Abstract
In cell extracts of Pseudaminobacter salicylatoxidans strain BN12, an enzymatic activity was detected which converted salicylate in an oxygen-dependent but NAD(P)H-independent reaction to a product with an absorbance maximum at 283 nm. This metabolite was isolated, purified, and identified by mass spectrometry and 1H and 13C nuclear magnetic resonance spectroscopy as 2-oxohepta-3,5-dienedioic acid. This metabolite could be formed only by direct ring fission of salicylate by a 1,2-dioxygenase reaction. Cell extracts from P. salicylatoxidans also oxidized 5-aminosalicylate, 3-, 4-, and 5-chlorosalicylate, 3-, 4-, and 5-methylsalicylate, 3- and 5-hydroxysalicylate (gentisate), and 1-hydroxy-2-naphthoate. The dioxygenase was purified and shown to consist of four identical subunits with a molecular weight of about 45,000. The purified enzyme showed higher catalytic constants with gentisate or 1-hydroxy-2-naphthoate than with salicylate. It was therefore concluded that P. salicylatoxidans synthesized a gentisate 1,2-dioxygenase with an extraordinary substrate range, which also allowed the oxidation of salicylate.
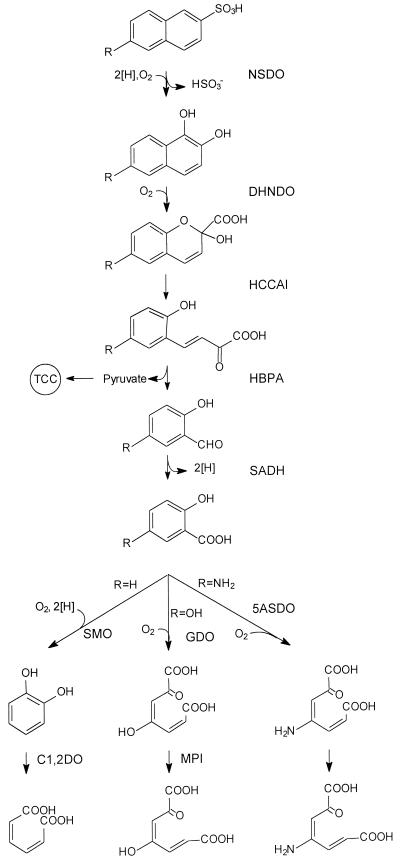
Naphthalenesulfonic acids are produced on a large scale as industrial detergents, dispersive materials, and intermediates for the production of azo dyes (34). Some pure and mixed bacterial cultures which grow with naphthalene-1-sulfonic acid, naphthalene-2-sulfonic acid (2NS), 2-aminonaphthalene-1-sulfonic acid, 6-aminonaphthalene-2-sulfonic acid (6A2NS), or naphthalenedisulfonic acids as the sole source of carbon and energy have been isolated. The initial reaction in the microbial degradation of (substituted) naphthalenesulfonic acids is catalyzed by desulfonating dioxygenases, which catalyze the formation of (substituted) 1,2-dihydroxynaphthalene(s) and the release of sulfite (4, 5, 26–30, 32, 44). Thus, the degradative pathway for naphthalenesulfonic acids converges at the level of 1,2-dihydroxynaphthalene (1,2-DHN) with the well-known pathway for the degradation of naphthalene (Fig. 1). The reactions which are catalyzed by the “upper” naphthalenesulfonic acid pathway result in the formation of (substituted) salicylate(s). For Sphingomonas xenophaga BN6, it was found that the enzymes participating in the upper naphthalenesulfonic acid pathway convert a wide range of substituted substrates and that therefore a single set of enzymes is sufficient for the conversion of various substituted naphthalenesulfonic acids (19–21, 26, 27, 35). In contrast to the degradation of (substituted) naphthalenesulfonic acids to the corresponding salicylates, the productive metabolism of (substituted) salicylates requires rather different metabolic pathways. Thus, salicylate is usually converted by a salicylate 1-monooxygenase to catechol, 3-hydroxysalicylate (2,3-dihyroxybenzoate) is directly oxidized by an extradiol ring fission reaction to a nonaromatic product, 4-hydroxysalicylate (2,4-dihydroxybenzoate) is converted by a monooxygenase to the ring fission substrate 1,2,4-trihydroxybenzene, and gentisate (5-hydroxysalicylate) and 5-aminosalicylate are substrates for ring fission dioxygenases (2, 36, 37, 41, 42).
FIG. 1.
Proposed pathways for the degradation of (substituted) naphthalenesulfonates. NSDO, naphthalenesulfonate dioxygenase; DHNDO, 1,2-dihydroxynaphthalene dioxygenase; HCCAI, 2-hydroxychromene-2-carboxylate isomerase; HBPA, 2′-hydroxybenzalpyruvate aldolase(hydratase); SADH, salicylaldehyde dehydrogenase; SMO, salicylate 1-monooxygenase; GDO, gentisate 1,2-dioxygenase; 5ASDO, 5-aminosalicylate 1,2-dioxygenase; C1,2DO, catechol 1,2-dioxygenase; MPI, maleylpyruvate isomerase; TCC, tricarboxylic acid cycle (19, 20, 26, 37).
So far, only one bacterial strain (Pseudaminobacter salicylatoxidans BN12) is known to completely degrade 6A2NS. In the course of attempts to characterize the metabolism of different substituted naphthalenesulfonates by this strain, a new enzymatic ring fission reaction for salicylate was detected, and it is described here.
MATERIALS AND METHODS
Bacterial strains and media.
The isolation and characterization of P. salicylatoxidans strain BN12 have been described previously (16, 25). Mineral media were prepared as described by Dorn et al. (9) and were supplemented with 100 mg of yeast extract per liter. Heat-labile and autoxidizable substrates were sterilized by membrane filtration (pore size, 0.2 μm; Sartorius, Göttingen, Germany); all other substrates were autoclaved at 121°C.
Preparation of cell extracts.
Cell suspensions in 20 mM Tris-HCl buffer, pH 8.0, were disrupted by using a French press (Aminco, Silver Spring, Md.) at 80 MPa. Cell debris was removed by centrifugation at 100,000 × g for 30 min at 4°C. The protein content of the cell extracts was determined by the method of Bradford (3) with bovine serum albumin as a standard.
Enzyme assays.
One unit of enzyme activity was defined as the amount of enzyme that converts 1 μmol of substrate per min. 1,2-Dihydroxynaphthalene dioxygenase and 2′-hydroxybenzalpyruvate aldolase activities were determined as described previously (19, 20).
Salicylate-1-monooxygenase was measured by a modification of the method of Lee et al. (23). Cell extract was added to a solution (final volume, 1 ml) containing 20 μmol of Tris-HCl buffer (pH 8.0), 0.1 μmol of salicylate, 1.5 nmol of flavin adenine dinucleotide, and 0.03 μmol of NADH. The decrease in absorption at 340 nm was measured spectrophotometrically.
The oxidation of 1-hydroxy-2-naphthoate was determined spectrophotometrically at 300 nm (molar reaction coefficient [ɛ300] = 11.5 mM−1 cm−1) as previously described by Iwabuchi and Harayama (15).
Gentisate 1,2-dioxygenase and 5-aminosalicylate 1,2-dioxygenase activities were measured spectrophotometrically at 340 nm (ɛ340 = 10.2 mM−1 cm−1) or 350 nm (ɛ350 = 10.2 mM−1 cm−1) (substrate concentration, 0.1 mM each) (37, 41).
For salicylate 1,2-dioxygenase, the cuvettes contained, in 1 ml, 20 μmol of Tris-HCl buffer (pH 8.0) and 0.1 μmol of salicylate. The reaction was started by the addition of cell extract, and the increase in absorbance was recorded at 283 nm. ɛ283 was experimentally determined as 13.6 mM−1 cm−1.
The kinetic parameters were determined in a concentration range of 0.01 to 0.5 mM. The enzymatic reactions were recorded for 1 min. Standard deviations were calculated by nonlinear regression using the Sigma-Plot program package, version 5.0.
The pH optimum of the salicylate 1,2-dioxygenase activity was determined using a universal buffer as described by Rauscher et al. The buffer consisted of a mixture of citric acid, phosphoric acid, boric acid, and NaOH, which was titrated with HCl to obtain the desired pH (31).
Purification of the salicylate 1,2-dioxygenase activity.
Protein was purified at 4°C by use of a fast-performance liquid chromatography system consisting of a LCC 500 controller, 500 pump, UV-1 monitor, REC-482 recorder, and FRAC autosampler from Pharmacia (Uppsala, Sweden).
Crude extract was applied to a Q Sepharose FF column (XK 16/10; Pharmacia). Protein was eluted with 240 ml of a linear gradient of Tris-HCl (20 mM, pH 8.0) into Tris-HCl (20 mM, pH 8.0) plus 0.35 M NaCl at a flow rate of 1.5 ml/min. Fractions (5 ml each) were collected, and enzyme activity was determined spectrophotometrically. The salicylate-converting enzyme was eluted as a single peak at a concentration of about 0.27 M NaCl. The active fractions were pooled and transferred to a hydroxyapatite column (XK 16/10, ceramic hydroxyapatite type I, 20-μm particle size; Bio-Rad) and eluted with a linear gradient (5 to 120 mM) of potassium phosphate buffer (pH 7.5) at a flow rate of 1.5 ml/min, and fractions of 2 ml were collected. The enzyme activity eluted when a potassium phosphate concentration of 20 mM was reached. The active fractions were pooled (30 ml) and applied to a Mono Q column (HR 5/5; Pharmacia) in order to concentrate the protein solution. The proteins were initially eluted (flow rate, 1 ml/min) with 5 ml of Tris-HCl (20 mM, pH 8.0) plus 0.3 M NaCl. The dioxygenase activity was eluted in 1 ml by an abrupt increase in the NaCl concentration to 1 M (reached by a linear gradient in 1 ml). From a saturated solution, (NH4)2SO4 was added to the active fraction to 20% saturation, and the protein solution was applied to a Phenyl Superose column (HR10/10; Pharmacia). The proteins were eluted with 10 ml of Tris-HCl (20 mM, pH 8.0) plus 0.8 M (NH4)2SO4 (flow rate, 0.75 ml/min), followed by 1 ml of a linear gradient to Tris-HCl (20 mM, pH 8.0) plus 0.45 M (NH4)2SO4 and a further elution with 10 ml of Tris-HCl (20 mM, pH 8.0) plus 0.45 M (NH4)2SO4. Fractions (1 ml each) were collected, and the dioxygenase activity was recovered in fractions which were eluted with about 0.53 M (NH4)2SO4. The pooled fractions (3 ml) were concentrated by ultrafiltration (Vivaspin 2; Sartorius) to a volume of 0.3 ml, and proteins were chromatographed on a Pharmacia Hiload Superdex 200 column (HR 10/30) by using 20 mM Tris-HCl (pH 8.0) with 150 mM NaCl as the elution buffer.
The enzyme preparations from the Mono Q, Phenyl Superose, and gel filtration columns were routinely incubated with 2 mM (NH4)2Fe(SO4)2 and 2 mM l-ascorbic acid for 30 min prior to assaying enzyme activity.
PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed by the method of Laemmli (22). Gels were silver stained by the method of Shevchenko et al. (33).
Analytical methods.
Salicylate and its oxidation product were analyzed by reverse-phase high-pressure liquid chromatography (HPLC). A reverse-phase column (Grom, Herrenberg, Germany; 125 by 4.0 mm [internal diameter], packed with 5-μm particles of Grom-Sil 100 Octyl) was used. The separated compounds were detected photometrically by using a photodiode array detector. The following solvent systems were used: 84.7% (vol/vol) water, 15% (vol/vol) methanol, and 0.3% H3PO4 or 90.0% (vol/vol) water, 10% (vol/vol) methanol, and an ion pair reagent according to the instructions of the manufacturer (PIC A; Waters). The usual flow rate was 0.7 ml/min.
Naphthalene-2-sulfonates and metabolites were analyzed by reverse-phase HPLC as described previously (26, 27).
Oxygen uptake experiments.
Oxygen uptake was measured at 25°C with an oxygen electrode (YSI 5350; Yellow Springs Instrument Co., Yellow Springs, Colo.). The reaction mixtures (total volume of 3 ml) contained 50 mM AMPSO {[1,1-dimethyl-2-hydroxyethyl)-amino]-2-hydroxypropane-1-sulfonic acid} buffer (pH 9.0) and cell extract (approximately 3 mg of protein). The endogenous respiration was determined for 2 min, and then different amounts of salicylate (0.03 to 0.6 μmol) were added. The oxygen uptake was recorded and corrected for endogenous respiration.
Isolation of the ring fission product from salicylate.
P. salicylatoxidans BN12 was grown in 3-liter Erlenmeyer flasks (600 ml of culture volume) with 6A2NS plus yeast extract (100 mg/liter) to an optical density at 546 nm of about 1.2. Cells were harvested by centrifugation, and cell extracts were prepared. This cell extract (about 100 mg of protein) was incubated in 50 ml of a sodium carbonate buffer (50 mM, pH 9.0) plus 5 mM of salicylate until the salicylate was completely converted. The resulting pool was separated from macromolecular contaminations by ultrafiltration (Diaflo PM10 membrane with a 10,000-molecular-weight cutoff; Amicon) and the filtrate was transferred to a Q Sepharose column (15 by 1.6 cm). Individual compounds were eluted from the column with a linear gradient of NaCl (from 0 to 1 M) in sodium carbonate buffer (30 mM, pH 9.0). Fractions which showed a significant absorbance at 283 nm eluted in the concentration range of 0.4 M NaCl and were pooled. This purified aqueous solution of the ring fission product (about 50 ml) was concentrated by evaporation to dryness. Methanol (3 ml) was added to the precipitate, and the methanolic phase containing most of the ring fission product was separated from the insoluble inorganic salts. The methanolic phase was evaporated, and the procedure was repeated. This resulted (according to HPLC and spectroscopic analysis) in the isolation of an almost pure preparation of the ring fission product.
Spectrometry.
Mass spectra were obtained on a Quattro LC triple-stage quadrupole mass spectrometer (Micromass, Manchester, United Kingdom) with electrospray ionization in the negative mode. The compound was introduced in a water-methanol solution by infusion with a syringe pump. The nuclear magnetic resonance (NMR) spectra were recorded with a Bruker ARX500 spectrometer in deuterated methanol. Chemical shifts (δ) are given in parts per million relative to tetramethylsilane as the internal standard.
Chemicals.
The sources of the chemicals used have been described (19, 20, 26, 27).
RESULTS
Growth phenotype.
P. salicylatoxidans BN12 grew in liquid media with 2NS, 6A2NS, or 6-hydroxynaphthalene-2-sulfonate (6H2NS) (2.5 mM each) supplemented with yeast extract (100 mg/liter). HPLC analysis indicated a complete conversion of 2NS, 6H2NS, and 6A2NS, because the substrates completely disappeared and only small amounts (<0.5 mM) of salicylate, gentisate, or 5-aminosalicylate were found transiently in the culture supernatants. Strain BN12 also grew with gentisate and 5-aminosalicylate (in the presence of yeast extract). No or very faint growth was observed with salicylate, 3-, 4-, or 6-hydroxysalicylate, 3- or 4-aminosalicylate, or 1-hydroxy-2-naphthoate.
Enzyme measurements with cell extracts.
The growth characteristics of strain BN12 suggested that the “upper” naphthalene pathway, which converts the naphthalenesulfonates to the corresponding salicylates, and the “lower” pathways for the metabolism of salicylate, 5HS, and 5AS both occurred in this strain (Fig. 1). This is in contrast to the 6A2NS-degrading mixed culture which was previously studied in our laboratory. In this coculture, S. xenophaga BN6 converts 2NS, 6A2NS, or 6H2NS into salicylate, 5AS, or 5HS. These products are not further degraded by strain BN6 but are excreted in almost stoichiometric amounts into the culture supernatant and are metabolized by other strains of the mixed culture (26).
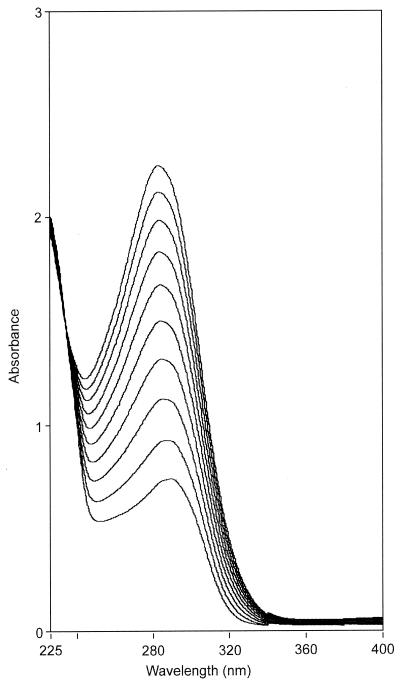
Therefore, we tested whether the enzymes for the upper naphthalene pathway and those for the oxidation of the salicylates (Fig. 1) occur together in P. salicylatoxidans BN12. The strain was grown with 6A2NS, 2NS, nutrient broth, or acetate and various enzymes involved in the degradation of naphthalenesulfonates and (substituted) salicylates assayed. Thus, 1,2-dihydroxynaphthalene dioxygenase, 2′-hydroxybenzalpyruvate aldolase, 5-aminosalicylate 1,2-dioxygenase, gentisate 1,2-dioxygenase, and maleylpyruvate isomerase activities were found in cell extracts. The enzymes of the upper naphthalene pathway and the gentisate pathway could be detected in extracts from cells which had been grown under nonselective conditions but showed some enzyme induction after growth with 2NS or 6A2NS (Table 1). Surprisingly, almost no salicylate 1-monoooxygenase activity was found after growth with 2NS or 6A2NS (<0.01 U/mg of protein). However, salicylate was converted without the addition of a cofactor to a product with an absorption maximum at 283 nm (Fig. 2). The oxidation of salicylate to a product with an absorption maximum at 283 nm has been observed for a different strain (BN11) which was isolated from the same mixed bacterial culture as P. salicylatoxidans BN12 but did not degrade naphthalenesulfonates (25).
TABLE 1.
Specific enzyme activities for the catabolism of naphthalenesulfonates from P. salicylatoxidans BN12 grown on different substrates
| Enzyme | Activity (U/mg) after growth with:
|
||
|---|---|---|---|
| 6A2NS | 2NS | Acetate | |
| 1,2-Dihydroxynaphthalene dioxygenase | 3.08 | 4.23 | 0.46 |
| 2′-Hydroxybenzalpyruvate aldolase | 0.23 | 0.15 | 0.03 |
| 5-Aminosalicylate 1,2-dioxygenase | 0.07 | 0.05 | 0.01 |
| Gentisate 1,2-dioxygenase | 2.26 | 2.20 | 0.31 |
| Maleylpyruvate isomerase | 0.13 | 0.07 | 0.02 |
FIG. 2.
Cofactor-independent conversion of salicylate by cell extracts from strain BN12. The reaction mixture (total volume, 3 ml) contained 20 mM Tris-HCl buffer (pH 8.0) and 0.1 mM salicylate. The reaction was started by the addition of 7 μl of cell extract (29.4 mg of protein per ml). Every 5 min a spectrum was recorded.
Analysis of the conversion of salicylate by HPLC.
The absorption spectrum of the product did not change significantly between pHs 5.0 and 12.0. The turnover of salicylate could be analyzed by HPLC (solvent system: 84.7% [vol/vol] water, 15% [vol/vol] methanol, and 0.3% [vol/vol] H3PO4). This reaction analysis demonstrated that salicylate (retention time [Rt] = 22.1 min) was converted to a single product (Rt = 3.3 min), which also demonstrated in the acidic solvent system the characteristic absorption maximum at 283 nm. Since we found no evidence in the relevant literature of a cofactor-independent conversion of salicylate to a product with an absorption maximum at 283 nm, the retention behavior and UV-visible (UV-Vis) spectra of several possible reaction products were analyzed. Thus, the putative products 3-, 4-, 5-, and 6-hydroxysalicylate, catechol, phenol, and benzoate were excluded according to their UV-Vis spectra and retention times. This suggested that salicylate was indeed converted by a novel reaction by cell extracts from P. salicylatoxidans strain BN12.
Oxygen-dependent conversion of salicylate.
Different amounts of salicylate were incubated in an oxygen electrode with cell extracts of P. salicylatoxidans, and the salicylate-dependent oxygen uptake was determined. The total amount of oxygen consumed at the expense of salicylate was strictly proportional to the initial concentration of the organic substrate: 1 mol of oxygen was consumed per mol of salicylate added.
Isolation of the oxidation product of salicylate.
All results suggested that salicylate was oxidized by cell extracts of P. salicylatoxidans by a direct 1,2-dioxygenolytic cleavage of the aromatic ring. We therefore attempted to isolate the product of the reaction for spectroscopic characterization. Thus, 250 μmol of salicylate was incubated overnight in 50 ml of sodium carbonate buffer (pH 9.0) with about 100 mg of cell extract. After the complete turnover of salicylate, the reaction product was isolated and purified by ultrafiltration and anion-exchange chromatography as described in Materials and Methods.
Identification of the ring cleavage product of salicylate.
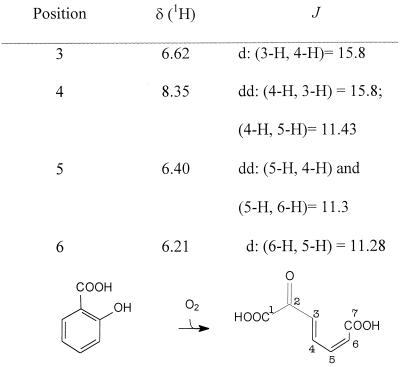
The purified product was dissolved in deuterated methanol, and 1H NMR (500 MHz) and 13C NMR (126 MHz) spectra were recorded. The 13C NMR spectrum clearly demonstrated the presence of seven different carbon atoms. Besides two carboxylate resonances (δ = 172.15 and 174.86 ppm), one signal (δ = 197.09 ppm) was observed, which suggested the presence of a keto group. The additional four signals of the 13C NMR spectrum resulted from olefinic carbon atoms (δ = 130.52, 133.94, 138.24, and 144.58).
The 1H NMR spectrum (Fig. 3) showed a set of four olefinic protons which, from the coupling pattern (two doublets and two doublets of doublets), are in adjacent positions. A chemical shift of 8.35 ppm must be assigned to the proton in the β position to the keto group (C-4) with its coupling constant (3J = 15.8 Hz) indicating a trans configuration for the double bond between C-3 and C-4. The double bond between C-5 and C-6 has a cis configuration (3J = 11.3 Hz). All NMR data were in accordance with the 2-oxohepta-3,5-dienedioic acid as a ring fission product of salicylate (Fig. 3).
FIG. 3.
1H NMR data for the oxidation product of salicylate.
The purified compound was further analyzed by mass spectrometry (MS). The sample was introduced by infusion and ionized by electrospray ionization, and the spectra were acquired in the negative mode. Two major signals with m/z values of 191 (M-2H + Na)− and 169 (M-H)− were observed. The molecular anion with an m/z value of 169 was further fragmented by collision induced dissociation (MS/MS). Thus, fragments with m/z values of 125 (M-H-CO2)−, 97 (M-H-CO2-CO)−, and 81 (M-H-CO2-CO2)− were detected. This fragmentation pattern could be explained by the loss of two carboxyl groups and one carbonyl group from the parent ion. The MS data thus agree with the oxidation of salicylate to 2-oxohepta-3,5-dienedioic acid.
Characterization of the salicylate-converting enzyme activity in cell extracts.
Cell extracts of strain BN12 were incubated in different buffer systems at different temperatures. The remaining activity of the salicylate-converting enzyme was assayed at different intervals. The enzyme was rather stable at −20°C in Na-K phosphate buffer (pH 7.0) or Tris-HCl buffer (pH 8.0). After 1 week of storage (7 mg of protein/ml) at −20°C, about 60% of the original activity was recovered. In contrast, within 2 days at 4°C in both buffer systems, the enzyme lost its activity completely.
Enzymatic activity with salicylate was determined within a pH range from 5 to 11 by using a universal buffer as described by Rauscher et al. (31) and measuring the increase in absorbance at 283 nm. The enzyme showed maximal activity at about pH 8.0 to 9.0. The enzyme showed the highest activity at about 40°C and 80 to 90% of this maximal activity at 30 or 50°C. ɛ283 of the product formed from salicylate was experimentally determined at pH 9.0 as 18.5 mM−1 cm−1. Because of the absorbance of the substrate salicylate at 283 nm (ɛ283 = 4.9 mM−1 cm−1), a molar reaction coefficient of 13.6 mM−1 cm−1 was used for the determination of the enzyme activity.
After the incubation of cell extracts with the Fe2+-chelating agent 8-hydroxyquinoline, o-phenanthroline, or 2,2′-dipyridyl (each 0.1 mM, 15 min), only 9, 2, or 28%, respectively, of the activity in an untreated control was recovered. This suggested that the salicylate-converting oxygenase activity required Fe2+ ions as previously described for gentisate 1,2-dioxygenases and 1-hydroxy-2-naphthoate 1,2-dioxygenase (14, 15).
Oxidation of different substituted salicylates by cell extracts.
The results presented above clearly demonstrated that cell extracts from P. salicylatoxidans BN12 were able to oxidize salicylate by a dioxygenolytic cleavage between carbon atoms 1 and 2. Similar cleavage reactions of aromatic 2-hydroxycarboxylic acids have been described previously for gentisate, 5-aminosalicylate, 5-chlorosalicylate, and 1-hydroxy-2-naphthoate (1, 7, 13, 15, 37). The cell extracts from P. salicylatoxidans BN12 oxidized (apart from salicylate) gentisate and 5-aminosalicylate (Table 1). Therefore, cell extracts were also incubated with 5-chlorosalicylate and 1-hydroxy-2-naphthoate, and the reactions were analyzed by UV-Vis overlay spectra. The UV-Vis spectra of the product(s) formed from 5-chlorosalicylate (λmax = 332 nm) and 1-hydroxy-2-naphthoate (λmax = 300 nm) were very similar to those of the corresponding 1,2-dioxygenation products, which had been described previously (1, 7). The UV-Vis spectrum of the ring fission product of 1-hydroxy-2-naphthoate (trans-2-carboxybenzalpyruvate) significantly changed after acidification from a λmax of 300 nm to λmax of 274 and 281 nm (1). The same observation was made with the product formed from 1-hydroxy-2-naphthoate by the salicylate-oxidizing enzyme from strain BN12. It was therefore concluded that 1-hydroxy-2-naphthoate was converted by P. salicylatoxidans BN12 to the same product as that for Nocardioides sp. strain KP7.
Furthermore, an increase in absorbance between 280 and 400 nm was also observed with cell extracts from P. salicylatoxidans with 3- and 4-chlorosalicylate, 3-hydroxysalicylate, and 3-, 4-, and 5-methylsalicylate. This suggested that the salicylate-oxidizing enzyme activity also cleaves several substituted salicylates.
Purification of the enzyme exhibiting salicylate 1,2-dioxygenase activity.
The ability of the cell extracts to oxidize gentisate, 5-aminosalicylate, and 1-hydroxy-2-naphthoate in addition to salicylate suggested that a single enzyme could be responsible for the conversion of these substrates. The enzyme was therefore purified according to the procedure shown in Table 2. The enzyme was purified 62-fold. The overall yield was 1.5% of the activity present in cell extract. This enzyme preparation gave one clearly dominant band after SDS-gel electrophoresis (>90% purity). The molecular weight of the purified dioxygenase as determined by gel filtration was estimated to be 185,000. The molecular weight of one subunit was determined by SDS-gel electrophoresis to be 45,000. Therefore, it can be assumed that the enzyme consists of four identical subunits.
TABLE 2.
Purification of the salicylate 1,2-dioxygenase activity from P. salicylatoxidans BN12
| Purification step | Vol (ml) | Total protein (mg) | Sp act (U/mg) | Total activity (U) | Recovery (%) | Purification (fold) |
|---|---|---|---|---|---|---|
| Crude extract | 16.5 | 760 | 0.019 | 14.4 | 100 | 1.00 |
| Q Sepharose FF | 14.0 | 70 | 0.038 | 2.7 | 19 | 2.0 |
| Hydroxyapatite | 30.0 | 19.5 | 0.089 | 1.74 | 12 | 4.7 |
| Mono Q | 1.0 | 6.4 | 0.45 | 2.9 | 20 | 24 |
| Phenyl Superose | 3.0 | 4.5 | 0.42 | 1.9 | 13 | 22 |
| Gel filtration | 1.0 | 0.19 | 1.18 | 0.22 | 1.5 | 62 |
Substrate specificity of the dioxygenase.
The purified enzyme was incubated with salicylate (0.1 mM), gentisate, 5-aminosalicylate, or 1-hydroxy-2-naphthoate (0.5 mM each), and the reaction products were analyzed spectrophotometrically. The enzyme preparation oxidized all four substrates. The purified enzyme converted the four substrates with almost the same relative activities as the cell extract (relative activities with salicylate, 5-aminosalicylate, 1-hydroxy-2-naphthoate, and gentisate with both preparations were about 1:10:40:120). This suggested that a single dioxygenase was responsible in P. salicylatoxidans BN12 for the oxidation of these substrates. Substrate inhibition kinetics were observed with all four substrates when the substrate concentrations exceeded about 1 mM. Therefore, the oxidation of the substrates was compared in a substrate range of 10 to 500 μM. Thus, it was found that the highest specific activities were observed with gentisate and the lowest activities were found with salicylate (Table 3). A comparison of the specificity constants (kcat/Km) suggested that gentisate and 1-hydroxy-2-naphthoate were the preferred substrates of the enzyme. This suggested that a gentisate dioxygenase with an extremely relaxed substrate specificity was responsible for the unusual oxidation of salicylate by P. salicylatoxidans BN12.
TABLE 3.
Kinetic constants of the dioxygenase with different substrates
| Substrate | Vmax (U mg−1) | Km (μM) | kcat (S−1) | kcat/Km (s−1 M−1) (104) |
|---|---|---|---|---|
| Salicylate | 1.4 ± 0.1 | 12 ± 4 | 1 | 8 |
| 5-Aminosalicylate | 79 ± 34 | 3,100 ± 1,500 | 59 | 2 |
| Gentisate | 125 ± 5 | 79 ± 10 | 94 | 119 |
| 1-Hydroxy-2-naphthoate | 59 ± 3 | 134 ± 17 | 44 | 33 |
DISCUSSION
Salicylate is degraded by most bacteria by a salicylate-1-monooxygenase, which oxidatively decarboxylates salicylate in a NAD(P)H-dependent reaction to catechol (42, 45, 46). In contrast, the results obtained during the present study unequivocally demonstrate the 1,2-dioxygenolytic cleavage of salicylate by cells and cell extracts from P. salicylatoxidans BN12. This extraordinary ring fission reaction was first suggested by the oxygen dependence but NAD(P)H independence of the reaction. This indicated a ring fission dioxygenase and contradicted a possible involvement of hydroxylating monooxygenases or dioxygenases. The involvement of a monooxygenase was also excluded by the spectrophotometric and HPLC analysis of the reaction product, which clearly excluded all possible hydroxylated salicylates and also catechol as reaction products. The definitive proof for the suggested novel ring fission reaction of salicylate to 2-oxohepta-3,5-dienedioic acid was obtained after purification of the reaction product. The results of the 1H and 13C NMR spectra clearly indicated the presence of two carboxylate groups, a further oxo function, and a set of four conjugated olefinic carbon atoms. Furthermore, these 1H and 13C NMR spectra clearly resembled the spectra previously reported for 6-oxohepta-2,4-dienoic acid (a compound studied in connection with the cis-trans isomerization of maleylpyruvate, the cleavage product of gentisate) (10) and 2-hydroxymuconate (an intermediate in the degradation of catechol by the meta cleavage pathway) (43). The evidence for the proposed ring fission reaction was further substantiated by the information obtained by MS. The 1H NMR spectrum indicated a trans configuration for the double bond between C-3 and C-4. It was expected that this double bond should conserve its cis configuration originating from the aromatic nucleus of the substrate, because there is no obvious mechanism by which this double bond could easily isomerize in the absence of an attached hydroxy group which would allow a keto-enol tautomerization and a facilitated isomerization at this position. This suggests that during the enzymatic ring fission of salicylate some transient rearrangements of the double bonds take place, as has been suggested previously for the reaction of a gentisate 1,2-dioxygenase with its substrate (14), or that some (reversible) lactonization reactions may occur, as previously suggested for the ring fission product of 5-chlorosalicylate (7).
The oxygenolytic 1,2-cleavage of salicylate contradicts a generally accepted paradigm that the enzymatic ring fission of the aromatic nucleus by bacteria requires the presence of two hydroxy groups attached to the aromatic ring. Nevertheless, a few examples have been described previously in which monohydroxylated aromatics, e.g., 5-aminosalicylate or 2-aminophenol, were cleaved by ring fission dioxygenases (8, 24, 37, 39). The ability of “traditional” ring fission dioxygenases to oxidize aminohydroxybenzene derivatives is mechanistically easily explained, because the amino group activates the aromatic nucleus similarly to the hydroxy group for an electrophilic attack of the ring-cleaving dioxygenases (37). There are also a few reports which describe the ring fission of monohydroxylated aromatic compounds which do not possess a second electron-donating substituent; this has been described for the oxidation of 5-chlorosalicylate by a Bacillus sp. and for the conversion of 1-hydroxy-2-naphthoate by several gram-negative (e.g., Aeromonas) and gram-positive (Nocardioides) bacteria (1, 7, 15, 17, 18). From these two reactions, only the oxidation of 1-hydroxy-2-naphthoate by Nocardioides sp. strain KP7 has been analyzed on an enzymatic and genetic level and the ring fission product has been isolated and characterized by various spectroscopic techniques (1, 15).
The results of the present study suggested that P. salicylatoxidans oxidized salicylate by a ring fission dioxygenase similar to gentisate 1,2-dioxygenase or 1-hydroxy-2-naphthoate dioxygenase. This was indicated by the ability of the enzyme to convert gentisate and 1-hydroxy-2-naphthoate, the size of the subunits, the structure of the holoenzyme, and the dependence of the enzyme on Fe2+ ions. The ring fission dioxygenase from P. salicylatoxidans was clearly different from currently known gentisate 1,2-dioxygenases or 1-hydroxy-2-naphthoate dioxygenases because of its unique ability to oxidatively cleave salicylate and also the ability to cleave gentisate and 1-hydroxy-2-naphthoate with high catalytic efficiencies. In contrast, the 1-hydroxy-2-naphthoate dioxygenase from Nocardioides sp. strain KP7 does not oxidize gentisate or salicylate (15), and gentisate 1,2-dioxygenases do not oxidize salicylate (14). (Apparently 1-hydroxy-2-naphthoate has not been tested as a substrate for gentisate 1,2-dioxygenases.)
From the kinetic data it is currently not possible to distinguish if the enzyme is more closely related to gentisate 1,2-dioxygenases or to 1-hydroxy-2-naphthoate dioxygenases. Furthermore, gentisate 1,2-dioxygenases and 1-hydroxy-2-naphthoate dioxygenases may be evolutionarily related, because the gentisate 1,2-dioxygenase from Sphingomonas sp. strain RW5 has been shown to have a low degree of sequence similarity (27%) to the 1-hydroxy-2-naphthoate dioxygenase from Nocardioides sp. strain KP7 (40). A closer relationship of the enzyme from P. salicylatoxidans with gentisate 1,2-dioxygenases was suggested by the enzyme purification experiments, which demonstrated that the enzyme is composed of four identical subunits with a size of about 45 kDa. Similar results have been obtained with gentisate 1,2-dioxygenases from different microorganisms (Comamonas testosteroni, Comamonas acidovorans, Haloferax sp., Klebsiella pneumoniae, Moraxella osloensis OA3, Pseudomonas alcaligenes, and Sphingomonas sp. strain RW5) which are composed of four identical subunits with a size of about 40 kDa (6, 11, 12, 13, 38, 40, 47). In contrast, it was suggested that the 1-hydroxy-2-naphthoate dioxygenase from Nocardioides sp. strain KP7 was composed of six subunits with a molecular mass of about 43 kDa (15). In order to obtain conclusive information about the evolution of the extraordinary salicylate 1,2-dioxygenase activity from strain BN12, we are attempting to clone the encoding gene.
REFERENCES
- 1.Adachi K, Iwabuchi T, Sano H, Harayama S. Structure of the ring-cleavage product of 1-hydroxy-2-naphthoate, an intermediate of the phenanthrene-degradative pathway of Nocardioides sp. strain KP7. J Bacteriol. 1999;181:757–763. doi: 10.1128/jb.181.3.757-763.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andreoni V, Canonica L, Galli E, Gennari C, Treccani V. 2,3-Dihydroxybenzoate pathway in Pseudomonas putida. Biochem J. 1981;194:607–610. doi: 10.1042/bj1940607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Brilon C, Beckmann W, Hellwig M, Knackmuss H-J. Enrichment and isolation of naphthalenesulfonic acid-utilizing pseudomonads. Appl Environ Microbiol. 1981;42:39–43. doi: 10.1128/aem.42.1.39-43.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brilon C, Beckmann W, Knackmuss H-J. Catabolism of naphthalenesulfonic acids by Pseudomonas sp. A3 and Pseudomonas sp. C22. Appl Environ Microbiol. 1981;42:44–55. doi: 10.1128/aem.42.1.44-55.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crawford R L, Hutton S W, Chapman P J. Purification and properties of gentisate 1,2-dioxygenase from Moraxella osloensis. J Bacteriol. 1975;121:794–799. doi: 10.1128/jb.121.3.794-799.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crawford R L, Olson P E, Frick T D. Catabolism of 5-chlorosalicylate by a Bacillus isolated from the Mississippi river. Appl Environ Microbiol. 1979;38:379–384. doi: 10.1128/aem.38.3.379-384.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis J K, He Z, Somerville C C, Spain J C. Genetic and biochemical comparison of 2-aminophenol 1,6-dioxygenase of Pseudomonas pseudoalcaligenes JS45 to meta-cleavage dioxygenases: divergent evolution of 2-aminophenol meta-cleavage pathway. Arch Microbiol. 1999;172:330–339. doi: 10.1007/s002030050787. [DOI] [PubMed] [Google Scholar]
- 9.Dorn E, Hellwig M, Reineke W, Knackmuss H J. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch Microbiol. 1974;99:61–70. doi: 10.1007/BF00696222. [DOI] [PubMed] [Google Scholar]
- 10.Feliu A L, Seltzer S. Synthesis and interconversion of the four isomeric 6-oxo-2,4-heptadienoic acids. J Org Chem. 1985;50:447–451. [Google Scholar]
- 11.Feng Y, Khoo H E, Poh C L. Purifcation and characterization of gentisate 1,2-dioxygenases from Pseudomonas alcaligenes NCIB 9867 and Pseudomonas putida NCIB 9869. Appl Environ Microbiol. 1999;65:946–950. doi: 10.1128/aem.65.3.946-950.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu W, Oriel P. Gentisate 1,2-dioxygenase from Haloferax sp. D1227. Extremophiles. 1998;2:439–446. doi: 10.1007/s007920050090. [DOI] [PubMed] [Google Scholar]
- 13.Harpel M R, Lipscomb J D. Gentisate 1,2-dioxygenase from Pseudomonas. Purification, characterization, and comparison of the enzymes from Pseudomonas testosteroni and Pseudomonas acidovorans. J Biol Chem. 1990;265:6301–6311. [PubMed] [Google Scholar]
- 14.Harpel M R, Lipscomb J D. Gentisate 1,2-dioxygenase from Pseudomonas. Substrate coordination to active site Fe2+ and mechanism of turnover. J Biol Chem. 1990;265:22187–22196. [PubMed] [Google Scholar]
- 15.Iwabuchi T, Harayama S. Biochemical and molecular characterization of 1-hydroxy-2-naphthoate dioxygenase from Nocardioides sp. KP7. J Biol Chem. 1998;273:8332–8336. doi: 10.1074/jbc.273.14.8332. [DOI] [PubMed] [Google Scholar]
- 16.Kämpfer P, Müller C, Mau M, Neef A, Auling G, Busse H-J, Osborn A M, Stolz A. Description of Pseudaminobacter gen. nov. with two new species, Pseudaminobacter salicylatoxidans sp. nov. and Pseudaminobacter defluvii sp. nov. Int J Syst Bacteriol. 1999;49:887–897. doi: 10.1099/00207713-49-2-887. [DOI] [PubMed] [Google Scholar]
- 17.Kiyohara H, Nakao K. Enzymatic conversion of 1-hydroxy-2-naphthoate in phenanthrene-grown Aeromonas sp. S45P1. Agric Biol Chem. 1977;41:705–707. [Google Scholar]
- 18.Kiyohara H, Nakao K. The catabolism of phenanthrene and naphthalene by bacteria. J Gen Microbiol. 1978;105:69–75. [Google Scholar]
- 19.Kuhm AE, Stolz A, Ngai K-L, Knackmuss H-J. Purification and characterization of a 1,2-dihydroxynaphthalene dioxygenase from a bacterium that degrades naphthalenesulfonic acids. J Bacteriol. 1991;173:3795–3802. doi: 10.1128/jb.173.12.3795-3802.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhm A E, Knackmuss H-J, Stolz A. Purification and properties of 2′-hydroxybenzalpyruvate aldolase from a bacterium that degrades naphthalenesulfonates. J Biol Chem. 1993;268:9484–9489. [PubMed] [Google Scholar]
- 21.Kuhm A E, Knackmuss H-J, Stolz A. 2-Hydroxychromene-2-carboxylate isomerase from bacteria that degrade naphthalenesulfonates. Biodegradation. 1993;4:155–162. [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Lee J, Oh J, Min K R, Kim Y. Nucleotide sequence of salicylate hydroxylase gene and its 5′-flanking region of Pseudomonas putida KF715. Biochem Biophys Res Commun. 1996;218:544–548. doi: 10.1006/bbrc.1996.0097. [DOI] [PubMed] [Google Scholar]
- 24.Lendenmann U, Spain J C. 2-Aminophenol 1,6-dioxygenase: a novel aromatic ring cleavage enzyme purified from Pseudomonas pseudoalcaligenes JS45. J Bacteriol. 1996;178:6227–6232. doi: 10.1128/jb.178.21.6227-6232.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nörtemann B. Bakterieller Abbau von Amino- und Hydroxynaphthalin-sulfonsäuren. Thesis. Stuttgart, Germany: University of Stuttgart; 1987. [Google Scholar]
- 26.Nörtemann B, Baumgarten J, Rast H G, Knackmuss H-J. Bacterial communities degrading amino- and hydroxynaphthalenesulfonates. Appl Environ Microbiol. 1986;52:1195–1202. doi: 10.1128/aem.52.5.1195-1202.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nörtemann B, Kuhm A E, Knackmuss H-J, Stolz A. Conversion of substituted naphthalenesulfonates by Pseudomonas sp. BN6. Arch Microbiol. 1994;161:320–327. [Google Scholar]
- 28.Ohe T, Watanabe Y. Degradation of 2-naphthylamine-1-sulfonic acid by Pseudomonas strain TA-1. Agric Biol Chem. 1986;50:1419–1426. [Google Scholar]
- 29.Ohe T, Watanabe Y. Microbial degradation of 1,6- and 2,6-naphthalenedisulfonic acid by Pseudomonas sp. S-1. Agric Biol Chem. 1988;52:2409–2414. [Google Scholar]
- 30.Ohe T, Ohmoto T, Kobayashi Y, Sato A, Watanabe Y. Metabolism of naphthalenesulfonic acids by Pseudomonas sp. TA-2. Agric Biol Chem. 1990;54:669–675. [Google Scholar]
- 31.Rauscher K, Voigt J, Wilke I, Wilke K-T. Chemische Tabellen und Rechentafeln für die analytische Praxis, 7. überarbeitete Auflage. Thun, Germany: Verlag Harri Deutsch; 1982. [Google Scholar]
- 32.Rozgaj R, Glancer-Äoljan M. Total degradation of 6-aminonaphthalene-2-sulphonic acid by a mixed culture consisting of different bacterial genera. FEMS Microbiol Ecol. 1992;86:229–235. [Google Scholar]
- 33.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 34.Shultz A. Sulfonic acids. In: Grayson V M, editor. Kirk-Othmer encyclopedia of chemical technology. 3rd ed. Vol. 22. New York, N.Y: John Wiley & Sons; 1983. pp. 45–63. [Google Scholar]
- 35.Stolz A. Degradation of substituted naphthalenesulfonic acids by Sphingomonas xenophaga BN6. Int J Ind Microbiol Biotechnology. 1999;23:391–399. doi: 10.1038/sj.jim.2900725. [DOI] [PubMed] [Google Scholar]
- 36.Stolz A, Knackmuss H-J. Degradation of 2,4-dihydroxybenzoate by Pseudomonas sp. BN9. FEMS Microbiol Lett. 1993;108:219–224. doi: 10.1111/j.1574-6968.1993.tb06102.x. [DOI] [PubMed] [Google Scholar]
- 37.Stolz A, Nörtemann B, Knackmuss H-J. Bacterial degradation of 5-aminosalicylic acid. Initial ring cleavage. Biochem J. 1992;282:675–680. doi: 10.1042/bj2820675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suárez M, Ferrer E, Martín M. Purification and biochemical characterization of gentisate 1,2-dioxygenase from Klebsiella pneumoniae M5a1. FEMS Microbiol Lett. 1996;143:89–95. doi: 10.1111/j.1574-6968.1996.tb08466.x. [DOI] [PubMed] [Google Scholar]
- 39.Takenaka S, Murakami S, Shinke R, Hatakeyama K, Yukawa H, Aoki K. Novel genes encoding 2-aminophenol 1,6-dioxygenase from Pseudomonas species AP-3 growing on 2-aminophenol and catalytic properties of the purified enzyme. J Biol Chem. 1997;272:14727–14732. doi: 10.1074/jbc.272.23.14727. [DOI] [PubMed] [Google Scholar]
- 40.Werwath J, Arfmann H-A, Pieper D H, Timmis K N, Wittich R-M. Biochemical and genetic characterization of a gentisate 1,2-dioxygenase from Sphingomonas sp. strain RW5. J Bacteriol. 1998;180:4171–4176. doi: 10.1128/jb.180.16.4171-4176.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wheelis M L, Palleroni N J, Stanier R J. The metabolism of aromatic acids by Pseudomonas testosteroni and P. acidovorans. Arch Mikrobiol. 1967;59:302–314. doi: 10.1007/BF00406344. [DOI] [PubMed] [Google Scholar]
- 42.White-Stevens R H, Kamin H. Studies of a flavoprotein, salicylate hydroxylase. I. Preparation, properties, and the uncoupling of oxygen reduction from hydroxylation. J Biol Chem. 1972;247:2358–2370. [PubMed] [Google Scholar]
- 43.Whitman C P, Aird B A, Gillespie W R, Stolowich N J. Chemical and enzymatic ketonization of 2-hydroxymuconate, a conjugated enol. J Am Chem Soc. 1991;113:3154–3162. [Google Scholar]
- 44.Wittich R-M, Rast H G, Knackmuss H-J. Degradation of naphthalene-2,6- and naphthalene-1,6-disulfonic acid by a Moraxella sp. Appl Environ Microbiol. 1988;54:1842–1847. doi: 10.1128/aem.54.7.1842-1847.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamamoto S, Katagiri M, Maeno H, Hayaishi O. Salicylate hydroxylase, a monooxygenase requiring flavin adenine dinucleotide. J Biol Chem. 1965;240:3408–3413. [PubMed] [Google Scholar]
- 46.You I-S, Murray R I, Jollie D, Gunsalus I C. Purification and characterization of salicylate hydroxylase from Pseudomonas putida PpG7. Biochem Biophys Res Commun. 1990;169:1049–1054. doi: 10.1016/0006-291x(90)92000-p. [DOI] [PubMed] [Google Scholar]
- 47.Zhou N-Y, Fuenmayor S L, Williams P A. nag genes of Ralstonia (formerly Pseudomonas) sp. strain U2 encoding enzymes for gentisate catabolism. J Bacteriol. 2001;183:700–708. doi: 10.1128/JB.183.2.700-708.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]