Abstract
We present a clinical case where a complex abnormal non‐invasive prenatal test (NIPT) result in a research project revealed carcinoma of the breast in the pregnant woman. Furthermore, the NIPT result did not demonstrate the same fetal chromosomal aberration as the chorion villus sample. A literature search for similar cases was performed identifying 43 unique cases, where abnormal NIPT results were related to maternal malignancy. Malignancy is a rare but important cause of complex abnormal non‐invasive prenatal test (NIPT) results and should be considered when fetal karyotype and abnormal NIPT results are discordant. Furthermore, a follow‐up invasive sample is essential for correct fetal diagnosis when abnormal NIPT results are found.
Keywords: cell‐free DNA, fetal diagnostics, genetics, malignancy, NIPT, prenatal diagnostics
Maternal malignancy is a rare but important cause of complex abnormal NIPT results and should not be overlooked.

1. INTRODUCTION
Non‐invasive prenatal test (NIPT) is a method validated to screen for the three most common fetal trisomies, trisomy 13, 18, and 21 (T13, T18, T21), by analysis of a maternal blood sample containing cell‐free DNA (cfDNA) from both mother and fetus. The fetal cfDNA stems primarily from placental cytotrophoblasts. 1 Some NIPTs may also detect increased risk of sex chromosome aberrations and some genome wide NIPTs may detect pathogenic copy number variations of considerable size. 2 Specific NIPT methods vary, but the overall point of the test is that the amount of DNA from different locations in the genome is “counted” (both maternal and fetal). Trisomy in a fetus is suspected, when there is surplus of DNA from one of the chromosomes 13, 18, and 21, as compared to what would be expected (normal regions/bins as reference). 3
Non‐invasive prenatal test is relatively recently implemented as part of prenatal screening programs in several countries. 4 It was introduced into clinical practice in some countries around 2011 5 , 6 /2012, 7 but in Denmark it became an integrated part of the national free‐of‐charge screening program in 2017. Women with a risk of Down Syndrome above 1:300 (high risk women) at the combined first trimester screening (CFTS) may choose between invasive testing with high‐resolution chromosomal microarray or NIPT validated for trisomy 13, 18, 21 and sex chromosome anomalies only.
Non‐invasive prenatal test has a high sensitivity and specificity for the trisomies 13, 18, and 21. 8 , 9 In some settings, NIPT is also used to screen for sex chromosome abnormalities. 8
Non‐invasive prenatal test results positive for fetal chromosomal abnormalities should always be confirmed by invasive testing, as the NIPT method itself is not diagnostic for fetal aneuploidy. For low‐risk women the positive predictive value for T21 may be below 50%, 10 and even for a high‐risk woman, a high negative predictive value may not exclude T21. 5
Discordant NIPT results are reported due to maternal genetic abnormalities such as copy number variations (CNV), 11 , 12 confined placental mosaicism, 12 , 13 , 14 , 15 a vanished twin pregnancy 16 or maternal malignancy 5 although the last complication is rare. 5
The present clinical case report is, to our knowledge, the first known case of a maternal malignancy in combination with a complex abnormal NIPT result from a Scandinavian site.
As other cases reported in the literature, it demonstrates that sometimes circulating tumor DNA (ctDNA) can interfere with and complicate a NIPT result.
2. CASE HISTORY
In January 2014, a +30‐year‐old pregnant woman underwent invasive testing by chorion villous sampling (CVS), as the nuchal translucency was 7.5 mm and the risk of T21 was above 1:50 at CFTS. Just prior to the CVS, a blood sample was drawn from the woman in the clinical setting. The CVS showed chromosomal abnormalities involving chromosome 21, 17 and after counseling the pregnancy was terminated on parental request.
In a later research project at “Center for Fetal Diagnostics” (CFFD) at Aarhus University Hospital, the blood sample drawn from the same woman in her pregnancy, was included in a cfDNA cohort for NIPT validation of atypical samples. Ethical approval for this cfDNA cohort was provided by The Regional Research Ethics Committee Central Denmark Region by request no 267/2015. The Danish Data Protection Agency approved the data handling (1‐16‐02‐659‐16). This NIPT, analyzed at Erasmus MC, Rotterdam in the Netherlands, revealed a complex abnormal result, and raised a suspicion of maternal neoplasia. The medical records of the woman were further investigated and it turned out that she had been diagnosed with a unilateral solid tumor, carcinoma of the breast, just 1 month after the original CVS was performed and the NIPT research sample was collected. She had had the genes BRCA1 and −2 screened and no pathogenic variations were detected. The woman had died from her breast cancer before the NIPT research project was initiated.
3. METHODS
3.1. Clinical case
3.1.1. Chorion villous sampling
Chromosomal microarray (CMA, Agilent oligoarray, 180 kb) was performed on uncultured cells from whole, untrypsinated chorionic villi selected by visual inspection of the sample. Furthermore, karyotyping and multicolor mBAND21 fluorescence in situ hybridization (FISH) was done on cultured cells.
3.1.2. NIPT analysis
The DNA from 2–3 ml plasma (from the pregnant woman presented in the clinical case) was extracted with the QIAamp Circulating Nucleic Acid kit (Qiagen). The concentration of the DNA was measured with the Quant‐iT dsDNA High‐Sensitivity assay kit (Life Technologies). The sequence library was created from 10 μl cfDNA of at least 0.05 ng/μl of cfDNA using the Thruplex plasma‐SEQ sample preparation kit (Rubicon Genomics). Two unique synthetic DNA “barcodes” (dual index) were attached to each sample and shallow whole genome sequencing was performed on a HiSeq2500 sequencer yielding 50‐bp reads in a 96‐plex reaction. Genome wide analysis (excluding sex chromosomes) was performed by using Wisecondor software after achieving 26.6mln reads. 18
The fetal fraction was only indirectly calculated by use of SeqFF algorithm. The SeqFF method published by Kim and colleagues 19 determines the fraction based on the assumption that fetal and maternal reads are not equally distributed over the human genome due to fragment size differences and short read lengths. The SeqFF is a multivariate regression model trained with read counts over specific autosomal regions in a large sample set used to predict the “fetal” fraction of the test samples. The SeqFF method is independent of the fetal gender. 19 The fetal fraction was estimated to be 11%, but it does not distinguish fetal cfDNA from tumor cfDNA.
3.2. Review
A semi‐systematic search of the online database PubMed was done in March 2019 by MHM, resulting in 141 possible relevant articles (see search strings in Table S2). Additionally, 3 records were found by mention of co‐authors. After removal of duplicates, 79 articles were screened by MHM and IV on title and abstract. Of these, 27 were found relevant for full article screening by MHM. IV did a 2nd screening of the 79 articles on title and abstract and found 22 articles relevant for full article screening. These 22 articles were all represented in the 27 chosen by MHM. MHM did full article screening of the 27 articles, to determine which articles presented relevant case reports.
4. RESULTS
4.1. Clinical case
4.1.1. Chorion villous sampling
Chromosomal microarray on uncultured CVS‐DNA detected a 5,5 Mb terminal deletion on 21q22.2q22.3 (Figure 1A). Karyotyping and multicolor mBAND21 FISH on cultured CVS‐cells revealed one normal chromosome 21 and one iso‐chromosome 21 in all metaphases analyzed (Figure 1B). CMA on cultured CVS confirmed duplication of chromosome 21 (q11.2q22.2) and a terminal chromosome 21 (q22.2q22.3) deletion (Figure 1B). When re‐evaluating the microarray result the duplication could still not be seen in the uncultured sample, but a potential low‐grade mosaicism below 10% could not be excluded.
FIGURE 1.

(A) UNCULTURED CVS. Array‐CGH revealed a 5.6 Mb terminal deletion on chromosome 21q (arr[GRCh37] 21q22.2q22.3(42538181‐48090317)x1). (BI) CULTURED CVS. Array‐CGH revealed a 27 Mb duplication and the terminal deletion of 5.6 Mb of chromosome 21q (arr[GRCh37] 21q11.2q22.2(15390816‐42482129)x3,21q22.2q22.3(42518800‐48090317)x1). (BII) Multicolor mBAND21 confirmed the suspicion of duplication of chromosome 21 (q11.2q22.2) and terminal deletion of chromosome 21 (q22.2q22.3) in cultured CVS. Karyotyping, not shown, revealed one normal chromosome 21 and one iso‐chromosome 21 in all metaphases analyzed.
4.1.2. NIPT analysis
Unexpectedly, instead of an aberration of chromosome 21, the cfDNA showed a complex abnormal chromosome profile. Multiple genomic aberrations of several chromosomes were found in the cfDNA that were discordant to the CVS karyotype. Figure 2 shows the cfDNA profile with multiple structural and numerical aberrations: interstitial 1q gain, terminal 1q gain, 3q gain, monosomy 5, loss in 8p and 8q gain, 12p gain and 12q loss, 13q gain, monosomy 14, and possible 19p and 19q gain. Chromosome 21 was not called as abnormal by the software but had an unusual pattern with a subtle gain and terminal loss. This was likely noticed only thanks to a priori knowledge of the CVS results.
FIGURE 2.
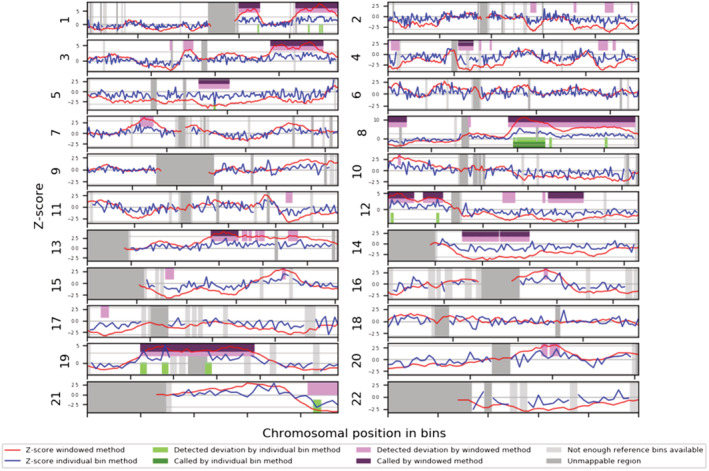
The whole genome Wisecondor results showing multiple events called by the sliding window method. The cfDNA profile shows the following aberrations: interstitial gain in 1q, terminal gain in 1q, gain in 3q, monosomy 5, loss in 8p and gain in 8q,gain in 12p and loss in 12q, gain in 13q, monosomy 14, and possible gain in 19p and 19q. Chromosome 21 was not called by window method, but it has an unusual pattern with a subtle interstitial gain and possible terminal loss. This was noticed likely only thanks to a priori knowledge of the CVS results.
4.2. Review
Of the 27 full articles screened, 13 articles were identified as relevant, presenting case reports of malignant disease in pregnant women with relation to aberrant NIPT results. 20 , 21 , 22 , 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 From these, 43 unique case reports of malignancy related to an unusual NIPT were found. Of the 13 articles, 10 were case reports, some of which were combined with literature reviews. The remaining 3 articles were retrospective follow‐ups, where pregnant women who underwent NIPT were the study populations.
A total of 45 cases were reported in the 13 articles included, but 2 cases were represented in 2 articles each, why the number of unique cases identified ultimately was 43.
Many of the cases were identified retrospectively, and often the authors were not physicians treating the women, limiting access to background information (see Table S1 for details of each of the cases, as they were stated in the published articles).
For some of the cases presented, the aberrant NIPT result lead to further clinical evaluations and cancer diagnosis. In others, cancer was diagnosed unrelated to the NIPT result.
In most of the cases included, the NIPT results showed multiple chromosomal aberrations, discordant with known fetal karyotypes. In a few cases, only one abnormality (e.g., monosomy or trisomy) was found, which were proven discordant because of invasive tests showing normal fetal karyotype.
Many different cancer types were represented (see Table S1). Cancer stage was only reported in some and ranged from early stages to late‐stage metastatic disease.
5. DISCUSSION
We present this unusual case of abnormal NIPT to facilitate the recognition of malignancy in a NIPT sample. Another important lesson from this case is that the malignancy may cause fetal aberrations to be overlooked. Most probably the fetal aberration of 21q in the present case was either too small to be detected (5 Mb) or masked by the presence of abnormal tumor DNA. Without the CVS result it is very unlikely that we would have diagnosed the true fetal aberration among the many cfDNA aberrations induced by the neoplastic cell line. We therefore suggest that also NIPT profiles raising the suspicion of malignancy are followed up by invasive sampling to investigate fetal genotype. Furthermore, this unusual case demonstrates discordant results from DNA extracted from uncultured, untrypsinated chorionic villus cells and cultured, trypsinated cells from other villi.
Malignancy profiles in NIPT samples have been described previously. Most of the identified published cases of maternal malignancy related to unusual NIPT results, including this clinical case, showed multiple genomic aberrations. Multiple, large deletions and duplications are unlikely to be compatible with a viable fetus, which of course offers reason for scrutiny, when a pregnancy otherwise appears to include a viable fetus. Such a complex abnormal NIPT result is highly suggestive of acquired chromosome aberrations in the pregnant woman. It is therefore necessary to look for malignancy (or a benign neoplasia such as a leiomyoma), which is also pointed out by Carlson et al. 30 and Dharajiya et al. 31 Snyder et al. 33 presents a follow‐up study on multiple aneuploidy and single monosomy NIPT results in a cohort of 113,415 NIPT cases, and has follow‐up information on 26 cases with multiple genomic aberrations, 5 of which turned out to have cancer.
Not all malignancies found in women with discordant NIPT results were linked to multiple chromosomal aberrations on NIPT—a few of the published cases mentioned only had single autosomal monosomies or trisomies (e.g., case 2,3 and 6 mentioned in Bianchi et al. 21 ). This underlines that malignancy is a rare differential diagnosis in all discordant NIPT results, not only the ones with multiple aneuploidies. It is, however, clear from Snyder et al. 33 that several of the reported cases 20 , 21 , 22 , 24 , 26 , 27 , 28 , 30 , 31 , 32 and our present case, that a NIPT with multiple aberrations is the common finding when maternal malignancy is actually present.
6. CONCLUSION
Malignancies are rare in pregnant women, 34 but NIPT for fetal aneuploidy may reveal complex abnormal patterns, when cancer is present. Because of this, when a complex abnormal pattern is detected on a NIPT, follow‐up analyses for neoplasia in the mother must be undertaken. It is, however, also important to initiate follow‐up invasive testing, as maternal neoplasia and ctDNA may have masked fetal aneuploidy.
AUTHOR CONTRIBUTIONS
Initial idea for this article was developed by Ida Vogel and Ida Charlotte Bay Lund, during a research co‐work with Anne‐Bine Skytte, Naja Becher, Lotte Andreasen, and Malgorzata Ilona Srebniak. The review of similar articles and initial article draft was written by Maria Hammer Møllgaard. Screening of possible review articles on abstract and title was also done by Ida Vogel, full text analysis by MHM as summarized in Tables S1 and S2. Alterations and proof reading of the article and especially contributions to the Methods, Results, and Conclusion sections of the article was a joint work between all co‐authors.
FUNDING INFORMATION
Ida Vogel is funded by a grant from the Novo Nordic Foundation: NNF16OC0018772.
CONFLICT OF INTEREST
No authors declared any conflicts of interest.
CONSENT
Written consent from the patient could not be obtained as the patient is deceased. See Tables S1 and S2 for further information on consent.
Supporting information
Tables S1‐S2
ACKNOWLEDGMENTS
The authors would like to thank their families for continuous support.
Moellgaard MH, Lund ICB , Becher N, et al. Incidental finding of maternal malignancy in an unusual non‐invasive prenatal test and a review of similar cases. Clin Case Rep. 2022;10:e06280. doi: 10.1002/ccr3.6280
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study are available in the Supplementary Material of this article.
REFERENCES
- 1. Alberry M, Maddocks D, Jones M, et al. Free fetal DNA in maternal plasma in anembryonic pregnancies: confirmation that the origin is the trophoblast. Prenat Diagn. 2007;27(5):415‐418. [DOI] [PubMed] [Google Scholar]
- 2. Kucharik M, Gnip A, Hyblova M, et al. Non‐invasive prenatal testing (NIPT) by low coverage genomic sequencing: detection limits of screened chromosomal microdeletions. PLoS One. 2020;15(8):e0238245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chiu RW, Chan KC, Gao Y, et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc Natl Acad Sci U S A. 2008;105(51):20458‐20463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gadsbøll K, Petersen OB, Gatinois V, et al. Current use of noninvasive prenatal testing in Europe, Australia and the USA: a graphical presentation. Acta Obstet Gynecol Scand. 2020;99(6):722‐730. [DOI] [PubMed] [Google Scholar]
- 5. Hartwig TS, Ambye L, Sorensen S, et al. Discordant non‐invasive prenatal testing (NIPT) – a systematic review. Prenat Diagn. 2017;37(6):527‐539. [DOI] [PubMed] [Google Scholar]
- 6. Bianchi DW, Wilkins‐Haug L. Integration of noninvasive DNA testing for aneuploidy into prenatal care: what has happened since the rubber met the road? Clin Chem. 2014;60(1):78‐87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gray KJ, Wilkins‐Haug LE. Have we done our last amniocentesis? Updates on cell‐free DNA for down syndrome screening. Pediatr Radiol. 2018;48(4):461‐470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gil MM, Quezada MS, Revello R, Akolekar R, Nicolaides KH. Analysis of cell‐free DNA in maternal blood in screening for fetal aneuploidies: updated meta‐analysis. Ultrasound Obstet Gynecol. 2015;45(3):249‐266. [DOI] [PubMed] [Google Scholar]
- 9. Iwarsson E, Jacobsson B, Dagerhamn J, Davidson T, Bernabé E, Heibert Arnlind M. Analysis of cell‐free fetal DNA in maternal blood for detection of trisomy 21, 18 and 13 in a general pregnant population and in a high risk population – a systematic review and meta‐analysis. Acta Obstet Gynecol Scand. 2017;96(1):7‐18. [DOI] [PubMed] [Google Scholar]
- 10. Petersen AK, Cheung SW, Smith JL, et al. Positive predictive value estimates for cell‐free noninvasive prenatal screening from data of a large referral genetic diagnostic laboratory. Am J Obstet Gynecol. 2017;217(6):691.e1‐691.e6. [DOI] [PubMed] [Google Scholar]
- 11. Snyder MW, Simmons LE, Kitzman JO, et al. Copy‐number variation and false positive prenatal aneuploidy screening results. N Engl J Med. 2015;372(17):1639‐1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang H, Gao Y, Jiang F, et al. Non‐invasive prenatal testing for trisomies 21, 18 and 13: clinical experience from 146,958 pregnancies. Ultrasound Obstet Gynecol. 2015;45(5):530‐538. [DOI] [PubMed] [Google Scholar]
- 13. Choi H, Lau TK, Jiang FM, et al. Fetal aneuploidy screening by maternal plasma DNA sequencing: ‘false positive’ due to confined placental mosaicism. Prenat Diagn. 2013;33(2):198‐200. [DOI] [PubMed] [Google Scholar]
- 14. Dugo N, Padula F, Mobili L, et al. Six consecutive false positive cases from cell‐free fetal DNA testing in a single referring Centre. J Prenat Med. 2014;8(1–2):31‐35. [PMC free article] [PubMed] [Google Scholar]
- 15. Huijsdens‐van Amsterdam K, Page‐Christiaens L, Flowers N, et al. Isochromosome 21q is overrepresented among false‐negative cell‐free DNA prenatal screening results involving down syndrome. Eur J Hum Genet. 2018;26(10):1490‐1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gromminger S, Yagmur E, Erkan S, et al. Fetal aneuploidy detection by cell‐free DNA sequencing for multiple pregnancies and quality issues with vanishing twins. J Clin Med. 2014;3(3):679‐692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Becher N, Skytte A. “Discrepancy between cytogenetic and molecular results in two cases with isochromosomes” 10th European cytogenetics conference 2015. Chromosome Res. 2015;23(1):1‐157. [Google Scholar]
- 18. Straver R, Sistermans EA, Reinders MJ. Introducing WISECONDOR for noninvasive prenatal diagnostics. Expert Rev Mol Diagn. 2014;14(5):513‐515. [DOI] [PubMed] [Google Scholar]
- 19. Kim SK, Hannum G, Geis J, et al. Determination of fetal DNA fraction from the plasma of pregnant women using sequence read counts. Prenat Diagn. 2015;35(8):810‐815. [DOI] [PubMed] [Google Scholar]
- 20. Osborne CM, Hardisty E, Devers P, et al. Discordant noninvasive prenatal testing results in a patient subsequently diagnosed with metastatic disease. Prenat Diagn. 2013;33(6):609‐611. [DOI] [PubMed] [Google Scholar]
- 21. Bianchi DW, Chudova D, Sehnert AJ, et al. Noninvasive prenatal testing and incidental detection of occult maternal malignancies. JAMA. 2015;314(2):162‐169. [DOI] [PubMed] [Google Scholar]
- 22. Amant F, Verheecke M, Wlodarska I, et al. Presymptomatic identification of cancers in pregnant women during noninvasive prenatal testing. JAMA Oncol. 2015;1(6):814‐819. [DOI] [PubMed] [Google Scholar]
- 23. Sun K, Jiang P, Chan KC, et al. Plasma DNA tissue mapping by genome‐wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci U S A. 2015;112(40):E5503‐E5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vandenberghe P, Wlodarska I, Tousseyn T, et al. Non‐invasive detection of genomic imbalances in Hodgkin/reed‐Sternberg cells in early and advanced stage Hodgkin's lymphoma by sequencing of circulating cell‐free DNA: a technical proof‐of‐principle study. Lancet Haematol. 2015;2(2):e55‐e65. [DOI] [PubMed] [Google Scholar]
- 25. Luskin MR, Discenza MN, Easter SR, et al. Maternal iAMP21 acute lymphoblastic leukemia detected on prenatal cell‐free DNA genetic screening. Blood Adv. 2017;1(19):1491‐1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Imbert‐Bouteille M, Chiesa J, Gaillard JB, et al. An incidental finding of maternal multiple myeloma by non invasive prenatal testing. Prenat Diagn. 2017;37(12):1257‐1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smith J, Kean V, Bianchi DW, et al. Cell‐free DNA results lead to unexpected diagnosis. Clin Case Rep. 2017;5(8):1323‐1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ji X, Chen F, Zhou Y, et al. Copy number variation profile in noninvasive prenatal testing (NIPT) can identify co‐existing maternal malignancies: case reports and a literature review. Taiwan J Obstet Gynecol. 2018;57(6):871‐877. [DOI] [PubMed] [Google Scholar]
- 29. Cordeiro Mitchell CN, Murdock T, Fader AN, Stone RL. Advanced ovarian cancer treated in pregnancy and detected by cell‐free DNA aneuploidy screening. Gynecol Oncol Rep. 2018;24:48‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carlson LM, Hardisty E, Coombs CC, Vora NL. Maternal malignancy evaluation after discordant cell‐free DNA results. Obstet Gynecol. 2018;131(3):464‐468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dharajiya NG, Grosu DS, Farkas DH, et al. Incidental detection of maternal neoplasia in noninvasive prenatal testing. Clin Chem. 2018;64(2):329‐335. [DOI] [PubMed] [Google Scholar]
- 32. Suzumori N, Sekizawa A, Takeda E, et al. Classification of factors involved in nonreportable results of noninvasive prenatal testing (NIPT) and prediction of success rate of second NIPT. Prenat Diagn. 2019;39(2):100‐106. [DOI] [PubMed] [Google Scholar]
- 33. Snyder HL, Curnow KJ, Bhatt S, Bianchi DW. Follow‐up of multiple aneuploidies and single monosomies detected by noninvasive prenatal testing: implications for management and counseling. Prenat Diagn. 2016;36(3):203‐209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van der Meij KRM, Sistermans EA, Macville MVE, et al. TRIDENT‐2: National Implementation of genome‐wide non‐invasive prenatal testing as a first‐tier screening test in The Netherlands. Am J Hum Genet. 2019;105(6):1091‐1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1‐S2
Data Availability Statement
The data that supports the findings of this study are available in the Supplementary Material of this article.


