Abstract
Despite routine analysis of a large panel of genes, pathogenic variants are only detected in approximately 20% of families with hereditary breast and/or ovarian cancer. Mobile element insertions (MEI) are known to cause genetic diseases in humans, but remain challenging to detect. Retrospective analysis of targeted next-generation sequencing (NGS) data from 359 patients was performed using a dedicated MEI detection pipeline. We detected one MEI in exon 9 of the PALB2 gene in a woman with a family history of breast cancer. The pathogenic variant, c.2872_2888delins114AluL2, disrupts the PALB2 coding sequence and leads to the production of a truncated protein, p.(Gln958Valfs*38). This is the first report of a pathogenic MEI in PALB2. This study illustrates that MEI analysis may help to improve molecular diagnostic yield and can be performed from targeted NGS data used for routine diagnosis.
Subject terms: Cancer genetics, Next-generation sequencing
Introduction
Hereditary breast and ovarian cancer (HBOC) syndrome is mainly caused by pathogenic variants in the BRCA1 [1] and BRCA2 [2] genes. Several additional genes, such as PALB2 [3], RAD51C [4], and RAD51D [5] are now recognized as susceptibility genes for HBOC, although they are associated with a lower risk of cancer than are BRCA1 and BRCA2. The French Genetic and Cancer Group (GGC)—Unicancer guidelines recommend testing 13 genes (BRCA1, BRCA2, PALB2, TP53, CDH1, PTEN, RAD51C, RAD51D, MLH1, MSH2, MSH6, PMS2, and EPCAM) in HBOC individuals [6]. However, the mutation detection rate remains low, at around 15–20%, even in high-risk families. Identifying pathogenic variants in known responsible genes that are not detected by current screening methods could account for some of the cases that still lack a molecular diagnosis. One potential class of variants is mobile element insertions (MEI). MEI occur when an active mobile element inserts into a new genomic location [7]. MEI could then cause disease by disrupting a gene coding sequence, altering gene regulation, or altering splicing, depending on the insertion site. One example in familial breast cancer is the c.156_157insAlu in exon 3 of BRCA2, which represents 27% of all deleterious BRCA1 and BRCA2 mutations in northern/central Portugal, due to a founder effect [8]. Pathogenic MEI have been shown to be involved in various conditions and are estimated to account for approximately 0.03–0.1% of pathogenic variants responsible for genetic diseases [9]. However, this is likely to be a substantial underestimation, since it is challenging to detect this type of variant using routine molecular tests.
Here we report the detection of a pathogenic Alu insertion in the PALB2 gene by using a dedicated MEI detection pipeline. This variant c.2872_2888delinsGTGTCCCCAATACGTCTAATAAATAGGCCTGCAGGTCTAGAGCTCAAGAAAGAGCTCAGAGGTAGAAATGTATGTTTGCAAGTAGTCAGCACCAGATTTTATATTTCCAGATGA (or c.2872_2888delins114AluL2), was detected in a patient with a strong hereditary predisposition for breast cancer for whom no pathogenic variant had previously been identified by NGS data analysis.
Methods
Patients
All patients included in this study were seen for genetic consultation or are being treated at a specialist center for the disease of interest. Blood samples were sent to our laboratory for molecular analysis because of their clinical presentation and/or family history. All patients provided written informed consent for genetic analysis.
Target capture sequencing method
Libraries were prepared according to the Kapa sample preparation protocol (Kapa Biosystems®). Libraries were pooled and captured using the SeqCap EZ Choice Library (Roche/NimbleGen, Madison, WI) according to the manufacturer’s protocol and sequenced using an Illumina Platform (Illumina, San Diego, CA).
Data analysis
Reads were mapped to hg19 using BWA (0.7.12). Only alignments with at least 10 soft clipped bases were kept. Coverage for all bases was assessed using GATK (3.5) and was used to detect significant differences between two consecutive positions (breakpoint) using an in-house R script (R 3.5.3). The detection step is based on differences in read depth (all reads vs. only soft clipped reads) and depth ratios between the coordinates. Some putative breakpoints are filtered using mismatch ratio and indel ratio parameters, which excludes events that appear in repeat sequences or regions with low mappability. Consensus sequences around breakpoint were predicted using Trinity (2.8.4) from soft clipped reads and their mates. Rebuilt contigs were filtered out based on the read number used for Trinity reconstruction and the CIGAR alignment pattern. Selected sequences were annotated using Blast (2.0.9+) and the RepeatMasker track from UCSC (hits with p value < 0.01). Finally, a depiction of the region was created using the IGV-snapshot-automator script.
Dedicated mobile element insertion polymerase chain reaction and Sanger sequencing
Each detected event was validated by dedicated MEI PCR and Sanger sequencing. Based on the rebuilt sequence generated bioinformatically, we were able to design MEI-specific primers located within or at one breakpoint of the MEI. DNA was PCR-amplified using primers specific for the gene of interest or Alu insertion–specific primers, where one primer (forward or reverse) annealed to the Alu insertion breakpoint or within the Alu insertion, and the second primer was specific for the gene of interest. All amplifications were performed using the following conditions: 95 °C for 10 min, (95 °C for 30 s, 60 °C for 45 s, 72 °C for 1 min) ×40, and 72 °C for 20 min. The PCR products were then subjected to Sanger sequencing analysis using an ABI 3730DNA Analyzer (Life Technologies).
Results
We performed retrospective analysis of targeted NGS data from 362 cases referred to our laboratory for molecular diagnosis of HBOC (n = 197) or angiogenetic diseases (n = 162), and for whom no pathogenic variants (single nucleotide variants or copy number variant) were previously identified. Analysis of MEI was performed using a dedicated pipeline that involves the following key steps: (1) selection of clipped reads; (2) preselection of events based on coverage (depth ratio); (3) a first filtering step based on alignment quality; (4) reconstruction of the inserted sequence; (5) a second filtering step based on the reconstructed sequence; and (6) annotation of the inserted sequence (Supplementary Fig. 1).
Fifteen distinct MEI were detected in 55 samples (Table 1). We also analyzed three samples with known Alu insertions as positive controls (supplementary Table 1). Two samples had the c.156 157insAlu in exon 3 of BRCA2, previously detected by MLPA. The third sample had an Alu insertion within MSH2 exon 8 previously described [10].
Table 1.
New mobile element insertions (MEI) detected in the retrospective cohort (n = 362).
| Event | Gene | Location | Coordinates (hg19) | Annotation | No. of samples | Molecular confirmation | Classification |
|---|---|---|---|---|---|---|---|
| 1 | RASA1 | Intron 8 | chr5:86647920 | L1 | 2 | Yes | VUS |
| 2 | RASA1 | Intron 1 | chr5:86597280 | AluYA5 | 8 | Yes | VUS |
| 3 | RASA1 | Intron 1 | chr5:86604030 | LIME3A | 20 | No | ND |
| 4 | ENG | Intron 2 | chr9:130597114 | AluYA5 | 13 | Yes | VUS |
| 5 | ENG | Intron 2 | chr9:130600053 | – | 2 | Yes | VUS |
| 6 | ACVRL1 | Intron 6 | chr12:52308548 | – | 1 | Yes | VUS |
| 7 | RASA1 | Intron 6 | chr5:86640516 | – | 1 | No | ND |
| 8 | CAV1 | Exon 2 | chr7:116166603 | – | 1 | No | ND |
| 9 | FGFR1 | Exon 6 | chr8:38283711 | MIRc | 1 | No | ND |
| 10 | ENG | Intron 3 | chr9:130591270 | LIME4a | 1 | No | ND |
| 11* | SMAD4 | Exon 1 | chr18:48556623 | LIME3E | 1 | Yes | Benign |
| 12* | SMAD4 | Intron 1 | chr18:48573287 | – | 1 | Yes | Benign |
| 13* | SMAD4 | Intron 11 | chr18:48604625 | – | 1 | Yes | Benign |
| 14 | AXIN2 | Exon 6 | chr17:63533653 | – | 1 | No | ND |
| 15 | PALB2 | Exon 9 | chr16:23634397 | – | 1 | Yes | Pathogenic |
Annotation was carried out by performing a Blast comparison between the rebuilt sequences and Repeatmasker data. * indicates MEI that were detected in the same sample. Molecular confirmation was performed by dedicated MEI PCR followed by Sanger sequencing, except for events 12 and 13, which were confirmed by next-generation sequencing analysis.
VUS variant of unknown significance, ND Not determined.
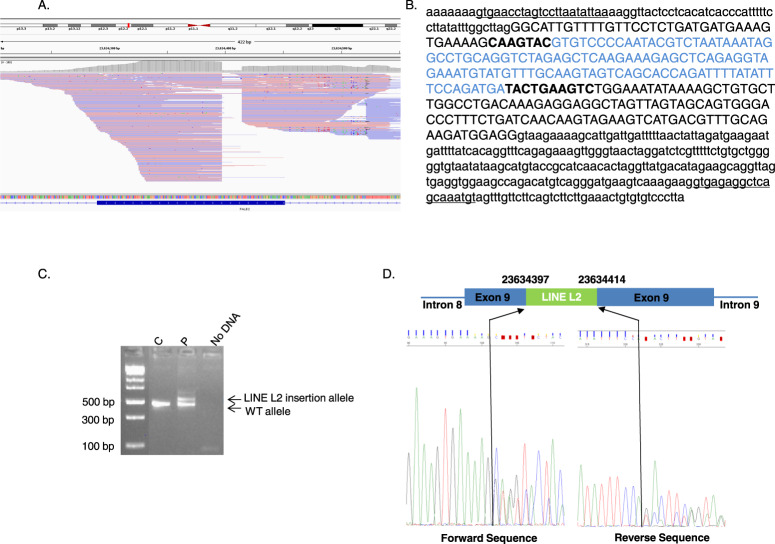
Each event was analyzed by dedicated MEI PCR followed by Sanger sequencing. We molecularly confirmed the presence of nine MEI. According to American College of Medical Genetics and Genomics (ACMG) criteria [11, 12], six events (1, 2, 4, 5, 6, and 15) were classified as variant of unknown significance based on the frequency of their detection and three events (11, 12 and 13) detected on the same sample were classified as benign since these events correspond to the integration of a SMAD4 processed pseudogene [13]. NoPCR amplification of the mutated allele was observed for six MEI whereas wild-type allele amplification was obtained using specific primers for the gene of interest (Supplementary figure 2). This suggests that these events were false positives. One MEI was detected in exon 9 of PALB2 (Fig. 1A). The rebuilt sequence predicted a 114-bp LINE type L2 element insertion associated with deletion of 17 bp of the PALB2 sequence at the insertion site, (Fig. 1B). Molecular analysis confirmed the presence of this insertion. This variant, c.2872_2888delins114AluL2, disrupts the PALB2 coding sequence and leads to the production of a truncated protein, p.(Gln958Valfs*38) (Fig. 1C, D). This variant was classified as pathogenic according to ACMG criteria [11, 12]. This variant was detected in a women diagnosed with breast cancer at 55 years of age. This woman had a familial history of breast cancer: her mother and maternal grandmother had both died from breast cancer at the age of 42 and before the age of 50, respectively. No DNA was available to perform a segregation study.
Fig. 1. Mobile element insertion (MEI) in exon 9 of the PALB2 gene.
A Integrative Genomics Viewer visualization of reads used for bioinformatics detection. B MEI rebuilt sequence generated by bioinformatic analysis. The PALB2 reference sequence (NM_024675.3) is indicated in black, with intronic sequences shown in lowercase letters and exonic sequence shown in uppercase letters. The detected LINE type L2 insertion sequence is indicated in blue. The PALB2 fragment that was deleted at the insertion breakpoints is indicated in bold. The primer sequences used for Sanger sequencing confirmation are underlined. C Gel electrophoresis of PALB2 exon 9 polymerase chain reaction (PCR) products from one control individual (C) and the patient (P) showing an additional band in the P sample compared to the C sample that corresponds to the allele with the insertion. D Schematic representation of the LINE type L2 element inserted into exon 9 of PALB2 and Sanger sequencing of the PCR product shown in C that confirmed the in silico rebuilt sequence.
Discussion
MEI are known to cause genetic disease, but detecting them in the context of routine diagnosis remains a challenge. We used a dedicated MEI detection pipeline to analyze targeted NGS data generated from the routine molecular diagnosis of 359 patients. We were able to identify a pathogenic MEI in exon 9 of PALB2 gene, suggesting that this type of variant may be responsible for cancers in some high-risk families for whom a pathogenic variant has not yet been identified. PALB2 (partner and localizer of BRCA2; also known as FANCN) was originally identified as a BRCA2-interacting protein that mediates BRCA2 recruitment to DNA damage sites and is therefore essential for BRCA2 function in double-strand break repair by homologous recombination [14]. Subsequently, it was also shown to interact with BRCA1, acting as a bridge between these two proteins [15]. PALB2 bi-allelic germline loss-of-function variants cause Fanconi’s anemia [16], whereas mono-allelic loss-of-function variants are associated with an increased risk of breast cancer [17]. The frequency of PALB2 pathogenic variants is variable among different populations, ranging from 0.73% to 3.40%. This is the first report of an MEI in this gene, even though it is known that large rearrangements can occur when initial breakpoints occur near Alu elements, and Alu-related tandem duplications have been reported in PALB2 [18, 19].
This case demonstrates that, even if MEI only account for a small proportion of pathogenic variants in HBOC syndrome, detecting these events could resolve some cases for which no molecular diagnosis was available. Identification of a genetic cause is essential for clinical management and genetic counseling in HBOC families. This study shed in light that MEI detection in routine diagnosis, using a dedicated pipeline that could be easily integrated into current diagnostic pipelines, could improve molecular diagnostic yield and the assessment of the prevalence of MEI in human disease.
Supplementary information
Acknowledgements
We would like to thank Dr. Stéphanie Baert-Desurmont for providing the MSH2 Alu insertion fastq files.
Author contributions
FC supervised the study and contributed to analyzing data, interpreting result, and writing the manuscript. ME designed the experiments and contributed to analyzing data, interpreting result, and writing the manuscript. OA, PDLG conceived and realized bioinformatic analyses and contributed to writing the manuscript. GL performed technical experiments. NB, EG, AP contributed to analyzing data and interpreting results. CD, OC-H collected and analyzed clinical data. All authors reviewed the manuscript.
Funding
No specific funding support this study.
Data availability
All additional data not provided in this article are available from the corresponding authors upon reasonable request. If you are interested in using the MEI detection pipeline, please contact pierre.delagrange@genodiag.com.
Competing interests
The authors declare no competing interests.
Ethical approval
All patients gave their informed consent for genetic research, which was approved by the “Comité de Protection des Personnes Ile de France-VI”, decision ID RCB2007-AO1347-46.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41431-022-01064-3.
References
- 1.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 2.Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–92. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 3.Antoniou AC, Casadei S, Heikkinen T, Barrowdale D, Pylkäs K, Roberts JK, et al. Breast-cancer risk in families with mutations in PALB2. N Engl J Med. 2014;371:497–506. doi: 10.1056/NEJMoa1400382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meindl A, Hellebrand H, Wiek C, Erven V, Wappenschmidt B, Niederacher D, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet. 2010;42:410–14. doi: 10.1038/ng.569. [DOI] [PubMed] [Google Scholar]
- 5.Loveday C, Turnbull C, Ramsay E, Hughes D, Ruark E, Frankum JR, et al. Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat Genet. 2011;43:879–82. doi: 10.1038/ng.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moretta J, Berthet P, Bonadona V, Caron O, Cohen-Haguenauer O, Colas C, et al. The French Genetic and Cancer Consortium guidelines for multigene panel analysis in hereditary breast and ovarian cancer predisposition. Bull Cancer. 2018;105:907–17. doi: 10.1016/j.bulcan.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 7.Kazazian HH, Moran JV. Mobile DNA in health and disease. N Engl J Med. 2017;377:361–70. doi: 10.1056/NEJMra1510092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Machado PM, Brandão RD, Cavaco BM, Eugénio J, Bento S, Nave M, et al. Screening for a BRCA2 rearrangement in high-risk breast/ovarian cancer families: evidence for a founder effect and analysis of the associated phenotypes. J Clin Oncol. 2007;25:2027–34. doi: 10.1200/JCO.2006.06.9443. [DOI] [PubMed] [Google Scholar]
- 9.Hancks DC, Kazazian HH. Roles for retrotransposon insertions in human disease. Mob DNA. 2016;7:9. doi: 10.1186/s13100-016-0065-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baert-Desurmont S, Coutant S, Charbonnier F, Macquere P, Lecoquierre F, Schwartz M, et al. Optimization of the diagnosis of inherited colorectal cancer using NGS and capture of exonic and intronic sequences of panel genes. Eur J Hum Genet. 2018;11:1597–602. doi: 10.1038/s41431-018-0207-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med J Am Coll Med Genet. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amendola LM, Jarvik GP, Leo MC, McLaughlin HM, Akkari Y, Amaral MD, et al. Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the Clinical Sequencing Exploratory Research Consortium. Am J Hum Genet. 2016;98:1067–76. doi: 10.1016/j.ajhg.2016.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watson CM, Camm N, Crinnion LA, Antanaviciute A, Adlard J, Markham AF, et al. Characterization and genomic localization of a SMAD4 processed pseudogene. J Mol Diagn. 2017;6:933–40. doi: 10.1016/j.jmoldx.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–29. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 15.Sy SM-H, Huen MSY, Zhu Y, Chen J. PALB2 regulates recombinational repair through chromatin association and oligomerization. J Biol Chem. 2009;284:18302–10. doi: 10.1074/jbc.M109.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia B, Dorsman JC, Ameziane N, de Vries Y, Rooimans MA, Sheng Q. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–61. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 17.Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–7. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richardson ME, Chong H, Mu W, Conner BR, Hsuan V, Willett S, et al. DNA breakpoint assay reveals a majority of gross duplications occur in tandem reducing VUS classifications in breast cancer predisposition genes. Genet Med J Am Coll Med Genet. 2019;21:683–93. doi: 10.1038/s41436-018-0092-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang C, Arnold AG, Trottier M, Sonoda Y, Abu-Rustum NR, Zivanovic O, et al. Characterization of a novel germline PALB2 duplication in a hereditary breast and ovarian cancer family. Breast Cancer Res Treat. 2016;160:447–56. doi: 10.1007/s10549-016-4021-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All additional data not provided in this article are available from the corresponding authors upon reasonable request. If you are interested in using the MEI detection pipeline, please contact pierre.delagrange@genodiag.com.