Abstract
Allogeneic CD19-specific chimeric antigen receptor (CAR) T cells with inactivated donor T cell receptor (TCR) expression can be used as an “off-the-shelf” therapeutic modality for lymphoid malignancies, thus offering an attractive alternative to autologous, patient-derived T cells. Current approaches for T cell engineering mainly rely on the use of viral vectors. Here, we optimized and validated a non-viral genetic modification platform based on Sleeping Beauty (SB) transposons delivered with minicircles to express CD19-28z.CAR and CRISPR-Cas9 ribonucleoparticles to inactivate allogeneic TCRs. Efficient TCR gene disruption was achieved with minimal cytotoxicity and with attainment of robust and stable CD19-28z.CAR expression. The CAR T cells were responsive to CD19+ tumor cells with antitumor activities that induced complete tumor remission in NALM6 tumor-bearing mice while significantly reducing TCR alloreactivity and GvHD development. Single CAR signaling induced the similar T cell signaling signatures in TCR-disrupted CAR T cells and control CAR T cells. In contrast, TCR disruption inhibited T cell signaling/protein phosphorylation compared with the control CAR T cells during dual CAR/TCR signaling. This non-viral SB transposon-CRISPR-Cas9 combination strategy serves as an alternative for generating next-generation CD19-specific CAR T while reducing GvHD risk and easing potential manufacturing constraints intrinsic to viral vectors.
Keywords: CRISPR-Cas9, transposon, chimeric antigen receptor, Sleeping Beauty, T cell receptor, leukemia
Graphical abstract

Non-viral SB transposon-CRISPR-Cas9 T cell engineering results in efficient CD19-specific CAR/TCR knockout T cell production. These universal CD19-CAR/TCR-negative T cells exhibited potent antitumor effects in vitro and in vivo while reducing TCR alloreactivity and GvHD development. CD19-CAR/TCR-negative T cells exhibit reduced TCR/CD3 signaling upon dual CAR/TCR co-activation.
Introduction
Chimeric antigen receptor (CAR) T cell adoptive immunotherapy has emerged as a promising therapeutic modality for lymphoid malignancies.1,2 The US Food and Drug Administration and the European Medicines Agency have approved CAR T cell products for refractory cell precursor acute lymphoblastic leukemia and large B cell lymphoma.3,4 Nevertheless, specific challenges hamper widespread use of CAR T cell therapy, including dependency on autologous patient-specific T cells and substantial inter-patient variability. Particularly for pediatric and heavily treated patients who previously received chemotherapies and/or stem cell transplantation, the resulting lymphopenic conditions may lead to inadequate T cell numbers, suboptimal CAR T cell functions, and unsuccessful CAR T cell production.5, 6, 7, 8 Although use of allogeneic CAR T cells could overcome some of the limitations of autologous, patient-specific CAR T cells,9,10 allogeneic T cell transplantation triggers graft-versus-host disease (GvHD) because of HLA-mismatched α/β T cells.11, 12, 13, 14, 15 Of note, depletion of α/β T cells alleviates GvHD effects,16,17 and TCR ablation in T cells prevents GvHD in mice.18, 19, 20
Several strategies of endogenous TCR inactivation in allogeneic CAR T cells have been reported to augment the safety and efficacy of adoptive immunotherapy and minimize GvHD risk. The current approaches largely rely on the combination of clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) technology and viral vector transduction for TCR disruption and CAR expression in allogeneic T cells, respectively.9,19,21,22 Despite the successes of CAR T cell manufacturing and therapeutic applications, there is an unmet need to overcome the limitations caused by viral vectors including complex large-scale manufacturing of viral vectors and rigorous biosafety testing to exclude the presence of replication-competent retroviruses.23, 24, 25, 26 As an alternative for gene delivery, the use of transposons offers a number of distinct advantages over viral vector-based approaches, including larger cargo capacity to deliver multiple transgenes, reliance on cost-effective non-viral transfection strategies and their relatively random integration profiles compared with the biased integration pattern of integrating viral vectors.27, 28, 29, 30, 31, 32, 33 These advantages could greatly facilitate vector manufacturing and simplify regulatory approval on the path to clinical translation.34 Nevertheless, to our knowledge, combining transposons with CRISPR-Cas9 technologies for the manufacturing of allogeneic TCR knockout (TCR KO) CAR T cells has not yet been explored.35
Here, we describe the optimization and validation of non-viral system based on (1) Sleeping Beauty (SB) transposon31,36-mediated expression of CD19-targeted CAR and (2) targeted inactivation of endogenous TCR genes using CRISPR-Cas9-ribonucleoparticles (RNPs) for engineering human “off-the-shelf” CAR T cell devoid of endogenous TCR. The resulting CAR T cells are highly functional against CD19-expressing target cells and CD19+ tumor-bearing mice for tumor eradication with reduced TCR alloreactivity and GvHD development. Moreover, a comprehensive molecular analysis was performed to examine the downstream signaling effects in the TCR-disrupted CAR T cells.
Results
CRISPR-Cas9 RNP for single- and dual-gene disruption in human T cells
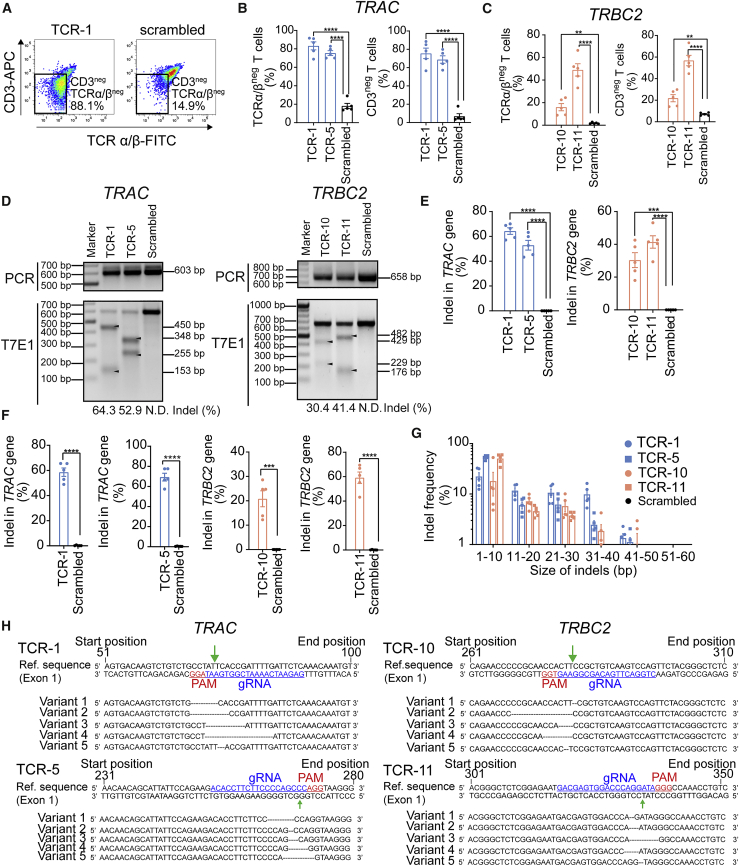
We first screened different single-guide RNAs (sgRNAs) specifically designed to target the TRAC and TRBC2 genes in the HEK293 cell line using a T7 endonuclease 1 assay (Tables S1 and S2). TRAC and TRBC2 gene disruption was readily detectable in most TCR-targeted guide RNA (gRNA) groups (4/5 TRAC-targeting sgRNAs; 6/6 TRBC2-targeting sgRNAs) (Figure S1). The indel percentages of TCR-1 and TCR-5 gRNAs targeting the TRAC gene and TCR-8, TCR-9, TCR-10, and TCR-11 gRNAs targeting TRBC2 gene were higher compared with those achieved using the other gRNAs (Figures S1C–S1D). We therefore selected the TCR-1, TCR-5, TCR-8, TCR-9, TCR-10, and TCR-11 gRNAs for subsequent TCR disruption in human CD8+ T cells.
CRISPR-Cas9 RNP complexes composed of TCR-targeting gRNA and purified Cas9 protein (TCR RNP) were nucleofected into T cells. TRAC-targeting TCR-1 and TCR-5 RNPs led to ∼80% TCR-negative (TCRneg) T cells (Figures 1A and 1B). TRBC2-targeting TCR-10 and TCR-11 RNPs unexpectedly outperformed TCR-8 and TCR-9 (in contrast to the results obtained in HEK293 cells) and resulted in ∼20%–50% TCRneg T cells for TCR-10/TCR-11 as opposed to 15%–38% for TCR-8/TCR-9 (Figures 1C and S2A). Loss of TCR expression after TCR disruption is consistent with a significant increase in CD3-negative (CD3neg) T cells (Figures 1A–1C). TCR downregulation impairs TCR/CD3 complex formation, which in turn results in decreased CD3 expression.37 The loss of TCR expression following TCR RNP delivery correlated with 50%–70% and ∼30%–40% indel percentages in TRAC and TRBC2 genes, respectively (Figures 1D, 1E, S2C, and S2D). Deep sequencing confirmed on-target TCR disruption, corresponding to 60%–70% and 20%–60% for TRAC and TRBC2 targeting, respectively (Figure 1F). Most indels ranged from 1 to 10 nucleotides (Figures 1G and 1H), resulting in frameshift variants and in-frame deletions (Table S3). Overall cell viability was comparable among all conditions, indicating that TCR disruption after CRISPR-Cas9 RNP transfection did not lead to any discernable cytotoxicity (Figure S3).
Figure 1.
CRISIPR-Cas9-mediated single-gene KO in human CD8+ T cells using RNP particles
After 48 h post-CD3/CD28 stimulation, 5 × 105 human CD8+ T cells were subjected to nucleofection of 70 pmol TRAC- or TRBC2-targeting RNP particles (Cas9:sgRNA ratio of 1:1). (A) Schematic representation of TCRα/β and CD3 coexpression in TRAC-targeting TCR-1 (left) or scrambled control (right) RNP-transfected cells. (B) Percentages of TCRα/βneg (left) and CD3neg (right) T cell population by flow cytometry after 48 h post-TRAC-targeting RNP nucleofection. (C) Percentages of TCRα/βneg (left) and CD3neg (right) T cell population by flow cytometry analysis 48 h post-nucleofection with TRBC2 RNP. (D) Schematic representatives of indel mutation detected by T7 endonuclease 1 (T7E1) assay after 48 h of TRAC-targeting (left) or TRBC2-targeting (right) RNP nucleofection. The indel percentage values are the averages of five biologically independent donors. (E) Quantitative analysis of indel mutation percentage in TRAC (left) and TRBC2 (right) genes detected by T7E1 assay after 48 h post-nucleofection. (F) Quantitative analysis of indel mutation percentage in TRAC and TRBC2 genes detected by deep sequencing analysis after 48 h post-RNP nucleofection. (G) Indel mutation frequency relative to the size of indels in TRAC and TRBC2 genes detected by deep sequencing analysis after 48 h post-nucleofection. Data are representative of five biologically independent donors. (H) Schematic representation of mutated DNA sequences in TRAC (left) and TRBC2 (right) after TCR-targeting RNP nucleofection. Data are shown as means ± SEM (n = 5 biologically independent donors); Student’s t test: ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
To broaden the versatility of the approach, we investigated whether the CRISPR-Cas9 RNP platform allows for efficient simultaneous disruption of multiple target genes in human CD8+ T cells. Simultaneous targeting of both the TRAC and TRBC2 loci did not significantly increase the percentages of TCRneg or CD3neg T cells compared with using only a single TCR-11 RNP (Figure S4). Based on this finding, we used a single TCR-specific gRNA (TCR-1-targeting TRAC) for TCR gene inactivation in subsequent experiments.
Non-viral transfection of a transgene of interest in the TCRneg cells
We subsequently optimized the platform to efficiently deliver any transgene of interest in the CRISPR-Cas9-modified TCR/CD3neg T cells. The commercially available pmaxGFP plasmid was selected for the platform optimization owing to its robust GFP reporter expression in a broad range of cells, including T cells, and its availability for instant use. The use of pmaxGFP described in previous studies highlights its convenience for platform optimization and serves as a benchmark to compare study outcomes by using the same standardized vector design and reporter gene.38, 39, 40, 41 The first strategy was based on a single co-transfection of CRISPR-Cas9 RNP and a GFP reporter plasmid (pmaxGFP) (Figure S5A, upper), which led to only ∼8.3% ± 2.27% GFP+CD3neg T cells in the pmaxGFP/TCR-1 RNP-transfected group (Figure S5B, left). The total percentage of CD3neg T cells was not significantly different between the pmaxGFP/TCR-1 RNP-treated groups and TCR-1 RNP controls, suggesting that pmaxGFP co-transfection did not interfere with the efficiency of TCR-1 RNP-mediated gene disruption (Figure S5C, left). In contrast, the total GFP+ fraction in the pmaxGFP/scrambled RNP group was 5.4-fold greater than in the pmaxGFP/TCR-1 RNP group (Figure S5D, left), indicating that the TCR CRISPR-Cas9 components attenuated the efficacy of GFP transfection and/or expression during nucleofection.
Since the major difference between the TCR-1 and the scrambled gRNA is the targeting sequence (TCR-targeting versus non-targeting sequences), we tested the hypothesis of whether it is restricted to TCR-targeting gRNA or whether it represents a more general phenomenon, irrespective of the gRNA and its target sequence. Therefore, an additional experiment was carried out by co-transfection of (1) 70 pmol TRAC-targeting TCR-1, programmed cell death protein 1 (PD1)-targeting PD1-5 (i.e., the PD1 KO activity was experimentally validated previously; representing a gene-targeting gRNA that is not relevant to TCR KO), or non-targeting scramble RNP (gRNA:Cas9 = 1:1), and (2) 500 ng of pmaxGFP into human T cells (Figures S6A–S6C). Forty-eight hours post-transfection, both PD1/pmaxGFP- and TCR/pmaxGFP-transfected cells revealed a pronounced decrease in total GFP+ population compared with the scrambled control/pmaxGFP group (Figures S6A and S6C). This indicates that the reduction of GFP transfection efficiency is not restricted to TCR targeting per se but may represent a more general phenomenon associated with CRISPR-Cas9-mediated gene targeting, irrespective of the gRNA and its target sequence. The exact reason is not clear and beyond the scope of this study but may be related to the cellular response following induction of dsDNA breaks.42
Then we investigated whether the Cas9 dosage titration can improve the GFP transfection efficiency while maintaining a relatively robust TCR KO efficiency (Figure S6D). Our result showed that a reduction of Cas9 to 35 and 17.5 pmol during a single nucleofection enhanced the total GFP+ population to ∼60% (Figure S6D, circular orange line) but inevitably compromised ∼20% of TCR gene disruption (Figure S6D, squared blue line).
The second strategy was based on sequential delivery of CRISPR-Cas9 RNP and pmaxGFP (24 h later) to generate GFP+TCR/CD3neg T cells (Figure S5A, middle). This approach yielded 53.7% ± 5.49% GFP+CD3neg T cells in the pmaxGFP/TCR-1 RNP-transfected group, which was 7.8-fold higher compared with the pmaxGFP/scrambled RNP-transfected group (Figure S5B, middle). Similar levels of total CD3neg T cells (Figure S5C, middle) and total GFP+ T cells (Figure S5D, middle) were observed between the TCR-1 RNP/pmaxGFP and control groups. This pattern suggests that pmaxGFP transfection did not interfere with the efficiency of TCR-1 RNP-mediated gene disruption and vice versa.
To further increase the overall yield of GFP+TCR/CD3neg T cell production, CD3neg cells were specifically enriched by CD3+ T cell depletion using magnetic activated cell sorting (MACS) in TCR-1 RNP-transfected T cells before pmaxGFP transfection (Figure S5A, lower). Up to 80% GFP+CD3neg T cells could be obtained following pmaxGFP/TCR-1 RNP transfection (Figure S5B, right). We also consistently observed equivalent percentages of total CD3+ T cells and GFP+ T cells between TCR-1 RNP/pmaxGFP and the corresponding control groups (Figures S5C, right and S5D, right). Depletion of CD3+ T cells led to a ∼1.5-fold increase in GFP+TCR/CD3neg T cell purity compared with sequential transfection of the bulk heterogeneous CD3+/CD3neg T cell population (Figures S5E and S5F). The overall transfection efficiency of pmaxGFP was unexpectedly enhanced ∼1.5-fold after CD3neg enrichment (Figure S5G). Despite the comparable cell viability (Figure S7), a gradual decrease in total GFP+CD3neg T cell numbers after CD3+ T cell depletion was apparent, indicating a loss of cells during cell processing (Figure S5H).
Rapid production of TCR KO CAR T cells targeting CD19 antigen for transient CAR expression
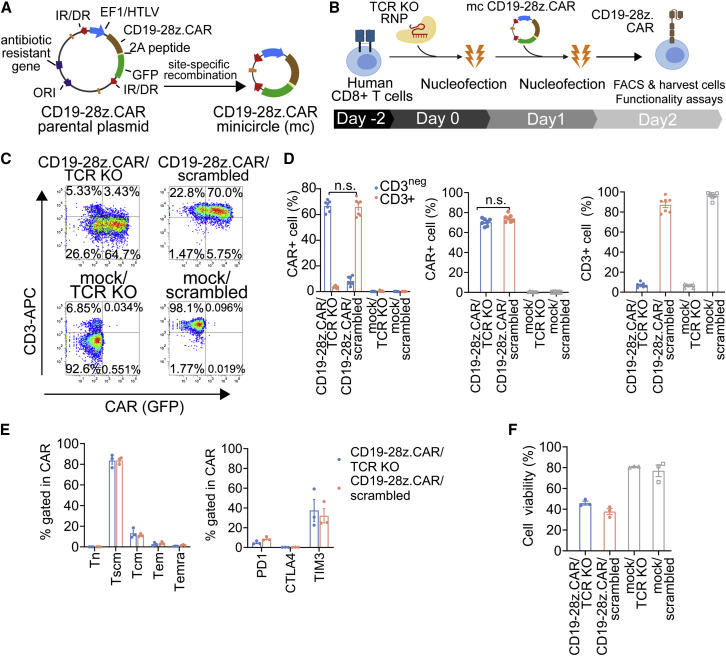
We employed the optimized sequential transfection approach to generate CD19-specific CAR T cells devoid of TCR expression (CD19-28z.CAR/TCR KO). For this purpose, we replaced the pmaxGFP plasmid with a minicircle (mc) DNA encoding a CD19-specific CAR fused to a GFP reporter gene via a T2A self-cleaving peptide, under the control of the hybrid elongation factor 1/human T cell leukemia virus promoter. The resulting mc plasmid, which was devoid of bacterial backbone DNA, was designated as mcCD19-28z.CAR (Figure 2A). The main advantages of mcDNA are its superior transfection efficiency and amelioration of the cellular toxicity caused by the bacterial backbone in conventional plasmids.43 Due to the differences in DNA size, vector design, and/or promoter strength when comparing mcCD19-28z.CAR vector with pmaxGFP, a dose-dependent transfection study was therefore carried out to assess the optimal DNA amount of the mcCD19-28z.CAR vector for achieving high transfection efficiency (Figure S8A). The results showed that, to obtain similar transfection efficiencies as pmaxGFP (i.e., ∼60%), the mcCD19-28z.CAR vector required a 2-fold higher DNA dose compared with the amount of pmaxGFP plasmid used (i.e., 1 μg mcCD19-28z.CAR versus 500 ng pmaxGFP; Figure S8A, orange line). We therefore selected the dose of 1 μg mcCD19-28z.CAR vector per 5 × 105 T cells for CAR T cell manufacturing in the subsequent experiments. As expected, CAR expression correlated directly with GFP (Figure S8B), supporting use of GFP as a surrogate marker for CAR expression in the subsequent experiments.
Figure 2.
Production and characterization of CD19-specific CAR/TCR KO T cells using the non-viral CRISPR-Cas9 RNP-mc vector platform for transient CAR expression
(A) Schematic representation of the mcDNA vector generated from the parental plasmid via site-specific recombination, which further removed the bacterial backbone and minimized the overall vector size. Specifically, the mcCD19-28z.CAR vector contains a single multicistronic construct under the regulation of elongation factor 1 alpha-HTLV (EF1/HTLV). The CD19-28z.CAR and GFP reporter genes are linked via a self-cleavable 2A peptide. The expression cassette is flanked with IR/DR inverted repeats. (B) Schematic representation of the CAR T cell production method: 5 × 105 stimulated human CD8+ T cells were subjected to nucleofection of 70 pmol of TCR-1 RNP. Twenty-four hours later, the cells were electroporated with 1 μg of mcCD19-28z.CAR. Next day, the cells were harvested for flow cytometry analysis and functionality tests. (C) Schematic flow cytometric plot. (D) Percentages of (left) fractioned CAR/CD3neg and CAR/CD3+ cells, (middle) total CAR+ cells, and (right) total CD3+ cells in CD19-28z.CAR/TCR KO, CD19-28z.CAR/scrambled control, mock/TCR KO, and mock/scrambled control T cells analyzed by flow cytometry at 24 h post-transfection. (E) Immunophenotyping analysis of (left) T cell memory markers and (right) PD1, CTLA4, and TIM3 immune checkpoint markers in CD19-28z.CAR/TCR KO and CD19-28z.CAR/scrambled control T cells at 24 h post-transfection. (F) Cell viability of CD19-28z.CAR/TCR KO, CD19-28z.CAR/scrambled control, mock/TCR KO, and mock/scrambled control T cells at 24 h after production. Data are shown as means ± SEM (n = 3–6 biologically independent donors); Student’s t test; n.s., not significant.
Consistent with our result obtained from the platform optimization experiment using pmaxGFP plasmid (Figures S5B–S5D, left), the single nucleofection of mcCD19-28z.CAR/TCR-1 RNP components is relatively inefficient to generate the CD19-28z.CAR/TCR KO T cells (Figures S9A–S9C). This justifies the use of sequential nucleofection platform for robust CAR/TCR KO production in the subsequent experiments (Figure 2B). Sequential transfection of T cells with TCR-1 or scrambled control RNP followed by mcCD19-28z.CAR resulted in ∼60%–70% of the target cell population (i.e., CAR+CD3neg population for mcCD19-28z.CAR/TCR-1 transfection; CAR+CD3+ population for mcCD19-28z.CAR/scrambled control transfection) (Figures 2C and 2D, left) and comparable amounts of total CAR T cells (both CD3neg and CD3+) (Figure 2D, middle). Similar percentages of total CD3neg T cells were obtained from the mcCD19-28z.CAR/TCR-1 RNP and mock/TCR-1 RNP control transfection, suggesting that TCR disruption was unaffected by mcCD19-28z.CAR DNA transfection (Figure 2D, right). Interestingly, in the CD19-28z.CAR/TCR KO T cells, which are mostly CD3neg, the percentage of CAR expression gated in the CD3neg fraction was significantly higher than that in the CD3+ fraction (Figure S9D). Conversely, in the CD19-28z.CAR/scrambled control T cells, which are predominately CD3+, a significant increase in CAR expression was observed in the CD3+ fraction compared with the CD3neg fraction (Figure S9D). CAR T cells consisted mainly of the stem cell-like memory (Tscm) (CD45RA+CD45RO+/negCD62L+CD95+) subset, and less central memory (CD45RO+CD62L+CD95+), effector memory (Tem) (CD45RO+CD62LnegCD95+), and terminally differentiated effector (CD45RA+CD62LnegCD95+) subsets (Figure 2E, left). In both CD19-28z.CAR/TCR KO and CD19-28z.CAR/scrambled control T cells, expression of PD1 and CTLA4 was barely discernible, whereas TIM3 expression was moderate (Figure 2E, right). Cell viability declined to 32.3%–49.6% in the CD19-28z.CAR/TCR KO and CD19-28z.CAR/scrambled groups, consistent with DNA-induced toxicity (Figure 2F).
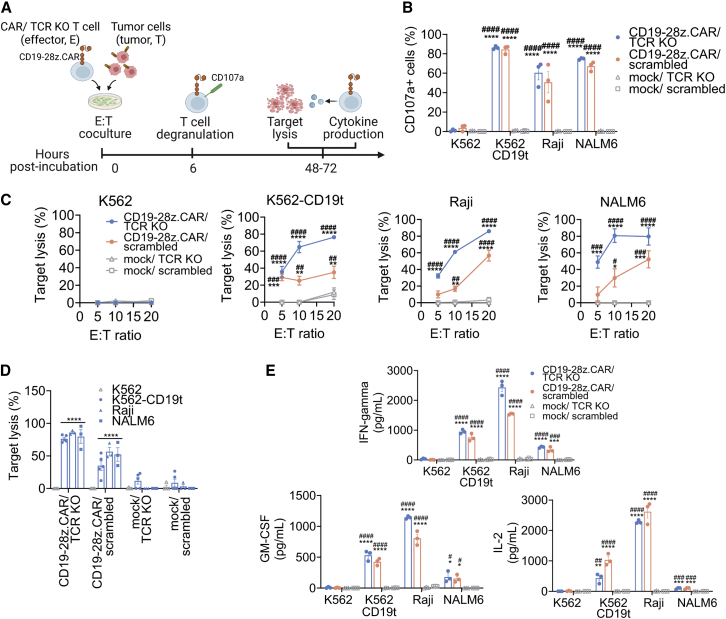
Transiently expressed CD19-28z.CAR/TCR KO T cells with antitumor functions in vitro
To characterize the functionality of CD19-28z.CAR/TCR KO and CD19-28z.CAR/scrambled T cells, we assessed their upregulation of the CD107a degranulation marker, oncolytic effect, and cytokine secretion pattern (Figure 3A). After a short incubation with CD19-expressing K562-CD19t, Raji, and NALM6 tumor cells (Figure S10), CD107a expression markedly increased in the CD19-28z.CAR/TCR KO and CD19-28z.CAR/scrambled groups compared with K562 control tumor cells (Figures 3B, S11A, and S11B). In contrast, CD107a expression was similar among all mock/TCR KO and mock/scrambled groups (Figures 3B, S11A, and S11B). Both CD19-28z.CAR/TCR KO and CD19-28z.CAR/scrambled CAR T cells effectively eliminated CD19+ tumor cells in an effector cell dose-dependent fashion (Figure 3C). Prolonged incubation to 72 h led to a 30%–60% increase in tumor lysis (Figure S11C). CD19-negative K562 control cells were resistant to the cytolytic effects of the CD19-28z.CAR/TCR KO or CD19-28z.CAR/scrambled T cells, indicating that the tumor lysis was CD19 antigen specific (Figure 3D). Concordantly, co-incubation with CD19+ tumor cells increased cytokine production (interferon γ [IFN-γ], granulocyte-macrophage colony-stimulating factor [GM-CSF], and interleukin-2 [IL-2]) in CD19-28z.CAR/TCR KO and CD19-28z.CAR/scrambled CAR T (Figure 3E).
Figure 3.
Transiently expressed CD19-redirected CAR/TCR KO T cells demonstrated a potent antitumor effect in response to CD19 antigen
(A) Schematic representation of experimental design. CD19-CAR/TCR KO, CD19-CAR/scrambled, mock/TCR KO, or mock/scrambled T cells were cocultured with CD19+ tumor cells (i.e., K562-CD19t, Raji, or NALM6) or CD19neg tumor cells (i.e., K562) at specific effector:tumor cell ratios based on the assays for 6–72 h and were subjected for subsequent analyses. (B) Percentage of CD107a+ cells upon 6 h of incubation with CD19+ and control tumor cells (∗compared with mock/TCR KO group; #compared with mock/scrambled control group). (C) Effector dose-response-specific lysis of effector cells (∗compared with mock/TCR KO group; #compared with mock/scrambled control group). (D) Comparison of specific lysis of effector cells between incubation with CD19+ and K562 control tumor cells (∗compared with K562 control cells). (E) Secretion of human IFN-γ, GM-CSF, and IL-2 cytokines from effector cells in response to target tumor cells (∗compared with mock/TCR KO group; #compared with mock/scrambled control group) after 72 h post-coculture. Data are shown as means ± SEM (n = 3 biologically independent donors); one-way ANOVA with Dunnett’s post hoc; n.s., not significant; ∗/#p < 0.05, ∗∗/##p < 0.01, ∗∗∗/###p < 0.001, ∗∗∗∗/####p < 0.0001.
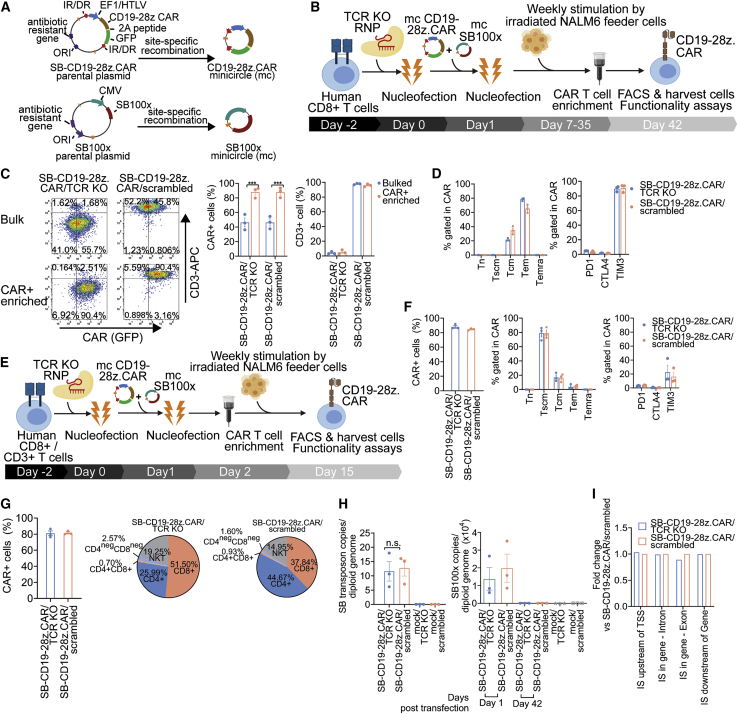
SB-mediated manufacture of CD19-specific CAR/TCR KO T cells for stable CAR expression
We next adapted our method to generate CD19-28z.CAR/TCR KO T cells stably expressing CAR using the SB transposon system. The mcDNA vectors mc-CD19-28z.CAR and mcSB100x harboring hyperactive SB100x transposase were co-transfected into CD8+ T cells initially transfected with TCR-1 or with scrambled RNPs as control (Figures 4A and 4B). The resulting transfected cells (designated as SB-CD19-28z.CAR/TCR KO and SB-CD19-28z.CAR/scrambled) were then expanded using irradiated NALM6 feeder cells in the presence of human IL-2, IL-7, IL-15, and IL-21. After 6 weeks of expansion, CAR T cells were enriched by MACS (Figure 4B). This approach resulted in a ∼10% weekly increase in the CAR+ T cell fraction (Figure S12B, left). An average 94- ± 28-fold expansion of CAR T cells in the bulk SB-CD19-28z.CAR/TCR KO and SB-CD19-28z.CAR/scrambled control T cell populations was observed at the end of expansion protocol (Figure S12B, right). Following selective expansion of CAR T cells and CAR enrichment with MACS, up to 89.7% and 90.6% of the SB-CD19-28z.CAR/TCR KO and SB-CD19-28z.CAR/scrambled groups expressed CAR (Figure 4C, left and middle). The SB-CD19-28z.CAR/TCR KO T cells exhibited only very low TCR expression levels compared with the SB-CD19-28z.CAR/scrambled T cells (Figure 4C, right), consistent with efficient TCR disruption in the CAR T cells. Similar results were obtained in the CAR-enriched and bulk CAR T cell populations (Figure 4C, right). Both the SB-CD19-28z.CAR/TCR KO and SB-CD19-28z.CAR/scrambled T cells predominately displayed the Tem phenotype (Figure 4D, left). PD1 and CTLA4 expression was barely detectable, whereas relatively high TIM 3 expression was observed in both effector cells (Figure 4D, right). Hence, the prolonged culture duration led to a more differentiated memory T cell phenotype and elevated TIM3 expression. To limit the cell expansion period, an early CAR+ T cell enrichment step was implemented at 24 h post-transfection. Subsequently, only the CAR+ T cell fraction was subjected to cell expansion for 14 days (Figure 4E). This was expected to delay terminal T cell differentiation and exhaustion caused by repeated NALM6 stimulation while attaining a high purity of CAR T cell production. Notably, using this method, an average of 88.1% ± 3.34% and 84.6% ± 1.42% of CAR expression was achieved after 14 days culture in SB-CD19-28z.CAR/TCR KO and SB-CD19-28z.CAR/scrambled cells, respectively (Figure 4F, left). The extent of the T cell expansion was comparable in both conditions (Figure S12C). In contrast to the previous approach based on the 6-week culture protocol, the phenotypes of these CAR T cells were mainly Tscm, and the percentage of the Tem subset was reduced (Figure 4F, middle). In addition to minimal PD1 and CTLA4 expression, the current method also decreased TIM3 expression by ∼70% compared with the 6-week culture protocol (Figure 4F, right). Similar results were obtained from CD19 CAR T cells generated from human CD3+ T cells (Figures 4G and S12D–S12F). Notably, the expansion of CAR T cells obtained from CD3+ T cells was significantly higher compared with CAR T cells produced from purified CD8+ T cells (Figure S12F). SB-CD19-28z.CAR/TCR KO cells showed a slight difference with respect to CD8+ and CD4+ composition compared with SB-CD19-28z.CAR/scrambled cells, whereas the distribution of other T cell subsets was comparable among the groups (Figure 4G, right). Analysis of genomic DNA from SB-CD19-28z.CAR/TCR KO and SB-CD19-28z.CAR/scrambled cells (Table S4) indicated similar SB transposon copies (Figure 4H, left) and gradual loss of the transposase-encoding SB100x plasmid toward the end of the culture (Figure 4H, right). This disappearance of the transposase-encoding plasmid is a built-in self-limiting safety feature preventing continuous transposition in the transfected T cells.
Figure 4.
Production and characterization of CD19-specific CAR/TCR KO T cells using the non-viral CRISPR-Cas9 RNP-mcSB transposon platform for sustained CAR expression
(A) Schematic representation of the mcDNA vector generated from the parental plasmid via site-specific recombination, which further removed the bacterial backbone and minimized the overall vector size. The production of mcCD19-28z.CAR vector was described previously (Figure 2A).The mcSB100x vector harbors the gene encoding hyperactive SB100x transposase to modulate SB transposition driven by the cytomegalovirus (CMV) promotor. (B) Schematic representation of CAR T cell production approach without early CAR enrichment: 5 × 105 stimulated human CD8+ T cells were subjected to nucleofection of 70 pmol of TCR-1 RNP. Twenty-four hours later, the cells were transfected with 1 μg of mcCD19-28z.CAR and 0.5 μg of mcSB100x. Subsequently, the CD19-specific CAR T cells were selectively expanded by weekly addition of 100-Gy-irradiated NALM6 feeder cells. Recombinant IL-2, IL-7, and IL-15 cytokines were supplemented to the culture on a Monday/Wednesday/Friday schedule. On week 6 post-culture, the CAR+ T cells were enriched using the MACS method by incubating the cells with APC-conjugated recombinant human CD19 protein and magnetic bead-conjugated anti-APC antibody. The enriched CAR+ cells were rested in complete T cell medium and harvested for flow cytometry analysis and functionality tests. (C) (Left) Schematic flow cytometric plot, (middle) total CAR+ cell percentage, and (right) total CD3+ cell percentage of CAR T cells before and after CAR+ enrichment by MACS. (D) Immunophenotyping analysis of (left) T cell memory markers and (right) PD1, CTLA4, and TIM3 immune checkpoint markers of SB-CD19-28z.CAR/TCR KO and SB-CD19-28z.CAR/scrambled control T cells at 6 weeks post-culture. Data are shown as means ± SEM (n = 3 biologically independent donors); Student’s t test; ∗∗∗p < 0.001. (E) Schematic representation of CAR T cell production approach with early CAR enrichment: 5 × 105 stimulated human CD8+ or CD3+ T cells were subjected to nucleofection of 70 pmol of TCR-1 RNP. Twenty-four hours later, the cells were transfected with 1 μg of mcCD19-28z.CAR and 0.5 μg of mcSB100x. Next day, only CAR+ T cells were enriched using the MACS method and subsequently expanded by weekly addition of 100-Gy-irradiated NALM6 feeder cells. Recombinant IL-2, IL-7, and IL-15 cytokines were supplemented to the culture on a Monday/Wednesday/Friday schedule. On week 2 post-culture, the CAR T cells were harvested for flow cytometry analysis and functionality tests. (F) (Left) Total CAR+ cell percentage, immunophenotyping analysis of (middle) T cell memory markers, and (right) PD1, CTLA4, and TIM3 immune checkpoint markers in SB-CD19-28z.CAR/TCR KO and SB-CD19-28z.CAR/scrambled control T cells generated from human CD8+ T cells at 2 weeks post-culture. Data are shown as means ± SEM (n = 3 biologically independent donors). (G) (Left) Total CAR+ cell percentage and (right) cellular composition of CD4+ and CD8+ CAR T cells generated from human CD3+ T cells at 2 weeks post-culture. (H) Quantitative analysis of (left) SB transposon copy number and (right) SB100x transposase after 6 weeks of CAR T cell expansion by qPCR assay (Table S4). Data are shown as means ± SEM (n = 3 biologically independent donors); Student’s t test; n.s., not significant. (I) Fold difference of SB transposon integration site (IS) frequency in the different genomic features between SB-CD19-28z.CAR/TCR KO versus SB-CD19-28z.CAR/scrambled control T cells.
The method of CAR T cell manufacturing described in our study is based on sequential electroporation of TRAC-targeted CRISPR RNP for TCR ablation and SB transposon/transposase DNA for CAR expression. Since the time interval between the CRISPR-mediated gene targeting and the SB transposon transfection is ∼24 h, we agree with the reviewer that it cannot be excluded a priori that CRISPR-Cas9 activity may alter the overall SB transposon integration pattern. Moreover, CRISPR-Cas9-mediated double-stranded DNA breaks at TRAC loci (TRAC DSB) may potentially favor preferential SB transposon integration into these TRAC DSB regions since DSBs are known to “capture” exogenous DNA.42,44 Hence, this could lead to differences in SB transposon integration profiles in the SB-CD19-28z.CAR/TCR KO T cells compared with the SB-CD19-28z.CAR/scrambled control T cells. To determine the effect of CRISPR-Cas9 activity on SB transposon insertion profiles in T cells, an unbiased genome-wide integration site analysis (ISA) of genomic DNA samples obtained from the SB-CD19-28z.CAR/TCR KO T cells and the SB-CD19-28z.CAR/scrambled control T cells was carried out. The ISA data showed that the relative frequencies of transgene insertion into different genomic features in the SB-CD19-28z.CAR/TCR KO T cells and the SB-CD19-28z.CAR/scrambled T cells control are highly similar (Figures 4I and S13A). The integration profile of the SB transposon vector in both samples displayed 51% of integration site (IS) in gene coding regions, of which 49% of ISs are located within intronic and 2% within exonic regions. The remaining 49% of ISs were found outside of the gene body with highly similar percentages of IS found either upstream of a transcriptional start site (TSS) (24%) or downstream of a gene (25%) (Figure S13A, upper). When compared with typical gammaretroviral and lentiviral profiles,45, 46, 47 these SB transposon-modified CAR T cells have lower insertion site proportions in gene coding regions, indicating lower preference toward transcription active parts of the genome. Consistently, ISs were detected across all chromosomes and are found to be in line with chromosome size, showing comparable frequencies in the SB-CD19-28z.CAR/TCR KO T cells and the SB-CD19-28z.CAR/scrambled T cells (Figure S13A, middle). The integration in both samples was distributed similarly throughout the gene relative to TSS, showing a reduced frequency upstream of the TSS (Figure S13A, lower). Both CAR T cells are highly polyclonal (Figure S13B) and absent of any integration site showing a relative contribution of >30% (of total retrieved integration sites) (Figure S13C), suggesting no biased insertion into any specific loci in the genome and clonal expansion. In particular, we did not observe an integration event within or nearby TRAC gene in the SB-CD19-28z.CAR/TCR KO T cells (Table S5), ruling out the possibility of preferential SB transposon insertion at TRAC DSBs.
The lack of SB transposon integration at TRAC DSBs was further confirmed using PCR amplification and DNA Sanger sequencing analysis. First, the plasmid library containing DNA variants derived from the TRAC DSBs was generated from genomic DNA of the SB-CD19-28z.CAR/TCR KO and the SB-CD19-28z.CAR/scrambled control T cells (Figure S14A). Next, we screened at least 100 plasmid clones to assess the presence of the SB transposon at the TRAC DSB library using PCR amplification specific to CAR DNA. The results showed that the SB transposon integration was undetectable at TRAC DSB regions in the SB-CD19-28z.CAR/TCR KO T cells (Figures S14B and S14C). Consistently, the DNA sequencing analysis indicated the absence of an SB transposon insertion in the TRAC DSBs (Figure S14D).
To evaluate the potential risks of insertional oncogenesis induced by SB transposon integration, the frequency of insertion sites harbored in 100 kb proximity to a TSS of the selected cancer-associated genes was determined. The results demonstrated that, in concordance with the polyclonal composition of SB-derived CAR T cells, the relative frequencies of these insertion sites that were found within or close to the proto-oncogenes, including CCND2 (AS1), HMGA2, LMO2, MECOM, or MN1, in previous gene therapy trials,29,30,48, 49, 50, 51 were very low (i.e., 0.0004%–0.0195%) (Table S6).
In vitro antitumor effects of SB-modified CD19-redirected CAR T cells devoid of TCR expression
We then explored whether SB-CD19-28z.CAR/TCR KO T cells were specifically responsive to CD19 antigen and exerted antitumor functions. CD19-expressing tumor cells induced ∼20%–40% higher CD107a expression than K562 control cells in both SB-CD19-28z.CAR/TCR KO and SB-CD19-28z.CAR/scrambled groups (Figures 5A, S15A, and S15B). After 48 and 72 h of effector/CD19+ tumor cell coculture, an increase in target lysis was uniquely identified in the SB-CD19-28z.CAR/TCR KO and SB-CD19 28z.CAR/scrambled control T cells (Figures 5B and S15C) and positively correlated with a higher effector-to-target ratio (Figure 5B). Consistent with these findings, cytolytic functions of SB-CD19-28z.CAR/TCR KO and SB-CD19-28z.CAR/scrambled T cells were restricted to CD19+ tumor cells (Figure 5C). In the absence of CAR expression, minimal or no lysis of CD19+ target cells was apparent, confirming that the specificity of cell killing was determined by CAR expression (Figure 5C). This result corresponded with increased IFN-γ, GM-CSF, and tumor necrosis factor alpha (TNF-α) cytokine levels in the SB-CD19-28zCAR/TCR KO and SB-CD19-28z.CAR/scrambled groups upon exposure to CD19 antigen (Figure 5D). Following co-incubation with CD19-depleted allogeneic PBMCs (allo-PBMCs), the TCR ablation significantly decreased CAR T cell proliferation compared with the SB-CD19-28z.CAR/scrambled control T cells in a time-dependent manner (Figure 5E).
Figure 5.
SB-CD19-28z.CAR/TCR KO were responsive to CD19-expressing tumor cells and displayed antigen-dependent effector functions
(A) Percentage of CD107a+ cells upon 6 h of incubation with CD19+ or K562 control tumor cells (∗compared with mock/TCR KO group; #compared with mock/scrambled control group). (B) Effector dose-response-specific lysis of effector cells after 48 h of incubation with CD19+ or K562 control tumor cells. One-way ANOVA with Dunnett’s post-hoc (∗compared with mock/TCR KO group; #compared with mock/scrambled control group). (C) Comparison of specific lysis of effector cells between incubation with CD19+ and K562 control tumor cells. One-way ANOVA with Dunnett’s post-hoc (∗compared with K562 control group). (D) Secretion of human IFN-γ, GM-CSF, and TNF-α cytokines from effector cells in response to target tumor cells after 72 h post-coculture. One-way ANOVA with Dunnett’s post-hoc (∗compared with mock/TCR KO group; #compared with mock/scrambled control group). (E) (Left) Schematic flow cytometric plot on day 4 post-incubation and (right) percentages of proliferating effector cells after contact with CD19-depleted allogeneic PBMCs (allo-PBMCs) at different time points. Student’s t test (∗compared with SB-CD19-28z.CAR/TCR KO at each time point). Data are shown as means ± SEM (n = 3–4 biologically independent donors); ∗∗/##p < 0.01, ∗∗∗/###p < 0.001, ∗∗∗∗/####p < 0.0001.
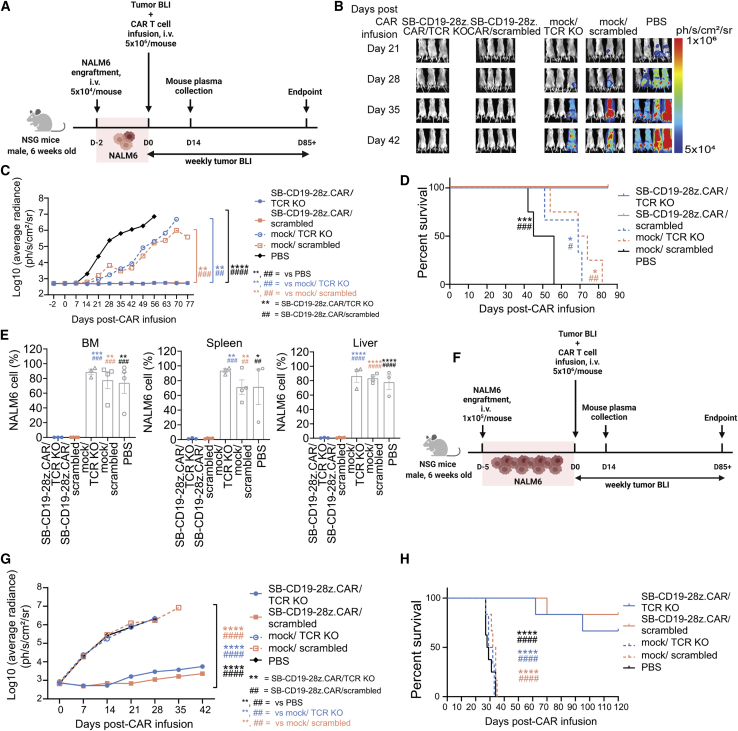
Potent in vivo antitumor functions of SB-engineered CD19-redirected CAR/TCR KO T cells
Next, we assessed the effect of SB-CD19-28z.CAR/TCR KO on CD19+ tumor progression in a xenograft model. For a low tumor burden setting, we administered 5 × 104 luciferase-expressing NALM6 cells by intravenous injection into NOD/SCID/IL2r−/− (NSG) male mice, followed by 5 × 106 SB-CD19-28z.CAR/TCR KO, SB-CD19-28z.CAR/scrambled, mock/TCR KO, or mock/scrambled T cell engraftment at 2 days post-NALM6 inoculation. As a negative control, NALM6-bearing mice were injected with phosphate-buffered saline (PBS) (Figure 6A). All PBS-injected tumor-bearing mice had a tumor burden from day 14, whereas most of the mock/TCR KO and mock/scrambled-treated groups developed tumors on day 42 (Figures 6B and 6C). In contrast, mice receiving the SB-CD19-28z.CAR/TCR KO CAR T cells or the SB-CD19-28z.CAR/scrambled control CAR T cells maintained complete tumor remission over an 11-week period (Figures 6B and 6C). The absence of tumor burden is consistent with a significantly prolonged median survival observed in SB-CD19-28z.CAR/TCR KO or SB-CD19-28z.CAR/scrambled control-injected groups compared with the control mice (Figure 6D). Moreover, endpoint examination of CAR T cell-treated mice showed that NALM6 cells were undetectable in the major lymphoid and non-lymphoid organs including bone marrow, spleen, and liver tissue (Figures 6E and S16A). In contrast, a marked accumulation of infiltrating NALM6 cells was identified in these organs obtained from all control groups (Figure S16A). In the next experiment, the same therapeutic dose of CD19 CAR T cells was tested in the NALM6-bearing mice with a higher tumor load (i.e., inoculation of 1 × 105 luciferase-expressing NALM6 cells per mouse; tumor was established for 5 days before CAR T cell infusion) (Figure 6F). Both SB-CD19-28z.CAR/TCR KO CAR T cells and SB-CD19-28z.CAR/scrambled control CAR T cells significantly reduced the leukemic burden in the mice with high tumor load and improved the survival rate compared with the control groups (Figures 6G, 6H, and S16B), consistent with the previous low tumor load experiment. Only few mice from these groups developed leukemia at later time points and had to be euthanized (i.e., two mice from SB-CD19-28z.CAR/TCR KO, euthanized on day 62 and 95 post-CAR infusion; one mouse from SB-CD19-28z.CAR/scrambled, euthanized on day 70 post-CAR infusion) (Figure 6H), indicating that CAR T cells exhibited antitumor effects in an effector cell dose-dependent manner. Collectively, both the SB-CD19-28z.CAR/TCR KO and the SB-CD19-28z.CAR/scrambled control T cells were equally efficient at suppressing NALM6 tumor progression and improving mouse survival compared with the non-CAR T cells (i.e., mock/TCR KO and mock/scrambled) and PBS control groups (Figure 6). Tumor growth of the mock/TCR KO and mock/scrambled groups was slightly reduced compared with the PBS control group, due to a CAR-independent T cell-mediated antitumor effect.
Figure 6.
SB-CD19-28z.CAR/TCR KO T cells efficiently rejected CD19+ leukemic cells in vivo
(A) Schematic representation of experimental design with low tumor burden. Six-week-old male NSG mice received 5 × 104 NALM6 tumor cells expressing luciferase via intravenous tail vein injection. Two days post-tumor inoculation, 5 × 106 SB-CD19-28z.CAR/TCR KO or SB-CD19-28z.CAR/scrambled control T cells were intravenously infused into xenografted mice. For the control groups, the equivalent numbers of mock/TCR KO or mock/scrambled control T cells or equivalent volumes of PBS were administered to the mice. Non-invasive bioluminescence imaging (BLI) was performed weekly to monitor the tumor progression over time. (B) Representative BLI of tumor development from all groups at different time points. The BLI signals were expressed in average radiance (ph/s/cm2/sr). (C) Quantitative tumor BLI signals from the whole mouse body at different time points. The BLI signals were expressed in log10(average radiance) (ph/s/cm2/sr). (D) Kaplan-Meier survival curve analysis of mice from all groups. (E) Quantitative analysis showing the presence of NALM6 tumor cells in bone marrow, spleen, and liver tissues of all treated groups at endpoint. For the low tumor burden experiment, data are shown as means ± SEM (n = 3–4 mice); one-way ANOVA with Dunnett’s post-hoc or log rank test for Kaplan-Meier survival analysis; ∗/#p < 0.05, ∗∗/## p < 0.01, ∗∗∗/###p < 0.001, ∗∗∗∗/#### p< 0.0001. (F) Schematic representation of experimental design with high tumor burden. Six-week-old male NSG mice received 1 × 105 NALM6 tumor cells expressing luciferase via intravenous tail vein injection. Five days post-tumor inoculation, 5 × 106 SB-CD19-28z.CAR/TCR KO or SB-CD19-28z.CAR/scrambled control T cells were intravenously infused into xenografted mice. For the control groups, the equivalent numbers of mock/TCR KO or mock/scrambled control T cells or equivalent volumes of PBS were administered to the mice. (G) Quantitative tumor BLI signals from the whole mouse body at different time points. The BLI signals were expressed in log10(average radiance) (ph/s/cm2/sr). (H) Kaplan-Meier survival curve analysis of mice from all groups. For high tumor burden experiment, data are shown as means ± SEM (n = 6–8 mice); one-way ANOVA with Dunnett’s post-hoc or log rank test for Kaplan-Meier survival analysis; ∗/#p < 0.05, ∗∗/##p < 0.01, ∗∗∗/###p < 0.001, ∗∗∗∗/####p < 0.0001.
TCR disruption in SB-derived CAR T cell-alleviated xenogeneic GvHD development
We further evaluated whether the SB-CD19-28z.CAR/TCR KO cells with reduced TCR alloreactivity could lower the risk of GvHD development in mice. To induce GvHD, we administered 10 × 106 SB-CD19-28z.CAR/TCR KO or SB-CD19-28z.CAR/scrambled control T cells into 2-Gy-irradiated NSG mice. Mice receiving only PBS served as a “healthy” control (Figure 7). Consistent with our results obtained from the alloreactivity experiment (Figure 5E) and previous studies,20,22,60 most of the mice (4/6 mice) receiving the SB-CD19-28z.CAR/scrambled control T cells developed clinical signs of GvHD and died after 2 months, whereas all SB-CD19-28z.CAR/TCR KO-treated mice survived and remained disease-free over 3 months (Figure 7A–7C; Table S7). This corresponds with the presence of characteristic GvHD histopathology findings, including high infiltration of mononuclear cells in liver and lung, shortening and/or structural abnormality of intestinal villi, and bronchial luminal narrowing (Figure 7D) in the SB-CD19-28z.CAR/scrambled control mice, as opposed to the lack of any such pathologies in the SB-CD19-28z.CAR/TCR KO-treated mice.
Figure 7.
SB-CD19-28z.CAR/TCR KO T cells inhibited GvHD development in vivo
Six-week-old male NSG mice were irradiated at a dose of 2 Gy. After 6 h of irradiation, the mice received 10 × 106 SB-CD19-28z.CAR/TCR KO or SB-CD19-28z.CAR/scrambled control T cell through intravenous administration. For the healthy control group, equivalent volumes of PBS were administered to the mice. (A and B) (A) Mouse body weight change and (B) GvHD clinical scoring were recorded over time. The GvHD clinical scoring criteria used in the experiment are described in Table S7.52, 53, 54 (C) Kaplan-Meier survival analysis of mice from all groups (n = 4–6 mice); log rank test; ∗p < 0.05. (D) Histopathology of intestine, liver, and lung tissues obtained from all groups. Solid black arrows indicate GvHD pathology, including high infiltration of mononuclear cells in hepatic and lung tissues, shortening and/or structural abnormality of intestinal villi, and bronchial luminal narrowing.55, 56, 57, 58, 59 The corresponding number at each scale bar indicates an image size (μm).
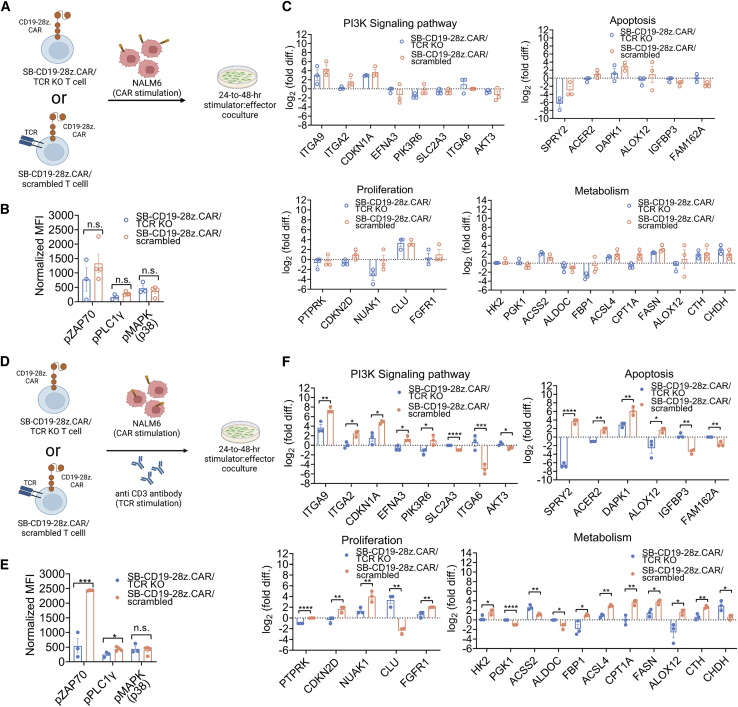
Analysis of signaling pathways in T cells engineered with SB-CD19-28z.CAR in the presence and absence of endogenous TCR signaling
Generally, the antileukemic activities of CAR T cells mainly associate with CAR signaling upon specific tumor antigen recognition. However, this may not fully recapitulate the current allogeneic CAR treatment where endogenous TCR of donor-derived CAR T cells can be reactive to viral antigens and/or allo-antigens.61,62 Hence, we believe that it is strongly necessary and relevant to explore the alteration of signaling profiles in the CAR/TCR KO cells compared with the CAR/scrambled control T cells under two different T cell signaling conditions: single CAR signaling (i.e., by stimulating the cells with NALM6) (Figure 8A) and dual CAR/endogenous TCR signaling (i.e., by stimulating the cells with NALM6 cell and anti CD3 antibody) (Figure 8D). This is aimed at providing a better molecular insight into allogeneic CAR T cell signaling in the absence and presence of endogenous TCR reactivity, respectively. As a negative control, the effector cells were cultured without stimulus to measure the background signals from the resting state.
Figure 8.
Effects of single CAR signaling and concomitant CAR/endogenous TCR co-signaling on molecular signature of SB-CD19-28z.CAR/TCR KO and SB-CD19-28zCAR/scrambled T cells
(A) Schematic representative of experimental design for analysis of single CAR-induced molecular signaling profile. A total of 3 × 106 SB-CD19-28z.CAR/TCR KO or SB-CD19-28z.CAR/scrambled T cells was stimulated with irradiated NALM6 cell (effector-to-target [E:T] = 1:1) for inducing CAR signaling for 24–48 h. As a negative control of resting state, CD19 CAR T cells were cultured in the absence of stimulus. (B) Quantitative analysis of ZAP70, PLCγ1, and p38 protein phosphorylation in CAR T cells after 24 h of stimulation. (C) Quantitative analysis of RNA expression profiling of selected genes associated with the PI3K signaling pathway, apoptosis, metabolism, and cell proliferation in SB-CD19-28z.CAR/TCR KO and SB-CD19-28z.CAR/scrambled T cells at 48 h post-stimulation. (D) Schematic representative of experimental design for analysis of simultaneous CAR/endogenous TCR-induced molecular signaling profile. A total of 3 × 106 SB-CD19-28z.CAR/TCR KO or SB-CD19-28z.CAR/scrambled T cells was stimulated with irradiated NALM6 cell (E:T = 1:1) and anti-CD3 antibody (bead:cell = 1:1) for inducing CAR and endogenous TCR co-signaling for 24–48 h. As a negative control of resting state, CD19 CAR T cells were cultured in the absence of stimulus. (E) Quantitative analysis of ZAP70, PLCγ1, and p38 protein phosphorylation in CAR T cells after 24 h of stimulation. (F) Quantitative analysis of RNA expression profiling of selected genes associated with the PI3K signaling pathway, apoptosis, metabolism, and cell proliferation in SB-CD19-28z.CAR/TCR KO and SB-CD19-28z.CAR/scrambled T cells at 48 h post-stimulation. Differentially expressed genes (DEGs) are defined as the genes showing p < 0.05 for differential expression. Data are shown as means ± SEM (n = 3 biologically independent donors); Student’s t test; n.s., not significant; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
First we assessed the phosphorylation states of essential protein kinases including tyrosine kinase ZAP70 (ZAP70),63 phospholipase C-γ1 (PLCγ1),63 and p38 mitogen-activated protein kinase (MAPK, p38),64 which are responsible for the initiation of T cell signaling (Table S8). We found that a single CAR signaling induced by NALM6 stimulation (Figure 8A) enhanced the similar levels of ZAP70, PLCγ1, and p38 protein phosphorylation in the SB-CD19-28z.CAR/TCR KO and SB-CD19-28z.CAR/scrambled control T cells (Figures 8B and S17A). The engagement of CAR and/or TCR to antigens is known to trigger several signaling transduction pathways that are essential for T cell function. In particular, the phosphoinositide 3-kinase (PI3K) signaling pathway plays a crucial role by promoting effector T cell differentiation and regulating cell proliferation, apoptosis, and metabolic activity.65,66 Therefore, the transcriptional profiles of genes associated with the PI3K pathway (total 29 genes) were compared in SB-CD19-28z.CAR/TCR KO and SB-CD19-28z.CAR/scrambled T cells in response to a signal CAR stimulation. As expected, the large majority of PI3K pathway genes (i.e., 24/29 = 82%) were not differentially expressed between SB-CD19-28z.CAR/TCR KO and SB-CD19-28z.CAR/scrambled control T cells, although some differences were apparent. In particular, only 5/29 differentially expressed genes (DEGs) belonging to the PI3K pathway were identified in the SB-CD19-28z.CAR/TCR KO compared with the SB-CD19-28z.CAR/scrambled control T cells following NALM6 stimulation (Figures 8C, S17B, and S17C). This suggests that, following single CAR signaling when exposed to CD19 antigen (on NALM6 cells), the SB-CD19-28z.CAR/TCR KO and the SB-CD19-28z.CAR/scrambled control T cells display comparable molecular signatures of T cell signaling (Figures 8A–8C), consistent with similar antitumor responses (Figure 5).
Nevertheless, dual CAR/endogenous TCR signaling (Figure 8D) led to a higher ZAP70 and PLCγ1 phosphorylation in SB-CD19-28z.CAR/scrambled T cells versus SB-CD19-28z.CAR/TCR KO T cells (Figures 8E and S17A), as opposed to a single CAR signaling. Consistently, comprehensive RNA analysis identified 14/29 DEGs (10 downregulated genes and 4 upregulated genes) belonging to the PI3K pathway in the SB-CD19-28z.CAR/TCR KO compared with the SB-CD19-28z.CAR/scrambled cells after simultaneous CAR/endogenous TCR signaling (Figures 8F, S17B, and S17D). This corresponded with the transcriptional alteration of the downstream processes that are associated with the PI3K pathway and influence apoptosis (10/19 DEGs), cell proliferation (12/19 DEGs), and metabolic activity (8/16 DEGs for glycolysis, 5/5 DEGs for fatty acid synthesis/oxidation, and 4/4 DEGs for amino acid metabolism) (Figures 8F, S17B, and S17D). Interestingly, the RNA expression fold changes of DEGs identified during single NALM6 stimulation were further augmented by additional anti-CD3 stimulation (Figures S17B and S17D). This suggests that TCR disruption in CAR T cells inhibits T cell signaling/protein phosphorylation and differential gene expression that is linked to the PI3K signaling pathway and some of its downstream processes compared with the control CAR T cells when endogenous TCR reactivation occurs (Figures 8D–8F and S18).
Discussion
In this study, we validated a non-viral T cell engineering platform based on SB transposons and CRISPR-Cas9 as a potential universal off-the-shelf allogeneic CAR T cell therapy. For this purpose, we stably expressed CD19-CAR genes in purified human T cells using hyperactive SB transposons. The use of mc-harboring SB transposons allowed for superior transfection and transposition rates and reduced DNA-mediated toxicity compared with the plasmid-based SB transposon.33,67 To minimize GvHD risk, endogenous TCR expression was inactivated using TCR gene-specific CRISPR-Cas9 RNPs. With optimized transfection and enrichment schemes, relatively robust CD19-CAR expression (90%) and TCR inactivation (99% TCRneg) was achieved. Compared with the sequential transfection approach used in this study, a simultaneous co-transfection of CRISPR-Cas9 and SB transposon components would be more convenient by reducing the steps of CAR T cell manufacturing compared with a sequential transfection method. Simultaneous co-transfection of CRISPR-Cas9 RNP and linear dsDNA template in human T cells was demonstrated in a previous study,68 suggesting the feasibility of this approach. Nevertheless, our study, which is based on a circular dsDNA (plasmid and mcDNA vector) transfection, clearly showed that simultaneous transfection of CRISPR-Cas9 RNP and transgene-encoding plasmid/mcDNA was relatively inefficient compared with a sequential transfection to produce the transgene-expressing TCR KO T cells (Figures S5B–S5D, left and S9A–S9C). In fact, co-electroporation of pmaxGFP plasmid and any CRISPR-Cas9 RNPs resulted in a significant reduction of the total GFP transfection efficiency compared with when the cells were transfected with only the pmaxGFP plasmid (Figure S5D, left). This suggests that the cellular uptake of the relatively large circular dsDNA during transfection could be hampered by the CRISPR-Cas9 RNP components, thereby compromising the overall efficiency of CAR/TCR KO T cell production. Most important, transplantation of CD19-specific CAR/TCRneg T cells into a xenograft tumor model resulted in efficient CD19-specific oncolysis in vitro and complete tumor remission in vivo, consistent with increased IFN-γ, GM-CSF, and TNF-α production. Most of the SB-CD19-28z.CAR/TCR KO T cells exhibited a predominant memory phenotype. Clinical trials have shown that T cell memory phenotypes correlated with antileukemic response and CAR-T cell persistence in patients with B cell acute lymphoblastic leukemia.69 Extended periods of CAR-T cell coculture with repeated antigen stimulation led to a relatively robust T cell expansion (100- to 1,000-fold expansion during a 6-week culture protocol and 10- to 30-fold expansion during a 2-week culture protocol with early CAR+ T cell enrichment. However, prolonged expansion may potentially contribute to T cell exhaustion.70 Nevertheless, it is encouraging that these in-vitro-expanded CAR/TCR KO T cells remained fully functional, consistent with the absence or low expression of exhaustion markers such as PD1 and CTLA4. Although relatively high TIM3 expression was apparent in the CAR T cells based on the 6-week expansion protocol, this likely resulted from the supplement of IL-2, IL-7, IL-15, and IL-21 cytokines in the culture medium.71 In accordance with our results, increased TIM3 expression without or only minimal PD1 expression in CAR T cells did not compromise CAR T cell functionality,60,72 supporting the notion that TIM3 is dispensable for T cell exhaustion.73 The prevailing memory phenotype and absence of a typical T cell exhaustion profile correlated with the antitumor effects and sustained and complete tumor remission in the xenograft model.60,74,75 Reduction of TCR alloreactivity in CAR/TCR KO T cells observed in our study exhibited its therapeutic potentials to minimize the risk of GvHD in mice, which was demonstrated in previous studies.22,60
To our knowledge, this study is the first to demonstrate that the combination of SB transposons and CRISPR-Cas9 enables efficient manufacturing of allogeneic TCR-disrupted CD19-CAR T cells, underscoring its novelty. Previous studies using transposon-based engineering of T cells relied on zinc finger nucleases (ZFNs) to disrupt the TCR genes.35 Here, we obtained significantly higher TCR targeting efficiencies (99%) with our optimized platform compared with the use of ZFNs, which usually yields ∼30% CD3/TCRneg CAR T cells, requiring an additional CD3+ T cell depletion step.35 The superior efficiency with the current approach may be due to more efficient delivery, expression, and/or stability of CRISPR-Cas9 RNPs compared with ZFN mRNA. However, we cannot exclude a contribution of differences in T cell activation status to these different targeting efficiencies. The relatively efficient nonviral platform described in this study supports its potential broad implications, not only for single TRAC gene disruption but also to achieve (simultaneous) multiple genetic ablations associated with HLA I/II expression including β-2 microglobulin (B2M) and class II major histocompatibility complex transactivator (CIITA) genes to generate universal off-the-shelf CAR T cells with minimal risks of GvHD and T cell graft rejection.19,22 This could be achieved by a co-transfection of TRAC-, B2M-, and CIITA-targeting CRISPR-Cas9 RNPs into T cells followed by subsequent transfection of SB transposon vector for stable CAR expression (Figure S5A, middle and lower). Additional depletion of HLA I, HLA II, and TCR/CD3+ T cells could be carried out before SB transposon transfection to augment the overall efficiency of universal CAR T cell production (Figure S5A, lower).19,22 The high and sustained CAR expression levels achieved with the SB transposon-CRISPR-Cas9 platform also compares favorably with previous findings typically attained with use of γ-retroviral or lentiviral vectors for CAR-T cell generation.5,26,35, 36, 37, 38, 39, 40 Moreover, the use of the 2-week expansion protocol with early CAR+ enrichment still yielded up to 70-fold expansion of the transposon-engineered CAR/TCR KO T cells with predominant memory phenotypes, consistent with the CAR/TCR KO T cell profiles generated from viral vector-based systems.60 This improved protocol is also highly relevant for current clinical-grade CAR T cell manufacturing methods, which typically require 2 weeks of cell expansion before harvest.34,76 However, it cannot be excluded that this platform allows for further shortening of CAR T cell production time by increasing higher numbers of input cells. For allogeneic treatment, ∼90 × 106 CAR T cells/patient (i.e., the dose ranges from 106/m2 to 108/m2 of recipient’s calculated body surface area) is typically required.34 By assuming (1) ∼40% of cell viability (Figure S12A) and (2) ∼50% of transfection efficiency (Figure S12B) upon gene transfer, we would need 56.3 × 106 allogenic CD3 T cells as a starting material to primarily obtain 11.25 × 106 CAR T cells, which would undergo ∼8-fold CAR expansion after 7 days of culture (Figure S12F) and result in 90 × 106 CAR T cells. Given a high abundance of T cells (∼60%) in PBMCs,77 this cell number is achievable from a single donor using lymphocyte apheresis. Interestingly, a recent study demonstrated that the SB transposon-mediated CAR T cells can be generated as early as 8 days and display cytotoxicity against CD19+ tumors,72 thus supporting the feasibility of our approach. Although non-viral DNA transfection can trigger some acute cytotoxicity, using mcDNA vector rather than conventional plasmids to deliver transposons reduced this effect.60
Interaction of the CAR-CD3 zeta domain with the endogenous TCR/CD3 complex may be required to optimize CAR T cell reactivity toward target antigens.78 Consequently, endogenous TCR disruption may perturb CAR signaling upon antigen engagement and compromise the oncolytic potential of CAR T cells. This scenario is supported by recent data showing that TCR ablation impairs CAR T cell function, accounting for incomplete tumor suppression in vivo.60 Nevertheless, it is particularly encouraging that we observed no significant impact of TCR disruption on the antileukemic function of CD19 CAR T cells in response to CD19+ tumor cells and in NALM6-bearing mice, consistent with similar protein kinase phosphorylation and transcriptional profiles in both T cells following CD19-mediated single CAR signaling. The comparative RNA expression analysis revealed only few DEGs involved with the PI3K signaling pathway. To our knowledge, studies reporting the roles of these DEGs (i.e., RXRA, IL3, COL6A5, PPP2R2B, and EFNA4) in CAR T cell antitumor functions are currently limited, warranting further investigation. Among these DEGs, retinoid X receptor alpha (RXRA) gene disruption limits T cell proliferation and promotes apoptosis following antigen stimulation. Moreover, deficiency of RXRA activity drives T cell differentiation to secrete more Th1 cytokines, including IFN-γ and TNF-α.79,80 Interestingly, we found that RXRA downregulation in the SB-CD19-28z.CAR/TCR KO cells compared with the SB-CD19-28z.CAR/scrambled control T cells does not significantly enhance IFN-γ and TNFα secretion upon CAR signaling in response to CD19+ tumor cells (Figure 5D).
It has been shown that dual CAR and TCR signaling contributes to differential expression of genes responsible for apoptosis, effector functions, and cellular metabolism compared with CAR activation in the mouse system.81 Our current findings complement these murine studies and provide further insights into the molecular interplay between CAR and endogenous TCR/CD3 signaling in human T cells, which alters the protein phosphorylation and RNA expression of genes associated with the PI3K pathway and its downstream processes (Figures S17 and S18). ZAP70 and PLC1γ proteins play key roles in signal transduction in T cells by inducing protein phosphorylation cascades and stabilizing the linker for activation of T cell protein complex during T cell signaling.82 In addition, a loss of ZAP70 activity impairs T cell activation, proliferation, and effector and memory T cell responses to target antigens.83 Suboptimal ZAP70 phosphorylation triggered by CAR-low-density antigen engagement leads to poor proximal receptor signaling and attenuation of T cell functions.84 Hence, an increase of ZAP70 and/or PLC1γ phosphorylation mediated by reactive endogenous TCR during CAR signaling may promote the overall sensitivity of CAR T cells to antigens and enhance antitumor response. Activation of PI3K pathway signaling is crucial to activate T cells and enhances CAR T cell function in vivo.85 However, the constitutive signaling of the PI3K pathway drives the terminal differentiation of T cells into short-lived effector T cells, which is less favorable toward achieving CAR T cell persistence.65 Furthermore, inhibition of the PI3K pathway reduces GvHD development by promoting tolerance of alloreactive T cells.86 Hence, the temporal regulation of the PI3K signaling pathway in response to CD19-specific T cell activation may not only improve CAR T cell persistence but also minimize GvHD risk. Our study is the first to confirm that TCR-disrupted CD19 CAR T cells are relatively resistant to endogenous TCR/CD3 signaling. Moreover, they maintain a stable molecular signaling profile. This may potentially minimize undesirable T cell signaling and/or adverse events caused by TCR reactivation.81,87
Based on the previous studies, it is possible that most DEGs identified in T cells following dual CAR/endogenous TCR signaling may enhance CAR T cell cytolytic functions. For example, upregulation of integrin subunit alpha 2 in activated endogenous TCR/CAR T cells could improve T cell growth and increase cytokine production.88 Cyclin-dependent kinase inhibitor 1A serves as an essential mediator to regulate an optimal proliferation and a homeostasis of activated/memory T cells.89,90 Death-associated protein kinase 1 (DAPK1) plays a crucial role in CD8+ T cell activation, trafficking, and potent antitumor effects.91 In addition, DAPK1-mediated mTORC1 pathway signaling enhances CD8+ T cell response and memory differentiation in a PI3K/AKT-independent manner.92 An increased fibroblast growth factor receptor 1 expression induced by CAR/endogenous TCR activation in T cells could augment IL-2 production and cell proliferation through NFAT and AP-1 activity.93 Upregulation of carnitine palmitoyltransferase 1A may facilitate mitochondrial fatty acid oxidation to support CD8+ memory T cell phenotype.94 Cystathionine gamma-lyase overexpression in the dual CAR/endogenous TCR-activated T cells may improve antitumor effects by controlling amino acid abundance in tumor microenvironment.95 Protein tyrosine phosphatase receptor type K regulates the ERK phosphorylation pathway, which is required for CD4+ T cell development.96 Nevertheless, for some DEGs observed upon CAR/endogenous TCR activation, it remains unclear whether their expression alteration provides biological advantages to CAR T cells. Hexokinase 2 (HK2) expression is essential for glycolysis and oxidative phosphorylation in activated T cells and supports antitumor effects through IFN-γ secretion,97 consistent with HK2 upregulation in CAR/TCR-activated T cells observed in our study. However, inhibition of HK2-directed glycolysis may enhance memory T cell formation and long-term persistence.98 Acyl-CoA synthetase long-chain family member 4 expression in T cell is indispensable for effective antitumor response in mice but may compromise T cell survival through iron-dependent programmed cell death pathway activation.99
This study takes advantage of the versatility of the SB and CRISPR-Cas9 technology and further expands their potential in an allogeneic TCR KO setting to minimize the risk of GvHD and open the way to a universal, off-the-shelf immunotherapy for lymphoid cancers. Autologous transposon-modified CAR T cells have been explored in clinical trials (NCT03389035, NCT04289220, and NCT04284254), demonstrating therapeutic efficacy toward leukemia and solid tumors.34,69,100,101 SB transposon-based vectors typically integrate close to random into the target cell genome, which sets them apart from γ-retroviral and lentiviral vectors that exhibit a biased integration into (active) genes, as also demonstrated in our current result (Figures 4I and S13A). We also found that CRISPR-Cas9 activity does not alter the SB transposon insertion patterns and does not skew the transposon insertions toward on-target DSBs in T cells (Figure S14), consistent with the previous study showing the absence of transposon insertion into CRISPR-Cas9-mediated DSBs.102 The current SB transposon-CRISPR-Cas9 RNP technology enables generation of universal allogenic CAR T cells with a relatively safe integration profile consistent with (1) lower integration bias toward transcriptionally active regions of the genome compared with integrating viral vectors,45, 46, 47 (2) polyclonal integration pattern and no dominant insertion preference, and (3) extremely low occurrence of insertion sites in the vicinity of proto-oncogenes. Although insertional oncogenesis cannot be excluded, the current available data support the safety advantages and feasibility of this approach for clinical translation. Nevertheless, caution is warranted in the face of recent reports that showed two out of ten patients developed T cell lymphomas derived from CD19-CAR T cells that were generated with the piggyBac transposon system.103, 104, 105, 106 Although no transposon insertions into typical oncogenes were apparent in both patients, lymphoma cells harbored structural variants (SVs) and copy number variations (CNVs), including gains and losses. In contrast to SB, the piggyBac transposase is known to interact with the bromodomain-containing protein 4, a keeper of genome stability. Whether this could have contributed to the observed high incidence of SVs and CNVs warrants further investigation. Of interest, both lymphomas shared ISs in the BACH2 locus, which encodes a transcription factor that regulates T cell plasticity, but it is unclear whether this is a contributing factor in lymphomagenesis. Although long-term follow-up of SB-based CAR T cell therapy revealed no evidence of malignant transformation,107 future studies are aimed at further reducing the risk of insertional oncogenesis either by reducing the number of integrations per cell by SB transposase mRNA delivery33,108 or ultimately by achieving targeted transposition.
Materials and methods
The design of CAR-expressing vector and guided RNA, detection of CRISPR-Cas9 on-target effects, primary human T cell isolation and culture, cell lines, flow cytometry, cytotoxicity assays, selective CAR T cell expansion, and other relevant information are described in the supplemental information.
Delivery of CRISPR-Cas9 RNP into human CD8+ T cells by nucleofection
The P3 Primary Cell 4D-Nucleofector X Kit was used in all nucleofection experiments. 5 × 105 cells were washed with 1× Dulbecco’s PBS and resuspended in 20 μL of kit P3 Primary Cell 4D-Nucleofector X (Lonza). The prepared 70 pmol of CRISPR-Cas9 RNP particles was added to the cell-buffer mixture and mixed well. Nucleofection was carried out in a 16-well strip using the EH-115 program. Immediately after nucleofection, 80 μL of pre-warmed complete T cell medium containing 50 U/mL of human rIL-2 was immediately added to cells, and the cells were rested for 30 min in 5% CO2 at 37°C. The transfected cells were transferred to 1 mL of complete T cell medium and culture for 48 h at 5% CO2 and 37°C.
Generation of CD19-28z.CAR/TCR KO T cells and SB-CD19-28z.CAR/TCR KO T cells
Sequential nucleofection of TRAC-targeting CRISPR-Cas9 RNP and mcDNA vector encoding CD19-28z.CAR was used to engineer CAR/TCRneg T cells for transient and sustained CAR expression. To generate CD19-28z.CAR/TCR KO T cells for transient CAR expression, 700 pmol of TCR-1 RNP particles was transfected into 5 × 106 CD8+ T cells using the EH-115 program to obtain TCRneg T cells. TCR-1 RNP was substituted with scrambled gRNA RNP to generate the scrambled control T cells. Twenty-four hours later, 10 μg of mcCD19-28z.CAR was transfected into 5 × 106 TCRneg or scrambled control T cells using the EO-115 program. Alternatively, 10 μg of mcCD19-28z.CAR and 5 μg of mcSB100x were transfected into 5 × 106 TCRneg or scrambled control T cells using the EO-115 program to produce SB-CD19-28z.CAR/TCR KO and SB-CD19-28z.CAR/scrambled T cells for stable CAR expression, respectively. As a control devoid of CAR expression, mock transfection of TCRneg and scrambled control T cells was carried out to generate mock/TCR KO and mock/scrambled control T cells, respectively. After 24 h of transfection, the surface CAR and TCR expression was examined by flow cytometry analysis, and the cells were harvested for functionality assays or CAR T cell culture.
Animal experiments
All animal experiments were carried out under the procedures approved by the Institutional Animal Ethics Committee of the Vrije Universiteit Brussel (Brussels, Belgium). The experimental details are described in the corresponding figure legends and supplemental information. The development of tumor burden was assessed weekly using non-invasive in vivo bioluminescence imaging. At several weeks post-NALM6 tumor engraftment, some mice developed the clinical signs of significant weight loss, severely hunched back, poor grooming, hindlimb paralysis, and/or limited mobility. These mice were euthanized as an endpoint by CO2 inhalation or cervical dislocation.
Statistical analysis and software
Data analysis was performed using GraphPad Prism 8 software. Schematic representatives were generated using BioRender software. Comparison of two mean values from two experimental groups was performed using two-tailed independent Student’s t tests. Alternatively, a one-way analysis of variance (ANOVA) test was used, with Dunnett’s post-hoc applied for multiple comparison of mean values from more than two experimental cohorts. Results are reported as the mean ± standard error of mean (SEM), and p < 0.05 was taken to indicate significance.
Data availability statement
The data that support the findings of this study are available from the corresponding author on reasonable request.
Acknowledgments
We thank Red Cross Flanders (Rode Kruis-Vlaanderen, Belgium) for supplying buffy coat samples for the study. This work was supported by the Fonds Wetenschappelijk Onderzoek (FWO, project nos. G041616N, G019114N), Stichting Tegen Kanker (STK, project no. C/2014/239), and the UPGRADE (Unlocking Precision Gene Therapy) project, that has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement no. 825825, to M.K.C. and T.V., and J.T. was supported by an FWO Aspirant and Research Grant (project no. 1531116N), the Award Cancer Research – Oncology Center Vrije Universiteit Brussel, funded by the bequests of the late Ms. Esther Desmedt and the late Ms. Irma Noë, and Wetenschappelijk Fonds Willy Gepts (WFWG) foundation – VUB.
Author contributions
J.T. designed and performed all experiments, analyzed data, and wrote the paper. E.S.-K. provided essential assistance in animal experiments. T.G. provided essential assistance in irradiation experiments. T.V. and M.K.C. designed the experiments, analyzed data, wrote and approved the paper, and led the study. T.V. and M.K.C. applied for and obtained financial support for this study. All authors approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2022.06.006.
Contributor Information
Marinee K. Chuah, Email: thierry.vandendriessche@vub.be.
Thierry VandenDriessche, Email: marinee.chuah@vub.be.
Supplemental information
References
- 1.Brown C.E., Mackall C.L. CAR T cell therapy: inroads to response and resistance. Nat. Rev. Immunol. 2019;19:73–74. doi: 10.1038/s41577-018-0119-y. [DOI] [PubMed] [Google Scholar]
- 2.Majzner R.G., Mackall C.L. Clinical lessons learned from the first leg of the CAR T cell journey. Nat. Med. 2019;25:1341–1355. doi: 10.1038/s41591-019-0564-6. [DOI] [PubMed] [Google Scholar]
- 3.Zheng P.P., Kros J.M., Li J. Approved CAR T cell therapies: ice bucket challenges on glaring safety risks and long-term impacts. Drug Discov. Today. 2018;23:1175–1182. doi: 10.1016/j.drudis.2018.02.012. [DOI] [PubMed] [Google Scholar]
- 4.Townsend M.H., Bennion K., Robison R.A., O'Neill K.L. Paving the way towards universal treatment with allogenic T cells. Immunol. Res. 2020;68:63–70. doi: 10.1007/s12026-020-09119-7. [DOI] [PubMed] [Google Scholar]
- 5.Graham C.E., Jozwik A., Quartey-Papafio R., Ioannou N., Metelo A.M., Scala C., Dickson G., Stewart O., Almena-Carrasco M., Peranzoni E., et al. Gene-edited healthy donor CAR T cells show superior anti-tumour activity compared to CAR T cells derived from patients with lymphoma in an in vivo model of high-grade lymphoma. Leukemia. 2021;35:3581–3584. doi: 10.1038/s41375-021-01324-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maude S.L., Laetsch T.W., Buechner J., Rives S., Boyer M., Bittencourt H., Bader P., Verneris M.R., Stefanski H.E., Myers G.D., et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 2018;378:439–448. doi: 10.1056/nejmoa1709866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuster S.J., Svoboda J., Chong E.A., Nasta S.D., Mato A.R., Anak Ö., Brogdon J.L., Pruteanu-Malinici I., Bhoj V., Landsburg D., et al. Chimeric antigen receptor T cells in refractory B-cell lymphomas. N. Engl. J. Med. 2017;377:2545–2554. doi: 10.1056/nejmoa1708566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neelapu S.S., Locke F.L., Bartlett N.L., Lekakis L.J., Miklos D.B., Jacobson C.A., Braunschweig I., Oluwole O.O., Siddiqi T., Lin Y., et al. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N. Engl. J. Med. 2017;377:2531–2544. doi: 10.1056/NEJMoa1707447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benjamin R., Graham C., Yallop D., Jozwik A., Mirci-Danicar O.C., Lucchini G., Pinner D., Jain N., Kantarjian H., Boissel N., et al. UCART19 Group Genome-edited, donor-derived allogeneic anti-CD19 chimeric antigen receptor T cells in paediatric and adult B-cell acute lymphoblastic leukaemia: results of two phase 1 studies. Lancet. 2020;396:1885–1894. doi: 10.1016/S0140-6736(20)32334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Depil S., Duchateau P., Grupp S.A., Mufti G., Poirot L. 'Off-the-shelf' allogeneic CAR T cells: development and challenges. Nat. Rev. Drug Discov. 2020;19:185–199. doi: 10.1038/s41573-019-0051-2. [DOI] [PubMed] [Google Scholar]
- 11.Anwer F., Shaukat A.A., Zahid U., Husnain M., McBride A., Persky D., Lim M., Hasan N., Riaz I.B. Donor origin CAR T cells: graft versus malignancy effect without GVHD, a systematic review. Immunotherapy. 2017;9:123–130. doi: 10.2217/imt-2016-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai B., Guo M., Wang Y., Zhang Y., Yang J., Guo Y., Dai H., Yu C., Sun Q., Qiao J., et al. Co-infusion of haplo-identical CD19-chimeric antigen receptor T cells and stem cells achieved full donor engraftment in refractory acute lymphoblastic leukemia. J. Hematol. Oncol. 2016;9:131. doi: 10.1186/s13045-016-0357-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallardo D., García-López J., SuredA A., SuredA A., Ferra C., Cancelas J., Berlanga J., Brunet S., Boqué C., Picón M., et al. Low-dose donor CD8+ cells in the CD4-depleted graft prevent allogeneic marrow graft rejection and severe graft-versus-host disease for chronic myeloid leukemia patients in first chronic phase. Bone Marrow Transplant. 1997;20:945–952. doi: 10.1038/sj.bmt.1701008. [DOI] [PubMed] [Google Scholar]
- 14.Champlin R., Ho W., Gajewski J., Feig S., Burnison M., Holley G., Greenberg P., Lee K., SchmId I., Giorgi J. Selective depletion of CD8+ T lymphocytes for prevention of graft-versus-host disease after allogeneic bone marrow transplantation. Blood. 1990;76:418–423. doi: 10.1182/blood.v76.2.418.bloodjournal762418. [DOI] [PubMed] [Google Scholar]
- 15.Shlomchik W.D. Graft-versus-host disease. Nat. Rev. Immunol. 2007;7:340–352. doi: 10.1038/nri2000. [DOI] [PubMed] [Google Scholar]
- 16.Bertaina A., Merli P., Rutella S., Pagliara D., Bernardo M.E., Masetti R., Pende D., Falco M., Handgretinger R., Moretta F., et al. HLA-haploidentical stem cell transplantation after removal of αβ+ T and B cells in children with nonmalignant disorders. Blood. 2014;124:822–826. doi: 10.1182/blood-2014-03-563817. [DOI] [PubMed] [Google Scholar]
- 17.Balashov D., Shcherbina A., Maschan M., Trakhtman P., Skvortsova Y., Shelikhova L., Laberko A., Livshits A., Novichkova G., Maschan A. Single-center experience of unrelated and haploidentical stem cell transplantation with TCRαβ and CD19 depletion in children with primary immunodeficiency syndromes. Biol. Blood Marrow Transpl. 2015;21:1955–1962. doi: 10.1016/j.bbmt.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 18.Morris G.P., Allen P.M. Cutting edge: highly alloreactive dual TCR T cells play a dominant role in graft-versus-host disease. J. Immunol. 2009;182:6639–6643. doi: 10.4049/jimmunol.0900638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren J., Liu X., Fang C., Jiang S., June C.H., Zhao Y. Multiplex genome editing to generate universal CAR T cells resistant to PD1 inhibition. Clin. Cancer Res. 2017;23:2255–2266. doi: 10.1158/1078-0432.ccr-16-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poirot L., Philip B., Schiffer-Mannioui C., Le Clerre D., Chion-Sotinel I., Derniame S., Potrel P., Bas C., Lemaire L., Galetto R., et al. Multiplex genome-edited T-cell manufacturing platform for "Off-the-Shelf" adoptive T-cell immunotherapies. Cancer Res. 2015;75:3853–3864. doi: 10.1158/0008-5472.can-14-3321. [DOI] [PubMed] [Google Scholar]
- 21.Eyquem J., Mansilla-Soto J., Giavridis T., van der Stegen S.J.C., Hamieh M., Cunanan K.M., Odak A., Gönen M., Sadelain M. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543:113–117. doi: 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kagoya Y., Guo T., Yeung B., Saso K., Anczurowski M., Wang C.H., Murata K., Sugata K., Saijo H., Matsunaga Y., et al. Genetic ablation of HLA class I, class II, and the T-cell receptor enables allogeneic T cells to be used for adoptive T-cell therapy. Cancer Immunol. Res. 2020;8:926–936. doi: 10.1158/2326-6066.cir-18-0508. [DOI] [PubMed] [Google Scholar]
- 23.Levine B.L., Miskin J., Wonnacott K., Keir C. Global manufacturing of CAR T cell therapy. Mol. Ther. Methods Clin. Dev. 2017;4:92–101. doi: 10.1016/j.omtm.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang J., Hubbard-Lucey V.M., Pearce L., O'Donnell-Tormey J., Shalabi A. The global landscape of cancer cell therapy. Nat. Rev. Drug Discov. 2018;17:465–466. doi: 10.1038/nrd.2018.74. [DOI] [PubMed] [Google Scholar]
- 25.Gu Q., Cynader M.S. Immunocytochemical localization of enkephalin in the cat visual cortex. Brain Res. 1993;620:155–158. doi: 10.1016/0006-8993(93)90284-t. [DOI] [PubMed] [Google Scholar]
- 26.Wang X., Rivière I. Clinical manufacturing of CAR T cells: foundation of a promising therapy. Mol. Ther. Oncolytics. 2016;3:16015. doi: 10.1038/mto.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tipanee J., Chai Y.C., VandenDriessche T., Chuah M.K. Preclinical and clinical advances in transposon-based gene therapy. Biosci. Rep. 2017;37 doi: 10.1042/bsr20160614. BSR20160614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tipanee J., VandenDriessche T., Chuah M.K. Transposons: moving forward from preclinical studies to clinical trials. Hum. Gene Ther. 2017;28:1087–1104. doi: 10.1089/hum.2017.128. [DOI] [PubMed] [Google Scholar]
- 29.Deichmann A., Hacein-Bey-Abina S., Schmidt M., Garrigue A., Brugman M.H., Hu J., Glimm H., Gyapay G., Prum B., Fraser C.C., et al. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J. Clin. Invest. 2007;117:2225–2232. doi: 10.1172/jci31659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hacein-Bey-Abina S., Garrigue A., Wang G.P., Soulier J., Lim A., Morillon E., Clappier E., Caccavelli L., Delabesse E., Beldjord K., et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 2008;118:3132–3142. doi: 10.1172/jci35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walisko O., Schorn A., Rolfs F., Devaraj A., Miskey C., Izsvák Z., Ivics Z. Transcriptional activities of the Sleeping Beauty transposon and shielding its genetic cargo with insulators. Mol. Ther. 2008;16:359–369. doi: 10.1038/sj.mt.6300366. [DOI] [PubMed] [Google Scholar]
- 32.Mátés L., Chuah M.K.L., Belay E., Jerchow B., Manoj N., Acosta-Sanchez A., Grzela D.P., Schmitt A., Becker K., Matrai J., et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 33.Monjezi R., Miskey C., Gogishvili T., Schleef M., SchMeer M., Einsele H., Ivics Z., Hudecek M. Enhanced CAR T-cell engineering using non-viral Sleeping Beauty transposition from minicircle vectors. Leukemia. 2017;31:186–194. doi: 10.1038/leu.2016.180. [DOI] [PubMed] [Google Scholar]
- 34.Kebriaei P., Singh H., Huls M.H., Figliola M.J., Bassett R., Olivares S., Jena B., Dawson M.J., Kumaresan P.R., Su S., et al. Phase I trials using Sleeping Beauty to generate CD19-specific CAR T cells. J. Clin. Invest. 2016;126:3363–3376. doi: 10.1172/jci86721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torikai H., Reik A., Liu P.Q., Zhou Y., Zhang L., Maiti S., Huls H., Miller J.C., Kebriaei P., Rabinovitch B., et al. A foundation for universal T-cell based immunotherapy: T cells engineered to express a CD19-specific chimeric-antigen-receptor and eliminate expression of endogenous TCR. Blood. 2012;119:5697–5705. doi: 10.1182/blood-2012-01-405365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivics Z., Hackett P.B., Plasterk R.H., Izsvák Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 37.Berdien B., Mock U., Atanackovic D., Fehse B., Mock U. TALEN-mediated editing of endogenous T-cell receptors facilitates efficient reprogramming of T lymphocytes by lentiviral gene transfer. Gene Ther. 2014;21:539–548. doi: 10.1038/gt.2014.26. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Z., Qiu S., Zhang X., Chen W. Optimized DNA electroporation for primary human T cell engineering. BMC Biotechnol. 2018;18:4. doi: 10.1186/s12896-018-0419-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ingegnere T., Mariotti F.R., Pelosi A., Quintarelli C., De Angelis B., Tumino N., Besi F., Cantoni C., Locatelli F., Vacca P., Moretta L. Human CAR NK cells: a new non-viral method allowing high efficient transfection and strong tumor cell killing. Front. Immunol. 2019;10:957. doi: 10.3389/fimmu.2019.00957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang R.S., Lai M.C., Shih H.A., Lin S. A robust platform for expansion and genome editing of primary human natural killer cells. J. Exp. Med. 2021;218 doi: 10.1084/jem.20201529. e20201529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hutson T.H., Buchser W.J., Bixby J.L., Lemmon V.P., Moon L.D.F. Optimization of a 96-well electroporation assay for postnatal rat CNS neurons suitable for cost-effective medium-throughput screening of genes that promote neurite outgrowth. Front. Mol. Neurosci. 2011;4:55. doi: 10.3389/fnmol.2011.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Riepe C., Zelin E., Frankino P.A., Meacham Z.A., Fernandez S.G., Ingolia N.T., Corn J.E. Double stranded DNA breaks and genome editing trigger loss of ribosomal protein RPS27A. FEBS J. 2022;289:3101–3114. doi: 10.1111/febs.16321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kay M.A., He C.Y., Chen Z.Y. A robust system for production of minicircle DNA vectors. Nat. Biotechnol. 2010;28:1287–1289. doi: 10.1038/nbt.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mátrai J., Cantore A., Bartholomae C.C., Annoni A., Wang W., Acosta-Sanchez A., Samara-Kuko E., De Waele L., Ma L., Genovese P., et al. Hepatocyte-targeted expression by integrase-defective lentiviral vectors induces antigen-specific tolerance in mice with low genotoxic risk. Hepatology. 2011;53:1696–1707. doi: 10.1002/hep.24230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu X., Burgess S.M. Integration target site selection for retroviruses and transposable elements. Cell Mol. Life Sci. 2004;61:2588–2596. doi: 10.1007/s00018-004-4206-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Demeulemeester J., De Rijck J., Gijsbers R., Debyser Z. Retroviral integration: site matters: mechanisms and consequences of retroviral integration site selection. Bioessays. 2015;37:1202–1214. doi: 10.1002/bies.201500051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bougnères P., Hacein-Bey-Abina S., Labik I., Adamsbaum C., Castaignède C., Bellesme C., Schmidt M. Long-term follow-up of hematopoietic stem-cell gene therapy for cerebral adrenoleukodystrophy. Hum. Gene Ther. 2021;32:1260–1269. doi: 10.1089/hum.2021.053. [DOI] [PubMed] [Google Scholar]
- 48.Hacein-Bey-Abina S., Von Kalle C., Schmidt M., McCormack M.P., Wulffraat N., Leboulch P., Lim A., Osborne C.S., Pawliuk R., Morillon E., et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 49.Ott M.G., Schmidt M., Schwarzwaelder K., Stein S., Siler U., Koehl U., Glimm H., Kühlcke K., Schilz A., Kunkel H., et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat. Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 50.Howe S.J., Mansour M.R., Schwarzwaelder K., Bartholomae C., Hubank M., Kempski H., Brugman M.H., Pike-Overzet K., Chatters S.J., de Ridder D., et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Invest. 2008;118:3143–3150. doi: 10.1172/jci35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Braun C.J., Boztug K., Paruzynski A., Witzel M., Schwarzer A., Rothe M., Modlich U., Beier R., Göhring G., Steinemann D., et al. Gene therapy for Wiskott-Aldrich syndrome--long-term efficacy and genotoxicity. Sci. Transl. Med. 2014;6:227ra33. doi: 10.1126/scitranslmed.3007280. [DOI] [PubMed] [Google Scholar]
- 52.Naserian S., Leclerc M., Thiolat A., Pilon C., Le Bret C., Belkacemi Y., Maury S., Charlotte F., Cohen J.L. Simple, reproducible, and efficient clinical grading system for murine models of acute graft-versus-host disease. Front. Immunol. 2018;9:10. doi: 10.3389/fimmu.2018.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lai H.Y., Chou T.Y., Tzeng C.H., Lee O.K.S. Cytokine profiles in various graft-versus-host disease target organs following hematopoietic stem cell transplantation. Cell Transplant. 2012;21:2033–2045. doi: 10.3727/096368912x653110. [DOI] [PubMed] [Google Scholar]
- 54.Westphal S., McGeary A., Rudloff S., Wilke A., Penack O. The green tea catechin epigallocatechin gallate ameliorates graft-versus-host disease. PLoS One. 2017;12:e0169630. doi: 10.1371/journal.pone.0169630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morin F., Kavian N., Nicco C., Cerles O., Chéreau C., Batteux F. Improvement of sclerodermatous graft-versus-host disease in mice by niclosamide. J. Invest. Dermatol. 2016;136:2158–2167. doi: 10.1016/j.jid.2016.06.624. [DOI] [PubMed] [Google Scholar]
- 56.Pai C.C.S., Hsiao H.H., Sun K., Chen M., Hagino T., Tellez J., Mall C., Blazar B.R., Monjazeb A., Abedi M., Murphy W.J. Therapeutic benefit of bortezomib on acute graft-versus-host disease is tissue specific and is associated with interleukin-6 levels. Biol. Blood Marrow Transplant. 2014;20:1899–1904. doi: 10.1016/j.bbmt.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsukuma K.E., Wei D., Sun K., Ramsamooj R., Chen M. Diagnosis and differential diagnosis of hepatic graft versus host disease (GVHD) J. Gastrointest. Oncol. 2016;7:S21–S31. doi: 10.3978/j.issn.2078-6891.2015.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu Y., Sakamaki S., Kuroda H., Kusakabe T., Konuma Y., Akiyama T., Fujimi A., Takemoto N., Nishiie K., Matsunaga T., et al. Prevention of lethal acute graft-versus-host disease in mice by oral administration of T helper 1 inhibitor, TAK-603. Blood. 2001;97:1123–1130. doi: 10.1182/blood.v97.4.1123. [DOI] [PubMed] [Google Scholar]
- 59.Tago Y., Kobayashi C., Ogura M., Wada J., Yamaguchi S., Yamaguchi T., Hayashi M., Nakaishi T., Kubo H., Ueda Y. Human amnion-derived mesenchymal stem cells attenuate xenogeneic graft-versus-host disease by preventing T cell activation and proliferation. Sci. Rep. 2021;11:2406. doi: 10.1038/s41598-021-81916-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stenger D., Stief T.A., Kaeuferle T., Willier S., Rataj F., Schober K., Vick B., Lotfi R., Wagner B., Grünewald T.G.P., et al. Endogenous TCR promotes in vivo persistence of CD19-CAR-T cells compared to a CRISPR/Cas9-mediated TCR knockout CAR. Blood. 2020;136:1407–1418. doi: 10.1182/blood.2020005185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cruz C.R.Y., Micklethwaite K.P., Savoldo B., Ramos C.A., Lam S., Ku S., Diouf O., Liu E., Barrett A.J., Ito S., et al. Infusion of donor-derived CD19-redirected virus-specific T cells for B-cell malignancies relapsed after allogeneic stem cell transplant: a phase 1 study. Blood. 2013;122:2965–2973. doi: 10.1182/blood-2013-06-506741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang K.S., Im H., Hong S., Pergolini I., del Castillo A.F., Wang R., Clardy S., Huang C.H., Pille C., Ferrone S., et al. Multiparametric plasma EV profiling facilitates diagnosis of pancreatic malignancy. Sci. Transl. Med. 2017;9:eaal3226. doi: 10.1126/scitranslmed.aal3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Malissen B., Grégoire C., Malissen M., Roncagalli R. Integrative biology of T cell activation. Nat. Immunol. 2014;15:790–797. doi: 10.1038/ni.2959. [DOI] [PubMed] [Google Scholar]
- 64.Ashwell J.D. The many paths to p38 mitogen-activated protein kinase activation in the immune system. Nat. Rev. Immunol. 2006;6:532–540. doi: 10.1038/nri1865. [DOI] [PubMed] [Google Scholar]
- 65.Zheng W., O'Hear C.E., Alli R., Basham J.H., Abdelsamed H.A., Palmer L.E., Jones L.L., Youngblood B., Geiger T.L. PI3K orchestration of the in vivo persistence of chimeric antigen receptor-modified T cells. Leukemia. 2018;32:1157–1167. doi: 10.1038/s41375-017-0008-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lucas C.L., Chandra A., Nejentsev S., Condliffe A.M., Okkenhaug K. PI3Kδ and primary immunodeficiencies. Nat. Rev. Immunol. 2016;16:702–714. doi: 10.1038/nri.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cheng C., Tang N., Li J., Cao S., Zhang T., Wei X., Wang H. Bacteria-free minicircle DNA system to generate integration-free CAR-T cells. J. Med. Genet. 2019;56:10–17. doi: 10.1136/jmedgenet-2018-105405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roth T.L., Puig-Saus C., Yu R., Shifrut E., Carnevale J., Li P.J., Hiatt J., Saco J., Krystofinski P., Li H., et al. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature. 2018;559:405–409. doi: 10.1038/s41586-018-0326-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Magnani C.F., Gaipa G., Lussana F., Belotti D., Gritti G., Napolitano S., Matera G., Cabiati B., Buracchi C., Borleri G., et al. Sleeping Beauty-engineered CAR T cells achieve antileukemic activity without severe toxicities. J. Clin. Invest. 2020;130:6021–6033. doi: 10.1172/jci138473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Neal L.R., Bailey S.R., Wyatt M.M., Bowers J.S., Majchrzak K., Nelson M.H., Haupt C., Paulos C.M., Varela J.C. The basics of artificial antigen presenting cells in T cell-based cancer immunotherapies. J. Immunol. Res. Ther. 2017;2:68–79. [PMC free article] [PubMed] [Google Scholar]
- 71.Mujib S., Jones R.B., Lo C., Aidarus N., Clayton K., Sakhdari A., Benko E., Kovacs C., Ostrowski M.A. Antigen-independent induction of Tim-3 expression on human T cells by the common gamma-chain cytokines IL-2, IL-7, IL-15, and IL-21 is associated with proliferation and is dependent on the phosphoinositide 3-kinase pathway. J. Immunol. 2012;188:3745–3756. doi: 10.4049/jimmunol.1102609. [DOI] [PubMed] [Google Scholar]
- 72.Chicaybam L., Abdo L., Viegas M., Marques L.V.C., de Sousa P., Batista-Silva L.R., Alves-Monteiro V., Bonecker S., Monte-Mór B., Bonamino M.H. Transposon-mediated generation of CAR-T cells shows efficient anti B-cell leukemia response after ex vivo expansion. Gene Ther. 2020;27:85–95. doi: 10.1038/s41434-020-0121-4. [DOI] [PubMed] [Google Scholar]
- 73.Avery L., Filderman J., Szymczak-Workman A.L., Kane L.P. Tim-3 co-stimulation promotes short-lived effector T cells, restricts memory precursors, and is dispensable for T cell exhaustion. Proc. Natl. Acad. Sci. USA. 2018;115:2455–2460. doi: 10.1073/pnas.1712107115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Konduri V., Joseph S.K., Byrd T.T., Nawas Z., Vazquez-Perez J., Hofferek C.J., Halpert M.M., Liu D., Liang Z., Baig Y., et al. A subset of cytotoxic effector memory T cells enhances CAR T cell efficacy in a model of pancreatic ductal adenocarcinoma. Sci. Transl. Med. 2021;13:eabc3196. doi: 10.1126/scitranslmed.abc3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maiti S.N., Huls H., Singh H., Dawson M., Figliola M., Olivares S., Rao P., Zhao Y.J., Multani A., Yang G., et al. Sleeping beauty system to redirect T-cell specificity for human applications. J. Immunother. 2013;36:112–123. doi: 10.1097/cji.0b013e3182811ce9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kebriaei P., Huls H., Neel R.N.S., Olivares S., Orozco A., Su S., Maiti S., Smith A., Groot E.D., Kantarjian H., et al. Shortening the time to manufacture CAR+T cells with sleeping beauty system supports T-cell engraftment and anti-tumor effects in patients with refractory CD19+Tumors. Blood. 2017;130:2060. [Google Scholar]
- 77.Stroncek D.F., Lee D.W., Ren J., Sabatino M., Highfill S., Khuu H., Shah N.N., Kaplan R.N., Fry T.J., Mackall C.L. Elutriated lymphocytes for manufacturing chimeric antigen receptor T cells. J. Transl. Med. 2017;15:59. doi: 10.1186/s12967-017-1160-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bridgeman J.S., Hawkins R.E., Bagley S., Blaylock M., Holland M., Gilham D.E. The optimal antigen response of chimeric antigen receptors harboring the CD3ζ transmembrane domain is dependent upon incorporation of the receptor into the endogenous TCR/CD3 complex. J. Immunol. 2010;184:6938–6949. doi: 10.4049/jimmunol.0901766. [DOI] [PubMed] [Google Scholar]
- 79.Stephensen C.B., Borowsky A.D., Lloyd K.C.K. Disruption of Rxra gene in thymocytes and T lymphocytes modestly alters lymphocyte frequencies, proliferation, survival and T helper type 1/type 2 balance. Immunology. 2007;121:484–498. doi: 10.1111/j.1365-2567.2007.02595.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dawson H.D., Collins G., Pyle R., Key M., Taub D.D. The Retinoic Acid Receptor-alpha mediates human T-cell activation and Th2 cytokine and chemokine production. BMC Immunol. 2008;9:16. doi: 10.1186/1471-2172-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang Y., Kohler M.E., Chien C.D., Sauter C.T., Jacoby E., Yan C., Hu Y., Wanhainen K., Qin H., Fry T.J. TCR engagement negatively affects CD8 but not CD4 CAR T cell expansion and leukemic clearance. Sci. Transl. Med. 2017;9:eaag1209. doi: 10.1126/scitranslmed.aag1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zeng L., Palaia I., Šarić A., Su X. PLCγ1 promotes phase separation of T cell signaling components. J. Cell Biol. 2021;220 doi: 10.1083/jcb.202009154. e202009154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lo W.L., Shah N.H., Rubin S.A., Zhang W., Horkova V., Fallahee I.R., Stepanek O., Zon L.I., Kuriyan J., Weiss A. Slow phosphorylation of a tyrosine residue in LAT optimizes T cell ligand discrimination. Nat. Immunol. 2019;20:1481–1493. doi: 10.1038/s41590-019-0502-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gudipati V., Rydzek J., Doel-Perez I., Gonçalves V.D.R., Scharf L., Königsberger S., Lobner E., Kunert R., Einsele H., Stockinger H., et al. Inefficient CAR-proximal signaling blunts antigen sensitivity. Nat. Immunol. 2020;21:848–856. doi: 10.1038/s41590-020-0719-0. [DOI] [PubMed] [Google Scholar]
- 85.Zhong X.S., Matsushita M., Plotkin J., Riviere I., Sadelain M. Chimeric antigen receptors combining 4-1BB and CD28 signaling domains augment PI3kinase/AKT/Bcl-XL activation and CD8+ T cell-mediated tumor eradication. Mol. Ther. 2010;18:413–420. doi: 10.1038/mt.2009.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Herrero-Sánchez M.C., Rodríguez-Serrano C., Almeida J., San Segundo L., Inogés S., Santos-Briz Á., García-Briñón J., Corchete L.A., San Miguel J.F., del Cañizo C., Blanco B. Targeting of PI3K/AKT/mTOR pathway to inhibit T cell activation and prevent graft-versus-host disease development. J. Hematol. Oncol. 2016;9:113. doi: 10.1186/s13045-016-0343-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lapteva N., Gilbert M., Diaconu I., Rollins L.A., Al-Sabbagh M., Naik S., Krance R.A., Tripic T., Hiregange M., Raghavan D., et al. T-cell receptor stimulation enhances the expansion and function of CD19 chimeric antigen receptor-expressing T cells. Clin. Cancer Res. 2019;25:7340–7350. doi: 10.1158/1078-0432.ccr-18-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.He P., Wang B.H., Cao R.R., Zhu D.C., Ge B., Zhou X., Wu L.F., Lei S.F., Deng F.Y. ITGA2 protein is associated with rheumatoid arthritis in Chinese and affects cellular function of T cells. Clin. Chim. Acta. 2021;523:208–215. doi: 10.1016/j.cca.2021.09.024. [DOI] [PubMed] [Google Scholar]
- 89.Arias C.F., Ballesteros-Tato A., García M.I., Martín-Caballero J., Flores J.M., Martínez-A C., Balomenos D. p21CIP1/WAF1 controls proliferation of activated/memory T cells and affects homeostasis and memory T cell responses. J. Immunol. 2007;178:2296–2306. doi: 10.4049/jimmunol.178.4.2296. [DOI] [PubMed] [Google Scholar]
- 90.Daszkiewicz L., Vázquez-Mateo C., Rackov G., Ballesteros-Tato A., Weber K., Madrigal-Avilés A., Di Pilato M., Fotedar A., Fotedar R., Flores J.M., et al. Distinct p21 requirements for regulating normal and self-reactive T cells through IFN-gamma production. Sci. Rep. 2015;5:7691. doi: 10.1038/srep07691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wei Z., Du Q., Li P., Liu H., Xia M., Chen Y., Bi G., Tang Z.H., Cheng X., Lu Y., et al. Death-associated protein kinase 1 (DAPK1) controls CD8(+) T cell activation, trafficking, and antitumor activity. FASEB J. 2021;35:e21138. doi: 10.1096/fj.201903067rr. [DOI] [PubMed] [Google Scholar]
- 92.Wei Z., Li P., He R., Liu H., Liu N., Xia Y., Bi G., Du Q., Xia M., Pei L., et al. DAPK1 (death associated protein kinase 1) mediates mTORC1 activation and antiviral activities in CD8(+) T cells. Cell Mol. Immunol. 2021;18:138–149. doi: 10.1038/s41423-019-0293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Byrd V.M., Kilkenny D.M., Dikov M.M., Reich M.B., Rocheleau J.V., Armistead W.J., Thomas J.W., Miller G.G. Fibroblast growth factor receptor-1 interacts with the T-cell receptor signalling pathway. Immunol. Cell Biol. 2003;81:440–450. doi: 10.1046/j.1440-1711.2003.01199.x. [DOI] [PubMed] [Google Scholar]
- 94.van der Windt G.J., Everts B., Chang C.H., Curtis J., Freitas T., Amiel E., Pearce E., Pearce E. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 2012;36:68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lancien M., Gueno L., Salle S., Merieau G., Beriou K., Nguyen T.H., Abidi A., Solomon P., Poschmann J. Cystathionine-gamma-lyase overexpression in T cells enhances antitumor effect independently of cysteine autonomy. Cancer Sci. 2021;112:1723–1743. doi: 10.1111/cas.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Erdenebayar N., Maekawa Y., Nishida J., Kitamura A., Yasutomo K. Protein-tyrosine phosphatase-kappa regulates CD4+ T cell development through ERK1/2-mediated signaling. Biochem. Biophys. Res. Commun. 2009;390:489–493. doi: 10.1016/j.bbrc.2009.09.117. [DOI] [PubMed] [Google Scholar]
- 97.Gu M., Zhou X., Sohn J.H., Zhu L., Jie Z., Yang J.Y., Zheng X., Xie X., Yang J., Shi Y., et al. NF-κB-inducing kinase maintains T cell metabolic fitness in antitumor immunity. Nat. Immunol. 2021;22:193–204. doi: 10.1038/s41590-020-00829-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sukumar M., Liu J., Ji Y., Subramanian M., Crompton J.G., Yu Z., Roychoudhuri R., Palmer D.C., Muranski P., Karoly E.D., et al. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. J. Clin. Invest. 2013;123:4479–4488. doi: 10.1172/jci69589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Drijvers J.M., Gillis J.E., Muijlwijk T., Nguyen T.H., Gaudiano E.F., Harris I.S., LaFleur M.W., Ringel A.E., Yao C.H., Kurmi K., et al. Pharmacologic screening identifies metabolic vulnerabilities of CD8(+) T cells. Cancer Immunol. Res. 2021;9:184–199. doi: 10.1158/2326-6066.cir-20-0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang Y., Zhang Z., Ding Y., Fang Y., Wang P., Chu W., Jin Z., Yang X., Wang J., Lou J., Qian Q. Phase I clinical trial of EGFR-specific CAR-T cells generated by the piggyBac transposon system in advanced relapsed/refractory non-small cell lung cancer patients. J. Cancer Res. Clin. Oncol. 2021;147:3725–3734. doi: 10.1007/s00432-021-03613-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Prommersberger S., Reiser M., Beckmann J., Danhof S., Amberger M., Quade-Lyssy P., Einsele H., Hudecek M., Bonig H., Ivics Z. CARAMBA: a first-in-human clinical trial with SLAMF7 CAR-T cells prepared by virus-free Sleeping Beauty gene transfer to treat multiple myeloma. Gene Ther. 2021;28:560–571. doi: 10.1038/s41434-021-00254-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hu B., Zou Y., Zhang L., Tang J., Niedermann G., Firat E., Huang X., Zhu X. Nucleofection with plasmid DNA for CRISPR/Cas9-Mediated inactivation of programmed cell death protein 1 in CD133-specific CAR T cells. Hum. Gene Ther. 2019;30:446–458. doi: 10.1089/hum.2017.234. [DOI] [PubMed] [Google Scholar]
- 103.Schambach A., Morgan M., Fehse B. Two cases of T cell lymphoma following Piggybac-mediated CAR T cell therapy. Mol. Ther. 2021;29:2631–2633. doi: 10.1016/j.ymthe.2021.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bishop D.C., Clancy L.E., Simms R., Burgess J., Mathew G., Moezzi L., Street J.A., Sutrave G., Atkins E., McGuire H.M., et al. Development of CAR T-cell lymphoma in 2 of 10 patients effectively treated with piggyBac-modified CD19 CAR T cells. Blood. 2021;138:1504–1509. doi: 10.1182/blood.2021010813. [DOI] [PubMed] [Google Scholar]
- 105.Micklethwaite K.P., Gowrishankar K., Gloss B.S., Li Z., Street J.A., Moezzi L., Mach M.A., Sutrave G., Clancy L.E., Bishop D.C., et al. Investigation of product-derived lymphoma following infusion of piggyBac-modified CD19 chimeric antigen receptor T cells. Blood. 2021;138:1391–1405. doi: 10.1182/blood.2021010858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wilson M.H., Gottschalk S. Expect the unexpected: piggyBac and lymphoma. Blood. 2021;138:1379–1380. doi: 10.1182/blood.2021012349. [DOI] [PubMed] [Google Scholar]
- 107.Srour S.A., Singh H., McCarty J., de Groot E., Huls H., Rondon G., Qazilbash M., Ciurea S., Bardelli G., Buck J., et al. Long-term outcomes of Sleeping Beauty-generated CD19-specific CAR T-cell therapy for relapsed-refractory B-cell lymphomas. Blood. 2020;135:862–865. doi: 10.1182/blood.2019002920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Querques I., Mades A., Zuliani C., Miskey C., Alb M., Grueso E., Machwirth M., Rausch T., Einsele H., Ivics Z., et al. A highly soluble Sleeping Beauty transposase improves control of gene insertion. Nat. Biotechnol. 2019;37:1502–1512. doi: 10.1038/s41587-019-0291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author on reasonable request.