Abstract
The current Dutch Pharmacogenetics Working Group (DPWG) guideline, describes the gene–drug interaction between CYP2D6 and the opioids codeine, tramadol and oxycodone. CYP2D6 genotype is translated into normal metaboliser (NM), intermediate metaboliser (IM), poor metaboliser (PM) or ultra-rapid metaboliser (UM). Codeine is contraindicated in UM adults if doses >20 mg every 6 h (q6h), in children ≥12 years if doses >10 mg q6h, or with additional risk factors. In PMs, an alternative analgesic should be given which is not or to a lesser extent metabolised by CYP2D6 (not tramadol). In IMs with insufficient analgesia, a higher dose or alternative analgesic should be given. For tramadol, the recommendations for IMs and PMs are the same as the recommendation for codeine and IMs. UMs should receive an alternative drug not or to a lesser extent metabolised by CYP2D6 or the dose should be decreased to 40% of the commonly prescribed dose. Due to the absence of effect on clinical outcomes of oxycodone in PMs, IMs and UMs no action is required. DPWG classifies CYP2D6 genotyping for codeine “beneficial” and recommends testing prior to, or shortly after initiation of treatment in case of higher doses or additional risk factors. CYP2D6 genotyping is classified as “potentially beneficial” for tramadol and can be considered on an individual patient basis.
Subject terms: Pharmacogenetics, Personalized medicine
Introduction
The role of heritable genetic variation on drug response is referred to as pharmacogenetics (PGx). Personalised medicine, also known as precision medicine, can be achieved with the use of PGx information. Knowledge of an individual’s genetic composition for drug metabolising enzymes, drug transporters, receptors or effector proteins may be used to guide pharmacological treatment. To establish the use of PGx in a clinical setting, guidelines informing physicians are essential. In order to resolve this, the Royal Dutch Pharmacists Association (KNMP) has appointed the Dutch Pharmacogenetics Working Group (DPWG) in 2005, a group of 15 professionals consisting of (clinical) pharmacists, physicians, general practitioners, clinical pharmacologists, clinical chemists and epidemiologists [1]. The role of DPWG is to develop evidence-based PGx-guided therapeutic recommendations based on systematic literature review and to implement these into computerised systems used nationwide for medication prescription, dispensing and monitoring. In order to meet the public request for this information also outside the Dutch pharmacist and physician systems, the DPWG guidelines and future updates will be published [2]. In the current guideline the gene–drug interaction between CYP2D6 and the opioids codeine, tramadol and oxycodone is presented. The pharmacotherapeutic rational for use of these drugs as well as the cost-effectiveness of PGx-guided dosing is outside the scope of this guideline. This manuscript provides information on the development of this guideline and presents an overview of the PGx therapeutic recommendations. Firstly, background information of opioids and of the CYP2D6 gene and its genetic variation is provided. This genetic information is followed by the evidence from the literature on the gene–drug interaction between CYP2D6 with the previously mentioned opioids. Finally, the therapeutic recommendations for the clinic (see the corresponding table) and clinical decision support systems are provided. These DPWG PGx-guided recommendations are also compared to other international guidelines. The goal of these DPWG recommendations is to optimise the choice of and treatment with these opioids, thereby reducing the occurrence of toxicity and insufficient analgesia.
Drugs: opioids (codeine, tramadol and oxycodone)
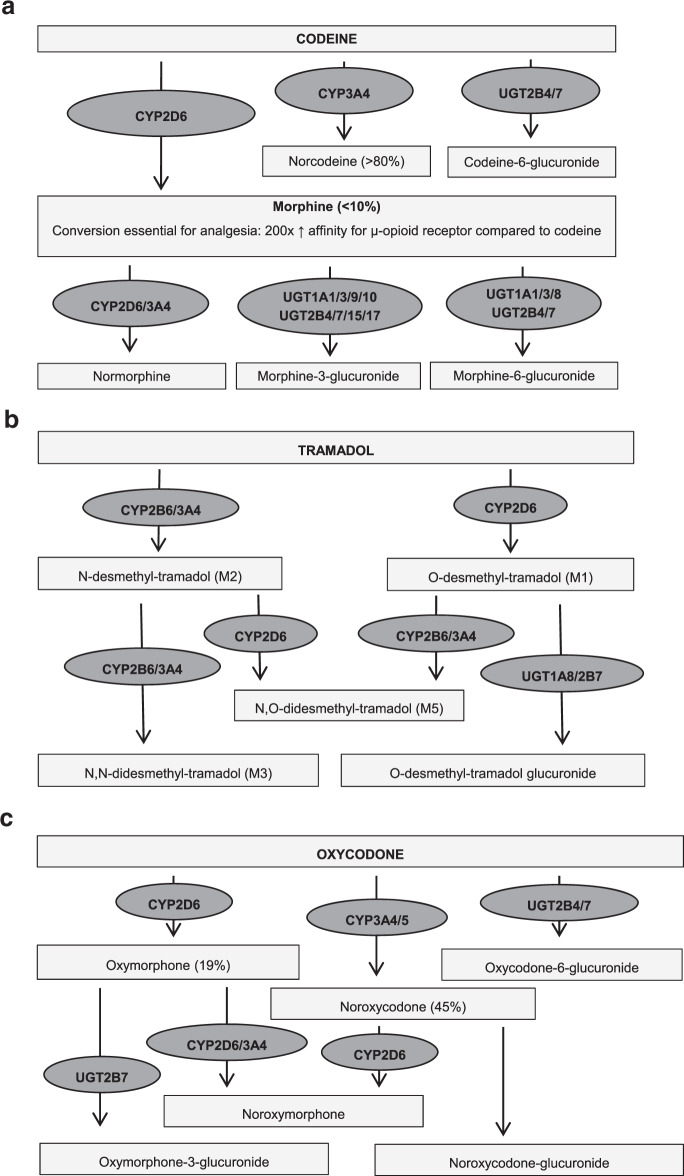
The opioids codeine and tramadol are prescribed for mild to moderate pain, in line with the WHO analgesic ladder. These compounds are typically used in combination with paracetamol or NSAIDs [3]. In addition to the analgesic potency, codeine is also prescribed for suppressing dry or painful cough. Both opioids are contraindicated in children below 18 years undergoing tonsillectomy and/or adenoidectomy for obstructive sleep apnoea syndrome due to the risk of life-threatening toxicity and in children with breathing problems in general [4]. Oxycodone is prescribed for moderate to severe pain. When used for postoperative pain, it is often used in combination with paracetamol. Oxycodone is always started at a low dose and is increased until analgesia is sufficient [3]. Codeine is metabolised by CYP2D6 into the active metabolite morphine through O-demethylation (10%), and by CYP3A4 to norcodeine and by UGT2B7 into codeine-6-glucuronide (50–70%). Morphine is further converted by UGT2B7 and UGT1A1 into morphine-3-glucuronide and morphine-6-glucuronide. The conversion of codeine into morphine is essential for the analgesic effect since the affinity of morphine for the µ-opioid receptor is 200× higher as compared to codeine [5, 6]. Tramadol is metabolised into two major metabolites, O-desmethyltramadol (M1) and N-desmethyltramadol (M2) by CYP2D6 and CYP3A4/CYP2B6, respectively. The conversion of tramadol into the active (+)-metabolite M1 leads to 700× higher affinity in vitro for μ-opioid receptor [5, 7]. Oxycodone is predominantly metabolised by CYP3A4 to the inactive metabolite noroxycodone (45%) and to a lesser extent by CYP2D6 to the active metabolite oxymorphone (19%). The remaining part of oxycodone is metabolised by primarily UGT2B7 and UGT2B4. Oxymorphone has a 30–40-fold stronger analgesic potency as compared to oxycodone. However, due to low brain concentrations, the contribution of the oxycodone metabolites to total analgesia seems very limited [5, 8]. For a schematic overview of codeine, tramadol and oxycodone metabolism see Fig. 1A–C.
Fig. 1. Metabolism opioids.
Schematic overview of codeine (a), tramadol (b) and oxycodone (c) metabolism [5–8]. CYP Cytochrome P450 isoenzyme; UGT UDP-glucuronosyltransferase isoenzyme.
Gene: Cytochrome P450 2D6 (CYP2D6)
CYP2D6 encodes the Cytochrome P450 family 2 subfamily D member 6 enzyme. CYP2D6 is located on chromosome 22q13.2. Transcription variant 1 (NC_000022.11) has 9 exons [9]. CYP2D6 is a highly polymorphic gene and contains 1536 variants [10], from which over 140 CYP2D6 alleles and 250 sub-alleles have been identified. An inactive enzyme activity in vivo has been found for, i.e. *3, *4, *5, *6, *7, *8, *11, *12, *13, *15, *18, *19, *20, *21, *31, *36, *38, *40, *42 and *114. Reduced activity in vivo was found for, i.e. *9, *10, *14, *17, *29 and *41. The activity of other *-alleles is either unknown or normal [11]. The prevalence of the majority of these alleles is low (<0.1%). Several duplications of these alleles (including wild-type allele *1) have been described. Another variation that adds to the complexity of the CYP2D6 gene is the occurrence of hybrid alleles with the pseudogene CYP2D7 [11]. Table 1 gives an overview of the most frequently occurring alleles translated to gene dose (gene activity score) and metabolic capacity. Also the HGVS nomenclature of these alleles is depicted in Table 1. The translation of genotype to phenotype and the corresponding gene dose (gene activity score) is depicted in Table 2. Major ethnic diversity exists in the frequency of the CYP2D6 alleles. The most common inactive allele in the European population is the *4-allele (frequency 16%). In the East Asian population, the decreased activity allele *10 is common (frequency 59%) and in the African population the *17-allele and *29-allele (frequency 9% and 20%). Gene duplications are mostly found in African populations (e.g. Ethiopia 29%) but are less frequent in Caucasians and Asians (1–2%) [12].
Table 1.
CYP2D6 alleles stratified by gene dose and predicted metabolic capacity.
| Metabolic capacity | Gene dose (gene activity score) | Allele number# |
|---|---|---|
| Increased functionality | ≥2 |
*1 duplication (2–13×) *2 duplication (2–13×) *35 duplication |
| Fully functional | 1 |
*1 (=wild-type, wt) *2 *33 *35 *39 |
| Reduced functionality | 0.5 |
*9 (also duplication) *10 (also duplication) *14 (previously 14B) *17 (also duplication) *29 (also duplication) *41 (also duplication) |
| Fully dysfunctional (null alleles) | 0 |
*3 through *8 (*3, *4 and *6 also duplication) *11 through *13 *15 *18 through *21 *31 *36 (also duplication) *38 *40 *42 *114 (previously *14A) |
Many variations exist for CYP2D6, more than 140 different allele variations have been identified/described in the literature [https://www.pharmvar.org/gene/CYP2D6; December 2020]. A number of these variations, including their functionality, are listed in Table 1. Genotyping usually screens for only the most common variant alleles. As a result, the reported genotype can differ from the actual genotype.
# The *-alleles mentioned in the table above are characterised by the following sequence variations:
*1: defined as the allele without variations affecting enzyme activity (in clinical practice as the allele without any of the determined variations).
*2: rs-numbers: rs16947 and rs1135840; NM_000106.6: c.[886C>T; 1457G>C]; NP_000097.3: p.(Arg296Cys; Ser486Thr); NC_000022.11: g.[42127941G>A; 42126611C>G].
*3: rs-number: rs35742686; NM_000106.6: c.775del; NP_000097.3: p.(Arg259fs); NC_000022.11: g.42128242del.
*4: rs-number: rs3892097; NG_008376.3(NM_000106.6): c.506-1G>A; protein sequence not available; NC_000022.11: g.42128945C>T.
*5: CYP2D6 full gene deletion.
*6: rs-number: rs5030655; NM_000106.6: c.454del; NP_000097.3: p.(Trp152fs); NC_000022.11: g.42129084del.
*7: rs-number: rs5030867; NM_000106.6: c.971A>C; NP_000097.3: p.(His324Pro); NC_000022.11: g.42127856T>G.
*8: rs-numbers: rs5030865, rs16947 and rs1135840; NM_000106.6: c.[505G>T; 886C>T; 1457G>C]; NP_000097.3: p.(Gly169Ter; Arg296Cys; Ser486Thr); NC_000022.11: g.[42129033C>A; 42127941G>A; 42126611C>G].
*9: rs-number: rs5030656; NM_000106.6: c.841_843del; NP_000097.3: p.(Lys281del); NC_000022.11: g.42128176_4218178del.
*10: rs-number: rs1065852 and rs1135840; NM_000106.6: c.[100C>T; 1457G>C]; NP_000097.3: p.(Pro34Ser; Ser486Thr); NC_000022.11: g.[42130692G>A; 42126611C>G].
*11: rs201377835, rs16947 and rs1135840; NG_008376.3(NM_000106.6): c.[181-1G>C; 886C>T; 1457G>C]; protein sequence not available; NC_000022.11: g.[42129910C>G; 42127941G>A; 42126611C>G].
*12: rs5030862, rs16947 and rs1135840; NM_000106.6: c.[124G>A; 886C>T; 1457G>C]; NP_000097.3: p.(Gly42Arg; Arg296Cys; Ser486Thr); NC_000022.11: g.[42130668C>T; 42127941G>A; 42126611C>G].
*13: CYP2D7/CYP2D6 hybrid gene.
*14: rs-numbers: rs5030865, rs16947 and rs1135840; NM_000106.6: c.[505G>A; 886C>T; 1457G>C]; NP_000097.3: p.(Gly169Arg; Arg296Cys; Ser486Thr); NC_000022.11: g.[42129033C>T; 42127941G>A; 42126611C>G].
*15: rs-number: rs774671100; NM_000106.6: c.137dup; NP_000097.3: p.(Leu47fs); NC_000022.11: g.42130655dup.
*17: rs28371706, rs16947 and rs1135840; NM_000106.6: c.[320C>T; 886C>T; 1457G>C]; NP_000097.3: p.(Thr107Ile; Arg296Cys; Ser486Thr); NC_000022.11: g.[42129770G>A; 42127941G>A; 42126611C>G].
*18: NM_000106.6: c. 1410_1411insGTGCCCACT; NP_000097.3: p.(Thr470_Gly471insValProThr); NC_000022.11: g.42126657_42126658insAGTGGGCAC.
*19: rs72549353, rs16947 and rs1135840; NM_000106.6: c.[765_768del; 886C>T; 1457G>C]; NP_000097.3: p.(Thr256fs); NC_000022.11: g.[42128251_42128254del; 42127941G>A; 42126611C>G].
*20: rs72549354, rs199535154, rs16947 and rs1135840; NM_000106.6: c.[635dup; 638T>C; 886C>T; 1457G>C]; NP_000097.3: p.(Leu213fs); NC_000022.11: g.[42128817dup; 42128812A>G; 42127941G>A; 42126611C>G].
*21: rs72549352, rs16947 and rs1135840; NM_000106.6: c.[805dup; 886C>T; 1457G>C]; NP_000097.3: p.(Arg269fs); NC_000022.11: g.[42128218dup; 42127941G>A; 42126611C>G].
*29: rs61736512, rs1058164, rs16947, rs59421388 and rs1135840; NM_000106.6: c.[406G>A; 408G>C; 886C>T; 1012G>A; 1457G>C]; NP_000097.3: p.(Val136Ile; Arg296Cys; Val338Met; Ser486Thr); NC_000022.11: g.[42129132C>T; 42129130C>G; 42127941G>A; 42127608C>T; 42126611C>G].
*31: rs-numbers: rs16947, rs267608319 and rs1135840; NM_000106.6: c.[886C>T; 1319G>A; 1457G>C]; NP_000097.3: p.(Arg296Cys; Arg440His; Ser486Thr); NC_000022.11: g.[42127941G>A; 42126749C>T; 42126611C>G].
*33: rs-number: rs28371717; NM_000106.6: c.709G>T; NP_000097.3: p.(Ala237Ser); NC_000022.11: g.42128308C>A.
*35: rs769258, rs16947 and rs1135840; NM_000106.6: c.[31G>A; 886C>T; 1457G>C]; NP_000097.3: p.(Val11Met; Arg296Cys; Ser486Thr); NC_000022.11: g.[42130761C>T; 42127941G>A; 42126611C>G].
*36: rs-number: rs1065852, rs1135833, rs1135835, rs28371735, rs766507177, rs74478221, rs75467367 and rs1135840; NM_000106.6: c.[100C>T; 1405C>G; 1408A>G; 1432C>T; 1433A>C; 1444G>A; 1445C>G; 1457G>C]; NP_000097.3: p.(Pro34Ser; Pro469Ala; Thr470Ala; His478Ser; Ala482Ser; Ser486Thr); NC_000022.11: g.[42130692G>A; 42126663G>C; 42126660T>C; 42126636G>A; 42126635T>G; 42126624C>T; 42126623G>C; 42126611C>G].
*38: rs-number: rs72549351; NM_000106.6: c.811_814del; NP_000097.3: p.(Thr272fs); NC_000022.11: g.42128199_42128202del.
*39: rs-number: rs1135840; NM_000106.6: c.1457G>C; NP_000097.3: p.(Ser486Thr); NC_000022.11: g.42126611C>G.
*40: rs-numbers: rs28371706, rs72549356, rs16947 and rs1135840; NM_000106.6: c.[320C>T; 522_523insTTTCGCCCCTTTCGCCCC; 886C>T; 1457G>C]; NP_000097.3: p.(Thr107Ile; Pro174_Asn175insPheArgProPheArgPro; Arg296Cys; Ser486Thr); NC_000022.11: g.[42129770G>A; 42127941G>A; 42128927_42128928insGGGGCGAAAGGGGCGAAA; 42126611C>G].
*41: rs-numbers: rs16947, rs28371725 and rs1135840;; NG_008376.3(NM_000106.6): c.[886C>T; 985+39G>A; 1457G>C]; NP_000097.3: p.(Arg296Cys; protein not available; Ser486Thr); NC_000022.11: g.[42127941G>A; 42127803C>T; 42126611C>G].
*42: rs-numbers: rs16947, rs72549346 and rs1135840; NM_000106.6: c.[886C>T; 1088_1089dup; 1457G>C]; NP_000097.3: p.(Arg296Cys; Gln364fs); NC_000022.11: g.[42127941G>A; 42127532_42127533dup; 42126611C>G].
*114: rs-numbers: rs1065852, rs5030865, rs16947 and rs1135840; NM_000106.6: c.[100C>T; 505G>A; 886C>T; 1457G>C]; NP_000097.3: p.(Pro34Ser; Gly169Arg; Arg296Cys; Ser486Thr); NC_000022.11: g.[42130692G>A; 42129033C>T; 42127941G>A; 42126611C>G].
Table 2.
The translation of genotypes to gene dose and predicted phenotypes.
| Patient genotype (given as functionality of both alleles) | Gene dose | Phenotype |
|---|---|---|
| Fully functional/fully functional | 2 | NM |
| Fully functional/reduced functionality | 1.5 | NM |
| Fully functional/inactive | 1 | IM |
| Reduced functionality/reduced functionality | 1 | IM |
| Reduced functionality/inactive | 0.5 | IM |
| Inactive/inactive | 0 | PM |
| (Fully functional/fully functional)+multiplication | ≥3 | UM |
| (Fully functional/reduced functionality)+duplication | 2.5 or 2 | NM |
| (Fully functional/reduced functionality)+multiplication reduced functional (3 alleles) | 2.5 | NM |
| (Fully functional/reduced functionality)+multiplication fully functional (≥3 alleles) | ≥3.5 | UM |
| (Fully functional/reduced functionality)+multiplication reduced functional (≥4 alleles) | ≥3 | UM |
| (Fully functional/inactive)+duplication fully functional allele | 2 | NM |
| (Fully functional/inactive)+multiplication fully functional allele (≥3 alleles) | ≥3 | UM |
| (Fully functional/inactive)+multiplication inactive allele | 1 | IM |
| (Reduced functionality/reduced functionality)+multiplication (resulting in 3–5 alleles in total) | 1.5–2.5 | NM |
| (Reduced functionality/reduced functionality)+multiplication (resulting in ≥6 alleles in total) | ≥3 | UM |
| (Reduced functionality/inactive)+multiplication inactive allele | 0.5 | IM |
| (Reduced functionality/inactive)+duplication reduced functional allele | 1 | IM |
| (Reduced functionality/inactive)+multiplication reduced functional allele (3–5 alleles) | 1.5–2.5 | NM |
| (Reduced functionality/inactive)+multiplication reduced functional allele (≥6 alleles) | ≥3 | UM |
| (Inactive/inactive) + multiplication | 0 | PM |
IM intermediate metaboliser, NM normal metaboliser, PM poor metaboliser, UM ultra-rapid metaboliser.
Translation of genotype to phenotype
Based on the CYP2D6 genotype, patients can be translated into four predicted phenotypes: normal metaboliser (NM), intermediate metaboliser (IM), poor metaboliser (PM) or ultra-rapid metaboliser (UM). The different predicted phenotypes are defined based on the total gene dose or gene activity score of the genotype, with the gene dose of a fully functional allele being 1, the gene dose of a reduced functional allele being 0.5 and the gene dose of an inactive allele being 0. NM is defined as having a gene dose of 1.5 through 2.5. IM is defined as having a gene dose of 0.5 or 1. PM is defined as having a gene dose of 0. UM is defined as having a gene dose ≥3. As previously mentioned the translation from genotype to phenotype is illustrated in Table 2. An extensive genotype to predicted phenotype translation can be found in Supplementary Table 1, which can be used to programme the translation of genotype results into predicted phenotypes in laboratory information systems.
Additional phenotyping test
Besides a CYP2D6 genotyping test, it is also possible to perform a CYP2D6 phenotyping test. A phenotyping test measures the actual CYP2D6 activity, in contrast with genotyping. This test is performed by administration of exogenous compounds exclusively metabolised via CYP2D6, such debrisoquine, metoprolol, dextromethorphan and sparteine [13]. The ratio between the parent compound and metabolite is indicative of the activity of the enzyme. Phenotyping tests are currently mainly performed by the pharmaceutical industry or in a research setting as in clinical practice safety issues (e.g. dosage errors, unexpected reactions) and practical reasons (e.g. delay between administration and sampling, not possible in vulnerable populations) hamper its use [14].
Gene-drug interaction
Pharmacological mechanism
CYP2D6 is essential for the analgesic effect of codeine and beneficial for the analgesic effect of tramadol and oxycodone since it is involved in metabolic activation. This enzyme is primarily expressed in the liver but is also found in other tissues such as the duodenum and small intestine [9]. CYP2D6 IM and PM groups are associated with decreased plasma concentrations of the active metabolite of codeine and the most active metabolites of tramadol and oxycodone. Theoretically, a less potent analgesic effect is expected with lower plasma concentrations of these active metabolites. On the contrary, in patients with gene duplications (CYP2D6 UM) higher levels of these active metabolites are observed, which are linked to an increased risk of adverse events. For the indication of cough, codeine instead of morphine is the main active substance. So, CYP2D6 activity is predicted to play a minor role in the effectiveness of codeine in the treatment of cough.
Supporting body of evidence
A detailed description of the methods used for literature collection, assessment and preparation of the gene–drug monograph has previously been published [1]. In brief, a systematic review of literature was performed and relevant articles were summarised by a scientist of the KNMP. The summary and scores for quality of evidence and for the clinical impact of each article were checked by two independent DPWG members and subsequently discussed with the entire group. The quality of evidence was scored on a 5-point scale ranging from 0 (lowest) to 4 (highest) and the impact of the clinical effect was scored on a 7-point scale ranging from AA# (positive effect) to F (highest negative effect). This clinical impact scale (AA#-F) runs parallel to the Common Terminology Criteria for Adverse Events (CTCAE); where CTCAE grade 5 severity is equal to clinical relevance score F (death) and CTCAE grade 1 severity is equal to clinical relevance score B. The clinical relevance score additionally includes the scores AA#, AA and A, since these do not exist in the CTCAE. These regard “Positive clinical effect”, “No clinical or kinetic effect”, and “Significant kinetic effect or not clinically relevant effect”, respectively. DPWG guidelines are checked for agreement with current evidence every 3–5 years in general. An updated version of the guideline will be published if recommendations are altered. The summaries and scores of the articles reviewed to devise this guideline can be found in Supplementary Tables 2, 3 and 4.
For codeine, the initial literature search was performed on 14 September 2006, followed by searches on 10 July 2009, 16 September 2013 and 18 October 2017. Studies in which only concentrations in urine were measured were not included. Articles from 2007 onwards that only mentioned ratios and not concentrations or AUCs of the individual substances, or which included insufficient data to distinguish between PM, IM, NM and UM in the study population, were not included. Articles from 2009 onwards with only kinetic endpoints were not included. For tramadol, the initial literature search was performed on 16 February 2006, followed by searches on 15 July 2009, 27 August 2013 and 19 October 2017. Studies in which only concentrations in urine were measured were not included. From 2009 onwards, kinetic studies were only included if the plasma concentration of O-desmethyl-tramadol was determined for UM and NM/NM. From 2012 onwards, studies on clinical effects in healthy volunteers were not included. Contrary to in healthy volunteers, tramadol is usually titrated and not the only analgesic drug in patients. For oxycodone, the initial literature search was performed on 28 June 2006, followed by searches on and 22 July 2009, 24 April 2013 and 18 October 2017. Studies in which clinical effects were modelled instead of measured and articles published after 2009 with only kinetic data were not included.
General conclusions of evidence
For codeine with the indication pain, four studies (36 healthy PM volunteers) found an association between reduced CYP2D6 activity with absence/strongly diminished analgesia [15–18]. This is confirmed in 1 clinical cohort, in which 2 PM individuals reported no analgesic effect [19]. On the contrary, 4 patient studies with a total number of 28 PM patients were unable to confirm this effect [17, 20–22]. However, in these studies, codeine was generally given with other analgesics (e.g. paracetamol or rescue medication). This could have masked the interaction between effect and genetic variation. As for PM, diminished formation of morphine was observed for IM. The DPWG concludes that there is a gene–drug interaction for PM and IM and that reduced CYP2D6 activity increases the risk of insufficient analgesia by codeine. Because of this, the DPWG recommends the use of another analgesic in PM and an enhanced vigilance for insufficient analgesia in IM. In case of insufficient analgesia in IM, the DPWG recommends a dose increase or an alternative analgesic. The alternative analgesic should not or to a lesser extent be metabolised by CYP2D6 (so not tramadol). For the indication suppressing dry or painful cough, there is no evidence available for a negative clinical effect due to reduced CYP2D6 activity. This indicates that therapy adjustment is not necessary.
Codeine use in breastfeeding mothers with CYP2D6 gene duplications increased the risk for central nervous system depression in the mother and infant [23]. This adverse event could be reduced if maternal codeine use was limited to a maximum of 4 days and the mothers were informed about this toxicity [21]. One fatal incident of an infant has been reported in a breastfeeding mother with the UM phenotype using codeine [24]. While in a case-control study two mothers receiving codeine, both with the UM phenotype, had symptomatic infants showing CNS depression [25]. In addition, two fatal cases in children with UM phenotype have been reported after the use of codeine [26, 27]. Furthermore, adverse events (e.g. obstipation, sedation and severe abdominal pain) have been also observed in UM patients [28, 29]. According to the drug label, the use of codeine is contraindicated for all patients with the UM phenotype [30]. The DPWG concludes that there is a gene–drug interaction and that individuals with UM phenotype have an increased risk of serious opioid toxicity of codeine. For the indication of a cough only with the co-occurrence of other risk factors (e.g. CYP3A4 inhibitors, impaired renal function), an increased risk of serious opioid toxicity was observed [29]. For this reason, the DPWG recommends the use of an alternative drug that is not or to a lesser extent metabolised by CYP2D6 in patients requiring higher doses (higher than 20 mg every 6 h in adults and 10 mg every 6 h in children) and/or with additional risk factors (concomitant use of CYP3A4 inhibitors and/or impaired kidney function). No therapy adjustment is required for other patients. The summaries of reviewed articles can be found in Supplementary Table 2.
The risk of reduced effectiveness is increased in healthy volunteers and patients with the IM and PM using tramadol [31–34]. In 2 UM cases life-threatening, morphine-like side effects have been reported [35, 36]. For this reason, DPWG concludes that IM and PM have an increased risk of reduced effectiveness and UM an increased risk of potentially serious opioid toxicity of tramadol. Because of this, the DPWG recommends enhanced vigilance for insufficient analgesia in IM and PM, and a dose increase or an alternative analgesic in case of insufficient analgesia. For UM, the DPWG recommends an alternative drug, or a dose decrease if an alternative is not possible. The alternative drug should not or to a lesser extent be metabolised by CYP2D6 (so not codeine). The summaries of reviewed articles can be found in Supplementary Table 3.
For oxycodone in the postoperative setting, two studies found no clinically significant effect of PM phenotype on drug use, duration of analgesia, the need for a new dose and side effects [37, 38]. However, another study showed an increase in cumulative oxycodone use until 12 h after surgery and the equianalgesic dose compared to piritramide with decreasing gene dose but no effect on pain score [39]. Within a cancer population, no effect was found of the PM phenotype (n = 27) on the occurrence of side effects and the median daily dose of oxycodone [40]. A case-control study with six PMs who used oxycodone and were breastfeeding showed no difference in the incidence of sleepiness and lethargy in mother and infant compared to NM + IM [41]. Two studies showed a smaller increase in pain threshold and pain tolerance to electrical stimulation of the calf nerve or in a cold pressor test in healthy volunteers with PM phenotype [42, 43]. The DPWG concludes that there is a gene–drug interaction. However, because none of the patient studies showed an effect of IM or PM phenotype on pain scores or adverse events, adjustment of therapy is not required. For the UM phenotype, clinical studies found no effect on (1) adverse events in cancer patients [40] or in breastfeeding mothers and infants [41], (2) the requirement of additional postoperative dose and pain score [39] and (3) the total amount of oxycodone required for analgesia, the duration of the analgesia and the plasma concentration of oxycodone during adequate analgesia and the need for a new dose [38]. One patient developed insomnia, anxiety and increased alertness on oxycodone 10 mg twice [44]. One study including healthy volunteers who were given a single dose per kg body weight showed increased sedation and decreased functioning in a psychomotor test (replacing numerical digits) with an increasing gene dose [42]. This study found also higher pain thresholds when exposed to ice water and electrical nerve stimulation in UMs compared to NMs (40%) and PMs (30-fold) [42]. Because none of the patient studies showed an effect of the UM phenotype on pain scores or adverse events and because no serious adverse events were observed in case reports, the DPWG concludes that adjustment of therapy is not required. The summaries of reviewed articles can be found in Supplementary Table 4.
Pharmacotherapeutic recommendations
The DPWG therapeutic recommendations to optimise therapy with codeine or tramadol in patients known to have a variant CYP2D6 phenotype are summarised in Table 3. The rationale, kinetic and clinical consequences of these therapeutic recommendations are depicted in Supplementary Tables 5 and 6. For oxycodone, no adjustment of therapy is required as summarised in Supplementary Table 7. See Supplementary Tables 8, 9 and 10 for an overview of suggested pop-up (or look-up) texts for electronic prescribing systems for pharmacists and physicians. These can be used to programme alerts into the clinical decision support system (CDSS).
Table 3.
Summary therapeutic recommendations based on CYP2D6 phenotype for codeine and tramadol.
| Drug | CYP2D6 phenotype | Therapeutic recommendation |
|---|---|---|
| Codeine | PM |
For PAIN: 1. choose an alternative Do not select tramadol, as this is also metabolised by CYP2D6. Morphine is not metabolised by CYP2D6. Oxycodone is metabolised by CYP2D6 to a limited extent, but this does not result in differences in analgesia in patients. 2. if an alternative is not an option: advise the patient to report inadequate analgesia. For COUGH: 1. no action required |
| IM |
For PAIN: 1. be alert to a reduced effectiveness 2. in the case of inadequate effectiveness: 1. try a dose increase 2. if this does not work: choose an alternative Do not select tramadol, as this is also metabolised by CYP2D6. Morphine is not metabolised by CYP2D6. Oxycodone is metabolised by CYP2D6 to a limited extent, but this does not result in differences in analgesia in patients. 3. if no alternative is selected: advise the patient to report inadequate analgesia For COUGH: 1. no action required |
|
| UM |
DOSES HIGHER THAN 20 mg every 6 h for adults and 10 mg every 6 h for children aged 12 years or older AND/OR ADDITIONAL RISK FACTORS, such as co-medication with CYP3A4 inhibitors and/or reduced kidney function: Codeine is contra-indicated 1. if possible, select an alternative 1. For PAIN: do not select tramadol, as this is also metabolised by CYP2D6. Morphine is not metabolised by CYP2D6 Oxycodone is metabolised by CYP2D6 to a limited extent, but this does not result in differences in side effects in patients. 2. For COUGH: noscapine is not metabolised by CYP2D6. DOSES LOWER THAN OR EQUAL TO 20 mg every 6 h for adults and 10 mg every 6 h for children aged 12 years or older AND NO ADDITIONAL RISK FACTORS, such as co-medication with CYP3A4 inhibitors and/or reduced kidney function: 1. no action required |
|
| Tramadol | PM |
1.be alert to a reduced effectiveness 2.in the case of inadequate effectiveness:1 1.try a dose increase1. 2. if this does not work: choose an alternative1.Do not select codeine, as this is also metabolised by CYP2D6. Morphine is not metabolised by CYP2D6. Oxycodone is metabolised by CYP2D6 to a limited extent, but this does not result in differences in analgesia in patients. 3.if no alternative is selected: advise the patient to report inadequate analgesia |
| IM |
1.be alert to a reduced effectiveness 2.in the case of inadequate effectiveness: 1.try a dose increase 2. if this does not work: choose an alternative Do not select codeine, as this is also metabolised by CYP2D6. Morphine is not metabolised by CYP2D6. Oxycodone is metabolised by CYP2D6 to a limited extent, but this does not result in differences in analgesia in patients. 3.if no alternative is selected: advise the patient to report inadequate analgesia |
|
| UM |
1. select an alternative Do not choose codeine, as it is contra-indicated for CYP2D6 UMs. Morphine is not metabolised by CYP2D6. Oxycodone is metabolised by CYP2D6 to a limited extent, but this does not result in differences in side effects in patients. 2. if an alternative is not possible: 1.use 40% of the standard dose 2.advise the patient to report side effects (e.g. drowsiness, confusion, constipation, nausea and vomiting, respiratory depression or urine retention) |
|
| Oxycodone | PM | NO action required |
| IM | NO action required | |
| UM | NO action required |
Implications for clinical practice
Ongoing debate persists whether and which single-drug gene pairs should be implemented into routine care. Points of debate include the amount of evidence that is necessary supporting the effectiveness of pre-emptive genotyping, cost-effectiveness of PGx testing in the pre-emptive setting and its reimbursement [45]. As a consequence, drug–gene pairs which are ready for implementation are hampered in application in clinical practice [46]. In an effort to overcome this inconclusiveness and to direct clinicians on whether or not to order relevant PGx genotyping tests before initiating therapy, the DPWG has developed the Clinical Implication Score. The DPWG Clinical Implication Score for a certain drug–gene pair can be scored as: essential, beneficial or potentially beneficial. These categories are clarified in Supplementary Table 11. The development of these categories and the systematic scoring criteria are discussed elsewhere [47]. In brief, the implications for clinical practice are based on four criteria: the clinical effect associated with gene–drug interaction; the level of evidence supporting the associated clinical effect; the number needed to genotype (NNG) in the Dutch population; and the availability of and type of PGx information in the drug label issued by the European Medicines Agency (EMA). The scores provided for each of these criteria by the DPWG can be found in Supplementary Table 11. Only gene–drug interactions which are actionable (codeine and tramadol) are subject to receiving a Clinical Implication Score. For codeine, cases of fatal adverse events have been observed at analgesic doses for UMs (grade 5 toxicity) and an UM with impaired renal function using two CYP3A4 inhibitors became comatose 4 days after starting codeine 25 mg three times daily to combat cough. No serious adverse events have been observed in patients without additional risk factors on low doses (≤20 mg every 6 h for adults and ≤10 mg every 6 h for children). In addition, studies confirming an increased risk for severe adverse events (CTCAE grade ≥3) are lacking and the number needed to genotype to prevent a severe adverse event is likely to be larger than 1000 (see Supplementary Table 11 for more details). The Dutch drug label of codeine indicates that use in patients who are known to be CYP2D6 ultra-rapid metabolisers (UM) is contra-indicated.
The Clinical Implication Score of the gene–drug interaction between CYP2D6 and codeine is 4 out of the maximum of 10 points for patients at higher doses and/or with additional risk factors. This indicates that genotyping before starting codeine is considered “beneficial” for adults with doses >20 mg every 6 h, for children ≥12 years with doses >10 mg every 6 h, and in patients with additional risk factors (e.g. co-medication with CYP3A4 inhibitors and/or impaired renal function). In order to guide drug and dose selection and improve drug safety these patients should ideally be genotyped before (or directly after) drug therapy has been initiated. For tramadol, serious life-threatening opioid toxicity (CTCAE grade 4) has been observed in ultra-rapid metabolisers. However, studies confirming an increased risk for severe adverse events (CTCAE grade ≥ 3) are lacking and the number needed to genotype to prevent a severe adverse event is likely to be larger than 1000 (see Supplementary Table 11 for more details). The Dutch drug label of tramadol does not mention any CYP2D6 genotype or phenotype. Based on this the Clinical Implication Score of the gene–drug interaction between CYP2D6 and tramadol is 1 out of a maximum of 10 points. This indicates that genotyping before starting tramadol is considered “potentially beneficial”. Genotyping before starting tramadol can be considered on an individual patient basis. In clinical practice genotyping could be considered in patients that experience problems as undersedation or oversedation.
Differences between available guidelines
To the best of our knowledge, other guidelines have only been found for codeine. Guidelines for codeine are available from the Clinical Pharmacogenetics Implementation Consortium (CPIC) [48] and the Canadian Pharmacogenomics Network for Drug Safety (CPNDS) [49]. Tramadol and oxycodone are briefly discussed in the CPIC guideline [48], but no therapeutic recommendations are provided for these opioids.
CPIC
The DPGW and CPIC guideline differ in translation from CYP2D6 genotype to phenotype and methodology of guideline development, as discussed earlier [50]. The CPIC guideline on codeine was recently updated [51]. The addition of evidence for other opioids (tramadol, hydrocodone, oxycodone and methadone) and the genes OPRM1 and Catechol-O-methyltransferase (COMT) was added to the previous guideline.
The advice for codeine in PMs and IMs is in agreement with the recommendations from this current DPGW guideline. On the contrary, the CPIC guideline provides rather general advice to avoid codeine in all UM patients without any further risk stratification. Although the risk factors in the UM group mentioned in this guideline are not included in the therapeutic advice of the CPIC guideline, it is stated that the risk of toxicity is present at ‘standard’ analgesic doses. This indirectly implies that at lower doses, as is the case with the use of codeine for cough, the risk probably is absent. No information is provided on the risk factors impaired kidney function and co-medication that inhibits CYP3A4.
The advice for tramadol in PMs and IMs is partly in agreement with the recommendations from this current DPWG guideline. The advice in this current DPWG guideline advises in both phenotype groups a dosage increase in case of inadequate effectiveness. Whereas an alternative opioid is advised if dosage increase did not result in adequate analgesia. In the CPIC guideline, no dosage increases are recommended and in the case of PMs, it is advised to immediately avoid tramadol in this group. In the UM group, both guidelines agree to avoid tramadol due to the risk of toxicity. However, in this current DPWG guideline also alternative advice is provided in the case where tramadol could not be switched. The DPWG recommends in that case a specific dosage (40% of standard dose).
Finally, the CPIC guideline also provides therapeutic recommendations for hydrocodone, whereas this is not included in the current DPWG guideline. Since hydrocodone is not available on the Dutch market the evidence supporting its interaction with CYP2D6 was not evaluated by the DPWG. This current DPWG guideline also has not evaluated the evidence of the OPRM1 and COMT genes. However, because CPIC does not recommend any therapy adjustments in the case of OPRM1 and COMT variants, this does not greatly affect the value of our guideline.
CPNDS
The therapeutic recommendations from CPNDS [49] are more in line with the CPIC guideline. They advise avoiding codeine in PMs and UMs, although the level of evidence is indicated as ‘strong’ for PMs ‘moderate’ for UMs. The lower classification of recommendation in the latter group could most likely be improved if further risk stratification occurs within the UM group, as is the case in the current guideline.
Disclaimer
The Pharmacogenetics Working Group of the KNMP (DPWG) formulates the optimal recommendations for each phenotype group based on the available evidence. If this optimal recommendation cannot be followed due to practical restrictions, e.g. therapeutic drug monitoring or a lower dose is not available, then the health care professional should consider the next best option.
Supplementary information
Genotype to predicted phenotype translation to be programmed into laboratory information system
Literature review of CYP2D6-codeine interactions to support the therapeutic guidelines to choose an alternative or optimise the dose
Literature review of CYP2D6-tramadol interactions to support the therapeutic guidelines to choose an alternative or optimise the dose
Literature review of CYP2D6-oxycodone interactions to support the absence of a need for therapy adjustment in patients with variant genotypes
Dutch Pharmacogenetics Working Group (DPWG) Guideline for CYP2D6 and codeine: the therapeutic recommendation and its rationale, and the kinetic and clinical consequences for each predicted phenotype
Dutch Pharmacogenetics Working Group (DPWG) Guideline for CYP2D6 and tramadol: the therapeutic recommendation and its rationale, and the kinetic and clinical consequences for each predicted phenotype
Dutch Pharmacogenetics Working Group (DPWG) Guideline for CYP2D6 and oxycodone: the therapeutic recommendation and its rationale, and the kinetic and clinical consequences for each predicted phenotype
Suggested clinical decision support texts for health care professionals for codeine.
Suggested clinical decision support texts for health care professionals for tramadol.
Suggested clinical decision support texts for health care professionals for oxycodone (preferably implemented as look-up texts only).
The Clinical Implication Score is “beneficial” for codeine in patients using higher doses and/or having additional risk factors and “potentially beneficial” for tramadol, based on the criteria and cor
Acknowledgments
Funding
The U-PGx consortium received funding from the European Community’s Horizon 2020 Programme under grant agreement No. 668353 (U-PGx). The DPWG received funding from the Royal Dutch Pharmacists Association.
Data availability
All data and material are either included in the supplementary information or publicly available (i.e. the published articles, PubMed). The guidelines and background information are available on the website of the Royal Dutch Pharmacists Association (KNMP) (Pharmacogenetic Recommendations. Available from: https://www.knmp.nl/). The guidelines and background information will be available on PharmGKB.org
COMPETING INTERESTS
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
9/27/2021
A Correction to this paper has been published: 10.1038/s41431-021-00969-9
Supplementary information
The online version contains supplementary material available at 10.1038/s41431-021-00920-y.
References
- 1.Swen JJ, Wilting I, de Goede AL, Grandia L, Mulder H, Touw DJ, et al. Pharmacogenetics: from bench to byte. Clin Pharm Ther. 2008;83:781–7. doi: 10.1038/sj.clpt.6100507. [DOI] [PubMed] [Google Scholar]
- 2.Guchelaar HJ. Pharmacogenomics, a novel section in the European. J Hum Genet Eur J Hum Genet. 2018;26:1399–400. doi: 10.1038/s41431-018-0205-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paineurope. WHO analgesic ladder (2018). http://www.paineurope.com/tools/who-analgesic-ladder. Accessed Oct 2017.
- 4.European Medicines Agency. Restrictions on use of codeine for pain relief in children—CMDh endorses PRAC recommendation (2018). http://www.ema.europa.eu/docs/en_GB/document_library/Referrals_document/Codeine_containing_medicinal_products/Position_provided_by_CMDh/WC500144850.pdf. Accessed Jun 2013.
- 5.Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, Sangkuhl K, Thorn CF, et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharm Ther. 2012;92:414–7. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thorn CF, Klein TE, Altman RB. Codeine and morphine pathway. Pharmacogenet Genom. 2009;19:556–8. doi: 10.1097/FPC.0b013e32832e0eac. [DOI] [PubMed] [Google Scholar]
- 7.Gong L, Stamer UM, Tzvetkov MV, Altman RB, Klein TE. PharmGKB summary: tramadol pathway. Pharmacogenet Genom. 2014;24:374–80. doi: 10.1097/FPC.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huddart R, Clarke M, Altman RB, Klein TE. PharmGKB summary: oxycodone pathway, pharmacokinetics. Pharmacogenet Genom. 2018;28:230–7. doi: 10.1097/FPC.0000000000000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.National Center for Biotechnology Information. CYP2D6 cytochrome P450 family 2 subfamily D member 6 [Homo sapiens (human)] (2018). https://www.ncbi.nlm.nih.gov/gene/1565. Accessed 4 Mar 2018.
- 10.gnomAD browser beta. Gene: CYP2D6. (2018). http://gnomad.broadinstitute.org/gene/ENSG00000100197.
- 11.Pharmacogene Variation Consortium. CYP2D6 allele nomenclature (2020). https://www.pharmvar.org/gene/CYP2D6.
- 12.Zhou Y, Ingelman-Sundberg M, Lauschke VM. Worldwide distribution of Cytochrome P450 alleles: a meta-analysis of Population-scale Sequencing Projects. Clin Pharm Ther. 2017;102:688–700. doi: 10.1002/cpt.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank D, Jaehde U, Fuhr U. Evaluation of probe drugs and pharmacokinetic metrics for CYP2D6 phenotyping. Eur J Clin Pharm. 2007;63:321–33. doi: 10.1007/s00228-006-0250-8. [DOI] [PubMed] [Google Scholar]
- 14.Magliocco G, Thomas A, Desmeules J, Daali Y. Phenotyping of human CYP450 enzymes by endobiotics: current knowledge and methodological approaches. Clin Pharmacokinet. 2019;58:1373–91. doi: 10.1007/s40262-019-00783-z. [DOI] [PubMed] [Google Scholar]
- 15.Sindrup SH, Brosen K, Bjerring P, Arendt-Nielsen L, Larsen U, Angelo HR, et al. Codeine increases pain thresholds to copper vapor laser stimuli in extensive but not poor metabolizers of sparteine. Clin Pharm Ther. 1990;48:686–93. doi: 10.1038/clpt.1990.212. [DOI] [PubMed] [Google Scholar]
- 16.Desmeules J, Gascon MP, Dayer P, Magistris M. Impact of environmental and genetic factors on codeine analgesia. Eur J Clin Pharm. 1991;41:23–6. doi: 10.1007/BF00280101. [DOI] [PubMed] [Google Scholar]
- 17.Poulsen L, Brosen K, Arendt-Nielsen L, Gram LF, Elbaek K, Sindrup SH. Codeine and morphine in extensive and poor metabolizers of sparteine: pharmacokinetics, analgesic effect and side effects. Eur J Clin Pharm. 1996;51:289–95. doi: 10.1007/s002280050200. [DOI] [PubMed] [Google Scholar]
- 18.Eckhardt K, Li S, Ammon S, Schanzle G, Mikus G, Eichelbaum M. Same incidence of adverse drug events after codeine administration irrespective of the genetically determined differences in morphine formation. Pain. 1998;76:27–33. doi: 10.1016/S0304-3959(98)00021-9. [DOI] [PubMed] [Google Scholar]
- 19.VanderVaart S, Berger H, Sistonen J, Madadi P, Matok I, Gijsen VM, et al. CYP2D6 polymorphisms and codeine analgesia in postpartum pain management: a pilot study. Ther Drug Monit. 2011;33:425–32. doi: 10.1097/FTD.0b013e3182272b10. [DOI] [PubMed] [Google Scholar]
- 20.Williams DG, Patel A, Howard RF. Pharmacogenetics of codeine metabolism in an urban population of children and its implications for analgesic reliability. Br J Anaesth. 2002;89:839–45. doi: 10.1093/bja/aef284. [DOI] [PubMed] [Google Scholar]
- 21.Kelly LE, Chaudhry SA, Rieder MJ, t Jong G, Moretti ME, Lausman A, et al. A clinical tool for reducing central nervous system depression among neonates exposed to codeine through breast milk. PLoS ONE. 2013;8:e70073. doi: 10.1371/journal.pone.0070073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baber M, Chaudhry S, Kelly L, Ross C, Carleton B, Berger H, et al. The pharmacogenetics of codeine pain relief in the postpartum period. Pharmacogenom J. 2015;15:430–5. doi: 10.1038/tpj.2015.3. [DOI] [PubMed] [Google Scholar]
- 23.Sistonen J, Madadi P, Ross CJ, Yazdanpanah M, Lee JW, Landsmeer ML, et al. Prediction of codeine toxicity in infants and their mothers using a novel combination of maternal genetic markers. Clin Pharm Ther. 2012;91:692–9. doi: 10.1038/clpt.2011.280. [DOI] [PubMed] [Google Scholar]
- 24.Koren G, Cairns J, Chitayat D, Gaedigk A, Leeder SJ. Pharmacogenetics of morphine poisoning in a breastfed neonate of a codeine-prescribed mother. Lancet. 2006;368:704. doi: 10.1016/S0140-6736(06)69255-6. [DOI] [PubMed] [Google Scholar]
- 25.Madadi P, Ross CJ, Hayden MR, Carleton BC, Gaedigk A, Leeder JS, et al. Pharmacogenetics of neonatal opioid toxicity following maternal use of codeine during breastfeeding: a case-control study. Clin Pharm Ther. 2009;85:31–5. doi: 10.1038/clpt.2008.157. [DOI] [PubMed] [Google Scholar]
- 26.Ciszkowski C, Madadi P, Phillips MS, Lauwers AE, Koren G. Codeine, ultrarapid-metabolism genotype, and postoperative death. N Engl J Med. 2009;361:827–8. doi: 10.1056/NEJMc0904266. [DOI] [PubMed] [Google Scholar]
- 27.Kelly LE, Rieder M, van den Anker J, Malkin B, Ross C, Neely MN, et al. More codeine fatalities after tonsillectomy in North American children. Pediatrics. 2012;129:e1343–7. doi: 10.1542/peds.2011-2538. [DOI] [PubMed] [Google Scholar]
- 28.Dalen P, Frengell C, Dahl ML, Sjoqvist F. Quick onset of severe abdominal pain after codeine in an ultrarapid metabolizer of debrisoquine. Ther Drug Monit. 1997;19:543–4. doi: 10.1097/00007691-199710000-00011. [DOI] [PubMed] [Google Scholar]
- 29.Gasche Y, Daali Y, Fathi M, Chiappe A, Cottini S, Dayer P, et al. Codeine intoxication associated with ultrarapid CYP2D6 metabolism. N Engl J Med. 2004;351:2827–31. doi: 10.1056/NEJMoa041888. [DOI] [PubMed] [Google Scholar]
- 30.Electronic Medicines Compendium. Codeine phosphate 15 mg tablets (2018). https://www.medicines.org.uk/emc/product/7030. Accessed Mar 2017.
- 31.Poulsen L, Arendt-Nielsen L, Brosen K, Sindrup SH. The hypoalgesic effect of tramadol in relation to CYP2D6. Clin Pharm Ther. 1996;60:636–44. doi: 10.1016/S0009-9236(96)90211-8. [DOI] [PubMed] [Google Scholar]
- 32.Fliegert F, Kurth B, Gohler K. The effects of tramadol on static and dynamic pupillometry in healthy subjects-the relationship between pharmacodynamics, pharmacokinetics and CYP2D6 metaboliser status. Eur J Clin Pharm. 2005;61:257–66. doi: 10.1007/s00228-005-0920-y. [DOI] [PubMed] [Google Scholar]
- 33.Matouskova O, Slanar O, Chytil L, Perlik F. Pupillometry in healthy volunteers as a biomarker of tramadol efficacy. J Clin Pharm Ther. 2011;36:513–7. doi: 10.1111/j.1365-2710.2010.01203.x. [DOI] [PubMed] [Google Scholar]
- 34.Stamer UM, Lehnen K, Hothker F, Bayerer B, Wolf S, Hoeft A, et al. Impact of CYP2D6 genotype on postoperative tramadol analgesia. Pain. 2003;105:231–8. doi: 10.1016/S0304-3959(03)00212-4. [DOI] [PubMed] [Google Scholar]
- 35.Stamer UM, Stuber F, Muders T, Musshoff F. Respiratory depression with tramadol in a patient with renal impairment and CYP2D6 gene duplication. Anesth Analg. 2008;107:926–9. doi: 10.1213/ane.0b013e31817b796e. [DOI] [PubMed] [Google Scholar]
- 36.Orliaguet G, Hamza J, Couloigner V, Denoyelle F, Loriot MA, Broly F, et al. A case of respiratory depression in a child with ultrarapid CYP2D6 metabolism after tramadol. Pediatrics. 2015;135:e753–5. doi: 10.1542/peds.2014-2673. [DOI] [PubMed] [Google Scholar]
- 37.Zwisler ST, Enggaard TP, Mikkelsen S, Brosen K, Sindrup SH. Impact of the CYP2D6 genotype on post-operative intravenous oxycodone analgesia. Acta Anaesthesiol Scand. 2010;54:232–40. doi: 10.1111/j.1399-6576.2009.02104.x. [DOI] [PubMed] [Google Scholar]
- 38.Cajanus K, Neuvonen M, Koskela O, Kaunisto MA, Neuvonen PJ, Niemi M, et al. Analgesic plasma concentrations of oxycodone after surgery for breast cancer-which factors matter? Clin Pharmacol Ther. 2017;103:653–62. doi: 10.1002/cpt.771. [DOI] [PubMed] [Google Scholar]
- 39.Stamer UM, Zhang L, Book M, Lehmann LE, Stuber F, Musshoff F. CYP2D6 genotype dependent oxycodone metabolism in postoperative patients. PLoS ONE. 2013;8:e60239. doi: 10.1371/journal.pone.0060239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andreassen TN, Eftedal I, Klepstad P, Davies A, Bjordal K, Lundstrom S, et al. Do CYP2D6 genotypes reflect oxycodone requirements for cancer patients treated for cancer pain? A cross-sectional multicentre study. Eur J Clin Pharm. 2012;68:55–64. doi: 10.1007/s00228-011-1093-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lam J, Kelly L, Matok I, Ross CJ, Carleton BC, Hayden MR, et al. Putative association of ABCB1 2677G>T/A with oxycodone-induced central nervous system depression in breastfeeding mothers. Ther Drug Monit. 2013;35:466–72. doi: 10.1097/FTD.0b013e318288f158. [DOI] [PubMed] [Google Scholar]
- 42.Samer CF, Daali Y, Wagner M, Hopfgartner G, Eap CB, Rebsamen MC, et al. Genetic polymorphisms and drug interactions modulating CYP2D6 and CYP3A activities have a major effect on oxycodone analgesic efficacy and safety. Br J Pharm. 2010;160:919–30. doi: 10.1111/j.1476-5381.2010.00709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zwisler ST, Enggaard TP, Noehr-Jensen L, Pedersen RS, Mikkelsen S, Nielsen F, et al. The hypoalgesic effect of oxycodone in human experimental pain models in relation to the CYP2D6 oxidation polymorphism. Basic Clin Pharm Toxicol. 2009;104:335–44. doi: 10.1111/j.1742-7843.2009.00378.x. [DOI] [PubMed] [Google Scholar]
- 44.de Leon J, Dinsmore L, Wedlund P. Adverse drug reactions to oxycodone and hydrocodone in CYP2D6 ultrarapid metabolizers. J Clin Psychopharmacol. 2003;23:420–1. doi: 10.1097/01.jcp.0000085421.74359.60. [DOI] [PubMed] [Google Scholar]
- 45.Pirmohamed M, Hughes DA. Pharmacogenetic tests: the need for a level playing field. Nat Rev Drug Discov. 2013;12:3–4. doi: 10.1038/nrd3921. [DOI] [PubMed] [Google Scholar]
- 46.Swen JJ, Huizinga TW, Gelderblom H, de Vries EG, Assendelft WJ, Kirchheiner J, et al. Translating pharmacogenomics: challenges on the road to the clinic. PLoS Med. 2007;4:e209. doi: 10.1371/journal.pmed.0040209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swen JJ, Nijenhuis M, van Rhenen M, de Boer-Veger NJ, Buunk AM, Houwink EJF, et al. Pharmacogenetic information in clinical guidelines: the European Perspective. Clin Pharm Ther. 2018;103:795–801. doi: 10.1002/cpt.1049. [DOI] [PubMed] [Google Scholar]
- 48.Crews KR, Gaedigk A, Dunnenberger HM, Leeder JS, Klein TE, Caudle KE, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin Pharm Ther. 2014;95:376–82. doi: 10.1038/clpt.2013.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Madadi P, Amstutz U, Rieder M, Ito S, Fung V, Hwang S, et al. Clinical practice guideline: CYP2D6 genotyping for safe and efficacious codeine therapy. J Popul Ther Clin Pharm. 2013;20:e369–96. [PubMed] [Google Scholar]
- 50.Bank PCD, Caudle KE, Swen JJ, Gammal RS, Whirl-Carrillo M, Klein TE, et al. Comparison of the guidelines of the clinical pharmacogenetics implementation Consortium and the Dutch Pharmacogenetics Working Group. Clin Pharm Ther. 2018;103:599–618. doi: 10.1002/cpt.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crews KR, Monte AA, Huddart R, Caudle KE, Kharasch ED, Gaedigk A, et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2D6, OPRM1, and COMT genotypes and select opioid therapy. Clin Pharmacol Ther. 2021. 10.1002/cpt.2149. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genotype to predicted phenotype translation to be programmed into laboratory information system
Literature review of CYP2D6-codeine interactions to support the therapeutic guidelines to choose an alternative or optimise the dose
Literature review of CYP2D6-tramadol interactions to support the therapeutic guidelines to choose an alternative or optimise the dose
Literature review of CYP2D6-oxycodone interactions to support the absence of a need for therapy adjustment in patients with variant genotypes
Dutch Pharmacogenetics Working Group (DPWG) Guideline for CYP2D6 and codeine: the therapeutic recommendation and its rationale, and the kinetic and clinical consequences for each predicted phenotype
Dutch Pharmacogenetics Working Group (DPWG) Guideline for CYP2D6 and tramadol: the therapeutic recommendation and its rationale, and the kinetic and clinical consequences for each predicted phenotype
Dutch Pharmacogenetics Working Group (DPWG) Guideline for CYP2D6 and oxycodone: the therapeutic recommendation and its rationale, and the kinetic and clinical consequences for each predicted phenotype
Suggested clinical decision support texts for health care professionals for codeine.
Suggested clinical decision support texts for health care professionals for tramadol.
Suggested clinical decision support texts for health care professionals for oxycodone (preferably implemented as look-up texts only).
The Clinical Implication Score is “beneficial” for codeine in patients using higher doses and/or having additional risk factors and “potentially beneficial” for tramadol, based on the criteria and cor
Data Availability Statement
All data and material are either included in the supplementary information or publicly available (i.e. the published articles, PubMed). The guidelines and background information are available on the website of the Royal Dutch Pharmacists Association (KNMP) (Pharmacogenetic Recommendations. Available from: https://www.knmp.nl/). The guidelines and background information will be available on PharmGKB.org