Abstract
Biomembrane order, dynamics, and other essential physicochemical parameters are controlled by cholesterol, a major component of mammalian cell membranes. Although cholesterol is well known to exhibit a condensing effect on fluid lipid membranes, the extent of stiffening that occurs with different degrees of lipid acyl chain unsaturation remains an enigma. In this review, we show that cholesterol locally increases the bending rigidity of both unsaturated and saturated lipid membranes, suggesting there may be a length-scale dependence of the bending modulus. We review our published data that address the origin of the mechanical effects of cholesterol on unsaturated and polyunsaturated lipid membranes and their role in biomembrane functions. Through a combination of solid-state deuterium NMR spectroscopy and neutron spin-echo spectroscopy, we show that changes in molecular packing cause the universal effects of cholesterol on the membrane bending rigidity. Our findings have broad implications for the role of cholesterol in lipid–protein interactions as well as raft-like mixtures, drug delivery applications, and the effects of antimicrobial peptides on lipid membranes.
Graphical Abstract
Keywords: Area per lipid, Cholesterol, Membrane elasticity, Neutron spin-echo, Rafts, Solid-state NMR spectroscopy
Introduction: Biophysical Functions of Sterols
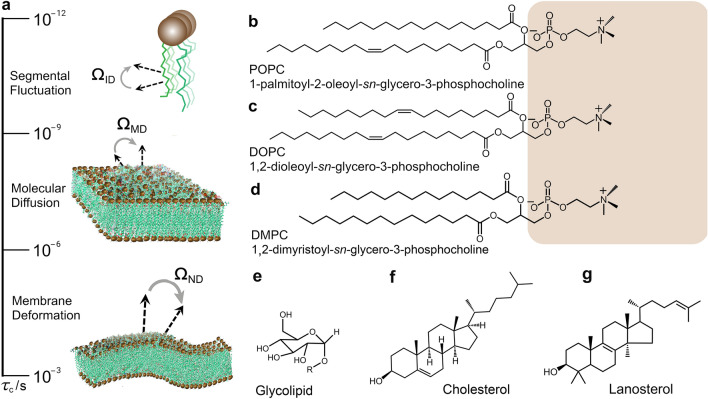
Biological membranes are the nexus of control and regulation of the cellular functions of life in which the lipid bilayer has a pivotal significance. Lipids form the matrix or scaffold of cellular membranes and have a remarkable diversity of molecular types that interact with proteins and peptides (Brown 2017; Lorent et al. 2020). The lipid bilayer is the primary platform for nutrient exchange, protein–cell interactions, and vesicular budding, among other vital cellular processes. Notably, the cell membranes of animals, plants, and microbes comprise mixtures of lipids and sterols, whose intermolecular associations play key roles in their physical properties at mesoscopic length scales, such as lipid packing and membrane rigidity. How the structural and dynamical properties of the phospholipids are controlled or regulated by steroids like cholesterol, lanosterol, or ergosterol are questions of high biological and biophysical significance (Chakraborty et al. 2020; Endress et al. 2002). Moreover, the lipid fluctuations are connected to the geometry of the interactions and the rates of motions, as described by the order parameters and correlation times. Phospholipid mobilities come with their own characteristic timescales, and various biophysical techniques can uncover how the molecular dynamics depend on time and distance by an energy landscape (Fig. 1a). The hierarchical dynamics in lipid bilayers include segmental fluctuations, molecular diffusion, and collective changes (membrane deformation), defining the topography of the motions as shown in Fig. 1a. Rapid segmental fluctuations with restricted amplitude are further averaged by molecular movements and collective lipid disturbances (Brown 1982, 2019), due to the force field of the bilayer membranes (Brown et al. 1983). Recently the dynamics landscape has been investigated for 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) in terms of multiple motions acting on various timescales and on multiple positions of the lipid molecules (Smith et al. 2022). In this review article, we address the collective motions of lipid membranes, and how sterols control their kinetics and mechanical properties such as the bending rigidity and spontaneous curvature.
Fig. 1.
Energy landscapes and mobilities of phospholipids in membranes entail characteristic timescales. a Fluctuations of 13C–1H or C–2H bonds are given by ΩID Euler angles for internal segmental frame (I) with respect to the membrane director axis (D). Phospholipid molecular motions entail anisotropic reorientation of the molecule-fixed frame (M) with respect to the membrane director axis (D) as described by ΩMD Euler angles. The liquid-crystalline bilayer lends itself to propagation of thermally excited, quasi-periodic fluctuations due to motion of the local membrane normal (N) relative to the average director axis (D) characterized by ΩND Euler angles. Examples are shown respectively for glycerophospholipids, b POPC, c DOPC, and d DMPC where the lipid headgroups are highlighted in brown; e glycolipids where R denotes the rest of the lipid molecule; f cholesterol which is the sterol component of animal biomembranes; and g lanosterol which is a precursor in the sterol biosynthesis pathway
Among sterols, it is currently thought that cholesterol is linked to the evolution of membrane biophysical functions (Bloom et al. 1991) as recapitulated by its metabolic pathway (Bloch 1965, 1983). Cholesterol is the most abundant sterol found in animal cells and is the precursor to steroid hormones, Vitamin D, bile acids, and other metabolites. It is implicated in key biological activities involving lysis, viral budding, and antibiotic resistance. By modifying the bending rigidity and spontaneous curvature of cell membranes, cholesterol imparts the ability to deform or withstand mechanical stresses. Cholesterol is derived in its metabolic pathway from its precursor lanosterol by removal of methyl substituents yielding a two-faced or Janus-like molecule with significantly altered properties (Bloom et al. 1991; Martinez et al. 2004). In eukaryotic membranes, cholesterol is present in varying amounts ranging from < 5 mol% in mitochondrial membranes up to ~ 40 mol% in cellular plasma membranes, while it is universally absent in prokaryotic membranes. It has essential regulatory functions involving protein activity, lipid permeability, and raft-like microdomains associated with trafficking and cell signaling, as pioneered by Erwin London and his colleagues (Brown and London 1998). Deviations from normal cholesterol homeostasis affects key membrane-related functions associated with cardiovascular disease, cancer, and various neurological conditions. Cholesterol is moreover implicated in viral infections due to influenza, HIV, and coronavirus (SARS-Cov-2) (Liao et al. 2001; Sun and Whittaker 2003; Wang et al. 2020). Understanding its functions requires knowledge of how cholesterol affects the biomembrane mechanics over relevant scales of length and time (Bloch 1983; Chakraborty et al. 2020; Trouard et al. 1999).
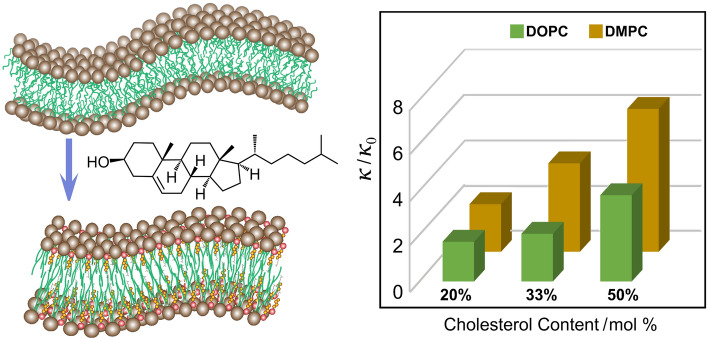
It has long been known that cholesterol yields a condensing effect on fluid (liquid-disordered, ld) lipid membranes, giving rise to the liquid-ordered (lo) phase (Ipsen et al. 1987; Mouritsen and Zuckermann 2004). Condensation of the area per lipid (packing) at the bilayer interface with water is accompanied by an increase in area elastic modulus (KA), which affects the energetics of membrane deformations. In accord with a polymer-brush model (Doktorova et al. 2019; Rawicz et al. 2000), the area elastic modulus is related to the Canham–Helfrich monolayer bending rigidity (κ) (elastic modulus). Cholesterol is thus expected to have a substantial influence on the curvature free energy of the membrane lipids, meaning the entire force field for the membrane deformation needs to be considered (Deserno 2015; Venable et al. 2015). Still, the question of whether the effects of cholesterol on membrane bending rigidity are universal for both saturated and (poly)unsaturated lipid membranes remains open and has attracted recent interest (Ashkar et al. 2021; Nagle 2021; Nagle et al. 2021). Knowing whether cholesterol stiffens both saturated and unsaturated lipid membranes to the same extent is important in the context of microdomains, line tension, and curvature free energy, which entail the force balance of the membrane lipids (Allender et al. 2019). Depending on the lipid polar head group, saturated and unsaturated lipid membranes differ in their intrinsic bending rigidity (Brown et al. 2002) and are the appropriate reference systems. Examples of biological functions that depend on membrane deformation include lipid–protein interactions (Brown 2017), endo/exocytosis and membrane biogenesis, and the budding of membrane-enveloped viruses (SARS-CoV-2, HIV).
Through combining studies of structure and dynamics, we can acquire insight regarding the interdependence of membrane lipid architecture, mechanics, and function (Brown 1997; Deserno 2015; Nagle 2013). Equilibrium properties of the lipids that render them suited to compartmentalize and affect membrane functions include: a very low critical micelle concentration (cmc) so that they self-assemble completely; a high resistance to tearing with a large area expansion modulus; an intermediate bending rigidity that allows them to be readily deformable yet with significant free energy; and a spontaneous monolayer curvature of the leaflets that introduces a free energy frustration due to the lipid composition. According to the flexible surface model (FSM), the functional lipid–protein interactions are affected by the energetics of membrane deformation by strong out-of-plane coupling (Brown 2017; Fried et al. 2021) as compared to the weaker in-plane couplings described by the fluid-mosaic model. By including equilibrium investigations with dynamical studies, we can formulate the forces and potentials that govern biomembrane structure, assembly, and function. In this context, how the properties of membrane lipids are modified and controlled by sterols takes on a central biological and pharmaceutical relevance.
Here we review our studies using a combination of solid-state nuclear magnetic resonance (NMR) and neutron spin-echo (NSE) spectroscopy that uniquely inform the physicochemical properties of cholesterol-containing lipid membranes. Both solid-state NMR and NSE spectroscopy are inherently sensitive to mesoscale bending fluctuations of lipid bilayers. These methods lead us to the idea of a local membrane stiffening effect of cholesterol over short length and time scales in conjunction with emergent bilayer material properties. Our observations are pertinent to the functional role of cholesterol in viral budding and membrane lipid–protein interactions as described by the flexible surface model (FSM) (Brown 1994). For 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) an approximately threefold increase in bending rigidity is seen with increasing cholesterol content up to a 50% mole fraction, which is supported by atomistic molecular dynamics (MD) simulations (Doktorova et al. 2019). Our results (Chakraborty et al. 2020) illuminate structure–property relations that connect the area compressibility modulus (KA) of saturated and unsaturated lipid bilayers to the curvature elastic modulus (bending rigidity, κ) using a polymer-brush model (Doktorova et al. 2019; Rawicz et al. 2000). Accordingly, we propose a scale-dependent manifestation of bilayer properties, highlighting how cholesterol has been evolutionarily selected to control membrane deformation over mesoscopic scales of time and space. A universal stiffening by cholesterol is evident based on our measurements that probe its local or mesoscopic influences on membrane energetics at an atomistic level (Ashkar et al. 2021). Our findings imply that there is a length- and time-scale dependence of cholesterol-induced influences on the membrane mechanical properties that are universal to both saturated and unsaturated lipid bilayers.
Cholesterol and Its Molecular Features
Illustrations of the chemical structures of the three major classes of membrane lipids, namely, phospholipids, glycolipids, and sterols, are provided in Fig. 1b–g. In cell membranes, lipids exist in various nanostructures that differ in their polar head group size, charge, and capacity for hydrogen bonding, as well as in their nonpolar acyl chain length and degree of unsaturation. Phospholipids are the main components of the lipid bilayers of mammalian cells. The second most abundant component is cholesterol, constituting up to ~ 40 mol% of the lipids of the plasma membrane (Huang et al. 1999; Krause and Regen 2014; van Meer et al. 2008) which has attracted significant interest. The fact that cholesterol is found in large amounts in mammalian cells but is absent in prokaryotic cells (Mouritsen and Zuckermann 2004) indicates its important role in cell evolution. Its mole fraction in various cell membranes is controlled through biosynthesis, efflux from cells, and influx of lipoprotein cholesterol into the cell (Simons and Ikonen 2000). Functionally, cholesterol is implicated in key biological processes such as cellular homeostasis, steroid and vitamin D synthesis, and the regulation of membrane order and dynamics (Craig et al. 2021). Well-known methods that describe how cholesterol modulates cellular functions include: (i) indirect effects on membrane properties and consequently on protein–membrane interactions (Brown 1994, 2012, 2017), and (ii) by direct effects on cholesterol–protein interactions (Liu et al. 2017; Sheng et al. 2012). Changes in cholesterol levels are commonly attributed to variations in membrane properties, stability, and pathology.
Structurally, cholesterol is a 27‑carbon compound characterized by a hydrocarbon tail, a central sterol nucleus comprising four hydrocarbon rings, and a hydroxyl group (Fig. 1f) (Craig et al. 2021). This structure enables incorporation of cholesterol into membranes and its alignment along the hydrocarbon chains of the lipids (Fig. 1b–d) (Bloch 1983). The orientation of cholesterol yields a more ordered state of the hydrocarbon chains, and thereby controls membrane structural, dynamical, and physical properties. In membranes with saturated and unsaturated lipids, cholesterol preferentially segregates into domains rich in saturated lipids called lipid rafts as shown by the work of Erwin London and coworkers (Brown and London 1998). These raft-like domains are thought to play an important role in cell signaling and pharmacology (Brown and London 1998; Klose et al. 2013; Levental and Veatch 2016; Sezgin et al. 2017; Simons and Gerl 2010), as well as maintaining membrane order (Zhang et al. 2018). Notably, the increase in lipid packing due to cholesterol can lower the membrane permeability, and it can affect the distribution and transport of anesthetics and other drugs in membranes (Auger et al. 1988; Siminovitch et al. 1984). Cholesterol also plays a significant regulatory role in many biophysical processes (Maxfield and van Meer 2010) including passive permeation (Corvera et al. 1992), protein and enzyme activity (Cornelius 2001; de Meyer et al. 2010), and viral infections, e.g., HIV (Prasad and Bukrinsky 2014), influenza (Sun and Whittaker 2003), and SARS-Cov-2 (coronavirus) (Meher et al. 2019).
Apart from its central role in membrane processes, cholesterol is also a key component in drug discovery applications. For instance, the presence of cholesterol in renal cell membranes is critical for improving the membrane resistance against damage by nephrotoxic antibiotics (Khondker et al. 2017). This is attributed to increased membrane stabilization and suppression of lipid and peptide mobility by cholesterol. Such modifications of membrane properties by cholesterol have been used in liposomal formulations, yielding significant advances in their use as drug carriers (Allen and Cullis 2013). Inclusion of cholesterol in liposomal membranes results in stable and long-circulating liposomes in vivo, which is needed for protection of the encapsulated drug from degradation and premature release (Allen and Cullis 2013; Leeb and Maibaum 2018). In addition, cholesterol reduces the activity of proteins and peptides of the innate immune system on liposomal bilayers (Zasloff 2002), thus prolonging the liposome circulation time. Such findings point to the importance of cholesterol-induced modifications of membrane structural, dynamical, and mechanical properties in controlling the efficacy of membrane-interacting drugs and in the developing of robust liposomal drug carriers.
From this perspective, the lipids are viewed as highly deformable materials which can be significantly altered by the presence of steroids such as cholesterol, lanosterol, or ergosterol. To further understand the effects of cholesterol on membrane mechanics, various biophysical techniques have been used, including flickering spectroscopy (Duwe and Sackmann 1990), micropipette aspiration (Bassereau et al. 2014; Dimova 2014; Evans and Rawicz 1990), electrodeformation (Gracià et al. 2010; Niggemann et al. 1995), X-ray diffusivity (Pan et al. 2009), molecular dynamics (MD) simulations (Doktorova et al. 2019; Johner et al. 2014; Khelashvili et al. 2013), deuterium nuclear magnetic resonance (2H NMR) (Brown et al. 1983; Molugu and Brown 2016), and neutron spin-echo (NSE) spectroscopy (Arriaga et al. 2009a; Chakraborty et al. 2020; Mell et al. 2013; Nagao et al. 2017). Among these techniques, solid-state 2H NMR and NSE spectroscopy together with MD simulations are uniquely able to access collective lipid fluctuations over length and time scales of biological processes, including protein–membrane interactions and signaling events. In this review article, we discuss the powerful combination of solid-state 2H NMR and NSE spectroscopy for investigating membrane mechanics using spin labeling methods. Figure 2 depicts the types of quasi-elastic bilayer deformations that may be considered for a free lipid membrane. The shape fluctuations are modeled as unconstrained splay, twist, and bend excitations, together with effective axial rotations of the lipids. Such a continuum picture represents a departure from the previous molecular theories for lipid bilayer relaxation, in which the absence of chemical details is both the strength and weakness. In solid-state 2H NMR spectroscopy, the relaxation of deuterium spin labels along the hydrocarbon chain of the lipid molecules is studied under the influence of a magnetic field. Experiments involving NSE spectroscopy characterize the change in spin polarization of an incident neutron beam as it exchanges energy with the lipid membrane. To illustrate the mode of operation of the two techniques, we first summarize the basic principles together with their application to studies of membrane fluctuations. Extension to lipid bilayers then gives a powerful and unique approach to determining their mechanical or viscous membrane properties, and how they emerge from atomistic-level forces that affect cholesterol–membrane interactions.
Fig. 2.
Excitations of a fluid lipid bilayer are described within the continuum elastic approximation. a Planar bilayer, b splay, c twist, and d bend deformations, together with axial rotations about the local director
Deuterium NMR Spectroscopy Applied to Lipid Membranes
Solid-state nuclear magnetic resonance (NMR) spectroscopy is an atomic-level method to investigate the structure and dynamics of solids, semi-solids, and liquid crystals (Reif et al. 2021). The reader will recall that lipid molecules are liquid crystals which fall in between a liquid and a solid. In a liquid, the molecules tumble over all space, which averages out the orientation-dependent interactions to zero, for example dipole–dipole, magnetic dipole, or electric quadrupolar interactions. Here you will observe the isotopic chemical shifts, together with the spin–spin couplings which are not dependent on orientation. In a solid, however, the angular dependent interactions are not averaged to zero, which means we can observe them in various ways. Solid-state NMR formulates these interactions in terms of order parameters capturing the solid-like properties of liquid crystals. One should also recognize that the term solid-state does not imply the system is a physical solid—only that it has an average or equilibrium structure despite significant molecular motion. Molecular solids are the limit, although considerable motions are still possible (Kinnun et al. 2013), and even isotropic liquids such as water can have a local structure. For chemical compounds and biomaterials, one thus needs to devise methods to characterize both their structure and dynamics, with lipid membranes and liquid crystals as prominent examples (Molugu et al. 2017; Reif et al. 2021). In this regard, solid-state 2H NMR spectroscopy gives a versatile technique for characterizing the molecular organization of lipids involving their structure, ordering, and rates of molecular motions. It provides information complementary to other physical techniques such as neutron spin-echo (NSE) spectroscopy, and is one of the premier biophysical tools applicable to lipid bilayers and biomembranes (Brown and Chan 1996; Molugu et al. 2017). By combining solid-state 2H NMR order parameter measurements with nuclear spin relaxation experiments, the collective membrane dynamics including their elastic deformations can be revealed at an atomically resolved level. Owing to their liquid-crystalline nature, lipid membranes clearly span the NMR time and length scales, which allows for analysis of their multiscale dynamics and elastic deformations in the presence of cholesterol.
A further important aspect is that the solid-state NMR technology is essential to validating the force fields in membrane simulations, as in the seminal work of Pastor et al. (Klauda et al. 2010; Marrink et al. 2019). It provides experimental insights into membrane properties that cannot otherwise be obtained. Because the coupling interactions in solid-state NMR are sensitive to orientation and/or distance, their values correspond with the average structure of the system. By introducing site-specific labels due to the individual C–2H bonds, atomic-level details for noncrystalline amorphous or liquid-crystalline systems can be acquired directly. In addition, the relaxation parameters of solid-state 2H NMR spectroscopy give us insight into the relevant molecular motions. In solid-state 2H NMR of biomolecular systems, one of the distinctive features is that both lineshape data (Molugu et al. 2017) and relaxation times (Brown 2019; Leftin and Brown 2011; Martinez et al. 2002c) are accessible to experimental measurements. An example using solid-state 2H NMR spectroscopy is shown in Fig. 3a and b, which yields information about the lipid structural dynamics over a range of timescales. Combined measurements of residual quadrupolar couplings (RQCs) and relaxation times thus provide information on the geometry and allow investigations of the multiscale molecular dynamics in the lipid systems of interest.
Fig. 3.
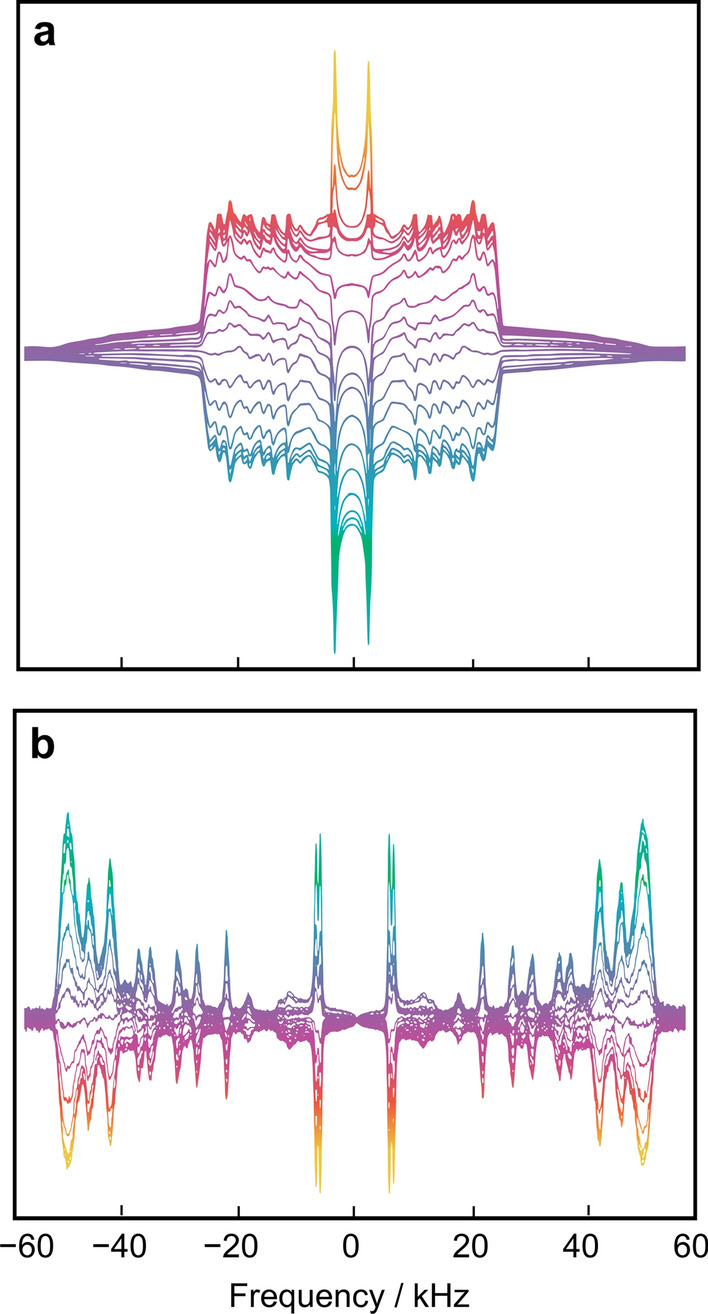
Solid-state 2H NMR spectroscopy of membrane lipids provides both lineshape data and relaxation times yielding information about structure and dynamics. a Inversion recovery of 2H nuclear magnetization for random DMPC-d54/cholesterol (1:1) multilamellar dispersion in the liquid-ordered phase at T = 44 °C showing partially relaxed 2H NMR spectra. b Deconvolved (de-Paked) 2H NMR spectra corresponding to bilayer normal parallel to magnetic field (θ = 0°). The sample contained 20 mM Tris buffer at pH 7.3 (50 wt% H2O). Data were acquired at 76.8 MHz using a phase-cycled, inversion-recovery quadrupolar-echo pulse sequence, π − t1 − (π /2)x − τ − (π/2)y − t2 (acquire), where t1 is a variable delay ranging from 5 ms (bottom) to 3 s (top). Adapted from Ref. (Martinez et al. 2002b)
Let us now briefly consider the general principles of how lipid membranes are studied with solid-state 2H NMR spectroscopy. This section includes a very concise explanation of what is essential for the reader to be able to understand the 2H NMR spectroscopic concepts. For further detailed explanation, general readers are referred to Refs. (Brown 1996; Kinnun et al. 2013; Molugu et al. 2017). Here, we note that the 2H nucleus has a spin of I = 1, which yields three Zeeman energy levels due to projection of its spin angular momentum onto the quantization axis, which is the main magnetic field direction. The three states are designated as and using quantum mechanical bra-ket notation, where the energy levels are due to the interaction Hamiltonian ĤZ for the nuclear magnetic moment with the static magnetic field. The allowed single-quantum nuclear spin transitions occur because the perturbation due to the electric quadrupolar coupling with the nuclear quadrupole moment removes their degeneracy in 2H NMR spectroscopy. The perturbing Hamiltonian ĤQ comes from the coupling of the quadrupole moment of the 2H nucleus with the electric field gradient (EFG) of the C–2H bond, yielding two spectral branches for each inequivalent site of the lipid molecules (Fig. 3).
To continue briefly, in solid-state 2H NMR spectroscopy the experimental quadrupolar coupling is given by the difference in the frequencies of the spectral lines coming from the perturbing Hamiltonian. The result for the quadrupolar frequencies () reads:
| 1 |
Here, is the static quadrupolar coupling constant, corresponds to the asymmetry parameter of the EFG tensor, is a Wigner rotation matrix element, and are the Euler angles (Rose 1957) that transform the principal axis system (PAS) of the EFG tensor (P) to the laboratory frame (L) (Brown 1996; Leftin et al. 2014b; Xu et al. 2014). Because the static EFG tensor of the C–2H bond is nearly axially symmetric (), the above result simplifies to yield:
| 2 |
The experimental quadrupolar splitting is thus obtained as:
| 3 |
where for convenience the various symbols are defined above.
Now for liquid-crystalline membranes the molecular motions are typically cylindrically symmetric about the bilayer normal, an axis called the director. The idea of a director is central to lipid biophysics. Rotation of the principal axis system of the coupling tensor to the laboratory frame, described by the Euler angles, can thus be represented by two consecutive rotations. First, the Euler angles represent the (time-dependent) rotation of the principal axis system of the C–2H bond to the director frame, and second the Euler angles correspond to the (static) rotation from the director to the laboratory frame. The use of the closure property of the rotation group (Xu et al. 2014) is very helpful in this regard, as explained in the Appendices of Refs. (Brown 1996; Molugu et al. 2017); see also Ref. (Kinnun et al. 2013) for an introductory description. Considering the cylindrical symmetry about the director, we can then expand Eq. (3), which now reads:
| 4a |
| 4b |
Here, is the angle of the bilayer normal to the static external magnetic field. The segmental order parameter SCD is provided by the formula:
| 5 |
where the angular brackets denote a time or ensemble average. Hence, it follows that
| 6 |
in which is the second-order Legendre polynomial (Arfken 1970; Varshalovich et al. 1988). The above formula describes how the quadrupolar splitting depends on the (Euler) angles that rotate the coupling tensor from its principal axes system (PAS) to the laboratory frame of the main magnetic field.
Next, we consider the process of NMR relaxation, which is due to fluctuations of the coupling Hamiltonian coming from the various motions of the lipid molecules within the membrane bilayer. The fluctuations lead to transitions between the various adjacent energy levels (Xu et al. 2014; Molugu et al. 2017). In solid-state 2H NMR spectroscopy of liquid-crystalline membrane lipids, we are often interested in the spin–lattice () relaxation rates. Experimental measurements of the relaxation rate typically involve inverting the magnetization followed by attainment of equilibrium through its recovery as a function of time. The observable relaxation rates depend on the spectral densities of motion in the laboratory frame by the relation:
| 7 |
In this formula, is the spin–lattice (longitudinal) relaxation rate, and designates the spectral densities of motion, where m = 1, 2, and is the deuteron Larmor frequency. The spectral densities describe the power spectra of the lipid motions as a function of frequency in terms of fluctuations of the Wigner rotation matrix elements that transform the coupling (EFG) tensor from its principal axis system to the laboratory coordinates frame (Varshalovich et al. 1988). They are the Fourier transform partners of the orientational correlation functions , which depend on time and thereby characterize the C–2H bond fluctuations of the lipids, as in the case of lipid molecular dynamics simulations (Klauda et al. 2010; Venable et al. 2015).
Clearly the segmental order parameters depend only on the amplitudes of the C–2H bond motions, while the relaxation rates also include the rates of the C–2H bond fluctuations. According to a generalized model-free (GMF) approach of relaxation rate analysis (Brown 1984; Xu et al. 2014), a functional dependence of the rates on the squared segmental order parameters () (square law) along the chain would result (Fig. 4). For short-wavelength excitations—on the order of the bilayer thickness and less—the spectral density can be written as (Nevzorov and Brown 1997):
| 8 |
Here, is the angular frequency, is the viscoelastic constant, is the dimensionality, and indicates the second-rank Wigner rotation matrix (Rose 1957). The irreducible spectral densities depend on the square of the observed order parameters, and the slope of the square-law plot is inversely related to the membrane softness. For 3D quasi-elastic bilayer fluctuations, the viscoelastic constant is given by as previously derived from first principles in Section IV.B of Ref. (Brown 1982). Here, K is the elastic (force) constant for splay, twist, and bend deformations in an one-elastic constant approximation, where is the viscosity coefficient, is the order parameter for the relatively slow motions, and the other symbols have their conventional meanings (Brown 1982). Because a single elastic constant is assumed, no distinction is made between splay, twist, and bend deformations, which is both a strength and weakness of our approach. In addition to the so-called bending modulus , the compression modulus comes into play (Nagle and Tristram-Nagle 2000). For splay deformations, in a one-elastic constant approximation the bending rigidity is , where is the bilayer thickness and DC is the acyl chain thickness per monolayer, giving a dependence of the rates as measured experimentally (Brown et al. 2001).
Fig. 4.
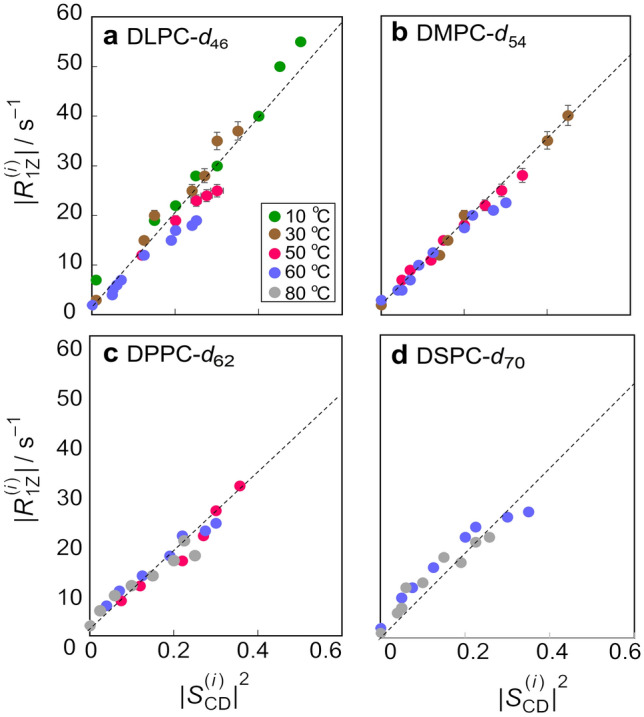
Functional dependence of relaxation times and order parameters from solid-state 2H NMR spectroscopy reveal dynamical bending rigidity of lipid membranes. Data are for unoriented lipid dispersions at 55.4 MHz and various temperatures (T ≥ Tm + 6 °C): a DLPC-d46, b DMPC-d54, c DPPC-d62, and d DSPC-d70, with acyl lengths of n = 12, 14, 16, and 18 carbons, respectively. Square-law functional relation of spin–lattice relaxation rates R1Z and order parameters |SCD| along the acyl chains (index i) for a homologous series of PCs in the Lα phase characterizes the influence of the acyl length (bilayer thickness). Adapted from Ref. (Brown et al. 2001)
The Energy Landscapes of Lipid Membranes
For membrane lipid bilayers, their deformations in response to external forces or thermal fluctuations are generally characterized by at least four material constants: (i) the surface tension (which is zero for a bilayer at equilibrium), (ii) the area expansion modulus or alternatively the lateral compressibility, (iii) the bending rigidity , and (iv) the monolayer spontaneous curvature. These structural quantities are inherent to the forces governing the nanoscopic structures of the lipid membrane assemblies. Representative applications of solid-state 2H NMR spectroscopy to lipid membranes include the influences of cholesterol (Brown and Seelig 1978; Brown et al. 2001; Trouard et al. 1999; Veatch et al. 2007; Vist and Davis 1990; Wassall et al. 2004) as well as acyl chain unsaturation (Huber et al. 2002; Huster et al. 1998; Salmon et al. 1987; Wassall et al. 2018). In this review, we aim to characterize how the average membrane structure, fluctuations, and elastic deformations are affected by cholesterol. The use of acyl chain-perdeuterated phospholipids (n = 12 to 18 carbons) in NMR relaxation studies allows one to observe nearly the entire hydrocarbon region of the bilayer simultaneously. As shown in Fig. 4, a square-law functional dependence of the rates on along the chains describes the data for the homologous PCs for °C above the main phase transition temperature (Fig. 4a–d), suggesting that the bending rigidity depends only weakly on temperature in the Lα state. These data are discussed here as a proof of concept for the square-law dependence theory and serve as reference samples for cholesterol/lipid systems to be discussed later in the paper. Still, it might be surprising to some readers that the NMR results for soft membrane bilayers—involving flexible lipid molecules with many degrees of freedom—can be interpreted simply by using concepts drawn from the physics of materials. In terms of our picture, the membrane lipids are effectively tethered to the aqueous interface via their polar head groups, and the bilayer interior is essentially liquid hydrocarbon. The reason why the R1Z relaxation is governed by collective order fluctuations is that the local segmental motions of the lipids are very fast (~ 10–11 s), with spectral densities extending to very high frequencies. With the above results in our hands, we are then able to study the influences of cholesterol on lipid membrane structural and dynamical properties at a mesoscopic level.
In what comes next, our focus is on the lipid membrane structural deformation and the emergent fluctuations that occur due to the presence of sterols. As a steroid molecule cholesterol is amphiphilic in nature, as in the case of other lipids. Solid-state 2H NMR studies clearly show the effect of cholesterol on the physical properties of bilayers of 1,2-diperdeuteriomyristoyl-sn-glycero-3-phosphocholine (DMPC-d54) at an atomic level (Fig. 5) consistent with the various other lipid NMR studies (Bartels et al. 2008; Bunge et al. 2008; Chakraborty et al. 2020; Morrison and Bloom 1994; Oldfield et al. 1978; Trouard et al. 1999; Veatch et al. 2007). Additional 2H NMR studies of the specifically deuterated cholesterol molecule in lipid membranes have been conducted (Bonmatin et al. 1990; Brown 1990). For acyl chain perdeuterated phospholipids, we can deconvolve or de-Pake the powder-type spectra of random multilamellar dispersions, as notably pioneered by the Canadian physicist Myer Bloom (Sternin et al. 1983) to yield more highly resolved subspectra of the θ = 0° bilayer orientation (Fig. 5a–d). Notice that a distribution of residual quadrupolar couplings (RQCs) is evident, corresponding to the profile of the SCD order parameters of the various C2H2 and C2H3 groups along the acyl chains. A striking increase in the residual quadrupolar splittings is seen for the acyl segments with progressively greater mole fraction of cholesterol, due to fewer degrees of freedom of the flexible lipids. Interaction of phospholipids with the rigid cholesterol molecule thus leads to greater magnitudes of the orientational order parameters of the lipid acyl chain segments versus the bilayer normal (Fig. 5a–d), which according to a mean-torque model indicates a greater bilayer thickness.
Fig. 5.
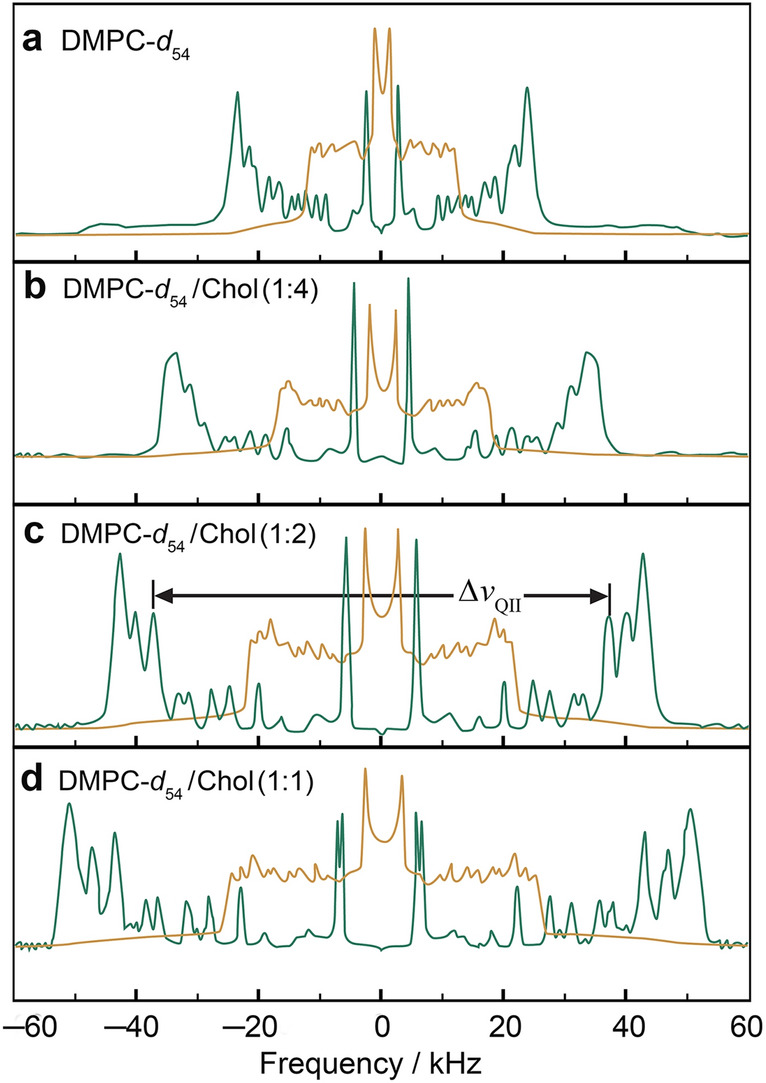
Cholesterol reduces the conformational degrees of freedom for lipid acyl chain segments yielding closer molecular packing. a DMPC-d54 in the liquid-disordered (ld) phase, and b–d DMPC-d54 containing various mole fractions of cholesterol (Chol) in the liquid-ordered (lo) phase. Powder type spectra (brown) of randomly oriented multilamellar dispersions were numerically inverted (de-Paked) to yield subspectra corresponding to the θ = 0° orientation (green). Note that a distribution of residual quadrupolar couplings (RQCs) corresponds to the various C2H2 and C2H3 groups with a progressive increase due to cholesterol. Adapted from Ref. (Martinez et al. 2004)
In addition, the reader should recall that because the lipid volume is almost constant to a first approximation (Petrache et al. 1997), i.e., the molecular volume is generally conserved, the increased bilayer thickness is accompanied by a reduction of the area at the aqueous interface. The well-known condensing effect of cholesterol is thus explained at the molecular level by a reduced area per phospholipid molecule at the aqueous interface. In turn, the closer lipid molecular packing with fewer degrees of freedom requires a greater energetic cost of deforming the bilayer, as shown by an increase in the bilayer elastic moduli for either area and/or curvature deformation (KA and κ, respectively). It follows that the solid-state 2H NMR order parameters are a direct measure of the lipid close packing, manifested by a decrease in the area per lipid and an increase in the bilayer thickness (Mallikarjunaiah et al. 2019; Petrache et al. 2000). Those in turn govern the area and curvature elasticity of the membrane (Chakraborty et al. 2020).
How Lipid Membranes are Stiffened by Cholesterol and Lanosterol
Further analysis of the solid-state 2H NMR spectral data obtained for lipid membranes can be accomplished by introducing a mean-torque model for the configurational statistics of the lipid acyl chains (Leftin et al. 2014a; Petrache et al. 2000, 2001). The first-order mean-torque model is based on the statistical mechanical precepts, and gives important structural parameters for lipid membranes: the mean area per lipid at the aqueous interface, the average bilayer thickness , the hydrophobic thickness DC of one bilayer leaflet, and the area elastic (or compressibility) modulus KA at an emergent atomistic level (Fig. 7c). Introduction of a polymer brush model then allows the values of KA to be related to the corresponding values of the bending rigidity (κ) (Doktorova et al. 2019; Rawicz et al. 2000). The mean-torque model has been recently reviewed and readers are guided to the previous studies (Kinnun et al. 2015; Mallikarjunaiah et al. 2019) for details of the theory, and how it is applied to bilayer deformation under an eternal force such as hydrostatic pressure or osmotic stress (Mallikarjunaiah et al. 2011).
Fig. 7.
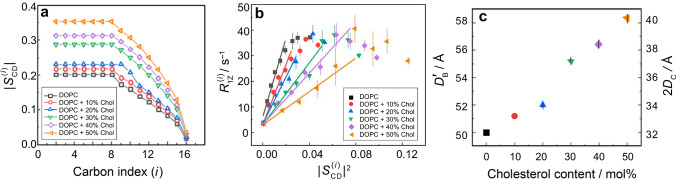
Results from solid-state 2H NMR spectroscopy shows cholesterol increases acyl chain ordering and bending stiffness of lipid membranes due to closer molecular packing in the bilayer. a Segmental order parameter versus acyl position for POPC-d31 probe lipid in DOPC/cholesterol membranes with different mol% cholesterol at T = 25 °C. b Dependence of spin–lattice relaxation rate on squared order parameters indicating a decrease in square-law slopes due to bilayer stiffening by cholesterol. c Structural parameters, i.e., steric thickness () and bilayer hydrocarbon thickness (2DC), obtained from 2H NMR lineshapes using the first-order mean-torque model. Adapted from Ref. (Chakraborty et al. 2020)
Notable applications of solid-state 2H NMR spectroscopy to lipid membranes include extensive studies of the role of cholesterol in raft-like lipid mixtures by Klaus Gawrisch and coworkers (Veatch et al. 2007) and by Stephen Wassall et al. (Brzustowicz et al. 2002; Wassall et al. 2004). In other related studies, solid-state 2H NMR spectroscopy has been used to investigate the effect of varying the mole fraction of cholesterol in raft-like binary and ternary mixtures with N-palmitoylsphingomyelin (PSM) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) for both aligned and unoriented multilamellar dispersions (Bartels et al. 2008; Bunge et al. 2008). Striking differences have been found in chain packing for monounsaturated POPC-d31 bilayers upon interaction with cholesterol in binary mixtures (Morrison and Bloom 1994), in which the acyl chains of the glycerophospholipid are less strongly affected than the sphingolipid by the rigid sterol backbone at low cholesterol concentrations (Bartels et al. 2008). In addition, a more continuous dependence of chain order on cholesterol concentration is seen in the case of POPC bilayers in the liquid-ordered phase. This behavior is evidently due to the less favorable interaction of the unsaturated acyl chain of POPC with cholesterol, as already shown for various unsaturated lipids (Brzustowicz et al. 2002; Shaikh et al. 2006; Wassall et al. 2004).
At this point we can conclude based on the solid-state 2H NMR spectroscopy that cholesterol leads to larger orientational order parameters of the lipids, due to a greater bilayer thickness and closer acyl chain packing (decreased interfacial area per lipid molecule with less free volume). The analysis can be taken a step further by recalling that the observables from solid-state 2H NMR spectroscopy include the SCD segmental order parameters and the corresponding NMR relaxation rates, e.g., the R1Z spin–lattice relaxation rates (Brown 2019). We next show that in addition to the greater segmental order parameters, the fewer degrees of freedom of the lipid acyl chains lead to a reduction in the area elastic modulus (KA) and an increase in the bending rigidity (κ), e.g., as described by a polymer brush model (Doktorova et al. 2019; Rawicz et al. 2000). For lipid membranes the contribution from so-called order-director fluctuations (ODF) to the relaxation has been interpreted in terms of short-wavelength excitations, on the order of the bilayer thickness and less (Brown 1982). In this case, the relaxation rates involve the spectral density (power spectra) and have a characteristic dependence on the resonance frequency and the SCD segmental order parameters. As a result, the solid-state 2H NMR spin–lattice relaxation times of lipid bilayers are often interpreted by a power law that includes the dependence of the R1Z relaxation rates on the square of the observed order parameters, where the slope of the square-law plot is inversely related to the viscoelastic constant as a measure of the membrane softness.
Following our generalized model-free (GMF) approach (Brown 1984; Xu et al. 2014), the relaxation time analysis then leads to a functional dependence of the rates on the squared segmental order parameters () (square law) along the chain (Fig. 4). In fact, there are two possibilities for interpreting the correspondence of the R1Z relaxation rates to the SCD order parameters that describe the order–director fluctuations (ODF) in liquid-crystalline membranes. Either one can introduce an approximate model that is exactly soluble, or one can use an exact model that is approximately soluble. In the first case, the measurements can be interpreted by an analytical chemistry approach, combining the and data as a metric for the bilayer stiffening, with the results for the canonical DMPC-d54 bilayer (Martinez et al. 2002b) as a refence (Méléard et al. 1997; Rawicz et al. 2000). Alternatively, the results can be interpreted together with theory (Brown 1982) to estimate the value of the bending rigidity κ (curvature elastic modulus). For 3D quasi-elastic bilayer fluctuations, because a single elastic constant is assumed, no distinction is made between splay, twist, and bend deformations. The so-called bending rigidity (splay elastic modulus) is thus , where is the bilayer thickness and DC is the acyl chain thickness per monolayer (Brown et al. 2001). With the single-elastic constant approximation, we obtain results in approximate quantitative agreement with other biophysical measurements, such a micropipette deformation and shape fluctuations of giant unilamellar vesicles (Brown et al. 2001). We can then combine the order parameters and relaxation rate profiles into a square-law functional dependence (Williams et al. 1985) in terms of plots of versus whose slope is inversely related to the bending rigidity (curvature elastic modulus) (Fig. 6).
Fig. 6.
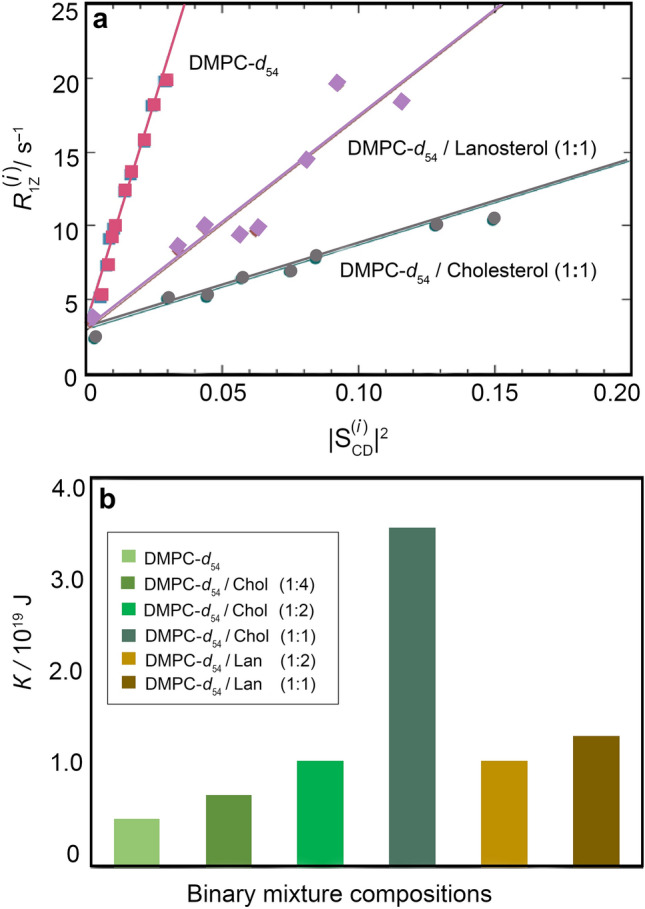
Square-law relations of NMR relaxation times and order parameters indicate bilayer stiffening by cholesterol and lanosterol. a Dependence of spin–lattice relaxation rates on squared order parameters for resolved 2H NMR splittings of DMPC-d54. Data were obtained at T = 44 °C and at 76.8 MHz (11.8 T). b Bending rigidity values calculated for DMPC-d54/cholesterol (DMPC-d54/Chol) and DMPC-d54/lanosterol (DMPC-d54/Lan) systems. Adapted from Ref. (Martinez et al. 2004)
In this way, it is found experimentally that in solid-state 2H NMR relaxation studies the effect of cholesterol on the area and curvature elasticity of DMPC-d54 lipid bilayers is characterized by a square-law dependence of the rates versus the order parameters along the entire acyl chain. The reader should note that the square law (Brown 1982) is a model-free correlation among the experimental observables as obtained by solid-state 2H NMR relaxation measurements. For multilamellar dispersions of DMPC-d54/cholesterol (Fig. 6a), an increase in bilayer stiffening at various cholesterol mole fractions is inferred from the solid-state 2H NMR relaxation analysis. Here cholesterol yields a large decrease in the square-law slopes, corresponding to a progressive reduction in bilayer elasticity (Fig. 6b). The square-law slope is a manifestation of the progressive increase in the bilayer rigidity of DMPC at various compositions of cholesterol. The reduction in the slope (Fig. 6a) reflects an increase in bending rigidity κ due to short-range cholesterol–phospholipid interactions, showing how membrane mechanical properties emerge for the local atomistic forces within the lipid bilayer. Hence, we conclude that the results for the DMPC-d54/cholesterol mixtures reveal stiffening in the case of saturated lipid bilayers, which provides a benchmark for the relaxation analysis that can be extended to mono and polyunsaturated lipids.
For most animal and fungal membranes, lanosterol is the steroid precursor and hence it is also of considerable biological and pharmacological interest in comparison to cholesterol (Bloom et al. 1991; Endress et al. 2002; Henriksen et al. 2006). How natural selection of biophysical function has occurred during the evolution of sterols has been explored from the vantage point of solid-state 2H NMR spectroscopy (Martinez et al. 2004; Miao et al. 2002) and quasielastic neutron scattering (Endress et al. 2002). Here we focus on solid-state NMR relaxation experiments, which inform the differences in behavior of cholesterol versus its metabolic precursor lanosterol in terms of the natural selection of membrane elasticity during biological evolution (Martinez et al. 2004). An example for DMPC-d54 binary mixtures (Fig. 6a) shows that the slope of the square-law plot is greater for lanosterol than for cholesterol. Additional studies for various molar ratios of sterols indicate that the reduction of the bilayer elasticity is less for lanosterol than for cholesterol (Martinez et al. 2004). Calculated values of the bending rigidity from the solid-state 2H NMR relaxation data analysis (Fig. 6b) are consistent with the results from video microscopy of large unilamellar vesicles (Méléard et al. 1997). The findings agree with comparative studies of sterols (Dahl et al. 1980; Yeagle 1985), which report that cholesterol increases the conformational order of the membrane lipid acyl chains more effectively than lanosterol and has a stronger ability to promote domain formation in membranes (Urbina et al. 1995; Yeagle 1985; Xu and London 2000). We thus conclude that removal of methyl groups from the α-face of lanosterol as it occurs in the biosynthetic pathway enhances the ability of cholesterol to stiffen the lipid membrane. Our results support the idea that natural selection of cholesterol to control the membrane bending rigidity has occurred during molecular evolution. According to this view, the biosynthetic pathway of cholesterol recapitulates its natural selection during biological evolution, with implications for synthetic biology and the development of artificial cells. Evidently an increase in the bilayer bending rigidity has co-evolved with the demethylation of lanosterol to yield cholesterol, suggesting the possibility of feedback control of cholesterol homeostasis in biomembranes.
Stiffening of Lipid Membranes by Cholesterol
The above results for saturated DMPC lipids give striking examples of the mesoscopic stiffening of bilayer membranes by cholesterol and lanosterol. Lipid systems with unsaturated acyl groups such as 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) (Pan et al. 2008; Wiener et al. 1991; Wiener and White 1991, 1992) or polyunsaturated lipid membranes (Eldho et al. 2003; Huster et al. 1998; Petrache et al. 2001; Salmon et al. 1987; Stillwell and Wassall 2003) are likewise of interest, e.g., as components of raft-like lipid mixtures (Honerkamp-Smith et al. 2009; Veatch et al. 2007; Wassall and Stillwell 2009; Wassall et al. 2018). Accordingly, the solid-state NMR approach (Fig. 5) has also been extended to include comparative studies of the interactions of cholesterol with unsaturated and polyunsaturated lipid bilayers (Huster et al. 1998; Wassall et al. 2004). Similarities and differences are evident versus saturated lipid bilayers in the liquid-crystalline state. For unsaturated POPC and DOPC lipids, an increase in the residual quadrupolar couplings and derived order parameter profiles occurs upon incorporation of cholesterol (Bartels et al. 2008; Chakraborty et al. 2020) as in the case of saturated DMPC-d54 lipids. For DOPC a small amount of 1-perdeuteriopalmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC-d31) is used as a tracer lipid (1–10 mol%) for its configurational properties, as it is otherwise challenging to chemically deuterate. This approach assumes additivity (sum rule) of the partial molar quantities of the components in the liquid-crystalline state (ld or lo phase) (Jansson et al. 1990, 1992; Otten et al. 2000). Because the tracer lipid is present only in very small amounts, it adequately captures the properties of the host bilayer (Doole et al. 2022). Notably, the smaller absolute values for the POPC-d31 tracer lipid in the DOPC bilayer versus saturated lipids like DMPC-d54 in the absence of cholesterol manifests the greater degrees of freedom of the acyl chains. According to Eq. (6) the observed residual quadrupolar couplings are directly related to the segmental order parameter SCD profile. Just as for the saturated DMPC-d54 bilayer (Fig. 5), the residual quadrupolar couplings give a profile of the absolute segmental order parameters as a function of chain position (index i), due to the effects of the bilayer packing on the trans–gauche isomerizations of the lipid acyl groups. An example of solid-state 2H NMR data for the DOPC bilayer using POPC-d31 as a tracer or probe lipid (Fig. 7a) shows that the resolved signals have a progressive increase in their quadrupolar splittings with greater mole fraction of cholesterol. In most aspects, the results for DOPC using the tracer lipid POPC-d31 are qualitatively like other lipid systems such as DMPC with cholesterol. Clearly there is a striking increase in the order parameter profiles due to cholesterol (Fig. 7a) that must be explained by any model of the phospholipid–sterol interactions.
We next turn to the influence of cholesterol on the dynamics of the di-monounsaturated DOPC bilayer as detected from the combined relaxation rate and order parameter profiles in solid-state 2H NMR spectroscopy (Chakraborty et al. 2020). The unsaturated lipid DOPC has attracted considerable attention as a prominent component of model raft-like lipid mixtures, as shown by the pioneering work of Klaus Gawrisch et al. (Veatch et al. 2007). For the unsaturated DOPC bilayer the presence of cholesterol yields an appreciable influence on its elastic deformation, revealed by a decrease in the square-law slopes (Fig. 7b) just as seen for the saturated DPMC-d54 bilayer (Fig. 6a). By contrast, local trans–gauche isomerizations along the chains do not predict such a square-law dependence as seen for the DOPC/cholesterol membranes. For DOPC as well as DMPC, addition of cholesterol yields a large decrease in the square-law slopes, which we interpret as a progressive reduction in bilayer elasticity (Fig. 7b). The increased bending rigidity is due to short-range cholesterol–phospholipid interactions, showing that membrane mechanical properties emerge from the local atomistic interactions within the lipid bilayer over nanoscopic distances for both unsaturated and saturated bilayers (Ashkar et al. 2021). Our findings differ from the previous diffusive X-ray scattering experiments (Nagle 2021; Pan et al. 2009), and suggest the membrane properties depend on the length and time scales of the measurements (Chakraborty et al. 2020). Based on the solid-state 2H NMR spectroscopy, the progressively smaller square-law slopes (Fig. 7b) correspond to a reduction in bilayer elasticity (Fig. 6b), which we suggest is due to the universal increase in the bending rigidity of saturated and unsaturated lipid bilayers due to cholesterol (Ashkar et al. 2021).
Further interpretation of the energetic cost of the membrane remodeling considers how the bilayer packing and structural properties affect the bending energy of the lipids. Here the bilayer deformation as deduced from the NMR relaxation model is compared with the lipid packing obtained from the mean-torque analysis for cholesterol-containing binary and ternary systems (Bartels et al. 2008). The order parameter profiles (or the spectral moments) from 2H NMR spectroscopy are interpreted by applying the first-order mean-torque model of Petrache and coworkers (Petrache et al. 2000) in terms of the area per lipid at the aqueous interface and the corresponding bilayer thickness (Fig. 7c). Notably, for both unsaturated and saturated lipid bilayers, the changes in the segmental order parameter profile due to cholesterol manifest the decrease in area per lipid and increase in bilayer thickness owing to closer packing of the lipids (less free volume, higher density) (Heerklotz and Tsamaloukas 2006). The matching decrease in square-law slope (Fig. 7b) thus implies a positive correlation of the NMR-derived bending rigidity with the area per lipid and the bilayer thickness obtained from the mean-torque model—that is to say, with the close lipid packing. From the combined solid-state 2H NMR analysis a self-consistent picture emerges, in which the acyl chain packing obtained from the segmental order parameter profiles (Mallikarjunaiah et al. 2019; Petrache et al. 2000) directly corresponds with the bilayer deformation obtained from the NMR relaxation analysis, i.e., bilayer stiffness or stretching (Figs. 6b and 7c). Accordingly, we propose that cholesterol-induced stiffening of saturated and unsaturated bilayers is driven by the same lipid-packing considerations. At the level of a continuum elastic picture, the bilayer properties are averaged over the solid-state 2H NMR time scale (lifetime ~ 10–5 s depending on the quadrupolar coupling) (Molugu et al. 2017). Reduction of the area per lipid at the aqueous interface leads to greater bilayer thickness, in which tighter acyl chain packing yields an increased energetic penalty for separating or bending the lipids (increased area elastic modulus KA and bending rigidity κ). The energy cost of curving the more tightly packed bilayer is thus increased by close packing of the lipids (condensing effect), in which the area and curvature elastic moduli (KA and κ, respectively) are related, e.g., as described by a polymer brush model (Doktorova et al. 2019; Rawicz et al. 2000) (Figs. 6b and 7c).
One should also keep in mind that the observables from 2H NMR spectroscopy are averaged by lipid motions over the relevant spectroscopic time scale, which for sake of illustration we can take as ~ 10–5 s (Molugu et al. 2017). Assuming a value for the lipid translational diffusion coefficient of DT ≈ 0.5 × 10–11 m2 s–1 taken from the pioneering work of Göran Lindblom et al. (Lindblom and Orädd 1994), together with an area per lipid of ⟨A⟩ = 70 Å2 and using the relation ⟨r2⟩ = 4DTt, we can conclude that the results are averaged over ~ 2 × 300 ≈ 600 lipid molecules (considering both monolayers). Although a bilayer average, the value does not specify the actual length scale of the deformations, which depends on the distribution function of the microdomains. Little evidence of cholesterol-induced phase separation is seen in the systems studied here on the 2H NMR timescale (~ 10–5 s); still, nonideal mixing and phase separations with fewer than ~ 600 lipid molecules are not precluded by the motional averaging (Molugu et al. 2017). Lipids would need to exchange between raft-like microdomains and nonraft regions with lifetimes of ≲ 10–5 s to account for the solid-state 2H NMR spectra. In addition, the relaxation times are on the order of ~ 30–400 ms and so they are averaged over even larger distances (Brown and Davis 1981). It follows that 2H NMR spectroscopy only places limits on the lifetimes of lipids in the microdomains—local stiffening is one possibility that could contribute to the averaged bilayer properties. Below, we will see that essentially a very similar picture is arrived at from analysis of the results of neutron spin-echo experiments.
Application of Neutron Spin-Echo Spectroscopy to Lipid–Cholesterol Interactions
The introduction of neutron spin-echo (NSE) spectroscopy to lipid membranes is highly complementary to solid-state NMR spectroscopy, because both methods detect the mean-square amplitudes and rates of the lipid motions (Ashkar et al. 2021). These techniques inform the structural dynamics of lipid bilayers and their response to perturbations from additives such as drugs and sterols, and external forces. In the case of NSE spectroscopy, the Larmor precession of the neutron spins, contained within the magnetic coils of the spectrometer, serves as a timer for each neutron, and allows the detection of tiny velocity changes ( 10–5) in a scattering event, measured as a change in the final beam polarization (Mezei 1972; Mezei et al. 2002; Monkenbusch and Richter 2007). Specifically, the precession angle is given by:
| 9 |
Here, s–1 T–1 is the gyromagnetic ratio of the neutrons with being the mass of the neutron, h the Planck constant, and λ, the neutron wavelength. In addition l is the length of the precession coil, B is the magnetic field, and x is the direction of the neutron beam. Experimentally, the change in polarization is measured at the sample position, which is directly related to the normalized intermediate scattering function, , as a function of Fourier time.
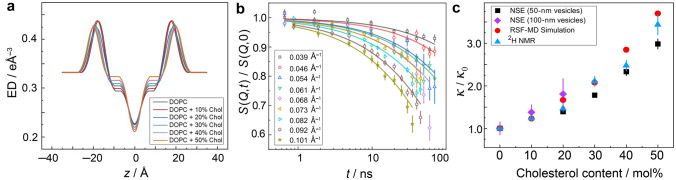
Applying an integrative approach, by combining the NSE data together with small-angle X-ray scattering (SAXS) results and solid-state NMR spectroscopy, we can obtain a comprehensive picture of how sterols control or affect the bending elasticity of lipid membranes. Initially the electron density profiles (Fig. 8a) obtained from the SAXS data are fit to yield the phosphate-to-phosphate (p–p) thickness, giving a direct measure of the bilayer structural dimensions. For di-monounsaturated DOPC bilayers, the addition of cholesterol yields a progressive increase in the p–p distance, showing the occurrence of a greater bilayer thickness (Fig. 8a). The concomitant reduction in the area per phospholipid indicates bilayer condensation due to cholesterol that has been extensively documented for saturated lipid bilayer systems (vide supra). Above we have concluded from solid-state 2H NMR data that closer bilayer packing (less free volume), corresponds to a larger bilayer area elastic modulus (KA) and greater bending rigidity (κ) for the unsaturated DOPC bilayer. Hence for the unsaturated DOPC and POPC bilayers, the findings are generally consistent with the previous results for saturated lipids (Martinez et al. 2002b).
Fig. 8.
Neutron spin-echo (NSE) spectroscopy and solid-state 2H NMR relaxometry show nearly identical increases in bending modulus of unsaturated lipid membranes with cholesterol mole fraction. a Electron density (ED) profiles along the membrane normal (z-axis) obtained from SAXS data for DOPC/cholesterol membranes, such that z = 0 denotes the center of the membrane. b NSE-measured intermediate scattering functions on 50-nm vesicles of protiated DOPC membranes with 20 mol% cholesterol. Error bars represent ± 1 standard deviation. The lines are fits to the data using a stretched exponential function. c Relative bending rigidity moduli (κ/κ0) calculated from nanoscale bending fluctuations sampled by NSE spectroscopy, solid-state 2H NMR relaxometry, and analysis of MD simulations. Adapted from Ref. (Chakraborty et al. 2020)
In neutron spin-echo spectroscopy, the relaxation rates are directly related to the bending elasticity through the Zilman–Granek theory, in which model-free observations of slower NSE decays are associated with bilayer stiffening. The reduced NSE data set yields the normalized intermediate scattering function S(Q,t)/S(Q,0) for discrete q-values within the accessed q-range, where t is the Fourier time (Fig. 8b). The time-independent (yet q-dependent) relaxation rate is analyzed in terms of bending fluctuations, as predicted by Zilman and Granek (ZG). Signatures of such undulatory motions are manifested in relaxation spectra with stretched exponential decays given by (Zilman and Granek 1996):
| 10 |
In the above formula, A is the amplitude of the fluctuations and the parameter is a Q-dependent decay rate which directly relates to the bending rigidity modulus. Stretched exponential decays of this form are typically observed in NSE relaxation spectra of vesicular lipid membranes. The underlying physical reason has to do with the collective membrane dynamics, e.g., these deformations are governed by membrane elastic properties, such as the bending rigidity modulus, . Early NSE measurements have provided direct experimental evidence that membrane bending undulations follow an elastic sheet model (Yamada et al. 2005).
Extraction of the bending rigidity modulus in these measurements uses refinements of the original ZG theory (Watson and Brown 2010), which take into account the interleaflet friction to interpret the relaxation rates in terms of the effective bending modulus, . In this notation, is related to the bilayer bending rigidity, , by where is the monolayer area compressibility and d is the distance between the neutral surface and the bilayer midplane (Watson and Brown 2010). Fits of the intermediate scattering functions using Eq. (10) yield the decay rate, at the various individual -values (Fig. 8b). With increasing cholesterol mole fraction the NSE decays become progressively slower (Chakraborty et al. 2020), which is indicative of bilayer stiffening due to the presence of cholesterol, consistent with the results of NMR spectroscopy.
For the case of bending fluctuations, the ZG relaxation rate (Hoffmann et al. 2014; Nagao et al. 2017) reads:
| 11 |
where is the solvent viscosity, is the Boltzmann constant, and T is the temperature on an absolute scale. In this formula, the neutral surface (where the cross-sectional area per lipid molecule remains constant) is considered to be at the interface between the hydrophilic head group and the hydrophobic fatty acid chains. With this approach, values of the bending rigidity moduli have been reported for various phospholipid membranes in liquid-disordered (ld) and liquid-ordered (lo) states. A direct measurement of membrane mechanics on nanoscopic scales thus becomes possible (Arriaga et al. 2009b; De Mel et al. 2020; Gupta et al. 2019; Lee et al. 2010; Sharma et al. 2017; Woodka et al. 2012). Following this method, the calculated bending-rigidity moduli shows ~ 3.5-fold relative increase for DOPC/cholesterol membranes with 50 mol% cholesterol, i.e., ∼ 3.5 (Fig. 8c). Analogous results have been obtained for unsaturated POPC lipid membranes by neutron spin-echo measurements (Arriaga et al. 2009a, 2010), as seen for interactions of cholesterol with POPC-d31 bilayers using solid-state 2H NMR spectroscopy (Martinez et al. 2002a; Molugu and Brown 2016). Excellent agreement is found with the results of different biophysical techniques: solid-state 2H NMR spectroscopy, neutron spin-echo (NSE) spectroscopy, and molecular dynamics (MD) simulations of saturated and unsaturated lipid bilayers (Ashkar et al. 2021; Chakraborty et al. 2020) all support the essential features of the analysis (Ashkar et al. 2021).
Accordingly, from the results of neutron spin-echo (NSE) spectroscopy, solid-state NMR spectroscopy, and molecular dynamics simulations (Chakraborty et al. 2020), we infer that cholesterol stiffens both saturated and unsaturated lipid membranes by well-established structure–property relations of self-assemblies. Although solid-state NMR and NSE spectroscopy measure the temporal decay of the measured quantities, they both include the mean-square amplitudes and decay rates (Ashkar et al. 2021). Clearly it is necessary to separate the mean-square amplitudes from the membrane dynamics in the data reduction and analysis. The relaxations measured by NMR and NSE spectroscopy both depend on both structure and dynamics and are related to elastic membrane constants, where the stiffening effects of cholesterol are consistent with the previous studies of saturated and unsaturated lipid membranes (Bartels et al. 2008; Chakraborty et al. 2020).The results are explicable by a modified polymer brush theory that relates the bending rigidity κ to the area compressibility modulus KA in agreement with molecular dynamics simulations (Doktorova et al. 2019). Our interpretation differs from the previous experiments on microscopic scales that have led to somewhat different conclusions regarding the mechanical effects of cholesterol for lipids with different acyl chain unsaturation (Ashkar et al. 2021; Pan et al. 2009). As an explanation, we suggest there may be a length-scale and time-scale dependence of the membrane elastic properties for unsaturated versus saturated lipid membranes (Chakraborty et al. 2020). Effectively the persistence length of the cholesterol-induced stiffening may be less in the case of more unsaturated lipids, e.g., due to nonideal mixing of multicomponent lipid systems. We have thus proposed that membrane elastic properties including bending rigidity κ scale with molecular packing, give rise to a universal stiffening of lipid membranes by sterols due to the natural selection of membrane biophysical properties (Ashkar et al. 2021).
Discussion
Cellular membrane properties entail lipid interactions with proteins, peptides, and sterols, and are deeply connected to the evolution of biophysical functions. Knowledge of how incorporation of these essential molecules affects acyl chain relaxations and collective lipid dynamics is required on the ps–ns timescales of protein conformational changes in relating biomembrane structure to function. Our integrative approach combining solid-state NMR measurements of lipid acyl chain relaxations and neutron spin-echo (NSE) studies of ps–ns membrane fluctuations brings out essential knowledge of the functional dynamics underlying a wide range of biological phenomena. For instance, the ability to correlate segmental acyl chain relaxations near the interleaflet plane or near the water interface (accessed through solid-state NMR) with emergent membrane fluctuations (acquired through NSE) can shed light on the molecular mechanisms by which biological inclusions like proteins and sterols regulate collective membrane dynamics and resultant functions. In the NMR studies, probe molecules that differentially sequester in different membrane compartments can be used to correlate local submolecular lipid relaxations with NSE investigations of collective fluctuations within selectively contrasted membrane compartments (Nickels et al. 2015). Such synergistic investigations can provide much needed insights into the fundamental dynamic processes that are vital to our understanding of biological functions going forward.
Our recent studies combining these two approaches have illuminated the need for understanding the effects of cholesterol on the local elastic and viscoelastic properties of lipid membranes with various degrees of acyl chain unsaturation. Such investigations are key to comprehending the role of local membrane mechanics in a biological setting involving functions that occur on mesoscopic length and time scales of lipid–protein interactions, cell and organelle replication, viral budding, and membrane shape deformations. From the existing literature, there is a clear correlation in favor of an overall increase in membrane rigidity and molecular packing due to lipid interactions with cholesterol. The cholesterol-induced variations in membrane mechanics as detected from various studies point us toward a general explanation of the underlying molecular interactions. Taken together, the various biophysical results give a more complete picture than with any single method, because of the likelihood of scale-dependent properties (Ashkar et al. 2021). Closer lipid packing (e.g., condensing effect due to cholesterol, less free volume) yields a greater energy cost of stretching or bending the bilayer. It is interesting that the stiffening effect due cholesterol found for both saturated and unsaturated lipid membranes follows the pathway of biosynthesis from lanosterol and retraces the molecular evolutionary history. The simultaneous increase in cholesterol-induced molecular packing, membrane viscosity, and bending rigidity (Chakraborty et al. 2020) indicates a molecular-level suppression of elastic fluctuations that alters the local viscoelastic properties of the membrane for both saturated and unsaturated bilayers (Molugu and Brown 2016).
Local stiffening due to cholesterol in both saturated and unsaturated lipid membranes has clear biophysical significance with regards to lipid mixing in biological membranes, as originally considered by Erwin London et al. (Brown and London 1998; Veatch et al. 2008). We have proposed that the condensing effect of cholesterol together with its influences on both area and curvature elastic deformation all have their origin in closer molecular packing (less free volume and a reduction of molecular area at the lipid/water interface) (Chakraborty et al. 2020). Future extension to membranes with varying degrees of lipid unsaturation or multicomponent lipid mixtures can inform differences in the mechanical response of asymmetric lipid membranes or domain-forming (raft-like) bilayers (Brown and London 1998; Simons and Gerl 2010). Because of the abundance of cholesterol in eukaryotic cellular membranes, these studies take on importance with regard to the actions of cholesterol in disorders of lipid metabolism associated with cardiovascular disease, cancer, and dysfunctions of G-protein–coupled receptors (GPCRs) and ion channels, as well the selective actions of antimicrobial peptides (Doole et al. 2022). In turn, biophysical properties due to lipid packing and membrane stiffening may have been selected during the molecular evolution of cholesterol from lanosterol, as recapitulated in its biosynthetic pathway. The bilayer stiffening due to cholesterol may also be relevant to the formation of membrane lipid rafts and the selectivity of antimicrobial peptides. In future applications to artificial cells and synthetic biology, we envision that demethylation of sterols affords a way to regulate bilayer curvature and flexibility and thus control cellular function. In addition, how cholesterol affects the actions of antimicrobial peptides as well as the maturation of viruses such as HIV and SARS-CoV-2 (coronavirus) remain as timely questions of urgent biomedical and biophysical relevance.
Author contributions
FTD, TK, RA, and MFB wrote and edited the manuscript with discussion, review, and contributions from all authors.
Funding
We gratefully acknowledge support from the U.S. National Science Foundation (CHE 1904125 to M.F.B. and MCB 2137154 to R.A.) and the U.S. National Institutes of Health (R01EY026041 to M.F.B.).
Data availability
The datasets generated during and/or analyzed during the current study are available in the University of Arizona Data Repository (https://doi.org/10.25422/azu.data.12712856.v4) or in the Virginia Tech Data Repository (VTechData; https://doi.org/10.7294/v8w6-7760). The data sharing for additional figures are not applicable as no datasets were generated or analyzed during the current study.
Declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare that they have no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Rana Ashkar, Email: ashkar@vt.edu.
Michael F. Brown, mfbrown@email.arizona.edu
References
- Allen TM, Cullis PR. Liposomal drug delivery systems: from concept to clinical applications. Adv Drug Deliv Rev. 2013;65:36–48. doi: 10.1016/j.addr.2012.09.037. [DOI] [PubMed] [Google Scholar]
- Allender DW, Sodt AJ, Schick M. Cholesterol-dependent bending energy is important in cholesterol distribution of the plasma membrane. Biophys J. 2019;116:2356–2366. doi: 10.1016/j.bpj.2019.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arfken G. Mathematical methods for physicsts. New York: Academic Press; 1970. [Google Scholar]
- Arriaga LR, López-Montero I, Monroy F, Orts-Gil G, Hellweg T. Stiffening effect of cholesterol on disordered lipid phases: a combined neutron spin echo + dynamic light scattering analysis of the bending elasticity of large unilamellar vesicles. Biophys J. 2009;96:3629–3637. doi: 10.1016/j.bpj.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arriaga LR, López-Montero I, Orts-Gil G, Farago B, Hellweg T, Monroy F. Fluctuation dynamics of spherical vesicles: frustration of regular bulk dissipation into subdiffusive relaxation. Phys Rev E. 2009;80:031908. doi: 10.1103/PhysRevE.80.031908. [DOI] [PubMed] [Google Scholar]
- Arriaga LR, Rodríguez-García R, López-Montero I, Farago B, Hellweg T, Monroy F. Dissipative curvature fluctuations in bilayer vesicles: coexistence of pure-bending and hybrid curvature-compression modes. Eur Phys J E. 2010;31:105–113. doi: 10.1140/epje/i2010-10551-1. [DOI] [PubMed] [Google Scholar]
- Ashkar R, Doktorova M, Heberle FA, Scott HL, Barrera FN, Katsaras J, Khelashvili G, Brown MF. Reply to Nagle et al.: The universal stiffening effects of cholesterol on lipid membranes. Proc Natl Acad Sci USA. 2021;118:e2102845118. doi: 10.1073/pnas.21028451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auger M, Jarrell HC, Smith IC. Interactions of the local anesthetic tetracaine with membranes containing phosphatidylcholine and cholesterol: a 2H NMR study. Biochemistry. 1988;27:4660–4667. doi: 10.1021/bi00413a012. [DOI] [PubMed] [Google Scholar]
- Bartels T, Bittman R, Beyer K, Brown MF. Raft-like mixtures of sphingomyelin and cholesterol investigated by solid-state 2H NMR spectroscopy. J Am Chem Soc. 2008;130:14521–14532. doi: 10.1021/ja801789t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassereau P, Sorre B, Lévy A. Bending lipid membranes: experiments after W. Helfrich's model. Adv Colloid Interface Sci. 2014;208:47–57. doi: 10.1016/j.cis.2014.02.002. [DOI] [PubMed] [Google Scholar]
- Bloch K. The biological synthesis of cholesterol. Science. 1965;150:19–28. doi: 10.1126/science.150.3692.19. [DOI] [PubMed] [Google Scholar]
- Bloch K. Sterol structure and membrane function. CRC Crit Rev Biochem. 1983;14:47–92. doi: 10.3109/10409238309102790. [DOI] [PubMed] [Google Scholar]
- Bloom M, Evans E, Mouritsen OG. Physical properties of the fluid lipid-bilayer component of cell membranes: a perspective. Q Rev Biophys. 1991;24:293–397. doi: 10.1017/S0033583500003735. [DOI] [PubMed] [Google Scholar]
- Bonmatin J-M, Smith ICP, Jarrell HC, Siminovitch DJ. Use of a comprehensive approach to molecular dynamics in ordered lipid systems: cholesterol reorientation in ordered lipid bilayers. A 2H NMR relaxation case study. J Am Chem Soc. 1990;112:1697–1704. doi: 10.1021/ja00161a007. [DOI] [Google Scholar]
- Brown D, London E. Functions of lipid rafts in biological membranes. Annu Rev Cell Dev Biol. 1998;14:111–136. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- Brown MF. Theory of spin-lattice relaxation in lipid bilayers and biological membranes. 2H and 14N quadrupolar relaxation. J Chem Phys. 1982;77:1576–1599. doi: 10.1063/1.443940. [DOI] [Google Scholar]
- Brown MF. Unified picture for spin-lattice relaxation of lipid bilayers and biomembranes. J Chem Phys. 1984;80:2832–2836. doi: 10.1063/1.447031. [DOI] [Google Scholar]
- Brown MF. Anisotropic nuclear spin relaxation of cholesterol in phospholipid bilayers. Mol Phys. 1990;71:903–908. doi: 10.1080/00268979000102201. [DOI] [Google Scholar]
- Brown MF. Modulation of rhodopsin function by properties of the membrane bilayer. Chem Phys Lipids. 1994;73:159–180. doi: 10.1016/0009-3084(94)90180-5. [DOI] [PubMed] [Google Scholar]
- Brown MF. Membrane structure and dynamics studied with NMR spectroscopy. In: Merz K Jr, Roux B, editors. Biological membranes. A molecular perspective from computation and experiment. Birkhäuser, Basel; 1996. [Google Scholar]
- Brown MF. Influence of nonlamellar-forming lipids on rhodopsin. In: Epand RM, editor. Current topics in membranes. San Diego: Academic Press; 1997. [Google Scholar]
- Brown MF. Curvature forces in membrane lipid–protein interactions. Biochemistry. 2012;51:9782–9795. doi: 10.1021/bi301332v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MF. Soft matter in lipid–protein interactions. Annu Rev Biophys. 2017;46:379–410. doi: 10.1146/annurev-biophys-070816-033843. [DOI] [PubMed] [Google Scholar]
- Brown MF. Collective dynamics in lipid membranes. In: Nieh M-P, Heberle FA, Katsaras J, editors. Characterization of biological membranes: structure and dynamics. Berlin: De Gruyter; 2019. [Google Scholar]
- Brown MF, Chan SI. Bilayer membranes: deuterium & carbon-13 NMR. In: Grant DM, Harris RK, editors. Encyclopedia of nuclear magnetic resonance. New York: Wiley; 1996. [Google Scholar]
- Brown MF, Davis JH. Orientation and frequency dependence of the deuterium spin-lattice relaxation in multilamellar phospholipid dispersions: implications for dynamic models of membrane structure. Chem Phys Lett. 1981;79:431–435. doi: 10.1016/0009-2614(81)85008-7. [DOI] [Google Scholar]
- Brown MF, Ribeiro AA, Williams GD. New view of lipid bilayer dynamics from 2H and 13C NMR relaxation time measurements. Proc Natl Acad Sci USA. 1983;80:4325–4329. doi: 10.1073/pnas.80.14.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MF, Seelig J. Influence of cholesterol on the polar region of phosphatidylcholine and phosphatidylethanolamine bilayers. Biochemistry. 1978;17:381–384. doi: 10.1021/bi00595a029. [DOI] [PubMed] [Google Scholar]
- Brown MF, Thurmond RL, Dodd SW, Otten D, Beyer K. Composite membrane deformation on the mesoscopic length scale. Phys Rev E. 2001;64:010901. doi: 10.1103/PhysRevE.64.010901. [DOI] [PubMed] [Google Scholar]
- Brown MF, Thurmond RL, Dodd SW, Otten D, Beyer K. Elastic deformation of membrane bilayers probed by deuterium NMR relaxation. J Am Chem Soc. 2002;124:8471–8484. doi: 10.1021/ja012660p. [DOI] [PubMed] [Google Scholar]
- Brzustowicz MR, Cherezov V, Caffrey M, Stillwell W, Wassall SR. Molecular organization of cholesterol in polyunsaturated membranes: microdomain formation. Biophys J. 2002;82:285–298. doi: 10.1016/S0006-3495(02)75394-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunge A, Müller P, Stöckl M, Herrmann A, Huster D. Characterization of the ternary mixture of sphingomyelin, POPC, and cholesterol: support for an inhomogeneous lipid distribution at high temperatures. Biophys J. 2008;94:2680–2690. doi: 10.1529/biophysj.107.112904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Doktorova M, Molugu TR, Heberle FA, Scott HL, Dzikovski B, Nagao M, Stingaciu LR, Standaert RF, Barrera FN, Katsaras J, Khelashvili G, Brown MF, Ashkar R. How cholesterol stiffens unsaturated lipid membranes. Proc Natl Acad Sci U S A. 2020;117:21896–21905. doi: 10.1073/pnas.2004807117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelius F. Modulation of Na, K-ATPase and Na-ATPase activity by phospholipids and cholesterol. I. Steady-state kinetics. Biochemistry. 2001;40:8842–8851. doi: 10.1021/bi010541g. [DOI] [PubMed] [Google Scholar]
- Corvera E, Mouritsen OG, Singer MA, Zuckermann MJ. The permeability and the effect of acyl-chain length for phospholipid bilayers containing cholesterol: theory and experiment. Biochim Biophys Acta. 1992;1107:261–270. doi: 10.1016/0005-2736(92)90413-G. [DOI] [PubMed] [Google Scholar]
- Craig M, Yarrarapu SNS, Dimri M. Biochemistry, cholesterol. Treasure Island (FL): StatPearls Publishing LLC; 2021. [PubMed] [Google Scholar]
- Dahl JS, Dahl CE, Bloch K. Sterols in membrane: growth characteristics and membrane properties of Mycoplasma capricolum cultured on cholesterol and lanosterol. Biochemistry. 1980;19:1467–1472. doi: 10.1021/bi00548a032. [DOI] [PubMed] [Google Scholar]
- De Mel JU, Gupta S, Perera RM, Ngo LT, Zolnierczuk P, Bleuel M, Pingali SV, Schneider GJ. Influence of external NaCl salt on membrane rigidity of neutral DOPC vesicles. Langmuir. 2020;36:9356–9367. doi: 10.1021/acs.langmuir.0c01004. [DOI] [PubMed] [Google Scholar]
- De Meyer FJM, Rodgers JM, Willems TF, Smit B. Molecular simulation of the effect of cholesterol on lipid-mediated protein-protein interactions. Biophys J. 2010;99:3629–3638. doi: 10.1016/j.bpj.2010.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deserno M. Fluid lipid membranes: from differential geometry to curvature stresses. Chem Phys Lipids. 2015;185:11–45. doi: 10.1016/j.chemphyslip.2014.05.001. [DOI] [PubMed] [Google Scholar]
- Dimova R. Recent developments in the field of bending rigidity measurements on membranes. Adv Colloid Interface Sci. 2014;208:225–234. doi: 10.1016/j.cis.2014.03.003. [DOI] [PubMed] [Google Scholar]
- Doktorova M, Levine MV, Khelashvili G, Weinstein H. A New computational method for membrane compressibility: bilayer mechanical thickness revisited. Biophys J. 2019;116:487–502. doi: 10.1016/j.bpj.2018.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doole FT, Chun CK, Streitwieser E, Sarkar D, Struts AV, Singharoy A, Brown MF. Rivalry of cholesterol and antimicrobial peptides as seen by molecular simulations and NMR spectroscopy. Biophys J. 2022;121:161a–162a. doi: 10.1016/j.bpj.2021.11.1922. [DOI] [Google Scholar]
- Duwe H, Sackmann E. Bending elasticity and thermal excitations of lipid bilayer vesicles: modulation by solutes. Physica A. 1990;163:410–428. doi: 10.1016/0378-4371(90)90349-W. [DOI] [Google Scholar]
- Eldho NV, Feller SE, Tristram-Nagle S, Polozov IV, Gawrisch K. Polyunsaturated docosahexaenoic vs docosapentaenoic acid—differences in lipid matrix properties from the loss of one double bond. J Am Chem Soc. 2003;125:6409–6421. doi: 10.1021/ja029029o. [DOI] [PubMed] [Google Scholar]
- Endress E, Heller H, Casalta H, Brown MF, Bayerl T. Anisotropic motion and molecular dynamics of cholesterol, lanosterol, and ergosterol in lecithin bilayers studied by quasielastic neutron scattering. Biochemistry. 2002;41:13078–13086. doi: 10.1021/bi0201670. [DOI] [PubMed] [Google Scholar]
- Evans E, Rawicz W. Entropy-driven tension and bending elasticity in condensed-fluid membranes. Phys Rev Lett. 1990;64:2094–2097. doi: 10.1103/PhysRevLett.64.2094. [DOI] [PubMed] [Google Scholar]
- Fried SDE, Lewis JW, Szundi I, Martínez-Mayorga K, Mahalingam M, Vogel R, Kliger DS, Brown MF. Membrane curvature revisited—the archetype of rhodopsin studied by time-resolved electronic spectroscopy. Biophys J. 2021;120:440–452. doi: 10.1016/j.bpj.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracià RS, Bezlyepkina N, Knorr RL, Lipowsky R, Dimova R. Effect of cholesterol on the rigidity of saturated and unsaturated membranes: fluctuation and electrodeformation analysis of giant vesicles. Soft Matter. 2010;6:1472–1482. doi: 10.1039/B920629A. [DOI] [Google Scholar]
- Gupta S, De Mel JU, Schneider GJ. Dynamics of liposomes in the fluid phase. Curr Opin Colloid Interface Sci. 2019;42:121–136. doi: 10.1016/j.cocis.2019.05.003. [DOI] [Google Scholar]
- Heerklotz H, Tsamaloukas A. Gradual change or phase transition: characterizing fluid lipid-cholesterol membranes on the basis of thermal volume changes. Biophys J. 2006;91:600–607. doi: 10.1529/biophysj.106.082669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksen J, Rowat AC, Brief E, Hsueh YW, Thewalt JL, Zuckermann MJ, Ipsen JH. Universal behavior of membranes with sterols. Biophys J. 2006;90:1639–1649. doi: 10.1529/biophysj.105.067652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann I, Michel R, Sharp M, Holderer O, Appavou MS, Polzer F, Farago B, Gradzielski M. Softening of phospholipid membranes by the adhesion of silica nanoparticles—as seen by neutron spin-echo (NSE) Nanoscale. 2014;6:6945–6952. doi: 10.1039/c4nr00774c. [DOI] [PubMed] [Google Scholar]
- Honerkamp-Smith AR, Veatch SL, Keller SL. An introduction to critical points for biophysicists; observations of compositional heterogeneity in lipid membranes. Biochim Biophys Acta. 2009;1788:53–63. doi: 10.1016/j.bbamem.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Buboltz JT, Feigenson GW. Maximum solubility of cholesterol in phosphatidylcholine and phosphatidylethanolamine bilayers. Biochim Biophys Acta. 1999;1417:89–100. doi: 10.1016/s0005-2736(98)00260-0. [DOI] [PubMed] [Google Scholar]
- Huber T, Rajamoorthi K, Kurze VF, Beyer K, Brown MF. Structure of docosahexaenoic acid-containing phospholipid bilayers as studied by 2H NMR and molecular dynamics simulations. J Am Chem Soc. 2002;124:298–309. doi: 10.1021/ja011383j. [DOI] [PubMed] [Google Scholar]
- Huster D, Arnold K, Gawrisch K. Influence of docosahexaenoic acid and cholesterol on lateral lipid organization in phospholipid mixtures. Biochemistry. 1998;37:17299–17308. doi: 10.1021/bi980078g. [DOI] [PubMed] [Google Scholar]
- Ipsen JH, Karlström G, Mouritsen OG, Wennerström H, Zuckermann MJ. Phase equilibria in the phosphatidylcholine-cholesterol system. Biochim Biophys Acta. 1987;905:162–172. doi: 10.1016/0005-2736(87)90020-4. [DOI] [PubMed] [Google Scholar]
- Jansson M, Thurmond RL, Barry JA, Brown MF. Deuterium NMR study of intermolecular interactions in lamellar phases containing palmitoyllysophosphatidylcholine. J Phys Chem. 1992;96:9532–9544. doi: 10.1021/j100202a083. [DOI] [Google Scholar]
- Jansson M, Thurmond RL, Trouard TP, Brown MF. Magnetic alignment and orientational order of dipalmitoylphosphatidylcholine bilayers containing palmitoyllysophosphatidylcholine. Chem Phys Lipids. 1990;54:157–170. doi: 10.1016/0009-3084(90)90009-g. [DOI] [PubMed] [Google Scholar]
- Johner N, Mondal S, Morra G, Caffrey M, Weinstein H, Khelashvili G. Protein and lipid interactions driving molecular mechanisms of in meso crystallization. J Am Chem Soc. 2014;136:3271–3284. doi: 10.1021/ja4129839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khelashvili G, Kollmitzer B, Heftberger P, Pabst G, Harries D. Calculating the bending modulus for multicomponent lipid membranes in different thermodynamic phases. J Chem Theory Comput. 2013;9:3866–3871. doi: 10.1021/ct400492e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khondker A, Alsop RJ, Dhaliwal A, Saem S, Moran-Mirabal JM, Rheinstädter MC. Membrane cholesterol reduces polymyxin B nephrotoxicity in renal membrane analogs. Biophys J. 2017;113:2016–2028. doi: 10.1016/j.bpj.2017.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnun JJ, Leftin A, Brown MF. Solid-state NMR spectroscopy for the physical chemistry laboratory. J Chem Ed. 2013;90:123–128. doi: 10.1021/ed2004774. [DOI] [Google Scholar]
- Kinnun JJ, Mallikarjunaiah KJ, Petrache HI, Brown MF. Elastic deformation and area per lipid of membranes: atomistic view from solid-state deuterium NMR spectroscopy. Biochim Biophys Acta. 2015;1848:246–259. doi: 10.1016/j.bbamem.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauda JB, Venable RM, Freites JA, O’Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, Mackerell AD, Jr, Pastor RW. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J Phys Chem B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose C, Surma MA, Simons K. Organellar lipidomics—background and perspectives. Curr Opin Cell Biol. 2013;25:406–413. doi: 10.1016/j.ceb.2013.03.005. [DOI] [PubMed] [Google Scholar]
- Krause MR, Regen SL. The structural role of cholesterol in cell membranes: from condensed bilayers to lipid rafts. Acc Chem Res. 2014;47:3512–3521. doi: 10.1021/ar500260t. [DOI] [PubMed] [Google Scholar]
- Lee JH, Choi SM, Doe C, Faraone A, Pincus PA, Kline SR. Thermal fluctuation and elasticity of lipid vesicles interacting with pore-forming peptides. Phys Rev Lett. 2010;105:038101. doi: 10.1103/PhysRevLett.105.038101. [DOI] [PubMed] [Google Scholar]
- Leeb F, Maibaum L. Spatially resolving the condensing effect of cholesterol in lipid bilayers. Biophys J. 2018;115:2179–2188. doi: 10.1016/j.bpj.2018.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leftin A, Brown MF. An NMR data base for simulations of membrane dynamics. Biochim Biophys Acta. 2011;1808:818–839. doi: 10.1016/j.bbamem.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leftin A, Molugu TR, Job C, Beyer K, Brown MF. Area per lipid and cholesterol interactions in membranes from separated local-field 13C NMR spectroscopy. Biophys J. 2014;107:2274–2286. doi: 10.1016/j.bpj.2014.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leftin A, Xu X, Brown MF. Phospholipid bilayer membranes: deuterium and carbon-13 NMR spectroscopy. eMagRes. 2014;3:199–214. doi: 10.1002/9780470034590.emrstm1368. [DOI] [Google Scholar]
- Levental I, Veatch S. The continuing mystery of lipid rafts. J Mol Biol. 2016;428:4749–4764. doi: 10.1016/j.jmb.2016.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao A, Cimakasky LM, Hampton R, Nguyen DH, Hildreth JEK. Lipid rafts and HIV pathogenesis: host membrane cholesterol is required for infection by HIV type 1. AIDS Res Hum Retroviruses. 2001;17:1009–1019. doi: 10.1089/088922201300343690. [DOI] [PubMed] [Google Scholar]
- Lindblom G, Orädd G. NMR studies of translational diffusion in lyotropic liquid crystals and in lipid membranes. Prog Nucl Magn Reson Spectrosc. 1994;26:483–515. doi: 10.1016/0079-6565(94)80014-6. [DOI] [Google Scholar]
- Liu S-L, Sheng R, Jung JH, Wang L, Stec E, O’Connor MJ, Song S, Bikkavilli RK, Winn RA, Lee D. Orthogonal lipid sensors identify transbilayer asymmetry of plasma membrane cholesterol. Nat Chem Biol. 2017;13:268–274. doi: 10.1038/nchembio.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorent JH, Levental KR, Ganesan L, Rivera-Longsworth G, Sezgin E, Doktorova M, Lyman E, Levental I. Plasma membranes are asymmetric in lipid unsaturation, packing and protein shape. Nat Chem Bio. 2020;16:644–652. doi: 10.1038/s41589-020-0529-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallikarjunaiah KJ, Kinnun JJ, Petrache HI, Brown MF. Flexible lipid nanomaterials studied by NMR spectroscopy. Phys Chem Chem Phys. 2019;21:18422–18457. doi: 10.1039/C8CP06179C. [DOI] [PubMed] [Google Scholar]
- Mallikarjunaiah KJ, Leftin A, Kinnun JJ, Justice MJ, Rogozea AL, Petrache HI, Brown MF. Solid-state 2H NMR shows equivalence of dehydration and osmotic pressures in lipid membrane deformation. Biophys J. 2011;100:98–107. doi: 10.1016/j.bpj.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrink SJ, Corradi V, Souza PCT, Ingólfsson HI, Tieleman DP, Sansom MSP. Computational modeling of realistic cell membranes. Chem Rev. 2019;119:6184–6226. doi: 10.1021/acs.chemrev.8b00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez GV, Castro-Sanchez J, Dykstra EM, Slavish J, Polt R, Brown MF. Structural and dynamic aspects of sphingomyelin rafts as studied by solid state deuterium NMR spectroscopy. Biophys J. 2002;82:506a. [Google Scholar]
- Martinez GV, Dykstra EM, Lope-Piedrafita S, Brown MF. Lanosterol and cholesterol-induced variations in bilayer elasticity probed by 2H NMR relaxation. Langmuir. 2004;20:1043–1046. doi: 10.1021/la036063n. [DOI] [PubMed] [Google Scholar]
- Martinez GV, Dykstra EM, Lope-Piedrafita S, Job C, Brown MF. NMR elastometry of fluid membranes in the mesoscopic regime. Phys Rev E. 2002;66:050902. doi: 10.1103/PhysRevE.66.050902. [DOI] [PubMed] [Google Scholar]
- Martinez GV, Dykstra EM, Lope-Piedrafita S, Job C, Brown MF. Solid state deuterium NMR study of the effects of sterols on membrane elasticity. Biophys J. 2002;82:544a. [Google Scholar]
- Maxfield FR, Van Meer G. Cholesterol, the central lipid of mammalian cells. Curr Opin Cell Biol. 2010;22:422–429. doi: 10.1016/j.ceb.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meher G, Bhattacharjya S, Chakraborty H. Membrane cholesterol modulates oligomeric status and peptide-membrane interaction of severe acute respiratory syndrome coronavirus fusion peptide. J Phys Chem B. 2019;123:10654–10662. doi: 10.1021/acs.jpcb.9b08455. [DOI] [PubMed] [Google Scholar]
- Méléard P, Gerbaud C, Pott T, Fernandez-Puente L, Bivas I, Mitov MD, Dufourcq J, Botherel P. Bending elasticities of model membranes: influences of temperature and sterol content. Biophys J. 1997;72:2616–2629. doi: 10.1016/S0006-3495(97)78905-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mell M, Moleiro LH, Hertle Y, Fouquet P, Schweins R, López-Montero I, Hellweg T, Monroy F. Bending stiffness of biological membranes: what can be measured by neutron spin echo? Eur Phys J E. 2013;36:75. doi: 10.1140/epje/i2013-13075-2. [DOI] [PubMed] [Google Scholar]
- Mezei F. Neutron spin echo: a new concept in polarized thermal neutron techniques. Z Physik. 1972;255:146–160. doi: 10.1007/bf01394523. [DOI] [Google Scholar]
- Mezei F, Pappas C, Gutberlet T. Neutron spin echo spectroscopy basics, trends and applications. Berlin: Springer-Verlag; 2002. [Google Scholar]
- Miao L, Nielsen M, Thewalt J, Ipsen JH, Bloom M, Zuckermann MJ, Mouritsen OG. From lanosterol to cholesterol: structural evolution and differential effects on lipid bilayers. Biophys J. 2002;82:1429–1444. doi: 10.1016/S0006-3495(02)75497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molugu TR, Brown MF. Cholesterol-induced suppression of membrane elastic fluctuations at the atomistic level. Chem Phys Lipids. 2016;199:39–51. doi: 10.1016/j.chemphyslip.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molugu TR, Lee S, Brown MF. Concepts and methods of solid-state NMR spectroscopy applied to biomembranes. Chem Rev. 2017;117:12087–12132. doi: 10.1021/acs.chemrev.6b00619. [DOI] [PubMed] [Google Scholar]
- Monkenbusch M, Richter D. High resolution neutron spectroscopy—a tool for the investigation of dynamics of polymers and soft matter. C R Phys. 2007;8:845–864. doi: 10.1016/j.crhy.2007.10.001. [DOI] [Google Scholar]
- Morrison C, Bloom M. Orientation dependence of 2H nuclear magnetic resonance spin-lattice relaxation in phospholipid and phospholipid: cholesterol systems. J Chem Phys. 1994;101:749–763. doi: 10.1063/1.468491. [DOI] [Google Scholar]
- Mouritsen OG, Zuckermann MJ. What's so special about cholesterol? Lipids. 2004;39:1101–1113. doi: 10.1007/s11745-004-1336-x. [DOI] [PubMed] [Google Scholar]
- Nagao M, Kelley EG, Ashkar R, Bradbury R, Butler PD. Probing elastic and viscous properties of phospholipid bilayers using neutron spin echo spectroscopy. J Phys Chem Lett. 2017;8:4679–4684. doi: 10.1021/acs.jpclett.7b01830. [DOI] [PubMed] [Google Scholar]
- Nagle JF. Introductory lecture: basic quantities in model biomembranes. Faraday Discuss. 2013;161:11–29. doi: 10.1039/C2FD20121F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagle JF. Measuring the bending modulus of lipid bilayers with cholesterol. Phys Rev E. 2021;104:44405. doi: 10.1103/PhysRevE.104.044405. [DOI] [PubMed] [Google Scholar]
- Nagle JF, Evans EA, Bassereau P, Baumgart T, Tristram-Nagle S, Dimova R. A needless but interesting controversy. Proc Natl Acad Sci USA. 2021;118:e2025011118. doi: 10.1073/pnas.2025011118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagle JF, Tristram-Nagle S. Structure of lipid bilayers. Biochim Biophys Acta. 2000;1469:159–195. doi: 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevzorov AA, Brown MF. Dynamics of lipid bilayers from comparative analysis of 2H and 13C nuclear magnetic resonance relaxation data as a function of frequency and temperature. J Chem Phys. 1997;107:10288–10310. doi: 10.1063/1.474169. [DOI] [Google Scholar]
- Nickels JD, Cheng X, Mostofian B, Stanley C, Lindner B, Heberle FA, Perticaroli S, Feygenson M, Egami T, Standaert RF, Smith JC, Myles DAA, Ohl M, Katsaras J. Mechanical properties of nanoscopic lipid domains. J Am Chem Soc. 2015;137:15772–15780. doi: 10.1021/jacs.5b08894. [DOI] [PubMed] [Google Scholar]
- Niggemann G, Kummrow M, Helfrich W. The bending rigidity of phosphatidylcholine bilayers: dependences on experimental method, sample cell sealing and temperature. J. Phys II (Paris) 1995;5:413–425. doi: 10.1051/jp2:1995141. [DOI] [Google Scholar]
- Oldfield E, Meadows M, Rice D, Jacobs R. Spectroscopic studies of specifically deuterium labeled membrane systems. Nuclear magnetic resonance investigation of the effects of cholesterol in model systems. Biochemistry. 1978;17:2727–2740. doi: 10.1021/bi00607a006. [DOI] [PubMed] [Google Scholar]
- Otten D, Brown MF, Beyer K. Softening of membrane bilayers by detergents elucidated by deuterium NMR spectroscopy. J Phys Chem B. 2000;104:12119–12129. doi: 10.1021/jp001505e. [DOI] [Google Scholar]
- Pan J, Tristram-Nagle S, Kučerka N, Nagle JF. Temperature dependence of structure, bending rigidity, and bilayer interactions of dioleoylphosphatidylcholine bilayers. Biophys J. 2008;94:117–124. doi: 10.1529/biophysj.107.115691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Tristram-Nagle S, Nagle JF. Effect of cholesterol on structural and mechanical properties of membranes depends on lipid chain saturation. Phys Rev E. 2009;80:021931. doi: 10.1103/PhysRevE.80.021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrache HI, Dodd SW, Brown MF. Area per lipid and acyl length distributions in fluid phosphatidylcholines determined by 2H NMR spectroscopy. Biophys J. 2000;79:3172–3192. doi: 10.1016/S0006-3495(00)76551-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrache HI, Feller SE, Nagle JF. Determination of component volumes of lipid bilayers from simulations. Biophys J. 1997;70:2237–2242. doi: 10.1016/S0006-3495(97)78867-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrache HI, Salmon A, Brown MF. Structural properties of docosahexaenoyl phospholipid bilayers investigated by solid-state 2H NMR spectroscopy. J Am Chem Soc. 2001;123:12611–12622. doi: 10.1021/ja011745n. [DOI] [PubMed] [Google Scholar]
- Prasad VR, Bukrinsky MI. New clues to understanding HIV nonprogressors: low cholesterol blocks HIV trans infection. mBio. 2014;5:e01396-14. doi: 10.1128/mBio.01396-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawicz W, Olbrich KC, Mcintosh T, Needham D, Evans E. Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophys J. 2000;79:328–339. doi: 10.1016/S0006-3495(00)76295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif B, Ashbrook SE, Emsley L, Hong M. Solid-state NMR spectroscopy. Nat Rev Methods Primers. 2021;1:2. doi: 10.1038/s43586-020-00002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose ME. Elementary theory of angular momentum. New York: Wiley & Sons; 1957. [Google Scholar]
- Salmon A, Dodd SW, Williams GD, Beach JM, Brown MF. Configurational statistics of acyl chains in polyunsaturated lipid bilayers from 2H NMR. J Am Chem Soc. 1987;109:2600–2609. doi: 10.1021/ja00243a010. [DOI] [Google Scholar]
- Sezgin E, Levental I, Mayor S, Eggeling E. The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat Rev Mol Cell Biol. 2017;18:361–374. doi: 10.1038/nrm.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh SR, Cherezov V, Caffrey M, Soni SP, Locascio D, Stillwell W, Wassall SR. Molecular organization of cholesterol in unsaturated phosphatidylethanolamines: X-ray diffraction and solid state 2H NMR reveal differences with phosphatidylcholines. J Am Chem Soc. 2006;128:5375–5383. doi: 10.1021/ja057949b. [DOI] [PubMed] [Google Scholar]
- Sharma VK, Mamontov E, Ohl M, Tyagi M. Incorporation of aspirin modulates the dynamical and phase behavior of the phospholipid membrane. Phys Chem Chem Phys. 2017;19:2514–2524. doi: 10.1039/c6cp06202d. [DOI] [PubMed] [Google Scholar]
- Sheng R, Chen YH, Gee HY, Stec E, Melowic HR, Blatner NR, Tun MP, Kim Y, Källberg M, Fujiwara TK. Cholesterol modulates cell signaling and protein networking by specifically interacting with PDZ domain-containing scaffold proteins. Nat Commun. 2012;3:1–9. doi: 10.1038/ncomms2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siminovitch DJ, Brown MF, Jeffrey KR. 14N NMR of lipid bilayers: effects of ions and anesthetics. Biochemistry. 1984;23:2412–2420. doi: 10.1021/bi00306a015. [DOI] [PubMed] [Google Scholar]
- Simons K, Gerl MJ. Revitalizing membrane rafts: new tools and insights. Nat Rev Mol Cell Biol. 2010;11:688–699. doi: 10.1038/nrm2977. [DOI] [PubMed] [Google Scholar]
- Simons K, Ikonen E. How cells handle cholesterol. Science. 2000;290:1721–1726. doi: 10.1126/science.290.5497.1721. [DOI] [PubMed] [Google Scholar]
- Smith AA, Vogel A, Engberg O, Hildebrand PW, Huster D. A method to construct the dynamic landscape of a bio-membrane with experiment and simulation. Nat Commun. 2022;13:108. doi: 10.1038/s41467-021-27417-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternin E, Bloom M, Mackay AL. De-Pake-ing of NMR spectra. J Magn Reson. 1983;55:274–282. doi: 10.1016/0022-2364(83)90239-1. [DOI] [Google Scholar]
- Stillwell W, Wassall SR. Docosahexaenoic acid: membrane properties of a unique fatty acid. Chem Phys Lipids. 2003;126:1–27. doi: 10.1016/s0009-3084(03)00101-4. [DOI] [PubMed] [Google Scholar]
- Sun X, Whittaker GR. Role for influenza virus envelope cholesterol in virus entry and infection. J Virol. 2003;77:12543–12551. doi: 10.1128/JVI.77.23.12543-12551.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouard TP, Nevzorov AA, Alam TM, Job C, Zajicek J, Brown MF. Influence of cholesterol on dynamics of dimyristoylphosphatidylcholine as studied by deuterium NMR relaxation. J Chem Phys. 1999;110:8802–8818. doi: 10.1063/1.478787. [DOI] [Google Scholar]
- Urbina JA, Pekerar S, Le H, Patterson J, Montez B, Oldfield E. Molecular order and dynamics of phosphatidylcholine bilayer membranes in the presence of cholesterol, ergosterol and lanosterol: a comparative study using 2H-, 13C- and 31P-NMR spectroscopy. Biochim Biophys Acta. 1995;1238:163–176. doi: 10.1016/0005-2736(95)00117-L. [DOI] [PubMed] [Google Scholar]
- Van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshalovich DA, Moskalev AN, Khersonskii VK. Quantum theory of angular momentum. Singapore: World Scientific Publishing Company; 1988. [Google Scholar]
- Veatch SL, Cicuta P, Sengupta P, Honerkamp-Smith A, Holowka D, Baird B. Critical fluctuations in plasma membrane vesicles. ACS Chem Biol. 2008;3:287–293. doi: 10.1021/cb800012x. [DOI] [PubMed] [Google Scholar]
- Veatch SL, Soubias O, Keller SL, Gawrisch K. Critical fluctuations in domain-forming lipid mixtures. Proc Natl Acad Sci USA. 2007;104:17650–17655. doi: 10.1073/pnas.070351310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venable RM, Brown FLH, Pastor RW. Mechanical properties of lipid bilayers from molecular dynamics simulation. Chem Phys Lipids. 2015;192:60–74. doi: 10.1016/j.chemphyslip.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vist MR, Davis JH. Phase equilibria of cholesterol/dipalmitoylphosphatidylcholine mixtures: 2H nuclear magnetic resonance and differential scanning calorimetry. Biochemistry. 1990;29:451–464. doi: 10.1021/bi00454a021. [DOI] [PubMed] [Google Scholar]
- Wang S, Li W, Hui H, Tiwari SK, Zhang Q, Croker BA, Rawlings S, Smith D, Carlin AF, Rana TM. Cholesterol 25-Hydroxylase inhibits SARS-CoV-2 and other coronaviruses by depleting membrane cholesterol. EMBO J. 2020;39:e106057. doi: 10.15252/embj.2020106057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassall SR, Brzustowicz MR, Shaikh SR, Cherezov V, Caffrey M, Stillwell W. Order from disorder, corralling cholesterol with chaotic lipids. The role of polyunsaturated lipids in membrane raft formation. Chem Phys Lipids. 2004;132:79–88. doi: 10.1016/j.chemphyslip.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Wassall SR, Leng X, Canner SW, Pennington ER, Kinnun JJ, Cavazos AT, Dadoo S, Johnson D, Heberle FA, Katsaras J, Shaikh SR. Docosahexaenoic acid regulates the formation of lipid rafts: a unified view from experiment and simulation. Biochim Biophys Acta. 2018;1860:1985–1993. doi: 10.1016/j.bbamem.2018.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassall SR, Stillwell W. Polyunsaturated fatty acid–cholesterol interactions: domain formation in membranes. Biochim Biophys Acta. 2009;1788:24–32. doi: 10.1016/j.bbamem.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Watson MC, Brown FL. Interpreting membrane scattering experiments at the mesoscale: the contribution of dissipation within the bilayer. Biophys J. 2010;98:L9–L11. doi: 10.1016/j.bpj.2009.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener MC, King GI, White SH. Structure of a fluid dioleoylphosphatidylcholine bilayer determined by joint refinement of x-ray and neutron diffraction data. I. Scaling of neutron data and the distributions of double bonds and water. Biophys J. 1991;60:568–576. doi: 10.1016/S0006-3495(91)82086-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener MC, White SH. Fluid bilayer structure determination by the combined use of x-ray and neutron diffraction. II. "Composition-space" refinement method. Biophys J. 1991;59:174–185. doi: 10.1016/S0006-3495(91)82209-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener MC, White SH. Structure of a fluid dioleoylphosphatidycholine bilayer determined by joint refinement of x-ray and neutron diffraction data III. Complete structure. Biophys J. 1992;61:434–447. doi: 10.1016/S0006-3495(92)81849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GD, Beach JM, Dodd SW, Brown MF. Dependence of deuterium spin–lattice relaxation rates of multilamellar phospholipid dispersions on orientational order. J Am Chem Soc. 1985;107:6868–6873. doi: 10.1021/ja00310a021. [DOI] [Google Scholar]
- Woodka AC, Butler PD, Porcar L, Farago B, Nagao M. Lipid bilayers and membrane dynamics: insight into thickness fluctuations. Phys Rev Lett. 2012;109:058102. doi: 10.1103/PhysRevLett.109.058102. [DOI] [PubMed] [Google Scholar]
- Xu X, London E. The effect of sterol structure on membrane lipid domains reveals how cholesterol can induce lipid domain formation. Biochemistry. 2000;39:843–849. doi: 10.1021/bi992543v. [DOI] [PubMed] [Google Scholar]
- Xu X, Struts AV, Brown MF. Generalized model-free analysis of nuclear spin relaxation experiments. eMagRes. 2014;3:275–286. doi: 10.1002/9780470034590.emrstm1367. [DOI] [Google Scholar]
- Yamada LN, Seto H, Takeda T, Nagao M, Kawabata Y, Inoue K. SAXS, SANS and NSE studies on “unbound state” in DPPC/Water/CaCl2 System. J Phys Soc Japan. 2005;74:2853–2859. doi: 10.1143/jpsj.74.2853. [DOI] [Google Scholar]
- Yeagle PL. Lanosterol and cholesterol have different effects on phospholipid acyl chain ordering. Biochim Biophys Acta. 1985;815:33–36. doi: 10.1016/0005-2736(85)90470-5. [DOI] [PubMed] [Google Scholar]
- Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- Zhang X, Barraza KM, Beauchamp JL. Cholesterol provides nonsacrificial protection of membrane lipids from chemical damage at air–water interface. Proc Natl Acad Sci U S A. 2018;115:3255–3260. doi: 10.1073/pnas.1722323115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilman AG, Granek R. Undulations and dynamic structure factor of membranes. Phys Rev Lett. 1996;77:4788–4791. doi: 10.1103/PhysRevLett.77.4788. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available in the University of Arizona Data Repository (https://doi.org/10.25422/azu.data.12712856.v4) or in the Virginia Tech Data Repository (VTechData; https://doi.org/10.7294/v8w6-7760). The data sharing for additional figures are not applicable as no datasets were generated or analyzed during the current study.