Abstract
Macrolides represent a clinically important class of antibiotics that block protein synthesis by interacting with the large ribosomal subunit. The macrolide binding site is composed primarily of rRNA. However, the mode of interaction of macrolides with rRNA and the exact location of the drug binding site have yet to be described. A new class of macrolide antibiotics, known as ketolides, show improved activity against organisms that have developed resistance to previously used macrolides. The biochemical reasons for increased potency of ketolides remain unknown. Here we describe the first mutation that confers resistance to ketolide antibiotics while leaving cells sensitive to other types of macrolides. A transition of U to C at position 2609 of 23S rRNA rendered E. coli cells resistant to two different types of ketolides, telithromycin and ABT-773, but increased slightly the sensitivity to erythromycin, azithromycin, and a cladinose-containing derivative of telithromycin. Ribosomes isolated from the mutant cells had reduced affinity for ketolides, while their affinity for erythromycin was not diminished. Possible direct interaction of ketolides with position 2609 in 23S rRNA was further confirmed by RNA footprinting. The newly isolated ketolide-resistance mutation, as well as 23S rRNA positions shown previously to be involved in interaction with macrolide antibiotics, have been modeled in the crystallographic structure of the large ribosomal subunit. The location of the macrolide binding site in the nascent peptide exit tunnel at some distance from the peptidyl transferase center agrees with the proposed model of macrolide inhibitory action and explains the dominant nature of macrolide resistance mutations. Spatial separation of the rRNA residues involved in universal contacts with macrolides from those believed to participate in structure-specific interactions with ketolides provides the structural basis for the improved activity of the broader spectrum group of macrolide antibiotics.
Macrolide antibiotics inhibit protein synthesis in a wide range of pathogenic bacteria. The drugs bind to a single site in the large ribosomal subunit located near the entrance to the nascent peptide tunnel and are thought to sterically hinder the growth of the polypeptide chain and cause dissociation of peptidyl-tRNA from the ribosome (1, 10, 18, 33). Macrolide antibiotics were also shown to affect ribosome assembly (9).
The macrolide binding site is composed primarily of RNA. Two segments of 23S rRNA, the central loop of domain V and the loop of hairpin 35 from domain II, are believed to be the major components of the drug binding site on the ribosome. Mutations of A2058 (Escherichia coli numeration) and several neighboring positions in the central loop of domain V confer resistance to macrolides (27), as reviewed in references 34 and 35. Methylation of A2058 by Erm-type methyl-transferases drastically reduces the affinity of macrolides for the ribosome, suggesting the direct interaction between this RNA residue and the drug (11, 17, 28). The interaction of macrolides with this region of rRNA is further supported by RNA footprinting: all of the macrolide antibiotics investigated strongly protect A2058 and A2059 from chemical modification (5, 16, 20, 37; L. Xiong and A. Mankin, unpublished results). The accessibility of several other positions within the central loop of domain V is also affected by macrolide binding (11, 25). Genetic and biochemical evidence also revealed interaction of macrolides with the helix 35 in domain II of 23S rRNA (16, 37). Though rRNA appears to be the major component of the macrolide binding site, mutations in two ribosomal proteins, L4 and L22 (1, 8, 23, 31, 36), were shown to confer resistance. It remains unclear, however, whether these proteins directly participate in the drug binding or the mutations allosterically affect the conformation of rRNA in the drug binding site (12, 13).
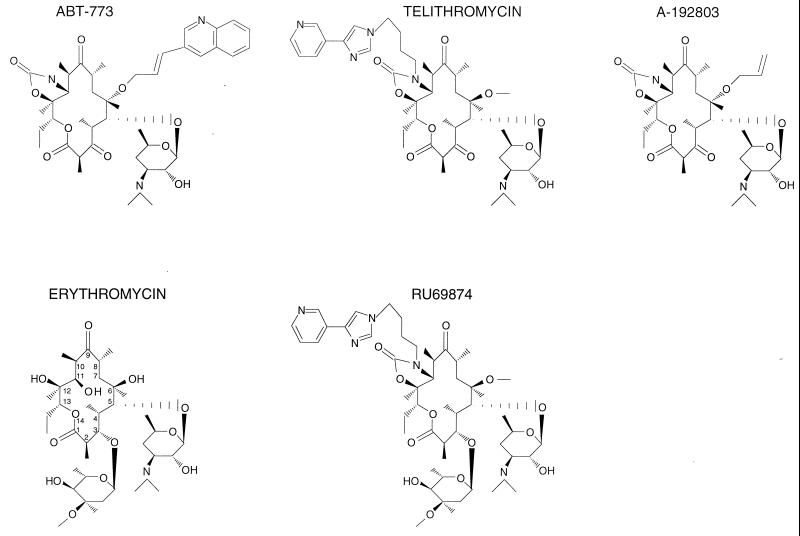
Erythromycin A, which was introduced in clinical practice in 1953, represented the first generation of the 14-member-ring macrolides (Fig. 1). The later developed drugs, such as clarithromycin, roxithromycin, and the 15-member-ring azithromycin, were characterized by better stability and a broader spectrum of action. The subsequent spread of resistant strains prompted a search for newer macrolide derivatives. The new class group of macrolides—ketolides, which contain a keto function instead of the 3-cladinose residue and carry an 11,12 carbamate and an additional alkyl-aryl side chain (Fig. 1) exhibit an improved activity profile and, importantly, show a significant activity against some macrolide-resistant isolates (21, 26). (Note that, in this paper, we will refer to cladinose-containing macrolides as cladinosolides, while drugs with the keto function at position 3 of the lacton ring will be referred to as ketolides.) The enhanced potency of ketolides correlates with their higher affinity for the ribosome, which depends, at least in part, on a stronger interaction with the helix 35 loop (11, 16, 37). Ketolides may interact idiosyncratically with the other rRNA segments as well: some rRNA mutations that render cells resistant to cladinosolides produce little or no effect on cell sensitivity to ketolides (11).
FIG. 1.
Chemical structures of macrolide antibiotics used in this study. Ketolides, containing the 3-keto group, are represented by ABT-773, telithromycin, and A-192803. Erythromycin and RU69874 are examples of cladinosolides that contain a 3-cladinose residue.
In this paper, we characterize the first ketolide-specific resistance mutation in rRNA. The mutation at position 2609 of rRNA confers resistance to ketolides, but not to cladinose-containing macrolides. The location of this and other RNA residues involved in interaction with macrolides in the ribosome tertiary structure positions the macrolide binding site in the nascent peptide exit tunnel. Structural information provides the rationale for the improved activity of ketolide antibiotics and illuminates a possible mode of macrolide action.
MATERIALS AND METHODS
Reagents, strains, and plasmids. (i) Antibiotics.
ABT-773 and A-192803 were from Abbott Laboratories, telithromycin, and RU 69874 from Aventis Pharma. Erythromycin was purchased from Sigma.
(ii) Chemicals and enzymes.
Dimethyl sulfate (DMS) and 1-cyclohexyl-3-(2-morpholinoethyl) carbodiimide metho-p-toluene sulfonate (CMCT), used in RNA probing experiments, were from Aldrich. All the other chemicals were purchased from either Fisher or Sigma. Restriction enzymes were from MBI Fermentas.
(iii) Plasmids.
Plasmid pKK3535 (Table 1) contains a complete wild-type rrnB operon of E. coli cloned in the BamHI site of pBR322 vector (7). A closely related pKK3535 derivative, pNK, lacking a 903-bp NaeI-BamHI fragment from the vector portion, provided a more favorable restriction site configuration for the fragment exchange (38). pKK754A is identical to pKK3535, except for the presence of a T754A mutation in the 23S rRNA gene (37). pSTL102 contains the rrnB operon with a C1192T spectinomycin resistance mutation in the 16S rRNA gene and an A2058G mutation in the 23S rRNA gene (32).
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Resistance markera | Source or reference |
|---|---|---|---|
| pKK3535 | pBR322 containing complete rrnB operon inserted in the BamHI site | Ampr | 7 |
| pNK | pKK3535 derivative in which a 903-bp NaeI-BamHI fragment has been removed from the inactivated tetracycline resistance gene of the vector | Ampr | 38 |
| pSTL102 | pKK3535 derivative containing spectinomycin resistance mutation, C1192U in 16S rRNA and Eryr mutation A2058G in 23S rRNA gene. | Ampr | 32 |
| pKK754A | Identical to pKK3535, except for the presence of the T754A mutation in the 23S rRNA gene | Ampr | 37 |
| pKK2609C | Mutant pKK3535 plasmid containing the T2609C mutation in the 23S rRNA gene isolated from the selected Ketr clones | Ampr | This work |
| pNK2609C | Identical to pNK except for the presence of the T2609C mutation in the 23S rRNA gene | Ampr | This work |
| pHK-rrnC+ | pSC101 derivative containing complete rrnC operon | Kanr | 3 |
Antibiotic resistance gene present on the plasmid and used for transformant selection.
(iv) E. coli strains.
Strain JM109 (39) [endA1 recA1 gyrA96 thi hsdR17(rK− mK+) relA1 supE44 Δ(lac-proAB) (F′ traD36, proAB lacIqZΔM15)] was from Promega. Strain TA531 (ΔEBHGzADcCΔcrecA/pTRNA66, pHK-rrnC+) (3) was a gift of C. Squires (Tufts University).
Selection of ketolide resistance mutations.
The E. coli strain TA531 was used for selection of the resistant mutants (3). In this strain, all seven chromosomal rRNA operons are deleted and the only source of rRNA is a pHK-rrnC+ plasmid carrying the rrnC operon as well as a kanamycin resistance marker. The plasmid pKK3535 was randomly mutagenized by passing through the E. coli mutator strain XL1-Red as described previously (38). TA531 cells were transformed with the mutant pKK3535 library and grown overnight in liquid culture in the presence of ampicillin to promote loss of the pHK-rrnC+ plasmid. A total of 106 Ampr cells were plated onto agar plates containing 100 μg of ampicillin per ml and 10 μg of ABT-773 per ml. About 50 ketolide-resistant (Ketr) clones appeared after a 24-h incubation. They were replica plated onto Luria-Bertani agar containing ampicillin (100 μg/ml) or kanamycin (30 μg/ml). Two clones that exhibited an Ampr Ketr Kans phenotype, containing only mutant pKK3535 as a rRNA source, were chosen for further analyses.
Plasmids isolated from these two clones were used to re-transform fresh TA531 cells and the plasmid-exchange procedure was repeated. The Ketr phenotype was cotransferable with the mutant pKK3535 plasmids, indicating that the resistance determinant was plasmid borne.
Mapping the site of the Ketr mutation.
DNA sequencing revealed the presence of the U2609C mutation in the 23S rRNA gene in plasmids isolated from both analyzed Ketr clones. Fragment exchange was used to test whether the presence of this only mutation was sufficient for the ketolide resistance. A Bpu1102I-PaeI 2.2-kbp restriction fragment from mutant pKK3535 plasmids containing the U2609C mutation was used to replace the corresponding fragment in the wild-type rrnB operon on plasmid pNK (38). The resulting plasmid, pNK2609C, conferred the same level of ketolide resistance as the originally selected mutant pKK3535. DNA sequencing confirmed the presence of only the U2609C mutation in the exchanged Bpu1102I-PaeI 2.2-kbp fragment, demonstrating that the presence of this mutation was sufficient to confer ketolide resistance.
Determining antibiotic MICs.
To determine the MICs of various macrolide and ketolide antibiotics, cultures of Ampr Kans TA531 cells carrying one of the following plasmids—wild-type or mutant pKK3535, pNK, or pSTL102—were grown overnight in the presence of 100 μg of ampicillin per ml, diluted 1,000-fold into a fresh ampicillin-containing medium supplemented with various drug concentrations, and grown overnight with constant shaking. The optical densities of the cultures were measured at 650 nm and plotted.
RNA footprinting.
E. coli ribosomes (200 nM) were incubated for 10 min at 37°C, followed by 10 min at 20°C in 50 μl of the modification buffer (30) containing 5 μM macrolide antibiotics. DMS or CMCT modification was carried out at 37°C essentially as described previously (19), except that the CMCT concentration was increased twofold. Primer extension was performed according to the method of Stern et al. (30).
Antibiotic binding.
One A260 unit (23 pmol) of ribosomes was incubated with [14C]ABT-773 (27.2 mCi/mmol) (prepared by Abbott Laboratories) or [14C]erythromycin (55.1 mCi/mmol) (NEN Life Science) at concentrations ranging from 2 nM up to 200 nM. The reactions were carried out at room temperature for 4.5 h in buffers containing 10 mM Tris-HCl (pH 7.2), 10 mM NH4Cl, 4 mM MgCl2, and 100 mM KCl in a final volume of 0.5 ml in sterile microcentrifuge tubes. At the end of the reactions, ribosomes were pelleted by centrifugation at 100,000 × g for 2 h in a Beckman TL-100 centrifuge (rotor TL-45). The amount of radioactive drug associated with the ribosomes was determined by scintillation counting. Data were analyzed by using KaleidaGraph software (Synergy Software). Kd values were determined with the formula: Kd = D1/2 − 1/2Rtotal, where D1/2 = concentration of the drugs at which 50% ribosomes bind antibiotic and Rtotal = total concentration of active ribosomes.
RESULTS
Selection of the ketolide-resistant mutant.
To identify rRNA mutations that confer resistance to ketolides, the plasmid pKK3535 containing a complete copy of E. coli rrnB operon and an ampicillin resistance marker was mutagenized by passing through a mutator E. coli strain. For mutant selection, we used an E. coli strain, TA531, with seven chromosomal rRNA operons deleted. rRNA in TA531 cells is transcribed from the wild-type rrn operon contained on a plasmid pHK-rrnC+ carrying a kanamycin resistance marker (2, 3). Growth of TA531 cells transformed with the wild-type pKK3535 was significantly inhibited at 5 μg of ketolide antibiotic ABT-773 per ml and completely abolished on agar plates containing 10 μg of ABT-773 per ml. The mutagenized pKK3535 plasmid library was introduced into the TA531cells and Ketr clones were selected on agar plates containing 10 μg of ABT-773 per ml. Replica plating allowed identification of two clones that exhibited the Kans phenotype, indicating that they had lost the pHK-rrnC+ plasmid. rRNA in these clones was expressed exclusively from the mutant pKK3535, and the ribosomes were expected to contain only mutant rRNA. The ABT-773 MIC for both clones was 75 μg/ml, 15 times higher than the ABT-773 MIC for the Ampr Kans TA531 cells transformed with the wild-type pKK3535 (Table 2). The ketolide resistance was cotransferable with the mutant pKK3535 plasmids from these clones, demonstrating the presence of the ketolide resistance marker on the plasmid-borne genes.
TABLE 2.
Effect of 23S rRNA mutations on E. coli sensitivity to macrolide antibiotics
| Mutation | Plasmid | MIC (μM) of a:
|
||||
|---|---|---|---|---|---|---|
| Erythromycin | Azithromycin | Telithromycin | ABT-773 | RU69874 | ||
| None | pKK3535 or pNKb | 150 | 10 | 10 | 5 | 75 |
| U2609C | pKK2609C or pNK2609Cc | 75 | 5 | 75 | 75 | 50 |
| U754A | pKK754A | 150 | 10 | 25 | 5 | NDd |
| A2058Ge | pSTL102 | >500 | >50 | >100 | >100 | ND |
MIC for Kans Ampr TA531 E. coli cells expressing rRNA exclusively from the mutant plasmid.
Cells transformed with wild-type pKK3535 or pNK exhibited the same sensitivity to the antibiotics tested.
Cells transformed with the originally selected pKK2609C or engineered pNK2609C exhibited the same sensitivity to the antibiotics tested.
ND, not determined.
The pSTL102 plasmid contains spectinomycin resistance mutation in 16S rRNA gene (C1192T) in addition to the A2058G mutation in 23S rRNA gene. The C1192T mutation does not contribute to macrolide resistance.
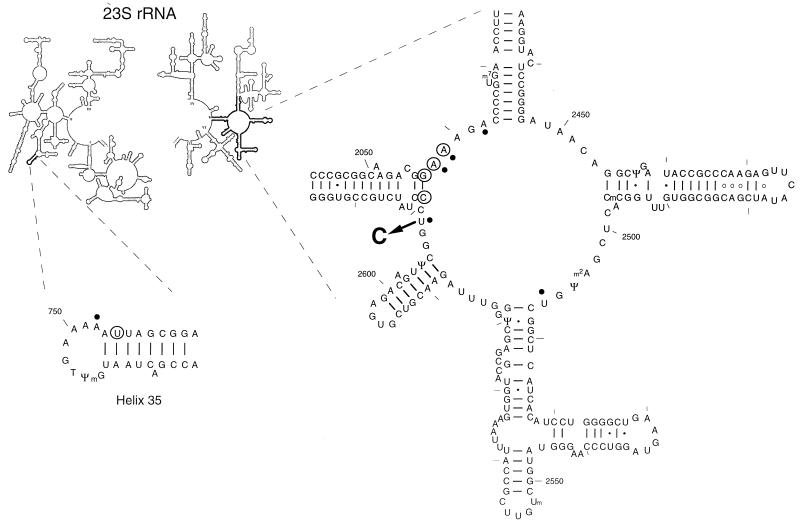
Two sites in the 23S rRNA were previously implicated in macrolide binding: the central loop of domain V and helix 35 in domain II of 23S rRNA. Sequencing domains II and V in the plasmid-borne 23S rRNA gene from two Ampr Kans Ketr clones revealed the presence of the same mutation, U2609 to C (Fig. 2), in both of them. To our best knowledge, this mutation was never previously recovered in antibiotic-resistant isolates. To test whether a single U2609C mutation was sufficient to confer ABT-773 resistance, a 2.2-kbp Bpu1102I-PaeI restriction fragment containing the U2609C mutation and no other nucleotide changes was excised from the mutant pKK3535 and used to replace an analogous fragment in the plasmid pNK (38) (Table 1). TA531 cells transformed with the resulting plasmid, pNK2609C, and cured from the wild-type pHK-rrnC+ plasmid, exhibited the same level of ketolide resistance as the originally selected Ketr mutants—hence proving that a single U2609C mutation in the 23S rRNA gene can confer resistance to ABT-773.
FIG. 2.
E. coli 23S rRNA positions involved in interaction with 14-member macrolide antibiotics. A newly identified ketolide resistance mutation, U2609 to C, is shown by an arrow. Sites of the previously characterized macrolide-resistance mutations are circled. For references, see reference 35. Nucleotide residues whose accessibility to chemical modification is affected by macrolides are marked by dots. The secondary structure is presented following those in references 4 and 14.
Peculiar spectrum of resistance of the U2609C mutant.
The spectrum of macrolide resistance conferred by the newly selected U2609C mutation was compared with that of the previously characterized mutation, A2058G (29). TA531 cells were transformed with pNK2609C or with pSTL102 carrying the A2058G mutations. Both of these plasmids contained an ampicillin resistance marker. The results of antibiotic sensitivity testing of the Ampr Kans transformants expressing exclusively mutant 23S rRNA are shown in Table 2. The A2058G mutation rendered cells resistant to all of the macrolides tested. In contrast, the U2609C mutation conferred considerable resistance to two ketolides, telithromycin and ABT-773, but made cells slightly more sensitive to the cladinosolides, erythromycin and azithromycin. Comparison of the effect of the U2609C mutation on cell sensitivity to telithromycin or its cladinose derivative, HMR69874, showed no resistance to the latter compound. Therefore, the U2609C mutation represents the first example of a nucleotide change in 23S rRNA that confers resistance specifically to ketolides.
Footprinting of ketolides and cladinosolides on the ribosome reveal structure-specific interactions.
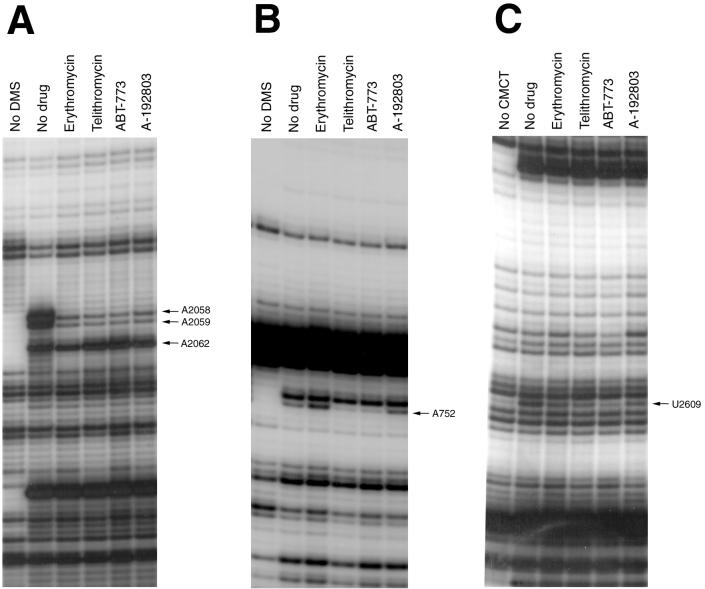
Ketolide resistance of the U2609C mutant suggested that U2609, as well as the neighboring rRNA sites, might be involved in direct interaction with the drug. To explore such possible interaction more directly, we used RNA footprinting (19). All of the macrolides tested strongly protected A2058 and A2059 in domain V from DMS modification (Fig. 3A), confirming the previous conclusion that these nucleotides may directly participate in macrolide binding (16, 20, 37). The ketolides ABT-773 and telithromycin enhance somewhat the accessibility of A2062 to chemical modification; similar effects were reported previously for telithromycin (11). Cladinosolides only marginally affected modification of A2062 (a minor protection of this position by erythromycin seen in Fig. 3A could not be reproduced in other experiments). It has been noted, however, that mutations of A2062 do not affect cell sensitivity to 14-member-ring macrolides (both cladinosolides and ketolides). Therefore, changes in the accessibility of A2062 to DMS modification in the presence of ketolides are probably the result of allosteric effects rather than an indication of a direct drug-RNA interaction.
FIG. 3.
Effect of macrolides (cladinosolides and ketolides) on accessibility of 23S rRNA nucleotides to chemical modification. (A) Central loop of domain V, DMS modification. (B) Hairpin 35, DMS modification. (C) Central loop domain V, CMCT modification. “No DMS” represents the unmodified control. “No drug” represents ribosomes modified in the absence of antibiotics. All of the other samples contained respective antibiotics present at a 5 μM concentration (see Materials and Methods for experimental details). Nucleotides with modification affected by bound antibiotics are indicated by arrows.
It has been reported previously that cladinosolides and ketolides with the 11,12 carbamate-linked alkyl-aryl side chain, had different effect on the accessibility of A752 in the loop of helix 35: ketolides (telithromycin) protected A752, while cladinosolides (erythromycin, clarithromycin, or roxithromycin) enhanced its modification by DMS (11, 16, 37; Xiong and Mankin, unpublished). Our RNA probing experiments showed that A752 is shielded from chemical modifications, not only by ketolides with the 11,12 side chain, like telithromycin, but also by ABT-773, which contains an alkyl-aryl side chain attached at position 6 of the lactone ring (Fig. 3B). On the other hand, binding of the ketolide A-192803, which has an 11,12 carbamate but lacks the side chain, did not change the accessibility of A752, indicating that it is not the carbamate group, but rather the side chain that is involved in interaction with the helix 35 loop.
CMCT probing revealed protection of U2609 by cladinosolides and ketolides. Interestingly, ABT-773 and, to a slightly lesser extent, telithromycin afforded stronger protection of U2609 than the other drugs tested (Fig. 3C), indicating possible idiosyncratic contacts between U2609 and therapeutically potent ketolide molecules. The U2609 protection suggests a close proximity of this residue to the macrolide binding site and its possible direct involvement in the drug binding. Both ketolides telithromycin and ABT-773 also appeared to protect U2506, although these effects were significantly weaker than protection of U2609 (data not shown).
The U2609C mutation reduces the affinity of ketolides for the ribosome.
rRNA mutations can confer resistance by reducing drug binding to the ribosome or by interfering with the drug action without perturbing its affinity (24). In order to verify the mode of resistance rendered by the U2609C mutation, binding studies were performed with radioactive erythromycin or ABT-773 and either wild-type or mutant (U2609C) ribosomes. Antibiotic binding was performed under equilibrium conditions, and the ribosome-drug complex was separated from the unbound antibiotic by high-speed centrifugation. The data showed that the U2609C mutation reduced binding of the ketolide, whereas the affinity of erythromycin, a cladinosolide, was slightly increased (Table 3), thus indicating that the U2609C mutation renders cells ketolide resistant by interfering with antibiotic binding. The results of the binding experiments were in excellent agreement with the resistance data (Table 2), in which the U2609C mutation rendered cells resistant to ABT-773 but slightly increased sensitivity to erythromycin.
TABLE 3.
Affinity of cladinosolides erythromycin and ketolide ABT-773 for E. coli ribosomes carrying different mutations in 23S rRNA
| Mutant or wild type |
Kd (nM)a
|
KdEry/KdABT-773 ratio | |
|---|---|---|---|
| Erythromycin | ABT-773 | ||
| Wild type | 7.8 | 1.3 | 6.0 |
| U754A | 6.7 | 2.8 | 2.4 |
| U2609C | 5.1 | 7.9 | 0.6 |
| A2058G | NDb | 1,500 | |
An average error in Kd measurements was 20%.
ND, not determined.
DISCUSSION
Dominant nature of macrolide resistance mutations.
In this paper, we described the selection of a ketolide-resistant mutant of E. coli. The presence of a single mutation, U2609C, in 23S rRNA was shown to be sufficient to confer resistance to ketolide antibiotics, but not to cladinose-containing macrolides.
During mutant selection, the resistance phenotype conferred by the U2609C mutation was initially manifested in a mixed-ribosome environment, in which a portion of the ribosome population contained mutant rRNA encoded in mutagenized pKK3535 plasmid, while the rest of the ribosomes contained wild-type rRNA transcribed from the pHK-rrnC+ plasmid. Hence, the U2609C-mediated resistance must be dominant or codominant. This conclusion is further supported by the fact that the pKK2609C plasmid rendered JM109 cells ketolide resistant in spite of the presence of the chromosome-encoded wild-type ribosomes (data not shown). The previously studied macrolide resistance mutations in rRNA were also shown to be dominant (see reference 34 for references). The dominant nature of these mutations is in agreement with the proposed mode of macrolide action according to which the drugs block nascent peptide growth and cause dissociation of peptidyl-tRNA (18). The peptidyl-tRNA loss should facilitate disengagement of sensitive ribosomes from mRNA, allowing resistant ribosomes to continue translation. The dominant nature of the U2609C mutation, as well as other rRNA mutations conferring resistance to macrolides, has an unfortunate consequence: the appearance of the mutation in just one out of several chromosomal rRNA operons may already provide a selective advantage for the mutant during antibiotic treatment. This may result in the subsequent spread of the mutation between other rRNA loci, rendering the pathogen resistant to even higher concentrations of the drug (31).
Macrolide binding site and specificity of ketolide interaction with the ribosome.
Several mutations conferring resistance to cladinose-containing macrolides have been found previously in the central loop of domain V of 23S rRNA (34, 35). The U2609C mutation is unique because it is the first mutation that confers resistance exclusively to ketolides without increasing cell resistance to cladinosolides. It is likely that the interaction of the nucleotide residue 2609 with the ketolide molecule differs from its interaction with the cladinose-containing macrolides. Such interaction can contribute to a tighter binding of ketolides to the ribosome and should be important for their activity.
The recently solved crystal structure of the 50S ribosomal subunit of the archaeon Halobacterium marismortui (4) provides a useful framework for modeling antibiotic binding sites in the ribosome (6). It should be kept in mind, however, that archaeal ribosomes differ from their bacterial counterparts both in protein composition and in rRNA structure. In particular, a number of nucleotide residues that are involved in macrolide binding are different in Bacteria and Archaea (15). Nevertheless, even though the nature and precise orientation of the nucleotide residues in Archaea and Bacteria can be idiosyncratic, their general ribosomal localization should be the same, allowing the use of the available crystallographic data for characterization of the macrolide binding site on the ribosome.
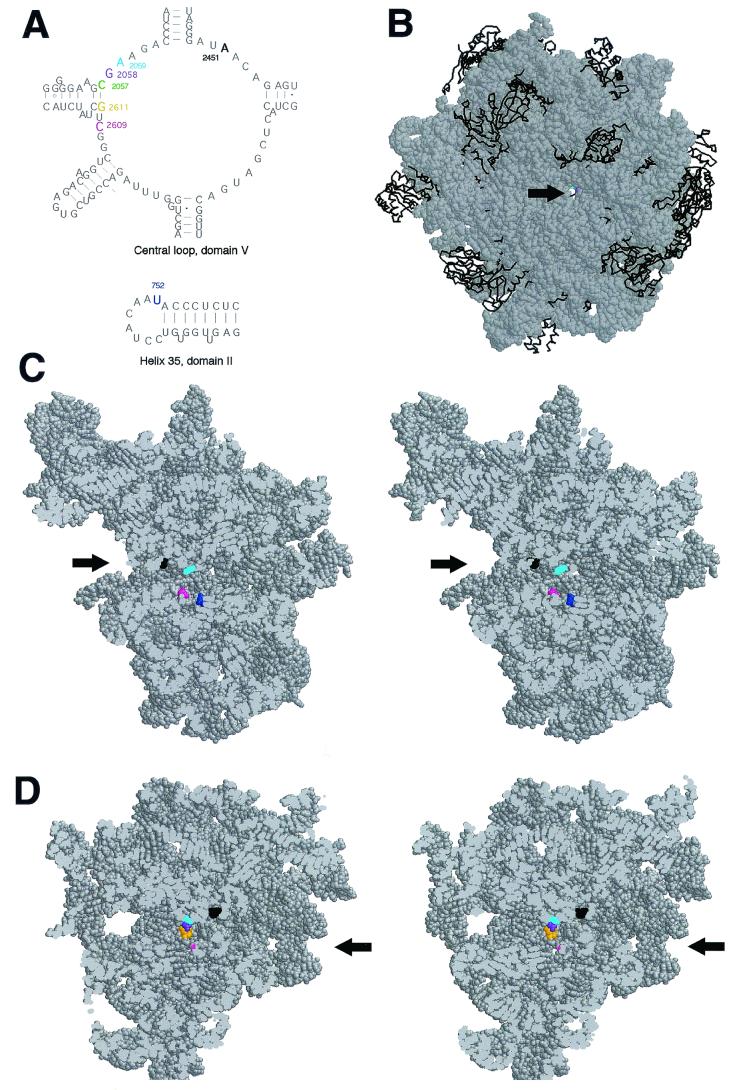
We used coordinates of nucleotides, according to genetic or biochemical criteria involved in macrolide binding, to map macrolide binding site in the ribosome. Residues 2057 to 2059, 2609, and 2611 in domain V and the residue at position 752 in domain II were included in our reconstructions (11, 16, 20, 25, 34, 37).
All of the nucleotides implicated in interactions with macrolides form a relatively tight cluster located at approximately one-third the distance from the interface side of the 50S subunit (Fig. 4 A to C). The drug binding site is located near the bottom of a cavity containing the peptidyl transferase center, at the entrance into the nascent peptide exit tunnel. The easiest way for the drug molecule to access this site in the isolated 50S subunit is from the interface side. However, given a relatively wide size of the exit tunnel and its hypothetical “nonstick properties” (22), it is also possible for the drug to diffuse to its binding site through the tunnel. This may be a preferred or even the only way for the drug to reach its site of action in 70S ribosome during early rounds of translation when the peptidyl transferase cavity is occupied by peptidyl- and aminoacyl-tRNAs.
FIG. 4.
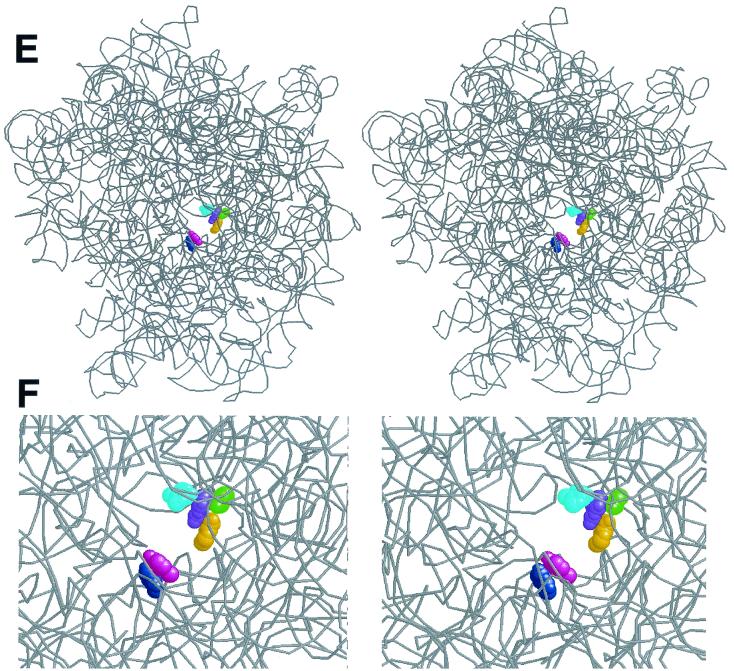
Relative position of nucleotides involved in macrolide binding in the crystallographic structure of the H. marismortui 50S ribosomal subunit. (A) Secondary structure of the central loop of domain V and hairpin 35 of H. marismortui 23S rRNA. Nucleotide positions in domain V at which mutations (in E. coli ribosome) confer macrolide resistance are shown in color. The nucleotide position in helix 35 corresponding to the adenine base in the E. coli ribosome for which modification is differentially affected by ketolides and cladinosolides is shown in blue. A2451 located within the active site of the peptidyl transferase center is shown in black. The nucleotide numbering is that of E. coli 23S rRNA. (B) “Front” view of the 50S subunit from the interface side. The opening of the peptidyl transferase center leading into the exit tunnel is indicated by an arrow. To make the tunnel visible, the subunit was rotated by 35° versus the y axis and tilted by 35° versus the z axis. rRNA is shown in a space-fill representation (gray), and protein backbones are shown in black. Positions 2058 and 2059 important for macrolide binding are seen at the bottom of the peptidyl transferase cavity. (C) Stereo view of a vertical cross-cut of the 50S subunit through the exit tunnel. (D) Same as panel C, but rotated by 180° versus the vertical axis. Only the rRNA moiety of the 50S subunit is shown. The entrance into the peptidyl transferase cavity (located at the interface surface of the subunit), which leads to the nascent peptide exit tunnel, is shown by arrows. Nucleotides involved in interaction with macrolide antibiotics are shown in color (same as in panel A), and A2451 located in the peptidyl transferase active site is shown in black. (E) Spatial arrangement of the nucleotides involved in macrolide binding in the exit tunnel seen from the peptidyl transferase (subunit interface) side. rRNA is shown as a gray backbone, while nucleotides involved in interaction with macrolide antibiotics are colored in a space-fill representation. Proteins were removed for clarity. (F) Close-up view of the macrolide binding site (approximately fourfold enlargement compared to panel E).
The nucleotide residues involved in macrolide binding form two clusters on the walls of the nascent peptide exit tunnel (Fig. 4B to E). The domain V residues, bp 2057 to 2611 (green and orange, in Fig. 4) and unpaired bp 2058 (purple) and 2059 (cyan) are grouped on one wall of the tunnel. These nucleotides appear to be involved in a similar type of interaction with different classes of macrolides: mono- or dimethylation of A2058 dramatically reduces the affinity of various macrolides (35), while all macrolides tested protect A2058 and A2059 from chemical modification (11, 16, 20, 25, 37). Mutations at these positions were shown to confer macrolide resistance (reviewed in reference 34). The “universal” contacts of macrolides with these domain V RNA residues should involve the regions in the drug structure that are shared by cladinosolides and ketolides. Since the lactone ring structure is rather variable between different macrolides, it is reasonable to think that at least some of the universal macrolide contacts should involve the desosamine sugar, the functional groups of which can participate in hydrogen bonding with RNA residues. The N1 position of G2058 in the 50S subunit of H. marismortui (corresponding to N1 of A2058 in Bacteria, which is protected by macrolides from chemical modification) is located at a distance of approximately 15 Å from A2451 (black in Fig. 4C and D), which demarcates the peptidyl transferase active site. This relatively large distance explains why the 14-member-ring macrolides do not interfere with the peptide bond formation, but start inhibiting the elongation of protein synthesis only when the nascent peptide reaches the size of 3 to 5 amino acids (33).
Position 2609 (magenta in Fig. 4) is located on the opposite wall of the exit tunnel (Fig. 4E and F). Both cladinosolides and ketolides protect U2609 from CMCT modification (25; this work). However, the U2609C mutation confers resistance exclusively to ketolides while increasing cell sensitivity to cladinosolides. Thus, the mode of interaction of ketolides with U2609 is probably different from that of cladinosolides. We think that the U2609 protection rendered by ketolides is likely to reflect a direct contact between U2609 and a ketolide molecule (involving probably the 3-keto group). In this case, changing the chemical identity of the RNA base is expected to disrupt contacts with the drug and should reduce affinity of the drug for the ribosome, which was exactly what was observed in our binding studies. The direct contact of ketolides with U2609 would explain in the most straightforward way the observed ketolide resistance conferred by the U2609 mutation. Cladinosolides, on the other hand, may cause protection of U2609 allosterically, or, alternatively, the cladinose residue may shield U2609 from chemical modification without making direct contact with the RNA base.
The residues at positions 752 (dark blue in Fig. 4) (U, in H. marismortui, but A in E. coli) in the loop of helix 35 is located behind U2609 if seen from the peptidyl transferase center (Fig. 4C). Similar to the interactions of macrolides with U2609, their contacts with A752 are also structure specific, since ketolides protect while cladinosolides increase exposure of A752 to modification with DMS (16, 37). The protection of A752 is affected by the 3-keto group and by the presence of an extended alkyl-aryl side chain (11). The location of the side chain is not critical, and ketolides with the side chain attached to either the 11,12 carbamate group (as in telithromycin) or the C6 oxygen atom (as in ABT-773) protect A752 equally well (Fig. 3B). Interestingly, however, protection of A752 depends dramatically on the chemical nature of the ketolide side chain, suggesting that it may form a direct contact with residue 752 (Xiong and Mankin, unpublished data). Interaction of the 3-keto group with U2609 can place the ketolide molecule in a position favorable for the protection of A752 from the chemical modification.
Spatial separation of the nucleotide residues involved in universal interactions with different macrolides (positions 2057, 2058, 2059, and 2611) and those that form structure-specific contacts (752 and 2609) explains the improved activity profile of ketolides and illuminates venues for future drug development. Specific interactions of the 3-keto group with U2609 and of the alkyl-aryl side chain with A752 may increase the affinity of ketolides and therefore provide enough binding energy, even when interaction with the “universal” macrolide binding site on the other side of the exit tunnel is impaired due to methylation of A2058 by Erm-type methylases or mutations. Thus, it appears that one of the promising ways to further improve drug activity would be through strengthening or extending drug contacts in the vicinity of residues 2609 and 752.
The position of the macrolide binding site in the ribosome is in very good agreement with the proposed mechanism of drug action in which the drug blocks translation by sterically hindering growth of the nascent polypeptide. Located in the narrow portion of the exit tunnel, the bound macrolide molecule should efficiently barricade it and prevent the nascent peptide from entering the tunnel. Since translation is blocked prior to insertion of the nascent peptide into the tunnel, the peptidyl-tRNA eventually dissociates from the ribosome with its subsequent hydrolysis by peptidyl-tRNA hydrolase. This mechanism of drug action explains why resistant mutations are dominant: sensitive ribosomes do not block mRNA for a long time and therefore allow a resistant ribosome to translate it. It is obvious that the drugs would be more potent if blockage of mRNA translation would be irreversible so that stalled ribosomes would not dissociate from mRNA. Theoretically, this could be achieved if compounds are found that bind “deeper” in the exit tunnel, close to its orifice at the “back” of the 50S subunit. A relatively long nascent peptide threaded through the tunnel will increase the affinity of peptidyl-tRNA to the ribosome, thus preventing its dissociation and extending the time during which translation of mRNA is blocked.
ACKNOWLEDGMENTS
We thank S. Douthwaite, F. Francheschi, J. Sutcliffe, and P. Mauvais for stimulating discussion and advice; N. Polacek for advice regarding the manuscript, H. F. Noller, C. Squires, and S. Douthwaite for making their plasmid constructs available to us, and M. Gomez for help in preparing the manuscript. We also thank Aventis Pharma for providing telithromycin and RU 69847.
This work was supported by research grants from Abbott Laboratories and the National Institutes of Health (GM53762).
REFERENCES
- 1.Arevalo M A, Tejedor F, Polo F, Ballesta J P. Protein components of the erythromycin binding site in bacterial ribosomes. J Biol Chem. 1988;263:58–63. [PubMed] [Google Scholar]
- 2.Asai T, Condon C, Voulgaris J, Zaporojets D, Shen B, Al-Omar M, Squires C, Squires C L. Construction and initial characterization of Escherichia coli strains with few or no intact chromosomal rRNA operons. J Bacteriol. 1999;181:3803–3809. doi: 10.1128/jb.181.12.3803-3809.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asai T, Zaporojets D, Squires C, Squires C L. An Escherichia coli strain with all chromosomal rRNA operons inactivated: complete exchange of rRNA genes between bacteria. Proc Natl Acad Sci USA. 1999;96:1971–1976. doi: 10.1073/pnas.96.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ban N, Nissen P, Hansen J, Moore P B, Steitz T A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 5.Beauclerk A A, Cundliffe E. Sites of action of two ribosomal RNA methylases responsible for resistance to aminoglycosides. J Mol Biol. 1987;193:661–671. doi: 10.1016/0022-2836(87)90349-4. [DOI] [PubMed] [Google Scholar]
- 6.Belova L, Tenson T, Xiong L, McNicholas P M, Mankin A S. A novel site of antibiotic action in the ribosome: interaction of evernimicin with the large ribosomal subunit. Proc Natl Acad Sci USA. 2001;98:3726–3731. doi: 10.1073/pnas.071527498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 8.Chittum H S, Champney W S. Ribosomal protein gene sequence changes in erythromycin-resistant mutants of Escherichia coli. J Bacteriol. 1994;176:6192–6198. doi: 10.1128/jb.176.20.6192-6198.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chittum H S, Champney W S. Erythromycin inhibits the assembly of the large ribosomal subunit in growing Escherichia coli cells. Curr Microbiol. 1995;30:273–279. doi: 10.1007/BF00295501. [DOI] [PubMed] [Google Scholar]
- 10.Cundliffe E. Antibiotic inhibitors of ribosome function. In: Gale E F, Cundliffe E, Reynolds P E, Richmond M H, Waring M J, editors. The molecular basis of antibiotic action. London, United Kingdom: John Wiley & Sons; 1981. pp. 402–545. [Google Scholar]
- 11.Douthwaite S, Hansen L H, Mauvais P. Macrolide-ketolide inhibition of MLS-resistant ribosomes is improved by alternative drug interaction with domain II of 23S rRNA. Mol Microbiol. 2000;36:183–193. doi: 10.1046/j.1365-2958.2000.01841.x. [DOI] [PubMed] [Google Scholar]
- 12.Gabashvili I S, Gregory S T, Valle M, Grassucci R, Worbs M, Wahl M C, Dahlberg A E, Frank J. The polypeptide tunnel system in the ribosome and its gating in erythromycin resistance mutants of L4 and L22. EMBO J. 2001;8:181–188. doi: 10.1016/s1097-2765(01)00293-3. [DOI] [PubMed] [Google Scholar]
- 13.Gregory S T, Dahlberg A E. Erythromycin resistance mutations in ribosomal proteins L22 and L4 perturb the higher order structure of 23 S ribosomal RNA. J Mol Biol. 1999;289:827–834. doi: 10.1006/jmbi.1999.2839. [DOI] [PubMed] [Google Scholar]
- 14.Gutell R R. Comparative sequence analysis and the structure of 16S and 23S rRNA. In: Zimmermann R A, Dahlberg A E, editors. Ribosomal RNA: structure, evolution, processing, and function in protein biosynthesis. Boca Raton, Fla: CRC Press; 1996. pp. 111–128. [Google Scholar]
- 15.Gutell R R, Weiser B, Woese C R, Noller H F. Comparative anatomy of 16S-like ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- 16.Hansen L H, Mauvais P, Douthwaite S. The macrolide-ketolide antibiotic binding site is formed by structures in domains II and V of 23S ribosomal RNA. Mol Microbiol. 1999;31:623–632. doi: 10.1046/j.1365-2958.1999.01202.x. [DOI] [PubMed] [Google Scholar]
- 17.Lai C J, Weisblum B. Altered methylation of ribosomal RNA in an erythromycin-resistant strain of Staphylococcus aureus. Proc Natl Acad Sci USA. 1971;68:856–860. doi: 10.1073/pnas.68.4.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menninger J R, Otto D P. Erythromycin, carbomycin, and spiramycin inhibit protein synthesis by stimulating the dissociation of peptidyl-tRNA from ribosomes. Antimicrob Agents Chemother. 1982;21:811–818. doi: 10.1128/aac.21.5.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merryman C, Noller H F. Footprinting and modification-interference analysis of binding sites on RNA. In: Smith C W J, editor. RNA:protein interactions, a practical approach. Oxford, United Kingdom: Oxford University Press; 1998. pp. 237–253. [Google Scholar]
- 20.Moazed D, Noller H F. Chloramphenicol, erythromycin, carbomycin and vernamycin B protect overlapping sites in the peptidyl transferase region of 23S ribosomal RNA. Biochimie. 1987;69:879–884. doi: 10.1016/0300-9084(87)90215-x. [DOI] [PubMed] [Google Scholar]
- 21.Nilius A M, Bui M H, Almer L, Hensey-Rudloff D, Beyer J, Ma Z, Or Y S, Flamm R K. Comparative in vitro activity of ABT-773, a novel antibacterial ketolide. Antimicrob Agents Chemother. 2001;45:2163–2168. doi: 10.1128/AAC.45.7.2163-2168.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nissen P, Hansen J, Ban N, Moore P B, Steitz T A. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 23.Pardo D, Rosset R. Genetic studies of erythromycin resistant mutants of Escherichia coli. Mol Gen Genet. 1974;135:257–268. doi: 10.1007/BF00268620. [DOI] [PubMed] [Google Scholar]
- 24.Porse B T, Leviev I, Mankin A S, Garrett R A. The antibiotic thiostrepton inhibits a functional transition within protein L11 at the ribosomal GTPase centre. J Mol Biol. 1998;276:391–404. doi: 10.1006/jmbi.1997.1541. [DOI] [PubMed] [Google Scholar]
- 25.Poulsen S M, Kofoed C, Vester B. Inhibition of the ribosomal peptidyl transferase reaction by the mycarose moiety of the antibiotics carbomycin, spiramycin and tylosin. J Mol Biol. 2000;304:471–481. doi: 10.1006/jmbi.2000.4229. [DOI] [PubMed] [Google Scholar]
- 26.Reinert R R, Bryskier A, Lütticken R. In vitro activities of the new ketolide antibiotics HMR 3004 and HMR 3647 against Streptococcus pneumoniae in Germany. Antimicrob Agents Chemother. 1998;42:1509–1511. doi: 10.1128/aac.42.6.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sigmund C D, Morgan E A. Erythromycin resistance due to mutation in a ribosomal RNA operon of Escherichia coli. Proc Natl Acad Sci USA. 1982;79:5602–5606. doi: 10.1073/pnas.79.18.5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skinner R, Cundliffe E, Schmidt F J. Site of action of a ribosomal RNA methylase responsible for resistance to erythromycin and other antibiotics. J Biol Chem. 1983;258:12702–12706. [PubMed] [Google Scholar]
- 29.Sor F, Fukuhara H. Identification of two erythromycin resistance mutations in the mitochondrial gene coding for the large ribosomal RNA in yeast. Nucleic Acids Res. 1982;10:6571–6577. doi: 10.1093/nar/10.21.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stern S, Moazed D, Noller H F. Analysis of RNA structure using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- 31.Tait-Kamradt A, Davies T, Cronan M, Jacobs M R, Appelbaum P C, Sutcliffe J. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob Agents Chemother. 2000;44:2118–2125. doi: 10.1128/aac.44.8.2118-2125.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Triman K, Becker E, Dammel C, Katz J, Mori H, Douthwaite S, Yapijakis C, Yoast S, Noller H F. Isolation of temperature sensitive mutants of 16S rRNA in Escherichia coli. J Mol Biol. 1989;209:645–653. doi: 10.1016/0022-2836(89)92000-7. [DOI] [PubMed] [Google Scholar]
- 33.Vazquez D. The macrolide antibiotics. In: Corcoran J W, Hahn F E, editors. Antibiotics III. Mechanism of action of antimicrobial and antitumor agents. New York, N.Y: Springer-Verlag; 1975. pp. 459–479. [Google Scholar]
- 34.Vester B, Douthwaite S. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob Agents Chemother. 2001;45:1–12. doi: 10.1128/AAC.45.1.1-12.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wittmann H G, Stoffler G, Apirion D, Rosen L, Tanaka K, Tamaki M, Takata R, Dekio S, Otaka E. Biochemical and genetic studies on two different types of erythromycin resistant mutants of Escherichia coli with altered ribosomal proteins. Mol Gen Genet. 1973;127:175–189. doi: 10.1007/BF00333665. [DOI] [PubMed] [Google Scholar]
- 37.Xiong L, Shah S, Mauvais P, Mankin A S. A ketolide resistance mutation in domain II of 23S rRNA reveals proximity of hairpin 35 to the peptidyl transferase centre. Mol Microbiol. 1999;31:633–639. doi: 10.1046/j.1365-2958.1999.01203.x. [DOI] [PubMed] [Google Scholar]
- 38.Xiong L, Kloss P, Douthwaite S, Andersen N M, Swaney S, Shinabarger D L, Mankin A S. Oxazolidinone resistance mutations in 23S rRNA of Escherichia coli reveal the central region of domain V as the primary site of drug action. J Bacteriol. 2000;182:5325–5331. doi: 10.1128/jb.182.19.5325-5331.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]