Abstract
The involvement of LexA in induction of RecA was investigated in Deinococcus radiodurans. As in the wild-type strain, an increase in RecA protein synthesis following γ irradiation was detected in a lexA disruptant, indicating that LexA is not involved in the induction of RecA in D. radiodurans.
Deinococcus radiodurans is characterized by its extraordinary radiation resistance phenotype, which is considered to be due to a highly proficient DNA repair capacity (3, 25, 34). The most striking feature of D. radiodurans is that it can mend over 100 double-strand breaks (DSBs) of genomic DNA during postirradiation incubation (2, 18). As the rejoining of DSBs can be prevented by adding chloramphenicol to the incubation mixture, proteins induced by irradiation are necessary for the rejoining of DNA breakages (18). Several DNA damage-inducible proteins that may be required for DNA repair have been detected in the cell extract of D. radiodurans by two-dimensional polyacrylamide gel electrophoresis (PAGE) (12, 35). These observations suggest that D. radiodurans possesses a DNA damage response mechanism. However, little is known about the molecular basis for the control of the inducible proteins.
In Escherichia coli, the inducible DNA repair system (the SOS system) is regulated by two key proteins; RecA and LexA (8, 38). E. coli RecA is activated by DNA damage to mediate the proteolytic cleavage of the E. coli LexA repressor, resulting in derepression of the SOS regulon. The SOS response in Bacillus subtilis progresses in a similar manner, with B. subtilis RecA having an identical role in controlling the SOS regulon together with a cellular repressor protein that is functionally homologous to the E. coli LexA repressor (42). The B. subtilis repressor (termed DinR) binds the promoter regions of several din genes and B. subtilis recA (20, 23, 40, 41) and undergoes autodigestion under alkaline conditions and RecA-mediated cleavage under more physiological conditions (23, 41). It has also been shown that the intracellular level of intact DinR is significantly reduced following DNA damage (23). Thus, the basic mechanism of the SOS response seems to be conserved between E. coli and B. subtilis. Deinococcus species form a coherent phylogenetic cluster related to the Thermus-Meiothermus lineage (30), indicating that the Deinococcus lineage is distinct from the lineages of both proteobacteria and gram-positive bacteria. Although SOS-like processes have been documented in a wide variety of eubacterial species (24, 32), the involvement of RecA and LexA in the SOS response is poorly understood in Deinococcus and closely related bacterial species.
As expression of the deinococcal recA gene is enhanced after γ irradiation (4), the recA gene seems to be a member of a DNA damage response regulon in D. radiodurans. In the present study, D. radiodurans LexA was purified from E. coli cells and its ability to cleave itself was examined. The changes in intracellular levels of the LexA and RecA proteins following γ irradiation were also investigated by using lexA and recA disruptant strains to gain insight into the DNA damage response mechanism in D. radiodurans. Our results indicated that deinococcal LexA undergoes RecA-mediated cleavage but is not involved in the regulation of deinococcal RecA.
Expression plasmid construction.
pDC144 is a cosmid clone in a genomic library of D. radiodurans strain KD8301 (Table 1). pZA8 (16) contains D. radiodurans lexA in a 6,005-bp SalI-SacI fragment from pDC144. The nucleotide sequence of this region was confirmed to be perfectly consistent with the corresponding region of D. radiodurans (39). To isolate the lexA coding region, PCR was carried out by using pZA8 DNA with the specific oligonucleotides 5′-GGCAAACTGCGCGCATATGCCGCCTGAACTG-3′ and 5′-GTCGGGATCCTACTCGGTCACGCGGTGGCTCACG-3′, containing restriction sites (NdeI and BamHI, which are underlined). PCR products were then digested with NdeI and BamHI to adapt the termini for the in-frame insertion of lexA into the NdeI-BamHI sites in the pET3a vector. The resultant expression plasmid was designated pET3lexAwt (Table 1). The DNA sequence of the expression plasmid was checked to confirm the lack of introduction of errors by PCR.
TABLE 1.
Strains, cosmids, and plasmids used in this study
| Designation | Relevant description | Source or reference |
|---|---|---|
| D. radiodurans | ||
| KR1 | Wild type (γ rayr) | 17 |
| KD8301 | KR1 Ade− DNase− Smr (γ rayr) | 28 |
| XE1 | KD8301 lexA166::cat; generated by insertional mutagenesis with pXKE6 | This work |
| RN201 | KR1recA229::cat; generated by insertional mutagenesis with pKSCR3 | This work |
| E. coli | ||
| JM109 | Host of pUC- and pET3a-based plasmids | Takara |
| BL21 (DE3) | Host for gene expression | Novagen |
| Cosmids | ||
| SuperCos 1 | Cosmid vector; 7.6 kb; Apr Kmr | Stratagene |
| pDC144 | SuperCos 1 with 34.4-kb D. radiodurans DNA; lexA+ | 28 |
| Plasmids | ||
| pUC18 | E. coli cloning vector; 2.7 kb; Apr | Takara |
| pUC19 | E. coli cloning vector; 2.7 kb; Apr | Takara |
| pET3a | E. coli expression vector; 4.6 kb; Apr | Novagen |
| pLysS | E. coli plasmid containing T7 lysozyme gene; 4.9 kb; Cmr | Novagen |
| pKatCAT | pUC19 containing cat and D. radiodurans katAp; 3.6 kb; Apr Cmr | 9 |
| pZA8 | pUC19 with 6,005-bp SalI-SacI fragment from pDC144 | 16 |
| pET3lexAwt | pET3a NdeI-BamHI::635-bp PCR product from pZA8 | This work |
| pXKE6 | pZA8 EagI::915-bp HincII fragment from pKatCAT; lexA166::cat | This work |
| pKS1 | pUC18 with 4,402-bp ApaLI-SphI fragment containing recA | 29 |
| pKSCR3 | pKS1 BglII::915-bp HincII fragment from pKatCAT; recA229::cat | This work |
Protein purification.
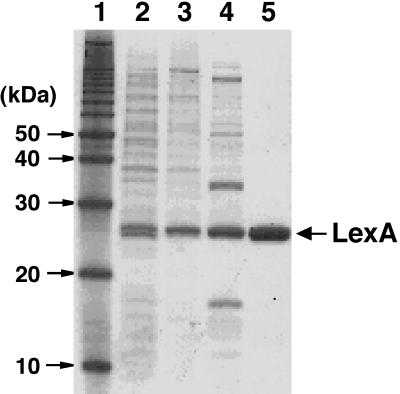
D. radiodurans LexA was induced by isopropyl-β-d-thiogalactopyranoside (IPTG) in E. coli strain BL21(DE3) carrying pLysS and pET3lexAwt. Cells were harvested, washed, and resuspended in a buffer containing 20 mM sodium phosphate (pH 7.4), 1 mM phenylmethylsulfonyl fluoride, and 0.1% (wt/vol) protease inhibitor cocktail. The suspension was sonicated for 10 min, and debris was removed by centrifugation. Ammonium sulfate was added to the supernatant to give 30% saturation. The suspension was stirred for 1 h and then centrifuged for 30 min. The pellets were resuspended in a buffer containing 20 mM sodium phosphate (pH 7.4) and 0.1 mM EDTA, and the suspension was dialyzed for 18 h. The protein was further purified to apparent homogeneity by column chromatography on HiTrap Heparin HP and Resource S (Fig. 1). The N-terminal amino acid sequence of the purified protein was found to be Pro-Pro-Glu-Leu-Thr-Pro-Thr-Arg-Arg-Ser-Ile-Leu-Gln-Ala-. This was completely consistent with the sequence from residue 2 to residue 15 of the primary structure predicted from the DNA sequence data. Thus, the purified protein was confirmed to be D. radiodurans LexA.
FIG. 1.
Purification of D. radiodurans LexA protein. Samples were subjected to sodium dodecyl sulfate–15% PAGE and stained with Coomassie brilliant blue. Lanes: 1, 10-kDa protein ladder (Invitrogen); 2, total cellular proteins from E. coli BL21(DE3)/pLysS/pET3lexAwt induced by IPTG; 3, resuspension from 30% ammonium sulfate precipitation; 4, pooled LexA fractions from a HiTrap Heparin HP column; 5, pooled LexA fractions from a Resource S column. The position of the 25-kDa band of LexA is indicated on the right.
Autodigestion and RecA-mediated cleavage.
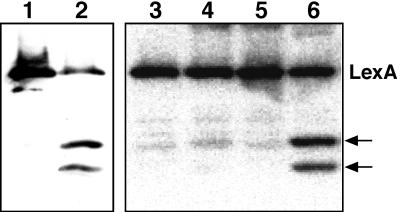
First, whether the purified protein has autodigestion and RecA-mediated cleavage activity was investigated. For this purpose, antiserum raised against the purified D. radiodurans LexA protein was generated. The autodigestion reaction was assayed by incubating 0.4 μM LexA in 50 mM Tris-HCl (pH 10) at 37°C and monitored by Western analysis. As shown in Fig. 2, LexA was autodigested to yield two breakdown products. For RecA-mediated cleavage, reactions were carried out in a buffer consisting of 20 mM Tris-HCl (pH 7.4) and 10 mM MgCl2 with 6 μM oligonucleotide (35-mer) and 1 mM adenosine-5′-O-[γ-thio]triphosphate (ATPγS). LexA (0.4 μM) was incubated with D. radiodurans RecA (4.2 μM) at 37°C for 1 h and sampled for Western analysis. The purification of the RecA protein will be reported elsewhere. LexA was cleaved by incubation with RecA to yield two breakdown products the sizes of which were identical to those observed in autodigestion. When the RecA, oligonucleotide, or ATPγS was omitted from the reaction mixture, no breakdown product was observed (Fig. 2). From these results, we concluded that D. radiodurans LexA maintains proteolytic activity and that D. radiodurans RecA can promote the proteolytic activity of LexA.
FIG. 2.
Autodigestion and RecA-mediated cleavage of purified LexA protein visualized by Western analysis with D. radiodurans LexA antiserum (diluted 1:10,000). LexA (0.4 μM) was incubated in 50 mM Tris-HCl (pH 10) for 0 h (lane 1) and 8 h (lane 2). For RecA-mediated cleavage, LexA was incubated with D. radiodurans RecA under the conditions described in the text. Lanes: 3, RecA omitted; 4, oligonucleotide omitted; 5, ATPγS omitted; 6, complete reaction mixture. The arrows on the right indicate the positions of breakdown products of LexA.
Construction of gene disruptant strains.
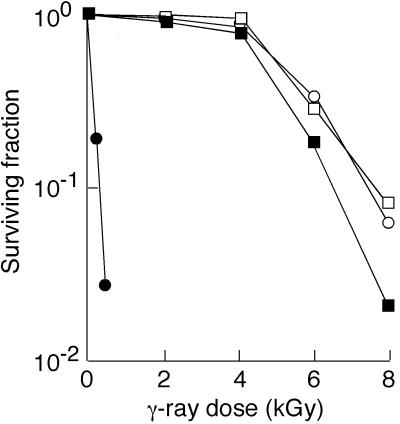
To test our assumption mentioned above, we investigated the in vivo interaction of LexA and RecA by detecting changes in intracellular protein levels following γ irradiation. For this purpose, we generated a lexA disruptant strain and a recA disruptant strain by the direct insertional mutagenesis technique (9). The disruptant strains were isolated on TGY plates (28) containing chloramphenicol at 3 μg/ml and designated XE1 (carrying lexA166::cat) and RN201 (carrying recA229::cat), respectively. Disruption of the genes was confirmed by amplifying the targeted allele by PCR. The DNA damage sensitivity phenotype of strain RN201 was confirmed by measuring cell survival following γ irradiation (Fig. 3). RN201 exhibited extreme γ ray sensitivity, as observed in the recA disruptant strain 1R1A constructed previously (10) and mutant strain rec30 (10, 26, 35) carrying a recA670 mutation (29). In addition, strain RN201 had a slow growth rate compared with that of its parental strain. On the other hand, the growth rate of strain XE1 carrying the lexA166::cat mutation was almost the same as that of the parental strain. Although XE1 cells showed a slightly higher rate of cell death than the parental strain at high doses (6 and 8 kGy) of γ rays, the disruption of lexA did not severely affect γ ray resistance (Fig. 3).
FIG. 3.
Sensitivity of lexA and recA disruptant strains to γ rays. Cells grown to early stationary phase were resuspended in 10 mM sodium phosphate buffer (pH 7), challenged with 60Co γ irradiation, spread on TGY plates, and incubated at 30°C. After 3 days, surviving colonies were counted. Each point represents the average result of three independent experiments. Symbols: open squares, strain KD8301; filled squares, strain XE1 (lexA166::cat); open circles, strain KR1; filled circles, strain RN201 (recA229::cat).
Changes in intracellular LexA and RecA levels following irradiation.
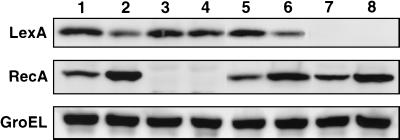
The changes in LexA and RecA levels following γ irradiation were compared among the disruptants and their parental strains. Early stationary phase cells were resuspended in 10 mM sodium phosphate buffer (pH 7) and divided into two fractions. One fraction was irradiated at a dose of 2 kGy, and the other fraction was not irradiated. The cells were then incubated in fresh TGY broth for 2 h at 30°C with agitation. The protein extracts were subjected to Western analysis with D. radiodurans LexA antiserum (diluted 1:10,000) and E. coli RecA antiserum (1:500). The E. coli RecA antiserum was raised against purified E. coli RecA protein (Promega). As a control, D. radiodurans GroEL (22) was detected by using E. coli GroEL antiserum (diluted 1:2,000) (StressGen Biotechnologies Corp.) (Fig. 4).
FIG. 4.
Changes in intracellular LexA, RecA, and GroEL levels following irradiation. Each sample contained 10 μg of protein. Lanes: 1 and 2, strain KR1; 3 and 4, strain RN201 (recA229::cat); 5 and 6, strain KD8301; 7 and 8, strain XE1 (lexA166::cat). Odd- and even-numbered lanes contained nonirradiated samples and those irradiated with 2 kGy, respectively.
In wild-type strain KR1, the level of LexA was decreased (2.7-fold) and the level of RecA was, in contrast, increased after γ irradiation (2.5-fold) (Fig. 4, lanes 1 and 2). These results suggested that RecA is activated during postirradiation incubation and promotes the proteolytic cleavage of LexA. In strain RN201, the RecA signal completely disappeared (Fig. 4, lanes 3 and 4), confirming the disruption of recA. Importantly, γ irradiation did not affect the level of LexA in RN201. These results supported our observation of in vitro RecA-mediated LexA cleavage and further suggested that RecA is the sole protein required for LexA cleavage. As in strain KR1, a decrease in LexA (2.5-fold) and an increase in RecA (2.5-fold) were observed in strain KD8301 after irradiation (Fig. 4, lanes 5 and 6). In strain XE1, a derivative of strain KD8301, the LexA signal disappeared because of disruption of lexA (Fig. 4, lanes 7 and 8). If RecA represses the expression of recA, constitutive production of LexA at an elevated level can be seen in unirradiated lexA disruptant cells. However, this was not the case in strain XE1. The level of RecA in unirradiated XE1 cells was comparable to those in unirradiated KR1 and KD8301 cells, and RecA induction following irradiation (2.0-fold) was observed in XE1 as in KR1 and KD8301. The level of GroEL was constant irrespective of irradiation in all of the strains tested. Thus, the results of our experiments did not support the involvement of LexA in the induction of RecA in D. radiodurans.
Discussion.
The results obtained in this study indicate that D. radiodurans LexA undergoes RecA-mediated cleavage (Fig. 2) and RecA is the sole protein responsible for cleavage of LexA in vivo (Fig. 4). E. coli RecA mediates the proteolytic cleavage of the bond between Ala-84 and Gly-85 of LexA (13). Hydrolysis of the LexA Ala-Gly bond proceeds similarly to that of serine proteases, with Ser-119 acting as a nucleophile and Lys-156 acting as an activator (33). Alignment of the amino acid sequences of E. coli LexA, B. subtilis DinR, and D. radiodurans LexA (Fig. 5) revealed that the amino acid residues involved in the cleavage reaction are also conserved in D. radiodurans LexA (Ala-83, Gly-84, Ser-119, and Lys-158). Therefore, we assumed that the two breakdown products observed in the in vitro cleavage assays (Fig. 2) were the N-terminal and C-terminal fragments of LexA cleaved between Ala-83 and Gly-84.
FIG. 5.
Multiple amino acid sequence alignment of E. coli RecA (14), B. subtilis DinR (31), and D. radiodurans LexA. Multiple alignment was achieved with the CLUSTAL W program (37). Dashes indicate gaps in the alignment. Numbers on the right are the coordinates of the proteins. Asterisks indicate identical residues. The conserved Ala and Gly residues in the cleavage site and the Ser and Lys residues required for cleavage are indicated by filled triangles. The locations of three α helices (H1, H2, and H3) found in E. coli LexA are indicated according to the description of Fogh et al. (7).
The N-terminal domain of E. coli LexA is involved in DNA binding (15), and the LexA DNA binding domain contains three α helices, of which helices 2 and 3 form a variant helix-turn-helix DNA binding motif (7). The N-terminal region of the D. radiodurans LexA sequence, however, showed very limited similarity to analogous regions in E. coli LexA and B. subtilis DinR (Fig. 5). If the N-terminal region of D. radiodurans LexA serves as a DNA binding domain, D. radiodurans possesses a distinct LexA binding motif that is different from the E. coli SOS box (38) and the B. subtilis DinR box (40). For the purification of D. radiodurans LexA, we found a heparin column to be very effective (Fig. 1), suggesting that the LexA protein retains DNA binding ability. In the preliminary gel mobility shift assay, we also found that the purified LexA protein could bind to the upstream region of its own gene (data not shown). A detailed analysis of the D. radiodurans lexA operator is ongoing in our laboratory.
Under our experimental conditions (irradiation with 2 kGy, followed by 2 h of incubation), the RecA induction ratio was 2.0- to 2.5-fold (Fig. 4). This induction ratio was much smaller than that observed in a previous study (50- to 100-fold; irradiation with 5 kGy, followed by 2 h of incubation) (4). It has been shown that, after irradiation at 3 kGy, a cell's recovery from DSBs is complete within 3 h (2). Therefore, one explanation for the induction ratio difference is the different γ ray doses used in the two experiments. It is conceivable that most DSB repair is completed under our experimental conditions; thereby, the induction ratio was smaller than that in the previous study. However, the 50- to 100-fold RecA induction ratio seems to be an overestimate because such radiation-induced protein was not detected by two-dimensional PAGE analysis in which cells were irradiated at 6 kGy and incubated for 2 h (35).
It has been shown that several eubacterial species lack a lexA gene (1, 6). A lack of inducibility of RecA has also been demonstrated in some genera (24). Thus, while the recA gene is clearly conserved in a wide variety of eubacterial species, the control mechanism of its expression is not. Our results indicated that the recA gene is part of a DNA damage response regulon in D. radiodurans. However, our findings did not support the suggestion that D. radiodurans LexA is involved in the induction of RecA. This, in turn, led us to speculate that D. radiodurans has an alternative DNA damage response mechanism with which to control recA expression. In E. coli, the sulA gene, whose products inhibit cell division, is under the direct control of E. coli LexA (38). Consequently, E. coli lexA (Def) mutants are viable only if they contain an additional mutation on the sulA gene. In addition, it has been shown that B. subtilis lexA (Def) mutants exhibit a strong filamentation phenotype, accompanied by significant loss of viability (11). On the other hand, the D. radiodurans lexA disruptant generated in this study was viable. This different behavior has been reported previously for lexA disruptants of Xanthomonas campestris (43) and Rhodobacter sphaeroides (36) in which LexA functions as a repressor of recA expression. The D. radiodurans genome does not encode a homolog of the E. coli sulA gene (39). However, it has been shown that the DNA damage-induced delay in D. radiodurans chromosomal DNA replication is dose dependent and that the length of the delay always exceeds the time required for repair of the DNA damage that caused the inhibition (5, 19, 27). Based on these observations, it has been proposed that there is a regulatory mechanism in D. radiodurans that controls chromosome replication and, as a consequence, controls cell division (2, 3). We suggest that D. radiodurans LexA is probably not involved in such a regulatory mechanism because of the behavior of LexA disruptant cells.
Interestingly, it has recently been shown that D. radiodurans encodes a second but diverged copy of LexA (DRA0074) that retains the potential DNA binding domain and the autocleavage domain (21). It would be interesting to determine whether DRA0074 is involved in the control of the DNA damage response in D. radiodurans. Recently, we succeeded in overproducing DRA0074 in E. coli. Characterization of this recombinant protein will provide useful information about control of the DNA damage response.
Nucleotide sequence accession number.
The nucleotide sequence reported here (the Dienococcus radiodurans gene for aldehyde dehydrogenase, succinic semialdehyde dehydrogenase, partial and complete cds) has been deposited in the DDBJ/EMBL/GenBank databases under accession no. AB003475.
Acknowledgments
We thank Xiang-Rong Kong for determining preliminary conditions for the purification of LexA. We are also grateful to Rieko Nakano for constructing strain RN201.
This work was performed as part of an Atomic Energy Crossover Project of the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan, and supported by a Grant-in-Aid for Scientific Research from MEXT.
REFERENCES
- 1.Aravind L, Walker D R, Koonin E V. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Battista J R. Against all odds: the survival strategies of Deinococcus radiodurans. Annu Rev Microbiol. 1997;51:203–224. doi: 10.1146/annurev.micro.51.1.203. [DOI] [PubMed] [Google Scholar]
- 3.Battista J R. DNA repair in Deinococcus radiodurans. In: Nickoloff J A, Hoekstra M F, editors. DNA damage and repair. I. DNA repair in prokaryotes and lower eukaryotes. Totowa, N.J: Humana Press; 1998. pp. 287–303. [Google Scholar]
- 4.Carroll J D, Daly M J, Minton K W. Expression of recA in Deinococcus radiodurans. J Bacteriol. 1996;178:130–135. doi: 10.1128/jb.178.1.130-135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dean C J, Feldschreiber P, Lett J T. Repair of X-ray damage to the deoxyribonucleic acid in Micrococcus radiodurans. Nature. 1966;209:49–52. doi: 10.1038/209049a0. [DOI] [PubMed] [Google Scholar]
- 6.Eisen J A, Hanawalt P C. A phylogenomic study of DNA repair genes, proteins, and processes. Mutat Res. 1999;435:171–213. doi: 10.1016/s0921-8777(99)00050-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fogh R H, Ottleben G, Rüterjans H, Schnarr M, Boelens R, Kaptein R. Solution structure of the LexA repressor DNA binding domain determined by 1H NMR spectroscopy. EMBO J. 1994;13:3936–3944. doi: 10.1002/j.1460-2075.1994.tb06709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C.: American Society for Microbiology; 1995. [Google Scholar]
- 9.Funayama T, Narumi I, Kikuchi M, Kitayama S, Watanabe H, Yamamoto K. Identification and disruption analysis of the recN gene in the extremely radioresistant bacterium Deinococcus radiodurans. Mutat Res. 1999;435:151–161. doi: 10.1016/s0921-8777(99)00044-0. [DOI] [PubMed] [Google Scholar]
- 10.Gutman P D, Carroll J D, Masters C I, Minton K W. Sequencing, targeted mutagenesis and expression of a recA gene required for the extreme radioresistance of Deinococcus radiodurans. Gene. 1994;141:31–37. doi: 10.1016/0378-1119(94)90124-4. [DOI] [PubMed] [Google Scholar]
- 11.Haijema B J, Sinderen D, Winterling K, Kooistra J, Venema G, Hamoen L W. Regulated expression of the dinR and recA genes during competence development and SOS induction in Bacillus subtilis. Mol Microbiol. 1996;22:75–85. doi: 10.1111/j.1365-2958.1996.tb02657.x. [DOI] [PubMed] [Google Scholar]
- 12.Hansen M T. Four proteins synthesized in response to deoxyribonucleic acid damage in Micrococcus radiodurans. J Bacteriol. 1980;141:81–86. doi: 10.1128/jb.141.1.81-86.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horii T, Ogawa T, Nakatani T, Hase T, Matsubara H, Ogawa H. Regulation of SOS functions: purification of E. coli LexA protein and determination of its specific site cleaved by the RecA protein. Cell. 1981;27:515–522. doi: 10.1016/0092-8674(81)90393-7. [DOI] [PubMed] [Google Scholar]
- 14.Horii T, Ogawa T, Ogawa H. Nucleotide sequence of the lexA gene of E. coli. Cell. 1981;23:689–697. doi: 10.1016/0092-8674(81)90432-3. [DOI] [PubMed] [Google Scholar]
- 15.Hurstel S, Granger-Schnarr M, Daune M, Schnarr M. In vitro binding of LexA repressor to DNA: evidence for the involvement of the amino-terminal domain. EMBO J. 1986;5:793–798. doi: 10.1002/j.1460-2075.1986.tb04283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kikuchi M, Narumi I, Kitayama S, Watanabe H, Yamamoto K. Genomic organization of the radioresistant bacterium Deinococcus radiodurans: physical map and evidence for multiple replicons. FEMS Microbiol Lett. 1999;174:151–157. [Google Scholar]
- 17.Kitayama S, Asaka S, Totsuka K. DNA double-strand breakage and removal of cross-links in Deinococcus radiodurans. J Bacteriol. 1983;155:1200–1207. doi: 10.1128/jb.155.3.1200-1207.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitayama S, Matsuyama A. Double-strand scissions in DNA of gamma-irradiated Micrococcus radiodurans and their repair during postirradiation incubation. Agric Biol Chem. 1971;35:644–652. [Google Scholar]
- 19.Lett J T, Feldschreiber P, Little J G, Steele K, Dean C J. The repair of X-ray damage to the deoxyribonucleic acid in Micrococcus radiodurans: a study of the excision process. Proc R Soc Lond Ser B. 1967;167:184–201. doi: 10.1098/rspb.1967.0023. [DOI] [PubMed] [Google Scholar]
- 20.Lovett C M, Jr, Cho K C, O'Gara T M. Purification of an SOS repressor from Bacillus subtilis. J Bacteriol. 1993;175:6842–6849. doi: 10.1128/jb.175.21.6842-6849.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makarova K S, Aravind L, Wolf Y I, Tatusov R L, Minton K W, Koonin E V, Daly M J. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol Mol Biol Rev. 2001;65:44–79. doi: 10.1128/MMBR.65.1.44-79.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meima R, Rothfuss H M, Gewin L, Lidstrom M E. Promoter cloning in the radioresistant bacterium Deinococcus radiodurans. J Bacteriol. 2001;183:3169–3175. doi: 10.1128/JB.183.10.3169-3175.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller M C, Resnick J B, Smith B T, Lovett C M., Jr The Bacillus subtilis dinR gene codes for the analogue of Escherichia coli LexA. Purification and characterization of the DinR protein. J Biol Chem. 1996;271:33502–33508. [PubMed] [Google Scholar]
- 24.Miller R V, Kokjohn T A. General microbiology of recA. Environmental and evolutionary significance. Annu Rev Microbiol. 1990;44:365–394. doi: 10.1146/annurev.mi.44.100190.002053. [DOI] [PubMed] [Google Scholar]
- 25.Moseley B E B. Photobiology and radiobiology of Micrococcus (Deinococcus) radiodurans. Photochem Photobiol Rev. 1983;7:223–275. [Google Scholar]
- 26.Moseley B E B, Copland H J R. Isolation and properties of a recombination-deficient mutant of Micrococcus radiodurans. J Bacteriol. 1975;121:422–428. doi: 10.1128/jb.121.2.422-428.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moseley B E B, Copland H J R. The rate of recombination repair and its relationship to the radiation-induced delay in DNA synthesis in Micrococcus radiodurans. J Gen Microbiol. 1976;93:251–258. doi: 10.1099/00221287-93-2-251. [DOI] [PubMed] [Google Scholar]
- 28.Narumi I, Cherdchu K, Kitayama S, Watanabe H. The Deinococcus radiodurans uvrA gene: identification of mutation sites of two mitomycin-sensitive strains and the first discovery of insertion sequence element from deinobacteria. Gene. 1997;198:115–126. doi: 10.1016/s0378-1119(97)00301-6. [DOI] [PubMed] [Google Scholar]
- 29.Narumi I, Satoh K, Kikuchi M, Funayama T, Kitayama S, Yanagisawa T, Watanabe H, Yamamoto K. Molecular analysis of the Deinococcus radiodurans recA locus and identification of a mutation site in a DNA repair-deficient mutant, rec30. Mutat Res. 1999;435:233–243. doi: 10.1016/s0921-8777(99)00048-8. [DOI] [PubMed] [Google Scholar]
- 30.Rainey F A, Nobre M F, Schumann P, Stackebrandt E, da Costa M S. Phylogenetic diversity of the deinococci as determined by 16S ribosomal DNA sequence comparison. Int J Syst Bacteriol. 1997;47:510–514. doi: 10.1099/00207713-47-2-510. [DOI] [PubMed] [Google Scholar]
- 31.Raymond-Denise A, Guillen N. Identification of dinR, a DNA damage-inducible regulator gene of Bacillus subtilis. J Bacteriol. 1991;173:7084–7091. doi: 10.1128/jb.173.22.7084-7091.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roca A I, Cox M M. The RecA protein: structure and function. Crit Rev Biochem Mol Biol. 1990;25:415–456. doi: 10.3109/10409239009090617. [DOI] [PubMed] [Google Scholar]
- 33.Slilaty S N, Little J W. Lysine-156 and serine-119 are required for LexA repressor cleavage: a possible mechanism. Proc Natl Acad Sci USA. 1987;84:3987–3991. doi: 10.1073/pnas.84.12.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith M D, Masters C I, Moseley B E B. Molecular biology of radiation-resistant bacteria. In: Herbert R A, Sharp R J, editors. Molecular biology and biotechnology of extremophiles. New York, N.Y: Chapman & Hall; 1992. pp. 258–280. [Google Scholar]
- 35.Tanaka A, Hirano H, Kikuchi M, Kitayama S, Watanabe H. Changes in cellular proteins of Deinococcus radiodurans following γ-irradiation. Radiat Environ Biophys. 1996;35:95–99. doi: 10.1007/BF02434031. [DOI] [PubMed] [Google Scholar]
- 36.Tapias A, Campoy S, Barbé J. Analysis of the expression of the Rhodobacter sphaeroides lexA gene. Mol Gen Genet. 2000;263:957–965. doi: 10.1007/pl00008696. [DOI] [PubMed] [Google Scholar]
- 37.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walker G C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984;48:60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White O, Eisen J A, Heidelberg J F, Hickey E K, Peterson J D, Dodson R J, Haft D H, Gwinn M L, Nelson W C, Richardson D L, Moffat K S, Qin H, Jiang L, Pamphile W, Crosby M, Shen M, Vamathevan J J, Lam P, McDonald L, Utterback T, Zalewski C, Makarova K S, Aravind L, Daly M J, Minton K W, Fleischmann R D, Ketchum K A, Nelson K E, Salzberg S, Smith H O, Venter J C, Fraser C M. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science. 1999;286:1571–1577. doi: 10.1126/science.286.5444.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winterling K W, Chafin D, Hayes J J, Sun J, Levine A S, Yasbin R E, Woodgate R. The Bacillus subtilis DinR binding site: redefinition of the consensus sequence. J Bacteriol. 1998;180:2201–2211. doi: 10.1128/jb.180.8.2201-2211.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winterling K W, Levine A S, Yasbin R E, Woodgate R. Characterization of DinR, the Bacillus subtilis SOS repressor. J Bacteriol. 1997;179:1698–1703. doi: 10.1128/jb.179.5.1698-1703.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wojciechowski M F, Peterson K R, Love P E. Regulation of the SOS response in Bacillus subtilis: evidence for a LexA repressor homolog. J Bacteriol. 1991;173:6489–6498. doi: 10.1128/jb.173.20.6489-6498.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang M-K, Wu P-I, Yang Y-C. Identification of a lexA gene in, and construction of a lexA mutant of, Xanthomonas campestris pv. citri. Curr Microbiol. 2000;40:233–238. doi: 10.1007/s002849910047. [DOI] [PubMed] [Google Scholar]