Abstract
Background
The aetiologic role of circulating proteins in the development of breast cancer subtypes is not clear. We aimed to examine the potential causal effects of circulating proteins on the risk of breast cancer by intrinsic-like subtypes within the Mendelian randomisation (MR) framework.
Methods
MR was performed using summary statistics from two sources: the INTERVAL protein quantitative trait loci (pQTL) Study (1890 circulating proteins and 3301 healthy individuals) and the Breast Cancer Association Consortium (BCAC; 106,278 invasive cases and 91,477 controls). The inverse-variance (IVW)-weighted method was used as the main analysis to evaluate the associations between genetically predicted proteins and the risk of five different intrinsic-like breast cancer subtypes and the weighted median MR method, the Egger regression, the MR-PRESSO, and the MRLocus method were performed as secondary analysis.
Results
We identified 98 unique proteins significantly associated with the risk of one or more subtypes (Benjamini–Hochberg false discovery rate < 0.05). Among them, 51 were potentially specific to luminal A-like subtype, 14 to luminal B/Her2-negative-like, 11 to triple negative, 3 to luminal B-like, and 2 to Her2-enriched-like breast cancer (ntotal = 81). Associations for three proteins (ICAM1, PLA2R1 and TXNDC12) showed evident heterogeneity across the subtypes. For example, higher levels of genetically predicted ICAM1 (per unit of increase) were associated with an increased risk of luminal B/HER2-negative-like cancer (OR = 1.06, 95% CI = 1.03–1.08, BH-FDR = 2.43 × 10−4) while inversely associated with triple-negative breast cancer with borderline significance (OR = 0.97, 95% CI = 0.95–0.99, BH-FDR = 0.065, Pheterogeneity < 0.005).
Conclusions
Our study found potential causal associations with the risk of subtypes of breast cancer for 98 proteins. Associations of ICAM1, PLA2R1 and TXNDC12 varied substantially across the subtypes. The identified proteins may partly explain the heterogeneity in the aetiology of distinct subtypes of breast cancer and facilitate the personalised risk assessment of the malignancy.
Subject terms: Risk factors, Breast cancer
Introduction
Breast cancer is the leading cause of global cancer incidence in women with an estimated 2.3 million new cases being diagnosed worldwide in 2020 [1]. Although the aetiology of breast cancer is not fully understood, it is widely recognised that breast cancer is a heterogeneous disease with distinct histological and molecular characteristics [2]. However, the majority of studies did not account for this heterogeneity when investigating the aetiology or risk factors for breast cancer.
Circulating proteins have been linked to breast cancer risk. For example, a large pooling study showed that levels of circulating insulin-like growth factor-1 (IGF-1) were associated with a 30% increased risk of breast cancer (highest versus the lowest fifth of IGF-1 levels) [3], suggesting the insulin/IGF-1 axis plays in a critical role in breast carcinogenesis [4]. We previously conducted a genetic instrumental analysis to search for novel circulating protein biomarkers for breast cancer risk [5]. A panel of 56 proteins was found significant, many of which are involved in the oestrogen receptor (ER) signalling and insulin resistance-related pathways. However, in these studies, the associations were not examined according to specific subtypes. Thereby important findings might be missed, and it remains unclear whether the identified biomarkers are subtype-specific or shared by different subtypes.
In this study, we evaluated the associations between 1890 circulating proteins and breast cancer risk within five distinct intrinsic-like subtypes by conducting Mendelian randomisation (MR) analysis. MR was developed as an effective tool to evaluate the causal relationship between an exposure and outcome of interest when randomised clinical trials (RCT) are not feasible. Similar to RCT, it is designed to minimise the impact of confounding, reverse causation, and other biases, providing more definitive evidence for causal inference [6]. Through the analysis, we aimed to identify potential subtype-specific proteins and those shared by different subtypes of breast cancer, which may help explain the heterogeneity in the disease aetiology and ultimately facilitate an effective risk assessment of breast cancer.
Materials and methods
Data source and study population
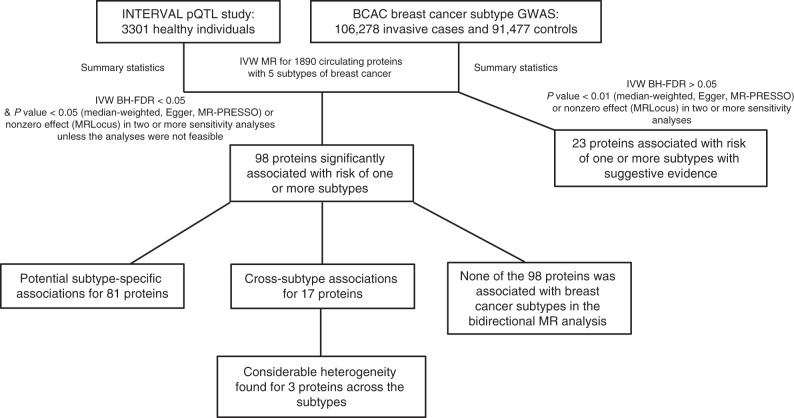
The study flowchart is shown in Fig. 1. We obtained summary statistics of the associations between genetic variants and circulating protein concentrations from a large-scale protein quantitative trait loci (pQTL) study conducted in 3301 healthy subjects of European descent [7]. Circulating proteins were quantified using the SOMAscan platform. The original GWAS reported 1927 significant pQTL associations for 1478 circulating proteins [7]. We extracted all the genetic variants associated with a specific protein with a P < 5.0 × 10−8. We excluded genetic variants with an imputation quality score (R2) < 0.8 and a minor allele frequency <0.05 from the current analysis. Summary statistics of selected pQTL variants in associations with risks of breast cancer intrinsic-like subtypes were obtained from a recent genome-wide association study (GWAS) conducted in the BCAC (106,278 invasive cases and 91,477 controls) [8]. Both data, from the pQTL study and BCAC, were used previously to identify the 56 proteins associated with overall breast cancer risk [5]. To obtain independent genetic variants associated with a specific protein, linkage disequilibrium (LD) pruning was then performed to filter out those in LD > 0.1 based on the data of CEU populations in the 1000 Genomes Project. Breast cancer intrinsic-like subtypes were determined based on the status of hormone receptors (i.e., ER, progesterone receptor [PR], and human epidermal growth factor receptor 2 [HER2]) and grade of primary breast cancer. Invasive cases were categorised into five distinct subtypes, including luminal A-like (ER+ and/or PR+, HER2−, Grades 1 and 2), luminal B/Her2-negative-like (ER+ and/or PR+, HER2−, grade 3), luminal B-like (ER+ and/or PR+, HER2+), HER2-enriched-like (ER− and PR−, HER2+), and triple-negative (ER−, PR−, HER2−). Summary statistics of GWAS of breast cancer intrinsic-like subtypes were downloaded from the BCAC website (http://bcac.ccge.medschl.cam.ac.uk/bcacdata/). Our analyses were limited to the women of European ancestry included in the BCAC as the participants of the consortium were predominantly white. Details of the genotyping protocols in the BCAC have been published elsewhere [8–10]. All the BCAC data were imputed by IMPUTE version 2 [11], using the 1000 Genomes Project (October 2014 version 3 release) dataset as the reference panel. All participating studies of the BCAC were approved by their corresponding ethics review boards and all subjects provided informed consent.
Fig. 1. The flowchart of the current study.
This flowchart describes what has been done for the study and summarises the main findings.
Statistical analysis
The inverse-variance (IVW)-weighted method [12] was performed as the main analysis. Three additional MR approaches, i.e., the weighted median MR method [13], the Egger regression [14], the MR-PRESSO [15] and MRLocus, were conducted as secondary analyses. The IVW approach assumes that all genetic variants used as instruments are valid. To address the potential violation of this assumption, we performed the weighted median approach, which accepts that up to half of the genetic instruments included are invalid [13]. In addition, the Egger regression and MR-PRESSO were applied to detect and correct for pleiotropic effects [14, 15] (another common violation of assumption in MR analysis).
The number of genetic variants used as instruments for proteins ranged from 1 to 51. Approximately 49.3% (932/1890) of the instruments were constructed using three or more pQTL variants. Odds ratios (ORs), 95% confidence intervals (CIs), and corresponding p-values were obtained for all four approaches unless the number of genetic variants were under the minimum requirement for certain methods (e.g., a minimum of three variants is required for median/Egger regression method). Associations with a Benjamini–Hochberg false discovery rate (BH-FDR) of <0.05 for the IVW method within each intrinsic-like subtype were considered significant in a two-sided test. These associations also had a P < 0.05 (weighted median, MR-Egger, or MR-PRESSO) or nonzero effect (MRLocus) in two of the four remaining MR approaches. Suggestive associations were defined as those had a BH-FDR >0.05 using IVW method, while their P < 0.01 (weighted median, MR-Egger and MR-PRESSO) or nonzero effect (MRLocus) in two MR approaches in the secondary analysis. Full results of the four MR approaches were presented in Supplementary Tables S2 and 3. We further conducted bidirectional MR analysis [16] with the genetic instruments associated with breast cancer subtypes (P < 5.0 × 10−8 & LD < 0.1) for the proteins found significant in the MR analysis mentioned above. Test of heterogeneity was performed to detect potential subtype-specific associations. Proteins identified with a Pheterogeneity < 0.05 and effect estimates showing opposite directions across subtypes were visualised by scatter plots. The web-based tool Panther [17] was used to assess whether the evaluated and identified proteins were overrepresented/enriched at the pathway level. STRING [18] was used to show reported interactions between the identified proteins. The linkage disequilibrium (LD) patterns between the pQTL variants used as instrumental variables in this study for the identified proteins and previously reported breast cancer susceptibility variants in the European populations of the 1000 Genomes reference panel (1000G Phase3 v5, EUR) were assessed [8, 10, 19, 20]. All the statistical analyses were completed using R 4.1.1 (R packages: MendelianRandomization, version 0.5.1; MR-PRESSO, version 1.0; mrlocus).
Results
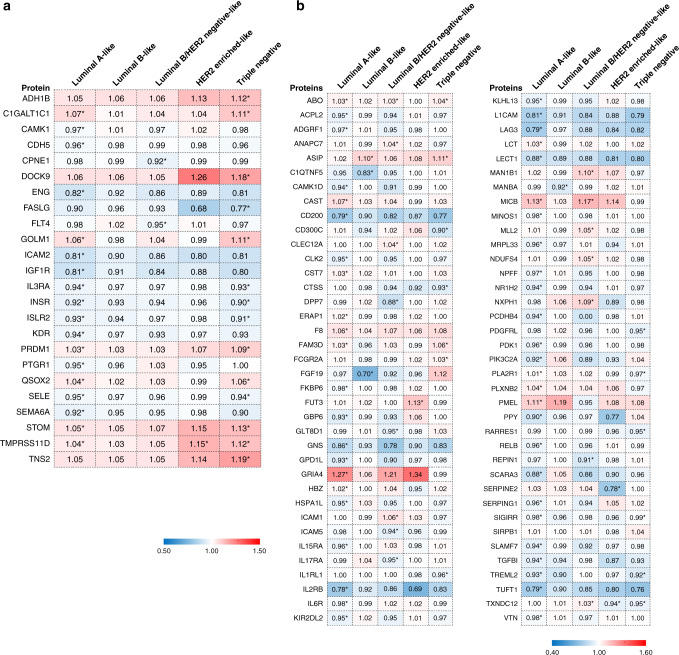
We first performed pathway enrichment analysis for the 1890 circulating proteins and found that they were enriched in 204 and 31 pathways in Reactome and PANTHER database, respectively (FDR <0.05). The top enriched ones were the immune system, cytokine signalling, interleukin signalling, innate immune system, and angiogenesis-related pathways (Supplementary Table S1). We then re-evaluated the associations of 56 previously reported proteins [5] with risk of breast cancer by intrinsic-like subtypes (Fig. 2a and Supplementary Table S2). After adjustment of multiple comparisons within each subtype, 24 of the 56 proteins were found to be significantly associated with the risk of one or more subtypes using IVW method (BH-FDR <0.05). The majority of the significant associations were driven by luminal A-like and/or triple-negative breast cancer (22/24, Supplementary Fig. S1). We also found that two proteins, Fms-related receptor tyrosine kinase 4 (FLT4, alias VEGFR3) and copine 1 (CPNE1), were exclusively associated with risk of luminal B/Her2-negative-like breast cancer. Associations shared by two or more subtypes were identified for nine proteins with risk of luminal A-like and triple-negative breast cancer, and one with luminal A-like, Her2-enriched-like, and triple-negative breast cancer. The association direction was consistent for these associations (Fig. 2a and Supplementary Table S2). In addition, suggestive associations were found for six proteins with risk luminal A-like breast cancer; two for luminal B/Her2-negative-like breast cancer; two with Her2-enriched-like breast cancer; and three within triple-negative breast cancer (Supplementary Table S2).
Fig. 2. Heatmaps of MR estimates of proteins significantly associated with risk of one or more breast cancer subtypes.
a Associations for previously reported proteins in Shu et al. [5]. b Newly identified associations. *BH-FDR < 0.05. MR estimates obtained from the IVW approach were presented in the figure. The full results of the four MR approaches are shown in Supplementary Tables S2 and S3.
We next evaluated the associations of the remaining 1834 proteins with the risk of breast cancer intrinsic-like subtypes. In total, we found significant associations between 74 additional proteins and risk of one or more subtypes (BH-FDR < 0.05, Fig. 2b and Supplementary Table S3), of which 67 were potentially subtype-specific and dominated by those associated with risk of luminal A-like (n = 43), luminal B/Her2-negative-like triple (n = 12), or triple-negative breast cancer only (n = 7) (Supplementary Fig. S2). Associations shared across subtypes were also found. For example, the association between agouti signalling protein (ASIP) and risk of triple-negative breast cancer (OR = 1.11, 95% CI = 1.08–1.14, BH-FDR = 4.51 × 10−10, per unit of increase) was the most significant association identified in this analysis. The protein was not associated with the risk of luminal A-like, luminal B/HER2-negative-like, or HER2-enriched-like breast cancer but was associated with luminal B-like breast cancer (OR = 1.10, 95% CI = 1.06–1.14, BH-FDR = 8.93 × 10−5). The summary of identified proteins in the current study and their overlapping with previously reported associations with overall breast cancer risk were shown in Table 1. A total of 118 associations were identified for 98 unique proteins with risk of one or more subtypes. Among them, 81 were specific to one subtype.
Table 1.
Summary of identified associations between proteins and risk of breast cancer intrinsic-like subtypes.
| BC subtypes | Significant associations with risks of overall BC and BC subtypes* | Novel associations with risk of BC subtypes only* | Total |
|---|---|---|---|
| Luminal A-like relevant (specific) | 18 (8) | 48 (43) | 66 (51) |
| Luminal B-like relevant (specific) | 0 (0) | 4 (3) | 4 (3) |
| Luminal B/HER2-negative-like relevant (specific) | 2 (2) | 15 (12) | 17 (14) |
| HER2-enriched-like relevant (specific) | 1 (0) | 3 (2) | 4 (2) |
| Triple-negative relevant (specific) | 14 (4) | 13 (7) | 27 (11) |
*The significance of the associations with risk of breast cancer subtype was determined if BH-FDR < 0.05, based on the P values from the inverse-variance weighted MR method. The associations between circulating proteins and risk of overall breast cancer were retrieved from a previous study conducted by Shu et al. [5]. In all, 98 unique proteins were associated with one or more subtypes among the totalled 118 significant associations identified.
The number in parentheses indicates the associations specific to the subtype of interest (also see Supplementary Figs. S1 and S2). A total of 81 subtype-specific protein associations were identified.
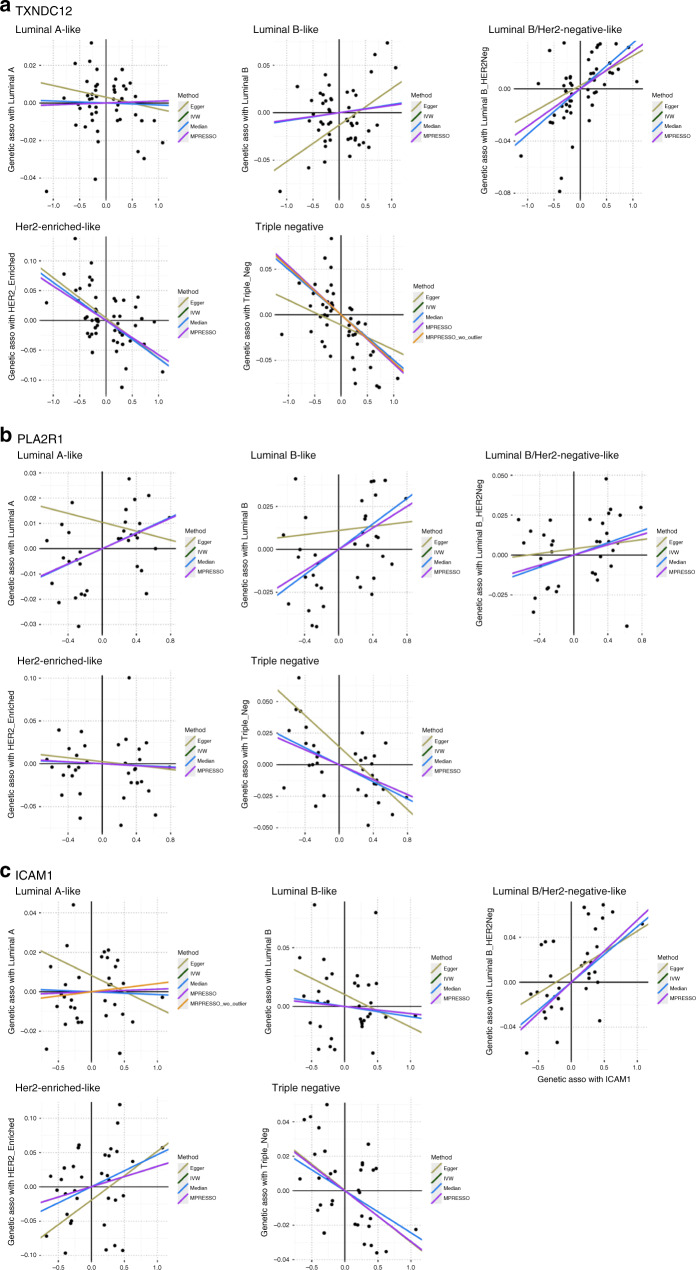
Three proteins, thioredoxin Domain Containing 12 (TXNDC12), phospholipase A2 Receptor 1 (PLA2R1), and intercellular adhesion molecule 1 (ICAM1), showed strong heterogeneity for their association estimates across the intrinsic-like subtypes (Fig. 3, Supplementary Table S3 and Supplementary Fig. S3). For example, higher levels of genetically predicted ICAM1 were associated with an increased risk of luminal B/HER2-negative-like cancer (OR = 1.06, 95% CI = 1.03–1.08, BH-FDR = 2.43 × 10−4) while inversely associated with triple-negative cancer with borderline significance (OR = 0.97, 95% CI = 0.96–0.99, BH-FDR = 0.065). Furthermore, the genetically predicted levels of PLA2R1 were positively associated with luminal A-like cancer while inversely with triple-negative cancer; and TXNDC12 was positively associated with luminal B/HER2-negative-like cancer while inversely with HER2-enriched-like and triple-negative cancer.
Fig. 3. Scatter plots of MR associations of TXNDC12, PLA2R1 and ICAM1 with risk of five breast cancer intrinsic-like subtypes.
Heterogeneity test was performed based on the MR estimates from the four approaches on each of the five subtypes.
The bidirectional MR analysis found no significant association for the 98 identified proteins (Supplementary Table S4). The identified proteins associated with the risk of luminal A-like breast cancer were found statistically significantly enriched in the immune system and insulin signalling pathways based on the overrepresentation analysis using data from the Reactome and PANTHER database (Table 2, see 'Methods'). Protein–protein interaction analysis highlighted interaction hubs for luminal A-like breast cancer, including clusters of cytokine receptors, cell-cell adhesion molecules, and growth factor receptors (Supplementary Fig. S4). We also compared LD patterns between the pQTL variants used in this study for the 98 significantly associated proteins and those reported breast cancer susceptibility variants. We found that the instrumental variables of eight proteins showed a moderate LD (r2: 0.25–0.63) with the known breast cancer susceptibility variants (Supplementary Table S5).
Table 2.
Significant pathways identified from the overrepresentation test of the significantly associated proteins.
| Breast cancer subtypes | Pathway | Proteins involved | Fold enrichment | FDR |
|---|---|---|---|---|
| Reactome pathway | ||||
| Luminal A-like specific | Immune system | KLHL13, SERPING1, ILRB, SLAMF7, IL6R, ERAP1, CD200, SIGIRR, GBP6, RELB, IL15RA, ICAM2, KIR2DL2, GNS, PDK1, VTN, LAG3 | 3.28 | 1.79 × 10−2 |
| Luminal A-like relevant | Immune system | S/A+ MICB, IL2RB, TREML2, IL3RA, STOM | 3.24 | 1.21 × 10−3 |
| PANTHER pathway | ||||
| Luminal A-like specific | Insulin/IGF pathway—protein kinase B signalling cascade | IPK3C2A, IGF1R, PDK1 | 29.70 | 1.39 × 10−2 |
| Interleukin signalling pathway | IL2RB, IL6R, IL15RA, PDK1 | 17.80 | 1.42 × 10−2 | |
| Luminal A-like relevant | Insulin/IGF pathway—protein kinase B signalling cascade | IPK3C2A, IGF1R, PDK1, INSR | 31.68 | 1.76 × 10−3 |
| Interleukin signalling pathway | IL2RB, IL6R, IL13R, IL15RA, PDK1 | 17.80 | 9.34 × 10−4 | |
Luminal A-like specific: proteins specifically associated with luminal A-like breast cancer.
Luminal A-like relevant: proteins specifically associated with luminal A-like breast cancer and those associated with luminal A-like breast cancer and any other intrinsic-like subtypes.
S/A: Same as above.
Discussion
In this study, we evaluated the relationships between circulating proteins and risks of breast cancer intrinsic-like subtypes by conducting a MR analysis. Through the analyses, we identified 74 novel associations of proteins with the risk of one or more intrinsic-like subtypes of breast cancer and confirmed 24 proteins that were also previously reported to be associated with overall breast cancer risk. Only a small proportion of the identified associations were driven by known breast cancer susceptibility loci (Supplementary Table S5). Among the identified proteins, three (ICAM1, PLA2R1 and TXNDC12) showed strong evidence of heterogeneity among intrinsic-like subtypes.
It is well recognised that breast cancer is highly heterogeneous, as it consists of subtypes with distinct pathological and molecular features [21, 22]. Previous studies have reported differences in risk factors for breast cancer molecular subtypes. For example, body mass index was reported to be inversely associated with luminal A breast cancer but positively associated with basal-like breast cancer in premenopausal women [23]. The greatest association of family history of breast cancer was found for basal-like breast cancer compared to that for other subtypes [23, 24]. Heterogeneity for the distinct subtypes of breast cancer was also consistently reported for reproductive risk factors [25, 26]. These findings support that different subtypes of breast cancer have distinguished etiologies, and identifying risk factors for breast cancer subtypes has important implications for the prevention of more aggressive subtypes such as luminal B, HER2 and basal-like cancers.
We previously conducted a genetic instrument analysis and showed significant associations of 56 proteins with risk of overall breast cancer [5]. This study is the first to extensively examine the potential causal role of circulating proteins played in the development of molecular subtypes of breast cancer. According to our findings, most of the identified associations were luminal A-like, luminal B/Her2-negative-like, or triple-negative breast cancer-specific. Associations shared by subtypes were also identified. As an example, ASIP was significantly associated with increased risk of both triple-negative and luminal B-like breast cancer. A meta-analysis of GWAS of breast cancer identified a susceptibility locus at 20q11 where ASIP and another two genes reside closely, showing the variant was more strongly associated with ER-negative breast cancer especially triple-negative breast cancer than overall breast cancer [27]. While the previous GWAS was unable to discern the potential causal gene player in the region [27], our findings serve as strong evidence supporting the potential causal role of ASIP in this locus. The same locus was also previously linked to pigmentation traits and risk of both cutaneous melanoma and basal cell carcinoma [28], suggesting a possible shared genetic susceptibility between triple-negative breast cancer and skin cancers.
Among the identified associations, estimates for three proteins varied substantially across the subtypes. The exact biological mechanisms that underlie these associations especially regarding subtype heterogeneity are not clear; thus, further investigations are warranted. For example, the biological function of ICAM1 in breast cancer remains controversial. ICAM1 is a cell surface transmembrane glycoprotein receptor, belonging to the immunoglobulin superfamily. The protein was reported to be involved in T-cell priming, transendothelial trafficking, and facilitating lymphocyte adhesion with tumour cells [29]. Ogawa et al. also reported that expression of ICAM1 was negatively associated with tumour infiltration, nuclear pleomorphism, as well as lymph node metastasis in breast cancer [30]. Conversely, it has been proposed that the downregulation of ICAM1 could attenuate the metastatic ability of MCF-7 cells, leading to a decreased migration and invasiveness of the cancer cells [31, 32]. In an in vitro experiment, Guo et al. also demonstrated that ICAM1 might be an effective therapeutic target by delivering small interfering RNA to triple-negative breast cancer MDA-MB-231 cells, resulting in an inhibition of cancer progression [33].
The overrepresented test indicated that the identified proteins were enriched for immune-related and insulin signalling pathways for luminal A-like breast cancer. Pathway analyses of GWAS data have highlighted the involvement of immune-response pathways in susceptibility to overall breast cancer [10]. Our findings provided new evidence at protein level, supporting their role in breast cancer aetiology, especially in the development of luminal A-like breast cancer. Whether the proteins enriched in immune-related pathways in the current study having clinical implication in patient management or treatment decision deserves future investigations.
Our study had several strengths. To our knowledge, no study has examined the relationship between circulating proteins and risk of breast cancer intrinsic-like subtypes via MR approaches. We employed different MR approaches to address potential issues of pleiotropy and invalid instruments in our analyses. Further, the large sample size included in the current study could improve the precision of association estimates. The bidirectional MR analysis provided further evidence that reverse causality was unlikely to have a strong impact on our findings. Nevertheless, we also recognise several limitations in our approach. Although Egger regression and MR-PRESSO were applied to detect and address potential pleiotropic effects, we cannot completely rule out the possibility of residual pleiotropic effects for the genetic instruments used in the analysis. Pleiotropic effects could be more appropriately addressed if individual-level data is available. In addition, the surrogate intrinsic-like subtypes of breast cancer were defined by histopathological information on ER, PR, HER2 and grade status instead of on the basis of actual molecular profiles [34], which may introduce misclassifications. This misclassification is also expected to be non-differential, leading to a null association if exists, given the data used in our two-sample MR analysis were collected from two independent populations. Another limitation was that the sample size for the subtypes other than luminal A-like in the BCAC was still relatively small. It is possible that proteins identified to be associated with luminal A-like breast cancer only in this study may also be associated with other subtypes if the sample size of other subtypes increased. Moreover, we lacked information on certain risk factors which precluded analysis in subgroups such as stratification by menopausal status or age at diagnosis. Also, as the circulating protein levels in whole blood may not accurately reflect its levels in the relevant issues such as breast tissues, additional investigations focused on tissue pQTLs are warranted to further study the relationship between proteins and breast cancer risk. Furthermore, our study could not evaluate other important circulating proteins that are not included in the SomaScan panel. Further investigation should be conducted once more comprehensive pQTL data became available.
In conclusion, this MR study investigated the potential causal relationship between circulating proteins and the risk of breast cancer intrinsic-like subtypes and identified 98 proteins associated with the risk of one or more subtypes. Levels of three proteins, ICAM1, PLA2R1 and TXNDC12, showed a strong heterogeneity for their associations, as the estimates varied significantly across the subtypes. These findings revealed the importance of accounting for subtype heterogeneity when investigating risk factors and searching for biomarkers for breast cancer, which in turn may be instrumental in effective risk classification and personalised screening.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
The top 20 enriched pathways among 1890 tested proteins from overrepresentation test
Previously reported proteins, comparisons between the associations with risk of overall breast cancer and associations with risk of breast cancer subtypes: Full results from five MR approaches
Significant associations with risk of breast cancer subtypes (not previously reported for risk of overall breast cancer): full results from five MR approaches.
Bidirectional MR analysis for the identified proteins in the main analysis: from breast cancer subtype to proteins.
The pQTL of eight proteins showed a moderate linkage disequilibrium (LD) with previously identified breast cancer variants
Acknowledgements
The authors would like to thank the researchers of the Breast Cancer Association Consortium (BCAC), as well as Benjamin B Sun and colleagues for sharing summary statistics of GWAS.
Author contributions
XS (Xiang Shu) designed the study. QZ performed the statistical analyses. XS (Xiaohui Sun) created the figures. XS (Xiang Shu) drafted the manuscript. XS (Xiang Shu), MF, XG, JL, MER, X-OS, WZ and JLB interpreted the data and edited the manuscript. All authors have given final approval of the version to be published.
Funding
XS (Xiang Shu) is supported, in part, by R00CA230205; XS (Xiaohui Sun) is supported by R00CA230205 & China Scholarship Council (CSC) (202108330197); MF is supported by the Quantitative Sciences Undergraduate Research Experience through R25CA214255; WZ is supported, in part by R01CA202981 and R01CA235553 for breast cancer research. We also acknowledge the Memorial Sloan Kettering P30 Cancer Center Support Grant (P30CA008748) for the statistical support.
Data availability
All data used in this study are publicly available summary-level data, with the relevant studies cited.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All included datasets were approved by respective ethics/institutional review committees, in accordance with the Declaration of Helsinki.
Consent to publish
Not applicable.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-022-01923-2.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011;5:5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Endogenous H, Breast Cancer Collaborative, G. Key TJ, Appleby PN, Reeves GK, Roddam AW. Insulin-like growth factor 1 (IGF1), IGF binding protein 3 (IGFBP3), and breast cancer risk: pooled individual data analysis of 17 prospective studies. Lancet Oncol. 2010;11:530–42. doi: 10.1016/S1470-2045(10)70095-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christopoulos PF, Msaouel P, Koutsilieris M. The role of the insulin-like growth factor-1 system in breast cancer. Mol Cancer. 2015;14:43. doi: 10.1186/s12943-015-0291-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shu X, Bao J, Wu L, Long J, Shu XO, Guo X, et al. Evaluation of associations between genetically predicted circulating protein biomarkers and breast cancer risk. Int J Cancer. 2020;146:2130–8. doi: 10.1002/ijc.32542. [DOI] [PubMed] [Google Scholar]
- 6.Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362:k601. doi: 10.1136/bmj.k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J, et al. Genomic atlas of the human plasma proteome. Nature. 2018;558:73–9. doi: 10.1038/s41586-018-0175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H, Ahearn TU, Lecarpentier J, Barnes D, Beesley J, Qi G, et al. Genome-wide association study identifies 32 novel breast cancer susceptibility loci from overall and subtype-specific analyses. Nat Genet. 2020;52:572–81. doi: 10.1038/s41588-020-0609-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45:353–61. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michailidou K, Lindstrom S, Dennis J, Beesley J, Hui S, Kar S, et al. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551:92–4. doi: 10.1038/nature24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37:658–65. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40:304–14. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–25. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693–8. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carreras-Torres R, Johansson M, Haycock PC, Relton CL, Davey Smith G, Brennan P, et al. Role of obesity in smoking behaviour: Mendelian randomisation study in UK Biobank. BMJ. 2018;361:k1767. doi: 10.1136/bmj.k1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–86. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–13. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milne RL, Kuchenbaecker KB, Michailidou K, Beesley J, Kar S, Lindstrom S, et al. Identification of ten variants associated with risk of estrogen-receptor-negative breast cancer. Nat Genet. 2017;49:1767–78. doi: 10.1038/ng.3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coignard J, Lush M, Beesley J, O’Mara TA, Dennis J, Tyrer JP, et al. A case-only study to identify genetic modifiers of breast cancer risk for BRCA1/BRCA2 mutation carriers. Nat Commun. 2021;12:1078. doi: 10.1038/s41467-020-20496-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360:790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 22.Reis-Filho JS, Weigelt B, Fumagalli D, Sotiriou C. Molecular profiling: moving away from tumor philately. Sci Transl Med. 2010;2:47ps43. doi: 10.1126/scitranslmed.3001329. [DOI] [PubMed] [Google Scholar]
- 23.Yang XR, Sherman ME, Rimm DL, Lissowska J, Brinton LA, Peplonska B, et al. Differences in risk factors for breast cancer molecular subtypes in a population-based study. Cancer Epidemiol Biomark Prev. 2007;16:439–43. doi: 10.1158/1055-9965.EPI-06-0806. [DOI] [PubMed] [Google Scholar]
- 24.Tamimi RM, Colditz GA, Hazra A, Baer HJ, Hankinson SE, Rosner B, et al. Traditional breast cancer risk factors in relation to molecular subtypes of breast cancer. Breast Cancer Res Treat. 2012;131:159–67. doi: 10.1007/s10549-011-1702-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson KN, Schwab RB, Martinez ME. Reproductive risk factors and breast cancer subtypes: a review of the literature. Breast Cancer Res Treat. 2014;144:1–10. doi: 10.1007/s10549-014-2852-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaudet MM, Gierach GL, Carter BD, Luo J, Milne RL, Weiderpass E, et al. Pooled analysis of nine cohorts reveals breast cancer risk factors by tumor molecular subtype. Cancer Res. 2018;78:6011–21. doi: 10.1158/0008-5472.CAN-18-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siddiq A, Couch FJ, Chen GK, Lindstrom S, Eccles D, Millikan RC, et al. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum Mol Genet. 2012;21:5373–84. doi: 10.1093/hmg/dds381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gudbjartsson DF, Sulem P, Stacey SN, Goldstein AM, Rafnar T, Sigurgeirsson B, et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nat Genet. 2008;40:886–91. doi: 10.1038/ng.161. [DOI] [PubMed] [Google Scholar]
- 29.Blank C, Brown I, Kacha AK, Markiewicz MA, Gajewski TF. ICAM-1 contributes to but is not essential for tumor antigen cross-priming and CD8+ T cell-mediated tumor rejection in vivo. J Immunol. 2005;174:3416–20. doi: 10.4049/jimmunol.174.6.3416. [DOI] [PubMed] [Google Scholar]
- 30.Ogawa Y, Hirakawa K, Nakata B, Fujihara T, Sawada T, Kato Y, et al. Expression of intercellular adhesion molecule-1 in invasive breast cancer reflects low growth potential, negative lymph node involvement, and good prognosis. Clin Cancer Res. 1998;4:31–6. [PubMed] [Google Scholar]
- 31.Di D, Chen L, Wang L, Sun P, Liu Y, Xu Z, et al. Downregulation of human intercellular adhesion molecule-1 attenuates the metastatic ability in human breast cancer cell lines. Oncol Rep. 2016;35:1541–8. doi: 10.3892/or.2016.4543. [DOI] [PubMed] [Google Scholar]
- 32.Figenschau SL, Knutsen E, Urbarova I, Fenton C, Elston B, Perander M, et al. ICAM1 expression is induced by proinflammatory cytokines and associated with TLS formation in aggressive breast cancer subtypes. Sci Rep. 2018;8:1–12. doi: 10.1038/s41598-018-29604-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo P, Yang J, Di Jia MAM, Auguste DT. ICAM-1-targeted, Lcn2 siRNA-encapsulating liposomes are potent anti-angiogenic agents for triple negative breast cancer. Theranostics. 2016;6:1. doi: 10.7150/thno.12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai X, Li T, Bai Z, Yang Y, Liu X, Zhan J, et al. Breast cancer intrinsic subtype classification, clinical use and future trends. Am J Cancer Res. 2015;5:2929–43. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The top 20 enriched pathways among 1890 tested proteins from overrepresentation test
Previously reported proteins, comparisons between the associations with risk of overall breast cancer and associations with risk of breast cancer subtypes: Full results from five MR approaches
Significant associations with risk of breast cancer subtypes (not previously reported for risk of overall breast cancer): full results from five MR approaches.
Bidirectional MR analysis for the identified proteins in the main analysis: from breast cancer subtype to proteins.
The pQTL of eight proteins showed a moderate linkage disequilibrium (LD) with previously identified breast cancer variants
Data Availability Statement
All data used in this study are publicly available summary-level data, with the relevant studies cited.