Abstract
Background
Hepatocellular carcinoma (HCC) is one of the most malignant tumors worldwide. The ST6 β-galactoside α-2, 6-sialyltransferase 1 (ST6GAL1) has been found aberrantly expressed in a variety of cancers including HCC, but its function and mechanism in regulating liver inflammation remain to be investigated. This study aimed to explore the role of ST6GAL1 in HCC. The data of ST6GAL1 expression, prognosis, and clinical parameters were collected and further analyzed from the public databases including The Cancer Genome Atlas (TCGA), Human Protein Atlas (HPA), and Gene Expression Omnibus (GEO). The HCC rat model was constructed by intraperitoneal injection of diethylnitrosamine. The mRNA and protein expression levels of ST6GAL1 in rat liver tissues were detected by real-time quantitative polymerase chain reaction, capillary electrophoresis, and Western blot.
Results
The ST6GAL1 mRNA and protein expression levels were both lower in HCC tissues compared with normal liver tissues in the public databases and HCC rat model. The survival analysis showed that upregulation of ST6GAL1 was an independent prognostic factor for good prognosis in HCC patients. The ST6GAL1 mRNA expression showed a negative correlation with ST6GAL1 methylation levels. Enrichment analysis showed that ST6GAL1 expression was most associated with metabolic, cancer, estrogen, axon guidance, cAMP, and PI3K-AKT signaling pathways. The ST6GAL1 mRNA expression negatively correlated with liver inflammation status and proportion of NK CD56bright, NK CD56dim, pDC, and CD8+ T cells in liver.
Conclusion
Compared with normal tissues, ST6GAL1 was lower expressed in HCC tumor tissues, and the downregulation of ST6GAL1 was associated with a poor prognosis in HCC patients. ST6GAL1 could further affect the infiltration of immune cells to exert anti-inflammation function in liver. Our study indicated that ST6GAL1 could be a potential biomarker and therapeutic target to assess the prognosis and regulate the immune cells infiltration level of HCC.
Keywords: hepatocellular carcinoma, ST6GAL1, prognosis, immune infiltration, biomarker
Background
Hepatocellular carcinoma (HCC) is one of the most malignant tumors in clinical practice, leading to at least 600,000 deaths each year.1 However, the pathological characteristics of HCC are not easily detected, making late radical treatment extremely difficult.2 Therefore, there is an urgent need to find steady and reliable biomarkers to screen patients with poor prognoses and provide more aggressive treatment earlier.
HCC usually arises from chronic inflammation, and the immune response plays a crucial role in cancer occurrence and development.3 Chronic liver inflammation causes repeated damage and regeneration of hepatocytes, which in turn induces immune cell tolerance, resulting in a dysregulated immune response and eventual progression to HCC.4 The immune cells, a major component of the tumor microenvironment (TME), can migrate from hematopoietic organs and peripheral blood to the liver, establishing an active immune ecological niche that interacts with parenchymal hepatic cells, and influences differentiation, tumorigenesis, and progression.5 A related study has shown that insufficient crosstalk between dendritic cells (DCs) and T cells was one of the main mechanisms of tumor tolerance in HCC.6 In addition, the immune cell infiltration level has a significant impact on the prognosis of HCC.7 Meanwhile, the immune therapeutics could achieve a good therapeutic effect by modulating the TME and maintaining the balance of immune homeostasis in solid tumors.8 Therefore, it is important to explore the molecular changes associated with oncogenic inflammation in tumor tissue or adjacent lymphocytes.
Beta-galactoside alpha-2, 6-sialyltransferase 1 (ST6GAL1) is a sialyltransferase that mediates the glycosylation of proteins and lipids by transferring alpha2-6-linked sialic acid to the glycosyl terminus of glycoproteins and glycolipids.9 Altered expression of sialyltransferase usually causes abnormalities in salivary acidification and change in cellular glycosylation, which are closely associated with tumorigenesis.10–14 Furthermore, the ST6GAL1 has been found that exerted an important role in tissue inflammation damage. It has been shown that ST6GAL1 could prevent intestinal inflammation by regulating T cell immunity levels and play a protective effect on IgA nephropathy by inhibiting the production of proinflammatory cytokines in patients.15,16 While the deficiency of ST6GAL1 could cause exaggerated acute neutrophilic inflammation, increase radiation-induced gastrointestinal damage, and promote the transformation of synovial fibroblasts into a pro-inflammatory phenotype in mice.17–19 For HCC, it has been reported that serum ST6GAL1 levels were positively correlated with tumor FGF19 expression in patients with surgically resected HCC, and could be a novel serum biomarker for lenvatinib-susceptible FGF19-driven HCC.20 However, the comprehensive mechanism of how ST6GAL1 regulates immune response and liver inflammation in HCC remains to be elucidated.
In this study, we first analyzed the ST6GAL1 expression and the relationships between ST6GAL1 expression and clinicopathological indicators in HCC through public databases. In addition, we validated the mRNA and protein expression of ST6GAL1 in rats’ normal and HCC liver tissues. Finally, we performed immune infiltration and functional enrichment analysis to further explore the potential mechanisms of ST6GAL1 in the development and progression of HCC.
Methods
Databases
The RNA-seq and clinicopathologic data of HCC were downloaded from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) module of the Xena Public Data Hubs in the UCSC Xena platform (http://xena.ucsc.edu/)38 for further analysis. The ST6GAL1 immunohistochemical staining data of liver tissues were obtained from the Human Protein Atlas (HPA) database (http://www.proteinatlas.org/). The gene expression profile data of GSE83148, GSE54238, GSE84044, GSE54236, and GSE22508 datasets were downloaded from the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/). These databases were further used for ST6GAL1 expression and clinicopathologic data analyses.
Survival Analysis
The Kaplan-Meier mapper platform39 was used as a meta-analysis tool to analyze the survival profile of HCC patients. The Kaplan-Meier mapper platform splits patients into high- and low-risk groups by automatically selecting the best cut-off value. All possible cutoff values between the lower and upper quartiles were computed, and the best performing threshold was used as a cutoff. Common clinical risk factors were used as a basis for patient stratification. A nomogram was constructed to predict the correlation between clinical factors associated with patients with hepatocellular carcinoma and disease prognosis.
ST6GAL1 Gene Copy Number Variation and Methylation Analysis
The copy number variation (CNV) and methylation level data of the ST6GAL1 gene were downloaded through the MEXPRESS platform.40 We selected the probe cg16751732, the most predominant ST6GAL1 CpG location, from the probes of interest for further analysis. The prognostic value of ST6GAL1 methylation levels in HCC was appraised using the MethSurv platform.41 The patients were stratified into high- and low-risk groups based on the q25 value of their risk scores.
Gene Enrichment Analysis
The DESeq2 R package (version 1.26.0) was used to calculate the correlation of ST6GAL1 expression with all human genes and to compare the expression profiles between high- and low-ST6GAL1 mRNA expression groups.42 |log2Fold Change| = 2 and adjusted P = 0.05 were considered as cut-off criteria for differential expression genes (DEGs), log2Fold Change > 2 and adjusted P < 0.05 were defined as up-regulated genes, and log2Fold Change < −2 and adjusted P < 0.05 were defined as down-regulated genes. The DEGs encoding proteins were used for gene enrichment analysis of gene annotation and Kyoto encyclopedia of genes and genomes (KEGG) by the Sangerbox platform (http://sangerbox.com/).43–45 Adjusted P < 0.1 and q value < 0.2 are used as thresholds for gene enrichment analysis. Sorting was also performed based on the magnitude of Count Values. The ggplot2 R package (version 3.3.3) was applied for DEGs correlation analysis and sorted in descending order using the absolute R value as the criterion.
Immune Infiltration Analysis
The GSVA R package (version 1.34.0) was utilized to appraise the relationship between 24 tumor-infiltrating immune cell markers and different ST6GAL1 mRNA expression groups in HCC samples.46 Spearman correlation between HCC immune cells infiltration and ST6GAL1 expression levels was calculated using the ssGSEA algorithm on 424 samples from TCGA. A P value < 0.05 was used as a criterion for screening immune cells that might be affected by ST6GAL1 expression. The TISIDB platform was used to explore the correlations between ST6GAL1 expression and immunoinhibitor or immunostimulator from the “Immunomodulator” module in HCC.47
Animals
Twelve 6-week-old male Wistar rats, weighing 200g ± 20g, were purchased from Beijing Vital River Laboratory Animal Technology Co. Ltd (license: SCXK (Beijing) 2016–0006). All animals were raised in the Barrier Environmental Animal Laboratory of Dongzhimen Hospital of Beijing University of Chinese Medicine (license: SYXK (Beijing) 2015–0001) and maintained under the National Standards for Laboratory Animals of China (GB14925-2010). Our study was carried out in compliance with the ARRIVE guidelines. This study was approved by the Ethics Committee of Laboratory Animals of Dongzhimen Hospital of Beijing University of Chinese Medicine (No.21–10). Animals were kept separately in an SPF laboratory, with the breeding environment: temperature 25 ± 1°C, humidity 50 ± 10%, free of food and drinking water, 12-hour day and night alternation, as well as adaptable feeding for 5 days. Rats were anesthetized by intraperitoneal injection using a 3% sodium pentobarbital solution. After anesthesia, blood was taken from the rats as well as the material, and if necessary, euthanasia by cervical dislocation was performed.
HCC Rat Model
The HCC rat model was constructed by intraperitoneal injection of diethylnitrosamine (Psaitong, N60001, 50 mg/kg/week, CN). After sixteen weeks, the rats were sacrificed and livers were obtained for the next experiments.
Real-Time Quantitative Polymerase Chain Reaction Analysis
The total RNA was extracted from the biopsy tissue of rat liver using the RaPure Total RNA Mini Kit (Magen, R4011, CN) according to the manufacturer’s instructions. Total RNA reverse transcription to cDNA was performed with the All-in-One First-Strand Synthesis MasterMix (with dsDNase) (BioMed, BM60501S, CN). Real-time quantitative polymerase chain reaction (RT-qPCR) was performed using the Real-time PCR Detection System (Agilent Technologies, US) with the Taq SYBR® Green qPCR Premix (BioMed, BM60304S, CN). The primers were provided as follows:
ST6GAL1-F: AAGGACAGTTTGTACACCGA;
ST6GAL1-R: CTGATACCACTTTGGGATATCTG;
GAPDH-F: AGACAGCCGCATCTTCTTGT;
GAPDH-R: CTTGCCGTGGGTAGAGTCAT.
Capillary Electrophoresis and Western Blot
The total protein was extracted from the biopsy tissue of the rat liver using the RIPA Lysis Buffer (Beyotime, P0013B, CN) according to the manufacturer’s instructions. The capillary electrophoresis was performed on the Wes instrument according to the manufacturer’s instructions (ProteinSimple, San Jose, CA; SM-W004). For Western blot, the protein lysates were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (Epizyme Biomedical Technology, PG112, CN) and then electrophoretically transferred onto the polyvinylidene fluoride membranes (Epizyme Biomedical Technology, WJ001, CN). The primary antibodies we used were rabbit anti-ST6GAL1 (Proteintech, 14355-1-AP, 1:1000, US); mouse anti-GAPDH (MBL, M171-3, 1:5000, JPN). The Image J software was used to analyze the integrated density of the protein bands.
Statistics
Statistical analysis of all data was performed using R (version 3.6.3) and GraphPad Prism (version 9.1.0). The Wilcoxon rank sum test was used to analyze the expression of ST6GAL1 in unpaired samples. The Kruskal–Wallis test and Dunn’s test were utilized to evaluate the relationship between ST6GAL1 expression and clinicopathological characteristics. Kaplan-Meier method was applied for survival curve plotting. Differences between groups were determined by an unpaired t-test. P value < 0.05 was considered statistically significant in this study. Spearman and statistical significance were used to analyze the gene expression correlations. |R| > 0.1 were considered as correlations and P value < 0.05 were considered statistically significant.
Results
ST6GAL1 Expression Was Downregulated in HCC
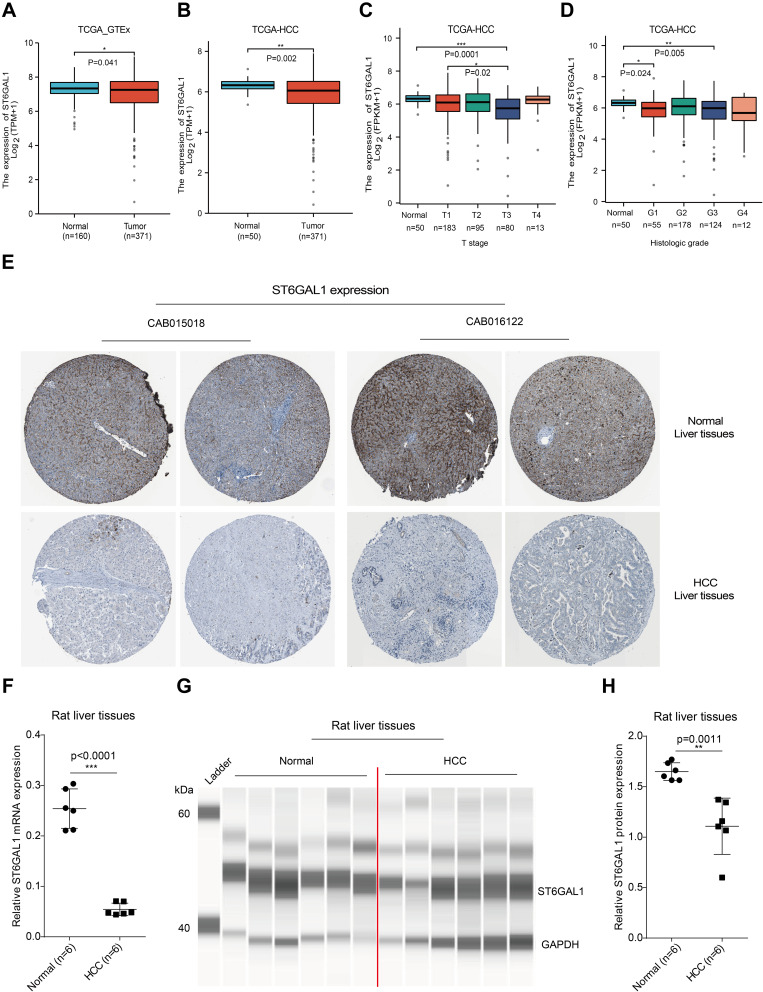
To clear ST6GAL1 expression and its correlations with clinicopathological parameters in HCC, we first analyzed data from the TCGA and GTEx databases. The results showed that ST6GAL1 mRNA expression was significantly lower in HCC compared with non-tumor liver tissues (Figure 1A). Meanwhile, ST6GAL1 mRNA expression was also lower in HCC tissues compared with adjacent non-tumor tissues in TCGA database (Figure 1B). We also analyzed ST6GAL1 mRNA expression in paired non-tumor and HCC tissues. The results showed that, compared with paired adjacent non-tumor liver tissues, ST6GAL1 expression was significantly downregulated in HCC tissues of TCGA database, GSE54236 dataset, and GSE22508 dataset (Figure S1). In addition, according to different clinicopathological parameters, we found that compared to the normal, ST6GAL1 mRNA expression was significantly decreased at different T stages (Figure 1C) and histological grades (Figure 1D). Next, we analyzed the ST6GAL1 protein expression in liver tissues in the HPA database, and the results showed that ST6GAL1 was also lower expressed in HCC liver tissues compared to the normal (Figure 1E). Through the RT-PCR and capillary electrophoresis approaches, we further detected the ST6GAL1 expression in liver tissues of the DEN-induced rat HCC model. The RT-PCR result showed that the ST6GAL1 mRNA expression in HCC-rat liver tissues was significantly lower than that in normal tissues (Figure 1F). The capillary electrophoresis experiment (CEE) demonstrated that ST6GAL1 protein level in HCC-rat liver tissues was also significantly lower than those in normal tissues (Figure 1G and H). And we also performed SDS-PAGE to detect ST6GAL1 protein expression, and the result was coincident with the CEE (Figure S2A and B). These results indicated that ST6GAL1 was downregulated in HCC on both mRNA and protein levels.
Figure 1.
ST6GAL1 expression in normal and HCC liver tissues. (A) The ST6GAL1 mRNA expression of HCC (n = 371) and normal tissues (n = 160) in the TCGA and GTEx databases; (B) The ST6GAL1 mRNA expression of HCC (n = 371) and adjacent non-tumor tissues (n = 50) in the TCGA database; (C and D) The ST6GAL1 mRNA expression of different T stages (C) or histologic grades (D) in the TCGA database; (E) The ST6GAL1 protein expression in the HPA database; (F) The ST6GAL1 mRNA expression levels in liver tissues of normal (n = 6) and HCC (n = 6) rats, detected by qRT-PCR and GAPDH as the internal reference; (G) The ST6GAL1 protein expression levels in liver tissues of normal (n = 6) and HCC (n = 6) rats, detected by capillary electrophoresis analysis and GAPDH as the internal reference; (H) The integrated density analysis of the protein bands. *p < 0.05, **p < 0.01, ***p < 0.001.
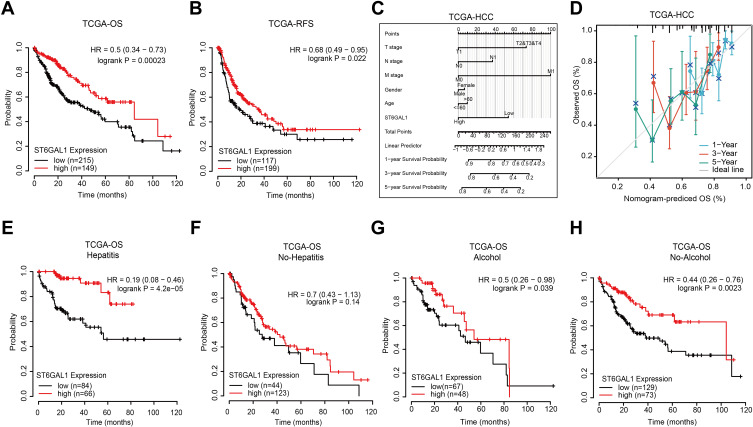
Lower ST6GAL1 mRNA Expression is Associated with Poor Prognosis in HCC
We next analyzed the correlation between ST6GAL1 mRNA expression and HCC patients’ survival. The Kaplan-Meier survival analysis showed that patients with low ST6GAL1 expression had a worse overall survival (OS) and recurrence-free survival (RFS) than those in the high expression group (Figure 2A and B). Next, we investigated the correlations between ST6GAL1 mRNA expression and prognosis in different clinical subgroups of HCC. As we could see from the results, the upregulation of ST6GAL1, decreasing T stage, and negative M stage were independent prognostic factors for a good prognosis (Figure 2C). Moreover, we evaluated the nomogram performance according to ST6GAL1 expression, and the C-index of OS was 0.642, which also presented the good prognostic predictive efficacy of ST6GAL1 expression (Figure 2D). Meanwhile, because viral hepatitis is one of the most important pathogenic factors of HCC, we further analyzed the prognostic relationship between viral hepatitis (VH), alcohol hepatitis (AH), and ST6GAL1 expression in HCC. The results showed that in VH-related HCC, patients with higher ST6GAL1 expression had better OS than those in the low expression group (Figure 2E), while there was no significant difference in patients without VH (Figure 2F). For AH or not, patients with higher ST6GAL1 expression had better OS than those in the lower expression group (Figure 2G and H), while the difference was more significant in the no AH-related HCC group. These results indicated the potential of ST6GAL1 expression to predict the prognosis of HCC, especially in the viral hepatitis-related patient population.
Figure 2.
The correlation between ST6GAL1 mRNA expression and HCC patients’ prognosis in TCGA database. (A and B) The overall survival (n = 364, (A)) and relapse-free survival (n = 316, (B)) analysis between ST6GAL1 expression and HCC; (C) A nomogram that combines ST6GAL1 and other prognostic factors in HCC; (D) The calibration curve of the nomogram; (E–H) The overall survival analysis of ST6GAL1 expression in HCC patients with viral hepatitis (n = 150, (E)), no-viral hepatitis (n = 167, (F)), alcohol consumption (n = 115, (G)), and no-alcohol consumption (n = 202, (H)).
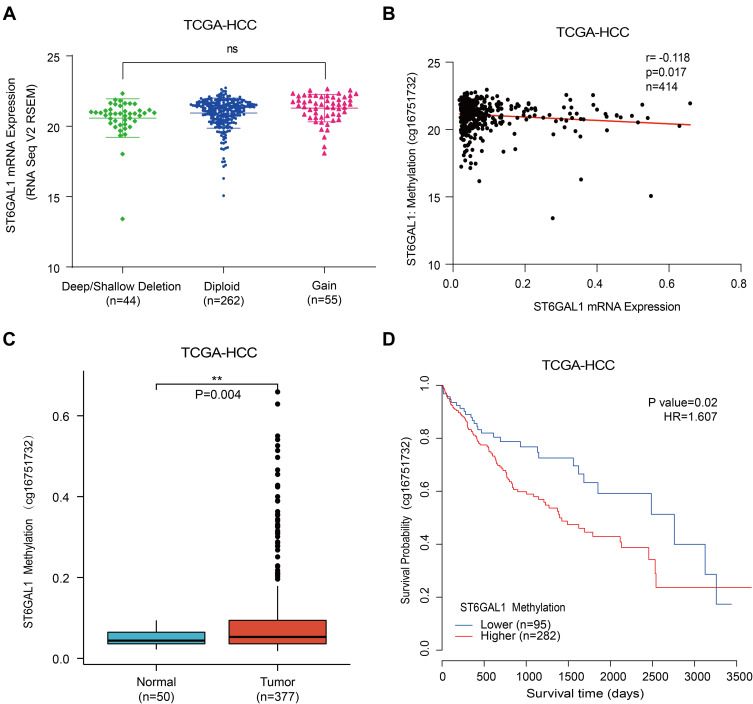
ST6GAL1 mRNA Expression Was Correlated with DNA Methylation
To further explore the expression of ST6GAL1, we analyzed the association of ST6GAL1 mRNA expression with its copy number variation (CNV) and methylation level in the TCGA database. As Figure 3A showed, patients with ST6GAL1 gene amplification or deletion were found to have a similar proportion and there was no significant difference between these two groups. Subsequently, we analyzed and found there existed a significant difference between ST6GAL1 mRNA expression and ST6GAL1 gene methylation level, and they showed a negative correlation (Figure 3B). In addition, methylation of ST6GAL1 promoter was higher in tumor tissues than in normal tissues (Figure 3C). We further performed the prognostic analysis of ST6GAL1 methylation in HCC, and the result showed that patients with high ST6GAL1 methylation had poor OS than those with low ST6GAL1 methylation (Figure 3D). These results suggested that ST6GAL1 expression was mainly regulated by gene methylation in HCC.
Figure 3.
The associations of ST6GAL1 mRNA expression with gene copy number variation and methylation in HCC. (A) The ST6GAL1 mRNA expression in tumor tissues of gene copy neutral (n = 262), deletion (n = 44), and duplication (n = 55) groups in the TCGA-HCC database; (B) The correlation between ST6GAL1 mRNA expression and gene methylation level of liver tissues in the TCGA-HCC database; (C) The ST6GAL1 methylation levels in HCC (n = 377) and normal liver tissues (n = 50) in the TCGA-HCC database; (D) The overall survival analysis between ST6GAL1 methylation level and HCC in the TCGA-HCC database. **p < 0.01.
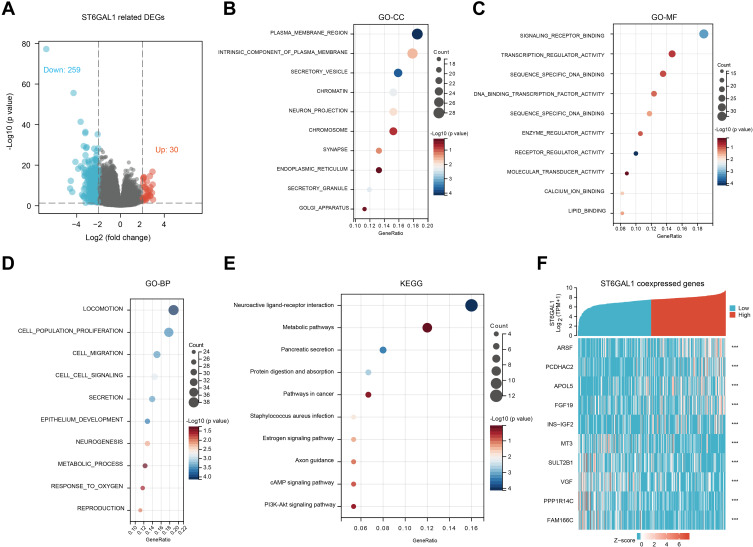
Functional Enrichment Analysis of ST6GAL1 Related Genes
To further explore the functions of ST6GAL1 in HCC, we analyzed differential expression genes (DEGs) according to ST6GAL1 expression level in the TCGA-HCC database. Compared to the ST6GAL1-low expression group, we obtained 289 DEGs, of which 30 genes were upregulated and 259 genes were downregulated (Figure 4A). Among the DEGs, 209 encoding protein genes were analyzed using gene ontology (GO) and KEGG functional enrichment analysis. The enrichment results showed that the most relevant cellular components (CC) of ST6GAL1-related DEGs were the plasma membrane region, an intrinsic component of the plasma membrane, and secretory vesicle (Figure 4B); the most relevant molecular function (MF) of ST6GAL1-related DEGs were signaling receptor binding, transcription regulator activity, and sequence-specific DNA binding (Figure 4C); the most relevant biological process (BP) of ST6GAL1-related DEGs were locomotion, cell population proliferation, cell migration, and cell-cell signaling (Figure 4D). The most relevant pathways of ST6GAL1-related DEGs were neuroactive ligand-receptor interaction, metabolic pathways, pancreatic secretion, protein digestion and absorption, pathways in cancer, estrogen signaling pathway, axon guidance, cAMP signaling pathway, and PI3K-Akt signaling pathway (Figure 4E). From the details, the top 5 up-regulated DEGs and top 5 down-regulated DEGs of ST6GAL1 were ARSF, PCDHAC2, APOL5, FGF19, INS-IGF2, MT3, SULT2B1, VGF, PPP1R14C, and FAM166C (Figure 4F). These ST6GAL1-related genes and pathways might play an important role during hepatocarcinogenesis.
Figure 4.
The ST6GAL1-related differential expression genes (DEGs) and functional enrichment analysis in HCC. (A) Volcano plot of ST6GAL1 related DEGs; (B–E) The GO-CC (B), GO-MF (C), GO-BP (D), and KEGG (E) enrichment analysis of ST6GAL1 related DEGs; (F) The expression heatmap of the ST6GAL1-related DEGs.
ST6GAL1 Expression Correlates with Liver Inflammation
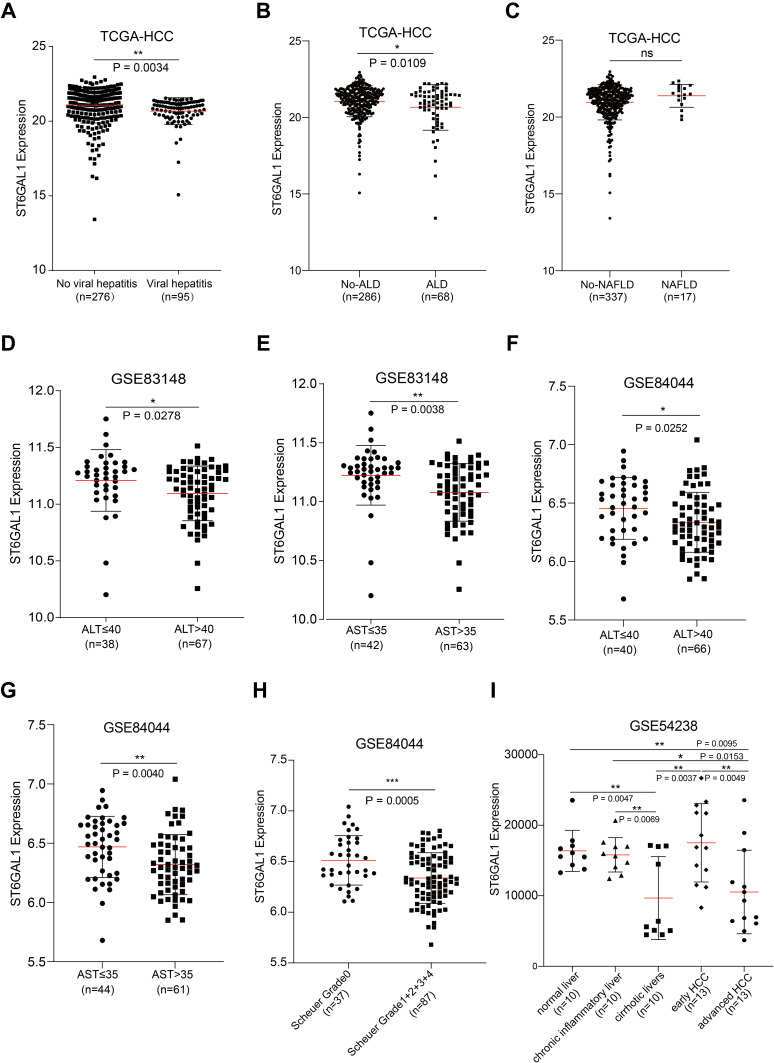
Considering the inflammatory regulation function of ST6GAL1,21 we analyzed the effects of VH, AH, and steatohepatitis (SH) on ST6GAL1 expression in the TCGA-HCC database. The results showed that ST6GAL1 was downregulated in VH-related HCC compared to the no-VH HCC group (Figure 5A). And lower expressed in the AH-related HCC group compared to the no-AH-related HCC group (Figure 5B). While, no difference was found between SH-related HCC and no-SH-related HCC (Figure 5C). Next, in the GSE83148 dataset which contained sequencing data of 122 chronic hepatitis B (CHB) liver samples22 and in the GSE84044 dataset which contained sequencing data of 124 CHB patients,23 we analyzed the correlation between the ST6GAL1 mRNA expression and levels of serum liver inflammation-related indicators including alanine aminotransferase (ALT) and aspartate aminotransferase (AST). The elevated ALT and AST levels are the earliest indications of liver inflammation and tissue damage.24 The results showed that in GSE83148 dataset, compared with the ALT- and AST-low level group, ST6GAL1 was lower expressed in the ALT- and AST-high level group (Figure 5D and E). The GSE84044 dataset also confirmed the above results (Figure 5F and G). Moreover, in GSE84044, according to Scheuer grades, we further analyzed the ST6GAL1 expression in different subgroups. The results showed that with the level of inflammatory activity increased, the ST6GAL1 expression downregulated (Figure 5H). Given that HCC is a progressive process, we further analyzed the differences in ST6GAL1 expression in different stages of liver disease in the GSE54238 dataset, which contained 10 normal livers (NL), 10 chronic inflammatory livers (IL), 10 cirrhotic livers (CL), 13 early HCC (eHCC) and 13 advanced HCC (aHCC) samples.25 The results showed that there was no difference in ST6GAL1 expression between NL and IL groups. While compared to NL or IL groups, ST6GAL1 expression was significantly downregulated in CL and aHCC groups (Figure 5I). These results indicated that ST6GAL1 expression was correlated with liver inflammation status, and ST6GAL1 had an anti-inflammation function during hepatocarcinogenesis.
Figure 5.
The association between ST6GAL1 mRNA expression and liver inflammation status. (A–C) ST6GAL1 expression of HCC tissues in no-viral hepatitis (VH) and VH groups (A), no-alcoholic liver disease (ALD) and ALD groups (B), and no-non-alcoholic fatty liver disease (NAFLD) and NAFLD groups (C) in the TCGA-HCC database; (D and E) Correlation between ST6GAL1 mRNA expression and alanine aminotransferase (ALT, (D)), and aspartate aminotransferase (AST, (E)) levels in the GSE83148 dataset; (F and G) Correlation between ST6GAL1 mRNA expression and ALT (F) and AST (G) levels in the GSE84044 dataset; (H) Expression of ST6GAL1 among hepatitis patients with Scheuer grades of G0, G1-G4 in the GSE84044 dataset; (I) ST6GAL1 expression of normal liver, chronic inflammatory liver, cirrhotic livers, early HCC, and advanced HCC in GSE54238 dataset. *p < 0.05, **p < 0.01, ***p < 0.001.
ST6GAL1 Expression Correlates with Immune Cell Infiltration in Liver Tissues
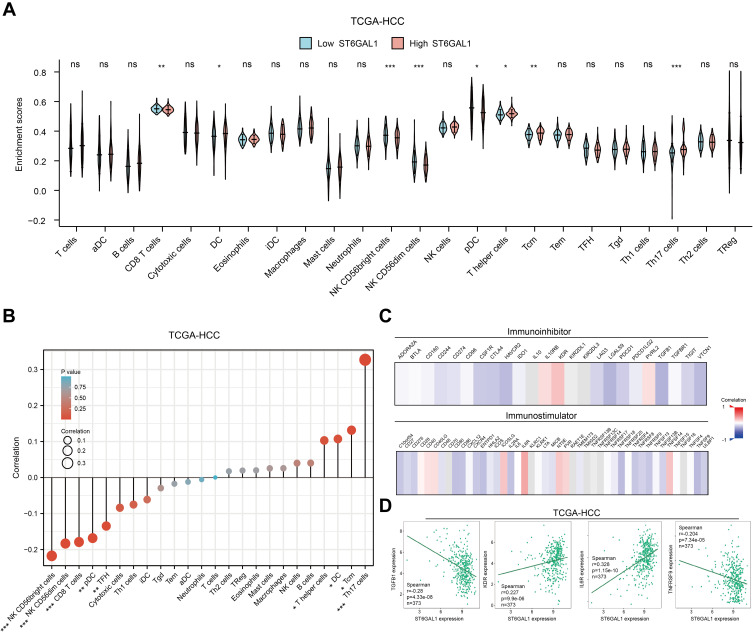
The above results suggested that ST6GAL1 might be involved in liver immune response, so we then analyzed the relationship between ST6GAL1 expression and the level of immune cell infiltration (ICI) in the TCGA database. The results showed that in the ST6GAL1-high expression group, the enrichment scores of dendritic cells (DC), T helper cells (Th), central memory T cells (Tcm), and Th17 cells were significantly higher, while the enrichment scores of CD8+ T cells, NK CD56bright cells, NK CD56dim cells and plasmacytoid dendritic cell (pDC) were decreased (Figure 6A). And the correlation analysis between the ICI and ST6GAL1 expression showed that there were significant negative correlations between ST6GAL1 expression and NK CD56bright cells, NK CD56dim cells, CD8+ T cells, pDC, and T follicular helper (TFH) cells, and significant positive correlations between ST6GAL1 expression and Th, Tcm, DC and Th17 cells (Figure 6B). Furthermore, we analyzed the correlations between ST6GAL1 expression and immunomodulators. The results showed that ST6GAL1 expression correlated to kinds of immunomodulators including immunoinhibitors and immunostimulators (Figure 6C). And among the immunoinhibitors, the ST6GAL1 had the highest negative correlation with TGFB1, and had the highest positive correlation with KDR; while among the immunostimulators, the ST6GAL1 had the highest positive correlation with IL6R, had the highest negative correlation with TNFRSF9 (Figure 6D). These results demonstrated that ST6GAL1 might exert an anti-inflammation function via regulating the infiltration of immune cells in the liver.
Figure 6.
The association of ST6GAL1 mRNA expression and immune cell infiltration. (A) Immune cell infiltration levels of high- and low-ST6GAL1 expression groups in HCC; (B) Correlation between ST6GAL1 mRNA expression and immune cell infiltration in HCC; (C) Correlation between ST6GAL1 mRNA expression and immunoinhibitors or immunostimulators in HCC; (D) Correlation between ST6GAL1 mRNA expression and TGFB1, KDR, IL6R, and TNFRSF9 in HCC. *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
HCC is one of the globally recognized malignancies with suboptimal OS rates, and the mechanisms of its pathogenesis and progression remain unclear.26,27 Therefore, exploring the pathogenesis behind HCC and evaluating its prognosis has become a priority. In this study, we found that ST6GAL1 was lower expressed in HCC tissues. And the lower ST6GAL1 expression, the worse of HCC prognosis. Furthermore, we found that the downregulation of ST6GAL1 was associated with aberration of immune cells infiltration in HCC liver tissues.
ST6GAL1 is a sialyltransferase that mediates the glycosylation of proteins and lipids to form functionally important glycoproteins and glycolipids in the Golgi apparatus, which plays an important role in cell proliferation and metastasis.28 The aberrant expression of ST6GAL1 has been reported in many human malignancies,14,29 indicating its different effects on different cancer types. As to HCC, there have been different views about ST6GAL1 as a prognostic indicator. Chen et al reported that ST6GAL1 expression was upregulated in HCC tissues and was associated with adverse prognosis,12 while Souady et al reported no difference in ST6GAL1 expression in cancerous and adjacent noncancerous tissues.30 However, these two studies were based on a small number of clinical HCC patients, only 78 and 35 cases respectively, resulting in some limitations and uncertainties. In our study, by analyzing the data in the public databases, we found that both ST6GAL1 mRNA and protein expressions were significantly lower in HCC compared with normal liver tissues. Subsequently, the same results were also seen in vivo experiments of the DEN-induced HCC rat model. And patients with lower ST6GAL1 mRNA expression had significantly poorer OS than those with high expression in the TCGA-HCC dataset. These results were consistent with previous reports of Poon et al and Cao et al.31,32 In addition, ST6GAL1 expression was also associated with clinical T stages and histological grades, as the HCC progresses, the expression of ST6GAL1 decreases. The CNV and methylation level of genes are two important factors affecting genes’ expression. And the ST6GAL1 expression has been reported to be regulated by DNA methylation in human gliomas and bladder cancer.33,34 Our results showed that decreased ST6GAL1 mRNA expression was associated with ST6GAL1 gene hypermethylation, but not with ST6GAL1 CNV status.
ST6GAL1 has been reported to participate in multiple signaling pathways during tumorigenesis including PI3K-AKT, TGF, EGFR, and HIF/VEGF signaling pathways.13 We here analyzed ST6GAL1 and its related DEGs in HCC. And the KEGG enrichment analysis showed that ST6GAL1 and its related DEGs may participate in the metabolic pathway, estrogen signaling pathway, axon guidance, and cAMP signaling pathway in HCC. In addition, among the top ten ST6GAL1-related DEGs we screened out, FGF19 has been proven to have an upregulating effect on ST6GAL1 expression.20 While relationships between ST6GAL1 and other genes needed further research.
More than that, ST6GAL1 is thought to play key roles in homeostasis and communication of immune cells. A previous study has found that knocking out the ST6GAL1 gene of hepatocytes could promote immune response and inflammation in liver.35 Our analysis also found that ST6GAL1 was lower expressed in VH-related HCC and AH-related HCC. And downregulation of ST6GAL1 was associated with liver inflammation levels, which indicated its anti-inflammation function in liver. Meanwhile, through the immune cells infiltration analysis, we found that ST6GAL1 could inhibit NK, pDC, and CD8+ T cells infiltrating in liver tissues. According to the previous reports, Danilo et al found that CD8+ T cells were heterogeneous. And although they were key mediators of antitumor immunity, they became dysfunctional after effector differentiation, thereby suppressing the anti-tumor immune response.36 And increasing evidences indicated that the tumor microenvironment (TME)-mediated suppression and modulation of tumor-infiltrated DCs impaired their function in initiating potent anti-tumor immunity and even promoted tumor progression.8 More evidence showed that NK cells were modulated and impaired in HCC to activate the elimination of tumor cells, which further involved in the pathogenesis of liver injury and inflammation.37 These studies suggested that high infiltration levels of NK CD56bright, NK CD56dim, pDC, and CD8+ T cells in liver could exacerbate HCC progression, which was consistent with the results of the present study. And the way that ST6GAL1 released liver inflammation might relied on regulating immune cell infiltration, which might be a therapeutic target in the clinic. However, more studies are needed to explore the mechanism of ST6GAL1 in regulating the TME and inflammation in HCC.
Conclusions
The results of this study showed that the decreased expression of ST6GAL1 in tumor tissues was associated with a poor prognosis for HCC patients. Its downregulation in liver tissues could promote intrahepatic inflammation. Our study suggested that ST6GAL1 could be a potential biological marker and therapy target for HCC. And provided some ideas to reveal the mechanism of ST6GAL1 on the occurrence and development of HCC by regulating the ICI and inhibiting liver inflammation.
Acknowledgments
We sincerely acknowledge the support from the Institute of Liver Diseases and the Key Laboratory of Dongzhimen Hospital, Beijing University of Chinese Medicine.
Funding Statement
This research was supported by [the Fundamental Research Funds for the Central Universities, Beijing University of Chinese Medicine] grant number [No.2021-JYB-XJSJJ-055 and No.2020-JYB-XJSJJ-060].
Abbreviations
HCC, Hepatocellular carcinoma; TME, Tumor microenvironment; DCs, dendritic cells; ST6GAL1, Beta-galactoside alpha-2, 6-sialyltransferase 1; TCGA, The Cancer Genome Atlas; GTEx, Genotype-Tissue Expression; HPA, Human protein atlas; GEO, Gene Expression Omnibus; CNV, Copy number variation; DEGs, Differential expression genes; KEGG, Kyoto encyclopedia of genes and genomes; RT-qPCR, Real-time quantitative polymerase chain reaction; SDS-PAGE, Sodium dodecyl sulfate polyacrylamide gel electrophoresis; CEE, capillary electrophoresis experiment; OS, Overall survival; RFS, Recurrence-free survival; VH, Viral hepatitis; AH, Alcohol hepatitis; GO, Gene ontology; CC, Cellular components; MF, Molecular function; BP, Biological process; SH, Steatohepatitis; CHB, Chronic hepatitis B; ALT, Alanine aminotransferase; AST, Aspartate aminotransferase; NL, Normal livers; IL, Chronic inflammatory livers; CL, Cirrhotic livers; eHCC, Early HCC; aHCC, Advanced HCC; ICI, Immune cell infiltration; DC, Dendritic cell; Th, T helper cell; Tcm, Central memory T cell; pDC, Plasmacytoid dendritic cell; TFH, T follicular helper.
Data Sharing Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories can be found in the article. The datasets generated or analyzed during the current study are available in the zenodo repository, [https://doi.org/10.5281/zenodo.6408057].
Ethics Approval and Consent to Participate
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Animal Research Ethics Board of Beijing University of Chinese medicine Dongzhimen Hospital (Approval No: 21-10). The study was carried out in compliance with the ARRIVE guidelines.
Disclosure
All authors have no conflict of interest to disclose.
References
- 1.Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589–604. doi: 10.1038/s41575-019-0186-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pinto Marques H, Gomes da Silva S, De Martin E, Agopian VG, Martins PN. Emerging biomarkers in HCC patients: current status. Int J Surg. 2020;82S:70–76. doi: 10.1016/j.ijsu.2020.04.043 [DOI] [PubMed] [Google Scholar]
- 3.Ruf B, Heinrich B, Greten TF. Immunobiology and immunotherapy of HCC: spotlight on innate and innate-like immune cells. Cell Mol Immunol. 2021;18(1):112–127. doi: 10.1038/s41423-020-00572-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ringelhan M, Pfister D, O’Connor T, Pikarsky E, Heikenwalder M. The immunology of hepatocellular carcinoma. Nat Immunol. 2018;19(3):222–232. doi: 10.1038/s41590-018-0044-z [DOI] [PubMed] [Google Scholar]
- 5.Lei X, Lei Y, Li J-K, et al. Immune cells within the tumor microenvironment: biological functions and roles in cancer immunotherapy. Cancer Lett. 2020;470:126–133. doi: 10.1016/j.canlet.2019.11.009 [DOI] [PubMed] [Google Scholar]
- 6.Lurje I, Hammerich L, Tacke F. Dendritic cell and T cell crosstalk in liver fibrogenesis and hepatocarcinogenesis: implications for prevention and therapy of liver cancer. Int J Mol Sci. 2020;21(19):7378. doi: 10.3390/ijms21197378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurebayashi Y, Ojima H, Tsujikawa H, et al. Landscape of immune microenvironment in hepatocellular carcinoma and its additional impact on histological and molecular classification. Hepatology. 2018;68(3):1025–1041. doi: 10.1002/hep.29904 [DOI] [PubMed] [Google Scholar]
- 8.Fu C, Jiang A. Dendritic cells and CD8 T cell immunity in tumor microenvironment. Front Immunol. 2018;9:3059. doi: 10.3389/fimmu.2018.03059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hedlund M, Ng E, Varki A, Varki NM. alpha 2-6-Linked sialic acids on N-glycans modulate carcinoma differentiation in vivo. Cancer Res. 2008;68(2):388–394. doi: 10.1158/0008-5472.CAN-07-1340 [DOI] [PubMed] [Google Scholar]
- 10.Han Y, Liu Y, Fu X, et al. miR-9 inhibits the metastatic ability of hepatocellular carcinoma via targeting beta galactoside alpha-2,6-sialyltransferase 1. J Physiol Biochem. 2018;74(3):491–501. doi: 10.1007/s13105-018-0642-0 [DOI] [PubMed] [Google Scholar]
- 11.Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15(9):540–555. doi: 10.1038/nrc3982 [DOI] [PubMed] [Google Scholar]
- 12.Chen X, Wang L, Yu X, Wang S, Zhang J. Caveolin-1 facilitates cell migration by upregulating nuclear receptor 4A2/retinoid X receptor α-mediated β-galactoside α2,6-sialyltransferase I expression in human hepatocarcinoma cells. Int J Biochem Cell Biol. 2021;137:106027. doi: 10.1016/j.biocel.2021.106027 [DOI] [PubMed] [Google Scholar]
- 13.Garnham R, Scott E, Livermore KE, Munkley J. ST6GAL1: a key player in cancer. Oncol Lett. 2019;18(2):983–989. doi: 10.3892/ol.2019.10458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorsett KA, Marciel MP, Hwang J, Ankenbauer KE, Bhalerao N, Bellis SL. Regulation of ST6GAL1 sialyltransferase expression in cancer cells. Glycobiology. 2021;31(5):530–539. doi: 10.1093/glycob/cwaa110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morosi LG, Cutine AM, Cagnoni AJ, et al. Control of intestinal inflammation by glycosylation-dependent lectin-driven immunoregulatory circuits. Sci Adv. 2021;7(25). doi: 10.1126/sciadv.abf8630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Yu H, Wu S, et al. Plasma ST6GAL1 regulates IgG sialylation to control IgA nephropathy progression. Ther Adv Chronic Dis. 2021;12:20406223211048644. doi: 10.1177/20406223211048644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Punch PR, Irons EE, Manhardt CT, Marathe H, Lau JTY. The sialyltransferase ST6GAL1 protects against radiation-induced gastrointestinal damage. Glycobiology. 2020;30(7):446–453. doi: 10.1093/glycob/cwz108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nasirikenari M, Segal BH, Ostberg JR, Urbasic A, Lau JT. Altered granulopoietic profile and exaggerated acute neutrophilic inflammation in mice with targeted deficiency in the sialyltransferase ST6Gal I. Blood. 2006;108(10):3397–3405. doi: 10.1182/blood-2006-04-014779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Khan A, Antonopoulos A, et al. Loss of α2-6 sialylation promotes the transformation of synovial fibroblasts into a pro-inflammatory phenotype in arthritis. Nat Commun. 2021;12(1):2343. doi: 10.1038/s41467-021-22365-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myojin Y, Kodama T, Maesaka K, et al. ST6GAL1 is a novel serum biomarker for Lenvatinib-susceptible FGF19-driven hepatocellular carcinoma. Clin Cancer Res. 2021;27(4):1150–1161. doi: 10.1158/1078-0432.CCR-20-3382 [DOI] [PubMed] [Google Scholar]
- 21.Holdbrooks AT, Ankenbauer KE, Hwang J, Bellis SL. Regulation of inflammatory signaling by the ST6Gal-I sialyltransferase. PLoS One. 2020;15(11):e0241850. doi: 10.1371/journal.pone.0241850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou W, Ma Y, Zhang J, et al. Predictive model for inflammation grades of chronic hepatitis B: large-scale analysis of clinical parameters and gene expressions. Liver Int. 2017;37(11):1632–1641. doi: 10.1111/liv.13427 [DOI] [PubMed] [Google Scholar]
- 23.Wang M, Gong Q, Zhang J, et al. Characterization of gene expression profiles in HBV-related liver fibrosis patients and identification of ITGBL1 as a key regulator of fibrogenesis. Sci Rep. 2017;7:43446. doi: 10.1038/srep43446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sookoian S, Pirola CJ. Liver enzymes, metabolomics and genome-wide association studies: from systems biology to the personalized medicine. World J Gastroenterol. 2015;21(3):711–725. doi: 10.3748/wjg.v21.i3.711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan SX, Wang J, Yang F, et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology. 2016;63(2):499–511. doi: 10.1002/hep.27893 [DOI] [PubMed] [Google Scholar]
- 26.Llovet JM, Montal R, Villanueva A. Randomized trials and endpoints in advanced HCC: role of PFS as a surrogate of survival. J Hepatol. 2019;70(6):1262–1277. doi: 10.1016/j.jhep.2019.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X, Li C, Zhang L, et al. The significance of exosomes in the development and treatment of hepatocellular carcinoma. Mol Cancer. 2020;19(1):1. doi: 10.1186/s12943-019-1085-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li F, Ding J. Sialylation is involved in cell fate decision during development, reprogramming and cancer progression. Protein Cell. 2019;10(8):550–565. doi: 10.1007/s13238-018-0597-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holdbrooks AT, Britain CM, Bellis SL. ST6Gal-I sialyltransferase promotes tumor necrosis factor (TNF)-mediated cancer cell survival via sialylation of the TNF receptor 1 (TNFR1) death receptor. J Biol Chem. 2018;293(5):1610–1622. doi: 10.1074/jbc.M117.801480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Souady J, Hülsewig M, Distler U, et al. Differences in CD75s- and iso-CD75s-ganglioside content and altered mRNA expression of sialyltransferases ST6GAL1 and ST3GAL6 in human hepatocellular carcinomas and nontumoral liver tissues. Glycobiology. 2011;21(5):584–594. doi: 10.1093/glycob/cwq200 [DOI] [PubMed] [Google Scholar]
- 31.Poon TCW, Chiu CHS, Lai PBS, et al. Correlation and prognostic significance of beta-galactoside alpha-2,6-sialyltransferase and serum monosialylated alpha-fetoprotein in hepatocellular carcinoma. World J Gastroenterol. 2005;11(42):6701–6706. doi: 10.3748/wjg.v11.i42.6701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao Y, Merling A, Crocker PR, Keller R, Schwartz-Albiez R. Differential expression of beta-galactoside alpha2,6 sialyltransferase and sialoglycans in normal and cirrhotic liver and hepatocellular carcinoma. Lab Invest. 2002;82(11):1515–1524. doi: 10.1097/01.LAB.0000038503.34655.98 [DOI] [PubMed] [Google Scholar]
- 33.Kroes RA, Moskal JR. The role of DNA methylation in ST6Gal1 expression in gliomas. Glycobiology. 2016;26(12):1271–1283. doi: 10.1093/glycob/cww058 [DOI] [PubMed] [Google Scholar]
- 34.Antony P, Rose M, Heidenreich A, Knüchel R, Gaisa NT, Dahl E. Epigenetic inactivation of ST6GAL1 in human bladder cancer. BMC Cancer. 2014;14:901. doi: 10.1186/1471-2407-14-901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oswald DM, Zhou JY, Jones MB, Cobb BA. Disruption of hepatocyte Sialylation drives a T cell-dependent pro-inflammatory immune tone. Glycoconj J. 2020;37(3):395–407. doi: 10.1007/s10719-020-09918-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Danilo M, Chennupati V, Silva JG, Siegert S, Held W. Suppression of Tcf1 by Inflammatory cytokines facilitates effector CD8 T cell differentiation. Cell Rep. 2018;22(8):2107–2117. doi: 10.1016/j.celrep.2018.01.072 [DOI] [PubMed] [Google Scholar]
- 37.Liu P, Chen L, Zhang H. Natural killer cells in liver disease and hepatocellular carcinoma and the NK cell-based immunotherapy. J Immunol Res. 2018;2018:1206737. doi: 10.1155/2018/1206737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vivian J, Rao AA, Nothaft FA, et al. Toil enables reproducible, open source, big biomedical data analyses. Nat Biotechnol. 2017;35(4):314–316. doi: 10.1038/nbt.3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Menyhárt O, Á N, Győrffy B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R Soc Open Sci. 2018;5(12):181006. doi: 10.1098/rsos.181006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koch A, De Meyer T, Jeschke J, Van Criekinge W. MEXPRESS: visualizing expression, DNA methylation and clinical TCGA data. BMC Genomics. 2015;16:636. doi: 10.1186/s12864-015-1847-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Modhukur V, Iljasenko T, Metsalu T, Lokk K, Laisk-Podar T, Vilo J. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics. 2018;10(3):277–288. doi: 10.2217/epi-2017-0118 [DOI] [PubMed] [Google Scholar]
- 42.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28(11):1947–1951. doi: 10.1002/pro.3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49(D1):D545–D551. doi: 10.1093/nar/gkaa970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. doi: 10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35(20):4200–4202. doi: 10.1093/bioinformatics/btz210 [DOI] [PubMed] [Google Scholar]