ABSTRACT
Riemerella anatipestifer is an important bacterial pathogen in the global duck industry and causes heavy economic losses. In our previous study, we demonstrated that R. anatipestifer type IX secretion system components GldK and GldM, and the secretion protein metallophosphoesterase, acted as virulence factors. In this study, R. anatipestifer AS87_RS02955 was investigated for virulence and enzymatic activity properties. We constructed AS87_RS02955 mutation and complementation strains to assess bacterial virulence. In vivo bacterial loads showed a significantly reduced bacterial loads in the blood of ducks infected with mutant strain Yb2Δ02955, which was recovered in the blood of ducks infected with the complementation strain cYb2Δ02955, demonstrating that AS87_RS02955 was associated with virulence. Further studies showed AS87_RS02955 was a novel nonspecific endonuclease with no functionally conserved domain, but enzymatic activity toward DNA and RNA was indicated. DNase activity was activated by Zn2+, Cu2+, Mg2+, Ca2+, and Mn2+ ions but inhibited by ethylenediaminetetraacetic acid. RNase activity was independent of metal cations, but stimulated by Mg2+, Ca2+, and Mn2+. RAS87_RS02955 enzymatic activity was active across a broad pH and temperature range. Moreover, we identified four sites in rAS87_RS02955, F39, F92, I134, and F145, which were critical for enzymatic activity. In summary, we showed that R. anatipestifer AS87_RS02955 encoded a novel endonuclease with important roles in bacterial virulence.
IMPORTANCE R. anatipestifer AS87_RS02955 was identified as a novel T9SS effector and displayed a nonspecific endonuclease activity in this study. The protein did not contain a conserved His-Asn-His motif structure, which is similar to the endonuclease from Prevotella sp. Its mutant strain Yb2Δ02955 demonstrated significantly attenuated virulence, suggesting AS87_RS02955 is an important virulence factor. Moreover, AS87_RS02955 displayed nonspecific endonuclease activity to cleave λ DNA and MS2 RNA, while four protein sites were critical for endonuclease activity. In conclusion, R. anatipestifer AS87_RS02955 plays important roles in bacterial virulence.
KEYWORDS: Riemerella anatipestifer, AS87_RS02955, virulence, endonuclease
INTRODUCTION
Riemerella anatipestifer is a Gram-negative, rod-shaped, non-spore-forming bacterium (1), and is classified as a member of the Flavobacteriaceae family in the rRNA superfamily V based on 16S rRNA sequence analysis (2). R. anatipestifer causes acute and chronic lethal septicemia characterized by fibrinous polyserositis and meningitis, and is transmitted in ducks, waterfowl, turkey, and other birds (3–5). Several R. anatipestifer virulence factors have been identified, including outer membrane protein A, glycosyltransferase VapD, CAMP cohemolysin, nicotinamidase PncA, and iron acquisition protein SprA (6–12).
Protein secretion systems are crucial for bacterial growth and survival and include nutrient acquisition, responses to different extracellular milieu, and antibiotic resistance (13). The type IX (T9SS) or Por secretion system is a novel system and has been described in members of the Bacteroidetes phylum (14). The presence of an N-terminal signal peptide and a conserved C-terminal domain (CTD) in most T9SS secreted proteins is a common topological characteristic (15). The N-terminal signal peptide allows protein transport across the plasma membrane via the secretion (Sec) system. The T9SS recognizes the CTD signal which allows proteins across the outer membrane (16). T9SS is required for the secretion of gliding motility apparatus components, adhesins, and various enzymes (13).
R. anatipestifer is a member of the Bacteroidetes phylum and was recently reported to contain a T9SS (17). In our previous study, 49 virulence genes from the R. anatipestifer strain Yb2 were reported using random transposon mutagenesis (18). Of these, three genes (AS87_08795, AS87_00505, and AS87_08785) encoded gliding-motility proteins (18) which are common features of T9SS function. Further studies also confirmed that R. anatipestifer AS87_RS08795 and AS87_RS08785 were T9SS component proteins and associated with bacterial gliding motility, protein secretion, and virulence (19, 20). Moreover, we demonstrated that R. anatipestifer AS87_RS00980 encoded a T9SS secretory protein which functioned as a metallophosphoesterase and virulence factor (21). We have also shown that R. anatipestifer AS87_RS02955 is a T9SS secretory protein in our previous study (22).
In this study, to clarify R. anatipestifer AS87_RS02955 functions, we used bioinformatics methods to analyze its conserved domains and sequence homology. Then, we constructed deletion and complementation strains to investigate functions related to bacterial adherence/invasion, and in vivo survival. Additionally, recombinant AS87_RS02955 and associated mutants were generated to clarify enzymatic activity and identify critical activity sites. Our study updates the literature on R. anatipestifer T9SS, and provides a new theoretical basis for R. anatipestifer pathogenesis research.
RESULTS
Characterization of R. anatipestifer mutant Yb2Δ02955 and complementation Yb2Δ02955 (cYb2Δ02955) strains.
PCR amplifications of AS87_RS02955, 16S rRNA, and Ermr were performed to confirm mutant and complementation strains. Data shown in Fig. S1A indicates that mutant Yb2Δ02955 and complementation cYb2Δ02955 strains were successfully constructed. In addition, His-tagged recombinant AS87_RS02955 (rAS87_RS02955) was successfully expressed in Escherichia coli BL21(DE3) cells and successfully purified using HisTrap affinity columns (Fig. S1B).
AS87_RS02955 expression in Yb2, Yb2Δ02955, and cYb2Δ02955 strains was determined by Western blotting. As shown in Fig. 1A, a 28-kDa protein was identified in whole protein extracts in Yb2 (lane 1) and cYb2Δ02955 (lane 3) strains, but not in the Yb2Δ02955 strain (lane 2). This analysis clearly demonstrated that AS87_RS02955 was expressed in Yb2 and cYb2Δ02955 strains, but not the Yb2Δ02955 strain, indicating that AS87_RS02955 was successfully deleted in the mutant strain Yb2Δ02955, but rescued in the complementation cYb2Δ02955 strain. A mouse antiserum directed against the TonB-dependent receptor of R. anatipestifer (generated in our laboratory) was used to control for protein loading.
FIG 1.
Western blotting. (A) The identification of the mutant Yb2Δ02955 and complementation strain cYb2Δ02955. Lane M, PageRuler prestained protein ladder; lanes 1 to 3, whole-cell proteins from Yb2, Yb2Δ02955, and cYb2Δ02955 strains, respectively. A mouse antiserum directed against the TonB-dependent receptor of R. anatipestifer was used to control for protein loading. (B) The identification of the bacterial secretion proteins. Lane M, PageRuler prestained protein ladder; lanes 1 to 3, secretion proteins from Yb2, Yb2Δ02955, and cYb2Δ02955 strains, respectively. The TonB-dependent receptor was used to control for protein loading.
Proteins in the cell-free culture medium of strains Yb2, Yb2Δ02955, and cYb2Δ02955 were also identified by Western blotting. As shown (Fig. 1B), a 17.1-kDa band of the AS87_RS02955 was detected in Yb2 (lane 1) but not in Yb2Δ02955 (lane 2) while this band was recovered in cYb2Δ02955 (lane 3). Combined, these data suggested AS87_RS02955 was a secretion protein.
AS87_RS02955 effects on bacterial growth and virulence.
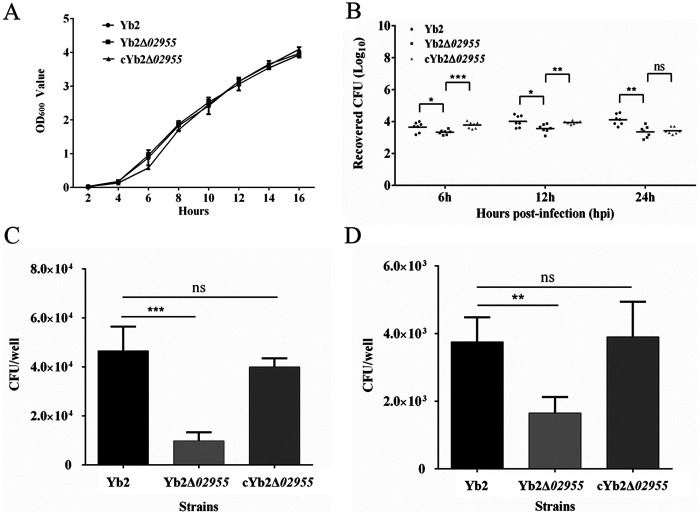
Optical density values at 600 nm (OD600) were used to analyze the growth rates of Yb2, Yb2Δ02955, and cYb2Δ02955 strains. Our results showed no significant growth rate differences among strains (Fig. 2A), suggesting AS87_RS02955 was not associated with R. anatipestifer growth.
FIG 2.
Bacterial growth curves and virulence. (A) Bacterial growth curves. No significant differences were detected between Yb2, Yb2Δ02955, and cYb2Δ02955 strains. (B) Bacterial loads in the blood of ducks infected with R. anatipestifer Yb2 and Yb2Δ02955 strains were determined at 6, 12, and 24 hpi. Bacterial CFU were presented as the mean ± standard deviations of eight infected ducks, then analyzed using a two-tailed independent Student’s t test. (C) Bacterial adherence assay. (D) Bacterial invasion assay. Assays were performed in DEFs at an MOI = 100. Data indicated bacteria which had bound to or invaded DEFs in wells of a 12-well plate. Error bars represented the standard deviation calculated from three independent experiments performed in triplicate. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05.
We further measured bacterial loads in the blood of ducks infected with Yb2, Yb2Δ02955, and cYb2Δ02955 strains. As shown (Fig. 2B), bacterial loads were significantly decreased at 6, 12, and 24 h postinfection (hpi) in Yb2Δ02955 mutant strain, compared with the wild-type strain Yb2. Complementation strain cYb2Δ02955 recovered most of the bacterial loads at 6 and 12 hpi, and partial loads at 24 hpi. Thus, AS87_RS02955 deletion significantly attenuated R. anatipestifer virulence.
Deleted AS87_RS02955 reduces bacterial adherence and invasion capacity.
The ability of Yb2, Yb2Δ02955, and cYb2Δ02955 strains to adhere and invade duck embryo fibroblasts (DEFs) was demonstrated by counting cell-adherent and cell-invasive bacterial numbers postinfection. As shown in Fig. 2C and D, cell-adherent and cell-invasive numbers for the Yb2Δ02955 strain were significantly lower than in cells infected with Yb2. The complementation strain cYb2Δ02955 restored adherence and invasion capacities. Thus, AS87_RS02955 had critical roles in bacterial adherence and invasion.
AS87_RS02955 affects IL-1β, IL-6, IL-8, and tumor necrosis factor-α expression in HD-11 cells infected with R. anatipestifer strains.
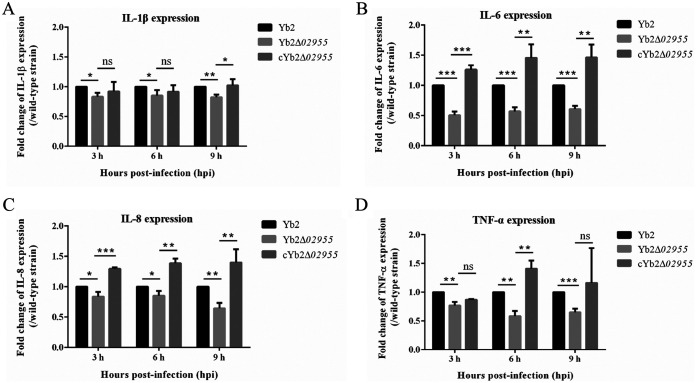
Quantitative real-time PCR (qRT-PCR) was used to analyze IL-1β, IL-6, IL-8, and tumor necrosis factor (TNF)-α gene expression profiles in HD-11 cells at 3, 6, and 9 hpi with Yb2, Yb2Δ02955, and cYb2Δ02955 strains. Relative expression levels were normalized to β-actin transcripts. Compared with Yb2, significantly reduced IL-1β, IL-6, IL-8, and TNF-α gene expression levels were observed at 3, 6, and 9 hpi in cells postinfection with Yb2Δ02955. The complementation strain cYb2Δ02955 restored IL-6 and IL-8 expression at 3, 6, and 9 hpi, with IL-1β at 9 hpi, and TNF-α at 6 hpi (Fig. 3A-D). Thus, the AS87_RS02955-encoded protein stimulated IL-1β, IL-6, IL-8, and TNF-α gene expression in HD-11 cells infected with different R. anatipestifer strains.
FIG 3.
Inflammatory cytokine expression levels in HD-11 cells infected with R. anatipestifer strains. HD-11 cells were separately infected with Yb2, Yb2Δ02955, and cYb2Δ02955 strains. IL-1β (A), IL-6 (B), IL-8 (C), and TNF-α (D) transcription levels were significantly decreased in HD-11 cells infected with the Yb2Δ02955 mutant strain compared with the wild-type strain Yb2 at 3, 6, and 9 hpi. Statistical significance was assessed using two-way analysis of variance *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05.
AS87_RS02955 encodes an endonuclease.
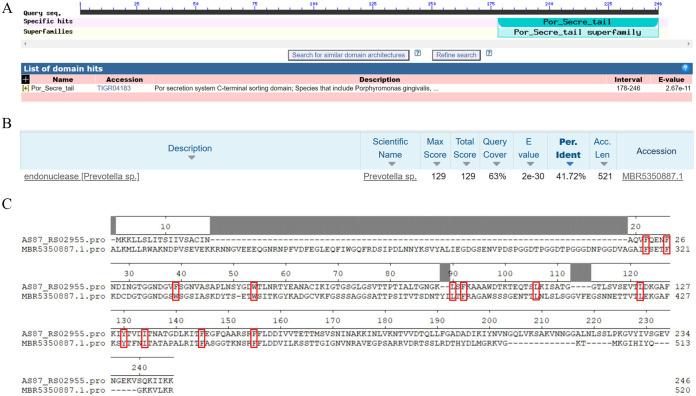
From the NCBI Conserved Domain Database, AS87_RS02955 contained a Por secretion system C-terminal sorting domain, but not a functional conserved domain (Fig. 4A). BLAST analyses showed that AS87_RS02955 shared a 41.72% sequence homology with an endonuclease from a Prevotella sp. in the His-Asn-His (HNH) superfamily (Fig. 4B). HNH endonucleases normally contain a His-Asn-His motif (23), but no conserved HNH motif structure was observed in R. anatipestifer AS87_RS02955, which is similar to the Prevotella sp. HNH endonuclease sequence (Fig. 4C).
FIG 4.
Bioinformatics analysis of R. anatipestifer AS87_RS02955. (A) R. anatipestifer AS87_RS02955 gene comprised 741 bp and encoded a polypeptide of 246 amino acid residues containing a Por secretion system C-terminal sorting domain. (B) BLAST analysis showed AS87_RS02955 shared 41.72% sequence homology with a Prevotella sp. (sequence ID: MBR5350887.1) endonuclease. (C) AS87_RS02955 sequence alignment with the Prevotella sp. endonuclease sequence (215 to 520 amino acids were displayed). Twelve potential critical sites for enzymatic activity are indicated by red boxes.
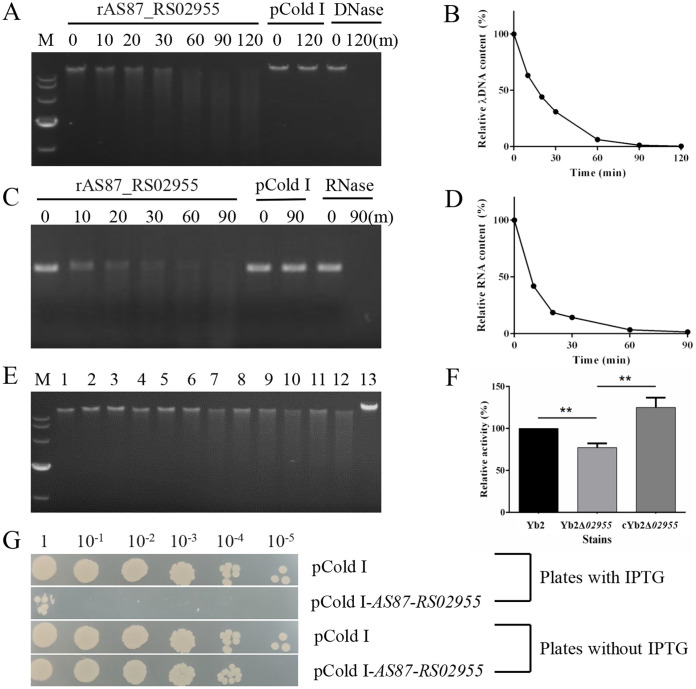
To ascertain if AS87_RS02955 encoded an endonuclease, a His-tagged recombinant AS87_RS02955 protein (rAS87_RS02955) was efficiently expressed in E. coli BL21(DE3) cells (Fig. S1B). According to previous reports, nonspecific endonucleases that cleave DNA will also cleave RNA in the presence of Mg2+ (24–26). The enzymatic activity of rAS87_RS02955 was measured using the substrates λ DNA or MS2 RNA in the presence of Mg2+. Recombinant AS87_RS02955 cleaved λ DNA (Fig. 5A and B) and MS2 RNA (Fig. 5C and D) in a reaction time-dependent pattern. DNase and RNase were both used as positive controls and pCold I as a negative control.
FIG 5.
Enzymatic activity assays of R. anatipestifer recombinant AS87_RS02955 (rAS87_RS02955). (A) Enzymatic activity of rAS87_RS02955 in cleaving λ DNA. (B) λ DNA cleavage curve. (C) Enzymatic activity of rAS87_RS02955 in cleaving MS2 RNA. (D) MS2 RNA cleavage curve. Purified rAS87_RS02955 was incubated with substrates at 37°C for 0, 10, 20, 30, 60, 90, and 120 min (except 120 min for RNA) in the presence of Mg2+. Purified pCold I was used as a negative control. DNase and RNase were used as positive controls. Cleavage curves were generated using Image J software. (E) Enzymatic activity of R. anatipestifer secretory proteins. Lane M, DL10,000 DNA marker; lanes 1 to 3, 1 h reactions; lanes 4 to 6, 3 h reactions; lanes 7 to 9, 6 h reactions; lanes 10 to 12, 9 h reactions. Lanes 1, 4, 7, and 10, secretory proteins from Yb2; lanes 2, 5, 8, and 11, secretory proteins from Yb2Δ02955; lanes 3, 6, 9, and 12, secretory proteins from cYb2Δ02955; lane 13, no protein reaction for 9 h at 37°C. (F) Relative enzymatic activity of secretory proteins after incubation for 9 h. Yb2 activity toward λ DNA was 100%: Yb2Δ02955 showed 70% relative activity compared with Yb2. Complementation with AS87_RS02955 recovered enzymatic activity. Error bars represent the standard deviation of data from three independent experiments (**, P < 0.01). (G) AS87_RS02955 activity in E. coli BL21(DE3) cells. Cells were transformed with pCold I-AS87_RS02955, grown to logarithmic phase, washed twice in PBS, adjusted to OD600 0.8, 10-fold serially diluted, and 2 μL spotted onto LB agar plates with/without IPTG for overnight culture at 37°C. Cells transformed with the pCold I vector were used as a control strain. “1” means 2 μL bacteria at OD600 = 0.8; 10−1, 10−2, 10−3, 10−4, and 10−5 indicated 10-fold series dilution of the bacteria at OD600 = 0.8.
The enzymatic activity of cell-free bacterial culture medium was assayed using a λ DNA substrate to confirm the endonuclease activity of the AS87_RS02955-encoded protein. As shown (Fig. 5E), cleavage activity of the mutant strain Yb2Δ02955 was lower than Yb2 (lanes 2, 5, 8, 11 versus lanes 1, 4, 7, 10, respectively); cYb2Δ02955 restored this activity (lanes 3, 6, 9, 12). After analysis in Image J software, Yb2Δ02955 generated 70% relative endonuclease activity compared with Yb2 in a 9 h reaction, but activity was restored in cYb2Δ02955 (Fig. 5F).
Recombinant AS87_RS02955 function toward bacterial survival was also investigated. Proteins that only exhibit exonuclease activity do not reduce host cell survival rates as previously reported (27, 28). After induction with isopropylb-D-1-thiogalactopyranoside (IPTG), a decreased survival rate of E. coli BL21(DE3) cells transformed with pCold I-AS87_RS02955 was observed compared with empty vector pCold I transformed cells (Fig. 5G). Thus, rAS87_RS02955 was a nonspecific endonuclease.
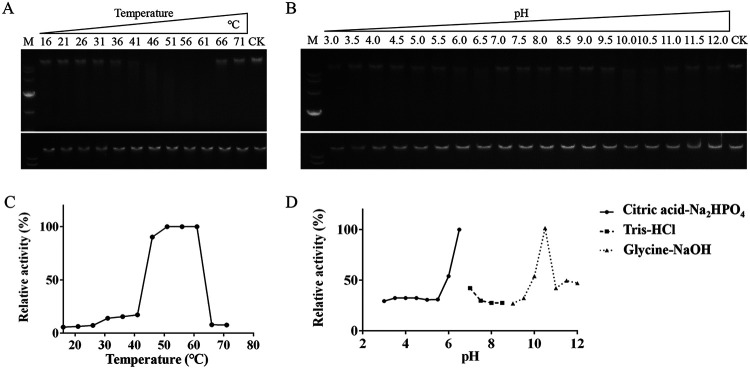
The effects of temperature and pH on rAS87_RS02955 enzymatic activity.
The optimal temperature for rAS87_RS02955 cleavage of λ DNA was determined over the 16 to 71°C temperature range, while optimal pH was determined after establishing relative activity versus pH values over the 3 to 12 range. Purified pCold I under the same conditions was used as a negative control (Fig. 6A and B). We observed rAS87_RS02955 generated > 90% relative activity between 46 and 61°C (Fig. 6C). This suggested that rAS87_RS02955 functioned at high temperatures while most enzymes operate at moderate temperatures. This characteristic could permit enzymatic reactions at high temperatures, thereby increasing conversion rates to improve bacterial thermal tolerance (29). As shown (Fig. 6D), rAS87_RS02955 generated enzymatic activity over a broad pH range; relative activity peaked at pH 6.5 and 10.5. The isoelectric point of AS87_RS02955 was 8.9 using Expasy (30), thus confirming the basic nature of the enzyme. From these data, pH 6.5 was chosen as the optimal working pH of the recombinant protein.
FIG 6.
The effects of temperature and pH on recombinant AS87_RS02955 (rAS87_RS02955) activity. (A) The effects of various temperatures on λ DNA cleavage activity of rAS87_RS02955. Optimal temperature effects were measured by incubating rAS87_RS02955 with λ DNA in citric acid-Na2HPO4 buffer (pH 6.5) for 30 min at 5°C intervals from 16 to 71°C. (B) The effects of different pH’s on the DNA cleavage activity of rAS87_RS02955. Optimal pH effects were measured for 30 min at 37°C in citric acid-Na2HPO4 buffer, Tris-HCl buffer, or glycine-NaOH buffer. CK was the reaction without enzyme. Gels in bottom panels indicated negative controls and contained purified pCold I plus λ DNA similarly incubated as for rAS87_RS02955. (C) Curve showing temperature effects. (D) Curve showing pH effects. Curves were determined by comparing the integrated density of each band with the CK using Image J software.
The effects of metal cations on rAS87_RS02955 enzymatic activity.
The effects of Zn2+, Cu2+, Mg2+, Ca2+, Mn2+, and Fe3+ cations on the ability of rAS87_RS02955 to cleave λ DNA and MS2 RNA suggested good relative activity (Table 1 and 2, respectively). As shown in Table 1, rAS87_RS02955 generated 97.45% to 61.69% relative activity upon the addition of Mn2+, Zn2+, Cu2+, Ca2+, and Mg2+ at 1 mM, while Fe3+ (1 mM) generated only 1.47% relative activity, suggesting poor Fe3+ effects with respect to rAS87_RS02955 DNase activity. However, rAS87_RS02955 generated the highest λ DNA cleaving activity with Mn2+, similar to other endonuclease HNH superfamily members (23, 31–34). Moreover, rAS87_RS02955 cleaved RNA in the absence of metal cations; however, activity was stimulated by Mn2+, Mg2+, and Ca2+, but inhibited by Zn2+, Cu2+, and Fe3+ (Table 2).
TABLE 1.
Metal cations effect on λ DNA cleavage activity of the rAS87_RS02955
| Metal cations | Concn (mM) | Relative activity (%)a |
|---|---|---|
| None | 0 | |
| Mn2+ | 1 | 97.45 |
| Zn2+ | 1 | 80.82 |
| Cu2+ | 1 | 79.74 |
| Ca2+ | 1 | 78.27 |
| Mg2+ | 1 | 61.69 |
| Fe3+ | 1 | 1.47 |
Relative activity was calculated as: (integrated density of gel without a metal cation − integrated density of gel with a metal cation)/(integrated density of reaction without a metal cation) × 100%.
TABLE 2.
Metal cations effect on MS2 RNA cleavage activity of rAS87_RS02955
| Metal cations | Concn (mM) | Relative activity (%)a |
|---|---|---|
| None | 100 | |
| Mn2+ | 1 | 206.06 |
| Mg2+ | 1 | 135.71 |
| Ca2+ | 1 | 111.75 |
| Zn2+ | 1 | 79.65 |
| Cu2+ | 1 | 61.33 |
| Fe3+ | 1 | 50.48 |
Relative activity was calculated as: integrated density of gel without a metal cation/integrated density of reaction with a metal cation × 100%.
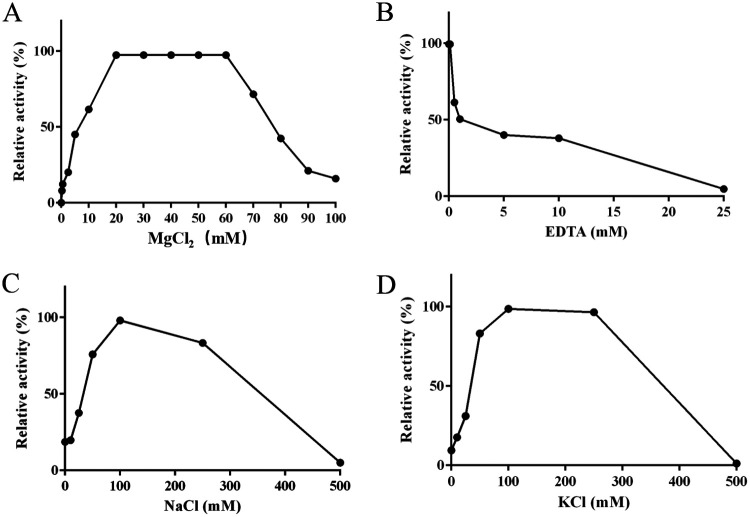
Recombinant AS87_RS02955 showed optimal activity with Mg2+ in the 20 mM to 60 mM range, but was almost completely inhibited at 100 mM (Fig. 7A). The effects of EDTA on recombinant activity were also analyzed (Fig. 7B); enzymatic activity was partially inhibited at 0.5 mM EDTA and completely inhibited at 25 mM.
FIG 7.
The effects of metal cations on λ DNA activity of recombinant AS87_RS02955 (rAS87_RS02955). Optimal cation concentrations were measured by incubating rAS87_RS02955 with λ DNA in citric acid-Na2HPO4 buffer (pH 6.5) for 30 min at 37°C. (A) The effects of Mg2+ on λ DNA cleavage activity of rAS87_RS02955. Optimal activity ranged from 20 to 60 mM. (B) The effects of EDTA on λ DNA cleavage activity of rAS87_RS02955. Enzymatic activity was inhibited by 0.5 mM EDTA and completely inhibited at 25 mM. (C) The effects of Na+ on λ DNA cleavage activity of rAS87_RS02955. Optimal rAS87_RS02955 activity was determined at 100 mM. (D) The effects of K+ on λ DNA cleavage activity of rAS87_RS02955. Optimal rAS87_RS02955 activity ranged from 100 to 250 mM.
The effects of ionic strength on rAS87_RS02955 activity were also evaluated by various NaCl and KCl concentrations in the 0 to 500 mM range. Recombinant AS87_RS02955 activity was promoted by NaCl and KCl, peaked at 100 mM, and maintained at over 80% relative activity in the 50 to 250 mM range, but completely inhibited at 500 mM NaCl or KCl (Fig. 7C and D). Agarose gel data are shown in Fig. S2.
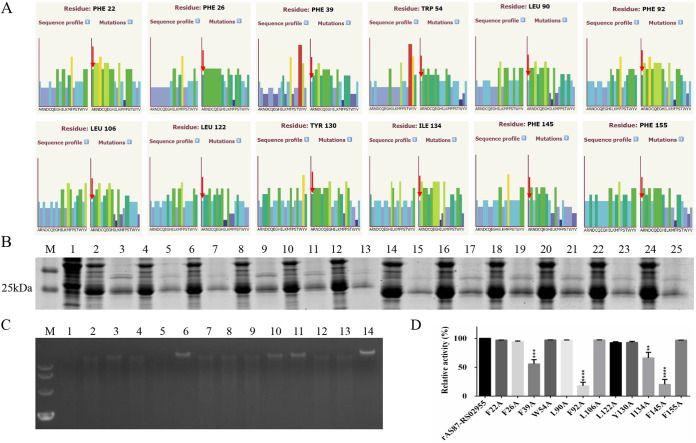
Positions F39, F92, I134, and F145 are critical for rAS87_RS02955 enzymatic activity.
Based on predictions from the Phyre2 Investigator web interface, mutational analysis graphs indicated the predictive effects of mutations at particular amino acid positions; tall, green or blue bars above residues were predicted as critical. As shown in Fig. 8A, 12 amino acid residues were predicted to affect rAS87_RS02955 activity when switched to alanine. Using in vitro site-directed mutagenesis, 12 His-tagged mutant proteins were efficiently expressed in E. coli BL21(DE3) cells. Cells transformed with pCold I were used as negative-expression controls (Fig. 8B).
FIG 8.
Expression and enzymatic activity of 12 mutant recombinant AS87_RS02955 (rAS87_RS02955) proteins. (A) Mutational analysis graphs of predicted residue positions affecting rAS87_RS02955 enzymatic activity. Tall, green or blue bars above alanine were predicted to affect rAS87_RS02955 enzymatic activity when alanine was substituted for different amino acid residues. (B) SDS-PAGE analysis of mutant protein expression. Lane M, PageRuler prestained protein ladder; lane 1, E. coli BL21(DE3) cells transformed with pCold I plus IPTG; lanes 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, and 24, supernatants from E. coli BL21(DE3) cells transformed with mutant pCold I-F22A, pCold I-F26A, pCold I-F39A, pCold I-W54A, pCold I-L90A, pCold I-F92A pCold I-L106A, pCold I-L122A, pCold I-Y130A, pCold I-I134A, pCold I-F145A, pCold I-F155A plus IPTG, respectively; lanes 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, and 25, purified F22A, F26A, F39A, W54A, L90A, F92A, L106A, L122A, Y130A, I134A, F145A, and F155A, respectively. (C) Enzymatic activity of mutant proteins. Lane M, DL10,000 DNA marker; lanes 1 to 12, purified F22A, F26A, F39A, W54A, L90A, F92A, L106A, L122A, Y130A, I134A, F145A, and F155A, respectively; lane 13, purified rAS87_RS02955; lane 14, purified pCold I. (D) Relative enzymatic activity of mutant proteins compared with rAS87_RS02955. Relative activity was determined by comparing the integrated density of each protein band with rAS87_RS02955 in Image J software. Error bars represent the standard deviation of data from three independent experiments. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
The effects of site-directed mutagenesis on endonuclease activity in mutant proteins are shown in Fig. 8C. After analysis in Image J software, the mutant proteins F39A and I134A retained approximately 50% relative activity, while F92A and F145A retained approximately 20% compared with rAS87_RS02955 (Fig. 8D). These data suggest positions F39, F92, I134, and F145 were required to maintain rAS87_RS02955 activity, while F92 and F145 were more critical.
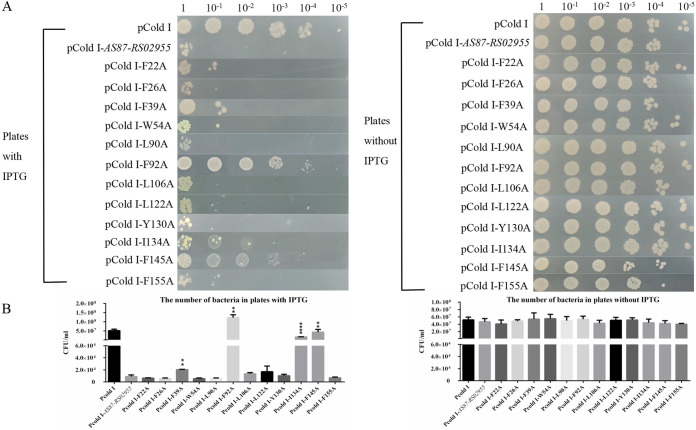
Bacterial survival assays were conducted to confirm the effects of critical amino acid residues. As shown in Fig. 9A and B, when positions F39, F92, I134, or F145 were mutated, bacterial numbers increased compared with E. coli BL21(DE3) cells transformed with pCold I-AS87_RS02955 after IPTG induction. Cells transformed with pCold I-F92A or pCold I-F145A were almost equal to pCold I empty vector cells. All E. coli BL21(DE3) with a transformer were spotted and plated to the plants containing 100 μg/mL ampicillin without IPTG as a negative control, the numbers of bacteria were also counted and analyzed as shown in the right panel.
FIG 9.
Bacterial survival assays. (A) Mutant protein activity in E. coli BL21(DE3) cells. Cells were transformed with a pCold I-mutant protein, grown to logarithmic phase, washed twice in PBS, adjusted to OD600 = 0.8, 10-fold serially diluted, 2 μL spotted onto LB agar plates with/without IPTG (left and panel, respectively), and cultured overnight at 37°C. Cells transformed with the pCold I vector were used as controls. “1” means 2 μL bacteria at OD600 = 0.8; 10−1, 10−2, 10−3, 10−4, and 10−5 indicate 10-fold series dilution of the bacteria at OD600 = 0.8. (B) Bacterial counting. E. coli BL21(DE3) colonies transformed with different mutant protein-plasmids were counted on IPTG (left panel) or without IPTG (right panel) plates. Error bars represent the standard deviation of data from three independent experiments. **, P < 0.01; ****, P < 0.0001.
DISCUSSION
Secreted bacterial proteins participate in many important tasks via protein secretion systems, and play key roles in antagonistic and mutualistic processes within and between species, including detoxification, antibiotic resistance, and scavenging (35–37). T9SS or the Por secretion system was discovered in several Bacteroidetes species, including Flavobacterium johnsoniae, Porphyromonas gingivalis, Tannerella forsythia, and R. anatipestifer (17). In a recent study, enzymes secreted by T9SS were implicated in the degradation of complex biopolymers and suggested as possible virulence factors, e.g., proteinases, glycosidases, nucleases, and lipases (15, 38, 39). In our previous study, we identified two R. anatipestifer T9SS component proteins associated with protein secretion, gliding motility, and virulence (19, 20). We also characterized a R. anatipestifer T9SS effector metallophosphoesterase which generated phosphatase activity and played important roles in bacterial virulence (21). In the current study, we characterized the R. anatipestifer T9SS effector AS87_RS02955 as an endonuclease; it functioned as a DNase and RNase and played key roles in bacterial virulence. As inflammatory cytokine expression and bacterial loads in the blood of ducks infected with Yb2Δ02955 mutant strains were significantly reduced, it was possible that AS87_RS02955 promoted inflammatory reactions and extracellular bacterial survival and spread in vivo.
AS87_RS02955 contains a Por secretion system C-terminal sorting domain, which we confirmed was secreted outside cells by Western blotting. AS87_RS02955 does not have a functionally conserved domain. However, by BLAST, it showed sequence homology with an HNH endonuclease, but did not contain a conserved HNH motif; thus, it may be a novel endonuclease. Recombinant AS87_RS02955 cleaved λ DNA and functioned optimally at pH 6.5 and 10.5 over a wide temperature range (40 to 60°C), which was very different to reported endonucleases (23, 31, 40, 41). Recombinant AS87_RS02955 also displayed activity toward λ DNA in the presence of metal cations; more than 60% relative activity was identified in the presence of Zn2+, Cu2+, Mg2+, Ca2+, and Mn2+ cations, but not Fe3+. Also, rAS87_RS02955 showed the highest activity in the presence of Mn2+, similar to other HNH endonucleases (23, 31–34). Recombinant AS87_RS02955 also demonstrated activity toward RNA; it cleaved RNA in the presence or absence of metal cations, but in previous research it required metal cations to cleave DNA like Aspergillus (42). Both Mg2+ and Ca2+ are the most abundant divalent cations in living organisms (43), while Mg2+ is the most abundant divalent cation inside cells (44). Other ions such as Fe2+, Zn2+, and Cu2+ are widespread, while Mn2+ and Ni2+ are essential but found at low concentrations (45). As Mn2+ concentrations inside cells are low, and Zn2+, Cu2+, and Ca2+ may cause protein precipitation, Mg2+ was chosen to explore enzymatic activity in vitro. The optimal Mg2+ concentration for rAS87_RS02955 activity ranged from 20 to 60 mM, which was higher than other endonucleases (29, 41, 46). Also, recombinant enzymatic activity was increased by low Na+ or K+ concentrations and suppressed by high concentrations. But rAS87_RS02955 was more tolerant to Na+ or K+ than other reported endonucleases (29, 45). Taken together, R. anatipestifer rAS87_RS02955 appeared independent of Na+ or K+ with respect to DNA cleavage activity.
We also confirmed that F39, F92, I134, and F145 positions were required to maintain rAS87_RS02955 activity, and that F92 and F145 were more critical. These positions are not conserved amino acid residues, but they may be involved in binding metal cofactors or substrates; thus, their altering may have changed the enzyme structure. Bacterial survival assays confirmed that AS87_RS02955 expression affected bacterial function, indicating it may be related to the competition of heterologous bacteria.
AS87_RS02955 is a novel nonspecific endonuclease with important roles in bacterial cell invasion and infection. From a previous report, most nonspecific nucleases are secreted to break down nucleotides and phosphates for bacterial survival, e.g., Serratia nuclease from Serratia marcescens, Staphylcoccal nuclease from Staphylococcus aureus, and P1 nuclease from Penicillium citrinum (47). Some nonspecific nucleases are involved in DNA repair and recombination, including Neurospora crassa endonuclease, yeast Rad52, and Nuc1. Mutations in genes encoding these nucleases cause incomplete DNA repair or genetic recombination functions, which affect bacterial virulence (48–50). These secreted toxins induce target cell death by randomly digesting nucleic acids in these cells, thus improving host cell survival chances during stress (51). R. anatipestifer may also adopt an aggressive and offensive approach for cell defense and survival by secreting AS87_RS02955. The secreted nucleases could not only provide nutrients for cells by degrading extracellular DNA, but also mediate bacterial escape from neutrophil extracellular traps (NETs) during infection (52, 53) or involve in the competition between bacteria and exogenous or endogenous bacteria (54). We searched the homologous endonuclease in Prevotella sp. (sequence ID: MBR5350887.1); unfortunately, the characteristic and function has not been determined. Because AS87_RS02955 is a secreted nonspecific endonuclease and involved in the survival of E. coli, we think AS87_RS02955 may help R. anatipestifer cleave DNA or RNA in the living environment for nutrients, or compete with other organisms through some systems to kill competitors and survive. However, the molecular mechanisms, whereby AS87_RS02955 acts during R. anatipestifer infection, require further investigation.
In conclusion, we propose that R. anatipestifer AS87_RS02955 is a novel endonuclease and virulence factor. The protein showed a nonspecific DNA cleavage activity; the highest efficiency occurred at 40 to 60°C and optimal pH’s of 6.5 and 10.5. AS87_RS02955 also displayed a divalent metal ion-dependent DNase activity and a metal ion-independent RNase activity. AS87_RS02955 was closely associated with cell infection, extracellular survival, and in vivo bacterial spread. However, AS87_RS02955 potential functions in protecting R. anatipestifer from infection of other organisms (bacteriophage, bacteria, and virus) via DNA or RNA destruction require further clarification. Our study facilitates the further investigation of bacterial pathogenesis mechanisms for AS87_RS02955 and the development of effective strategies to control R. anatipestifer infection.
MATERIALS AND METHODS
Animals and ethics statement.
One-day-old Cherry Valley ducks were obtained from JinHu Duck Farm (Jiangsu, China). Ducks were raised in cages in a temperature-controlled environment with unlimited access to antibiotic-free feed, water, and a 12-h light/day cycle for the study duration. Duck maintenance and experimental procedures were performed in accordance with the Guide for the Care and Use of Laboratory Animals of the Institutional Animal Care and Use Committee guidelines set out by Shanghai Veterinary Research Institute, the Chinese Academy of Agricultural Sciences. The study protocol was approved by the Institutional Animal Care and Use Committee of our institution (approval no. SHVRI-SZ-20210930-Y01).
Bacterial strains, plasmids, and culture conditions.
Bacterial strains, plasmids, and primers are listed (Table 3). The virulent wild-type R. anatipestifer strain Yb2 (18) was used in this study. The mutant Yb2Δ02955 and complementation cYb2Δ02955 strains were established in this study. All R. anatipestifer strains were grown in tryptic soy broth medium (TSB; Difco, NJ, USA) or on solid tryptic soy agar (TSA; Difco) plates at 37°C in 5% CO2. E. coli strains were grown at 37°C on Luria-Bertani (LB) plates or in LB broth. The pCP29 plasmid and E. coli strain S17-1 were kindly provided by Mark J. McBride (University of Wisconsin-Milwaukee, Milwaukee, WI, USA). Antibiotics were used at the following concentrations: ampicillin = 100 μg/mL; erythromycin = 1.0 μg/mL; kanamycin = 50 μg/mL; streptomycin = 50 μg/mL; and cefoxitin = 5 μg/mL.
TABLE 3.
Strains, plasmids, and primers used in this study
| Strains, plasmids, or primers | Characteristics |
|---|---|
| Strains, and plasmids | |
| Yb2 | Riemerella anatipestifer serotype 2 strain |
| Yb2Δ02955 | AS87_RS02955 gene mutant of R. anatipestifer Yb2 |
| cYb2Δ02955 | Mutant Yb2Δ02955 carrying plasmid pCP29-AS87_RS02955 |
| pCP29 | ColE1 ori (pCP1 ori), Apr (Emr); E. coli-F. johnsoniae shuttle plasmid |
| pCP29-AS87_RS02955 | pCP29 containing ompA promoter and AS87_RS02955 ORF,a cfxAb (Apr) |
| pCold I | ColE1 ori, Apr, N-terminal His-tag |
| Primers | |
| AS87_RS02955-F | 5′-GGTACAGGTGGTAATGATGG-3′ |
| AS87_RS02955-R | 5′-GAAACTTTCTCACCGTTTAC-3′ |
| AS87_RS02955 P1-F | 5′-CATAATGCACAACGGGTAATAACTTTA-3′ |
| AS87_RS02955 P1-R | 5′-GAACGGGCAATTTCTTTTTTGTCATGAGAGAGTAATTTTTTCATTTTTAA-3′ |
| Erm-P-F | 5′-TTAAAAATGAAAAAATTACTCTCTCATGACAAAAAAGAAATTGCCCGTTC-3′ |
| Erm-P-R | 5′-AAAAATAAATTTGACTTATTTCTTGCTACGAAGGATGAAATTTTTCAGGG-3′ |
| AS87_RS02955 P2-F | 5′-CCCTGAAAAATTTCATCCTTCGTAGCAAGAAATAAGTCAAATTTATTTTT-3′ |
| AS87_RS02955 P2-R | 5′-TTGTTTTTGTACTTGCTCAAAATGATT-3′ |
| Erm-F | 5′-ATGACAAAAAAGAAATTGCCCGTT-3′ |
| Erm-R | 5′-CTACGAAGGATGAAATTTTTCAGGG-3′ |
| 16S rRNA-F | 5′-GAGCGGTAGAGTATCTTCGGATACT-3′ |
| 16S rRNA-R | 5′-TATTACCGCGGCTGCTGGCA-3′ |
| AS87_RS02955-orf-F | 5′-CGACTCGAGATGAAAAAATTACTCTCTC-3′ (XhoI site underlined) |
| AS87_RS02955-orf-R | 5′-CATGCATGCTTATTTCTTGATAATTTTTTGAG-3′ (SphI site underlined) |
| AS87_RS02955-pCold-F | 5′-TCGGTACCCTCGAGGGATCCTGCATCAATGCTCAAGTTTTC-3′ |
| AS87_RS02955-pCold-R | 5′-TCTAGACTGCAGGTCGACTTTCTTGATAATTTTTTGAG-3′ |
| Chβ-actin-F | 5′-GAGAAATTGTGCGTGACATCA-3′ |
| Chβ-actin-R | 5′-CCTGAACCTCTCATTGCCA-3′ |
| ChTNF-α-F | 5′-CGCTCAGAACGACGTCAA-3′ |
| ChTNF-α-R | 5′-GTCGTCCACACCAACGAG-3′ |
| ChIL-1β-F | 5′-GGGCATCAAGGGCTACA-3′ |
| ChIL-1β-R | 5′-TCGGGTTGGTTGGTGATG-3′ |
| ChIL-6-F | 5′-GTTCGCCTTTCAGACCTAC-3′ |
| ChIL-6-R | 5′-ACCACTTCATCGGGATTTA-3′ |
| ChIL-8-F | 5′-TTGGAAGCCACTTCAGTCAGAC-3′ |
| ChIL-8-R | 5′-GGAGCAGGAGGAATTACCAGTT-3′ |
| AS87_RS02955-F22-F | 5′-TCGGTACCCTCGAGGGATCCTGCATCAATGCTCAAGTTGCCCAAG-3′ |
| AS87_RS02955-F26-F | 5′-TCGGTACCCTCGAGGGATCCTGCATCAATGCTCAAGTTTTCCAAGAAAACGCTAATGACATTAATGG-3′ |
| AS87_RS02955-F39-F | 5′-GCTAGTGGTAATGTAGCCTCTGCACCTCTCAATTCTTA-3′ |
| AS87_RS02955-F39-R | 5′-GAGGCTACATTACCACTAGCAACACCATCATTACCACCT-3′ |
| AS87_RS02955-W54-F | 5′-GCGACTCTAAACAGAACTTACGAAGCTAATGCGTGTATTA-3′ |
| AS87_RS02955-W54-R | 5′-CGTAAGTTCTGTTTAGAGTCGCGTCTCCATAAGAATTGAG-3′ |
| AS87_RS02955-L90-F | 5′-GCTTCCTTTAAAGCAGCCGCTTGGGATACTAAAACAGAA-3′ |
| AS87_RS02955-L90-R | 5′-GCGGCTGCTTTAAAGGAAGCTTTTCCGTTTCCTGTTAAT-3′ |
| AS87_RS02955-F92-F | 5′-GCTAAAGCAGCCGCTTGGGATACTAAAACAGAACAAACCT-3′ |
| AS87_RS02955-F92-R | 5′-CAAGCGGCTGCTTTAGCGGAAAGTTTTCCGTTTCCTGTTA-3′ |
| AS87_RS02955-L106-F | 5′-GCTAAAATTTCAGCAACTGGAGGAACTCTAAGTGTTTCAGA-3′ |
| AS87_RS02955-L106-R | 5′-GTTGCTGAAATTTTAGCAGAGGTTTGTTCTGTTTTAGTATCCC-3′ |
| AS87_RS02955-L122-F | 5′-GCAGACAAAGGAGCTTTTAAGATCTATACAGTAGATATCACTAACG-3′ |
| AS87_RS02955-L122-R | 5′-AAAGCTCCTTTGTCTGCTGTAACTTCTGAAACACTTAGAGTTC-3′ |
| AS87_RS02955-Y130-F | 5′-GCTACAGTAGATATCACTAACGCAACGGGAGATTTAAAG-3′ |
| AS87_RS02955-Y130-R | 5′-GCGTTAGTGATATCTACTGTAGCGATCTTAAAAGCTCC-3′ |
| AS87_RS02955-I134-F | 5′-GCCACTAACGCAACGGGAGATTTAAAGATTACCTTTGAAGG-3′ |
| AS87_RS02955-I134-R | 5′-CTCCCGTTGCGTTAGTGGCATCTACTGTATAGATCTTA-3′ |
| AS87_RS02955-F145-F | 5′-GCTGAAGGTTTTCAGGCTGCTAGATCAAGATTTTTTTTGGACG-3′ |
| AS87_RS02955-F145-R | 5'-GCAGCCTGAAAACCTTCAGCGGTAATCTTTAAATCTCC-3′ |
| AS87_RS02955-F155-F | 5′-GCTTTTTTGGACGATATAGTAGTTACAGAAACCACTATGTCTG-3′ |
| AS87_RS02955-F155-R | 5′-CTATATCGTCCAAAAAAGCTCTTGATCTAGCAGCCTGAAAACC-3′ |
ORF, open reading frame.
cfxA, cefoxitin resistance gene.
Constructing mutant Yb2Δ02955 and complementation cYb2Δ02955 strains.
The mutant strain Yb2Δ02955 was constructed using the natural transformation method (55). Briefly, upstream (~800 bp) and downstream sequences (~800 bp) of AS87_RS02955 were amplified from Yb2 genomic DNA using the primer pairs AS87_RS02955 P1-F/AS87_RS02955 P1-R and AS87_RS02955 P2-F/AS87_RS02955 P2-R (Table 3). The erythromycin resistance (Ermr) gene was amplified from the R. anatipestifer strain HXb2 using Erm-P-F and Erm-P-R primers (Table 3). The resulting PCR fragments were ligated by overlap PCR using AS87_RS02955 P1-F and AS87_RS02955 P2-R primers (56), then purified, and introduced into Yb2 by natural transformation. Transformants were selected on TSA plates containing 1.0 μg/mL erythromycin.
A complementation strain cYb2Δ02955 was constructed using the shuttle plasmid pCP29, which carried the R. anatipestifer ompA promoter (57). Briefly, the open reading frame (ORF) of AS87_RS02955 was amplified from R. anatipestifer Yb2 genomic DNA using AS87_RS02955-orf-F/AS87_RS02955-orf-R primers (Table 3), digested with SphI and XhoI, and ligated into pCP29 at corresponding enzyme sites to generate the pCP29-AS87_RS02955 plasmid. The complementation strain cYb2Δ02955 was generated by transferring the plasmid into the mutant strain via conjugation.
To confirm Yb2Δ02955 mutant and cYb2Δ02955 complement strains, AS87_RS02955-F/AS87_RS02955-R, 16S rRNA-F/16S rRNA-R, and Erm-F/Erm-R (Table 3) primer pairs were used to amplify AS87_RS02955, R. anatipestifer 16S rRNA, and Ermr, respectively.
Recombinant protein expression and Western blotting.
R. anatipestifer AS87_RS02955 was amplified from Yb2 using AS87_RS02955-pCold-F and AS87_RS02955-pCold-R primers (Table 3). After purification, the amplicon was cloned into the expression vector pCold I to construct the pCold I-AS87_RS02955 plasmid. After confirmation by DNA sequencing, the plasmid was transformed into E. coli BL21(DE3) cells (Invitrogen, Carlsbad, CA, USA). His-tagged recombinant AS87_RS02955 protein (rAS87_RS02955) expression was induced by supplementing cells with 1 mM IPTG (Sigma, St. Louis, MO, USA) for 24 h at 16°C with shaking. Then, cells were harvested by centrifugation for 5 min at 10,000 × g and 4°C, resuspended in lysis buffer (20 mM Na3PO4, 0.5 M NaCl, pH 7.4), and purified on HisTrap affinity columns (GE Healthcare, Uppsala, Sweden) according to manufacturer’s instructions. Purified fractions were confirmed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), followed by Coomassie brilliant blue staining.
To identify Yb2Δ02955 mutant and complementation strains, whole-cell and secretion proteins from Yb2, the mutant strain Yb2Δ02955, and the complementation strain cYb2Δ02955, were extracted and characterized by Western blotting as previously described (21). Samples (same concentration) were separated by SDS-PAGE and electrophoretically transferred onto nitrocellulose membranes (Millipore, Billerica, MA, USA). Then, membranes were probed using the primary antibody mouse anti-AS87_RS02955 polyclonal antibody (generated in our laboratory) and the secondary antibody horseradish peroxidase-conjugated goat anti-mouse IgG polyclonal antibody (Abcam, Cambridge, UK). A mouse anti-TonB-dependent receptor antibody was used as the protein loading control. Assays were performed in triplicate and replicated three times.
Bacterial growth, adherence, and invasion assays.
Growth curves for Yb2, Yb2Δ02955, and cYb2Δ02955 were conducted as previously described (58). Briefly, bacterial strains at the logarithmic growth phase in TSB were optically measured at OD600, then diluted to same bacterial CFU values. Equal bacterial counts were then transferred to fresh TSB at a 1:100 (vol/vol) ratio and cultured at 37°C with shaking at 220 rpm. Growth was monitored by measuring OD600 values at 2-h intervals for 16 h.
Adhesion and invasion assays were performed using DEFs as previously described, with some modifications (7). DEFs were obtained from 9-day-old healthy duck embryos using a standard method (59) and maintained in Dulbecco's modified Eagle medium (DMEM; Gibco, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA), 5,000 U/mL streptomycin, and 5,000 μg/mL penicillin (Gibco, USA). Briefly, wells in a 12-well tissue culture plate were seeded with 2.5 × 105 DEFs in complete fresh DMEM without antibiotics. Bacteria were pelleted, washed twice in phosphate-buffered saline (PBS), and resuspended in fresh medium without antibiotics, at 2.5 × 107 CFU/mL. After incubation at 37°C in 5% CO2, cell monolayers were rinsed in PBS and infected with bacteria at a multiplicity of infection (MOI) = 100. Infected cells were incubated at 37°C in 5% CO2 for 2 h, washed three times in PBS, and lysed in 0.1% trypsin (100 μL/well). Cell suspensions were serially 10-fold diluted and spread onto TSA plates for bacterial colony counting after placing in the 37°C, 5% CO2 incubator for overnight incubation.
For invasion assays, infected cells were incubated for an additional 1 h in DMEM supplemented with 300 μg/mL gentamicin to kill extracellular bacteria. Infected cells were then washed three times in PBS, lysed, serially 10-fold diluted and spread onto TSA plates, incubated, and intracellular bacterial numbers determined at indicated time point. Assays were performed in triplicate.
Determining bacterial loads in infected duck blood.
To determine bacterial survival in infected duck blood, 16 21-day-old ducks were randomly divided into two groups and intramuscularly inoculated with Yb2 or Yb2Δ02955 strain at 2.5 × 107 CFU. Blood samples were collected at 6, 12, and 24 hpi, serially 10-fold diluted, and spread on TSA plates. For bacterial counting, plates were incubated overnight at 37°C in 5% CO2 (10).
Quantitative real-time PCR of inflammatory cytokines.
To determine if AS87_RS02955 influenced inflammatory cytokine expression in cells, interleukin (IL)-1β, IL-6, IL-8, and TNF-α mRNA levels in HD-11 cells (ATCC, Rockefeller, MD, USA) infected with Yb2, Yb2Δ02955, or cYb2Δ02955 strains were measured by qRT-PCR as previously described, but with some modifications (60). Briefly, HD-11 cells were infected with bacteria at an MOI = 100, after which samples were collected at 3, 6, and 9 hpi. Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and stored at −80°C. Total RNA was measured using a spectrophotometer and the Turbo DNA-free kit (Thermo Scientific, Waltham, MA, USA) used to remove genomic DNA. Then, single-strand cDNA was synthesized from total RNA using random hexamer primers and the PrimeScript RT reagent kit (TaKaRa, China). We performed qRT-PCR in duplicate using SYBR Premix Ex Taq (TaKaRa) and gene-specific primers (Table 3). A melting curve was generated at the end of each amplification program to confirm the presence of a single amplification product without primer dimers. Gene expression was quantified using the comparative 2−ΔΔCT method, with β-actin acting as a normalization reference. All assays were performed in triplicate and replicated three times.
Bioinformatics and enzymatic activity analyses.
The AS87_RS02955 amino acid sequence was retrieved from Universal Protein Resource (https://www.uniprot.org/uniprot). Pfam domains and conserved amino acids were predicted by searching the Conserved Domain Database of the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Proteins homologous with AS87_RS02955 were identified by BLAST analysis of AS87_RS02955 amino acid sequences in the NCBI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
The enzymatic activity of purified rAS87_RS02955 was measured using λ DNA (Sangon Biotech, China) cleavage as previously described, with some modifications (41). The reaction mixture (20 μL) contained 200 ng λ DNA, 20 mM MgCl2, 10 mM citric acid-Na2HPO4 buffer (pH 6.5), 100 mM NaCl, and 2 μg rAS87_RS02955. The reaction proceeded for 0, 10, 20, 30, 60, 90, and 120 min at 37°C. Purified pCold I and DNase (Invitrogen) were used as negative and positive controls, respectively. Secretory proteins from Yb2, Yb2Δ02955, and cYb2Δ02955 strains were extracted and quantified as previously described (21). Enzymatic activity was measured using 8 μg secretory proteins from each strain, and reaction mixtures were incubated for 1, 3, 6, and 9 h at 37°C.
To identify RNA cleavage activity in rAS87_RS02955, MS2 RNA (Sigma) was used as a substrate. The reactions contained 800 ng MS2 RNA, 2 μg rAS87_RS02955, and 20 mM MgCl2 and proceeded for 0, 10, 20, 30, 60, and 90 min at 37°C. Purified pCold I and RNase A (Solarbio, Beijing, China) were used as negative and positive controls, respectively. Reactions were analyzed on 0.8% agarose gels and quantified in ImageJ software (National Institutes of Health, MD, USA). All data were collected in triplicate.
The effects of temperature and pH on rAS87_RS02955 enzymatic activity.
We performed assays to determine the effects of temperature on rAS87_RS02955 DNA cleavage. Reaction mixtures were incubated for 30 min at 16, 21, 26, 31, 36, 41, 46, 51, 56, 61, 66, and 71°C.
The effects of pH on rAS87_RS02955 activity were evaluated using a pH range of 3.0 to 12.0 in reactions (20 μL) containing 200 ng λ DNA, 20 mM MgCl2, 10 mM buffers with various pH’s, 100 mM NaCl, and 2 μg rAS87_RS02955. Three different buffers (all at 10 mM concentrations) were used to prepare different pH’s: citric acid-Na2HPO4 (pH 3.0 to 6.5), Tris-HCl (pH 7.0 to 8.5), and glycine-NaOH (pH 9.0 to 12.0). Reactions were maintained for 30 min at 37°C. Purified pCold I was used as a negative control. Data were collected in triplicate and the results analyzed as described above.
The effects of metal cations on rAS87_RS02955 enzymatic activity.
Zn2+, Cu2+, Mg2+, Ca2+, Mn2+, and Fe3+ metal cations were examined for their effects on rAS87_RS02955 enzymatic activity. Reactions were performed using 200 ng λ DNA or 800 ng MS2 RNA as the substrate plus 2 μg rAS87_RS02955 for 30 min at 37°C in different metal cation concentrations. The optimal Mg2+ concentration was measured by incubating rAS87_RS02955 with different Mg2+ concentrations and λ DNA in 10 mM citric acid-Na2HPO4 buffer (pH 6.5) for 30 min at 37°C. The effects of EDTA on enzymatic activity were measured by adding several EDTA concentrations, ranging from 0 to 25 mM, to reactions.
To determine the effects of NaCl and KCl on rAS87_RS02955 endonuclease activity, we performed DNA cleavage assays, which contained different NaCl and KCl concentrations ranging from 0 to 500 mM in 20 μL reactions with rAS87_RS02955. Purified pCold I was used as a negative control. Reactions were performed as described above. Experiments were performed in triplicate.
Site-directed mutagenesis of rAS87_RS02955.
The AS87_RS02955 protein sequence was subjected to comparative homology modeling using the Phyre2 Investigator web interface (http://www.sbg.bio.ic.ac.uk/phyre2) (61, 62). According to a mutational analysis graph which represented the predicted effects of mutations at a particular position, tall, green or blue bars above a residue were predicted to exert a phenotypic effect and could be classified as conserved sites (61). To determine if these predicted residues affected enzymatic activity, site-directed mutagenesis, using the two-stage PCR-based overlap extension method (63), was used to generate the following mutant proteins: rAS87_RS02955 F22A, F26A, F39A, W54A, L90A, F92A, L106A, L122A, Y130A, I134A, F145A, and F155A. Complementary oligodeoxyribonucleotide primers (Table 3) were used to generate mutant proteins in E. coli BL21(DE3) cells, followed by HisTrap affinity column purification, and SDS-PAGE to confirm expression.
Determining mutant protein enzymatic activity and performing bacterial survival assays.
Mutant protein enzymatic activity was measured by incubating 2 μg protein with 200 ng λ DNA for 30 min at 37°C. We used rAS87_RS02955 as a positive control and purified pCold I as a negative control.
To determine if rAS87_RS02955 expression affected bacterial function, transformant expressing rAS87_RS02955, mutant proteins, or pCold I was cultured to logarithmic phase, washed three times in PBS, and diluted to the same OD600 value. Suspensions were then spotted onto LB plates containing 100 μg/mL ampicillin and 0.1 mM IPTG. Plates containing 100 μg/mL ampicillin without IPTG were used as negative controls. Plates were incubated overnight at 37°C.
Statistical analysis.
Statistical analyses were conducted in GraphPad Software version 6.0 (La Jolla, CA, USA). One-way analysis of variance was used for growth curve, adhesion and invasion, qPCR, and enzymatic activity analyses. Two-tailed independent Student’s t tests were used to analyze bacterial loads in blood. P < 0.05 values were considered statistically significant.
ACKNOWLEDGMENTS
This work was supported by the Shanghai Science and Technology Innovation Action Plan (Grant no.19391902800).
We thank all the authors who contributed to this study: M.Z. prepared the manuscript; M.Z., Z.C., R.S., P.N., Y.F., and D.L. performed the experiments and analyzed the data; S.Y. designed the study and revised the manuscript.
We declare that they have no competing interests.
We also thank the Institutional Animal Care and Use Committee of Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences for supporting the animal experiments.
Footnotes
Supplemental material is available online only.
Contributor Information
Shengqing Yu, Email: yus@shvri.ac.cn.
Christopher A. Elkins, Centers for Disease Control and Prevention
REFERENCES
- 1.Segers P, Mannheim W, Vancanneyt M, De Brandt K, Hinz KH, Kersters K, Vandamme P. 1993. Riemerella anatipestifer gen. nov., comb. nov., the causative agent of septicemia anserum exsudativa, and its phylogenetic affiliation within the Flavobacterium-Cytophaga rRNA homology group. Int J Syst Bacteriol 43:768–776. 10.1099/00207713-43-4-768. [DOI] [PubMed] [Google Scholar]
- 2.Subramaniam S, Chua KL, Tan HM, Loh H, Kuhnert P, Frey J. 1997. Phylogenetic position of Riemerella anatipestifer based on 16S rRNA gene sequences. Int J Syst Bacteriol 47:562–565. 10.1099/00207713-47-2-562. [DOI] [PubMed] [Google Scholar]
- 3.Leavitt S, Ayroud M. 1997. Riemerella anatipestifer infection of domestic ducklings. Can Vet J 38:113. [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, Yue J, Ding C, Wang S, Liu B, Tian M, Yu S. 2016. Deletion of AS87_03730 gene changed the bacterial virulence and gene expression of Riemerella anatipestifer. Sci Rep 6:22438. 10.1038/srep22438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hess C, Enichlmayr H, Jandreski-Cvetkovic D, Liebhart D, Bilic I, Hess M. 2013. Riemerella anatipestifer outbreaks in commercial goose flocks and identification of isolates by MALDI-TOF mass spectrometry. Avian Pathol 42:151–156. 10.1080/03079457.2013.775401. [DOI] [PubMed] [Google Scholar]
- 6.Subramaniam S, Huang B, Loh H, Kwang J, Tan HM, Chua KL, Frey J. 2000. Characterization of a predominant immunogenic outer membrane protein of Riemerella anatipestifer. Clin Diagn Lab Immunol 7:168–174. 10.1128/CDLI.7.2.168-174.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Q, Han X, Zhou X, Ding C, Zhu Y, Yu S. 2011. OmpA is a virulence factor of Riemerella anatipestifer. Vet Microbiol 150:278–283. 10.1016/j.vetmic.2011.01.022. [DOI] [PubMed] [Google Scholar]
- 8.Chang CF, Hung PE, Chang YF. 1998. Molecular characterization of a plasmid isolated from Riemerella anatipestifer. Avian Pathol 27:339–345. 10.1080/03079459808419349. [DOI] [PubMed] [Google Scholar]
- 9.Crasta KC, Chua KL, Subramaniam S, Frey J, Loh H, Tan HM. 2002. Identification and characterization of CAMP cohemolysin as a potential virulence factor of Riemerella anatipestifer. J Bacteriol 184:1932–1939. 10.1128/JB.184.7.1932-1939.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Liu B, Dou Y, Fan H, Wang S, Li T, Ding C, Yu S. 2016. The Riemerella anatipestifer AS87_01735 gene encodes nicotinamidase PncA, an important virulence factor. Appl Environ Microbiol 82:5815–5823. 10.1128/AEM.01829-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang M, Zhang P, Zhu D, Wang M, Jia R, Chen S, Sun K, Yang Q, Wu Y, Chen X, Biville F, Cheng A, Liu M. 2017. Identification of the ferric iron utilization gene B739_1208 and its role in the virulence of R. anatipestifer CH-1. Vet Microbiol 201:162–169. 10.1016/j.vetmic.2017.01.027. [DOI] [PubMed] [Google Scholar]
- 12.Hu D, Guo Y, Guo J, Wang Y, Pan Z, Xiao Y, Wang X, Hu S, Liu M, Li Z, Bi D, Zhou Z. 2019. Deletion of the Riemerella anatipestifer type IX secretion system gene sprA results in differential expression of outer membrane proteins and virulence. Avian Pathol 48:191–203. 10.1080/03079457.2019.1566594. [DOI] [PubMed] [Google Scholar]
- 13.Abby SS, Cury J, Guglielmini J, Neron B, Touchon M, Rocha EP. 2016. Identification of protein secretion systems in bacterial genomes. Sci Rep 6:23080. 10.1038/srep23080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McBride MJ, Nakane D. 2015. Flavobacterium gliding motility and the type IX secretion system. Curr Opin Microbiol 28:72–77. 10.1016/j.mib.2015.07.016. [DOI] [PubMed] [Google Scholar]
- 15.Veith PD, Nor Muhammad NA, Dashper SG, Likic VA, Gorasia DG, Chen D, Byrne SJ, Catmull DV, Reynolds EC. 2013. Protein substrates of a novel secretion system are numerous in the Bacteroidetes phylum and have in common a cleavable C-terminal secretion signal, extensive post-translational modification, and cell-surface attachment. J Proteome Res 12:4449–4461. 10.1021/pr400487b. [DOI] [PubMed] [Google Scholar]
- 16.de Diego I, Ksiazek M, Mizgalska D, Koneru L, Golik P, Szmigielski B, Nowak M, Nowakowska Z, Potempa B, Houston JA, Enghild JJ, Thogersen IB, Gao J, Kwan AH, Trewhella J, Dubin G, Gomis-Ruth FX, Nguyen KA, Potempa J. 2016. The outer-membrane export signal of Porphyromonas gingivalis type IX secretion system (T9SS) is a conserved C-terminal beta-sandwich domain. Sci Rep 6:23123. 10.1038/srep23123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McBride MJ, Zhu Y. 2013. Gliding motility and Por secretion system genes are widespread among members of the phylum bacteroidetes. J Bacteriol 195:270–278. 10.1128/JB.01962-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Ding C, Wang S, Han X, Yu S. 2015. Whole-genome sequence analysis and genome-wide virulence gene identification of Riemerella anatipestifer strain Yb2. Appl Environ Microbiol 81:5093–5102. 10.1128/AEM.00828-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malhi KK, Wang X, Chen Z, Ding C, Yu S. 2019. Riemerella anatipestifer gene AS87_08785 encodes a functional component, GldK, of the type IX secretion system. Vet Microbiol 231:93–99. 10.1016/j.vetmic.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 20.Chen Z, Wang X, Ren X, Han W, Malhi KK, Ding C, Yu S. 2019. Riemerella anatipestifer GldM is required for bacterial gliding motility, protein secretion, and virulence. Vet Res 50:43. 10.1186/s13567-019-0660-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niu P, Chen Z, Ren X, Han W, Dong H, Shen R, Ding C, Zhu S, Yu S. 2021. A Riemerella anatipestifer metallophosphoesterase that displays phosphatase activity and is associated with virulence. Appl Environ Microbiol 87. 10.1128/AEM.00086-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Z, Niu P, Ren X, Han W, Shen R, Zhu M, Yu Y, Yu S. 2022. Genome-wide analysis and characterization of the Riemerella anatipestifer putative T9SS secretory proteins with a conserved C-terminal domain. J Bacteriol 204:e0007322. 10.1128/jb.00073-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L, Huang Y, Xu D, Yang L, Qian K, Chang G, Gong Y, Zhou X, Ma K. 2016. Biochemical characterization of a thermostable HNH endonuclease from deep-sea thermophilic bacteriophage GVE2. Appl Microbiol Biotechnol 100:8003–8012. 10.1007/s00253-016-7568-7. [DOI] [PubMed] [Google Scholar]
- 24.Meiss G, Franke I, Gimadutdinow O, Urbanke C, Pingoud A. 1998. Biochemical characterization of Anabaena sp. strain PCC 7120 non-specific nuclease NucA and its inhibitor NuiA. Eur J Biochem 251:924–934. 10.1046/j.1432-1327.1998.2510924.x. [DOI] [PubMed] [Google Scholar]
- 25.Friedhoff P, Meiss G, Kolmes B, Pieper U, Gimadutdinow O, Urbanke C, Pingoud A. 1996. Kinetic analysis of the cleavage of natural and synthetic substrates by the Serratia nuclease. Eur J Biochem 241:572–580. 10.1111/j.1432-1033.1996.00572.x. [DOI] [PubMed] [Google Scholar]
- 26.Iwasaki M, Igarashi H, Yutsudo T. 1997. Mitogenic factor secreted by Streptococcus pyogenes is a heat-stable nuclease requiring His122 for activity. Microbiology (Reading) 143:2449–2455. 10.1099/00221287-143-7-2449. [DOI] [PubMed] [Google Scholar]
- 27.Handa N, Morimatsu K, Lovett ST, Kowalczykowski SC. 2009. Reconstitution of initial steps of dsDNA break repair by the RecF pathway of E. coli. Genes Dev 23:1234–1245. 10.1101/gad.1780709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dianov G, Sedgwick B, Daly G, Olsson M, Lovett S, Lindahl T. 1994. Release of 5'-terminal deoxyribose-phosphate residues from incised abasic sites in DNA by the Escherichia coli RecJ protein. Nucleic Acids Res 22:993–998. 10.1093/nar/22.6.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mankai H, Wende W, Slama N, Ayed A, Roberts RJ, Limam F. 2020. Biochemical and molecular characterization of a restriction endonuclease Tvu2HI from Thermoactinomyces vulgaris 2H and study of its R-M system. Int J Biol Macromol 164:3105–3113. 10.1016/j.ijbiomac.2020.08.151. [DOI] [PubMed] [Google Scholar]
- 30.Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. 1999. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol 112:531–552. 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 31.Zhou H, Li P, Wu D, Ran T, Wang W, Xu D. 2015. EheA from Exiguobacterium sp. yc3 is a novel thermostable DNase belonging to HNH endonuclease superfamily. FEMS Microbiol Lett 362:fnv204. 10.1093/femsle/fnv204. [DOI] [PubMed] [Google Scholar]
- 32.Xu SY, Gupta YK. 2013. Natural zinc ribbon HNH endonucleases and engineered zinc finger nicking endonuclease. Nucleic Acids Res 41:378–390. 10.1093/nar/gks1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu SY, Kuzin AP, Seetharaman J, Gutjahr A, Chan SH, Chen Y, Xiao R, Acton TB, Montelione GT, Tong L. 2013. Structure determination and biochemical characterization of a putative HNH endonuclease from Geobacter metallireducens GS-15. PLoS One 8:e72114. 10.1371/journal.pone.0072114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen BW, Landthaler M, Shub DA, Stoddard BL. 2004. DNA binding and cleavage by the HNH homing endonuclease I-HmuI. J Mol Biol 342:43–56. 10.1016/j.jmb.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 35.Ruhe ZC, Low DA, Hayes CS. 2013. Bacterial contact-dependent growth inhibition. Trends Microbiol 21:230–237. 10.1016/j.tim.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Viprey V, Del Greco A, Golinowski W, Broughton WJ, Perret X. 1998. Symbiotic implications of type III protein secretion machinery in Rhizobium. Mol Microbiol 28:1381–1389. 10.1046/j.1365-2958.1998.00920.x. [DOI] [PubMed] [Google Scholar]
- 37.Wandersman C, Delepelaire P. 2004. Bacterial iron sources: from siderophores to hemophores. Annu Rev Microbiol 58:611–647. 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- 38.Declercq AM, Haesebrouck F, Van den Broeck W, Bossier P, Decostere A. 2013. Columnaris disease in fish: a review with emphasis on bacterium-host interactions. Vet Res 44:27. 10.1186/1297-9716-44-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Veith PD, Chen YY, Gorasia DG, Chen D, Glew MD, O'Brien-Simpson NM, Cecil JD, Holden JA, Reynolds EC. 2014. Porphyromonas gingivalis outer membrane vesicles exclusively contain outer membrane and periplasmic proteins and carry a cargo enriched with virulence factors. J Proteome Res 13:2420–2432. 10.1021/pr401227e. [DOI] [PubMed] [Google Scholar]
- 40.Zhang L, Wang L, Wu L, Jiang D, Tang C, Wu Y, Wu M, Chen M. 2021. Biochemical characterization and mutational studies of a thermostable endonuclease III from Sulfolobus islandicus REY15A. Int J Biol Macromol 193:856–865. 10.1016/j.ijbiomac.2021.10.143. [DOI] [PubMed] [Google Scholar]
- 41.Wang W, Ma L, Wang L, Zheng L, Zheng M. 2020. RecJ from Bacillus halodurans possesses endonuclease activity at moderate temperature. FEBS Lett 594:2303–2310. 10.1002/1873-3468.13809. [DOI] [PubMed] [Google Scholar]
- 42.Endo Y, Huber PW, Wool IG. 1983. The ribonuclease activity of the cytotoxin alpha-sarcin. The characteristics of the enzymatic activity of alpha-sarcin with ribosomes and ribonucleic acids as substrates. J Biol Chem 258:2662–2667. 10.1016/S0021-9258(18)32977-6. [DOI] [PubMed] [Google Scholar]
- 43.Cowan JA. 2002. Structural and catalytic chemistry of magnesium-dependent enzymes. Biometals 15:225–235. 10.1023/a:1016022730880. [DOI] [PubMed] [Google Scholar]
- 44.Maguire ME, Cowan JA. 2002. Magnesium chemistry and biochemistry. Biometals 15:203–210. 10.1023/a:1016058229972. [DOI] [PubMed] [Google Scholar]
- 45.Yang W. 2011. Nucleases: diversity of structure, function and mechanism. Q Rev Biophys 44:1–93. 10.1017/S0033583510000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma L, Wang W, Hao C, Zheng L, Wang L, Zheng M. 2021. Coexistence of endonuclease and exonuclease activities in a novel RecJ from Bacillus cereus. Biotechnol Lett 43:1349–1355. 10.1007/s10529-021-03107-z. [DOI] [PubMed] [Google Scholar]
- 47.Rangarajan ES, Shankar V. 2001. Sugar non-specific endonucleases. FEMS Microbiol Rev 25:583–613. 10.1111/j.1574-6976.2001.tb00593.x. [DOI] [PubMed] [Google Scholar]
- 48.Alani E, Padmore R, Kleckner N. 1990. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell 61:419–436. 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- 49.Mishra NC, Forsthoefel AM. 1983. Biochemical genetics of Neurospora nuclease II: mutagen sensitivity and other characteristics of the nuclease mutants. Genet Res 41:287–297. 10.1017/s0016672300021340. [DOI] [PubMed] [Google Scholar]
- 50.Chow TY, Resnick MA. 1988. An endo-exonuclease activity of yeast that requires a functional RAD52 gene. Mol Gen Genet 211:41–48. 10.1007/BF00338391. [DOI] [PubMed] [Google Scholar]
- 51.Hsia KC, Li CL, Yuan HS. 2005. Structural and functional insight into sugar-nonspecific nucleases in host defense. Curr Opin Struct Biol 15:126–134. 10.1016/j.sbi.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 52.Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, Tsang LH, Smeltzer MS, Horswill AR, Bayles KW. 2009. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One 4:e5822. 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kiedrowski MR, Kavanaugh JS, Malone CL, Mootz JM, Voyich JM, Smeltzer MS, Bayles KW, Horswill AR. 2011. Nuclease modulates biofilm formation in community-associated methicillin-resistant Staphylococcus aureus. PLoS One 6:e26714. 10.1371/journal.pone.0026714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Michel-Briand Y, Baysse C. 2002. The pyocins of Pseudomonas aeruginosa. Biochimie 84:499–510. 10.1016/s0300-9084(02)01422-0. [DOI] [PubMed] [Google Scholar]
- 55.Liu M, Zhang L, Huang L, Biville F, Zhu D, Wang M, Jia R, Chen S, Sun K, Yang Q, Wu Y, Chen X, Cheng A. 2017. Use of natural transformation to establish an easy knockout method in Riemerella anatipestifer. Appl Environ Microbiol 83. 10.1128/AEM.00127-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiong AS, Yao QH, Peng RH, Duan H, Li X, Fan HQ, Cheng ZM, Li Y. 2006. PCR-based accurate synthesis of long DNA sequences. Nat Protoc 1:791–797. 10.1038/nprot.2006.103. [DOI] [PubMed] [Google Scholar]
- 57.Hu Q, Zhu Y, Tu J, Yin Y, Wang X, Han X, Ding C, Zhang B, Yu S. 2012. Identification of the genes involved in Riemerella anatipestifer biofilm formation by random transposon mutagenesis. PLoS One 7:e39805. 10.1371/journal.pone.0039805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu Q, Han X, Zhou X, Ding S, Ding C, Yu S. 2010. Characterization of biofilm formation by Riemerella anatipestifer. Vet Microbiol 144:429–436. 10.1016/j.vetmic.2010.02.023. [DOI] [PubMed] [Google Scholar]
- 59.Guo Y, Shen C, Cheng A, Wang M, Zhang N, Chen S, Zhou Y. 2009. Anatid herpesvirus 1 CH virulent strain induces syncytium and apoptosis in duck embryo fibroblast cultures. Vet Microbiol 138:258–265. 10.1016/j.vetmic.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang S, Xu X, Liu X, Wang D, Liang H, Wu X, Tian M, Ding C, Wang G, Yu S. 2017. Escherichia coli type III secretion system 2 regulator EtrA promotes virulence of avian pathogenic Escherichia coli. Microbiology (Reading) 163:1515–1524. 10.1099/mic.0.000525. [DOI] [PubMed] [Google Scholar]
- 61.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rafeeq CM, Vaishnav AB, Manzur Ali PP. 2021. Characterisation and comparative analysis of hydrophobin isolated from Pleurotus floridanus (PfH). Protein Expr Purif 182:105834. 10.1016/j.pep.2021.105834. [DOI] [PubMed] [Google Scholar]
- 63.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59. 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aem.01276-22-s0001.pdf, PDF file, 0.2 MB (192.7KB, pdf)