ABSTRACT
Pectobacterium carotovorum subsp. carotovorum is a necrotrophic plant pathogen that secretes plant cell wall-degrading enzymes (PCWDEs) that cause soft rot disease in various crops. Bacteriophages have been under consideration as harmless antibacterial agents to replace antibiotics and copper-based pesticides. However, the emergence of bacteriophage resistance is one of the main concerns that should be resolved for practical phage applications. In this study, we developed a phage cocktail with three lytic phages that recognize colanic acid (phage POP12) or flagella (phages POP15 and POP17) as phage receptors to minimize phage resistance. The phage cocktail effectively suppressed the emergence of phage-resistant P. carotovorum subsp. carotovorum compared with single phages in in vitro challenge assays. The application of the phage cocktail to napa cabbage (Brassica rapa subsp. pekinensis) resulted in significant growth retardation of P. carotovorum subsp. carotovorum (P < 0.05) and prevented the symptoms of soft rot disease. Furthermore, phage cocktail treatments of young napa cabbage leaves in a greenhouse environment indicated effective prevention of soft rot disease compared to that in the nonphage negative control. We isolated 15 phage-resistant mutants after a phage cocktail treatment to assess the virulence-associated phenotypes compared to those of wild-type (WT) strain Pcc27. All mutants showed reduced production of four different PCWDEs, leading to lower levels of tissue softening. Ten of the 15 phage-resistant mutants additionally exhibited decreased swimming motility. Taken together, these results show that the phage cocktail developed here, which targets two different types of phage receptors, provides an effective strategy for controlling P. carotovorum subsp. carotovorum in agricultural products, with a potential ability to attenuate P. carotovorum subsp. carotovorum virulence.
IMPORTANCE Pectobacterium carotovorum subsp. carotovorum is a phytopathogen that causes soft rot disease in various crops by producing plant cell wall-degrading enzymes (PCWDEs). Although antibiotics and copper-based pesticides have been extensively applied to inhibit P. carotovorum subsp. carotovorum, the emergence of antibiotic-resistant bacteria and demand for harmless antimicrobial products have emphasized the necessity of finding alternative therapeutic strategies. To address this problem, we developed a phage cocktail consisting of three P. carotovorum subsp. carotovorum-specific phages that recognize colanic acids and flagella of P. carotovorum subsp. carotovorum. The phage cocktail treatments significantly decreased P. carotovorum subsp. carotovorum populations, as well as soft rot symptoms in napa cabbage. Simultaneously, they resulted in virulence attenuation in phage-resistant P. carotovorum subsp. carotovorum, which was represented by decreased PCWDE production and decreased flagellum-mediated swimming motility. These results suggested that preparations of phage cocktails targeting multiple receptors would be an effective approach to biocontrol of P. carotovorum subsp. carotovorum in crops.
KEYWORDS: Pectobacterium carotovorum subsp. carotovorum, bacteriophage cocktail, phage receptor, soft rot disease, virulence attenuation
INTRODUCTION
Pectobacterium carotovorum subsp. carotovorum is a ubiquitous necrotrophic phytopathogen that is responsible for soft rot disease in various crops and ornamental plants during their cultivation, transport, and storage (1). P. carotovorum subsp. carotovorum produces and secretes plant cell wall-degrading enzymes (PCWDEs), including pectate lyase (Pel), polygalacturonase (Peh), cellulase (Cel), and protease (Prt), which are the major virulence factors of P. carotovorum subsp. carotovorum (2). These pathogens primarily degrade cell walls by using Pel and penetrate host tissue through natural pores (e.g., lenticels and stomata) or wounds, which consequently leads to tissue maceration in tubers, rhizomes, and leaves (1, 3). P. carotovorum subsp. carotovorum has been reported as a causative agent of many infectious crop disease outbreaks and is ranked among the top 10 pathogens of importance in agriculture (4–7). Copper-based pesticides and antibiotics have been applied to protect agricultural products from Pectobacterium spp. (former Erwinia spp.) infections (8–11). However, a novel therapeutic strategy for the biocontrol of P. carotovorum subsp. carotovorum needs to be developed due to the spread of bacterial resistance against chemotherapeutics and the public’s desire to use safer and ecologically friendly biological agents (12).
Bacteriophages (phages) are viruses that specifically infect host bacteria. They hijack the host machinery for self-replication and subsequently lyse their host bacterium to release progeny viral particles (13). Phages have several advantages for agricultural application as alternative antimicrobial agents compared to conventional chemical biocides: harmlessness to humans and symbiotic bacteria, relatively lower development and production costs than antibiotics, and self-dosing (14, 15). Several therapeutic uses of bacteriophages for the biocontrol of Pectobacterium have been studied. Phages PP1, DU_PP13B, ϕA38, and ϕ41 were characterized and successfully protected lettuce and potato tubers from P. carotovorum subsp. carotovorum infections (16–18). However, these cases of single-phage treatments have several limitations, such as narrow host ranges and the easy development of bacterial resistance (19).
Antiphage mechanisms in bacteria, including spontaneous mutations, restriction-modification systems, and CRISPR-Cas adaptive immunity, have been studied (20). Bacteria may alter or eliminate their cell surface components that function as phage receptors through spontaneous mutations, which block the adsorption of phages. For instance, we recently revealed that a spontaneous mutant resistant to colanic acid (CA)-recognizing phage POP72 contained a missense mutation within a putative wzc gene that is involved in the biosynthesis of the capsular polysaccharide CA (21). Salmonella enterica serovar Typhimurium strain LT2(c) could block phage SPC35 adsorption through phase-variable glucosylation of the receptor O antigen (22), and phage 117-resistant Klebsiella prevented phage adsorption by introducing mobile genetic elements into the coding region of the wcaJ gene, which is associated with CA biosynthesis (23). The use of phage cocktails has been considered to be an effective strategy to broaden host ranges and to overcome phage resistance mechanisms in the biocontrol of foodborne pathogens or phytopathogens. Kim et al. and Bai et al. developed phage cocktails targeting different phage receptors, and they successfully retarded the emergence of phage-resistant Salmonella Typhimurium in in vitro challenge assays (24, 25). Czajkowski et al. and Zaczek-Moczydłowska et al. reported the efficacy of phage mixtures that were able to suppress Pectobacterium growth in semi-in planta experiments on potato tubers (26, 27).
Phage resistance induced by strong selective pressure may trigger trade-off costs, including attenuated virulence and reduced fitness within heterogeneous populations (28). Phage-mediated selection induced a modified lipopolysaccharide (LPS) composition in Pectobacterium atrosepticum, which resulted in reduced motility and virulence while unexpectedly improving abiotic-surface adhesion (29). Phage-resistant Ralstonia solanacearum strains that were isolated from phage combination-treated tomatoes exhibited defective growth rates and reduced competitive abilities compared to those of the phage-susceptible ancestral pathogen (30). In contrast, phenotypic changes beneficial to bacterial survival, such as increased antibiotic resistance and exopolysaccharide production, were reported in Escherichia coli and Pseudomonas fluorescens (31, 32). These various trade-offs associated with phage resistance inevitably occur in pathogens during the arms race with predator phages. Therefore, it is important to understand the fitness costs associated with phage treatments when formulating phage cocktails.
In this study, we developed a P. carotovorum subsp. carotovorum phage cocktail consisting of three phages that recognize different phage receptors (CA and flagella). The phage cocktail treatment more effectively retarded both P. carotovorum subsp. carotovorum growth and soft rot disease progression in napa cabbage (Brassica rapa subsp. pekinensis) compared to single-phage treatments. The attenuated virulence of phage-resistant P. carotovorum subsp. carotovorum due to phage cocktail treatment could indicate the usefulness of phage cocktails as alternative biocontrol agents against P. carotovorum subsp. carotovorum infections.
RESULTS
Receptor analysis of Pectobacterium phages to develop phage cocktails.
Phage cocktails, which consist of multiple phages that recognize various phage receptors, can retard the development of phage-resistant bacteria (25, 33). Six Pectobacterium phages with clear plaques were isolated from sewage by using WT P. carotovorum subsp. carotovorum strain Pcc27 as the host bacterium. As all Pectobacterium phages that were previously isolated by our group recognize CA as a phage receptor (21), we attempted to obtain phages that target phage receptors other than CA. We conducted spot assays to determine the phage receptors by using three Pcc27 mutants with insertions of transposon Tn5 (cpsG::Tn5, wcaA::Tn5, and gmd::Tn5) that could not synthesize CA (21, 34). Among the six phages, four phages (POP11, POP12, POP13, and POP14) could not infect the three transposon mutants (Table S1 in the supplemental material). The sensitivity of Pcc27 against the four phages was restored upon plasmid complementation with the wcaA gene even without IPTG (isopropyl-β-d-thiogalactopyranoside) induction (Fig. 1; Fig. S1). This result suggested that POP11, POP12, POP13, and POP14 recognized CA as a phage receptor. However, POP15 and POP17 formed single plaques on the lawns of the three transposon mutants, which indicated that these two phages recognized an apparatus other than CA on the Pcc27 cell surface.
FIG 1.
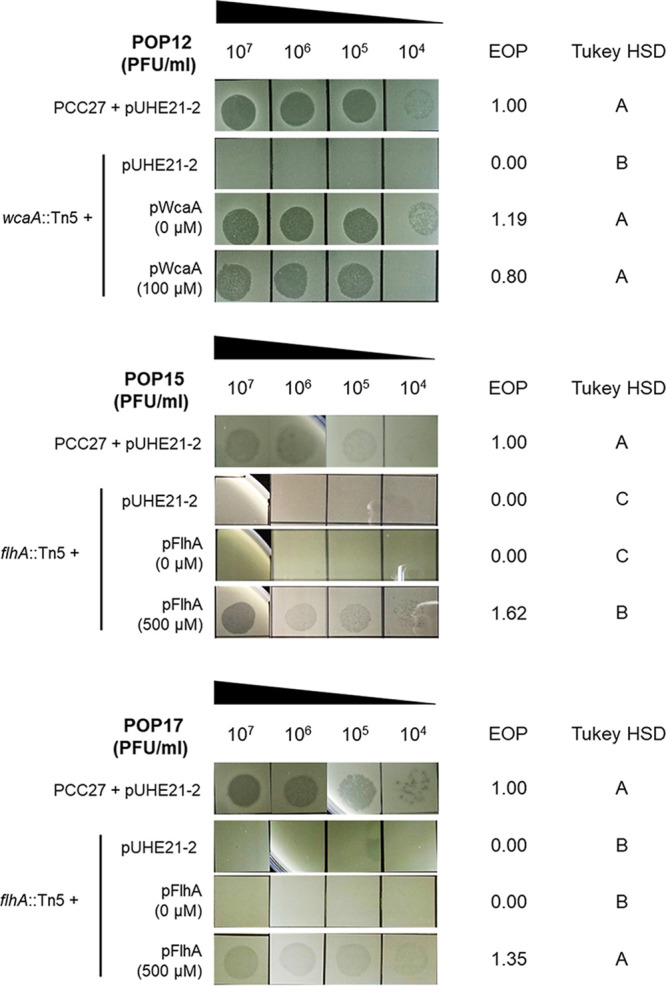
Determination of phage receptors for Pectobacterium-targeting phages. Transposon mutants with wcaA or flhA genes disrupted by Tn5 insertion did not form plaques in the phage spot assays. Complementation of each gene in the Tn5 mutants restored phage susceptibility. The IPTG concentrations are indicated in parentheses. Efficiency of plating (EOP) was calculated by dividing the titer of the phage on each indicated strain by the titer of the same phage on WT strain Pcc27 harboring pUHE21-2. Significant differences among the experimental groups are marked with letters to the right. One representative result from triplicate experiments is shown. HSD, honestly significant difference.
A Tn5 insertional mutant library in the Pcc27 background was constructed and screened for phage POP15 and POP17 resistance to identify phage receptor(s) (35, 36). We acquired two POP17-resistant bacterial strains from the screening. Through partial DNA sequencing, we found that these two mutants had a Tn5 insertion in a putative flhA (homology to Pcc21_RS13355) or putative flhD (homology to Pcc21_13415) gene, respectively. The putative flhA gene in Erwinia amylovora is associated with flagellum biosynthesis, and the putative flhD gene encodes a master regulator that controls the expression of flagellum genes in Pectobacterium carotovorum subsp. carotovorum (formerly Erwinia carotovora subsp. carotovora) (37, 38). When the flhA gene was complemented under the control of the IPTG-inducible promoter, the susceptibility of the flhA::Tn5 mutant against phages POP15 and POP17 was restored to the WT level (Fig. 1). These results suggested that phages POP15 and POP17 recognize flagella as phage receptors.
Host range determination of Pectobacterium phages.
The host ranges of six phages were determined by using 70 Pectobacterium strains that were isolated in South Korea (39, 40). Phage POP12 had the broadest host range compared to the other CA-recognizing phages and showed clear plaques or bacterial growth inhibition zones on P. carotovorum subsp. carotovorum isolates (43 of 47 strains), P. carotovorum subsp. brasiliensis isolates (12 of 17 strains), and P. carotovorum subsp. odoriferum isolates (4 of 4 strains) (Fig. S2). Phages POP15 and POP17 showed relatively narrower host ranges than POP12, but both flagellum-dependent phages could form plaques or inhibition zones against P. carotovorum subsp. carotovorum strains (e.g., Pcc19 and Pcc92) and two Pectobacterium atrosepticum strains that are insusceptible to POP12. Phage POP15 could infect a P. carotovorum subsp. carotovorum strain, Pcc22, that is resistant to POP12 and POP17, and phage POP17 could form plaques or inhibition zones against P. carotovorum subsp. brasiliensis strains (e.g., E10, E12, and E42) that are resistant to POP12 and POP15 (Fig. S2). The results of the host range assays of POP15 and POP17 indicated that the use of flagellum-dependent phages together with POP12 could broaden the host spectrum. Thus, we mixed POP12, POP15, and POP17 to construct a phage cocktail in which CA- and flagellum-recognizing phages were mixed at the same concentration.
Genomic and morphological characterization of POP12, POP15, and POP17.
We conducted whole-genome sequencing of the three phages chosen for the phage cocktail to investigate whether the genes associated with bacterial virulence, antibiotic resistance, and phage lysogen decisions were present in the phage genomes. Bioinformatic analysis revealed that phages POP12, POP15, and POP17 contained 170,838 bp, 153,445 bp, and 155,176 bp of double-stranded DNA, respectively, and that the %GC contents were 36.3%, 51.7%, and 51.9%, respectively (Fig. S3). Genome annotation by GeneMark.hmm and the RAST annotation engine suggested that phages POP12, POP15, and POP17 had 257, 301, and 308 putative open reading frames (ORFs), respectively (41, 42). No genes associated with lysogen formation, toxins, or bacterial virulence factors were identified, which implies the lytic nature and safety of these three phages.
Morphological observations by transmission electron microscopy (TEM) revealed that the three phages belonged to the family Myoviridae (Fig. S4). Phage POP12 had an elongated icosahedral head (125.6 ± 2.6 nm long and 85.6 ± 0.9 nm wide) and contractile tail (122.2 ± 1.9 nm). Phages POP15 and POP17 had isometric heads and contractile, long tails. The head diameter of POP15 was 69.5 ± 2.7 nm, and that of POP17 was 94.9 ± 0.7 nm. The tail length of POP15 was 100.7 ± 5.0 nm, whereas that of POP17 was 129.0 ± 5.7 nm.
Bacterial challenge assays with phage cocktails.
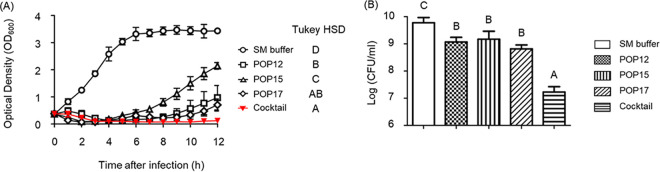
We conducted in vitro bacterial challenge assays to evaluate the efficacy of single phages or phage cocktails in the inhibition of bacterial growth. Single phages or phage cocktails were added to the strain Pcc27 culture at an MOI of 1. The growth of Pcc27 was rapidly suppressed by phage POP15 but resumed at 4 h postinfection. Phages POP12 and POP17 each inhibited bacterial growth until 8 h postinfection. In contrast, treatment with phage cocktails inhibited the emergence of phage-resistant mutants for at least 12 h postinfection (Fig. 2A). The numbers of viable P. carotovorum subsp. carotovorum cells 12 h postinfection with the phage cocktail were also significantly lower than those of the single-phage treatments, as well as the negative control (P < 0.001) (Fig. 2B). These results indicated that the phage cocktail could effectively inhibit host growth and delay the emergence of phage-resistant mutants.
FIG 2.
Bacterial challenge assay with phage POP12, POP15, or POP17 or the phage cocktail comprised of all three phages. Wild-type Pcc27 was infected with phages at MOIs of 1. (A) The bacterial growth was monitored hourly by measuring the optical densities (OD600). (B) The viable bacterial populations were enumerated at 12 h postinfection. SM buffer was added to the Pcc27 culture as a negative control. The mean values with standard deviations (SD) from triplicate experiments are shown. Statistical analysis was conducted by one-way analysis of variance (ANOVA) with Tukey’s multiple-comparison tests among the experimental groups 12 h postinfection. Significant differences among the experimental groups are marked with letters (P < 0.001).
Application of phage cocktails to prevent soft rot in napa cabbage.
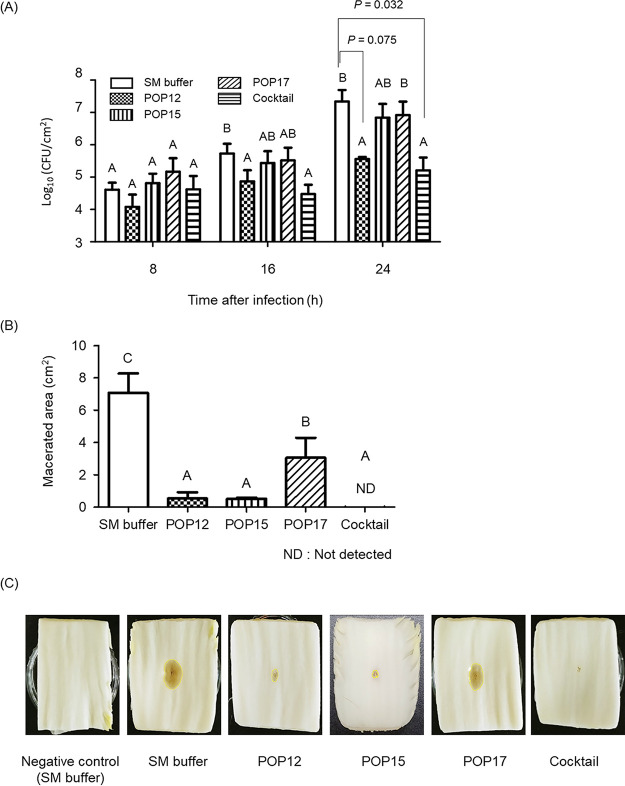
The bactericidal effects of single phages and the phage cocktail against P. carotovorum subsp. carotovorum infection were evaluated by using napa cabbage. Each sample was artificially inoculated with 3 × 105 CFU/mL of rifampicin-resistant Pcc27 (Pcc27RifR) and was then treated with single phages or a phage cocktail at a multiplicity of infection (MOI) of 103. When phages were not applied, the number of Pcc27RifR cells in napa cabbage increased up to approximately 7-log CFU/cm2 within 24 h, and consequently, the sample exhibited typical soft rot symptoms. In contrast, the phage cocktail treatment, rather than single-phage treatments, significantly inhibited the growth of Pcc27RifR (P = 0.032) and the progression of disease in napa cabbage after 24 h of incubation (Fig. 3). The growth of Pcc27RifR was also retarded by a phage POP12 treatment up to 16 h of incubation, but the number of recovered Pcc27RifR populations at 24 h was higher than that of the cocktail-treated group and showed no significant difference from the negative control (P = 0.075) (Fig. 3A). In addition, soft rot symptoms began after 24 h of incubation in POP12-treated napa cabbage (Fig. 3C). A single-phage treatment with phage POP15 or POP17 did not significantly affect the growth of Pcc27RifR cells in napa cabbage (Fig. 3A), and the symptoms of soft rot were also noticeable (Fig. 3C). Comparable protective effects were also achieved against soft rot disease in napa cabbage artificially inoculated with P. carotovorum subsp. carotovorum strain Pcc19 or Pcc21 (Fig. S5).
FIG 3.
Retardation of soft rot disease development in napa cabbage by phage treatment. Each artificially inoculated crop sample was treated with or without phages at an MOI of 103 and incubated in a humid chamber. (A) The numbers of P. carotovorum subsp. carotovorum cells were determined at the indicated time points. (B) Macerated areas of napa cabbage were measured after 24 h of incubation using ImageJ. (C) The symptoms of soft rot were monitored after 24 h of incubation. A sample inoculated with SM buffer was used as a negative control, and strain Pcc27RifR treated with SM buffer was used as a nonphage control. Each column in panels A and B represents the mean value from triplicate experiments, and error bars indicate the standard deviations. One-way analysis of variance (ANOVA) with Tukey’s multiple-comparison test was performed for comparing the CFU values and macerated areas among the groups at the indicated time points. Significant differences among the experimental groups are marked with letters (P < 0.001). Representative results from triplicate experiments are shown in panel C.
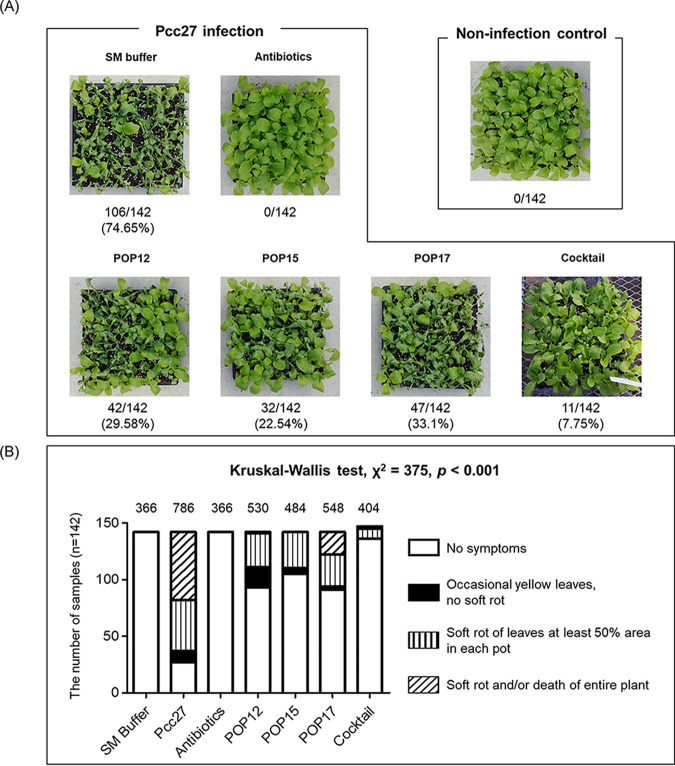
Next, single phages or the phage cocktail were applied by foliar spraying onto young napa cabbage leaves in a greenhouse to assess their antibacterial effects against Pcc27. The three-phage cocktail treatment protected young leaves from soft rot disease better than the single-phage treatments (Fig. 4A). The percentage of rotten leaves with the phage cocktail treatment was 7.75%, but the values obtained after applying single phages ranged from 22.54% to 33.1% (Fig. 4A). Moreover, the qualitative ordinal scale of the soft rot disease indicated that the disease severity of the phage cocktail-treated samples was similar to that of antibiotic-treated samples (Fig. 4B). These results suggested that phage cocktail treatments exhibited better antimicrobial effects in preventing soft rot disease than single-phage treatments, at least in the laboratory and greenhouse tests performed.
FIG 4.
Effect of the phage cocktail in preventing soft rot in young leaves of napa cabbage grown in the greenhouse. (A) Each phage or phage cocktail was sprayed onto young leaves at an MOI of 100. P. carotovorum subsp. carotovorum Pcc27 was added to experimental groups at 1 × 106 CFU/pot 1 day after phage treatments, and the samples were incubated for 30 h in a humidity chamber. The disease symptoms were examined 2 days after transfer of the samples to the greenhouse. Antibiotics were used as a positive control, and SM buffer was used as a nonphage negative control. The rates of rotten napa cabbage are indicated below the images. One representative result from the triplicate experiments is shown. (B) The extent of soft rot was evaluated based on visual assessment of symptom severity in leaves using a four-point scale. Statistical analysis was performed by using nonparametric Kruskal-Wallis analysis. The chi-square (χ2) test result is presented above each bar. A lower chi-square value indicates weaker symptoms of soft rot.
Virulence evaluation of phage-resistant mutants.
Attenuation of bacterial virulence caused by phage infections has been well reported in various foodborne pathogens (43–45). However, studies on the association of bacterial virulence with phage resistance have been reported for only a few plant pathogens, such as P. atrosepticum and R. solanacearum (29, 46, 47). To determine whether the P. carotovorum subsp. carotovorum virulence was altered by phage infection, we obtained 80 colonies from the phage cocktail treatment and purified them by sequential streaking on fresh LB plates at least three times. The susceptibility of each clone against single phages was tested with spot assays. Fifteen clones (PccR1 to PccR15) were insusceptible to phage POP12, and five of the clones (PccR11 to PccR15) were additionally resistant to phage POP15 and POP17 infections (Table S2). In addition to the 15 mutants isolated after phage cocktail treatment (PccR1 to PccR15; also referred to as “phage-resistant mutants” below), we obtained POP12- or POP17-resistant mutants (PccPOP12 and PccPOP17) from single-phage treatments for use as controls in the virulence comparison.
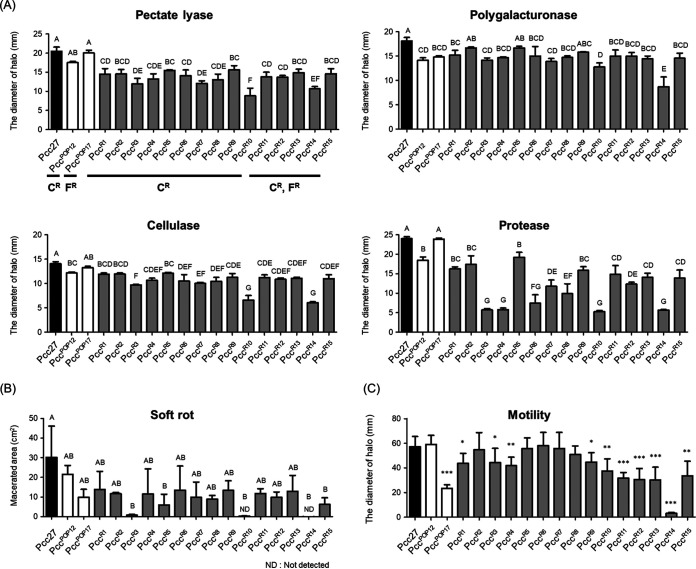
Among the important virulence factors of P. carotovorum subsp. carotovorum are PCWDEs, which are secreted through the type II secretion system (48). We evaluated the production of four different PCWDEs in the 15 phage-resistant mutants by performing extracellular enzyme assays and virulence assays using napa cabbage. Interestingly, all 15 mutants (PccR1 to PccR15) and PccPOP12 produced smaller amounts of PCWDEs than WT Pcc27 (Fig. 5A; Fig. S6 and Table S3). We then compared the PCWDE activities of the 15 phage-resistant mutants to those of PccPOP12, which presented lower PCWDE activities than PccPOP17. Ten mutants exhibited lower Pel activities than PccPOP12, and the levels of Cel and Prt production were also reduced in 6 and 11 of the 15 phage-resistant P. carotovorum subsp. carotovorum strains, respectively (Fig. 5A). However, all of the 15 mutants except PccR14 had Peh activities similar to that of PccPOP12 (Table S3). In accordance with the much lower production of PCWDEs than for the other phage-resistant mutants (Fig. 5A), napa cabbage inoculated with the PccR10 and PccR14 strains showed no symptoms of soft rot (Fig. 5B; Fig. S6). Three more mutants (PccR3, PccR5, and PccR15) also showed significant reduction in soft rot symptoms compared to those in napa cabbage inoculated with PccPOP17, and the rotten-tissue areas of napa cabbage inoculated with the other 10 phage-resistant P. carotovorum subsp. carotovorum strains were smaller than those of the WT Pcc27- or PccPOP12-inoculated controls (Fig. 5B; Fig. S6).
FIG 5.
Attenuated virulence of phage-resistant mutants evaluated by extracellular enzyme assay (A), virulence assay in napa cabbage (B), and motility assay (C). (A) Each column indicates the diameters of the haloes around the wells that were measured to represent the enzyme activities (details in Materials and Methods). (B) Macerated areas of napa cabbage were measured after 24 h of incubation using ImageJ. (C) The swimming haloes of bacterial growth on 0.3% agar plates were measured after 24 h of incubation. The mean values with SD from triplicate experiments are shown. Significant differences among the experimental groups are marked with letters and asterisks. CR, resistant to CA-recognizing phage; FR, resistant to flagellum-recognizing phage; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The group of mutants that were resistant to all three phages (PccR11 to PccR15) and the group of mutants that were resistant only to phage POP12 (PccR1 to PccR10) both exhibited similarly low levels of PCWDE production and soft rot symptoms. These results indicated that phage cocktail treatments could impair the production of PCWDEs more than single-phage treatments, leading to the alleviation of soft rot symptoms.
Because flagellum-mediated motility was recognized as another essential virulence determinant in P. carotovorum subsp. carotovorum (2, 49, 50), we also assessed the swimming motilities of WT strain Pcc27 and phage-resistant mutants. Among 15 mutants resistant to the phage cocktail, 10 showed significant reductions in motility compared to that of the WT Pcc27 (Fig. 5C). In particular, all five mutants (PccR11 to PccR15) that evaded infection by flagellum-recognizing phages POP15 and POP17 exhibited further decreases in motility compared with that of the other mutants that were resistant only to POP12 (particularly PccR1, PccR3, PccR4, PccR9, and PccR10). The PccR14 strain was even nonmotile (Fig. 5C; Table S2). This result implied that phage cocktail treatments could confer strong selection pressure on WT Pcc27 and increase fitness costs, such as the accumulation of phage resistance mutations. Taken together, the virulence assessments of phage-resistant mutants revealed that the use of a phage cocktail that targeted two different receptors could effectively attenuate the virulence of P. carotovorum subsp. carotovorum.
DISCUSSION
P. carotovorum subsp. carotovorum is a widespread plant pathogen that causes quality depreciation and loss of agricultural products. Phages have been considered to be attractive antimicrobial agents for the biocontrol of P. carotovorum subsp. carotovorum, but the emergence of phage resistance should be overcome to provide practical phage therapies. In this study, we developed a phage cocktail by considering the host range spectra and types of phage receptors to constrain the emergence of phage resistance. The CA-targeting phage POP12, with the broadest host range, and two flagellum-recognizing phages that can complement the host spectrum of POP12 were selected to formulate the cocktail.
We tested napa cabbage, which is a major isolation source of Pectobacterium spp. in South Korea (4, 51). The results of the semi-in planta bioassay suggested that our phage cocktail, which targeted two phage receptors, reduced the pathogen population in napa cabbage and alleviated soft rot symptoms more significantly than single-phage treatments. Interestingly, no symptoms of soft rot were observed in phage cocktail-treated napa cabbage (Fig. 3C), even though the average number of Pcc27RifR cells that were recovered from the phage cocktail-treated napa cabbage was more than 105 CFU/cm2 (Fig. 3A), which is sufficient for disease progression (52), suggesting that phage-resistant P. carotovorum subsp. carotovorum produces smaller amounts of PCWDEs (Fig. 5A).
Gill and Abedon proposed several factors that are associated with successful phage therapy in plants, such as the location or niche where target bacteria exist, density of target bacteria, adequate solution for phage diffusion, and environmental conditions that can allow phage amplification (53). In the in vitro challenge assay performed in liquid broth, treatment with phage POP15 or POP17 efficiently prevented the growth of P. carotovorum subsp. carotovorum cells for up to 4 or 8 h, respectively (Fig. 2A). However, the same treatment with 100 times more PFU of phage was not able to restrict the growth of P. carotovorum subsp. carotovorum cells in napa cabbage, and consequently, noticeable soft rot symptoms were observed (Fig. 3C). This might be due to the optimized conditions, such as nutrition and temperature, for P. carotovorum subsp. carotovorum in the in vitro assay, which allowed superior propagation and consequently better bactericidal activities of the phages than in napa cabbage.
Considering that the three phages were applied under the same experimental conditions, we speculated that the more efficient control of P. carotovorum subsp. carotovorum cells by POP12 in napa cabbage would originate from the characteristics of POP12, such as the efficiency of adsorption to Pcc27. Indeed, phage POP12, recognizing CA, was adsorbed to WT Pcc27 more rapidly than the flagellotropic phages, POP15 and POP17 (Fig. S7A).
A mixture of validamycin and streptomycin was used as a positive control in the greenhouse test, which resulted in effective prevention of the disease (Fig. 4). The overuse of aminoglycoside antibiotics like validamycin and streptomycin is, however, a serious problem due to its negative impacts on plants, food, and humans (54). Over 40,000 tons of validamycin have been produced annually to prevent sheath blight disease in rice plants. Streptomycin, which is recognized as critically and highly important for human medication by the World Health Organization (WHO), is used at a rate of approximately 80,000 pounds per year to prevent phytopathogen infections (54–57). Because the preventive effect of the phage cocktail was comparable to those of the antibiotics tested in the present study (Fig. 4B), phage cocktails are suggested as important alternatives and/or adjuvants to minimize the use of antibiotics.
An understanding of how simultaneous application of multiple phages affects the acquisition of phage resistance in bacteria is necessary to formulate effective phage combinations. Intriguingly, we isolated 15 phage-resistant mutants that exhibited different patterns of susceptibility against the three phages. All mutants were resistant to the CA-recognizing phage POP12. Similar to our previously reported study, in which a spontaneous mutant resistant to another CA-targeting phage, POP72, produced a smaller amount of CA than WT Pcc27 (21), this result suggested that the 15 mutants might be deficient in CA biosynthesis. Among them, 5 mutant strains (PccR11 to PccR15) that were also resistant to flagellum-dependent phages had decreased motility (Fig. 5C; Table S2). Wright et al. reported that simultaneous treatments with multiple phages targeting different receptors often led to single receptor-specific mutations in host bacteria (58), because it is a burden for bacteria to acquire resistance against all phage components at once. Interestingly, two flagellotropic phages, POP15 and POP17, adsorbed better to Pcc27 when CA was absent (Fig. S7B), which indicated that CA might act as a physical obstacle for phage POP15 and POP17 infections. These results implied that POP12 first affected WT Pcc27 extensively and had a primary impact on the development of phage resistance, and then POP15 and POP17 affected the resultant CA-lacking resistant mutants and WT host as secondary or auxiliary components of the phage cocktail.
The malfunction or altered functions of phage receptor molecules as a cost for phage resistance often lead to virulence attenuation in bacterial pathogens (59). In the present study, all 15 mutants (PccR1 to PccR15), which had in common resistance to the CA-targeting phage POP12, exhibited decreased PCWDE production and showed reduced soft rot symptoms in napa cabbage compared to the soft rot symptoms caused by infection with WT Pcc27 (Fig. 5A; Fig. S6 and Table S3). Because the PCWDE activity was also lowered in PccPOP12, the POP12 resistance that was probably driven by the loss of CA might have had a large effect on the virulence attenuation in the 15 mutants. Many genes are involved in the production of CA and PCWDEs (34, 60), but their known regulation mechanisms are not enough to elucidate the regulatory link between CA and PCWDE synthesis. Thus, the causal relationship between P. carotovorum subsp. carotovorum virulence and CA production needs to be investigated in further studies by using mutants with targeted gene deletions.
The two flagellotropic phages, POP15 and POP17, also played roles as components of the phage cocktail, not only by lysing the susceptible P. carotovorum subsp. carotovorum cells but also by contributing to further virulence attenuation in the resistant mutants by inducing restricted motility, similar to PccPOP17 (Fig. 5C). Flagellum-mediated motility is reported to be an important determinant for P. carotovorum subsp. carotovorum virulence because flagellum deficiencies affect the adhesion to and penetration of phytopathogens into plant tissue (2, 61). Although both PccPOP12 and 10 of the 15 phage-resistant mutants obtained after phage cocktail treatment were resistant to phage POP12 only, the mutant PccPOP12 obtained after phage POP12 treatment alone was fully functional in terms of its swimming motility, whereas five of the resistant mutants (PccR1, PccR3, PccR4, PccR9, and PccR10) that were isolated after phage cocktail treatment exhibited significantly impaired motilities (Fig. 5C; Table S2). This result further supported the concept that phage cocktail treatments impose strong selective pressure on WT Pcc27, which leads to greater attenuation of bacterial virulence. Phenotypic variations among phage-resistant bacteria can originate from genetic changes like stochastic gene expression related to receptor biosynthesis (62) and/or phase variations (63). Through these diverse evolutionary trajectories, phage cocktail treatments could trigger the attenuation of P. carotovorum subsp. carotovorum virulence at various levels. Further genetic and physiological studies are needed to elucidate reasons for the attenuated virulence of the phage-resistant mutants.
In conclusion, we designed a bacteriophage cocktail that consists of three phages that recognize two different phage receptors (CA and flagella) in P. carotovorum subsp. carotovorum to delay the emergence of bacterial resistance. The cocktail could efficiently retard the growth of P. carotovorum subsp. carotovorum and the subsequent progress of tissue softening in napa cabbage. Moreover, phage cocktail treatment attenuated P. carotovorum subsp. carotovorum’s virulence, including PCWDE production and bacterial motility. These results suggest that the preparation of phage cocktails that target multiple phage receptors and an understanding of the fitness trade-offs that are generated by phage resistance would be helpful for the development of effective alternative strategies to control P. carotovorum subsp. carotovorum with phages instead of using conventional antibiotics.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Pectobacterium carotovorum subsp. carotovorum isolates were provided by the Rural Development Administration (RDA) at Wanju-gun, South Korea. All bacterial strains and plasmids used in this study are listed in Table 1. P. carotovorum subsp. carotovorum strains were grown in LB (Luria-Bertani) broth and plates (1.5% [wt/vol] agar) at 30°C. Escherichia coli strain MFDpir was grown at 37°C in LB broth and plates that were supplemented with 0.3 mM diaminopimelic acid (DAP). Antibiotics were used at the following concentrations: ampicillin (Amp), 50 μg/mL; carbenicillin (Car), 100 μg/mL; kanamycin (Kan), 50 μg/mL; and rifampicin (Rif), 50 μg/mL. IPTG (isopropyl β-d-1-thiogalactopyranoside) was added at a concentration of 100 or 500 μM.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Pectobacterium carotovorum subsp. carotovorum strains | ||
| Pcc27 | P. carotovorum subsp. carotovorum isolate Pcc27; wild type, host for phages POP12, POP15, and POP17 | 39 |
| Pcc27RifR | Spontaneous rifampicin-resistant mutant of Pcc27 | 21 |
| Pcc27/pUHE21-2 | Pcc27 with pUHE21-2 lacIq | 21 |
| wcaA::Tn5 strain | Pcc27 with transposon insertion in putative wcaA | 21 |
| gmd::Tn5 strain | Pcc27 with transposon insertion in putative gmd | 21 |
| cpsG::Tn5 strain | Pcc27 with transposon insertion in putative cpsG | 21 |
| flhA::Tn5 strain | Pcc27 with transposon insertion in putative flhA | This study |
| flhD::Tn5 strain | Pcc27 with transposon insertion in putative flhD | This study |
| wcaA::Tn5/pUHE21-2 | wcaA::Tn5 strain with pUHE21-2 lacIq | 21 |
| wcaA::Tn5/pWcaA | wcaA::Tn5 strain complemented with wcaA gene from Pcc27 | 21 |
| flhA::Tn5/pUHE21-2 | flhA::Tn5 strain with pUHE21-2 lacIq | This study |
| flhA::Tn5/pFlhA | flhA::Tn5 strain complemented with flhA gene from Pcc27 | This study |
| Escherichia coli strains | ||
| DH5α/λpir | ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 hsdR17 deoR thi-1 supE44 gyrA96 relA1/λpir | 76 |
| MFDpir | MG1655 RP4-2-Tc::[ΔMu1::Δaac(3)IV-ΔaphA-Δnic35-ΔMu2::zeo] ΔdapA::(erm-pir) ΔrecA | 36 |
| Plasmids | ||
| pUHE21-2 lacIq | reppMB1 lacIq; inducible Lac promoter, Ampr | 77 |
| pWcaA | pUHE21-2 lacIq::PCC21_RS06680; Ampr | 21 |
| pFlhA | pUHE21-2 lacIq::PCC21_RS13355; Ampr | This study |
Ampr, ampicillin resistant.
Bacteriophage isolation and propagation.
Bacteriophages were isolated from sewage by using P. carotovorum subsp. carotovorum strain Pcc27 as a bacterial host. Five milliliters of sewage was mixed with 5 mL of 2× LB broth, and the mixture was incubated overnight with the host strain at 30°C. The culture was centrifuged at 13,000 × g for 10 min and filtered to exclude bacterial cells. The filtered supernatants were serially diluted 10-fold and spotted onto host bacterial lawns (see below). The plates were incubated overnight at 30°C, and the phage plaques that formed were observed. Single plaques were picked with a sterile tip and eluted in a sodium chloride-magnesium sulfate (SM) buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 8 mM MgSO4·7H2O). Dilution, spotting, and plaque picking were sequentially repeated at least three times to purify a single phage. For phage propagation, the phage lysate was added to the exponentially growing host bacterial culture at a multiplicity of infection (MOI) of 1 and incubated for 3 h. The propagated phages were precipitated with polyethylene glycol (PEG) 6000 and concentrated by using CsCl density gradient ultracentrifugation (78,500 × g for 2 h at 4°C) (64).
Bacteriophage spot assay.
The bacterial lawn was prepared as described elsewhere (64). Briefly, 100 μL of cultured host cells was inoculated into 5 mL of LB soft agar (0.4% agar), supplemented with the appropriate antibiotic and IPTG if necessary. This mixture was poured onto LB agar plates and solidified for 30 min. Ten microliters of serially diluted (10-fold) phage lysates was spotted on the bacterial lawn and dried for 20 min at room temperature. The plates were incubated for 12 h at 30°C, and the phage plaques were monitored.
Screening phage-resistant mutants from the transposon Tn5 insertional-mutant library of P. carotovorum subsp. carotovorum.
Phage-insensitive P. carotovorum subsp. carotovorum mutants were screened from the Tn5 transposon insertional mutant library of P. carotovorum subsp. carotovorum as previously described, with some modifications (35, 36). The suicide vector pRL27 containing Tn5 was transferred from donor E. coli MFDpir cells to recipient Pcc27 cells by conjugation as follows. Donor and recipient strains at the early log phase were harvested and washed three times with 10 mM MgSO4. A mixture of donor and recipient cells (3:1, vol/vol) was spotted onto LB agar plates supplemented with DAP and incubated for 24 h at 30°C for conjugation. The transconjugants were resuspended in LB broth, and the cell dilutions were mixed with phage POP17 (109 PFU/mL). After incubation at room temperature for 15 min, the mixture was plated on LB/Kan plates and incubated for 24 h at 30°C. Each emerging colony was isolated, and the absence of remaining phages in the cells was verified by spotting the culture supernatant on a bacterial lawn. Determining transposon insertion sites is similar to cloning of plasposons (65). The genomic DNA that was extracted from the phage-resistant Tn5 mutants was digested with the restriction enzyme BamHI and circularized by T4 ligase (Roche). E. coli strain DH5α/λpir was transformed with a portion of the ligated mixture by heat shock, and the transformants were selected on LB agar/Kan plates. Plasmid DNA containing Tn5 was extracted from the selected transformants, and the locus of the transposon insertion site was determined by sequencing with transposon-specific primers tpnRL17-1 and tpnRL 13-2 (Table 2) (35). The nucleotide sequence was compared to the sequence of Pectobacterium carotovorum subsp. carotovorum strain Pcc21 (GenBank accession number CP003776) as a reference for the analysis.
TABLE 2.
Primers used in this study
| Purpose, primer | Sequence (5′–3′)a |
|---|---|
| Plasmid construction | |
| Pcc27_wcaA_F_BamHI | ATAGGATCCATGTCAACAAATAATTTAGTCAGTGTTATTATT |
| Pcc27_wcaA_R_HindIII | ATAAAGCTTTGAACGCAAGTCAATCATTTTATTTTTTCC |
| Pcc27_flhA_F_EcoRI | ATAGAATTCCCGGATGCACTGGATTTTGCT |
| Pcc27_flhA_R_HindIII | ATAAAGCTTCTGCAGCCAGAGAATGCATCG |
| Sequence confirmation | |
| Pcc27_flhA_F_confirm | ACCGACTTCAGCAATACGTC |
| Pcc27_flhA_R_confirm | CGGTGCAACCAGATCCTTAT |
| pUHE21-2_F1 | AGATTCAATTGTGAGCGGATAAC |
| pUHE21-2_R3 | GGTCATTACTGGATCTATCAACA |
| tpnRL17–1 | AACAAGCCAGGGATGTAACG |
| tpnRL13–2 | CAGCAACACCTTCTTCACGA |
Restriction enzyme sites are underlined.
Sequencing of phage DNA and bioinformatics analysis.
Phage DNA was extracted by the phenol-chloroform method as previously described (66). The purified phage DNA was sequenced using a Genome Sequencer FLX titanium sequencer (Roche, Mannheim, Germany) and assembled with GS De Novo Assembler software (Roche) at LabGenomics, Inc., and Sanigen Co. Ltd., South Korea. The ORFs were predicted by using Glimmer3 (67), GeneMarkS (41), Fgenesb software (Softberry, Inc., Mount Kisco, NY), and the RAST annotation server (http://rast.nmpdr.org/) (42, 68). The annotated data were assorted using Artemis (69). The tRNA sequence in the phage genome was analyzed with the tRNAscan-SE program (70). The predictions of protein functions were performed with NCBI BLASTp and InterProScan (71, 72).
TEM analysis.
The three purified phages were morphologically characterized by transmission electron microscopy (TEM) analysis as described by Kim and Ryu (64). Briefly, 5 μL of high-titer phage stock (approximately 1 × 1011 PFU/mL) was placed on carbon-coated copper grids and negatively stained with 2% aqueous uranyl acetate (pH 4.0). Phage POP12 was observed with TEM (Carl Zeiss LEO 913AB) at a 100-kv acceleration voltage at the National Institute of Agricultural Sciences (Wanju-gun, South Korea). Phages POP15 and POP17 were observed with TEM (energy-filtered [EF]-TEM) (JEM-1010; JEOL, Japan) at an acceleration voltage of 80 kV at the NICEM (Seoul, South Korea). These phages were classified morphologically by using the International Committee on Taxonomy of Viruses (ICTV) classification (73).
Bacterial challenge assays.
A phage cocktail consisting of three phages (POP12, POP15, and POP17) was prepared at a ratio of 2:1:1. Host Pcc27 cultures at the early exponential phase were infected with each phage or phage cocktail at an MOI of 1 (3 × 109 PFU/mL). The optical densities at 600 nm were measured every hour for 16 h. The cultures were sampled at 12 h post-phage infection and plated onto LB agar medium to count the numbers of viable cells. SM buffer instead of phages was added to the negative control. The experiments were conducted in triplicate.
Biocontrol assays on crop models using a phage cocktail.
The napa cabbage used in this study was purchased from a local market in Seoul, South Korea. Samples were cut into equal sizes (approximately 10 cm by 7 cm) and sanitized with 1% sodium hypochlorite for 5 min. After washing with sterilized water for 5 min and air drying, the samples were stabbed with a sterilized needle. Ten microliters of Pcc27RifR bacterial culture (107 CFU/mL) or sterilized water (negative control) was inoculated into the wounds. After air drying for 10 min, single phages or the prepared phage cocktail was spotted on the Pcc27RifR-inoculated wound at an MOI of 103. The samples were stored in a plastic box to maintain humidity and incubated at 30°C for 24 h. At the indicated time points, the numbers of Pcc27RifR cells were measured as described by Bai et al. (25). The crop samples were transferred to sterile bags (Filtra-Bag with open top, code SCTO7012A; Labplas) containing 100 mL of sterilized buffered peptone water (BPW) and homogenized with a stomacher (BagMixer 400 Laboratory Blender, Interscience) for 1 min. Sample debris was removed, and the bacterial cells in the supernatants were collected via centrifugation (13,000 × g for 1 min at 4°C). The cell pellets were resuspended in phosphate-buffered saline (PBS). A serially diluted (10-fold) cell suspension was plated onto LB agar/Rif plates, and the colonies were counted after 12 h of incubation at 30°C. The macerated areas of napa cabbage samples were measured using ImageJ.
Greenhouse trials.
The greenhouse trials to determine the prophylactic effect of phages as biocontrol agents were kindly supported by the Highland Agriculture Research Institute (Pyeongchang-gun, South Korea). Seeds of napa cabbage (Brassica rapa subsp. pekinensis) were purchased in a local market in Pyeongchang-gun. These seeds were planted in plastic pots (2 cm by 2 cm) and cultivated for 1 month. The young napa cabbage leaves were sprayed with a single phage or phage cocktail (1 × 108 PFU/pot). SM buffer instead of phages was used as a nonphage negative control, and antibiotics (validamycin-A [15%] plus streptomycin [5%]) purchased from the local market were sprayed as a positive control. After air drying for 1 day, serially diluted (10-fold) Pcc27 cells (1 × 106 CFU/pot) with 10 mM MgSO4 were sprayed onto the young leaves. The inoculated samples were placed in a humidity chamber (28°C and 90% relative humidity) for 30 h and then transferred to the greenhouse. The extent of soft rot was evaluated 2 days after incubation based on visual assessment of symptom severity in leaves using the following four-point scale: 0, no symptoms; 1, occasional yellow leaves, no soft rot; 2, soft rot of leaves of at least 50% of area in each pot; and 3, soft rot and/or death of entire plant. Leaves that turned dark green over at least 50% of their total area were considered diseased. The day/night temperatures in the greenhouse ranged from 15 to 30°C.
Isolation of phage-resistant P. carotovorum subsp. carotovorum mutants.
Mutants with resistance to the phage cocktail were isolated by high-titer overlay as previously described (64). The WT Pcc27 (107 CFU/mL) culture was mixed with the phage cocktail at an MOI of 103 in 5 mL of soft LB soft agar. The mixture was poured onto LB agar plates and solidified, and the plates were incubated at 30°C for 24 h. The emerging colonies were streaked three times onto LB plates to isolate a single colony. At each streaking step, the newly formed colonies were tested by spot assays to identify phage resistance.
Extracellular enzyme assays.
Extracellular enzyme assays were conducted as described by Chatterjee et al. and Lee et al., with some modifications (2, 74). The media were prepared as follows. The Pel assay plate contained 1 % polygalacturonic acid (PGA), 1 % yeast extract, 0.38 μM calcium chloride, and 100 mM Tris/HCl, pH 8.5; the Peh assay plate contained 1 % PGA, 1 % yeast extract, 2.2 mM EDTA and 110 mM sodium acetate, pH 5.5; the Cel assay plate contained 1 % carboxymethyl cellulose and 25 mM sodium phosphate, pH 7.0; and the Prt assay plate contained 1 % skim milk and 0.1 % yeast extract. All plates were supplemented with 0.8% agarose and 0.2% sodium azide. In each plate, wells were created with a no. 2 cork borer (Sigma-Aldrich), and the bottoms of the wells were covered with 0.8% (vol/vol) molten agarose containing 0.2% sodium azide. The P. carotovorum subsp. carotovorum cell cultures at the stationary phase grown in LB medium were centrifuged (16,000 × g for 1 min), and 30 μL of the supernatant containing extracellular enzymes was applied to each well. Then, the plates were incubated at 30°C for 16 h. For the Peh and Pel assay plates, 4 mL of 4 N HCl was poured, and the diameters of the clear zones were measured. The Cel assay plates were stained with 0.1% Congo red solution for 30 min and washed three times with 1 M NaCl until clear zones were visible. The haloes on the Prt assay plates were measured after 36 h of incubation without any further treatment.
Infection assay of phage cocktail-resistant bacteria in napa cabbage.
Napa cabbage samples were prepared by using the same method as described for the biocontrol assays. Amounts of 10 μL of P. carotovorum subsp. carotovorum bacterial culture and each phage-resistant-mutant culture (5 × 107 CFU/mL) were inoculated into the wounds. After air drying for 10 min, the samples were stored in a plastic box to maintain humidity and incubated at 30°C for 36 h. Macerated areas of napa cabbage were measured using ImageJ.
Motility assay.
The swimming motilities of the P. carotovorum subsp. carotovorum strains were evaluated as described by Choi et al. with some modifications (75). Amounts of 1 μL of the overnight-cultured P. carotovorum subsp. carotovorum cells were inoculated onto semisolid LB agar plates (0.3% agar) by stabbing. The diameters of the bacterial growth zones were measured after plate incubation at 30°C for 24 h.
Adsorption assays.
The phage adsorption assays were conducted as previously described with some modifications (22). Briefly, P. carotovorum subsp. carotovorum cells were harvested at an optical density at 600 nm (OD600) of 1.0 (5 × 108 CFU/mL) and washed with LB medium. Each phage was added to a bacterial suspension at an MOI of 0.01 and the suspension aliquoted in equal volumes into five microtubes. During phage adsorption for 15 min at 30°C, each tube was centrifuged (16,000 × g for 1 min at 4°C) at the indicated time points, and the supernatants were immediately filtered (0.22-μm pore size; Millipore). The numbers of unadsorbed phage particles in the filtrates were determined by spot assays using WT Pcc27 and the cpsG::Tn5 mutant as the indicator strains.
Statistical analysis.
Statistical Package for Social Science (SPSS) 25 software was used for statistical analysis. Statistical analysis was conducted by one-way analysis of variance (ANOVA) with Tukey’s multiple-comparison tests among the experimental groups. The visual assessment of the extent of soft rot whose results are shown in Fig. 4B was statistically analyzed by nonparametric Kruskal-Wallis test. Statistical analysis for the adsorption assay whose results are shown in Fig. S7B was performed by using the unpaired t test.
Data availability.
The complete genome information of the three phages chosen for the phage cocktail was registered in the NCBI GenBank database under accession numbers MT560058 for POP12, MT560059 for POP15, and MT552976 for POP17.
ACKNOWLEDGMENTS
This study received support from LG Chem and a National Research Foundation of Korea (NRF) grant (grant no. NRF-2020R1A2B5B03094303). This work was carried out with the support of the Cooperative Research Program for Agriculture Science and Technology Development (project no. PJ01574702), Rural Development Administration, Republic of Korea, with a Yonsei University Future-leading Research Initiative of 2019 grant (grant no. 2019-22-0083). H.K. was supported by the BK21 Plus Program of Department of Agricultural Biotechnology, Seoul National University, Seoul, South Korea.
H.K., M.K., and S.R. conceived and designed the experiments. H.K. performed the experiments and analyzed the data. H.K., M.K., and S.R. wrote the paper. S.-N.J. supported the greenhouse experiments. S.H. provided H.K. and M.K. with the Pectobacterium carotovorum strains.
We declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Sangryeol Ryu, Email: sangryu@snu.ac.kr.
Martha Vives, Universidad de los Andes.
REFERENCES
- 1.Charkowski AO. 2018. The changing face of bacterial soft-rot diseases. Annu Rev Phytopathol 56:269–288. 10.1146/annurev-phyto-080417-045906. [DOI] [PubMed] [Google Scholar]
- 2.Lee DH, Lim J-A, Lee J, Roh E, Jung K, Choi M, Oh C, Ryu S, Yun J, Heu S. 2013. Characterization of genes required for the pathogenicity of Pectobacterium carotovorum subsp. carotovorum Pcc21 in Chinese cabbage. Microbiology (Reading) 159:1487–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Des Essarts YR, Cigna J, Quêtu-Laurent A, Caron A, Munier E, Beury-Cirou A, Hélias V, Faure D. 2016. Biocontrol of the potato blackleg and soft rot diseases caused by Dickeya dianthicola. Appl Environ Microbiol 82:268–278. 10.1128/AEM.02525-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jee S, Choi J-G, Lee Y-G, Kwon M, Hwang I, Heu S. 2020. Distribution of Pectobacterium species isolated in South Korea and comparison of temperature effects on pathogenicity. Plant Pathol J 36:346–354. 10.5423/PPJ.OA.09.2019.0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi O, Kim J. 2013. Pectobacterium carotovorum subsp. brasiliense causing soft rot on paprika in Korea. J Phytopathol 161:125–127. 10.1111/jph.12022. [DOI] [Google Scholar]
- 6.Hahm S-S, Han K-S, Shim M-Y, Park J-J, Kwon K-H, Park J-E. 2003. Occurrence of bacterial soft rot of lily bulb caused by Pectobacterium carotovorum subsp. carotovorum and Pseudomonas marginalis in Korea. Plant Pathol J 19:43–45. 10.5423/PPJ.2003.19.1.043. [DOI] [Google Scholar]
- 7.Clark C, Hoy M, Bond J, Chen C, Goh Y-K, Liang X, Liu X, Lotrakul P. 1998. First report of Erwinia carotovora subsp. carotovora causing bacterial root rot of sweetpotato (Ipomoea batatas) in Louisiana. Plant Dis 82:129. 10.1094/PDIS.1998.82.1.129A. [DOI] [PubMed] [Google Scholar]
- 8.Blom TJ, Brown W. 1999. Preplant copper-based compounds reduce Erwinia soft rot on calla lilies. Horttech 9:56–59. 10.21273/HORTTECH.9.1.56. [DOI] [Google Scholar]
- 9.Gracia-Garza J, Allen W, Blom T, Brown W. 2002. Pre-and post-plant applications of copper-based compounds to control Erwinia soft rot of calla lilies. Can J Plant Pathol 24:274–280. 10.1080/07060660209507009. [DOI] [Google Scholar]
- 10.Elphinstone J, Perombelon M. 1987. Control of contamination of potatoes with air-borne Erwinia carotovora by foliar application of copper oxychloride. Ann Appl Biol 110:535–544. 10.1111/j.1744-7348.1987.tb04171.x. [DOI] [Google Scholar]
- 11.Jones AL, Schnabel EL. 2000. The development of streptomycin-resistant strains of Erwinia amylovora, p 235–251. In Vanneste JL (ed), Fire blight: the disease and its causative agent, Erwinia amylovora. CABI Publishing, Wallingford, Oxfordshire, United Kingdom. [Google Scholar]
- 12.Buttimer C, McAuliffe O, Ross RP, Hill C, O’Mahony J, Coffey A. 2017. Bacteriophages and bacterial plant diseases. Front Microbiol 8:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drulis-Kawa Z, Majkowska-Skrobek G, Maciejewska B, Delattre A-S, Lavigne R. 2012. Learning from bacteriophages—advantages and limitations of phage and phage-encoded protein applications. Curr Protein Pept Sci 13:699–722. 10.2174/138920312804871193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuzaki S, Rashel M, Uchiyama J, Sakurai S, Ujihara T, Kuroda M, Ikeuchi M, Tani T, Fujieda M, Wakiguchi H, Imai S. 2005. Bacteriophage therapy: a revitalized therapy against bacterial infectious diseases. J Infect Chemother 11:211–219. 10.1007/s10156-005-0408-9. [DOI] [PubMed] [Google Scholar]
- 15.Meaden S, Koskella B. 2013. Exploring the risks of phage application in the environment. Front Microbiol 4:358. 10.3389/fmicb.2013.00358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim JA, Jee S, Lee DH, Roh E, Jung K, Oh C, Heu S. 2013. Biocontrol of Pectobacterium carotovorum subsp carotovorum using bacteriophage PP1. J Microbiol Biotechnol 23:1147–1153. 10.4014/jmb.1304.04001. [DOI] [PubMed] [Google Scholar]
- 17.Lee J, Kim S, Park T. 2017. Diversity of bacteriophages infecting Pectobacterium from potato fields. J Plant Pathol 99:453–460. [Google Scholar]
- 18.Smolarska A, Rabalski L, Narajczyk M, Czajkowski R. 2018. Isolation and phenotypic and morphological characterization of the first Podoviridae lytic bacteriophages ϕA38 and ϕA41 infecting Pectobacterium parmentieri (former Pectobacterium wasabiae). Eur J Plant Pathol 150:413–425. 10.1007/s10658-017-1289-3. [DOI] [Google Scholar]
- 19.Loc-Carrillo C, Abedon ST. 2011. Pros and cons of phage therapy. Bacteriophage 1:111–114. 10.4161/bact.1.2.14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat Rev Microbiol 8:317–327. 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 21.Kim H, Kim M, Bai J, Lim J-A, Heu S, Ryu S. 2019. Colanic acid is a novel phage receptor of Pectobacterium carotovorum subsp. carotovorum phage POP72. Front Microbiol 10:143. 10.3389/fmicb.2019.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim M, Ryu S. 2012. Spontaneous and transient defence against bacteriophage by phase-variable glucosylation of O-antigen in Salmonella enterica serovar Typhimurium. Mol Microbiol 86:411–425. 10.1111/j.1365-2958.2012.08202.x. [DOI] [PubMed] [Google Scholar]
- 23.Tan D, Zhang Y, Qin J, Le S, Gu J, Chen L-k, Guo X, Zhu T. 2020. A frameshift mutation in wcaJ associated with phage resistance in Klebsiella pneumoniae. Microorganisms 8:378. 10.3390/microorganisms8030378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim M, Kim S, Park B, Ryu S. 2014. Core lipopolysaccharide-specific phage SSU5 as an auxiliary component of a phage cocktail for Salmonella biocontrol. Appl Environ Microbiol 80:1026–1034. 10.1128/AEM.03494-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bai J, Jeon B, Ryu S. 2019. Effective inhibition of Salmonella Typhimurium in fresh produce by a phage cocktail targeting multiple host receptors. Food Microbiol 77:52–60. 10.1016/j.fm.2018.08.011. [DOI] [PubMed] [Google Scholar]
- 26.Czajkowski R, Ozymko Z, de Jager V, Siwinska J, Smolarska A, Ossowicki A, Narajczyk M, Lojkowska E. 2015. Genomic, proteomic and morphological characterization of two novel broad host lytic bacteriophages ΦPD10.3 and ΦPD23.1 infecting pectinolytic Pectobacterium spp. and Dickeya spp. PLoS One 10:e0119812. 10.1371/journal.pone.0119812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zaczek-Moczydłowska MA, Young GK, Trudgett J, Plahe C, Fleming CC, Campbell K, O’Hanlon R. 2020. Phage cocktail containing Podoviridae and Myoviridae bacteriophages inhibits the growth of Pectobacterium spp. under in vitro and in vivo conditions. PLoS One 15:e0230842. 10.1371/journal.pone.0230842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mangalea MR, Duerkop BA. 2020. Fitness trade-offs resulting from bacteriophage resistance potentiate synergistic antibacterial strategies. Infect Immun 88:e00926-19. 10.1128/IAI.00926-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evans T, Ind A, Komitopoulou E, Salmond G. 2010. Phage-selected lipopolysaccharide mutants of Pectobacterium atrosepticum exhibit different impacts on virulence. J Appl Microbiol 109:505–514. 10.1111/j.1365-2672.2010.04669.x. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Wei Z, Yang K, Wang J, Jousset A, Xu Y, Shen Q, Friman V-P. 2019. Phage combination therapies for bacterial wilt disease in tomato. Nat Biotechnol 37:1513–1520. 10.1038/s41587-019-0328-3. [DOI] [PubMed] [Google Scholar]
- 31.Kortright KE, Doss-Gollin S, Chan BK, Turner PE. 2021. Evolution of bacterial cross-resistance to lytic phages and albicidin antibiotic. Front Microbiol 12:658374. 10.3389/fmicb.2021.658374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scanlan PD, Buckling A. 2012. Co-evolution with lytic phage selects for the mucoid phenotype of Pseudomonas fluorescens SBW25. ISME J 6:1148–1158. 10.1038/ismej.2011.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oechslin F. 2018. Resistance development to bacteriophages occurring during bacteriophage therapy. Viruses 10:351. 10.3390/v10070351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stevenson G, Andrianopoulos K, Hobbs M, Reeves PR. 1996. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol 178:4885–4893. 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larsen RA, Wilson MM, Guss AM, Metcalf WW. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch Microbiol 178:193–201. 10.1007/s00203-002-0442-2. [DOI] [PubMed] [Google Scholar]
- 36.Ferrieres L, Hemery G, Nham T, Guerout AM, Mazel D, Beloin C, Ghigo JM. 2010. Silent mischief: bacteriophage Mu insertions contaminate products of Escherichia coli random mutagenesis performed using suicidal transposon delivery plasmids mobilized by broad-host-range RP4 conjugative machinery. J Bacteriol 192:6418–6427. 10.1128/JB.00621-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chatterjee A, Cui Y, Chatterjee AK. 2009. RsmC of Erwinia carotovora subsp. carotovora negatively controls motility, extracellular protein production, and virulence by binding FlhD and modulating transcriptional activity of the master regulator, FlhDC. J Bacteriol 191:4582–4593. 10.1128/JB.00154-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamber T, Pothier JF, Pelludat C, Rezzonico F, Duffy B, Smits TH. 2017. Role of the type VI secretion systems during disease interactions of Erwinia amylovora with its plant host. BMC Genomics 18:628. 10.1186/s12864-017-4010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roh E-J, Lee S-D, Lee Y-H, Ra D-S, Choi J-H, Moon E-P, Heu S-G. 2009. Diverse antibacterial activity of Pectobacterium carotovorum subsp. carotovorum isolated in Korea. J Microbiol Biotechnol 19:42–50. [PubMed] [Google Scholar]
- 40.Lee DH, Kim JB, Lim JA, Han SW, Heu S. 2014. Genetic diversity of Pectobacterium carotovorum subsp brasiliensis isolated in Korea. Plant Pathol J 30:117–124. 10.5423/PPJ.OA.12.2013.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lukashin AV, Borodovsky M. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res 26:1107–1115. 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia FF, Stevens R. 2014. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:D206–D214. 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Capparelli R, Nocerino N, Lanzetta R, Silipo A, Amoresano A, Giangrande C, Becker K, Blaiotta G, Evidente A, Cimmino A, Iannaccone M, Parlato M, Medaglia C, Roperto S, Roperto F, Ramunno L, Iannelli D. 2010. Bacteriophage-resistant Staphylococcus aureus mutant confers broad immunity against staphylococcal infection in mice. PLoS One 5:e11720. 10.1371/journal.pone.0011720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gu J, Liu X, Li Y, Han W, Lei L, Yang Y, Zhao H, Gao Y, Song J, Lu R, Sun C, Feng X. 2012. A method for generation phage cocktail with great therapeutic potential. PLoS One 7:e31698. 10.1371/journal.pone.0031698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oechslin F, Piccardi P, Mancini S, Gabard J, Moreillon P, Entenza JM, Resch G, Que Y-A. 2016. Synergistic interaction between phage therapy and antibiotics clears Pseudomonas aeruginosa infection in endocarditis and reduces virulence. J Infect Dis 215:703–712. 10.1093/infdis/jiw632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Evans T, Trauner A, Komitopoulou E, Salmond G. 2010. Exploitation of a new flagellatropic phage of Erwinia for positive selection of bacterial mutants attenuated in plant virulence: towards phage therapy. J Appl Microbiol 108:676–685. 10.1111/j.1365-2672.2009.04462.x. [DOI] [PubMed] [Google Scholar]
- 47.Addy HS, Askora A, Kawasaki T, Fujie M, Yamada T. 2012. The filamentous phage φRSS1 enhances virulence of phytopathogenic Ralstonia solanacearum on tomato. Phytopathology 102:244–251. 10.1094/PHYTO-10-11-0277. [DOI] [PubMed] [Google Scholar]
- 48.Fan J, Ma L, Zhao C, Yan J, Che S, Zhou Z, Wang H, Yang L, Hu B. 2020. Transcriptome of Pectobacterium carotovorum subsp. carotovorum PccS1 infected in calla plants in vivo highlights a spatiotemporal expression pattern of genes related to virulence, adaptation, and host response. Mol Plant Pathol 21:871–891. 10.1111/mpp.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hossain MM, Shibata S, Aizawa S-I, Tsuyumu S. 2005. Motility is an important determinant for pathogenesis of Erwinia carotovora subsp. carotovora. Physiol Mol Plant Pathol 66:134–143. 10.1016/j.pmpp.2005.06.001. [DOI] [Google Scholar]
- 50.Cui Y, Chatterjee A, Yang H, Chatterjee AK. 2008. Regulatory network controlling extracellular proteins in Erwinia carotovora subsp. carotovora: FlhDC, the master regulator of flagellar genes, activates rsmB regulatory RNA production by affecting gacA and hexA (lrhA) expression. J Bacteriol 190:4610–4623. 10.1128/JB.01828-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park H, Lim C, Hur J. 2011. Phylogeny of the Korean Erwinia species as determined by comparison of 16S rDNA sequences. J Agric Life Environ Sci 23:62–69. [Google Scholar]
- 52.Liu H, Coulthurst SJ, Pritchard L, Hedley PE, Ravensdale M, Humphris S, Burr T, Takle G, Brurberg MB, Birch PRJ, Salmond GPC, Toth IK. 2008. Quorum sensing coordinates brute force and stealth modes of infection in the plant pathogen Pectobacterium atrosepticum. PLoS Pathog 4:e1000093. 10.1371/journal.ppat.1000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gill J, Abedon ST. 2003. Bacteriophage ecology and plants. APSnet Feature 2003:1103. [Google Scholar]
- 54.Serwecińska L. 2020. Antimicrobials and antibiotic-resistant bacteria: a risk to the environment and to public health. Water 12:3313. 10.3390/w12123313. [DOI] [Google Scholar]
- 55.Liu H-W, Begley T (ed). 2020. Comprehensive natural products III: chemistry and biology. Elsevier Health Sciences, London, United Kingdom. [Google Scholar]
- 56.Donley N. 2019. The USA lags behind other agricultural nations in banning harmful pesticides. Environ Health 18:1–12. 10.1186/s12940-019-0488-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.World Health Organization. 2017. Critically important antimicrobials for human medicine: 5th revision. Ranking of antimicrobial agents for risk management of antimicrobial resistance due to non-human use. Antimicrobial Resistance Division, Global Coordination and Partnership, WHO, Geneva, Switzerland.
- 58.Wright RC, Friman V-P, Smith MC, Brockhurst MA. 2019. Resistance evolution against phage combinations depends on the timing and order of exposure. mBio 10:e01652-19. 10.1128/mBio.01652-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brockhurst MA, Koskella B, Zhang Q-G. 2021. Bacteria-phage antagonistic coevolution and the implications for phage therapy, p 231–251. In Harper D, Abedon S, Burrowes B, McConville M (ed), Bacteriophages. Springer, Cham, Switzerland. [Google Scholar]
- 60.Li X, Ma Y, Liang S, Tian Y, Yin S, Xie S, Xie H. 2018. Comparative genomics of 84 Pectobacterium genomes reveals the variations related to a pathogenic lifestyle. BMC Genomics 19:1–22. 10.1186/s12864-018-5269-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leonard S, Hommais F, Nasser W, Reverchon S. 2017. Plant–phytopathogen interactions: bacterial responses to environmental and plant stimuli. Environ Microbiol 19:1689–1716. 10.1111/1462-2920.13611. [DOI] [PubMed] [Google Scholar]
- 62.Chapman-McQuiston E, Wu X. 2008. Stochastic receptor expression allows sensitive bacteria to evade phage attack. Part I: experiments. Biophys J 94:4525–4536. 10.1529/biophysj.107.120212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bikard D, Marraffini LA. 2012. Innate and adaptive immunity in bacteria: mechanisms of programmed genetic variation to fight bacteriophages. Curr Opin Immunol 24:15–20. 10.1016/j.coi.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 64.Kim M, Ryu S. 2011. Characterization of a T5-like coliphage, SPC35, and differential development of resistance to SPC35 in Salmonella enterica serovar typhimurium and Escherichia coli. Appl Environ Microbiol 77:2042–2050. 10.1128/AEM.02504-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dennis JJ, Zylstra GJ. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl Environ Microbiol 64:2710–2715. 10.1128/AEM.64.7.2710-2715.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sambrook J, Russell D. 2001. Molecular cloning: a laboratory manual, 3rd ed, p 6.4–6.12. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 67.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res 27:4636–4641. 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carver T, Berriman M, Tivey A, Patel C, Böhme U, Barrell BG, Parkhill J, Rajandream M-A. 2008. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24:2672–2676. 10.1093/bioinformatics/btn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic Local Alignment Search Tool. J Mol Biol 215:403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 72.Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong S-Y, Lopez R, Hunter S. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed). 2011. Virus taxonomy. Classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 74.Chatterjee A, Cui Y, Liu Y, Dumenyo CK, Chatterjee AK. 1995. Inactivation of rsmA leads to overproduction of extracellular pectinases, cellulases, and proteases in Erwinia carotovora subsp. carotovora in the absence of the starvation/cell density-sensing signal, N-(3-oxohexanoyl)-L-homoserine lactone. Appl Environ Microbiol 61:1959–1967. 10.1128/aem.61.5.1959-1967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Choi Y, Shin H, Lee J-H, Ryu S. 2013. Identification and characterization of a novel flagellum-dependent Salmonella-infecting bacteriophage, iEPS5. Appl Environ Microbiol 79:4829–4837. 10.1128/AEM.00706-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Platt R, Drescher C, Park S-K, Phillips GJ. 2000. Genetic system for reversible integration of DNA constructs and lacZ gene fusions into the Escherichia coli chromosome. Plasmid 43:12–23. 10.1006/plas.1999.1433. [DOI] [PubMed] [Google Scholar]
- 77.Soncini FC, Vescovi EG, Groisman EA. 1995. Transcriptional autoregulation of the Salmonella-Typhimurium phoPQ operon. J Bacteriol 177:4364–4371. 10.1128/jb.177.15.4364-4371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aem.00761-22-s0001.pdf, PDF file, 1.5 MB (1.5MB, pdf)
Data Availability Statement
The complete genome information of the three phages chosen for the phage cocktail was registered in the NCBI GenBank database under accession numbers MT560058 for POP12, MT560059 for POP15, and MT552976 for POP17.