Abstract
Resistance to β-lactam antibiotics in staphylococci is mediated by mecA and blaZ, genes encoding a penicillin-binding protein (PBP2a) with low β-lactam affinity and β-lactamase, respectively. The mec and bla regulators, mecR1-mecI and blaR1-blaI, respectively, encode inducer-repressors with sufficient amino acid homology to suggest that they could coregulate PBP2a production. In order to test this hypothesis, plasmids containing mec and bla regulatory sequences were introduced into Staphylococcus aureus containing a chromosomal mecA-lacZ transcriptional fusion. Corepression was confirmed by demonstrating a gene dosage-dependent reduction in β-galactosidase activity by either MecI or BlaI and additive repression when both were present. Both MecI-MecI and BlaI-BlaI homodimer and MecI-BlaI heterodimer interactions were demonstrated in the yeast two-hybrid assay, and purified MecI and BlaI protected the same mec promoter-operator sequences. However, MecI was approximately threefold more effective at mecA-lacZ transcriptional repression than was BlaI. While MecI and BlaI displayed similar activity as repressors of mecA transcription, there was a marked difference between MecR1 and BlaR1 in the rate and specificity of induction. Induction through BlaR1 by a β-lactam was 10-fold greater than through MecR1 at 60 min and was 81% of maximal by 2 h, while induction through MecR1 never exceeded 20% of maximal. Furthermore, complementation studies showed that MecI- or BlaI-mediated mecA transcriptional repression could be relieved by induction through homologous but not heterologous sensor-inducer proteins, demonstrating the repressor specificity of induction.
The targets for β-lactam antibiotics are penicillin-binding proteins (PBPs), enzymes that function to cross-link peptidoglycan in the bacterial cell wall (7). Staphylococci can resist the action of β-lactam antibiotics by producing a new PBP, PBP2a, with reduced β-lactam affinity that is able to mediate essential cell wall construction when β-lactam-susceptible PBPs have been inactivated (14, 15). The gene that encodes PBP2a, mecA, was acquired from an unknown donor bacterium, together with 30 to 50 kb of additional DNA, and inserted at a specific chromosomal site (1, 6, 19). The mec complex also contains a two-gene operon, mecR1-mecI, that is divergently transcribed from and has overlapping promoter-operator regions with mecA (31, 34). mecR1 encodes a signal sensor/transducer with antirepressor activity, while mecI encodes a repressor of mecA transcription (21). In the absence of induction via MecR1 by a β-lactam antibiotic, mecA transcription is tightly repressed by mecI (31, 33).
Staphylococci can also resist the action of certain β-lactam antibiotics by producing an enzyme, β-lactamase, that inactivates these antibiotics by hydrolyzing the β-lactam ring. The gene encoding β-lactamase, blaZ, is usually plasmid encoded. β-Lactamase production is also regulated by two divergently transcribed genes, blaR1 and blaI, whose gene products have amino acid homology to mecR1 and mecI, respectively (9, 17, 38). There are similarities between the promoter-operator regions of the mecA and blaZ regulons, and both purified MecI and BlaI have been shown to bind to bla promoter-operator sequences in DNase protection assays (9).
On the basis of the similarities between blaZ and mecA regulatory sequences, it is reasonable to assume that each regulon may modulate the transcriptional expression of mecA. Previous studies provide evidence that mecA transcription and PBP2a production can be affected by BlaI. Hackbarth and Chambers (10) reported that PBP2a production was converted from unregulated and constitutive to inducibly repressed by the introduction of a plasmid containing blaR1 and blaI. In addition, Ryffel et al. (32)found that bla-mediated repression of mecA transcription was less stringent at baseline and more rapidly inducible than mec-mediated repression. However, the staphylococcal strains used in these studies contained uncharacterized mec regulatory sequences or were nonisogenic, and mecA transcription was not quantified.
In this study, we used constructs that differed only in terms of defined mutations in mec and bla regulatory sequences in the presence of mec-reporter gene fusions to quantify the effect of these regulatory sequences on mecA expression. In addition, we investigated the biochemical basis of mecA coregulation by comparing the binding of purified MecI and BlaI to mecA promoter-operator sequences and by assessing MecI-BlaI protein-protein interactions.
MATERIALS AND METHODS
All strains and plasmids used in this study are listed in Table 1. For routine culture, S. aureus strains were grown at 37°C in either Trypticase soy or brain heart infusion broth. Escherichia coli strains were grown in Luria broth. For solid media, agar-agar was added to broth at a final concentration of 1.5%. All media used for growth and maintenance of bacteria strains were purchased from Difco Laboratories, Detroit, Mich. For the culture of yeast strain Y190 (Clontech, Palo Alto, Calif.) and its derivatives, yeast extract-peptone-dextrose (YPD) or dropout base supplemented with complete synthetic medium lacking leucine, tryptophan, or both (Bio 101, Vista, Calif.) was used. Antibiotics were used at the following concentrations: ampicillin (Amp), 50 μg/ml for E. coli; chloramphenicol, 10 μg/ml; erythromycin, 10 μg/ml; minocycline, 1 μg/ml; and gentamicin, 1 μg/ml. All antibiotics and chemicals were purchased from Sigma Chemical Company, St. Louis, Mo. Restriction endonucleases and other enzymes were purchased from New England Biolabs, Beverly, Mass.
TABLE 1.
Plasmids and strains
| Plasmid or strain | Genotype or phenotype | Remarks and reference |
|---|---|---|
| Plasmids | ||
| pUC19 | Apr; 2.7 kb | General cloning vector (35) |
| pBluescript SK+ | Apr; 3.0 kb | General cloning vector (Stratagene) |
| pRN5542 | Cmr; 3 kb | Staphylococcal cloning vector, derivative of pC194 (25) |
| pJIM2246 | Cmr; 6.0 kb | Shuttle vector (27) |
| pGO621 | Apr; 5.2 kb | mecR1-mecI ligated into puc19 at EcoRI-BamHI sites (this study) |
| pGO621C | Apr Cmr; 8.2 kb | pGO621 with pSK265 ligated at BamHI site (this study) |
| pGO630 | Spr Tcr Emr ts; 16 kb | Promoterless lacZ gene from E. coli fused to first 50 bp of mecA, intact mecR1-mecI ligated into pSK950 (31) |
| pSK950 | Emr Spr Tcr ts; 10.4 kb | pCL84 with pE194ts at PstI site (24); integration vector |
| pGO640 | Apr; 5.2 kb | PstI-EaeI fragment containing intact blaZ, blaR1, and truncated blaI from pCH2278 ligated into PstI-NotI sites of pBluescript KS+ |
| pGO641 | Apr Emr; 9.5 kb | pE194 ligated into ClaI site of pGO640 (this study) |
| pCH2278 | Cmr; 6.2 kb | 3.2-kb EcoRI-XhoI fragment containing bla locus ligated into EcoRI-SalI sites of pRN5542 (10) |
| pCH1988 | Cmr; 6.2 kb | Same as pCH2278; contains linker insertion mutation in blaR1 (10) |
| pGO630ΔR1 | Spr Tcr Emr ts; 16 kb | mecR1 mutant of pGO630 created by filling in ClaI site present in mecR1 (this study) |
| pGO681 | Cmr; 8.0 kb | 2.0-kb BamHI-PstI fragment containing first 50 bp of mecA, intact mecR1, and mecI truncated at PstI site ligated into BamHI-PstI sites of pJIM2246 (this study) |
| pGO682 | Cmr; 8.8 kb | 2.8-kb PstI-EaeI fragment containing intact blaZ, blaR1, and blaI truncated at EaeI site ligated into PstI-NotI sites of pJIM2246 (this study) |
| pACT2 | Apr; 8.1 kb | Activation domain hybrid cloning vector (Clonetech) |
| pAS1 | Apr; 7.3 kb | DNA-binding domain hybrid vector (Clonetech) |
| S. aureus | ||
| RN450 | Cms Ems Gms Tcsbla | ATCC 8235-4 |
| RN4220 | Cms Ems Gms Tcsbla | Restriction-deficient mutagenized RN450 shuttle plasmid host (22) |
| GO450TF | Cms Ems Gms Tcsbla | Provided by Tim Foster |
| GO450TF::630 | Cms Emr Gms Tcrbla geh | GO450TF with integrated pGO630 (this study) |
| GO450TF::630ΔI | Cms Emr Gms Tcrbla geh | mecR1 mecI mutant derivative of GO450TF::630 created by allelic replacement (this study) |
| GO450TF::630ΔRI | Cms Emr Gms Tcrbla geh | mecR1 mecI+ derivative of GO450TF::630 created by allelic replacement (this study) |
| E. coli | ||
| DH5α | λ−φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 phoA hsdR17(r−K m+K) supE441 thi-1 gyrA96 relA91 glnV44 (7) | |
| Able C | lac (LacZw−) (Kanr McrA− McrB− McrF− Mrr− | |
| hsdR(rK− mK−) (F′ proAB laclqZΔM15 Tn10(Tetr) (Stratagene) | ||
| S. cerevisiae | ||
| Y190 | MATa ura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu2-3 gal4Δ gal80Δ cyhr2 LYS::GAL1uas−HIS3tata−HIS3 URA3::GAL1uas−GAL1tata−lacZ (Clontech) |
Cloning, transformations, and DNA manipulations.
All restriction endonuclease digestions and ligations were performed as directed by the manufacturer. E. coli DH5α and Able K competent cells, obtained from Gibco-BRL (Grand Island, N.Y.) and Stratagene (La Jolla, Calif.), respectively, were transformed according to the manufacturer's directions. Yeast strain Y190 was transformed by the lithium acetate method as described by Clontech (Clontech Matchmaker System Manual; Clontech Laboratories, Palo Alto, Calif.). Small-scale isolation of plasmid DNA from E.coli and S. aureus was done by the method of Birnboim and Doly (3) and the hexadecyltrimethylammonium bromide extraction method of Townsend et al. (35), respectively. Large-scale plasmid DNA isolations were obtained using Qiagen Midi Prep affinity columns (Qiagen, Chatsworth, Calif.). Plasmid DNA was isolated from Saccharomyces cerevisiae using the RPM yeast plasmid isolation kit (Bio 101, Vista, Calif.). Recombinant plasmids were transferred between S. aureus strains by transduction as described by Sharma et al. (33).
β-Galactosidase activity.
An overnight culture diluted in warmed BHI or BHI supplemented with β-lactam inducer was adjusted to an optical density at 600 nm (OD600) of approximately 0.050 and shaken at 37°C until an OD600 of ≈0.600 was reached (≈2 h). Cells were harvested by centrifugation and stored overnight at −80°C. The thawed pellet was resuspended in 1 ml of Z buffer (0.06 M Na2HPO4, 0.04 M NaH2PO4, 0.01 M KCl, 0.001 M MgSO4, 0.05 M β-mercaptoethanol, pH 7.0), and 500 μl of the suspension plus an additional 500 μl of Z buffer was added to 2.5 g of 1-mm Zirconia beads (Biospec Products, Bartlesville, Okla.). Cell lysates were produced by bead beating for 5 min at 4°C using the Biospec Products (Bartlesville, Okla.) bead beater, and lysates were centrifuged for 10 min at 26,895 × g. A 50- to 100-μl sample was taken for assay of β-galactosidase activity using ONPG (o-nitrophenyl-β-d-galactoside) as the substrate. Activity was expressed as Miller units per milligram of protein. Protein concentration was determined with the Bio-Rad protein assay (Bio-Rad, Hercules, Calif.)
DNase I footprinting.
DNA fragments used for DNase I footprinting were prepared by PCR amplification, and the procedure was performed as described by Sharma et al. (33).
Construction of Gal4 protein fusion plasmids and production of yeast extracts.
A 422-bp BamHI fragment from plasmid pGO443 containing the mecI coding sequence was cloned into the BamHI site of pACT2 and pAS1. The blaI coding sequence was obtained from plasmid pI6187 by PCR amplification with the following primers: blaI upstream primer, 5′AA ATG GGT GTT TCA AAT G 3′ (bold C denotes a T in the wild-type sequence, change made to prevent early termination), and blaI downstream primer, 5′GAC ACT GAA TTT GCT C 3′. The blaI PCR products were purified by gel electrophoresis and cloned into the SmaI site of pAS1 and pACT2. To confirm the presence of the in-frame junction from both fusion constructs, recombinant plasmids were sequenced using the Dye Terminator cycle sequencing kit (Applied Biosystems, Foster City, Calif.) and the ABI 377 DNA sequencer.
Cell extracts were prepared by a modification of the method of Rose and Botstein (30). Yeast cells grown at 30°C to stationary phase in dropout base supplemented with complete synthetic medium lacking leucine and tryptophan were diluted 1:10 into fresh medium and grown at 30°C to an OD600 of ≈0.4 to 0.5. Then 5-ml aliquots were pelleted by centrifuging for 5 min at 4,066 × g. Cells were washed twice with equal volumes of ice-cold water. Pellets were stored frozen overnight at −80°C. The frozen pellet was resuspended in 250 μl of breaking buffer (100 mM Tris-HCl, 1 mM dithiothreitol, and 20% glycerol). The suspension was added to 250 μl of 0.5-mm glass beads. Cell lysates were produced by five cycles of 1-min pulses followed by 1 min of incubation on ice using the Biospec Products bead beater. We used 100 μl of sample for assay of β-galactosidase activity, using ONPG as the substrate. Activity was expressed as nanomoles of ONPG hydrolyzed per minute per milligram of protein.
Western blot analysis.
For Western immunoblot analysis, S. aureus whole-cell lysates were prepared as described by Gregory et al. (9).We resolved 80 μg of total protein in 16.5% Tris–tricine minigels by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. The membranes were blocked with 5% skim milk in TBST (20 mM Tris-HCl, 500 mM NaCl, 0.05% Tween 20, pH 7.5) for 1 h and incubated with rabbit anti-MecI polyclonal antiserum diluted 1:100 with the blocking solution overnight at 4°C. The secondary antibody was horseradish peroxidase-conjugated donkey anti-rabbit immunoglobulin (Amersham Pharmacia Biotech Inc., Piscataway, N.J.). Bound antibodies were detected by color development using nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate (Bio-Rad Laboratories, Richmond, Calif.) or by chemiluminescence with the ECL Western blot detection system (Amersham Pharmacia Biotech Inc., Piscataway, N.J.).
Constructs used for mecA transcriptional analysis.
mecA transcription was assessed by analysis of β-galactosidase activity from a mecA-lacZ fusion. The previously described plasmid pGO630 (33) contains the 5′ 50 bp of mecA transcriptionally fused to lacZ. mecA is preceded by the mecA-mecR1 intergenic region, which contains both the mecA and mecR1 promoter-operator sequences, followed by the mecR1 and mecI regulatory genes. This plasmid was introduced into the chromosomal phage L54a att site of methicillin-susceptible S. aureus strain GO450TF, interrupting the lipase (geh) gene. This strain was designated GO450TF::630.
For complementation studies, a mecR1 mutant of pGO630 was created by filling in at the ClaI site of mecR1, producing a frameshift mutation. This plasmid, pGO630ΔR1, was introduced into the chromosome of strain GO450TF, and the strain was designated GO450TF::630ΔR1. Plasmid pCH2278 and derivatives (10), containing blaZ, blaR1, and blaI on a high-copy staphylococcal replicon (pRN5542), provided bla regulation.
The studies reported below were performed in a methicillin-susceptible S. aureus strain in order to avoid the possible confounding effects of having more than one copy of mec regulatory targets or other mec regulatory genes in the same cell. However, most of the studies reported for GO450TF were duplicated in GO450M, a derivative of GO450 containing the chromosomal mec region (26). Differences in β-galactosidase activity in the presence of mec and bla regulatory sequences were the same as those generated in GO450TF (data not shown). Likewise, the β-lactamase regulatory genes were removed from their usual wild-type plasmid location and ligated into another replicon both to avoid the possible confounding effect of other genes contained on the naturally occurring penicillinase plasmids and to afford comparable gene dosages when different plasmid-borne regulatory genes were compared. However, results obtained with pCH2278 were repeated with the naturally occurring penicillinase plasmid pI6187 (28) and found to be similar despite the higher copy number of pCH2278 (data not shown).
RESULTS
Binding of MecI and BlaI to mec promoter-operator sequences.
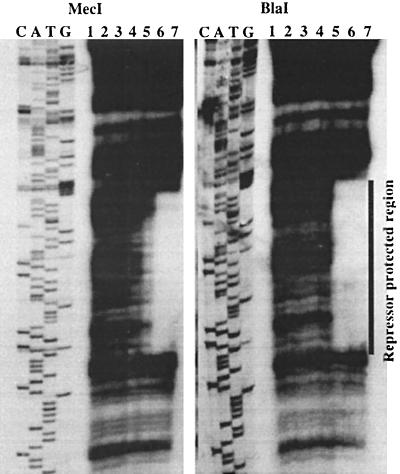
MecI and BlaI were purified as glutathione-S-transferase fusion proteins and cleaved from the fusion as previously described (33). Equal amounts of cleaved, purified MecI and BlaI proteins, as estimated by Coomassie blue staining of polyacrylamide gels, were used in DNase protection assays. A 210-bp DNA fragment containing the mecA-mecR1 intergenic region, 16 bp of the 5′ end of mecA, and 6 bp of the 5′ end of mecR1 was used as the mec target. As previously described (33), MecI bound to a 43-bp region of mec DNA that included a 30-bp palindrome with 15 bp of dyad symmetry, the mecA −10 promoter sequence, and the mecR1 −35 sequence (Fig. 1). As shown in Fig. 1, BlaI protected the same sequences as MecI.
FIG. 1.
DNase I protection analysis. DNase I footprint of the 210-bp target fragment containing the region between the mecA and mecR1 translational start sites. MecI and BlaI protected regions are shown. Lanes 1 through 7 contain 0, 0.001, 0.01, 0.05, 0.15, 0.4, and 1 mg of MecI or BlaI, respectively. C, A, T, and G are the labeled nucleotides of the sequencing ladder generated from each target fragment by using primers complementary to the 5′ end. The sequence is read from bottom (5′end) to ttop (3′ end). This image was scanned from the original film by using an AlphaImager 1000 (Alpha Innotech) and was cropped and labeled by using Canvas 5 graphics software (Deneba, Miami, Fla). The same systems were used to prepare Fig. 2.
Repression of mecA transcription.
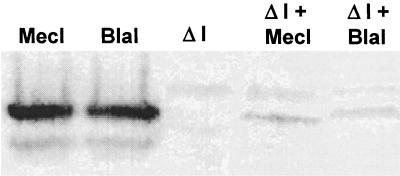
Maximal unregulated transcription from a chromosomal mecA-lacZ fusion was determined after inactivation of mecR1 and mecI by allelic replacement with the tetM gene, as described previously (26). As shown in Table 2, β-galactosidase activity from a mecR1 mecI mutant (GO450TF::630ΔΙ) was 66-fold greater than the activity from the regulated chromosomal gene (GO450TF::630). A gene dosage effect of MecI-mediated repression was shown when plasmid-encoded MecI (pGO621C) was introduced into the strain containing the regulated chromosomal mecA-lacZ fusion (GO450TF::630). β-Galactosidase activity decreased more than sixfold in the presence of plasmid-encoded MecI. Similarly, there was an almost twofold decrease in transcription in the presence of high-copy blaI (pCH2278). Thus, there was additive repression provided by the increased gene dosage of plasmid-encoded repressors over that seen when only the chromosomal copy of mecI was present. However, there was more repression mediated by the MecI-containing plasmid constructs than by those containing BlaI. This was despite the observation, shown in the Western blot in Fig. 2, that approximately the same amounts of BlaI and MecI were produced by each of the plasmids. The increased repression of MecI over BlaI was also seen when a plasmid encoding each of the repressors was introduced into the strain containing an unregulated chromosomal mecA-lacZ fusion (GO450TF::630ΔI; Table 2).
TABLE 2.
Determination of mec- and bla-mediated repression and induction of mecA expression
| Strain | Relevant genotypea | β-Galactosidase activityb with CBAP at:
|
Fold inductionc | |
|---|---|---|---|---|
| 0 μg/ml | 10 μg/ml | |||
| GO450TF::630 | mecR1+mecI+ (c) | 355 ± 94 | 4,404 ± 386 | 12 |
| GO450TF::630 + pGO621C | mecR1+mec1+ (c) | 54 ± 12.5 | 4,291 ± 459 | 80 |
| mecR1+mec1+ (p) | ||||
| GO450TF::630 + pCH2278 | mecR1+mecI (c) | 187 ± 87.5 | 10,597 ± 893 | 57 |
| blaR1+blaI+ (p) | ||||
| GO450TF::630ΔI | mecR1 ΔmecI (c) | 23,265 ± 1,972 | 21,461 ± 2,691 | 0 |
| GO450TF::630ΔI + pGO621C | mecR1 ΔmecI (c) | 195 ± 95 | 5,676 ± 124 | 29 |
| mecR1+mec1+ (p) | ||||
| GO450TF::630ΔI + pCH2278 | mecR1 ΔmecI (c) | 431 ± 174 | 18,811 ± 182 | 44 |
| blaR1+blaI+ (p) | ||||
c, chromosomal; p, plasmid borne.
Miller units per milligram of protein per hour. Means of at least three experiments ± standard deviation.
Fold increase in activity as calculated by the activity obtained in the presence of mecI following a 2-h exposure to CBAP divided by the activity in the absence of mecI and/or blaI.
FIG. 2.
Western immunoblot analysis showing relative affinity of anti-MecI polyclonal antiserum for MecI and BlaI. In the first and second lanes, equal amounts of purified MecI and BlaI, as determined by Coomassie staining, were probed to demonstrate the equal affinity of anti-MecI antiserum for both repressors. The other lanes illustrate the production of repressor protein from plasmid constructs: GO450::630ΔI alone, third lane; GO450::630ΔI plus pGO621C (MecI), fourth lane; and GO450::630ΔI plus pCH2278 (BlaI), last lane.
Analysis of MecI and BlaI interactions.
The gene dosage effect, by which increasing amounts of repressor led to increasing transcriptional repression, may have been due in part to protein-protein interactions. A previous study suggested that optimal repression was dependent on the ability of MecI to form dimers and higher-0order oligomers (33). We used the yeast two-hybrid assay to assess the formation of MecI-MecI homodimers and MecI-BlaI heterodimers.
mecI and blaI were cloned in the correct orientation and reading frame to generate a hybrid protein consisting of the repressor protein and either the activation or binding domain of the yeast protein Gal4. The resulting plasmids were transformed into yeast strain Y190. Protein-protein interactions were detected by an increase in β-galactosidase activity and by growth on synthetic medium lacking histidine and containing 25 to 50 mM 3-aminotriazole, an inhibitor of yeast HIS3-encoded imidazoleglycerol-phosphate dehydratase. As shown in Table 3, MecI and BlaI interacted with both homologous (MecI-MecI and BlaI-BlaI homodimers) and heterologous (MecI-BlaI heterodimers) proteins. Interestingly, the strength of the interactions, as measured by β-galactosidase activity, was as great between heterodimers as between homodimers. Neither mecI nor blaI cloned into either the activation or binding domains alone produced a significant increase in β-galactosidase activity or allowed growth on His auxotrophic medium.
TABLE 3.
Analysis of MecI and BlaI interactions with the yeast two-hybrid systema
| Plasmids | β-Galactosidase activityb |
|---|---|
| pAS1-mecI + pACT2-mecI | 276 ± 69 |
| pAS1-blaI + pACT2-blaI | 287 ± 127 |
| pAS1-mecI + pACT2-blaI | 300 ± 70 |
| pAS1-mecI + pACT2 | 55 ± 19 |
| pAS1 + pACT2-blaI | 47 ± 17 |
| pAS1 + pACT | 11 ± 4 |
Reporter strain Y190 was transformed with the indicated plasmids.
Nanomoles of ONPG hydrolyzed per minute per milligram of protein. Means of at least two assays ± standard deviation.
Induction of mecA transcription.
MecI-repressed mecA transcription was induced from the chromosomal copy of mec regulators (GO450TF::630) with CBAP (2-[2′-carboxyphenyl] benzoyl-β-aminopenicillanic acid) at concentrations of 1, 5, 10, and 20 μg/ml. Maximal induction without growth inhibition was achieved at 10 μg/ml. As shown in Table 2, after 2 h of induction, CBAP at 10 μg/ml produced only a 12-fold induction of MecI-repressed mecA transcription, a value that represented only 19% of unregulated transcription seen in the mecI mutant (GO450TF::630ΔI). There was a much greater induction from the plasmid-encoded copy of mec regulators (pGO621C) due to increased baseline repression, but induction still reached only 18% of the unregulated value. In contrast, induction by CBAP at 10 μg/ml of BlaI-repressed mecA transcription in the mecR1 mecI mutant resulted in a 44-fold increase in mecA transcription that was 81% of maximal.
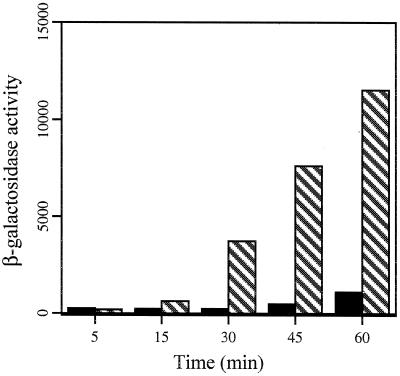
The relative induction through BlaR1 or MecR1 by two different β-lactam antibiotics was also assessed. CBAP (FW 440.8), a penicillin, was compared to cefoxitin (FW 449.4), a cephalosporin, at concentrations that did not inhibit growth (1 μg/ml). As shown in Table 4, both CBAP and cefoxitin were from 5- (cefoxitin) to 20-fold (CBAP) better inducers through BlaR1 than MecR1. In addition, cefoxitin was an eightfold better inducer of mecA than was CBAP through MecR1 while cefoxitin was only twofold better than CBAP through BlaR1, demonstrating differential responses to β-lactams of different classes. Induction over time was also assessed in MecI- and BlaI-repressed systems by measuring β-galactosidase activity at 15-min intervals for 1 h after induction with CBAP (10 μg/ml). As shown in Fig. 3, induction through BlaR1 occurred more rapidly and completely than through MecR1, beginning at 15 min for BlaR1 versus 45 min for MecR1, and achieving 50% of maximal at 60 min for BlaR1 versus only 4.8% for MecR1.
TABLE 4.
Differential induction of mecA expression
| Strain | Genotypea | β-Galactosidase activityb with:
|
Ratio, cefoxitin/CBAPd | ||
|---|---|---|---|---|---|
| No addition | CBAP (1 μg/ml) | Cefoxitin (1 μg/ml) | |||
| GO450TF::630ΔI + pGO621C | mecR1 ΔmecI (c) | 226 ± 103 | 263 ± 76 (1)c | 2,197 ± 809 (10) | 8 |
| mecR1+mecI+ (p) | |||||
| GO450TF::630ΔI + pCH2278 | mecR1 ΔmecI (c) | 350 ± 139 | 7,812 ± 831 (22) | 17,055 ± 1,419 (49) | 2 |
| blaR1+blaI+ (p) | |||||
c, chromosomal; p, plasmid borne.
Miller units per milligram of protein per hour. Means of at least three experiments ± standard deviation.
Fold increase in activity as calculated by the activity in the presence of a β-lactam divided by the activity in the absence of β-lactams is shown in parentheses.
Fold increase in activity for cefoxitin versus CBAP induction as calculated by the activity in the presence of cefoxitin divided by the activity in the presence of CBAP.
FIG. 3.
Time course comparing induction of mecA expression via MecR1 and BlaR1. β-Galactosidase activity is in Miller units per milligram of protein per hour. Samples were taken at the indicated times after induction with 10 μg of CBAP per ml. The solid bars represent CBAP induction of strain GO450TF::630ΔI (mecR1 mecI) plus pGO621C (mecR1+ mecI+); the striped bars represent induction of strain GO450TF::630ΔI (mecR1 mecI) plus pCH2278 (blaR1+ blaI+).
We next sought to assess whether induction of repressed mecA transcription could be achieved through heterologous sensor signal transducers. The following strains were constructed (see Table 1 for detailed descriptions): strain GO450TF::630ΔR1, chromosomal mecR1 mecI+ strain containing plasmid pGO682 (blaR1+ blaI) created by deletion of blaI at the EaeI site; GO450TF::630ΔR1 containing plasmid pGO681 (mecR1+ mecI), created by deletion of mecI at the PstI site; and GO450TF::630ΔI containing pCH1988 (blaR1 blaI+) (10) and pGO641 (blaR1+ blaI). Constructs were confirmed by DNA sequence analysis. The bla sequences on pGO641 were identical to those on pGO682. The two plasmids differed by staphylococcal replicons and antibiotic resistance markers to allow complementation to be performed in different backgrounds.
Complementation of mutations was assessed by repression and induction of β-galactosidase production from the chromosomal mecA-lacZ fusion. Results are shown in Table 5. Induction of mecA transcription with CBAP at 10 μg/ml was achieved only by complementation with the homologous sensor-signal transducer, mecR1. No induction was achieved when the mecR1 mutation was complemented by blaR1, yet blaR1 could complement the homologous plasmid-encoded blaR1 mutation. Thus, there was no cross-induction by a sensor-signal transducer for the heterologous repressor.
TABLE 5.
Complementation of mecA repression and induction with homologous and heterologous mec regulatorsa
| Strain | Relevant genotype | β-Galactosidase activity
|
Fold inductionb | |
|---|---|---|---|---|
| No inducer | CBAP (10 μg/ml) | |||
| GO450TF::630ΔRI | mecR1 mecI+ (c) | 254 ± 40 | 285 ± 24 | 1 |
| GO450TF::630ΔRI + pGO681 | mecR1 mecI+ (c) | 286 ± 37 | 2,824 ± 630 | 10 |
| mecR1+ ΔmecI (p) | ||||
| GO450TF::630ΔRI + pGO682 | mecR1 mecI+ (c) | 318 ± 45 | 345 ± 31 | 1 |
| blaR1+ ΔblaI (p) | ||||
| GO450TF::630ΔI + pGO641 + pCH1988 | mecR1 ΔmecI (c) | 198 ± 43 | 15,639 ± 5,016 | 79 |
| blaR1+ ΔblaI (p) | ||||
| blaR1 blaI+ (p) | ||||
See Table 4, footnotes a and b.
Value with no inducer divided by value with CBAP at 10 μg/ml.
DISCUSSION
Investigators have previously shown that the genes regulating production of β-lactamase can also regulate PBP2a production or mecA transcription (10, 31, 36). We have also recently demonstrated, using Northern blots in clinical isolates containing β-lactamase plasmids and mecI allelic replacement knockouts, that BlaI also regulates mecA transcription in Staphylococcus epidermidis. (5). We extended these data by demonstrating that purified MecI and BlaI also bound to the same mecA-mecR1 promoter-operator sequences. Gregory et al. (9) demonstrated that both MecI and BlaI also bound the same bla promoter-operator sequences, and Lewis and Dyke (25) showed that both could regulate β-lactamase production. While the overall sequence homology of the mecA-mecR1 and blaZ-blaR1 intergenic regions is not high (59%), there is 81% homology between the arms of the respective mec and bla promoter-operator dyads.
In order to compare the activities of MecI and BlaI as repressors, we assessed the ability of plasmid-encoded regulators to affect transcription of a mecA-lacZ transcriptional fusion. We have previously shown that mecA can be regulated by MecI and BlaI in methicillin-resistant S. aureus isolates using Northern blots (T. Dickinson and G. Archer, unpublished observations). All comparisons were made in the same genetic background, and both plasmid-encoded repressors were provided in similar amounts, as shown by Western blot analysis. Several observations were notable. First, there was a gene dosage effect of MecI, so that when repressor was provided on a high-copy plasmid there was an increase in mecA transcriptional repression over that seen when MecI was encoded on the chromosome. Furthermore, there was additive repression between either plasmid-encoded MecI or BlaI and chromosomal MecI.
The explanation for the direct association of mecA transcription with repressor quantity may be progressive saturation of unsaturated binding sites. The amount of MecI produced from the chromosomal gene may be insufficient, due to autoregulation, to completely saturate binding sites. This provides sufficient low-level transcription of mecA to mediate β-lactam resistance and of mecR1 to mediate induction. As repressor quantity increases, protein-protein interactions may also promote target saturation. Repressors with amino-terminal helix-turn-helix motifs like MecI and BlaI bind to DNA as dimers (13). Dimerization is energetically coupled to operator binding, and monomers and dimers are in dynamic equilibrium (2, 20). We used the yeast two-hybrid assay to demonstrate the ability of MecI and BlaI to form both homodimers and heterodimers with equal efficiency. In addition, we have previously associated formation of higher-order protein oligomers with optimum MecI repression (33), a finding also noted for the lambda cI repressor (20). Thus, increasing quantities of either MecI or BlaI could shift the equilibrium from monomers to dimers and higher-order oligomers, promoting more rapid and complete saturation of repressor binding sites.
Whereas the MecI and BlaI repressors were virtually interchangeable with regard to target interactions and activity, there was a marked difference between the two inducers. First, induction of mecA transcription was more rapid and more complete through BlaR1 than through MecR1. Second, there was a difference in the relative responses of the two inducers to low concentrations of structurally different β-lactam antibiotics: CBAP, a penicillin, and cefoxitin, a cephalosporin. Finally, there was a lack of cross-induction by inducer (BlaR1) for the heterologous repressor (MecI).
The differential response of the two inducers may be due to a difference in amino acid composition between the sensor regions of BlaR1 and MecR1. BlaR1 from Bacillus licheniformis is a 606-amino-acid transmembrane protein with its carboxy terminus external to the cell membrane. The terminal 250 amino acids contain motifs for β-lactam binding and have been shown to complex with β-lactam antibiotics (22, 39). While the β-lactam binding motifs are identical between sequences from Bacillus and staphylococcal BlaR1 and MecR1, there is only 48% amino acid identity between the entire staphylococcal BlaR1 and MecR1 carboxy-terminal sensor regions. This low homology could easily explain differences in the rapidity of induction and β-lactam selectivity of the two proteins.
The specificity of each inducer for its homologous repressor may also be related to sequence differences between the predicted cytoplasmic portions of BlaR1 and MecR1. Zhang et al. (38) recently showed that induction through BlaR1 produced a series of reactions whereby BlaR1 first underwent autoproteolysis, converting itself into a protease. Activated BlaR1 then inactivated BlaI by site-specific proteolytic cleavage. We have evidence that MecI is cleaved at a site very similar to the one in BlaI following β-lactam induction through MecR1 (J. M. Bosilevac and G. Archer, unpublished data). Signal transduction is thought to occur through four membrane-spanning segments, inducing conformational change in two cytoplasmic domains, one of which contains the zinc metalloprotease site (38). The existence of additional chromosomal genes that are required for induction has also been postulated (4). Since there is only 13% homology between the MecR1 and BlaR1 cytoplasmic domains outside the metalloprotease motif, the specificity for induction by homologous inducers is probably due to specific interactions between inducer and repressor or inducer and an intermediate molecule. While there are examples of proteolytic cleavage as a mechanism of gene regulation, such as in the inactivation of alternative sigma factors in recovery from the heat shock response (16), response regulation through repressor-specific proteolytic inactivation, as exemplified by BlaR1 and MecR1, constitutes a unique mechanism for signal transduction that may have homologues among other bacteria.
ACKNOWLEDGMENT
This work was supported in part by USPHS grant R37 AI35705 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Archer G L, Niemeyer D M, Thanassi J A, Pucci M J. Dissemination among staphylococci of DNA sequences associated with methicillin resistance. Antimicrob Agents Chemother. 1994;38:447–454. doi: 10.1128/aac.38.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beckett D, Koblan K S, Ackers G K. Quantitative study of protein association at picomolar concentration: the lambda phage cI repressor. Anal Biochem. 1991;196:69–75. doi: 10.1016/0003-2697(91)90118-d. [DOI] [PubMed] [Google Scholar]
- 3.Birnboim H, Doly J. A rapid extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1525. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen S, Sweeney H M. Constitutive penicillinase formation in Staphylococcus aureus owing to mutations unlinked to the penicillinase plasmid. J Bacteriol. 1968;95:1368–1374. doi: 10.1128/jb.95.4.1368-1374.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dickinson T M, Archer G L. Phenotypic expression of oxacillin resistance in Staphylococcus epidermidis: roles of mecA transcriptional regulation and resistant-subpopulation selection. Antimicrob Agents Chemother. 2000;44:1616–1623. doi: 10.1128/aac.44.6.1616-1623.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fey P D, Climo M W, Archer G L. Determination of the chromosomal relationship between mecA and gyrA in methicillin-resistant coagulase-negative staphylococcus. Antimicrob Agents Chemother. 1998;42:306–312. doi: 10.1128/aac.42.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghuysen J M. Molecular structures of penicillin binding proteins and beta-lactamases. Trends Microbiol. 1994;2:372–380. doi: 10.1016/0966-842x(94)90614-9. [DOI] [PubMed] [Google Scholar]
- 8.Grant S G N, Jesse J, Bloom F R, Hanahan D. Differential plasmid rescue from transgenic mouse DNA into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gregory P D, Lewis R A, Curnock S P, Dyke K G H. Studies of the repressor (BlaI) of β-lactamase synthesis in Staphylococcus aureus. Mol Microbiol. 1997;24:1025–1037. doi: 10.1046/j.1365-2958.1997.4051770.x. [DOI] [PubMed] [Google Scholar]
- 10.Hackbarth C J, Chambers H F. blaI and blaR1 regulate β-lactamase and PBP2a production in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1993;37:1144–1149. doi: 10.1128/aac.37.5.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hackbarth C J, Mick C, Chambers H F. Altered production of penicillin-binding protein 2a can affect phenotypic expression of methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1994;38:2568–2571. doi: 10.1128/aac.38.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardt K, Joris B, Lepage S, Brasseur R, Lampen J O, Frere J M, Fink A L, Ghuysen J M. The penicillin sensory transducer, BlaR, involved in the inducibility of β-lactamase synthesis in Bacillus licheniformis is embedded in the plasma membrane via a four-α-helix bundle. Mol Microbiol. 1997;23:935–944. doi: 10.1046/j.1365-2958.1997.2761642.x. [DOI] [PubMed] [Google Scholar]
- 13.Harrison S C, Aggarwal A K. DNA recognition by proteins with the helix-turn-helix motif. Annu Rev Biochem. 1990;59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- 14.Hartman B J, Tomasz A. Expression of methicillin resistance in heterogeneous strains of Staphylococcus aureus. Antimicrob Agents Chemother. 1986;29:85–92. doi: 10.1128/aac.29.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartman B J, Tomasz A. Low-affinity penicillin-binding protein associated with β-lactam resistance in Staphylococcus aureus. J Bacteriol. 1984;158:513–516. doi: 10.1128/jb.158.2.513-516.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herman C, Thevant P, D'Ari R, Bouloc P. Degradation of ς32, the heat shock regulator in Escherichia coli, is governed by HflB. Proc Natl Acad Sci USA. 1995;92:3516–3520. doi: 10.1073/pnas.92.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiramatsu K. Molecular evolution of MRSA. Microbiol Immunol. 1995;39:531–543. doi: 10.1111/j.1348-0421.1995.tb02239.x. [DOI] [PubMed] [Google Scholar]
- 18.Horinouchi S, Weisblum B. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibiotics. J Bacteriol. 1982;150:804–814. doi: 10.1128/jb.150.2.804-814.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito T, Katayama Y, Hiramatsu K. Cloning and nucleotide sequence determination of entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315P. Antimicrob Agents Chemother. 1999;43:1449–1458. doi: 10.1128/aac.43.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson A D, Poteete A R, Laucr G, Sauer R T, Ackers G K, Ptashne M. Lambda repressor and Cro: components of an efficient switch. Nature. 1981;294:217–223. doi: 10.1038/294217a0. [DOI] [PubMed] [Google Scholar]
- 21.Joris B, Hardt K, Ghuysen J M. Induction of β-lactamase and low-affinity penicillin-binding protein 2′ synthesis in gram-positive bacteria. In: Ghuysen J M, Hackenbeck R, editors. Bacterial cell wall. New York, N.Y: Elsevier Science; 1994. pp. 505–515. [Google Scholar]
- 22.Joris B, Ledent P, Kobayashi T, Lampen J O, Ghuysen J M. Expression in Escherichia coli of the Met346-Arg601 carboxyl terminal protein of Bacillus licheniformis as a water-soluble 26000 Mr penicillin-binding protein. FEMS Microbiol Lett. 1990;70:107–113. doi: 10.1016/0378-1097(90)90111-3. [DOI] [PubMed] [Google Scholar]
- 23.Kreiswirth B N, Lofdahl A, Betley M J, O'Reilly M, Schlievert P M, Bergdoll M S, Novick R P. The toxic shock exotoxin structural gene is not detectably transmitted by a phage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 24.Lee C Y, Buranen S L, Ye Z. Construction of a single-copy integration vector for Staphylococcus aureus. Gene. 1991;103:101–105. doi: 10.1016/0378-1119(91)90399-v. [DOI] [PubMed] [Google Scholar]
- 25.Lewis R A, Dyke K G H. MecI represses synthesis from the β-lactamase operon of Staphylococcus aureus. J Antimicrob Chemother. 2000;45:139–144. doi: 10.1093/jac/45.2.139. [DOI] [PubMed] [Google Scholar]
- 26.Niemeyer D M, Pucci M J, Thanassi J A, Sharma V K, Archer G L. Role of mecA transcriptional regulation in the phenotypic expression of methicillin resistance in Staphylococcus aureus. J Bacteriol. 1996;178:5464–5471. doi: 10.1128/jb.178.18.5464-5471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novick R P. Genetic systems of staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- 28.Novick R P, Murphy E, Gryczan T J, Baron E, Edelman I. Penicillinase plasmids of Staphylococcus aureus: restriction-deletion maps. Plasmid. 1979;2:109–129. doi: 10.1016/0147-619x(79)90010-6. [DOI] [PubMed] [Google Scholar]
- 29.Renault P, Corthier G, Goupil N, Delorme C, Ehrlich S D. Plasmid vectors for Gram-positive bacteria switching from high to low copy number. Gene. 1996;183:175–182. doi: 10.1016/s0378-1119(96)00554-9. [DOI] [PubMed] [Google Scholar]
- 30.Rose M, Botstein D. Construction and use of gene fusions with lacZ (β-galactosidase) which are expressed in yeast. Methods Enzymol. 1983;101:167–180. doi: 10.1016/0076-6879(83)01012-5. [DOI] [PubMed] [Google Scholar]
- 31.Ryffel C, Kayser F H, Berger-Bächi B. Correlation between regulation of mecA transcription and expression of methicillin resistance in staphylococci. Antimicrob Agents Chemother. 1992;36:25–31. doi: 10.1128/aac.36.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryffel C, Strassle A, Kayser F, Berger-Bächi B. Mechanisms of heteroresistance in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1994;38:724–728. doi: 10.1128/aac.38.4.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharma V K, Hackbarth C J, Dickinson T M, Archer G L. Interaction of native and mutant MecI repressors with sequences that regulate mecA, the gene encoding penicillin-binding protein 2a in methicillin-resistant staphylococci. J Bacteriol. 1998;180:2160–2166. doi: 10.1128/jb.180.8.2160-2166.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki E, Kuwahara-Arai K, Richardson J F, Hiramatsu K. Distribution of mec regulator genes in methicillin-resistant Staphylococcus clinical strains. Antimicrob Agents Chemother. 1993;37:1219–1226. doi: 10.1128/aac.37.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Townsend D E, Ashdown N, Bolton J, Grubb W B. The use of cetyltrimethylammoniumbromide for the rapid isolation from Staphylococcus aureus of relaxable and nonrelaxed plasmid DNA suitable for in vitro manipulation. Lett Appl Microbiol. 1985;1:87–94. [Google Scholar]
- 36.Ubukata K, Nonoguchi R, Matsuhashi M, Konno M. Expression and inducibility in Staphylococcus aureus of the mecA gene, which encodes a methicillin-resistant S. aureus-specific penicillin-binding protein. J Bacteriol. 1989;171:2882–2885. doi: 10.1128/jb.171.5.2882-2885.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13 mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 38.Zhang H Z, Hackbarth C J, Chansky K M, Chambers H F. A proteolytic transmembrane signaling pathway and resistance to β-lactams in staphylococci. Science. 2001;291:1962–1965. doi: 10.1126/science.1055144. [DOI] [PubMed] [Google Scholar]
- 39.Zhu Y F, Englebert S S, Joris B, Ghuysen J M, Kobayashi T, Lampen J O. Structure, function, and fate of the BlaR signal transducer involved in induction of β-lactamase in Bacillus licheniformis. J Bacteriol. 1992;174:6171–6178. doi: 10.1128/jb.174.19.6171-6178.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]