Sir,
The infection of SARS-CoV-2 is characterized by respiratory symptoms, which indicates droplet transmission. The clinical spectrum of SARS-CoV-2 infection ranged from asymptomatic, mild infection to severe pneumonia with severe acute respiratory syndrome or multi-organ failure, which may result in a deadly outcome1,2. It was reported in 2002-2003 SARS outbreak that about 16-73 per cent of patients had diarrhoea as one of the symptoms within the first week of the infection usually3. RNA of SARS-CoV was detected in stool from the fifth day of infection onwards. The proportion of positivity from stool specimens for viral RNA gradually started increasing and peaked at day 11th of the infection. A small proportion of patients showed the presence of viral RNA in the faeces even after 30 days of illness4. In the present pandemic of SARS-CoV-2, gastrointestinal (GI) symptoms have been commonly reported among the patients5. The presence of angiotensin converting enzyme 2 (ACE2) cell receptors in the enterocytes of small intestine contributes to the gut infection of SARS-CoV-26. The detection of SARS-CoV-2 RNA in the stool remained longer after respiratory specimens became negative in a group of patients with COVID-197,8. However, the reports are scanty and need wider confirmations.
RNA viruses have high mutation rates and undergo rapid evolution, due to which their virulence, infectivity and transmissibility are enhanced. Monitoring changes/mutations in the SARS-CoV-2 genome at population level are important for tracing the outbreak origin, tracking transmission chains and understanding the evolution of virus9. Since the viral RNA is detected in the stool even after the respiratory specimens test negative, stool specimens provide an opportunity to characterize genomic changes/mutations specific to that particular patient. This study was aimed to sequence the full genome of the SARS-CoV-2 by next generation sequencing (NGS) from stool samples of both symptomatic and asymptomatic individuals with SARS-CoV-2 infection.
Stool samples were collected from patients, positive for SARS-CoV-2 by naso/oropharyngeal swab by reverse transcription (RT)-PCR. Written informed consent was taken from all the participants. This study was a part of an ongoing study approved by the Ethics Committee of Indian Council of Medical Research (ICMR)-National Institute of Virology (NIV, 20-2-2 R), Pune, India and conducted from April 2020-2021. The viral RNA was extracted from 30 per cent (w/v) suspensions of stool specimens using QIAmp Viral RNA kits (Qiagen, Hilden, Germany) as per the manufacturer’s instructions. All the stool specimens were tested for SARS-CoV-2 by real-time RT-PCR targeting the genes, E, RNA-dependent RNA-polymerase (RdRp), ORF-1b-nsp14 and RNaseP (internal control) as per the NIV protocol10. A few representative samples with the highest threshold cycle (Ct) values were selected for NGS analysis. Sequencing of SARS-CoV-2 genome was performed using the Ion AmpliSeq platform11. cDNA was synthesized with the SuperScript VILO reverse transcriptase kit (Invitrogen, Thermo Fisher Scientific, MA, USA), and the libraries were prepared according to the manufacturer’s instructions (Thermo Fisher Scientific, MA, USA). The enriched templates were loaded onto an Ion 316 Chip for sequencing on Ion PGM machine (Thermo Fisher Scientific, MA, USA). Sequence data were compared with the complete genome of the SARS-CoV-2 Wuhan-Hu-1 isolate (GenBank accession number MN908947.3). The entire sequence database was submitted in EpiFlu of GISAID (https://www.gisaid.org/epiflu-applications/submitting-data-to-epiflutm/) global database.
In this study, (12M, 5F) 17 patients were included. All patients were positive for SARS-CoV-2 in the stool specimens. All the 17 specimens were processed for genome sequencing. According to their clinical conditions, three patients were admitted in intensive care unit, two were asymptomatic and six were having mild symptoms with/without comorbidities. One patient had diarrhoea only. RT-PCR from throat/nasal swabs was done 3-7 days before stool collection (Table).
Table.
Clinical, demographic and mutations details of patients
| Sample ID | Age/sex | Symptoms | Mode of infection | Comorbidities | Geographical location | Date of oropharyngeal swab tested | Date of stool sample received in the lab | Ct values for E genes in stool | Treated | Lineage | Spike mutations of interest |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2003583 | 46/male | Asymptomatic (high containment zone) | No | Chinchawad, Pune | December 24, 2020 | December 26, 2020 | 30.08 | No | B.1.36.16 | D614G | |
| 2001729 | 16/male | Sore throat | No | Nanapeth, Pune | May 15, 2020 | May 21, 2020 | 21.96 | No | B.1.1.306 | D614G | |
| 2001676-3 (repeat samples) | 37/male | Fever, breathing problem, in ICU | Symptomatic health care worker | Diabetes | Vadagaon, Pune | May 4, 2020 | June 9, 2020 | 29.18 | Yes* | B.1.1.281 | D614G: Q677R |
| 2001676-4 (repeat samples) | 37/male | Fever, breathing problem, in ICU | Symptomatic health care worker | Diabetes | Vadagaon, Pune | May 4, 2020 | June 19, 2020 | 31.55 | Yes* | B.1.1.281 | D614G: Q677R |
| 2001692-1 | 22/male | Mild fever, sore throat | Symptomatic health care worker | No | Dhole Patil Road, Pune | May 1, 2020 | May 26, 2020 | 27.8 | No | B.1.1.281 | D614G: F1220L |
| 2001884 | 67/female | Diarrhoea only | Hypertension | Shivaji Nagar, Pune | May 27, 2020 | June 12, 2020 | 28.02 | No | B.1.1.306 | D614G | |
| 2002247 | 30/male | Asymptomatic | Healthcare worker | No | Sangli | August 2, 2020 | August 5, 2020 | 26.5 | No | B.1.1.306 | D614G |
| 2002157 | 80/male | ICU, severely ill | Hypertension | Kothrud | July 23, 2020 | July 27, 2020 | 26.13 | No | B.1.210 | D614G | |
| 2002181 | 65/male | Cough, headache | No | Chikali | August 1, 2020 | August 4, 2020 | 28.75 | No | B.1.1.306 | D614G: Y789H | |
| 210357 | 60/female | Symptomatic, breathlessness | No | Jalgaon | February 12, 2021 | February 11, 2021 | 19.13 | No | B.1.36.29 | D614G: N440K, S929I | |
| 210354 | 70/male | Fever, weakness | Diabetes mellitus, hypertension, traumatic bleed Epilepsy Anaemia |
Kothrud, Pune | February 7, 2021 | February 11, 2021 | 29.59 | No | B.1.617.1# | P681R: D614G L452R: E484Q G142D: S94T E154K: D1153Y Q1071H: V382L | |
| 210014 | 33/male | Sore throat, fever, nasal discharge | No | Ravet, Pune | January 1, 2021 | January 7, 2021 | 28.89 | No | B.1.1.421 | D614G: Y351P T345C: N343R, F342Y, F347L, N481H, A348C, S349T, S816T, A344C |
* Treated: Repeat sample collected after discharge from hospital after treatment completion; # RT-PCR not done at NIV. Ct, cycle threshold; RT-PCR, reverse transcription polymerase chain reaction; ICU, intensive care unit; NIV, National Institute of Virology
NGS was successful in all samples but complete only in 12 samples. The coverage for five samples was <26,000 bp and these samples were not included in the analysis. The final sequence was 29,508 bp long with a GC content of 37.98 per cent. All the sequences from the samples were compared with the complete genome of the Wuhan-Hu-1 isolate of SARS-CoV-2 using the BLAST tool (https://www.ncbi.nlm.nih.gov/nuccore/1798174254; GenBank accession number MN908947.3) and 99.8 per cent similarity was observed with Wuhan-Hu-1 isolate. The phylogenetic tree was constructed using MEGA v.6 software12, employing the Neighbor-Joining method with “Maximum Composite Likelihood” as the substitution model for the analysis of these genome sequences (Figure). Our results revealed that these sequences belonged to clade GR except three with GH clade. All these sequences were submitted in Global Initiative on Sharing All Influenza Data (GISAID) database (https://www.gisaid.org/epiflu-applications/submitting-data-to-epiflutm/).
Figure.
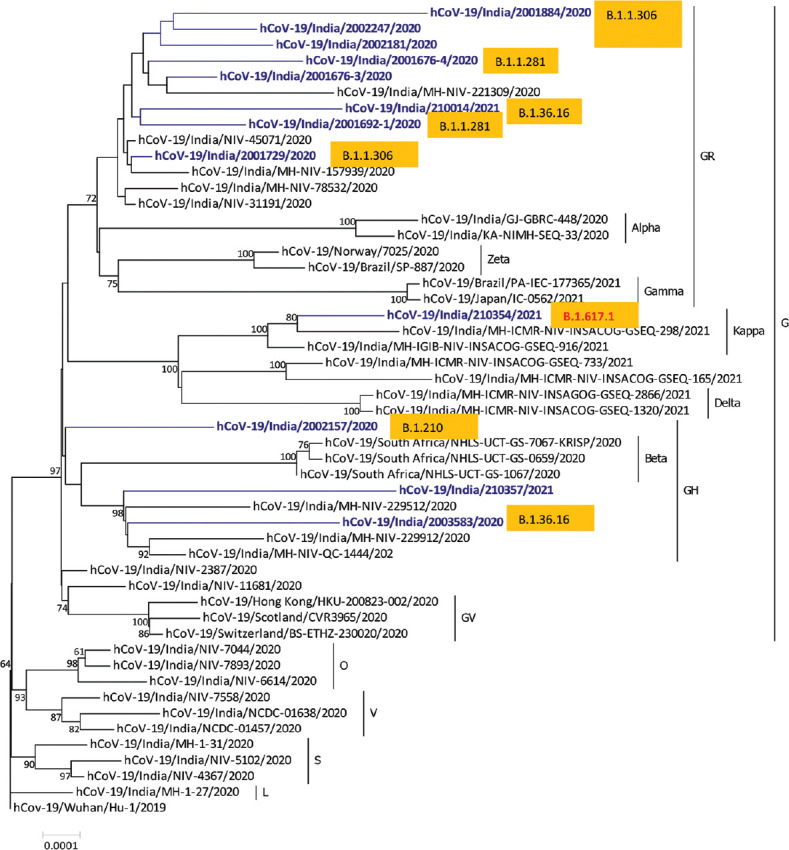
Phylogeny of SARS-CoV-2 genomes from stool samples of COVID-19–positive patients from Pune, Maharashtra (in blue text), according to lineages.
Sequence analysis identified a number of nucleotide variants at position 241 (C-T), 3037 (C-T), 23403 (A-G) and 14408 (C-T) in all patients and mutations at positions 313 (C-T), 5700 (C-A), 28881–28882–28883. Many silent mutations (241, 3037) and non-synonymous mutations (14408, 23403 and 28881–28882–28883) were also observed. Among the non-synonymous mutations, an already observed mutation at position 14408, which is located in the viral RdRp gene, a key component of the replication/transcription machinery, was also noticed. Mutations at 241, 3037, 14408 and 23403 were observed in all the sequences. The non-synonymous mutations were observed in ORF1b, ORF3a and ORF8 (nucleocapsid phosphoprotein) genes, resulting in the amino acid mutation Q57H (glutamine to histidine), P227L (proline to leucine) and S194L (serine to leucine), respectively. D614G, one of the pre-dominant mutations which is located in the spike glycoprotein, was observed in all the 12 genomes. One sample (210354) showed mutations at E154K, L452R, E484Q, P681R and Q1071H along with D614G (Table).
Detection of SARS-CoV-2 by RT-PCR and NGS13,14,15 from stool specimens has been reported. In this study, SARS-CoV-2 was detected in patients who were positive by RT-PCR and complete genomes were sequenced in 12 stool samples. NGS identified the whole-genome sequence of SARS-CoV-2 from stool specimens from patients with positive oropharyngeal RT-PCR. One patient (2001676-4) tested positive for SARS-CoV-2 by NGS from stool even after 44 days of positive nasopharyngeal RT-PCR test. Among the globally sequenced strains of SARS-CoV-2, this D614G substitution was not common before March 2020, but the frequency for this variant increased to over 75 per cent after June16. This mutation has been suggested to be associated with enhanced transmission in populations with lower ACE2 expression17,18. In another sample (210354), multiple mutations were observed in spike protein which characterized that as lineage B.1.617.1.
Findings from our study suggest that SARS-CoV-2 is constantly detectable and continuously showing infectivity in the intestine in spite of the negative RT-PCR result from the respiratory pathway. The infectivity and transcriptional activity of SARS-CoV-2 may decrease gradually19. As per the emergence of new variants from throat samples20, we also observed similar trend from the stool samples (Figure). The prolonged existence of SARS-CoV-2 in the stool of patients without the involvement of gut as well as in recovered patients emphasizes a potential for faecal-oral transmission. This observation suggests that the virus may stay for a longer time period in the gut than predicted. Since sequences obtained from stool samples represent the last evolved virus within a patient, it can be compared with viral genome sequences obtained using a respiratory specimen to understand the evolution of virus within a patient. However, in the present study, since whole-genome sequences were not available from respiratory specimens, it was not possible to compare both the sequences and this aspect needs further explorations. For the better understanding of the infectivity of SARS-CoV-2 in the human intestine, there is a need for the future research. Active and prolonged activity of SARS-CoV-2 in the GI tract of COVID-19–positive patients, even in the absence of GI-related symptoms and after the recovery, reflects the importance of long-term surveillance of SARS-CoV-2 and the threat of potential faecal-oral transmission. Approaches that include neutralizing gut SARS-CoV-2 activity and modulating gut microbiome should be explored in a prospective manner for better treatment of SARS-CoV-2 infection.
Acknowledgment:
Authors thank Dr Priya Abraham, Director ICMR-NIV, Pune, for the support during the study. The assistance provided by Servshree P.S. Jadhav & Santosh (Deenanath Mangeshkar Hospital, Pune) during sample collection from the hospital is duly acknowledged. Shri Manohar Shinde and Ms Nutan S. Chavan are acknowledged for providing technical support for processing of samples.
Footnotes
Financial support & sponsorship: The authors acknowledge the ICMR-National Institute of Virology and ICMR, New Delhi, for financial support.
Conflicts of Interest: None.
References
- 1.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–20. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. WHO issues consensus document on the epidemiology of SARS. Wkly Epidemiol Rec. 2003;78:373–5. [PubMed] [Google Scholar]
- 4.Chan KH, Poon LL, Cheng VC, Guan Y, Hung IF, Kong J, et al. Detection of SARS coronavirus in patients with suspected SARS. Emerg Infect Dis. 2004;10:294–9. doi: 10.3201/eid1002.030610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vuille-Dit-Bille RN, Liechty KW, Verrey F, Guglielmetti LC. SARS-CoV-2 receptor ACE2 gene expression in small intestine correlates with age. Amino Acids. 2020;52:1063–5. doi: 10.1007/s00726-020-02870-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–7. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu CL, Raval M, Schnall JA, Kwong JC, Holmes NE. Duration of respiratory and gastrointestinal viral shedding in children with SARS-CoV-2:A systematic review and synthesis of data. Pediatr Infect Dis J. 2020;39:e249–56. doi: 10.1097/INF.0000000000002814. [DOI] [PubMed] [Google Scholar]
- 8.Zuo T, Zhang F, Lui GC, Yeoh YK, Li AY, Zhan H, et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology. 2020;159:944–55. doi: 10.1053/j.gastro.2020.05.048. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forster P, Forster L, Renfrew C, Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc Natl Acad Sci U S A. 2020;117:9241–3. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choudhary ML, Vipat V, Jadhav S, Basu A, Cherian S, Abraham P, et al. Development of in vitro transcribed RNA as positive control for laboratory diagnosis of SARS-CoV-2 in India. Indian J Med Res. 2020;151:251–4. doi: 10.4103/ijmr.IJMR_671_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Potdar V, Cherian SS, Deshpande GR, Ullas PT, Yadav PD, Choudhary ML, et al. Genomic analysis of SARS-CoV-2 strains among Indians returning from Italy, Iran &China, &Italian tourists in India. Indian J Med Res. 2020;151:255–60. doi: 10.4103/ijmr.IJMR_1058_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6:Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen C, Gao G, Xu Y, Pu L, Wang Q, Wang L, et al. SARS-CoV-2-positive sputum and feces after conversion of pharyngeal samples in patients with COVID-19. Ann Intern Med. 2020;172:832–4. doi: 10.7326/M20-0991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu J, Liu J, Li S, Peng Z, Xiao Z, Wang X, et al. Detection and analysis of nucleic acid in various biological samples of COVID-19 patients. Travel Med Infect Dis. 2020;37:101673. doi: 10.1016/j.tmaid.2020.101673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papoutsis A, Borody T, Dolai S, Daniels J, Steinberg S, Barrows B, et al. Detection of SARS-CoV-2 from patient fecal samples by whole genome sequencing. Gut Pathog. 2021;13:7. doi: 10.1186/s13099-021-00398-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, et al. Tracking changes in SARS-CoV-2 spike:Evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–27. doi: 10.1016/j.cell.2020.06.043. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang SW, Miller SO, Yen CH, Wang SF. Impact of genetic variability in ACE2 expression on the evolutionary dynamics of SARS-CoV-2 spike D614G mutation. Genes (Basel) 2020;12:E16. doi: 10.3390/genes12010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi AC, Xie X. Making sense of spike D614G in SARS-CoV-2 transmission. Sci China Life Sci. 2021;64:1062–7. doi: 10.1007/s11427-020-1893-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ranshing S, Lavania M, Potdar V, Patwardhan S, Prayag PS, Jog S, et al. Transmission of COVID-19 infection within a family cluster in Pune, India. Indian J Med Res. 2021;153:555–8. doi: 10.4103/ijmr.IJMR_3378_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cherian S, Potdar V, Jadhav S, Yadav P, Gupta N, Das M, et al. SARS-CoV-2 spike mutations, L452R, T478K, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. Microorg. 2021;9:1542. doi: 10.3390/microorganisms9071542. [DOI] [PMC free article] [PubMed] [Google Scholar]


