Abstract
Previously, we reported that heterologous expression of an embryonic transcription factor, Tbx18, reprograms ventricular cardiomyocytes into induced pacemaker cells (Tbx18-iPMs), though the key pathways are unknown. Here, we have used a tandem mass tag proteomic approach to characterize the impact of Tbx18 on neonatal rat ventricular myocytes. Tbx18 expression triggered vast proteome remodeling. Tbx18-iPMs exhibited increased expression of known pacemaker ion channels, including Hcn4 and Cx45 as well as upregulation of the mechanosensitive ion channels Piezo1, Trpp2 (PKD2), and TrpM7. Metabolic pathways were broadly downregulated, as were ion channels associated with ventricular excitation–contraction coupling. Tbx18-iPMs also exhibited extensive intracellular cytoskeletal and extracellular matrix remodeling, including 96 differentially expressed proteins associated with the epithelial-to-mesenchymal transition (EMT). RNAseq extended coverage of low abundance transcription factors, revealing upregulation of EMT-inducing Snai1, Snai2, Twist1, Twist2, and Zeb2. Finally, network diffusion mapping of >200 transcriptional regulators indicates EMT and heart development factors occupy adjacent network neighborhoods downstream of Tbx18 but upstream of metabolic control factors. In conclusion, transdifferentiation of cardiac myocytes into pacemaker cells entails massive electrogenic, metabolic, and cytostructural remodeling. Structural changes exhibit hallmarks of the EMT. The results aid ongoing efforts to maximize the yield and phenotypic stability of engineered biological pacemakers.
Keywords: proteomics, systems biology, network, heart, pacemaker, cardiomyocytes, epithelial−mesenchymal transition
Introduction
Cardiac pacing originates from the sinoatrial node (SAN). Cardiac defects in the SAN and/or the conduction system cause bradycardia and irregular heartbeats.1 Current therapy relies on the implantation of electronic pacing devices. However, the indwelling hardware has drawbacks, including the need to change batteries at regular intervals, which entails a surgical procedure that comes with the risk of infection. Likewise, pediatric patients must undergo multiple surgeries to reposition or replace the pacemaker as they outgrow their devices. The development of biological pacemakers promises to transcend these issues.2 We previously showed that somatic gene transfer of an embryonic transcription factor, Tbx18, suffices to reprogram quiescent neonatal rat ventricular myocytes (NRVMs) in vitro and adult ventricular myocytes (VMs) in vivo to induced pacemaker cells.3 The Tbx18-induced pacemaker cells (Tbx18-iPMs) faithfully recapitulated native SAN pacemaker cells’ automaticity. Remarkably, Tbx18-iPMs exhibited the unique morphology of the nodal pacemaker cells, indicating that the reprogramming was not only limited to generating rhythmic action potentials, but it also reconfigured the cellular architecture of the iPMs. Yet, how Tbx18 effectuates this cellular transition is not understood. A mechanistic understanding of Tbx18-induced reprogramming would inform both gene regulatory models of the factors driving the conversion of VMs to PMs, with implications for refined bio-pacemaker engineering as well as the mechanisms of sinoatrial node development in cardiogenesis.
To define the molecular distinctions between VMs and Tbx18-induced pacemakers (Tbx18-iPMs) and elucidate plausible models underlying transdifferentiation, we analyzed global proteomic and select transcript changes. Collectively, these data indicate that Tbx18 rewires the metabolic and electrogenic landscapes while extensively remodeling intracellular and extracellular structures, conferring a biosignature that bears many of the hallmarks of the epithelial-to-mesenchymal transition.
Materials and Methods
Animal Care and NRVM Isolation
Timed-pregnancy Sprague Dawley rats were purchased from Charles River and maintained in accordance with the Emory University Institutional Animal Care and Use Committee (IACUC). NRVMs were isolated from 1–3-day-old pups and cultured as a monolayer, as described previously.3 Only the lower area (from the apex to the midline) was excised to minimize contaminating atrioventricular nodal cells. A monolayer of NRVMs was transduced with either Ad-Tbx18-IRES-zsGreen1 on the day of isolation by incubating them on the shaker for 2 h. Adenoviral vector transduction was performed at a multiplicity of infection (MOI) of 1 fluorescence forming unit (ffu) per cell. Transduction was performed in a routine NRVM culture media based on M199 with the following components: 10 mM HEPES, 0.1 mM nonessential amino acids, 3.5 mg/mL glucose, 2 mM L-glutamine, 4 μg/mL vitamin B12, 100 U/mL penicillin, and heat-inactivated fetal bovine serum at 10% (first two days of culture) or 2% (after two days of culture) final concentration.
RNA-Sequencing Analysis
Total RNA was prepared from GFP-NRVMs and Tbx18-iPMs. After the library was prepared, the sequencing was done with Illumina HiSeq. 3000. RNASeq reads were aligned to the rn5 reference assembly of the rat genome from the UCSC genome browser website (http://genome.ucsc.edu/). Alignment was performed using the STAR aligner with the annotation as a splice junction reference.4 Transcripts were annotated using the corresponding rn5 annotation, and transcript abundance was estimated using htseq-count (version 0.6.1p1).5 Library size normalization and differential expression analyses were performed using the DESeq2. R package.6 A negative binomial generalized linear model was fitted, and the Wald test was used to assess differential expression, including the Benjamini–Hochberg procedure to calculate an adjusted p-value to control the false discovery rate (FDR). Pathways, upstream regulators, and cellular functions were generated using Ingenuity Pathway Analysis (IPA).7
Experimental Design for Mass Spectrometry
NRVMS were isolated and distributed among 10 plates. Five plates are individually virally transduced with the Ad-Tbx18-IRES-zsGreen1, and five are transduced with Ad-GFP, which serve as controls. The relative abundance of proteins from each plate is determined by a method that involves labeling proteolytically digested peptides with isobaric tandem mass tags (TMTs). All samples were subsequently pooled prior to extensive peptide fractionation and mass spectrometry. Peptides and proteins are identified by matching to a rat protein sequence database. Peptide-conjugated mass tags are fragmented in the MS2 phase of MS to yield characteristic reporter ions whose currents are quantified to determine relative protein abundance and analyzed statistically.
Protein Sample Preparation, Proteolysis, and TMT Labeling
Prior to cell harvest, media was removed, and the wells of each plate were washed three times with ice-cold phosphate-buffered saline. Cells were subsequently harvested by addition of 200 μL of ice-cold RIPA buffer and scraping each well before transferring the cell lysate to a conical tube. Samples were flash-frozen in liquid nitrogen and stored at −80 °C until further use. Prior to digestion, cell lysates were thawed quickly and subjected to methanol/chloroform/water extraction and protein precipitation by the method of Wessel and Flugge.8 Samples were dried under nitrogen gas to remove residual chloroform before resuspending protein in filtered and deionized 9 M urea. Samples were allowed to solubilize by vortexing for 30 min. After 30 min, residual protein aggregates were disrupted by brief bursts of sonication (<30 s total). Peptides were diluted to 6-fold into 60 mM HEPES, 0.6 M thiourea, 0.6 mM DTT, pH 7.5, such that the final reaction buffer contained 50 mM HEPES, 1.5 M urea, 0.5 M thiourea, 0.5 mM DTT, pH 7.5. All samples were diluted further with reaction buffer to a common final protein concentration of 1mg/mL. Samples (about 500 μg) were subjected to proteolytic digestion with proteomics grade Trypsin (10 μg; Promega) at room temperature overnight. The following morning, samples were supplemented with 10 μg of Trypsin. At that time, DTT was added to samples at a concentration of 5 mM, and the digest was allowed to proceed for 1 h prior to peptide alkylation by adding iodoacetamide to a final concentration of 15 mM. Alkylation was allowed to proceed for 1 h at room temperature in the dark. Peptides were subsequently acidified by the addition of trifluoroacetic acid to a final concentration of 0.5% (v/v) and purified by solid-phase extraction using SepPak tC18 cartridges (Waters) on a vacuum manifold. Purified peptides were eluted with 60% (v/v) acetonitrile in aqueous 0.1% (v/v) formic acid. Peptides were evaporated to dryness on an Eppendorf Vacufuge. Peptide samples (approximately 500 μg) were re-dissolved in triethylammonium bicarbonate (TEAB) pH 8.5 and labeled with TMT reagents according to the manufacturer’s instructions.
Chromatography and Mass Spectrometry
Following TMT labeling of individual samples, peptides were pooled and subjected to high-pH reversed-phase high-pressure liquid chromatography (bRP-HPLC).9 The apparatus consisted of an offline Agilent capillary LC operating in normal flow mode with a Waters XBridge 5 μm C18, 4.6 mm × 250 mm column. Mobile phase solvents were 10 mM TEAB (pH 8.5) for Buffer A, and 7 mM TEAB, 90% Acetonitrile (pH 8.5) for buffer B. Elution of the peptides was performed at 1 ml/min with a biphasic gradient (5–30% B over 55 min followed by a 35–45% B over 10 min). Seventy-five fractions were collected in Fisher DeepWell plates with a Probot fraction collector (LC Packings). Fractions were pooled according to a concatenation strategy (i.e., pooling samples 1 + 26 + 51, 2 + 27 + 52, etc.) to maximize the orthogonality of peptide separation with respect to subsequent low-pH RP-HPLC.9 Following concatenation, each sample was analyzed by low-pH RP-HPLC coupled to tandem mass spectrometry (LC-MS/MS). Peptides were injected onto a 2 cm trap column at 5 μL/min for 6 min before being eluted onto a 75 μm × 15 cm in-house packed column (Michrom Magic C18AQ, 5 μm, 100Å) using a nanoAquity nanoLC system (Waters) operating at 300 nL/min. Each sample was run on a 90 min gradient. Solvent A consisted of 0.1% (v/v) formic acid and solvent B contained 0.1% (v/v) formic acid and 90% (v/v) acetonitrile. Peptides were eluted and ionized into a Q-Exactive Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher) at a spray voltage of 2.0 kV using a data-dependent “Top 15” method operating in FT-FT acquisition mode. The survey full-scan MS (350–1800 Th) was performed at a resolution of 70,000 with an automatic gain control setting of 3 × 106, while MS2 scans were performed at a resolution of 35,000 with a target value of 1 × 105. Maximum injection times were 100 ms for MS1 and 250 ms for MS2. The MS2 ion selection threshold was set to 4 × 104, and an isolation window of 1.5 Daltons was used to perform HCD fragmentation with a normalized collision energy of 30%. Persistent systemic polysiloxane produced a background peak at 371.101230, which was used as an internal calibrant for each survey scan. Spectra whose charges were unassigned or +1 were not tabulated. Dynamic exclusion was set to 30 s.
Protein Identification
Peak list files (.RAW) were searched against a rat sequence database (NCBI RefSeq, taxonomy: Rattus Norvegicus, 2012, 29 449 sequences) using Mascot Version: 2.2.0 (Matrix Science) interfaced through Proteome Discoverer 1.4 (Thermo). Spectra were searched with a parent ion mass tolerance of 30 ppm and an MS2 mass tolerance of 0.05 Da. Trypsin was specified as the enzyme, and 2 missed cleavage sites were allowed. Cysteine carbamidomethylation and N-terminal amine labeling with TMT reagent were specified as fixed modifications for the database search. Dynamic modifications included N-pyroglutamate from glutamine, oxidized methionine, deamidation of asparagine and glutamine, as well as TMT labeling of lysine. All searches were conducted with the reversed database search mode engaged. Percolator software was used for peptide FDR (q-value) calculations. Mascot output files (.dat) tabulated were in Proteome Discoverer. Proteins were identified on the basis of high-confidence peptides only (q < 0.01).
Quantification of TMT Reporter Ions and Estimation of Relative Protein Abundance
Analysis was confined to uniquely- and unambiguously assigned spectra (1% peptide FDR). Proteins identified by a single peptide were quantified, provided that 2 spectra were observed. Reporter ion intensities were integrated over 20 ppm by the most confident centroid method. Missingness of ion intensities for a single spectrum across reporter channels was low (<2% of unique spectra), indicative of efficient labeling and fragmentation. Spectra with missing channel data were omitted from the analysis. TMT signals were quantified using the median sweep algorithm originally described by Herbrich et al.,10 as implemented in Foster et al.11 Briefly, TMT reporter ion signals were logarithmically transformed (base 2) before each spectrum was median-centered. The estimated relative protein abundance within a channel is considered to be the median value of all log-transformed, median-centered spectra assigned to a protein. Total protein in each TMT channel was normalized by median-centering all relative protein abundances within that channel.
The choice of TMT-based MS2 quantitation has the documented limitation that derived fold changes tend to be compressed, owing largely to peptide co-isolation interference, which we have minimized by extensive peptide fractionation. Nevertheless, the fold changes reported here are best considered as conservative estimates of the actual fold change.
Differential Protein Regulation and Statistics
Differential protein abundance between GFP-transduced and Tbx18-transduced experimental groups was assessed with an empirical Bayes-modified 2-sample t-test. This procedure is essentially equivalent to the linear modeling of microarrays (LIMMA12), modified for quantitative proteomic experiments,11,13−15 essentially as described in the online primer, http://www.biostat.jhsph.edu/~kkammers/software/eupa/R_guide.html.(13) The resulting modified p-values were further corrected for multiple hypothesis testing by Storey’s q-value method16−18 using the R package, “q-values”.19 In the interest of proper accounting for differential regulation false discovery (q), and given the use of moderated p-values, a fold-change threshold was not considered.14 However, for reference, the minimum fold change at q < 0.0001 was ±1.15-fold.
Principal Component Analysis and Hierarchical Clustering
Principal component analysis was performed on log-transformed median swept protein abundances (unfiltered) using Partek software (Genomic Solutions). Unsupervised agglomerative hierarchical clustering was performed using Ward’s clustering method, where the distance metric is defined as half of the square of the Euclidean distance as implemented in Spotfire (Qiagen). Though precise cluster composition is specific to the clustering method, similar results were obtained by clustering using complete linkage and Euclidean distance.
Pathway and Upstream Regulator Analysis
Protein accessions were mapped to their corresponding genes before Pathway analysis was performed using Ingenuity (Qiagen). Pathway overrepresentation was determined using the full proteome as the reference protein set. Specifically, proteins (mapped to their respective genes) from each of the 6 protein clusters in Figure 2 were analyzed for pathway member overrepresentation using Fisher’s exact test. P-values <0.05 were considered statistically significant, which does not necessarily correlate with biological significance. Where the information in the Ingenuity knowledge base permitted, the extent of pathway activation or inhibition is inferred from the direction and magnitude of fold changes among pathway member proteins, which is given by the z-score.7 Pathways are curated by Ingenuity and can be visualized by entering the pathway name at https://targetexplorer.ingenuity.com/index.htm. Overrepresentation can also be applied to the identification of likely upstream regulators, be they transcriptional regulators, kinases, or growth factors. The tenet is that if the identity of the target genes of a given upstream regulator is known from prior experimentation (e.g., microarrays or proteome studies in data repositories), one may infer its likely involvement based on the extent of target gene coregulation in a new data set represented by the p-value (Fisher’s exact test). Moreover, as in pathway analysis, the direction and magnitude of the coregulation permit inference about the likely activation or inhibition given by the z-score. Upstream regular analysis was performed in Ingenuity on proteins (mapped as above) that were differentially regulated between Tbx18 and GFP-transduced groups at an FDR threshold of q < 0.0001.
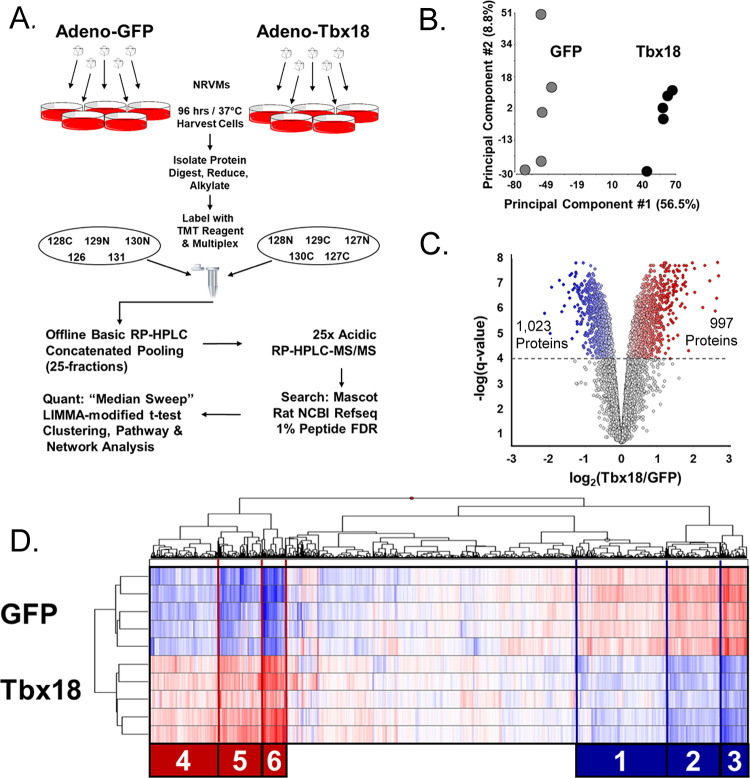
Figure 2.
Overview of the Tbx18-iPM Proteome. (A) Experimental design for Tbx18 viral transduction of induction of NRVMs, protein isolation, and quantitative mass spectrometric analysis. (B) PCA analysis of proteomics from GFP or Tbx18-transduced NRVMs. Each dot represents the proteome arising from independent viral GFP or Tbx18 transduction replicates. (C) Volcano plots depicting significant protein changes between myocytes transduced with GFP or Tbx18. For the purposes of upstream regulator analysis that informs subsequent analyses, differential regulation was defined as q < 0.0001. (D) Hierarchically clustered heat map (Ward’s method, Euclidean distance) of the whole proteome. The most highly regulated proteins fell into six major clusters, three downregulated and three upregulated in Tbx18-iPMs. Each coregulated cluster was used as input for IPA analysis in Figures 3 and 4.
Network Analysis
Functional protein association/interaction networks were constructed by loading the gene identifiers of up and downregulated proteins into stringApp 1.4.2,20 embedded in Cytoscape 3.7.1,21 and then searching the STRING v11 database.22 The default association/interaction threshold (STRING score > 0.4) was used to map relationships between proteins. Network modularity was assessed with the Markov clustering function in the clusterMaker2 1.3.1 app23 using the STRING score (>0.6) for edge weighting. The granularity parameter (inflation value) was set empirically. The final networks are presented in an edge-weighted, spring-embedded layout using the Markov cluster (module) number for edge weighting, with modules rearranged for clarity.
Immunoblot Analysis
GFP and Tbx18-transduced NRVM cell lysates were collected in ice-cold RIPA buffer (Thermo Scientific #PI89900) combined with Halt Protease and Phosphatase inhibitor cocktail (Thermo Scientific #P178442). Lysate concentrations were measured by BCA assay (Thermo Scientific #J63283.QA). Reduced protein samples were run on a 4–12% Bolt Bis-Tris gel (Invitrogen #NW04125BOX) at 10 μg per well. Transferred PVDF membranes were incubated with primary antibodies against total Notch1 (Cell Signaling Technology #3608, 1:1000) and Snail (Invitrogen #MA5-14801, 1:1000) overnight at 4 °C. Secondary staining was performed for 1 h at room temperature with Li-Cor IRDye 800CW and 680RD secondary antibodies (Li-Cor #926-32211 and #926-68071, 1:10000). Stained membranes were digitally scanned with a Li-Cor Odyssey DLx Imaging System and quantified with Image Studio Lite (Li-Cor Biosciences). All measurements were normalized to total protein stain by REVERT 700 Total Protein Stain kit (Li-Cor Biosciences #926-11010).
Results
Preparation of Tbx18-induced Pacemaker Cells
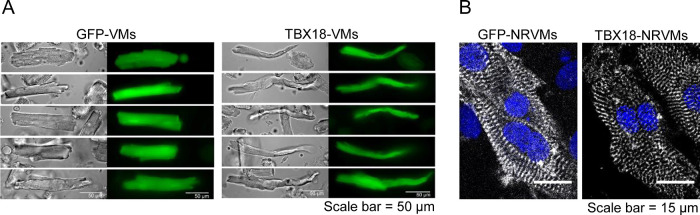
Our previous work indicated that phenotypic conversion of ventricular myocytes to the iPMs was complete by D3–D5 after Tbx18 gene transfer in vitro and in vivo.(3,24) Reprogramming of adult rat ventricular myocytes to the iPMs in vivo by direct myocardial injection of Tbx18 demonstrated that myocytes expressing Tbx18 underwent significant changes in cell morphology, adopting the cell shape of the native SAN pacemaker cells (Figure 1A). The change in cell shape was accompanied by disorganization of sarcomere structure, as illustrated by sarcomeric α-actinin staining in Tbx18-NRVMs compared to GFP-NRVMs (Figure 1B). To gain a general understanding of the proteome changes during Tbx18-induced reprogramming of the cardiomyocytes without contaminating non-myocytes and blood serum, we chose NRVMs as the cardiomyocyte model. Hence, Tbx18-induced pacemaker cells (Tbx18-iPMs) were created by somatic gene transfer of Tbx18 on NRVMs cultured in serum-free media and harvesting the iPMs 4 days after gene transfer. NRVMs transduced with GFP served as the control.
Figure 1.
Overexpression of Tbx18 in ventricular myocytes changes cell morphology accompanied by the disarray of sarcomere structure. (A) Adenoviral vector expressing only a GFP reporter (left) or Tbx18 as well as a zsGreen1 reporter (right) was injected directly into the apex of the rat heart in vivo. Four days later, the heart was harvested, and ventricular myocytes from the apex were isolated ex vivo. Tbx18-expressing ventricular myocytes (Tbx18-VMs) showed spindle-shaped morphology compared to typical brick-like shapes of GFP-expressing ventricular myocytes (GFP-VMs). Scale bar: 50 μm. (B) Neonatal rat ventricular myocytes (NRVMs) were transduced with Ad-GFP (left) or Ad-Tbx18-IRES-zsGreen1 (right) in vitro. Four days later, the myocytes were fixed and stained with an antibody against α-sarcomeric actinin (A7811, Sigma-Aldrich). The sarcomere structure is disorganized in Tbx18-NRVMs compared to that in GFP-NRVMs. Scale bar: 15 μm.
Global Proteomic Trends in the Tbx18-iPMs
To understand the extent of coordinated protein remodeling evoked by Tbx18-induced NRVM/iPM transdifferentiation at a comprehensive level, we undertook a gel-free, peptide-labeled, 2D-LC-MS/MS proteomic identification and quantification strategy (Figure 2A). NRVM culture plates were independently virally transduced GFP (n = 5) or Tbx18 (n = 5) for 96 h prior to cell collection and downstream protein and peptide processing. Principal component analysis (Figure 2B) shows that Tbx18 treatment is the primary contributor to variance (nearly 57%) in these experiments and that iPMs share proteomes that are distinct from those of control, GFP-NRVMs. In all, 6661 proteins were quantified and analyzed statistically. The breadth of protein remodeling was notable. Indeed, at a conventional significance threshold of p < 0.05, 4475 of 6661 proteins (67%) are differentially regulated at a differential expression false discovery rate (q) of 1.6%. For this study, we imposed a threshold q < 0.0001 for significance to keep network analyses manageable. At this threshold, 997 proteins were upregulated, while 1023 were downregulated (Figure 2C). Hierarchical clustering of the entire proteome of TBX18-iPMs vs GFP-NRVMs yielded six major clusters (three downregulated and three upregulated, Figure 2D) encompassing proteins with the largest fold changes, or just over 50% of the proteome (3422 proteins), which we leveraged for pathway analysis.
Protein Downregulation by Tbx18 Implicates Metabolic and Mitochondrial Pathways
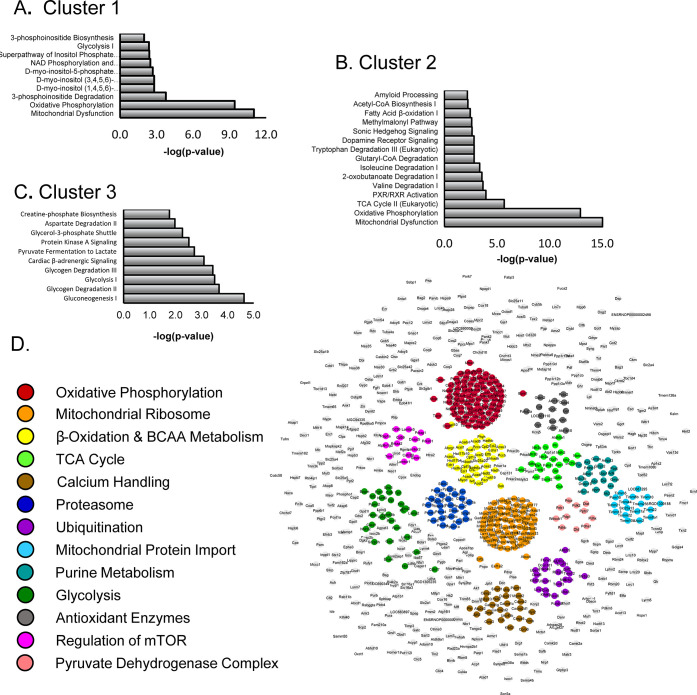
Clusters 1–3 (Figure 2D) encompass the most downregulated proteins in Tbx-iPM proteome. In cluster 1, the average fold change was −1.21, and the top associated pathways (by descending −log(p-value)) are depicted in Figure 3A. Oxidative phosphorylation (OxPhos) was implicated based on changes in 36 proteins from respiratory complexes I–V and their associated assembly factors (p = 1 × 10–11). Next, several pathways associated with inositol phosphate metabolism were implicated. Other pathways such as heme and cardiolipin synthesis are encompassed by cluster 1 (see Online Supplement S1).
Figure 3.
Pathway analysis of downregulated clusters in Tbx18-iPMs indicates broad attenuation of metabolic pathways. (A–C) Clusters 1–3 (Figure 2D), encompassing downregulated proteins, were subjected to pathway analysis (Ingenuity). Pathways are ranked by −log(p-value), which reflects the overlap between the data set and the curated pathways, not the relative importance to the phenotype. Full tables are provided in the Online Supplement. Pathways indicate extensive mitochondrial protein downregulation and broad downregulation of enzymes involved in glucose, fatty acid, and amino acid metabolism. (D) STRING functional protein association network comprised of proteins differentially regulated at a threshold of q < 0.0001. The largest multinode modules are colored. The network uniquely highlights the downregulation of mitochondrial ribosome and protein import as well as the ubiquitin-proteasome machinery and antioxidant proteins, among others.
In cluster 2, the average fold change was −1.42. The remainder of proteins involved in mitochondrial respiration or OxPhos fall into this cluster (Figure 3B), as do the enzymes of the TCA cycle. Amino acid degradation is a dominant theme, particularly of the branched-chain amino acids, valine and isoleucine, due to downregulation of Auh, Acad8, Dld, Hadhb, and Ehhadh, but also tryptophan degradation due to Hsd17b10, L3hypdh, Hadhb, Ehhadh, and Hsd17b4. Other pathways, such as the metabolism of phenylalanine, tyrosine, methionine, glutamine, and aspartate, were also noted (Online Supplement S1). Finally, β-oxidation of long-chain fatty acids (Hsd17b10, scp2, Auh, Hadhb, Ehhadh, Hsd17b4, Eci1; p = 0.004) and the odd-chained fatty acid pathway (Ehhadh, Eci1; p = 0.04) are enriched in this cluster.
Cluster 3 encompasses the most downregulated proteins, with an average fold change of −1.89. Gluconeogenesis, primarily a function of the liver, emerged as a significant pathway (Figure 3C) due to the overlap of enzymes primarily involved in glycolysis in NRVMs, which was, likewise, significant (Eno3, Eno2, Pgam2, Tpi1, Aldoc; p = 3 × 10–4). Glycogen degradation is likely also reduced (PYGM, PGM1, PYGB, AGl; p = 2 × 10–4). The cardiac β-adrenergic signaling pathway is implicated since several protein kinase A scaffolds and substrates involved in excitation–contraction coupling are substantially downregulated (e.g., Pde2a, Akap6, and Ryr2, among others). Finally, creatine phosphate biosynthesis and pyrimidine ribonucleotide interconversion via adenylate kinases were also downregulated in Tbx18-iPMs (p = 0.017 and p = 0.047 respectively).
Since cluster analysis was performed on coregulated clusters, irrespective of significance, we sought to corroborate dominant themes among significantly downregulated proteins. To keep the data set manageable, we imposed an FDR threshold of q < 0.0001. A STRING functional protein association network (i.e., direct interactions, coexpression, gene ontology, conserved domains; see methods) was constructed in Cytoscape 3.7.1 and depicted in Figure 3D. The network contains 920 nodes connected by 7717 edges or medium- to high-confidence relationships (STRING score ≥ 0.6), which is more than what would be expected from a similarly sized, randomly chosen set of proteins (p < 10–16). Thirteen network clusters are highlighted, several of which are concordant with pathway analysis (e.g., OxPhos, TCA cycle, β-oxidation, and branched-chain amino acid metabolism), whereas others figure more prominently in the network analysis. Notably, other mitochondrial processes, including mitochondrial protein synthesis via the mitochondrial ribosomal subunits, as well as mitochondrial protein import via the TOM/TIM complexes, are profoundly downregulated by Tbx18. The Ca2+ handling module encompasses both sarcolemmal and sarcoplasmic reticulum proteins associated with Ca2+-induced Ca2+ release such as L-type Ca2+ channel subunits, ryanodine receptors, SERCA2, phospholamban, and calsequestrin. Other modules highlight the impact of Tbx18 on ubiquitination and proteasomal protein degradation.
Protein Upregulation by Tbx18 Spans Nuclear, Cytoskeletal, and Extracellular Pathways
Upregulated proteins, as with the downregulated ones, were parsed by cluster, as depicted in Figure 2D. Several significant pathways were also associated with z-scores that provide a measure of likely pathway activation or inhibition and are included in Figure 4. For cluster 4 (Figure 4A), the average fold change was 1.31. As shown in Figure 1A,B, during Tbx18-mediated pacemaker cell reprogramming, normal ventricular cardiomyocytes undergo dramatic morphological changes.3 Consistent with this observation, actin cytoskeletal pathways, including Rho and Rac GTPase signaling, is upregulated. These elements are shared with other pathways such as focal adhesion kinase (FAK) signaling, hepatocyte growth factor (HGF) signaling, and regulation of cellular mechanics by calpain. Other elements of the Rho and Rac pathways are shared with NFκB-related pathways, including NFκB activation and Cd27 activation in lymphocytes (e.g., RelA, NFκBIE, NFκB2). The upregulation of NFκB subunits, as well as disheveled1–3 and frizzled1 and 2, suggest activation of either the Wnt/Ca2+ pathway or possibly the Wnt/PCP pathway.
Figure 4.
Pathway analysis of upregulated clusters reveals extensive remodeling of the cytoskeleton, ECM, and signaling. (A–C) Clusters 4–6 (Figure 2D) encompassing the substantially upregulated proteins were subjected to pathway analysis (Ingenuity). Pathway overrepresentation is ranked by −log(p-value). Some pathways have an associated z-score as a metric of inferred pathway inhibition or activation. Full tables are provided in the Online Supplement. Dominant themes include upregulation of matrix, cytoskeletal, membrane trafficking proteins, and all of the small GTPases associated with the processes. DNA replication and repair enzymes are implicated, and several signaling pathways are inferred to be activated. (D) STRING functional protein association network comprised of proteins differentially regulated at a threshold of q < 0.0001. The largest multinode modules are colored. The network uniquely highlights the number of upregulated proteins involved in ribosomal RNA processing.
Cluster 5 comprises 498 proteins with an average fold change of 1.6 (Figure 4B). This cluster partially overlaps with those of cluster 4, particularly activation of the Rho-family GTPases. An acute inflammatory response program is inferred from the upregulation of tumor necrosis factor (TNF) receptor signaling components (Tnfrs11b, Tradd, Irak). Interleukin-8 (Il-8) signaling is also upregulated with an instance of p38 MAPK signaling. Given that the same multiplicity of infection was used for adenoviral gene transfer of Tbx18 or GFP, we interpret the inflammatory response as a bona fide feature of the Tbx18-iPMs phenotype.
Cluster 6 harbors a small group of the most upregulated proteins with the average fold change of 2.42 in Tbx18-iPMs (Figure 4C). Interferon signaling dominates this cluster, which includes interferon-inducible proteins (e.g., Ifit3, Ifitm2, Stat2). Many of the proteins are also common to the pathway of interferon regulatory factors (IRF) by pattern recognition receptors. Overrepresented in this cluster is upregulation of extracellular matrix remodeling, with collagen regulation and fibrosis as the central pathway. Again, more Rho-signaling and cytoskeletal remodeling proteins are upregulated in cluster 6, as are proteins involved in integrin-linked kinase (Ilk) signaling, including keratin and vimentin (Kr18, Vim). Finally, Notch signaling is inferred from the upregulation of Notch2, 3, and numb-like endocytic adapter protein (Numbl).
STRING network analysis of the significantly upregulated proteins (q < 0.0001; Figure 4D) highlighted 751 nodes and 5263 moderate-to high-confidence edges (score ≥ 0.6; p < 10–16). These modules corroborate the pathway analysis, particularly with respect to interferon signaling, the extracellular matrix, adhesion molecules, cytoskeletal remodeling, and the prominence of small GTPase regulation. The network modules also highlight the extensive upregulation of ribosomal RNA processing enzymes as well as DNA synthesis and repair enzymes. Finally, two of the modules reveal the prominence of endoplasmic reticulum/Golgi/vesicle processes. Vesicular proteins, in particular, were clustered with growth factor receptors (e.g., Egfr and Pdgfrb), presumably an indication of their association, as the receptors are shuttled to the sarcolemma.
Hallmark of Tbx18-iPMs #1—Loss of Ventricular and Gain of Pacemaker Electrogenic Proteins
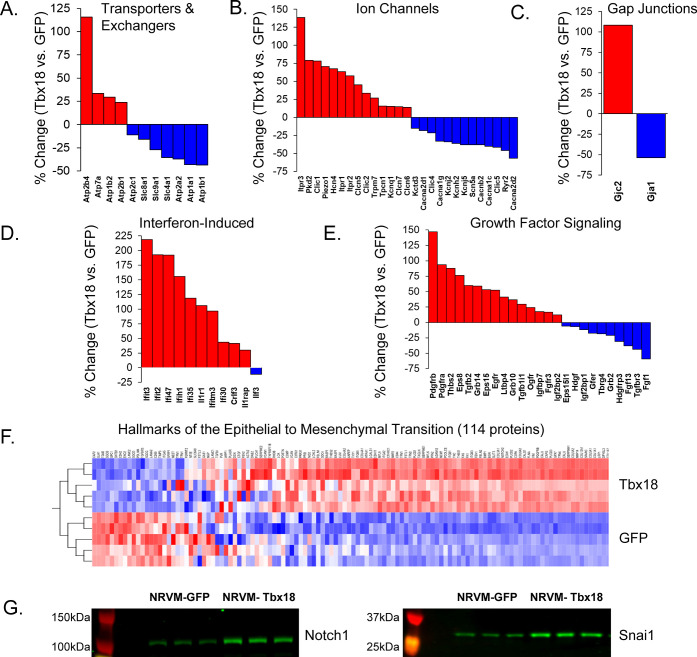
The hallmark of the pacemaker cells is automaticity, which is associated with the emergence of a distinct ion channel program that permits spontaneous and rhythmic diastolic depolarizations that initiate the action potential.25 Tbx18 overexpression in NRVMs led to extensive modulation of ion channels and gap junctions, resembling the native pacemaker cells (Figure 5A–C). Key ion channels that figure prominently in ventricular myocytes’ action potential were downregulated (Figure 5A). These include Na+ channels that underpin the fast upstroke velocity (SCN5A downregulated by 1.61-fold), voltage-gated L-type Ca2+ channels that provide the long plateau phase (Cacna1c, Cacnb2, and Cacnd2 downregulated 1.62-, 1.67-, and 2.31-fold respectively), and the inward rectifier K+ channel that is responsible for stabilizing the resting membrane potential (Kcnj2 downregulated by 1.5-fold). In cardiac myocytes, preservation of the Na+ gradient is the function of the Na+-K+ATPase, both subunits of which (Atp1a1, Atp1b1; Figure 5B) are downregulated by about 1.8-fold. Gap junctions allow for electrical and metabolic coupling between cells and are composed of connexins.26 The dominant ventricular myocyte connexin, Cx43, was downregulated 2.2-fold (Figure 5C). The release of Ca2+ from the myocyte sarcoplasmic reticulum (SR) in response to sarcolemmal depolarization is mediated by the ryanodine receptor, which is down by 1.9-fold in Tbx18-iPMs. Sequestration of SR calcium is achieved by the sarco/endoplasmic reticulum Ca2+ ATPase (Serca2 or Atp2a2) whose activity is modulated by phospholamban (Plb).27 These proteins were downregulated 1.6-fold and 1.8-fold, respectively, further demonstrating loss of chamber cardiomyocyte phenotype.
Figure 5.
Hallmarks of NRVM/PM transdifferentiation: electrogenic remodeling and EMT biosignature. (A) Differentially regulated (p < 0.05) ion channels. (B) Differentially regulated active transporters and exchangers. (C) Gap junctions in the Tbx18-iPMs. (D) Interferon-induced proteins. (E) Growth factor Signaling. (F) Heat map of EMT-associated proteins found in MSigDB (hierarchically clustered by sample, proteins ranked by inter-group variance). (G) Immunoblot analysis of key regulators of the EMT, Notch1, and Snai1.
The emerging Tbx18-iPM ion channel profile was characterized by 1.7-fold upregulation of HCN4 (Figure 5B), the molecular correlate of the funny current, If,28 sarco/endoplasmic reticulum Ca2+ channels, and the inositol trisphosphate receptors have been reported to positively regulate the spontaneous firing rate of the SAN pacemaker cells29,30 and interstitial cells of Cajal.31 Indeed, Itpr1, Itpr2, and Itpr3 were highly upregulated (1.6-, 1.6-, and 2.4-fold, respectively) in Tbx18-iPMs compared to control myocytes. Other notably upregulated channels fall within the class of mechanosensitive cation channels, including Piezo1 (1.7-fold), Pkd2 (aka Trpp2, 1.8-fold), and TrpM7 (1.3-fold). Intracellular voltage-gated Cl– channels involved in endosomal/lysosomal acidification were also among the upregulated electrogenic proteins (Figure 5B). Together, this proteomic data validates our previous findings that Tbx18 expression is sufficient to set the pacemaker electrical program.
Hallmark of Tbx18-iPMs #2–Transdifferentiation Recapitulates Features of the Epithelial–Mesenchymal Transition
Pathway analysis of clusters 4–6 showed inferred activation of several signaling pathways, including the Integrin, Wnt, Notch, and Acute Phase Response (e.g., IL1, IL6, TNFa). These four pathways drew our attention as they are known to be key modulators of another well-studied pathway of cellular plasticity, namely, the epithelial–mesenchymal transition (EMT). Other signaling pathways known to modulate the EMT are TGFβ/p38 and receptor tyrosine kinases. p38 pathway activation was inferred from cluster 4, while TGFβ2 was upregulated substantially in the iPMs’ proteome (Figure 5E). Receptor tyrosine kinases such as EGF receptor (Egfr) and PDGF receptors A and B (Pdgfra, Pdgfrb) are highly upregulated in Tbx18-iPMS.
We also saw that many of the matrix, cell surface, and cytostructural proteins, considered to be biomarkers of EMT, were upregulated in Tbx18-iPMs, including collagens a1 and a3 (Col1a1 and Col3a1), fibronectin (Fn1), and OB-cadherin (Cdh11). A broader molecular signature of the EMT consisting of 200 genes has been compiled from prior transcriptomic studies (Gene Set: Hallmark_Epithelial_Mesenchymal_Transition), of which 114 were quantified in the Tbx18-iPM proteome (Figure 5F). Of these, 96 were differentially regulated at p < 0.05 (q < 0.011), while 73 met the more stringent threshold of q < 0.0001.
An EMT is typically associated with activation of specific transcription factors, including Snai1 (snail), Snai2 (slug), Zeb1, Zeb2, Twist1, Twist2, Ets1, and Foxc2. In our proteome, Zeb2 was upregulated 1.49-fold (p = 9.38 × 10–5) while Zeb1 was unchanged (1.03-fold, NS; Online Supplement S1). To extend our coverage of transcription factors that are often underrepresented in proteomic data sets, we turned to RNAseq analysis (Online Supplement S2). Notwithstanding that this constituted an independent data set, the overall correlation between protein fold change and RNA fold changes was high (Pearson’s r = 0.62; Figure S1). For factors unrepresented in the proteome, the mRNA changes represent a reasonable indication of the likely direction of protein changes, though, for several reasons, inferences about the likely magnitude should be tempered. Nevertheless, examining the other EMT transcription factors, we found upregulation of Snai1 (2.2-fold), Snai2 (3.1-fold), Twist1 (1.9-fold), Twist2 (1.8-fold), and Foxc2 (1.35-fold). Ets1 was not significantly regulated. Taken together, the broad remodeling of matrix and cytoskeletal EMT hallmarks, as well as the upregulation of EMT-associated transcription factors suggest that Tbx18 induces morphologic transdifferentiation of ventricular myocyte to pacemaker cells by recruiting the EMT program.
Gene Regulatory Modules Observed in and Inferred From Proteomic and RNAseq Data
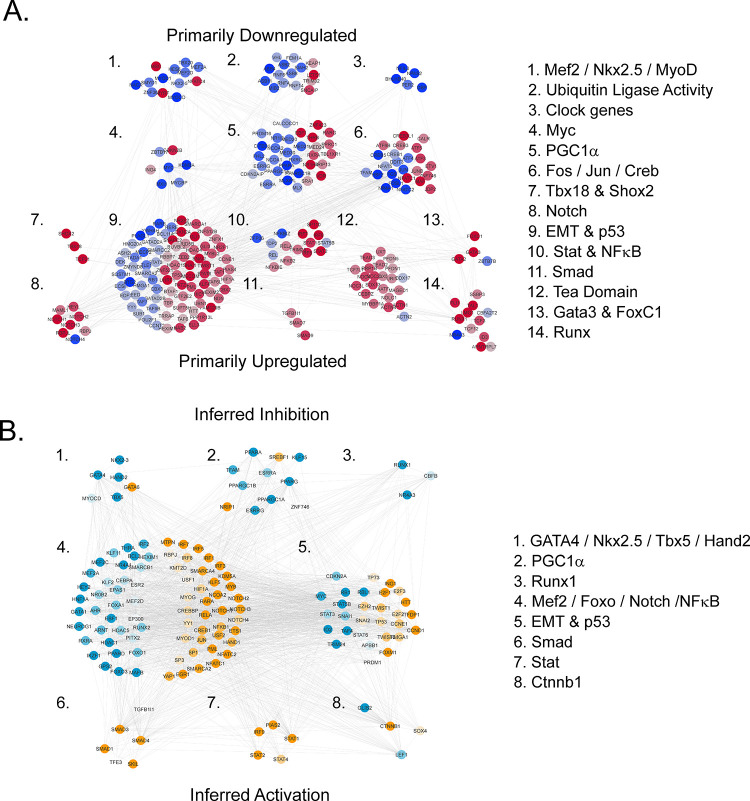
To investigate how Tbx18 might orchestrate the global changes in electrogenic, structural, and signaling protein levels, we compiled a STRING functional association network of transcription factors from the proteome and RNAseq data sets (Figure 6A, see methods). Markov clustering parsed the network into 14 distinct modules, which are ordered according to the number of module members that were down (blue) or upregulated (red). The top 6 modules contain downregulated members, which suggests inhibition of the module. Downregulation of Module 1 (Gata4, NKx2.5, and MyoD) is consistent with trends identified in the pathway analysis (Figure 2) in that ventricular ion channels are transcriptionally regulated by Gata4, Nkx2.5, and MyoD. Downregulation of glucose and fatty acid metabolism, as well as mitochondrial biogenesis, is consistent with downregulation of modules 5 and 6, which include Pgc1α and Esrra and Esrrg. Modules 9–14 are composed primarily of upregulated transcription factors. Tbx18 expression has been shown to correlate with Shox2, a transcriptional regulator of the pacemaker channel, Hcn4. Upregulation of modules 8, 10, and 11 are consistent with upregulated pathways identified in Figure 3, including Notch, TGFβ, and acute phase response signaling. The association network also clusters functionally similar molecules exemplified by module 2, composed of downregulated ubiquitin ligases of the transcriptional machinery, whereas module 12 highlights the broad upregulation of proteins with DNA-binding TEA domains.
Figure 6.
Observed and inferred networks of transcriptional regulators. (A) STRING functional association network composed of transcriptional regulators from both proteome and RNAseq data sets that were differentially regulated at an FDR threshold of q < 0.005. Functional modules were obtained by Markov clustering. Node color represents relative expression (log2(Tbx18/GFP)) and spans from blue (downregulated) to red (upregulated). Modules composed primarily of downregulated factors are near the top, while those composed of upregulated factors are near the bottom. (B) STRING network composed of transcriptional regulators inferred to be implicated in the coordinate regulation of proteins differentially regulated at q < 0.0001 (URA analysis, p < 0.05). Node color represents the range of z-scores and spans from turquoise (inferred inhibited) to yellow-orange (inferred activated). Network modules are ordered from primarily inhibited, at the top, to primarily activated, at the bottom. Module names in A and B capture modular themes.
Noting that the expression of transcription factors is not equivalent to their activity and that some factors may be regulated via post-translational modification, we compiled a functional association network of transcription factors from Ingenuity’s upstream regulator analysis (URA), inferred from coordinate changes in protein levels (q < 0.0001; Figure 6B). Modules are ordered by likelihood of inhibition (top, blue-coded) to the likelihood of activation (bottom, orange-coded). The major modules largely fit with many of the modules in Figure 6A. A new module involves Ctnnb1 (module 8), whose activation is inferred despite the fact that the protein itself was not differentially regulated. The activity of Ctnnb1, which encodes β-catenin downstream in the Wnt pathway, is governed by phosphorylation-dependent nucleus-cytosol shuttling. It is possible that membrane-bound β-catenin translocates into the nucleus, thus activating canonical Wnt signaling in Tbx18-iPMs, a notion that warrants further investigation.
Candidate Links between Tbx18 and EMT Program Prioritized by Network Mapping
To clarify how Tbx18 expression might elicit an EMT-like gene expression program, a “heat-diffusion” algorithm was used to map information flow through the transcriptional network (Figure 6A,6B). Orange nodes are proximal (fewer edges back) to Tbx18, while yellow nodes and modules are more distal (Figure 7A). Under this heat map, modules 1–5 are proximal to Tbx18, while modules 6–11 are distal. This indicates that key EMT transcription factors in module 2 are closer to Tbx18 than the proteins of the PGC1α, NFκB, Fos/Jun, or STAT modules in the network, whose perturbation would likely be a consequence of EMT activation rather than its proximal cause.
Figure 7.
Proximity of the EMT program to Tbx18 in the transcription factor network. The nodes from the networks in Figure 6A,6B were merged and re-clustered. Module 1 corresponds to what the PathCards pathway unification database (https://pathcards.genecards.org/) calls the “heart development superpathway,” which is divided into submodules (a) GMTH, (b) Ctnnb1, and (c) Notch, for clarity. (A) 250 nodes were ranked by proximity to Tbx18 (see Online Supplement S1), of which the top 125 are depicted. Orange nodes are closer within the network (fewer edges back) to Tbx18, while yellow nodes and modules are more distal. (B) Top-ranked Tbx18-proximal nodes (Top 10, 20, 40, and 60; Online Supplement S1) are highlighted, in turn, across network modules 1–5 to illustrate candidate routes of information flow between Tbx18 and key EMT factors in module 2. (C) Nodes from modules 1–5 in panel (A) were recolored to integrate expression log2(Tbx18/GFP) (node color; blue, white, red) and inferred activity (border color; turquoise, white, orange). Tbx18 acts directly on module 1, likely inhibiting the action of the GMTH submodule. Communication between Tbx18 and Snai2 of the EMT module may be direct or indirect.
Figure 7B further illustrates the flow of network information among the topmost proximal modules based on currently known associations. Among the top 10 closest network associations with Tbx18 are proteins Shox2, Tcf21, and Scx, whose expression correlate with it, albeit by unknown mechanisms. Tbx18 is known to interact with the proteins of module 1a (GMTH). The influence of Tbx18 on module 1b is supported by the mutually exclusive expression patterns of Tbx18 and Pitx2.32 Among the 20 closest neighbors to Tbx18, the heat map extends to Hey2 in the module 1c and to a key EMT transcription factor, Snai2, in module 2 (EMT and p53). This is supported by evidence for direct binding of Tbx18 to the promoter of Snai2 in murine primary epicardial cells.33 Other EMT transcription factors, including Zeb2, Snai1, Twist1, and Twist2 are among the top 40 nodes closest to Tbx18 in module 3. The nodes of modules 4 (activators/repressors) and 5 (Smad) round out the top 60 closest factors to Tbx18 in the network but are downstream of the EMT module (module 2) based on currently known associations.
Expression and inferred activity are superimposed on modules 1–5 in Figure 7C. Specifically, Tbx18 acts as a suppressor of GATA4/Tbx5, which would have an immediate impact on most nodes in module 1a, including Nkx2.5, myocardin (Myocd), and Tbx20, among others. Inhibited factors in module 1a are complemented by coincident activation of module 1b, the EMT factors in module 2, and activation of Notch programs. Thus, the networks of Figure 7 and the network proximity rankings in Online Supplement S1 provide a model of prioritized targets to validate the gene regulatory program underlying Tbx18-induced reprogramming of ventricular cardiomyocytes to pacemaker cells.
Discussion
Ectopic expression of Tbx18 in the ventricular myocytes causes vast and profound protein remodeling consistent with changing cellular identity, specifically, the emergence of pacemaker cells. Key observations from our study include (1) significant metabolic remodeling in the induced pacemaker cells, (2) upregulation of mechanically gated ion channels in Tbx18-iPMs that may contribute to the Bainbridge reflex (atrial reflex) and, (3) upregulation of proteins and transcripts which point to the recruitment of the EMT program to effect changes in the cell morphology in the induced pacemaker cells. We present a transcriptional network model to help evaluate and prioritize mechanisms underlying the Tbx18-mediated reprogramming of ventricular myocytes to pacemaker cells.
Diminution of Cardiomyocyte Identity by Tbx18
Cardiac myocytes are characterized by a protein biosignature that includes expression of specific sarcolemmal and sarcoplasmic reticulum ion channels and pumps that participate in excitation–contraction (EC), sarcomeric contractile proteins, and a robust metabolic program to meet the energetic demands of contraction. In this study, we demonstrate that ectopic Tbx18 expression in myocytes has the most profound impact on EC proteins and the metabolic program. The transcriptional programs that underlie these two facets of the cardiomyocyte function may be regarded as distinct but communicating modules: (1) a minimal myocyte identity module similar to module 1a in Figure 7A/C and (2) A metabolic/bioenergetic hub similar to module 5 in Figure 6A.
The minimum myocyte identity program has emerged from studies showing that the transcription factors Gata4, Mef2c, and Tbx5 alone (GMT) or with Hand2 (GMTH) suffice to convert cardiac fibroblasts into cardiac myocytes.34,35 These factors constitute the core of negatively regulated ones in module 1a (Figure 7A,C), which also includes Nkx2.5 and Tbx20. Inhibition of a transcriptional module involving the GMTH factors is consistent with the robust and significant downregulation of proteins that affect electrical excitation and mechanical contraction of ventricular cardiomyocytes, including Scn5A, Kcnj2, and Ryr2 (Figure S2). The network in Figure 7A/C shows that they are among the proximal targets of Tbx18. Specifically, Tbx5 and Tbx20 act with Gata4 and Nkx2.5 to drive myocyte gene expressions such as atrial natriuretic peptide and Scn5a.36−38 Tbx18, as in other T-box proteins, Tbx2, and Tbx3, suppresses the program by competing with Tbx5 for Nkx2.5 or GATA4.39 While Gata4 and Hand2 expression levels do not change in response to Tbx18, the activity of these factors from URA was inferred to be inhibited nonetheless. Repression of Tbx5, in turn, would downregulate Mef2c expression,40 consistent with model prediction in Figure 6B (z-score: −3.02). Mef2a and Mef2d were detected in the proteome and were down −1.49 and −1.47-fold, respectively.
Downregulation of metabolism and bioenergetics genes is the dominant theme across clusters 1–3 in Figure 3. Glycolysis, glycogen metabolism, fatty acid oxidation, TCA cycle, and oxidative phosphorylation were nearly universally downregulated. The core components of the underlying transcriptional program include many of the proteins and transcripts in module 5 of Figure 6A, including Pgc1a, Ppara, Pparg, Esrra, and Esrrg, among others. There is currently no evidence that Tbx18 is directly associated with these factors. Based on heat diffusion through the transcription factor network (Figure 7A), the members of the bioenergetic transcriptional hub are further away from Tbx18 than the inhibited GMTH module and the activated EMT factors. Since the GMTH module controls the expression of energetically expensive ATPases such as myosin, Serca2a, and the Na+-K+ ATPase, this suggests that downregulation of metabolism is a secondary or adaptive, response to lower energetic demands of the pacemaker cell.
Emergent Electrogenic Identity Conferred by Tbx18
The emergence of pacemaker identity was associated with two prominent features at the protein level: (1) upregulation of a distinct pacemaker ion channel and Ca2+-handling profile and (2) substantial remodeling of the cytoskeleton, matrix, and signaling pathways consistent with activation of a canonical EMT. Expectedly, Tbx18 caused upregulation (1.7-fold) of Hcn4, the molecular correlate of If that destabilizes the diastolic sarcolemmal membrane potential and confers the early (linear) rise of phase-4 diastolic depolarization.41 Inspection of cluster 1 pathways (Figure 3A and Online Supplement S1) shows that IP3-catabolic pathways are downregulated, which would favor accumulation of intracellular IP3. This would, in turn, activate IP3 receptors (Itpr), which are also substantially upregulated in Tbx18-iPMs. This is in line with the recent reports that the IP3-mediated Ca2+ release mechanism regulates native SA nodal pacemaker cells’ automaticity,30 though interestingly, Itpr1–3 were not differentially expressed in a proteomic study comparing the sinoatrial node with atrial myocardium.42
The other significantly upregulated channels are classified as mechanosensitive and may be pertinent to atrial mechano-electric feedback, including the classic Bainbridge (atrial) reflex.43 Briefly, increased stretching of the right atrium, in response to elevated central venous pressure, increases heart rate. The underlying mechanism is thought to have both extrinsic (innervation-dependent) and intrinsic (innervation-independent) components (reviewed in44). Evidence for the latter comes from studies showing the reflex can be triggered in denervated hearts, including in heart transplant patients,45 isolated mammalian heart models,46,47 isolated atrial tissues, and even isolated pacemaker cells.48 Mechanically gated ion channels (MGCs) are candidate mediators of the atrial reflex, though the precise MGC complement whose expression correlates with pacemaker phenotype is not known.44,49 Our data indicate that the stretch-activated nonselective cation channel, Piezo1 (Fam38a),50,51 is higher in Tbx18-iPMs at both protein (1.7-fold) and transcript levels (1.7-fold). Piezo1 can sense both cell stretch and cell crowding.52 Piezo1 further mediates flow-sensitive vasoconstriction in the endothelial cells of mesenteric arteries53 and is elevated in cardiomyocytes in response to Angiotensin II and in heart failure.54 Polycystin2 (Pkd2 or Trpp2) is implicated in Ca2+-dependent autophagy and intracellular Ca2+ handling in cardiomyocytes, (because there are several "and"s in the sentence that require some help55 and mediates flow-induced extracellular Ca2+-influx in the primary cilium of kidney cells. Our data indicate that Pkd2 was significantly upregulated at the protein level (1.8-fold) while Pkd1 transcripts were up 1.9-fold. TrpM7 protein, which has been reported to be important for pacemaker cells’ automaticity,56 was upregulated (1.3-fold). Other Trp channels such as TrpC3, TrpC6, and TrpV2, all of which have documented roles modulating sarcolemmal Ca2+-flux, were significantly upregulated at the transcript level. The fact that Piezo1, Pkd2, and TrpM7 are upregulated in cardiac fibroblasts relative to myocytes57 provides further impetus to examine the roles of these channels in the mechano-sensitivity of the sinus rhythm.
Transcriptional regulators of Hcn4 are implicated in module 1a of Figure 7A–7C. Evidence from knockout mouse studies indicates that Nkx2.5 is a transcriptional repressor of Hcn4.58 Tbx18 may inhibit Nkx2.5 directly or indirectly through increased expression of Shox2, which has been shown to suppress the expression of Nkx2.5.58,59 Alternatively, Notch signaling may also play a role, as overexpression of Notch is sufficient to increase Hcn4 expression and subsequently If density in NRVMs.60 In this regard, Notch2 and Notch3 are upregulated by 1.8- and 2-fold, respectively, in the Tbx18-iPM proteome. These mechanisms need not be mutually exclusive and may all contribute to the pacemaker electrogenic profile in Tbx18-iPMs.
Transdifferentiation into Pacemaker Cell Morphology by Tbx18
EMT figures prominently in the morphological plasticity of many cell types, including epicardial cells, and is integral to cardiac development.61 Briefly, epithelial cells undergoing EMT are characterized by structural polarity and tight cell coupling. They undergo a transformation to a mesenchymal fate whose morphology is well suited for motility and migration, all without dedifferentiating to a pluripotent state.61,62 Tbx18 is expressed in both epicardial progenitors and SA nodal cells. Interestingly, Tbx18 expression is required to mediate TGFβ-initiated epicardial transition in vitro,33 yet knocking out Tbx18 in mice did not prevent the epicardial EMT in vivo.(63) Here, we show that Tbx18 expression suffices to recruit the EMT transcriptional program (upregulation of at least 6 EMT transcription factors) and elicit differential regulation of 96 cytostructural EMT hallmarks in ventricular myocytes (Figure 5D). The process is orchestrated by specific transcription factors, including Snai1, Snai2, Twist1, Twist2, Zeb1, and Zeb2. These factors bind and redistribute epigenetic modulators, including histone acetylases/deacetylases and methyltransferase/demethylases, serving as both transcriptional repressors and activators. We have shown previously that conversion of ventricular myocytes to pacemaker cells by Tbx18 involves durable epigenetic reprogramming, increasing H3K27Me3-mediated repression of the promoters of Kcnj2, Gja1, and Actn2 genes while increasing H3K4Me3-mediated activation of Hcn4.3
The network models in Figure 7A–7C offer testable hypotheses for how Tbx18 activates the EMT program in myocytes. Heat diffusion indicates that EMT transcription factors and minimal myocyte identity factors (GMTH) are positioned close to each other in the network neighborhood, suggesting that effects of Tbx18 are direct, or need only a few intermediaries. Direct binding of Tbx18 on the promoter region of Snai2 in epicardial cells33 is in line with our network model. Recently reported Tbx18-binding partners in HEK293 cells64 have little overlap with the top candidates from our network analysis. This is likely attributable to differences between primary cardiomyocytes and the immortalized embryonic kidney cells. Future ChIP-Seq and ChIP-MS studies in Tbx18-treated iPMs will inform and help refine the Tbx18/EMT model.
Acknowledgments
The authors thank Jun Li for preparing the RNA samples subjected to RNAseq. The authors thank Gregory K. Tharp and Nirav B. Patel in Yerkes National Primate Research Center Nonhuman Primate Genomic Core for performing RNAseq and data analysis. The table of contents graphic was created with BioRender.com.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.2c00133.
Author Contributions
∇ D.B.F. and J.-m.G. contributed equally to this work. J.-m.G., D.B.F., R.N.C., and H.C.C. designed the research. D.B.F., J.-m.-G., E.H.K., D.W.W., and R.O. performed the experiments. J.-m.G., D.B.F., and H.C.C. analyzed the data and wrote the manuscript.
D.B.F. was supported by AHA 12SDG12060056, AHA 18TPA34170575, and NIH R01HL134821. J.-m.G. was supported by NSF 1609831 to H.C.C. H.C.C. was supported by R01HL111646-01A1.
The authors declare no competing financial interest.
Notes
The MS proteomics raw data (.raw), complete search results (.msf), and spectra (.mzidentML &.mgf) have been deposited to the ProteomeXchange Consortium (http://www.proteomexchange.org/;65) via the PRIDE partner repository66 with the data set identifier PXD013351 and 10.6019/PXD013351. The RNAseq data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus67 and are accessible through GEO Series accession number GSE129545 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE129545). Further requests for resources and reagents should be directed to and will be fulfilled by, Dr. Hee Cheol Cho (hcho21@jhu.edu)
Supplementary Material
References
- John R. M.; Kumar S. Sinus Node and Atrial Arrhythmias. Circulation 2016, 133, 1892–1900. 10.1161/CIRCULATIONAHA.116.018011. [DOI] [PubMed] [Google Scholar]
- Cho H. C. Pacing the Heart with Genes: Recent Progress in Biological Pacing. Curr. Cardiol. Rep. 2015, 17, 65. 10.1007/s11886-015-0620-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor N.; Liang W.; Marban E.; Cho H. C. Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat. Biotechnol. 2013, 31, 54–62. 10.1038/nbt.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A.; Davis C. A.; Schlesinger F.; Drenkow J.; Zaleski C.; Jha S.; Batut P.; Chaisson M.; Gingeras T. R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S.; Pyl P. T.; Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I.; Huber W.; Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome Biol. 2014, 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A.; Green J.; Pollard J. Jr; Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel D.; Flugge U. I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984, 138, 141–143. 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Yang F.; Gritsenko M. A.; Wang Y.; Clauss T.; Liu T.; Shen Y.; Monroe M. E.; Lopez-Ferrer D.; Reno T.; Moore R. J.; Klemke R. L.; Camp D. G. 2nd; Smith R. D. Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics 2011, 11, 2019–2026. 10.1002/pmic.201000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbrich S. M.; Cole R. N.; West K. P. Jr; Schulze K.; Yager J. D.; Groopman J. D.; Christian P.; Wu L.; O’Meally R. N.; May D. H.; McIntosh M. W.; Ruczinski I. Statistical inference from multiple iTRAQ experiments without using common reference standards. J. Proteome Res. 2013, 12, 594–604. 10.1021/pr300624g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster D. B.; Liu T.; Kammers K.; O’Meally R. N.; Yang N.; Papanicolaou K. N.; Talbot C.; Cole R. N.; O’Rourke B. Integrated Omic Analysis of a Guinea Pig Model of Heart Failure and Sudden Cardiac Death. J. Proteome Res. 2016, 15, 3009. 10.1021/acs.jproteome.6b00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G. K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3, 1–25. 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Kammers K.; Cole R. N.; Tiengwe C.; Ruczinski I. Detecting Significant Changes in Protein Abundance. EuPA Open Proteomics 2015, 7, 11–19. 10.1016/j.euprot.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammers K.; Foster D. B.; Ruczinski I.. Analysis of Proteomic Data. In In Manual of Cardiovascular Proteomics Agnetti G.; Lindsey M. L.; Foster D. B., Eds.; Springer: UK, 2016. [Google Scholar]
- Schwämmle V.; Leon I. R.; Jensen O. N. Assessment and improvement of statistical tools for comparative proteomics analysis of sparse data sets with few experimental replicates. J. Proteome Res. 2013, 12, 3874–3883. 10.1021/pr400045u. [DOI] [PubMed] [Google Scholar]
- Storey J. D. A direct approach to false discovery rates. J. R. Stat. Soc.: Series B 2002, 64, 479–498. 10.1111/1467-9868.00346. [DOI] [Google Scholar]
- Storey J. D. The positive false discovery rate: a Bayesian interpretation and the q-value. Ann. Stat. 2003, 31, 2013–2035. 10.1214/aos/1074290335. [DOI] [Google Scholar]
- Storey J. D.; Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 9440–9445. 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J. D.Qvalue: Q-value Estimation for False Discovery Rate Control. R Package Version 2.0.0., 2015.
- Doncheva N. T.; Morris J. H.; Gorodkin J.; Jensen L. J. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2019, 18, 623–632. 10.1021/acs.jproteome.8b00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su G.; Morris J. H.; Demchak B.; Bader G. D. Biological network exploration with Cytoscape 3. Curr. Protoc. Bioinf. 2014, 47, 1–24. 10.1002/0471250953.bi0813s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D.; Gable A. L.; Lyon D.; Junge A.; Wyder S.; Huerta-Cepas J.; Simonovic M.; Doncheva N. T.; Morris J. H.; Bork P.; Jensen L. J.; Mering C. V. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. H.; Apeltsin L.; Newman A. M.; Baumbach J.; Wittkop T.; Su G.; Bader G. D.; Ferrin T. E. clusterMaker: a multi-algorithm clustering plugin for Cytoscape. BMC Bioinf. 2011, 12, 436. 10.1186/1471-2105-12-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor N.; Galang G.; Marban E.; Cho H. C. Transcriptional suppression of connexin43 by TBX18 undermines cell-cell electrical coupling in postnatal cardiomyocytes. J. Biol. Chem. 2011, 286, 14073–14079. 10.1074/jbc.M110.185298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler N. J.; Greener I. D.; Tellez J. O.; Inada S.; Musa H.; Molenaar P.; Difrancesco D.; Baruscotti M.; Longhi R.; Anderson R. H.; Billeter R.; Sharma V.; Sigg D. C.; Boyett M. R.; Dobrzynski H. Molecular architecture of the human sinus node: insights into the function of the cardiac pacemaker. Circulation 2009, 119, 1562–1575. 10.1161/CIRCULATIONAHA.108.804369. [DOI] [PubMed] [Google Scholar]
- van Kempen M. J.; Fromaget C.; Gros D.; Moorman A. F.; Lamers W. H. Spatial distribution of connexin43, the major cardiac gap junction protein, in the developing and adult rat heart. Circ. Res. 1991, 68, 1638–1651. 10.1161/01.RES.68.6.1638. [DOI] [PubMed] [Google Scholar]
- Akin B. L.; Hurley T. D.; Chen Z.; Jones L. R. The structural basis for phospholamban inhibition of the calcium pump in sarcoplasmic reticulum. J. Biol. Chem. 2013, 288, 30181–30191. 10.1074/jbc.M113.501585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFrancesco D. The role of the funny current in pacemaker activity. Circ. Res. 2010, 106, 434–446. 10.1161/CIRCRESAHA.109.208041. [DOI] [PubMed] [Google Scholar]
- Ju Y. K.; Liu J.; Lee B. H.; Lai D.; Woodcock E. A.; Lei M.; Cannell M. B.; Allen D. G. Distribution and functional role of inositol 1,4,5-trisphosphate receptors in mouse sinoatrial node. Circ. Res. 2011, 109, 848–857. 10.1161/CIRCRESAHA.111.243824. [DOI] [PubMed] [Google Scholar]
- Kapoor N.; Tran A.; Kang J.; Zhang R.; Philipson K. D.; Goldhaber J. I. Regulation of calcium clock-mediated pacemaking by inositol-1,4,5-trisphosphate receptors in mouse sinoatrial nodal cells. J. Physiol. 2015, 593, 2649–2663. 10.1113/JP270082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M. H.; Sung T. S.; O’Driscoll K.; Koh S. D.; Sanders K. M. Intracellular Ca(2+) release from endoplasmic reticulum regulates slow wave currents and pacemaker activity of interstitial cells of Cajal. Am. J. Physiol.: Cell Physiol. 2015, 308, C608–20. 10.1152/ajpcell.00360.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlueter J.; Brand T. A right-sided pathway involving FGF8/Snai1 controls asymmetric development of the proepicardium in the chick embryo. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 7485–7490. 10.1073/pnas.0811944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi M.; Nimura K.; Mori M.; Nakagami H.; Kaneda Y. The transcription factors Tbx18 and Wt1 control the epicardial epithelial-mesenchymal transition through bi-directional regulation of Slug in murine primary epicardial cells. PLoS One 2013, 8, e57829 10.1371/journal.pone.0057829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M.; Tsuchihashi T.; Ivey K. N.; Ross R. S.; Hong T. T.; Shaw R. M.; Srivastava D. Cardiac fibroblasts regulate myocardial proliferation through beta1 integrin signaling. Dev. Cell 2009, 16, 233–244. 10.1016/j.devcel.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y. J.; Song K.; Luo X.; Daniel E.; Lambeth K.; West K.; Hill J. A.; DiMaio J. M.; Baker L. A.; Bassel-Duby R.; Olson E. N. Reprogramming of human fibroblasts toward a cardiac fate. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 5588–5593. 10.1073/pnas.1301019110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg V.; Kathiriya I. S.; Barnes R.; Schluterman M. K.; King I. N.; Butler C. A.; Rothrock C. R.; Eapen R. S.; Hirayama-Yamada K.; Joo K.; Matsuoka R.; Cohen J. C.; Srivastava D. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 2003, 424, 443–447. 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- Bruneau B. G.; Nemer G.; Schmitt J. P.; Charron F.; Robitaille L.; Caron S.; Conner D. A.; Gessler M.; Nemer M.; Seidman C. E.; Seidman J. G. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 2001, 106, 709–721. 10.1016/S0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- Hiroi Y.; Kudoh S.; Monzen K.; Ikeda Y.; Yazaki Y.; Nagai R.; Komuro I. Tbx5 associates with Nkx2-5 and synergistically promotes cardiomyocyte differentiation. Nat. Genet. 2001, 28, 276–280. 10.1038/90123. [DOI] [PubMed] [Google Scholar]
- Farin H. F.; Bussen M.; Schmidt M. K.; Singh M. K.; Schuster-Gossler K.; Kispert A. Transcriptional repression by the T-box proteins Tbx18 and Tbx15 depends on Groucho corepressors. J. Biol. Chem. 2007, 282, 25748–25759. 10.1074/jbc.M703724200. [DOI] [PubMed] [Google Scholar]
- Hiroi Y.; Kudoh S.; Monzen K.; Ikeda Y.; Yazaki Y.; Nagai R.; Komuro I. Tbx5 associates with Nkx2-5 and synergistically promotes cardiomyocyte differentiation. Nat. Genet. 2001, 28, 276–280. 10.1038/90123. [DOI] [PubMed] [Google Scholar]
- Lakatta E. G.; Maltsev V. A.; Vinogradova T. M. A coupled SYSTEM of intracellular Ca2+ clocks and surface membrane voltage clocks controls the timekeeping mechanism of the heart’s pacemaker. Circ. Res. 2010, 106, 659–673. 10.1161/CIRCRESAHA.109.206078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linscheid N.; Logantha S. J. R. J.; Poulsen P. C.; Zhang S.; Schrölkamp M.; Egerod K. L.; Thompson J. J.; Kitmitto A.; Galli G.; Humphries M. J.; Zhang H.; Pers T. H.; Olsen J. V.; Boyett M.; Lundby A. Quantitative proteomics and single-nucleus transcriptomics of the sinus node elucidates the foundation of cardiac pacemaking. Nat. Commun. 2019, 10, 2889 10.1038/s41467-019-10709-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge F. A. The influence of venous filling upon the rate of the heart. J. Physiol. 1915, 50, 65–84. 10.1113/jphysiol.1915.sp001736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhtz-Buschbeck J. P.; Schaefer J.; Wilder N. Mechanosensitivity: From Aristotle’s sense of touch to cardiac mechano-electric coupling. Prog. Biophys. Mol. Biol. 2017, 130, 126–131. 10.1016/j.pbiomolbio.2017.05.001. [DOI] [PubMed] [Google Scholar]
- Bernardi L.; Salvucci F.; Suardi R.; Solda P. L.; Calciati A.; Perlini S.; Falcone C.; Ricciardi L. Evidence for an intrinsic mechanism regulating heart rate variability in the transplanted and the intact heart during submaximal dynamic exercise?. Cardiovasc. Res. 1990, 24, 969–981. 10.1093/cvr/24.12.969. [DOI] [PubMed] [Google Scholar]
- Blinks J. R. Positive chronotropic effect of increasing right atrial pressure in the isolated mammalian heart. Am. J. Physiol.-Legacy Content 1956, 186, 299–303. 10.1152/ajplegacy.1956.186.2.299. [DOI] [PubMed] [Google Scholar]
- Keatinge W. R. The effect of increased filling pressure on rhythmicity and atrioventricular conduction in isolated hearts. J. Physiol. 1959, 149, 193–208. 10.1113/jphysiol.1959.sp006334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper P. J.; Lei M.; Cheng L. X.; Kohl P. Selected contribution: axial stretch increases spontaneous pacemaker activity in rabbit isolated sinoatrial node cells. J. Appl. Physiol. 2000, 89, 2099–2104. 10.1152/jappl.2000.89.5.2099. [DOI] [PubMed] [Google Scholar]
- Peyronnet R.; Nerbonne J. M.; Kohl P. Cardiac Mechano-Gated Ion Channels and Arrhythmias. Circ. Res. 2016, 118, 311–329. 10.1161/CIRCRESAHA.115.305043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste B.; Mathur J.; Schmidt M.; Earley T. J.; Ranade S.; Petrus M. J.; Dubin A. E.; Patapoutian A. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science 2010, 330, 55–60. 10.1126/science.1193270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste B.; Xiao B.; Santos J. S.; Syeda R.; Grandl J.; Spencer K. S.; Kim S. E.; Schmidt M.; Mathur J.; Dubin A. E.; Montal M.; Patapoutian A. Piezo proteins are pore-forming subunits of mechanically activated channels. Nature 2012, 483, 176–181. 10.1038/nature10812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudipaty S. A.; Lindblom J.; Loftus P. D.; Redd M. J.; Edes K.; Davey C. F.; Krishnegowda V.; Rosenblatt J. Mechanical stretch triggers rapid epithelial cell division through Piezo1. Nature 2017, 543, 118–121. 10.1038/nature21407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rode B.; Shi J.; Endesh N.; Drinkhill M. J.; Webster P. J.; Lotteau S. J.; Bailey M. A.; Yuldasheva N. Y.; Ludlow M. J.; Cubbon R. M.; Li J.; Futers T. S.; Morley L.; Gaunt H. J.; Marszalek K.; Viswambharan H.; Cuthbertson K.; Baxter P. D.; Foster R.; Sukumar P.; Weightman A.; Calaghan S. C.; Wheatcroft S. B.; Kearney M. T.; Beech D. J. Piezo1 channels sense whole body physical activity to reset cardiovascular homeostasis and enhance performance. Nat. Commun. 2017, 8, 350 10.1038/s41467-017-00429-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J.; Huang B.; Yuan G.; Chen Y.; Liang F.; Zeng H.; Zheng S.; Cao L.; Geng D.; Zhou S. Stretch-activated channel Piezo1 is up-regulated in failure heart and cardiomyocyte stimulated by AngII. Am. J. Transl. Res. 2017, 9, 2945–2955. [PMC free article] [PubMed] [Google Scholar]
- Criollo A.; Altamirano F.; Pedrozo Z.; Schiattarella G. G.; Li D. L.; Rivera-Mejias P.; Sotomayor-Flores C.; Parra V.; Villalobos E.; Battiprolu P. K.; Jiang N.; May H. I.; Morselli E.; Somlo S.; de Smedt H.; Gillette T. G.; Lavandero S.; Hill J. A. Polycystin-2-dependent control of cardiomyocyte autophagy. J. Mol. Cell. Cardiol. 2018, 118, 110–121. 10.1016/j.yjmcc.2018.03.002. [DOI] [PubMed] [Google Scholar]
- Sah R.; Mesirca P.; Van den Boogert M.; Rosen J.; Mably J.; Mangoni M. E.; Clapham D. E. Ion channel-kinase TRPM7 is required for maintaining cardiac automaticity. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, E3037–46. 10.1073/pnas.1311865110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen P. C.; Schrölkamp M.; Bagwan N.; Leurs U.; Humphries E. S. A.; Bomholzt S. H.; Nielsen M. S.; Bentzen B. H.; Olsen J. V.; Lundby A. Quantitative proteomics characterization of acutely isolated primary adult rat cardiomyocytes and fibroblasts. J. Mol. Cell. Cardiol. 2020, 143, 63–70. 10.1016/j.yjmcc.2020.04.021. [DOI] [PubMed] [Google Scholar]
- Espinoza-Lewis R. A.; Liu H.; Sun C.; Chen C.; Jiao K.; Chen Y. Ectopic expression of Nkx2.5 suppresses the formation of the sinoatrial node in mice. Develop. Biol. 2011, 356, 359–369. 10.1016/j.ydbio.2011.05.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.; Chen C. H.; Espinoza-Lewis R. A.; Jiao Z.; Sheu I.; Hu X.; Lin M.; Zhang Y.; Chen Y. Functional redundancy between human SHOX and mouse Shox2 genes in the regulation of sinoatrial node formation and pacemaking function. J. Biol. Chem. 2011, 286, 17029–17038. 10.1074/jbc.M111.234252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentschler S.; Yen A. H.; Lu J.; Petrenko N. B.; Lu M. M.; Manderfield L. J.; Patel V. V.; Fishman G. I.; Epstein J. A. Myocardial Notch signaling reprograms cardiomyocytes to a conduction-like phenotype. Circulation 2012, 126, 1058–1066. 10.1161/CIRCULATIONAHA.112.103390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainock M.; Toubat O.; Danopoulos S.; Beckham A.; Warburton D.; Kim R. Epicardial Epithelial-to-Mesenchymal Transition in Heart Development and Disease. J. Clinical Med. 2016, 5, 27. 10.3390/jcm5020027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacic J. C.; Dimmeler S.; Harvey R. P.; Finkel T.; Aikawa E.; Krenning G.; Baker A. H. Endothelial to Mesenchymal Transition in Cardiovascular Disease: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 2019, 73, 190–209. 10.1016/j.jacc.2018.09.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greulich F.; Farin H. F.; Schuster-Gossler K.; Kispert A. Tbx18 function in epicardial development. Cardiovasc. Res. 2012, 96, 476–483. 10.1093/cvr/cvs277. [DOI] [PubMed] [Google Scholar]
- Rivera-Reyes R.; Kleppa M. J.; Kispert A. Proteomic analysis identifies transcriptional cofactors and homeobox transcription factors as TBX18 binding proteins. PLoS One 2018, 13, e0200964 10.1371/journal.pone.0200964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch E. W.; Csordas A.; Sun Z.; Jarnuczak A.; Perez-Riverol Y.; Ternent T.; Campbell D. S.; Bernal-Llinares M.; Okuda S.; Kawano S.; Moritz R. L.; Carver J. J.; Wang M.; Ishihama Y.; Bandeira N.; Hermjakob H.; Vizcaino J. A. The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2017, 45, D1100–D1106. 10.1093/nar/gkw936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y.; Csordas A.; Bai J.; Bernal-Llinares M.; Hewapathirana S.; Kundu D. J.; Inuganti A.; Griss J.; Mayer G.; Eisenacher M.; Perez E.; Uszkoreit J.; Pfeuffer J.; Sachsenberg T.; Yilmaz S.; Tiwary S.; Cox J.; Audain E.; Walzer M.; Jarnuczak A. F.; Ternent T.; Brazma A.; Vizcaino J. A. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.; Domrachev M.; Lash A. E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.