Abstract
Background
Anti–programmed cell death 1 (anti–PD-1) and PD ligand 1 (PD-L1) immune checkpoint therapies (ICTs) provided durable responses only in a subset of cancer patients. Thus, biomarkers are needed to predict nonresponders and offer them alternative treatments. We recently implicated discoidin domain receptor tyrosine kinase 2 (DDR2) as a contributor to anti–PD-1 resistance in animal models; therefore, we sought to investigate whether this gene family may provide ICT response prediction.
Methods
We assessed mRNA expression of DDR2 and its family member DDR1. Transcriptome analysis of bladder cancer (BCa) models in which DDR1 and 2 were perturbed was used to derive DDR1- and DDR2-driven signature scores. DDR mRNA expression and gene signature scores were evaluated using BCa–The Cancer Genome Atlas (n = 259) and IMvigor210 (n = 298) datasets, and their relationship to BCa subtypes, pathway enrichment, and immune deconvolution analyses was performed. The potential of DDR-driven signatures to predict ICT response was evaluated and independently validated through a statistical framework in bladder and lung cancer cohorts. All statistical tests were 2-sided.
Results
DDR1 and DDR2 showed mutually exclusive gene expression patterns in human tumors. DDR2high BCa exhibited activation of immune pathways and a high immune score, indicative of a T-cell–inflamed phenotype, whereas DDR1high BCa exhibited a non–T-cell–inflamed phenotype. In IMvigor210 cohort, tumors with high DDR1 (hazard ratio [HR] = 1.53, 95% confidence interval [CI] = 1.16 to 2.06; P = .003) or DDR2 (HR = 1.42, 95% CI = 1.01 to 1.92; P = .04) scores had poor overall survival. Of note, DDR2high tumors from IMvigor210 and CheckMate 275 (n = 73) cohorts exhibited poorer overall survival (HR = 1.56, 95% CI = 1.20 to 2.06; P < .001) and progression-free survival (HR = 1.77 95%, CI = 1.05 to 3.00; P = .047), respectively. This result was validated in independent cancer datasets.
Conclusions
These findings implicate DDR1 and DDR2 driven signature scores in predicting ICT response.
Immune checkpoint therapies (ICTs) are emerging as an important pillar of anticancer therapy (1-3). ICTs target co-inhibitory receptors on T cells, such as programmed cell death 1 (PD-1), PD-1 ligand 1 (PD-L1), and cytotoxic T lymphocyte–associated protein 4 (1). Despite many durable clinical responses in melanoma, non-small cell lung carcinoma (NSCLC), bladder cancer (BCa), and renal cell cancer (4), many patients are nonresponders (5,6). In an attempt to stratify response, biomarkers including PD-L1 overexpression on cancer cells or immune cells, tumor mutational burden (TMB), microsatellite instability, and neoantigen load have been studied (4). Although there are associations between these biomarkers and ICT outcomes, neither PD-L1 overexpression nor TMB as a single marker can distinguish ICT responders from nonresponders (7,8). A meta-analysis of 45 studies using ICTs showed that PD-L1 overexpression in cancer or immune cells was predictive in only 28.9% cases and not predictive or not tested in the remaining 53.3% and 17.8% cases, respectively (9). These studies highlight the need to improve or identify new biomarkers for stratifying ICT responses, which is the objective of the current study.
We recently identified discoidin domain receptor 2 (DDR2) expression as a determinant of ICT response in murine models of BCa and then used genetic and pharmacologic methods to show that DDR2 inhibition can sensitize murine BCa ICT response (10). Interestingly, DDR2 inhibition alone did not influence murine BCa growth. Therefore, we reasoned that signatures composed of genes modulated by DDR2, or its family member DDR1, can possibly stratify patient response to ICT. Here, we show that scores generated from such gene signatures can stratify response to anti–PD-L1 therapy in BCa and NSCLC.
Methods
Institutional Review Board Approval
All human data were obtained from previously reported sources and references in publications.
Statistical Analysis
We performed a secondary analysis of existing published transcriptome datasets from The Cancer Genome Atlas (TCGA) and IMvigor210 studies, which are publicly available. Signature score computation using deidentified transcriptome data to validate the signature performance without direct access to identifiable patient clinical data on 2 NSCLC datasets (from authors from Caris and Tempus) and CheckMate 275 dataset (from authors from Icahn School of Medicine) was performed. A χ2 test was used to test inverse relationship of DDR1 and DDR2 (DDR1/2) gene expression in BCa. Association of clinical outcomes was assessed using Kaplan-Meier survival curves and log-rank and/or Cox statistics. To test whether DDR signature performance was prognostic independent of other clinical variables, multivariable analyses were performed adjusting for the age, sex, grade, and stage when it is available. A P value less than .05 was considered statistically significant. We used the MATLAB package including the Statistics toolbox (Mathworks, Natick, MA, USA), the R package (v.4.1 http://www.r-project.org/) for all statistical tests, and computational analysis.
Results
Gene Expression of DDR1/2 in BCa
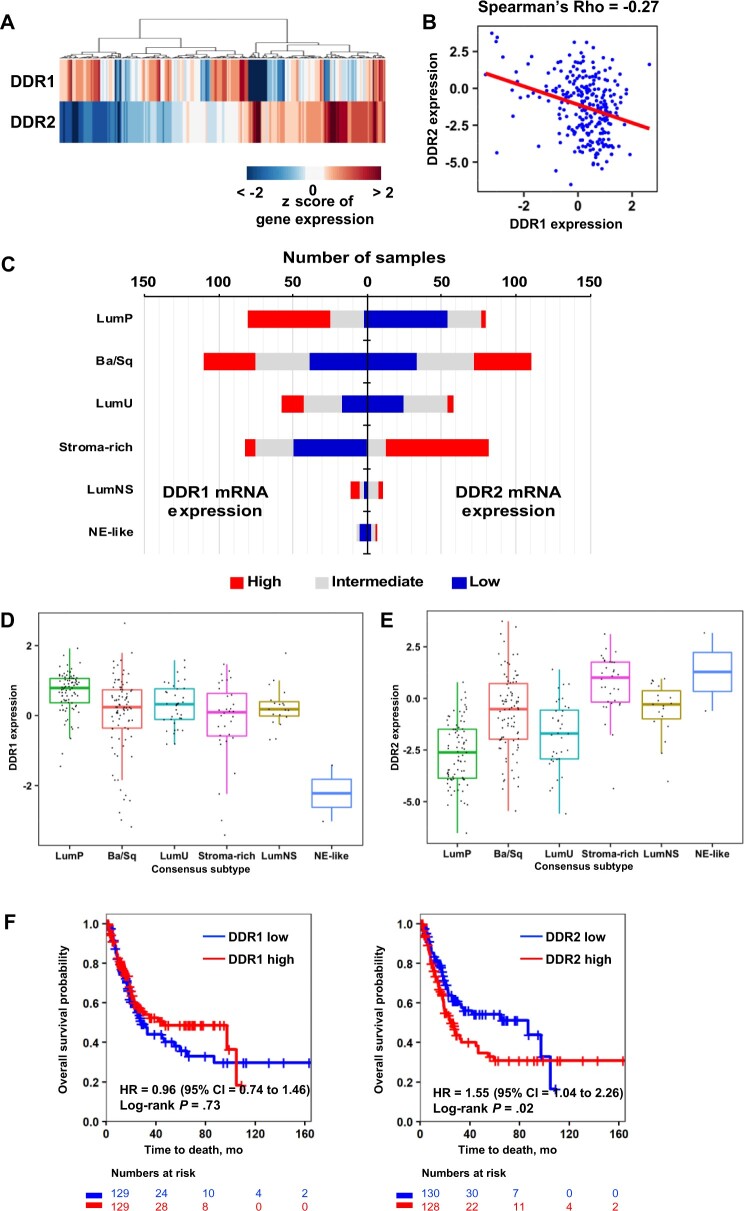
We have reported the roles of the DDR family, DDR1/2, in modulating BCa metastasis and ICT response (10,11). Unsupervised clustering of DD1/2 expression from the TCGA-BCa cohort (n = 259) (Table 1) (12) suggested that tumors could be divided into DDR1highDDR2low and DDR1lowDDR2high groups (Figure 1, A; χ2 P < .001), with DDR1/2 expression exhibiting an inverse relationship (Figure 1, B; Spearman ρ = −0.27).
Table 1.
Clinicopathological characteristics of TCGA BCa cohort by DDR expression levels (n = 259a)
| Characteristics | Level of DDR1 and DDR2 gene expression |
|||
|---|---|---|---|---|
| DDR1high and DDR2high | DDR1high and DDR2low | DDR1low and DDR2high | DDR1low and DDR2low | |
| Sex | ||||
| Female | 10 | 22 | 26 | 12 |
| Male | 39 | 58 | 54 | 38 |
| Pathologic stage | ||||
| II | 10 | 34 | 12 | 12 |
| III | 17 | 27 | 37 | 24 |
| IV | 22 | 19 | 31 | 14 |
| T stage | ||||
| T2 | 11 | 39 | 13 | 14 |
| T3 | 28 | 33 | 54 | 28 |
| T4 | 10 | 8 | 13 | 8 |
| Recurrent | ||||
| No | 17 | 40 | 28 | 18 |
| Recurrent | 2 | 10 | 5 | 5 |
| Not specified | 30 | 30 | 47 | 27 |
| Overall survival | ||||
| Living | 24 | 53 | 40 | 29 |
| Deceased | 25 | 27 | 40 | 29 |
| Histological grade | ||||
| High grade | 49 | 80 | 80 | 50 |
| Low grade | 0 | 0 | 0 | 0 |
From a total of 407 samples, 259 samples were selected by filtering with the following criteria: 1) any stage other than T2-T4; 2) 52 (13%) had urothelial carcinoma with variant histology, including 42 squamous, 4 small cell and/or neuroendocrine, 2 micropapillary, and 4 plasmacytoid; 5 additional tumors that met screening criteria were included: 3 pure squamous cell bladder carcinomas, 1 squamous cell carcinoma of nonbladder origin and 1 bladder adenocarcinoma; 3) from a total of 57 patients, 35 had received prior intravesical immunotherapy with Bacille Calmette-Guerin; 4) 12 patients had received neoadjuvant chemotherapy. BCa = bladder cancer; DDR = discoidin domain receptor; TCGA = The Cancer Genome Atlas.
Figure 1.
Expression of DDR genes in human bladder tumors as a function of molecular subtype and clinical outcome. A) Heatmap depicts expression pattern of DDR1 and DDR2 in the TCGA-BCa cohort described in “Materials” and “Methods” (n = 259). B) Scatter plot and regression line (red) shows an inverse relationship between DDR1 and DDR2 expression. Hierarchical clustering was performed using the Manhattan distance and ward linkage method. C) Stacked bar graphs depict the distribution of tumors from the TCGA-BCa cohort by bladder cancer consensus subtypes (13). Tumor samples in each subtype were stratified into 3 groups by DDR expression at tercile values. D and E) Box plots show DDR1 (D) and DDR2 (E) expression in BCa consensus subtypes. F) Kaplan-Meier survival curves for DDR1 and DDR2 expression in the TCGA BCa cohort. Tumors were stratified into high and low groups at median expression of DDR. Statistical significance of differential survival between the groups were tested by log-rank test. Ba/Sq = basal and squamous; BCa = bladder cancer; CI = confidence interval; DDR = discoidin domain receptor; HR = hazard ratio; LumNS = luminal nonspecified; LumP = luminal papillary; LumU = luminal unstable; NE = neuroendocrine; TCGA = The Cancer Genome Atlas.
Because gene expression–based BCa subtypes associate with different clinical behaviors (12,13), we investigated the distribution of high DDR1 and DDR2 tumors among luminal papillary (LumP), basal and squamous (Ba/Sq), luminal unstable, stromal-rich, luminal nonspecified, and neuroendocrine-like subtypes (Figure 1, C). The LumP subtype has a high proportion of DDR1high tumors and overall DDR1 expression (Figure 1, C, D; χ2 P = .002), whereas the stroma-rich subtype has a higher proportion of DDR2high tumors (Figure 1, C, E). The stark difference between these 2 subtypes is intriguing and may reflect the expression of DDR2 in both cancer cells and stromal fibroblasts in the tumor microenvironment (TME) (14).
These findings suggest that the classical BCa subtypes are more heterogeneous than previously thought, and their clinical behavior may in part be driven by DDR1/2 expression. Given this, we examined if DDR1/2 expression stratified patient outcomes in TCGA-BCa. DDR1high tumors trended toward a better overall survival (OS), but this was not statistically significant (hazard ratio [HR] = 0.96, 95% confidence interval [CI] = 0.74 to 1.46; P = .73), whereas DDR2high tumors had statistically significantly poorer OS (HR = 1.55, 95% CI = 1.04 to 2.26; P = .02; Figure 1, F).
The Bladder Tumor Microenvironment in DDR1high and DDR2high Tumors
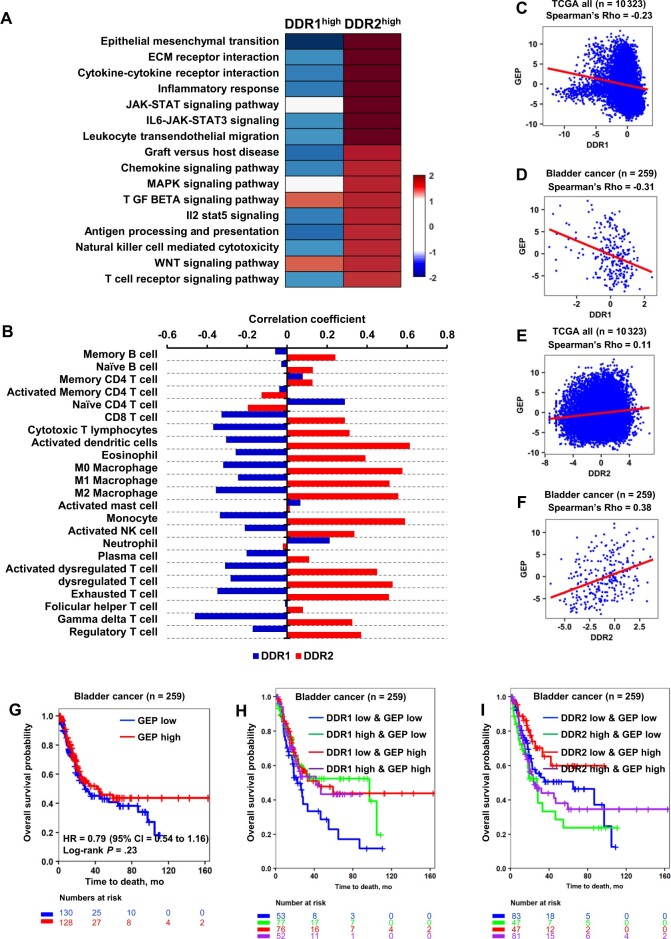
Gene set enrichment analysis (GSEA) identified 16 statistically significantly enriched cellular processes in DDR2high compared with DDR2low tumors, which were not enriched in DDR1high tumors (Figure 2, A). Epithelial mesenchymal transition (EMT) and Extracellular matrix (ECM) receptor interaction were statistically significantly enriched in DDR2high tumors (Figure 2, A), echoing the higher prevalence of DDR2high tumors in the stroma-rich subtype (Figure 1, C). Intriguingly, transforming growth factor–β signaling pathway and wingless/integrated (WNT) signaling pathway were enriched in both DDR1high and DDR2high tumors (Figure 2, A). Another key finding is the enrichment of immune cell–related processes, such as cytokine-cytokine receptor interaction, inflammatory response, and T-cell receptor signaling in DDR2high tumors (Figure 2, A), indicative of a DDR2 role in immune regulation and suggesting a T-cell–inflamed phenotype that is ineffectual in immune rejection of the tumors (15). In contrast, DDR1high tumors have statistically significant enrichment in P53 pathway (Normalized Enrichment Score [NES] = 1.51; P = .003), estrogen response early (NES = 1.48; P = .002), and protein secretion (NES = 1.45; P = .009). This suggests that DDR1high tumors are positive in distinct cellular functions from DDR2high tumors and negatively associated with these immune cell–related processes (Figure 2, A), proposing that they are non–T-cell inflamed.
Figure 2.
Impact of DDR expression on gene set enrichment analysis, molecularly defined cellular composition, and T-cell–inflamed GEP in human bladder tumors. A) Heatmap displays differentially enriched hallmark gene sets between DDR1high and DDR2high bladder tumors. Color bar represents the log-twofold change of the DDR1 or DDR2 high group compared with the DDR1 or DDR2 low group. B) Bar graphs depict Spearman correlation coefficient of DDR1/2 expression and 23 immune infiltration scores described in “Materials” and “Methods.” C-F) TCGA pan-cancer patients (n = 10,323) and TCGA-BCa patients (n = 259) analysis of DDR1/2 expression and T-cell–inflamed GEP score shows negative correlation with DDR1 expression (C, D) and positive correlation with DDR2 expression (E, F). Spearman method was used to estimate the correlation coefficient. The red line indicates the regression line. G) Tumors were stratified by T-cell–inflamed GEP score at the median of the TCGA-BCa patients (n = 259). H) Survival curves shows survival patterns of the 4 groups by DDR1 expression and T-cell–inflamed GEP score. Multiple log-rank tests were performed with the DDR1low and GEPlow group as a base line. I) Survival curves show survival patterns of the 4 groups by DDR2 and T-cell–inflamed GEP score. BCa = bladder cancer; CI = confidence interval; DDR = discoidin domain receptor; GEP = gene-expression profile; HR = hazard ratio; TCGA = The Cancer Genome Atlas; ECM = extracellular matrix; JAK = Janus kinase; STAT = signal transducer and activator of transcription; IL6 = interleukin 6; NK = natural killer; MAPK = mitogen-activated protein kinase; TGF = transforming growth factor; IL2 = interleukin 2; WNT = wingless/integrated.
Next, we assessed the correlation between DDR1/2 expression levels and 23 immune cell signatures (Supplementary Table 1, available online). Consistent with the GSEA results (Figure 2, A), the correlation coefficients of immune cell types demonstrated reciprocal values between DDR1high and DDR2high tumors in TCGA-BCa (Figure 2, B). DDR2high tumors were positively correlated with 19 immune cells (Figure 2, B), indicative of an immunologically “hot” TME. Conversely, DDR1high tumors were negatively correlated with most immune cells (Figure 2, B), indicative of an immunologically “cold” TME. These indicate that distinct TMEs are present in DDR1high and DDR2high BCa, suggesting these 2 genes have differential biological roles despite their structural similarity (16-18).
We also found that DDR1 expression was inversely correlated with the T-cell–inflamed gene-expression profile (GEP) score in both the TCGA–pan-cancer cohort (n = 10 323) (Figure 2, C; ρ = −0.23, P < .001) and TCGA-BCa (Figure 2, D; ρ = −0.31, P < .001), whereas DDR2 expression was positively correlated (Figure 2, E; ρ = 0.11, P < .001; Figure 2, F; ρ = 0.38, P < .001). DDR2 expression was also statistically significantly correlated with the presence of CD8+ T cells (ρ = 0.29, P < .001) and cytotoxic T lymphocytes (ρ = 0.31, P < .001) in the TCGA-BCa (Figure 2, B).
Because GEP score is known to be associated with ICT responsiveness, we tested whether DDR expression in addition to GEP score has an additional benefit in identifying patients with distinct survival. GEP score alone had no statistically significant association with OS in TCGA-BCa (Figure 2, G). However, GEP scores stratified by DDR1/2 expression exhibited differences. The DDR1lowGEPlow group showed worse OS compared with the DDR1highGEPlow and DDR1lowGEPhigh groups (Figure 2, H, Table 2). Additionally, the DDR2low groups (ie, DDR2lowGEPlow, DDR2lowGEPhigh) exhibited better OS compared with DDR2high groups (ie, DDR2high and GEPlow and DDR2high and GEPhigh) (Figure 2, I; Table 2). This indicates that DDR expression offers better predictive information compared with GEP scores.
Table 2.
Multiple log-rank tests were performed with DDRlow and GEPhigh group as baseline in TCGA BCa and IMvigora
| Comparison | HR (95% CI) | P | Cohort |
|---|---|---|---|
| DDR1 high and GEPlow vs DDR1low and GEPlow | 0.65 (0.39 to 1.10) | .11 | TCGA BCa |
| DDR1 low and GEPhigh vs DDR1low and GEPlow | 0.58 (0.35 to 0.97) | .04 | TCGA BCa |
| DDR1 high and GEPhigh vs DDR1low and GEPlow | 0.73 (0.42 to 1.27) | .26 | TCGA BCa |
| DDR2 low and GEPlow vs DDR2low and GEPhigh | 1.86 (0.98 to 3.53) | .06 | TCGA BCa |
| DDR2 high and GEPlow vs DDR2low and GEPhigh | 2.61 (1.33 to 5.12) | .005 | TCGA BCa |
| DDR2 high and GEPhigh vs DDR2low and GEPhigh | 2.19 (1.16 to 4.13) | .02 | TCGA BCa |
| DDR1 high and GEPlow vs DDR1low and GEPlow | 0.79 (0.55 to 1.14) | .20 | IMvigor |
| DDR1 low and GEPhigh vs DDR1low and GEPlow | 0.76 (0.53 to 1.10) | .15 | IMvigor |
| DDR1 high and GEPhigh vs DDR1low and GEPlow | 1.13 (0.80 to 1.60) | .48 | IMvigor |
| DDR2 low and GEPlow vs DDR2low and GEPhigh | 0.76 (0.53 to 1.12) | .17 | IMvigor |
| DDR2 high and GEPlow vs DDR2low and GEPhigh | 1.05 (0.74 to 1.51) | .78 | IMvigor |
| DDR2 high and GEPhigh vs DDR2low and GEPhigh | 0.89 (0.62 to 1.29) | .53 | IMvigor |
BCa = bladder cancer; CI = confidence interval; DDR = discoidin domain receptor; GEP = gene-expression profile; HR = hazard ratio; TCGA = The Cancer Genome Atlas.
Association of DDR Expression and Response to ICT
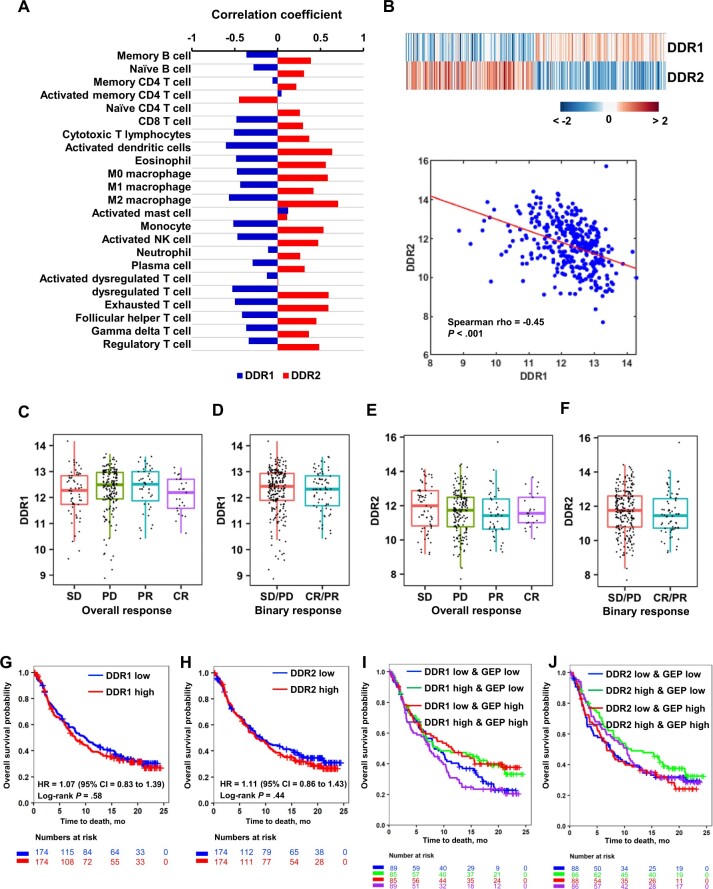
DDR1 high and DDR2high tumors appear to associate with immunologically cold and hot TMEs, respectively, whereas DDR2high tumors have worse patient outcome following surgery. Given these associations, we investigated the relationship of DDR1/2 expression in patients undergoing ICT (19). Using the IMvigor210 dataset, where BCa patients were treated with atezolizumab anti–PD-L119, we assessed if DDR1/2 expression associates with immune infiltration. Consistent with the TCGA-BCa results, IMvigor210 showed similar patterns of correlation between DDR1/2 expression and immune infiltration scores of immune cells (Figure 3, A). Unsupervised clustering analysis of DDR1/2 expression showed separation of DDR1highDDR2low vs DDR1lowDDR2high tumors with an inverse linear relationship (ρ = -0.45, P < .001) (Figure 3, B). DDR1/2 expression had no correlation with ICT response (Figure 3, C-F) or patient survival (Figure 3, G, H), even when DDR expression was further stratified by GEP scores (Figure 3, I and J; Table 2).
Figure 3.
Association of DDR expression with TME features and immune checkpoint therapy response in human BCa. A) Bar graph depicts Spearman correlation coefficients of DDR1 and DDR2 expression with immune cell type scores (n = 23). B) Heatmap and scatter plot show an inverse relationship of DDR1 and DDR2 expression in the IMvigor cohort. Color bar represents the z score of gene expression. C, D) Box plots depict expression distribution of DDR1 by immunotherapy response groups in the IMvigor210 cohort (n = 298). E, F) Box plots depict expression distribution of DDR2 by immunotherapy response groups in IMvigor (n = 298). G, H) Kaplan-Meier (KM) survival curves for DDR1 (G) and DDR2 (H) expression in IMvigor. Tumors were stratified into high and low groups at median expression of DDR. Significance of differential survival between the groups was tested by log-rank test. I) KM curves show survival patterns of the 4 groups by DDR1 expression and T-cell–inflamed GEP score. Multiple log-rank tests were performed with DDR1low and GEPlow group as a baseline. J) KM curves show survival patterns of the 4 groups by DDR2 and T-cell–inflamed GEP score. Multiple log-rank tests were performed with DDR2low and GEPhigh group as a baseline. Table 2 show hazard ratio, significance level (P value), and confidence interval for each comparison. BCa = bladder cancer; CI = confidence interval; CR = complete response; DDR = discoidin domain receptor; GEP = gene-expression profile; HR = hazard ratio; PD = partial disease; PR = partial response; SD = stable disease; TCGA = The Cancer Genome Atlas; TGF = transforming growth factor; TME = tumor microenvironment.
Development of DDR-Driven Gene Signatures
The lack of outcome stratification by DDR1/2 expression led us to explore a broader evaluation of DDR-regulated gene expression. We hypothesized expression changes would represent biologically active DDR signaling rather than just high DDR expression with minimal downstream transcriptional consequences. We developed gene signatures that were sensitive to changes in DDR1/2 expression (20). We either overexpressed DDR1 (DDR1-OE) or depleted DDR2 (DDR2-KD) in BCa models. Subcutaneous tumors in mice generated from either DDR1-OE T24 human BCa cells and controls. In these experiments, female Rag2 and Il2rg double-knockout mice (Taconic) were used. Four mice per group were used with tumors removed 6 weeks after initial tumor cell inoculation and profiled. The details of the DDR2 experiments were described in our prior publication (10,21) and carried out on tumors derived from a murine cell line NA13 BCa (10,21) transduced with shDDR2 or scrambled shRNA, subjected to RNA-sequencing (Supplementary Figure 1, A, available online). Differential expression analysis was performed (Supplementary Figure 1, B, C, available online). A total of 225 upregulated and 367 downregulated genes (Supplementary Table 2, available online) by DDR1-OE, and 211 upregulated and 69 downregulated genes (Supplementary Table 3, available online) by DDR2-KD were identified with a false discovery rate less than .05 and log2-fold-change of at least 1. Functional enrichment analysis using DAVID (22) of 225 upregulated genes by DDR1-OE and 69 downregulated genes by DDR2-KD revealed statistically significant and specific enrichment of epithelial development and excretion by DDR1-OE (Supplementary Figure 1, D, available online) and chemokine signaling and immune and/or inflammatory responses by DDR2-KD (Supplementary Figure 1, E, available online). Regulation of cell proliferation and wound response were similarly enriched by both DDR1 and DDR2 manipulations (Supplementary Figure 1, F, available online).
From the 225 upregulated genes by DDR1-OE and 69 downregulated genes by DDR2-KD (Supplementary Figure 2, A, available online), we first selected the top 50 most differentially expressed genes (Supplementary Figure 2, B, available online) because recent data from a large-scale transcriptome analysis revealed that the top 50 ranked genes usually provided markers that discriminate between experimental groups with high confidence (23,24). These 2 sets of genes had no overlap. We used them to compute DDR1/2 activity scores and examined their association with molecular functions, clinical, and therapeutic outcomes (Supplementary Figure 2, B, available online).
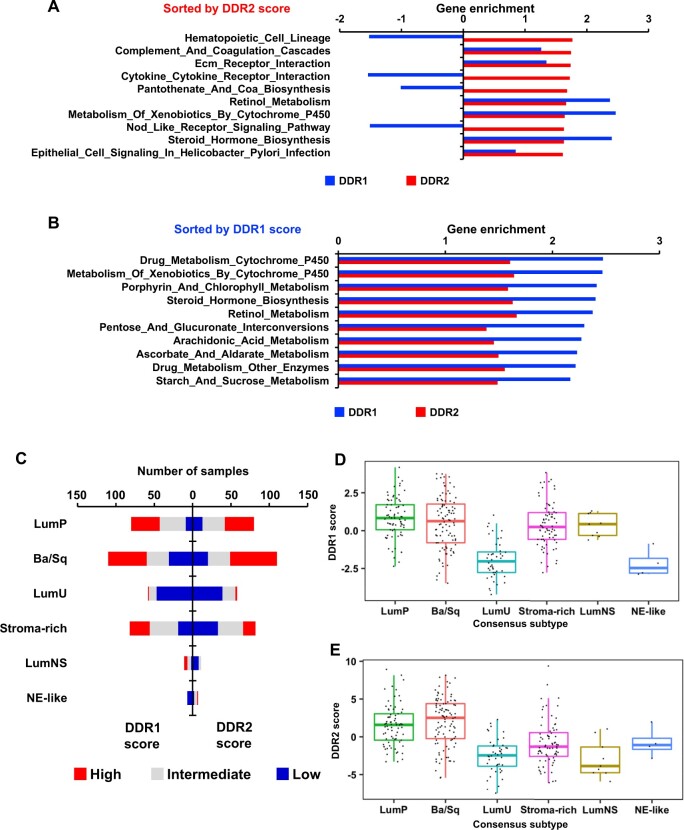
Next, we stratified IMvigor210 patients into DDR1/2–high and –low median score groups (25). GSEA were performed, and the top 10 KEGG pathways statistically significantly enriched in DDR1- or DDR2-high score tumors are presented in Figure 4, A and B. Consistent with the enriched gene sets in Figure 2, A, DDR2high tumors exhibited statistically significant enrichment of immune-related pathways (Figure 4, A), whereas DDR1high tumors enriched for metabolic pathways (Figure 4, B). LumP and Ba/Sq subtypes had the largest fractions of tumors with high DDR1/2 scores (Figure 4, C).
Figure 4.
Relationship of DDR gene signature scores with molecular pathways and tumor subtypes in bladder cancer. A, B) Bar plots depict enrichment of KEGG pathways by DDR2 (A) or DDR1 (B) active tumors. DDR1/2 gene expression score was computed by a z score method and used to stratify tumors into 2 groups with high and low DDR1 or DDR2 scores. Pathways were sorted by their DDR score, and the top 10 pathways were selected. C) Stacked bar graph depicts distribution of the tumors from IMvigor by BCa consensus subtypes. Tumor samples in each subtype were stratified into 3 groups by DDR score at tercile values. D, E) Box plot shows DDR1 (D) and DDR2 (E) scores in BCa consensus subtypes in IMvigor. Ba/Sq = basal and squamous; BCa = bladder cancer; DDR = discoidin domain receptor; LumNS = luminal nonspecified; LumU = luminal unstable; NE = neuroendocrine.
Evaluation of DDR Signature Scores as Predictors of Anti–PD-L1 Response in Patients
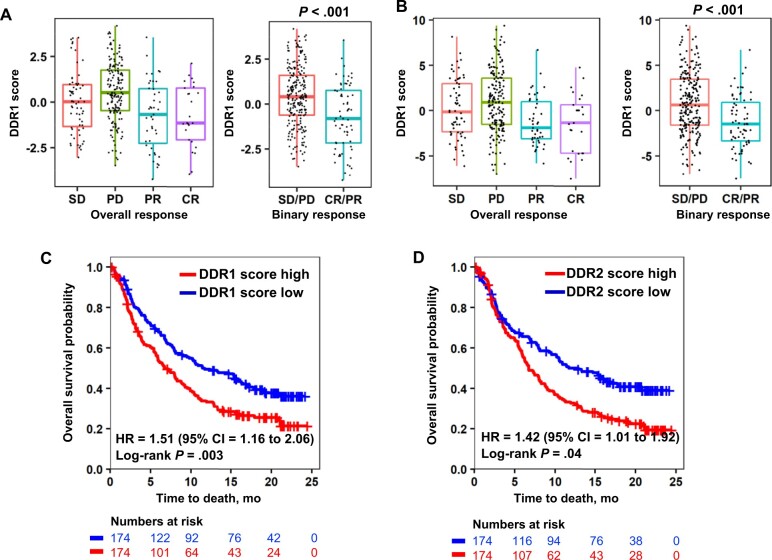
Next, we examined the ability of the DDR signature scores to stratify ICT response and survival from IMvigor210. DDR scores were statistically significantly different between stable disease or partial disease and complete response or partial response (P < .001 for DDR1 and P < .001 for DDR2) (Figure 5, A and B). Furthermore, tumors with high DDR1 (HR = 1.51, 95% CI = 1.16 to 2.06; P = .003) (Figure 5, C) or high DDR2 (HR = 1.42, 95% CI = 1.01 to 1.92; P = .04) (Figure 5, D) scores had poorer OS, suggesting that DDR scores have the potential to discriminate between patients with differential responses to anti–PD-L1 therapy.
Figure 5.
Association of DDR gene signature scores with immune checkpoint therapy response of bladder cancer patients. A, B) Boxplots depicts DDR1 (A) and DDR2 (B) scores as a function of the clinical response (stable disease [SD], progressive disease [PD], partial response [PR] and complete response [CR]) from IMvigor. C, D) Survival curves for DDR1 (C) and DDR2 (D) scores in IMvigor. Tumors were stratified into high and low groups at median score levels of IMvigor. Statistical significance of differential survival between the groups was tested by log-rank test. CI = confidence interval; DDR = discoidin domain receptor; HR = hazard ratio.
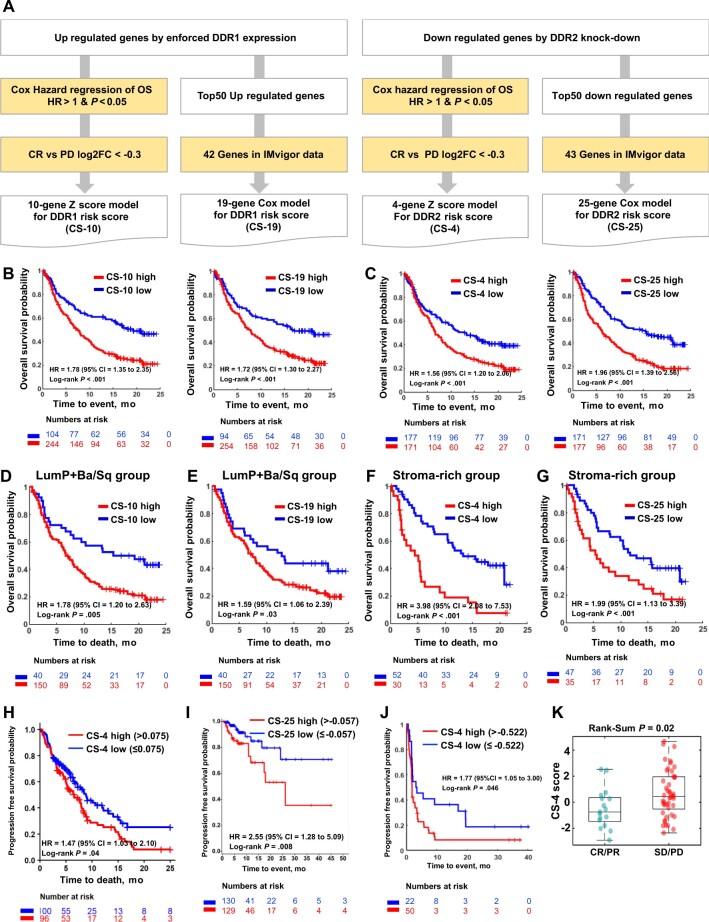
To optimize the composition of the DDR gene signature, we selected core genes among the 50 that were highly associated with OS in IMvigor210 through a statistical framework (Figure 6, A; see Supplementary Methods, available online), resulting in gene signatures for risk stratification of patients: a 10-gene signature based on a z score model (CS-10) and a 19-gene signature based on a Cox model (CS-19) for DDR1 and a 4-gene signature based on a z score model (CS-4) and a 25-gene signature based on a Cox model (CS-25) for DDR2.
Figure 6.
Optimization and validation of DDR gene signature scores as predictors of tumor response to anti–PD-L1 therapy. A) Schematic diagram of the gene selection process for model construction. Given the upregulated or downregulated genes from DDR murine models, 2 branches of subtractive approaches were employed to make 2 different gene models based on the gene signatures for DDR1 and DDR2, respectively, which correspond to the z score model and Cox model. B) Clinical association of DDR1 gene models. Left and right graphs show survival analysis results for 10-gene DDR1 z score model (CS-10) and 19-gene DDR1 Cox risk score model (CS-19), respectively. C) Clinical association of DDR2 gene models. Left and right graphs show survival analysis results for 4-gene DDR2 z score model (CS-4) and 25-gene DDR2 Cox risk score model (CS-25), respectively. D) Survival curves of CS-10 high and low groups in LumP and Ba/Sq subtypes in IMvigor. E) Survival curves of CS-19 high and low groups in LumP and Ba/Sq subtypes in IMvigor. F) Survival curves of CS-4 high and low groups in stroma-rich subtype in IMvigor. G) Survival curves of CS-25 high and low groups in stroma-rich subtype in IMvigor. H) Survival curves of CS-4 high and low groups in NSCLC cohort treated with anti–PD-L1 therapy from the Tempus cohort. I) Survival curves of CS-25 high and low groups in the NSCLC cohort treated with anti–PD-L1 therapy from the Caris cohort. J) Survival curves of CS-4 high and low groups in the cohort treated with anti–PD-L1 therapy from the CheckMate 275 cohort. K) Boxplots depict CS-4 scores as a function of the clinical response as defined in Figure 5, A, from the CheckMate 275 cohort. Ba/Sq = basal and squamous; CI = confidence interval; CR = complete response; DDR = discoidin domain receptor; HR = hazard ratio; LumP = luminal papillary; NSCLC = non-small cell lung carcinoma; OS = overall survival; PD = partial disease; PD-L1 = programmed cell death ligand 1; PR = partial response; SD = stable disease.
We also surveyed optimal cut points of the signature score using IMvigor210 data by assessing hazard ratios across all DDR signature scores for OS. We identified optimal cut points of CS-10, CS-19, CS-4, and CS-25, which are −0.77, −0.079, 0.039, and −0.059, respectively. Survival differences at these cut points are all statistically significant (Figure 6, B, C). We also assessed survival association of the DDR signature scores in BCa consensus subtypes. The LumP and Ba/Sq subtypes with high CS-10 (Figure 6, D) and CS-19 (Figure 6, E) scores were associated with worse OS, whereas the stroma-rich subtype with high CS-4 (Figure 6, F) or high CS-25 (Figure 6, G) scores were associated with worse OS. Of note, CS-25 showed the best performance in the stroma-rich subtype and statistically significant stratification of OS in all subtypes. We further made 2 combined models by union of CS-10 and CS-4 models for combined DDR z score model (CS-14) and union of CS-19 and CS-25 Cox models for combined DDR Cox model (CS-44). CS-14 and CS-44 gene models have better association with OS compared with DDR1 or DDR2 models before combining with a better hazard ratio of 2.03 (95% CI = 1.53 to 2.71; P < .001) and 2.48 (95% CI = 1.85 to 3.33; P < .001), respectively (Supplementary Figure 3, available online).
Next, we identified RNA-sequencing and outcome data from 2 NSCLC cohorts (ie, Tempus and Caris) treated with anti–PD-L1 in routine practice. We applied our scoring of the 4-gene signatures to these. This analysis was done in a double-blinded fashion. The survival curves were computed by investigators with the patient DDR signature scores but no information on gene composition or how the score was derived. Clinical characteristics are described in Supplementary Table 4 (available online). Notably, the 2 NSCLC cohorts had differences in the proportion of PD-L1–stained tumor-infiltrating lymphocyte (TIL) immune cells, with 29% of tumors (n = 59) in the Tempus cohort having more than 5% of PD-L1–positive cells compared with 50% of tumors (n = 129) in the Caris cohort. This may explain why CS-4 high and low groups exhibited statistically significant survival differences in the Tempus cohort (HR = 1.47, 95% CI = 1.03 to 2.10; P = .04; Figure 6, H), whereas CS-25 high and low groups exhibited statistically significant survival differences in the Caris cohort (HR = 2.55, 95% CI = 1.28 to 5.09; P = .008; Figure 6, I). Both CS-4 and CS-25 are DDR2 signature scores, and the survival difference with CS-4 shows a higher hazard ratio compared with CS-25 in the stroma-rich subtype (Figure 6, F and G). We also evaluate the performance of CS-4 with an independent cohort from CheckMate 275 study (26), a dataset comprised of patients with metastatic urothelial cancer treated with the PD-1 inhibitor nivolumab, and found statistically significant poorer survival in CS-4 high tumors (HR = 1.77, 95% CI = 1.05 to 3.00; P = .046; Figure 6, J) consistent with the statistically significant difference of CS-4 scores between complete response or partial response and stable disease or partial disease groups (P = .02; Figure 6, K). Along with this, we further assessed DDR2 signature scores in 6 independent cohorts (4 melanoma, 1 glioblastoma, and 1 with metastatic urothelial carcinoma) with checkpoint inhibitors responses (Supplementary Figure 4, available online).
Discussion
Biomarkers for stratifying ICT responses can be broadly categorized into 2 subsets: biomarkers indicative of a T-cell–inflamed TME and genomic biomarkers. The expression of PD-L1 is associated with clinical response to ICT in multiple cancers including melanoma, NSCLC, renal cell carcinoma, and colon cancer (27-30). However, PD-L1 can be expressed on tumor and immune cells, and the predictive value of PD-L1 expression on either tumor or immune cells differs based on the cancer type (29,31-33). Additionally, challenges exist interpreting PD-L1 expression by immunohistochemistry, because of variability in antibodies and scoring methods (34). More important, some studies show that patients with PD-L1–negative tumors can also benefit from ICT (35,36), suggesting the presence of biological drivers of response unrelated to PD-L1.
Other parameters evaluated as predictors of ICT response include 1) the presence of TILs (29), 2) T-cell–inflamed GEP (37), and 3) interferon (IFN)–γ gene signature (38). In general, high densities of TILs (ie, CD8+ T cells) have been associated with better clinical outcome after surgical removal of primary tumors (39). In the context of ICT, targeting PD-1 and PD-L1 signaling is thought to reinvigorate preexisting antitumor response of TILs, which express immune checkpoint molecules (40). IFN-γ is a cytokine released by activated T cells, nature killer cells, and natural killer T cells and is important for both antitumor response and adaptive immune resistance mechanisms. IFN-γ signaling also upregulates expression of PD-L1 on tumor, stromal, and other immune infiltrating cells, which can interact with PD-1 on TILs (41). Genomic biomarkers such as TMB and microsatellite instability have also been associated with ICT response (42). High TMB has been associated with improved clinical outcome in NSCLC (43), melanoma (44), bladder, and colorectal cancers (42,45-47). The TCGA-BCa and pan-cancer cohorts have shown that DDR2 gene expression has positive association with PD-L1 gene expression, whereas DDR1 gene expression does not (Supplementary Figure 5, available online). Association of DDR gene expression with human leukocyte antigens (HLA) gene mutation could not be confirmed to be statistically significant because of the small number of tumor samples harboring HLA defects in the TCGA data (Supplementary Figure 6 and 7, available online). We also checked the association of DDR gene expression with TMB, resulting in no statistical significance in the TCGA data (Supplementary Figure 8, available online).
The above markers have pan-tumor US Food and Drug Administration approval; however, further improvements are needed in BCa, which motivated the current study. Our work indicates that DDR2 and DDR1 scores have value in stratifying PD-L1 response not only in BCa but also other tumor types, indicating possibly broader utility. These findings reveal intriguing differential biology of DDR1/2 high expression BCa, exhibiting a non–T-cell–inflamed and a T-cell–inflamed phenotype, respectively. Although these are from the same family, our findings revealed their differential involvement regulating immune cell infiltration and a role in the stromal microenvironment in BCa. For instance, in the CheckMate 275 BCa cohort, a high CD8+ T-cell infiltration together with a low EMT and stromal core signature was associated with the highest response and longest progression-free survival and OS following treatment with nivolumab (26). Conversely, patients with a high CD8+ T-cell infiltration but also a high EMT and stromal core signature showed a statistically significantly worse progression-free survival and OS (26), implicating a role of the stromal microenvironment in impeding T-cell function and driving ICT resistance (48,49). Future studies will be important to validate the utility of the DDR scores in predicting ICT response in prospective clinical trials. In these studies, when tumor tissues are evaluated, it is particularly important to do so using single cell technologies. Recently, it has been shown that single cell expression profiling of human bladder cancer provides striking novel information that will likely change how we classify tumor subtypes and other established markers in the future (50). If such gene expression scores are predictive, it would be important to determine if this translates to superior outcomes for patients with high DDR1 and/or DDR2 scores treated with DDR1/2 inhibitors such as sitravatinib (48) and dasatinib (10) in combination with ICT in bladder (NCT03606174) and other cancers (lung: NCT02750514, hematologic NCT02819804, NCT02011945).
In conclusion, these results suggest that DDR gene signatures define differential T-cell tumor-infiltration biology and stratify patient survival following PD-L1 checkpoint inhibitor therapy in urothelial cancer. Notably, some signatures also stratified outcome of other cancer types such as melanoma. Further research is needed to understand the importance of tumor type in the utility of these various DDR-based signatures. Overall, our findings suggest that the DDR2- and DDR1-dependent transcriptional program defines differential tumor biology, intriguingly, both linked with immunotherapy response.
Funding
This work was supported by National Institutes of Health CA075115 (DT) and CA175397 (KSC).
Notes
Role of the funder: The study funders had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
Disclosures: DS, JA, DM, PJ are employees of commercial entities (Caris and Tempus) as indicated. All other authors declare no competing interests.
Authors contributions: Conceptualization: DT. Methodology: KSC, YCL, and DT. Software and Formal analysis: SY and MK. Validation: SY and MK. Investigation: KSC, SY, YCL, and DT. Resources and Data Curation: KSC, DT, DS, JA, DM, PJ, and MDG. Writing—Original Draft: SY, KSC, XPH, MDG, and DT. Writing—Review & Editing: All authors. Visualization: SY and MK. Supervision: DT. Project Administration: DT. Funding Acquisition: DT. and KSC.
Supplementary Material
Contributor Information
Sungyong You, Department of Surgery, Cedars-Sinai Medical Center, Los Angeles, CA, USA; Samuel Oschin Comprehensive Cancer Institute, Los Angeles, CA, USA.
Minhyung Kim, Department of Surgery, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Xen Ping Hoi, Samuel Oschin Comprehensive Cancer Institute, Los Angeles, CA, USA; Department of Pathology and Laboratory Medicine, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Yu Cheng Lee, Graduate Institute of Medical Sciences, Taipei Medical University, Taipei, Taiwan.
Li Wang, Department of Medicine, Division of Hematology Oncology, Icahn School of Medicine at Mount Sinai, Tisch Cancer Institute, New York, NY, USA.
David Spetzler, Caris Life Sciences, Irving, TX, USA.
Jim Abraham, Caris Life Sciences, Irving, TX, USA.
Dan Magee, Caris Life Sciences, Irving, TX, USA.
Prerna Jain, Tempus, Chicago, IL, USA.
Matthew D Galsky, Department of Medicine, Division of Hematology Oncology, Icahn School of Medicine at Mount Sinai, Tisch Cancer Institute, New York, NY, USA.
Keith Syson Chan, Samuel Oschin Comprehensive Cancer Institute, Los Angeles, CA, USA; Department of Pathology and Laboratory Medicine, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Dan Theodorescu, Department of Surgery, Cedars-Sinai Medical Center, Los Angeles, CA, USA; Samuel Oschin Comprehensive Cancer Institute, Los Angeles, CA, USA; Department of Pathology and Laboratory Medicine, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Data Availability
The RNAseq data generated from the DDR1 (GSE191097) and DDR2 (GSE190698) experiments can be found in Gene Expression Omnibus (GEO).
References
- 1. Sharma P, Allison JP. Dissecting the mechanisms of immune checkpoint therapy. Nat Rev Immunol. 2020;20(2):75-76. doi: 10.1038/s41577-020-0275-8. [DOI] [PubMed] [Google Scholar]
- 2. Tran L, Xiao JF, Agarwal N, Duex JE, Theodorescu D. Advances in bladder cancer biology and therapy. Nat Rev Cancer. 2021;21(2):104-121. doi:10.1038/s41568-020-00313-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348(6230):56-61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 4. Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer. 2019;19(3):133-150. doi: 10.1038/s41568-019-0116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schneider AK, Chevalier MF, Derré L. The multifaceted immune regulation of bladder cancer. Nat Rev Urol. 2019;16(10):613-630. doi:10.1038/s41585-019-0226-y. [DOI] [PubMed] [Google Scholar]
- 6. Rouanne M, Roumiguié M, Houédé N, et al. Development of immunotherapy in bladder cancer: present and future on targeting PD(L)1 and CTLA-4 pathways. World J Urol. 2018;36(11):1727-1740. doi: 10.1007/s00345-018-2332-5. [DOI] [PubMed] [Google Scholar]
- 7. Motzer RJ, Rini BI, McDermott DF, et al. Nivolumab for metastatic renal cell carcinoma: results of a randomized phase II trial. J Clin Oncol. 2015;33(13):1430-1437. doi:10.1200/J Clin Oncol.2014.59.0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Robert C, Long GV, Brady B, et al. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2015;372(4):320-330. doi: 10.1056/NEJMoa1412082. [DOI] [PubMed] [Google Scholar]
- 9. Davis AA, Patel VG. The role of PD-L1 expression as a predictive biomarker: an analysis of all US Food and Drug Administration (FDA) approvals of immune checkpoint inhibitors. J Immunother Cancer. 2019;7(1):278.doi: 10.1186/s40425-019-0768-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tu MM, Lee FYF, Jones RT, et al. Targeting DDR2 enhances tumor response to anti-PD-1 immunotherapy. Sci Adv. 2019;5(2):eaav2437.doi:10.1126/sciadv.aav2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee YC, Kurtova AV, Xiao J, et al. Collagen-rich airway smooth muscle cells are a metastatic niche for tumor colonization in the lung. Nat Commun. 2019;10(1):2131.doi: 10.1038/s41467-019-09878-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Robertson AG, Kim J, Al-Ahmadie H, et al. ; for the TCGA Research Network. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell. 2017;171(3):540-556.e25. doi: 10.1016/j.cell.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kamoun A, de Reyniès A, Allory Y, et al. ; for the Bladder Cancer Molecular Taxonomy Group. A consensus molecular classification of muscle-invasive bladder cancer. Eur Urol. 2020;77(4):420-433. doi: 10.1016/j.eururo.2019.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Corsa CAS, Brenot A, Grither WR, et al. The action of discoidin domain receptor 2 in basal tumor cells and stromal cancer-associated fibroblasts is critical for breast cancer metastasis. Cell Rep. 2016;15(11):2510-2523. doi:10.1016/j.celrep.2016.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Iafolla MAJ, Selby H, Warner K, Ohashi PS, Haibe-Kains B, Siu LL. Rational design and identification of immuno-oncology drug combinations. Eur J Cancer. 2018;95:38-51. doi: 10.1016/j.ejca.2018.02.027. [DOI] [PubMed] [Google Scholar]
- 16. Agarwal G, Smith AW, Jones B. Discoidin domain receptors: micro insights into macro assemblies. Biochim Biophys Acta Mol Cell Res. 2019;1866(11):118496.doi: 10.1016/j.bbamcr.2019.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lafitte M, Sirvent A, Roche S. Collagen kinase receptors as potential therapeutic targets in metastatic colon cancer. Front Oncol. 2020;10:125.doi: 10.3389/fonc.2020.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Auguste P, Leitinger B, Liard C, et al. Meeting report - first discoidin domain receptors meeting. J Cell Sci. 2020;133(4):jcs243824. doi: 10.1242/jcs.243824. [DOI] [PubMed] [Google Scholar]
- 19. Mariathasan S, Turley SJ, Nickles D, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554(7693):544-548. doi: 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Smith SC, Baras AS, Owens CR, Dancik G, Theodorescu D. Transcriptional signatures of Ral GTPase are associated with aggressive clinicopathologic characteristics in human cancer. Cancer Res. 2012;72(14):3480-3491. doi: 10.1158/0008-5472.CAN-11-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tu MM, Abdel-Hafiz HA, Jones RT, et al. Inhibition of the CCL2 receptor, CCR2, enhances tumor response to immune checkpoint therapy. Commun Biol. 2020;3(1):720.doi: 10.1038/s42003-020-01441-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44-57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 23. Vandenbon A, Diez D. A clustering-independent method for finding differentially expressed genes in single-cell transcriptome data. Nat Commun. 2020;11(1):4318.doi: 10.1038/s41467-020-17900-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hou D, Koyutürk M. Comprehensive evaluation of composite gene features in cancer outcome prediction. Cancer Inform. 2014;13(suppl 3):93-104. doi: 10.4137/CIN.S14028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Levine DM, Haynor DR, Castle JC, et al. Pathway and gene-set activation measurement from mRNA expression data: the tissue distribution of human pathways. Genome Biol. 2006;7(10):R93.doi: 10.1186/gb-2006-7-10-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang L, Saci A, Szabo PM, et al. EMT- and stroma-related gene expression and resistance to PD-1 blockade in urothelial cancer. Nat Commun. 2018;9(1):3503.doi: 10.1038/s41467-018-05992-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443-2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Weber JS, D’Angelo SP, Minor D, et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 2015;16(4):375-384. doi: 10.1016/S1470-2045(15)70076-8. [DOI] [PubMed] [Google Scholar]
- 29. Taube JM, Klein A, Brahmer JR, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res. 2014;20(19):5064-5074. doi:10.1158/1078-0432.CCR-13-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Daud AI, Wolchok JD, Robert C, et al. Programmed death-ligand 1 expression and response to the anti-programmed death 1 antibody pembrolizumab in melanoma. J Clin Oncol. 2016;34(34):4102-4109. doi:10.1200/J Clin Oncol.2016.67.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Faraj SF, Munari E, Guner G, et al. Assessment of tumoral PD-L1 expression and intratumoral CD8+ T cells in urothelial carcinoma. Urology. 2015;85(3):703.e1-703.e6. doi: 10.1016/j.urology.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Powles T, Eder JP, Fine GD, et al. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature. 2014;515(7528):558-562. doi: 10.1038/nature13904 [DOI] [PubMed] [Google Scholar]
- 33. Llosa NJ, Cruise M, Tam A, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5(1):43-51. doi:10.1158/2159-8290.CD-14-0863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Patel SP, Kurzrock R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol Cancer Ther. 2015;14(4):847-856. doi:10.1158/1535-7163.MCT-14-0983 [DOI] [PubMed] [Google Scholar]
- 35. Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. 2015;373(2):123-135. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Motzer RJ, Escudier B, McDermott DF, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med. 2015;373(19):1803-1813. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ayers M, Lunceford J, Nebozhyn M, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127(8):2930-2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Grasso CS, Tsoi J, Onyshchenko M, et al. Conserved interferon-γ signaling drives clinical response to immune checkpoint blockade therapy in melanoma. Cancer Cell. 2020;38(4):500-515.e3. doi: 10.1016/j.ccell.2020.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fridman WH, Pagès F, Sautès-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12(4):298-306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 40. Tumeh PC, Harview CL, Yearley JH, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515(7528):568-571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Taube JM, Anders RA, Young GD, et al. Colocalization of inflammatory response with B7-H1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med. 2012;4(127):127ra37.doi: 10.1126/scitranslmed.3003689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51(2):202-206. doi: 10.1038/s41588-018-0312-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124-128. doi: 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roh W, Chen PL, Reuben A, et al. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci Transl Med. 2017;9(379):eaah3560. doi:10.1126/scitranslmed.aah3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rosenberg JE, Hoffman-Censits J, Powles T, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387(10031):1909-1920. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Powles T, Durán I, van der Heijden MS, et al. Atezolizumab versus chemotherapy in patients with platinum-treated locally advanced or metastatic urothelial carcinoma (IMvigor211): a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2018;391(10122):748-757. doi:10.1016/S0140-6736(17)33297-X. [DOI] [PubMed] [Google Scholar]
- 47. Patel VG, Oh WK, Galsky MD. Treatment of muscle‐invasive and advanced bladder cancer in 2020. CA Cancer J Clin. 2020;70(5):404-423. doi: 10.3322/caac.21631. [DOI] [PubMed] [Google Scholar]
- 48. Du W, Huang H, Sorrelle N, Brekken RA. Sitravatinib potentiates immune checkpoint blockade in refractory cancer models. JCI Insight. 2018;3(21):e124184. doi: 10.1172/jci.insight.124184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lee YC, Lam HM, Rosser C, Theodorescu D, Parks WC, Chan KS. The dynamic roles of the bladder tumour microenvironment [published online ahead of print June 28, 2022]. Nat Rev Urol. 2022. doi:10.1038/s41585-022-00608-y. https://www.nature.com/articles/s41585-022-00608-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gouin KH, Ing N, Plummer JT, et al. An N-Cadherin 2 expressing epithelial cell subpopulation predicts response to surgery, chemotherapy and immunotherapy in bladder cancer. Nat Commun. 2021;12(1):4906. doi:10.1038/s41467-021-25103-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNAseq data generated from the DDR1 (GSE191097) and DDR2 (GSE190698) experiments can be found in Gene Expression Omnibus (GEO).