Abstract
Sulfation is a common modification of glycans and glycoproteins. Sulfated N-glycans have been identified in various glycoproteins and implicated for biological functions, but in vitro synthesis of structurally well-defined full length sulfated N-glycans remains to be described. We report here the first in vitro enzymatic sulfation of biantennary complex type N-glycans by recombinant human CHST2 (GlcNAc-6-O-sulfotransferase 1, GlcNAc6ST-1). We found that the sulfotransferase showed high antennary preference and could selectively sulfate the GlcNAc moiety located on the Manα1,3Man arm of the biantennary N-glycan. The glycan chain was further elongated by bacterial β1,4 galactosyltransferase from Neiserria meningitidis and human β1,4 galactosyltransferase IV(B4GALT4), which led to the formation of different sulfated N-glycans. Using rituximab as a model IgG antibody, we further demonstrated that the sulfated N-glycans could be efficiently transferred to an intact antibody by using a chemoenzymatic Fc glycan remodeling method, providing homogeneous sulfated glycoforms of antibodies. Preliminary binding analysis indicated that sulfation did not affect the apparent affinity of the antibody for FcγIIIa receptor.
Keywords: Sulfated N-glycans, Sulfation, Sulfotransferase, Sulfated antibody, Chemoenzymatic synthesis, Glycoforms
1. Introduction
Sulfated N-glycans are a group of biologically important oligosaccharides, which are commonly found on glycoproteins formed by enzymatic modifications of N-glycans. Sulfation of N-glycans in glycoproteins occurs in the Golgi apparatus, where sulfotransferases catalyze the transfer of a sulfate group from 3′-phosphoadenosine-5′-phosphosulfate (PAPS) to glycoprotein substrates. Among the N-glycans, complex type structures are the most commonly sulfated, where the sulfate group is attached on the C-3 or C-6 position of galactose (Gal), the C-6 position of N-acetylglucosamine (GlcNAc), or the C-4 position of N-acetylgalactosamine (GalNAc) of the non-reducing terminal Galβ1,4GlcNAc (LacNAc) and GalNAcβ1,4GlcNAc (LacdiNAc) disaccharide moieties [1].
A variety of biologically important glycoproteins have been shown to possess sulfated N-glycans. For example, 20 sulfated N-glycans of IgG antibodies as trace species were identified from human sera, which are implicated as biomarkers of rheumatoid arthritis [2]. Although the influence of sulfated N-glycans on IgG functions remains to be characterized, it is speculated that sulfated N-glycans might alter the ligand specificity and biological functions of IgG antibodies [2]. Large structural diversity of sulfated N-glycans have been found on influenza glycoproteins, e.g., the viral hemagglutinin (HA) and neuraminidase (NA) [3], which might significantly affect viral replication and receptor binding [4–6]. Moreover, sulfated N-glycans play a critical role in lymphocyte homing and recruitment mediated by L-selectins [7]. The half-life of pituitary hormone is regulated by sulfated N-glycans [8]. Besides, sulfated N-glycans participate in peripheral nervous system (PNS) myelination both in porcines and murine tissues [9]. Sulfated di-, tri- and tetra-antennary complex type N-glycans are identified in human Tamm-Horsfall glycoprotein [10], which are involved in the binding to neutrophils [11] and immunosuppressive properties [12]. Sulfated N-glycans have also been reported in porcine thyroglobulin, which are the major iodinated glycoprotein formed in the thyroid gland [13]. Additionally, sulfated N-glycans have been reported in erythropoietin expressed in baby hamster kidney (BHK) cells [14], hen egg albumin [15], lysosomal enzyme [16], urokinase [17], and recombinant L-selectins produced in HEK293F cells [18].
The diverse biological functions of sulfated glycans have stimulated tremendous interest in the synthesis of sulfated carbohydrates. For example, sulfated oligosaccharides containing HNK-1 epitope, sulfated core 2O-GalNAc glycans and O-sulfated sialyl Lewis X with O-sulfation at different sites have been synthesized by a chemoenzymatic approach, in which respective sulfate group is added on the glucuronic acid, N-acetylglucosamine, galactose and N-acetylglucosamine moieties chemically followed by enzymatic sugar chain elongation [19–21]. Nevertheless, in vitro chemoenzymatic synthesis of structurally well-defined sulfated full-length N-glycans has not been described so far. For the biosynthesis of sulfated complex type N-glycans in mammalian systems, three types of sulfotransferases, including the galactose-3-O-sulfotransferases, the N-acetylglucosamine-6-O-sulfotransferases and the N-acetylgalactosamine-4-O-sulfotransferases, are responsible for the enzymatic sulfation. In human, four galactose-3-O-sulfotransferases have been identified and characterised, i.e., Gal3ST1, Gal3ST2, Gal3ST3 and Gal3ST4) [22–26], and two homologous enzymes of N-acetylgalactosamine-4-O-sulfotransferases have been cloned and expressed, which are able to transfer sulfate group to a GalNAc β1,4 GlcNAc (LacdiNAc) moiety [27]. In addition, five isoenzymes of GlcNAc-6-O-sulfotransferases have been produced and characterized, i.e., CHST2 (GlcNAc6ST-1), CHST4 (GlcNAc6ST-2), CHST5 (GlcNAc6ST-3), CHST7 (GlcNAc6ST-4), and CHST6 (GlcNAc6ST-5) (gene names followed by alternate names in parenthesis), which transfer a sulfate group to terminal GlcNAc leading to the formation of different cell surface epitopes including biologically important 6-sulfo sLex [28–33]. However, none of those five GlcNAc-6-O-sulfotransferases has been examined for in vitro sulfation of full-length intact N-glycans. We report here the first in vitro enzymatic sulfation of biantennary complex type N-glycans by recombinant human GlcNAc-6-O-sulfotransferase 1 (CHST2). We found that sulfotransferase CHST2 showed antennary preference for sulfation and could selectively add a sulfate group on the terminal GlcNAc of the Man-α1,3-Man arm. Further elongation by two different β1,4 galactosyltransferases gave additional sulfated N-glycans. Using rituximab as a model IgG antibody, we further demonstrated that the sulfated N-glycans could be successfully transferred to deglycosylated rituximab using an endoglycosidase mutant, Endo-S2 D184M, to provide homogeneous sulfated glycoforms of antibodies. We have previously shown that the Endo-S2 mutant (D184M) could use different glycan oxazolines as donor substrates for antibody Fc glycan remodeling [34–37]. The present study provided the first example showing that the Endo-S2 mutant is equally efficient to transfer sulfated N-glycans to antibodies to give structurally well-defined sulfated antibody glycoforms.
2. Results and discussion
2.1. Synthesis of sulfated N-glycans
To obtain biantennary complex type N-glycans, sialoglycopeptide (SGP) was isolated from egg yolk powder on a gram-scale according to the method reported previously [38–39]. SGP was treated with pronase to give the asparagine-linked (N-linked) bi-antennary N-glycan [40]. The free amino group in the asparagine moiety was protected by a fluorenylmethyloxycarbonyl (Fmoc) group to facilitate purification of products by preparative HPLC in each enzymatic transformation. The terminal sialic acid and galactose moieties were then removed by sialidase from Micromonospara Viridifaciens [41] and a β-1,4 galactosidase, respectively. The resulting product (G0-Asn-Fmoc, 1) was utilized as substrate to evaluate enzymatic sulfation by recombinant human GlcNAc-6-O-sulfotransferase 1 (CHST2) expressed in HEK293F cells (Scheme 1). Incubation of 1 with CHST2 yielded a new product (2) that eluted later than the starting material (1) on reverse phase analytical HPLC, and MALDI-TOF MS analysis indicated that it was a monosulfated N-glycan. Afterwards, preparative scale synthesis was executed, which resulted in 80 % isolated yield of product 2. To validate the sulfation position, we initially treated the product with commercial β-N-acetylglucosaminidase S and found that both sulfated and non-sulfated terminal GlcNAc moieties were removed, thus the β-N-acetylglucosaminidase S could not distinguish between the sulfated and free terminal GlcNAc moieties. It has been previously reported that the bacterial β-1,4-galactosyltransferase from Neisseria meningitidis was able to catalyze galactosylation specifically on free terminal GlcNAc rather than a sulfated GlcNAc [42]. Therefore, the free terminal GlcNAc could be temporarily blocked by galactosylation. Thus, treatment of 2 with the Neisseria meningitidis β-1,4-galactosyltransferase gave a monogalactosylated N-glycan (3), as shown by MALDI-TOF MS analysis (Fig. S1). Then the intermediate was treated with the commercial β-N-acetylglucosaminidase S and the MALDI-TOF MS indicated that a sulfated GlcNAc moiety was removed to give a truncated N-glycan (Fig. S2). Finally, the resulting product was treated with a commercial α-1,3-mannosidase, which could specifically remove free terminal α1,3-linked mannose. MALDI-TOF MS analysis showed that a mannose moiety was removed from the substrate upon treatment with the α-1,3-mannosidase to give a truncated N-glycan (4) (Fig. S3). The experimental data confirm that in product 2, the Man-1,3-Man arm carries a sulfated GlcNAc moiety that could not be galactosylated by the Neisseria meningitidis β-1,4-galactosyltransferase, and would be removed selectively by the β-N-acetylglucosaminidase S, thus freeing the mannose moiety on the α-1,3Man arm (the lower arm in the N-glycan). These results clearly show a high-regioselectivity of the CHST2 at the low-arm antennal GlcNAc moiety. Nevertheless, we also found that use of excess amount of donor substrate PAPS and enzyme CHST2 could slowly drive the addition of a second sulfate group on the other terminal GlcNAc, as shown by the MALDI-TOF MS monitoring of the reaction, although a complete double sulfation would need further optimization of the reaction condition (Fig. S4).
Scheme 1.
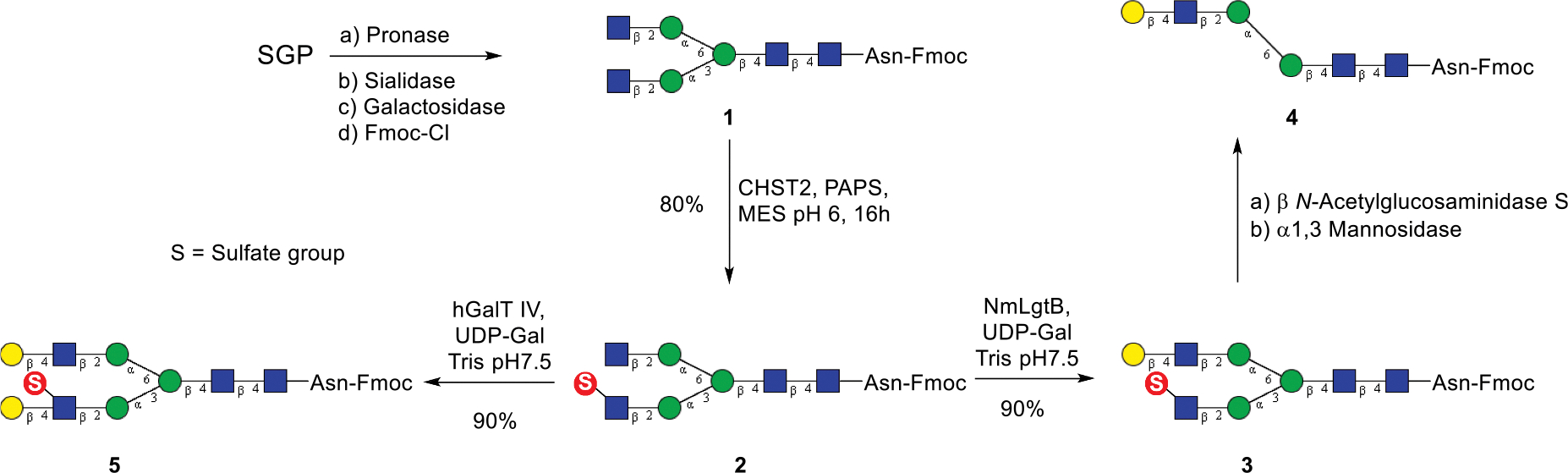
Chemoenzymatic synthesis of sulfated N-glycans.
It should be mentioned that treatment of the fully galactosylated N-glycan with sulfotransferase CHST2 did not give any sulfated N-glycan product (data not shown), suggesting that CHST2 only transfers sulfate group to a terminal instead of internal GlcNAc moiety in an N-glycan [43]. To synthesize additional sulfated N-glycan, the sulfated N-glycan (2) was elongated with human β1,4 galactosyltransferase IV (B4GALT4) to give a fully galactosylated N-glycan (5). This result suggests that the human B4GALT4 is promiscuous and efficient to galactosylate both non-sulfated and the 6-sulfated GlcNAc moieties. Thus, the use of the bacterial and human galactosyltransferases gave the two differentially galactosylated and sulfated N-glycans (3 and 5), respectively, in excellent yields. (Scheme 1). All the sulfated N-glycans were purified by preparative HPLC and their identity was confirmed by HRMS and NMR analysis.
2.2. Synthesis of sulfated N-glycan oxazolines as enzyme substrates
In order to synthesize novel antibody glycoforms carrying sulfated N-glycans, we sought to apply our chemoenzymatic Fc glycan remodeling method [44] to investigate whether an endoglycosynthase mutant such as Endo-S2 D184M could recognize sulfated glycan oxazolines for transglycosylation. We have previously demonstrated that the Endo-S2 mutant (D184M) can take different glycan oxazolines as donor substrates for antibody Fc glycan remodeling [34–37]. For that purpose, we first synthesized the corresponding sulfated N-glycan oxazolines. Treatment of the sulfated Fmoc-N-glycans (2, 3, and 5) with wild-type Endo-S2 gave the free N-glycans, which were subsequently treated with DMC and TEA in water to afford the sulfated glycan oxazolines (6, 7, and 8), respectively, in excellent yields (Scheme 2). The sulfated glycan oxazolines were desalted by G10 size exclusion chromatography and lyophilized. All three sugar oxazolines were analysed by 1H NMR. The anomeric protons at reducing termini of 6, 7, and 8 appeared as a doublet at ca. 6.0 ppm with a coupling constant (J1,2) of ca. 7.0 Hz, which are a signature signal of the anomeric proton of a glycan oxazoline (H-1 for 6, 5.98 ppm, 7.28 Hz; H-1 for 7, 5.96 ppm, 7.26 Hz; H-1 for 8, 5.96 ppm, 7.26 Hz. Figs. S5–7).
Scheme 2.
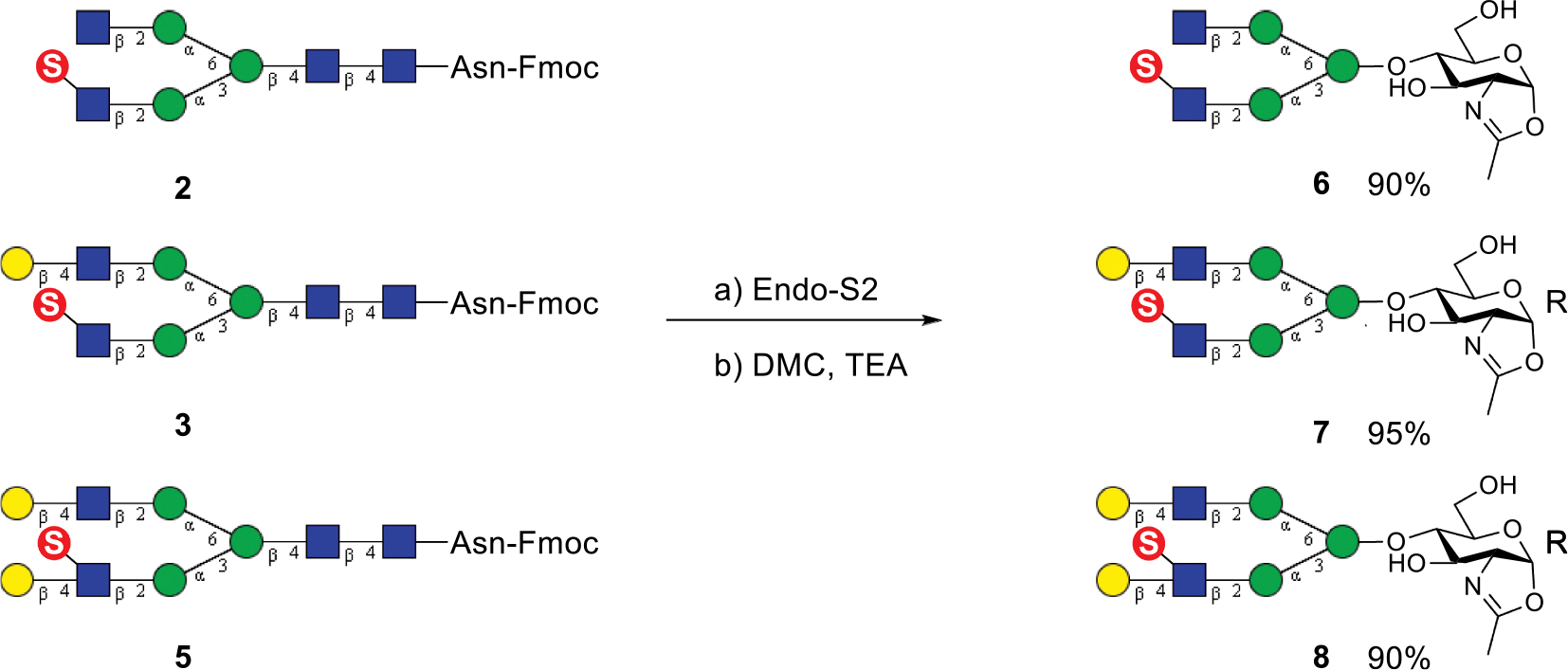
Synthesis of sulfated N-glycan oxazolines.
2.3. Chemoenzymatic synthesis of antibody glycoforms carrying sulfated N-glycans
To test the feasibility of the sulfated N-glycan oxazolines as substrates for enzymatic glycan remodeling of antibodies, we chose rituximab, a therapeutic monoclonal antibody, as a model for antibody Fc glycan remodeling. Thus, rituximab was first deglycosylated with Endo-S2 to give the Fucα1,6 GlcNAc-rituximab intermediate [34]. Then Endo-S2 mutant D184M from Streptococcus pyogenes was used for examining the transglycosylation with the sulfated N-glycan oxazolines, as the D184M mutant has shown excellent activity to transfer different types of N-glycans [34]. We found that all the three sulfated N-glycan oxazolines acted as excellent substrates of the D184M mutant to give the corresponding sulfated glycoforms of rituximab (9, 10, and 11), respectively, in high yields (Scheme 3). Interestingly, an attempt to perform direct sulfation of the terminal GlcNAc moiety in the G0F antibody glycoform (12) by CHST2 failed to give detectable sulfated antibody, as monitored by LC-MS analysis (Fig. S8), although the corresponding free N-glycan served as an excellent substrate of human CHST2. These results suggest that other human GlcNAc-6-O-sulfotransferases may be involved in the biosynthesis of such sulfated antibody glycoforms in vivo.
Scheme 3.
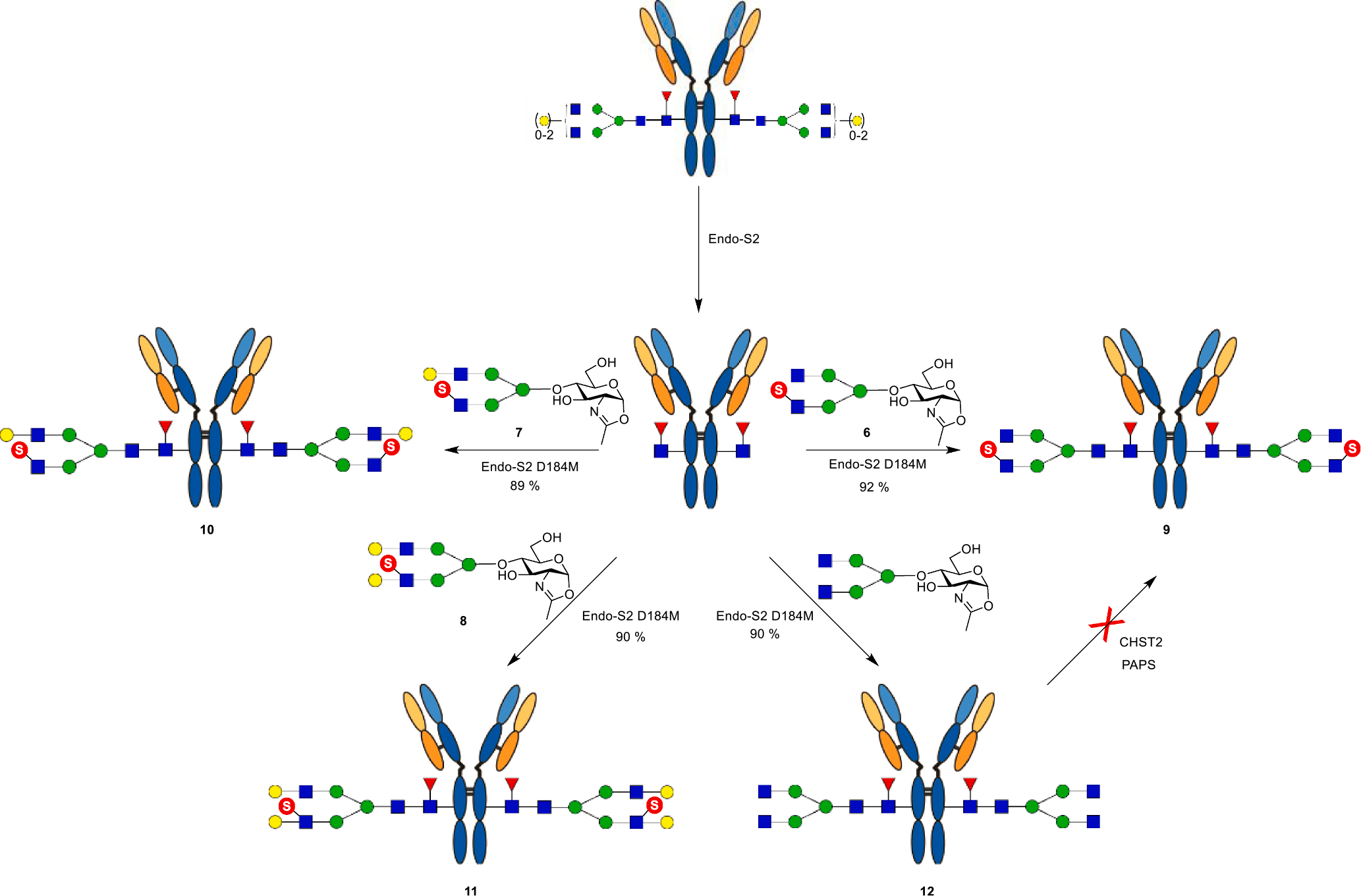
Chemoenzymatic synthesis of sulfated glycoforms of rituximab.
The antibody glycoforms were isolated by protein A affinity chromatography and their identity was confirmed by LC-ESI-MS analysis (Fig. 1). Calcd for intact antibody 9, M = 147234 Da, found, 147236 Da; calcd for intact sulfated antibody 10, M = 147558 Da, found, 147560 Da; calcd for intact sulfated antibody 11, M = 147882 Da, found, 147884 Da; calcd for antibody 12, M = 147076 Da, found, 147074 Da. All data were deconvoluted. To further confirm the structural homogeneity, the glycoengineered antibodies were treated with protease IdeS which can specifically digest immunoglobulin G to generate the monomeric Fc domain. The product Fc domain was analysed by LC-ESI-MS (Fig. S9), which indicate that the sulfated N-glycans have been transferred specifically to the Fc domain of antibody rituximab.
Fig. 1.
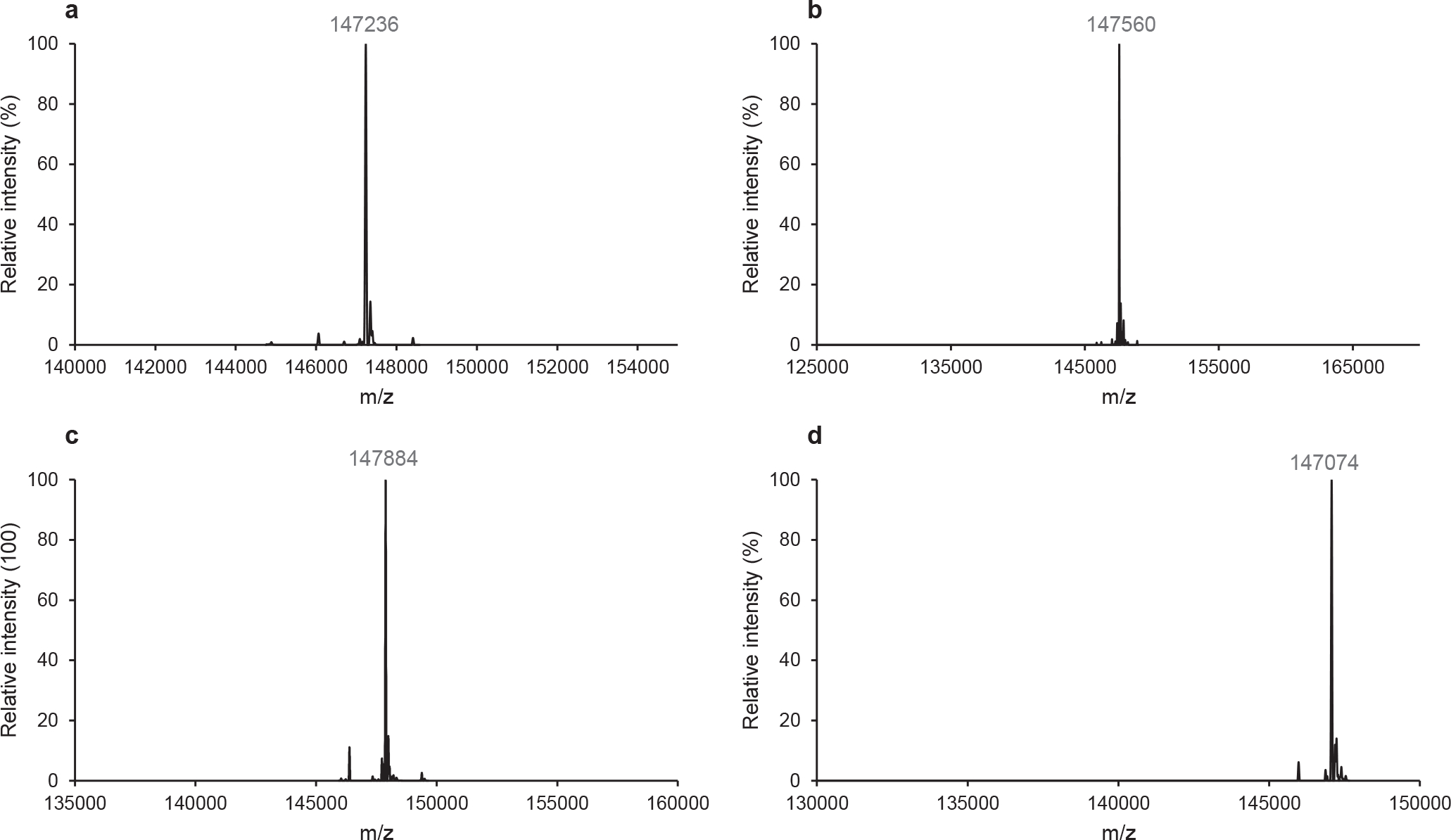
LC-ESI-MS analysis of glycoengineered rituximab. (a) Intact antibody 9. (b) Intact antibody 10. (c) Intact antibody 11. (d) Intact antibody 12. The spectra are the deconvoluted spectra.
2.4. Binding of the sulfated glycoforms of antibody with Fcγ receptor FcγRIIIa-V158
Next, we investigated the impact of sulfate group on the binding between glycans on antibody rituximab and Fc receptors. Surface plasmon resonance (SPR) analysis was utilized to assess the binding affinity of FcγRIIIa-V158 to sulfated glycoforms and corresponding non-sulfated glycoforms. Rituximab with different glycoforms were site-specifically immobilized on the CM5 sensor chip which was pre-functionalised with protein A. Different concentrations of FcγRIIIa-V158 were injected as analyte following the procedures reported previously [35]. The SPR sensorgrams for the interaction between different glycoforms of rituximab and FcγRIIIa-V158 were shown in Fig. 2. The dissociation constant (KD) between the binding of rituximab with various glycoforms to FcγRIIIa-V158 was estimated to be 198 nM (G0F-rituximab), 172 nM (SulfG0F-rituximab), 108 nM (G2F-rituximab) and 92 nM (SulfG2F-rituximab), respectively. Apparently, the complex-type glycoforms (G2F-rituximab, G0F-rituximab) showed the similar binding affinity compared to the corresponding sulfated glycoforms. Furthermore, the KD value of the binding between sulfated G1F-rituximab and FcγRIIIa-V158 was also obtained, which was 84 nM (Fig. S10). These results suggest that the introduction of a sulfate group at the lower arm GlcNAc moiety of the Fc glycans does not significantly affect the affinity of the antibody for the Fcγ receptor FcγRIIIa-V158. The present study represents the first in vitro enzymatic sulfation of full-length N-glycans by the human sulfotransferase CHST2. An expanded chemoenzymatic synthesis of diverse sulfated N-glycans using other sulfotransferases and the evaluation of the binding of the synthetic sulfated N-glycans with different lectins are currently underway in our laboratory and the results will be reported in due course.
Fig. 2.
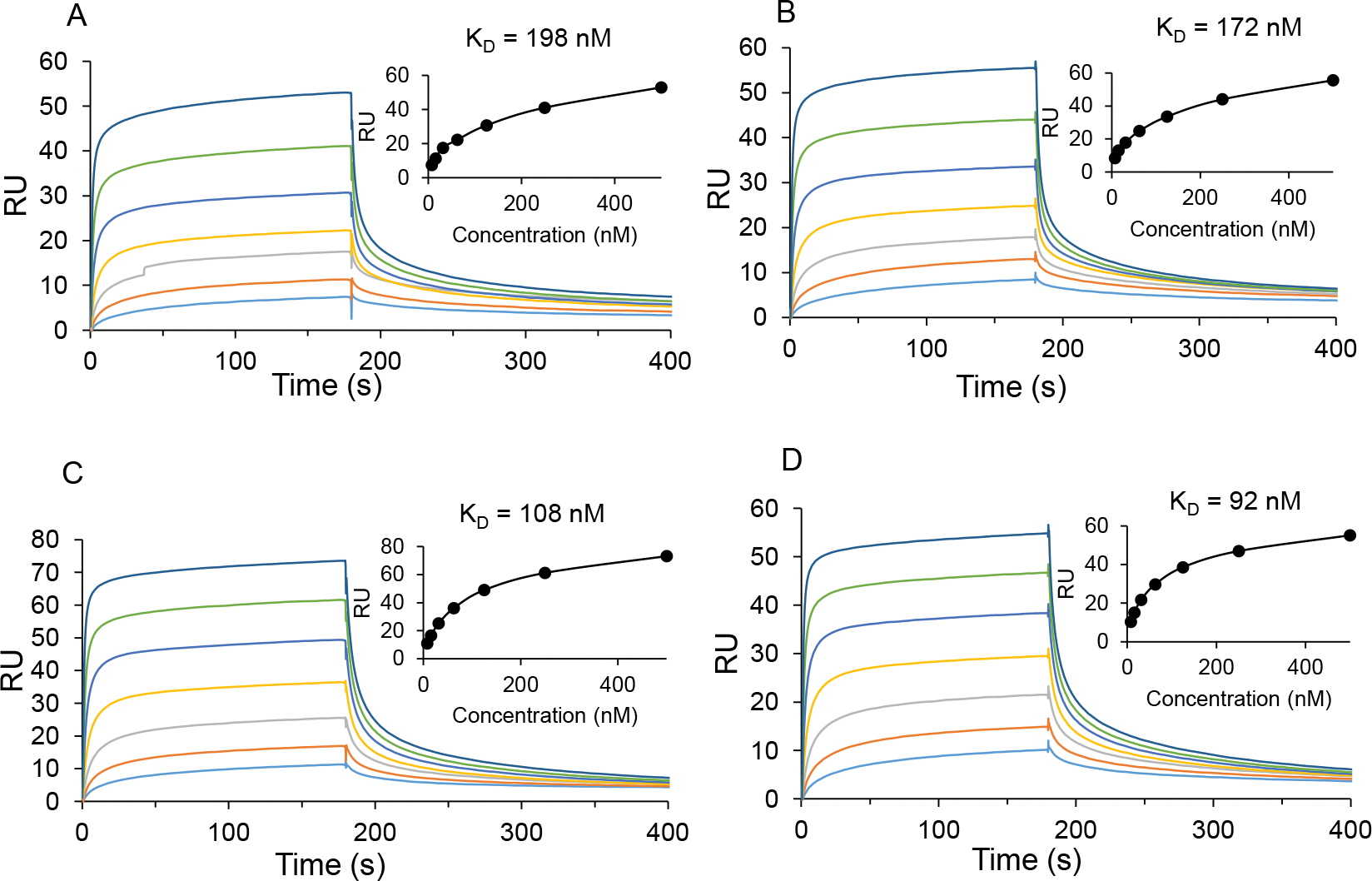
SPR sensorgrams of the binding of FcγRIIIa-V158 with different glycoforms of rituximab. A) G0F-rituximab (12); B) Sulfated G0F-rituximab (9); C) G2F-rituximab; D) Sulfated G2F-rituximab (11).
3. Conclusion
The in vitro enzymatic sulfation of biantennary complex type N-glycans by recombinant human GlcNAc-6-O-sulfotransferase 1 (CHST2) and the first chemoenzymatic synthesis of sulfated antibody glycoforms are described. The high site-selectivity of sulfation at the lower arm of N-glycans by human sulfotransferase CHST2 is an unexpected discovery. The availability of structurally well-defined sulfated N-glycans and sulfated glycoforms of antibodies provides an exciting opportunity to investigate their biological functions. The next step is to expand this approach to evaluation of other sulfotransferases for the construction of a library of specific N-glycans and antibody glycoforms with diverse sulfation patterns, which will be highly valuable for structural and functional studies.
4. Experimental section
4.1. Materials
The Endo-S2 and Endo-S2 D184M glycosynthase mutant were prepared by our lab as described previously [34]. The plasmids bearing human CHST2 and B4GALT4 genes were generated as described previously [45]. PAPS was kindly provided by Prof. Jian Liu (the University of North Carolina, Chapel Hill, US). Bacterial β1,4-galactosyltransferase from Neisseria meningitidis was expressed following the established procedure [46]. Polyethylenimine (PEI), Fmoc-Cl, formic acid and acetonitrile were purchased from Sigma-Aldrich. E.coli DH5α, the α-1,3 mannosidase, the β-N-acetylglucosaminidase S, and the β1,4-galactosidase were purchased from New England Biolab. The sialidase from Micromonospara Viridifaciens was expressed and purified following the previously described procedure [41]. 1 ml HisTrap™ FF column was purchased from GE Healthcare. NucleoBond®Xtra Midi EF kit was bought from MACHEREY-NAGEL.
4.2. Analytical methods
HPLC:
Analytical RP-HPLC was carried out on a Thermo Scientific instrument with a YMC C18 column (5 μm, 4.6 × 250 mm). The column was eluted with a linear gradient of 10–50 % acetonitrile with 0.1 % formic acid (v/v) for 30 min at a flow rate of 1 ml/min under UV 266 nm. The retention time of sulfated N-glycans (2, 3, 5) were 23.5 min, 22.6 min, and 22.2 min, respectively. Reverse phase preparative HPLC was executed on a Waters 600 HPLC instrument with a SymmetryPrep C18 column (7 μm, 19 × 300 mm). The column was eluted with a linear gradient of 20–60 % acetonitrile with 0.1 % formic acid (v/v) for 30 min at a flow rate of 10 ml/min under UV 266 nm.
LC-ESI-MS:
The core-fucosylated antibodies were analysed by liquid chromatography electrospray mass spectrometry (LC-ESI-MS). LC-ESI-MS was carried out on an Exactive Plus Orbitrap Mass Spectrometry (Thermo Scientific) with a C8 column (5 μm, 1.0 × 75 mm) for IdeS digested antibody and C4 column (3.5 μm, 2.1 × 50 mm) for intact antibody analysis.
HRMS:
High resolution mass spectrometry was executed on an Exactive Plus Orbitrap Mass Spectrometry (Thermo Scientific).
NMR:
1H, 13C, 1H–1H COSY and 1H–13C HSQC NMR spectra were recorded on a 600 MHz spectrometer (Bruker, Tokyo, Japan) with D2O as the solvent containing 1 % (v/v) DMSO-d6 as an inner standard.
4.3. Enzyme expression and purification of human CHST2 and B4GALT4
The coding region of human CHST2 and B4GALT4 genes were inserted into pGEn2 vector, which were utilized for HEK293F (ThermoFisher) cell transient expression as described previously [45,47]. For the purification, the culture supernatant was collected and used for Ni2+-NTA column purification. The column was equilibrated with the binding buffer (Tris 50 mM, NaCl 500 mM, imidazole 10 mM, pH 7.5). The culture supernatant was loaded and washed with 20 times column volumes of washing buffer (Tris 50 mM, NaCl 500 mM, Imidazole 20 mM, pH 7.5), and eluted with a buffer containing Tris 50 mM, NaCl 500 mM, imidazole 300 mM, pH 7.5. The fractions containing proteins were collected and buffer exchanged using buffer containing HEPES 20 mM, NaCl 100 mM, pH 7.0, sodium azide 0.05 %, and glycerol 10 %. The protein concentrations were measured by Nanodrop. The two enzymes were analysed by SDS-PAGE (Fig. S11). The aliquots of the two enzymes were stored in −80 °C freezer until use.
4.4. The sulfation position determination
A reaction mixture composed of monosulfated G1-Asn-Fmoc (100 μg), Glycobuffer 1 (CaCl2 5 mM, sodium acetate 50 mM), β-N-acetylglucosaminidase S was incubated at 37° C. The product was analysed by MALDI-ToF. When the sulfated N-acetylglucosamine was hydrolysed completely, BSA (100 μg) and α1–2,3 mannosidase (32 U) were added to the reaction mixture. The product was analysed by MALDI-ToF (Fig. S1–3).
4.5. Enzymatic synthesis of monosulfated G0-Asn-Fmoc
In total, 550 μl reaction mixture composed of G0-Asn-Fmoc (11 mg, 6.6 μmole), MES buffer (50 mM, pH 6), MgCl2 (5 mM), PAPS (10 μmole), NaF (10 mM), ATP (1 mM) and human CHST2 (221 μg) was incubated at 37 °C. The reaction was monitored by HPLC. After the reaction was completed, the reaction was quenched by heating at 95 °C for 5 min. The product was purified by preparative HPLC. After lyophilisation, the product (9.1 mg, 80 %) was obtained as white powder. The compound was characterised by NMR and HRMS. 1H NMR (D2O + 1 % DMSO-d6, 600 MHz): δ = 7.73 (m, 2H, FmocH), 7.54 (m, 2H, FmocH), 7.34 (m, 2H, FmocH), 7.29 (m, 2H, FmocH), 5.04 (m, 1H), 4.91 (d, J = 9.6 Hz, 1H), 4.83 (m, 1H), 4.50–4.30 (m, 7H), 4.26 (d, J = 10.8 Hz, 1H), 4.15–3.40 (m, 55H, rest protons on the sugar rings), 1.97–1.80 (m, 12H, –COCH3). 13C NMR (D2O + 1 % DMSO-d6, 151 MHz): δ = 174.3, 174.25, 174.16, 157.9, 143.3, 140.5, 127.6, 127.0, 124.7, 119.7, 100.7, 99.8, 99.3, 99.2, 99.1, 96.6, 80.1, 79.2, 78.3, 77.8, 76.3, 76.0, 75.8, 75.4, 73.9, 73.3, 73.1, 73.0, 72.7, 72.4, 72.2, 71.5, 69.7, 69.5, 69.1, 69.0, 66.9, 66.8, 66.6, 66.1, 65.6, 65.3, 61.2, 61.1, 59.5, 54.9, 54.8, 54.5, 53.4, 50.0, 46.5, 36.8, 21.9, 21.8, 21.5. HRMS (ESI) m/z [M + H]+ calcd for C69H101N6O43S+: 1733.5616 found: 1733.5661.
4.6. Enzymatic synthesis of monosulfated G1-Asn-Fmoc
In total, 260 μl reaction mixture containing monosulfated G0-Asn-Fmoc (5.22 mg, 3 μmol), Tris buffer (50 mM, pH 7.5), MnCl2 (10 mM), UDP-Gal (3.9 μmol), CIAP (5 U) and NmLgtB (109 μg) was incubated at 37 °C. The reaction was monitored by HPLC. After the reaction was completed, the reaction was quenched by heating at 95 °C for 5 min. The product (5.1 mg, 90 %) was purified by preparative HPLC, which was subsequently lyophilised as white powder. The compound was characterised by NMR and HRMS. 1H NMR (D2O + 1 % DMSO-d6, 600 MHz): δ = 7.78 (m, 2H, FmocH), 7.57 (m, 2H, FmocH), 7.37 (m, 2H, FmocH), 7.31 (m, 2H, FmocH), 5.03 (m, 1H), 4.90 (d, J = 9.6 Hz, 1H), 4.83 (m, 1H), 4.50–4.01 (m, 11H), 3.81 (d, J = 10.2 Hz, 1H), 3.68–3.41 (m, 45H, rest protons on the sugar rings), 1.98–1.95 (m, 12H, –COCH3). 13C NMR (D2O + 1 % DMSO-d6, 151 MHz): δ = 174.2, 174.1, 143.3, 140.5, 127.6, 127.0, 124.6, 119.7, 102.5, 100.8, 99.9, 99.3, 99.1, 96.6, 80.0, 79.1, 78.3, 78.1, 77.8, 76.3, 75.9, 75.8, 74.9, 74.3, 73.9, 73.3, 73.1, 72.8, 72.4, 72.2, 72.1, 71.7, 71.5, 70.5, 69.7, 69.1, 68.1, 66.9, 66.8, 66.5, 66.0, 65.5, 65.3, 61.2, 60.6, 59.6, 54.8, 54.5, 54.4, 53.4, 49.9, 46.5, 36.7, 21.9. HRMS (ESI) m/z [M + H]+ calcd for C75H111N6O48S+: 1895.6144 found: 1895.6190.
4.7. Enzymatic synthesis of monosulfated G2-Asn-Fmoc
In total, 260 μl reaction mixture composed of monosulfated G0-Asn-Fmoc (5.14 mg, 3 μmol), Tris buffer (100 mM, pH 7.5), MnCl2 (10 mM), UDP-Gal (6.9 μmol), CIAP (5 U) and human B4GALT4 (257 μg) was incubated at 37 °C. The reaction was monitored by HPLC. After the reaction was completed, the reaction was quenched by heating at 95 °C for 5 min. The product (5.6 mg, 90 %) was purified by preparative HPLC, which was obtained as white powder after lyophilisation. The compound was characterised by HRMS and NMR. 1H NMR (D2O + 1 % DMSO-d6, 600 MHz): δ = 7.79 (m, 2H, FmocH), 7.57 (m, 2H, FmocH), 7.38 (m, 2H, FmocH), 7.31 (m, 2H, FmocH), 5.03 (m, 1H), 4.91 (m, 1H), 4.83 (m, 1H), 4.45–4.10 (m, 12H), (d, J = 1.8 Hz, 1H), 3.83–3.41 (m, 65H, rest protons on the sugar rings), 1.98–1.80 (m, 12H, –COCH3). 13C NMR (D2O + 1 % DMSO-d6, 151 MHz): δ = 174.3, 174.2, 172.0, 143.2, 140.5, 127.6, 127.0, 124.6, 119.7, 102.5, 102.1, 100.8, 99.9, 99.1, 98.9, 96.6, 80.0, 79.0, 78.2, 78.1, 77.7, 77.0, 76.3, 75.9, 75.7, 74.8, 74.2, 73.9, 73.1, 72.4, 72.2, 71.6, 71.5, 71.4, 70.5, 69.7, 68.9, 68.1, 68.0, 66.9, 66.8, 66.0, 65.8, 65.4, 65.2, 61.2, 61.1, 60.5, 59.5, 54.5, 54.4, 54.3, 53.9, 49.9, 46.5, 21.9. HRMS (ESI) m/z [M + H]+ calcd for C81H121N6O53S +: 2057.6673 found: 2057.6723.
4.8. Synthesis of sugar oxazoline of monosulfated G0, G1 and G2 N-glycans
A solution containing monosulfated G0-Asn-Fmoc (11 mg) or monosulfated G1-Asn-Fmoc (5 mg) or monosulfated G2-Asn-Fmoc (5.9 mg) and Endo-S2 (300 μg) in PBS buffer (pH7.4) was incubated at 37 °C. The reaction was monitored by LC-MS. When no substrate was found in LC-MS, the reaction was quenched by heating at 95 °C for 5 min. Thereafter, the mixture was spun down at 12247 g for 5 min at room temperature. The supernatant was subsequently loaded on G15 column. The fractions containing N-glycans were pooled together and lyophilised. A solution composed of monosulfated glycans hydrolysed by Endo-S2, triethylamine (40 eq) and 2-chloro-1,3-dimethylimidazolinium chloride (20 eq) was incubated on ice for 30 min. Thereafter, the solution was loaded on size exclusion chromatography G10 column. The fractions containing glycans were collected and lyophilised. The sugar oxazolines were characterised by 1H NMR and HRMS. For monosulfated G0 oxazoline (6): 1H NMR (400 MHz, D2O) δ 5.98 (d, 1H, J = 7.28 Hz, H-1-GlcNAc-ox), 5.02 (s, 1H), 4.85–4.81 (m, 1H), 4.61 (s, 1H), 4.51–4.46 (m, 2H), 4.29–4.21 (m, 2H), 4.17–4.02 (m, 5H), 3.85–3.24 (m, 29H), 1.97–1.93 (m, 9H). HRMS (ESI) m/z [M + H]+ calcd for C42H70N3O33S+: 1176.3607, found: 1176.3529. For monosulfated G1 oxazoline (7): 1H NMR (600 MHz, D2O) δ 5.96 (d, 1H, J = 7.26 Hz, H-1-GlcNAc-ox), 5.00 (s, 1H), 4.83 (s, 1H), 4.62 (s, 1H), 4.50–4.44 (m, 2H), 4.34 (d, J = 7.86 Hz, 1H), 4.28–4.20 (m, 2H), 4.15–4.01 (m, 6H), 3.89–3.35 (m, 34H), 1.96–1.91 (m, 9H). HRMS (ESI) m/z [M + H]+ calcd for C48H80N3O38S+: 1338.4135, found: 1338.4068. For monosulfated G2 oxazoline (8): 1H NMR (600 MHz, D2O) δ 5.96 (d, 1H, J = 7.26 Hz, H-1-GlcNAc-ox), 5.00 (s, 1H), 4.84–4.79 (m, 1H), 4.62 (s, 1H), 4.52–4.45 (m, 2H), 4.41 (d, 1H, J = 7.8 Hz, 1H), 4.34 (d,1H, J = 7.86 Hz, 1H), 4.31–4.19 (m, 3H), 4.15–3.97 (m, 6H), 3.89–3.26 (m, 39H), 1.96–1.90 (m, 9H). HRMS (ESI) m/z [M + H]+ calcd for C54H90N3O43S+: 1500.4663, found: 1500.4611.
4.9. Enzymatic synthesis of rituximab with sulfated N-Glycans
Synthesis of 9.
The reaction mixture containing Fucα1,6 GlcNAc-rituximab (1.28 mg), sugar oxazoline of 6 (750 μg) and glycosynthase mutant Endo-S2 D184M (40 μg) in 1 × PBS buffer (pH7.4) was incubated at 30 °C. The reaction was monitored by Thermo LC-ESI-MS. After the reaction was completed, the product of 9 (1.19 mg, 92 %) was purified by protein A chromatography. LC-ESI-MS analysis: calcd for intact antibody of 9, M = 147234 Da, found, 147236 Da; after IdeS digestion of 9, LC-ESI-MS calcd for the monomeric Fc part 25283 Da, found, 25279 Da (deconvolution data).
Synthesis of 10.
The reaction mixture containing Fucα1,6 GlcNAc-rituximab (1.3 mg), sugar oxazoline of 7 (750 μg) and glycosynthase mutant Endo-S2 D184M (40 μg) in 1 × PBS buffer (pH7.4) was incubated at 30 °C. The reaction was monitored by Thermo LC-ESI-MS. After the reaction was completed, the product of 10 (1.18 mg, 89 %) was purified by protein A chromatography. LC-ESI-MS analysis: calcd for intact antibody of 10, M = 147558 Da, found, 147560 Da; after IdeS digestion of 10, LC-ESI-MS calcd for the monomeric Fc part 25445 Da, found, 25441 Da (deconvolution data).
Synthesis of 11.
The reaction mixture containing Fucα1,6 GlcNAc-rituximab (1.3 mg), sugar oxazoline of 8 (750 μg) and glycosynthase mutant Endo-S2 D184M (40 μg) in 1 × PBS buffer (pH7.4) was incubated at 30 °C. The reaction was monitored by Thermo LC-ESI-MS. After the reaction was completed, the product of 11 (1.19 mg, 90 %) was purified by protein A chromatography. LC-ESI-MS analysis: calcd for intact antibody of 11, M = 147882 Da, found, 147884 Da; after IdeS digestion of 11, LC-ESI-MS calcd for the monomeric Fc part 25607 Da, found, 25603 Da (deconvolution data).
Synthesis of 12.
The reaction mixture containing Fucα1,6 GlcNAc-rituximab (900 μg), the G0 N-glycan oxazoline [48] (750 μg), and glycosynthase mutant Endo-S2 D184M [34] (40 μg) in 1 × PBS buffer (pH7.4) were incubated at 30 °C. The reaction was monitored by Thermo LC-ESI-MS. After the reaction was completed, the product of 12 (822 μg, 90 %) was purified by protein A chromatography. LC-ESI-MS analysis: calcd for intact antibody of 12, M = 147076 Da, found, 147074 Da; after IdeS digestion of 12, LC-ESI-MS calcd for the monomeric Fc domain, M = 25204 Da, found, 25199 Da (deconvolution data).
4.10. Enzymatic activity of CHST2 with rituximab G0F glycoform (12).
The reaction mixture containing the rituximab G0F glycoform (12) (391 μg), MgCl2 (5 mM), PAPS (1 mM), NaF (10 mM), ATP (1 mM) and CHST2 (5.2 μg) in MES buffer (100 mM, pH 6) was incubated at 37 °C. The reaction was monitored by LC-ESI-MS. No enzymatic sulfation of the sulfation was observed after incubation for 16 h at 37 °C (Fig. S8).
4.11. Surface plasmon resonance assays
Biacore T200 (GE Healthcare, USA) surface plasmon resonance (SPR) was utilised to determine the binding affinity between various glycoforms of rituximab and FcγRIIIa-V158. To capture a variety of rituximab glycoforms, a standard primary amine coupling chemistry at pH4.5 was utilised to immobilise protein A on a CM5 biosensor chip (GE Healthcare, USA). A flow cell prepared with BSA was used as reference. Each rituximab glycoform in HBS-P buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 0.05 % surfactant P20) was injected individually at 10 μl/min. The antibodies were captured at 200 RU for FcγRIIIa-V158. 2 × serial dilutions of FcγRIIIa receptor in HBS-P buffer was injected at 30 μl/min. After each cycle, the surface was regenerated by injecting glycine HCl buffer (10 mM, pH 2.0). To obtain the equilibrium constant KD, the experimental data was fitted into a 1:1 Langmuir binding model utilising the BIAcore T200 evaluation software (GE Healthcare).
Supplementary Material
Acknowledgements
We thank Prof. Jian Liu (The University of North Carolina) for providing 3’-phosphoadenosine-5’-phosphosulfate (PAPS) and members of the Wang lab for technical assistance and helpful discussions. We thank Mr. Chin Huang (University of Georgia) for expressing the CHST2 enzyme. This work was supported by the National Institutes of Health (NIH grants R01 AI155716 to L.X.W and R01 GM130915 to K.W.M.).
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioorg.2022.106070.
Data availability
All the data and experimental procedures are described in the main text and the supporting information.
References
- [1].Klaassen CD, Boles JW, Sulfation and sulfotransferases 5: the importance of 3’-phosphoadenosine 5’-phosphosulfate (PAPS) in the regulation of sulfation, FASEB J. 11 (6) (1997) 404–418. [DOI] [PubMed] [Google Scholar]
- [2].Wang JR, Gao WN, Grimm R, Jiang S, Liang Y, Ye H, Li ZG, Yau LF, Huang H, Liu J, Jiang M, Meng Q, Tong TT, Huang HH, Lee S, Zeng X, Liu L, Jiang ZH, A method to identify trace sulfated IgG N-glycans as biomarkers for rheumatoid arthritis, Nat. Commun. 8 (2017) 631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].She Y-M, Li X, Cyr TD, Remarkable Structural Diversity of N-Glycan Sulfation on Influenza Vaccines, Anal. Chem. 91 (8) (2019) 5083–5090. [DOI] [PubMed] [Google Scholar]
- [4].Wang W, Hu T, Frantom PA, Zheng T, Gerwe B, Del Amo DS, Garret S, Seidel RD 3rd, Wu P, Chemoenzymatic synthesis of GDP-L-fucose and the Lewis X glycan derivatives, Proc. Natl. Acad. Sci. USA 106 (2009) 16096–16101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ichimiya T, Nishihara S, Takase-Yoden S, Kida H, Aoki-Kinoshita K, Frequent glycan structure mining of influenza virus data revealed a sulfated glycan motif that increased viral infection, Bioinformatics 30 (5) (2014) 706–711. [DOI] [PubMed] [Google Scholar]
- [6].Gambaryan AS, Tuzikov AB, Pazynina GV, Webster RG, Matrosovich MN, Bovin NV, H5N1 chicken influenza viruses display a high binding affinity for Neu5Acalpha2–3Galbeta1–4(6-HSO3)GlcNAc-containing receptors, Virology 326 (2004) 310–316. [DOI] [PubMed] [Google Scholar]
- [7].Mitoma J, Bao X, Petryanik B, Schaerli P, Gauguet J-M, Yu S-Y, Kawashima H, Saito H, Ohtsubo K, Marth JD, Khoo K-H, von Andrian UH, Lowe JB, Fukuda M, Critical functions of N-glycans in L-selectin-mediated lymphocyte homing and recruitment, Nat. Immunol. 8 (4) (2007) 409–418. [DOI] [PubMed] [Google Scholar]
- [8].Hemmerich S, Handbook of Neurochemistry and Molecular Neurobiology, in: Lajtha A, Banik N (Eds.), Handbook of Neurochemistry and Molecular Neurobiology, Springer US, Boston, MA, 2007, pp. 283–302, 10.1007/978-0-387-30379-6_9. [DOI] [Google Scholar]
- [9].Yoshimura T, Hayashi A, Handa-Narumi M, Yagi H, Ohno N, Koike T, Yamaguchi Y, Uchimura K, Kadomatsu K, Sedzik J, Kitamura K, Kato K, Trapp BD, Baba H, Ikenaka K, GlcNAc6ST-1 regulates sulfation of N-glycans and myelination in the peripheral nervous system, Sci. Rep. 7 (2017) 42257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].van Rooijen JJM, Kamerling JP, Vliegenthart JFG, Sulfated di-, tri- and tetraantennary N-glycans in human Tamm-Horsfall glycoprotein, Eur. J. Biochem. 256 (2) (1998) 471–487. [DOI] [PubMed] [Google Scholar]
- [11].Toma G, Bates JM, Kumar S, Uromodulin (Tamm-Horsfall protein) is a leukocyte adhesion molecule, Biochem. Biophys. Res. Commun. 200 (1) (1994) 275–282. [DOI] [PubMed] [Google Scholar]
- [12].Sathyamoorthy N, Decker JM, Sherblom AP, Muchmore A, Evidence that specific high mannose structures directly regulate multiple cellular activities, Mol. Cell. Biochem. 102 (1991) 139–147. [DOI] [PubMed] [Google Scholar]
- [13].Kamerling JP, Rijkse I, Maas AA, van Kuik JA, Vliegenthart JF, Sulfated N-linked carbohydrate chains in porcine thyroglobulin, FEBS Lett. 241 (1988) 246–250. [DOI] [PubMed] [Google Scholar]
- [14].Kawasaki N, Haishima Y, Ohta M, Itoh S, Hyuga M, Hyuga S, Hayakawa T, Structural analysis of sulfated N-linked oligosaccharides in erythropoietin, Glycobiology 11 (12) (2001) 1043–1049. [DOI] [PubMed] [Google Scholar]
- [15].Yamashita K, Ueda I, Kobata A, Sulfated asparagine-linked sugar chains of hen egg albumin, J. Biol. Chem. 258 (23) (1983) 14144–14147. [PubMed] [Google Scholar]
- [16].Freeze HH, Wolgast D, Structural analysis of N-linked oligosaccharides from glycoproteins secreted by Dictyostelium discoideum. Identification of mannose 6-sulfate, J. Biol. Chem. 261 (1) (1986) 127–134. [PubMed] [Google Scholar]
- [17].Bergwerff AA, Oostrum J, Kamerling JP, Vliegenthart JFG, The major N-linked carbohydrate chains from human urokinase. The occurrence of 4-O-sulfated, (alpha 2–6)-sialylated or (alpha 1–3)-fucosylated N-acetylgalactosamine(beta 1–4)-N-acetylglucosamine elements, Eur. J. Biochem. 228 (3) (1995) 1009–1019. [DOI] [PubMed] [Google Scholar]
- [18].Wedepohl S, Kaup M, Riese SB, Berger M, Dernedde J, Tauber R, Blanchard V, N-glycan analysis of recombinant L-Selectin reveals sulfated GalNAc and GalNAc-GalNAc motifs, J. Proteome Res. 9 (7) (2010) 3403–3411. [DOI] [PubMed] [Google Scholar]
- [19].Gao T, Yan J, Liu C-C, Palma AS, Guo Z, Xiao M, Chen X.i., Liang X, Chai W, Cao H, Chemoenzymatic Synthesis of O-Mannose Glycans Containing Sulfated or Nonsulfated HNK-1 Epitope, J. Am. Chem. Soc. 141 (49) (2019) 19351–19359. [DOI] [PubMed] [Google Scholar]
- [20].Xu Z, Deng Y, Zhang Z, Ma W, Li W, Wen L, Li T, Diversity-Oriented Chemoenzymatic Synthesis of Sulfated and Nonsulfated Core 2 O-GalNAc Glycans, J. Org. Chem. 86 (15) (2021) 10819–10828. [DOI] [PubMed] [Google Scholar]
- [21].Santra A, Yu H, Tasnima N, Muthana MM, Li Y, Zeng J, Kenyon NJ, Louie AY, Chen X.i., Systematic Chemoenzymatic Synthesis of O-Sulfated Sialyl Lewis x Antigens, Chem. Sci. 7 (4) (2016) 2827–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Honke K, Yamane M, Ishii A, Kobayashi T, Makita A, Purification and characterization of 3’-phosphoadenosine-5’-phosphosulfate:GalCer sulfotransferase from human renal cancer cells, J. Biochem. 119 (3) (1996) 421–427. [DOI] [PubMed] [Google Scholar]
- [23].El-Fasakhany FM, Uchimura K, Kannagi R, Muramatsu T, A novel human Gal-3-O-sulfotransferase: molecular cloning, characterization, and its implications in biosynthesis of (SO(4)-3)Galbeta1–4(Fucalpha1–3)GlcNAc, J. Biol. Chem. 276 (29) (2001) 26988–26994. [DOI] [PubMed] [Google Scholar]
- [24].Suzuki A, Hiraoka N, Suzuki M, Angata K, Misra AK, McAuliffe J, Hindsgaul O, Fukuda M, Molecular cloning and expression of a novel human beta-Gal-3-O-sulfotransferase that acts preferentially on N-acetyllactosamine in N- and O-glycans, J. Biol. Chem. 276 (2001) 24388–24395. [DOI] [PubMed] [Google Scholar]
- [25].Chandrasekaran EV, Lakhaman SS, Chawda R, Piskorz CF, Neelamegham S, Matta KL, Identification of physiologically relevant substrates for cloned Gal: 3-O-sulfotransferases (Gal3STs): distinct high affinity of Gal3ST-2 and LS180 sulfotransferase for the globo H backbone, Gal3ST-3 for N-glycan multiterminal Galbeta 1, 4GlcNAcbeta units and 6-sulfoGalbeta1, 4GlcNAcbeta, and Gal3ST-4 for the mucin core-2 trisaccharide, J. Biol. Chem. 279 (11) (2004) 10032–10041. [DOI] [PubMed] [Google Scholar]
- [26].Seko A, Hara-Kuge S, Yamashita K, Molecular cloning and characterization of a novel human galactose 3-O-sulfotransferase that transfers sulfate to gal beta 1–>3galNAc residue in O-glycans, J. Biol. Chem. 276 (2001) 25697–25704. [DOI] [PubMed] [Google Scholar]
- [27].Hiraoka N, Misra A, Belot F, Hindsgaul O, Fukuda M, Molecular cloning and expression of two distinct human N-acetylgalactosamine 4-O-sulfotransferases that transfer sulfate to GalNAc beta 1–>4GlcNAc beta 1–>R in both N- and O-glycans, Glycobiology 11 (2001) 495–504. [DOI] [PubMed] [Google Scholar]
- [28].Grunwell JR, Bertozzi CR, Carbohydrate sulfotransferases of the GalNAc/Gal/GlcNAc6ST family, Biochemistry 41 (44) (2002) 13117–13126. [DOI] [PubMed] [Google Scholar]
- [29].Uchimura K, El-Fasakhany FM, Hori M, Hemmerich S, Blink SE, Kansas GS, Kanamori A, Kumamoto K, Kannagi R, Muramatsu T, Specificities of N-acetylglucosamine-6-O-sulfotransferases in relation to L-selectin ligand synthesis and tumor-associated enzyme expression, J. Biol. Chem. 277 (6) (2002) 3979–3984. [DOI] [PubMed] [Google Scholar]
- [30].Lee JK, Bistrup A, van Zante A, Rosen SD, Activities and expression pattern of the carbohydrate sulfotransferase GlcNAc6ST-3 (I-GlcNAc6ST): functional implications, Glycobiology 13 (2003) 245–254. [DOI] [PubMed] [Google Scholar]
- [31].Hiraoka N, Petryniak B, Nakayama J, Tsuboi S, Suzuki M, Yeh J-C, Izawa D, Tanaka T, Miyasaka M, Lowe JB, Fukuda M, A novel, high endothelial venule-specific sulfotransferase expresses 6-sulfo sialyl Lewis(x), an L-selectin ligand displayed by CD34, Immunity 11 (1) (1999) 79–89. [DOI] [PubMed] [Google Scholar]
- [32].Akama TO, Misra AK, Hindsgaul O, Fukuda MN, Enzymatic synthesis in vitro of the disulfated disaccharide unit of corneal keratan sulfate, J. Biol. Chem. 277 (45) (2002) 42505–42513. [DOI] [PubMed] [Google Scholar]
- [33].Uchimura K, Fasakhany F, Kadomatsu K, Matsukawa T, Yamakawa T, Kurosawa N, Muramatsu T, Diversity of N-acetylglucosamine-6-O-sulfotransferases: molecular cloning of a novel enzyme with different distribution and specificities, Biochem. Biophys. Res. Commun. 274 (2) (2000) 291–296. [DOI] [PubMed] [Google Scholar]
- [34].Li T, Tong X, Yang Q, Giddens JP, Wang LX, Glycosynthase Mutants of Endoglycosidase S2 Show Potent Transglycosylation Activity and Remarkably Relaxed Substrate Specificity for Antibody Glycosylation Remodeling, J. Biol. Chem. 291 (2016) 16508–16518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Li T, DiLillo DJ, Bournazos S, Giddens JP, Ravetch JV, Wang L-X, Modulating IgG effector function by Fc glycan engineering, Proc. Natl. Acad. Sci. U S A 114 (13) (2017) 3485–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ou C, Li C, Zhang R, Yang Q, Zong G, Dai Y, Francis RL, Bournazos S, Ravetch JV, Wang LX, One-Pot Conversion of Free Sialoglycans to Functionalized Glycan Oxazolines and Efficient Synthesis of Homogeneous Antibody-Drug Conjugates through Site-Specific Chemoenzymatic Glycan Remodeling, Bioconjug. Chem. 32 (2021) 1888–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhang X, Liu H, He J, Ou C, Donahue TC, Muthana MM, Su L, Wang LX, Site-Specific Chemoenzymatic Conjugation of High-Affinity M6P Glycan Ligands to Antibodies for Targeted Protein Degradation, ACS Chem. Biol. 17 (2022), 10.1021/acschembio.1c00751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sun B, Bao W, Tian X, Li M, Liu H, Dong J, Huang W, A simplified procedure for gram-scale production of sialylglycopeptide (SGP) from egg yolks and subsequent semi-synthesis of Man3GlcNAc oxazoline, Carbohydr. Res. 396 (2014) 62–69. [DOI] [PubMed] [Google Scholar]
- [39].Liu L, Prudden AR, Bosman GP, Boons GJ, Improved isolation and characterization procedure of sialylglycopeptide from egg yolk powder, Carbohydr. Res. 452 (2017) 122–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kajihara Y, Suzuki Y, Yamamoto N, Sasaki K, Sakakibara T, Juneja LR, Prompt chemoenzymatic synthesis of diverse complex-type oligosaccharides and its application to the solid-phase synthesis of a glycopeptide with Asn-linked sialyl-undeca- and asialo-nonasaccharides, Chem. Eur. J. 10 (2004) 971–985. [DOI] [PubMed] [Google Scholar]
- [41].Watson JN, Dookhun V, Borgford TJ, Bennet AJ, Mutagenesis of the conserved active-site tyrosine changes a retaining sialidase into an inverting sialidase, Biochemistry 42 (43) (2003) 12682–12690. [DOI] [PubMed] [Google Scholar]
- [42].Lau K, Thon V, Yu H, Ding L, Chen Y, Muthana MM, Wong D, Huang R, Chen X, Highly efficient chemoenzymatic synthesis of beta1–4-linked galactosides with promiscuous bacterial beta1–4-galactosyltransferases, Chem. Commun. (Camb.) 46 (2010) 6066–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bowman KG, Cook BN, de Graffenried CL, Bertozzi CR, Biosynthesis of L-selectin ligands: sulfation of sialyl Lewis x-related oligosaccharides by a family of GlcNAc-6-sulfotransferases, Biochemistry 40 (2001) 5382–5391. [DOI] [PubMed] [Google Scholar]
- [44].Wang LX, Tong X, Li C, Giddens JP, Li T, Glycoengineering of Antibodies for Modulating Functions, Annu. Rev. Biochem. 88 (2019) 433–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Moremen KW, Ramiah A, Stuart M, Steel J, Meng L.u., Forouhar F, Moniz HA, Gahlay G, Gao Z, Chapla D, Wang S, Yang J-Y, Prabhakar PK, Johnson R, Rosa MD, Geisler C, Nairn AV, Seetharaman J, Wu S-C, Tong L, Gilbert HJ, LaBaer J, Jarvis DL, Expression system for structural and functional studies of human glycosylation enzymes, Nat. Chem. Biol. 14 (2) (2018) 156–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Park JE, Lee KY, Do SI, Lee SS, Expression and characterization of beta-1,4-galactosyltransferase from Neisseria meningitidis and Neisseria gonorrhoeae, J. Biochem. Mol. Biol. 35 (2002) 330–336. [DOI] [PubMed] [Google Scholar]
- [47].Meng L.u., Forouhar F, Thieker D, Gao Z, Ramiah A, Moniz H, Xiang Y, Seetharaman J, Milaninia S, Su M, Bridger R, Veillon L, Azadi P, Kornhaber G, Wells L, Montelione GT, Woods RJ, Tong L, Moremen KW, Enzymatic basis for N-glycan sialylation: structure of rat alpha2,6-sialyltransferase (ST6GAL1) reveals conserved and unique features for glycan sialylation, J. Biol. Chem. 288 (48) (2013) 34680–34698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zong G, Li C, Prabhu SK, Zhang R, Zhang X, Wang L-X, A facile chemoenzymatic synthesis of SARS-CoV-2 glycopeptides for probing glycosylation functions, Chem. Commun. (Camb.) 57 (55) (2021) 6804–6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data and experimental procedures are described in the main text and the supporting information.


