Abstract
Wastewater surveillance (WS) of SARS-CoV-2 currently requires multiple steps and suffers low recoveries and poor sensitivity. Here, we report an improved analytical method with high sensitivity and recovery to quantify SARS-CoV-2 RNA in wastewater. To improve the recovery, we concentrated SARS-CoV-2 viral particles and RNA from both the solid and aqueous phases of wastewater using an electronegative membrane (EM). The captured viral particles and RNA on the EM were incubated in our newly developed viral inactivation and RNA preservation (VIP) buffer. Subsequently, the RNA was concentrated on magnetic beads and inhibitors removed by washing. Without eluting, the RNA on the magnetic beads was directly detected using reverse transcription quantitative polymerase chain reaction (RT-qPCR). Analysis of SARS-CoV-2 pseudovirus (SARS-CoV-2 RNA in a noninfectious viral coat) spiked to wastewater samples showed an improved recovery of 80%. Analysis of 120 wastewater samples collected twice weekly between May 2021 and February 2022 from two wastewater treatment plants showed 100% positive detection, which agreed with the results independently obtained by a provincial public health laboratory. The concentrations of SARS-CoV-2 RNA in these wastewater samples ranged from 2.4×102 to 2.9×106 copies per 100 mL of wastewater. Our method's capability of detecting trace and diverse concentrations of SARS-CoV-2 in complex wastewater samples is attributed to the enhanced recovery of SARS-CoV-2 RNA and efficient removal of PCR inhibitors. The improved method for the recovery and detection of viral RNA in wastewater is important for wastewater surveillance, complementing clinical diagnostic tests for public health protection.
Keywords: Wastewater, Virus, Viral RNA, Environmental monitoring, Pathogens, Surveillance
Graphical abstract
Introduction
The Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) that caused the COVID-19 pandemic has created unprecedented impacts worldwide. SARS-CoV-2 is continuously mutating, and at least five variants of concerns (VOCs) (Alpha, Beta, Gamma, Delta, and Omicron) have been reported by World Health Organization (WHO, 2022a). The WHO has reported over 584 million total cases of SARS-CoV-2, and over 6.4 million deaths, with cases continuing to rise and new Omicron sub-variants dominating globally (WHO, 2022b). The emergence of these VOCs in combination with waning population immunity and risk of re-infection create a need for efficient community surveillance (Hrudey et al., 2022).
The conventional approach for community surveillance of SARS-CoV-2 heavily relies on clinical testing using nasopharyngeal swab (NPS) sample collection followed by reverse transcription quantitative polymerase chain reaction (RT-qPCR) detection. However, using the clinical diagnostic tests alone for public health surveillance can become overwhelmed during a pandemic situation. Furthermore, to manage the demand for testing and shortages in diagnostic resources, healthcare systems on a global scale are often limited to only testing symptomatic patients and close contacts. Thus, pre-symptomatic, asymptomatic, and mild symptomatic cases, which can significantly contribute to the spread of SARS-CoV-2, are often undetected by clinical diagnostics. As a result, clinical testing of SARS-CoV-2 alone underestimates the true scale of the pandemic, and public health officials must make decisions on community quarantine guidelines with inadequate surveillance data.
Wastewater surveillance has become a useful public health tool for assessing the status of community infections, and many organizations are adopting WS for national surveillance programs (Hrudey et al., 2022). For example, the U.S. Centers for Disease Control and Prevention (CDC, 2022), the International Water Association (IWA, 2022), the Global Institute for Water Security (2022), the Public Health Agency of Canada (NCCID, 2022), and the Canadian Water Network (CWN, 2022) have adopted WS to complement clinical testing for monitoring the status and trends of the COVID-19 pandemic in the community.
Wastewater is a composite sample of the entire community, containing fecal, urine, and other biological products from individuals in the community. Several studies have demonstrated that up to 89% of infected patients shed SARS-CoV-2 viral particles and RNA into feces as early as one-day post-infection (Wolfel et al., 2020; Zhang et al., 2020; Gupta et al., 2020; CDC, 2022). Additionally, respiratory secretions of SARS-CoV-2 through bath, shower, and laundry water are also captured in wastewater. Several studies have even reported successful detection of SARS-CoV-2 and variants in wastewater up to 14 days prior to clinically reported cases (Karthikeyan et al., 2022; Vo et al., 2022; Xie et al., 2022). Sensitivity analysis showed that the viral RNA could be detected in wastewater at 99% probability if there were higher than 38 new cases (range 17-97) per 100,000 people in the community (Le, 2023; Li et al., 2023).
Because of the need to respond rapidly to the COVID-19 pandemic, current WS of SARS-CoV-2 is based on individually accessible laboratory methods without national or international standardized procedures. WS of viruses generally involves multiple steps, including sample collection, viral particle concentration, RNA extraction, and RT-qPCR detection. To detect SARS-CoV-2 RNA accurately and sensitively in wastewater, a WS process with a high overall recovery is required. However, multi-laboratory studies have found poor recoveries of surrogates (enveloped viruses) or SARS-CoV-2 from wastewater, with recoveries ranging from 0.08% to 66% amongst the participating laboratories (Chik et al., 2021; Kantor et al., 2021; Kumblathan et al., 2021, Kumblathan et al., 2022; Pecson et al., 2021). Four major factors contribute to the poor recoveries and large variations: (1) the use of only the aqueous phase of wastewater samples while discarding the solid phase; (2) incomplete concentration of viral particles and viral RNA from wastewater; (3) inefficient RNA extraction and inadequate removal of RT-qPCR inhibitors; and (4) insufficient sample volume used for analysis, especially when viral loads are very low. The poor reproducibility within the studies can also be attributed to sample-to-sample matrix differences. Insufficient removal of matrix interference during the concentration and RNA extraction steps can contribute to inaccurate and inconsistent RT-qPCR results. Therefore, we aimed to develop a method for enhanced recovery and detection of viral RNA in wastewater. To achieve this, we will focus on three main components: (1) efficient capture and concentration of viral particles and RNA from both liquid and solid phases, (2) efficient removal of the sample matrices, particularly RT-qPCR inhibitors while ensuring efficient extraction of viral RNA, and (3) capability of handling large volume of samples.
Here we report an improved analytical method that can provide consistent and highly efficient concentration of viral particles, extraction of RNA, and removal of inhibitors for RT-qPCR detection, resulting in highly sensitive detection of SARS-CoV-2 RNA in wastewater. The method involves concentration of viral particles and RNA from the whole wastewater sample using electronegative membrane (EM) filtration, followed by viral inactivation and RNA preservation (VIP), magnetic capture of RNA, and RT-qPCR detection of the N1 gene of SARS-CoV-2 (VIP-Mag-RT-qPCR). The EM filtration used in this method allows for the concentration of viruses from a large volume of sample. Our improved method, EM-VIP-Mag-RT-qPCR, allows for enhanced recovery and detection of RNA of SARS-CoV-2 and variants in wastewater. The EM-VIP-Mag-RT-qPCR method is also inexpensive and broadly accessible, therefore it can be widely used for the detection of viral RNA in wastewater samples containing both high and low concentrations of SARS-CoV-2 viral particles and RNA.
1. Materials and methods
1.1. Reagents
Proteinase K and Buffer RLT lysis buffer were purchased from QIAgen (Germantown, MD, USA). TaqPath 1-Step RT-qPCR Master Mix, CG, UltraPure guanidine isothiocyanate, Gibco Beef Extract Powder, RNA-grade glycogen, and THE RNA Storage Solution were bought from ThermoFisher Scientific (Carlsbad, CA, USA). Polyethylene glycol (PEG) Bio Ultra 8000, glycine (electrophoresis grade), magnesium chloride hexahydrate, and the proteinase K Inhibitor (tetrapeptidyl chloromethyl ketone) were bought from MilliporeSigma (Oakville, Ontario, Canada). SPRIselect magnetic beads were bought from Beckman Coulter (Brea, CA, USA). 2-Mercaptoethanol (2-ME), biotechnology grade, was bought from BioShop Canada (Burlington, Ontario, Canada). RNasin Plus Rnase Inhibitor was bought from Promega (Madison, WI, USA). Pseudovirus (SARS-CoV-2 RNA targets in a noninfectious viral coat) solution, AccuPlex SARS-CoV-2 Verification Panel-Full Genome, was bought from Sera Care, LGC (Milford, MA, USA). Purified SARS-CoV-2 RNA was provided by our colleagues in the Department of Medical Microbiology and Immunology at the University of Alberta. CDC N1 and N2 primer-probes 2019-nCoV RUO kit was bought from Integrated DNA Technologies (Coralville, IA, USA).
1.2. Wastewater sample collection
Using appropriate personal protective equipment, wastewater samples were collected from two wastewater treatment plants (WWTPs) in Calgary and Edmonton, Alberta, Canada. Five hundred milliliters of post-grit raw influent wastewater samples from a 24 hr composite sampler were collected twice a week from May 2021 to October 2021. After October 2021, sampling frequency was increased to three times per week. All the samples were stored at 4°C after collection and shipped to Dr. Lilly (Xiaoli) Pang's research laboratory on a weekly basis. We refer to Dr. Pang's laboratory as the reference laboratory in the following discussion as they participate in the Pan Alberta WS program. An aliquot of the same sample was analyzed by Pang's lab using their method (Hasing et al., 2021; Qiu et al., 2022) and another aliquot was analyzed using our in-house method described in detail below.
1.3. Concentration of SARS-CoV-2 from wastewater samples
An aliquot of 80 mL of sample was transferred into a sterile DNase free and RNase free conical tube and centrifuged at 1510 ×g for 30 min. This allowed for the separation of the aqueous and solid phase. The resulting supernatant of the aqueous phase was transferred into another DNase and RNase free sterile conical tube and the solid pellet was re-suspended in beef extract solution (3% W/V beef extract in 0.05 mol/L glycine (pH 9.0)) at a ratio of 1:5. The pellet suspension was agitated at 800 r/min for 30 min at room temperature on a shaker followed by centrifugation at 1510 ×g for 10 min. The resulting supernatant was transferred into a new tube, neutralized with HCl and then was combined with the aqueous phase. Then MgCl2 (1 mol/L) was added into each sample to reach the final concentration of 25 mmol/L. The treated wastewater sample was then filtered through an electronegative membrane (EM) using a vacuum filtration set-up. The mixed cellulose ester (MCE) membrane filter had a diameter of 47 mm and a pore size of 0.45 µm (Millipore Sigma).
1.4. Extraction of viral RNA from wastewater samples using the Viral Inactivation and Preservation (VIP) buffer
The EMs containing the captured SARS-CoV-2 particles were directly used to extract viral RNA. The viral inactivation and preservation (VIP) buffer, developed in-house (Liu et al., 2022), was used for RNA extraction. The EMs were vortexed thoroughly in 600 µL of the VIP buffer, placed on a heating block for 10 min at 55°C, centrifuged for 2 min at 13,000 ×g, and the final supernatant was transferred to a new tube.
1.5. Concentration of extracted viral RNA on magnetic beads
The supernatant containing the extracted SARS-CoV-2 RNA is then incubated with the in-house developed magnetic beads mixture. The details on the preparation and optimization of the magnetic beads buffer can be found in our previous publication (Liu et al., 2022). 400 µL of the magnetic beads suspension and 200 µL of 100% ethanol (RNA grade) were added into the extracted RNA supernatant, vortexed, and incubated on a shaker at room temperature for 10 min. Samples were then centrifuged to collect the magnetic beads and the resulting beads were washed three times with 0.8 mL of 75% ethanol containing sodium citrate. The magnetic beads containing the extracted RNA were air-dried and resuspended in a solution containing 25 µL of RNase-free water, 4 µL proteinase K inhibitor, and 1 µL RNase inhibitor.
1.6. RT-qPCR analysis of concentrated viral RNA directly from magnetic beads
The TaqPath 1-Step RT-qPCR Master Mix (Thermo Fisher Scientific) and CDC N1 primer-probes from the 2019-nCoV RUO kit (IDT) were used according to the manufacturers’ instructions. The RT-qPCR assay for the N1 segment of the SARS-CoV-2 RNA was optimized as described previously (Liu et al., 2022). RT-qPCR was performed on a QuantStudio 3 Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific) using the QuantStudio Design and Analysis Software v1.5.1. The 1/6 portion of the extracted RNA was used as template in each RT-qPCR reaction. The RT-qPCR standard curve of the N1 gene segment (Appendix A Fig. S4) was used to convert the Ct values to viral RNA copies. The number of SARS-CoV-2 RNA copies per 100 mL of original wastewater sample was obtained using the following equation: RNA copies/100 mL = RNA copies × 6 × 100 mL/80 mL.
1.7. Recovery experiments
The recovery experiments were performed using a non-infectious SARS-CoV-2 pseudovirus solution containing both viral particles and free viral RNA (purchased from Sera Care, LGC, Milford, MA, USA). The copy number of total RNA in the solution was determined in our previous study (Liu et al., 2022). An aliquot of this solution containing 50200 copies of the viral RNA was spiked in two sets of wastewater samples for recovery experiments. In the first set of recovery experiments, three previously determined SARS-CoV-2 negative wastewater samples (1 L each) were pooled together. Aliquots of 80 mL of the pooled negative samples were used for spiking. In the second set of recovery experiments, six positive wastewater samples containing low SARS-CoV-2 RNA copies were pooled. Aliquots of 80 mL were spiked with the SARS-CoV-2 pseudovirus. The two sets of recovery experiments were repeated twice on different days with triplicate sample aliquots analyzed in each set of repeats. For spiked negative samples, the recovery of SARS-CoV-2 RNA in each sample was calculated as the amount of SARS-CoV-2 RNA measured divided by that of pseudovirus SARS-CoV-2 RNA spiked into the sample. For spiked wastewater samples with low concentrations of SARS-CoV-2 RNA, the amount of previously determined SARS-CoV-2 RNA was subtracted from the total SARS-CoV-2 RNA. The net measured amount divided by the amount of pseudovirus SARS-CoV-2 RNA spiked into the sample yielded the recovery value.
2. Results and discussion
2.1. Development of an EM-VIP-Mag-RT-qPCR method for enhanced detection of SARS-CoV-2 in wastewater
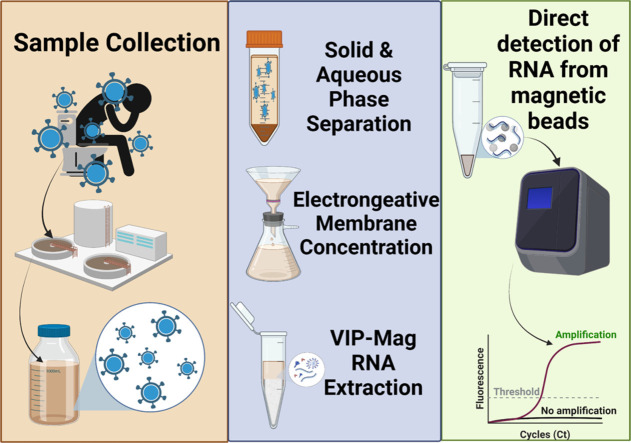
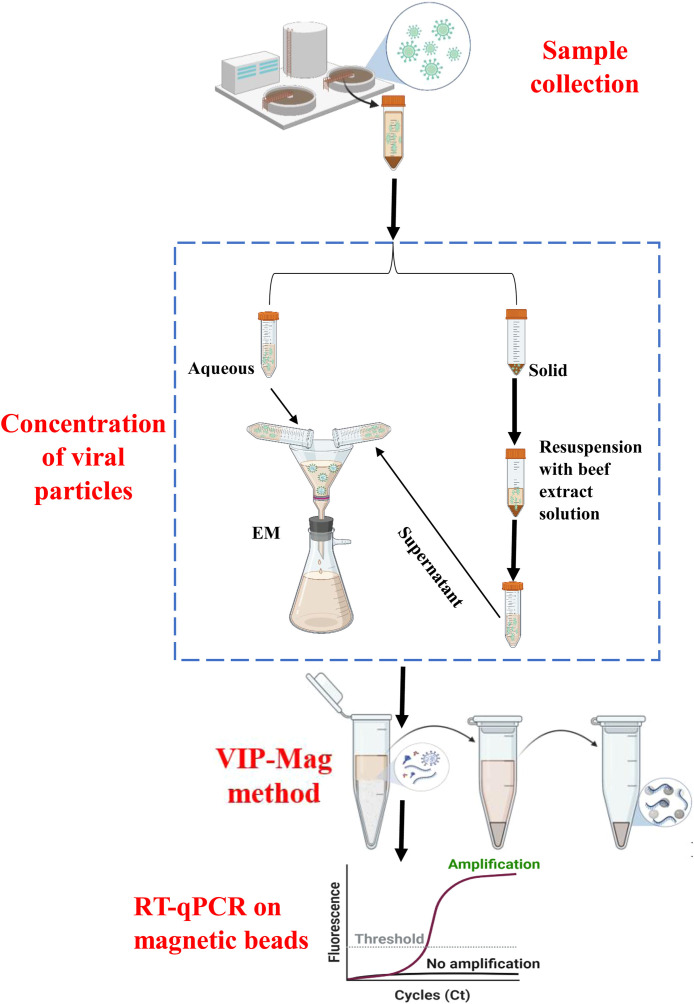
Fig. 1 shows the major steps of our method for the detection of SARS-CoV-2 in wastewater, including EM concentration of viral particles and RNA from the combined aqueous phase and supernatant collected from the solids pellet of each sample, RNA extraction and removal of inhibitors using the VIP-Mag method, and direct detection of RNA on magnetic beads by RT-qPCR. The methods and reagents for each step were carefully selected and optimized to improve recovery of SARS-CoV-2. The following sections describe the rationale and improvements of our method for the enhanced wastewater analysis of viral RNA.
Fig. 1.
Schematic showing steps of wastewater sample processing and analysis, including sample collection, concentration of viral particles and RNA using an electronegative membrane (EM), extraction of RNA onto magnetic beads along with viral inactivation and RNA preservation (VIP-Mag), and direct RT-qPCR detection of RNA on magnetic beads.
2.2. Variable distribution of viral RNA in solid and aqueous phase of wastewater samples
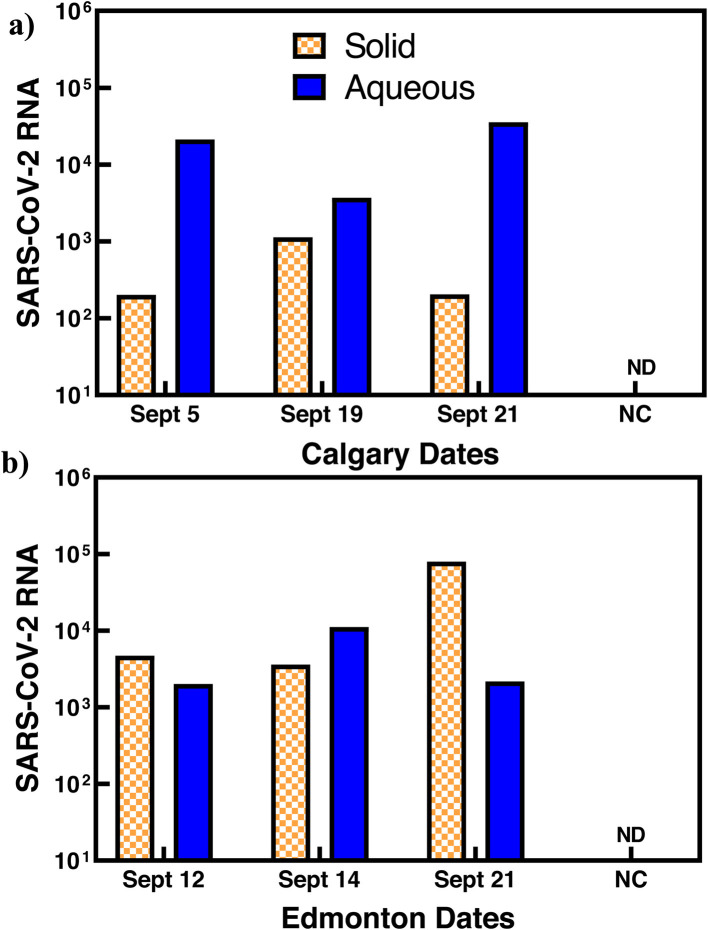
To improve the efficiency and reproducibility of the concentration of viral particles and RNA from wastewater samples, we aimed to collect all SARS-CoV-2 viral particles and RNA from both the solid and aqueous phases of the wastewater. We separately analyzed the aqueous and solid phases of three representative wastewater samples collected from WWTPs in Calgary and Edmonton (Appendix A Table S1). Fig. 2 presents the RNA copies detected in both the solid and aqueous phases of these three samples. These results show that significant amounts of viral RNA were detected in both the solid phase (402 – 159,226 RNA copies/100 mL) and aqueous phase (4,049 – 71,100 RNA copies/100 mL) of the samples. The detected copies of the viral RNA in the aqueous and solid phases varied between samples even when the samples were collected from the same sewer system. The percentage of SARS-CoV-2 RNA present in the solid phase varied from1%-23% in the Calgary samples and 24%-97% in the Edmonton samples. Three of the Calgary wastewater samples had higher viral RNA concentrations in the aqueous phase than in the solid phase (Fig. 2a). In the Edmonton wastewater samples, two samples had higher viral RNA concentrations in the solid phase than aqueous phase. The variable distribution of viral RNA in both solid and aqueous phases demonstrates that discarding either the solid or liquid phase of wastewater can result in large variations, lower recoveries, and falsely decreased or even negative results. Therefore, the use of both the solid and aqueous phase is important to improve recoveries and reduce variations resulting from unpredictable composition of wastewater.
Fig. 2.
Separate analysis of the aqueous and solid phases of wastewater samples collected from two wastewater treatment plants in the cities of Calgary (a) and Edmonton (b). NC indicates negative control and ND indicates not detectable. All wastewater samples collected from both wastewater treatment plants have detectable SARS-CoV-2 RNA in both the aqueous phase and solid phase. The relative amounts of SARS-CoV-2 RNA in the aqueous phase and solid phase vary with the samples.
The results in Fig. 2 clarify one of the major contributors to low recovery with large variations in previous WS of SARS-CoV-2 studies, as many of these studies excluded the solid phase and analyzed only the aqueous phase (Balboa et al., 2021; Chik et al., 2021; Hata et al., 2021; LaTurner et al., 2021; Pecson et al., 2021). While discarding solids may reduce inhibitors for the subsequent RT-qPCR analysis, this practice could underestimate the overall SARS-CoV-2 concentrations in wastewater samples. Several studies have demonstrated that SARS-CoV-2 particles could be detected in 2-3 magnitude higher concentrations in solids and sludge of some wastewater samples than in the aqueous phase (Peccia et al., 2020; D'Aoust et al., 2021; Graham et al., 2021; Carrillo-Reyes et al., 2021; Westhaus et al., 2021; Buonerba et al., 2021). Roldan-Hernandez et al. (2022) have shown persistence of SARS-CoV-2 in solids with a slower rate of decay than in the wastewater influent, further suggesting the importance of including the solid phase which can adsorb the viral particles and RNA. Our results suggest the need for a simple and efficient procedure for concentration of viral particles and RNA from the entire sample.
To achieve highly efficient and reproducible recovery, we designed our method using the 3% beef extract solution to release the viral particles from the solids. The resultant supernatant was then re-combined with the aqueous phase for subsequent concentration of viral particles and RNA (Fig. 1). This approach facilitates the efficient concentration of viral particles and RNA from wastewater samples, reduces loss of viral particles and RNA adsorbed on solids, and minimizes interference of solid matter on subsequent concentration and extraction steps.
2.3. EM filtration of wastewater and concentration of viral particles and RNA
A challenge of analyzing wastewater is the low abundance of target viral particles and RNA and complicated sample matrix that may inhibit RT-qPCR. Therefore, our objective is to efficiently concentrate the viral particles and RNA from wastewater while minimizing co-concentration of the inhibitory matrix. We chose to use filtration with electronegative membranes (EM) for the concentration of viral particles and RNA to achieve cost-effective, fast, and large volume sample processing. It is expected that co-concentration of inhibitors is minimal, compared to other wastewater processing methods (Lu et al., 2020).
The principle of EM capture is based on the idea that SARS-CoV-2 viral particles and RNA is negatively charged. The EM allows small particulate matter and debris smaller than the 0.45 µm pore size to pass through the membrane while capturing viral particles and RNA on the membrane via charge interactions. Adding MgCl2 allows for Mg2+ to serve as salt bridge to facilitate the adsorption of negatively charged viral particle and RNA to the EM (Shi et al., 2017). This technique is widely accessible as only a vacuum filtration set-up is needed. We tested two common types of EMs (MilliporeⓇ): mixed cellulose ester (MCE) membrane filters and sterilely packed (SPAK) gridded mixed cellulose ester membrane filters. We first prepared triplicate spiked samples containing an equivalent of 1941 RNA copies of SARS-CoV-2 pseudovirus solution into deionized water (diH2O). These spiked samples were analyzed to evaluate the capture of viral particles and RNA on the SPAK and MCE membranes. As shown in Appendix A Fig. S1, the MCE membranes captured more spiked SARS-CoV-2 RNA (1823 ± 131), while the SPAK membranes captured lower copies of RNA (849 ± 218). To better understand the performance of the membranes with real wastewater samples, we spiked SARS-CoV-2 pseudovirus solution (containing 1941 RNA copies) into previously confirmed SARS-CoV-2 negative wastewater samples and analyzed the samples in triplicate using both the SPAK and MCE membrane. The MCE membranes again captured more spiked SARS-CoV-2 RNA than SPAK, supported by RNA copies of 1560 ± 226 and 1063 ± 139 and recoveries of 80% and 55%, respectively in the wastewater samples (Appendix A Fig. S1). Thus, the MCE EM was used for all the subsequent experiments. Furthermore, Ahmed et al., 2020 has used MCE EM to concentrate Murine Hepatitis Virus, a surrogate of SARS-CoV-2, from wastewater and obtained a recovery of 65% ± 23%, indicating that EM are appropriate for concentration of viruses from wastewater.
2.4. VIP-Mag-RT-qPCR for efficient removal of RT-qPCR inhibitors and ultra-sensitive in-situ amplification on beads
Detection of SARS-CoV-2 by RT-qPCR is highly impacted by the purity and integrity of the RNA template extracted. Enzymes involved in the RT-qPCR can be partially or completely inhibited by the residual sample matrix components present in the RNA extract, possibly leading to false-negative results (Schrader et al., 2012; Graham et al., 2021; Kitajima et al., 2020). Wastewater contains a variety of compounds, such as ions, bile salts, urea, alcohols, and numerous proteins (e.g., collagen, myoglobin, hemoglobin, and RNases). Many of these can inhibit RT-qPCR while the RNases can degrade RNA (Kumblathan et al., 2021). We overcame this challenge by designing efficient steps of removing inhibitors while sufficiently degrading RNases. During the first solid phase separation step, we removed most of the RT-qPCR inhibitors, where the viral particles were released into the beef extract solution, and the remaining solids were discarded to remove the associated RT-qPCR inhibitors. Secondly, we used our in-house developed VIP-Mag method, which concentrated pure RNA with high integrity. Our VIP buffer, which includes reagents such as 2-Mercaptoethanol, guanidinium isothiocyanate, Triton X-100, proteinase K, and glycogen, effectively lysed the SARS-CoV-2 viral particles and denatured RNases (Liu et al., 2022; Ramón-Nuńez et al., 2017). The extracted RNA was then captured on magnetic beads, and any remaining RT-qPCR inhibitors were removed through repeated washing of the beads (Appendix A Fig. S2). Thirdly, final addition of the RNase inhibitor and proteinase K inhibitor to the extracted RNA minimized any degradation of RNA and protein enzymes.
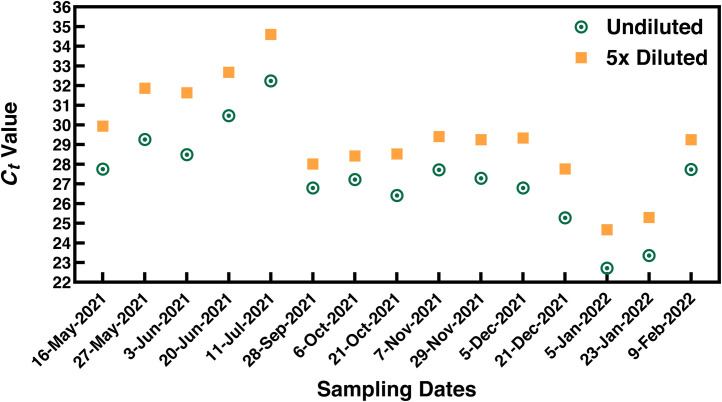
To confirm that there was no RT-qPCR inhibition, we analyzed both undiluted and five-fold diluted RNA on magnetic beads (Fig. 3 ). If RT-qPCR inhibition were present, the five-time dilution would minimize the inhibition, leading to a lower Ct for positive detections. Appendix A Table S2 shows that the differences (ΔC t) in the Ct values between the 5× diluted and undiluted beads samples ranged from 0.8 to 2.5. The mean ΔCt was 1.7±0.6, which is close to the excepted ΔCt value of 2.3 from a 5× dilution, suggesting that no apparent inhibitors are present in the undiluted magnetic beads suspension. These results demonstrate that our VIP-Mag method achieved sufficient removal of RT-qPCR inhibitors.
Fig. 3.
Analysis of undiluted and five-time diluted sample extracts. Monitoring of the presence of RT-qPCR inhibitors by monthly analyzing two samples. Two wastewater samples were analyzed every month from May 2021 to February 2022 alongside regular wastewater sample analysis using the EM-VIP-Meg-RT-qPCR protocol. Prior to RT-qPCR analysis, magnetic beads samples were diluted 5x and run simultaneously alongside the undiluted sample. The differences (ΔCt) in the Ct Values of the 5x diluted and undiluted beads samples are summarized in Appendix A Table S2 and range from 0.8 to 2.5. The mean ΔCt was 1.7 ± 0.6. Our results demonstrate that our VIP-Mag method allows for sufficient removal of RT-qPCR inhibitors without the need for dilution.
2.5. Improved recovery of viral RNA
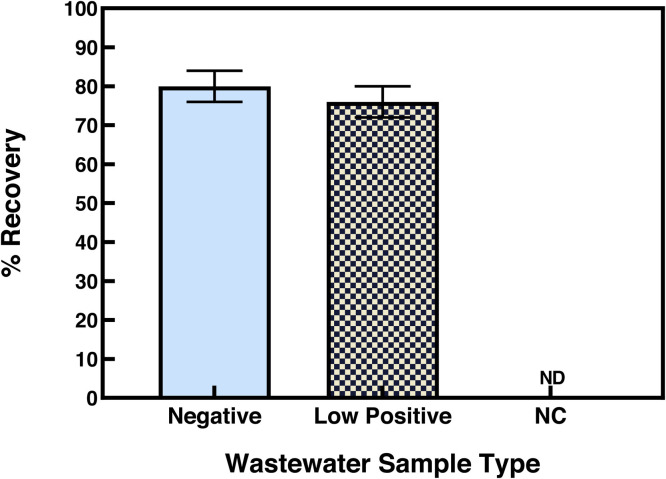
Having optimized our EM-VIP-Mag-RT-qPCR method, we evaluated the overall recovery of SARS-CoV-2 by analyzing two sets of wastewater samples spiked with SARS-CoV-2 pseudovirus. Prior to these spiking experiments, the two sets of the wastewater samples were confirmed to be negative or positive with low copies of SARS-CoV-2 RNA by the Alberta Public Health Laboratory. In the first set of recovery experiments using negative wastewater samples, the recovery was 80% ± 4% (Fig. 4 ). In the second set of recovery experiments using positive samples with low copies of RNA, the recovery was 76% ± 4% (Fig. 4). The second set experiment was performed to account for sample-to-sample variation. The recovery was calculated by taking the detected RNA copies and subtracting the RNA copies detected in the corresponding unspiked samples, and then dividing by the spiked amount. These results demonstrate that our method constantly provided high recovery of viral RNA in wastewater samples.
Fig. 4.
Recovery of viral RNA from wastewater samples. A previously confirmed SARS-CoV-2 negative wastewater sample and a wastewater sample with a low concentration of SARS-CoV-2 RNA were analyzed. RNA of SARS-CoV-2 pseudovirus (104 copies) was added to an aliquot of each sample. The N1 gene of SARS-CoV-2 was detected using RT-qPCR. The recovery was 80% ±4% and 76% ± 4% from the analysis of these wastewater samples spiked with 104 copies of the viral RNA. NC indicates negative control and ND indicates not detectable.
The EM-VIP-Mag-RT-qPCR method achieved consistently higher recovery than those previously reported. Several WS of SARS-CoV-2 studies using enveloped surrogate viruses have reported largely variable recoveries ranging from 0.08% to 66% between studies and even within the same study (Kumblathan et al., 2021). Furthermore, our method achieved higher recoveries than those of the previous studies (0.96%-65.7%) that also used EM for viral particle and RNA concentration (Ahmed et al., 2020, LaTurner et al., 2021, Gonzalez et al., 2020). The improved recovery provided by the EM-VIP-Meg-RT-qPCR method can be attributed to several strategies we incorporated. To maximize viral particle concentration and minimize RNA loss we released and isolated SARS-CoV-2 and RNA from solids using beef extract solution, efficiently extracted and maintained RNA integrity using the VIP buffer, and utilized magnetic beads to concentrate and directly detect RNA without an elution step. Additionally, to minimize co-concentration of RT-qPCR inhibitors we removed inhibitors by separating the solid and aqueous phase prior to the EM filtration step and used the VIP-Mag method with three wash steps. Thus, we successfully developed a method with enhanced recovery.
2.6. Blind test of composite wastewater samples with diverse SARS-CoV-2 RNA concentrations
In a blind inter-laboratory comparison format, we analyzed a set of composite wastewater samples previously collected from a long-term care facility (SARS-CoV-2 negative) and from the Edmonton WWTP (SARS-CoV-2 positive). Alberta Public Health Laboratory (APHL) diluted the SARS-CoV-2 positive wastewater sample with the SARS-CoV-2 negative wastewater sample by 10, 100, and 1000 folds. We received the undiluted and diluted composite wastewater samples without prior knowledge of the sample characteristics or their viral RNA concentrations. Composed of wastewater from two sources, these samples appeared to have a very dirty and complex matrix. Using our EM-VIP-Mag-RT-qPCR method, we were able to quantify the SARS-CoV-2 RNA concentrations in these composite wastewater samples. Our analyses showed that the concentrations of the SARS-CoV-2 RNA ranged from 1.9×102 copies per 100 mL to 4.1×105 copies per 100 mL wastewater. These results are consistent with the expected dilution factors and the concentrations in the composite wastewater samples, despite that this information was blind to us prior to our analysis.
Also in a blind inter-laboratory comparison format, we analyzed the composite wastewater sample that APHL prepared by diluting 0.1 mL Edmonton WWTP wastewater sample (SARS-CoV-2 positive) with 100 mL long-term care wastewater sample (SARS-CoV-2 negative). Our analysis showed that the concentration of SARS-CoV-2 RNA was 190 copies per 100 mL of wastewater. For comparison, APHL diluted 0.1 mL of the same Edmonton WWTP wastewater sample (SARS-CoV-2 positive) with 100 mL distilled deionized water (ddH2O) and analyzed this relatively clean sample using its routine method. APHL detected 102 copies per 100 mL of this diluted sample (mostly in ddH2O). Our results of detecting 190 copies of the SARS-CoV-2 RNA per 100 mL of wastewater, in agreement with APHL's result of detecting 102 copies per 100 mL of ddH2O, demonstrate the ability of our EM-VIP-Mag-RT-qPCR method for detecting trace and diverse concentrations of SARS-CoV-2 RNA in wastewater.
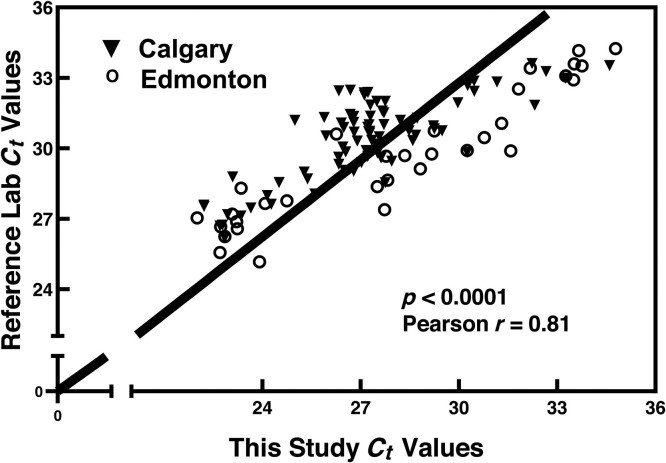
2.7. Detection of SARS-CoV-2 in 120 wastewater samples using EM-VIP-Mag-RT-qPCR
We successfully applied our EM-VIP-Mag-RT-qPCR method to detect SARS-CoV-2 in wastewater samples collected from two WWTPs in Calgary and Edmonton over a period of ten months (May 2021 to February 2022). Fig. 5 shows Ct values from the analysis of 120 samples (60 samples from each city) using our EM-VIP-Mag-RT-qPCR method and the results obtained independently by APHL (reference lab). Pearson correlation between the two sets of Ct values was r=0.81 and p<0.0001, indicating that our results are significantly correlated with those of the reference lab.
Fig. 5.
Pearson correlation analysis of results obtained using the EM-VIP-Mag-RT-qPCR method with the results provided by the reference laboratory from the analysis of 120 wastewater samples. The samples were collected from two wastewater treatment plants in Calgary and Edmonton.
Appendix A Fig. S3 shows the SARS-CoV-2 RNA concentrations in representative wastewaters collected from Calgary and Edmonton WWTPs. These results show that our EM-VIP-Mag-RT-qPCR method was able to detect the viral RNA in all 120 wastewater samples, with the lowest concentrations being 2.4×102 copies per 100 mL and the highest concentrations being 2.9×106 copies per 100 mL of wastewater. These results demonstrate excellent sensitivity and wide dynamic range of our method.
3. Conclusions
We successfully developed the EM-VIP-Mag-RT-qPCR method that enhanced the recovery and detection of SARS-CoV-2 in wastewater samples. The main features that contribute to the improvement include: recovery of SARS-CoV-2 viral particles and RNA from both solids and aqueous phase, processing with a large volume (80 mL) of wastewater, efficient RNA extraction and preservation using the VIP solution, removal of sample matrix and inhibitors using magnetic beads, and direct RT-qPCR detection on magnetic beads. This method provides reproducible recovery of (76%-80% ± 4%), representing a significant improvement compared to the previous studies (0.96%-65.7%). This method uses accessible equipment and reagents and is cost-effective. It can be applied for monitoring of SARS-CoV-2 in wastewater for community surveillance, complementing clinical testing. This method can also be adapted for detection of other pathogens in wastewater.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this article.
Acknowledgments
This work was supported by the Natural Sciences and Engineering Research Council of Canada, the Canada Research Chairs Program, Alberta Innovates, and Alberta Health. The authors would like to thank the wastewater treatment plants of EPCOR and the City of Calgary for the collection of wastewater samples analyzed in this study. Fig. 1 and TOC were created using BioRender.com, and other figures were created using GraphPad Prism 9.4.1.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jes.2022.10.006.
Appendix. Supplementary materials
References
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboa S., Mauricio-Iglesias M., Rodríguez S., Martínez Lamas L., Vasallo F.J., Regueiro B., et al. The fate of SARS-CoV-2 in wastewater treatment plants points out the sludge line as a suitable spot for incidence monitoring. Sci. Total Environ. 2021;772 doi: 10.1016/j.scitotenv.2021.145268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonerba A., Corpuz M.V., Ballesteros F., Choo K.H., Hasan S.W., Korshin G.V., et al. Coronavirus in water media: Analysis, fate, disinfection and epidemiological applications. J. Hazard. Mater. 2021;415 doi: 10.1016/j.jhazmat.2021.125580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canadian Water Network. COVID-19 wastewater coalition, 2022. Available: https://cwn-rce.ca/covid-19-wastewater-coalition/ Accessed August 10, 2022.
- Carrillo-Reyes J., Barragán-Trinidad M., Buitrón G. Surveillance of SARS-CoV-2 in sewage and wastewater treatment plants in Mexico. J. Water Process. Eng. 2021;40 doi: 10.1016/j.jwpe.2020.101815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention . 2022. National Wastewater Surveillance System. [Google Scholar]; Available: https://www.cdc.gov/healthywater/surveillance/wastewater-surveillance/wastewater-surveillance.html. Accessed August 10, 2022.
- Chik A.H.S., Glier M.B., Servos M., Mangat C.S., Pang X.L., Qiu Y., et al. Comparison of approaches to quantify SARS-CoV-2 in wastewater using RT-qPCR: results and implications from a collaborative inter-laboratory study in Canada. J. Environ. Sci. 2021;107:218–229. doi: 10.1016/j.jes.2021.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aoust P.M., Mercier E., Montpetit D., Jia J.J., Alexandrov I., Neault N., et al. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. 2021;188 doi: 10.1016/j.watres.2020.116560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Global Institute for Water Security. COVID-19 Early Indicators, 2022. Available: 10.1007/11744023 Data Accessed August 10, 2022. [DOI]
- Graham K.E., Loeb S.K., Wolfe M.K., Catoe D., Sinnott-Armstrong N., Kim S., et al. SARS-CoV-2 RNA in wastewater settled solids is associated with COVID-19 cases in a large urban sewershed. Environ. Sci. Technol. 2021;55:488–498. doi: 10.1021/acs.est.0c06191. [DOI] [PubMed] [Google Scholar]
- Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetka K., et al. COVID-19 surveillance in southeastern Virginia using wastewater based epidemiology. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Parker J., Smits S., Underwood J., Dolwani S. Persistent viral shedding of SARS-CoV-2 in faeces – a rapid review. Colorectal Dis. 2020;22:611–620. doi: 10.1111/codi.15138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasing M., Yu J., Qiu Y., Maal-Bared R., Bhavanam S., Lee B., et al. Comparison of detecting and quantitating SARS-CoV-2 in wastewater using moderate-speed centrifuged solids versus an ultrafiltration method. Water. 2021;13:2166. [Google Scholar]
- Hata A., Hara-Yamamura H., Meuchi Y., Imai S., Honda R. Detection of SARS-CoV-2 in wastewater in Japan during a COVID-19 outbreak. Sci. Total Environ. 2021;758 doi: 10.1016/j.scitotenv.2020.143578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrudey S.E., Bischel H.N., Charrois J., Chik A.H.S., Conant B., Delatolla R., et al. Wastewater surveillance for SARS-CoV-2 RNA in Canada. The Royal Society of Canada. 2022 [Google Scholar]; Available: https://rsc-src.ca/sites/default/files/WWS%20PB_EN_3.pdf. Accessed August 10, 2022.
- International Water Association . IWA; 2022. COVID-19 Wastewater-Based Epidemiology. [Google Scholar]; Available: https://iwa-network.org/learn/covid-19-wastewater-based-epidemiology/. Accessed August 10, 2022.
- Kantor R.S., Nelson K.L., Greenwald H.D., Kennedy L.C. Challenges in measuring the recovery of SARS-CoV-2 from wastewater. Environ. Sci. Technol. 2021;55:3514–3519. doi: 10.1021/acs.est.0c08210. [DOI] [PubMed] [Google Scholar]
- Karthikeyan S., Levy J.I., De Hoff P., Humphrey G., Birmingham A., Jepsen, et al. Wastewater sequencing reveals early cryptic SARS-CoV-2 variant transmission. Nature. 2022;609:101–108. doi: 10.1038/s41586-022-05049-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., et al. SARS-CoV-2 in wastewater: State of the knowledge and research needs. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumblathan T., Liu Y., Uppal G.K., Hrudey S.E., Li X.-F. Wastewater-based epidemiology for community monitoring of SARS-CoV-2: Progress and Challenges. ACS Environ. Au. 2021;1:18–31. doi: 10.1021/acsenvironau.1c00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumblathan T., Piroddi N., Hrudey S.E., Li X.-F. Wastewater based surveillance of SARS-CoV-2: challenges and perspective from a Canadian inter-laboratory study. J. Environ. Sci. 2022;116:229–232. doi: 10.1016/j.jes.2022.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaTurner Z.W., Zong D.M., Kalvapalle P., Gamas K.R., Terwilliger A., Crosby T., et al. Evaluating recovery, cost, and throughput of different concentration methods for SARS-CoV-2 wastewater-based epidemiology. Water Res. 2021;197 doi: 10.1016/j.watres.2021.117043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le C. Sensitivity of wastewater surveillance: What is the minimum COVID-19 cases required in population for SARS-CoV-2 RNA to be detected in wastewater? J. Environ. Sci. 2023;125:851–853. doi: 10.1016/j.jes.2022.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Lee B.E., Gao T., Qiu Y., Ellehoj E., Yu J., et al. Number of COVID-19 cases required in a population to detect SARS-CoV-2 RNA in wastewater in the province of Alberta, Canada: Sensitivity assessment. J. Environ. Sci. 2023;125:843–850. doi: 10.1016/j.jes.2022.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Kumblathan T., Feng W., Pang B., Tao J., Xu J., et al. On-site viral inactivation and RNA preservation of gargle and saliva samples combined with direct analysis of SARS-CoV-2 RNA on magnetic beads. ACS Meas. Sci. Au. 2022;2:224–232. doi: 10.1021/acsmeasuresciau.1c00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Huang Z., Luo J., Zhang X., Sha S. Primary concentration: The critical step in implementing the wastewater based epidemiology for the COVID-19 pandemic: A mini-review. Sci. Total Environ. 2020;747 doi: 10.1016/j.scitotenv.2020.141245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCCID (National Collaborating Centre for Infectious Disease) 2022. Public Health Agency of Canada Wastewater surveillance program for COVID-19. [Google Scholar]; Available: https://nccid.ca/wastewater-surveillance-for-covid-19/. Accessed August 10, 2022.
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecson B.M., Darby E., Haas C.N., Amha Y.M., Bartolo M., Danielson R., et al. Reproducibility and sensitivity of 36 methods to quantify the SARS-CoV-2 genetic signal in raw wastewater: findings from an interlaboratory methods evaluation in the U.S. Environ. Sci.: Water Res. Technol. 2021;7:504–520. doi: 10.1039/d0ew00946f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Yu J., Pabbaraju K., Lee B.E., Gao T., Ashbolt N.J., et al. Validating and optimizing the method for molecular detection and quantification of SARS-CoV-2 in wastewater. Sci. Total Environ. 2022;812 doi: 10.1016/j.scitotenv.2021.151434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramón-Nuńez L.A., Martos L., Fernández-Pardo Á., Oto J., Medina P., España F., Navarro S. Comparison of protocols and RNA carriers for plasma miRNA isolation. Unraveling RNA carrier influence on miRNA isolation. PLoS One. 2017;12 doi: 10.1371/journal.pone.0187005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldan-Hernandez L., Graham K., Duong D., Boehm A. Persistence of endogenous SARS-CoV-2 and pepper mild mottle virus RNA in wastewater settled solids. ACS EST Water. 2022 doi: 10.1021/acsestwater.2c00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader C., Schielke A., Ellerbroek L., Johne R. PCR inhibitors...occurrence, properties and removal. J. Appl. Microbiol. 2012;113:1014–1026. doi: 10.1111/j.1365-2672.2012.05384.x. [DOI] [PubMed] [Google Scholar]
- Shi H., Pasco E.V., Tarabara V.V. Membrane-based methods of virus concentration from water: A review of process parameters and their effects on virus recovery. Environ. Sci: Water Res. Technol. 2017;3:778–792. [Google Scholar]
- Vo V., Tillett R.L., Papp K., Shen S., Gu R., Gorzalski A., et al. Use of wastewater surveillance for early detection of Alpha and Epsilon SARS-CoV-2 variants of concern and estimation of overall COVID-19 infection burden. Sci. Total Environ. 2022;835 doi: 10.1016/j.scitotenv.2022.155410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhaus S., Weber F.A., Schiwy S., Linnemann V., Brinkmann M., Widera M., et al. Detection of SARS-CoV-2 in raw and treated wastewater in Germany–suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021;751 doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- World Health Organization . 2022. Tracking SARS-CoV-2 variants. [PubMed] [Google Scholar]; Available: https://www.who.int/activities/tracking-SARS-CoV-2-variants. Accessed August 10, 2022.
- World Health Organization . 2022. Coronavirus (COVID-19) Dashboard. [Google Scholar]; Available: https://covid19.who.int/. Accessed August 10, 2022.
- Xie Y., Challis J.K., Oloye F.F., Asadi M., Cantin J., Brinkmann M., et al. RNA in municipal wastewater reveals magnitudes of COVID-19 outbreaks across four waves driven by SARS-CoV-2 variants of concern. ACS EST Water. 2022 doi: 10.1021/acsestwater.1c00349. [DOI] [PubMed] [Google Scholar]
- Zhang W., Du R.H., Li B., Zheng X.S., Yang X.L., Hu B., et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microb. Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.