ABSTRACT
Biofilms are widespread in the environment, where they allow bacterial species to survive adverse conditions. Cells in biofilms are densely packed, and this proximity is likely to increase the frequency of horizontal gene transfer. Gene transfer agents (GTAs) are domesticated viruses with the potential to spread any gene between bacteria. GTA production is normally restricted to a small subpopulation of bacteria, and regulation of GTA loci is highly coordinated, but the environmental conditions that favor GTA production are poorly understood. Here, we identified a serine acetyltransferase gene, cysE1, in Rhodobacter capsulatus that is required for optimal receipt of GTA DNA, accumulation of extracellular polysaccharide, and biofilm formation. The cysE1 gene is directly downstream of the core Rhodobacter-like GTA (RcGTA) structural gene cluster and upregulated in an RcGTA overproducer strain, although it is expressed on a separate transcript. The data we present suggest that GTA production and biofilm are coregulated, which could have important implications for the study of rapid bacterial evolution and understanding the full impact of GTAs in the environment.
IMPORTANCE Direct exchange of genes between bacteria leads to rapid evolution and is the major factor underlying the spread of antibiotic resistance. Gene transfer agents (GTAs) are an unusual but understudied mechanism for genetic exchange that are capable of transferring any gene from one bacterium to another, and therefore, GTAs are likely to be important factors in genome plasticity in the environment. Despite the potential impact of GTAs, our knowledge of their regulation is incomplete. In this paper, we present evidence that elements of the cysteine biosynthesis pathway are involved in coregulation of various phenotypes required for optimal biofilm formation by Rhodobacter capsulatus and successful infection by the archetypal RcGTA. Establishing the regulatory mechanisms controlling GTA-mediated gene transfer is a key stepping stone to allow a full understanding of their role in the environment and wider impact.
KEYWORDS: gene transfer agent, Rhodobacter, amino acid biosynthesis, bacteriophage evolution, biofilms, capsular polysaccharide, cysteine
INTRODUCTION
Bacterial abundance in environmental water sources is routinely reported as millions to tens of millions of planktonic cells per mL (1–4). In addition, there are numerous abiotic and biotic surfaces available for bacterial attachment colonization. Biofilm formation is widespread in the environment and particularly in aquatic environments, where every solid surface is likely to be colonized by bacteria (5). Production of biofilm has numerous benefits, including protection from predation, resistance to starvation, and reduction of antibiotic susceptibility. Adherence to surfaces and establishment of biofilm architecture is known to be dependent on cell-cell communication and accumulation of cell-associated extracellular polysaccharides (EPS), DNA, and proteins. EPS itself is not usually essential for bacterial life; however, it is highly advantageous for survival and adaptation to challenging environments (6).
One of the major contributors to bacterial aquatic communities and biofilm is the alphaproteobacteria (7), of which Rhodobacterales species are key constituents (8, 9). Rhodobacter capsulatus is a metabolically versatile species capable of anaerobic photosynthetic and aerobic chemoheterotrophic growth (10). R. capsulatus biofilm production has not been extensively studied. However, one study has linked biofilm production to intracellular cyclic di-GMP (c-di-GMP) levels and the persulfide-responsive transcription factor SqrR (11). The EPS biosynthesis and export pathway has been identified, and this pathway is controlled by quorum sensing (12–14). EPS is also required for efficient gene transfer agent (GTA) binding to recipient cells in R. capsulatus (13, 15, 16).
GTAs are small virus-like particles that are produced by diverse species of bacteria and at least one archaeon (17), and they are thought to be descended from ancient bacteriophages that were domesticated by the host (18, 19). The genes encoding GTAs are embedded in the genomes of their host, often at multiple locations (20, 21), and coordinated expression is initiated from a small subset of the bacterial population (22–25). GTAs usually package and transfer “random” fragments of DNA from their host to compatible recipients (25–28), although Dinoroseobacter shibae GTAs do have biases to certain genomic regions (29). Unlike true viruses, GTAs do not preferentially transfer their own genes (25, 27). Timing and regulation of GTA production are tightly controlled by interlinked host regulatory circuits, including quorum sensing (14, 30), stringent response (31), SOS response (32), c-di-GMP (33, 34) and the pleiotropic transcription factor CtrA (35, 36). In Rhodobacter-like GTAs (RcGTAs), these complex pathways are integrated via a specific GTA transcriptional regulator, GafA (23), and an enigmatic secreted repressor encoded by rcc00280 (37).
In the alphaproteobacteria, GTAs are predicted to be present in at least 50% of sequenced genomes where transmission appears to be via vertical inheritance (18, 38, 39). The ecological role of GTAs in the environment and the evolutionary benefit of GTA production are not currently known (38, 40–42); however, the only in situ study of GTA activity reported extraordinary gene transfer frequencies far in excess of other transfer mechanisms (43). Here, we explored the roles of the two genes located immediately downstream of the core R. capsulatus GTA gene cluster, rcc01699 (referred to as g16 here) and rcc01700 (cysE1). Our findings indicate that cysE1 plays an important role in the regulation of biofilm production and RcGTA receipt, and we propose that biofilms could provide an ideal environment for GTA activity.
RESULTS
Extension of the core RcGTA gene cluster.
The core genes required for production of the R. capsulatus GTA particles (RcGTA), rcc01682 to rcc01698 (also known as g1 to g15), cluster in an ~14.5-kb region that is flanked on either side by host metabolic genes (36). As the RcGTA genes are not excised during propagation, and there are no signature sequences (att sites) to indicate the precise boundaries of the RcGTA gene cluster, the true limits of the operon are unknown (17). Previous transcriptome data indicated that several genes downstream of the RcGTA gene cluster are upregulated in a RcGTA overproducer strain (R. capsulatus DE442) to levels comparable to those of the preceding RcGTA genes (Fig. 1) (23, 44).
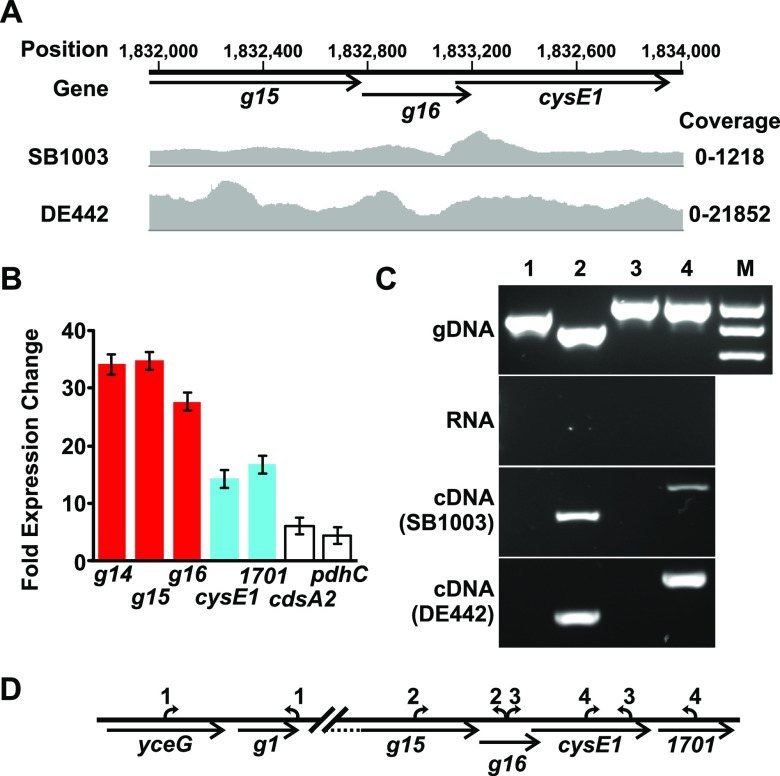
FIG 1.
Defining the RcGTA transcriptional unit. (A) Representative histograms showing RNA-seq transcript coverage from the R. capsulatus wild-type strain SB1003 and the GTA overproducer DE442. Genome position (in base pairs) is indicated at the top, and the location of each gene is depicted with arrows beneath. The RNA-seq data set was described in reference 23, and the raw data can be found in the NCBI GEO Database, accession no. GSE118116. (B) Chart showing fold changes in gene expression between R. capsulatus DE442 (n = 8) and wild-type SB1003 (n = 4) in the GSE118116 data set. Red bars indicate constituents of the core RcGTA gene cluster, cyan shows the cysE1 transcriptional unit, and white indicates the genes immediately downstream. Error bars represent standard deviations. (C) Agarose gel of a representative RT-PCR amplification. Templates were genomic DNA, DNA-free RNA, and cDNA isolated from SB1003 and DE442, as indicated. The 600- to 1,000-bp portion of the HyperLadder 1-kb DNA marker is included for reference (M) (Meridian Bioscience, Cincinnati, OH). PCR amplification occurred across the gene junctions indicated above each well and defined in panel D. (D) Schematic of the regions immediately before and after the RcGTA gene cluster. Primer pairs are numbered 1 to 4 and correspond to the wells shown in the preceding gel. The yceG gene immediately precedes the RcGTA small terminase gene, g1, which has previously been confirmed as the first gene of the operon and was used here as a negative control for genomic DNA carryover.
The first downstream gene, rcc01699 (or g16), begins 3 bases after the RcGTA g15 stop codon. The g16 gene encodes a hypothetical protein, and we were not able to assign a definitive predicted function through primary sequence or structural comparisons. The structure of g16 was predicted using the AlphaFold software (45), and the top-ranked model was submitted as a query to the Dali protein structure comparison server (see Table S1 in the supplemental material) (46). The best match was to the catalytic domain of the Bacillus cereus SleB protein (PDB code 4F55). SleB is a lytic transglycosylase that is involved in peptidoglycan remodeling during spore germination (47). It is conceivable that g16 has a role in ingress or egress of RcGTAs during lysis or infection, but no experimental data have been produced so far to support this. The second downstream gene, rcc01700 (cysE1), overlaps g16 by 59 bp. CysE1 is a putative serine O-acetyltransferase, an enzyme that produces O-acetyl-l-serine (OAS) from acetyl coenzyme A (acetyl-CoA) and serine (48), an early step in the production of cysteine and methionine (49). Other than amino acid biosynthesis, OAS and its racemer N-acetyl-serine (NAS) have also been implicated as signaling molecules in various cellular processes, including biofilm production by other species (50–55).
Raw transcriptome sequencing (RNA-seq) reads obtained from the wild-type R. capsulatus SB1003 strain and the DE442 RcGTA overproducer data sets were mapped onto the g15-to-cysE1 region of the genome, and no gaps in mRNA coverage were detected between the genes (Fig. 1A) (23). These data are consistent with a previous gene coexpression network analysis that indicated that rcc01688 to cdsA2 (rcc01702) form a single transcriptional unit (44). PCRs using cDNA template and primers located on either side of the RcGTA g15-g16 and cysE1-rcc01701 gene boundaries produced successful amplifications (Fig. 1C and D). However, no amplification was detected with primers spanning the g16-cysE1 junction (Fig. 1C and D). Control amplification of a genomic DNA template using identical conditions successfully amplified all target sequences (Fig. 1C). These data indicate that g16 is transcribed as part of the core RcGTA mRNA transcript and cysE1 and rcc01701 are located on a separate transcript, which is consistent with the relative expression levels observed in RNA-seq data (Fig. 1B) (23). Meanwhile, no amplification was detected across the boundary of the RcGTA small terminase gene (rcc01682 [RcGTA g1]) and the gene immediately upstream (yceG), which has never been associated with RcGTA production or regulation (Fig. 1C). The data presented indicate that RcGTA g1 is the first gene of the RcGTA cluster and g16 is likely to be the last. To assess the function of g16 and cysE1, individual gene knockouts were produced by GTA-mediated gene replacement.
There are two functional cysE genes in R. capsulatus.
The cysteine biosynthesis pathway is an essential biological process, but the R. capsulatus ΔcysE1 strains were not auxotrophic when grown on RCV minimal medium (Fig. 2). This unexpected viability can be explained by the presence of an additional serine O-acetyltransferase gene elsewhere in the genome, cysE2 (rcc02246; protein accession no. ADE85976). ClustalΩ alignment of the two cysE genes revealed 51.62% DNA sequence identity, and the protein products are 29.57% identical.
FIG 2.
Deletion of the R. capsulatus cysE1 gene does not lead to auxotrophy. (A) Serial dilutions of cultures of R. capsulatus SB1003 (WT), an isogenic cysE1 knockout containing empty pCM66T plasmid (ΔcysE1), and the cysE1 knockout complemented with pCM66T-cysE1 (ΔcysE1 + cysE1) were spotted onto either minimal-medium (RCV) or complex-medium agar plates (YPS). (B) Optical density growth curve of R. capsulatus SB1003 (black line; gray shading shows the standard deviation) and an isogenic cysE1 knockout (red line; pink shading shows the standard deviation) (n = 10). Cultures were grown in 200 μL of RCV liquid growth medium, and optical density at 600 nm was measured at 15-min intervals for 72 h.
Previous transcriptome data showed that cysE2 is expressed at lower levels than cysE1 in wild-type R. capsulatus SB1003, and this was confirmed here by qPCR; cysE2 transcripts were ~10-fold less abundant than cysE1 in wild-type cells (Fig. S1A). Interestingly, cysE2 transcript abundance increased by 2- to 3-fold in cysE1 null mutants, albeit with a final concentration that was still only ~25% of cysE1 transcript abundance in wild-type (WT) cells (Fig. S1A and B). Therefore, it is possible that limited upregulation of cysE2 expression can partially mitigate the loss of cysE1. Similar to the ΔcysE1 strain, deletion of cysE2 did not lead to an auxotrophic phenotype (Fig. S1C and D). A double cysE1 cysE2 mutant could not be made even using complex YPS medium, suggesting that loss of all serine O-acetyltransferase activity has wider implications than simply production of amino acids. In contrast to cysE1, cysE2 is not upregulated in the RcGTA overproducer DE442 compared to wild-type SB1003 (23, 44).
It is worth noting that growth of the ΔcysE1 and ΔcysE2 strains appears paler than the wild-type on solid media, and both produce slightly smaller colonies (Fig. 2A and Fig. S1C and D). This is particularly evident on minimal medium. The reason for this phenotype is unclear, but the number of colonies is equivalent to that of the wild type (Fig. 2A and Fig. S1C and D), indicating that the cultures reached the same cell density. The plates were left for several more days to see if the morphology of the cysE strains changed after longer incubation, as would be expected if they were simply growing more slowly. No substantial changes were observed with longer incubation times. The rate of logarithmic growth in liquid minimal medium was comparable between the wild-type and ΔcysE1 strains, but there was some divergence at the transition to stationary phase (Fig. 2B). Both growth curves began to plateau and then rose again shortly after, with the rise being more prominent for the wild-type (Fig. 2B). The numbers of CFU for both strains at stationary phase were comparable (Fig. 2A); therefore, it is unlikely that the increase in optical density was due to growth.
The CysE1 and CysE2 proteins contain most of the conserved amino acids that are predicted to be involved in catalytic activity (Fig. S2). To test whether both proteins are functional, plasmids containing the R. capsulatus cysE1 or cysE2 gene were transformed into an auxotrophic E. coli ΔcysE mutant from the Keio collection (56). As expected, the E. coli ΔcysE strain could grow on LB agar plates but no growth was observed on RCV minimal agar. The ability to grow on RCV was restored by in trans complementation with the R. capsulatus cysE1 or cysE2 gene (Fig. 3A and B). The R. capsulatus CysE1 and CysE2 proteins were also purified to homogeneity by affinity chromatography for use in an in vitro serine acetyltransferase assay. Both proteins possessed clear enzymatic activity (Fig. 3C). CysE2 also had low-level activity in the absence of a serine acceptor and slightly higher overall activity in the presence of serine, possibly indicating autoacetylation of one of the CysE2 serine residues or the presence of an intermediate state (Fig. 3C).
FIG 3.
The R. capsulatus CysE1 and CysE2 proteins are both functionally active. (A and B) Serial dilutions of cultures of an E. coli K-12 strain BW25113 cysE knockout containing empty pCM66T plasmid (ΔcysE), the cysE knockout complemented with pCM66T-R. capsulatus cysE1 (ΔcysE + cysE1), and the cysE knockout complemented with pCM66T-R. capsulatus cysE2 (ΔcysE + cysE2) were spotted onto either minimal-medium (RCV) or complex-medium agar plates (LB). (C) Purified R. capsulatus CysE1 and CysE2 proteins were tested for in vitro transfer of CoA from acetyl-CoA to serine in a fluorescence-based assay. Reactions were carried out in the presence or absence of the serine receiver (−Serine/+Serine). Bars represent means, and filled circles are the individual data points. Statistical significance is indicated above the chart (one-way analysis of variance [ANOVA] using the Holm-Šidák test; *, P < 0.05; ***, P < 0.001).
To our knowledge, the presence of multiple cysE gene copies in bacteria has been reported only for Lactobacillus (57, 58), while redundant serine acetyltransferase activity was shown in Rhizobium leguminosarum but the gene responsible was not identified (59). Sequence homology searches using BLASTp revealed that CysE1 has long been associated with the Rhodobacterales (196 hits out of 201 [Table S2]; database update, 16 May 2022), whereas CysE2 is likely to have been recently acquired by horizontal gene transfer: only 26 hits out of 186 against Rhodobacterales but 148 hits against betaproteobacterial species (Table S3) (database update, 16 May 2022). The GC content of the cysE1 gene is 64.7%, which is slightly closer to the genome average (66.6%) than that of cysE2 (70.0%). Furthermore, CysE2 is predicted to be part of a KEGG pathway with gene products encoded on the endogenous R. capsulatus plasmid pRCB133 rather than by chromosomal genes (44). A homologue of CysE2 was also identified in R. leguminosarum (Fig. S2), which is likely to be the source of the residual serine acetyltransferase activity observed by Parker et al. (59). Given the data presented above, it is probable that the two R. capsulatus cysE genes have a redundant function with regard to essential biosynthesis.
CysE1 affects the production of cell-associated EPS.
Visual inspection of cell pellets produced by cysE1 mutants revealed a decrease in the characteristic loose mucoidy phenotype of the wild-type, whereas deletion of g16 had no obvious effect (Fig. 4A and B). Loose-pellet formation has previously been attributed to accumulation of cell-associated extracellular polysaccharide (EPS) (13). The amount of EPS in the wild-type and mutant strains was quantified by EDTA extraction and phenol-sulfuric acid colorimetric quantification (13). This method does not distinguish between capsular EPS and lipopolysaccharides (LPS); however, LPS is not thought to be involved in the mucoidy pellet formation or with any RcGTA phenotypes (13). For simplicity, the extracted polysaccharides are referred to as EPS here. In agreement with the visual inspection, deletion of g16 had no effect on EPS production (Fig. 4A), but deletion of the cysE1 gene reduced EPS secretion to 47.0% of that of the wild type (Fig. 4B). Complementation in trans with a plasmid containing the cysE1 gene expressed from its native promoter partially restored cell-associated EPS production to 84.6% of that of the wild type, and expression from the puf photosynthesis promoter (see Materials and Methods) overrescued production to 120.8% (Fig. 4B). We hypothesize that the increase in optical density (OD) in early stationary phase shown in Fig. 2B was a result of EPS production and that the smaller increase in OD for the cysE1 knockout (KO) could reflect the impaired EPS production phenotype (Fig. 2B and 4). Deletion of cysE2 had a lesser effect on EPS production, ≥78.8% of the wild-type level (Fig. S4A).
FIG 4.
EPS and capsule production by R. capsulatus. (A and B) Quantification of cell-associated extracellular polysaccharide produced by the R. capsulatus derivatives annotated on the x axes, relative to the wild-type R. capsulatus parental strain. Images of the pellets formed after centrifugation of 1 mL of cells in the presence of 250 mM salt are provided above the respective bars. Bars represent means, and filled circles are the individual data points. Statistical significance is indicated above each chart (one-way ANOVA using the Holm-Šidák test [n = 3]; n.s., no significance; ***, P < 0.001). (C) Negative stain of the R. capsulatus capsule. Cells and background are stained dark with crystal violet, while the capsule itself is counterstained with copper sulfate and presents as a light halo around the cells. All images were taken using a 100× objective.
To determine whether the reduction in EPS was also associated with visibly impaired capsule formation, cells were negatively stained using crystal violet and copper sulfate (60). Consistent with the EPS data, substantial thinning of the capsule was observed for ΔcysE1 cells but not Δg16 cells (Fig. 4C). The correlation between decreased EPS production and impaired capsule formation is also supported by a previous study of a R. capsulatus polysaccharide biosynthesis cluster (13). Overexpression of either g16 or cysE1 also significantly increased the levels of EPS produced to 136.7% (P < 0.001) and 170.0% (P < 0.001), respectively, of the wild-type level (Fig. S5). The precise role of g16 is not clear and requires further investigation, but it does appear to directly or indirectly affect EPS production when expression levels are elevated. The connection between g16 and cysE1 is also supported by several examples in sequenced genomes where the two genes are fused (Fig. S6).
Effect of O-acetyl-l-serine on wild-type R. capsulatus B10 cells.
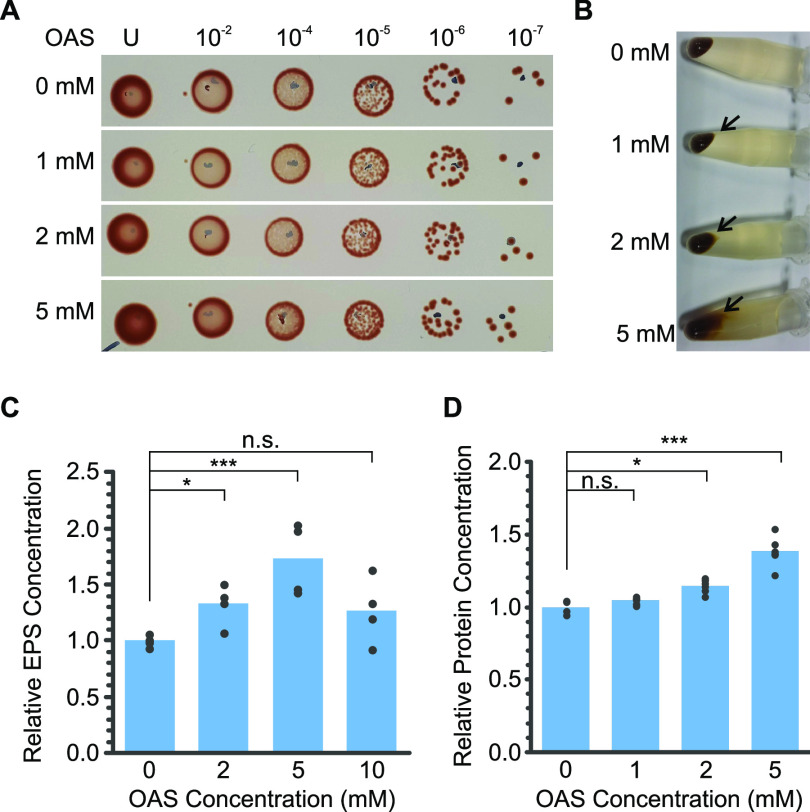
The CysE1 enzyme produces O-acetyl-l-serine (OAS) from acetyl-CoA and serine (48, 61). To test the effect of OAS on wild-type R. capsulatus cells, various concentrations were added directly to late-log-phase cultures (Fig. 5 and 6). OAS had no effect on the number of CFU (Fig. 5A) but did increase loose-pellet formation (Fig. 5B), cell-associated EPS (Fig. 5C), and protein accumulation (Fig. 5D). At 5 mM OAS, EPS quantities peaked at 172.8% compared to the buffer-only control (Fig. 5C). Meanwhile, extracellular protein concentration rose to 138.4% with 5 mM OAS (Fig. 5D). Interestingly, OAS also altered cell morphology from typical rods to truncated rods/cocci, reminiscent of the biofilm-type morphology (Fig. 6A). Quantification of the area of individual cells indicated that the median area decreased to 74.6% in the presence of 5 mM OAS compared to 0 mM (Fig. 6B). Taken together, the data presented here corroborate the hypothesis that deletion of cysE1 and subsequent reduction of OAS biosynthesis lead to the various phenotypes observed.
FIG 5.
Effect of OAS on EPS and protein production by wild-type R. capsulatus SB1003. In all panels, data were obtained from wild-type R. capsulatus cultures with or without OAS treatment (0, 1, 2, 5, or 10 mM, as indicated). (A) Ten-microliter spot dilution series to assess the effect of OAS on CFU. Dilution factors are given across the top. U, undiluted. (B) Cell pellets from 1 mL of culture show increased mucoidy appearance in the presence of OAS. NaCl (250 mM) was added prior to centrifugation to facilitate pellet formation. The black arrow highlights loose-pellet formation. (C) Quantification of cell-associated extracellular polysaccharide produced by wild-type cells in response to the concentrations of OAS annotated on the x axes (n = 5). (D) Quantification of cell-associated extracellular protein produced by wild-type cells in response to the concentrations of OAS annotated on the x axes (n = 6). Bars represent means (n = 4), and filled circles are the individual data points. Statistical significance is indicated above each chart (one-way ANOVA using the Holm-Šidák test; n.s., no significance; *, P < 0.05; ***, P < 0.001).
FIG 6.
Effects of AOS on cell morphology. (A) Phase-contrast microscopy of wild-type R. capsulatus SB1003 cells at ×100 magnification. The R. capsulatus cells are rod shaped when grown in RCV defined medium but truncate in the presence of OAS. Rhodobacter are usually found in chains of two cells; individual exemplar cells are highlighted with a white outline. Images of typical biofilm cells are shown for comparison. (B) Box plot of cell size (area) observed after OAS treatments. Measurements were made using ImageJ from a total of five fields of view from two independent cultures per condition. The ends of the boxes define the 25th and 75th percentiles, with a line at the median. Error bars define the 10th and 90th percentiles, and white circles define 5th and 95th percentiles. Statistical significance is indicated above the plots (one-way ANOVA using the Holm-Šidák test [0 mM, n = 4,488; 2 mM, n = 7,947; 5 mM, n = 6,115; 10 mM, n = 9,363]; ***, P < 0.001).
CysE1 is required for optimal RcGTA receipt.
It is known that EPS is required for production of the R. capsulatus capsule, which is in turn required for optimal RcGTA infection (13, 15). Because expression levels of g16 and cysE1 affect EPS production/capsule formation and transcription of both genes is coregulated with the RcGTA genes (Fig. 1), we hypothesized that there may also be an effect on RcGTA receipt. Standard GTA bioassays were carried out to monitor the transfer of antibiotic resistance from a rifampicin-resistant donor to a rifampicin-sensitive recipient (R. capsulatus B10). Wild-type strains of R. capsulatus produce RcGTAs at low frequency; therefore, the RcGTA overproducer strain DE442 was used as a donor in all assays to increase sensitivity (22, 37, 62). We found that deletion of g16 did not significantly decrease receipt of RcGTA DNA compared to that in wild-type B10 (98.2%, P = 0.837). The frequency of RcGTA receipt by the ΔcysE1 derivative was 47.7% compared to the control (Fig. 7). Complementation of the B10 ΔcysE1 mutant with a plasmid containing the cysE1 gene expressed from its native promoter restored RcGTA recipient efficiency to 119.5% of the wild type value, and expression from the puf promoter rescued it to 112.1% (Fig. 7B). Meanwhile, deletion of cysE2 had no significant effect on RcGTA receipt (Fig. S4B). Overall, these data indicate that CysE1 is required for optimal receipt of RcGTA DNA.
FIG 7.
Effect of g16 and cysE1 genes on RcGTA receipt capability. Representative bar charts of the frequencies of rifampicin resistance transfer between R. capsulatus DE442 and sensitive recipient strains in a GTA bioassay. All gene transfer frequencies were normalized to wild-type R. capsulatus B10 (WT). (A) The WT recipient (n = 5) was compared to an isogenic RcGTA g16 knockout strain (Δg16, n = 5). (B) The WT recipient (n = 3) was compared to an isogenic cysE1 knockout (ΔcysE1, n = 3). The cysE1 knockout was complemented in trans with either cysE1 expressed from its native promoter (+ cysE1; n = 3) or from the puf photosynthesis promoter (+ OX; n = 3). All other wild-type and mutant strains carried the empty pCM66T plasmid. In each case, bars represent means and filled circles are the individual data points. Statistical significance is indicated above the chart (one-way ANOVA using the Holm-Šidák test; n.s., no significance; **, P < 0.01; ***, P < 0.001).
Biofilm production is impaired in a cysE1 deletion strain.
EPS is also known to be a key prerequisite for bacterial adherence to solid surfaces and formation of three-dimensional biofilm architecture (6, 63), so we tested the ability of the ΔcysE1 strain to form biofilms on untreated polystyrene and glass surfaces. To assess biofilm production, cultures were incubated statically for 5 days in 96-well flat bottom polystyrene microtiter plates. Quantification of the adhered biomass agreed with the biochemical and histochemical assays for EPS/capsule. Biofilm production by the SB1003 ΔcysE1 mutant was 9% of that of the wild type (Fig. 8A). Complementation of the ΔcysE1 strain in trans restored biofilm production to 110% (Fig. 8A). Comparable complementation with the puf promoter alone or puf-g16 did not restore biofilm formation. Furthermore, deletion of the g16 gene did not decrease biofilm formation, and complementation of SB1003 Δg16 with puf-cysE1 did not further increase biofilm production beyond wild-type levels (Fig. 8A). Meanwhile, biofilms were allowed to form for 5 days on glass microscope coverslips and then imaged by phase-contrast microscopy. Wild-type R. capsulatus cultures readily produced evenly distributed, densely packed biofilms across the full surface of the coverslip (Fig. 8B). Although ΔcysE1 strains were capable of forming biofilm, the overall cell density was substantially lower than that of the wild type, with fewer close cell-cell contacts and frequent gaps visible throughout the colonized surface (Fig. 8B). Biofilms produced by Δg16 strains were indistinguishable from those of the wild type. Overall, the data presented clearly indicate that cysE1 is involved in regulation of extracellular polymeric substances and biofilm production.
FIG 8.
Effect of cysE1 deletion on biofilm. (A) Quantification of relative biofilm production on an uncoated polystyrene microtiter plate surface. Wild-type R. capsulatus SB1003 (WT) was compared to isogenic cysE1 (ΔcysE1) and g16 (Δg16) knockouts. Knockout strains were complemented in trans with empty pCM66T plasmid (66T) or pCM66T containing the puf photosynthesis promoter alone (puf), puf-cysE1 (cysE1), or puf-g16 (g16). Bars represent means (n = 6), and filled circles are the individual data points. Statistical significance is indicated above the chart (one-way ANOVA using the Holm-Šidák test; n.s., no significance; *, P < 0.05; ***, P < 0.001). (B) Representative phase-contrast images (×100 magnification) of R. capsulatus biofilm formed on an uncoated glass slide.
DISCUSSION
In this study, we investigated the roles of two genes, g16 and cysE1, immediately downstream of the R. capsulatus core GTA gene cluster. The impact of the CysE1 serine O-acetyltransferase on RcGTA receipt, secretion of polymeric substances to the cell surface, and formation of biofilm was clearly observed, but the role of RcGTA g16 is more enigmatic. Deletion of the g16 gene had no significant impact on any of the phenotypes tested in this study (production of EPS, biofilm and capsule formation, or RcGTA activity). However, overexpression did lead to an increase in EPS production. Attempts to identify a function for the g16 protein via sequence and structural homology searches yielded no high-confidence hits. Clearly, further work is required to definitively identify the role RcGTA g16 plays and the nature of its possible interaction with CysE1.
GTA production is known to be influenced by external nutrient availability (31, 64), and Westbye et al. (31) showed that chemical inhibition of histidine biosynthesis boosted RcGTA production. The data presented here are the first to link an amino acid biosynthesis pathway to regulation of RcGTA recipient ability. CysE catalyzes the production of OAS and its spontaneous racemer NAS (Fig. S7) (50, 65). In addition to being precursors to cysteine, OAS and NAS are also signaling molecules. NAS is a ligand for CysB, a LysR family transcriptional regulator that autoinhibits its own expression and activates expression of various sulfur metabolism and amino acid biosynthesis regulons (50, 51, 66, 67). CysB is not currently annotated in the R. capsulatus SB1003 genome, but there are at least 20 putative LysR-type regulators, including rcc02272, which is a strong HHPRED match to Klebsiella aerogenes CysB (PDB code 1AL3; E value, 1.3e−46) (Table S4).
Previous studies in other species have determined that CysE is involved in biofilm formation. The mechanism, however, is not fully understood, and there is some variation from species to species. Escherichia coli and Providencia stuartii cysE is required for production of an unidentified extracellular signal that promotes differential gene expression, and deletion of the cysE gene leads to increased biofilm formation (50). Addition of OAS but not NAS complemented the null phenotype, and this response was cysB dependent (50). In contrast, Vibrio fischeri cysE is an essential gene, and CysE is required for synthesis of an extracellular signal that promotes biofilm formation (68). It is also notable that a recent study identified two natural products as inhibitors of Staphylococcus aureus CysE, both of which also inhibited biofilm formation and even promoted dissolution of mature biofilm (69).
CysB is also known to regulate expression of diverse genes, such as those encoding acid tolerance (70, 71), antibiotic resistance (72, 73), type III secretion (74), and 4-hydroxy-2-alkylquinolones (HAQs). HAQs are core constituents of Pseudomonas aeruginosa cell-to-cell signaling that control various physiological functions, such as the secretion of extracellular DNA and biofilm formation (73, 75). PqsR is the direct regulator of HAQ synthesis, and pqsR expression is in turn regulated by CysB via a cysteine-independent mechanism (73). A similar mechanism could explain the phenotypes observed here, i.e., a reduction of OAS/NAS production in R. capsulatus could lead to impaired quorum sensing, which is known to be an important regulator of both EPS biosynthesis and RcGTA production/receipt (13, 14).
Under normal laboratory conditions, GTA genes are expressed in only a small subset of a given bacterial population; for example, ~10% of Bartonella henselae cells spontaneously produce GTAs (64), as do ~1 to 3% of R. capsulatus cells (22, 25). Moreover, wild-type strains produce modest in vitro transduction frequencies of no more than 10−4 (64, 76, 77). In contrast, the only large-scale in situ study to date reported frequencies of gene transfer that were orders of magnitude higher (43). Some of the discrepancy between the in vitro and in situ data could in part be due to poor replication of the optimal natural conditions for GTA transfer in the lab. Indeed, simply coculturing marine GTA producers from the Roseobacter clade with a natural symbiont, Synechococcus, led GTA proteins to constitute up to 13% of total extracellular protein (78). Mathematical modeling has raised doubts about whether GTA gene transfer, at the frequencies reported in vitro, would provide sufficient advantage to producer cells via recombination/allelic diversity alone, to outweigh the cost of lysis (41). Some of these reservations could be allayed by increased production of GTAs in a setting where potential recipients are at high density, receptor polysaccharide is enhanced, and loss of GTAs by diffusion is more constrained (Fig. S8).
We propose that an environmental niche for R. capsulatus GTA production could be in biofilm and that RcGTA activity is coregulated with biofilm production, although more work will be required to definitively prove this. We have shown that the two genes immediately adjacent to the known RcGTA cluster are coregulated with the RcGTA genes and one, g16, is also cotranscribed. Deletion of cysE1 leads to a substantial decrease in extracellular polysaccharide and substantial thinning of the capsule; EPS and capsule are both essential for successful adsorption to and infection of a target cell by RcGTAs (13–15). In a variety of species, polysaccharide secretion in particular is known to be a prerequisite for optimal biofilm formation (6, 79, 80), and here, we also observed substantial loss of biofilm formation by the R. capsulatus cysE1 knockout. This study focused on R. capsulatus, but GTAs are found in many species (17, 38), most of which are known to form biofilms, and therefore, our hypotheses could be tested in other species in the future.
We hypothesize that CysE1 is a constituent of interlocking biofilm and RcGTA regulatory circuits. For example, FliQ and CheY are known contributors to biofilm, and both are under the control of CtrA, a well-characterized pleiotropic regulator that is essential for RcGTA production (81–83). Furthermore, a recent global expression analysis of an Agrobacterium pdhS2 mutant identified a regulon that included control of motility, production of adhesive polysaccharides, surface attachment, and biofilm production via c-di-GMP signaling (84). Interestingly, homologues of most of the core components of the Agrobacterium biofilm regulation pathway (CtrA, CckA, PleC, and DivK) and c-di-GMP are all established regulators of RcGTA production (23, 32, 33, 35, 82, 85).
It is generally accepted that high levels of c-di-GMP are associated with reduced motility and increased biofilm formation in multiple species (75). For example, c-di-GMP levels in biofilm cells from the alphaproteobacterium Ruegeria mobilis are 20-fold higher than in planktonic cells (86). In contrast, there is only limited information about the role of c-di-GMP in GTA regulation. Two recent papers reported that increased c-di-GMP concentrations repress both RcGTA production and flagellar motility in R. capsulatus (33, 34); however, both high and low c-di-GMP concentrations appear to be important for different stages of RcGTA production and release (87). It is likely that multiple factors, such as the precise spatiotemporal levels of c-di-GMP, phosphorylation states of essential regulators (e.g., CtrA or CckA), and input from other signaling molecules (e.g., homoserine lactone [HSL], rcc00280, OAS/NAS, etc.), all combine to provide fine control of RcGTA expression and biofilm formation (14, 32, 35–37, 82, 87). More work is needed to establish the complex regulatory mechanisms, but our data provide the first link between regulation of GTA gene transfer and production of biofilm. Establishing the environmental niche for GTAs is essential to fully understand their impact on host communities and to effectively identify new GTAs in a broader range of species.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Three Rhodobacter capsulatus strains were used in this study: rifampicin-sensitive wild-type strain B10 (88), the rifampicin-resistant derivative SB1003 (ATCC BAA-309), and the RcGTA overproducer strain DE442 (23, 37, 62). All R. capsulatus cultures were grown at 30°C either with aeration in the dark or in anoxic sealed tubes under constant illumination. Two growth media were used, YPS complex medium (0.3% [wt/vol] yeast extract, 0.3% [wt/vol] peptone, 2 mM MgCl2, 2 mM CaCl2) and RCV defined medium (89); 1.5% (wt/vol) agar was added for solid media. E. coli strain S17-1, which contains chromosomally integrated tra genes, was used as a donor for all conjugations. NEB 10-beta competent E. coli (New England Biolabs [NEB], Ipswich, MA, USA) were used for standard cloning and plasmid maintenance. E. coli K-12 BW25113 ΔcysE was sourced from the Keio collection (56). All E. coli cultures were grown at 37°C in LB medium. Antibiotics were used for selection and counterselection as follows: for R. capsulatus, rifampicin at 100 μg/mL, gentamicin at 3 μg/mL, and kanamycin at 10 μg/mL; for E. coli, gentamicin at 10 μg/mL and kanamycin at 50 μg/mL. OAS (1 M solution in 10 mM Tris, pH 6; Sigma-Aldrich, Poole, UK) was added directly to cultures to the indicated final concentrations.
General cloning.
All cloning reactions were carried out with the In-Fusion cloning kit (TaKaRa Bio Europe, Saint-Germain-en-Laye, France) or NEBuilder (NEB) to produce the constructs listed in Table 1. Inserts were amplified using primers with 15-bp 5′ overhangs of complementary sequence to the DNA with which it was to be joined. Oligonucleotides were obtained from either Integrated DNA Technologies (IDT) or Sigma (Table 2). Q5 Polymerase (NEB) was used for all amplifications. pCM66T was used as a broad-host-range destination vector for all constructs and was linearized with BamHI (NEB).
TABLE 1.
Plasmids used in this studya
| Name | Description | Vector | Source or reference |
|---|---|---|---|
| pCM66T | Broad-host-range vector (gift from Mary Lidstrom); ptac IncP ColE1 | NA | Addgene plasmid 74738 |
| pETTF11 | Protein expression plasmid, His6-3c tag, T7 inducible promoter | NA | 94 |
| pmCRc | Codon-optimized mCherry | pEX | Eurofins Genomics |
| pCMF130 | RcGTA promoter fused to codon-optimized mCherry | pCM66T | This study |
| pCMF151 | puf promoter fused to SB1003 RcGTA g16 gene | pCM66T | This study |
| pCMF152 | puf promoter fused to SB1003 cysE1 gene | pCM66T | This study |
| pCMF278 | puf promoter only control | pCM66T | This study |
| pDS1 | cysE1 gene and flanking DNA | pCM66T | This study |
| pDS2 | g16 gene and flanking DNA | pCM66T | This study |
| pDS3 | cysE1::Gent | pCM66T | This study |
| pDS4 | g16::Gent | pCM66T | This study |
| pDS5 | g16-cysE1::Gent | pCM66T | This study |
| pDS6 | cysE2 gene and flanking DNA | pCM66T | This study |
| pDS7 | cysE2::Gent | pCM66T | This study |
| pDS8 | cysE2::Kan | pCM66T | This study |
| pCMF140 | His6-CysE1 | pETTF11 | This study |
| pDS9 | His6-CysE2 | pETTF11 | This study |
NA, not applicable.
TABLE 2.
Oligonucleotides used in this study
| Use and name | Sequence (5′-3′) | Construct produceda |
|---|---|---|
| RT-PCR | ||
| yceG RT1 F | GGGATGCGGCTGCAGACCGATCC | NA |
| g1 RT1 R | GTCAACCTCCTGCGGCGTC | NA |
| g15-16 RT2 F | GGGAGGTGTTTCAATTCGCC | NA |
| g15-16 RT2 R | GGTAAAGATCGGCCCAATGCG | NA |
| g16-cysE1 RT3 F | GCTGGCACAATGGTTGC | NA |
| g16-cysE1 RT3 R | GTGAATATTGCCCAGCA | NA |
| cyse1-1701 RT4 F | AGGACATGGCCTATTTCGTGC | NA |
| cyse1-1701 RT4 R | CATCACATGGTTCAGGTTCG | NA |
| qPCR | ||
| cysE1 qF | ATGCTGCATTCGGTGACA | NA |
| cysE1 qR | TGAATATTGCCCAGCACCTT | NA |
| cysE2 qF | CCAGCTTGACCAGGATGTC | NA |
| cysE2 qR | CGATGCGATGGTGGATCA | NA |
| uvrD qF | CAGAAGGAACACACGGTCAA | NA |
| uvrD qR | AAAGTGTCAGGCGGAATCTC | NA |
| Gene knockout constructs | ||
| Gent F | GATCCCCTGATTCCCTTTGT | pDS3, pDS4 and pDS5 |
| Gent R | CTTGAACGAATTGTTAGG | pDS3, pDS4 and pDS5 |
| cysE1 Up F | CGACTCTAGAGGATCCAGGTGTCGGATGTGTATGGC | pDS1 and pDS3 |
| cysE1 Up R | AACAATTCGTTCAAGCGGGTTTTGGACATGGGACT | pDS1 and pDS3 |
| cysE1 Down F | GGGAATCAGGGGATCCGGTCAACATGGATCAGCAG | pDS1 and pDS3 |
| cysE1 Down R | CGGTACCCGGGGATCCCAAAAACAGCCCGATCACCC | pDS1, pDS3, and pDS5 |
| cysE2 Up F | CGACTCTAGAGGATCCGATGCGCTGGCCCAGATCC | pDS6, pDS7, and pDS8 |
| cysE2 Up R-Gent | GGGAATCAGGGGATCACCGGAACCAGGGAAAACC | pDS7 |
| cysE2 Down F-Gent | AACAATTCGTTCAAGATGTCTGGCTGACCGAAAGC | pDS7 |
| cysE2 Down R | CGGTACCCGGGGATCTCGTCGTAATCGACGAAGG | pDS6, pDS7, and pDS8 |
| cysE2 Up R-Kan | GACACATGCAGCTCCACCGGAACCAGGGAAAACC | pDS8 |
| cysE2 Down F-Kan | ATTTTGAGACACAACATGTCTGGCTGACCGAAAGC | pDS8 |
| g16 Up F | CGACTCTAGAGGATCCGGGAGGTGTTTCAATTCGCC | pDS2, pDS4, and pDS5 |
| g16 Up R | AACAATTCGTTCAAGGTAAAGATCGGCCCAATGCG | pDS2 and pDS4 |
| g16 Down F | GGGAATCAGGGGATCCGACGATCTTGCAGAGGTGA | pDS2 and pDS4 |
| g16 Down R | CGGTACCCGGGGATCCCATCACATGGTTCAGGTTCG | pDS2 and pDS4 |
| Overexpression constructs | ||
| pPuf F | CGACTCTAGAGGATCCGAGCTTCGGAATCTGCG | pCMF151 and pCMF152 |
| pPuf R | CATAACAACCTCCGGATTGGCAGAC | pCMF151 and pCMF152 |
| cysE1 OX F | CCGGAGGTTGTTATGTCCAAAACCCGTGCG | pCMF152 |
| cysE1 OX R | CGGTACCCGGGGATCGTCAGCCCTCGCAGTCGC | pCMF152 |
| g16 OX F | CCGGAGGTTGTTATGCGGCCGATCCTGATC | pCMF151 |
| g16 OX R | CGGTACCCGGGGATCTCACAGATCCGCATCCAC | pCMF151 |
| puf con F | CGACTCTAGAGGATCCGAGCTTCGGAATCTGCG | pCMF278 |
| puf con R | CGGTACCCGGGGATCCATAACAACCTCCGGATT | pCMF278 |
| Reporter construct | ||
| pGTA F | CGACTCTAGAGGATCGATTGTCGATCAGATCAC | pCMF130 |
| pGTA R | GCTGACCATCGCCAGGGCCAGTTCC | pCMF130 |
| mCherryRc F | CCTGGCGATGGTCAGCAAGGGGGAG | pCMF130 |
| mCherryRc R | CGGTACCCGGGGATCCTGTTGTGTGGAATTGTGAGC | pCMF130 |
| Protein purification | ||
| H6-Cyse1 F | TCCAGGGACCAGCAATGATGTCCAAAACCCGTGCG | pCMF140 |
| H6-CysE1 R | TGAGGAGAAGGCGCGTCAGCCCTCGCAGTCGCAGC | pCMF140 |
| H6-Cyse2 F | TCCAGGGACCAGCAATGACGCTTGACACGCAACG | pDS9 |
| H6-CysE2 R | TGAGGAGAAGGCGCGTCGGTCATTCACGTTCTCC | pDS9 |
NA, not applicable.
Transformation/conjugation.
E. coli was transformed with 100 ng of plasmid DNA by standard heat shock transformation (90). For conjugation, 1-mL aliquots of overnight cultures of the E. coli S17-1 donor and Rhodobacter recipient strains were centrifuged at 5,000 × g for 1 min, washed with 1 mL YPS, centrifuged again, and resuspended in 100 μL YPS. Ten-microliter portions of concentrated donor and recipient cells were mixed and spotted onto YPS agar or spotted individually as negative controls. Plates were incubated overnight at 30°C. Spots were scraped, suspended in 100 μL YPS broth, and plated on YPS containing 100 μg/mL rifampicin (counterselection against E. coli) and 10 μg/mL kanamycin (plasmid selection). Plates were incubated overnight at 30°C and then restreaked onto fresh agar to obtain pure single colonies.
Gene knockouts.
Knockouts were created by GTA transfer. pCM66T plasmid constructs were created with a gentamicin resistance cassette flanked by 500 to 1,000 bp of DNA from either side of the target gene (Table 1). Assembly was achieved by a one-step, four-component NEBuilder (NEB) reaction and transformation into NEB 10-beta cells. Deletion constructs were introduced into the RcGTA overproducer strain, and a standard GTA transduction assay was carried out to replace the intact chromosomal gene with the deleted version.
Microtiter plate growth curves.
Two hundred microliters of growth medium was dispensed into the wells of a 96-well microtiter plate. For each replicate, 4 μL of independently grown cells was seeded into the well. The plates were assayed in a SpectroStar Nano device (BMG Labtech, Ortenberg, Germany). Plates were held at a constant 30°C, and optical density readings were taken at 15-min intervals. Plates were agitated at 150 rpm using an orbital pattern for 1 min before every reading.
Spot dilution assays.
Bacterial cultures were twice pelleted and resuspended in liquid minimal medium (RCV) to remove residual complex growth medium components or secondary metabolites. Washed cells were diluted in a 1-in-10 serial dilution series in RCV. Ten microliters from each dilution was spotted onto dry minimal-medium agar plates (RCV) or complex-medium agar plates (YPS or LB) and allowed to dry completely. Plates were incubated at 30°C for R. capsulatus or 37°C for E. coli.
Gene overexpression.
Overexpression in Rhodobacter was achieved by a transcriptional fusion of the genes of interest to the puf photosynthesis promoter. The promoter contains 1,002 bp of sequence upstream of the pufB gene, with the entire pufB start codon directly preceded by each gene of interest. The promoter and its use for expression of heterologous genes were described previously (22, 91). Growth and general maintenance of Rhodobacter strains containing overexpression plasmids was carried out at 30°C under aerated, chemotrophic growth conditions, where transcription from the puf promoter is strongly repressed. To establish overexpression conditions, cultures were grown in sealed glass tubes and incubated at 30°C with illumination to induce puf promoter activity.
Rhodobacter gene transfer assays.
Rhodobacter assays were carried out essentially as described in reference 92. RcGTA donor cultures were grown in YPS for ~72 h, and recipient cultures were grown in RCV for ~24 h. Cells were cleared from donor cultures by centrifugation, and the supernatant was filtered through a 0.45-μm syringe filter. Recipient cells were concentrated 3-fold by centrifugation at 5,000 × g and resuspension in 1/3 volume G buffer (10 mM Tris-HCl [pH 7.8], 1 mM MgCl2, 1 mM CaCl2, 1 mM NaCl, 0.5 mg/mL bovine serum albumin [BSA]). Reactions were carried out in polystyrene culture tubes (Starlab Ltd., Milton Keynes, UK) containing 400 μL G buffer, 100 μL recipient cells, and 100 μL filter donor supernatant, and then the mixtures were incubated at 30°C for 1 h. Nine hundred microliters of YPS was added to each tube and incubated for a further 3 h. Cells were harvested by centrifugation at 5,000 × g and plated on YPS containing 100 μg/mL rifampicin (for standard GTA assays) or 3 μg/mL gentamicin (for gene knockouts).
Reverse transcriptase PCR (RT-PCR).
Total RNA was extracted from stationary growth phase R. capsulatus cultures using the NucleoSpin RNA minikit (Macherey-Nagel GmbH & Co., Düren, Germany), including on-column DNase treatment. Trace DNA contamination was eliminated by using a Turbo DNase-free kit (Thermo Fisher Scientific, Waltham, MA, USA). RNA was quantified with a Nanodrop instrument (Thermo Fisher Scientific), and cDNA was synthesized from 1 μg of total RNA using the LunaScrip RT SuperMix kit (NEB). PCR amplification across gene boundaries was carried out using 1 μL of cDNA, standard Q5 polymerase conditions, 25 cycles of amplification, and primers as listed in Table 2.
Quantitative RT-PCR.
Dilutions (1:50) of the cDNA template were prepared in distilled water (dH2O), and 1 μL was used per reaction. Reactions contained Fast Sybr green Mastermix (Applied Biosystems, Waltham, MA), cDNA, and primers (500 nM). Standard conditions were used, with an annealing temperature of 60°C. All primer efficiencies were calculated as between 90 and 110%. Relative gene expression was determined using the ΔΔCT method (93). For each sample, variance was calculated for three independent biological replicates, which were each the mean for three technical replicates. A QuantStudio 3 real-time PCR system was used for all experiments (Applied Biosystems).
Protein purification.
For His6-tagged proteins, 500-mL cultures of E. coli containing the relevant expression plasmid were induced at mid-exponential growth phase with 0.2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) overnight at 20°C (94). Concentrated cells were lysed in 20 mL binding buffer (0.5 M NaCl, 75 mM Tris; pH 7.75) plus 0.2 mg mL−1 lysozyme (Avantor-VWR, Radnor, PA) and 500 U Basemuncher endonuclease (Abcam, Cambridge, UK) for 30 min on ice and then sonicated. Cleared supernatant was applied to a 5-mL HisTrap FF crude column (Cytiva, Marlborough, MA), and the bound, His-tagged protein was eluted with 125 mM imidazole. Eluted protein was desalted on a HiPrep 26/10 desalting column (Cytiva) and then further separated by size exclusion chromatography on a HiLoad 16/60 Superdex 200 preparative-grade gel filtration column. All chromatography steps were carried out on an AKTA Prime instrument (Cytiva). Purified proteins were concentrated in a Spin-X UF centrifugal concentrator (Corning Inc., Corning, NY) and quantified by the extinction coefficient method and a NanoDrop 1000 Spectrophotometer (Thermo Scientific, Waltham, MA). Samples were stored at −80°C in binding buffer plus 50% glycerol.
Transacetylation assay.
An endpoint assay for transacetylation was performed using a fluorometric acetyltransferase activity assay kit (Abcam) according to the manufacturer’s instructions. CysE1 and CysE2 proteins were used at a final concentration of 100 nM and l-serine at a final concentration of 0.25 mM. All reaction mixtures were incubated for 30 min at room temperature. Negative-control reactions were carried out in the absence of protein and/or l-serine. Fluorescence output was measured in an Infinite M200 microplate reader (Tecan, Männedorf, Switzerland) at excitation and emission wavelengths of 380 and 520 nm.
Quantification of cell-associated extracellular substances.
Extracellular polysaccharide and protein were extracted from R. capsulatus cells grown in RCV medium under photosynthetic conditions to stationary growth phase. In brief, filtered NaCl solution was added to each culture to a final concentration of 250 mM, and 1-mL samples were harvested by centrifugation at 14,000 × g for 10 min. Addition of NaCl was found to improve the formation of tight cell pellets, which allowed the supernatant to be carefully removed and discarded. To remove residual free polysaccharides that may have been present in the supernatant, cell pellets were resuspended in 1 mL of 250 mM NaCl and harvested again by centrifugation. This wash step was sufficient to remove >90% of loosely associated or free polysaccharides (Fig. S3). Washed cell pellets were resuspended in 1 mL of 50 mM EDTA and incubated at 37°C for 3 h. The samples were cleared by centrifugation at 14,000 × g for 10 min, and the supernatant was carefully transferred to a fresh tube. EPS quantification was carried out essentially as previously described (13) using a modified phenol-sulfuric acid method and a calibration curve constructed using an equal-concentration mixture of sucrose and fructose. Each quantification reaction mixture consisted of 100 μL sample supernatant, 100 μL 5% (wt/vol) phenol, and 500 μL 93% (vol/vol) sulfuric acid. Tubes were incubated at room temperature for 10 min before the absorbance at 490 nm was measured in a spectrophotometer. Extracellular protein concentration was determined by bicinchoninic acid assay (BCA) and an albumin standard curve according to the manufacturer’s instructions (Abcam).
Capsule staining.
Capsule formation was measured by negative staining (13, 60). In brief, 1-mL samples of stationary-phase R. capsulatus cultures were harvested by centrifugation, and the pellets were resuspended in 1 mL of 1% (wt/vol) Carnation skim milk powder. The mixture was smeared onto a microscope slide and allowed to air dry. The cells were stained with 1% (wt/vol) crystal violet for 5 min and then washed with 20% (wt/vol) copper sulfate. Slides were allowed to air dry before being imaged by oil immersion phase-contrast microscopy at ×100 magnification. Original images were black and white, and post factum false color was added using ImageJ to approximate the original image.
Biofilm assays.
R. capsulatus cultures were grown in 200 μL YPS medium in uncoated, polystyrene 96-well plates. Plates were incubated at 30°C, in the dark and without agitation, for 5 days. The culture medium was decanted and the wells were washed 5 times with dH2O to remove any unadhered cells. The cells were stained with 0.1% (wt/vol) crystal violet for 10 min, washed 5 times with dH2O to remove excess dye, and allowed to air dry. Bound crystal violet was resolubilized with 250 μL of 30% (vol/vol) acetic acid, and absorbance was measured at 590 nm. For coverslip assays, R. capsulatus cells were inoculated into 10 mL of YPS medium in a 50-mL Falcon tube containing a partially submerged glass coverslip and incubated for 5 days statically at 30°C in the dark or with illumination, as indicated. The coverslip was removed from the tube and washed thoroughly in dH2O, and excess liquid was wicked away with blotting paper. The biofilm was then sandwiched between a microscope slide and a fresh coverslip. Imaging of the mature biofilm was carried out using an Axioscope inverted microscope (Carl Zeiss Ltd., Cambridge, UK) and mounted charge-coupled device (CCD) camera.
Data analysis.
The RNA-seq data set in the NCBI Gene Expression Omnibus (GEO) database, accession number GSE118116, was analyzed to assess transcriptional units and compared to the meta-analysis of GSE18149, GSE33176, GSE41014, and GSE53636 carried out by Peña-Castillo et al. (44). Transcript abundance was visualized using the Broad Institute’s IGV viewer (95). Protein alignments were made in ClustalΩ (96) and then visualized in Jalview (97). FIJI software (98) was used to analyze microscopy images. Statistical analysis was carried out using SigmaPlot software version 13 (Systat Software Inc., Slough, UK; www.systatsoftware.com), and for each use, the test parameters are indicated in the text and/or figure legends. All data presented in figures correspond to one representative experiment consisting of at least three biological replicates; all experimental data were confirmed on at least two different occasions. CorelDraw 2018 (Corel Corporation, Ottawa, Canada) was used for figure preparation and ChemDraw (Perkin Elmer, Waltham, MA, USA) for chemical structure diagrams.
Sequence similarity analysis.
NCBI BLASTp searches to identify homologues of R. capsulatus SB1003 proteins were performed using the default parameters: expect threshold = 0.05, word size = 6, blosum62 similarity matrix, gap costs of existence = 11, and extension = 1. Queries were made against the nonredundant protein sequences database (nr; posted 5 May 2022, accessed 16 May 2022). A summary of the full outputs for CysE1 and CysE2 can be found in Tables S2 and S3. HHPRED analysis of the R. capsulatus SB1003 rcc02272 gene product was carried out using the pdb_mmcif70_14_Apr and NCBI Conserved Domains (CD) (v3.19) databases, accessed on 20 May 2022 (Table S4) (99, 100). The default parameters were used, i.e., the HHBlits UniRef30 MSA generation method, maximal generation steps = 3, and an E-value threshold of 1e−3. Minimum coverage of MSA hits was 20%, and minimum sequence identity of MSA hits with the query was 0%. Secondary structure scoring was done during alignment.
Protein structure and function prediction.
Three-dimensional structures for the R. capsulatus RcGTA g16 protein was predicted using the AlphaFold colab server (https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/beta/AlphaFold2_advanced.ipynb#scrollTo=bQe3KeyTcv0n) using msa_method:jackhammer and all other parameters set to default (45). The top-ranked AlphaFold model was submitted to the Dali protein structure comparison server to predict possible protein function; the full output can be found in Table S1.
Data availability.
All data required to assess the findings are available in the main text or the supplemental information files. Reagents and materials used in this paper are publicly available. Any unique bacterial strains or recombinant DNA constructs described in this paper will be provided by the corresponding author upon request.
ACKNOWLEDGMENTS
We thank the University of York Technology Facility for providing access to specialist equipment and expert technical assistance as required. We also thank Jelena Kusakina for critical reading of the manuscript.
This research was funded in whole by a Wellcome Trust and Royal Society independent fellowship grant (grant 109363/Z/15/A).
Conceptualization, P.C.M.F.; Methodology, D.S. and P.C.M.F.; Investigation, D.S. and P.C.M.F.; Writing, P.C.M.F.; Visualization, P.C.M.F.; Funding Acquisition, P.C.M.F.; Resources, P.C.M.F.; Supervision, P.C.M.F.
We declare no competing interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Paul C. M. Fogg, Email: paul.fogg@york.ac.uk.
Arpita Bose, Washington University in St. Louis.
REFERENCES
- 1.Rowe JM, Saxton MA, Cottrell MT, DeBruyn JM, Berg GM, Kirchman DL, Hutchins DA, Wilhelm SW. 2008. Constraints on viral production in the Sargasso Sea and North Atlantic. Aquat Microb Ecol 52:233–244. 10.3354/ame01231. [DOI] [Google Scholar]
- 2.Moran MA. 2015. The global ocean microbiome. Science 350:aac8455. 10.1126/science.aac8455. [DOI] [PubMed] [Google Scholar]
- 3.Heidelberg JF, Heidelberg KB, Colwell RR. 2002. Seasonality of Chesapeake Bay bacterioplankton species. Appl Environ Microbiol 68:5488–5497. 10.1128/AEM.68.11.5488-5497.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rowe JM, DeBruyn JM, Poorvin L, LeCleir GR, Johnson ZI, Zinser ER, Wilhelm SW. 2012. Viral and bacterial abundance and production in the Western Pacific Ocean and the relation to other oceanic realms. FEMS Microbiol Ecol 79:359–370. 10.1111/j.1574-6941.2011.01223.x. [DOI] [PubMed] [Google Scholar]
- 5.Dang H, Lovell CR. 2016. Microbial surface colonization and biofilm development in marine environments. Microbiol Mol Biol Rev 80:91–138. 10.1128/MMBR.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Decho AW, Gutierrez T. 2017. Microbial extracellular polymeric substances (epss) in ocean systems. Front Microbiol 8:922. 10.3389/fmicb.2017.00922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang W, Ding W, Li Y-X, Tam C, Bougouffa S, Wang R, Pei B, Chiang H, Leung P, Lu Y, Sun J, Fu H, Bajic VB, Liu H, Webster NS, Qian P-Y. 2019. Marine biofilms constitute a bank of hidden microbial diversity and functional potential. Nat Commun 10:517. 10.1038/s41467-019-08463-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vavourakis CD, Ghai R, Rodriguez-Valera F, Sorokin DY, Tringe SG, Hugenholtz P, Muyzer G. 2016. Metagenomic insights into the uncultured diversity and physiology of microbes in four hypersaline soda lake brines. Front Microbiol 7:211. 10.3389/fmicb.2016.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kimes NE, Callaghan AV, Aktas DF, Smith WL, Sunner J, Golding B, Drozdowska M, Hazen TC, Suflita JM, Morris PJ. 2013. Metagenomic analysis and metabolite profiling of deep-sea sediments from the Gulf of Mexico following the Deepwater Horizon oil spill. Front Microbiol 4:50. 10.3389/fmicb.2013.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McEwan AG. 1994. Photosynthetic electron transport and anaerobic metabolism in purple non-sulfur phototrophic bacteria. Antonie Van Leeuwenhoek 66:151–164. 10.1007/BF00871637. [DOI] [PubMed] [Google Scholar]
- 11.Shimizu T, Aritoshi T, Beatty JT, Masuda T. 2022. Persulfide-responsive transcription factor SqrR regulates gene transfer and biofilm formation via the metabolic modulation of cyclic di-GMP in Rhodobacter capsulatus. Microorganisms 10:908. 10.3390/microorganisms10050908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schaefer AL, Taylor TA, Beatty JT, Greenberg EP. 2002. Long-chain acyl-homoserine lactone quorum-sensing regulation of Rhodobacter capsulatus gene transfer agent production. J Bacteriol 184:6515–6521. 10.1128/JB.184.23.6515-6521.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brimacombe CA, Stevens A, Jun D, Mercer R, Lang AS, Beatty JT. 2013. Quorum-sensing regulation of a capsular polysaccharide receptor for the Rhodobacter capsulatus gene transfer agent (RcGTA). Mol Microbiol 87:802–817. 10.1111/mmi.12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leung MM, Brimacombe CA, Spiegelman GB, Beatty JT. 2012. The GtaR protein negatively regulates transcription of the gtaRI operon and modulates gene transfer agent (RcGTA) expression in Rhodobacter capsulatus. Mol Microbiol 83:759–774. 10.1111/j.1365-2958.2011.07963.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Westbye AB, Kuchinski K, Yip CK, Beatty JT. 2016. The gene transfer agent RcGTA contains head spikes needed for binding to the Rhodobacter capsulatus polysaccharide cell capsule. J Mol Biol 428:477–491. 10.1016/j.jmb.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 16.Bárdy P, Füzik T, Hrebík D, Pantůček R, Thomas Beatty J, Plevka P. 2020. Structure and mechanism of DNA delivery of a gene transfer agent. Nat Commun 11:3034. 10.1038/s41467-020-16669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lang AS, Zhaxybayeva O, Beatty JT. 2012. Gene transfer agents: phage-like elements of genetic exchange. Nat Rev Microbiol 10:472–482. 10.1038/nrmicro2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shakya M, Soucy SM, Zhaxybayeva O. 2017. Insights into origin and evolution of α-proteobacterial gene transfer agents. Virus Evol 3:vex036. 10.1093/ve/vex036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lang AS, Westbye AB, Beatty JT. 2017. The distribution, evolution, and roles of gene transfer agents in prokaryotic genetic exchange. Annu Rev Virol 4:87–104. 10.1146/annurev-virology-101416-041624. [DOI] [PubMed] [Google Scholar]
- 20.Hynes AP, Shakya M, Mercer RG, Grüll MP, Bown L, Davidson F, Steffen E, Matchem H, Peach ME, Berger T, Grebe K, Zhaxybayeva O, Lang AS. 2016. Functional and evolutionary characterization of a gene transfer agent’s multilocus “genome.” Mol Biol Evol 33:2530–2543. 10.1093/molbev/msw125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Motro Y, La T, Bellgard MI, Dunn DS, Phillips ND, Hampson DJ. 2009. Identification of genes associated with prophage-like gene transfer agents in the pathogenic intestinal spirochaetes Brachyspira hyodysenteriae, Brachyspira pilosicoli and Brachyspira intermedia. Vet Microbiol 134:340–345. 10.1016/j.vetmic.2008.09.051. [DOI] [PubMed] [Google Scholar]
- 22.Fogg PCM, Westbye AB, Beatty JT. 2012. One for all or all for one: heterogeneous expression and host cell lysis are key to gene transfer agent activity in Rhodobacter capsulatus. PLoS One 7:e43772. 10.1371/journal.pone.0043772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fogg PCM. 2019. Identification and characterization of a direct activator of a gene transfer agent. Nat Commun 10:595. 10.1038/s41467-019-08526-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Québatte M, Dehio C. 2019. Bartonella gene transfer agent: evolution, function, and proposed role in host adaptation. Cell Microbiol 21:e13068. 10.1111/cmi.13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hynes AP, Mercer RG, Watton DE, Buckley CB, Lang AS. 2012. DNA packaging bias and differential expression of gene transfer agent genes within a population during production and release of the Rhodobacter capsulatus gene transfer agent, RcGTA. Mol Microbiol 85:314–325. 10.1111/j.1365-2958.2012.08113.x. [DOI] [PubMed] [Google Scholar]
- 26.Berglund EC, Frank AC, Calteau A, Vinnere Pettersson O, Granberg F, Eriksson A-S, Näslund K, Holmberg M, Lindroos H, Andersson SGE. 2009. Run-off replication of host-adaptability genes is associated with gene transfer agents in the genome of mouse-infecting Bartonella grahamii. PLoS Genet 5:e1000546. 10.1371/journal.pgen.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherlock D, Leong JX, Fogg PCM. 2019. Identification of the first gene transfer agent (GTA) small terminase in Rhodobacter capsulatus, its role in GTA production and packaging of DNA. J Virol 93:e01328-19. 10.1128/JVI.01328-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esterman ES, Wolf YI, Kogay R, Koonin EV, Zhaxybayeva O. 2021. Evolution of DNA packaging in gene transfer agents. Virus Evol 7:veab015. 10.1093/ve/veab015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomasch J, Wang H, Hall ATK, Patzelt D, Preusse M, Petersen J, Brinkmann H, Bunk B, Bhuju S, Jarek M, Geffers R, Lang AS, Wagner-Döbler I. 2018. Packaging of Dinoroseobacter shibae DNA into gene transfer agent particles is not random. Genome Biol Evol 10:359–369. 10.1093/gbe/evy005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koppenhöfer S, Wang H, Scharfe M, Kaever V, Wagner-Döbler I, Tomasch J. 2019. Integrated transcriptional regulatory network of quorum sensing, replication control, and SOS response in Dinoroseobacter shibae. Front Microbiol 10:803. 10.3389/fmicb.2019.00803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Westbye AB, O'Neill Z, Schellenberg-Beaver T, Beatty JT. 2017. The Rhodobacter capsulatus gene transfer agent is induced by nutrient depletion and the RNAP omega subunit. Microbiology (Reading) 163:1355–1363. 10.1099/mic.0.000519. [DOI] [PubMed] [Google Scholar]
- 32.Kuchinski KS, Brimacombe CA, Westbye AB, Ding H, Beatty JT. 2016. The SOS response master regulator LexA regulates the gene transfer agent of Rhodobacter capsulatus and represses transcription of the signal transduction protein CckA. J Bacteriol 198:1137–1148. 10.1128/JB.00839-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pallegar P, Peña-Castillo L, Langille E, Gomelsky M, Lang AS. 2020. Cyclic di-GMP-mediated regulation of gene transfer and motility in Rhodobacter capsulatus. J Bacteriol 202:e00554-19. 10.1128/JB.00554-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pallegar P, Canuti M, Langille E, Peña-Castillo L, Lang AS. 2020. A two-component system acquired by horizontal gene transfer modulates gene transfer and motility via cyclic dimeric GMP. J Mol Biol 432:4840–4855. 10.1016/j.jmb.2020.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Westbye AB, Kater L, Wiesmann C, Ding H, Yip CK, Beatty JT. 2018. The protease ClpXP and the PAS domain protein DivL regulate CtrA and gene transfer agent production in Rhodobacter capsulatus. Appl Environ Microbiol 84:e00275-18. 10.1128/AEM.00275-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lang AS, Beatty JT. 2000. Genetic analysis of a bacterial genetic exchange element: the gene transfer agent of Rhodobacter capsulatus. Proc Natl Acad Sci USA 97:859–864. 10.1073/pnas.97.2.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding H, Grüll MP, Mulligan ME, Lang AS, Beatty JT. 2019. Induction of Rhodobacter capsulatus gene transfer agent (RcGTA) gene expression is a bistable stochastic process repressed by an extracellular calcium-binding RTX protein homologue. J Bacteriol 201:e00430-19. 10.1128/JB.00430-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kogay R, Neely TB, Birnbaum DP, Hankel CR, Shakya M, Zhaxybayeva O. 2019. Machine-learning classification suggests that many alphaproteobacterial prophages may instead be gene transfer agents. Genome Biol Evol 11:2941–2953. 10.1093/gbe/evz206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kogay R, Wolf YI, Koonin EV, Zhaxybayeva O. 2020. Selection for reducing energy cost of protein production drives the GC content and amino acid composition bias in gene transfer agents. mBio 11:e01206-20. 10.1128/mBio.03139-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Price MN, Wetmore KM, Waters RJ, Callaghan M, Ray J, Liu H, Kuehl JV, Melnyk RA, Lamson JS, Suh Y, Carlson HK, Esquivel Z, Sadeeshkumar H, Chakraborty R, Zane GM, Rubin BE, Wall JD, Visel A, Bristow J, Blow MJ, Arkin AP, Deutschbauer AM. 2018. Mutant phenotypes for thousands of bacterial genes of unknown function. Nature 557:503–509. 10.1038/s41586-018-0124-0. [DOI] [PubMed] [Google Scholar]
- 41.Redfield RJ, Soucy SM. 2018. Evolution of bacterial gene transfer agents. Front Microbiol 9:2527. 10.3389/fmicb.2018.02527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lang AS, Taylor TA, Beatty JT. 2002. Evolutionary implications of phylogenetic analyses of the gene transfer agent (GTA) of Rhodobacter capsulatus. J Mol Evol 55:534–543. 10.1007/s00239-002-2348-7. [DOI] [PubMed] [Google Scholar]
- 43.McDaniel LD, Young E, Delaney J, Ruhnau F, Ritchie KB, Paul JH. 2010. High frequency of horizontal gene transfer in the oceans. Science 330:50. 10.1126/science.1192243. [DOI] [PubMed] [Google Scholar]
- 44.Peña-Castillo L, Mercer RG, Gurinovich A, Callister SJ, Wright AT, Westbye AB, Beatty JT, Lang AS. 2014. Gene co-expression network analysis in Rhodobacter capsulatus and application to comparative expression analysis of Rhodobacter sphaeroides. BMC Genomics 15:730. 10.1186/1471-2164-15-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holm L. 2020. Using DALI for protein structure comparison. Methods Mol Biol 2112:29–42. 10.1007/978-1-0716-0270-6_3. [DOI] [PubMed] [Google Scholar]
- 47.Li Y, Jin K, Setlow B, Setlow P, Hao B. 2012. Crystal structure of the catalytic domain of the Bacillus cereus SleB protein, important in cortex peptidoglycan degradation during spore germination. J Bacteriol 194:4537–4545. 10.1128/JB.00877-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pye VE, Tingey AP, Robson RL, Moody PCE. 2004. The structure and mechanism of serine acetyltransferase from Escherichia coli. J Biol Chem 279:40729–40736. 10.1074/jbc.M403751200. [DOI] [PubMed] [Google Scholar]
- 49.Kredich NM. 2008. Biosynthesis of cysteine. EcoSal Plus 3. 10.1128/ecosalplus.3.6.1.11. [DOI] [PubMed] [Google Scholar]
- 50.Sturgill G, Toutain CM, Komperda J, O'Toole GA, Rather PN. 2004. Role of CysE in production of an extracellular signaling molecule in Providencia stuartii and Escherichia coli: loss of CysE enhances biofilm formation in Escherichia coli. J Bacteriol 186:7610–7617. 10.1128/JB.186.22.7610-7617.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jovanovic M, Lilic M, Savic DJ, Jovanovic G. 2003. The LysR-type transcriptional regulator CysB controls the repression of hslJ transcription in Escherichia coli. Microbiology (Reading) 149:3449–3459. 10.1099/mic.0.26609-0. [DOI] [PubMed] [Google Scholar]
- 52.Wiethaus J, Schubert B, Pfänder Y, Narberhaus F, Masepohl B. 2008. The GntR-like regulator TauR activates expression of taurine utilization genes in Rhodobacter capsulatus. J Bacteriol 190:487–493. 10.1128/JB.01510-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ostrowski J, Kredich NM. 1989. Molecular characterization of the cysJIH promoters of Salmonella typhimurium and Escherichia coli: regulation by cysB protein and N-acetyl-L-serine. J Bacteriol 171:130–140. 10.1128/jb.171.1.130-140.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neuenschwander U, Suter M, Brunold C. 1991. Regulation of sulfate assimilation by light and O-acetyl-l-serine in Lemna minor L. Plant Physiol 97:253–258. 10.1104/pp.97.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim H, Hirai MY, Hayashi H, Chino M, Naito S, Fujiwara T. 1999. Role of O-acetyl-l-serine in the coordinated regulation of the expression of a soybean seed storage-protein gene by sulfur and nitrogen nutrition. Planta 209:282–289. 10.1007/s004250050634. [DOI] [PubMed] [Google Scholar]
- 56.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wüthrich D, Wenzel C, Bavan T, Bruggmann R, Berthoud H, Irmler S. 2018. Transcriptional regulation of cysteine and methionine metabolism in Lactobacillus paracasei FAM18149. Front Microbiol 9:1261. 10.3389/fmicb.2018.01261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bogicevic B, Irmler S, Portmann R, Meile L, Berthoud H. 2012. Characterization of the cysK2-ctl1-cysE2 gene cluster involved in sulfur metabolism in Lactobacillus casei. Int J Food Microbiol 152:211–219. 10.1016/j.ijfoodmicro.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 59.Parker G, Walshaw D, O'Rourke K, Broad S, Tingey A, Poole PS, Robson RL. 2001. Evidence for redundancy in cysteine biosynthesis in Rhizobium leguminosarum RL3841: analysis of a cysE gene encoding serine acetyltransferase. Microbiology (Reading) 147:2553–2560. 10.1099/00221287-147-9-2553. [DOI] [PubMed] [Google Scholar]
- 60.Anthony EE. 1931. A note on capsule staining. Science 73:319–320. 10.1126/science.73.1890.319. [DOI] [PubMed] [Google Scholar]
- 61.Kredich NM, Tomkins GM. 1966. The enzymic synthesis of L-cysteine in Escherichia coli and Salmonella typhimurium. J Biol Chem 241:4955–4965. 10.1016/S0021-9258(18)99657-2. [DOI] [PubMed] [Google Scholar]
- 62.Ding H, Moksa MM, Hirst M, Beatty JT. 2014. Draft genome sequences of six Rhodobacter capsulatus strains, YW1, YW2, B6, Y262, R121, and DE442. Genome Announc 2:e00050-14. 10.1128/genomeA.00050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Davey ME, O'toole GA. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev 64:847–867. 10.1128/MMBR.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Québatte M, Christen M, Harms A, Körner J, Christen B, Dehio C. 2017. Gene transfer agent promotes evolvability within the fittest subpopulation of a bacterial pathogen. Cell Syst 4:611–621.E6. 10.1016/j.cels.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Flavin M, Slaughter C. 1965. Synthesis of the succinic ester of homoserine, a new intermediate in the bacterial biosynthesis of methionine. Biochemistry 4:1370–1375. 10.1021/bi00883a022. [DOI] [PubMed] [Google Scholar]
- 66.Turnbull AL, Surette MG. 2008. L-cysteine is required for induced antibiotic resistance in actively swarming Salmonella enterica serovar Typhimurium. Microbiology (Reading) 154:3410–3419. 10.1099/mic.0.2008/020347-0. [DOI] [PubMed] [Google Scholar]
- 67.Parry J, Clark DP. 2002. Identification of a CysB-regulated gene involved in glutathione transport in Escherichia coli. FEMS Microbiol Lett 209:81–85. 10.1111/j.1574-6968.2002.tb11113.x. [DOI] [PubMed] [Google Scholar]
- 68.Singh P, Brooks JF, Ray VA, Mandel MJ, Visick KL. 2015. CysK plays a role in biofilm formation and colonization by Vibrio fischeri. Appl Environ Microbiol 81:5223–5234. 10.1128/AEM.00157-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen C, Yan Q, Tao M, Shi H, Han X, Jia L, Huang Y, Zhao L, Wang C, Ma X, Ma Y. 2019. Characterization of serine acetyltransferase (CysE) from methicillin-resistant Staphylococcus aureus and inhibitory effect of two natural products on CysE. Microb Pathog 131:218–226. 10.1016/j.micpath.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 70.Rowbury RJ. 1997. Regulatory components, including integration host factor, CysB and H-NS, that influence pH responses in Escherichia coli. Lett Appl Microbiol 24:319–328. 10.1046/j.1472-765x.1997.00065.x. [DOI] [PubMed] [Google Scholar]
- 71.Shi X, Bennett GN. 1994. Effects of rpoA and cysB mutations on acid induction of biodegradative arginine decarboxylase in Escherichia coli. J Bacteriol 176:7017–7023. 10.1128/jb.176.22.7017-7023.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thulin E, Andersson DI. 2019. Upregulation of PBP1B and LpoB in cysB mutants confers mecillinam (amdinocillin) resistance in Escherichia coli. Antimicrob Agents Chemother 63:e00612-19. 10.1128/AAC.00612-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Farrow JM, Hudson LL, Wells G, Coleman JP, Pesci EC. 2015. CysB negatively affects the transcription of pqsR and Pseudomonas quinolone signal production in Pseudomonas aeruginosa. J Bacteriol 197:1988–2002. 10.1128/JB.00246-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Song Y, Yang C, Chen G, Zhang Y, Seng Z, Cai Z, Zhang C, Yang L, Gan J, Liang H. 2019. Molecular insights into the master regulator CysB-mediated bacterial virulence in Pseudomonas aeruginosa. Mol Microbiol 111:1195–1210. 10.1111/mmi.14200. [DOI] [PubMed] [Google Scholar]
- 75.Valentini M, Filloux A. 2016. Biofilms and cyclic di-GMP (c-di-GMP) signaling: lessons from Pseudomonas aeruginosa and other bacteria. J Biol Chem 291:12547–12555. 10.1074/jbc.R115.711507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Biers EJ, Wang K, Pennington C, Belas R, Chen F, Moran MA. 2008. Occurrence and expression of gene transfer agent genes in marine bacterioplankton. Appl Environ Microbiol 74:2933–2939. 10.1128/AEM.02129-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Solioz M, Marrs B. 1977. The gene transfer agent of Rhodopseudomonas capsulata. Purification and characterization of its nucleic acid. Arch Biochem Biophys 181:300–307. 10.1016/0003-9861(77)90508-2. [DOI] [PubMed] [Google Scholar]
- 78.Christie-Oleza JA, Scanlan DJ, Armengaud J. 2015. “You produce while I clean up”, a strategy revealed by exoproteomics during Synechococcus-Roseobacter interactions. Proteomics 15:3454–3462. 10.1002/pmic.201400562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Danese PN, Pratt LA, Kolter R. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J Bacteriol 182:3593–3596. 10.1128/JB.182.12.3593-3596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dickschat JS. 2010. Quorum sensing and bacterial biofilms. Nat Prod Rep 27:343–369. 10.1039/b804469b. [DOI] [PubMed] [Google Scholar]
- 81.Porter SL, Wadhams GH, Armitage JP. 2008. Rhodobacter sphaeroides: complexity in chemotactic signalling. Trends Microbiol 16:251–260. 10.1016/j.tim.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 82.Mercer RG, Callister SJ, Lipton MS, Pasa-Tolic L, Strnad H, Paces V, Beatty JT, Lang AS. 2010. Loss of the response regulator CtrA causes pleiotropic effects on gene expression but does not affect growth phase regulation in Rhodobacter capsulatus. J Bacteriol 192:2701–2710. 10.1128/JB.00160-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mercer RG, Quinlan M, Rose AR, Noll S, Beatty JT, Lang AS. 2012. Regulatory systems controlling motility and gene transfer agent production and release in Rhodobacter capsulatus. FEMS Microbiol Lett 331:53–62. 10.1111/j.1574-6968.2012.02553.x. [DOI] [PubMed] [Google Scholar]
- 84.Heindl JE, Crosby D, Brar S, Pinto JF, Singletary T, Merenich D, Eagan JL, Buechlein AM, Bruger EL, Waters CM, Fuqua C. 2019. Reciprocal control of motility and biofilm formation by the PdhS2 two-component sensor kinase of Agrobacterium tumefaciens. Microbiology (Reading) 165:146–162. 10.1099/mic.0.000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Westbye AB, Leung MM, Florizone SM, Taylor TA, Johnson JA, Fogg PC, Beatty JT. 2013. Phosphate concentration and the putative sensor kinase protein CckA modulate cell lysis and release of the Rhodobacter capsulatus gene transfer agent. J Bacteriol 195:5025–5040. 10.1128/JB.00669-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.D'Alvise PW, Magdenoska O, Melchiorsen J, Nielsen KF, Gram L. 2014. Biofilm formation and antibiotic production in Ruegeria mobilis are influenced by intracellular concentrations of cyclic dimeric guanosinmonophosphate. Environ Microbiol 16:1252–1266. 10.1111/1462-2920.12265. [DOI] [PubMed] [Google Scholar]
- 87.Farrera-Calderon RG, Pallegar P, Westbye AB, Wiesmann C, Lang AS, Beatty JT. 2021. The CckA-ChpT-CtrA phosphorelay controlling Rhodobacter capsulatus gene transfer agent production is bidirectional and regulated by cyclic di-GMP. J Bacteriol 203:e00525-20. 10.1128/JB.00525-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wall JD, Weaver PF, Gest H. 1975. Gene transfer agents, bacteriophages, and bacteriocins of Rhodopseudomonas capsulata. Arch Microbiol 105:217–224. 10.1007/BF00447140. [DOI] [PubMed] [Google Scholar]
- 89.Weaver PF, Wall JD, Gest H. 1975. Characterization of Rhodopseudomonas capsulata. Arch Microbiol 105:207–216. 10.1007/BF00447139. [DOI] [PubMed] [Google Scholar]
- 90.Maniatis T, Fritsch EF, Sambrook J. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 91.Adams CW, Forrest ME, Cohen SN, Beatty JT. 1989. Structural and functional analysis of transcriptional control of the Rhodobacter capsulatus puf operon. J Bacteriol 171:473–482. 10.1128/jb.171.1.473-482.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Leung M, Beatty J. 2013. Rhodobacter capsulatus gene transfer agent transduction assay. Bio Protoc 3:e334. [Google Scholar]
- 93.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 94.Fogg MJ, Wilkinson AJ. 2008. Higher-throughput approaches to crystallization and crystal structure determination. Biochem Soc Trans 36:771–775. 10.1042/BST0360771. [DOI] [PubMed] [Google Scholar]
- 95.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zimmermann L, Stephens A, Nam S-Z, Rau D, Kübler J, Lozajic M, Gabler F, Söding J, Lupas AN, Alva V. 2018. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J Mol Biol 430:2237–2243. 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 100.Gabler F, Nam S-Z, Till S, Mirdita M, Steinegger M, Söding J, Lupas AN, Alva V. 2020. Protein sequence analysis using the MPI bioinformatics toolkit. Curr Protoc Bioinformatics 72:e108. 10.1002/cpbi.108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S8. Download aem.00944-22-s0001.pdf, PDF file, 1.2 MB (1.2MB, pdf)
Tables S1 to S4. Download aem.00944-22-s0002.xlsx, XLSX file, 0.3 MB (257.9KB, xlsx)
Data Availability Statement
All data required to assess the findings are available in the main text or the supplemental information files. Reagents and materials used in this paper are publicly available. Any unique bacterial strains or recombinant DNA constructs described in this paper will be provided by the corresponding author upon request.