Abstract
An ongoing healthcare debate is whether controlling hospital-acquired infection (HAI) from methicillin-resistant Staphylococcus aureus (MRSA) will result in lowering the global HAI rate, or if MRSA will simply be replaced by another pathogen and there will be no change in overall disease burden. With surges in drug-resistant hospital-acquired pathogens during the COVID-19 pandemic, this remains an important issue. Using a dataset of more than 1 million patients in 51 acute care facilities across the USA, and with the aid of a threshold model that models the nonlinearity in outbreaks of diseases, we show that MRSA is additive to the total burden of HAI, with a distinct ‘epidemiological position’, and does not simply replace other microbes causing HAI. Critically, as MRSA is reduced it is not replaced by another pathogen(s) but rather lowers the overall HAI burden. The analysis also shows that control of MRSA is a benchmark for how well all non-S. aureus nosocomial infections in the same hospital are prevented. Our results are highly relevant to healthcare epidemiologists and policy makers when assessing the impact of MRSA on hospitalized patients. These findings further stress the major importance of MRSA as a unique cause of nosocomial infections, as well as its pivotal role as a biomarker in demonstrating the measured efficacy (or lack thereof) of an organization’s Infection Control program.
Subject terms: Infectious diseases, Statistics, Epidemiology
Introduction
Much effort has been directed at reducing the acquisition of methicillin-resistant Staphylococcus aureus (MRSA) disease in healthcare settings1–6. Such efforts have been prompted by data suggesting that MRSA disease is associated with substantial morbidity and cost7–9, and that the most important risk for MRSA disease is being colonized in the nares with MRSA10. During the COVID-19 pandemic, several drug-resistant pathogens, including MRSA, have been recognized as increasing problems during the pandemic11,12 . In a recent study, MRSA is identified to be a leader in global deaths as far as antimicrobial resistance is concerned12. This indicates a need for ongoing prevention efforts, even during the SARS-CoV-2 epidemic. MRSA accounts for a sufficiently large proportion of hospital-acquired infection (HAI) disease, thus making it “worth” targeting13–15. Because of its impact and societal concerns, MRSA is one of a growing range of pathogens (along with organisms such as CRE, Acinetobacter, ESBL-producing bacteria, Klebsiella and Clostridium difficile) that have a visible ‘public health niche,’ in the sense that Infection Control departments are highly tuned to pick up signals of these pathogens, resulting in ongoing surveillance for changes in infection rates, followed by deployment of various control measures when infections rise. We postulate that, for this reason, MRSA may be able to act as an indicator for overall success of HAI control. An open question is whether MRSA also occupies a distinct ‘epidemiological position’ in a hospital setting, interpreted here as a unique combination of factors determining transmissibility and persistence. If so, then this would imply that if MRSA were eliminated, new, similarly virulent organisms would not simply replace it as a cause of nosocomial disease. Hence, these conjectures suggest that if MRSA control efforts succeed in reducing MRSA disease, all-cause HAIs are reduced—and reduced to a substantial extent. These arguments could, in the absence of supporting data, be challenged. It has widely been reported that MRSA accounts for a large proportion of total nosocomial S. aureus infection in many hospitals16–18. Also, there is evidence that in- and outpatient MRSA disease is adding to rather than simply substituting for methicillin-sensitive Staphylococcus aureus infection19.
In this study, we present and analyze data to support our hypothesis that MRSA has its distinct epidemiological position with respect to other nosocomial infections in the acute care hospital setting. We also hypothesize that, in such nosocomial settings, MRSA acts as an indicator in explaining trend changes in all non-S. aureus infections. For this, we provide a quantitative marker and a threshold level that could be used to gauge hospital performance. We aim to demonstrate that MRSA is additive to the total burden of healthcare-associated all-organism disease and that its control significantly reduces HAI. Our goal is to help infection control programs focus on organisms that impact their overall hospital acquired infection issues.
Methods
Data
Our national dataset comes from the database of a medical information technology company (MedMined Services, Care Fusion, Inc., Birmingham, AL) and at the time of collection was one of the largest datasets compiled investigating hospital-acquired infections with over one million patients from 51 different hospitals throughout the United States. MedMined Services is a third-party provider that offers proprietary electronic automated data mining surveillance tool to monitor HAIs. The dataset is diverse and comprises both academic and community hospitals. Our study is based on monthly data (August 2005–September 2008) on the number of HAI caused by various bacteria (MRSA, methicillin-susceptible S. aureus [MSSA], and non-S. aureus [all other organisms]), along with the number of admissions to each of the 51 hospitals. None of the data was collected at the patient level. The data was aggregated by hospital and on a monthly basis over the 36-month study period; and all the hospitals were de-identified before we received the data. The 51 hospitals included in the study represent all hospitals for which the MedMined data was available for the entire 36-month study period. The dataset covers eight regions as identified by MedMined services over the entire country. No further information about the 51 hospitals (such as geographical location, socio-economic status, urbanicity) was provided.
We use the measured system-wide rates of HAI and MRSA from our four hospitals at NorthShore University HealthSystem for the year 2013, to validate the results of the analysis conducted using the MedMined data above. We also used data from the same network of NorthShore University HealthSystem hospitals for the years 2013–2016, comprising rates of nosocomial MRSA blood stream infections, rates of nosocomial vancomycin resistant enterococci (VRE) blood stream infections, and rates of nosocomial multidrug resistant Gram-negative organism blood stream infections in hospital intensive-care units (ICUs).
HAI was identified based on a validated indicator, namely ‘Nosocomial Infection Marker’ (NIM), as previously investigated20. The NIM rates are adjusted for comparison between hospitals based on the case-mix index that is specifically calculated for each facility. The sensitivity and specificity of marker analysis is 0.86 [95% confidence interval: (0.76, 0.96)] and 0.984 [95% confidence interval: (0.976, 0.992)] as previously demonstrated20.
Statistical analysis
We assess the relationship between the number of non-S. aureus infections per 1000 admissions (called the rate of non-S. aureus infections) and the number of S. aureus infections per 1000 admissions (called the rate of non-S. aureus infections) and we find it to be nonlinear; see Fig. 1a and the Results section below. As a result of this, we model the nonlinear behavior of the rate of non-S. aureus infections explained by changes in the MRSA and MSSA infection rates, using a threshold model for count data known as the Generalized Threshold Model21. For this, denotes the rate of non-S. aureus infections in hospital h in a given month t, and is the number of admissions in hospital h in a given month t. Then, the number of non-S. aureus infections in hospital h at time t (measured in months), is assumed to be a Poisson random variable whose mean = is given by
| 1 |
where and is the number of S. aureus (i.e., MSSA + MRSA) infections per 1000 admissions d months earlier in hospital h (which we will refer to as the lag-d S. aureus rate for that hospital), and is number of MRSA infections per 1000 admissions in hospital h in a given month t (which we will refer to as the rate of MRSA infections in month t for hospital h). The parameter is referred to as the threshold and is a non-negative integer referred to as the delay or threshold lag, both of which are unknown and being estimated. The variation across hospitals is accounted for in the model through the regression parameters and which vary across the 51 hospitals.
Figure 1.
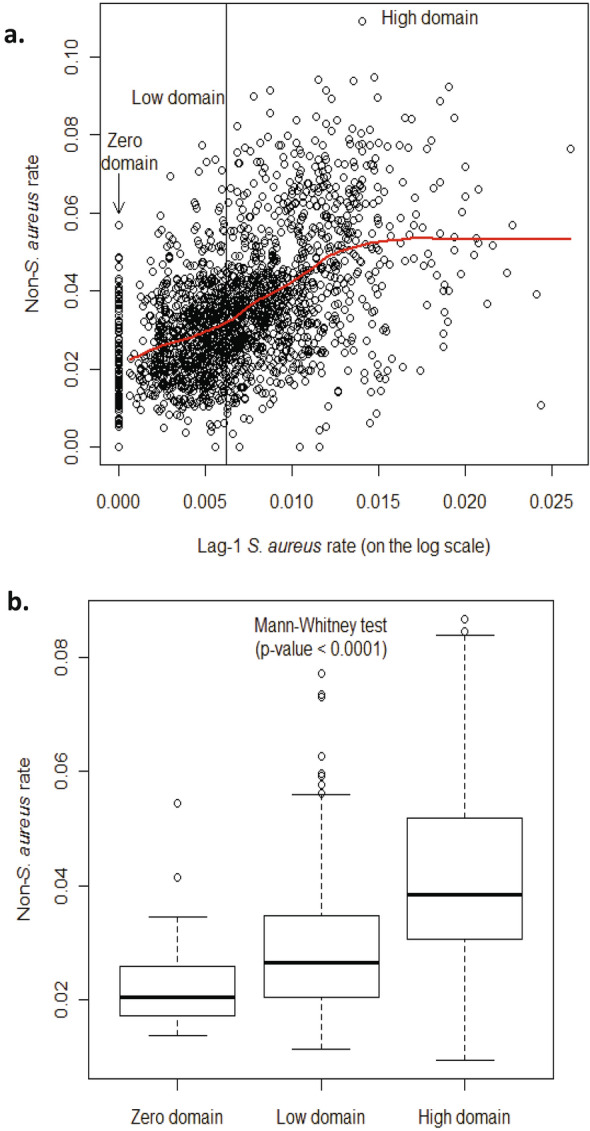
(a) Plot of the rate of non-S. aureus infections versus the threshold variable, namely the (log-transformed) lag-1 S. aureus rate. The zero domain corresponds to a domain such that the lag-1 S. aureus rate being equal to zero. The vertical line shows the location of the threshold estimate that defines two non-zero regimes: the low domain and the high domain. The red curve explores nonparametrically the mean of the non-S. aureus rate as a function of the lag-1 S. aureus rate, by fitting a local regression model. The shape of this red curve attests that the underlying process is nonlinear. (b) Boxplots of the fitted non-S. aureus rate in each of the 3 domains (zero, low, and high domains). The zero-domain median of the fitted non-S. aureus rate is significantly lower than the low-domain median (Mann–Whitney test; p value < 0.0001); the low-domain median of the fitted non-S. aureus rate is significantly lower than the high-domain median (Mann–Whitney test; p value < 0.0001).
We make the following underlying assumptions in our modeling approach. We assume that infections longer than 3 months ago no longer influence the rate of non-S. aureus infections. We also assume that hospitals are well-mixed in the sense that every new admission has the same probability to get infected with non-S. aureus.
The above fitted model using a Poisson distribution suggests the presence of overdispersion which occurs due to unmeasured factors (e.g., virulence of bacteria) and missing covariates (e.g., number of cultures performed to document infection per hospital and per admission). Hence, we modify the above model to account for this over dispersion by building a hierarchical model such that there is an unobserved random variable having a Gamma()/ distribution with mean 1 and variance . Then, conditional on , the number of non-S. aureus infections is Poisson distributed with mean . The marginal distribution of the number of non-S. aureus infections is then negative binomial with mean (with the corresponding infection rate modeled as in Eq. [1] above) and an inflated variance , where represents the over dispersion parameter (see Venables and Ripley22 and McCullagh and Nelder23 for a further discussion of modeling using the negative binomial distribution). Using a likelihood-based estimation approach21, the threshold delay d and the threshold r are estimated using a grid search between the 10th percentile and the 90th percentile of the lagged threshold variable
This study was approved by the Institutional Review Board of NorthShore University HealthSystem (Project EH-03–150). As reported in this approval by Institutional Review Board of NorthShore University HealthSystem is the statement 'It is noted that a consent form is not required for this study’. All methods were performed in accordance with the relevant guidelines and regulations.
Ethics approval
This study was approved by the Institutional Review Board of NorthShore University HealthSystem (Project EH-03–150). As reported in this approval by Institutional Review Board of NorthShore University HealthSystem is the statement ‘It is noted that a consent form is not required for this study’. All methods were performed in accordance with relevant guidelines and regulations.
Results
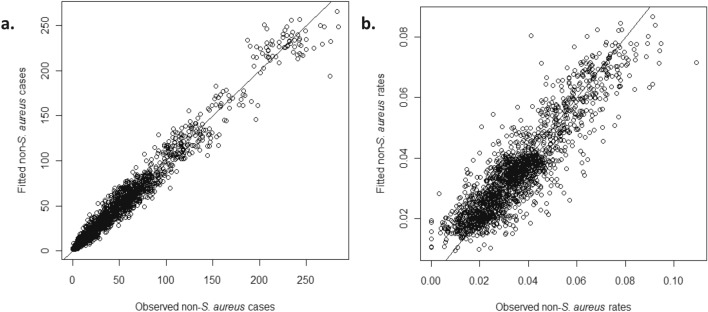
Figure 1a shows that the rate of non-staphylococcal (i.e., HAI minus MSSA and MRSA) infection varies nonlinearly with the log-transformed S. aureus rate of the previous month, i.e., the lag-1 S. aureus rate: the strong departure from linearity is unveiled by fitting a nonparametric local regression function (displayed in red in Fig. 1a), confirming that the underlying process is nonlinear. Hence, we model the nonlinear behavior of the rate of non-S. aureus infections explained by changes in the MRSA and MSSA rates, using a threshold model for count data24, specifically the Generalized Threshold Model21, where the threshold variable is the log-transformed lag-1 S. aureus rate; see “Methods”. We model the number of non-S. aureus infections using a negative binomial distribution that accounts for the over dispersion that occurs due to unmeasured factors (e.g., virulence of bacteria) as well as missing covariates (e.g., microbiological documentation policies)25,26; as described in the “Methods”. Table 1 summarizes the maximum likelihood estimates of the parameters in the model given by Eq. [1] in “Methods”. The good agreement between the observed values and the fitted values of the number of non-S. aureus cases in Fig. 2a and of the non-S. aureus rates in Fig. 2b, attests the credentials and importance of the fitted model in explaining the data; see Supplementary Materials (SM) for details on the model diagnostics.
Table 1.
Maximum likelihood estimates of the parameters in the fitted threshold model.
| Variable | Estimated value | Asymptotic | Asymptotic |
|---|---|---|---|
| Standard error | 95% Confidence interval | ||
| When the lag-1 S. aureus rate is zero | |||
| Intercept | − 4.33 | 0.098 | (− 4.53, − 4.14) |
| Lag-1 non-S. aureus rate | 17.7 | 3.5 | (10.7, 24.6) |
| Lag-2 non-S. aureus rate | 6.76 | 3.3 | (0.251, 13.2) |
| When the lag-1 S. aureus rate is positive and ≤ r | |||
| Hospital-specific intercept (See Table S1 in Supplementary Material) | |||
| MRSA rate | 8.63 | 4.2 | (0.376, 16.8) |
| Lag-1 non-S. aureus rate | 6.24 | 1.2 | (3.86, 8.62) |
| Lag-2 non-S. aureus rate | 4.62 | 1.3 | (2.08, 7.16) |
| Lag-3 non-S. aureus rate | 3.01 | 1.2 | (0.667, 5.35) |
| When the lag-1 S. aureus rate is positive and > r | |||
| Hospital-specific intercept (See Table S1 in Supplementary Material) | |||
| MRSA rate | 10.5 | 2.4 | (5.83, 15.23) |
| Lag-1 non-S. aureus rate | 4.47 | 0.75 | (3.00, 5.95) |
| Lag-2 non-S. aureus rate | 2.96 | 0.76 | (1.46, 4.45) |
| Lag-3 non-S. aureus rate | 2.12 | 0.77 | (0.615, 3.62) |
Figure 2.
Plot of the fitted values versus the observed values of (a) the number of non-S. aureus cases and (b) the rate of non-S. aureus infections.
The vertical line in Fig. 1a shows the location of the threshold estimate (0.62%) that defines two non-zero regimes: the low domain and the high domain. As displayed in the box plots of Fig. 1b, the distribution of the fitted non-S. aureus rates is shown to be statistically different across the three domains (i.e., the zero, low, and high domains) defined by the lag-1 S. aureus threshold variable. In particular, the zero-domain median of the fitted non-S. aureus rate is significantly lower than the low-domain median (Mann–Whitney test; p value < 0.0001); the low-domain median of the fitted non-S. aureus rate is significantly lower than the high-domain median (Mann–Whitney test; p value < 0.0001). In addition, Fig. 1b shows that the non-S. aureus rate is least variable in the zero domain, with increasing variability from zero to high domains (across the three domains).
The set of explanatory variables used to fit the model in Eq. [1] and their statistical significance in the fitted model reported for each regime in Table 1 show that the trend of total S. aureus nosocomial infection of the previous month (i.e., the threshold variable or lag-1 S. aureus rate), as well as total non-S. aureus nosocomial infection 1, 2, and 3 months earlier, is a benchmark for how well all non-S. aureus nosocomial infections are controlled. The lag-1 S. aureus rate determines whether or not all non-S. aureus nosocomial infections will be moving toward a high or low nosocomial infection domain. These domains are separated by a threshold determined by the lag-1 S. aureus rate that indicates whether or not changes in healthcare infection control practices are likely to occur (e.g., practice is likely to change if above the threshold). Below the threshold, the nosocomial infections would appear to be ‘under control’ using current practice; and above the threshold, they would appear ‘out of control’ and require some practice modification or intervention in order to re-enter the low domain. Furthermore, Table 1 shows that the lag-1, -2, and -3 non-S. aureus rates are somewhat more dominant in the low nosocomial infection domain (as shown by their parameter estimates and their corresponding 95% CI’s in each domain), where all hospitals with low nosocomial infection rates are tightly clustered as compared to the high domain (which shows more nosocomial infection diversity and variability; see Fig. 1a and b) suggesting that the lagged non-S. aureus nosocomial infections are more important in the low domain (along with lag-1 S. aureus) and that lag-1 S. aureus nosocomial infections are more dominant in the high infection domain. In addition, the fitted model in Eq. [1] (“Methods”) and Table 1 show that the actual rate of MRSA nosocomial infection is a significant predictor of non-S. aureus nosocomial infection, and that MRSA is more important in high domain hospitals and may thus dominate all nosocomial infections when these rates are high (Fig. S3 in SM).
Therefore, MRSA at any time was a good marker for identifying the status of overall rate of nosocomial infection at a given healthcare organization. A high overall rate of infection could represent shifts in risk of the underlying population (e.g., changes in use of invasive devices, or changes in performance of certain high-risk procedures, changes in prevalence or ‘colonization pressure’ of certain high-risk pathogens, etc.). Thus, a high rate of MRSA can serve as a trigger to investigate the underlying causes and examine whether or not this increased change in MRSA rate is due to inadequate infection control practice.
The threshold for lag-1 total S. aureus nosocomial infection separating the two domains is 0.62% or 6.2 nosocomial S. aureus infections per 1000 admissions (see “Methods” for the estimation of the threshold); of these 3.7 to 3.9 are MRSA nosocomial infections per 1000 admissions (mean and median are 0.6 and 0.63 MRSA percentage of S. aureus, respectively). In the dataset, the S. aureus HAI rate is approximately 17% of the overall HAI rate (mean and median). Thus, one can calculate that the total HAI/S. aureus HAI ratio is 5.88. If the threshold variable for separating the domains is 0.62 S. aureus infections, then this would suggest that a total HAI rate above 3.65% would serve as a trigger to investigate current infection control practices and possible changes thereof. Thus, hospitals with overall total nosocomial infection proportion less than 3.65% can be classified as ‘Good’ performers in their infection control program, as opposed to being classified as ‘Poor’ performers for having an overall total nosocomial infection proportion above 3.65%. This designation is model informed and is based on the entire realm of hospitals enrolled with MedMined and under consideration in this study. A core component of the model used is its benchmarking capacity that is key to infection prevention and control required to reduce HAI and plays an important role in the absence of national benchmarking designations.
Thus, in summary, the data implies that the MRSA HAI rate can be used as a surrogate to determine how well the overall Infection Control program is performing. If it is ≤ 0.37–0.39%, then the program is likely doing well and is governed by the low domain model where all HAIs are more equal in driving changes in HAI rate, but if it is > 0.39% there is room for improvement (some change in infection control practice may be warranted) and then the total HAI rate is governed by the upper domain model where MRSA disease is most important and likely should be the first target of any intervention.
Dividing the data into three domains (zero, low, and high domain) plays a key role in demonstrating that MRSA is additive to nosocomial infections, where movements in all-organism infection rates from one month to the next are not explained by the monthly fluctuations in non-S. aureus infection rates, but by the monthly movements in MRSA (and MSSA) infection rates. We studied the behavior of the monthly change of non-S. aureus rates (between two consecutive months), the monthly change in MRSA rates, and the monthly change in MSSA rates. Figure 3a shows that by combining all observations from the low and high domains, the distributions of the monthly changes in non-S. aureus rates, MRSA rates, and MSSA rates are all centered around zero. Therefore, distinguishing between data from the low and high domains (by modeling this nonlinearity using the threshold model) is required to study the impact of the fluctuations in MRSA rates with infections caused by all organisms. It is only when we separate the data between the low and high domains, that we observe that the monthly change in non-S. aureus rates is centered around zero but that the monthly changes in MRSA rates and MSSA rates are distributed around a median that is significantly different than zero (Fig. 3b and c). These findings suggest that MRSA adds to the total burden of all-organism nosocomial infections rather than simply replacing other organisms as a cause of infection.
Figure 3.
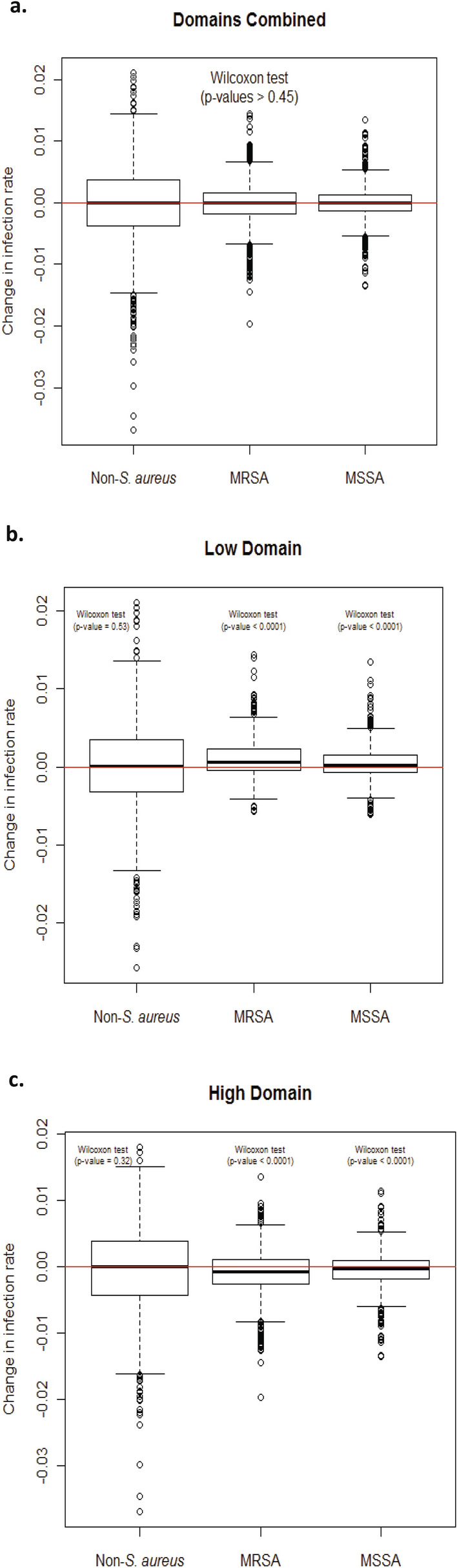
(a) Boxplots of the monthly change in the non-S. aureus infection rate (i.e., the difference in non-S. aureus rate between two consecutive months), the monthly change in the MRSA infection rate, and the monthly change in the MSSA infection rate for all observations corresponding to the low and high domains. The red horizontal line refers to a monthly change of zero between two consecutive months. The medians of the monthly change in non-S. aureus rate, the monthly change in MRSA rate, and the monthly change in MSSA rate are all not statistically different than zero (Wilcoxon test; p values > 0.45). (b) Boxplots of the monthly change in the non-S. aureus infection rate, the monthly change in the MRSA infection rate, and the monthly change in the MSSA infection rate corresponding to the observations in the low domain (when lag-1 S. aureus rate is less than or equal to 0.62%). The red horizontal line refers to a monthly change of zero between two consecutive months. The median change in non-S. aureus rate (median = 0.000086) is not statistically different than zero (Wilcoxon test; p value = 0.53); however, the median change in MRSA rate (median = 0.00054) and the median change in MSSA rate (median = 0.00019) are both statistically different than zero (Wilcoxon test; p values < 0.0001). (c) Boxplots of the monthly change in the non-S. aureus infection rate, the monthly change in the MRSA infection rate, and the monthly change in the MSSA infection rate corresponding to the observations in the high domain (when lag-1 S. aureus rate is greater than 0.62%). The red horizontal line refers to a monthly change of zero between two consecutive months. The median change in non-S. aureus rate (median = − 0.000086) is not statistically different than zero (Wilcoxon test; p value = 0.32); however, the median change in MRSA rate (median = − 0.00077) and the median change in MSSA rate (median = − 0.00032) are both statistically different than zero (Wilcoxon test; p values < 0.0001).
To validate the estimation of the threshold and the corresponding domains, using data not used in fitting the threshold model for count data, we use our own four-hospital system-wide data at NorthShore University HealthSystem in the year 2013 and we find that the measured system-wide rate of MRSA is 0.15% and the measured system-wide rate of HAI is 2.7%. Table S2 gives the rates of MRSA, VRE, and multidrug-resistant Gram-negative infections, as measured in the network of the NorthShore University HealthSystem for 2013–2016. Further commentary on these findings is included in the Discussion section.
Discussion
Others have argued that MRSA adds to the total burden of HAI disease in the outbreak setting27–32, but we are not aware of any other work that looks at MRSA in general (outbreak and endemic MRSA disease), which rigorously demonstrates using a national dataset that as MRSA goes up so do all HAI and as MRSA goes down so do all HAI. Our results are very relevant to healthcare epidemiologists and policy makers when assessing the impact of MRSA on hospitalized patients. In view of the fact that our study group represents a broad cross section of (community and academic) healthcare facilities, we believe the results are generalizable to most acute care centers in the developed world. First and foremost, we show that MRSA occupies its distinct epidemiological position; hence confronting MRSA on its own as a cause of HAI is worthwhile, as it is unlikely to be replaced by another pathogen, at least in the short term14. Additionally, those organizations that succeed in lowering their MRSA rate, should expect the total nosocomial infection rate to decrease, because of the indicator effect of MRSA and its public health niche.
We have tested the threshold hypothesis using our own HAI data for 2013. During that year our 4-hospital, system-wide MRSA rate was 0.15% (lower than the low domain threshold rate of 0.37–0.39%), which would suggest the overall HAI rate being less than 3.65% based on the model; our measured system-wide rate for 2013 was 2.7%. Furthermore, MRSA did not dominate as a pathogen and represented only 5.6% of all organisms causing HAI, again as predicted for an organization in the low domain model. This is also consistent with the model whereby being in the low domain suggests that non-S. aureus HAI are more important–in 2013 there was more than 30 microbial species contributing to HAI, with Escherichia coli being most common, followed by enterococci, Clostridium difficile, yeast, and then S. aureus.
Evolutionary speaking, we believe that the MRSA-niche in a hospital setting is also not likely to be filled in the long term by another pathogen since MRSA is an adaptive change in the parent microbe, S. aureus, which has always been an aggressive pathogen33. When antibiotics became widely available that could suppress or treat S. aureus, the organism adapted and became more antibiotic resistant (MRSA)33,34. In doing so, it added to or replaced the parent (MSSA) as a hospital pathogen35. Our results illustrate that if MRSA is controlled, it will indeed not be replaced at least in the short term. This is corroborated by data from the Netherlands and the Scandinavian countries, which have a (relatively) long history of controlling and monitoring MRSA, and where no ‘new’ organism has stepped up to fill the void for at least 25 years36,37. Furthermore, S. aureus behaves similarly in that if one controls it (via decolonization) before surgery, the post-operative infection rates fall four to six-fold and S. aureus is not replaced by another pathogen38,39.
Our study has limitations. Since the cost of the MedMined Services program is not inconsequential, it could be argued that our hospital cohort is willing to dedicate more resources than most to their infection control program and are thus not representative of most U.S. facilities. While this may be true, this weakness is countered by the strength of the data that is completely objective since information is drawn directly from laboratory information systems and electronic admission-discharge-transfer records with no human ‘interpretation’ that could bias the reported HAI results. Furthermore, other large healthcare systems have invested large resources in MRSA control40, so we believe our hospital data set is representative of the broad HAI model patterns in U.S. hospitals. The finding that there was a spread of MRSA rates that correlated with overall nosocomial infection trends does not contradict the representativeness of the data we used, despite this potential bias. Since this study was completed, MedMined was acquired by BD integrated analytics and the authors can no longer access additional data. This would not alter the investigation reported but would impair any further data investigation.
Further research of the underlying factors involved in the MRSA epidemiological position is warranted. This, however, would require a different study design. For such a study, one should look at the entire hospital, and its interaction with the outside world (society), as an ecosystem in which pathogens encounter favorable and unfavorable circumstances for their survival, reproduction and spread, and where different pathogens are under different pressure. Likely factors are number of patients (i.e., potential susceptible hosts for the nosocomial pathogen species), turnover rate of patients, intensity of treatment of individual patients, differences in susceptibility among patients (e.g., age-related or related to cause of hospitalization), the level of infection control and hand hygiene measures, compartmentalization of patients (metapopulation structure), and the connectivity of the contact network of patients and healthcare workers, in addition to antibiotic stewardship.
Microbiology laboratory data from hospitals are critical for following the evolution of multidrug resistant organisms41. Unfortunately, the hospital laboratory cannot detect potential nosocomial infections that are sometimes not well microbiologically documented (e.g., surgical site infections) or occur after discharge of the patient41.
The database used is relatively old (2005–2008). At that time, other pathogens, in particular, extended-spectrum β-lactamase producing Enterobacteriacae and carbapenemase-producing Enterobacteriacae (CPE), caused fewer nosocomial infections compared to MRSA. While many countries have reported an improvement in MRSA infection rates during these last 10 years, studies have also highlighted the progression of infections caused by multidrug resistant Gram negative organisms42. An effective overall infection prevention program is multifaceted including antimicrobial stewardship, interventions targeted on problematic pathogens (such as CPE surveillance), and monitoring of hand hygiene. While data about other emerging pathogens was not available to use in this study, we reviewed four years of data (2013–2016) from the NorthShore University HealthSystem network of four hospitals to give insight into the observed pattern of these other critical pathogens. The rates of the nosocomial blood stream infections caused by MRSA and VRE, and the rates of nosocomial blood stream infections caused by multidrug resistant Gram negative organisms in hospital intensive-care units showed that other infections were not replacing MRSA. Interestingly, VRE and resistant Gram negatives were actually either decreasing (VRE) or simply not emerging (the Gram negatives); see Table S2 in SM, supporting our hypothesis.
In summary, the proposed MRSA indicator in this study should be seen as an important part of a set of indicators. One could argue that the dataset used in this study is very elaborate from a time where a possible signal to be derived from MRSA control is not yet masked by the much larger number of important bacterial threats we have today. We recognize how the world of healthcare associated infections has changed and what this could mean for the use of this MRSA marker. Thus, follow-up investigation should be conducted to include these organisms using a large dataset in contemporary time.
In conclusion, while healthcare-associated MRSA infection has been recognized as an important adverse event, our analysis shows its major importance as a unique cause of nosocomial infection, as well as its pivotal role as a biomarker in demonstrating the measured efficacy (or lack thereof) for a given Infection Control program in the acute care setting. Until current electronic databases that keep track of all HAIs become widely used, our data suggest that to lower the HAI rate, hospitals can benefit from an infection control program that includes monitoring MRSA disease rates and enhances comprehensive patient safety efforts when these rates are rising.
Supplementary Information
Acknowledgements
The authors would like to thank MedMined Services, Care Fusion-BD for providing the data.
Author contributions
N.I.S., A.R., and L.R.P. designed research. N.I.S., H.H., and L.R.P. performed research. N.I.S. analyzed the data. N.I.S., A.R., H.H., and L.R.P. wrote the paper.
Funding
There was no external support for this work, and only the authors had a role in the study design, data analysis, data interpretation, or writing of this article.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-21300-6.
References
- 1.Jinno S, Chang S, Donskey CJ. A negative nares screen in combination with absence of clinical risk factors can be used to identify patients with very low likelihood of methicillin-resistant Staphylococcus aureus infection in a veterans affairs hospital. Am. J. Infect. Control. 2012;40(9):782–786. doi: 10.1016/j.ajic.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 2.Hanberger H, Walther S, Leone M, et al. Increased mortality associated with methicillin-resistant Staphylococcus aureus (MRSA) infection in the intensive care unit: results from the EPIC II study. Int. J. Antimicrob. Agents. 2011;38(4):331–335. doi: 10.1016/j.ijantimicag.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 3.Johnson AP, Davies J, Guy R, et al. Mandatory surveillance of methicillin-resistant Staphylococcus aureus (MRSA) bacteraemia in England: The first 10 years. J. Antimicrob. Chemother. 2012;67(4):802–809. doi: 10.1093/jac/dkr561. [DOI] [PubMed] [Google Scholar]
- 4.Milstone AM, Goldner BW, Ross T, Shepard JW, Carroll KC, Perl TM. Methicillin-resistant Staphylococcus aureus colonization and risk of subsequent infection in critically ill children: Importance of preventing nosocomial methicillin-resistant Staphylococcus aureus transmission. Clin. Infect. Dis. 2011;53(9):853–859. doi: 10.1093/cid/cir547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jain R, Kralovic SM, Evans ME, et al. Veterans affairs initiative to prevent methicillin-resistant Staphylococcus aureus infections. N. Engl. J. Med. 2011;364(15):1419–1430. doi: 10.1056/NEJMoa1007474. [DOI] [PubMed] [Google Scholar]
- 6.Robicsek A, Beaumont JL, Paule SM, et al. Universal surveillance for methicillin-resistant Staphylococcus aureus in 3 affiliated hospitals. Ann. Intern. Med. 2008;148(6):409–418. doi: 10.7326/0003-4819-148-6-200803180-00003. [DOI] [PubMed] [Google Scholar]
- 7.Cosgrove SE, Sakoulas G, Perencevich EN, Schwaber MJ, Karchmer AW, Carmeli Y. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: A meta-analysis. Clin. Infect. Dis. 2003;36(1):53–59. doi: 10.1086/345476. [DOI] [PubMed] [Google Scholar]
- 8.Cosgrove SE, Qi Y, Kaye KS, Harbarth S, Karchmer AW, Carmeli Y. The impact of methicillin resistance in Staphylococcus aureus bacteremia on patient outcomes: Mortality, length of stay, and hospital charges. Infect. Control Hosp. Epidemiol. 2005;26(2):166–174. doi: 10.1086/502522. [DOI] [PubMed] [Google Scholar]
- 9.Turner NA, Sharma-Kuinkel BK, Maskarinec SA, et al. Methicillin-resistant Staphylococcus aureus: An overview of basic and clinical research. Nat. Rev. Microbiol. 2019;17(4):203–218. doi: 10.1038/s41579-018-0147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robicsek A, Beaumont JL, Thomson RB, Jr, Govindarajan G, Peterson LR. Topical therapy for methicillin-resistant Staphylococcus aureus colonization: Impact on infection risk. Infect. Control Hosp. Epidemiol. 2009;30(7):623–632. doi: 10.1086/597550. [DOI] [PubMed] [Google Scholar]
- 11.Segala FV, Bavaro DF, Di Gennaro F, et al. Impact of SARS-CoV-2 epidemic on antimicrobial resistance: A literature review. Viruses. 2021;13(11):2110. doi: 10.3390/v13112110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antimicrobial RC. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet. 2022;399(10325):629–655. doi: 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jernigan JA. Is the burden of Staphylococcus aureus among patients with surgical-site infections growing? Infect. Control Hosp. Epidemiol. 2004;25(6):457–460. doi: 10.1086/502421. [DOI] [PubMed] [Google Scholar]
- 14.David MZ, Medvedev S, Hohmann SF, Ewigman B, Daum RS. Increasing burden of methicillin-resistant Staphylococcus aureus hospitalizations at US academic medical centers, 2003–2008. Infect. Control Hosp. Epidemiol. 2012;33(8):782. doi: 10.1086/666640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magill SS, Edwards JR, Bamberg W, et al. Multistate point-prevalence survey of health care-associated infections. N. Engl. J. Med. 2014;370(13):1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livermore DM, Pearson A. Antibiotic resistance: Location, location, location. Clin. Microbiol. Infect. 2007;13(Suppl 2):7–16. doi: 10.1111/j.1469-0691.2007.01724.x. [DOI] [PubMed] [Google Scholar]
- 17.McDonald LC. Trends in antimicrobial resistance in health care-associated pathogens and effect on treatment. Clin. Infect. Dis. 2006;42(Suppl 2):S65–71. doi: 10.1086/499404. [DOI] [PubMed] [Google Scholar]
- 18.Naimi TS, LeDell KH, Como-Sabetti K, et al. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA. 2003;290(22):2976–2984. doi: 10.1001/jama.290.22.2976. [DOI] [PubMed] [Google Scholar]
- 19.Hacek DM, Paule SM, Thomson RB, Jr, Robicsek A, Peterson LR. Implementation of a universal admission surveillance and decolonization program for methicillin-resistant staphylococcus aureus (MRSA) reduces the number of MRSA and total number of S. aureus isolates reported by the clinical laboratory. J. Clin. Microbiol. 2009;47(11):3749–52. doi: 10.1128/JCM.01223-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brossette SE, Hacek DM, Gavin PJ, et al. A laboratory-based, hospital-wide, electronic marker for nosocomial infection: The future of infection control surveillance? Am. J. Clin. Pathol. 2006;125(1):34–39. doi: 10.1309/502AUPR8VE67MBDE. [DOI] [PubMed] [Google Scholar]
- 21.Samia NI, Chan KS. Maximum likelihood estimation of a generalized threshold stochastic regression model. Biometrika. 2011;98(2):433–448. doi: 10.1093/biomet/asr008. [DOI] [Google Scholar]
- 22.Venables WN, Ripley BD. Modern applied statistics with S. 4. Springer; 2002. [Google Scholar]
- 23.McCullagh P, Nelder JA. Generalized linear models. 2. Chapman and Hall; 1989. [Google Scholar]
- 24.Tong H. Non-linear time series: A dynamical system approach. Clarendon Press, Oxford University Press; 1990. [Google Scholar]
- 25.Cox DR. Some remarks on overdispersion. Biometrika. 1983;70(1):269–274. doi: 10.1093/biomet/70.1.269. [DOI] [Google Scholar]
- 26.Agresti A. Categorical data analysis. 2. Wiley-Interscience; 2002. [Google Scholar]
- 27.Stamm AM, Long MN, Belcher B. Higher overall nosocomial infection rate because of increased attack rate of methicillin-resistant Staphylococcus aureus. Am. J. Infect. Control. 1993;21(2):70–74. doi: 10.1016/0196-6553(93)90227-u. [DOI] [PubMed] [Google Scholar]
- 28.Boyce JM, White RL, Spruill EY. Impact of methicillin-resistant Staphylococcus aureus on the incidence of nosocomial staphylococcal infections. J. Infect. Dis. 1983;148(4):763. doi: 10.1093/infdis/148.4.763. [DOI] [PubMed] [Google Scholar]
- 29.Chaix C, Durand-Zaleski I, Alberti C, Brun-Buisson C. Control of endemic methicillin-resistant Staphylococcus aureus: A cost-benefit analysis in an intensive care unit. JAMA. 1999;282(18):1745–1751. doi: 10.1001/jama.282.18.1745. [DOI] [PubMed] [Google Scholar]
- 30.Jernigan JA, Clemence MA, Stott GA, et al. Control of methicillin-resistant Staphylococcus aureus at a university hospital: One decade later. Infect. Control Hosp. Epidemiol. 1995;16(12):686–696. doi: 10.1086/647042. [DOI] [PubMed] [Google Scholar]
- 31.Harbarth S, Martin Y, Rohner P, Henry N, Auckenthaler R, Pittet D. Effect of delayed infection control measures on a hospital outbreak of methicillin-resistant Staphylococcus aureus. J. Hosp. Infect. 2000;46(1):43–49. doi: 10.1053/jhin.2000.0798. [DOI] [PubMed] [Google Scholar]
- 32.Ammerlaan HS, Harbarth S, Buiting AG, et al. Secular trends in nosocomial bloodstream infections: Antibiotic-resistant bacteria increase the total burden of infection. Clin. Infect. Dis. 2013;56(6):798–805. doi: 10.1093/cid/cis1006. [DOI] [PubMed] [Google Scholar]
- 33.Turner NA, Sharma-Kuinkel BK, Maskarinec SA, et al. Methicillin-resistant Staphylococcus aureus: An overview of basic and clinical research. Nat. Rev. Microbiol. 2019;17(4):203–218. doi: 10.1038/s41579-018-0147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chambers HF, Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009;7(9):629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mostofsky E, Lipsitch M, Regev-Yochay G. Is methicillin-resistant Staphylococcus aureus replacing methicillin-susceptible S aureus? J. Antimicrob. Chemother. 2011;66(10):2199–214. doi: 10.1093/jac/dkr278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bode LG, Wertheim HF, Kluytmans JA, et al. Sustained low prevalence of meticillin-resistant Staphylococcus aureus upon admission to hospital in The Netherlands. J Hosp Infect. 2011;79(3):198–201. doi: 10.1016/j.jhin.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 37.Bocher S, Skov RL, Knudsen MA, et al. The search and destroy strategy prevents spread and long-term carriage of methicillin-resistant Staphylococcus aureus: Results from the follow-up screening of a large ST22 (E-MRSA 15) outbreak in Denmark. Clin. Microbiol. Infect. 2010;16(9):1427–1434. doi: 10.1111/j.1469-0691.2009.03137.x. [DOI] [PubMed] [Google Scholar]
- 38.Hacek DM, Robb WJ, Paule SM, Kudrna JC, Stamos VP, Peterson LR. Staphylococcus aureus nasal decolonization in joint replacement surgery reduces infection. Clin. Orthop. Relat. Res. 2008;466(6):1349–1355. doi: 10.1007/s11999-008-0210-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bode LG, Kluytmans JA, Wertheim HF, et al. Preventing surgical-site infections in nasal carriers of Staphylococcus aureus. N. Engl. J. Med. 2010;362(1):9–17. doi: 10.1056/NEJMoa0808939. [DOI] [PubMed] [Google Scholar]
- 40.Jones M, Ying J, Huttner B, et al. Relationships between the importation, transmission, and nosocomial infections of methicillin-resistant Staphylococcus aureus: An observational study of 112 veterans affairs medical centers. Clin. Infect. Dis. 2014;58(1):32–39. doi: 10.1093/cid/cit668. [DOI] [PubMed] [Google Scholar]
- 41.Peterson LR, Brossette SE. Hunting health care-associated infections from the clinical microbiology laboratory: Passive, active, and virtual surveillance. J. Clin. Microbiol. 2002;40(1):1–4. doi: 10.1128/jcm.40.1.1-4.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jernigan JA, Hatfield KM, Wolford H, et al. Multidrug-resistant bacterial infections in U.S. hospitalized patients, 2012–2017. N. Engl. J. Med. 2020;382(14):1309–19. doi: 10.1056/NEJMoa1914433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.