Abstract
This review reflects upon our own as well as other investigators’ studies on the role of receptor for advanced glycation end-products (RAGE), bringing up the latest information on RAGE in physiology and pathology of the nervous system. Over the last ten years, major progress has been made in uncovering many of RAGE-ligand interactions and signaling pathways in nervous tissue; however, the translation of these discoveries into clinical practice has not come to fruition yet. This is likely, in part to be the result of our incomplete understanding of this crucial signaling pathway. Clinical trials examining the therapeutic efficacy of blocking RAGE-external ligand interactions by genetically engineered soluble RAGE or an endogenous RAGE antagonist, has not stood up to its promise; however, other trials with different blocking agents are being considered with hope for therapeutic success in diseases of the nervous system.
Keywords: Receptor for advanced glycation end-products, Nervous system, Neurological disorders, Sensorimotor disorders
Introduction
Neurodegenerative disorders are characterized by progressive loss of neurons causing disturbances in cognitive, psychomotor, and/or autonomic functions. The mechanism of these disorders in most cases remains unknown and that fact has hampered the identification of appropriate interventional strategies through the years. Molecules that may play a role in the exacerbation of these conditions are likely to be good candidates for therapeutic intervention in neurodegeneration. Receptor for advanced glycation end-products (RAGE) belongs to the group of so-called pattern recognition receptors that are part of the innate immune response signaling system and interact with a multitude of ligands involved in the pathogen- or damage-associated signaling pathways [1]. RAGE was first cloned from the bovine lung cDNA library in the early 1990s [2]. Apart from lung, RAGE was found to have a broad tissue distribution including vascular tissue, cardiac tissue, renal tissue, immune cells, and neural tissue [3].
Diabetic hyperglycemia-induced non-enzymatic glycation is recognized as a common source of advanced glycation end products (AGEs) within the body, Thus, the role of RAGE was initially studied in the context of pathology associated with diabetes mellitus (DM) such as diabetic neuropathy [4]. Apart from non-enzymatic glycation resulting from hyperglycemia, there is a second source of glycotoxins and AGEs, which is diet. Up to 10% of AGEs in the diet is taken up within the blood stream [5]. AGEs can be produced in food products via the Maillard reaction between amino-acids and reducing sugars [6]. Dietary AGEs may be an independent environmental risk factor in diabetic neuropathy [7]. High levels of AGEs are found in high-heat processed nuts, grains, and canned meats [8]. Similar to AGEs derived within the body, dietary AGEs can induce signaling through RAGE and contribute not only to diabetic neuropathy but modify the course of all the disorders in which RAGE participates. Thus, dietary AGEs in processed food may be an important contributor to the chronic ailments of modern times. In addition to diabetic neuropathy, by the beginning of this century the role of RAGE in neurodegenerative conditions also became apparent [9].
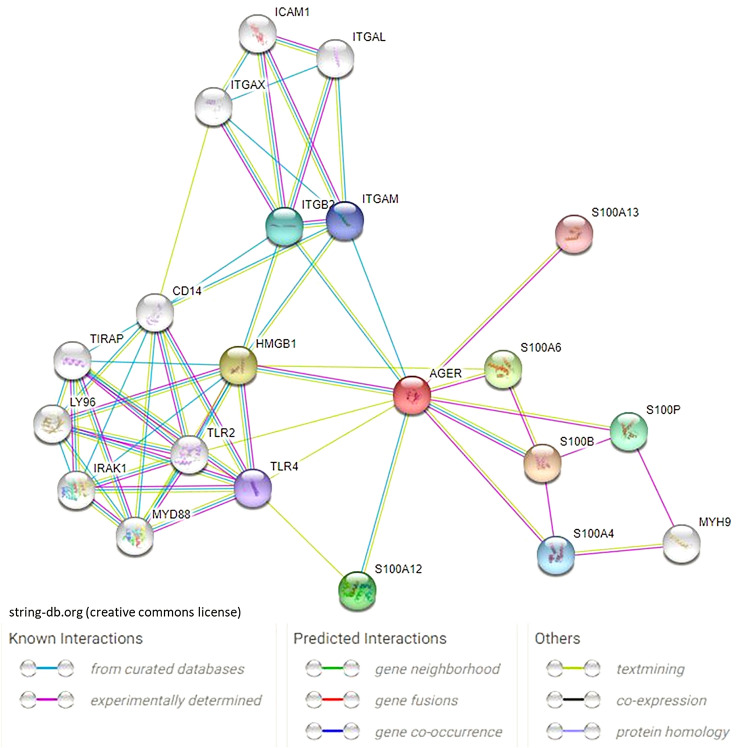
In recent years, major progress in deciphering RAGE-ligand interactions has been made, identifying several novel, previously unknown binding partners and signaling pathways. At the time of manuscript preparation, >25 biding partners have been reported: besides its first described ligands—advanced glycation end products (AGEs) [10]—RAGE interacts with advanced oxidation protein products (AOPP) [11], the S100/calgranulin protein family [12] (S100A1, S100A12, S100A14, and S100B), amphoterin/high mobility group box 1 [13] (HMGB1), protein diaphanous homolog 1 (DIAPH1) [14], epidermal growth factor receptor [15], growth factor receptor-bound protein 2 [16], transforming protein RhoA [17] Toll/interleukin-1 receptor domain-containing adapter protein (TIRAP) [18] , TPA-induced transmembrane protein, transthyretin [19], complement components C3a and C1q [20], amyloid β precursor protein [21], brain-derived neurotrophic factor [22] β-2 Mac1 [23], nucleic acids [24], phosphatidylserine [25], lysophosphatidic acid [26], and heat shock protein 70 (HSP70), [27] and is predicted to interact with: chromosome 2 open reading frame (C2orf15), chemokine-like factor superfamily member 7, cyclic AMP-responsive element-binding protein 3, MAP kinase-activated protein kinase 5, mesoderm induction early response protein 1, cytoplasmic protein NCK1, protein kinase C zeta type—predications sourced from the Uniprot (https://www.uniprot.org/) [28] and HitPredict databases (http://www.hitpredict.org/) [29] (Fig. 1). The RAGE extracellular domain via its multifarious ligands is likely to participate in numerous signal transduction processes that may have both physiological and pathological implications. Indeed, most recent studies have implied a role of RAGE in a variety of neurodegenerative conditions that may not only be linked with just the accumulation of glycation end-products but also may involve the interaction of RAGE with other ligands. Therefore, RAGE signaling has attracted a multiplicity of investigations. In contrast to the large number of interactions that occur with RAGE via its extracellular domain, RAGE via its cytosolic domain usually interacts only with actin regulating protein - DIAPH1 (formerly mDia1) [1, 14]. These biochemical findings raise important questions regarding the ability of the RAGE-Diaph1 signaling axis to distinguish between signaling by the multifarious ligands in the physiological or pathological process. A key challenge for the future in RAGE research is to identify how the otherwise physiological function of RAGE is co-opted by pathological processes.
Fig. 1.
String depiction of RAGE-ligand interactions. For clarity, only the most prominent and well-studied protein interactions are shown; AGER (RAGE), S100 family of Ca2+-binding molecules –S100A4, S100A6, S100A12, S100A13, S100B, S100P; TLR2, TLR, MyD88, IRAK1, TIRAP, LY96 and CD14 parts of the innate immune response pathways; ITGAM, ITGB2, ICAM1, ITGAL and ITGAX—involved in the adaptive immune response, participate in antigen presentation, leukocyte migration, and T cell cytotoxicity.
Source: https://string-db.org/ [134]
RAGE Structure
Human RAGE is encoded by a gene called AGER, located within the Major Histocompatibility Complex class III region on chromosome 6 [30], while in mice the gene encoding RAGE is located on chromosome 17. There are >30 known polymorphisms of human AGER, most of them classified as single nucleotide polymorphisms, ten of which are considered to be clinically relevant [31, 32].
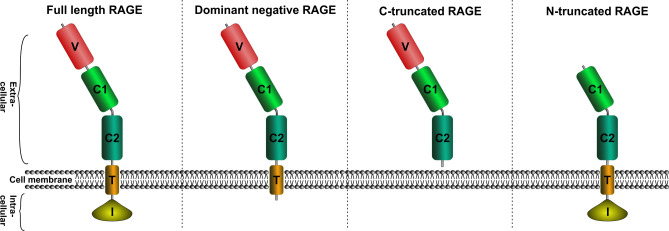
Structurally, RAGE is a single-pass transmembrane protein belonging to the immunoglobulin superfamily [33]. RAGE is comprised of three regions: an extracellular region made up of three distinct domains, V, C1, and C2; a single transmembrane domain; and a short intracellular tail also known as a cytosolic domain [34, 35] (Fig. 2). RAGE is known to self-associate and it oligomerizes on the plasma membrane. The self-association of RAGE is primarily mediated by the C1 domain. While the C2 domain of RAGE is structurally independent, the V and C1 domains have been shown to combine into a VC1 supradomain. RAGE interactions with its ligands are mediated by a basic surface formed in the VC1 domain which interacts with the acidic motifs within the ligand in glycation end-products [36]. An isothermal calorimetry study indicated that the S100B binding may be different and non-competitive to AGEs and mediated by a hydrophobic region [37]. These differences in the binding mechanism of different ligands with RAGE may differently transduce the signals via Diaph1 or lead to the recruitment of different intracellular signaling partners (see next section). Changes in the levels of different ligands of RAGE are therefore likely to profoundly alter RAGE-mediated signaling.
Fig. 2.
RAGE forms and structural domains: V—variable, biding site for extracellular ligands; C—constant, conserved domain with structural similarities to the C domains in other immunoglobulins; T—transmembrane, I—intracellular, binding site for Diaph1 and other, not yet identified intracellular ligands.
So far, a few different RAGE mRNA splice variants of physiological importance have been described: full-length RAGE, N-RAGE (N-truncated RAGE, also called RAGE splice variant 2, RAGE_v2) devoid of the V domain, DN-RAGE (dominant-negative RAGE) devoid of the cytosolic domain, and a C-truncated splice variant called endogenous secretory RAGE (esRAGE, also known as RAGE splice variant 1, RAGE_v1), missing both transmembrane and cytosolic domains [38]. In addition to these variants, an array of other isoforms has been identified or genetically engineered such as RAGE splice variant 3 thru 13 of unknown functions or sRAGE (soluble RAGE) —a genetically-engineered RAGE inhibitor [32]. Both naturally-occurring esRAGE and genetically-engineered sRAGE inhibit RAGE signaling by acting as RAGE substitutes and binding to its ligands [35]. These splicing variants are likely produced by a highly-regulated process and play a critical role in fine-tuning RAGE signaling under physiological conditions. Breakdown of this regulation may thus lead to aberrant RAGE signaling and to the development of pathological states.
The expression levels of RAGE vary depending on the developmental stage. A high level of RAGE expression occurs during embryonic development, promoting neuronal differentiation from undifferentiated embryonic stem cells both in vivo and in vitro [39, 40]; however, the overall expression level of RAGE declines over time, remaining physiologically high only in lung tissue in the postnatal period [35]. Apart from lungs, postnatally, physiological levels of RAGE expression are found in endocrine glands, kidneys, and the brain. At the cellular level, RAGE expression has been reported in numerous cells of the immune system, i.e. macrophages, monocytes, neutrophils, lymphocytes, and dendritic cells; and in podocytes, cardiomyocytes, vascular endothelial and vascular smooth muscle cells, as well as on glial and neuronal cells of the central nervous system (CNS) [34, 35]. The temporo-spatial regulation of RAGE expression in a variety of tissues underscores its physiologically relevant role. Altered expression of RAGE is therefore often associated with pathological states.
RAGE-ligand Interactions and Signaling Patterns
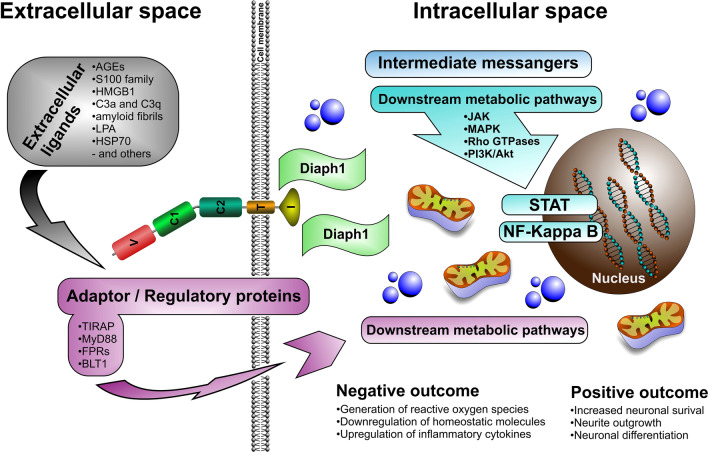
Studies have shown that, upon binding to its ligands via its extracellular domain, RAGE often undergoes biochemical and structural changes such as PKC phosphorylation and homo-, hetero-, or oligo-dimerization. These structural changes prompt RAGE to bind to various adaptor and/or regulatory proteins such as TIRAP and MyD88 (myeloid differentiation primary response gene 88), FPRs (formyl peptide receptors), and BLT1 (leukotriene B4 receptor 1) and triggers a cascade of intracellular responses, activating different metabolic pathways [1, 34, 41]. RAGE-ligand interactions via its extracellular domain also have effects on its intracellular, cytosolic domain, driving RAGE interactions with its cytosolic partner, DIAPH1, another type of regulator protein, and activating second messengers, thus triggering a set of differential metabolic signaling pathways within cells [34, 42].
The results of RAGE-ligand interactions vary depending on the cell and ligand type as well as ligand and RAGE concentrations on the cell surface. Evidence shows that, in many cases, the results of RAGE-ligand interactions lead to pathological changes; however, some studies show that depending on the developmental stage (pre- versus postnatal) and the aforementioned factors, RAGE interaction might be beneficial.
Studies have revealed that in the neuromuscular system during prenatal development, RAGE signaling is beneficial, promoting neuronal and myotubular differentiation, neurite outgrowth, myotube formation, and neuron and muscle cell proliferation [43, 44]. Furthermore, it has been demonstrated that, at low concentrations, RAGE-ligand interaction are beneficial after spinal cord injury by targeting Schwann cells and promoting axonal myelination and regeneration, counteracting to some extent the detrimental effects of neuroinflammation caused by excessive RAGE activation present on microglia and astrocytes [45]. Another study examining RAGE signaling in neuroblastoma cells demonstrated that RAGE, upon binding to one of its ligands, S100B, at low concentration offers neuronal protection against Aβ-driven neurotoxicity, while at high doses it accelerates neuronal death in conjunction with detrimental effects of Amyloid-β1-42 (Aβ1-42) neurotoxicity on neuronal cells [46, 47].
Similarly, low-level expression of RAGE seems to benefit migration, maturation, and proliferation of at least some cells of the immune system such as T cells, dendritic cells, and granulocytes [48], while high RAGE expression leads to exacerbated immune response, driven by overly reactive monocytes and macrophages/microglia [49].
RAGE in the Nervous System —A Janus-faced Receptor
Positive Effects of RAGESignaling in the Nervous System
The presence of RAGE in the nervous system was first described by Brett and colleagues [3] in a survey study examining the distribution of a novel (at the time) receptor for advanced glycation end-products in various mature bovine tissues. In that study, the presence of RAGE was detected in a subset of neurons, ependymal cells, and microvessels of the cerebral cortex and motor neurons of the spinal cord, as well as in neurite outgrowths and cell bodies of NGF-stimulated neuroendocrine PC12 cells [3]. Subsequent studies showed that RAGE, through its ligand interaction, is involved in neuronal differentiation, neurite outgrowth, and elongation, and post-injury nerve regeneration [45, 50–54] both in the embryonic and mature nervous system, underscoring its mediatory role in the developing and mature, adult, nervous system (Fig. 3)
Fig. 3.
RAGE signaling pathways: Ligands (such as AGEs, S100, HMGB1, and others) along with regulatory/adaptor proteins (such as TIRAM, MyD88, and others) by interaction with RAGE trigger a cascade of intracellular signaling pathways (such as JAK-STAT, MAPK, NF-κB, and others) that destabilize neuronal cells and lead to their dysfunction with negative consequences: such as: induction of reactive oxygen, pro-inflammatory cytokines, and downregulation of homeostatic molecules and positive consequences such as: increased neuronal survival, differentiation, and neurite outgrowth.
Many of the physiological or positive effects of RAGE in the nervous system were discovered during early studies of its interactions with a couple of its ligands, i.e. S100s and HMGB1. For example, it has been reported that RAGE activation by both HMGB1 and S100 in neuroblastoma and glioma cells promotes neuronal cell survival via an NF-κB induced increase in expression of the anti-apoptotic protein Bcl-2 [40]. Similar effects were also reported upon RAGE-S100B-HMGB1 binding, when both ligands, in a seemingly coordinated manner, bound to RAGE, triggering metabolic pathways that lead to neurite outgrowth [40]. This finding was later confirmed in dorsal root ganglion cell culture, where RAGE, upon simultaneous binding to S100B and HMGB1, activated neurite outgrowth promoting signaling pathways [55].
The role of RAGE in neuronal differentiation was documented in mouse-derived adult neuronal progenitor cells [53]. Here, as in the case of RAGE-mediated neurite outgrowth, stimulation of neuronal differentiation was mediated via RAGE interactions with S100B, HMGB1, and glycated bovine serum albumin coupled with NF-κB signaling [53]. The results of another set of experiments conducted in embryonic carcinoma cell lines on the role of RAGE in neuronal differentiation showed that all three proteins, RAGE, HMGB1, and S100B, are upregulated throughout the entire differentiation period in these cell lines. Furthermore, it was shown that the RAGE-ligand interaction is coupled with the upregulation of chromogranin, a structural component of secretory vesicles involved in neurosecretion at chemical synapses [50].
Studies have shown that positive RAGE signaling in the nervous system is not only driven by its interactions with S100B or HMGB1 but also, surprisingly, by its binding to monomeric Aβ1-42. In neuroblastoma cells, RAGE and Aβ1-42 interaction leads to upregulation of AMIGO (amphoterin-induced gene and ORF) proteins involved in neurite elongation and fasciculation, thus leading to neuronal differentiation [52].
The positive role of RAGE in the nervous system has also been reported under non-physiological, post-injury circumstances, as demonstrated by studies on the role of RAGE in the spinal cord and peripheral nerve injury models [45, 54]. It was demonstrated that in the spinal cord injury model, modulatory, pro-regenerative RAGE-HMGB1 signaling outweighs its negative, pro-inflammatory signaling, promoting neuronal stem cell differentiation, and accelerating spinal cord recovery [56]; while in peripheral nerve injury, RAGE-S100B interaction triggers pathways mediating Schwann cell migration, thus promoting post-injury peripheral nerve recovery [57].
Negative Effects of RAGE Signaling in the Nervous System
While the number of reports of positive effects of RAGE signaling in the nervous system is limited, the reports on the negative impact of RAGE signaling in the nervous system are abundant and the negative effects are well documented (Fig. 3). It is assumed that the negative impact of RAGE results from its role as a transduction receptor through its binding to multiple, pro-inflammatory and pro-oxidative stress ligands that activate inflammatory and oxidative stress pathways, leading to cellular dysfunction and degeneration resulting in cell death [58, 59].
Normally, in a non-disease setting, RAGE expression in adult cells of the nervous system is low, however, it increases as the concentration of its inflammatory and oxidative stress triggering ligands such as AGEs, HMGB1, S100B or when Aβ1-42 increases excessively [60].
As discussed earlier, HMGB1, S100B, and/or Aβ1-42 can trigger positive changes in the nervous system via RAGE signaling, but to do so, their concentration has to be low, slightly above their baseline. The higher the concentration, the more detrimental the effects of their interaction with RAGE [61, 62]. This kind of dichotomy is not true for AGEs, as the results of their binding to RAGE are always negative, triggering a cascade of metabolic changes leading to oxidative stress and excessive inflammatory responses [63, 64].
Similarly, negative outcomes of RAGE binding on cells and/or tissues has also been reported in the case of its interactions with its other ligands such as AOPP, complement factors, HSP70 or Mac-1, traditionally considered to be a part of the oxidative stress/inflammatory signaling pathways [63]. In addition to its increased activity upon ligand binding, the RAGE signaling pathway might be further enhanced by its upregulation in different nervous system cell types, such as microglia, resident macrophages, and astrocytes, further accelerating the accumulation of pathological processes in nerve cells and tissue [61, 65].
Though detrimental effects of RAGE signaling have been reported in several neurodegenerative diseases, its exact role in the pathogenesis of neurodegeneration remains elusive [66]. Based on current knowledge, it is generally accepted that RAGE on its own does not trigger neurodegeneration but exacerbates it by increasing oxidative stress and neuroinflammation in disease. According to the latest research models of RAGE signaling in the nervous system, immune cells of the nervous system (microglia of the CNS and resident macrophages of the peripheral nervous system) are the first responders to excessive accumulation of RAGE pro-inflammatory and pro-oxidative stress ligands. RAGE on the surface of these immune cells becomes activated upon binding to its ligands and, along with triggering downstream pathological pathways, stimulates the expression of neuronal and likely astrocytic RAGE, entering a positive expression loop and accelerating the progression of pathological changes and leading to neuronal dysfunction and degeneration [61, 65]. The dual nature of RAGE may thus result from its rather broad tissue distribution.
RAGE and its Role in Central Nervous System Diseases
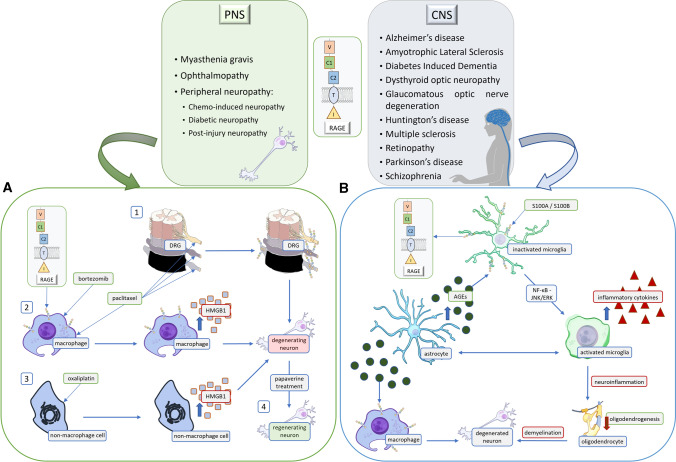
Since its first discovery, RAGE has been implicated in the processes of inflammation and neurodegeneration [65, 67]. The presence of this receptor was detected in neuroinflammatory, neurovascular, and neurodegenerative disorders of the CNS such as multiple sclerosis, diabetes-induced dementia, amyotrophic lateral sclerosis (ALS), Alzheimer’s disease, Parkinson’s disease, and Huntington’s disease. Here we discuss the latest evidence on the role of RAGE in the pathogenesis of CNS disorders, providing the most recent data since our last publications reviewing RAGE and its contribution to neurodegeneration (Figs 4, 5).
Fig. 4.
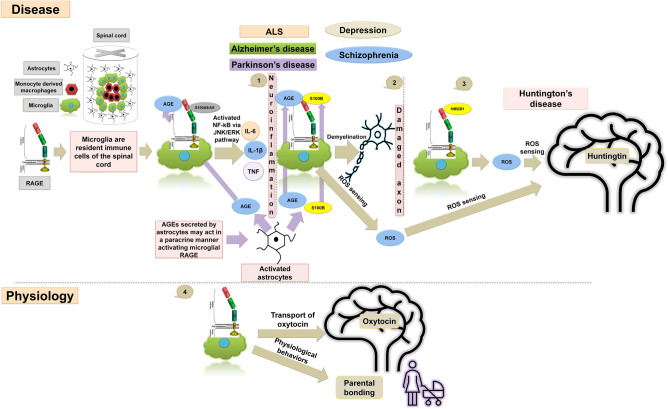
RAGE-activation-dependent signaling is present in numerous disorders of the nervous system. The diseases noted in the text are divided into two tables based on the location of the pathological changes. The RAGE ligands listed are assigned to specific conditions. Pro-inflammatory RAGE ligands such as S100A, S100B, and HMGB1 participate in the development of disease by binding to RAGE. Although in some circumstances, low and slightly raised levels of a given molecule may have a neuroprotective, pro-regenerative effect on the neuronal system, excessive levels lead to pathological pathway activation. A Chemo-induced neuropathy is an exemplary peripheral nervous system (PNS) disease. The figure presents multiple mechanisms that lead to neurodegeneration. Paclitaxel is an anti-cancer drug that causes an increased expression of RAGE in the DRG (1). At the same time, it increases the expression of HMGB1 in macrophages (2). Oxaliplatin has a similar effect on non-macrophage cells (3), as it increases the production of HMGB1. Interestingly, recent research provides data suggesting that papaverine treatment increases neuronal recovery, which helps in alleviating neuropathic pain (4). B Multiple sclerosis is an exemplary CNS neurodegenerative disease. Microglial RAGE binds to S100A8/A9, activating NF-κB via JNK/ERK pathway, which then increases the production of inflammatory cytokines. Moreover, due to metabolic changes, astrocytes produce excessive AGEs, which also bind to RAGE on microglia and macrophages within the lesion. Subsequently, this leads to demyelination along with inflammation, to further neurodegeneration and health decline.
Fig. 5.
RAGE and its role in CNS diseases. Schematic of RAGE signal transduction and its role in CNS diseases. The presence of RAGE has been detected in many nervous system disorders (1) RAGE on the surface of microglia acts as a receptor for the Ca2+-binding protein S100A8/A9 to activate NF-kB via the JNK/ERK pathway to enhance the production of inflammatory cytokines, such as TNF, IL-6, and IL-1β. Neuroinflammatory diseases of the CNS are characterized by the presence of inflammatory infiltrates in the brain and/or spinal cord parenchyma; simultaneously, the S100B-RAGE interaction may enhance the axonal demyelination in neuroinflammatory disorders such as ALS, Alzheimer’s disease, Parkinson’s disease, schizophrenia, and depression. Not only microglia, but also astrocytes may produce excessive AGEs. (2) Evidence suggests that these astrocyte-secreted AGEs act in a paracrine manner to activate microglial RAGE. AGE-RAGE interaction may impact NF-kB via the JNK/ERK pathway and enhance the production of inflammatory cytokines resulting in demyelination. (3) HMGB1, another ligand of RAGE, may be involved in ROS sensing by huntingtin and its nuclear transport. (4) Moreover, RAGE plays a critical role in transport of the hormone oxytocin that is important for physiological behaviors like maternal bonding. Overall, RAGE may participate in a large number of neurodegenerative and mental health conditions.
It is well-accepted that oxidative stress, inflammation, and apoptosis are implicated in the pathogenesis and progression of a number of neurodegenerative diseases, including ALS [68, 69]. Multiple studies implicate ligand-RAGE interaction in the genesis of reactive oxygen species (ROS) and amplification of inflammatory stressors [65, 70]. The key to the biology of RAGE is that its activity is driven by enhanced generation and accumulation of its ligands; our and others’ studies revealed that the ligands of RAGE are upregulated in human and murine models of ALS [71–78]. In the CNS, RAGE has been found in neurons, microglia, and astrocytes. Many studies indicate a central and crucial role played by RAGE in the CNS [79]. The signal transduction pathways activated downstream from the RAGE-ligand interaction depend on the stimulation of different cell types and ligands and elicit cell-type-specific effects [51, 65, 79]. One of the novel effects by which microglial RAGE works in a murine model of ALS is by disrupting cell-cell communication leading to the generation of a pro-inflammatory microglial phenotype [80]. The effects of RAGE activation on neurons, astrocytes, and microglia vary, depending on the specific isoform as well as the concentration level of RAGE on the cell surface [79].
Neuroinflammatory diseases are another group of CNS diseases whose main and most characteristic feature is the presence of inflammatory infiltrates in the brain and/or spinal cord parenchyma. Microglial cells are thought to be the resident immune cells within the CNS and play a crucial role in neuroinflammation via the secretion of pro-inflammatory cytokines and other factors. RAGE on the surface of microglia acts as a receptor for the Ca2+-binding protein S100A8/A9 to activate NF-κB via the JNK/ERK pathway to enhance the production of inflammatory cytokines [81]. Neuroinflammatory disorders such as multiple sclerosis [82] are often associated with demyelination. Another Ca2+-binding protein, S100B, may signal through RAGE in downregulating oligodendrogenesis required for myelin repair. Thus, RAGE may act as a double-edged sword in these conditions by increasing inflammation and at the same time inhibiting myelination. In fact, inhibition of the S100B-RAGE axis may improve remyelination in lysophosphatidylcholine-mediated demyelination [83]. Indeed, downregulation of membrane-bound RAGE expression is often seen in multiple sclerosis, and a higher level of sRAGE has been reported to be associated with responsiveness to IFN-β therapy and lower levels of disability in this disease [84]. Besides Ca2+-binding proteins, in multiple sclerosis lesions, astrocytes may produce excessive AGEs due to a switch in metabolism. Evidence suggests that these AGEs secreted by astrocytes may act in a paracrine manner to activate RAGE on microglia and macrophages present within the lesions [85]. Thus, RAGE may play a dynamic role in conditions like multiple sclerosis due to signaling via multiple different ligands including Ca2+-binding proteins and AGEs.
Together with RAGE, HMGB1 may also function in triggering inflammation in the murine model of multiple sclerosis via astrocytic SHH release in a RAGE-dependent manner via p38, JNK and STAT3 phosphorylation [86].
Inflammation in the CNS is, however, not limited to neuroimmunological disorders. In fact, inflammation is a hallmark of many neurodegenerative conditions such as Alzheimer’s disease and Huntington’s disease. Most neurodegenerative disorders are accompanied by the formation of protein aggregates and defects in protein homeostasis. In Huntington’s disease, protein aggregation is a result of CAG repeat expansion; however glycation may contribute to defective proteostasis [87]. In fact, there is increased expression and co-localization of RAGE with its ligands in the striatum of patients with Huntington’s disease [88]. HMGB1, another ligand for RAGE may be involved in ROS sensing by huntingtin and its nuclear transport [89]. However, despite these reports suggesting that RAGE signaling contributes to Huntington’s disease, a clear picture regarding role of RAGE in this pathology is yet to emerge.
Similarly, there are indications that RAGE may also play a role in Alzheimer’s disease.
Studies have shown that RAGE on endothelial cells mediates the influx of amyloid beta through the blood-brain barrier, triggering release of inflammatory cytokines and simultaneous vasoconstriction, as well as increased T cell infiltration leading to amyloid accumulation and increased inflammation in the brain [90–92]. Further, altered metabolism may affect RAGE cleavage in patients with Alzheimer’s disease, but the pathophysiological relevance of this finding remains to be investigated [93]. Experimentally, intracranial injection of AGE in murine models enhances many of the hallmarks of Alzheimer’s disease [94]. It is also known that inhibition of RAGE signaling in microglia may be beneficial in a mouse model of Alzheimer’s disease since some synaptic plasticity can be regained by inactivating RAGE [95]. Consistent with this notion, it has been found that placement of mesenchymal cells engineered to secrete sRAGE protects against neuronal loss in an Alzheimer’s mouse model [96].There have been three clinical trials first testing the safety and efficacy and later followed up by examining therapeutic effects of RAGE blockers in Alzheimer’s disease; while they were safe and effective, the therapeutic effects were less promising (for more information, see clinicaltrials.gov), indicating that in the pathogenesis of the disease some other factors besides RAGE–Aβ1-42 interactions might play a more prominent role. It should be noted though that the neuroinflammatory aspect of RAGE signaling has not been tested, hence it might provide a new venue for Alzheimer’s disease treatment options.
Parkinson’s disease is the second most common later age-onset neurodegenerative condition. In toxin-induced Parkinson’s disease mouse models it has been reported that pharmacological inhibition of RAGE protects dopaminergic neurons in the substantia nigra from 6-hydroxydopamine mediated cell-death [97]. Further, MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine) treatment (another toxin producing parkinsonism) has been shown to cause alterations in molecules belonging to the RAGE signaling pathway [98]. Downregulation of RAGE in MPTP-treated mice decreases the associated inflammation by suppression of NF-κB [99]. Overall, available data indicate that RAGE signaling plays a significant role in pathophysiology of both major neurodegenerative conditions, Alzheimer’s and Parkinson’s disease, and is likely to be a common molecular target for therapeutic intervention. Aside from the role in neuroinflammatory and neurodegenerative diseases, new evidence has revealed that the contribution of RAGE may also extend to neurovascular disorders such as diabetes-induced dementia affecting brain vasculature, leading to disturbances of cerebrovascular homeostasis and cognitive decline via its interactions with brain-derived neurotrophic factor [22]).
Finally, the role of RAGE has also been noted in ALS and some mental disorders including schizophrenia. Both pharmacological and genetic suppression of RAGE produces notable improvement in the muscle power of a murine model of ALS hSODG93A [100]. Despite the symptomatic improvement in muscle power these manipulations of RAGE fail to produce any significant improvement in the course of the disease and survival of these mice [100], indicating that the role of RAGE is downstream of the pathogenic mechanism and most likely involved in inflammation. In support of this idea, complete abolition of RAGE expression in hSODG93A mice reduces microgliosis and inflammation, significantly slows the progression of the phenotypes, and extends the survival [80]. Mechanistically, these benefits may be due to the suppression of inflammatory genes that is induced in this murine model due to aberrant expression of RAGE and its ligand S100B in astrocytes [101].
RAGE also plays a critical role in transport of the hormone oxytocin in the brain and is important for physiological behaviors like maternal bonding [102, 103]. Contradictorily, increased RAGE expression in microglia may contribute to depressive behavior in stress [104]. In fact, polymorphisms within the RAGE gene have been found to be associated with the risk of schizophrenia, a prevalent mental disorder [105]. One possible mechanism that has been put forward is that a decrease in the soluble splice variant esRAGE in schizophrenia causes an increase in carbonyl stress [106].
Overall data suggest that, due to its role in triggering inflammation and oxidative stress, RAGE participates in a large number of neurodegenerative and mental health conditions. In addition, RAGE performs valuable functions in the brain, including hormone transport and essential behaviors such as maternal bonding. This Ying and Yang nature of RAGE function needs further investigation before RAGE can be safely used as a target for therapeutic intervention.
RAGE and Pathogenesis of Sensorimotor Disorders
The presence of RAGE on immune cells as well as endothelium indicates that RAGE participates both in inflammation and vasculopathy, hallmarks of peripheral neuropathies. In addition, RAGE signaling defects in neurons also may produce axonopathy, the third arm of peripheral neuropathy [107]. Thus, aberrant RAGE signaling may participate in peripheral neuropathy pathogenesis in many different manners (Fig. 4, 6).
Fig. 6.
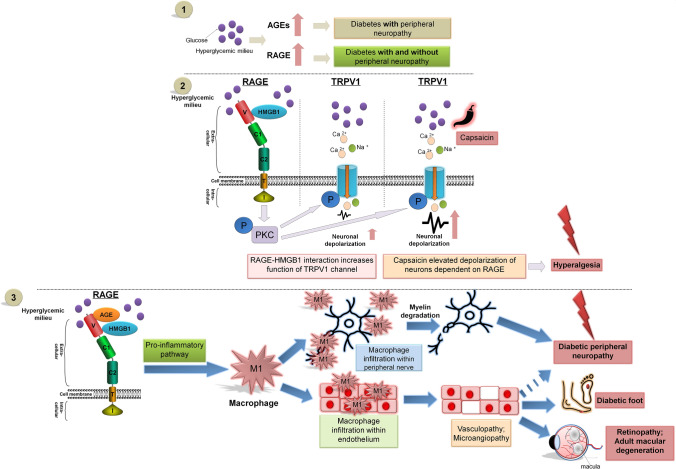
RAGE and pathogenesis of peripheral nervous system diseases. Schematic of RAGE signal transduction and its role in the pathogenesis of peripheral nervous system diseases. (1) RAGE and AGE are both elevated in the hyperglycemic milieu. However, while RAGE is increased in both diabetics with and without peripheral neuropathy, a higher amount of AGEs is found only in diabetics with neuropathy. Hence, pathways that downregulate RAGE may be important in the pathogenesis of peripheral nervous system diseases. The extracellular domains of RAGE (V, C1, and C2) have the ability to bind ligands, i.e. AGEs, HMGB1 and others. RAGE-HMGB1 interaction triggers signal transduction via the PKC pathway. (2) RAGE-HMGB1 interplay may increase the activity of TRPV1 channels affecting neuronal depolarization in peripheral nerve, amplified by the presence of capsaicin modulation of pain perception in patients with diabetic peripheral neuropathy. (3) RAGE is connected to many pro-inflammatory pathways. Hence, RAGE signaling is important for macrophage infiltration within peripheral nerves and the endothelium of vessels. These phenomena may be crucial for the progression of diabetic peripheral neuropathy, diabetic foot, retinopathy, adult macular degeneration, and other peripheral nervous system diseases.
In the last four decades, the global incidence of DM and the resulting hyperglycemia has doubled and is nearly 8.5% of the population [108]. RAGE, as a multi-ligand messenger receptor, is present in many tissues sensitive to hyperglycemia and is associated with many pathological processes in the initiation as well as the progression of diabetic neuropathy, such as increased inflammation, oxidative stress, protein glycation, and increased apoptosis. In fact, sensorimotor neuropathy is a common consequence of uncontrolled DM and hyperglycemia. Although the pathology of diabetic neuropathy begins insidiously, it can rapidly progress to conditions like diabetic foot with ulcerations and Charcot’s neuropathy with the need for amputation of the lower limb. Besides diabetic foot, diabetic neuropathy affects the quality of life and increases mortality in several other manners including chronic neuropathic pain and increased risk of falling. Besides control of blood sugar level and foot care there are no disease-modifying agents for diabetic neuropathy at the moment, making it crucial that molecules such as RAGE and their signaling mechanisms be studied intensively. Further, such studies are likely to inform us about other neuropathies.
The role of RAGE in the pathogenesis of peripheral neuropathy, especially diabetic neuropathy, is well studied and has been well described by us and others [109–112]. Studies on the role of RAGE in the pathogenesis of diabetic neuropathy, conducted in murine models of this disease, have shown that RAGE contributes to the exacerbated local inflammation and the increase of oxidative stress in hyperglycemia-sensitive tissues.
Intriguingly, the expression of RAGE as well as its activation has been reported not only in diabetic but also in non-diabetic neuropathies in patients [109, 113] and animal models. For example, peripheral neuropathies are common adverse effects of chemotherapy against neoplasms. Commonly-used drugs like paclitaxel are known to increase the expression levels of RAGE in dorsal root ganglia and cause peripheral neuropathies [114]. At the same time, paclitaxel also causes the release of the RAGE interactor molecule HMGB1 from macrophages within peripheral nerves [115]. A similar increase in capsaicin-mediated HMGB1 release from macrophages has also been recently reported in response to the proteasome-inhibiting chemotherapeutic drug, bortezomib, in a mouse model, suggesting that changes in RAGE signaling is modified in most chemotherapy-induced peripheral neuropathies [116]. Interestingly, there is evidence that HMGB1 is released from non-macrophage cells and participates in chemotherapy-induced peripheral neuropathy as seen with oxaliplatin in rodent models [116]. Depending on the nature of the pathology, peripheral neuropathy can be either painful or painless in nature. Painful peripheral neuropathies may produce allodynia (aberrant pain), and it has been reported that RAGE antagonists are capable of reducing paclitaxel-induced allodynia in an animal model, suggesting a direct role of RAGE signaling in this form of neuropathy [115]. Similarly, degradation of HMGB1 by thrombomodulin also reduces allodynia in oxaliplatin-induced peripheral neuropathy.
AGEs and RAGE are both elevated in the diabetic population when compared to non-diabetics [117]. However, while RAGE is increased in both diabetics with and without peripheral neuropathy, higher amounts of AGEs are found in diabetics with neuropathy [117]. This may lead to higher signaling through RAGE in neuropathy. In fact, culturing neurons under hyperglycemic conditions is sufficient to increase capsaicin-evoked depolarization that is dependent on RAGE. Comparable results have also been obtained from the nerves of diabetic animals [118] hence supporting the notion that increased RAGE signaling in hyperglycemia directly affects the function of sensory neurons, producing conditions such as hyperalgesia (increased pain) and allodynia. Besides AGEs, disturbed sensory neurons via AGE signaling could also be due to RAGE signaling via HMGB1, which increases the activity of TRPV1 channels [119]. Experimentally, deleting RAGE in an animal model of DM indeed partially rescues functional deficits in nerves, further supporting a role of RAGE in diabetic peripheral neuropathy. Besides directly affecting neurons, RAGE signaling is also likely to affect inflammatory cells. In animal model experiments, RAGE signaling is important for the infiltration of macrophage within peripheral nerves, with more macrophages exhibiting a pro-inflammatory M1 phenotype [120]. RAGE has been associated with vasculopathy and may be specifically involved in microangiopathy-associated diabetic neuropathy [121]. The importance of such RAGE signaling on overall nerve function extends beyond diabetic or chemotherapeutic neuropathy; it has been reported that in animal models of spinal nerve ligation, RAGE plays a role in hyperalgesia, thus providing evidence of RAGE signaling in injury-mediated neuropathies. The precise molecular pathway is uncertain, but it is known that the inflammatory pathway is upregulated [122]. Altogether, RAGE may be a target for intervention in both diabetic and non-diabetic peripheral neuropathies due to its functional importance in endothelial cells, as well as both neurons and immune cells. A recent report on sepsis-induced neuropathy indicated that using anti-inflammatory and antioxidant drugs such as papaverine effectively blocks RAGE-HMGB1 signaling and increases the chance of recovery [123]. Thus, RAGE blockers could be used in the management of neuropathic pain induced by a variety of conditions, including DM, chemotherapy, sepsis, and even injury.
RAGE is also upregulated in optic nerve neuropathy in Alzheimer’s patients [124]. Furthermore, RAGE-mediated accelerated aging has also been suggested to participate in glaucomatous optic nerve degeneration [125]. A recent investigation found increases in the levels of both RAGE and HMGB1 in Grave’s ophthalmopathy, and RAGE expression in these cases are directly correlated with dysthyroid optic neuropathy [126]. Apart from peripheral and cranial neuropathies, microangiopathy is also a hallmark of various retinopathies, and indeed RAGE has also been implicated in diabetic retinopathy as well as adult macular degeneration [127, 128]. Besides peripheral sensation and vision, RAGE signaling also likely affects other sensory nerves and pathways. Deletion of RAGE in mice increases their sensitivity to auditory stimuli [129], indicating a physiological role of RAGE in auditory sensation. RAGE may also play a pathological role in the auditory pathway, especially damage to cochlear hair cells under hyperglycemic conditions [130]. RAGE expression increases within the auditory cortex in animal models of noise-induced hearing loss, although the implications of this are not known [131]. Finally, RAGE expression increases with aging in the spiral ganglion cells of the cochlea and may play a role in the pathogenesis of presbycusis (age-dependent hearing loss) [132]. Overall, the literature indicates that, besides influencing sensory neurons in periphery, RAGE also play physiological and pathological roles in the sensory modalities of cranial nerves.
Interestingly, RAGE signaling has also been reported in diseases of neuromuscular junctions such as myasthenia gravis (MG), demonstrating lower levels of soluble RAGE, acting as a RAGE decoy, while levels of its pro-inflammatory ligands such as S100A, S100B, and HMGB1 are increased in the blood serum of MG patients, revealing biomarker and therapeutic potential [133].
Summary
In conclusion, RAGE, as a transmembrane multifaceted pattern-recognizing molecule, is capable of interacting with multiple endogenous ligands. It participates in both physiological and pathophysiological pathways in the peripheral and central nervous systems. The multidirectional involvement of RAGE in neuropathies makes it a promising target for interventional treatment. The major unknown to resolve is whether it is possible to determine the therapeutic dose for the pharmaceutical manipulation of RAGE. Further research is therefore required to gain a more in-depth understanding of the receptor's signaling modulation and to develop potent therapies of the pathologies in which RAGE has a detrimental effect.
Conflict of interest
All authors claim that there are no conflicts of interest
Footnotes
Judyta Juranek and Konark Mukherjee have contributed equally to this work.
Contributor Information
Judyta Juranek, Email: judytajuranek@gmail.com.
Marta Banach, Email: martabanach@yahoo.com.
References
- 1.Teissier T, Boulanger É. The receptor for advanced glycation end-products (RAGE) is an important pattern recognition receptor (PRR) for inflammaging. Biogerontology. 2019;20:279–301. doi: 10.1007/s10522-019-09808-3. [DOI] [PubMed] [Google Scholar]
- 2.Neeper M, Schmidt AM, Brett J, Yan SD, Wang F, Pan YC, et al. Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. J Biol Chem. 1992;267:14998–15004. doi: 10.1016/S0021-9258(18)42138-2. [DOI] [PubMed] [Google Scholar]
- 3.Brett J, Schmidt AM, Yan SD, Zou YS, Weidman E, Pinsky D, et al. Survey of the distribution of a newly characterized receptor for advanced glycation end products in tissues. Am J Pathol. 1993;143:1699–1712. [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt AM, Hori O, Chen JX, Li JF, Crandall J, Zhang J, et al. Advanced glycation endproducts interacting with their endothelial receptor induce expression of vascular cell adhesion molecule-1 (VCAM-1) in cultured human endothelial cells and in mice. A potential mechanism for the accelerated vasculopathy of diabetes. J Clin Invest. 1995;96:1395–1403. doi: 10.1172/JCI118175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peiretti PG, Medana C, Visentin S, Giancotti V, Zunino V, Meineri G. Determination of carnosine, anserine, homocarnosine, pentosidine and thiobarbituric acid reactive substances contents in meat from different animal species. Food Chem. 2011;126:1939–1947. doi: 10.1016/j.foodchem.2010.12.036. [DOI] [PubMed] [Google Scholar]
- 6.Liu LC, Liu L, Xie JH, Shen MY. Formation mechanism of AGEs in Maillard reaction model systems containing ascorbic acid. Food Chem. 2022;378:132108. doi: 10.1016/j.foodchem.2022.132108. [DOI] [PubMed] [Google Scholar]
- 7.Koschinsky T, He CJ, Mitsuhashi T, Bucala R, Liu C, Buenting C, et al. Orally absorbed reactive glycation products (glycotoxins): An environmental risk factor in diabetic nephropathy. Proc Natl Acad Sci USA. 1997;94:6474–6479. doi: 10.1073/pnas.94.12.6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheijen JLJM, Clevers E, Engelen L, Dagnelie PC, Brouns F, Stehouwer CDA, et al. Analysis of advanced glycation endproducts in selected food items by ultra-performance liquid chromatography tandem mass spectrometry: presentation of a dietary AGE database. Food Chem. 2016;190:1145–1150. doi: 10.1016/j.foodchem.2015.06.049. [DOI] [PubMed] [Google Scholar]
- 9.Sousa MM, Saraiva MJ. Neurodegeneration in familial amyloid polyneuropathy: from pathology to molecular signaling. Prog Neurobiol. 2003;71:385–400. doi: 10.1016/j.pneurobio.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt AM, Hori O, Cao R, Yan SD, Brett J, Wautier JL, et al. RAGE: a novel cellular receptor for advanced glycation end products. Diabetes. 1996;45:S77–S80. doi: 10.2337/diab.45.3.S77. [DOI] [PubMed] [Google Scholar]
- 11.Guo ZJ, Niu HX, Hou FF, Zhang L, Fu N, Nagai R, et al. Advanced oxidation protein products activate vascular endothelial cells via a RAGE-mediated signaling pathway. Antioxid Redox Signal. 2008;10:1699–1712. doi: 10.1089/ars.2007.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hofmann MA, Drury S, Fu C, Qu W, Taguchi A, Lu Y, et al. RAGE mediates a novel proinflammatory axis: a central cell surface receptor for S100/calgranulin polypeptides. Cell. 1999;97:889–901. doi: 10.1016/S0092-8674(00)80801-6. [DOI] [PubMed] [Google Scholar]
- 13.Nogueira-Machado JA, de Oliveira Volpe CM. HMGB-1 as a target for inflammation controlling. Recent Pat Endocr Metab Immune Drug Discov. 2012;6:201–209. doi: 10.2174/187221412802481784. [DOI] [PubMed] [Google Scholar]
- 14.Rai V, Maldonado AY, Burz DS, Reverdatto S, Yan SF, Schmidt AM, et al. Signal transduction in receptor for advanced glycation end products (RAGE): solution structure of C-terminal rage (ctRAGE) and its binding to mDia1. J Biol Chem. 2012;287:5133–5144. doi: 10.1074/jbc.M111.277731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aboushousha T, Hammam O, Safwat G, Eesa A, Ahmed S, Esmat ME, et al. Differential expression of RAGE, EGFR and ki-67 in primary tumors and lymph node deposits of breast carcinoma. Asian Pac J Cancer Prev. 2018;19:2269–2277. doi: 10.22034/APJCP.2018.19.8.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakaguchi M, Murata H, Aoyama Y, Hibino T, Putranto EW, Ruma IMW, et al. DNAX-activating protein 10 (DAP10) membrane adaptor associates with receptor for advanced glycation end products (RAGE) and modulates the RAGE-triggered signaling pathway in human keratinocytes. J Biol Chem. 2014;289:23389–23402. doi: 10.1074/jbc.M114.573071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rao JL, Ye ZC, Tang H, Wang C, Peng H, Lai WY, et al. The RhoA/ROCK pathway ameliorates adhesion and inflammatory infiltration induced by AGEs in glomerular endothelial cells. Sci Rep. 2017;7:39727. doi: 10.1038/srep39727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakaguchi M, Murata H, Yamamoto KI, Ono T, Sakaguchi Y, Motoyama A, et al. TIRAP, an adaptor protein for TLR2/4, transduces a signal from RAGE phosphorylated upon ligand binding. PLoS One. 2011;6:e23132. doi: 10.1371/journal.pone.0023132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sousa MM, du Yan S, Fernandes R, Guimaraes A, Stern D, Saraiva MJ. Familial amyloid polyneuropathy: receptor for advanced glycation end products-dependent triggering of neuronal inflammatory and apoptotic pathways. J Neurosci. 2001;21:7576–7586. doi: 10.1523/JNEUROSCI.21-19-07576.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma WC, Rai V, Hudson BI, Song F, Schmidt AM, Barile GR. RAGE binds C1q and enhances C1q-mediated phagocytosis. Cell Immunol. 2012;274:72–82. doi: 10.1016/j.cellimm.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 21.Sturchler E, Galichet A, Weibel M, Leclerc E, Heizmann CW. Site-specific blockade of RAGE-Vd prevents amyloid-beta oligomer neurotoxicity. J Neurosci. 2008;28:5149–5158. doi: 10.1523/JNEUROSCI.4878-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim OY, Song J. The importance of BDNF and RAGE in diabetes-induced dementia. Pharmacol Res. 2020;160:105083. doi: 10.1016/j.phrs.2020.105083. [DOI] [PubMed] [Google Scholar]
- 23.Chavakis T, Bierhaus A, Al-Fakhri N, Schneider D, Witte S, Linn T, et al. The pattern recognition receptor (RAGE) is a counterreceptor for leukocyte integrins: a novel pathway for inflammatory cell recruitment. J Exp Med. 2003;198:1507–1515. doi: 10.1084/jem.20030800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sirois CM, Jin TC, Miller AL, Bertheloot D, Nakamura H, Horvath GL, et al. RAGE is a nucleic acid receptor that promotes inflammatory responses to DNA. J Exp Med. 2013;210:2447–2463. doi: 10.1084/jem.20120201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He M, Kubo H, Morimoto K, Fujino N, Suzuki T, Takahasi T, et al. Receptor for advanced glycation end products binds to phosphatidylserine and assists in the clearance of apoptotic cells. EMBO Rep. 2011;12:358–364. doi: 10.1038/embor.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ray R, Jangde N, Singh SK, Sinha S, Rai V. Lysophosphatidic acid-RAGE axis promotes lung and mammary oncogenesis via protein kinase B and regulating tumor microenvironment. Cell Commun Signal. 2020;18:170. doi: 10.1186/s12964-020-00666-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grunwald MS, Ligabue-Braun R, Souza CS, Heimfarth L, Verli H, Gelain DP, et al. Putative model for heat shock protein 70 complexation with receptor of advanced glycation end products through fluorescence proximity assays and normal mode analyses. Cell Stress Chaperones. 2017;22:99–111. doi: 10.1007/s12192-016-0746-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Consortium U UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.López Y, Nakai KT, Patil A. HitPredict version 4: Comprehensive reliability scoring of physical protein-protein interactions from more than 100 species. Database (Oxford) 2015, 2015: bav117. [DOI] [PMC free article] [PubMed]
- 30.Sugaya K, Fukagawa T, Matsumoto K, Mita K, Takahashi E, Ando A, et al. Three genes in the human MHC class III region near the junction with the class II: gene for receptor of advanced glycosylation end products, PBX2 homeobox gene and a Notch homolog, human counterpart of mouse mammary tumor gene int-3. Genomics. 1994;23:408–419. doi: 10.1006/geno.1994.1517. [DOI] [PubMed] [Google Scholar]
- 31.Serveaux-Dancer M, Jabaudon M, Creveaux I, Belville C, Blondonnet R, Gross C, et al. Pathological implications of receptor for advanced glycation end-product (AGER) gene polymorphism. Dis Markers. 2019;2019:2067353. doi: 10.1155/2019/2067353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hudson BI, Carter AM, Harja E, Kalea AZ, Arriero M, Yang H, et al. Identification, classification, and expression of RAGE gene splice variants. FASEB J. 2008;22:1572–1580. doi: 10.1096/fj.07-9909com. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt AM, Stern DM. Receptor for age (RAGE) is a gene within the major histocompatibility class III region: implications for host response mechanisms in homeostasis and chronic disease. Front Biosci. 2001;6:D1151–D1160. doi: 10.2741/schmidt. [DOI] [PubMed] [Google Scholar]
- 34.MacLean M, Derk J, Ruiz HH, Juranek JK, Ramasamy R, Schmidt AM. The receptor for advanced glycation end products (RAGE) and DIAPH1: implications for vascular and neuroinflammatory dysfunction in disorders of the central nervous system. Neurochem Int. 2019;126:154–164. doi: 10.1016/j.neuint.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalea AZ, Schmidt AM, Hudson BI. Alternative splicing of RAGE: Roles in biology and disease. Front Biosci (Landmark Ed) 2011;16:2756–2770. doi: 10.2741/3884. [DOI] [PubMed] [Google Scholar]
- 36.Allmen EUV, Koch M, Fritz G, Legler DF. V domain of RAGE interacts with AGEs on prostate carcinoma cells. Prostate. 2008;68:748–758. doi: 10.1002/pros.20736. [DOI] [PubMed] [Google Scholar]
- 37.Leclerc E, Fritz G, Vetter SW, Heizmann CW. Binding of S100 proteins to RAGE: an update. Biochim Biophys Acta. 2009;1793:993–1007. doi: 10.1016/j.bbamcr.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 38.Lee EJ, Park JH. Receptor for advanced glycation endproducts (RAGE), its ligands, and soluble RAGE: potential biomarkers for diagnosis and therapeutic targets for human renal diseases. Genom Inform. 2013;11:224–229. doi: 10.5808/GI.2013.11.4.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huttunen HJ, Kuja-Panula J, Rauvala H. Receptor for advanced glycation end products (RAGE) signaling induces CREB-dependent chromogranin expression during neuronal differentiation. J Biol Chem. 2002;277:38635–38646. doi: 10.1074/jbc.M202515200. [DOI] [PubMed] [Google Scholar]
- 40.Huttunen HJ, Kuja-Panula J, Sorci G, Agneletti AL, Donato R, Rauvala H. Coregulation of neurite outgrowth and cell survival by amphoterin and S100 proteins through receptor for advanced glycation end products (RAGE) activation. J Biol Chem. 2000;275:40096–40105. doi: 10.1074/jbc.M006993200. [DOI] [PubMed] [Google Scholar]
- 41.Riuzzi F, Sorci G, Sagheddu R, Chiappalupi S, Salvadori L, Donato R. RAGE in the pathophysiology of skeletal muscle. J Cachexia Sarcopenia Muscle. 2018;9:1213–1234. doi: 10.1002/jcsm.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Egaña-Gorroño L, López-Díez R, Yepuri G, Ramirez LS, Reverdatto S, Gugger PF, et al. Receptor for advanced glycation end products (RAGE) and mechanisms and therapeutic opportunities in diabetes and cardiovascular disease: insights from human subjects and animal models. Front Cardiovasc Med. 2020;7:37. doi: 10.3389/fcvm.2020.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chou DKH, Zhang JH, Smith FI, McCaffery P, Jungalwala FB. Developmental expression of receptor for advanced glycation end products (RAGE), amphoterin and sulfoglucuronyl (HNK-1) carbohydrate in mouse cerebellum and their role in neurite outgrowth and cell migration. J Neurochem. 2004;90:1389–1401. doi: 10.1111/j.1471-4159.2004.02609.x. [DOI] [PubMed] [Google Scholar]
- 44.Hori O, Brett J, Slattery T, Cao R, Zhang J, Chen JX, et al. The receptor for advanced glycation end products (RAGE) is a cellular binding site for amphoterin. Mediation of neurite outgrowth and co-expression of rage and amphoterin in the developing nervous system. J Biol Chem. 1995;270:25752–25761. doi: 10.1074/jbc.270.43.25752. [DOI] [PubMed] [Google Scholar]
- 45.Song J, Lee WT, Park KA, Lee JE. Receptor for advanced glycation end products (RAGE) and its ligands: focus on spinal cord injury. Int J Mol Sci. 2014;15:13172–13191. doi: 10.3390/ijms150813172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Businaro R, Leone S, Fabrizi C, Sorci G, Donato R, Lauro GM, et al. S100B protects LAN-5 neuroblastoma cells against Abeta amyloid-induced neurotoxicity via RAGE engagement at low doses but increases Abeta amyloid neurotoxicity at high doses. J Neurosci Res. 2006;83:897–906. doi: 10.1002/jnr.20785. [DOI] [PubMed] [Google Scholar]
- 47.Huttunen HJ, Fages C, Rauvala H. Receptor for advanced glycation end products (RAGE)-mediated neurite outgrowth and activation of NF-κB require the cytoplasmic domain of the receptor but different downstream signaling pathways. J Biol Chem. 1999;274:19919–19924. doi: 10.1074/jbc.274.28.19919. [DOI] [PubMed] [Google Scholar]
- 48.Kierdorf K, Fritz G. RAGE regulation and signaling in inflammation and beyond. J Leukoc Biol. 2013;94:55–68. doi: 10.1189/jlb.1012519. [DOI] [PubMed] [Google Scholar]
- 49.Sorci G, Riuzzi F, Giambanco I, Donato R. RAGE in tissue homeostasis, repair and regeneration. Biochim Biophys Acta. 2013;1833:101–109. doi: 10.1016/j.bbamcr.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 50.Kim J, Wan CK, O'Carroll SJ, Shaikh SB, Nicholson LFB. The role of receptor for advanced glycation end products (RAGE) in neuronal differentiation. J Neurosci Res. 2012;90:1136–1147. doi: 10.1002/jnr.23014. [DOI] [PubMed] [Google Scholar]
- 51.Wang LY, Li ST, Jungalwala FB. Receptor for advanced glycation end products (RAGE) mediates neuronal differentiation and neurite outgrowth. J Neurosci Res. 2008;86:1254–1266. doi: 10.1002/jnr.21578. [DOI] [PubMed] [Google Scholar]
- 52.Piras S, Furfaro AL, Piccini A, Passalacqua M, Borghi R, Carminati E, et al. Monomeric Aβ1-42 and RAGE: key players in neuronal differentiation. Neurobiol Aging. 2014;35:1301–1308. doi: 10.1016/j.neurobiolaging.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 53.Meneghini V, Francese MT, Carraro L, Grilli M. A novel role for the receptor for advanced glycation end-products in neural progenitor cells derived from adult SubVentricular Zone. Mol Cell Neurosci. 2010;45:139–150. doi: 10.1016/j.mcn.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 54.Rong LL, Yan SF, Wendt T, Hans D, Pachydaki S, Bucciarelli LG, et al. RAGE modulates peripheral nerve regeneration via recruitment of both inflammatory and axonal outgrowth pathways. FASEB J. 2004;18:1818–1825. doi: 10.1096/fj.04-1900com. [DOI] [PubMed] [Google Scholar]
- 55.Saleh A, Smith DR, Tessler L, Mateo AR, Martens C, Schartner E, et al. Receptor for advanced glycation end-products (RAGE) activates divergent signaling pathways to augment neurite outgrowth of adult sensory neurons. Exp Neurol. 2013;249:149–159. doi: 10.1016/j.expneurol.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 56.Wang HY, Mei XF, Cao Y, Liu C, Zhao ZM, Guo ZP, et al. HMGB1/Advanced Glycation End Products (RAGE) does not aggravate inflammation but promote endogenous neural stem cells differentiation in spinal cord injury. Sci Rep. 2017;7:10332. doi: 10.1038/s41598-017-10611-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sbai O, Devi TS, Melone MAB, Feron F, Khrestchatisky M, Singh LP, et al. RAGE-TXNIP axis is required for S100B-promoted Schwann cell migration, fibronectin expression and cytokine secretion. J Cell Sci. 2010;123:4332–4339. doi: 10.1242/jcs.074674. [DOI] [PubMed] [Google Scholar]
- 58.Tóbon-Velasco JC, Cuevas E, Torres-Ramos MA. Receptor for AGEs (RAGE) as mediator of NF-kB pathway activation in neuroinflammation and oxidative stress. CNS Neurol Disord Drug Targets. 2014;13:1615–1626. doi: 10.2174/1871527313666140806144831. [DOI] [PubMed] [Google Scholar]
- 59.Daffu G, del Pozo CH, O'Shea KM, Ananthakrishnan R, Ramasamy R, Schmidt AM. Radical roles for RAGE in the pathogenesis of oxidative stress in cardiovascular diseases and beyond. Int J Mol Sci. 2013;14:19891–19910. doi: 10.3390/ijms141019891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kalea AZ, Reiniger N, Yang H, Arriero M, Schmidt AM, Hudson BI. Alternative splicing of the murine receptor for advanced glycation end-products (RAGE) gene. FASEB J. 2009;6:1766–1774. doi: 10.1096/fj.08-117739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ray R, Juranek JK, Rai V. RAGE axis in neuroinflammation, neurodegeneration and its emerging role in the pathogenesis of amyotrophic lateral sclerosis. Neurosci Biobehav Rev. 2016;62:48–55. doi: 10.1016/j.neubiorev.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 62.Hudson BI, Lippman ME. Targeting RAGE signaling in inflammatory disease. Annu Rev Med. 2018;69:349–364. doi: 10.1146/annurev-med-041316-085215. [DOI] [PubMed] [Google Scholar]
- 63.Corica D, Aversa T, Ruggeri RM, Cristani M, Alibrandi A, Pepe G, et al. Could AGE/RAGE-related oxidative homeostasis dysregulation enhance susceptibility to pathogenesis of cardio-metabolic complications in childhood obesity? Front Endocrinol (Lausanne) 2019;10:426. doi: 10.3389/fendo.2019.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Davis KE, Prasad C, Vijayagopal P, Juma S, Imrhan V. Advanced glycation end products, inflammation, and chronic metabolic diseases: links in a chain? Crit Rev Food Sci Nutr. 2016;56:989–998. doi: 10.1080/10408398.2012.744738. [DOI] [PubMed] [Google Scholar]
- 65.Derk J, MacLean M, Juranek J, Schmidt AM. The receptor for advanced glycation endproducts (RAGE) and mediation of inflammatory neurodegeneration. J Alzheimers Dis Parkinsonism. 2018;8:421. doi: 10.4172/2161-0460.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Juranek J, Ray R, Banach M, Rai V. Receptor for advanced glycation end-products in neurodegenerative diseases. Rev Neurosci. 2015;26:691–698. doi: 10.1515/revneuro-2015-0003. [DOI] [PubMed] [Google Scholar]
- 67.Clynes R, Moser B, Yan SF, Ramasamy R, Herold K, Schmidt AM. Receptor for AGE (RAGE): weaving tangled webs within the inflammatory response. Curr Mol Med. 2007;7:743–751. doi: 10.2174/156652407783220714. [DOI] [PubMed] [Google Scholar]
- 68.Mitchell JD, Borasio GD. Amyotrophic lateral sclerosis. Lancet. 2007;369:2031–2041. doi: 10.1016/S0140-6736(07)60944-1. [DOI] [PubMed] [Google Scholar]
- 69.Rowland LP, Shneider NA. Amyotrophic lateral sclerosis. N Engl J Med. 2001;344:1688–1700. doi: 10.1056/NEJM200105313442207. [DOI] [PubMed] [Google Scholar]
- 70.Richardson K, Allen SP, Mortiboys H, Grierson AJ, Wharton SB, Ince PG, et al. The effect of SOD1 mutation on cellular bioenergetic profile and viability in response to oxidative stress and influence of mutation-type. PLoS One. 2013;8:e68256. doi: 10.1371/journal.pone.0068256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Casula M, Iyer AM, Spliet WGM, Anink JJ, Steentjes K, Sta M, et al. Toll-like receptor signaling in amyotrophic lateral sclerosis spinal cord tissue. Neuroscience. 2011;179:233–243. doi: 10.1016/j.neuroscience.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 72.Lo Coco D, Veglianese P, Allievi E, Bendotti C. Distribution and cellular localization of high mobility group box protein 1 (HMGB1) in the spinal cord of a transgenic mouse model of ALS. Neurosci Lett. 2007;412:73–77. doi: 10.1016/j.neulet.2006.10.063. [DOI] [PubMed] [Google Scholar]
- 73.Hoyaux D, Boom A, van den Bosch L, Belot N, Martin JJ, Heizmann CW, et al. S100A6 overexpression within astrocytes associated with impaired axons from both ALS mouse model and human patients. J Neuropathol Exp Neurol. 2002;61:736–744. doi: 10.1093/jnen/61.8.736. [DOI] [PubMed] [Google Scholar]
- 74.Hoyaux D, Alao J, Fuchs J, Kiss R, Keller B, Heizmann CW, et al. S100A6, a calcium- and zinc-binding protein, is overexpressed in SOD1 mutant mice, a model for amyotrophic lateral sclerosis. Biochim Biophys Acta. 2000;1498:264–272. doi: 10.1016/S0167-4889(00)00101-4. [DOI] [PubMed] [Google Scholar]
- 75.Kamo H, Haebara H, Akiguchi I, Kameyama M, Kimura H, McGeer PL. A distinctive distribution of reactive astroglia in the precentral cortex in amyotrophic lateral sclerosis. Acta Neuropathol. 1987;74:33–38. doi: 10.1007/BF00688335. [DOI] [PubMed] [Google Scholar]
- 76.Kawaguchi M, Shibata N, Horiuchi S, Kobayashi M. Glyoxal inactivates glutamate transporter-1 in cultured rat astrocytes. Neuropathology. 2005;25:27–36. doi: 10.1111/j.1440-1789.2004.00579.x. [DOI] [PubMed] [Google Scholar]
- 77.Kato S, Horiuchi S, Liu J, Cleveland DW, Shibata N, Nakashima K, et al. Advanced glycation endproduct-modified superoxide dismutase-1 (SOD1)-positive inclusions are common to familial amyotrophic lateral sclerosis patients with SOD1 gene mutations and transgenic mice expressing human SOD1 with a G85R mutation. Acta Neuropathol. 2000;100:490–505. doi: 10.1007/s004010000226. [DOI] [PubMed] [Google Scholar]
- 78.Shibata N, Hirano A, Kato S, Nagai R, Horiuchi S, Komori T, et al. Advanced glycation endproducts are deposited in neuronal hyaline inclusions: a study on familial amyotrophic lateral sclerosis with superoxide dismutase-1 mutation. Acta Neuropathol. 1999;97:240–246. doi: 10.1007/s004010050980. [DOI] [PubMed] [Google Scholar]
- 79.Ding QX, Keller JN. Evaluation of rage isoforms, ligands, and signaling in the brain. Biochim Biophys Acta. 2005;1746:18–27. doi: 10.1016/j.bbamcr.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 80.Lee JD, McDonald TS, Fung JNT, Woodruff TM. Absence of receptor for advanced glycation end product (RAGE) reduces inflammation and extends survival in the hSOD1 G93A mouse model of amyotrophic lateral sclerosis. Mol Neurobiol. 2020;57:4143–4155. doi: 10.1007/s12035-020-02019-9. [DOI] [PubMed] [Google Scholar]
- 81.Ma L, Sun P, Zhang JC, Zhang Q, Yao SL. Proinflammatory effects of S100A8/A9 via TLR4 and RAGE signaling pathways in BV-2 microglial cells. Int J Mol Med. 2017;40:31–38. doi: 10.3892/ijmm.2017.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sternberg Z, Chiotti A, Tario J, Chichelli T, Patel N, Chadha K, et al. Reduced expression of membrane-bound (m)RAGE is a biomarker of multiple sclerosis disease progression. Immunobiology. 2016;221:193–198. doi: 10.1016/j.imbio.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 83.Santos G, Barateiro A, Brites D, Fernandes A. S100B impairs oligodendrogenesis and myelin repair following demyelination through RAGE engagement. Front Cell Neurosci. 2020;14:279. doi: 10.3389/fncel.2020.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rahimi M, Aghabozorg Afjeh SS, Omrani MD, Arsang-Jang S, Ganji M, Noroozi R, et al. Soluble receptor for advanced glycation end products (sRAGE) is up-regulated in multiple sclerosis patients treated with interferon β-1a. Cell Physiol Biochem. 2018;46:561–567. doi: 10.1159/000488622. [DOI] [PubMed] [Google Scholar]
- 85.Wetzels S, Vanmierlo T, Scheijen JLJM, van Horssen J, Amor S, Somers V, et al. Methylglyoxal-derived advanced glycation endproducts accumulate in multiple sclerosis lesions. Front Immunol. 2019;10:855. doi: 10.3389/fimmu.2019.00855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xiao YF, Sun Y, Liu W, Zeng FF, Shi JY, Li J, et al. HMGB1 promotes the release of sonic hedgehog from astrocytes. Front Immunol. 2021;12:584097. doi: 10.3389/fimmu.2021.584097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brás IC, König A, Outeiro TF. Glycation in Huntington's disease: a possible modifier and target for intervention. J Huntingtons Dis. 2019;8:245–256. doi: 10.3233/JHD-190366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim J, Waldvogel HJ, Faull RLM, Curtis MA, Nicholson LFB. The RAGE receptor and its ligands are highly expressed in astrocytes in a grade-dependant manner in the striatum and subependymal layer in Huntington's disease. J Neurochem. 2015;134:927–942. doi: 10.1111/jnc.13178. [DOI] [PubMed] [Google Scholar]
- 89.Son S, Bowie LE, Maiuri T, Hung CLK, Desmond CR, Xia JR, et al. High-mobility group box 1 links sensing of reactive oxygen species by huntingtin to its nuclear entry. J Biol Chem. 2019;294:1915–1923. doi: 10.1074/jbc.RA117.001440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Deane R, Yan SD, Submamaryan RK, LaRue B, Jovanovic S, Hogg E, et al. RAGE mediates amyloid-beta peptide transport across the blood-brain barrier and accumulation in brain. Nat Med. 2003;9:907–913. doi: 10.1038/nm890. [DOI] [PubMed] [Google Scholar]
- 91.Li M, Shang DS, Zhao WD, Tian L, Li B, Fang WG, et al. Amyloid beta interaction with receptor for advanced glycation end products up-regulates brain endothelial CCR5 expression and promotes T cells crossing the blood-brain barrier. J Immunol. 2009;182:5778–5788. doi: 10.4049/jimmunol.0803013. [DOI] [PubMed] [Google Scholar]
- 92.Liu LP, Hong H, Liao JM, Wang TS, Wu J, Chen SS, et al. Upregulation of RAGE at the blood-brain barrier in streptozotocin-induced diabetic mice. Synapse. 2009;63:636–642. doi: 10.1002/syn.20644. [DOI] [PubMed] [Google Scholar]
- 93.Fuller KNZ, Miranda ER, Thyfault JP, Morris JK, Haus JM. Metabolic derangements contribute to reduced sRAGE isoforms in subjects with Alzheimer's disease. Mediators Inflamm. 2018;2018:2061376. doi: 10.1155/2018/2061376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kong YY, Wang FS, Wang J, Liu CP, Zhou YP, Xu ZQ, et al. Pathological mechanisms linking diabetes mellitus and Alzheimer's disease: The receptor for advanced glycation end products (RAGE) Front Aging Neurosci. 2020;12:217. doi: 10.3389/fnagi.2020.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Criscuolo C, Fontebasso V, Middei S, Stazi M, Ammassari-Teule M, Yan SS, et al. Entorhinal Cortex dysfunction can be rescued by inhibition of microglial RAGE in an Alzheimer's disease mouse model. Sci Rep. 2017;7:42370. doi: 10.1038/srep42370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Son M, Oh S, Park H, Ahn H, Choi J, Kim H, et al. Protection against RAGE-mediated neuronal cell death by sRAGE-secreting human mesenchymal stem cells in 5xFAD transgenic mouse model. Brain Behav Immun. 2017;66:347–358. doi: 10.1016/j.bbi.2017.07.158. [DOI] [PubMed] [Google Scholar]
- 97.Gasparotto J, Ribeiro CT, Bortolin RC, Somensi N, Rabelo TK, Kunzler A, et al. Targeted inhibition of RAGE in substantia nigra of rats blocks 6-OHDA-induced dopaminergic denervation. Sci Rep. 2017;7:8795. doi: 10.1038/s41598-017-09257-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Viana SD, Valero J, Rodrigues-Santos P, Couceiro P, Silva AM, Carvalho F, et al. Regulation of striatal astrocytic receptor for advanced glycation end-products variants in an early stage of experimental Parkinson's disease. J Neurochem. 2016;138:598–609. doi: 10.1111/jnc.13682. [DOI] [PubMed] [Google Scholar]
- 99.Wang XL, Sun XX, Niu MY, Zhang XN, Wang J, Zhou C, et al. RAGE silencing ameliorates neuroinflammation by inhibition of p38-NF-κB signaling pathway in mouse model of Parkinson's disease. Front Neurosci. 2020;14:353. doi: 10.3389/fnins.2020.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu LP, Killoy KM, Vargas MR, Yamamoto Y, Pehar M. Effects of RAGE inhibition on the progression of the disease in hSOD1 G93A ALS mice. Pharmacol Res Perspect. 2020;8:e00636. doi: 10.1002/prp2.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Serrano A, Donno C, Giannetti S, Perić M, Andjus P, D'Ambrosi N, et al. The astrocytic S100B protein with its receptor RAGE is aberrantly expressed in SOD1 G93A models, and its inhibition decreases the expression of proinflammatory genes. Mediators Inflamm. 2017;2017:1626204. doi: 10.1155/2017/1626204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yamamoto Y, Liang MK, Munesue S, Deguchi K, Harashima A, Furuhara K, et al. Vascular RAGE transports oxytocin into the brain to elicit its maternal bonding behaviour in mice. Commun Biol. 2019;2:76. doi: 10.1038/s42003-019-0325-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yamamoto Y, Higashida H. RAGE regulates oxytocin transport into the brain. Commun Biol. 2020;3:70. doi: 10.1038/s42003-020-0799-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Franklin TC, Wohleb ES, Zhang Y, Fogaça M, Hare B, Duman RS. Persistent increase in microglial RAGE contributes to chronic stress-induced priming of depressive-like behavior. Biol Psychiatry. 2018;83:50–60. doi: 10.1016/j.biopsych.2017.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fu JW, Zuo X, Yin JW, Luo XD, Li Z, Lin JD, et al. Association of polymorphisms of the receptor for advanced glycation endproducts gene with schizophrenia in a Han Chinese population. Biomed Res Int. 2017;2017:6379639. doi: 10.1155/2017/6379639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Miyashita M, Watanabe T, Ichikawa T, Toriumi K, Horiuchi Y, Kobori A, et al. The regulation of soluble receptor for AGEs contributes to carbonyl stress in schizophrenia. Biochem Biophys Res Commun. 2016;479:447–452. doi: 10.1016/j.bbrc.2016.09.074. [DOI] [PubMed] [Google Scholar]
- 107.Zglejc-Waszak K, Mukherjee K, Juranek JK. The cross-talk between RAGE and DIAPH1 in neurological complications of diabetes: a review. Eur J Neurosci. 2021;54:5982–5999. doi: 10.1111/ejn.15433. [DOI] [PubMed] [Google Scholar]
- 108.Sloan G, Selvarajah D, Tesfaye S. Pathogenesis, diagnosis and clinical management of diabetic sensorimotor peripheral neuropathy. Nat Rev Endocrinol. 2021;17:400–420. doi: 10.1038/s41574-021-00496-z. [DOI] [PubMed] [Google Scholar]
- 109.Juranek JK, Kothary P, Mehra A, Hays A, Brannagan THI, Schmidt AM. Increased expression of the receptor for advanced glycation end-products in human peripheral neuropathies. Brain Behav. 2013;3:701–709. doi: 10.1002/brb3.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Juranek JK, Geddis MS, Song F, Zhang JH, Garcia J, Rosario R, et al. RAGE deficiency improves postinjury sciatic nerve regeneration in type 1 diabetic mice. Diabetes. 2013;62:931–943. doi: 10.2337/db12-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Juranek JK, Aleshin A, Rattigan EM, Johnson L, Qu W, Song F, et al. Morphological changes and immunohistochemical expression of RAGE and its ligands in the sciatic nerve of hyperglycemic pig (Sus scrofa) Biochem Insights. 2010;2010:47–59. doi: 10.4137/BCI.S5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lukic IK, Humpert PM, Nawroth PP, Bierhaus A. The RAGE pathway: activation and perpetuation in the pathogenesis of diabetic neuropathy. Ann N Y Acad Sci. 2008;1126:76–80. doi: 10.1196/annals.1433.059. [DOI] [PubMed] [Google Scholar]
- 113.Bekircan-Kurt CE, Tan E, Erdem ÖS. The activation of RAGE and NF-KB in nerve biopsies of patients with axonal and vasculitic neuropathy. Noro Psikiyatr Ars. 2015;52:279–282. doi: 10.5152/npa.2015.8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Klein I, Lehmann HC. Pathomechanisms of paclitaxel-induced peripheral neuropathy. Toxics. 2021;9:229. doi: 10.3390/toxics9100229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sekiguchi F, Domoto R, Nakashima K, Yamasoba D, Yamanishi H, Tsubota M, et al. Paclitaxel-induced HMGB1 release from macrophages and its implication for peripheral neuropathy in mice: Evidence for a neuroimmune crosstalk. Neuropharmacology. 2018;141:201–213. doi: 10.1016/j.neuropharm.2018.08.040. [DOI] [PubMed] [Google Scholar]
- 116.Tsubota M, Miyazaki T, Ikeda Y, Hayashi Y, Aokiba Y, Tomita S, et al. Caspase-dependent HMGB1 release from macrophages participates in peripheral neuropathy caused by bortezomib, a proteasome-inhibiting chemotherapeutic agent, in mice. Cells. 2021;10:2550. doi: 10.3390/cells10102550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wafa A, El-Nahas M, Al Biaumy A, Mansour Y. Study of advanced glycation endproducts and their receptors in Egyptian type 2 diabetic individuals with peripheral neuropathy. Egypt J Obes Diabet Endocrinol. 2017;3:15. doi: 10.4103/2356-8062.205209. [DOI] [Google Scholar]
- 118.Lam D, Momeni Z, Theaker M, Jagadeeshan S, Yamamoto Y, Ianowski JP, et al. RAGE-dependent potentiation of TRPV1 currents in sensory neurons exposed to high glucose. PLoS One. 2018;13:e0193312. doi: 10.1371/journal.pone.0193312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bestall SM, Hulse RP, Blackley Z, Swift M, Ved N, Paton K, et al. Sensory neuronal sensitisation occurs through HMGB-1-RAGE and TRPV1 in high-glucose conditions. J Cell Sci. 2018;131:jcs215939. doi: 10.1242/jcs.215939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Osonoi S, Mizukami H, Ogasawara S, Takahashi K, Sango K, Yagihashi S. Suppression of neuropathy development in diabetic rage-deficient mice is associated with absence of M1/M2 macrophage skewing in the sciatic nerve. Diabetes. 2018 doi: 10.2337/db18-575-P. [DOI] [Google Scholar]
- 121.Wautier JL, Guillausseau PJ. Advanced glycation end products, their receptors and diabetic angiopathy. Diabet Metab. 2001;27:535–542. [PubMed] [Google Scholar]
- 122.Li XN, Yang HQ, Ouyang Q, Liu FT, Li J, Xiang ZH, et al. Enhanced RAGE expression in the dorsal root ganglion may contribute to neuropathic pain induced by spinal nerve ligation in rats. Pain Med. 2016;17:803–812. doi: 10.1093/pm/pnv035. [DOI] [PubMed] [Google Scholar]
- 123.Solmaz V, Kaya M, Uslu FB, Atasoy O, Erbaş O. Papaverine has therapeutic potential for Sepsis-induced neuropathy in rats, possibly via the modulation of HMGB1-RAGE axis and its antioxidant prosperities. J Invest Surg. 2022;35:7–13. doi: 10.1080/08941939.2020.1809751. [DOI] [PubMed] [Google Scholar]
- 124.Wang MY, Ross-Cisneros FN, Aggarwal D, Liang CY, Sadun AA. Receptor for advanced glycation end products is upregulated in optic neuropathy of Alzheimer's disease. Acta Neuropathol. 2009;118:381–389. doi: 10.1007/s00401-009-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tezel G, Luo C, Yang XJ. Accelerated aging in glaucoma: Immunohistochemical assessment of advanced glycation end products in the human retina and optic nerve head. Invest Ophthalmol Vis Sci. 2007;48:1201–1211. doi: 10.1167/iovs.06-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Łacheta D, Poślednik KB, Czerwaty K, Ludwig N, Molińska-Glura M, Kantor I, et al. RAGE and HMGB1 expression in orbital tissue microenvironment in Graves' ophthalmopathy. Mediators Inflamm. 2021;2021:8891324. doi: 10.1155/2021/8891324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hudson BI, Stickland MH, Futers TS, Grant PJ. Effects of novel polymorphisms in the RAGE gene on transcriptional regulation and their association with diabetic retinopathy. Diabetes. 2001;50:1505–1511. doi: 10.2337/diabetes.50.6.1505. [DOI] [PubMed] [Google Scholar]
- 128.Hammes HP, Hoerauf H, Alt A, Schleicher E, Clausen JT, Bretzel RG, et al. N(Epsilon)(carboxymethyl)lysin and the AGE receptor RAGE colocalize in age-related macular degeneration. Invest Ophthalmol Vis Sci. 1999;40:1855–1859. [PubMed] [Google Scholar]
- 129.Sakatani S, Yamada K, Homma C, Munesue S, Yamamoto Y, Yamamoto H, et al. Deletion of RAGE causes hyperactivity and increased sensitivity to auditory stimuli in mice. PLoS One. 2009;4:e8309. doi: 10.1371/journal.pone.0008309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Xing Y, Ji QH, Li XM, Ming J, Zhang NN, Zha DJ, et al. Asiaticoside protects cochlear hair cells from high glucose-induced oxidative stress via suppressing AGEs/RAGE/NF-κB pathway. Biomed Pharmacother. 2017;86:531–536. doi: 10.1016/j.biopha.2016.12.025. [DOI] [PubMed] [Google Scholar]
- 131.Lee CH, Kim KW, Lee DH, Lee SM, Kim SY. Overexpression of the receptor for advanced glycation end-products in the auditory cortex of rats with noise-induced hearing loss. BMC Neurosci. 2021;22:38. doi: 10.1186/s12868-021-00642-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gao X, Lin C, Lu L, Liang G, Chen Z, Zhang X. RAGE, NF-kappaB, p21 expressions in mouse spiral ganglion cells. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2014;28:265–268. [PubMed] [Google Scholar]
- 133.Angelopoulou E, Paudel YN, Piperi C. Unraveling the role of receptor for advanced glycation end products (RAGE) and its ligands in myasthenia gravis. ACS Chem Neurosci. 2020;11:663–673. doi: 10.1021/acschemneuro.9b00678. [DOI] [PubMed] [Google Scholar]
- 134.Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]