Abstract
Background
Phthalates and bisphenols are non-persistent endocrine disrupting chemicals that are ubiquitously present in our environment and may have long-lasting health effects following fetal exposure. A potential mechanism underlying these exposure–outcome relationships is differential DNA methylation. Our objective was to examine the associations of maternal phthalate and bisphenol concentrations during pregnancy with DNA methylation in cord blood using a chemical mixtures approach.
Methods
This study was embedded in a prospective birth cohort study in the Netherlands and included 306 participants. We measured urine phthalates and bisphenols concentrations in the first, second and third trimester. Cord blood DNA methylation in their children was processed using the Illumina Infinium HumanMethylation450 BeadChip using an epigenome-wide association approach. Using quantile g-computation, we examined the association of increasing all mixture components by one quartile with cord blood DNA methylation.
Results
We did not find evidence for statistically significant associations of a maternal mixture of phthalates and bisphenols during any of the trimesters of pregnancy with DNA methylation in cord blood (all p values > 4.01 * 10–8). However, we identified one suggestive association (p value < 1.0 * 10–6) of the first trimester maternal mixture of phthalates and bisphenols and three suggestive associations of the second trimester maternal mixture of phthalates and bisphenols with DNA methylation in cord blood.
Conclusions
Although we did not identify genome-wide significant results, we identified some suggestive associations of exposure to a maternal mixture of phthalates and bisphenols in the first and second trimester with DNA methylation in cord blood that need further exploration in larger study samples.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13148-022-01345-0.
Keywords: Bisphenols, Phthalates, Prenatal, Mixture, DNA methylation
Background
Endocrine disrupting chemicals (EDCs) such as phthalates and bisphenols are used in many consumer products such as cosmetics and plastic food packaging [1, 2]. As a result, throughout their lives, humans are continuously exposed to a mixture of these endocrine disruptors. Previous studies have shown associations of exposure to endocrine disruptors with cardiometabolic health, insulin resistance and fertility in humans [3–6]. As phthalates and bisphenols are able to cross the placenta, exposure begins in utero [7, 8]. Fetal life has been suggested to be a sensitive period for exposure to environmental factors that determine life-long patterns of health and disease, as suggested in the Developmental Origins of Health and Disease framework [9]. Previous studies of EDC exposure during fetal life with childhood health have also shown an association of these exposures with birth weight, neurodevelopment and cardiometabolic risk factors during childhood, among others [3, 10–13].
One of the proposed mechanisms by which phthalates and bisphenols could affect health is by influencing DNA methylation [14]. Results from studies that have examined the association between phthalate or bisphenol exposure during fetal life and DNA methylation in humans are not consistent. Varying associations of fetal exposure to phthalates or bisphenol A were found with DNA methylation of the insulin-like growth factor 2 (IGF2) gene in candidate-gene studies [15–19]. These studies assessed DNA methylation in different tissues. In the study by Montrose et al., DNA methylation was assessed in cord blood, while in two other studies, placental tissue was used [16, 17, 19]. Finally, in two other studies, whole blood during childhood was used [15, 18]. As DNA methylation is tissue- and age-specific, results of these studies are not directly comparable. Several other studies of prenatal bisphenol and phthalate exposure, using an epigenome-wide approach, have identified differential DNA methylation at multiple CpG sites in cord blood in relation to exposure to different phthalates or bisphenols [20–25]. In addition, one study explored the associations of prenatal exposure to phthalates with DNA methylation in peripheral blood and buccal epithelial cells during childhood [26]. When exploring the associations with DNA methylation in cord blood, only the studies by Miura et al. and Petroff et al. included more than 70 mother–child pairs and the studies by Chen et al. and Petroff et al. were the only studies that included a phthalate [20, 22, 24, 25]. Most previous studies that assessed the associations of maternal phthalate or bisphenol urine concentrations with DNA methylation focused on one or a few phthalates or bisphenols and did not consider joint effects of the exposures [15–30]. This is an oversimplification, since humans are exposed to a mixture of different chemicals that could influence each other and thus could have synergistic or antagonistic effects. Focusing on only one single exposure might not fully elucidate the total mixture-effect of endocrine disruptors during pregnancy.
To overcome this limitation, we investigated the associations of fetal exposure to a mixture of phthalates and bisphenols, using a novel statistical approach, with DNA methylation in offspring [31]. We measured phthalate and bisphenol concentrations at three time points during pregnancy in spot urine samples obtained from 306 participants in a Dutch population-based cohort study and measured DNA methylation in an epigenome-wide association study (EWAS) in cord blood in their children.
Methods
Design
This study was embedded in the Generation R Study, a population-based prospective cohort study starting during early fetal life in Rotterdam, the Netherlands [32]. The study has been approved by the Medical Ethical Committee of the Erasmus MC, University Medical Center Rotterdam. Written informed consent was obtained for all participants.
Mother–child pairs were included in this exploratory study if bisphenols and phthalates urine concentrations were available at all three time points during pregnancy, which was in a subsample of 1379 mothers of the full Generation R Study, and if DNA methylation was measured in cord blood collected at birth. DNA methylation data were collected in a subsample of children participating in the full Generation R Study, consisting of participants with parents born in the Netherlands. A total of 306 mothers and their children were included in the current analyses (Additional file 1: Fig. S1).
Phthalate and bisphenol measurements
Between February 2004 and July 2005, women were invited to our research facility during early (median 12.6 weeks, interquartile range (IQR) 2.0 weeks), mid (median 20.4 weeks, IQR 1.0 weeks) and late pregnancy (median 30.2 weeks, IQR 1.3 weeks), at which time they provided a spot urine sample. The analyses of the phthalate, bisphenol and creatinine concentrations were performed at the Wadsworth Center, New York State Department of Health, Albany, New York, USA. Collection, transportation and analysis of these urine samples have been previously described [33]. Of all measured phthalates and bisphenols, we included those in the assessment that had less than 25% of their concentrations during all trimesters below the limit of detection (LOD). Phthalates and bisphenols with more than 25% of the samples below the limit of detection were thus not included in the further analysis. Concentrations below the LOD were substituted by LOD divided by the square root of 2 (LOD/√2) [34]. To account for urinary dilution in the analysis, all urine concentrations of phthalates and bisphenols were converted to µmol/g creatinine. An overview of the concentrations of included phthalates and bisphenols is presented in Table 1. (Non-participants are shown in Additional file 1: Table S1.) Finally, to reduce skewedness of the distributions, the phthalate and bisphenol urine concentrations were natural log transformed. As a sensitivity analysis, all analyses were repeated using the averaged concentrations over pregnancy.
Table 1.
Urine concentrations of phthalates and bisphenols, specified per trimester
| LOD (nmol/L) | First trimester | Second trimester | Third trimester | ||||
|---|---|---|---|---|---|---|---|
| Median (25–75th percentile) | Percentage < LOD | Median (25–75th percentile) | Percentage < LOD | Median (25–75th percentile) | Percentage < LOD | ||
| Phthalic acid (PA) (nmol/L) | 6.68 | 349.2 (195.4–844.3) | 0.3 | 953.2 (381.6–1558.6) | 0 | 345.7 (187.1–715.6) | 0.3 |
| Monomethylphthalate (mMP) (nmol/L) | 0.33 | 28.3 (14.8–52.4) | 0.3 | 18.5 (9.2–34.1) | 0.3 | 16.8 (9.4–36.6) | 1.0 |
| Monoethylphthalate (mEP) (nmol/L) | 0.31 | 671.5 (198.0–2412.9) | 0 | 330.5 (123.2–1057.6) | 0 | 591.3 (207.8–1775.7) | 0 |
| Mono-isobutylphthalate (mIBP) (nmol/L) | 0.40 | 84.2 (38.3–157.8) | 0 | 35.6 (18.9–66.4) | 0 | 58.9 (33.6–115.0) | 0.3 |
| Mono-n-butylphthalate (mBP) (nmol/L) | 0.63 | 68.5 (30.8–124.5) | 0.3 | 41.0 (24.4–75.1) | 0 | 45.7 (23.9–78.8) | 0 |
| Monobenzylphthalate (mBzBP) (nmol/L) | 0.59 | 24.6 (8.8–43.6) | 6.9 | 17.4 (7.2–33.6) | 2.9 | 9.9 (3.3–19.1) | 3.6 |
| Mono-(2-ethyl-5-carboxy-pentyl)phthalate (mECPP) (nmol/L) | 0.94 | 46.8 (26.3–92.9) | 0 | 33.2 (18.7–59.2) | 0 | 51.7 (26.8–90.3) | 0 |
| Mono-(2-ethyl-5-hydroxy-hexyl)phthalate (mEHHP) (nmol/L) | 0.27 | 35.1 (16.5–73.8) | 0 | 19.1 (10.6–36.8) | 0 | 33.0 (15.9–58.0) | 0 |
| Mono-(2-ethyl-5oxohexyl)phthalate (mEOHP) (nmol/L) | 0.14 | 22.6 (10.4–45.7) | 0 | 26.5 (14.2–56.7) | 0 | 22.5 (12.1–43.4) | 0 |
| Mono-[(2-carboxymethyl)-hexyl]phthalate (mCMHP) (nmol/L) | 0.13 | 42.4 (23.3–74.3) | 0 | 12.3 (7.1–23.4) | 0.3 | 9.1 (5.0–17.4) | 0 |
| Mono(3-carboxypropyl)phthalate (mCPP) (nmol/L) | 0.03 | 5.2 (3.0–10.4) | 0 | 3.6 (2.0–6.6) | 0 | 6.6 (3.7–12.1) | 0 |
| Bisphenol A (BPA) (nmol/L) | 0.66 | 6.2 (1.6–14.8) | 18.0 | 5.2 (2.4–12.2) | 8.2 | 6.0 (2.9–11.0) | 9.2 |
Values represent medians (25–75th percentiles). Absolute urine concentration of the limit of detection (in nmol/L urine) and individual exposures (in nmol/L urine) with concentrations below the limit of detection imputed as limit of detection/square root of 2. Only phthalates and bisphenols that have at least 75% detection in all trimesters are presented in this table. Individual exposures assessed but not included in the analysis in this study due to less than 75% of concentrations above the limit of detection in all trimesters include monoisononylphthalate, monocyclohexylphthalate, monooctylphthalate, mono-(8-methyl-1-nonyl)phthalate, mono-hexylphthalate, mono-2-heptylphthalate, mono-(7-carboxy-n-heptyl)phthalate, bisphenol S, bisphenol Z, bisphenol B, bisphenol F, bisphenol AP, bisphenol AF and bisphenol P
LOD limit of detection
DNA methylation measurement
After birth, cord blood was drawn by the attending physician or midwife. From these samples, DNA was extracted using the salting-out method. After bisulfite conversion of 500 ng DNA using the EZ-96 DNA Methylation kit (Shallow) (Zymo Research Corporation, Irvine, USA), samples were processed with the Illumina Infinium HumanMethylation450 BeadChip (Illumina Inc., San Diego, USA). At the time of processing the cord blood samples from the Generation R Study, the MethylationEPIC BeadChip was not available yet and whole-genome bisulfite sequencing was not feasible for high-throughput analysis in population studies. Beta values, which represent the ratio of methylated signal relative to the total (methylated and unmethylated) signal per CpG, were calculated. Quality control and normalization were performed using the CPACOR workflow [35]. Probes with a detection p value ≥ 1.0 * 10–16 were set to missing. Intensity values were quantile normalized. Arrays with observed technical problems or with a mismatch between sex of the proband and sex determined by the intensities of the X- and Y-chromosome were removed from the analysis. Only arrays with a call rate > 95% per sample were processed further, and DNA methylation beta values outside the range of (25th percentile – 3 * IQR, 75th percentile + 3 * IQR) were set to missing. Probes on the X and Y chromosomes were excluded from the dataset. Additionally, we removed cross-reactive probes, leaving information on 415,786 CpGs at birth [36, 37]. Probes that map to DNA containing a single nucleotide polymorphism (SNP), repetitive sequence elements or DNA harboring an insertion or deletion were flagged, but not removed [36, 37].
Covariates
Information on potential confounders was collected using questionnaires during pregnancy. Potential confounders were chosen based on their known association with both phthalate and bisphenol exposure and with DNA methylation. Included covariates were maternal age at inclusion, maternal pre-pregnancy body mass index (BMI), maternal educational level and maternal smoking habits (sustained versus non-sustained smoking during pregnancy). Child sex was obtained from midwife and hospital records. Sample plate number was included in the analysis to correct for batch effects. Plates with fewer than two participants were grouped together, which was done for six plates, as not all mother–child pairs from the Generation R Study in whom DNA methylation was measured in cord blood had information on the maternal phthalate and bisphenol urine concentrations during pregnancy available. White blood cell composition was estimated with the Salas method for cord blood, which included B-lymphocytes, CD4+ T-lymphocytes, CD8+ T-lymphocytes, granulocytes, monocytes, natural killer cells and nucleated red blood cells. Ethnicity and use of folic acid supplements were not assessed as potential confounders in this study, since all participants were of European ancestry and almost all participants (93.6%) used folic acid supplements.
Statistical analysis
Missing data for covariates (ranging between 0.3 and 11.4%) were imputed ten times by the Multivariate Imputation by Chained Equations (MICE) method in R. Imputation was successful for all covariates, and the last imputed dataset was used for all analysis. When all association analyses were repeated with a random other dataset as a sensitivity analysis, there were no differences in the reported associations. To assess the joint effects of the phthalate and bisphenol mixture in a specific trimester, we used the quantile-based g-computation approach from the qgcomp package in R [31]. In quantile g-computation, the exposures of interest are quantized (e.g., transformed into categories of exposure), after which the effect of increasing all exposures by one quantile simultaneously is evaluated by estimating the parameters of a marginal structural model given the joint intervention on the exposures. The main advantages of this method are the easy interpretation of the association and the absence of a need for directional homogeneity. Using this method, we were able to estimate the joint effect of increasing all mixture components by one quartile.
To examine associations of the chemical mixture with DNA methylation in cord blood, we first ran basic linear models adjusting for child sex, estimated cell types and batch. We then ran fully adjusted linear models adjusting for child sex, maternal education, maternal smoking during pregnancy, maternal age at inclusion, maternal pre-pregnancy BMI, estimated cell types and batch.
We used Bonferroni correction (p value cutoff < 4.01 * 10–8 based on an original p value cutoff of 0.05 and 415,786 tests per trimester, giving a total of 1,247,358 tests for the three trimesters) as the primary cutoff to assess statistical significance. Additionally, we defined suggestive associations based on a p value cutoff of < 1.0 * 10–6, as we feared to be too rigorous in dismissing potential associations that did not reach statistical significance due to the exploratory nature of this study. To provide a more comprehensive overview of the results, we present all associations with a p value cutoff of < 1.0 * 10–5 in the supplemental tables. We performed a priori defined exploratory analyses stratified on sex, as it has been hypothesized that exposure to endocrine disruptors could have different effects based on sex [38].
Results
Subject characteristics
Compared to non-participants, participating mothers in the present study were more often of European ancestry, highly educated and were less likely to sustain smoking during pregnancy (Table 2). Almost all participants used folic acid supplementation during early pregnancy. Most phthalate concentrations were higher among non-participants than among participants, but bisphenol A concentrations during first trimester were lower among non-participants (Table 1 and Additional file 1: Table S1).
Table 2.
Participant and non-participant characteristics
| Participants (three trimesters) | Non-participants | |
|---|---|---|
| n = 306 | n = 1 073 | |
| Maternal characteristics | ||
| Age at enrollment, mean (SD) (years) | 32.1 (3.9)* | 30.1 (5.0)* |
| Ethnicity, n (%) | ||
| European ancestry | 298 (97.4%)* | 455 (42.9%)* |
| Non-European ancestry | 8 (2.6%)* | 605 (57.1%)* |
| Education, n (%) | ||
| Low-middle | 92 (30.2%)* | 564 (55.6%)* |
| High | 213 (69.8%)* | 450 (44.4%)* |
| Pre-pregnancy BMI, median (95% range) (kg/m2) | 22.6 (18.7–34.4) | 22.7 (18.4–35.1) |
| Folic acid supplementation, n (%), yes | 233 (93.6%)* | 654 (76.9%)* |
| Smoking sustained during pregnancy, n (%), yes | 24 (8.9%)* | 155 (15.9%)* |
| Alcohol consumption sustained during pregnancy (any), n (%), yes | 202 (55.8%)* | 340 (35.1%)* |
| Child characteristics | ||
| Gender (boys), n (%) | 160 (52.3%) | 536 (50.0%) |
| Birth weight, mean (SD) (g) | 3556 (468)* | 3425 (503)* |
| Gestational age at birth, mean (SD) (weeks) | 40.3 (1.3)* | 40.0 (1.5)* |
Values represent numbers (valid percent), mean (SD) or median (95% range)
SD standard deviation
*p value < 0.05
Associations of exposure to a mixture of endocrine disruptors and DNA methylation in cord blood
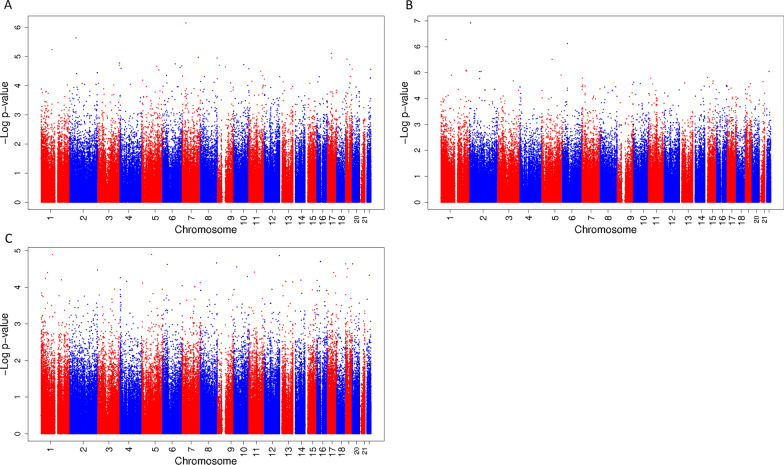
In the total study population, there were no significant associations of fetal exposure to a mixture of phthalates and bisphenols during either first, second or third trimester with DNA methylation in cord blood. (Figure 1A–C shows the Manhattan plots, CpGs with a p value < 1.0 * 10–6 are presented in Table 3, and CpGs with a p value < 1.0 * 10–5 are presented in Additional file 1: Table S2.) There were a few suggestive associations. The strongest association in the first trimester was found with decreased DNA methylation of cg05058973 (effect − 1.20 * 10–2 (standard error (SE) 2.37 * 10–3) per quartile increase in the mixture, p value 7.08 * 10–7), which maps to the growth hormone-releasing hormone receptor (GHRHR) gene. In the second trimester, we found a suggestive association with an increase in DNA methylation of cg00141688, located near the hippocalcin-like 1 (HPCAL1) gene, and cg15961211, which is close to the family with sequence similarity 183 member A (FAM183A) gene (effect per quartile increase in the mixture: 1.59 * 10–2 (SE 2.93 * 10–3), p value 1.21 * 10–7 and 3.65 * 10–3 (SE 7.11 * 10–4), p value 5.32 * 10–7, respectively) and with a decrease in DNA methylation of cg20840540, which is close to transcriptional-regulating factor 1 (TRERF1) (effect per quartile increase in the mixture − 1.28 * 10–2 (SE 2.52 * 10–3), p value 7.54 * 10–7). The analysis for the third trimester mixture showed no suggestive associations. Results for the models unadjusted for demographic covariates were comparable to those from the fully adjusted model (Additional file 1: Table S3 and Fig. S2).
Fig. 1.
Manhattan plot of associations between a mixture of phthalates and bisphenols during first, second and third trimester with DNA methylation at birth. Manhattan plot of associations between a mixture of phthalates and bisphenols during first (A), second (B) and third (C) trimester with DNA methylation at birth in the total population. In all Manhattan plots, the x-axis represents the autosomal chromosomes, the y-axis represents the −log10 of the p value and the dots represent CpGs
Table 3.
CpGs with p values < 1.0 * 10–6 from epigenome-wide association study of a mixture of phthalates and bisphenols in maternal urine during first, second and third trimester and DNA methylation in cord blood
| CpG | Chr | Position | Gene | Effect | SE | p value | Flag# | |
|---|---|---|---|---|---|---|---|---|
| First trimester | cg05058973 | 7 | 31002599 | GHRHR | − 1.20 * 10–2 | 2.37 * 10–3 | 7.08 * 10–7 | 0 |
| Second trimester | cg00141688 | 2 | 10517352 | HPCAL1 | 1.59 * 10–2 | 2.93 * 10–3 | 1.21 * 10–7 | 0 |
| cg15961211 | 1 | 43613440 | FAM183A | 3.65 * 10–3 | 7.11 * 10–4 | 5.32 * 10–7 | 0 | |
| cg20840540 | 6 | 42363749 | TRERF1 | − 1.28 * 10–2 | 2.52 * 10–3 | 7.54 * 10–7 | 0 |
There were no CpGs presented for third trimester, as none reached our uncorrected p value cutoff of < 1.0 * 10–6. There were no associations that reached significance (p value < 0.05) after further FDR-adjustment of the p value for multiple testing including the three trimesters
#We have indicated probes that map to DNA containing a single nucleotide polymorphism (SNP), repetitive sequence elements or DNA harboring an insertion or deletion with a ‘1’ in this column
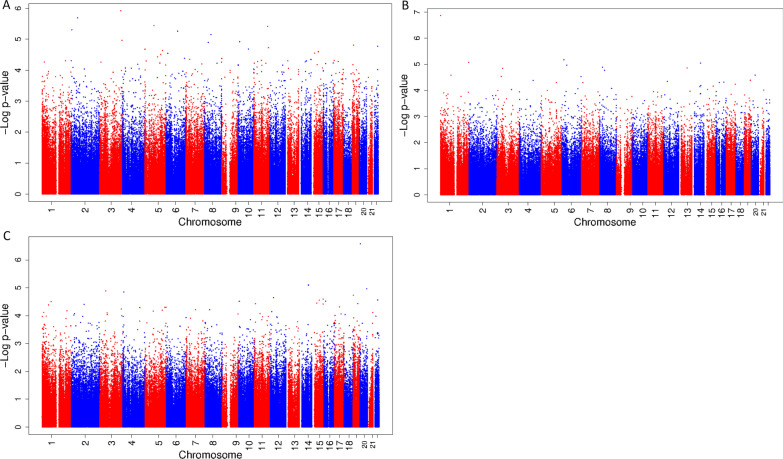
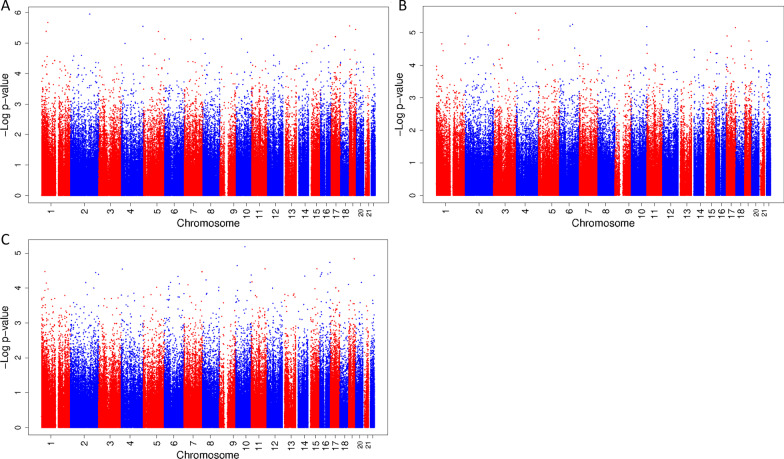
In the explorative stratified analyses among boys, there were no statistically significant associations of exposure to a mixture of phthalates and bisphenols during pregnancy with cord blood DNA methylation at birth. (Figure 2A–C shows the Manhattan plots, CpGs with a p value < 1.0 * 10–6 are presented in Table 4, and CpGs with a p value < 1.0 * 10–5 are presented in Additional file 1: Table S4.) However, there were some suggestive associations of exposure to the mixture during second trimester with a decrease in DNA methylation of cg03764767, which is located in the peroxisomal biogenesis factor 10 (PEX10) gene, and exposure to the mixture during third trimester with an increase in DNA methylation of cg23462052, located close to the small nuclear ribonucleoprotein polypeptides B and B1 (SNRPB) gene. (Figure 2A–C shows the Manhattan plots, CpGs with a p value < 1.0 * 10–6 are presented in Table 4, and CpGs with a p value < 1.0 * 10–5 are presented in Additional file 1: Table S4.) Among girls, there were no statistically significant or suggestive associations of exposure to the mixture during any of the trimesters with cord blood DNA methylation. (Figure 3A–C shows the Manhattan plots, and CpGs with a p value < 1.0 * 10–5 are presented in Additional file 1: Table S5.)
Fig. 2.
Manhattan plot of associations between a mixture of phthalates and bisphenols during first, second and third trimester with DNA methylation at birth among boys. Manhattan plot of associations between a mixture of phthalates and bisphenols during first (A), second (B) and third (C) trimester with DNA methylation at birth among boys. In all Manhattan plots, the x-axis represents the autosomal chromosomes, the y-axis represents the −log10 of the p value and the dots represent CpGs
Table 4.
CpGs with p values < 1.0 * 10–6 from epigenome-wide association study of a mixture of phthalates and bisphenols in maternal urine during first, second and third trimester and DNA methylation in cord blood among boys
| CpG | Chr | Position | Gene | Effect | SE | p value | Flag# | |
|---|---|---|---|---|---|---|---|---|
| Second trimester | cg03764767 | 1 | 2338210 | PEX10 | − 1.85 * 10–2 | 3.33 * 10–3 | 1.36 * 10–7 | 1 |
| Third trimester | cg23462052 | 20 | 2452871 | SNRPB | 1.27 * 10–2 | 2.36 * 10–3 | 2.65 * 10–7 | 1 |
There were no CpGs presented for first trimester, as none reached our uncorrected p value cutoff of < 1.0 * 10–6. There were no associations that reached significance (p value < 0.05) after further FDR-adjustment of the p value for multiple testing including the three trimesters
#We have indicated probes that map to DNA containing a single nucleotide polymorphism (SNP), repetitive sequence elements or DNA harboring an insertion or deletion with a ‘1’ in this column
Fig. 3.
Manhattan plot of associations between a mixture of phthalates and bisphenols during first, second and third trimester with DNA methylation at birth among girls. Manhattan plot of associations between a mixture of phthalates and bisphenols during first (A), second (B) and third (C) trimester with DNA methylation at birth among girls. In all Manhattan plots, the x-axis represents the autosomal chromosomes, the y-axis represents the −log10 of the p value and the dots represent CpGs
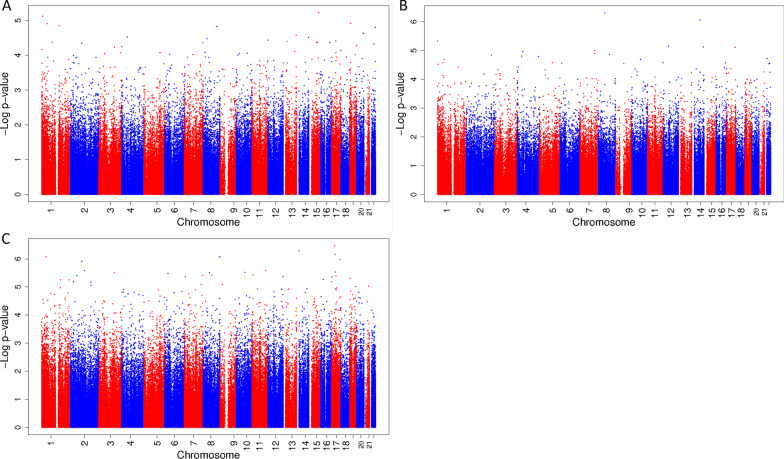
In the sensitivity analysis, in which we used the averaged mixture during pregnancy, there were no significant associations for fetal exposure to the mixture of phthalates and bisphenols in the total study population. (Figure 4A–C shows the Manhattan plots, CpGs with a p value < 1.0 * 10–6 are presented in Table 5, and CpGs with a p value < 1.0 * 10–5 are presented in Additional file 1: Table S6.) Among boys, there were two CpGs that had a suggestive association and among girls, five CpGs had a suggestive association. These associations did not overlap with the suggestive associations we identified in the trimester-specific analyses.
Fig. 4.
Manhattan plot of associations between a mixture of phthalates and bisphenols averaged over pregnancy with DNA methylation at birth in the total population and among boys and girls specifically. Manhattan plot of associations between a mixture of phthalates and bisphenols averaged over pregnancy in the total population (A), among boys (B) and among girls (C) with DNA methylation at birth. In all Manhattan plots, the x-axis represents the autosomal chromosomes, the y-axis represents the −log10 of the p value and the dots represent CpG
Table 5.
CpGs with p values < 1.0 * 10–6 from epigenome-wide association study of a mixture of phthalates and bisphenols in maternal urine averaged over pregnancy and DNA methylation in cord blood among boys and girls specifically
| CpG | Chr | Position | Gene | Effect | SE | p value | Flag# | |
|---|---|---|---|---|---|---|---|---|
| Boys | cg20400361 | 8 | 55,014,040 | LYPLA1 | 9.50 * 10–3 | 1.80 * 10–3 | 4.93 * 10–7 | 0 |
| cg00025138 | 14 | 71,275,917 | MAP3K9 | − 2.57 * 10–3 | 5.01 * 10–4 | 8.64 * 10–7 | 0 | |
| Girls | cg13344757 | 17 | 26904381 | ALDOC | 1.20 * 10–2 | 2.24 * 10–3 | 3.40 * 10–7 | 0 |
| cg04913443 | 14 | 21566084 | ZNF219;C14orf176 | − 7.00 * 10–3 | 1.33 * 10–3 | 5.13 * 10–7 | 0 | |
| cg11723896 | 17 | 34136427 | TAF15 | − 1.05 * 10–2 | 2.02 * 10–3 | 6.95 * 10–7 | 1 | |
| cg10074813 | 8 | 144637872 | GSDMD | 1.51 * 10–2 | 2.93 * 10–3 | 8.43 * 10–7 | 1 | |
| cg15504459 | 1 | 38478291 | UTP11L | − 8.02 * 10–3 | 1.55 * 10–3 | 8.60 * 10–7 | 0 |
There were no CpGs presented for the total group, as none reached our uncorrected p value cutoff of < 1.0 * 10–6
#We have indicated probes that map to DNA containing a single nucleotide polymorphism (SNP), repetitive sequence elements or DNA harboring an insertion or deletion with a ‘1’ in this column
Discussion
Main findings
In this relatively small population-based cohort study including children of European ancestry, using a novel statistical method for the analysis of joint mixture effects of exposure to phthalates and bisphenols during each trimester of pregnancy, we found that exposure to a mixture of phthalates and bisphenols was not significantly associated with DNA methylation in cord blood. However, there were some suggestive associations for exposure in the first and second trimester.
Comparison to the literature
In the total group, we found that exposure to the mixture of phthalates and bisphenols during the first trimester has a suggestive association with a decrease in DNA methylation of cg05058973, which is close to the GHRHR gene. This receptor is mainly expressed in the anterior pituitary gland, and in vitro studies have shown that it plays a role in the regulation of growth hormone, which influences protein and fat metabolism [39]. From animal studies, we know that expression of the GHRHR gene could be influenced by estrogen [40]. Exposure to the mixture during second trimester had a suggestive association with an increase in DNA methylation of cg00141688, which is located near HPCAL1. It has been reported that neonatal exposure to estrogens or BPA led to an increase in DNA methylation of the promoter of HPCAL1 in the prostate gland of rats with persisting effects during life [41]. We additionally found a suggestive association of exposure to the mixture during second trimester with an increase in DNA methylation of cg15961211, which is located near the FAM183A gene. This gene has no known function in relation to exposure to phthalates or bisphenols. Exposure to the mixture during the second trimester also had a suggestive association with a decrease in DNA methylation of cg20840540, which is located close to TRERF1, a gene that interacts with the progesterone receptor after its activation by progesterone [42]. Among boys, second trimester exposure to the mixture showed a suggestive association with a decrease in DNA methylation of cg03764767, which is within the PEX10 gene, which has been indicated as having a role in male fertility as it is important for spermatocyte development [43]. Also among boys, there was some indication that exposure to the mixture during third trimester could be associated with an increase in DNA methylation of cg23462052, which is near the SNRPB gene that has no known function related to our exposures.
The differentially methylated CpG sites identified in this study have not been associated with prenatal BPA and phthalate exposure in previous studies in humans, although a direct comparison of this study with previous literature is difficult, as none of the previous studies studied mixture effects using quantile g-imputation. Apart from implementing a mixtures model in this analysis, this study also differed from previous studies in other ways. First, several of the studies that have shown associations between fetal phthalate and bisphenol exposure and DNA methylation at birth have conducted targeted analyses using pyrosequencing [19, 44, 45]. Different methods have also been used to model chemical exposure, including exposure in the 25th percentile versus the 75th percentile, comparing subjects above and below the 75th percentile, or including the chemicals separately as continuous variables [17, 20–24, 27, 29, 44, 46]. To our knowledge, the only other study that used a chemicals mixture in their model was by Goodrich and colleagues, which used principal components to model third trimester BPA and phthalates in conjunction with targeted pyrosequencing of LINE-1 repetitive elements and IGF2, H19 and Hydroxysteroid 11-Beta Dehydrogenase 2 (HSD11B2) among children aged 8–14 years [15]. In that study, it was found that children from mothers with higher third trimester urine BPA concentrations had increased DNA methylation of IGF2 during peri-adolescence and that higher third trimester maternal urine monobenzylphthalate (mBzBP) and mono-isobutylphthalate (mIBP) concentrations were associated with increased DNA methylation of H19 during peri-adolescence. In the primary components analysis, higher third trimester maternal urine mono-n-butylphthalate (mBP), mBzBP, mIBP and mono(3-carboxypropyl)-phthalate (mCPP) concentrations predicted increased H19 methylation. We are unaware of any studies that have analyzed bisphenol or phthalate exposure from all three trimesters in relation to DNA methylation.
Strengths and limitations
To our knowledge, we have conducted the first study exploring the associations of exposure to a mixture of phthalates and bisphenols at multiple time points during pregnancy with DNA methylation in cord blood. Using this novel mixture approach, we ensured that possible synergistic or antagonistic effects of the components of the mixture are taken into account.
We recognize that our ability to find associations was limited due to the relatively small study population. In addition, the Illumina Infinium Human Methylation450 BeadChip only covers 2% of all CpG sites in the DNA and we only assessed DNA methylation in cord blood, while other tissues could be more informative. The generalizability of our results could be limited due to the fact that the study population was relatively highly educated and only of European ancestry. However, even in this small population, we found potentially promising results that warrant further exploration of these associations in larger studies. As we assessed exposure during all three trimesters, we were able to explore possible vulnerable periods. Based on the number of associations, there is some indication that exposure during first and second trimester might be more relevant. It is known that early pregnancy in particular is a sensitive period for environmental exposures impacting DNA methylation [47]. However, due to the short biological half-lives of phthalates and bisphenols, it could be that our measurements do not accurately represent exposure during the whole trimester. These results should therefore be seen as exploratory and hypothesis-generating.
Future perspectives
Our results support further exploration of exposure to a mixture of phthalates and bisphenols during pregnancy, preferably in a contemporary, larger, multi-ethnic cohort with multiple measures of exposure at different times during pregnancy.
Conclusions
In this exploratory study, we found some suggestive associations for exposure to a mixture of non-persistent endocrine disruptors during first and second trimester of pregnancy with DNA methylation in cord blood. This study underscores the need for larger contemporary studies to further explore the association of mixtures of phthalates and bisphenols with DNA methylation.
Supplementary Information
Additional file 1. Additional file containing supplemental figures and tables. Fig. S1. Flowchart of participants included in the study. Fig. S2. Manhattan plot of associations between a mixture of phthalates and bisphenols during first, second and third trimester with DNA methylation at birth. Table S1. Urine concentrations of phthalates and bisphenols during pregnancy in non-participants. Table S2. CpGs with p-values <1.0 * 10–5 from epigenome-wide association study of a mixture of phthalates and bisphenols in maternal urine during first, second and third trimester and DNA methylation in cord blood. Table S3. CpGs with p-values <1.0 * 10–5 from epigenome-wide association study of a mixture of phthalates and bisphenols in maternal urine during first, second and third trimester and DNA methylation in cord blood. Table S4. CpGs with p-values <1.0 * 10–5 from epigenome-wide association study of a mixture of phthalates and bisphenols in maternal urine during first, second and third trimester and DNA methylation in cord blood among boys. Table S5. CpGs with p-values <1.0 * 10–5 from epigenome-wide association study of a mixture of phthalates and bisphenols in maternal urine during first, second and third trimester and DNA methylation in cord blood among girls. Table S6. CpGs with p-values <1.0 * 10–5 from epigenome-wide association study of a mixture of phthalates and bisphenols in maternal urine averaged over pregnancy and DNA methylation in cord blood in the total group and among boys and girls specifically.
Acknowledgements
The Generation R Study is conducted by Erasmus MC, University Medical Center Rotterdam, in close collaboration with the School of Law and Faculty of Social Sciences of the Erasmus University Rotterdam, the Municipal Health Service Rotterdam area, Rotterdam, the Rotterdam Homecare Foundation, Rotterdam and the Stichting Trombosedienst & Artsenlaboratorium Rijnmond (STAR-MDC), Rotterdam. We gratefully acknowledge the contribution of participating children and parents, general practitioners, hospitals, midwives and pharmacies in Rotterdam. The study protocol was approved by the Medical Ethical Committee of the Erasmus Medical Centre, Rotterdam. Written informed consent was obtained for all participants. The generation and management of the Illumina 450K methylation array data (EWAS data) for the Generation R Study was executed by the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, the Netherlands. We thank Mr. Michael Verbiest, Ms. Mila Jhamai, Ms. Sarah Higgins, Mr. Marijn Verkerk and Dr. Lisette Stolk for their help in creating the EWAS database. We thank Dr. A. Teumer for his work on the quality control and normalization scripts.
Abbreviations
- EDCs
Endocrine disrupting chemicals
- EWAS
Epigenome-wide association study
- FAM183A
Family with sequence similarity 183 member A
- GHRHR
Growth hormone-releasing hormone receptor
- HPCAL1
Hippocalcin-like 1
- HSD11B2
Hydroxysteroid 11-beta dehydrogenase 2
- IGF2
Insulin-like growth factor 2
- IQR
Interquartile range
- LOD
Limit of detection
- mBP
Mono-n-butylphthalate
- mBzBP
Monobenzylphthalate
- mCPP
Mono(3-carboxypropyl)-phthalate
- mIBP
Mono-isobutylphthalate
- MICE
Multivariate imputation by chained equations
- PEX10
Peroxisomal biogenesis factor 10
- SD
Standard deviation
- SE
Standard error
- SNP
Single nucleotide polymorphism
- SNRPB
Small nuclear ribonucleoprotein polypeptides B and B1
- TRERF1
Transcriptional-regulating factor 1
Author contributions
JF and LT contributed to the conceptualization and performed the supervision of the study. VJ, JF and LT have acquired the funding for this project. CS performed the formal analysis. CS and AG wrote the original draft. CS, AG, SS, VJ, JF and LT reviewed and edited the original draft and approved the final manuscript.
Funding
The general design of the Generation R Study is made possible by financial support from the Erasmus MC, University Medical Center Rotterdam, Rotterdam, the Netherlands, Erasmus University Rotterdam, the Organization for Health Research and Development (ZonMw) and the Ministry of Health, Welfare and Sport. The EWAS data were funded by a grant to VWVJ from the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) Netherlands Consortium for Healthy Aging (NCHA; project nr. 050-060-810), by funds from the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, and by a grant from the National Institute of Child and Human Development (R01HD068437). This study was supported by grants R01ES-022972 and R01ES-029779 from the National Institutes of Health, USA. The content is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health. This project received funding from the European Union’s Horizon 2020 research and innovation programme (733206, LifeCycle; 874739, LongITools; 874583, ATHLETE) and from the European Joint Programming Initiative ‘A Healthy Diet for a Healthy Life’ (JPI HDHL, NutriPROGRAM project, ZonMw the Netherlands no. 529051022). VWVJ received a grant from the European Research Council (ERC Consolidator Grant, ERC-2014-CoG-64916). The funding sources had no involvement in the study design, the collection, analysis and interpretation of data, the writing of the report and the decision to submit the article for publication.
Availability of data and materials
The data that support the findings of this study are not publicly available due to privacy or ethical restrictions but are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The study has been reviewed and approved by the Medical Ethical Committee of the Erasmus MC, University Medical Center Rotterdam. Informed consent has been received from all subjects participating in the study prior to conducting the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Janine F. Felix and Leonardo Trasande contributed equally
References
- 1.Liu J, Wattar N, Field CJ, Dinu I, Dewey D, Martin JW, et al. Exposure and dietary sources of bisphenol A (BPA) and BPA-alternatives among mothers in the APrON cohort study. Environ Int. 2018;119:319–326. doi: 10.1016/j.envint.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Zhu H, Kannan K. A review of biomonitoring of phthalate exposures. Toxics. 2019;7(2):21. doi: 10.3390/toxics7020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Golestanzadeh M, Riahi R, Kelishadi R. Association of exposure to phthalates with cardiometabolic risk factors in children and adolescents: a systematic review and meta-analysis. Environ Sci Pollut Res Int. 2019;26(35):35670–35686. doi: 10.1007/s11356-019-06589-7. [DOI] [PubMed] [Google Scholar]
- 4.Ranciere F, Lyons JG, Loh VH, Botton J, Galloway T, Wang T, et al. Bisphenol A and the risk of cardiometabolic disorders: a systematic review with meta-analysis of the epidemiological evidence. Environ Health. 2015;14:46. doi: 10.1186/s12940-015-0036-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao H, Chen D, Zang M. Association between phthalate exposure and insulin resistance: a systematic review and meta-analysis update. Environ Sci Pollut Res Int. 2021;28(40):55967–55980. doi: 10.1007/s11356-021-16252-9. [DOI] [PubMed] [Google Scholar]
- 6.Panagiotou EM, Ojasalo V, Damdimopoulou P. Phthalates, ovarian function and fertility in adulthood. Best Pract Res Clin Endocrinol Metab. 2021;35(5):101552. doi: 10.1016/j.beem.2021.101552. [DOI] [PubMed] [Google Scholar]
- 7.Nahar MS, Liao C, Kannan K, Harris C, Dolinoy DC. In utero bisphenol A concentration, metabolism, and global DNA methylation across matched placenta, kidney, and liver in the human fetus. Chemosphere. 2015;124:54–60. doi: 10.1016/j.chemosphere.2014.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mose T, Knudsen LE, Hedegaard M, Mortensen GK. Transplacental transfer of monomethyl phthalate and mono(2-ethylhexyl) phthalate in a human placenta perfusion system. Int J Toxicol. 2007;26(3):221–229. doi: 10.1080/10915810701352721. [DOI] [PubMed] [Google Scholar]
- 9.Gluckman PD, Hanson MA. Living with the past: evolution, development, and patterns of disease. Science. 2004;305(5691):1733–1736. doi: 10.1126/science.1095292. [DOI] [PubMed] [Google Scholar]
- 10.Engdahl E, Svensson K, Lin PD, Alavian-Ghavanini A, Lindh C, Rüegg J, et al. DNA methylation at GRIN2B partially mediates the association between prenatal bisphenol F exposure and cognitive functions in 7-year-old children in the SELMA study. Environ Int. 2021;156:106617. doi: 10.1016/j.envint.2021.106617. [DOI] [PubMed] [Google Scholar]
- 11.Jahreis S, Trump S, Bauer M, Bauer T, Thürmann L, Feltens R, et al. Maternal phthalate exposure promotes allergic airway inflammation over 2 generations through epigenetic modifications. J Allergy Clin Immunol. 2018;141(2):741–753. doi: 10.1016/j.jaci.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 12.Zhou A, Chang H, Huo W, Zhang B, Hu J, Xia W, et al. Prenatal exposure to bisphenol A and risk of allergic diseases in early life. Pediatr Res. 2017;81(6):851–856. doi: 10.1038/pr.2017.20. [DOI] [PubMed] [Google Scholar]
- 13.Minatoya M, Kishi R. A review of recent studies on bisphenol A and phthalate exposures and child neurodevelopment. Int J Environ Res Public Health. 2021;18(7):3585. doi: 10.3390/ijerph18073585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutta S, Haggerty DK, Rappolee DA, Ruden DM. Phthalate Exposure and Long-Term Epigenomic Consequences: A Review. Front Genet. 2020;11:405. doi: 10.3389/fgene.2020.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodrich JM, Dolinoy DC, Sanchez BN, Zhang Z, Meeker JD, Mercado-Garcia A, et al. Adolescent epigenetic profiles and environmental exposures from early life through peri-adolescence. Environ Epigenet. 2016;2(3):dvw018. doi: 10.1093/eep/dvw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao Y, Chen J, Wang X, Song Q, Xu HH, Zhang YH. Third trimester phthalate exposure is associated with DNA methylation of growth-related genes in human placenta. Sci Rep. 2016;6:33449. doi: 10.1038/srep33449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montrose L, Padmanabhan V, Goodrich JM, Domino SE, Treadwell MC, Meeker JD, et al. Maternal levels of endocrine disrupting chemicals in the first trimester of pregnancy are associated with infant cord blood DNA methylation. Epigenetics. 2018;13(3):301–309. doi: 10.1080/15592294.2018.1448680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi YJ, Lee YA, Hong YC, Cho J, Lee KS, Shin CH, et al. Effect of prenatal bisphenol A exposure on early childhood body mass index through epigenetic influence on the insulin-like growth factor 2 receptor (IGF2R) gene. Environ Int. 2020;143:105929. doi: 10.1016/j.envint.2020.105929. [DOI] [PubMed] [Google Scholar]
- 19.LaRocca J, Binder AM, McElrath TF, Michels KB. The impact of first trimester phthalate and phenol exposure on IGF2/H19 genomic imprinting and birth outcomes. Environ Res. 2014;133:396–406. doi: 10.1016/j.envres.2014.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miura R, Araki A, Minatoya M, Miyake K, Chen ML, Kobayashi S, et al. An epigenome-wide analysis of cord blood DNA methylation reveals sex-specific effect of exposure to bisphenol A. Sci Rep. 2019;9(1):12369. doi: 10.1038/s41598-019-48916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCabe CF, Padmanabhan V, Dolinoy DC, Domino SE, Jones TR, Bakulski KM, et al. Maternal environmental exposure to bisphenols and epigenome-wide DNA methylation in infant cord blood. Environ Epigenet. 2020;6(1):dvaa021. doi: 10.1093/eep/dvaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen CH, Jiang SS, Chang IS, Wen HJ, Sun CW, Wang SL. Association between fetal exposure to phthalate endocrine disruptor and genome-wide DNA methylation at birth. Environ Res. 2018;162:261–270. doi: 10.1016/j.envres.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 23.Huang YF, Chang CH, Chen PJ, Lin IH, Tsai YA, Chen CF, et al. Prenatal bisphenol a exposure, DNA methylation, and low birth weight: a pilot study in Taiwan. Int J Environ Res Public Health. 2021;18(11):6144. doi: 10.3390/ijerph18116144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miura R, Ikeda-Araki A, Ishihara T, Miyake K, Miyashita C, Nakajima T, et al. Effect of prenatal exposure to phthalates on epigenome-wide DNA methylations in cord blood and implications for fetal growth: the Hokkaido Study on Environment and Children's Health. Sci Total Environ. 2021;783:147035. doi: 10.1016/j.scitotenv.2021.147035. [DOI] [PubMed] [Google Scholar]
- 25.Petroff RL, Padmanabhan V, Dolinoy DC, Watkins DJ, Ciarelli J, Haggerty D, et al. Prenatal exposures to common phthalates and prevalent phthalate alternatives and infant DNA methylation at birth. Front Genet. 2022;13:793278. doi: 10.3389/fgene.2022.793278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.England-Mason G, Merrill SM, Gladish N, Moore SR, Giesbrecht GF, Letourneau N, et al. Prenatal exposure to phthalates and peripheral blood and buccal epithelial DNA methylation in infants: an epigenome-wide association study. Environ Int. 2022;163:107183. doi: 10.1016/j.envint.2022.107183. [DOI] [PubMed] [Google Scholar]
- 27.Junge KM, Leppert B, Jahreis S, Wissenbach DK, Feltens R, Grützmann K, et al. MEST mediates the impact of prenatal bisphenol A exposure on long-term body weight development. Clin Epigenetics. 2018;10:58. doi: 10.1186/s13148-018-0478-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alavian-Ghavanini A, Lin PI, Lind PM, Risén Rimfors S, Halin Lejonklou M, Dunder L, et al. Prenatal bisphenol A exposure is linked to epigenetic changes in glutamate receptor subunit gene Grin2b in female rats and humans. Sci Rep. 2018;8(1):11315. doi: 10.1038/s41598-018-29732-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Solomon O, Yousefi P, Huen K, Gunier RB, Escudero-Fung M, Barcellos LF, et al. Prenatal phthalate exposure and altered patterns of DNA methylation in cord blood. Environ Mol Mutagen. 2017;58(6):398–410. doi: 10.1002/em.22095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bowman A, Peterson KE, Dolinoy DC, Meeker JD, Sánchez BN, Mercado-Garcia A, et al. Phthalate exposures, DNA methylation and adiposity in mexican children through adolescence. Front Public Health. 2019;7:162. doi: 10.3389/fpubh.2019.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keil AP, Buckley JP, O'Brien KM, Ferguson KK, Zhao S, White AJ. A quantile-based g-computation approach to addressing the effects of exposure mixtures. Environ Health Perspect. 2020;128(4):47004. doi: 10.1289/EHP5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaddoe VW, van Duijn CM, Franco OH, van der Heijden AJ, van Iizendoorn MH, de Jongste JC, et al. The Generation R Study: design and cohort update 2012. Eur J Epidemiol. 2012;27(9):739–756. doi: 10.1007/s10654-012-9735-1. [DOI] [PubMed] [Google Scholar]
- 33.Philips EM, Jaddoe VWV, Asimakopoulos AG, Kannan K, Steegers EAP, Santos S, et al. Bisphenol and phthalate concentrations and its determinants among pregnant women in a population-based cohort in the Netherlands, 2004–5. Environ Res. 2018;161:562–572. doi: 10.1016/j.envres.2017.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hornung RWRL. Estimation of average concentration in the presence of nondetectable values. Appl Occup Environ Hyg. 1990;5(1):46–51. doi: 10.1080/1047322X.1990.10389587. [DOI] [Google Scholar]
- 35.Lehne B, Drong AW, Loh M, Zhang W, Scott WR, Tan ST, et al. A coherent approach for analysis of the Illumina HumanMethylation450 BeadChip improves data quality and performance in epigenome-wide association studies. Genome Biol. 2015;16(1):37. doi: 10.1186/s13059-015-0600-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen YA, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8(2):203–209. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naeem H, Wong NC, Chatterton Z, Hong MK, Pedersen JS, Corcoran NM, et al. Reducing the risk of false discovery enabling identification of biologically significant genome-wide methylation status using the HumanMethylation450 array. BMC Genomics. 2014;15(1):51. doi: 10.1186/1471-2164-15-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Padmanabhan V, Song W, Puttabyatappa M. Praegnatio perturbatio-impact of endocrine-disrupting chemicals. Endocr Rev. 2021;42(3):295–353. doi: 10.1210/endrev/bnaa035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bilezikjian LM, Vale WW. Stimulation of adenosine 3',5'-monophosphate production by growth hormone-releasing factor and its inhibition by somatostatin in anterior pituitary cells in vitro. Endocrinology. 1983;113(5):1726–1731. doi: 10.1210/endo-113-5-1726. [DOI] [PubMed] [Google Scholar]
- 40.Petersenn S, Rasch AC, Heyens M, Schulte HM. Structure and regulation of the human growth hormone-releasing hormone receptor gene. Mol Endocrinol. 1998;12(2):233–247. doi: 10.1210/mend.12.2.0057. [DOI] [PubMed] [Google Scholar]
- 41.Tang WY, Morey LM, Cheung YY, Birch L, Prins GS, Ho SM. Neonatal exposure to estradiol/bisphenol A alters promoter methylation and expression of Nsbp1 and Hpcal1 genes and transcriptional programs of Dnmt3a/b and Mbd2/4 in the rat prostate gland throughout life. Endocrinology. 2012;153(1):42–55. doi: 10.1210/en.2011-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gizard F, Robillard R, Gross B, Barbier O, Révillion F, Peyrat JP, et al. TReP-132 is a novel progesterone receptor coactivator required for the inhibition of breast cancer cell growth and enhancement of differentiation by progesterone. Mol Cell Biol. 2006;26(20):7632–7644. doi: 10.1128/MCB.00326-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gu X, Li H, Chen X, Zhang X, Mei F, Jia M, et al. PEX10, SIRPA-SIRPG, and SOX5 gene polymorphisms are strongly associated with nonobstructive azoospermia susceptibility. J Assist Reprod Genet. 2019;36(4):759–768. doi: 10.1007/s10815-019-01417-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kundakovic M, Gudsnuk K, Herbstman JB, Tang D, Perera FP, Champagne FA. DNA methylation of BDNF as a biomarker of early-life adversity. Proc Natl Acad Sci USA. 2015;112(22):6807–6813. doi: 10.1073/pnas.1408355111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tindula G, Murphy SK, Grenier C, Huang Z, Huen K, Escudero-Fung M, et al. DNA methylation of imprinted genes in Mexican-American newborn children with prenatal phthalate exposure. Epigenomics. 2018;10(7):1011–1026. doi: 10.2217/epi-2017-0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song X, Zhou X, Yang F, Liang H, Wang Z, Li R, et al. Association between prenatal bisphenol a exposure and promoter hypermethylation of CAPS2, TNFRSF25, and HKR1 genes in cord blood. Environ Res. 2020;190:109996. doi: 10.1016/j.envres.2020.109996. [DOI] [PubMed] [Google Scholar]
- 47.Felix JF, Cecil CAM. Population DNA methylation studies in the Developmental Origins of Health and Disease (DOHaD) framework. J Dev Orig Health Dis. 2019;10(3):306–313. doi: 10.1017/S2040174418000442. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Additional file containing supplemental figures and tables. Fig. S1. Flowchart of participants included in the study. Fig. S2. Manhattan plot of associations between a mixture of phthalates and bisphenols during first, second and third trimester with DNA methylation at birth. Table S1. Urine concentrations of phthalates and bisphenols during pregnancy in non-participants. Table S2. CpGs with p-values <1.0 * 10–5 from epigenome-wide association study of a mixture of phthalates and bisphenols in maternal urine during first, second and third trimester and DNA methylation in cord blood. Table S3. CpGs with p-values <1.0 * 10–5 from epigenome-wide association study of a mixture of phthalates and bisphenols in maternal urine during first, second and third trimester and DNA methylation in cord blood. Table S4. CpGs with p-values <1.0 * 10–5 from epigenome-wide association study of a mixture of phthalates and bisphenols in maternal urine during first, second and third trimester and DNA methylation in cord blood among boys. Table S5. CpGs with p-values <1.0 * 10–5 from epigenome-wide association study of a mixture of phthalates and bisphenols in maternal urine during first, second and third trimester and DNA methylation in cord blood among girls. Table S6. CpGs with p-values <1.0 * 10–5 from epigenome-wide association study of a mixture of phthalates and bisphenols in maternal urine averaged over pregnancy and DNA methylation in cord blood in the total group and among boys and girls specifically.
Data Availability Statement
The data that support the findings of this study are not publicly available due to privacy or ethical restrictions but are available from the corresponding author on reasonable request.