Abstract
Cotton is a major fiber crop grown worldwide. Nitrogen (N) is an essential nutrient for cotton production and supports efficient crop production. It is a crucial nutrient that is required more than any other. Nitrogen management is a daunting task for plants; thus, various strategies, individually and collectively, have been adopted to improve its efficacy. The negative environmental impacts of excessive N application on cotton production have become harmful to consumers and growers. The 4R’s of nutrient stewardship (right product, right rate, right time, and right place) is a newly developed agronomic practice that provides a solid foundation for achieving nitrogen use efficiency (NUE) in cotton production. Cropping systems are equally crucial for increasing production, profitability, environmental growth protection, and sustainability. This concept incorporates the right fertilizer source at the right rate, time, and place. In addition to agronomic practices, molecular approaches are equally important for improving cotton NUE. This could be achieved by increasing the efficacy of metabolic pathways at the cellular, organ, and structural levels and NUE-regulating enzymes and genes. This is a potential method to improve the role of N transporters in plants, resulting in better utilization and remobilization of N in cotton plants. Therefore, we suggest effective methods for accelerating NUE in cotton. This review aims to provide a detailed overview of agronomic and molecular approaches for improving NUE in cotton production, which benefits both the environment and growers.
Keywords: nitrogen use efficiency, cotton, nitrogen metabolism, molecular approaches, physiological approach
Introduction
Source of nitrogen and plant soil interaction
Nitrogen (N) is the key component of plant chlorophyll, nucleic acids, and amino acids, and, compared to other elements, plants acquire N in large amounts from the soil (Shafreen et al., 2021). It is important for plant growth, leaf area, biomass, and crop yield (Kumar et al., 2020). Approximately 78% of the atmospheric N is dinitrogen gas (N2), which is converted into various forms of NH4+ and NO3– by microorganisms. Undisturbed soil organic matter comprises almost 95% N (Walworth, 2013). From an agricultural perspective, certain N sources are major sources of available N in crop production. These are natural and organic N sources, some of which have been artificially developed. The conversion of N from one form to another greatly influences nitrogen use efficiency (NUE). At earlier stages, nitrate (NO3–) is essential, but it is not commonly used as a fertilizer alone, and the other forms are released into the atmosphere through the denitrification process. Although urea is the most widely used N fertilizer source, it is rapidly nitrified after its conversion to ammonium (NH4+) (Robinson et al., 2011). The application of urea to the soil results in NO3– and NH4+. However, urea’s uptake process and the plants’ metabolic changes are not yet clear (Witte, 2011). Soil N availability in the soil is an indicator to examine the N efficiency in crop fields (Cui et al., 2008). Various field studies of N-labeled fertilizers have shown that N uptake is primarily obtained from the soil (Franco et al., 2011).
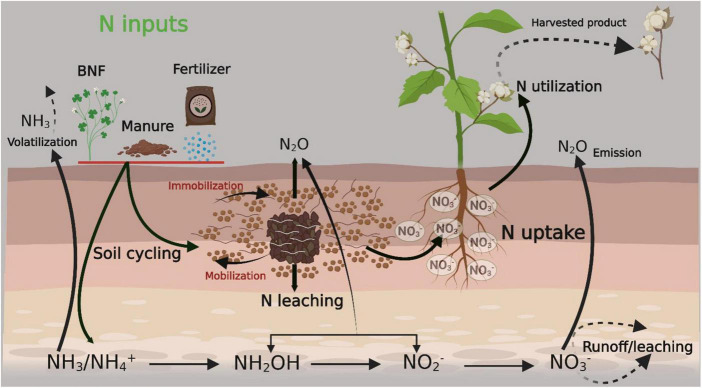
Mineralization and bacterial N fixation are natural sources of available N in the soil (Iqbal et al., 2020c; Gu et al., 2022). NH4+ and NO3– are the available forms of N for plant uptake. Nitrite (NO2–), nitrous oxide (NO), and atmospheric N are not readily available to plants unless they are converted into NH4+ and NO3– (Ma et al., 2015; Su et al., 2022). Microorganisms also degrade N, which is naturally available in the soil. When plants die, they decompose and deposit N into the soil. Legume crops contribute more N than other field crops (Hocking and Reynolds, 2012). Legumes are grown during crop rotation, help fix atmospheric N, and deposit it into the uppermost layers of the soil. Animal bones and bone meals are also an important source of higher levels of N than chemical ones (Sharma and Bali, 2017). A summary of N sources and their conversion, availability to plants, final harvested product, and losses within and outside of the soil are presented in Figure 1.
FIGURE 1.
Summary of nitrogen sources and their conversion, availability to plant along with final harvested product and losses within/outside of soil. The figure created with Biorender (https://biorender.com/). BNF, biological nitrogen fixation.
One N and three hydrogens (H) form anhydrous ammonia (NH3), an easily available and relatively cheap N fertilizer. However, it is explosive, caustic, and toxic. The use of this fertilizer is strictly regulated in the United States [Occupational Safety and Health Administration (OSHA), 2017]. Urea is another source of N and is widely used in crop production. It is readily available in a granular form and is better to use in windy conditions. It is degraded into NH4+ and carbon dioxide (CO2). It is an excellent source of N with appropriate attention for better crop production. Ammonium nitrate (NH4NO3) is chemically composed of NO3– and NH4+ cations. N uptake by plants is in the form of NH4+ and NO3–, and NH4NO3 is simply a mixture of both and is applied as a source of N. It has a low pH and is better for us in low wind conditions because of its structure (Kaiwen et al., 2015). One of the first N fertilizers produced 150 years ago was ammonium sulfate [(NH4)2SO4], which contains 21% N and 24% sulfur (S), which is most easiest to manage. (NH4)2SO4 is an important source of N and S, which are crucial for various plant functions, such as protein synthesis. (NH4)2SO4 is a well-known N fertilizer that lowers soil pH and enhances soil S. It is an excellent source of S in soil for better crop production (Khan et al., 2017b).
What is nitrogen use efficiency and the common factors contributing to low nitrogen use efficiency?
Nitrogen, a structural component of proteins and DNA, is essential for life and considered the most important crop-yield limiting nutrient (Mueller et al., 2012). Therefore, most farmers rely on N fertilizers to increase crop yields and economic returns (McLellan et al., 2018). However, N is prone to different types of losses, including ammonia (NH3) volatilization, nitrate (NO3–) leaching, denitrification losses as dinitrogen (N2) gas emissions, and nitrous oxide (N2O) emissions, which lead to environmental pollution and contribute to climate change (Fowler et al., 2013). Recent studies have reported that the agricultural sector is a major source of N loss to the environment (Ahmar and Gruszka, 2022). The rapid increase in population and ever-increasing food demand will further increase the demand for N fertilizers in the future, which may consequently increase N losses unless significant improvements are made in the whole food production-consumption chain, and more appropriate N management strategies are developed (Chen et al., 2020a).
Nitrogen uptake, translocation, assimilation, and remobilization are complex processes referred to as nitrogen use efficiency (NUE). It also shows the extent of cotton lint and seed yield in response to N application (Shafreen et al., 2021). Cotton NUE results from N uptake efficiency (UpE) and utilization efficiency (UtE). Cotton NUE was represented by lint yield recorded after N application. UpE is defined as the total N uptake by plants recorded after N application, and UtE is the cotton yield ratio divided by total plant N. Under N deficiency, plant N UpE is more important than UtE (Witcombe et al., 2008). In addition, N is a mobile nutrient in the soil and is more prone to leaching than other soil nutrients (Shimono et al., 2009; Tariq et al., 2019). Leaching, runoff, and volatilization result in more N loss, and crop plants take up less N (Cavigelli et al., 2012). Therefore, sustainable crop production requires an improvement in NUE by reducing N loss (Khan et al., 2017a).
Improvement in NUE is associated with several agronomic practices, such as improved irrigation methods, improved fertilizer application considering the 4Rs, and using hybrids with greater potential yields and lower N inputs (Venterea et al., 2012). Efficient management, that is, N source, rate, time, and placement, increases NUE in cropping systems (Snyder et al., 2014). N inhibitors, split application of N, irrigation time, and correct placement method of fertilizer that considers soil and crop type improve overall NUE (Afridi et al., 2014; Halvorson et al., 2014). Recently, Tang et al. (2012a) reported an increase in N accumulation during the boll-setting stage with late N application (Tang et al., 2012a). Moreover, NUE is considered an important factor for fertilizer inputs to any agricultural system, as it maintains the N balance between inputs and outputs without any economic or environmental loss (McAllister et al., 2012). Application of N fertilizer at the first bloom stage is another way to improve NUE because cotton plants utilize N more efficiently for reproduction (Ali, 2015).
Factors contributing to low nitrogen use efficiency
Nitrogen losses
Half of the applied N is usually lost through NO3– leaching, denitrification, or ammonia volatilization (Anthony and Armytage, 2008). Being negatively charged and highly soluble, NO3– is prone to leaching when the soil becomes saturated after heavy rains or flood irrigation. Anoxic conditions develop and denitrifying microorganisms start using nitrate as an alternate e–acceptor, reducing NO3– N to N2 (Shabbir et al., 2022). Weather conditions, irrigation patterns, and fertilizers also influence leaching (Follett, 2001; Riley et al., 2001). Bock and Kissel (1988) stated that alkaline soils resulting from NH3 volatilization lead to greater N loss because they have higher NH4+ concentrations on the soil surface. Various environmental factors, primarily higher temperature and wind speed, increase the risk of NH3 volatilization, which is a chemical process (Bock and Kissel, 1988). Soil characteristics, pH, cation exchange capacity (CEC), and moisture content also affect the volatilization rate (Jones, 2006).
Temperature and soil characteristics
The nitrification process increases with an increase in soil temperature, resulting in N loss and consequently influencing NUE (Engel et al., 2011). The availability and loss of N in the soil depend on the soil type (Chen et al., 2010). The higher the CEC, the higher the soil buffering capacity and the rate of NH4+ absorption will be greater than the loss rate. Soils with lower anion capacity lose negatively charged molecules, such as NO3– (Ferguson et al., 1984). Soil pH affects the activity of nitrifying and denitrifying bacteria (Ali et al., 2022a,c). The optimum pH for efficient N cycling is approximately 7 (Cameron et al., 2013). NUE is affected by the soil moisture content, which affects NO3– leaching and the nitrification or denitrification rates (Darwish et al., 2006). NUE is positively correlated with the efficiency of irrigation systems (Bronson, 2008).
Cropping systems, and C-N balance
The selection of cropping systems is crucial for balancing N input and output by improving N uptake and lowering the risk of N loss from the soil (Herridge et al., 2008). Several cropping systems tend to have a steady state of organic and inorganic soil N pools, with a slight change. Fast alteration in N pools in new soil management systems can influence the C-N balance as the soil organic matter remains constant. Overall, NUE in these cropping systems should incorporate changes in organic and inorganic soil N pools and N recovery efficiency (Lawlor, 2002). The C-N balance is an important factor unless adequate C is present, which improves the ability of plants to take up and utilize more N. Nitrogen levels can significantly affect C fixation and can be compromised (Castro et al., 1994). Photosynthetic rate and C level regulate N mineralization, uptake, assimilation, and immobilization. Hence, higher NUE can be attained through a higher photosynthetic rate (Lawlor, 2002).
Nitrogen fertilizer type
Several N fertilizers are used without considering the soil type, crop genetics, and fertilizer chemistry, whereas an appropriate type of fertilizer reduces the percentage losses of N (Spicer, 2002). NH3 (82% N) is gradually converted to NO3–, with a minimal risk of N loss due to leaching or denitrification, while urea (46% N) rapidly transformed to NO3–. Wet or compact soils have serious denitrification issues, while coarse or no-till soils have higher leaching or volatilization. (NH4)2SO4 (21% N) applications have less or no volatilization losses. NH4NO3 (34% N) contains 50% NH4+ and 50% NO3– when applied to the soil, and NH4+ N rapidly transforms into NO3– N. NH4NO3 should not be used in soils subjected to leaching and denitrification; however, it is more suitable for surface applications (Nielsen, 2006).
Nitrogen rates
The cotton crop requires almost 250–300 kg N ha–1 but utilizes only half to attain the maximum yield. Several studies have suggested that cotton uses N already available in the soil compared to applied N. On average, the cotton crop recuperates 33% of the applied N (Xu et al., 2012). In contrast, 25% of N remains in the soil until the maturity stage, with approximately 42% loss from the system (Tang et al., 2012b). Excessive N and N deficiency negatively affect plant growth and productivity (Liu et al., 2010; Yasmin et al., 2020). Moreover, extra N gradually leaches through underground water runoff, contaminating the groundwater with NO3–. Optimization of the N application rate depends on the soil type, climatic conditions, and several other soil and crop factors for better cotton production (Manghwar et al., 2021). However, the N rate for maximum economic yield depends on the N fertilizer cost and the commodity’s market price (Wajid et al., 2007). Owing to the complex chemical changes in N cycling that influence N loss, it is difficult to accurately predict the amount of N required. However, soil N availability, crop needs, and management practices should be considered to achieve greater NUE.
Nitrogen timing
The application of N had a significant effect on the overall NUE. Generally, N is applied during three growth stages: pre-plant, first bloom, and peak bloom. The pre-planting application provides sufficient time for N conversion into plant-available forms. However, it is subject to higher risks of N losses, especially due to young seedlings’ lower requirement of N. N remains exposed to heavy rains for more than 60 days and is more prone to leaching losses (Halkier and Gershenzon, 2006; Ayaz et al., 2021). Moreover, nitrification and urease inhibitors are recommended for delaying nitrification and urea hydrolysis for fall-applied anhydrous ammonia and early applied urea, respectively (Lasisi et al., 2020).
Approaches for improving nitrogen use efficiency
Improving NUE and guarding environmental quality are the two challenges cotton plant nutritionists face. NUE is described by several complicated interconnected factors that require close observation for improvement. Several field studies have revealed that site-specific N management tends to be more profitable and is considered sustainable. Several agronomic practices are also considered major factors influencing NUE.
Spatial variation
Different pedological processes and management practices lead to spatial variability within the field (Bouma and Finke, 1993). These spatial variations can be horizontal along the horizons or vertically across depth. Variations in horizons have made it difficult to determine the boundaries of the soil types, while vertical variation is an indicator of the layers in soil classification (Mulla and McBratney, 2002; Kong et al., 2021b). Several studies have revealed that soil spatial variability is usually the main factor causing variation in soil properties and crop yield (Ping et al., 2005) and spatial and temporal variability in cotton yield (Ward and Cox, 2000). Cotton yield was found to be spatially correlated with a range of 23–40 m, fiber quality measurement was found to be spatially correlated with a range of 15–106 m, and cotton quality measurements were found to be spatially correlated (Johnson and Bradow, 2000; Bronson et al., 2003).
Ping et al. (2007) reported that soil NO3– N highly depends on spatial variation. The spatial variability of soil properties suggests that site-specific cotton management is an appropriate option for improving cotton production with increased NUE (Ping et al., 2007). Site-specific management originated in efforts to adjust fertilizers to account for within-field variations in soil physical properties. Quantifying the spatial variability of soil properties and crop yield is important for decision-making in site-specific crop management because the spatial variability of soil and crop growth is the critical factor for determining variable-rate inputs of fertilizers and other chemicals. Improving the synchrony between demand and supply of N in the crop from all sources, that is, fertilizer application throughout the growing season, improves crop NUE (Cassman et al., 2003; Shankar et al., 2020; Ali et al., 2022b). However, site-specific N management is only possible following spatial variations. Recently, precision agriculture has provided an opportunity to address and measure spatial variability to promote sustainable agriculture and environmental stewardship (Ping et al., 2007; Ali Q. et al., 2021).
4R nutrient stewardship
An appropriate amount of N is required by crop plants for biomass production, and it helps restore soil organic carbon (C) levels (Lal, 2010). The best nutrient management practices (BNMPs) for N fertilizers play a key role in reducing nitrate leaching, lowering the risk of N2O emissions (a potent greenhouse gas), and improving overall NUE (Good and Beatty, 2011). Considering the weather and site-specific conditions when selection a suitable N fertilizer source can help improve NUE (Bruulsema et al., 2009; Ali M. et al., 2021). Intensive cropping systems meet the world’s need for food, fiber, and biofuel, which depend on better and sustainable crop production with improved NUE (Snyder et al., 2007; Solangi et al., 2021). Significant improvements in nutrient management practices may take several years to implement, as 4R practices are based on incremental progress and have an interim improvement/site-specific nature. However, implementing each management practice improves overall NUE and N2O emission mitigation (Wang et al., 2001).
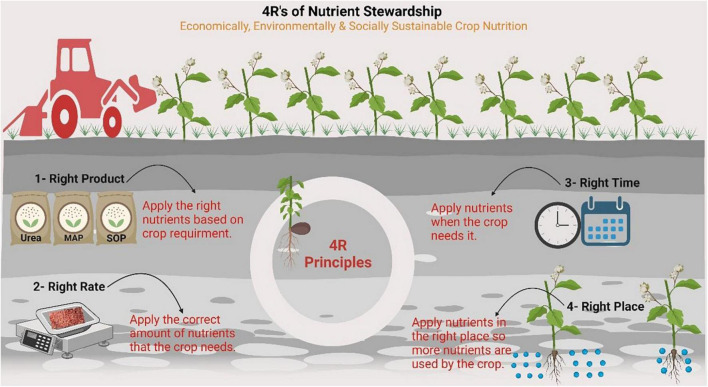
Nitrogen loss pathways include NH3 volatilization, denitrification, leaching, and runoff, leading to the formation of secondary aerosols, contamination of groundwater, and eutrophication, while N2O is emitted mainly from nitrification or denitrification processes (Ding et al., 2020; Manghwar et al., 2022). Thus, quantifying the different N loss pathways is important for developing proper N management strategies to improve overall NUE. One of the greatest factors leading to low N use efficiency and excess N losses in annual cropping systems is the mismatched timing of N availability with crop needs (Ranjan and Yadav, 2019). Recently, a 4R Nutrient Stewardship (4RNS) initiative has been developed and is being supported by the global fertilizer industry to achieve the basic economic (generation of more revenue per unit cropped area/input), social (production of better-quality food to meet global needs), and environmental elements (sustenance/improvement in soil quality and soil fertility, preservation of wildlife habitat, and biological diversity) of sustainability (Fixen, 2020). 4RNS is the adaptive management of mineral nutrients that has evolved through the refinement of interconnected practices and is site-specific (Bruulsema, 2018). The 4R practices that most clearly resulted in an improved NUE are presented in Figure 2.
FIGURE 2.
4Rs nutrient stewardship provides a framework to achieve cropping system goals, such as increased production, increased farmer profitability, enhanced environmental protection and improved sustainability. The figure created with Biorender (https://biorender.com/).
Right source, rate, time, and place
The key principle for selecting the right source is to ensure a balanced supply of nutrients, considering soil and crop plant characteristics, in addition to the rate, time, and placement method. Furthermore, we consider the synergism among nutrients and ensure the supply of nutrients in available forms to plants while selecting the right form. The use of enhanced N fertilizer sources with nitrification, urease, and dual inhibitors is a reliable means of reducing N losses and increasing NUE (Zhang et al., 2021). The greatest opportunities for increasing NUE were associated with lower rates of N fertilizer application. The key concept in selecting the correct rate is to assess the soil and crop needs in the source, time, and placement method. Rate-specific economics and fertilizer use efficiency (FUE) also play a key role in determining the correct rate. Better accounting for soil and residue N sources and targeting N rates for maximal N-use efficiency resulted in reduced overall N losses. To choose the appropriate fertilizer application time, split application of N fertilizers, especially for long-duration crops during the growing season, effectively reduces N losses with increased NUE. For example, the best time to apply N to late-sown cotton is at the appearance of the first flower. Afterward, cotton plants can utilize N much more efficiently in later stages (Zhang et al., 2021). Fertilizer placement can increase the efficiency of N fertilizer use by reducing NH3 emissions. The crop plant rooting pattern is the main deciding factor for the placement method, considering the source, rate, and time. It has also been shown that placement interacts with the tillage system, moisture, and temperature content to influence NUE. 4RNS is specifically built to achieve sustainability goals, including GHGs emission mitigation, particularly N2O, to reduce nutrient losses and produce more food per acre (Snyder et al., 2014). There are also opportunities to improve NUE associated with non-4R practices. Considering the effects of crop rotation on N availability, the impact of the tillage system, the use of tile drainage, and the inclusion of legumes in rotation are all important for improving NUE and developing 4R practices (Burton, 2018). Snyder et al. (2014) suggested that growers, crop advisors, researchers, policymakers, consumers, and the public could play a role in best management practices.
Metabolic and molecular pathways of nitrogen use in plants and specifically in cotton
The study of plant metabolism is a key aim in plant research. An improved understanding of the metabolic activities will improve production and our understanding of the influence on the environment (Weißenborn and Walther, 2017; Yu et al., 2021). Several basic metabolic pathways, including the shikimic acid, mevalonate, amino acid, glutamine, proline, aspartate, carbohydrate, nitrogen (N), and lipid pathways (Rojas et al., 2014), have been studied in plants. All enzymatic steps are specific to the metabolic process and act on several families of associated molecules. As a result of metabolism, metabolic pathways produce several products accumulated in the same cell or different compartments or exported where needed. These molecules are channeled through metabolic pathways called metabolic flux. Other metabolites that regulate the primary and secondary metabolic pathways are also produced. Thus, it is difficult to understand the metabolic pathways (Farré et al., 2014).
Nitrogen metabolism is a basic and vital process for optimum plant growth, stress tolerance, and normal physiological processes (Zhong et al., 2017; Johnson et al., 2022). It is also an essential component of nucleic acids, chlorophyll, photosynthesis, RuBisCO, and some hormones (Liao et al., 2019). In the second half of the 20th century, nitrogenous fertilizers significantly augmented crops worldwide. However, excessive and limited application of nitrogenous fertilizers causes stagnation of cotton yields and reduces NUE. Moreover, external cues, such as biotic and abiotic stresses, also reduce crop yield by decreasing gas exchange and chlorophyll fluorescence triggered by increased resistance to CO2 diffusion and metabolic constraints (Zhong et al., 2017; Ihtisham et al., 2020). Exposure to drought alters plant metabolic activities and biological functions, which are responsible for restricted growth (Mahmood et al., 2019).
In cotton, the excessive use of nitrogenous fertilizers, very high costs, and external cues, such as biotic and abiotic stresses (Khan et al., 2017a; Sarraf et al., 2022) have become challenging tasks to improve NUE (Zhang et al., 2018). Furthermore, half of the applied nitrogenous fertilizers are not absorbed by plants and are leached, polluting groundwater reservoirs, which ultimately threatens ecosystems (Cameron et al., 2013). Iqbal et al. (2020b) studied four cotton genotypes and found that N concentration and different N-metabolic enzymes are important regulators of NUE. Additionally, 48 candidate genes involved in nitrogen metabolism were identified in wheat (Liu et al., 2020). A recent review by Ueda et al. (2017) emphasized the importance of N-responsive genes involved in the efficient uptake of N (Ueda et al., 2017). A recent study identified that the TaPAP, TaUPS, and TaNMR genes were differentially expressed in wheat with varying N levels. Expression studies revealed their roles as conserved N-metabolism genes. TaNMR has been identified as a novel gene in N metabolism (Li et al., 2019).
Technical/metabolic pathway at the cellular, organ, and structural levels
Various studies on N metabolism have been conducted in wheat, rice, Arabidopsis, and cotton (Zhong et al., 2017; Vidal et al., 2020; The et al., 2021). N metabolism is a complex process that includes many physicochemical and biochemical processes, including N transportation, distribution, use, and reuse (Xing et al., 2018). Few studies have focused on N metabolism in cotton. However, even fewer studies describe the importance of N metabolism related to drought stress (Iqbal et al., 2020a), variation in N metabolism in response to NUE (Iqbal et al., 2020b), the effects of salt stress on N metabolism in roots and stems (Taliercio et al., 2010), the varying ratio of NH4+/NO3– and its impact on N metabolism, higher N application, and increased N metabolism (Iqbal et al., 2020a) and recent advancements in N metabolism in cotton (Baslam et al., 2020).
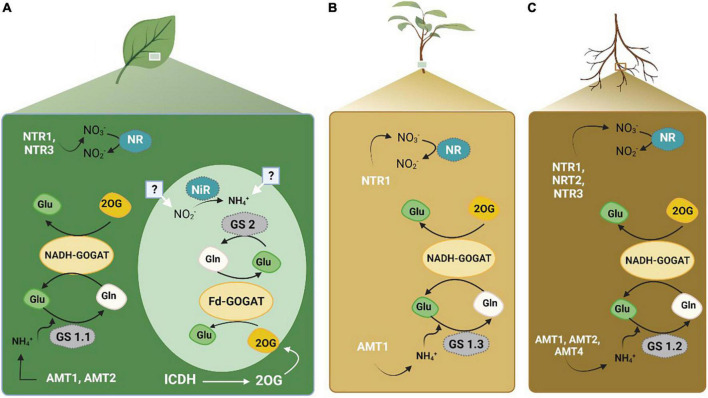
Nitrogen metabolism (Figure 3) is one of the critical processes in plants that reduces nitrate and convert it into amino acids. It has proven to be influential in determining the NUE in cotton (Iqbal et al., 2020b). During this process, key enzymes including nitrate reductase (NR), nitrite reductase (NiR), glutamine synthetase (GS), glutamate dehydrogenase (GDH), glutamine synthase (GOGAT), asparagine synthetase (AS), and aspartate aminotransferase (AspAT), are used to make the N available to the plants in the form of amino acids (Xu and Zhou, 2004). These enzymes were assessed in citrus to determine biochemical markers of N status (Singh et al., 2018). The conversion of input N as a raw material to final product amino acids is mediated by a series of enzymes. NO3– is converted to NH4+ by NR and NiR in the cytoplasm using one mole of NADPH or NADH, and NH4+ to glutamine in plastids using six moles of the reduced form of ferredoxin (Marcondes and Lemos, 2012; Ohyama et al., 2017). Initially, the enzyme GS, which consumes only one mole of ATP, assimilates the ammonium ion (NH4+) into Gln coupled with Glu. Furthermore, in plastids, 2 moles of ferredoxin are used by GOGAT to convert Gln into an organic acid, 2 oxoglutarate (2-OG) (Crawford, 2000). As a result of the transfer of Glu into organic acids, several amino acids (AA) are produced via transaminases. Previously, NH4+ was viewed as a factor responsible for the assimilation of Glu by GDH. However, recent research has validated the GS/GOGAT cycle as the principal route of ammonium assimilation in plants (Crawford, 2000; Crawford and Forde, 2002).
FIGURE 3.
Nitrogen transporters and key enzymes involved in N metabolism and acquisition in plants. Panel (A) represents the activity in leaves. Panel (B) represents the activity in plant. Panel (C) represents the activity within the roots. The figure created with Biorender (https://biorender.com/).
Previously, cotton was an orphan crop; many studies have been conducted to increase the NUE by focusing on the N concentrations and morphological and biochemical traits, but not much has been done on N metabolism and related enzymes, such as NR, NiR, GS, GOGAT, and GDH, which carry out the whole process from N uptake to use (Abenavoli et al., 2016; Luo and Zhou, 2019). Furthermore, N-containing compounds, such as amino acids and proteins, are key partners of N metabolism processes, including assimilation and metabolism, which determine genotypic responses to N supply (Quan et al., 2017). Therefore, these enzymes and N metabolism are considered the most vital biochemical factors for improving NUE in cotton (Xu et al., 2012). Another study was conducted to determine the contrasting NUE of six cotton genotypes. Biochemical and morpho-physiological traits such as N metabolic enzymes, shoot dry weight, and root traits were mostly affected in response to varying nitrate concentrations. NUE positively correlated with improved N uptake efficiency (Iqbal et al., 2020d).
In another study on cotton (Iqbal et al., 2020b), a close association between N uptake and utilization efficiency was demonstrated by applying various N doses. Because it is challenging to improve NUE by lowering N supply and selecting N efficient cotton genotypes, uptake, utilization, and remobilization of available N. Based on a contrasting N metabolic study, N uptake efficiency (NUpE), and N utilization efficiency (NUtE), CCRI-69 and XLZ-30 showed efficient NUE (Iqbal et al., 2020b). The N concentration and N-metabolizing enzymes were attributed as important traits that confer high NUpE. After applying higher N concentrations, shoot and root NR, GOGAT, and GDH enzyme activities increased. However, different genotypes exhibited contrasting behaviors (Iqbal et al., 2020b).
Nitrogen transporters and role in nitrogen use efficiency
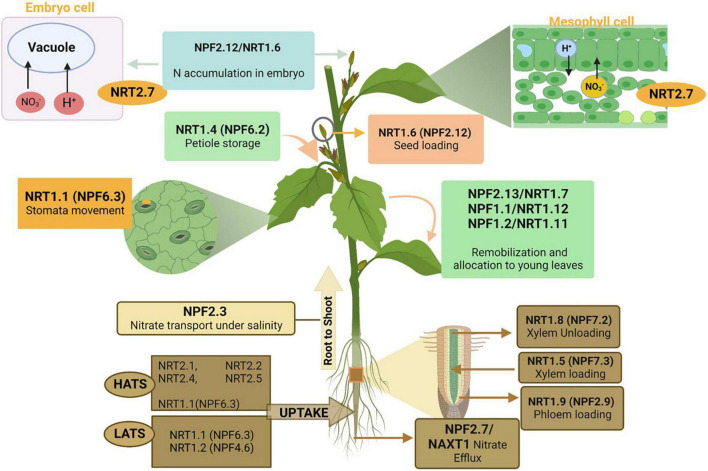
Plants absorb N in the form of nitrate at the root level by four nitrate transporter families: the nitrate peptide family NPF (previously known as NRT1/PTR), nitrate transporter 2 family NRT2/NNP (Nitrate-Nitrite Porter), chloride channel/transporter family (CLC-1), and slow anion associated channel homolog (SLAC1/SLAH) family (Iqbal et al., 2020d) (Figure 4). However, according to recent literature (Fan et al., 2017), the NRT1 (NPF) and NRT2 families are considered the main nitrate transporters for the uptake and transfer of nitrate in plant roots (Fan et al., 2017). NPF and NRT2 families are mainly responsible for low-affinity transport systems (LATS) and high-affinity transport systems (HATS), respectively (Williams and Miller, 2001; Tsay et al., 2007), whereas NRT1⋅1 (NPF6.3) and NRT1.3 (NPF6.4) (Morère-Le Paven et al., 2011) are responsible for both LATS and HATS (Liu et al., 1999). However, at high nitrate concentrations (>1 mM), it responds toward LATS (Raddatz et al., 2020). The NRT1 (NPF) nitrate transporters in other crops and transporters that behave differentially based on the varying nitrate concentration has been discussed in detail (Segonzac et al., 2007; Morère-Le Paven et al., 2011; Hawkesford, 2012; Hu et al., 2015; Chen et al., 2016).
FIGURE 4.
Roles of different nitrogen transporters in nitrate uptake and efflux from the soil, transportation from roots to shoots, allocation and assimilation in plant leaves, and seed development. As mentioned in the above section, these nitrogen transporters are linked to different families. The figure created with Biorender (https://biorender.com/).
In plants, multiple nitrate transporters (Table 1) perform NO3– uptake. Both NRT1 (NPF) and NRT2 transporter families affect plant growth and seed development because of the differences in NO3– uptake efficiency (Wang et al., 2020). NRT1 (NPF) was identified in Arabidopsis (Tsay et al., 1993) and is part of the NPF family (Léran et al., 2014). The NPF family includes 53 genes in Arabidopsis, 139 were identified in higher plants, and 93 genes in rice. These are further subdivided into 8–10 families as reviewed by Iqbal et al. (2020d); NPF6.3 (NRT1.1) and NPF4.6 (NRT1.2) are responsible for NO3– uptake in roots (Iqbal et al., 2020d). NRT1.1 (NPF6.3) is the first and most studied NO3– transporter in plants (Tsay et al., 1993). In the NRT1 (NPF) family, Arabidopsis includes 53 genes, of which 51 show differential expression patterns in the whole plant (Tsay et al., 2007). The NRT1 (NPF) family functions as NO3– transporters and a diverse range of substrates, including abscisic acid, nitrite, amino acids, peptides, chloride, glucosinolates, gibberellins, auxin, and jasmonoyl-isoleucine (Krouk et al., 2010; Kiba et al., 2012; Nour-Eldin et al., 2012; Saito et al., 2015; David et al., 2016; Tal et al., 2016).
TABLE 1.
Various nitrogen transporters involved in different functions of nitrate uptake, utilization, and remobilization.
| Sr. no. | N transporters | Function/s | References |
| (1) | NPF2.7/NAXT1 | Root efflux | O’Brien et al., 2016 |
| (2) | AMT1;1, AMT2;1, NPF6.2/NRT1.4, LHT1 | Leaf import | Noguero and Lacombe, 2016 |
| (3) | NPF2.12/NRT1.6, UmamiT11, UmamiT14 | Phloem unloading, and senescing | Noguero and Lacombe, 2016 |
| (4) | NRT2.7 | Seed vacuole | O’Brien et al., 2016 |
| (5) | NPF7.2/NRT1.8, NPF6.2/NRT1.4, NPF1.2/NRT1.11, NPF1.1/NRT1.12, AAP6, AAP2 | Xylem and phloem transfer | Fan et al., 2017 |
| (6) | CLCa, AVT3a | Leaf vacuole | Tegeder and Masclaux-Daubresse, 2018 |
| (7) | DiT2 | Chloroplast | Tegeder and Masclaux-Daubresse, 2018 |
| (8) | NPF2.13/NRT1.7, NPF1.2/NRT1.11, NPF1.1/NRT1.12, NRT2.4 NRT2.5, UmamiT18 AAP8 | Leaf export and phloem loading | Zhang et al., 2018 |
| (9) | NPF5.5, UmamiT28, UmamiT29, CAT6, AAP8, AAP1 | Loading to seed | Tegeder and Masclaux-Daubresse, 2018 |
| (10) | AMT1.1, AMT1.2, AMT1.3, AMT1.5, NRT2.1, NRT2.2, NRT2.4, NRT2.5, NPF6.3/NRT1.1, NPF4.6/NRT1.2, AAP1, AAP5, LTH1, LTH6, ProT2, ANR1 | Nitrogen uptake | Tegeder and Masclaux-Daubresse, 2018 |
| (11) | NPF7.3/NRT1.5, NPF2.3, UmamiT14, UmamiT18, UPS1-1, UPS1-2 | Movement from root nodule to xylem | Tegeder and Masclaux-Daubresse, 2018 |
| (12) | NPF7.2/NRT1.8, NPF2.9/NRT1.9 | Root reimport | Fiaz et al., 2021 |
Eight NRT2 transporters that respond to HATS have been identified in different plants (Von Wittgenstein et al., 2014). Although not many nitrate transporters have been identified in, we present the transporters from other plants such as Arabidopsis and rice. In Arabidopsis, several NRT2 transporters have been identified; four of them, including NRT2.1, NRT2.2, NRT2.4, and NRT2.5, function in nitrate influx and play a role in 95% nitrate uptake under low NO3– concentrations. However, NRT2.1 and NRT2.2 are the principal members of the NRT2 family for nitrate uptake (Lezhneva et al., 2014). Although NRT2.1 is a member of HATS but works efficiently under low nitrate availability (Li et al., 2007), its HATS activity is reduced, and plant growth is affected by low NO3– (Skopelitis et al., 2006) as demonstrated using mutants (Carvalho et al., 2003). However, compared to NRT2.1, NRT2.2 showed less expression for the uptake of NO3– (Suzuki and Knaff, 2005). Moreover, NRT2.4 and NRT2.5 are also responsible for NO3– uptake, but NRT2.4 was identified as a high-affinity transporter, as its mutant showed a reduction in NO3– uptake upon 0.025 mM nitrate (Kiba et al., 2012). Overall, NO3– acquisition depends on the specificity of NO3– transporters because NRT2.4 and NRT2.5 absorb NO3– from soil root hairs while NRT2.1 and NRT2.2 transport it from the apoplast to the apoplast cortex and endodermis (Suzuki and Knaff, 2005). In response to long-term starvation, NRT2.5 absorb NO3– efficiently from shoots and roots of adult plants, as its expression is increased along with several other NRT2 transporters (Lezhneva et al., 2014). In addition, NAR2(NRT3) from the NRT3 family is another transporter that develops a coupling relationship with NRT2 to transporters to NO3– in plants (Li et al., 2007). Many NRT2 transporters have been identified and studied in other plants, including Chlamydomonas reinhardtii (Zhou et al., 2000), barley (Marschner, 2011), and rice (Chen et al., 2016). These NRT transporters can be manipulated to improve NUE.
NH4+ uptake is carried out by ammonium transporters (AMTS) and other regulators such as cation channels or aquaporins (Glass et al., 2002). However, the overexpression of genes responsible for ammonium transporters has not yet been successful (Meister et al., 2014). Because of the excess availability of NH4+ in cells, it becomes toxic, a possible hindrance to targeting NH4+ transporters to improve N uptake (Bittsánszky et al., 2015). In addition to inorganic N acquisition, plants absorb organic nitrate in amino acids (AA) (Näsholm et al., 2009). Various root transporters, including AAP1 and AAP5, proline transporter ProT2, and lysine-histidine-type transporters LHT1 and LHT6, are responsible for the uptake of amino acids (The et al., 2021). However, this is only possible in fields that rely on manure or compost (Enggrob et al., 2019).
After NO3– uptake, it is assimilated in different parts of the shoot and loaded into the xylem vessels of roots using several transporters such as NRT1(NPF)/PTR and NRT (O’Brien et al., 2016). Upon assimilation, it is converted into AA (Meyer and Stitt, 2001). More assimilation takes place in shoots that in roots (O’Brien et al., 2016), using the same process discussed earlier. Different NO3– transporters have been shown to increase N assimilation in plants. According to a recent review (O’Brien et al., 2016), NRT1(NPF)/PTR and NRT2 members are expressed in the xylem and phloem. In Arabidopsis, NPF7.3 (NRT1.5) (Meng et al., 2016), NPF7.2 (NRT1.8) (Tong et al., 2005), and NPF2.9 (NRT1.9) (Wang and Tsay, 2011) play roles in influx/efflux, removal of NO3– from the xylem, and loading of NO3– into the root phloem, respectively. Besides these, many other genes are also responsible for the better uptake, assimilation, and remobilization of NO3– from roots to shoots (Fan et al., 2017).
According to the cited literature (Fang et al., 2013; Fan et al., 2016a; Feng et al., 2017; Wang Y. Y. et al., 2018), only NRT(NPF) transporters have been overexpressed in both leaves and roots to improve the NUE (The et al., 2021). However, it is yet to be determined whether overexpression of these NRT2 transporters in roots is sufficient to improve NUE. In the latest research on Arabidopsis, rice, and tobacco, overexpression of the hyperactive chimeric NO3– transporter AtNC4N in the phloem of old leaves increased N uptake and improved NUE under low N levels. Another study showed that the OsNRT1.1A (OsNPF6.3) NO3– transporter gene is involved in improving NUE, flowering, high yield, and early maturation in rice. Many other N transporters, such as NRT1.1B (NPF6.3B), NRT2.1, NRT2.3a, NRT2.3, NRT2.3b, PTR9, AMT1.1, and qNGR9, have been shown to increase NUE under high and low N concentrations (Fang et al., 2013; Ranathunge et al., 2014; Sun et al., 2014; Fu et al., 2015; Hu et al., 2015; Chen et al., 2016, 2017, 2020b; Fan et al., 2016a,b). Furthermore, several other genes in different plants have been shown to improve plant growth and NUE, including the nitrate transporter OsNPF4.5 (Sun et al., 2014), NAC42-activated nitrate transporter (Tang et al., 2019), and nitrate reductase gene OsNR2 (Gao et al., 2019).
Molecular and signaling pathways involved in nitrogen use efficiency
How the nitrate signaling and gene expression networks can be used to improve NUE in plants are not yet completely understood. Additionally, researchers have attempted to increase NUE by modulating the expression of key genes involved in NO3– uptake, assimilation, and remobilization in various plants. However, no significant success has been recorded (McAllister et al., 2012). Thus, success cannot be achieved until all other processes, including uptake, transport, assimilation, and remobilization, are understood. To improve NUE, it is also necessary to understand these processes and how they coordinate the expression of genes involved in the NO3– response. Apart from the N source, NO3– also modulates gene expression and plays a role in various developmental processes, including seed germination, shoot development, flowering, and root architecture (O’Brien et al., 2016; Lin and Tsay, 2017; Liu et al., 2017; Kant, 2018; Wang Y. Y. et al., 2018).
NO3– induces a primary NO3– response that regulates the transcriptional response without requiring de novo protein synthesis (Gowri et al., 1992). As a result of this rapid response, gene expression can be induced within minutes, reaching a peak at approximately 30 min. However, NO3– concentration determines the induction levels of PNR genes (Hu et al., 2009). Transcriptome analysis revealed the importance of NO3– in gene expression, as it controls 10% of the genome (Bouguyon et al., 2012). Various PNR gene families have been studied in plants, such as NRT1(NPF), NRT2, NIR, NIA1, NIA2, and other genes responsible for different metabolic processes, including the trehalose-6-P metabolism pentose phosphate pathway and glycolysis (Scheible et al., 1997). Several main NO3– signaling pathways, including transcription factors, peptides and proteins, kinases, NO3– transporters, and calcium signaling. Because cotton has not been well studied regarding NO3– molecular signaling pathways, we focused on these pathways in other crops to understand the potential mechanism in cotton.
NRT1.1 (NPF6.3) is involved in the uptake of NO3–. Two NO3– transporters, NRT1.1 (NPF6.3) and NRT2.1, are responsible for NO3– signaling and sensing. NRT1.1 (NPF6.3) also regulates the expression of NRT2.1 (Munþos et al., 2004). In the case of short-term and long-term NO3– supply, NRT1.1 (NPF6.3) upregulates NRT2.1, which involves the feedback repression of NRT2.1 (Bouguyon et al., 2015). Based on its dual-affinity property, it can switch between high- and low-level NO3– responses (Hu et al., 2009), which is caused by the phosphorylation status of the T101 residue (Hu et al., 2009). Furthermore, NO3– signaling is differentially regulated by the interaction of two CALCINEURIN B-LIKE (CBL)-INTERACTION PROTEIN KINASES (CIPKs) with NRT1.1 (NPF6.3), and CIPK8 and CIPK23 engage in low- and high-affinity responses, respectively (Ho et al., 2009). Except for PNR regulation, NRT1.1 (NPF6.3) is also involved in the root during the aging process. NRT1.1 (NPF6.3), which plays a role in sensing NO3– concentration as root growth depends on NO3– sensing; at high and low NO3– concentrations, lateral root growth is promoted and inhibited, respectively. At high NO3– concentrations, NRT1.1 (NPF6.3) upregulates Arabidopsis NO3– regulated 1 (ANR1) for root proliferation and lateral root growth development (Remans et al., 2006). While At low NO3– concentrations, NRT1.1 (NPF6.3) controls auxin levels and meristem activation for the repression of lateral root development (Mounier et al., 2014). An additional NO3– sensing system is also present, as the abolishment of NRT1.1 (NPF6.3) does not disrupt the PNR system, but its exact function in NO3– signaling is not understood (Sun et al., 2017).
After NO3– sensing by NRT1.1 (NPF6.3), the next step is to transmit the NO3– signals to the nucleus, which magnifies the cytosolic regulators. Calcium and various transcription factors are the main role players in signal transduction. Three CIPKs (CPK10, CPK30, and CPK32) and their partner CBL indicated that calcium also functions in NO3– signaling (Krouk, 2017). Calcium acts as a secondary messenger in the NO3– signaling pathway (Liu et al., 2017), responsible for changes in gene expression (Medici and Krouk, 2014). The first time, this study was conducted 30 years ago on barley and maize, where it showed that NO3– responsive genes show varying expression as a result of EGTA or LaCl3 pretreatment (Sakakibara et al., 1997). Furthermore, its function as a NO3– signaling pathway has been investigated in various studies (Riveras et al., 2015). Moreover, many other transcription factors (TFs), like NLP7, TCP20, LBD37, LBD38, LBD39, CIPK8, SLP9, TGA1, TGA4, CIPK23, bZIP1, BT1, and BT2 have been identified that interact with nitrate responsive genes, carry out the function of NO3– response, and improve NUE (Wang Y. Y. et al., 2018).
Based on recent bioinformatics analyses, BT1 and BT2 have proven to be great assets for improving NUE. BT1 was identified as the closest homolog of BT2. BT1 and BT2 improve NUE by repressing the expression of the nitrate transporters NRT2.1 and NRT2.4, which reduces NO3– uptake (Araus et al., 2016). Both TFs are involved in plant growth. However, it is yet to be determined whether BT1 and BT2 target multiple genes under N-sufficient and N-deficient conditions. The RWP-PK TFs family includes NLP7 TF. In a genome-wide analysis, NLP7 was identified as a player in modulating the expression of various genes involved in NO3– signaling, uptake, and assimilation (Marchive et al., 2013). Similarly, SPLN also regulates the expression of different NO3– transporter genes, including NPF6.3 (NRT1.1), NRT 2.1, NRT 2.2, NIA1, NIA2, and NIR (Krouk et al., 2010).
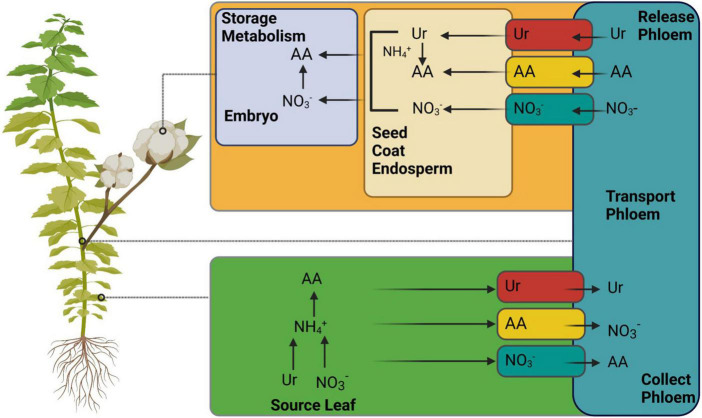
Cross talk on source/sink relationship and nitrogen use efficiency
Generally, N is recycled and remobilized in source leaves after uptake by the roots. It is delivered to the sinks via the phloem as amino acids, NO3–, and ureides (Figure 5) (Tegeder and Masclaux-Daubresse, 2018). However, different agronomic characteristics, such as NUE, grain filling, and yield, depend on better N remobilization in the source and its allocation to sink organs. The removal of N is initiated mainly during leaf senescence (The et al., 2021). In plants, senescence is a developmental process that regulates the nutrient requirements. Therefore, leaf senescence is also linked to the source/sink relationship (Guiboileau et al., 2010). In addition to the natural aging process, many other factors, such as nutritional starvation, pathogen infections, C-N ratio, photosynthetic activity, photoperiod, C accumulation, and various other cues, can initiate senescence. The limited availability of N also increases leaf senescence earlier than the sufficient availability in sunflowers (Bieker and Zentgraf, 2013).
FIGURE 5.
Illustrates the mechanism of nitrogen remobilization from source (leaf) to sink (seed). Various known and unknown transporters are involved in portioning, as mentioned in Tegeder and Masclaux-Daubresse (2018). AA, amino acids. The figure created with Biorender (https://biorender.com/).
In addition to N, some environmental factors, such as light quality and quantity and reactive oxygen species (ROS) (Zimmermann et al., 2006; Kong et al., 2021a), induce leaf senescence, which ultimately regulates nutrient mobility from source to sink organs where needed. Different sink organs and developmental processes require N. During anthesis, it varies according to the genotype. Varying concentrations of N are responsible for the differential uptake of N among different Arabidopsis accessions. Upon application of High N concentrations, most of the N is remobilized to the seeds, whereas at low N concentrations, it is allocated to rosette leaves (Masclaux-Daubresse and Chardon, 2011). During seed filling and flowering, the uptake of N is arrested in response to a reduction in HATS and HATS + LATS activities (Masclaux-Daubresse et al., 2010). Here, we will focus on natural-induced senescence accompanied by the movement of nitrogenous nutrients from source leaves to sink organs and various efforts to increase NUE in response to the source and sink relationship.
Generally, due to autophagy, proteins, organelles, and cytosolic macromolecules are degraded during leaf senescence, and N is translocated toward sink organs (Chen et al., 2019). Associated cytoplasmic components are dismantled and degraded during autophagy, which plays an important role in regulating and remobilizing nutrients from source to sink organs (Tegeder and Masclaux-Daubresse, 2018). ATGs genes are responsible for this autophagy process during leaf senescence, and are upregulated during autophagy (Chung et al., 2010; Breeze et al., 2011). Autophagy-related procedures can degrade the chloroplast and the most abundant protein, Rubisco, accounting for 80% of cellular N (Wada et al., 2009). This is further reinforced by the fact that leaf senescence is initially observed because of chloroplast dismantling and degradation. According to the cited literature (Martínez et al., 2008), enlisted protease enzymes, including chloroplastic DegP, FstH, and CND41 are responsible for the degradation of proteins D1 and Rubisco, respectively. Details regarding protein degradation mechanisms are provided by various studies (Tegeder and Masclaux-Daubresse, 2018; Gill et al., 2021). One of the efficient methods, the “apparent remobilization” method, which works based on N15 labeling, is used to orchestrate the amount of N present in different developmental stages of plants (Gallais et al., 2006).
Seeds are the major sink organs during N remobilization, which depends on different factors: strength of the seed sink, translocation processes in leaves, stems, and reproductive organs, and efficiency of phloem pathways (Manghwar et al., 2022). Based on acquired knowledge in different plants, it is well known that asparagine and glutamine are the major translocated amino acids from source to sink organs. Their concentrations also increased during leaf senescence. The amino acid permease (AAP) family is a candidate gene for improving the phloem loading efficiency. Different genes, including AAP4, AAP5, BnAAP1, and BnAAP2, have been implicated in phloem loading (Fischer et al., 2002; Koch et al., 2003; Chaffei et al., 2004; Tilsner et al., 2005). During leaf senescence, N remobilization and assimilation can improve the NUE of plants. Both GS1 and GS2 proved influential in improving NUE. Further improvement of N remobilization depends on the severity and severity of leaf senescence activity (Diaz et al., 2008).
There is a long list of protease genes responsible for protein degradation and N remobilization. Among these genes, SAG12 is a widely studied enzyme that catalyzes the cysteine protein in leaves. In addition, its homologs are also present in other crops and are responsible for N remobilization and improvement of NUE in oilseed and tobacco (Chung et al., 2010). Furthermore, glutamate dehydrogenase (GDH) is also important for improving NUE, as it functions to remobilize N (Lea and Miflin, 2011). However, overexpression of GDH did not result in N remobilization as in the case of tomato Slgdh-NAD: B1 in tobacco, GDH from Sclerotinia sclerotiorum and Magnaporthe grisea in rice (Purnell et al., 2005), and Nicotiana plumbaginifolia GDHA and GDHB in Nicotiana tabacum (Tercé-Laforgue et al., 2013). In contrast it improved NUE in other plants, such as overexpression of EcgdhA from Escherichia coli in tobacco and maize (Ameziane et al., 2000). In addition, overexpression of AngdhA from Aspergillus nidulans in potato (Egami et al., 2012) and fungal GDH from Cylindrocarpon ehrenbergii (CeGDH) in rice (Zhou et al., 2015) augmented NUE. Autophagy respondent genes such as AtATG8 in Arabidopsis, OsATG8b, OsATG8a, and OsATG8c in rice also proved valuable in improving NUE under sufficient conditions of N supply (Gallais et al., 2006; Zimmermann et al., 2006).
Other factors, such as N remobilization in the form of inorganic or ionic N, or combined with organic molecules, also play a differential role in improving NUE. Different studies have reinforced the significant role of inorganic N in N remobilization toward sink organs, but it also depends on the availability of sufficient N inputs (Masclaux-Daubresse et al., 2010). Various studies on NO3– transporters, including NRT2.5, NRT1.6 (NPF2.12), NRT1.5 (NPF7.3), and AMT1.5, could improve NO3– remobilization from source to sink organs, which ultimately also increases NUE (Wu et al., 2014; Havé et al., 2017). Another study was conducted on the NO3– transporter NRT1.7 (NPF2.13). They manipulated its remobilization, which improved NUE and increased plant growth, as it was involved in the meditation of stored NO3– from the source to sink organs (Chen et al., 2020c).
In addition to molecular-level studies, some common agronomic practices are also essential for better source-to-sink relationships in plants. Research on efficient and inefficient cotton genotypes, CCRI-69 and XLZ-30, showed that moderate to high N applications improve the remobilization of the source-to-sink relationship, which ultimately increases the NUE and yield (Iqbal et al., 2020c). Furthermore, different foliar applications also proved beneficial. Under treatment with different N foliar applications, such as NO3– and urea, winter wheat plants showed increased grain filling and source-to-sink relationships of N remobilization, as (Lyu et al., 2020; Liang et al., 2022). However, many N studies have been conducted on plants other than cotton plants. Currently, there is a dire need to study cotton plants to improve the NUE and source-to-sink relationship for better remobilization of N by employing molecular and agronomic studies.
Crop plants largely absorb N in the form of nitrate (NO3–) (Robinson et al., 2011; Manghwar et al., 2022), or ammonium (NH4+) when growing in acidic soils (Robinson et al., 2011; Wang L. et al., 2018). After uptake from the soil, plant roots assimilate NH4+ to avoid the toxicity of free ammonium, which would eliminate the transmembrane proton gradients pivotal for respiratory electron transport. Unlike ammonium, a large concentration of nitrate ions are assimilated in plant tissues and then translocated to plant shoots for assimilation. It is well established that N rates and sources significantly affect plant growth and developmental process (Irshad et al., 2008; Lu et al., 2009). Therefore, selecting the appropriate rate and method based on plant species, growth stage, soil, and environmental conditions is necessary to maximize plant growth and NUE (Marschner, 2011; Niu et al., 2011; Mendoza-Villarreal et al., 2015). Other farm management practices, including genotype selection, also significantly improved NUE. Genotype development and screening have been reported to be imperative for improving the uptake and utilization of N (Iqbal et al., 2020b).
In a recent study, Iqbal et al. (2020a) evaluated the performance of N-efficient and N-inefficient cotton genotypes supplied with nitrate and ammonium-N. They reported increased N uptake and utilization efficiency, better plant growth, chlorophyll content, and gas exchange in the N-efficient genotype when supplied with nitrate-N (Iqbal et al., 2020a). Improved root traits in nitrate-fed plants, including length, dry weight, and surface area, have been reported previously (Schortemeyer and Feil, 1996). Another study also reported better seedling growth in cotton when fed nitrate N than ammonium-fed seedlings, mainly due to better photosynthesis under increased translocation of nitrate to the photosynthesis system (Irshad et al., 2008). Ammonium application is also not recommended, as it reduces the uptake of potassium and cations to retard stomatal function and inhibit osmotic regulation, respectively (Lopes and Araus, 2006). Similarly, low NUE in cotton under ammonium application has also been attributed to inhibited ammonium metabolism and reduced protein synthesis (Pessarakli and Tucker, 1985). Chen et al. (2020a) studied different N application rates under field conditions. They reported that a moderate N application rate (240 kg ha–1) significantly increased seedling growth and cotton yield, thereby promoting NUE (Chen et al., 2020a). Similarly, in a recent study, Wang et al. (2021) reported that N application at 250 kg ha–1 considerably increased the N uptake and NUE in cotton (Wang et al., 2021).
Increasing plant nitrogen utilization and remobilization efficiency in plants
Nitrogen application rate, time of application, the N source, and other agronomic practices affect crop yield, N uptake, and its movement and metabolism within the plant, depending on the plant species (Ali, 2015; Ali et al., 2022a). Xu et al. (2012) studied the effects of different planting densities and reported that increasing planting densities in cotton enhanced N accumulation in plant parts (Xu et al., 2012). Similarly, Yao et al. (2015) reported that plant density alters N allocation. In partitioning, medium planting density results in high leaf N allocation to the P/S apparatus and its different partitioning components (Yao et al., 2015). Recently, Liu et al. (2022) demonstrated that N application significantly affects the N utilization efficiency. N at 180 kg ha–1 was applied at the flowering stage, promoting N utilization and efficiency in cotton grown under a wheat-cotton double cropping system (Liu et al., 2022).
In another study, Du et al. (2016) evaluated different cropping systems and reported reduced N accumulation rates and NUE in cotton in wheat-cotton rotations compared with monoculture (Du et al., 2016). Wei et al. (2012) studied the effects of different irrigation methods and N application rates. They reported that a moderate N application rate with drip irrigation promoted N uptake, translocation, and efficiency in cotton grown in arid regions (Wei et al., 2012). N application at 150 kg ha–1 and straw incorporation significantly improved N uptake and NUE, as reported by Wang et al. (2020). Niu et al. (2020) examined the responses of various cultivars and N application rates to NUE. They reported that CRI 69 and ZZM 1017, as N-inefficient cultivars, had increased N uptake and translocation into different parts and ultimately NUE in cotton (Niu et al., 2020). Plants inoculated with Azospirillum brasilense also show better N uptake, translocation into different tissues, and NUE (Saubidet et al., 2002).
Nitrogen use efficiency regulating enzymes and genes
Plants have evolved various mechanisms involving enzymes and activating genes to increase NUE (Table 2). However, only a few studies have discussed the key enzymes and genes directly or indirectly involved in enhancing NUE in cotton (Iqbal et al., 2020d). NUE highly depends on N uptake, utilization efficiency, and N assimilation rate (Garnett et al., 2015; Hawkesford, 2017). The involvement of the N-assimilation enzyme glutamine synthetase in promoting NUE through increased N harvest index (Brauer et al., 2011), increased number of grains (Martin et al., 2006), biomass production (Oliveira et al., 2002), and photorespiration (Hoshida et al., 2000) has been well documented in previous reports. Cai et al. (2009) documented that glutamine synthetase significantly promoted N uptake by crop plants (Cai et al., 2009). GS has been reported to enhance biomass production (Chichkova et al., 2001) and seed weight (Yamaya et al., 2002). NR and NiR are important enzymes for N assimilation; their role in enhancing NUE has been well reported in terms of enhancing dry weights (Abiko et al., 2010), nitrate content (Lillo et al., 2003), NiR activity (Crété et al., 1997), and NO2– assimilation (Takahashi et al., 2001). The GDH (NADH-GDH) has been reported to play a role in assimilating inorganic N to form glutamate by combining ammonium with 2-oxoglutarate (Fontaine et al., 2012).
TABLE 2.
Nitrogen use efficiency regulating enzymes and their specific roles in plants.
| Enzymes | Roles | References |
| Plastidic isoenzyme-GS2 | Involved in primary N assimilation |
Masclaux et al., 2000; Hirel et al., 2001 |
| Cytosolic GS isoenzyme-GS1 | Involved in the recycling of organic N | Masclaux et al., 2000; Hirel et al., 2001 |
| Glutamine synthetase | Increased N uptake and photorespiration | Habash et al., 2001 |
| Nitrate reductase and nitrite reductase | NO2– assimilation and reduced the nitrate levels | Takahashi et al., 2001; Lillo et al., 2003 |
| GS1.3 isoenzyme | Plays a putative role in controlling the yield under variable N conditions | Martin et al., 2006 |
| Alanine aminotransferase | Increased biomass and grain yield | Good et al., 2007 |
| NADH-GOGAT | Involved in glutamate synthesis to promote the growth of plants; promoted inorganic nitrogen assimilation notably in the roots | Tabuchi et al., 2007; Garnett et al., 2015 |
| Cytosolic asparagine synthetase and carbamoylphosphate synthase enzymes | Increased ammonium assimilation | Lam et al., 2003; Ranjan and Yadav, 2019 |
It is also well documented that NADH-GDH facilitates ammonium assimilation, which has an advantage over glutamine synthetase. The plastidic isoenzyme (GS2) is also involved in primary N assimilation, and the cytosolic GS isoenzyme (GS1) is involved in the recycling of organic N (Masclaux et al., 2000; Hirel et al., 2001). According to Martin et al. (2006), the GS1.3 isoenzyme plays a putative role in controlling yield under variable N conditions. NADH-GOGAT, a pyridine nucleotide-dependent GOGAT isoenzyme, is present in bundle-sheath cells and is involved in glutamate synthesis to promote plant growth (Tabuchi et al., 2007; Plett et al., 2016). The essential roles of another N-assimilation enzyme, alanine aminotransferase, in increasing biomass and seed yield have been well documented in previous reports (Good et al., 2007; Shrawat et al., 2008). According to Lam et al. (2003) and Ranjan and Yadav (2019), increased ammonium assimilation is also a result of the involvement of carbamoyl phosphate synthase and cytosolic asparagine synthetase enzymes (encoded by ASN1, ASN2, and ASN3).
Nitrogen use efficiency responsive genes manipulation
Different breeding approaches have been used in recent years to select the most appropriate traits for improving NUE. In plants, NUE is a complex trait that depends on the availability of soil N and various internal and external factors, including photosynthetic carbon fixation. NUE, the ratio of grain yield and N uptake, also varies with N application rates, where different genes are expressed in plants according to N rates (Hirel et al., 2001; Good and Beatty, 2011). According to Hirel et al. (2001), cytosolic GS genes, namely, gln1, gln2, and gln4, regulate leaf cytosolic enzyme activity. The GS gene gln4 has been reported as a housekeeping gene that controls NUE and influences yield by promoting ammonia assimilation (Hirel et al., 2001). A previous study reported enhanced NUE and yield upon overexpression of Gln1-3 and Gln1-4 (He et al., 2014). In an investigation by Castaings et al. (2009), it was concluded that the regulatory gene NLP7 was identified in Arabidopsis to regulate the nitrate content (Castaings et al., 2009). In a recent study, Cao et al. (2017) reported that regulating genes ZmNLP6 and ZmNLP8 help in nitrate signaling to increase the productivity of model plants (Cao et al., 2017). Table 3 lists the genes involved in NUE.
TABLE 3.
Genes directly or indirectly involved in nitrogen use efficiency.
| Genes | Direct/Indirect functions | References |
| GS2 | Increased the photorespiration and biomass production | Migge et al., 2000 |
| NiR | NO2– assimilation | Takahashi et al., 2001 |
| GOGAT | Improved plant biomass and seed weight | Yamaya et al., 2002 |
| AtDUR3 | Upregulation of AtDUR3 plays a putative role in organic N uptake |
Liu and Tsay, 2003
|
| GDHA | Increased ammonium assimilation, biomass, and dry weight | Mungur et al., 2006 |
| GS1 | A glutamine synthetase gene had higher grain yield, NUtE, and kernel number upon overexpression | Gallais et al., 2006 |
| AlaAT | Increased plant biomass and seed yield | Good et al., 2007 |
| AMT genes (AMT 1.1, 1.3, 1.5) | Help in ammonium uptake and its transport in plants | Yuan et al., 2007 |
| HvAlaAT | As alanine aminotransferase gene increased biomass and grain yield | Shrawat et al., 2008 |
| AtNRT1.1 | It can result in better nitrate uptake when acting as a nitrate sensor | Ho et al., 2009 |
| ENOD93–1 | Increased plant biomass and seed weight |
Bi et al., 2009
|
| NIA (Nia1 and Nia2) | As nitrate reductase genes significantly increased grain weight, dry biomass, and protein contents when overexpressed in model crops | Lillo et al., 2003; Zhao et al., 2013 |
| DOF1 | Promoted the growth and N uptake | Wang et al., 2013 |
| AGL21 | Increased plant biomass and seed weight | Yu et al., 2014 |
| Gln1-3 and Gln1-4 | Overexpression of these genes improved the yields and enhanced NUE | He et al., 2014 |
| OsAMT1;1 | As an ammonium transporter gene, helped in enhancing NUE | Ranathunge et al., 2014 |
| OsAMT1;1GS1 | Facilitate in achieving higher seed yield, NUE, and more numbers of seeds | Ranathunge et al., 2014 |
| TOND1 | Enhanced tolerance under N deficit condition | Zhang et al., 2015 |
| OsNRT2.1 | Increased biomass production | Chen et al., 2016 |
| NRT1.1B and OsNRT2.3b | As nitrate transporter genes helped in increasing the biomass production and seed yield to help in enhancing NUE | Gallais et al., 2006; Hu et al., 2015; Fan et al., 2016b |
| OsNRT2.3b and NRT1.1B | Worked as nitrate transporter genes to enhance NUE | Hu et al., 2015; Fan et al., 2016a |
| NLP7, ZmNLP6, and ZmNLP8 | NLP7 regulates the nitrate content, and ZmNLP6 and ZmNLP8 are helped in nitrate signaling to increase the productivity of model plants | Castaings et al., 2009; Cao et al., 2017 |
| GLN1 and GLN2 | GLN2 nuclear gene involved in the assimilation and re-assimilation of ammonium produced by nitrate reduction and from photorespiration, respectively. GLN1 helped in the recycling of ammonium during leaf senescence. | Bernard and Habash, 2009; Ranjan and Yadav, 2019 |
The glutamine synthetase gene GS1 and nitrate transporter genes OsNRT2.3b and NRT1.1B (NPF6.3B) have been reported to promote biomass production and seed yield, thereby enhancing NUE (Hu et al., 2015; Fan et al., 2016a). Similarly, according to Zhao et al. (2013), the expression of the nitrate reductase gene NIA promotes seed weight and protein content (Zhao et al., 2013). In another study, Shrawat et al. (2008) demonstrated that the alanine aminotransferase gene HvAlaAT significantly increases biomass production and seed yield (Shrawat et al., 2008). In addition, the ammonium transporter gene OsAMT1;1GS1 has been reported to have a higher seed yield, nitrogen use efficiency, and number of seeds (Ranathunge et al., 2014). In plants, the GLN2 nuclear gene is involved in the assimilation and re-assimilation of ammonium produced by nitrate reduction and photorespiration (Ranjan and Yadav, 2019). Similarly, according to Bernard and Habash (2009), the GLN1 gene facilitates the recycling of ammonium during leaf senescence (Bernard and Habash, 2009). The dual affinity transporter protein AtNRT1.1 (AtNPF6.3), which is expressed in root tips, can act as a nitrate sensor and improve nitrate uptake (Tsay et al., 2007; Ho et al., 2009), even under low N conditions (Liu and Tsay, 2003). Similarly, Li et al. (2006) demonstrated that AtNRT2.1 and AtNRT2.2 are involved in high-affinity nitrate uptake. Improved NUE via increased biomass has been well reported as a result of gene expression, including GS1 (Fuentes et al., 2001), GOGAt (Chichkova et al., 2001), AlaAT (Good et al., 2007), and ENOD93–1 (Bi et al., 2009) (Table 3).
Integrated approaches to enhance nitrogen use efficiency in cotton
Nitrogen use efficiency is defined as a plant’s ability to absorb nutrients from the soil, assimilate them, and utilize them to maximize crop yield (Hawkesford, 2017; Erisman et al., 2018). Increasing crop production is largely associated with fertilizer use and application rates. The optimum supply of nutrients plays a significant role in increasing crop production and NUE (Erisman et al., 2018; Fernie et al., 2020). In agricultural farming systems, N management should achieve higher crop productivity without polluting the environment (Klerkx et al., 2010). In the form of nitrate, N is a mobile nutrient that severely impacts the environment through GHGs emissions (Hristov et al., 2013). Plants obtain N from different sources, including residual N, organic matter decomposition, and biological N fixation, and it is imperative to understand the contribution of these sources to enhance NUE (Plett et al., 2020). Various approaches have been used in recent years to manage N and enhance the NUE. Soil and plant-based analysis, based on SOM, yield goal, N credit from the previous crop, manure, and irrigation water, is among the most commonly used techniques for N management in different crops (Dobermann and Cassman, 2002). According to a recent study by Guo et al. (2019), soil-based approaches are more suitable for areas with a homogeneous landscape and environmental and soil conditions with similar EC, crop productivity, and resource use efficiency (Benedetti et al., 2019; Guo et al., 2019). Variables such as slopes, soil depth, and drainage within a landscape significantly affected seed yield. Higher N fertility status has been reported in foot slopes due to higher SOM content and flow of water, while it is also well established that soils with upper landscape positions are poor with SOM (Sharma et al., 2017).
Fertilizer placement and timing
Compared to other nutrients, N is more prone to many soil transformations that occur within the soil, particularly above the soil layer, and can influence NUE (Yadav et al., 2017). Significant N losses have been reported owing to leaching, soil runoff, and volatilization. Therefore, it is necessary to choose appropriate methods of N application to maximize N availability to crop plants and NUE (Sharma et al., 2017). The broadcasting method of N application is not recommended because it causes severe losses in crop yield, and N is applied through volatilization and immobilization (Lasisi et al., 2020; Nasielski et al., 2020).
Use of tissue analysis for nitrogen management
Sensitive plants have been used for a long time as a marker of soil nutrient status. Some crop species are good markers for overall growing conditions, as they are immediately correlated with climatic conditions and soil management techniques, as discussed recently by Sharma and Bali (2017). In a recent study, Iqbal et al. (2020b) demonstrated that increased availability of N to crop plants results in N accumulation in different tissues, thereby increasing the chlorophyll content and net photosynthetic rates (Iqbal et al., 2020b). A positive correlation between chlorophyll content, corn yield, and N accumulation has been reported previously (Han et al., 2016; Ali et al., 2020).
Breeding program
Breeding for improved NUE can be attained through an assortment of different components (Cormier et al., 2013); where compensation and regulation are abundant and dependent on N application rates, plant species, and growth stage, directing obstacles to create effective NUE phenotypes (Cormier et al., 2016). However, omics-based studies offer results that enable us to focus on routes for improvement (Cormier et al., 2016). In addition, high-throughput phenotyping using high-throughput genotyping techniques will promote research on variable environments and species, which could significantly enhance their nutrients (Banerjee et al., 2020).
Conclusion
This review critically assessed various agronomic and molecular approaches for quick and efficient NUE in cotton production. Here, we describe opportunities to increase NUE through new agronomic practices such as 4R’S nutrient stewardship and recently developed molecular strategies, including manipulating metabolic pathways and transport in cotton plants. However, several concerns remain to be addressed. These could improve metabolic activities, the role of nitrogen transporters, signaling and sensing of metabolic pathways, and the relationship between sources and sinks. Ultimately, we recognize that improving NUE in cotton will be equally beneficial for the environment and growers to maximize their profits. This also involves expanding the growers’ objectives from the primary emphasis on cotton production for profit, which is crucial for livelihoods, to adding to the stewardship of the land and surrounding environment, which is essential for sustainability in cotton production. Understanding and following these motives will assist in building better education and other activities to eliminate barriers to nitrogen use efficiency in cotton farming.
Author contributions
MC, QA, LZ, and LS-L: conceptualization, visualization, and writing—original draft. LZ and LS-L: writing—review and editing, validation, conceptualization, supervision, and funding acquisition. MH, SH, MA, TJ, TM, MS, AU, WA, and SN: writing—review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the high-talent introduction and continuous training fund to LZ (Grant No: 10300000021LL05) and Discipline construction funds (Grant No: 10407000019CC2213G), supported by Zhejiang Academy of Agricultural Sciences (ZAAS) and State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products (Grant No: 10417000022CE0601G/029).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Abenavoli M. R., Longo C., Lupini A., Miller A. J., Araniti F., Mercati F., et al. (2016). Phenotyping two tomato genotypes with different nitrogen use efficiency. Plant Physiol. Biochem. 107 21–32. 10.1016/j.plaphy.2016.04.021 [DOI] [PubMed] [Google Scholar]
- Abiko T., Wakayama M., Kawakami A., Obara M., Kisaka H., Miwa T., et al. (2010). Changes in nitrogen assimilation, metabolism, and growth in transgenic rice plants expressing a fungal NADP (H)-dependent glutamate dehydrogenase (gdhA). Planta 232 299–311. 10.1007/s00425-010-1172-3 [DOI] [PubMed] [Google Scholar]
- Afridi M. Z., Jan T. M., Munsif F., Khan A., Nabi G., Ahmad M., et al. (2014). Nitrogen partitioning and translocation in wheat under fertilizer-N levels, application time and decapitation stress. J. Biol. Agric. Healthc. 13 13–19. [Google Scholar]
- Ahmar S., Gruszka D. (2022). In-Silico study of brassinosteroid signaling genes in rice provides insight into mechanisms which regulate their expression. Front. Genet. 13:953458. 10.3389/fgene.2022.953458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali A. M., Ibrahim S. M.,, Singh B. (2020). Wheat grain yield and nitrogen uptake prediction using atLeaf and GreenSeeker portable optical sensors at jointing growth stage. Inf. Process. Agric. 7 375–383. 10.1016/j.inpa.2019.09.008 [DOI] [Google Scholar]
- Ali M., Ali Q., Sohail M. A., Ashraf M. F., Saleem M. H., Hussain S., et al. (2021). Diversity and taxonomic distribution of endophytic bacterial community in the rice plant and its prospective. Int. J. Mol. Sci. 22:10165. 10.3390/ijms221810165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali Q., Ahmar S., Sohail M. A., Kamran M., Ali M., Saleem M. H., et al. (2021). Research advances and applications of biosensing technology for the diagnosis of pathogens in sustainable agriculture. Environ. Sci. Pollut. Res. 28 9002–9019. 10.1007/s11356-021-12419-6 [DOI] [PubMed] [Google Scholar]
- Ali N. (2015). Nitrogen utilization features in cotton crop. Am. J. Plant Sci. 6:55897. 10.4236/ajps.2015.67105 [DOI] [Google Scholar]
- Ali Q., Ayaz M., Yu C., Wang Y., Gu Q., Wu H., et al. (2022a). Cadmium tolerant microbial strains possess different mechanisms for cadmium biosorption and immobilization in rice seedlings. Chemosphere 303:135206. 10.1016/j.chemosphere.2022.135206 [DOI] [PubMed] [Google Scholar]
- Ali Q., Yu C., Hussain A., Ali M., Ahmar S., Sohail M. A., et al. (2022b). Genome engineering technology for durable disease resistance: Recent progress and future outlooks for sustainable agriculture. Front. Plant Sci. 13:860281. 10.3389/fpls.2022.860281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali Q., Zheng H., Rao M. J., Ali M., Hussain A., Saleem M. H., et al. (2022c). Advances, limitations, and prospects of biosensing technology for detecting phytopathogenic bacteria. Chemosphere 296:133773. 10.1016/j.chemosphere.2022.133773 [DOI] [PubMed] [Google Scholar]
- Ameziane R., Bernhard K., Lightfoot D. (2000). Expression of the bacterial gdhA gene encoding a NADPH glutamate dehydrogenase in tobacco affects plant growth and development. Plant Soil 221 47–57. 10.1023/A:1004794000267 [DOI] [Google Scholar]
- Anthony D., Armytage P. (2008). Cotton Catchment Communities CRC Annual Report 2007-2008. Available online at: http://www.insidecotton.com/xmlui/handle/1/77 [Google Scholar]
- Araus V., Vidal E. A., Puelma T., Alamos S., Mieulet D., Guiderdoni E., et al. (2016). Members of BTB gene family of scaffold proteins suppress nitrate uptake and nitrogen use efficiency. Plant Physiol. 171 1523–1532. 10.1104/pp.15.01731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayaz M., Ali Q., Farzand A., Khan A. R., Ling H., Gao X. (2021). Nematicidal volatiles from Bacillus atrophaeus gbsc56 promote growth and stimulate induced systemic resistance in tomato against Meloidogyne incognita. Int. J. Mol. Sci. 22:5049. 10.3390/ijms22095049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee B. P., Joshi S., Thoday-Kennedy E., Pasam R. K., Tibbits J., Hayden M., et al. (2020). High-throughput phenotyping using digital and hyperspectral imaging-derived biomarkers for genotypic nitrogen response. J. Exp. Bot. 71 4604–4615. 10.1093/jxb/eraa143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baslam M., Mitsui T., Sueyoshi K., Ohyama T. (2020). Recent advances in carbon and nitrogen metabolism in C3 plants. Int. J. Mol. Sci. 22:318. 10.3390/ijms22010318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti I., Branca G., Zucaro R. (2019). Evaluating input use efficiency in agriculture through a stochastic frontier production: An application on a case study in Apulia (Italy). J. Clean. Prod. 236:117609. 10.1016/j.jclepro.2019.117609 [DOI] [Google Scholar]
- Bernard S. M., Habash D. Z. (2009). The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New Phytol. 182 608–620. 10.1111/j.1469-8137.2009.02823.x [DOI] [PubMed] [Google Scholar]
- Bi Y., Kant S., Clark J., Gidda S., Ming F., Xu J., et al. (2009). Increased nitrogen-use efficiency in transgenic rice plants over-expressing a nitrogen-responsive early nodulin gene identified from rice expression profiling. Plant Cell Environ. 32 1749–1760. 10.1111/j.1365-3040.2009.02032.x [DOI] [PubMed] [Google Scholar]
- Bieker S., Zentgraf U. (2013). Plant senescence and nitrogen mobilization and signaling. Senescence Senescence Relat. Disord. 104 53–83. 10.5772/54392 [DOI] [Google Scholar]
- Bittsánszky A., Pilinszky K., Gyulai G., Komives T. (2015). Overcoming ammonium toxicity. Plant Sci. 231 184–190. 10.1016/j.plantsci.2014.12.005 [DOI] [PubMed] [Google Scholar]
- Bock B. R., Kissel D. E. (1988). Ammonia volatilization from urea fertilizers. Knoxville, TN: Tennessee Valley Authority. [Google Scholar]
- Bouguyon E., Brun F., Meynard D., Kubeš M., Pervent M., Leran S., et al. (2015). Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nat. Plants 1:15015. 10.1038/nplants.2015.15 [DOI] [PubMed] [Google Scholar]
- Bouguyon E., Gojon A., Nacry P. (2012). Nitrate sensing and signaling in plants. Semin. Cell Dev. Biol. 23 648–654. 10.1016/j.semcdb.2012.01.004 [DOI] [PubMed] [Google Scholar]
- Bouma J., Finke P. A. (1993). “Origin and nature of soil resource variability,” in Proceedings of soil specific crop management: A workshop on research and development issues, eds Robert P. C., Rust R. H., Larson W. E. (Hoboken, NJ: Wiley; ), 1–13. 10.2134/1993.soilspecificcrop.c1 [DOI] [Google Scholar]
- Brauer E. K., Rochon A., Bi Y., Bozzo G. G., Rothstein S. J., Shelp B. J. (2011). Reappraisal of nitrogen use efficiency in rice overexpressing glutamine synthetase1. Physiol. Plant. 141 361–372. 10.1111/j.1399-3054.2011.01443.x [DOI] [PubMed] [Google Scholar]
- Breeze E., Harrison E., McHattie S., Hughes L., Hickman R., Hill C., et al. (2011). High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23 873–894. 10.1105/tpc.111.083345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronson K. F. (2008). Nitrogen use efficiency of cotton varies with irrigation system. Better Crop Plant Food 92 20–22. 10.2134/jeq2017.10.0389 [DOI] [PubMed] [Google Scholar]
- Bronson K. F., Keeling J. W., Booker J. D., Chua T. T., Wheeler T. A., Boman R. K., et al. (2003). Influence of landscape position, soil series, and phosphorus fertilizer on cotton lint yield. Agron. J. 95 949–957. 10.2134/agronj2003.9490 [DOI] [Google Scholar]
- Bruulsema T. (2018). Managing nutrients to mitigate soil pollution. Environ. Pollut. 243 1602–1605. 10.1016/j.envpol.2018.09.132 [DOI] [PubMed] [Google Scholar]
- Bruulsema T., Lemunyon J., Herz B. (2009). Know your fertilizer rights. Crop Soils 42 13–18. [Google Scholar]
- Burton D. (2018). A review of the recent scientific literature documenting the impact of 4R management on N2O emissions relevant to a canadian context. Available Online at: https://fertilizercanada.ca/wp-content/uploads/2018/08/NERP-Science-Review-Paper-.pdf (assessed July 8, 2022). [Google Scholar]
- Cai H., Zhou Y., Xiao J., Li X., Zhang Q., Lian X. (2009). Overexpressed glutamine synthetase gene modifies nitrogen metabolism and abiotic stress responses in rice. Plant Cell Rep. 28 527–537. 10.1007/s00299-008-0665-z [DOI] [PubMed] [Google Scholar]
- Cameron K. C., Di H. J., Moir J. L. (2013). Nitrogen losses from the soil/plant system: A review. Ann. Appl. Biol. 162 145–173. 10.1111/j.1469-8137.2012.04300.x [DOI] [PubMed] [Google Scholar]
- Cao H., Qi S., Sun M., Li Z., Yang Y., Crawford N. M., et al. (2017). Overexpression of the maize ZmNLP6 and ZmNLP8 can complement the Arabidopsis nitrate regulatory mutant nlp7 by restoring nitrate signaling and assimilation. Front. Plant Sci. 8:1703. 10.3389/fpls.2017.01703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho H. G., Lopes-Cardoso I. A., Lima L. M., Melo P. M., Cullimore J. V. (2003). Nodule-specific modulation of glutamine synthetase in transgenic Medicago truncatula leads to inverse alterations in asparagine synthetase expression. Plant Physiol. 133 243–252. 10.1104/pp.102.017830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassman K. G., Dobermann A. R., Walters D. T., Yang H. (2003). Meeting cereal demand while protecting natural resources and improving environmental quality. Annu. Rev. Environ. Resour. 28 315–358. 10.1146/annurev.energy.28.040202.122858 [DOI] [Google Scholar]
- Castaings L., Camargo A., Pocholle D., Gaudon V., Texier Y., Boutet-Mercey S., et al. (2009). The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 57 426–435. 10.1111/j.1365-313X.2008.03695.x [DOI] [PubMed] [Google Scholar]
- Castro M. S., Peterjohn W. T., Melillo J. M., Steudler P. A., Gholz H. L., Lewis D. (1994). Effects of nitrogen fertilization on the fluxes of N2O, CH4, and CO2 from soils in a Florida slash pine plantation. Can. J. For. Res. 24 9–13. 10.1139/x94-002 [DOI] [Google Scholar]
- Cavigelli M. A., Del Grosso S. J., Liebig M. A., Snyder C. S., Fixen P. E., Venterea R. T., et al. (2012). US agricultural nitrous oxide emissions: Context, status, and trends. Front. Ecol. Environ. 10 537–546. 10.1890/120054 [DOI] [Google Scholar]
- Chaffei C., Pageau K., Suzuki A., Gouia H., Ghorbel M. H., Masclaux-Daubresse C. (2004). Cadmium toxicity induced changes in nitrogen management in Lycopersicon esculentum leading to a metabolic safeguard through an amino acid storage strategy. Plant Cell Physiol. 45 1681–1693. 10.1093/pcp/pch192 [DOI] [PubMed] [Google Scholar]
- Chen J., Fan X., Qian K., Zhang Y., Song M., Liu Y., et al. (2017). pOsNAR 2.1: Os NAR 2.1 expression enhances nitrogen uptake efficiency and grain yield in transgenic rice plants. Plant Biotechnol. J. 15 1273–1283. 10.1111/pbi.12714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Liu L., Wang Z., Zhang Y., Sun H., Song S., et al. (2020a). Nitrogen fertilization increases root growth and coordinates the root–shoot relationship in cotton. Front. Plant Sci. 11:880. 10.3389/fpls.2020.00880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Liu X., Liu S., Fan X., Zhao L., Song M., et al. (2020b). Co-Overexpression of OsNAR2.1 and OsNRT2.3a increased agronomic nitrogen use efficiency in transgenic rice plants. Front. Plant Sci. 11:1245. 10.3389/fpls.2020.01245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K.-E., Chen H.-Y., Tseng C.-S., Tsay Y.-F. (2020c). Improving nitrogen use efficiency by manipulating nitrate remobilization in plants. Nat. Plants 6 1126–1135. 10.1038/s41477-020-00758-0 [DOI] [PubMed] [Google Scholar]
- Chen J., Zhang Y., Tan Y., Zhang M., Zhu L., Xu G., et al. (2016). Agronomic nitrogen-use efficiency of rice can be increased by driving OsNRT2.1 expression with the OsNAR2.1 promoter. Plant Biotechnol. J. 14 1705–1715. 10.1111/pbi.12531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Soulay F., Saudemont B., Elmayan T., Marmagne A., Masclaux-Daubresse C. (2019). Overexpression of ATG8 in Arabidopsis stimulates autophagic activity and increases nitrogen remobilization efficiency and grain filling. Plant Cell Physiol. 60 343–352. 10.1093/pcp/pcy214 [DOI] [PubMed] [Google Scholar]
- Chen W., Hou Z., Wu L., Liang Y., Wei C. (2010). Effects of salinity and nitrogen on cotton growth in arid environment. Plant Soil 326 61–73. 10.13227/j.hjkx.201910137 [DOI] [PubMed] [Google Scholar]
- Chichkova S., Arellano J., Vance C. P., Hernández G. (2001). Transgenic tobacco plants that overexpress alfalfa NADH-glutamate synthase have higher carbon and nitrogen content. J. Exp. Bot. 52 2079–2087. 10.1093/jexbot/52.364.2079 [DOI] [PubMed] [Google Scholar]
- Chung T., Phillips A. R., Vierstra R. D. (2010). ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J. 62 483–493. 10.1111/j.1365-313X.2010.04166.x [DOI] [PubMed] [Google Scholar]
- Cormier F., Faure S., Dubreuil P., Heumez E., Beauchêne K., Lafarge S., et al. (2013). A multi-environmental study of recent breeding progress on nitrogen use efficiency in wheat (Triticum aestivum L.). Theor. Appl. Genet. 126 3035–3048. 10.1007/s00122-013-2191-9 [DOI] [PubMed] [Google Scholar]
- Cormier F., Foulkes J., Hirel B., Gouache D., Moënne-Loccoz Y., Le Gouis J. (2016). Breeding for increased nitrogen-use efficiency: A review for wheat (T. aestivum L.). Plant Breed. 135 255–278. 10.1111/pbr.12371 [DOI] [Google Scholar]
- Crawford N. M. (2000). “Nitrogen and sulfur,” in Biochemistry & molecular biology of plants, eds Buchanan B. B., Gruissem W., Jones R. L. (Rockville, MD: American Society of Plant Biologists; ), 824–849. [Google Scholar]
- Crawford N. M., Forde B. G. (2002). Molecular and developmental biology of inorganic nitrogen nutrition. Arabidopsis Book 1:e0011. 10.1199/tab.0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crété P., Caboche M., Meyer C. (1997). Nitrite reductase expression is regulated at the post-transcriptional level by the nitrogen source in Nicotiana plumbaginifolia and Arabidopsis thaliana. Plant J. 11 625–634. 10.1046/j.1365-313x.1997.11040625.x [DOI] [PubMed] [Google Scholar]
- Cui Z., Zhang F., Chen X., Miao Y., Li J., Shi L., et al. (2008). On-farm evaluation of an in-season nitrogen management strategy based on soil Nmin test. Field Crops Res. 105 48–55. 10.1016/j.fcr.2007.07.008 [DOI] [Google Scholar]
- Darwish T. M., Atallah T. W., Hajhasan S., Haidar A. (2006). Nitrogen and water use efficiency of fertigated processing potato. Agric. Water Manag. 85 95–104. 10.1016/j.agwat.2006.03.012 [DOI] [Google Scholar]
- David L. C., Berquin P., Kanno Y., Seo M., Daniel-Vedele F., Ferrario-Méry S. (2016). N availability modulates the role of NPF3. 1, a gibberellin transporter, in GA-mediated phenotypes in Arabidopsis. Planta 244 1315–1328. 10.1007/s00425-016-2588-1 [DOI] [PubMed] [Google Scholar]
- Diaz C., Lemaître T., Christ A., Azzopardi M., Kato Y., Sato F., et al. (2008). Nitrogen recycling and remobilization are differentially controlled by leaf senescence and development stage in Arabidopsis under low nitrogen nutrition. Plant Physiol. 147 1437–1449. 10.1104/pp.108.119040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W., He P., Zhang J., Liu Y., Xu X., Ullah S., et al. (2020). Optimizing rates and sources of nutrient input to mitigate nitrogen, phosphorus, and carbon losses from rice paddies. J. Clean. Prod. 256:120603. 10.1016/j.jclepro.2020.120603 [DOI] [Google Scholar]
- Dobermann A., Cassman K. G. (2002). Plant nutrient management for enhanced productivity in intensive grain production systems of the United States and Asia. Plant Soil 247 153–175. 10.1023/A:1021197525875 [DOI] [Google Scholar]
- Du X., Chen B., Zhang Y., Zhao W., Shen T., Zhou Z., et al. (2016). Nitrogen use efficiency of cotton (Gossypium hirsutum L.) as influenced by wheat–cotton cropping systems. Eur. J. Agron. 75 72–79. 10.1016/j.eja.2016.01.001 [DOI] [Google Scholar]
- Egami T., Wakayama M., Aoki N., Sasaki H., Kisaka H., Miwa T., et al. (2012). The effects of introduction of a fungal glutamate dehydrogenase gene (gdhA) on the photosynthetic rates, biomass, carbon and nitrogen contents in transgenic potato. Plant Biotechnol. 29 57–64. 10.5511/plantbiotechnology.12.0127a [DOI] [Google Scholar]
- Engel R., Jones C., Wallander R. (2011). Ammonia volatilization from urea and mitigation by NBPT following surface application to cold soils. Soil Sci. Soc. Am. J. 75 2348–2357. 10.2136/sssaj2011.0229 [DOI] [Google Scholar]
- Enggrob K. L., Jakobsen C. M., Pedersen I. F., Rasmussen J. (2019). Newly depolymerized large organic N contributes directly to amino acid uptake in young maize plants. New Phytol. 224 689–699. 10.1111/nph.16070 [DOI] [PubMed] [Google Scholar]
- Erisman J. W., Leach A., Bleeker A., Atwell B., Cattaneo L., Galloway J. (2018). An integrated approach to a nitrogen use efficiency (NUE) indicator for the food production–consumption chain. Sustainability 10:925. 10.3390/su10040925 [DOI] [Google Scholar]
- Fan X., Feng H., Tan Y., Xu Y., Miao Q., Xu G. (2016a). A putative 6-transmembrane nitrate transporter OsNRT1. 1b plays a key role in rice under low nitrogen. J. Integr. Plant Biol. 58 590–599. 10.1111/jipb.12382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Tang Z., Tan Y., Zhang Y., Luo B., Yang M., et al. (2016b). Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proc. Natl. Acad. Sci. U.S.A. 113 7118–7123. 10.1073/pnas.1525184113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Naz M., Fan X., Xuan W., Miller A. J., Xu G. (2017). Plant nitrate transporters: From gene function to application. J. Exp. Bot. 68 2463–2475. 10.1093/jxb/erx011 [DOI] [PubMed] [Google Scholar]
- Fang Z., Xia K., Yang X., Grotemeyer M. S., Meier S., Rentsch D., et al. (2013). Altered expression of the PTR/NRT 1 homologue Os PTR 9 affects nitrogen utilization efficiency, growth and grain yield in rice. Plant Biotechnol. J. 11 446–458. 10.1111/pbi.12031 [DOI] [PubMed] [Google Scholar]
- Farré G., Blancquaert D., Capell T., Van Der Straeten D., Christou P., Zhu C. (2014). Engineering complex metabolic pathways in plants. Annu. Rev. Plant Biol. 65 187–223. 10.1146/annurev-arplant-050213-035825 [DOI] [PubMed] [Google Scholar]
- Feng H., Li B., Zhi Y., Chen J., Li R., Xia X., et al. (2017). Overexpression of the nitrate transporter, OsNRT2. 3b, improves rice phosphorus uptake and translocation. Plant Cell Rep. 36 1287–1296. 10.1007/s00299-017-2153-9 [DOI] [PubMed] [Google Scholar]
- Ferguson R. B., Kissel D. E., Koelliker J. K., Basel W. (1984). Ammonia volatilization from surface-applied urea: Effect of hydrogen ion buffering capacity. Soil Sci. Soc. Am. J. 48 578–582. 10.2136/sssaj1984.03615995004800030022x [DOI] [Google Scholar]
- Fernie A. R., Bachem C. W. B., Helariutta Y., Neuhaus H. E., Prat S., Ruan Y.-L., et al. (2020). Synchronization of developmental, molecular and metabolic aspects of source–sink interactions. Nat. Plants 6 55–66. 10.1038/s41477-020-0590-x [DOI] [PubMed] [Google Scholar]
- Fiaz S., Wang X., Khan S. A., Ahmar S., Noor M. A., Riaz A., et al. (2021). Novel plant breeding techniques to advance nitrogen use efficiency in rice: A review. GM Crops Food 12 627–646. 10.1080/21645698.2021.1921545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer W., Loo D. D. F., Koch W., Ludewig U., Boorer K. J., Tegeder M., et al. (2002). Low and high affinity amino acid H+-cotransporters for cellular import of neutral and charged amino acids. Plant J. 29 717–731. 10.1046/j.1365-313x.2002.01248.x [DOI] [PubMed] [Google Scholar]
- Fixen P. E. (2020). A brief account of the genesis of 4R nutrient stewardship. Agron. J. 112 4511–4518. 10.1002/agj2.20315 [DOI] [Google Scholar]
- Follett R. F. (2001). “Nitrogen transformation and transport processes,” in Nitrogen in the environment: Sources, problems, and solutions, eds Follett R. F., Hatfield J. (Amsterdam: Elsevier Science Publishers; ), 17–44. 10.1016/B978-044450486-9/50004-2 [DOI] [Google Scholar]
- Fontaine J.-X., Tercé-Laforgue T., Armengaud P., Clément G., Renou J.-P., Pelletier S., et al. (2012). Characterization of a NADH-dependent glutamate dehydrogenase mutant of Arabidopsis demonstrates the key role of this enzyme in root carbon and nitrogen metabolism. Plant Cell 24 4044–4065. 10.1105/tpc.112.103689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler D., Coyle M., Skiba U., Sutton M. A., Cape J. N., Reis S., et al. (2013). The global nitrogen cycle in the twenty-first century. Philos. Trans. R. Soc. B Biol. Sci. 368:20130164. 10.1098/rstb.2013.0164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco H. C. J., Otto R., Faroni C. E., Vitti A. C., de Oliveira E. C. A., Trivelin P. C. O. (2011). Nitrogen in sugarcane derived from fertilizer under Brazilian field conditions. Field Crops Res. 121 29–41. 10.1016/j.fcr.2010.11.011 [DOI] [Google Scholar]
- Fu Y., Yi H., Bao J., Gong J. (2015). LeNRT2. 3 functions in nitrate acquisition and long-distance transport in tomato. FEBS Lett. 589 1072–1079. 10.1016/j.febslet.2015.03.016 [DOI] [PubMed] [Google Scholar]
- Fuentes S. I., Allen D. J., Ortiz-Lopez A., Hernández G. (2001). Over-expression of cytosolic glutamine synthetase increases photosynthesis and growth at low nitrogen concentrations. J. Exp. Bot. 52 1071–1081. 10.1093/jexbot/52.358.1071 [DOI] [PubMed] [Google Scholar]
- Gallais A., Coque M., Quilléré I., Prioul J., Hirel B. (2006). Modelling postsilking nitrogen fluxes in maize (Zea mays) using 15N-labelling field experiments. New Phytol. 172 696–707. 10.1111/j.1469-8137.2006.01890.x [DOI] [PubMed] [Google Scholar]
- Gao Z., Wang Y., Chen G., Zhang A., Yang S., Shang L., et al. (2019). The indica nitrate reductase gene OsNR2 allele enhances rice yield potential and nitrogen use efficiency. Nat. Commun. 10:5207. 10.1038/s41467-019-13110-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett T., Plett D., Conn V., Conn S., Rabie H., Rafalski J. A., et al. (2015). Variation for N uptake system in maize: Genotypic response to N supply. Front. Plant Sci. 6:936. 10.3389/fpls.2015.00936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill R. A., Ahmar S., Ali B., Saleem M. H., Khan M. U., Zhou W., et al. (2021). The role of membrane transporters in plant growth and development, and abiotic stress tolerance. Int. J. Mol. Sci. 22:12792. 10.3390/ijms222312792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass A. D. M., Britto D. T., Kaiser B. N., Kinghorn J. R., Kronzucker H. J., Kumar A., et al. (2002). The regulation of nitrate and ammonium transport systems in plants. J. Exp. Bot. 53 855–864. 10.1093/jexbot/53.370.855 [DOI] [PubMed] [Google Scholar]
- Good A. G., Beatty P. H. (2011). Fertilizing nature: A tragedy of excess in the commons. PLoS Biol. 9:e1001124. 10.1371/journal.pbio.1001124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good A. G., Johnson S. J., De Pauw M., Carroll R. T., Savidov N., Vidmar J., et al. (2007). Engineering nitrogen use efficiency with alanine aminotransferase. Botany 85 252–262. 10.1139/B07-019 [DOI] [Google Scholar]
- Gowri G., Kenis J. D., Ingemarsson B., Redinbaugh M. G., Campbell W. H. (1992). Nitrate reductase transcript is expressed in the primary response of maize to environmental nitrate. Plant Mol. Biol. 18 55–64. 10.1007/BF00018456 [DOI] [PubMed] [Google Scholar]
- Gu Q., Qiao J., Wang R., Lu J., Wang Z., Li P., et al. (2022). The role of pyoluteorin from Pseudomonas protegens Pf-5 in suppressing the growth and pathogenicity of Pantoea ananatis on maize. Int. J. Mol. Sci. 23:6431. 10.3390/ijms23126431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiboileau A., Sormani R., Meyer C., Masclaux-Daubresse C. (2010). Senescence and death of plant organs: Nutrient recycling and developmental regulation. C. R. Biol. 333 382–391. 10.1016/j.crvi.2010.01.016 [DOI] [PubMed] [Google Scholar]
- Guo X., Dias D., Pan Q. (2019). Probabilistic stability analysis of an embankment dam considering soil spatial variability. Comput. Geotech. 113:103093. 10.1016/j.compgeo.2019.103093 [DOI] [Google Scholar]
- Habash D. Z., Massiah A. J., Rong H. L., Wallsgrove R. M., Leigh R. A. (2001). The role of cytosolic glutamine synthetase in wheat. Ann. Appl. Biol. 138 83–89. [Google Scholar]
- Halkier B. A., Gershenzon J. (2006). Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57 303–333. 10.1146/annurev.arplant.57.032905.105228 [DOI] [PubMed] [Google Scholar]
- Halvorson A. D., Snyder C. S., Blaylock A. D., Del Grosso S. J. (2014). Enhanced-efficiency nitrogen fertilizers: Potential role in nitrous oxide emission mitigation. Agron. J. 106 715–722. 10.2134/agronj2013.0081 [DOI] [Google Scholar]
- Han M., Wong J., Su T., Beatty P. H., Good A. G. (2016). Identification of nitrogen use efficiency genes in barley: Searching for QTLs controlling complex physiological traits. Front. Plant Sci. 7:1587. 10.3389/fpls.2016.01587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havé M., Marmagne A., Chardon F., Masclaux-Daubresse C. (2017). Nitrogen remobilization during leaf senescence: lessons from Arabidopsis to crops. J. Exp. Bot. 68 2513–2529. 10.1093/jxb/erw365 [DOI] [PubMed] [Google Scholar]
- Hawkesford M. J. (2012). “Improving nutrient use efficiency in crops,” in eLS, (Chichester: John Wiley & Sons; ). 10.1002/9780470015902.a0023734 [DOI] [Google Scholar]
- Hawkesford M. J. (2017). Genetic variation in traits for nitrogen use efficiency in wheat. J. Exp. Bot. 68 2627–2632. 10.1093/jxb/erx079 [DOI] [PubMed] [Google Scholar]
- He C., Liu C., Liu Q., Gao X., Li N., Zhang J., et al. (2014). Over-expression of glutamine synthetase genes Gln1-3/Gln1-4 improved nitrogen assimilation and maize yields. Maydica 59 250–256. [Google Scholar]
- Herridge D. F., Peoples M. B., Boddey R. M. (2008). Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil 311 1–18. 10.1007/s11104-008-9668-3 [DOI] [Google Scholar]
- Hirel B., Bertin P., Quilleré I., Bourdoncle W., Attagnant C., Dellay C., et al. (2001). Towards a better understanding of the genetic and physiological basis for nitrogen use efficiency in maize. Plant Physiol. 125 1258–1270. 10.1104/pp.125.3.1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C.-H., Lin S.-H., Hu H.-C., Tsay Y.-F. (2009). CHL1 functions as a nitrate sensor in plants. Cell 138 1184–1194. 10.1016/j.cell.2009.07.004 [DOI] [PubMed] [Google Scholar]
- Hocking M. D., Reynolds J. D. (2012). Nitrogen uptake by plants subsidized by Pacific salmon carcasses: A hierarchical experiment. Can. J. For. Res. 42 908–917. 10.1139/x2012-045 [DOI] [Google Scholar]
- Hoshida H., Tanaka Y., Hibino T., Hayashi Y., Tanaka A., Takabe T., et al. (2000). Enhanced tolerance to salt stress in transgenic rice that overexpresses chloroplast glutamine synthetase. Plant Mol. Biol. 43 103–111. 10.1023/a:1006408712416 [DOI] [PubMed] [Google Scholar]
- Hristov A. N., Oh J., Firkins J. L., Dijkstra J., Kebreab E., Waghorn G., et al. (2013). Special topics—Mitigation of methane and nitrous oxide emissions from animal operations: I. A review of enteric methane mitigation options. J. Anim. Sci. 91 5045–5069. 10.2527/jas.2013-6583 [DOI] [PubMed] [Google Scholar]
- Hu B., Wang W., Ou S., Tang J., Li H., Che R., et al. (2015). Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 47 834–838. 10.1038/ng.3337 [DOI] [PubMed] [Google Scholar]
- Hu H.-C., Wang Y.-Y., Tsay Y.-F. (2009). AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J. 57 264–278. 10.1111/j.1365-313X.2008.03685.x [DOI] [PubMed] [Google Scholar]
- Ihtisham M., Liu S., Shahid M. O., Khan N., Lv B., Sarraf M., et al. (2020). The optimized N, P, and K fertilization for bermudagrass integrated turf performance during the establishment and its importance for the sustainable management of urban green spaces. Sustainability 12:10294. 10.3390/su122410294 [DOI] [Google Scholar]
- Iqbal A., Dong Q., Wang X., Gui H., Zhang H., Zhang X., et al. (2020a). High nitrogen enhance drought tolerance in cotton through antioxidant enzymatic activities, nitrogen metabolism and osmotic adjustment. Plants 9:178. 10.3390/plants9020178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal A., Dong Q., Wang X., Gui H., Zhang H., Zhang X., et al. (2020b). Variations in nitrogen metabolism are closely linked with nitrogen uptake and utilization efficiency in cotton genotypes under various nitrogen supplies. Plants 9:250. 10.3390/plants9020250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal A., Qiang D., Alamzeb M., Xiangru W., Huiping G., Hengheng Z., et al. (2020c). Untangling the molecular mechanisms and functions of nitrate to improve nitrogen use efficiency. J. Sci. Food Agric. 100 904–914. 10.1002/jsfa.10085 [DOI] [PubMed] [Google Scholar]
- Iqbal A., Qiang D., Zhun W., Xiangru W., Huiping G., Hengheng Z., et al. (2020d). Growth and nitrogen metabolism are associated with nitrogen-use efficiency in cotton genotypes. Plant Physiol. Biochem. 149 61–74. 10.1016/j.plaphy.2020.02.002 [DOI] [PubMed] [Google Scholar]
- Irshad M., Eneji A. E., Yasuda H. (2008). Comparative effect of nitrogen sources on maize under saline and non-saline conditions. J. Agron. Crop Sci. 194 256–261. 10.1111/j.1439-037X.2008.00310.x [DOI] [Google Scholar]
- Johnson R., Vishwakarma K., Hossen M. S., Kumar V., Shackira A. M., Puthur J. T., et al. (2022). Potassium in plants: Growth regulation, signaling, and environmental stress tolerance. Plant Physiol. Biochem. 172 56–69. 10.1016/j.plaphy.2022.01.001 [DOI] [PubMed] [Google Scholar]
- Johnson R. M., Bradow J. M. (2000). “Potential for precision management of cotton fiber quality,” in Proceedings of the 5th international conference on precision agriculture, (Bloomington, MN: American Society of Agronomy; ), 1–10. [Google Scholar]
- Jones C. (2006). Ammonia Volatilization: Process Ammonia Volatilization: Process, Amounts, and Yield Effects. In MABA/MGEA 2006 Convention. Available online at: https://landresources.montana.edu/soilfertility/ [Google Scholar]
- Kaiwen P. A. N., Pimin G., Jinchuang W., Yanjie W., Chenggang L. I. U., Wei L. I., et al. (2015). Applications of nitrate and ammonium fertilizers alter soil nematode food webs in a continuous cucumber cropping system in Southwestern Sichuan, China. Eurasian J. Soil Sci. 4 287–300. 10.18393/ejss.2015.4.287-300 [DOI] [Google Scholar]
- Kant S. (2018). Understanding nitrate uptake, signaling and remobilisation for improving plant nitrogen use efficiency. Semin. Cell Dev. Biol. 74 89–96. 10.1016/j.semcdb.2017.08.034 [DOI] [PubMed] [Google Scholar]
- Khan A., Tan D. K. Y., Afridi M. Z., Luo H., Tung S. A., Ajab M., et al. (2017a). Nitrogen fertility and abiotic stresses management in cotton crop: A review. Environ. Sci. Pollut. Res. 24 14551–14566. 10.1007/s11356-017-8920-x [DOI] [PubMed] [Google Scholar]
- Khan A., Tan D. K. Y., Munsif F., Afridi M. Z., Shah F., Wei F., et al. (2017b). Nitrogen nutrition in cotton and control strategies for greenhouse gas emissions: A review. Environ. Sci. Pollut. Res. 24 23471–23487. 10.1007/s11356-017-0131-y [DOI] [PubMed] [Google Scholar]
- Kiba T., Feria-Bourrellier A.-B., Lafouge F., Lezhneva L., Boutet-Mercey S., Orsel M., et al. (2012). The Arabidopsis nitrate transporter NRT2. 4 plays a double role in roots and shoots of nitrogen-starved plants. Plant Cell 24 245–258. 10.1105/tpc.111.092221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klerkx L., Aarts N., Leeuwis C. (2010). Adaptive management in agricultural innovation systems: The interactions between innovation networks and their environment. Agric. Syst. 103 390–400. 10.1016/j.agsy.2010.03.012 [DOI] [Google Scholar]
- Koch W., Kwart M., Laubner M., Heineke D., Stransky H., Frommer W. B., et al. (2003). Reduced amino acid content in transgenic potato tubers due to antisense inhibition of the leaf H+/amino acid symporter StAAP1. Plant J. 33 211–220. 10.1046/j.1365-313x.2003.01618.x [DOI] [PubMed] [Google Scholar]
- Kong M., Liang J., Ali Q., Wen W., Wu H., Gao X., et al. (2021a). 5-methoxyindole, a chemical homolog of melatonin, adversely affects the phytopathogenic fungus Fusarium graminearum. Int. J. Mol. Sci. 22:10991. 10.3390/ijms222010991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong M., Sheng T., Liang J., Ali Q., Gu Q., Wu H., et al. (2021b). Melatonin and its homologs induce immune responses via receptors trP47363-trP13076 in Nicotiana benthamiana. Front. Plant Sci. 12:691835. 10.3389/fpls.2021.691835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G. (2017). Nitrate signalling: Calcium bridges the nitrate gap. Nat. Plants 3:17095. 10.1038/nplants.2017.95 [DOI] [PubMed] [Google Scholar]
- Krouk G., Mirowski P., LeCun Y., Shasha D. E., Coruzzi G. M. (2010). Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biol. 11:R123. 10.1186/gb-2010-11-12-r123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N., Srivastava P., Vishwakarma K., Kumar R., Kuppala H., Maheshwari S. K., et al. (2020). “The rhizobium–plant symbiosis: State of the art,” in Plant microbe symbiosis, eds Varma A., Tripathi S., Prasad R. (Cham: Springer; ), 1–20. 10.1128/mr.59.1.124-142.1995 [DOI] [Google Scholar]
- Lal R. (2010). Beyond Copenhagen: Mitigating climate change and achieving food security through soil carbon sequestration. Food Secur. 2 169–177. 10.1007/s12571-010-0060-9 [DOI] [Google Scholar]
- Lam H., Wong H., Chan K., Yam L., Chen C., Chow M., et al. (2003). Overexpression of the ASN1 gene enhances nitrogen status in seeds of Arabidopsis. Plant Physiol. 132 926–935. 10.1104/pp.103.020123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasisi A. A., Akinremi O. O., Zhang Q., Kumaragamage D. (2020). Efficiency of fall versus spring applied urea-based fertilizers treated with urease and nitrification inhibitors I. Ammonia volatilization and mitigation by NBPT. Soil Sci. Soc. Am. J. 84 949–962. 10.1002/saj2.20062 [DOI] [Google Scholar]
- Lawlor D. W. (2002). Carbon and nitrogen assimilation in relation to yield: Mechanisms are the key to understanding production systems. J. Exp. Bot. 53 773–787. 10.1093/jexbot/53.370.773 [DOI] [PubMed] [Google Scholar]
- Lea P. J., Miflin B. J. (2011). Nitrogen assimilation and its relevance to crop improvement. Annu. Plant Rev. 42 1–40. 10.1002/9781444328608.ch1 [DOI] [Google Scholar]
- Léran S., Varala K., Boyer J.-C., Chiurazzi M., Crawford N., Daniel-Vedele F., et al. (2014). A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci. 19 5–9. 10.1016/j.tplants.2013.08.008 [DOI] [PubMed] [Google Scholar]
- Lezhneva L., Kiba T., Feria-Bourrellier A.-B., Lafouge F., Boutet-Mercey S., Zoufan P., et al. (2014). The Arabidopsis nitrate transporter NRT2.5 plays a role in nitrate acquisition and remobilization in nitrogen-starved plants. Plant J. 80 230–241. 10.1111/tpj.12626 [DOI] [PubMed] [Google Scholar]
- Li L., Gong H., Sun Z., Li T. (2019). Identification of conserved genes involved in nitrogen metabolic activities in wheat. PeerJ 7:e7281. 10.7717/peerj.7281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Chen F., Sun L., Zhang Z., Yang Y., He Z. (2006). Expression profiling of rice genes in early defense responses to blast and bacterial blight pathogens using cDNA microarray. Physiol. Mol. Plant Pathol. 68 51–60. 10.1016/j.pmpp.2006.06.002 [DOI] [Google Scholar]
- Li W., Wang Y., Okamoto M., Crawford N. M., Siddiqi M. Y., Glass A. D. M. (2007). Dissection of the AtNRT2. 1: AtNRT2. 2 inducible high-affinity nitrate transporter gene cluster. Plant Physiol. 143 425–433. 10.1104/pp.106.091223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z., Ali Q., Wang Y., Mu G., Kan X., Ren Y., et al. (2022). Toxicity of Bacillus thuringiensis strains derived from the novel crystal protein Cry31Aa with high nematicidal activity against rice parasitic nematode Aphelenchoides besseyi. Int. J. Mol. Sci. 23:8189. 10.3390/ijms23158189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao L., Dong T., Liu X., Dong Z., Qiu X., Rong Y., et al. (2019). Effect of nitrogen supply on nitrogen metabolism in the citrus cultivar ‘Huangguogan.’ PLoS One 14:e0213874. 10.1371/journal.pone.0213874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillo C., Lea U. S., Leydecker M., Meyer C. (2003). Mutation of the regulatory phosphorylation site of tobacco nitrate reductase results in constitutive activation of the enzyme in vivo and nitrite accumulation. Plant J. 35 566–573. 10.1046/j.1365-313x.2003.01828.x [DOI] [PubMed] [Google Scholar]
- Lin Y.-L., Tsay Y.-F. (2017). Influence of differing nitrate and nitrogen availability on flowering control in Arabidopsis. J. Exp. Bot. 68 2603–2609. 10.1093/jxb/erx053 [DOI] [PubMed] [Google Scholar]
- Liu A., Ma X., Zhang Z., Liu J., Luo D., Yang L., et al. (2022). Single dose fertilization at reduced nitrogen rate improves nitrogen utilization without yield reduction in late-planted cotton under a wheat–cotton cropping system. Ind. Crops Prod. 176:114346. 10.1016/j.indcrop.2021.114346 [DOI] [Google Scholar]
- Liu K., Tsay Y. (2003). Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J. 22 1005–1013. 10.1093/emboj/cdg118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K.-H., Huang C.-Y., Tsay Y.-F. (1999). CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11 865–874. 10.1105/tpc.11.5.865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K.-H., Niu Y., Konishi M., Wu Y., Du H., Sun Chung H., et al. (2017). Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature 545 311–316. 10.1038/nature22077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Zhang S., Zhang L. (2010). Above-ground dry matter accumulation of cotton genetics at different nitrogen applications. Cott. Sci. 22 77–82. [Google Scholar]
- Liu X., Yin C., Xiang L., Jiang W., Xu S., Mao Z. (2020). Transcription strategies related to photosynthesis and nitrogen metabolism of wheat in response to nitrogen deficiency. BMC Plant Biol. 20:448. 10.1186/s12870-020-02662-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M. S., Araus J. L. (2006). Nitrogen source and water regime effects on durum wheat photosynthesis and stable carbon and nitrogen isotope composition. Physiol. Plant. 126 435–445. 10.1111/j.1399-3054.2006.00595.x [DOI] [Google Scholar]
- Lu Y. L., Xu Y. C., Shen Q. R., Dong C. X. (2009). Effects of different nitrogen forms on the growth and cytokinin content in xylem sap of tomato (Lycopersicon esculentum Mill.) seedlings. Plant Soil 315 67–77. 10.1007/s11104-008-9733-y [DOI] [Google Scholar]
- Luo J., Zhou J.-J. (2019). Growth performance, photosynthesis, and root characteristics are associated with nitrogen use efficiency in six poplar species. Environ. Exp. Bot. 164 40–51. 10.1016/j.envexpbot.2019.04.013 [DOI] [Google Scholar]
- Lyu D., Backer R., Subramanian S., Smith D. L. (2020). Phytomicrobiome coordination signals hold potential for climate change-resilient agriculture. Front. Plant Sci. 11:634. 10.3389/fpls.2020.00634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F., Ma H. L., Qiu H., Yang H. Y. (2015). Effects of water levels and the additions of different nitrogen forms on soil net nitrogen transformation rate and N2O emission in subtropical forest soils. Ying Yong Sheng Tai Xue Bao 26 379–387. [PubMed] [Google Scholar]
- Mahmood T., Khalid S., Abdullah M., Ahmed Z., Shah M. K. N., Ghafoor A., et al. (2019). Insights into drought stress signaling in plants and the molecular genetic basis of cotton drought tolerance. Cells 9:105. 10.3390/cells9010105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manghwar H., Hussain A., Ali Q., Liu F. (2022). Brassinosteroids (BRs) role in plant development and coping with different stresses. Int. J. Mol. Sci. 23:1012. 10.3390/ijms23031012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manghwar H., Hussain A., Ali Q., Saleem M. H., Abualreesh M. H., Alatawi A., et al. (2021). Disease severity, resistance analysis, and expression profiling of pathogenesis-related protein genes after the inoculation of Fusarium equiseti in wheat. Agronomy 11:2124. 10.3390/agronomy11112124 [DOI] [Google Scholar]
- Marchive C., Roudier F., Castaings L., Bréhaut V., Blondet E., Colot V., et al. (2013). Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat. Commun. 4:1713. 10.1038/ncomms2650 [DOI] [PubMed] [Google Scholar]
- Marcondes J., Lemos E. G. M. (2012). “Nitrogen metabolism in citrus based on expressed tag analysis,” in Advances in Citrus Nutrition, ed. Srivastava A. (Dordrecht: Springer; ), 245–255. 10.1007/978-94-007-4171-3_17 [DOI] [Google Scholar]
- Marschner H. (2011). Marschner’s mineral nutrition of higher plants. Cambridge, MA: Academic Press. [Google Scholar]
- Martin A., Lee J., Kichey T., Gerentes D., Zivy M., Tatout C., et al. (2006). Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell 18 3252–3274. 10.1105/tpc.106.042689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez D. E., Costa M. L., Guiamet J. J. (2008). Senescence-associated degradation of chloroplast proteins inside and outside the organelle. Plant Biol. 10 15–22. 10.1111/j.1438-8677.2008.00089.x [DOI] [PubMed] [Google Scholar]
- Masclaux C., Valadier M.-H., Brugière N., Morot-Gaudry J.-F., Hirel B. (2000). Characterization of the sink/source transition in tobacco (Nicotiana tabacum L.) shoots in relation to nitrogen management and leaf senescence. Planta 211 510–518. 10.1007/s004250000310 [DOI] [PubMed] [Google Scholar]
- Masclaux-Daubresse C., Chardon F. (2011). Exploring nitrogen remobilization for seed filling using natural variation in Arabidopsis thaliana. J. Exp. Bot. 62 2131–2142. 10.1093/jxb/erq405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C., Daniel-Vedele F., Dechorgnat J., Chardon F., Gaufichon L., Suzuki A. (2010). Nitrogen uptake, assimilation and remobilization in plants: Challenges for sustainable and productive agriculture. Ann. Bot. 105 1141–1157. 10.1093/aob/mcq028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister C. H., Beatty P. H., Good A. G. (2012). Engineering nitrogen use efficient crop plants: The current status. Plant Biotechnol. J. 10 1011–1025. 10.1111/j.1467-7652.2012.00700.x [DOI] [PubMed] [Google Scholar]
- McLellan E. L., Cassman K. G., Eagle A. J., Woodbury P. B., Sela S., Tonitto C., et al. (2018). The nitrogen balancing act: Tracking the environmental performance of food production. Bioscience 68 194–203. 10.1093/biosci/bix164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medici A., Krouk G. (2014). The primary nitrate response: A multifaceted signalling pathway. J. Exp. Bot. 65 5567–5576. 10.1093/jxb/eru245 [DOI] [PubMed] [Google Scholar]
- Meister R., Rajani M. S., Ruzicka D., Schachtman D. P. (2014). Challenges of modifying root traits in crops for agriculture. Trends Plant Sci. 19 779–788. 10.1016/j.tplants.2014.08.005 [DOI] [PubMed] [Google Scholar]
- Mendoza-Villarreal R., Valdez-Aguilar L. A., Sandoval-Rangel A., Robledo-Torres V., Benavides-Mendoza A. (2015). Tolerance of lisianthus to high ammonium levels in rockwool culture. J. Plant Nutr. 38 73–82. 10.1080/01904167.2014.920379 [DOI] [Google Scholar]
- Meng S., Peng J.-S., He Y.-N., Zhang G.-B., Yi H.-Y., Fu Y.-L., et al. (2016). Arabidopsis NRT1. 5 mediates the suppression of nitrate starvation-induced leaf senescence by modulating foliar potassium level. Mol. Plant 9 461–470. 10.1016/j.molp.2015.12.015 [DOI] [PubMed] [Google Scholar]
- Meyer C., Stitt M. (2001). Nitrate reduction. In plant nitrogen. Heidelberg: Springer Verlag. 10.1007/978-3-662-04064-5_2 [DOI] [Google Scholar]
- Migge A., Carrayol E., Hirel B., Becker T. W. (2000). Leaf-specific overexpression of plastidic glutamine synthetase stimulates the growth of transgenic tobacco seedlings. Planta 210 252–260. 10.1007/PL00008132 [DOI] [PubMed] [Google Scholar]
- Morère-Le Paven M.-C., Viau L., Hamon A., Vandecasteele C., Pellizzaro A., Bourdin C., et al. (2011). Characterization of a dual-affinity nitrate transporter MtNRT1. 3 in the model legume Medicago truncatula. J. Exp. Bot. 62 5595–5605. 10.1093/jxb/err243 [DOI] [PubMed] [Google Scholar]
- Mounier E., Pervent M., Ljung K., Gojon A., Nacry P. (2014). Auxin-mediated nitrate signalling by NRT1.1 participates in the adaptive response of Arabidopsis root architecture to the spatial heterogeneity of nitrate availability. Plant Cell Environ. 37 162–174. 10.1111/pce.12143 [DOI] [PubMed] [Google Scholar]
- Mueller K. E., Eissenstat D. M., Hobbie S. E., Oleksyn J., Jagodzinski A. M., Reich P. B., et al. (2012). Tree species effects on coupled cycles of carbon, nitrogen, and acidity in mineral soils at a common garden experiment. Biogeochemistry 111 601–614. 10.1007/s10533-011-9695-7 [DOI] [Google Scholar]
- Mulla D. J., McBratney A. B. (2002). “Soil spatial variability,” in Soil physics companion, ed. Warrick A. W. (Boca Raton, FL: CRC Press; ), 343–373. 10.1201/9781420041651.ch9 [DOI] [Google Scholar]
- Mungur R., Wood A. J., Lightfoot D. A. (2006). Water potential is maintained during water deficit in Nicotiana tabacum expressing the Escherichia coli glutamate dehydrogenase gene. Plant Growth Regul. 50 231–238. 10.1007/s10725-006-9140-4 [DOI] [Google Scholar]
- Munþos S., Cazettes C., Fizames C., Gaymard F., Tillard P., Lepetit M., et al. (2004). Transcript profiling in the chl1-5 mutant of Arabidopsis reveals a role of the nitrate transporter NRT1. 1 in the regulation of another nitrate transporter, NRT2. 1. Plant Cell 16 2433–2447. 10.1105/tpc.104.024380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näsholm T., Kielland K., Ganeteg U. (2009). Uptake of organic nitrogen by plants. New Phytol. 182 31–48. 10.1111/j.1469-8137.2008.02751.x [DOI] [PubMed] [Google Scholar]
- Nasielski J., Grant B., Smith W., Niemeyer C., Janovicek K., Deen B. (2020). Effect of nitrogen source, placement and timing on the environmental performance of economically optimum nitrogen rates in maize. Field Crops Res. 246:107686. [Google Scholar]
- Nielsen R. L. (2006). “N loss mechanisms and nitrogen use efficiency,” in Purdue nitrogen management workshops, (West Lafayette, IN: Purdue University; ), 1–5. [Google Scholar]
- Niu G., Rodriguez D., Gu M. (2011). Response of Sophora secundiflora to nitrogen form and rate. HortScience 46 1303–1307. [Google Scholar]
- Niu J., Gui H., Iqbal A., Zhang H., Dong Q., Pang N., et al. (2020). N-use efficiency and yield of cotton (G. hirsutumn L.) are improved through the combination of N-fertilizer reduction and N-efficient cultivar. Agronomy 11:55. 10.3390/agronomy11010055 [DOI] [Google Scholar]
- Noguero M., Lacombe B. (2016). Transporters involved in root nitrate uptake and sensing by Arabidopsis. Front. Plant Sci. 7:1391. 10.3389/fpls.2016.01391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nour-Eldin H. H., Andersen T. G., Burow M., Madsen S. R., Jørgensen M. E., Olsen C. E., et al. (2012). NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature 488 531–534. 10.1038/nature11285 [DOI] [PubMed] [Google Scholar]
- O’Brien J. A., Vega A., Bouguyon E., Krouk G., Gojon A., Coruzzi G., et al. (2016). Nitrate transport, sensing, and responses in plants. Mol. Plant 9 837–856. 10.1016/j.molp.2016.05.004 [DOI] [PubMed] [Google Scholar]
- Occupational Safety and Health Administration (OSHA) (2017). Storage and handling of anhydrous ammonia. Washington, DC: Occupational Safety and Health Administration, 1–28. [Google Scholar]
- Ohyama T., Ohtake N., Sueyoshi K., Ono Y., Tsutsumi K., Ueno M., et al. (2017). “Amino acid metabolism and transport in soybean plants,” in Amino acid new insights roles plant animal, eds Asaduzzaman M., Asao T. (London: IntechOpen; ), 171–196. [Google Scholar]
- Oliveira I. C., Brears T., Knight T. J., Clark A., Coruzzi G. M. (2002). Overexpression of cytosolic glutamine synthetase. Relation to nitrogen, light, and photorespiration. Plant Physiol. 129 1170–1180. 10.1104/pp.020013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessarakli M., Tucker T. C. (1985). Ammonium (15N) metabolism in cotton under salt stress. J. Plant Nutr. 8 1025–1045. 10.1080/01904168509363404 [DOI] [Google Scholar]
- Ping J. L., Green C. J., Bronson K. F., Zartman R. E., Dobermann A. (2005). Delineating potential management zones for cotton based on yields and soil properties. Soil Sci. 170 371–385. 10.1631/jzus.B071379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping J. L., Green C. J., Zartman R. E., Bronson K. F., Morris T. F. (2007). Spatial variability of soil properties, cotton yield, and quality in a production field. Commun. Soil Sci. Plant Anal. 39 1–16. 10.1080/00103620701758840 [DOI] [Google Scholar]
- Plett D., Holtham L., Baumann U., Kalashyan E., Francis K., Enju A., et al. (2016). Nitrogen assimilation system in maize is regulated by developmental and tissue-specific mechanisms. Plant Mol. Biol. 92 293–312. 10.1007/s11103-016-0512-5 [DOI] [PubMed] [Google Scholar]
- Plett D. C., Ranathunge K., Melino V. J., Kuya N., Uga Y., Kronzucker H. J. (2020). The intersection of nitrogen nutrition and water use in plants: New paths toward improved crop productivity. J. Exp. Bot. 71 4452–4468. 10.1093/jxb/eraa049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purnell M. P., Skopelitis D. S., Roubelakis-Angelakis K. A., Botella J. R. (2005). Modulation of higher-plant NAD (H)-dependent glutamate dehydrogenase activity in transgenic tobacco via alteration of beta subunit levels. Planta 222 167–180. 10.1007/s00425-005-1510-z [DOI] [PubMed] [Google Scholar]
- Quan X., Zeng J., Han Z., Zhang G. (2017). Ionomic and physiological responses to low nitrogen stress in Tibetan wild and cultivated barley. Plant Physiol. Biochem. 111 257–265. 10.1016/j.plaphy.2016.12.008 [DOI] [PubMed] [Google Scholar]
- Raddatz N., Morales de los Ríos L., Lindahl M., Quintero F. J., Pardo J. M. (2020). Coordinated transport of nitrate, potassium, and sodium. Front. Plant Sci. 11:247. 10.3389/fpls.2020.00247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranathunge K., El-Kereamy A., Gidda S., Bi Y.-M., Rothstein S. J. (2014). AMT1; 1 transgenic rice plants with enhanced NH4+ permeability show superior growth and higher yield under optimal and suboptimal NH4+ conditions. J. Exp. Bot. 65 965–979. 10.1093/jxb/ert458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan R., Yadav R. (2019). Targeting nitrogen use efficiency for sustained production of cereal crops. J. Plant Nutr. 42 1086–1113. 10.1080/01904167.2019.1589497 [DOI] [Google Scholar]
- Remans T., Nacry P., Pervent M., Filleur S., Diatloff E., Mounier E., et al. (2006). The Arabidopsis NRT1. 1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc. Natl. Acad. Sci. U.S.A. 103 19206–19211. 10.1073/pnas.0605275103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley W. J., Ortiz-Monasterio I., Matson P. A. (2001). Nitrogen leaching and soil nitrate, nitrite, and ammonium levels under irrigated wheat in Northern Mexico. Nutr. Cycl. Agroecosyst. 61 223–236. 10.1023/A:1013758116346 [DOI] [Google Scholar]
- Riveras E., Alvarez J. M., Vidal E. A., Oses C., Vega A., Gutiérrez R. A. (2015). The calcium ion is a second messenger in the nitrate signaling pathway of Arabidopsis. Plant Physiol. 169 1397–1404. 10.1104/pp.15.00961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson N., Brackin R., Vinall K., Soper F., Holst J., Gamage H., et al. (2011). Nitrate paradigm does not hold up for sugarcane. PLoS One 6:e19045. 10.1371/journal.pone.0019045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas C. M., Senthil-Kumar M., Tzin V., Mysore K. S. (2014). Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front. Plant Sci. 5:17. 10.3389/fpls.2014.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H., Oikawa T., Hamamoto S., Ishimaru Y., Kanamori-Sato M., Sasaki-Sekimoto Y., et al. (2015). The jasmonate-responsive GTR1 transporter is required for gibberellin-mediated stamen development in Arabidopsis. Nat. Commun. 6:6095. 10.1038/ncomms7095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H., Kobayashi K., Deji A., Sugiyama T. (1997). Partial characterization of the signaling pathway for the nitrate-dependent expression of genes for nitrogen-assimilatory enzymes using detached maize leaves. Plant Cell Physiol. 38 837–843. 10.1093/oxfordjournals.pcp.a029242 [DOI] [Google Scholar]
- Sarraf M., Vishwakarma K., Kumar V., Arif N., Das S., Johnson R., et al. (2022). Metal/Metalloid-based nanomaterials for plant abiotic stress tolerance: An overview of the mechanisms. Plants 11:316. 10.3390/plants11030316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saubidet M. I., Fatta N., Barneix A. J. (2002). The effect of inoculation with Azospirillum brasilense on growth and nitrogen utilization by wheat plants. Plant Soil 245 215–222. 10.1023/A:1020469603941 [DOI] [Google Scholar]
- Scheible W. R., Gonzalez-Fontes A., Lauerer M., Muller-Rober B., Caboche M., Stitt M. (1997). Nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. Plant Cell 9 783–798. 10.1105/tpc.9.5.783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schortemeyer M., Feil B. (1996). Root morphology of maize under homogeneous or spatially separated supply of ammonium and nitrate at three concentration ratios. J. Plant Nutr. 19 1089–1097. 10.1242/jcs.45.1.147 [DOI] [PubMed] [Google Scholar]
- Segonzac C., Boyer J.-C., Ipotesi E., Szponarski W., Tillard P., Touraine B., et al. (2007). Nitrate efflux at the root plasma membrane: Identification of an Arabidopsis excretion transporter. Plant Cell 19 3760–3777. 10.1105/tpc.106.048173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabbir R., Javed T., Hussain S., Ahmar S., Naz M., Zafar H., et al. (2022). Calcium homeostasis and potential roles to combat environmental stresses in plants. S. Afr. J. Bot. 148 683–693. 10.1016/j.sajb.2022.05.038 [DOI] [Google Scholar]
- Shafreen M., Vishwakarma K., Shrivastava N., Kumar N. (2021). “Physiology and distribution of nitrogen in soils,” in Soil nitrogen ecology. Soil biology, Vol. 62 eds Cruz C., Vishwakarma K., Choudhary D. K., Varma A. (Cham: Springer; ), 3–31. 10.1007/978-3-030-71206-8_1 [DOI] [Google Scholar]
- Shankar A., Gupta R. K.,, Singh B. (2020). Site-specific fertilizer nitrogen management in Bt cotton using chlorophyll meter. Exp. Agric. 56 397–406. 10.1017/S0014479720000046 [DOI] [Google Scholar]
- Sharma L. K., Bali S. K. (2017). A review of methods to improve nitrogen use efficiency in agriculture. Sustainability 10:51. 10.3390/su10010051 [DOI] [Google Scholar]
- Sharma L. K., Bali S. K., Dwyer J. D., Plant A. B., Bhowmik A. (2017). A case study of improving yield prediction and sulfur deficiency detection using optical sensors and relationship of historical potato yield with weather data in maine. Sensors 17:1095. 10.3390/s17051095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono H., Okada M., Yamakawa Y., Nakamura H., Kobayashi K., Hasegawa T. (2009). Genotypic variation in rice yield enhancement by elevated CO2 relates to growth before heading, and not to maturity group. J. Exp. Bot. 60 523–532. 10.1093/jxb/ern288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrawat A. K., Carroll R. T., DePauw M., Taylor G. J., Good A. G. (2008). Genetic engineering of improved nitrogen use efficiency in rice by the tissue-specific expression of alanine aminotransferase. Plant Biotechnol. J. 6 722–732. 10.1111/j.1467-7652.2008.00351.x [DOI] [PubMed] [Google Scholar]
- Singh R., Parihar P., Prasad S. M. (2018). Sulfur and calcium simultaneously regulate photosynthetic performance and nitrogen metabolism status in As-challenged Brassica juncea L. seedlings. Front. Plant Sci. 9:772. 10.3389/fpls.2018.00772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skopelitis D. S., Paranychianakis N. V., Paschalidis K. A., Pliakonis E. D., Delis I. D., Yakoumakis D. I., et al. (2006). Abiotic stress generates ROS that signal expression of anionic glutamate dehydrogenases to form glutamate for proline synthesis in tobacco and grapevine. Plant Cell 18 2767–2781. 10.1105/tpc.105.038323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder C. S., Bruulsema T. W., Jensen T. L. (2007). Best management practices to minimize greenhouse gas emissions associated with fertilizer use. Better Crop. 91 16–18. [Google Scholar]
- Snyder C. S., Davidson E. A., Smith P., Venterea R. T. (2014). Agriculture: Sustainable crop and animal production to help mitigate nitrous oxide emissions. Curr. Opin. Environ. Sustain. 9 46–54. 10.1016/j.cosust.2014.07.005 [DOI] [Google Scholar]
- Solangi Z. A., Yao Y., Zhang Yani T. M., Rattar Q. A., Tahir S., Mustafa G., et al. (2021). QTL mapping for seed weight and seed yield-related traits in oil-seed crop rapeseed (Brassica sp.). Int. J. Agric. Res. 9 51–60. [Google Scholar]
- Spicer S. (2002). Fertilizers, manure, or biosolids. Water Environ. Technol. 14 32–37. [Google Scholar]
- Su C., Ding F., Wang W., Song Z., Ali Q., Ali M., et al. (2022). Time-resolved fluorescent microsphere lateral flow biosensors for rapid detection of Candidatus Liberibacter asiaticus. Plant Biotechnol. J. 20 1235–1237. 10.1111/pbi.13828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C.-H., Yu J.-Q., Hu D.-G. (2017). Nitrate: A crucial signal during lateral roots development. Front. Plant Sci. 8:485. 10.3389/fpls.2017.00485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Qian Q., Wu K., Luo J., Wang S., Zhang C., et al. (2014). Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Nat. Genet. 46 652–656. 10.1038/ng.2958 [DOI] [PubMed] [Google Scholar]
- Suzuki A., Knaff D. B. (2005). Glutamate synthase: Structural, mechanistic and regulatory properties, and role in the amino acid metabolism. Photosynth. Res. 83 191–217. 10.1007/s11120-004-3478-0 [DOI] [PubMed] [Google Scholar]
- Tabuchi M., Abiko T., Yamaya T. (2007). Assimilation of ammonium ions and reutilization of nitrogen in rice (Oryza sativa L.). J. Exp. Bot. 58 2319–2327. 10.1093/jxb/erm016 [DOI] [PubMed] [Google Scholar]
- Takahashi M., Sasaki Y., Ida S., Morikawa H. (2001). Nitrite reductase gene enrichment improves assimilation of NO2 in Arabidopsis. Plant Physiol. 126 731–741. 10.1104/pp.126.2.731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal I., Zhang Y., Jorgensen M. E., Pisanty O., Barbosa I. C., Zourelidou M., et al. (2016). Coordinated transport of nitrate, potassium, and sodium. Front. Plant Sci. 11:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taliercio E., Kwanyen P., Scheffler J. (2010). Nitrogen metabolism in cotton stems and roots during reproductive development. J. Cotton Sci. 14 107–112. [Google Scholar]
- Tang H. Y., Yang G. Z., Nie Y. C. (2012a). “Fertilization effect on N distribution in cotton plant,” in Proceedings of the 2012 international conference of agricultural engineering and food engineering, (Harbin: ), 93–96. [Google Scholar]
- Tang H. Y., Yang G. Z., Zhang X. L., Siddique K. (2012b). Improvement of fertilizer N recovery by allocating more N for later application in cotton (Gossypium hirsutum L.). Int. J. Basic Appl. Sci. 12 32–37. [Google Scholar]
- Tang W., Ye J., Yao X., Zhao P., Xuan W., Tian Y., et al. (2019). Genome-wide associated study identifies NAC42-activated nitrate transporter conferring high nitrogen use efficiency in rice. Nat. Commun. 10:5279. 10.1038/s41467-019-13187-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq A., Waqas M., Beihai T., Iqbal M., Ali S., Khan S. U. (2019). Hazardous impacts of pesticide usage on farmer’s health in cotton growing region of District Muzaffargarh, Pakistan. Pacific Int. J. 2 12–20. [Google Scholar]
- Tegeder M., Masclaux-Daubresse C. (2018). Source and sink mechanisms of nitrogen transport and use. New Phytol. 217 35–53. [DOI] [PubMed] [Google Scholar]
- Tercé-Laforgue T., Bedu M., Dargel-Grafin C., Dubois F., Gibon Y., Restivo F. M., et al. (2013). Resolving the role of plant glutamate dehydrogenase: II. Physiological characterization of plants overexpressing the two enzyme subunits individually or simultaneously. Plant Cell Physiol. 54 1635–1647. [DOI] [PubMed] [Google Scholar]
- The S. V., Snyder R., Tegeder M. (2021). Targeting nitrogen metabolism and transport processes to improve plant nitrogen use efficiency. Front. Plant Sci. 11:628366. 10.3389/fpls.2020.628366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilsner J., Kassner N., Struck C., Lohaus G. (2005). Amino acid contents and transport in oilseed rape (Brassica napus L.) under different nitrogen conditions. Planta 221 328–338. 10.1007/s00425-004-1446-8 [DOI] [PubMed] [Google Scholar]
- Tong Y., Zhou J., Li Z., Miller A. J. (2005). A two-component high-affinity nitrate uptake system in barley. Plant J. 41 442–450. 10.1111/j.1365-313X.2004.02310.x [DOI] [PubMed] [Google Scholar]
- Tsay Y.-F., Chiu C.-C., Tsai C.-B., Ho C.-H., Hsu P.-K. (2007). Nitrate transporters and peptide transporters. FEBS Lett. 581 2290–2300. 10.1016/j.febslet.2007.04.047 [DOI] [PubMed] [Google Scholar]
- Tsay Y.-F., Schroeder J. I., Feldmann K. A., Crawford N. M. (1993). The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72 705–713. 10.1016/0092-8674(93)90399-b [DOI] [PubMed] [Google Scholar]
- Ueda Y., Konishi M., Yanagisawa S. (2017). Molecular basis of the nitrogen response in plants. Soil Sci. Plant Nutr. 63 329–341. 10.1080/00380768.2017.1360128 [DOI] [Google Scholar]
- Venterea R. T., Halvorson A. D., Kitchen N., Liebig M. A., Cavigelli M. A., Del Grosso S. J., et al. (2012). Challenges and opportunities for mitigating nitrous oxide emissions from fertilized cropping systems. Front. Ecol. Environ. 10 562–570. 10.1111/gcb.13341 [DOI] [PubMed] [Google Scholar]
- Vidal E. A., Alvarez J. M., Araus V., Riveras E., Brooks M. D., Krouk G., et al. (2020). Nitrate in 2020: Thirty years from transport to signaling networks. Plant Cell 32 2094–2119. 10.1105/tpc.19.00748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Wittgenstein N. J. J. B., Le C. H., Hawkins B. J., Ehlting J. (2014). Evolutionary classification of ammonium, nitrate, and peptide transporters in land plants. BMC Evol. Biol. 14:11. 10.1186/1471-2148-14-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S., Ishida H., Izumi M., Yoshimoto K., Ohsumi Y., Mae T., et al. (2009). Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol. 149 885–893. 10.1104/pp.108.130013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajid A., Ghaffar A., Maqsood M., Hussain K., Nasim W. (2007). Yield response of maize hybrids to varying nitrogen rates. Pak. J. Agric. Sci. 44 217–220. [Google Scholar]
- Walworth J. (2013). AZ1591; College of agriculture and life sciences. Tucson, AZ: University of Arizona, 3. [Google Scholar]
- Wang G., Dobermann A., Witt C., Sun Q., Fu R. (2001). Performance of site-specific nutrient management for irrigated rice in southeast China. Agron. J. 93 869–878. 10.2134/agronj2001.934869x [DOI] [Google Scholar]
- Wang H., Wu L., Wang X., Zhang S., Cheng M., Feng H., et al. (2021). Optimization of water and fertilizer management improves yield, water, nitrogen, phosphorus and potassium uptake and use efficiency of cotton under drip fertigation. Agric. Water Manag. 245:106662. 10.1016/j.agwat.2020.106662 [DOI] [Google Scholar]
- Wang L., Xue C., Pan X., Chen F., Liu Y. (2018). Application of controlled-release urea enhances grain yield and nitrogen use efficiency in irrigated rice in the yangtze river basin, China. Front. Plant Sci. 9:999. 10.3389/fpls.2018.00999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Fu B., Pan L., Chen L., Fu X., Li K. (2013). Overexpression of Arabidopsis Dof1, GS1 and GS2 enhanced nitrogen assimilation in transgenic tobacco grown under low-nitrogen conditions. Plant Mol. Biol. Rep. 31 886–900. [Google Scholar]
- Wang Y.-Y., Cheng Y.-H., Chen K.-E., Tsay Y.-F. (2018). Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 69 85–122. 10.1146/annurev-arplant-042817-040056 [DOI] [PubMed] [Google Scholar]
- Wang W., Hu B., Li A., Chu C. (2020). NRT1. 1s in plants: Functions beyond nitrate transport. J. Exp. Bot. 71 4373–4379. 10.1093/jxb/erz554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-Y., Tsay Y.-F. (2011). Arabidopsis nitrate transporter NRT1. 9 is important in phloem nitrate transport. Plant Cell 23 1945–1957. 10.1105/tpc.111.083618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward B. D., Cox M. S. (2000). “Influences of soil chemical and physical properties on site-specific cotton production,” in Proceedings of the 5th international conference on precision agriculture, (Bloomington, MN: American Society of Agronomy; ), 1–11. [Google Scholar]
- Wei C., Ma T., Wang X. (2012). The fate of fertilizer N applied to cotton in relation to irrigation methods and N dosage in arid area. J. Arid Land 4 320–329. 10.3724/SP.J.1227.2012.00320 [DOI] [Google Scholar]
- Weißenborn S., Walther D. (2017). Metabolic pathway assignment of plant genes based on phylogenetic profiling–a feasibility study. Front. Plant Sci. 8:1831. 10.3389/fpls.2017.01831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L. E., Miller A. J. (2001). Transporters responsible for the uptake and partitioning of nitrogenous solutes. Annu. Rev. Plant Biol. 52 659–688. 10.1146/annurev.arplant.52.1.659 [DOI] [PubMed] [Google Scholar]
- Witcombe J. R., Hollington P. A., Howarth C. J., Reader S., Steele K. A. (2008). Breeding for abiotic stresses for sustainable agriculture. Philos. Trans. R. Soc. B Biol. Sci. 363 703–716. 10.1098/rstb.2007.2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte C.-P. (2011). Urea metabolism in plants. Plant Sci. 180 431–438. 10.1016/j.plantsci.2010.11.010 [DOI] [PubMed] [Google Scholar]
- Wu T., Qin Z., Fan L., Xue C., Zhou X., Xin M., et al. (2014). Involvement of CsNRT1. 7 in nitrate recycling during senescence in cucumber. J. Plant Nutr. Soil Sci. 177 714–721. 10.1002/jpln.201300665 [DOI] [Google Scholar]
- Xing Y., Guo S., Chen X., Du D., Liu M., Xiao Y., et al. (2018). Nitrogen metabolism is affected in the nitrogen-deficient rice mutant esl4 with a calcium-dependent protein kinase gene mutation. Plant Cell Physiol. 59 2512–2525. 10.1093/pcp/pcy169 [DOI] [PubMed] [Google Scholar]
- Xu G., Fan X., Miller A. J. (2012). Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 63 153–182. 10.1146/annurev-arplant-042811-105532 [DOI] [PubMed] [Google Scholar]
- Xu Z., Zhou G. (2004). Research advance in nitrogen metabolism of plant and its environmental regulation. Ying Yong Sheng Tai Xue Bao 15 511–516. [PubMed] [Google Scholar]
- Yadav M. R., Kumar R., Parihar C. M., Yadav R. K., Jat S. L., Ram H., et al. (2017). Strategies for improving nitrogen use efficiency: A review. Agric. Rev. 38 29–40. 10.18805/ag.v0iOF.7306 [DOI] [Google Scholar]
- Yamaya T., Obara M., Nakajima H., Sasaki S., Hayakawa T., Sato T. (2002). Genetic manipulation and quantitative-trait loci mapping for nitrogen recycling in rice. J. Exp. Bot. 53 917–925. 10.1093/jexbot/53.370.917 [DOI] [PubMed] [Google Scholar]
- Yao H., Zhang Y., Yi X., Hu Y., Luo H., Gou L., et al. (2015). Plant density alters nitrogen partitioning among photosynthetic components, leaf photosynthetic capacity and photosynthetic nitrogen use efficiency in field-grown cotton. Field Crops Res. 184 39–49. 10.1016/j.fcr.2015.09.005 [DOI] [Google Scholar]
- Yasmin H., Naeem S., Bakhtawar M., Jabeen Z., Nosheen A., Naz R., et al. (2020). Halotolerant rhizobacteria Pseudomonas pseudoalcaligenes and Bacillus subtilis mediate systemic tolerance in hydroponically grown soybean (Glycine max L.) against salinity stress. PLoS One 15:e0231348. 10.1371/journal.pone.0231348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Liu X., Zhang X., Zhang M., Gu Y., Ali Q., et al. (2021). Mycosubtilin produced by Bacillus subtilis ATCC6633 inhibits growth and mycotoxin biosynthesis of Fusarium graminearum and Fusarium verticillioides. Toxins 13:791. 10.3390/toxins13110791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Luo H., Xiong J., Zhou Q., Xia L., Sun M., et al. (2014). Bacillus thuringiensis C ry6 A exhibits nematicidal activity to Caenorhabditis elegans bre mutants and synergistic activity with C ry5 B to C. elegans. Lett. Appl. Microbiol. 58 511–519. 10.1111/lam.12219 [DOI] [PubMed] [Google Scholar]
- Yuan L., Loque D., Kojima S., Rauch S., Ishiyama K., Inoue E., et al. (2007). The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. Plant Cell 19 2636–2652. 10.1105/tpc.107.052134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Xiaoqiong F. U., Xiangru W., Huiping G. U. I., Qiang D., Nianchang P., et al. (2018). Identification and screening of nitrogen-efficient cotton genotypes under low and normal nitrogen environments at the seedling stage. J. Cotton Res. 1:6. 10.1186/s42397-018-0006-x [DOI] [Google Scholar]
- Zhang P., Wei T., Li Y., Wang K., Jia Z., Han Q., et al. (2015). Effects of straw incorporation on the stratification of the soil organic C, total N and C: N ratio in a semiarid region of China. Soil Tillage Res. 153 28–35. [Google Scholar]
- Zhang Z., Chattha M. S., Ahmed S., Liu J., Liu A., Yang L., et al. (2021). Nitrogen reduction in high plant density cotton is feasible due to quicker biomass accumulation. Ind. Crops Prod. 172:114070. 10.1016/j.indcrop.2021.114070 [DOI] [Google Scholar]
- Zhao X.-Q., Nie X.-L., Xiao X.-G. (2013). Over-expression of a tobacco nitrate reductase gene in wheat (Triticum aestivum L.) increases seed protein content and weight without augmenting nitrogen supplying. PLoS One 8:e74678. 10.1371/journal.pone.0074678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong C., Cao X., Hu J., Zhu L., Zhang J., Huang J., et al. (2017). Nitrogen metabolism in adaptation of photosynthesis to water stress in rice grown under different nitrogen levels. Front. Plant Sci. 8:1079. 10.3389/fpls.2017.01079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.-J., Trueman L. J., Boorer K. J., Theodoulou F. L., Forde B. G., Miller A. J. (2000). A high affinity fungal nitrate carrier with two transport mechanisms. J. Biol. Chem. 275 39894–39899. 10.1074/jbc.M004610200 [DOI] [PubMed] [Google Scholar]
- Zhou X., Lin J., Zhou Y., Yang Y., Liu H., Zhang C., et al. (2015). Overexpressing a fungal CeGDH gene improves nitrogen utilization and growth in rice. Crop Sci. 55 811–820. 10.1111/j.1469-8137.2011.03929.x [DOI] [PubMed] [Google Scholar]
- Zimmermann P., Heinlein C., Orendi G., Zentgraf U. (2006). Senescence-specific regulation of catalases in Arabidopsis thaliana (L.) Heynh. Plant Cell Environ. 29 1049–1060. 10.1111/j.1365-3040.2005.01459.x [DOI] [PubMed] [Google Scholar]