Abstract
The pituitary gland, as a nodal component of the endocrine system, is responsible for the regulation of growth, reproduction, metabolism, and homeostasis. Although pituitary formation though the hierarchical action of different transcription factors is well studied in mouse models, there is little evidence of the analogous developmental processes in humans. Herein, we present a female patient with a phenotype that includes blepharoptosis–ptosis–epicanthus syndrome and premature ovarian failure. Clinical exome sequencing revealed two heterozygous variants in two genes, LHX4 (pathogenic) and NR5A1 (VUS) genes and no mutation in FOXL2 gene. We propose a model of genetic interaction between LHX4 and NR5A1 during pituitary and ovarian development that may lead to a similar phenotype mediated by reduced FOXL2 expression.
Subject terms: Pituitary diseases, Gene regulation
Introduction
The pituitary gland is an integral part of the endocrine system for the regulation of growth, reproduction, metabolism, and homeostasis. Although the development of the pituitary gland in mouse models is well characterized, there is little evidence of the analogous developmental processes in humans. In this context, clinical cases with the specific characterization of the genotype–phenotype duality and mode of inheritance may give an insight for underlying interactions among human genes. Herein, we present a patient with premature ovarian failure (POF) and features of blepharophimosis–ptosis–epicanthus syndrome (BPES) (OMIM #110100) harboring two heterozygous variants in two genes, LHX4 and NR5A1. We propose the hypothesis of how genetic interaction between LHX4 and NR5A1 may reduce FOXL2 gene expression and lead to BPES.
Clinical report and genetic analysis
A 16-year-old girl was presented to our department with primary amenorrhea and short stature. Her height was 150.6 cm (<5th percentile, 2 SD below from her target mid-parental height) and her weight 37 kg (<3rd percentile). She had bilateral blepharophimosis, ptosis of the eyelids, and cubitus valgus. Both breast and pubic hair were Tanner III stage without any signs of further pubertal progression. She was the second child of a family of non-consanguineous parents, born full term but small for gestational age.
Her hormonal evaluation revealed hypergonadotropic hypogonadism with normal thyroid and adrenal function (Table 1). Her bone age was delayed by 3 years. A full panel of several autoantibodies was done, which was negative for autoimmunity. A pelvic ultrasound showed a uterus of early-pubertal size and small ovaries (2.5 cm3) containing a few follicles. The anti-Mullerian hormone (AMH) was almost undetectable (0.01 ng/mL). Magnetic resonance imaging of the pituitary gland showed normal morphology and size (6 mm height and 10 mm depth).
Table 1.
Hormonal profile of the 16-year-old female patient.
| TSH = 2.73 mIU/mL (0.270–4.2) | LH = 22.5 U/l (2.4–12.6) |
| FT4 = 1.35 ng/dL (0.8–2) | FSH = 64.4 U/l (3.5–12.5) |
| β-HCG = 0.4 mIU/mL (<5) | E2 = 8.91 pg/mL (26.7–1506) |
| 17-OHP = 0.56 ng/mL (0.1–1.1) | SHBG = 148.5 nmol/l (26.1–110) |
| DHEAS = 162.7 μg/dL (148–407) | ACTH = 50.9 pg/mL (5–60) |
| IGF-1 = 243 ng/mL (183–792) | Δ4 = 1.6 ng/mL (0.47–3.40) |
| AMH = 0.01 ng/mL (1.52–9.95) | Cortisol = 22 μg/dL (6.2–19.4) |
To convert to SI units: testosterone, nmol/L = ng/dL × 0.03467; FT4 pmol/L = pg/mL × 1.29; DHEAS, μmol/L = μg/dL × 0.027; Δ4, nmol/L = ng/mL × 3.479; 17-OHP, nmol/L = ng/mL × 3.026; ACTH, pmol/L = pg/mL × 0.2222; Cortisol, nmoL/L = μg/dL × 27.58; AMH, pmol/L = ng/mL × 7.14; IGF-1, nmol/L = ng/mL × 0.13.
17-OHP 17-OH-progesterone, Δ4 Δ4-androstenedione.
Estrogen replacement therapy with transdermal patches was initiated and during the pubertal induction period, her growth rate was increased substantially from the 3rd percentile to 20th percentile of height trajectory.
Upon taking written and signed informed consent from the patient and her parents, she underwent a full genetic analysis. Her karyotype was normal, while microarray-based copy number screening was normal for FOXL2 gene. Clinical exome sequencing (CES) was carried out employing the Clinical Exome Solution kit (4900 genes) by Sophia Genetics® carried out on an Illumina NextSeq 500. Computational analysis included read alignment to the human genome reference sequence (GRCh37/hg19), base quality score recalibration, indel realignment, and variant calling, using the SOPHiA DDM® platform (Sophia Genetics, SA) as previously described [1]. Variants were annotated with the ANNOVAR algorithm and filtrated using VarAFT v2.16 (http://varaft.eu) application. Variant classification was based on the American College of Medical Genetics and Genomics (ACMG) criteria [2] and evaluated with the in silico analysis tools by the VarSome platform.
Although FOXL2 gene was included in the CES panel employed, it was also Sanger-sequenced. Specifically, the single exon of the FOXL2 gene (NM_023067.4) was PCR amplified and sequenced using the Big Dye Terminator cycle sequencing kit on an ABI 3500 genetic analyzer (Applied Biosystems, CA, USA). FOXL2 gene PCR and sequencing primers are available on request. The CES findings were verified by Sanger-sequencing the coding regions and the intron-exon boundaries of exon 3 of the LHX4 and exon 4 of the NR5A1 genes.
Results
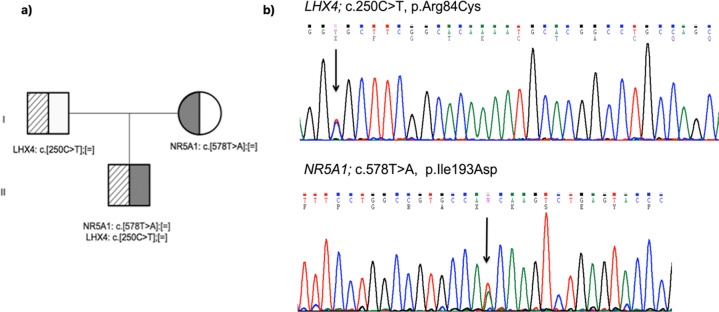
CES achieved 25× coverage of the 99.6% of the targeted sequences and mean coverage rate of 70×. Particularly, 100% of the FOXL2 gene was covered at least 50×. Neither CES, nor Sanger sequencing, found any pathogenic or likely pathogenic variant in FOXL2 gene. CES revealed in our patient two heterozygous variants in two genes; the, p.Arg84Cys in LHX4 gene (NM_033343.3: c.250C>T) and the p.Ile193Asn in NR5A1 gene (NM_004959.5: c.578T>A). These variants were confirmed by Sanger-sequencing patient’s and her parents’ DNA and it was shown that the LHX4 variant was paternal, whereas the NR5A1 variant of maternal inheritance (Fig. 1).
Fig. 1. Genetic analysis of the proband and her parents.
a Pedigree of the patient’s family and b parts of the sequencing chromatograms of LHX4 gene showing the exon 3 c.250C>T, p.Arg84Cys and NR5A1 gene showing the exon 4 c.578T>A, p.Ile193Asn variants detected in our patient.
The LHX4 gene variant p. Arg84Cys, was classified according to the ACMG criteria as Pathogenic (criteria applied: PVS1 very strong, PP3 and PP5 supporting,). Criterion PVS1 has been applied since the LHX4 variant c.250C>T is located within exon 3 acceptor splice site [acag|G(C/T)GC] and it is likely that disturbs normal splicing. It is a very rare variant with gnomAD exomes frequency 0.0000558 and gnomAD genomes frequency 0.0000131. Seventeen in silico tools of the VarSome platform as well as Polyphen 2, categorize this variant as damaging/pathogenic.
The LHX4 gene variant p.Arg84Cys, has been previously described in a male patient with short stature, growth hormone deficiency and hypothyroidism [3]. This variant lies within the zinc-binding 1 region of the LHX4 protein, impairing activity on CGA, POU1F1 and TSHB promoters. Interestingly, our patient’s father had a normal puberty and attained a normal final height.
The NR5A1 gene variant p.Ile193Asn is novel and it was classified according to the ACMG criteria as variant of unknown significance (criteria applied: PM2 moderate and PP2 and PP3 supporting). It is a highly conserved variant, not present in gnomAD exomes or gnomAD genomes databases. Her 56-year-old mother was a carrier of the NR5A1 gene variant, p.Ile193Asn, with menarche at the age of 14 years and no history of POF. A specific variant at the adjacent position (p.Arg192Phe), located at the hinge region, results in a weakly conserved protein with predicted intermediate pathogenicity [4]. Eighteen in silico tools of the VarSome platform as well as Polyphen 2, categorize this variant as damaging/pathogenic.
Discussion
Herein, we present a case of a girl with BPES phenotype, POF, and short stature. Because short stature is not a typical characteristic of the BPES phenotype we performed CES in addition to single FOXL2 gene sequencing. No intragenic variants, deletions or cytogenetic rearrangements of FOXL2, accountable for the BPES [5] were found, but CES revealed two heterozygous variants, each in LHX4 and NR5A1 genes, indicating digenic inheritance.
Based on the existent data, we propose, within the functional framework of the LHX4 and NR5A1 genes, a model for the explanation of our patient’s clinical phenotype. LHX3 and LHX4 proteins belong to the LIM-homeodomain transcription factors and play essential roles in the pituitary gland and nervous system development [6]. LHX3 binds to the promoter region of the alpha glycoprotein subunit (aGSU) and the beta subunits of FSH, TSH, and Pit-1 genes [7, 8]. This multitargeting action is critical, for both the early pituitary gland formation and the development of lactotrope, somatotrope, gonadotrope, and thyrotrope cell lineages. A recent study, using single-cell RNA sequencing from human fetal pituitaries, assigned LHX3 a major role in the early phase of pituitary developmental patterning [9]. The structurally related LHX4 protein is required for the proper expression of LHX3 [6]. Gene targeting studies in mice have elucidated the overlapping, but distinct roles of LHX3 and LHX4 in development with the LHX3 being essential for the emergence of all differentiated cell types in Rathke’s pouch [10]. Recently, six disease-causing variants of LHX4 gene in a heterozygous state were reported, in a large cohort of 417 unrelated patients with isolated growth hormone deficiency or combined pituitary hormone deficiency [11].
Steroidogenic factor-1 (SF-1 or NR5A1) is an orphan nuclear receptor that has multiple functions in steroidogenesis, sexual differentiation, and reproduction. Variants in NR5A1 gene have been found in a small percentage of women with POF, while many female carriers show no phenotype, indicating varying degrees of expression and incomplete penetrance [12]. At the hypophyseal level, PTX1 (Pituitary Homeobox-1) regulates the LHX3 gene expression [13] and this regulation is affected by NR5A1 and POU1F1 transcription factors.
Our patient’s clinical phenotype, described as BPES with or without POF [14], is usually encountered in individuals with FOXL2 gene mutations. FOXL2 is expressed in the ovary, the pituitary gland and the eye during early development [15] and requires the transcription factors LHX3 and LHX4 [16]. Recently, in a large cohort of patients with BPES only 67% harbored pathogenic FOXL2 variants [17] and some of them had coexistent hypopituitarism, supporting FOXL2 involvement in human pituitary development [18]. In addition, data from single-cell RNA sequencing from human fetal pituitaries indicate that FOXL2 acts with NR5A1 for the gonadotrope cell lineage development.
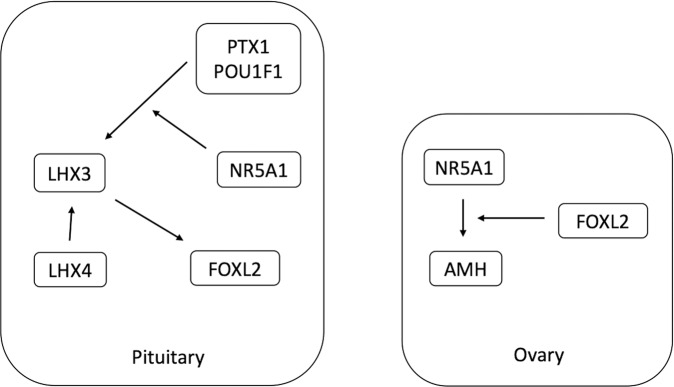
Based on our findings, we postulate that the additive effect of heterozygous variants in LHX4 and NR5A1, due to digenic inheritance, may lead to a BPES type I-like phenotype, by acting on the FOXL2 expression levels during development (Fig. 2). Firstly, the transcriptional balance of LHX3 and LHX4 is affected by the NR5A1 transcription levels because NR5A1 is needed for PITX1 and POU1F1 to regulate LHX3 gene expression [13]. Normal FOXL2 expression requires the transcription factor LHX3, because it is an upstream activator of FOXL2 [16]. LHX4 is indispensable for the proper expression of LHX3 and inductively affects FOXL2. These interactions may explain the BPES phenotype of our patient. Focusing on the ovaries, the inter-regulatory network of AMH, FOXL2, and NR5A1 is essential for developing the ovarian reserve during development. Experimental evidence supports that AMH is a target gene of NR5A1 and FOXL2 acts as an important factor in this transcriptional regulation [19]. Consequently, low AMH expression during ovarian development due to reduced drive by NR5A1 and FOXL2 may lead to POF (Fig. 2). The limitation of our study is the lack of functional studies on FOXL2 interaction with NR5A1 and LHX4 variants. However, we think that developmental tissue-specific and spatiotemporal gene interactions cannot be easily captured by in vitro cellular systems or the more informative transgenic mouse models due to the increased redundancy of gene networks. With an integrative approach of biological and computational data [20], we consider our patient’s phenotype to be the result of a natural gene perturbation experiment that provides valuable biologic data concerning the genes LHX4 and NR5A1 in the development of pituitary and ovary.
Fig. 2. Part of networks depicting gene interactions among LHX4, NR5A1, and FOXL2 in pituitary and ovary during development. Pituitary: NR5A1 co-regulates with PITX1 and POU1F1 the transcription of LHX3.
FOXL2 transcription needs LHX3 and is enhanced by LHX4 Ovary: SF-1 transactivates ovarian AMH in a FOXL2-dependent manner and FOXL2 is required for the binding of SF-1 to the AMH promoter.
In conclusion, we present a patient with the phenotype of BPES type I syndrome and heterozygous variants in LHX4 and NR5A1 genes. We propose a model of interaction between LHX4 and NR5A1 during pituitary and ovarian development that results to this phenotype by reduced FOXL2 expression.
Author contributions
AG conceived and drafted the manuscript. AS performed the genetic analysis and AS and DC critically revised the manuscript.
Data availability
The datasets generated during and/or analyzed during the current study are available in the LOVD v.3 repository https://www.lovd.nl/3.0/home.
Competing interests
The authors declare no competing interests.
Ethical approval
The study is approved by the ethics committee of the University Hospital of Patras.
Consent for publication
Full informed consent was obtained from the patient and her parents to publish the results of phenotyping and genetic studies.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Marinakis NM, Svingou M, Veltra D, Kekou K, Sofocleous C, Tilemis F-N, et al. Phenotype-driven variant filtration strategy in exome sequencing toward a high diagnostic yield and identification of 85 novel variants in 400 patients with rare Mendelian disorders. Am J Med Genet Part A. 2021;185:2561–71. doi: 10.1002/ajmg.a.62338. [DOI] [PubMed] [Google Scholar]
- 2.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pfaeffle RW, Hunter CS, Savage JJ, Duran-Prado M, Mullen RD, Neeb ZP, et al. Three novel missense mutations within the LHX4 gene are associated with variable pituitary hormone deficiencies. J Clin Endocrinol Metab. 2008;93:1062–71. doi: 10.1210/jc.2007-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janse F, De With LM, Duran KJ, Kloosterman WP, Goverde AJ, Lambalk CB, et al. Limited contribution of NR5A1 (SF-1) mutations in women with primary ovarian insufficiency (POI). Fertil Steril. 2012;97:141–6.e2. [DOI] [PubMed]
- 5.Beysen D, De Paepe A, De Baere E. FOXL2 mutations and genomic rearrangements in BPES. Hum Mutat. 2009;30:158–69. doi: 10.1002/humu.20807. [DOI] [PubMed] [Google Scholar]
- 6.Mullen RD, Colvin SC, Hunter CS, Savage JJ, Walvoord EC, Bhangoo APS, et al. Roles of the LHX3 and LHX4 LIM-homeodomain factors in pituitary development. Mol Cell Endocrinol. 2007;265-6:190–5. doi: 10.1016/j.mce.2006.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sloop KW, Meier BC, Bridwell JAL, Parker GE, Schiller AMC, Rhodes SJ. Differential activation of pituitary hormone genes by human LHx3 isoforms with distinct DNA binding properties. Mol Endocrinol. 1999;13:2212–25. doi: 10.1210/mend.13.12.0395. [DOI] [PubMed] [Google Scholar]
- 8.Bach I, Rhodes SJ, Pearse RV, Heinzel T, Gloss B, Scully KM, et al. P-Lim, a LIM homeodomain factor, is expressed during pituitary organ and cell commitment and synergizes with Pit-1. Proc Natl Acad Sci USA. 1995;92:2720–4. doi: 10.1073/pnas.92.7.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang S, Cui Y, Ma X, Yong J, Yan L, Yang M, et al. Single-cell transcriptomics identifies divergent developmental lineage trajectories during human pituitary development. Nat Commun. 2020;11:5275. [DOI] [PMC free article] [PubMed]
- 10.Sheng HZ, Moriyama K, Yamashita T, Li H, Potter SS, Mahon KA, et al. Multistep control of pituitary organogenesis. Science. 1997;278:1809–12. doi: 10.1126/science.278.5344.1809. [DOI] [PubMed] [Google Scholar]
- 11.Cohen E, Maghnie M, Collot N, Leger J, Dastot F, Polak M, et al. Contribution of LHX4 mutations to pituitary deficits in a cohort of 417 unrelated patients. J Clin Endocrinol Metab. 2017;102:290–301. doi: 10.1210/jc.2016-3158. [DOI] [PubMed] [Google Scholar]
- 12.Philibert P, Paris F, Lakhal B, Audran F, Gaspari L, Saâd A, et al. NR5A1 (SF-1) gene variants in a group of 26 young women with XX primary ovarian insufficiency. Fertil Steril. 2013;99:484–9. doi: 10.1016/j.fertnstert.2012.10.026. [DOI] [PubMed] [Google Scholar]
- 13.Tremblay JJ, Lanctôt C, Drouin J. The pan-pituitary activator of transcription, Ptx1 (Pituitary Homeobox 1), acts in synergy with SF-1 and Pit1 and is an upstream regulator of the LIM-homeodomain gene Lim3/Lhx3. Mol Endocrinol. 1998;12:428–41. doi: 10.1210/mend.12.3.0073. [DOI] [PubMed] [Google Scholar]
- 14.Crisponi L, Deiana M, Loi A, Chiappe F, Uda M, Amati P, et al. The putative forkhead transcription factor FOXL2 is mutated in blepharophimosis/ptosis/epicanthus inversus syndrome. Nat Genet. 2001;27:159–66. doi: 10.1038/84781. [DOI] [PubMed] [Google Scholar]
- 15.Cocquet J, Pailhoux E, Jaubert F, Servel N, Xia X, Pannetier M, et al. Evolution and expression of FOXL2 [1] J Med Genet. 2002;39:916–21. doi: 10.1136/jmg.39.12.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellsworth BS, Egashira N, Haller JL, Butts DL, Cocquet J, Clay CM, et al. FOXL2 in the pituitary: molecular, genetic, and developmental analysis. Mol Endocrinol. 2006;20:2796–805. doi: 10.1210/me.2005-0303. [DOI] [PubMed] [Google Scholar]
- 17.Bunyan DJ, Thomas NS. Screening of a large cohort of blepharophimosis, ptosis, and epicanthus inversus syndrome patients reveals a very strong paternal inheritance bias and a wide spectrum of novel FOXL2 mutations. Eur J Med Genet. 2019;62:103668. doi: 10.1016/j.ejmg.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 18.Castets S, Roucher-Boulez F, Saveanu A, Mallet-Motak D, Chabre O, Amati-Bonneau P, et al. Hypopituitarism in patients with blepharophimosis and FOXL2 mutations. Horm Res Paediatr. 2020;93:30–9. [DOI] [PubMed]
- 19.Jin H, Won M, Park SE, Lee S, Park M, Bae J. FOXL2 is an essential activator of SF-1-induced transcriptional regulation of anti-Müllerian hormone in human granulosa cells. PLoS One. 2016;11:e0159112. [DOI] [PMC free article] [PubMed]
- 20.Fernandez-Valverde SL, Aguilera F, Ramos-Díaz RA. Inference of developmental gene regulatory networks beyond classical model systems: new approaches in the post-genomic era. Integr Comp Biol. 2018;58:640–53.. doi: 10.1093/icb/icy061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available in the LOVD v.3 repository https://www.lovd.nl/3.0/home.