Abstract
In Saccharomyces cerevisiae, the rRNA Gm2270 methyltransferase, Pet56p, has an essential role in the maturation of the mitochondrial large ribosomal subunit that is independent of its methyltransferase activity. Here we show that the proposed Escherichia coli ortholog, RlmB (formerly YjfH), indeed is essential for the formation of Gm in position 2251 of 23S rRNA. However, a ΔrlmB mutant did not show any ribosome assembly defects and was not outgrown by a wild-type strain even after 120 cell mass doublings. Thus, RlmB has no important role in ribosome assembly or function in E. coli.
The assembly of the 50S and 30S ribosomal subunits has been proposed to be a self-assembly process as demonstrated by the ability to reconstitute in vitro fully active ribosomes from the isolated components (36). However, recent observations indicate that proteins that assist in the formation of ribosomes, besides the ribosomal components, rRNA-processing enzymes, and rRNA-modifying and ribosomal-protein-modifying enzymes, have to be present in vivo. Indeed, there are at least seven candidates in Escherichia coli: SrmB and DbpA, two DEAD box RNA helicases (14, 15, 26, 33, 37, 46); Era, an essential GTPase (30, 31–34, 41); RimM and RbfA, two proteins associated with free 30S subunits but not with 50S subunits or 70S ribosomes (5, 6, 10, 27); and DnaK and GroEL, two molecular chaperones that at least at high temperature seem important for ribosome maturation (1, 12, 42). Whether DnaK and GroEL are directly or indirectly involved in the assembly of ribosomes in E. coli is not known.
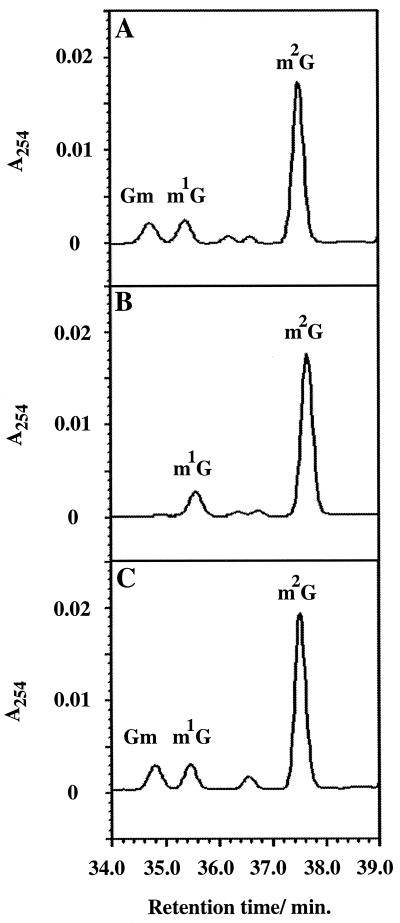
In Saccharomyces cerevisiae, the Pet56 protein catalyzes 2′-O-methylation at the universally conserved G at position 2270 (Gm2270) in the mitochondrial 21S rRNA (43), corresponding to position 2251 of 23S rRNA of E. coli. Further, pet56 null mutants lack functional mitochondrial ribosomes, indicating that Pet56p is essential for the in vivo assembly of the mitochondrial large ribosomal subunits (43). Recently it was demonstrated that Pet56p variants with amino acid substitutions in the SAM binding site, which abolished methyltransferase activity, could support the in vivo assembly of functional mitochondrial ribosomes, suggesting that Pet56p has a role in ribosome assembly that is independent of its methyltransferase activity (T. L. Mason, personal communication). In E. coli, the protein encoded by the yjfH gene is a putative 2′-O-methyltransferase, based on its similarities to other 2′-O-methyltransferases, especially Pet56p (21). The yjfH gene in E. coli encodes a hypothetical protein 243 amino acids in length and is downstream from the rnr gene, encoding RNase R (9). Just upstream from yjfH there are sequences that match those for promoters dependent on ς70 for transcription initiation, and downstream from yjfH there is a sequence characteristic of rho-independent transcriptional terminators. To investigate whether YjfH was the E. coli rRNA Gm2251 methyltransferase, a chromosomal deletion of yjfH was constructed. The 835-bp region upstream from yjfH was amplified by PCR using the oligonucleotides rnr-F1 and ΔyjfH-R1, trimmed with HindIII and SalI, and cloned into the temperature-sensitive plasmid vector pMAK705, yielding plasmid pMW458 (Table 1). The 917-bp region downstream from yjfH was amplified by PCR using the oligonucleotides ΔyjfH-F1 and yjfJ-R1, trimmed with SalI and BamHI, and inserted into plasmid pMW458. The resulting plasmid, pMW465, which contains an in-frame deletion that covers all except the first two and last three codons of yjfH replaced by a SalI site, was used to delete the yjfH gene on the chromosome of strain MW100 following the procedure described by Hamilton et al. (23). One of the resulting strains, MW244, was confirmed by PCR analyses to contain the yjfH deletion on the chromosome. A yjfH+ strain, MW245, was isolated together with MW244 and used as a control. Further, the yjfH+ gene was amplified by PCR using the oligonucleotides yjfH-F2 and yjfH-R2, trimmed with EcoRI and HindIII, and cloned into pBAD30, yielding plasmid pMW467. rRNA and tRNA from the yjfH+ strain MW245, the ΔyjfH mutant MW244, and the mutant harboring the yjfH+ plasmid pMW467 were prepared (13), degraded with nuclease P1 and alkaline phosphatase to nucleosides (17), and analyzed by high-pressure liquid chromatography (HPLC) (16). The ΔyjfH mutant MW244 did not show any deficiency in the modification of tRNA (data not shown); however, it was found to lack Gm in 23S rRNA (Fig. 1B). This modification deficiency was fully complemented by plasmid pMW467 (Fig. 1C). These findings and the similarity of YjfH to Pet56p in S. cerevisiae, together with the fact that in E. coli Gm is found in only one position in rRNA, suggest that YjfH is indeed the E. coli rRNA Gm2251 methyltransferase. Therefore, we rename YjfH RlmB (for “rRNA large-subunit methylation”).
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides
| Strain, plasmid, or oligonucleotide | Description | Referencea |
|---|---|---|
| Strains | ||
| MW100 | Hfr P4X | 47 |
| MW244 | MW100 ΔrlmB | |
| MW245 | MW100 rlmB+ | |
| Plasmids | ||
| pBAD30 | bla cat′ araC PBAD | 22 |
| pMAK705 | rep(Ts) cat | 23 |
| pMW458 | rep(Ts) cat ′rnr | |
| pMW465 | rep(Ts) cat ′rnr ΔrlmB yjfI yjfJ′ | |
| pMW467 | bla cat′ araC PBAD-rlmB | |
| Oligonucleotides | ||
| rnr-F1 | 5′-TTTTAAGCTTGGAAGAGATGCTGCAACTGG-3′ | |
| ΔyjfH-R1 | 5′-GCTGCGCTGGTCGACGCTCATTAATGTACTCGTTG-3′ | |
| ΔyjfH-F1 | 5′-CATTAATGAGCGTCGACCAGCGCAGCTAATTTCTCAG-3′ | |
| yjfJ-R1 | 5′-TTTTGGATCCAACACTTGCTCTTCAGCGG-3′ | |
| yjfH-F2 | 5′-TTTTGAATTCGGATAAACGTAGTGCATCAGG-3′ | |
| yjfH-R2 | 5′-TTTTAAGCTTACGTAATTATCTTACCAGCTATAG-3′ |
Unless otherwise noted, the origin was this study.
FIG. 1.
HPLC analysis of modified nucleosides in rRNA. Only the part of the chromatograms that showed a difference between the strains is shown. (A) Strain MW245 (rlmB+); (B) strain MW244 (ΔrlmB); (C) strain MW244 (ΔrlmB)/pMW467 (rlmB+). The indicated nucleosides are 2′-O-methyl guanosine (Gm), 1-methyl guanosine (m1G), and 2-methyl guanosine (m2G). The identity of Gm was confirmed by spectrum analysis and by comparing the chromatograms with that for a trmH::Kmr mutant lacking Gm in tRNA (data not shown; see also reference 38).
RlmB is dispensable for fast growth.
To examine whether RlmB is important for efficient growth, the growth rate at 37°C in Luria-Bertani (LB) medium (2) was determined for the ΔrlmB mutant MW244 and the wild-type strain MW245. The specific growth rate, k (= ln2/g, where g is the mass doubling time in hours), of the ΔrlmB mutant was identical to that of the rlmB+ strain (1.34; standard deviations of 0.021 and 0.031, respectively). Further, no difference between the two strains was observed in their ability to grow at 21, 30, 37, and 44°C on rich medium plates or at 30, 37, and 42.5°C on medium E plates containing glucose. Also, stationary-phase cell culture density, measured as optical density at 600 nm, and survival in stationary phase, monitored by viable count determinations, after 24 and 48 h of incubation at 37°C in LB medium did not differ between the two strains (data not shown). To test the ability of the ΔrlmB mutant to grow in competition with the rlmB+ strain over the whole range of the growth cycle, the two strains were grown separately in LB medium at 37°C with shaking to stationary phase, at which the optical density at 600 nm was 5.1 for both strains. Equal amounts of stationary-phase cultures of mutant and wild-type cells were mixed, diluted 106-fold in LB medium, and incubated for 24 h. The dilution and incubation steps of the mixed culture were repeated five times, and samples were taken at the start and after each cycle and plated on rich medium. To determine the ratio of ΔrlmB cells to rlmB+ cells in the mixed culture, 48 colonies from each sampling time were subjected to PCR with oligonucleotides rnr-F1 and yjfH-R2. Even after six cycles, corresponding to more than 120 cell doublings, approximately 50% of the cells in the mixed culture were still ΔrlmB mutants (data not shown). Thus, the lack of RlmB did not confer any disadvantage to the mutant cells in their competition with rlmB+ cells.
RlmB is not important for ribosome maturation.
Since pet56 mutants of S. cerevisiae that lack the rRNA Gm2270 methyltransferase are deficient in the maturation of mitochondrial large ribosomal subunits, we examined whether RlmB was essential for assembly of ribosomes in E. coli. Polysome extracts of strains MW244 (ΔrlmB) and MW245 (rlmB+) were prepared (40) and fractionated by sucrose gradient centrifugation (39). No differences between the two strains were observed with respect to the amounts of ribosomal subunits, 70S ribosomes, or polysomes (data not shown). To detect any subtle deficiency in ribosome maturation of the ΔrlmB mutant, the kinetics of ribosome assembly were studied by pulse-labeling techniques. Log-phase cultures of the two strains grown in rich MOPS medium (35) lacking uracil were labeled with [3H]uridine for 1 and 2 min. Cellular extracts were prepared and analyzed by sucrose gradient centrifugation under conditions that dissociated the 70S ribosomes into 50S and 30S subunits (29). The amounts of 50S subunit assembly intermediates in the two strains were indistinguishable at both 1 and 2 min of labeling (data not shown). Furthermore, when the maturation of the 50S subunits was probed by primer extension analysis of the 5′ end of 23S rRNA (6), no increased accumulation of precursors to 23S rRNA was observed in the ΔrlmB mutant (data not shown). These findings strongly suggest that RlmB is not important for ribosome maturation in E. coli.
In E. coli, 16S rRNA contains 11 modified nucleosides, of which 10 are methylated, while 23S rRNA contains 23 modified nucleosides, of which 14 are methylated (3). Some of the methylated nucleosides have ribose methylations (2′-O-methylations), whereas others have base methylations. Modified nucleosides are clustered at the functional domains of rRNA, such as the A and P sites of 16S rRNA and the peptidyltransferase center of 23S rRNA (3). It has been proposed that the modifications could affect rRNA maturation and structure, ribosomal subunit association, tRNA binding, and peptidyltransferase activity; however, little is known about their exact functions. In E. coli, three 16S rRNA methyltransferases (RsmA [KsgA], RsmB [RrmB], and RsmC) and two 23S rRNA methyltransferases (RrmA and RrmJ [FtsJ]) have been identified. A mutation in the E. coli ksgA (rsmA) gene, encoding the 16S rRNA m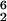 A1518,1519 methyltransferase (24), leads to a reduced growth rate in some media and a reduced polypeptide-synthetic activity in vitro (25), whereas a disruption of the gene for RsmB, the 16S rRNA m5C967 methyltransferase (19, 44), has no discernible effect on the growth rate of the cells (19). No mutation in rsmC, encoding the 16S rRNA m2G1207 methyltransferase (45), is available; however, RsmC cannot be essential for ribosome function, since functional 30S subunits can be prepared using 16S rRNA lacking all modifications (11, 28). A strain that lacks RrmA, the 23S rRNA m1G745 methyltransferase, shows a 1.4-fold-increased mass doubling time, an increased amount of free ribosomal subunits, and a decreased polypeptide chain elongation rate (20). Similarly, a mutant deficient in RrmJ, the 23S rRNA Um2552 methyltransferase (4, 7), has a severe growth disadvantage and a dramatically altered polysome profile (4, 8). Since the ΔrlmB mutant studied here showed no growth or ribosome assembly defects, Gm2251 cannot play any essential role in ribosome assembly or function, which is also supported by measurements of the peptidyltransferase activity of reconstituted ribosomes containing in vitro-transcribed 23S rRNA lacking Gm2251 (18). However, we cannot exclude the possibility that Gm2251 has some importance for ribosome function under conditions which we have not tested. Our results also suggest that RlmB itself has no important function in ribosome assembly. This contrasts with the situation in S. cerevisiae, where the RlmB counterpart, Pet56p, has an essential function in the maturation of the mitochondrial large ribosomal subunit that is independent of its methyltransferase activity (43; Mason, personal communication). In comparison to RlmB, Pet56p has an N-terminal extension of 143 amino acids. Conceivably, the maturation function of Pet56p might reside in this part of the protein, explaining why the ΔrlmB mutant was not deficient in ribosome maturation. This maturation function might be completely absent in E. coli or might be performed by another protein or by 23S rRNA.
A1518,1519 methyltransferase (24), leads to a reduced growth rate in some media and a reduced polypeptide-synthetic activity in vitro (25), whereas a disruption of the gene for RsmB, the 16S rRNA m5C967 methyltransferase (19, 44), has no discernible effect on the growth rate of the cells (19). No mutation in rsmC, encoding the 16S rRNA m2G1207 methyltransferase (45), is available; however, RsmC cannot be essential for ribosome function, since functional 30S subunits can be prepared using 16S rRNA lacking all modifications (11, 28). A strain that lacks RrmA, the 23S rRNA m1G745 methyltransferase, shows a 1.4-fold-increased mass doubling time, an increased amount of free ribosomal subunits, and a decreased polypeptide chain elongation rate (20). Similarly, a mutant deficient in RrmJ, the 23S rRNA Um2552 methyltransferase (4, 7), has a severe growth disadvantage and a dramatically altered polysome profile (4, 8). Since the ΔrlmB mutant studied here showed no growth or ribosome assembly defects, Gm2251 cannot play any essential role in ribosome assembly or function, which is also supported by measurements of the peptidyltransferase activity of reconstituted ribosomes containing in vitro-transcribed 23S rRNA lacking Gm2251 (18). However, we cannot exclude the possibility that Gm2251 has some importance for ribosome function under conditions which we have not tested. Our results also suggest that RlmB itself has no important function in ribosome assembly. This contrasts with the situation in S. cerevisiae, where the RlmB counterpart, Pet56p, has an essential function in the maturation of the mitochondrial large ribosomal subunit that is independent of its methyltransferase activity (43; Mason, personal communication). In comparison to RlmB, Pet56p has an N-terminal extension of 143 amino acids. Conceivably, the maturation function of Pet56p might reside in this part of the protein, explaining why the ΔrlmB mutant was not deficient in ribosome maturation. This maturation function might be completely absent in E. coli or might be performed by another protein or by 23S rRNA.
Acknowledgments
We thank Kerstin Jacobsson for analyzing RNA by HPLC. We are grateful to Thomas L. Mason for communicating data on Pet56p prior to publication. We thank Olof P. Persson, Glenn R. Björk, and Tord G. Hagervall for stimulating discussions and comments on the manuscript.
This work was supported by the Swedish Natural Science Research Council (B-BU 9911), the Carl Trygger Foundation, and the Magnus Bergvall Foundation.
REFERENCES
- 1.Alix J-H, Guérin M-F. Mutant DnaK chaperones cause ribosome assembly defects in Escherichia coli. Proc Natl Acad Sci USA. 1993;90:9725–9729. doi: 10.1073/pnas.90.20.9725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Björk G R. Modification of stable RNA. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 2. Washington, D.C.: American Society for Microbiology; 1987. pp. 719–731. [Google Scholar]
- 4.Bugl H, Fauman E B, Staker B L, Zheng F, Kushner S R, Saper M A, Bardwell J C, Jakob U. RNA methylation under heat shock control. Mol Cell. 2000;6:349–360. doi: 10.1016/s1097-2765(00)00035-6. [DOI] [PubMed] [Google Scholar]
- 5.Bylund G O, Persson B C, Lundberg L A C, Wikström P M. A novel ribosome-associated protein is important for efficient translation in Escherichia coli. J Bacteriol. 1997;179:4567–4574. doi: 10.1128/jb.179.14.4567-4574.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bylund G O, Wipemo L C, Lundberg L A C, Wikström P M. RimM and RbfA are essential for efficient processing of 16S rRNA in Escherichia coli. J Bacteriol. 1998;180:73–82. doi: 10.1128/jb.180.1.73-82.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caldas T, Binet E, Bouloc P, Costa A, Desgres J, Richarme G. The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23 S ribosomal RNA methyltransferase. J Biol Chem. 2000;275:16414–16419. doi: 10.1074/jbc.M001854200. [DOI] [PubMed] [Google Scholar]
- 8.Caldas T, Binet E, Bouloc P, Richarme G. Translational defects of Escherichia coli mutants deficient in the Um2552 23S ribosomal RNA methyltransferase RrmJ/FTSJ. Biochem Biophys Res Commun. 2000;271:714–718. doi: 10.1006/bbrc.2000.2702. [DOI] [PubMed] [Google Scholar]
- 9.Cheng Z F, Zuo Y, Li Z, Rudd K E, Deutscher M P. The vacB gene required for virulence in Shigella flexneri and Escherichia coli encodes the exoribonuclease RNase R. J Biol Chem. 1998;273:14077–14080. doi: 10.1074/jbc.273.23.14077. [DOI] [PubMed] [Google Scholar]
- 10.Dammel C S, Noller H F. Suppression of a cold-sensitive mutation in 16S rRNA by overexpression of a novel ribosome-binding factor, RbfA. Genes Dev. 1995;9:626–637. doi: 10.1101/gad.9.5.626. [DOI] [PubMed] [Google Scholar]
- 11.Denman R, Negre D, Cunningham P R, Nurse K, Colgan J, Weitzmann C, Ofengand J. Effect of point mutations in the decoding site (C1400) region of 16S ribosomal RNA on the ability of ribosomes to carry out individual steps of protein synthesis. Biochemistry. 1989;28:1012–1019. doi: 10.1021/bi00429a014. [DOI] [PubMed] [Google Scholar]
- 12.El Hage A, Sbaï M, Alix J H. The chaperonin GroEL and other heat-shock proteins, besides DnaK, participate in ribosome biogenesis in Escherichia coli. Mol Gen Genet. 2001;264:796–808. doi: 10.1007/s004380000369. [DOI] [PubMed] [Google Scholar]
- 13.Emilsson V, Kurland C G. Growth rate dependence of transfer RNA abundance in Escherichia coli. EMBO J. 1990;9:4359–4366. doi: 10.1002/j.1460-2075.1990.tb07885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuller-Pace F V. RNA helicases: modulators of RNA structure. Trends Cell Biol. 1994;4:271–274. doi: 10.1016/0962-8924(94)90210-0. [DOI] [PubMed] [Google Scholar]
- 15.Fuller-Pace F V, Nicol S M, Reid A D, Lane D P. DbpA: a DEAD box protein specifically activated by 23S rRNA. EMBO J. 1993;12:3619–3626. doi: 10.1002/j.1460-2075.1993.tb06035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gehrke C W, Kuo K C. Ribonucleoside analysis by reversed-phase high performance liquid chromatography. In: Gehrke C W, Kuo K C, editors. Chromatography and modification of nucleosides, part A: analytical methods for major modified nucleosides. Amsterdam, The Netherlands: Elsevier; 1990. pp. 3–72. [Google Scholar]
- 17.Gehrke C W, Kuo K C, McCune R A, Gerhardt K O, Agris P F. Quantitative enzymatic hydrolysis of tRNAs: reversed-phase high-performance liquid chromatography of tRNA nucleosides. J Chromatogr. 1982;230:297–308. [PubMed] [Google Scholar]
- 18.Green R, Noller H F. In vitro complementation analysis localizes 23S rRNA posttranscriptional modifications that are required for Escherichia coli 50S ribosomal subunit assembly and function. RNA. 1996;2:1011–1021. [PMC free article] [PubMed] [Google Scholar]
- 19.Gu X R, Gustafsson C, Ku J, Yu M, Santi D V. Identification of the 16S rRNA m5C967 methyltransferase from Escherichia coli. Biochemistry. 1999;38:4053–4057. doi: 10.1021/bi982364y. [DOI] [PubMed] [Google Scholar]
- 20.Gustafsson C, Persson B C. Identification of the rrmA gene encoding the 23S rRNA m1G745 methyltransferase in Escherichia coli and characterization of an m1G745-deficient mutant. J Bacteriol. 1998;180:359–365. doi: 10.1128/jb.180.2.359-365.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gustafsson C, Reid R, Greene P J, Santi D V. Identification of new RNA modifying enzymes by iterative genome search using known modifying enzymes as probes. Nucleic Acids Res. 1996;24:3756–3762. doi: 10.1093/nar/24.19.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guzman L M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helser T L, Davies J E, Dahlberg J E. Mechanism of kasugamycin resistance in Escherichia coli. Nat New Biol. 1972;235:6–9. doi: 10.1038/newbio235006a0. [DOI] [PubMed] [Google Scholar]
- 25.Igarashi K, Kishida K, Kashiwagi K, Tatokoro I, Kakegawa T, Hirose S. Relationship between methylation of adenine near the 3′ end of 16-S ribosomal RNA and the activity of 30-S ribosomal subunits. Eur J Biochem. 1981;113:587–593. doi: 10.1111/j.1432-1033.1981.tb05103.x. [DOI] [PubMed] [Google Scholar]
- 26.Iggo R, Picksley S, Southgate J, McPheat J, Lane D P. Identification of a putative RNA helicase in E. coli. Nucleic Acids Res. 1990;18:5413–5417. doi: 10.1093/nar/18.18.5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones P G, Mitta M, Kim Y, Jiang W, Inouye M. Cold shock induces a major ribosomal-associated protein that unwinds double-stranded RNA in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:76–80. doi: 10.1073/pnas.93.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krzyzosiak W, Denman R, Nurse K, Hellmann W, Boublik M, Gehrke C W, Agris P F, Ofengand J. In vitro synthesis of 16S ribosomal RNA containing single base changes and assembly into a functional 30S ribosome. Biochemistry. 1987;26:2353–2364. doi: 10.1021/bi00382a042. [DOI] [PubMed] [Google Scholar]
- 29.Lindahl L. Intermediates and time kinetics of the in vivo assembly of Escherichia coli ribosomes. J Mol Biol. 1975;92:15–37. doi: 10.1016/0022-2836(75)90089-3. [DOI] [PubMed] [Google Scholar]
- 30.Lu Q, Inouye M. The gene for 16S rRNA methyltransferase (ksgA) functions as a multicopy suppressor for a cold-sensitive mutant of era, an essential RAS-like GTP-binding protein in Escherichia coli. J Bacteriol. 1998;180:5243–5246. doi: 10.1128/jb.180.19.5243-5246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meier T I, Peery R B, Jaskunas S R, Zhao G. 16S rRNA is bound to era of Streptococcus pneumoniae. J Bacteriol. 1999;181:5242–5249. doi: 10.1128/jb.181.17.5242-5249.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nashimoto H. Non-ribosomal proteins affecting the assembly of ribosomes in Escherichia coli. In: Nierhaus K H, Franceschi F, Subramanian A R, Erdmann V A, Wittman-Liebold B, editors. The translational apparatus. New York, N.Y: Plenum Press; 1993. pp. 185–195. [Google Scholar]
- 33.Nashimoto H, Miura A, Saito H, Uchida H. Suppressors of temperature-sensitive mutations in a ribosomal protein gene, rpsL (S12), of Escherichia coli K12. Mol Gen Genet. 1985;199:381–387. doi: 10.1007/BF00330746. [DOI] [PubMed] [Google Scholar]
- 34.Nashimoto H, Uchida H. DNA sequencing of the Escherichia coli ribonuclease III gene and its mutations. Mol Gen Genet. 1985;201:25–29. doi: 10.1007/BF00397981. [DOI] [PubMed] [Google Scholar]
- 35.Neidhardt F C, Bloch P L, Pedersen S, Reeh S. Chemical measurement of steady-state levels of ten aminoacyl-transfer ribonucleic acid synthetases in Escherichia coli. J Bacteriol. 1977;129:378–387. doi: 10.1128/jb.129.1.378-387.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nierhaus K H. The assembly of prokaryotic ribosomes. Biochimie. 1991;73:739–755. doi: 10.1016/0300-9084(91)90054-5. [DOI] [PubMed] [Google Scholar]
- 37.Nishi K, Morel-Deville F, Hershey J W B, Leighton T, Schnier J. An eIF-4A-like protein is a suppressor of an Escherichia coli mutant defective in 50S ribosomal subunit assembly. Nature. 1988;336:496–498. doi: 10.1038/336496a0. [DOI] [PubMed] [Google Scholar]
- 38.Persson B C, Jäger G, Gustafsson C. The spoU gene of Escherichia coli, the fourth gene of the spoT operon, is essential for tRNA (Gm18) 2′-O-methyltransferase activity. Nucleic Acids Res. 1997;25:4093–4097. doi: 10.1093/nar/25.20.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Powers T, Noller H F. Dominant lethal mutations in a conserved loop in 16S rRNA. Proc Natl Acad Sci USA. 1990;87:1042–1046. doi: 10.1073/pnas.87.3.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ron E Z, Kohler R E, Davis B D. Polysomes extracted from Escherichia coli by freeze-thaw-lysozyme lysis. Science. 1966;153:1119–1120. doi: 10.1126/science.153.3740.1119. [DOI] [PubMed] [Google Scholar]
- 41.Sayed A, Matsuyama S, Inouye M. Era, an essential Escherichia coli small G-protein, binds to the 30S ribosomal subunit. Biochem Biophys Res Commun. 1999;264:51–54. doi: 10.1006/bbrc.1999.1471. [DOI] [PubMed] [Google Scholar]
- 42.Sbaï M, Alix J H. DnaK-dependent ribosome biogenesis in Escherichia coli: competition for dominance between the alleles dnaK756 and dnaK+ Mol Gen Genet. 1998;260:199–206. doi: 10.1007/s004380050886. [DOI] [PubMed] [Google Scholar]
- 43.Sirum-Connolly K, Mason T L. Functional requirement of a site-specific ribose methylation in ribosomal RNA. Science. 1993;262:1886–1889. doi: 10.1126/science.8266080. [DOI] [PubMed] [Google Scholar]
- 44.Tscherne J S, Nurse K, Popienick P, Michel H, Sochacki M, Ofengand J. Purification, cloning, and characterization of the 16S RNA m5C967 methyltransferase from Escherichia coli. Biochemistry. 1999;38:1884–1892. doi: 10.1021/bi981880l. [DOI] [PubMed] [Google Scholar]
- 45.Tscherne J S, Nurse K, Popienick P, Ofengand J. Purification, cloning, and characterization of the 16 S RNA m2G1207 methyltransferase from Escherichia coli. J Biol Chem. 1999;274:924–929. doi: 10.1074/jbc.274.2.924. [DOI] [PubMed] [Google Scholar]
- 46.Tsu C A, Uhlenbeck O C. Kinetic analysis of the RNA-dependent adenosinetriphosphatase activity of DbpA, an Escherichia coli DEAD protein specific for 23S ribosomal RNA. Biochemistry. 1998;37:16989–16996. doi: 10.1021/bi981837y. [DOI] [PubMed] [Google Scholar]
- 47.Wikström P M, Byström A S, Björk G R. Non-autogenous control of ribosomal protein synthesis from the trmD operon in Escherichia coli. J Mol Biol. 1988;203:141–152. doi: 10.1016/0022-2836(88)90098-8. [DOI] [PubMed] [Google Scholar]