Abstract
Alzheimer's disease (AD) is a chronic neurodegenerative disease, which leads to impairment of cognition and memory. The heat shock protein 70 (HSP70) family plays an important role in the pathogenesis of AD. It is known to regulate protein misfolding in a variety of diseases, including inhibition of Aβ aggregation and NFT formation in AD. As yet, the diagnostic molecular markers of AD remain unclear. Herein, we sought to investigate molecular markers of HSP70 family that can affect diagnosis and treatment in AD through computational analysis. In this study, the intersection between HSP70 family members and immune molecules was taken to screen immune-related HSP70 family genes. Based on the datasets from the NCBI-Gene Expression Omnibus (GEO) database, we found that the expression levels of HSPA1A and HSPA2 were significantly increased in AD samples, while HSPA8 significantly decreased. Surprisingly, the combination of the 3 hub genes had a good diagnosis of AD via receiver operating characteristic curve (ROC). Moreover, the clinical value of the 3 hub genes was further assessed by the Spearman correlation analysis with AD-related genes, β-secretase activity, and γ-secretase activity. In terms of immune cell infiltration, we showed that the distribution of seven immune cell types (macrophages M2, neutrophils, T cells CD4 memory activated, macrophages M0, NK cells activated, plasma cells, and T cells follicular helper) was associated with the occurrence of AD by CIBERSORT. Furthermore, our data suggested that EP300, MYC, TP53, JUN, CREBBP, and ESR1 might be key transcription factors (TFs) for the 3 hub genes. In general, these findings suggest that HSPA1A, HSPA2, and HSPA8 are potential molecular biomarkers for prognosis among HSP70 family in AD, and it provides a new perspective on diagnostic and therapeutic targets for AD.
1. Introduction
It is estimated that 47 million people live with dementia globally, and it will increase more than threefold (~131 million) by 2050 [1]. Alzheimer's disease (AD) is a common neurodegenerative disease, which is the major cause of dementia [2]. It will reduce the life quality of patients, affect the normal life of their families, and increase the cost burdens of care [3]. Currently, it has evolved into one of the great health-care challenges of the twenty-first century [4]. The pathogenesis of AD has been attributed to extracellular aggregates of amyloid β (Aβ) plaques and intracellular neurofibrillary tangles formed by hyperphosphorylated τ-protein (tau) in the human brain [1]. It is characterized by memory impairment and progressive neurocognitive dysfunction [5]. Some studies have shown that it also involves disturbances of mitochondria, abnormalities of lipid metabolism, oxidative stress, immune system and inflammatory responses, and other pathways [6, 7]. For instance, oxidative stress and neuroinflammation ultimately increase neuronal death through apoptosis or other mechanisms, and they are responsible for the behavioral deficits including AD [8]. They produce some of the insidious effects, including cognitive dysfunctions such as learning and memory impairment [9, 10]. In terms of AD-related molecular markers, APOE gene has been shown to be closely associated with the incidence of AD in most cases [11]. Other related genes include mutations in amyloid protein precursor (APP), presenilin-1 (PSEN1) and presenilin-2 (PSEN2) that make individuals susceptible to AD [12]. However, the pathogenesis and diagnostic molecular markers of AD and remains largely unclear. In particular, little is known about the immune aspects of AD. Therefore, it is of great significance to study the underlying molecular mechanisms and identify more reliable molecular markers of AD.
It is well known that misfolding protein aggregation in the human brain is one of the key features of many neurodegenerative diseases [13]. And the main hallmark in AD is the formation of Aβ plaques and tau protein aggregates [14]. The heat shock protein (HSP) family, particularly HSP70, plays an important role in this process and is known to regulate protein misfolding, including tau levels and toxicity in AD. The HSP70 family, consisting of 17 members, is a class of highly abundant and widely expressed chaperone proteins [13, 15]. They are encoded by a multigene family encompassing up to 17 genes and 30 pseudogenes [16]. In terms of function, the HSP70 family members are involved in numerous biological processes, including nascent polypeptide folding, protein trafficking, the refolding or degradation of aggregated peptides, and an immunomodulatory effect [17, 18]. In AD, it has been shown that HSP70 colocalizes with Aβ plaques and takes part in the neuroprotective response to suppress Aβ aggregation [19–21]. HSP70 also promotes solubility and the ability to bind microtubules of tau, hence inhibiting formation of neurofibrillary tangles [22]. In addition, an exon microarray data showed that HSPA1A was found to be upregulated in patients with AD, and it might be involved in protein folding abnormality and altered synaptic transmission on the pathomechanism of AD [23]. HSPA2 elevated Aβ40 and Aβ42 levels and phosphorylated-tau in two cell lines with HSPA2 overexpression and was nominated HSPA2 as a specific key regulator of late-onset Alzheimer's disease [24]. These evidences manifest that the HSP70 family plays a major role in AD's pathogenesis. However, at present, studies on the role of HSP70 family members in the pathogenesis of AD are scattered, only targeting a specific gene, such as HSPA1A. Considering the involvement of processes in AD etiology, targeting all members of the HSP70 family is a promising diagnostic and therapeutic prospect.
In this study, after integrating combined analysis between HSP70 family members and immune molecules in the ImmPort database and differential expression analysis, we preliminarily identified HSPA1A, HSPA2, and HSPA8 as hub genes of immune-related HSP70 family in AD. An in-depth analysis of the 3 hub genes functions in AD patients based on the datasets from the GEO database published online to determine their distinct diagnostic values and the potential functions in AD (Figure 1). To our knowledge, this is the first report that screens HSP70 family members to identify a series of hub genes in AD and performs clinical-related analysis, immune-related analysis, and biological function analysis of hub genes. Our data reveals a novel understanding of AD pathogenesis in HSP70 family and provides a set of potential useful targets for further research on molecular mechanisms and diagnostic biomarkers.
Figure 1.
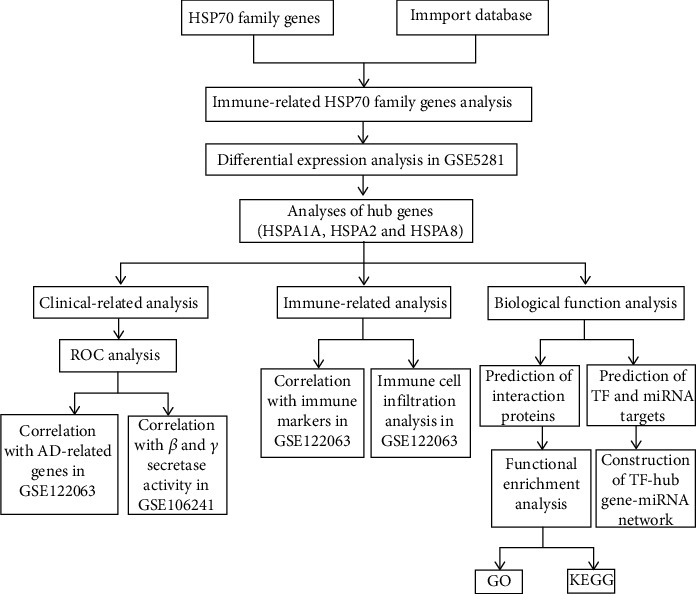
Workflow of computational analysis of immune-related HSP70 family members in AD.
2. Materials and Methods
2.1. Data Collection
Immune molecules data were retrieved from the ImmPort database [25] (https://www.immport.org/) (Supplementary Table 1). FunRich software [26] is user-friendly and provides graphical representation of the data. It was used to take intersection for screening candidate immune-related HSP70 family genes. The Gene Expression Omnibus [27] (GEO, http://www.ncbi.nlm.nih.gov/geo) is a public, free, and easy-to-use database with a large amount of data for researchers. We searched the GEO database for microarray datasets using the keyword “Alzheimer's disease.” Datasets were included if they met the following criteria: (1) were from humans; (2) included expression data from the brain regions of both AD and control samples and expression data from the brain regions of AD samples with β/γ secretase activity; (3) the number of AD samples was ≥50, and the number of control samples was ≥40; and (4) were downloaded to analysis. Finally, three datasets were obtained (GSE5281 [28], GSE132903 [29], and GSE122063 [30], including AD and control samples); one dataset (GSE106241 [31]) from AD samples with β/γ secretase activity was selected. Because GSE5281 (AD: 84, control: 74) and GSE132903 (AD: 97, control: 98) had far more samples, the array data of GSE5281 were used for differential expression analysis between the AD and control groups, and GSE132903 was used to validate further the differential expression of hub genes. The array data of GSE122063 was used for correlation with AD-related genes and immune-related analysis. The array data of GSE106241 was used for the correlation with β and γ secretase activity.
2.2. Disease-Diagnosing Ability and Correlation Analysis
To assess the diagnostic role of hub genes, we performed the receiver operating characteristic curve (ROC) analysis by SPSS 18.0 [32] (SPSS Inc., Chicago, USA). Moreover, we evaluated the Spearman correlation analysis between the 3 hub genes and AD-related genes, β-secretase activity and γ-secretase activity to understand whether the hub genes could reflect the clinical significance in AD.
2.3. Immune Cell Infiltration Analysis
To understand the status of the immune microenvironment in AD patients, we carried out CIBERSORT R package [33] of 110 samples in GSE122063, which was a robust algorithm for calculating the cellular composition of a tissue. The LM22 (22 immune cell types) was downloaded to act as a reference gene expression signature. Finally, Pearson correlation coefficient was utilized to evaluate the correlation with immune cell infiltration.
2.4. Prediction of Interaction Proteins and Functional Enrichment Analysis
GeneMANIA [34] (http://www.genemania.org) is a flexible, fast updated, and user-friendly website, which can provide interactions of proteins and genes and a variety of functions of submitted genes. To predict the interaction proteins of the 3 hub genes, we utilized GeneMANIA and exported the visualization results. Metascape [35] (http://metascape.org) is a practical, visual tool to perform gene annotation and pathway enrichment analysis. In terms of biological functions, the 3 hub genes, predicted interaction genes, and star molecules in AD were analyzed using Metascape, including the gene ontology (GO) enrichment and KOBAS-Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways analysis.
2.5. Construction of TF-Hub Gene-miRNA Network
The hTFtarget [36] (http://bioinfo.life.hust.edu.cn/hTFtarget#!/) was utilized to predict transcription factors (TFs) of hub genes. Then, the intersection of predicted TFs was taken via Jvenn website [37] (http://jvenn.toulouse.inra.fr/app/example.html), and the CytoHubba plug-in was used to select the TFs with high scores in Cytoscape [38]. The miRNAs of hub genes were predicted by using two online databases, TargetScan [39] (http://www.targetscan.org/) and miRDB [40] (http://mirdb.org/), which are commonly used to predict miRNAs. The miRNAs predicted by these two programs overlapped were identified as miRNA targets, in which the miRNA score predicted by the miRDB was over 80. Finally, the TF-hub gene-miRNA network was constructed using the Cytoscape software.
2.6. Statistical Analysis
Data obtained from GEO database were analyzed through Student's t-test. GraphPad Prism version 6.00 [41] was used for differential expression and correlation analysis, and values were expressed as Mean ± SD. P < 0.05 was considered statistically significant.
3. Results
3.1. Screening and Identification of Hub Genes by Computational Analysis
The 17 members of the HSP70 family (Table 1) and immune molecules in the ImmPort database were screened to take intersection for identifying immune-related HSP70 family genes using FunRich software. A total of 8 overlapped genes were obtained (Figure 2(a)). Furthermore, 87 AD samples and 74 control samples of GSE5281 were used for differential expression analysis of immune-related HSP70 family genes by GraphPad Prism 6.00 software. The results showed that HSPA1A and HSPA2 were significantly increased in AD group when compared with the control group (P < 0.0001 and P < 0.0001, respectively, Figure 2(b)), while HSPA8 decreased significantly (P < 0.0001, Figure 2(b)). It is worth mentioning that the HSPA1B gene was not clearly matched in the dataset, so it was excluded in the subsequent analysis. We have also analyzed GSE132903 dataset (AD: 97, Control: 98), and the verification results showed that the expression level trend of HSP1A, HSPA2, and HSPA8 gene was consistent (P < 0.05, P < 0.05, and P < 0.001, respectively, Supplementary Figure 1). Finally, we preliminary identified HSPA1A, HSPA2, and HSPA8 as hub genes of immune-related HSP70 family.
Table 1.
The 17 members of the HSP70 family in human.
| Gene | Gene ID | UniProt ID |
|---|---|---|
| HSPA1A | 3303 | P0DMV8 |
| HSPA1B | 3304 | P0DMV9 |
| HSPA1L | 3305 | P34931 |
| HSPA2 | 3306 | P54652 |
| HSPA4 | 3308 | P34932 |
| HSPA4L | 22824 | O95757 |
| HSPA5 | 3309 | P11021 |
| HSPA6 | 3310 | P17066 |
| HSPA7 | 3311 | P48741 |
| HSPA8 | 3312 | P11142 |
| HSPA9 | 3313 | P38646 |
| HSPA12A | 259217 | O43301 |
| HSPA12B | 116835 | Q96MM6 |
| HSPA13 | 6782 | P48723 |
| HSPA14 | 51182 | Q0VDF9 |
| HSPH1 | 10808 | Q92598 |
| HYOU1 | 10525 | Q9Y4L1 |
Figure 2.
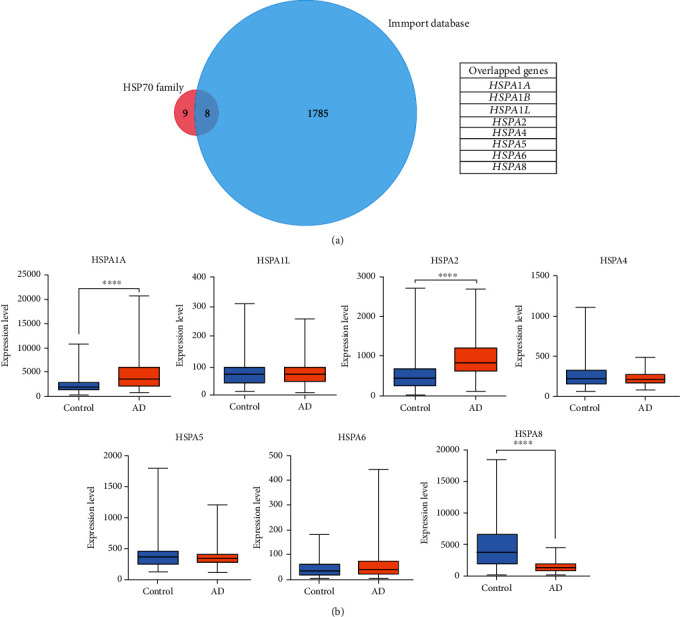
Screening and identification of hub genes by computational analysis. (a) Immune-related HSP70 family genes between the HSP70 family members and immune molecules in the ImmPort database were screened by Venn diagrams. The HSP70 family and the ImmPort database contain 17 and 1793 molecules, respectively, of which 8 are common between both groups. (b) The expression levels of seven immune-related HSP70 family members in GSE5281, except HSPA1B. The blue box indicates the control group, and the orange box indicates the AD group. Data were analyzed by Student's t-test and expressed as the Mean ± SD (AD: 84, control: 74). ∗∗∗∗ (P < 0.0001) represents significance compared to the control group.
3.2. ROC and Correlation Analyses Reveal Potential Clinical Values of Hub Genes
ROC analysis was performed to assess the diagnostic role of hub genes by SPSS 18.0. Surprisingly, we found that the combination of HSPA1A, HSPA2, and HSPA8 had a good ability to differentiate the AD (n = 56) and control (n = 44) groups in GSE122063 (Figure 3(a)). The area under the curve (AUC) for differentiating AD and control samples is 0.905 [95%confidence interval (CI) = 0.847–0.963]. Next, GSE122063 was also chosen for correlation analysis between hub genes and AD-related genes. Previous studies have shown that familial autosomal dominant AD genes amyloid protein precursor (APP) presenilin-1 (PSEN1), presenilin-2 (PSEN2), and major genetic risk factor APOE have been elucidated as the AD-related genes [42]. We found that HSPA1A and HSPA8 were negatively associated with APP (P = 0.0025 and P = 0.0089, respectively, Figure 3(b)), and HSPA2 was positively related to PSEN1 (P < 0.0001, Figure 3(b)). Moreover, the results also showed that HSPA1A and HSPA2 were negatively related to PSEN2 (P < 0.0001 and P = 0.0004, respectively, Figure 3(b)), and HSPA1A was negatively related to APOE (P = 0.0421, Figure 3(b)). In general, AD symptoms are associated with memory and cognition impairment. It has been suggested that cognitive deficit was associated with enhanced acetylcholinesterase activity in both cerebral cortex and hippocampus [10]. In view of this, we performed the correlation analysis between acetyl choline esterase (ACHE) and the 3 hub genes of HSP70 family in GSE122063 dataset. The results showed that HSPA2 was negatively related to ACHE (P < 0.0001, Figure 3(b)), and HSPA8 was positively related to ACHE (P = 0.0103, Figure 3(b)), but HSPA1A had no significant correlation (P = 0.2286, Figure 3(b)). Amyloid pathogenesis of AD reportedly starts with altered cleavage of APP by β-secretase and γ-secretase to produce insoluble Aβ fibrils [1, 43]. It suggests that β-secretase and γ-secretase play important roles in aggregates of Aβ plaques. Therefore, the correlation was also analyzed between the hub genes and the secretases. The results showed that HSPA2 was positively related to β-secretase activity (P = 0.0058, Figure 3(c)), and HSPA8 were negatively related to β-secretase activity (P < 0.0001, Figure 3(c)). Moreover, HSPA8 was negatively related to γ-secretase activity (P = 0.0117, Figure 3(c)).
Figure 3.
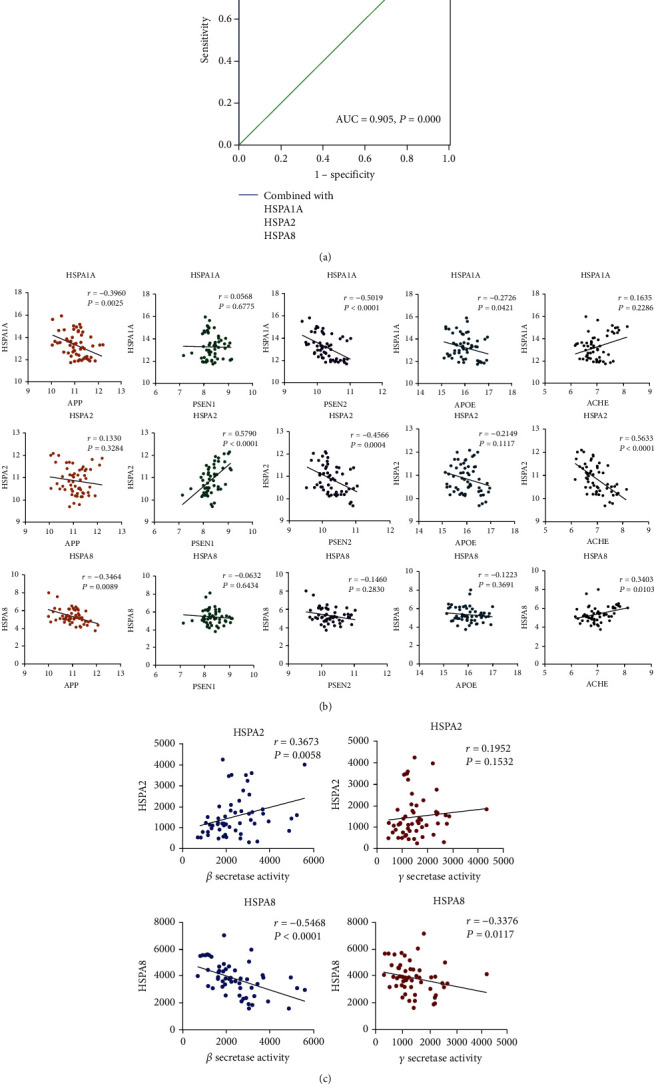
ROC and correlation analyses reveal potential clinical values of hub genes. (a) ROC curve in GSE122063 for distinction of AD and control based on the combination of HSPA1A, HSPA2, and HSPA8 (AD: 56, control: 44). (b) Correlation analysis between the 3 hub genes (HSPA1A, HSPA2, and HSPA8) and AD-related genes (APP, PSEN1, PSEN2, APOE, and ACHE) in GSE122063 (n = 56). Data were analyzed by the Spearman correlation analysis. (c) Correlation analysis to determine the clinical values between the hub genes (HSPA2 and HSPA8, except HSPA1A) and β-secretase activity and γ-secretase activity in GSE106241 (n = 60). Data were analyzed by the Spearman correlation analysis. For panels (b) and (c), P values in red are significant (P < 0.05).
3.3. Immune-Related Analysis Indicates the Relevance of Immune Cells and Hub Genes to AD
To investigate the relationship between 3 hub genes of immune-related HSP70 family in AD and the diverse immune infiltrating cells, we focused on the correlations between hub genes and immune cell type markers of various immune cells in AD samples of GSE122063. We analyzed the correlations between the expression of 3 hub genes and immune marker genes of different immune cells, such as B cells, neutrophils, and monocyte/macrophages. After the correlation analysis, the results suggested that there was a significant correlation between the expression of 3 hub genes and most immune markers (Table 2). We analyzed the immune infiltration of AD and control samples in GSE122063 by CIBERSORT algorithm. Figure 4(a) shows that macrophages account for most significant proportion among the 22 immune cells in the samples. As shown in Figures 4(b) and 4(t), cells CD4 naive had the strongest positive correlation with T cells gamma delta (r = 0.46); nevertheless, T cells CD4 memory resting had the strongest negative correlation with T cells CD8 (r = −0.64). We also carried out immune cell composition analysis to further compare the difference in the immune cells between the AD and control groups in GSE122063. The results suggested that the infiltration of macrophages M2, neutrophils, and T cells CD4 memory activated was higher in AD than in control group (P < 0.0001, P < 0.0001, and P < 0.01, respectively, Figure 4(c)), while the infiltration of macrophages M0, NK cells activated, plasma cells, and T cells follicular helper was opposite (P < 0.01, P < 0.01, P < 0.001, and P < 0.001, respectively, Figure 4(c)). These data indicated that the distribution of seven immune cell types was related to the occurrence of AD.
Table 2.
Correlation analysis between HSPA1A/HSPA2/HSPA8 and immune cell type markers in AD samples of GSE122063.
| Cell type | Gene markers | HSPA1A | HSPA2 | HSPA8 | |||
|---|---|---|---|---|---|---|---|
| COR | P value | COR | P value | COR | P value | ||
| CD8+ T cells | CD8A | -0.2879 | 0.0314 | -0.5776 | <0.0001 | 0.0861 | 0.5280 |
| Th1 cells | TBX21 | 0.0174 | 0.8985 | 0.3440 | 0.0094 | -0.3606 | 0.0063 |
| Treg | FOXP3 | -0.2967 | 0.0264 | 0.1456 | 0.2843 | -0.3557 | 0.0071 |
| CCR8 | 0.1485 | 0.2746 | 0.1529 | 0.2607 | -0.0801 | 0.5573 | |
| B cells | CD22 | 0.0331 | 0.8087 | 0.6700 | < 0.0001 | -0.2869 | 0.0321 |
| Neutrophils | CEACAM3 | 0.1321 | 0.3319 | 0.3057 | 0.0220 | 0.0054 | 0.9685 |
| FPR1 | 0.5884 | <0.0001 | -0.1791 | 0.1866 | 0.4027 | 0.0021 | |
| CSF3R | -0.0662 | 0.6280 | 0.0049 | 0.9713 | -0.1613 | 0.2349 | |
| S100A12 | 0.1714 | 0.2065 | 0.3556 | 0.0072 | -0.2007 | 0.1381 | |
| Monocyte/macrophages | CD86 | -0.0057 | 0.9667 | 0.4965 | < 0.0001 | -0.2304 | 0.0876 |
| CSF1R | 0.2432 | 0.0709 | 0.01097 | 0.9360 | 0.1407 | 0.3009 | |
| CD68 | 0.4656 | 0.0003 | -0.0192 | 0.8883 | 0.3343 | 0.0118 | |
| CD84 | 0.3693 | 0.0051 | -0.0025 | 0.9852 | 0.3444 | 0.0093 | |
| CD163 | 0.3700 | 0.0050 | 0.1468 | 0.2803 | 0.0874 | 0.5221 | |
| MS4A4A | 0.4919 | 0.0001 | -0.3283 | 0.0135 | 0.4726 | 0.0002 | |
| CD14 | 0.4918 | 0.0001 | -0.3242 | 0.0148 | 0.3908 | 0.0029 | |
| CD33 | 0.2943 | 0.0277 | -0.3928 | 0.0028 | 0.5218 | < 0.0001 | |
| NK cells | KIR3DL3 | -0.1883 | 0.1646 | 0.1963 | 0.1470 | -0.3442 | 0.0094 |
| Dendritic cells | CD83 | -0.1420 | 0.2964 | -0.5224 | < 0.0001 | 0.1111 | 0.4151 |
Figure 4.
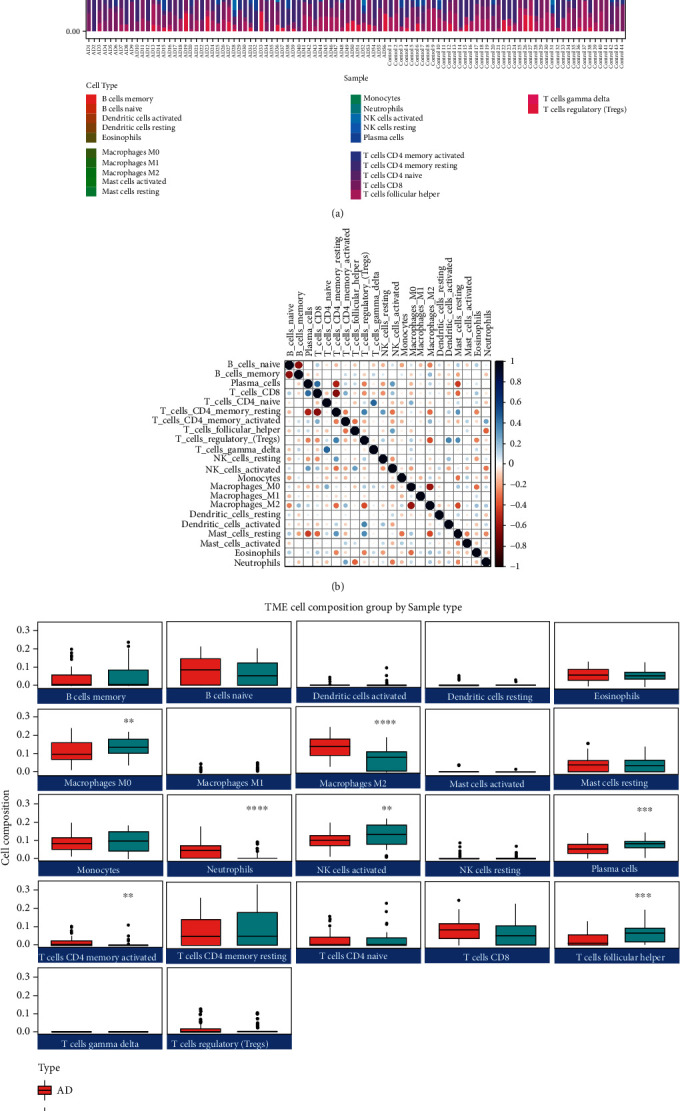
Immune-related analysis indicates the relevance of immune cells and hub genes to AD. (a) The proportion of the 22 immune cell types in AD and control samples in GSE122063 (AD: 56, control: 44). The horizontal coordinates indicate the samples, and the vertical coordinates indicate the proportion of immune cells. (b) Correlation matrix between the 22 immune cell types in GSE122063. Each row (column) represents one immune cell type. Blue represents positive correlations and red represents negative correlations. Darker colors indicate stronger correlations. (c) The total distribution of immune cells between the AD and control groups in GSE122063. The green box indicates the control group, and the red box indicates the AD group. Data were analyzed by Student's t-test and expressed as the Mean ± SD (AD: 56, control: 44). ∗∗ (P < 0.01), ∗∗∗ (P < 0.001), and ∗∗∗∗ (P < 0.0001) represent significance compared to the control group.
3.4. Functional Enrichment Analysis of Hub Genes, Interaction Genes, and Star Molecules in AD
Prediction of interaction proteins with the 3 hub genes of immune-related HSP70 family in AD was performed to explore the potential interactions using GeneMANIA. The results revealed that there were primarily 20 genes (HSPB1, HSPA6, STUB1, PSMD2, HSPH1, CITED1, BAG3, HSPBP1, HSPA4, BAG1, HSPA4L, DNAJB12, HSPA1L, EGFR, BAG2, BAG5, DNAJB14, PINK1, GAK, and METTL21A) associated with the 3 hub genes (Figure 5(a)). Then, the 3 hub genes, these interaction genes and star molecules in AD (APP, PSEN1, PSEN2, and APOE) were submitted in Metascape for analyzing the functions. Figure 5(b) shows the most highly enriched GO items. In the biological process (BP) category, protein folding, response to topologically incorrect protein, proteasomal protein catabolic process, regulation of protein stability, mitochondrial transport, response to oxidative stress, and positive regulation of amyloid fibril formation were associated with the occurrence and progression of AD. In the cellular component (CC) category, the inclusion body, blood microparticle, perinuclear region of cytoplasm, chaperone complex, focal adhesion, coated vesicle, and spindle were the most highly enriched items. These genes were mainly enriched in heat shock protein binding, adenyl-nucleotide exchange factor activity, ubiquitin protein ligase binding, misfolded protein binding, HSP70 protein binding, Tau protein binding, and other aspects in the molecular function (MF) category. Additionally, KEGG pathway analyses showed that protein processing in endoplasmic reticulum, antigen processing, and presentation and Alzheimer's disease were significantly implicated in the occurrence and progression of AD (Figure 5(b)).
Figure 5.
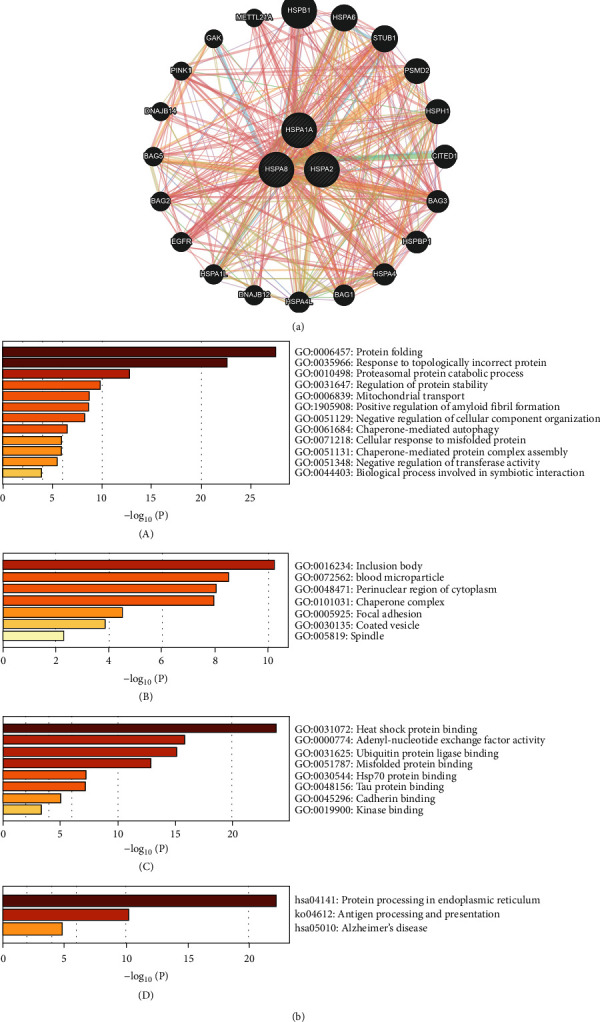
Functional enrichment analysis of hub genes, interaction genes, and star molecules in AD. (a) Prediction of interaction proteins with the 3 hub genes (HSPA1A, HSPA2, and HSPA8) of immune-related HSP70 family in AD are shown by protein–protein interaction network. (b) The GO and KEGG enrichment analysis of the 3 hub genes (HSPA1A, HSPA2, and HSPA8), 20 interaction genes (HSPB1, HSPA6, STUB1, PSMD2, HSPH1, CITED1, BAG3, HSPBP1, HSPA4, BAG1, HSPA4L, DNAJB12, HSPA1L, EGFR, BAG2, BAG5, DNAJB14, PINK1, GAK, and METTL21A), and 4 star molecules (APP, PSEN1, PSEN2, and APOE) in AD by Metascape. Bar plot of GO enrichment in biological process terms (A), cellular component terms (B), and molecular function terms (C). Bar plot of KEGG enriched terms (D).
3.5. Construction of TF-Hub Gene-miRNA Network May Be a New Potential Regulatory Network of AD
Due to the significantly different expression levels of the 3 hub genes of immune-related HSP70 family in the AD vs. control groups, the feasible transcription factor (TF) targets of the hub genes were explored by hTFtarget. Then, the intersection of predicted TF was taken using the Jvenn website, and 130 possible TFs that jointly regulated the 3 hub genes were obtained (Supplementary Table 2). CytoHubba plug-in was further used to select the TFs with high scores in Cytoscape focusing screening of 6 algorithms (MCC, degree, MNC, closeness, radiality, and EPC) and taken intersection, and the last 6 important TF targets (EP300, MYC, TP53, JUN, CREBBP, and ESR1) of hub genes were obtained. Subsequently, miRNA targets of the hub genes were predicted using the two online databases, TargetScan and miRDB. The miRNAs predicted by these two programs overlapped were identified as miRNA targets, in which the miRNA score predicted by the miRDB was ≥80, and we obtained 6 miRNA targets of HSPA1A, 13 miRNA targets of HSPA2, and 35 miRNA targets of HSPA8, respectively (Supplementary Table 3). Finally, the TF-hub gene-miRNA network was constructed using the Cytoscape software (Figure 6). It may be a new potential regulatory network of AD.
Figure 6.
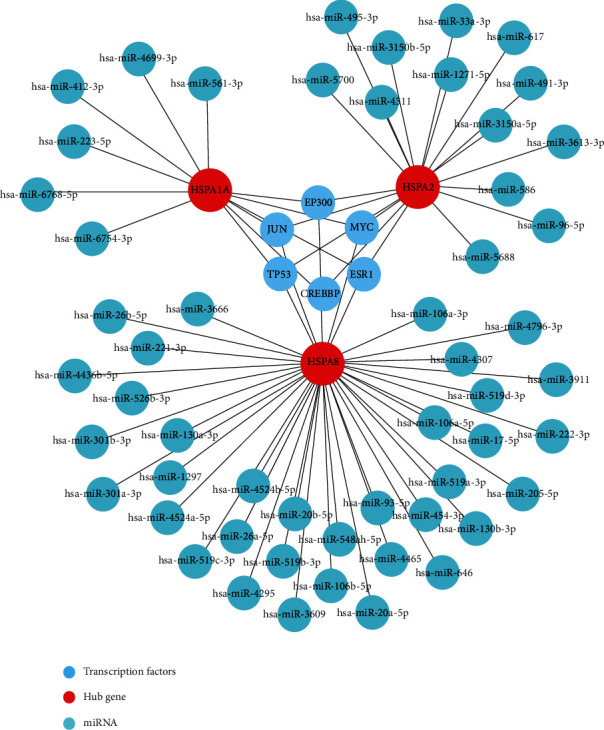
Construction of TF-hub gene-miRNA network may be a new potential regulatory network of AD. The red circle indicates the hub genes, the blue circle indicates the TF targets, and the brilliant blue circle indicates the miRNA targets.
4. Discussion
In this study, 3 hub genes of HSP70 family associated with AD were identified by computational analysis using gene expression data extracted from the GEO datasets. To our knowledge, this is the first report to screen HSP70 family members for identifying a series of hub genes as candidate biomarkers or therapeutic targets for AD and integrate clinical-related analysis, immune-related analysis, and biological function analysis of hub genes. According to ROC and correlation analyses, the 3 hub genes revealed potential clinical values. Moreover, immune cell infiltration analysis indicated the relevance of immune cells and hub genes to AD. Furthermore, our data suggested that EP300, MYC, TP53, JUN, CREBBP, and ESR1 might be key transcription factors (TFs) for the 3 hub genes and provided a new potential regulatory network of AD. Overall, we identified that HSPA1A, HSPA2, and HSPA8 among HSP70 family played vital roles in AD pathogenesis, and it provided a new perspective for AD diagnosis and treatment targets for AD. HSPA1A is a major heat shock protein in HSP70 family. A proteomics study of extracellular vesicles separated from cerebrospinal fluid (CSF) of AD showed that the expression level of HSPA1A was higher in the AD group than in the MCI and CTRL groups, and it might be used to monitor the progression of MCI to AD [44]. Another exon microarray data demonstrated that HSPA1A was found to be upregulated in patients with AD [23]. Our results showed increased expression of HSPA1A in AD, which is consistent with previous studies. This indicates HSPA1A may play an important part in AD progression. It was reported that HSPA2 was not only significantly highly expressed in late-onset Alzheimer's disease (LOAD), but also further elevated Aβ40 and Aβ42 levels and phosphorylated-tau in two cell lines with HSPA2 overexpression [24]. In our study, we found that HSPA2 was significantly overexpressed in AD. In addition, we also found that HSPA2 was significantly upregulated in Braak 5-6 when compared with Braak 0 in GSE131617. It is similar to the results of previous studies. This work further demonstrates HSPA2 as a specific key regulator of late-onset AD. In addition, HSPA8 was significantly downregulated in Braak 5-6 when compared with Braak 0 in our analysis. However, the expression level of HSPA1A decreased with different processes of AD. This suggests that the hub genes may not be a good predictor of the changes in different stages of AD, but limited by the dataset, no good research results could be obtained temporarily. Results of qRT-PCR revealed that a significant downregulation of HSPA8 was observed in AD across the three brain regions (entorhinal and auditory cortices and hippocampus) compared to the controls [45]. In this study, we found that HSPA8 decreased significantly in AD compared with control group in GSE5281. The results are consistent with previous studies. These findings indicate that HSPA1A, HSPA2, and HSPA8 play significant roles in the pathogenesis of AD.
A study revealed that the ROC result of HSPA1A mRNA in the diagnosis of myasthenia gravis was 0.830 [46]. Zhang et al. confirmed that HSPA8 as a potential serum biomarker for distinguishing renal cell carcinoma from benign urologic disease with ROC area under the curve (AUC) of 0.86 [47]. However, there is no assessment of the diagnostic ability of the hub genes in AD. Interestingly, we found that the combination of HSPA1A, HSPA2, and HSPA8 had a good ability to differentiate the AD and control groups in GSE122063. The AUC for predicting AD and control samples is 0.905 [95%confidence interval (CI) = 0.847–0.963]. The AUC of HSPA1A, HSPA2, and HSPA8 alone were 0.858 [95%confidence interval (CI) = 0.788–0.928], 0.821 [95%confidence interval (CI) = 0.739–0.902], and 0.667 [95%confidence interval (CI) = 0.562–0.772], respectively. This indicates that the predictive value of a single target is limited, while the combined target of the 3 hub genes has a higher predictive value.
Studies have shown that AD is a complex disorder with a relatively clear genetic component, involving several common AD-related genes. PSEN1, PSEN2, and APP have been identified as the cause of early onset familial AD (EOAD) [48]. In fact, early-onset autosomal dominant disease with age of onset younger than sixty years seems to be completely explained by pathogenic mutations in these three genes [49]. In LOAD, APOE is linked to the occurrence of AD and known to act in a dose-dependent manner. For instance, the effect of the ε4 allele in the risk for AD increases from 20 to 90% [50]. It was reported that the heat shock protein HSPA1A negatively regulated APP processing and Aβ production [51]. Our results showed that HSPA1A was negatively associated with APP. This demonstrated that HSPA1A was involved in the regulation mechanism of AD. In addition, a genetic association case-control study found that cross-talk in AD might be associated with multiway interactions between polymorphisms of candidate genes (HSPA1A, APOE, and so on) [52]. The correlation analysis in our study suggested that HSPA1A was negatively associated with APOE, further indicating a relationship between HSPA1A and susceptibility genes in AD. Interestingly, we also found that HSPA8 were negatively associated with APP, HSPA2 was positively associated with PSEN1, and HSPA1A and HSPA2 were negatively associated with PSEN2. These results have not been reported before. Moreover, AD causes varying degrees of memory and cognition impairment. Chonpathompikunlert et al. pointed out that acetyl choline esterase activity had played a role in the protection of neurodegeneration and improvement in memory impairment of AD's rats [53]. The results showed that HSPA2 was negatively related to ACHE and HSPA8 was positively related to ACHE. However, we have not found the appropriate dataset of GEO database which contains the level of acetyl choline to analyze the correlation with the 3 hub genes of HSP70 family. This may be due to the fact that the dataset related to AD in GEO database itself involves very few datasets of acetyl choline.
The main pathological process of AD is the accumulation of Aβ pathologic form which was caused by the continuous cleavage of APP via β- and γ-secretase in the brain [54]. Jo et al. reported that levels of β- and γ-secretase activities were greater in brain tissue samples from AD patients compared to nondemented control subjects [55]. It has been suggested that inhibition of the β- and γ-secretase was a valuable approach, which had been extensively tested in the clinic to prevent or delay the pathogenic effects of AD [56]. Based on the important role of β- and γ-secretase in Aβ formation, we analyzed the correlation in GSE106241 between the hub genes and β/γ-secretase activity, respectively. The results showed that HSPA2 was positively associated with β-secretase activity, HSPA8 was negatively associated with β-secretase activity, and HSPA8 was negatively associated with γ-secretase activity. However, due to the lack of HSPA1A data in this dataset, we did not confirm the correlation between HSPA1A and β- and γ-secretase activities and will add data later when available. Clinically, these results indicated that the 3 hub genes were closely relevant to the occurrence and development of AD. It may be possible to inhibit the activity of secretases by targeting the 3 hub genes, which in turn may have a therapeutic effect on AD.
There is evidence that immune system dysfunction in AD may be a major factor [57]. Many kinds of immune cells make up the immune system, including neutrophils and macrophages, to protect the body from external invasion. Hyperactivation of neutrophils is regarded as a characteristic of AD. The neutrophil formyl peptide receptors (FPR1 and FPR2) have become a therapeutic target that may be available for reducing injuries in inflammatory diseases including AD [58]. In our study, we found that HSPA1A and HSPA8 were significantly positive correlation with FPR1. Moreover, the infiltration of neutrophils in AD was significantly higher than in the control group. This suggests that these genes may play a part in neutrophils of immune system. It is worth mentioning that microglia had previously been reported to be associated with AD neuropathology and play a role in the etiology [59]. Microglia, as tissue-resident macrophages, play a fundamental role in the health of neurons [60]. Furthermore, the innate immune response may be modulated via polarizing microglia/macrophages in APP/PS1 mice [61]. In large genetic screens, key microglia/macrophages genes, such as CD33 and MS4As, have been associated with an increased risk of AD [59]. In this study, we found that HSPA1A, HSPA2, and HSPA8 were significantly correlated with CD33/MS4A4A through correlation analysis with immune cell type markers. In addition, the 3 hub genes have different levels of correlation with CD68, CD84, and other macrophages markers. The macrophages account for most significant proportion among the 22 immune cells in immune cell infiltration analysis, and we also found that infiltration of macrophages M2 was significantly increased in AD samples, while macrophages M0 decreased. These evidences may suggest that inflammatory activity of microglia in AD is increased and activated to promote the proliferation. This view is consistent with the research of Olmos-Alonso et al. [62]. NK cells have also been involved in AD [63], but controversial results have been published regarding NK cell number in AD patients. A study found no differences in the number of CD56 + CD16+ NK cells in AD patients [64], yet a decreased cells percentage of NK cells from 3xTg AD mice compared to NTg was reported by Maté et al. [65]. Our results are consistent with Ianire Maté's, that is, the infiltration of NK cells activated are reduced in AD, which may be due to the different functions of NK cells in different stages of AD. In general, our findings demonstrate an underlying immune role for HSPA1A/HSPA2/HSPA8 in AD.
The results of functional enrichment analysis indicated that 3 hub genes and their adjacent interaction genes mainly participated in biological process, such as protein folding, response to topologically incorrect protein, proteasomal protein catabolic process, regulation of protein stability, mitochondrial transport, response to oxidative stress, and positive regulation of amyloid fibril formation. The accumulation of misfolded protein aggregates is a common characteristic of AD, and it mainly includes an extensive aggregation of Aβpeptides and NFTs of hyper-phosphorylated tau protein [66]. There is the evidence that Aβ can be degraded through the proteasome to prevent its aggregation [67]. Several evidences suggest that the upregulation of molecular chaperones may inhibit NTFs' formation via dividing tau into productive folding pathways, thus preventing tau aggregation [68]. Moreover, a distinguishing feature of detrimental effects of Aβ peptides on neurons in AD is a significant decrease in mitochondria density and accumulation of mtDNA [69]. This may be due to an increase in mitochondrial degradation products by abnormal mitochondrial transport function, autophagy, or messed up proteolytic systems [70]. On the side, the enriched KEGG pathways include protein processing in endoplasmic reticulum (ER), antigen processing, and presentation and Alzheimer's disease. The ER is responsible for the proper folding or processing of nascent proteins, and the alterations in its homeostasis might be connected with accumulation of misfolded proteins in the brain of AD patients [71]. In addition, the analysis of TF-hub gene-miRNA network manifested that the 3 hub genes were related to TF targets EP300, MYC, TP53, JUN, CREBBP, and ESR1. Among them, it has been reported that MYC was increased in vulnerable neurons of AD patients, and the increased expression level of MYC might be in connection with cell death of neurons and the regulation of AD-related genes [72, 73]. An integrative analysis of hippocampus gene expression profiles also found dysregulation of transcription factors (CEBPB and MYC) in AD [74]. The miRNAs, a class of small noncoding RNAs (20-22 nucleotides), regulate the expression of key proteins and alter the expression of some miRNAs in AD pathogenesis. In our study, HSPA1A, HSPA2, and HSPA8 were predicted to be regulated by 6 miRNA targets, 13 miRNA targets, and 35 miRNA targets, respectively. It has been shown that hsa-miR-26b-5p can regulate the expression of the transporter vGLUT2, and a possible increased vGLUT2 level may lead to a higher intake of glutamate in the presynaptic vesicles to protect the nervous system from the excitatory toxicity of the glutamate itself, improving memory and cognition in AD [75]. We found that HSPA8 may be regulated by hsa-miR-26b-5p and then participate in the regulation of AD. However, studies on the regulation of miRNAs of HSP70 family members in AD are rare at present. Therefore, our findings may provide a new perspective to explore novel regulatory mechanism for research in this area.
The present study has some limitations. First, based on GEO datasets, some genes were lost due to inappropriate matching. The correlation analysis was lacking between the hub genes of HSP70 family and the definite AD biomarker including Aβ42, phospho-tau, and total tau due to the fact that the datasets in GEO database related to these AD biomarkers were very few. Second, the analysis of expression profile cannot reflect global changes, but only changes in transcription level. Third, further independent cohort with a larger sample size and molecular biological experiments are required to verify the roles of these hub genes in AD pathogenesis. To sum up, we hope our findings provide novel insights of AD pathogenesis and help to provide a set of potential helpful targets for further research on molecular mechanism and biomarkers in future.
5. Conclusion
We first reported that screens HSP70 family members to identify a series of hub genes in AD and performs clinical-related analysis, immune-related analysis, and biological function analysis of hub genes. Thus, HSPA1A, HSPA2, and HSPA8 may be 3 hub genes of immune-related HSP70 family in AD. Our findings might provide a novel understanding of AD pathogenesis and provide a set of potential useful targets for further studies on molecular mechanism and biomarkers.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 62027812). In addition, we thank Shengli Wang, Meijuan Li, and Yuying Qiu for helpful discussions on the study and manuscript.
Data Availability
The authors declare that all relevant data of this study are available within the article or from the corresponding author on reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest.
Authors' Contributions
Jie Li designed and supervised the study. Yeqing Dong, Tongxin Li, and Zhonghui Ma coordinated this study and analyzed the data. Chi Zhou performed part of the data analysis and Xinxu Wang supported a bioinformatics analysis. All authors participated in the draft writing and revising of the final manuscript. Yeqing Dong, Tongxin Li, and Zhonghui Ma contributed equally to this work and should be considered as co-first authors.
Supplementary Materials
Supplementary Fig.1 Expression of three immune-related HSP70 family members in GSE132903. The blue box indicates the control group, and the orange box indicates the AD group. Data were analyzed by Student's T-test and expressed as the Mean ± SD. ∗P < 0.05; ∗∗∗P < 0.001. Supplementary Table 1. Immune molecules in the Immport database. Supplementary Table 2. Common TFs of the 3 hub genes from hTFtarget by jvenn. Supplementary Table 3. The overlapped miRNAs of HSPA1A/HSPA2/HSPA8 predicted by TargetScan and miRDB
References
- 1.Tiwari S., Atluri V., Kaushik A., Yndart A., Nair M. Alzheimer’s disease: pathogenesis, diagnostics, and therapeutics. International Journal of Nanomedicine . 2019;Volume 14:5541–5554. doi: 10.2147/IJN.S200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballard C., Gauthier S., Corbett A., Brayne C., Aarsland D., Jones E. Alzheimer’s disease. Lancet . 2011;377(9770):1019–1031. doi: 10.1016/S0140-6736(10)61349-9. [DOI] [PubMed] [Google Scholar]
- 3.Wang Q. H., Wang X., Bu X. L., et al. Comorbidity burden of dementia: a hospital-based retrospective study from 2003 to 2012 in seven cities in China. Neuroscience Bulletin . 2017;33(6):703–710. doi: 10.1007/s12264-017-0193-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scheltens P., Blennow K., Breteler M. M., et al. Alzheimer’s disease. Lancet . 2016;388(10043):505–517. doi: 10.1016/S0140-6736(15)01124-1. [DOI] [PubMed] [Google Scholar]
- 5.Silva M. V. F., Loures C. M. G., Alves L. C. V., de Souza L. C., Borges K. B. G., Carvalho M. . G. Alzheimer’s disease: risk factors and potentially protective measures. Journal of Biomedical Science . 2019;26(1):p. 33. doi: 10.1186/s12929-019-0524-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guerreiro R., Hardy J. Genetics of Alzheimer’s disease. Neurotherapeutics . 2014;11(4):732–737. doi: 10.1007/s13311-014-0295-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozlov S., Afonin A., Evsyukov I., Bondarenko A. Alzheimer’s disease: as it was in the beginning. Reviews in the Neurosciences . 2017;28(8):825–843. doi: 10.1515/revneuro-2017-0006. [DOI] [PubMed] [Google Scholar]
- 8.Tiwari V., Kuhad A., Chopra K. Suppression of neuro-inflammatory signaling cascade by tocotrienol can prevent chronic alcohol-induced cognitive dysfunction in rats. Behavioural Brain Research . 2009;203(2):296–303. doi: 10.1016/j.bbr.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 9.Tiwari V., Chopra K. Resveratrol abrogates alcohol-induced cognitive deficits by attenuating oxidative-nitrosative stress and inflammatory cascade in the adult rat brain. Neurochemistry International . 2013;62(6):861–869. doi: 10.1016/j.neuint.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 10.Tiwari V., Chopra K. Attenuation of oxidative stress, neuroinflammation, and apoptosis by curcumin prevents cognitive deficits in rats postnatally exposed to ethanol. Psychopharmacology . 2012;224(4):519–535. doi: 10.1007/s00213-012-2779-9. [DOI] [PubMed] [Google Scholar]
- 11.Kanatsu K., Tomita T. Molecular mechanisms of the genetic risk factors in pathogenesis of Alzheimer disease. Front Biosci (Landmark Ed) . 2017;22(1):180–192. doi: 10.2741/4480. [DOI] [PubMed] [Google Scholar]
- 12.Van Cauwenberghe C., Van Broeckhoven C., Sleegers K. The genetic landscape of Alzheimer disease: clinical implications and perspectives. Genetics in Medicine . 2016;18(5):421–430. doi: 10.1038/gim.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lackie R. E., Maciejewski A., Ostapchenko V. G., et al. The Hsp70/Hsp90 chaperone machinery in neurodegenerative diseases. Frontiers in Neuroscience . 2017;11:p. 254. doi: 10.3389/fnins.2017.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng Q., Kebede M. T., Kemeh M. M., et al. Inhibition of the self-assembly of Aβ and of tau by polyphenols: mechanistic studies. Molecules . 2019;24(12):p. 2316. doi: 10.3390/molecules24122316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campanella C., Pace A., Caruso Bavisotto C., et al. Heat shock proteins in Alzheimer’s disease: role and targeting. International Journal of Molecular Sciences . 2018;19(9):p. 2603. doi: 10.3390/ijms19092603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brocchieri L., Conway de Macario E., Macario A. J. hsp70 genes in the human genome: conservation and differentiation patterns predict a wide array of overlapping and specialized functions. BMC Evolutionary Biology . 2008;8(1):p. 19. doi: 10.1186/1471-2148-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bukau B., Weissman J., Horwich A. Molecular chaperones and protein quality control. Cell . 2006;125(3):443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 18.Gastpar R., Gehrmann M., Bausero M. A., et al. Heat shock protein 70 surface-positive tumor exosomes stimulate migratory and cytolytic activity of natural killer cells. Cancer Research . 2005;65(12):5238–5247. doi: 10.1158/0008-5472.CAN-04-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magrané J., Smith R. C., Walsh K., Querfurth H. W. Heat shock protein 70 participates in the neuroprotective response to intracellularly expressed beta-amyloid in neurons. The Journal of Neuroscience . 2004;24(7):1700–1706. doi: 10.1523/JNEUROSCI.4330-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kakimura J., Kitamura Y., Takata K., et al. Microglial activation and amyloid-β clearance induced by exogenous heat-shock proteins. The FASEB Journal . 2002;16(6):601–603. doi: 10.1096/fj.01-0530fje. [DOI] [PubMed] [Google Scholar]
- 21.Hoshino T., Murao N., Namba T., et al. Suppression of Alzheimer’s disease-related phenotypes by expression of heat shock protein 70 in mice. The Journal of Neuroscience . 2011;31(14):5225–5234. doi: 10.1523/JNEUROSCI.5478-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dou F., Netzer W. J., Tanemura K., et al. Chaperones increase association of tau protein with microtubules. Proceedings of the National Academy of Sciences of the United States of America . 2003;100(2):721–726. doi: 10.1073/pnas.242720499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chi L. M., Wang X., Nan G. X. In silico analyses for molecular genetic mechanism and candidate genes in patients with Alzheimer’s disease. Acta Neurologica Belgica . 2016;116(4):543–547. doi: 10.1007/s13760-016-0613-6. [DOI] [PubMed] [Google Scholar]
- 24.Petyuk V. A., Chang R., Ramirez-Restrepo M., et al. The human brainome: network analysis identifies HSPA2 as a novel Alzheimer’s disease target. Brain . 2018;141(9):2721–2739. doi: 10.1093/brain/awy215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhattacharya S., Dunn P., Thomas C. G., et al. ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Scientific Data . 2018;5(1, article 180015) doi: 10.1038/sdata.2018.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pathan M., Keerthikumar S., Ang C. S., et al. FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics . 2015;15(15):2597–2601. doi: 10.1002/pmic.201400515. [DOI] [PubMed] [Google Scholar]
- 27.Liu Z., Li H., Pan S. Discovery and validation of key biomarkers based on immune infiltrates in Alzheimer’s disease. Frontiers in Genetics . 2021;12, article 658323 doi: 10.3389/fgene.2021.658323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang W. S., Dunckley T., Beach T. G., et al. Gene expression profiles in anatomically and functionally distinct regions of the normal aged human brain. Physiological Genomics . 2007;28(3):311–322. doi: 10.1152/physiolgenomics.00208.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piras I. S., Krate J., Delvaux E., et al. Transcriptome changes in the Alzheimer’s disease middle temporal gyrus: importance of RNA metabolism and mitochondria-associated membrane genes. Journal of Alzheimer's Disease . 2019;70(3):691–713. doi: 10.3233/JAD-181113. [DOI] [PubMed] [Google Scholar]
- 30.McKay E. C., Beck J. S., Khoo S. K., et al. Peri-infarct upregulation of the oxytocin receptor in vascular dementia. Journal of Neuropathology and Experimental Neurology . 2019;78(5):436–452. doi: 10.1093/jnen/nlz023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marttinen M., Paananen J., Neme A., et al. A multiomic approach to characterize the temporal sequence in Alzheimer’s disease-related pathology. Neurobiology of Disease . 2019;124:454–468. doi: 10.1016/j.nbd.2018.12.009. [DOI] [PubMed] [Google Scholar]
- 32.Zhang S., Hu C., Lu R., et al. Diagnostic accuracy of bronchodilator response for asthma in a population of South China. Advances in Therapy . 2018;35(10):1578–1584. doi: 10.1007/s12325-018-0783-0. [DOI] [PubMed] [Google Scholar]
- 33.Newman A. M., Liu C. L., Green M. R., et al. Robust enumeration of cell subsets from tissue expression profiles. Nature Methods . 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warde-Farley D., Donaldson S. L., Comes O., et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Research . 2010;38(suppl_2):W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Y., Zhou B., Pache L., et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nature Communications . 2019;10(1):p. 1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Q., Liu W., Zhang H. M., et al. hTFtarget: a comprehensive database for regulations of human transcription factors and their targets. Genomics, Proteomics & Bioinformatics . 2020;18(2):120–128. doi: 10.1016/j.gpb.2019.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bardou P., Mariette J., Escudié F., Djemiel C., Klopp C. jvenn: an interactive Venn diagram viewer. BMC Bioinformatics . 2014;15(1):p. 293. doi: 10.1186/1471-2105-15-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research . 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agarwal V., Bell G. W., Nam J. W., Bartel D. P. Predicting effective microRNA target sites in mammalian mRNAs. Elife . 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y., Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Research . 2020;48(D1):D127–d131. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romaniuk D., Smagur J., Cisek A., Antonik J., Romaniuk W. Does preservative content affect drug permeability? The permeability analysis in vitro of 3 ophthalmic Latanoprost formulations in a human epithelial cell culture model. Klinika Oczna . 2016;118(2):127–132. [PubMed] [Google Scholar]
- 42.Kim J. H. Genetics of Alzheimer’s disease. Dement Neurocogn Disord . 2018;17(4):131–136. doi: 10.12779/dnd.2018.17.4.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen J. X., Yan S. S. Role of mitochondrial Amyloid-β in Alzheimer’s disease. Journal of Alzheimer's Disease . 2010;20(Suppl 2):S569–S578. doi: 10.3233/JAD-2010-100357. [DOI] [PubMed] [Google Scholar]
- 44.Muraoka S., Jedrychowski M. P., Yanamandra K., Ikezu S., Gygi S. P., Ikezu T. Proteomic profiling of extracellular vesicles derived from cerebrospinal fluid of Alzheimer’s disease patients: a pilot study. Cells . 2020;9(9):p. 1959. doi: 10.3390/cells9091959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silva P. N., Furuya T. K., Braga I. L., et al. Analysis of HSPA8 and HSPA9 mRNA expression and promoter methylation in the brain and blood of Alzheimer’s disease patients. Journal of Alzheimer's Disease . 2014;38(1):165–170. doi: 10.3233/JAD-130428. [DOI] [PubMed] [Google Scholar]
- 46.Nong W., Huang F., Mao F., Lao D., Gong Z., Huang W. DCAF12 and HSPA1A may serve as potential diagnostic biomarkers for myasthenia gravis. BioMed Research International . 2022;2022:15. doi: 10.1155/2022/8587273.8587273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y., Cai Y., Yu H., Li H. iTRAQ-based quantitative proteomic analysis identified HSC71 as a novel serum biomarker for renal cell carcinoma. BioMed Research International . 2015;2015:6. doi: 10.1155/2015/802153.802153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rogaeva E. The solved and unsolved mysteries of the genetics of early-onset Alzheimer’s disease. Neuromolecular Medicine . 2002;2(1):01–10. doi: 10.1385/NMM:2:1:01. [DOI] [PubMed] [Google Scholar]
- 49.Guerreiro R. J., Gustafson D. R., Hardy J. The genetic architecture of Alzheimer’s disease: beyond APP, PSENs and APOE. Neurobiology of Aging . 2012;33(3):437–456. doi: 10.1016/j.neurobiolaging.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corder E. H., Saunders A. M., Strittmatter W. J., et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science . 1993;261(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 51.Gerber H., Mosser S., Boury-Jamot B., et al. The APMAP interactome reveals new modulators of APP processing and beta-amyloid production that are altered in Alzheimer’s disease. Acta Neuropathologica Communications . 2019;7(1):p. 13. doi: 10.1186/s40478-019-0660-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kitzlerová E., Fišar Z., Lelková P., et al. Interactions among polymorphisms of susceptibility loci for Alzheimer’s disease or depressive disorder. Medical Science Monitor . 2018;24:2599–2619. doi: 10.12659/MSM.907202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chonpathompikunlert P., Wattanathorn J., Muchimapura S. Piperine, the main alkaloid of Thai black pepper, protects against neurodegeneration and cognitive impairment in animal model of cognitive deficit like condition of Alzheimer’s disease. Food and Chemical Toxicology . 2010;48(3):798–802. doi: 10.1016/j.fct.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 54.Hardy J., Selkoe D. J. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science . 2002;297(5580):353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 55.Jo D. G., Arumugam T. V., Woo H. N., et al. Evidence that γ-secretase mediates oxidative stress-induced β-secretase expression in Alzheimer’s disease. Neurobiology of Aging . 2010;31(6):917–925. doi: 10.1016/j.neurobiolaging.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mehta D., Jackson R., Paul G., Shi J., Sabbagh M. Why do trials for Alzheimer’s disease drugs keep failing? A discontinued drug perspective for 2010-2015. Expert Opinion on Investigational Drugs . 2017;26(6):735–739. doi: 10.1080/13543784.2017.1323868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jevtic S., Sengar A. S., Salter M. W., McLaurin J. A. The role of the immune system in Alzheimer disease: etiology and treatment. Ageing Research Reviews . 2017;40:84–94. doi: 10.1016/j.arr.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 58.Forsman H., Kalderén C., Nordin A., Nordling E., Jensen A. J., Dahlgren C. Stable formyl peptide receptor agonists that activate the neutrophil NADPH- oxidase identified through screening of a compound library. Biochemical Pharmacology . 2011;81(3):402–411. doi: 10.1016/j.bcp.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 59.Stalder M., Deller T., Staufenbiel M., Jucker M. 3D-reconstruction of microglia and amyloid in APP23 transgenic mice: no evidence of intracellular amyloid. Neurobiology of Aging . 2001;22(3):427–434. doi: 10.1016/S0197-4580(01)00209-3. [DOI] [PubMed] [Google Scholar]
- 60.Bartels T., De Schepper S., Hong S. Microglia modulate neurodegeneration in Alzheimer’s and Parkinson’s diseases. Science . 2020;370(6512):66–69. doi: 10.1126/science.abb8587. [DOI] [PubMed] [Google Scholar]
- 61.Fu A. K., Hung K. W., Yuen M. Y., et al. IL-33 ameliorates Alzheimer’s disease-like pathology and cognitive decline. Proceedings of the National Academy of Sciences of the United States of America . 2016;113(19):E2705–E2713. doi: 10.1073/pnas.1604032113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olmos-Alonso A., Schetters S. T., Sri S., et al. Pharmacological targeting of CSF1R inhibits microglial proliferation and prevents the progression of Alzheimer’s-like pathology. Brain . 2016;139(3):891–907. doi: 10.1093/brain/awv379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poli A., Kmiecik J., Domingues O., et al. NK cells in central nervous system disorders. Journal of Immunology . 2013;190(11):5355–5362. doi: 10.4049/jimmunol.1203401. [DOI] [PubMed] [Google Scholar]
- 64.Richartz-Salzburger E., Batra A., Stransky E. Altered lymphocyte distribution in Alzheimer’s disease. Journal of Psychiatric Research . 2007;41(1-2):174–178. doi: 10.1016/j.jpsychires.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 65.Maté I., Cruces J., Giménez-Llort L., de la Fuente M. Function and redox state of peritoneal leukocytes as preclinical and prodromic markers in a longitudinal study of triple-transgenic mice for Alzheimer’s disease. Journal of Alzheimer's Disease . 2015;43(1):213–226. doi: 10.3233/JAD-140861. [DOI] [PubMed] [Google Scholar]
- 66.Braak H., Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathologica . 1991;82(4):239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 67.Tseng B. P., Green K. N., Chan J. L., Blurton-Jones M., LaFerla F. M. Aβ inhibits the proteasome and enhances amyloid and tau accumulation. Neurobiology of Aging . 2008;29(11):1607–1618. doi: 10.1016/j.neurobiolaging.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morawe T., Hiebel C., Kern A., Behl C. Protein homeostasis, aging and Alzheimer’s disease. Molecular Neurobiology . 2012;46(1):41–54. doi: 10.1007/s12035-012-8246-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brunk U. T., Jones C. B., Sohal R. S. A novel hypothesis of lipofuscinogenesis and cellular aging based on interactions between oxidative stress and autophagocytosis. Mutation Research . 1992;275(3-6):395–403. doi: 10.1016/0921-8734(92)90042-N. [DOI] [PubMed] [Google Scholar]
- 70.Pająk B., Kania E., Orzechowski A. Killing me softly: connotations to unfolded protein response and oxidative stress in Alzheimer’s disease. Oxidative Medicine and Cellular Longevity . 2016;2016:17. doi: 10.1155/2016/1805304.1805304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Plácido A. I., Pereira C. M., Duarte A. I., et al. The role of endoplasmic reticulum in amyloid precursor protein processing and trafficking: implications for Alzheimer’s disease. Biochimica et Biophysica Acta . 2014;1842(9):1444–1453. doi: 10.1016/j.bbadis.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 72.Lee H. P., Kudo W., Zhu X., Smith M. A., Lee H. G. Early induction of c-Myc is associated with neuronal cell death. Neuroscience Letters . 2011;505(2):124–127. doi: 10.1016/j.neulet.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ferrer I., Blanco R., Carmona M., Puig B. Phosphorylated c-MYC expression in Alzheimer disease, Pick’s disease, progressive supranuclear palsy and corticobasal degeneration. Neuropathology and Applied Neurobiology . 2001;27(5):343–351. doi: 10.1046/j.1365-2990.2001.00348.x. [DOI] [PubMed] [Google Scholar]
- 74.Lanke V., Moolamalla S. T. R., Roy D., Vinod P. K. Integrative analysis of hippocampus gene expression profiles identifies network alterations in aging and Alzheimer’s disease. Frontiers in Aging Neuroscience . 2018;10:p. 153. doi: 10.3389/fnagi.2018.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Capelli E., Torrisi F., Venturini L., et al. Low-frequency pulsed electromagnetic field is able to modulate miRNAs in an experimental cell model of Alzheimer’s disease. Journal of Healthcare Engineering . 2017;2017:10. doi: 10.1155/2017/2530270.2530270 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig.1 Expression of three immune-related HSP70 family members in GSE132903. The blue box indicates the control group, and the orange box indicates the AD group. Data were analyzed by Student's T-test and expressed as the Mean ± SD. ∗P < 0.05; ∗∗∗P < 0.001. Supplementary Table 1. Immune molecules in the Immport database. Supplementary Table 2. Common TFs of the 3 hub genes from hTFtarget by jvenn. Supplementary Table 3. The overlapped miRNAs of HSPA1A/HSPA2/HSPA8 predicted by TargetScan and miRDB
Data Availability Statement
The authors declare that all relevant data of this study are available within the article or from the corresponding author on reasonable request.


