Abstract
The world is facing rapid climate change and a fast-growing global population. It is believed that the world population will be 9.7 billion in 2050. However, recent agriculture production is not enough to feed the current population of 7.9 billion people, which is causing a huge hunger problem. Therefore, feeding the 9.7 billion population in 2050 will be a huge target. Climate change is becoming a huge threat to global agricultural production, and it is expected to become the worst threat to it in the upcoming years. Keeping this in view, it is very important to breed climate-resilient plants. Legumes are considered an important pillar of the agriculture production system and a great source of high-quality protein, minerals, and vitamins. During the last two decades, advancements in OMICs technology revolutionized plant breeding and emerged as a crop-saving tool in wake of the climate change. Various OMICs approaches like Next-Generation sequencing (NGS), Transcriptomics, Proteomics, and Metabolomics have been used in legumes under abiotic stresses. The scientific community successfully utilized these platforms and investigated the Quantitative Trait Loci (QTL), linked markers through genome-wide association studies, and developed KASP markers that can be helpful for the marker-assisted breeding of legumes. Gene-editing techniques have been successfully proven for soybean, cowpea, chickpea, and model legumes such as Medicago truncatula and Lotus japonicus. A number of efforts have been made to perform gene editing in legumes. Moreover, the scientific community did a great job of identifying various genes involved in the metabolic pathways and utilizing the resulted information in the development of climate-resilient legume cultivars at a rapid pace. Keeping in view, this review highlights the contribution of OMICs approaches to abiotic stresses in legumes. We envisage that the presented information will be helpful for the scientific community to develop climate-resilient legume cultivars.
Keywords: legumes, climate change, drought stress, marker-assisted breeding, marker-trait association
Introduction
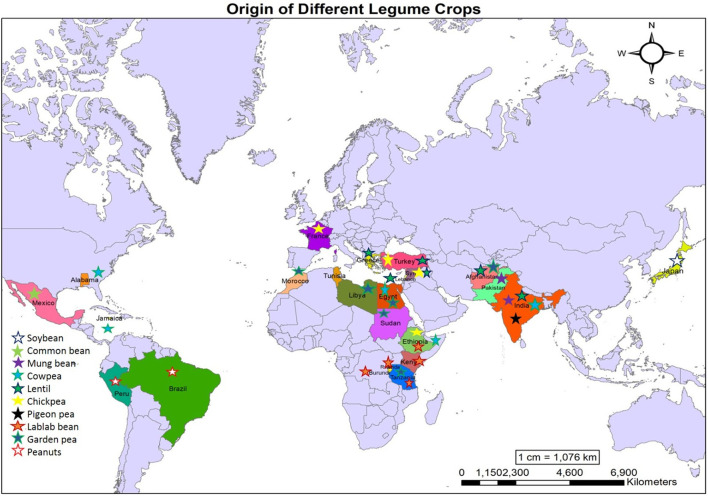
The grand challenge facing scientists in the twenty-first century is to optimize climate change adaptation, agricultural productivity, food security, and environmental protection. Climate-related changes will likely boost the severity of both sole and joint abiotic stresses, especially drought, cold, salinity, and heat (Pandey et al., 2017; Anwar et al., 2022). These climate-change vulnerabilities pose a high threat to global food and nutritional security. Crops that can help overcome the effects of climate change are urgently needed. Legumes may prove to be one such suitable group of crops with promising future possibilities. Legumes being the members of Fabaceae family have a great value for their versatile usage and demand for humans and livestock. Legumes are a staple food for millions of people all over the world and serve as a cheap source of high-quality protein. As a result of its higher global consumption, this family ranks second after Gramineae and is the third largest flowering plant family followed by Asteraceae and Orchidaceae (Çakir et al., 2019). Presently, 88% of species of these families are examined and 750 genera consisting of 17,000–18,000 species play a significant role as food grains, pasture, and agroforestry to maintain the environments (Gupta and Pandey, 2021). Legumes are nutritionally used as staples worldwide. Legumes are a low-cost and good source of vitamins, carbohydrates, protein, and fiber. Concerning global distribution, chickpeas, pigeon pea, and lentils are widely cultivated in South Asian countries; faba beans are primarily cultivated in North Africa, China, and the Mediterranean countries; cowpeas in West African (WA) countries; soybean and other oilseed crops predominantly found in Indonesia, China and Japan while the Central and South America (C&SA) is also famous for the cultivations of beans (Joshi and Rao, 2017). The origins of legume crops are shown in Figure 1.
Figure 1.
Origin centers of various legume crops.
The consumption of legumes in different forms is common, with a beneficial aspect for the human diet, aiding good health. Legumes are full of substantial elements of diets including necessary proteins, minerals, complex carbohydrates, amino acids, dietary fibers, and vitamins such as riboflavin, vitamins B complex, tocopherols, pyridoxine, thiamine, and folates (Mefleh et al., 2022). Phaseolin is the major globulin in domesticated Phaseolus beans. Phaseolin is a protein containing a neutral sugar, usually mannan, and it is organized into three subunits, each of which possesses different molecular weights, isoelectric points, and glycosylation degrees. The legumes contain low saturated fats, and for that reason can be considered cholesterol-free. Protein is the main element for good health and is found high in legumes (40%) as compared to cereals (17%) (Kamau et al., 2020). Legumes seeds contain manyproteins such as oligomeric globulins, albumins, glutelins, and prolamins. These proteins are generally synthesized in the seed, and provide support for seedling germination. The amino acid analysis of legumes revealed a moderate quantity of sulfur-containing L-tryptophan and amino acids but high content of the primary Lysine. The carbohydrate is present in the form of starch, galactose ribose, á-galactosides, glucose, maltose, sucrose, and fructose as well as fermentable fibers in legumes seeds, and plants. The presences of amylopectin and amylose in dry grains and lignin, oligosaccharides, hemicellulose, cellulose, and pectin in plants are a vital source of carbohydrates that help in digestion. A significant amount of micronutrients is also present in legume crops. The micronutrients such as calcium, potassium, selenium, iron, magnesium, copper, and zinc are reported in mung bean, common bean, faba beans, chickpeas, lupins, and lentils which collectively play a vital role in body development (Roorkiwal et al., 2021). The nutritional comparison among food legumes is given in Table 1.
Table 1.
Nutritional comparison among food legumes.
| Crops | Cowpea | Chickpea | Lentil | Mung bean | Kidney bean | Pigeon pea | Soybean | Moth-bean |
|---|---|---|---|---|---|---|---|---|
| Protein (%) | 24 | 21 | 25 | 24 | 24 | 22 | 37 | 23 |
| Dietary fiber (%) | 11 | 12 | 11 | 16 | 25 | 15 | 9 | – |
| Carbohydrate (%) | 60 | 63 | 63 | 63 | 60 | 63 | 30 | 62 |
| Lipids (%) | 2 | 6 | 1 | 1 | 1 | 2 | 20 | 2 |
| K (μg g−1) | 13,750 | 7,180 | 6,770 | 12,460 | 14,060 | 13,920 | 17,970 | 11,910 |
| Zn (μg g−1) | 61 | 28 | 33 | 27 | 51 | 28 | 50 | 19 |
| Ca (μg g−1) | 850 | 570 | 350 | 1,320 | 1,430 | 1,300 | 2,770 | 1,500 |
| Na (μg g−1) | 80 | 240 | 60 | 150 | 240 | 170 | 20 | 300 |
| Fe (μg g−1) | 100 | 43 | 65 | 67 | 82 | 52 | 157 | 109 |
| P (μg g−1) | 4,380 | 2,520 | 2,810 | 3,670 | 4,070 | 3,670 | 7,040 | 4,890 |
| Mg (μg g−1) | 3,330 | 790 | 470 | 1,890 | 1,400 | 1,830 | 2,800 | 3,810 |
| Vitamin A (IU) | 33 | 67 | 39 | 114 | 0 | 28 | 22 | 32 |
| Vitamin C (μg g−1) | 15 | 40 | 45 | 48 | 45 | 0 | 60 | 40 |
| vitamin B6 (μg g−1) | 4 | 5 | 4 | 4 | 4 | 3 | 4 | 4 |
| Vitamin E (μg g−1) | 0 | 8 | 5 | 5 | 2 | 0 | 9 | 0 |
| Thiamin (μg g−1) | 7 | 5 | 9 | 6 | 5 | 6 | 9 | 6 |
Despite their commercial and economic importance, legumes have not received the same level of attention as cereals to increase crop productivity. The legumes crop is threatened by a series of biotic diseases especially caused by fungi and nematodes. The fungal diseases such as ascochyta blight (Ascochyta rabiei), charcoal rot (Macrophomina phaseolina), powdery mildew (Erysiphcpoiygoni), alternaria blight (Alternaria alternate), botrytis gray mold (Botrytis cinerea), cercospora leaf spot (Cercospora canescens), collar rot (Sclerotium rolfsii), phytophthora blight (Phytophthora drechsleri), stemphylium blight (Stemphylium sp), fusarium wilt (Fusarium oxysporum f. sp. ciceris), wet root rot (Rhizoctonia solani), vascular wilt (F. oxysporum f. sp. lentis) are contributed to major yield loss in legumes crops as compared to all others (Sampaio et al., 2020; Mahmoud, 2021; Pandey and Basandrai, 2021; Urva, 2021). Nematodes (Cyst and root-knot) are also a major factor of yield loss in legumes (Zwart et al., 2019). Similarly, crops are susceptible to insects, weeds, and diseases, which are also responsible for devastating yield losses (Reddy et al., 2004).
Globally, 55 developing countries having semiarid and tropical areas are affected by abiotic stress where mainly cultivation consists of grain legumes. In abiotic stress, multiple stresses are involved like drought, waterlogging, chilling, salinity, and high temperature are the main cause of the decrease in crop production annually (Ozturk Gokce et al., 2020). The cultivated land faced about 90% environmental stresses. Temperature plays a significant role in the survival of plant and their growth along with root nodules development. The rise in high-temperature result in poor pollination, and cause a significant reduction in the crop yield (Settele et al., 2016). Drought is an important abiotic stress issue that terribly affects crop production. Drought hinders the legume symbiotic routine, slowing down the growth, and eventually leading to decreased crop yield (Anjum et al., 2017). However, the influence of drought on the exposed plant is mainly dependent on the long drought period and magnitude of the stress. Cold-induced stress negatively affects photosynthetic pigments and capacity, antioxidant defense mechanisms, and mineral nutrient uptake; it alters metabolic and hormonal processes, and it disturbs plant developmental stages from seed germination to maturation. Plants exposed to low temperatures can increase the accumulation of reactive oxygen species (ROS), including superoxide anion radicals (), singlet oxygen (1O2), hydrogen peroxide (H2O2), and hydroxyl ions (OH−), which damage mitochondria and chloroplasts and can lead to cell death (Gill and Tuteja, 2010). Salinity is also an important abiotic stress issue that is proved as harmful to the survival of legumes (Shabala and Munns, 2012). In newly emerging leaves, the abscisic acid (ABA) level rises in the transpiration portion due to sodium chloride (Badhan et al., 2018). In the leaf veins and root, an ATP binding cassette (ABC) carrier was expressed that play a fundamental role toward ABC by showing hypersensitivity in plants regarding seedling and germination phase. Genetically designed plants were upregulated for ABA expression from separated leave have displayed a reduction in water loss as well as higher lead temperature than the normal plants (Raza et al., 2019).
Enhancement of already existing germplasm through focused evolution needs tracking of the desired traits to combine them. Previously, phenotype assisted in tracking the qualities of a plant in better ways that are nowadays exchanged in more meaningful ways in sense of DNA markers. Combinations of molecular studies and other breeding tools have moved into deeper knowledge to study the possible traits of a selected plant, including genomic and gene regulatory traits (Dhaliwal et al., 2020). The whole bunch of genes quantitative trait loci (QTLs) are studied along with the molecular markers that are connected with traits for trustworthy marker-aided breeding. The marker technology development in legumes is relatively slower as compared to cereals, giving them the label of an orphan crop (Gresta et al., 2016). The gradual development of molecular markers proceeds includes first-generation markers i.e., Restriction fragment length polymorphism (RFLP), amplified fragment length polymorphism (AFLP), random amplification of polymorphic DNA (RAPD), producing data of various loci in a single turn, mainly used in the study of diversity in pigeon pea, winged bean, cowpea, cluster bean, and dolichos bean (Chapman, 2015; Kumar et al., 2020). Furthermore, sequence-oriented markers, specifically, single nucleotide polymorphism (SNPs), simple sequence repeats (SSRs), and their alternation to increase reproducibility and reliability, efficiently working in trait mapping, fine mapping as well as linkage mapping studies (Choi, 2019) have also been documented. Simple sequence repeats (SSRs)markers have been used for the assessment of genetic diversity.
SSR markers have been used in cluster bean, pigeon pea, cowpea, and winged bean, whereas inter-species SSRs have been employed in lablab for the assessment of diversity and other purposes. SSRs markers have also been used in many dolichos bean studies for the assessment of genetic diversity (Keerthi et al., 2018; Singh et al., 2021). SNPs are desired markers due to their pervasive nature and lavishness in the genetic material. SNPs have been recognized as effective and used in genetic material-aided breeding of cluster bean, pigeon pea, winged bean, cowpea, and dolichos bean. As the desired markers are recognized, it provides us a pathway for mapping traits of concern going toward cloning as well as fine mapping (Bohra et al., 2019; Nawaz and Chung, 2020). This review article will provide a brief overview of multi-omics techniques and their uses for abiotic stress research on legumes.
Legumes genetic resources
Genetic diversity is essential for fulfilling the basic dietary and nutritional needs of an ever-increasing population, and it also serves as the foundation for selection response. Information on crop genetic resources, including pulse crops conserved in national and international gene banks, is available online at global portals such as Genesys (https://www.genesys-pgr.org/) and GRIN (https://www.ars-grin.gov/), as well as in a 1996 FAO publication titled “The second report on the state of the world's plant genetic resources for food and agriculture” (https://www.fao.org/3/i1500e/i1500e00.htm). According to the FAO report, CG centers hold 35,891 common bean accessions, 33,359 chickpea accessions, 15,588 cowpea accessions, 13,289 pigeonpea accessions, 10,864 lentil accessions, 9,186 faba bean accessions, 6,129 field pea accessions, and 3,225 grass pea accessions in trust (Table 2). Since then, the ICARDA gene bank has grown to include 50,968 accessions of pulse crops, including 15,749 chickpeas, 14,597 lentils, 10,034 faba beans, 6,131 peas, and 4,457 grass peas. Similarly, ICRISAT holds 20,764 and 13,783 accessions of chickpea and pigeonpea, respectively. CIAT, IITA, and AVRDC hold 37,938, 16,460, and 10,946 accessions of Phaseolus beans, cowpea, and Vigna species, respectively. To facilitate accessibility and better use of the germplasm available with gene banks, various sets of core (Frankel, 1984; Brown, 1989), mini-core (Upadhyaya and Ortiz, 2001), focused identification of germplasm strategy (FIGS) (Mackay et al., 2004), and reference (Odong et al., 2011) germplasm have been developed. The FIGS strategy (https://www.icarda.org/research/innovations/focused-identification-germplasm-strategy-figs) is being pursued at ICARDA, which has proved effective for various adaptive traits such as heat, drought, cold, and salt tolerance, as well as insect pest and disease resistance. FIGS sets for chickpea, lentils, grass pea, and faba bean have recently been released, which can be used to find and apply valuable genes in preferred agronomic environments. Similarly, ICRISAT-developed mini-core sets of chickpea, pigeonpea, and groundnut provide a rich source of variety for desired traits in breeding operations (Upadhyaya et al., 2013). Except for a few features, the present germplasm of pulse crops contains sufficient variety for crucial economic traits. The creation of structured and representative collections of germplasm from the worldwide collection increases the efficiency with which beneficial alleles/traits can be identified and used.
Table 2.
Legume germplasm holdings in major gene banks.
| Crop | Global status | CG center | USDA | NBPGR,/India |
|---|---|---|---|---|
| Chickpea | 98,285 | 33,359 | 7,000 | 14,704 |
| Lentil | 58,405 | 10,864 | – | 9,989 |
| Vigna species | – | – | – | 5,549 |
| Common bean | 261,963 | 35,891 | – | 1,514 |
| Grass pea | 26,066 | 3,225 | – | 2,797 |
| Field pea | 94,001 | 6,129 | 6,161 | 3,070 |
| Cowpea | 65,323 | 15,588 | 1,287 | 3,317 |
| Pigeonpea | 40,820 | 13,289 | 4,806 | 12,859 |
| Faba bean | 43,695 | 9,186 | – | – |
| Others | 183,078 | 13,690 | – | 19,579 |
| Total | 1,069,897 | 141,221 | 73,378 |
CG, Crop Genebanks; NBPGR, National Bureau of Plant Genetic Resources, India; USDA, United States Department of Agriculture.
OMICS approaches in the technological era
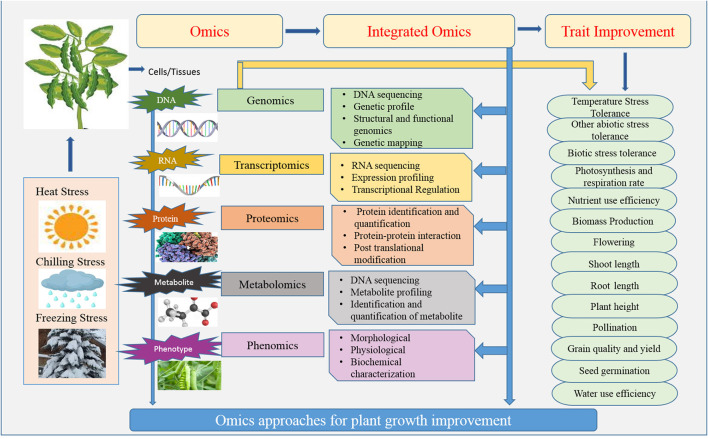
Omics is a fast-evolving technique that provides the methodology, technological proficiency, and interdisciplinary and transdisciplinary advancement needed to improve awareness and identification of all the plant's genomic and transcriptomic processes. Omics is a beneficial aid for understanding the change in plant metabolism as a consequence of contact with the external environment (Kumar et al., 2021). Transcriptomic, metabolomic, and genomic tools scan and give gene - expression and protein expression levels immediately, so in the advanced era these play a vital role in plant improvements (Jha et al., 2020). Omics technology not just increases our awareness, but also gives us new insights into how plants react when stressed. Whenever, a small change occurs in genetics, nutrients, or fluctuation in the external environment, a wide range of technologies is used to perceive these variations in plants. In the recent and modern era, these advanced tools are providing support to understand the whole plant genome, and the biggest example of this is Arabidopsis thaliana. Innovation in the modern era started with the complete genome sequencing of this model plant. Rice, maize, and soybeans are among the main crop plants whose genomes are complicated but have been fully sequenced. The introduction of high-throughput “Omics” methodologies has ushered in a time of successful plant molecular tools for responding to environmental changes. Rapid innovations and advancements in “omics” regarding the post-genomic epoch such as molecular characterization, next-generation sequencing, modeling of various molecular and physiological knowledge, and association of these assertions with plant establishment have contributed to a successful transition to resilience and efficiency under abiotic stress conditions (Pandey et al., 2021). With the invention of next-generation sequencing techniques, it become feasible and reliable to sequence plant species. Furthermore, current omics tools such as proteomics and transcriptomics aids to understand the level of genes and protein. Different results from reported studies showed that not all the genes are turned on or off at the same time; for that reason, the metabolism adopts a complex phenotype that cannot be determined by genotype (Jha et al., 2019; Singh et al., 2020). As a result, the successful integration of genomics, transcriptomics, phonemics, proteomics, epigenomics, metabolomics, interact omics, and ionomics would aid breeders in identifying promising candidates' genes and optimal features for generating and improving the productivity of legume crops under abiotic stresses (Kumar et al., 2022). Figure 2 is showing different omics approaches for the improvement of plant growth.
Figure 2.
Schematic visualizing the contribution of OMIC's approaches for legume plant improvement.
Contribution of OMICs approaches in legumes for abiotic stresses
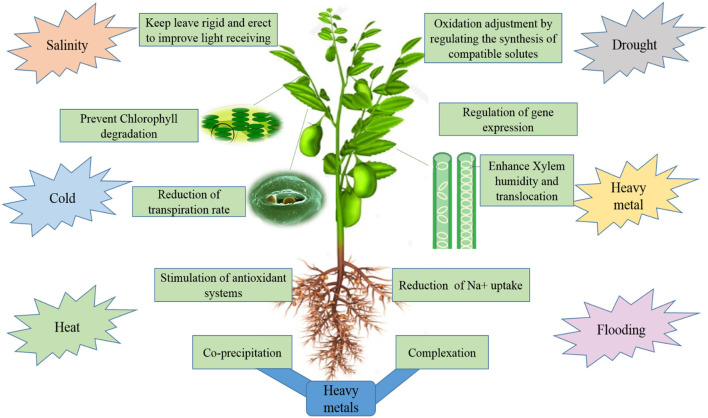
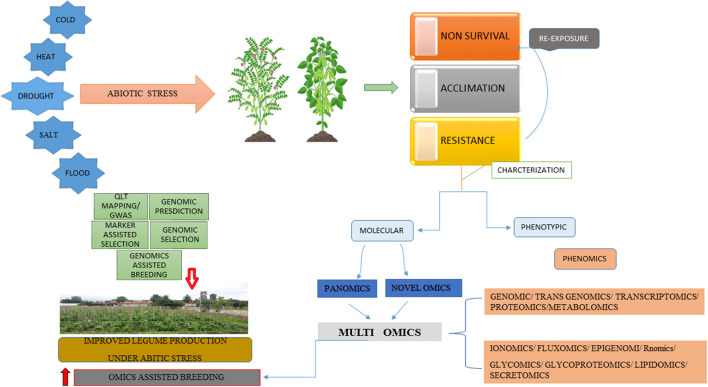
The genome size of the legumes is large and some of them are polyploidy (Stai et al., 2019). As a result, model systems have been established to better recognize the legume biology and their modeling system that is developed for genetics studies regarding nodule formation and some dynamic processes like active tolerance, resistance, and susceptibility toward numerous stresses. Figure 3 provide a brief information about the effects of various a biotic stress on the growth of legumes. The basic research on legumes is performed on plant model systems, most frequently Lotus japonicas and Medicago truncatula, because of their diploid small genome, short life cycle, and enormous production of seed (Cook, 1999; Cervantes et al., 2019). The implementation of all these models has significantly enhanced the genetic and genomic datasets regarding legumes, and the similarity between distinct legume genomes has indeed augmented the data's usefulness (Stracke et al., 2004). In addition, the genome sequencing of soybean and their higher similarity to model legume species may greatly assist many genetic methods for investigation, like positional cloning. The genome sequence data is extremely valuable and serve as a great initial point, it is insufficient to understand gene activity, and the numerous metabolic reactions and regulatory responses get stimulated as the stress condition prolongs in the legumes. New suitable approaches and methods for overcoming these barriers to biotic stresses including the study of quantitative and qualitative traits are essential for checking gene expression (Wan et al., 2017). These must be done through the use of transcriptome, proteomic, and metabolomic levels study with an understanding of the use of genetically modifying crops and marker-assisted screening (Dita et al., 2006). Genomic, metabolomics, transcriptomics, transgenomics, proteomics, phenomics, and functional genomics fall under 'omics' and are admired as omics tools. Significant advancements within those distinct omics (Figure 4 and Table 3) have played an important role in the better understanding of stress responses at the molecular as well as genetic levels (Langridge and Fleury, 2011).
Figure 3.
Exploring the effect of abiotic stresses on the growth of legume crops.
Figure 4.
Improved abiotic stress tolerance in legumes using integrated multi-omics techniques.
Table 3.
Novel omics approaches for legumes breeding.
| Legumes | Abiotic stress | Omics technology | Details | References |
|---|---|---|---|---|
| Cowpea | Drought | Phosphoproteomics | Protein phosphorylation is induced by a gradual water deficiency. | Subba et al., 2013 |
| Secretomics | Dehydration, the stress-responsive secretome, and the highly complex metabolic network activity in the extracellular matrix have all been studied in depth. | Gupta et al., 2015 | ||
| Oxidative | Secretomics | CaFer1's role in iron buffering and oxidative stress adaption under different weather conditions. | Parveen et al., 2016 | |
| Common bean | Chlorpyrifos | Lipidomics | Triacylglycerol levels in pods and seeds are decreasing. | Fernandes et al., 2018 |
| Soybean | Heat | Lipidomics | Reduced expression of fatty acid desaturase results in lower quantities of lipids with 18:3 acyl chains. | Narayanan et al., 2020 |
| Low phosphorus | Lipidomics | Under low-phosphorus circumstances, lipid remodeling occurs. | Okazaki et al., 2017 | |
| Flooding | Phosphoproteomics | During flood stress, the ethylene signaling pathway was critical for protein phosphorylation in root tips. | Yin et al., 2014 | |
| Glycoproteomics | Protein N-glycosylation was negatively affected by flooding. | Showalter et al., 2016 |
Genomics advances for abiotic stress tolerance in legumes
Genomics focuses on the physical integrity of the genome, with the goal of recognizing, diagnosing, and regulating genomic features throughout the chromosomes. Now we deliberate some genomic advancements to apprehend the abiotic stress endurance in legumes (Chandrashekharaiah et al., 2021).
Molecular marker resources
The advent of genomic research has opened up new possibilities for genetic improvement of complex traits like salinity and drought endurance. In comparison to traditional breeding, a combination of genomic techniques and MAS can contribute to the identification of individual-specific genes in breeding populations at a considerably faster rate (Saade et al., 2016). SNPs are excellent for distinguishing complex traits employing massively multiplexed marker oligonucleotides like the Affymetrix GeneChip, as these are fast, high-throughput, co-dominant, highly abundant, cost-effective, and sequence-tagged (Missanga et al., 2021; Thudi et al., 2021). High throughput Axiom® SNP array genotyping is an efficient and cost-effective method for genotyping and the development of high-density linkage maps. Recently “Axiom® CicerSNP Array” has been developed for genotyping of Recombinant inbred lines of chickpea (Roorkiwal et al., 2018).
Quantitative trait loci mapping for abiotic stresses in legumes
The heredity of abiotic stress resistance is a complex phenomenon, and QTL mapping is the most popular method of discovering QTL, as well as genetic and linkage mapping of genomic regions related to tolerance. QTL analysis allows researchers to examine the genetic structure of a characteristic. QTLs can identify genomic areas linked to the expression of the trait being researched (Kushwah et al., 2021). Strong linkage maps might be used for spatial replication of important genes. Because they are comprised of a sequence-tagged marker therefore these can also be used in functional genomics to analyze chromosomal arrangement and evolutionary purposes. Markers are successful in determining the QTLs for specific traits from various regions of chromosomes, for identifying genotypes against the different biotic stresses and multifunctional. QTLs were previously identified from the many legume crops by using linked markers (Luo et al., 2021; Zate, 2021). Advancements in phenomics and genomics now allowed us to better characterize the QTLs that influence a certain trait, admired as QTLome (Salvi and Tuberosa, 2015). Breeders have a responsibility to use QTLome's immense knowledge effectively because of its vastness. Improved QTL meta-analysis, estimations of QTL impact, and crop modeling will enable the QTLome to be used effectively against the abiotic stress of legumes. Several studies have been conducted to dissect QTLs in legumes crop for abiotic stresses. The identification of these QTLs against abiotic stress in various legumes are helpful for breeders during breeding activities. We have provided QTLs based studies in legumes against abiotic stress in Table 4.
Table 4.
QTLs identified against abiotic stresses for various legumes crops.
| Crop | Abiotic stress | Associated marker (s) | Population type | Parental lines | Linkage group(s)/QTLs | References |
|---|---|---|---|---|---|---|
| Chickpea | Salinity | SSR (135) | RILs | JG-62 × ICCV 2 | LG3, LG6, and LG4 | Vadez et al., 2012 |
| Salinity | SSRs (28) and SNPs (28) | RILs | JG 11 × ICCV 2 | CaLG05 and 07 | Pushpavalli et al., 2015 | |
| Salinity | SSRs (150) | F2 | Vignaluteola oblonga × Vignaluteola | LG1 | Chankaew et al., 2014 | |
| Heat | SNPs (271) | RILs | ICC 15,614 × ICC 4,567 | CaLG05 and 06 | Paul et al., 2018 | |
| Cold | SNPs (747) | RILs | PI 489,777 and ICC 4,958 | CTCa3.1 and 8.1 | Mugabe et al., 2019 | |
| Drought | SSRs (97) | RILs | ILC 3,279 × ILC 588 | Q1-1 and Q3-1 | Rehman et al., 2011 | |
| Salinity | SSRs (150) | F2 | Vignaluteola oblonga × Vignaluteola | LG1 | Chankaew et al., 2014 | |
| Cowpea | Heat | SNPs (8) | RILs | IT82E-18 × CB27 | Cht 5 | Lucas et al., 2013 |
| Drought | AFLP (306) | RILs | CB46 × IT93K503-1 | 10 QTL (Dro) | Muchero et al., 2009 | |
| Salinity | SSRs (32) and RFLPs (116) | F2:5 | Tokyo × S-100 | LG N | Lee et al., 2004 | |
| Aluminum toxicity | SSRs, FLP, and AFLP (2,639) | RILs | Huaxia 3 × Zhonghuang 24 | qAAC_04 and qRRE_04 | Wang et al., 2019a | |
| Soybean | Aluminum toxicity | DNA markers (14) | RILs | Forrest × Essex | LG F | Sharma et al., 2011 |
| Aluminum toxicity | SSRs (11) | RILs | NN1138-2 × KF No.1 | LG B1 | Korir et al., 2013 | |
| Drought | SNPs (4,117) | RILs | Magellan × PI 567,731, PI 567,690 × Pana | Gm09, 05, 10, 06, 19, and 12 | Ye et al., 2020 | |
| Pea | Salinity | SNPs (705) | RILs | Parafield × Parafield | Ps III and Ps VII | Leonforte et al., 2013 |
| Frost/cold | SNPs (258) | RILs | Terese × Champagne | LG5 and 6 | Dumont et al., 2009 | |
| Drought | SSRs (6) and SNPs (2) | RILs | cv Messire × P665 | AA175, AB141, PsAAP2_SNP4, and A6 | Iglesias-García et al., 2015 | |
| Heat | SSRs (7) | F2 | PDL-1 × E-153 and PDL-2 × JL-3 | qHt_ps and qHt_ss | Singh et al., 2017a | |
| Lentil | Frost/cold | AFLP (94), RAPD (56), and ISSR (106) | RILs | Precoz × WA8649090 | LG4 | Kahraman et al., 2004 |
| Drought | SNPs (220) and AFLPs (180) | RILs | ILL 5,888 × ILL 6,002 | QRSAVII: 21.94 QSL12IV: 103.83, QSL22VII: 21.94, QSPADVIII: 72.15 and QDRWVII: 21.93 | Idrissi et al., 2016 | |
| Faba bean | Frost/cold1 | SNPs (5) | RILs | Bean Gottingen Winter and Bean Pure Line 4628 | LG-01, LG-02, LG-03, LG-04, LG-08, and LG-10 | Sallam et al., 2016 |
| Mung Bean | Drought | SSRs (313) | RILs | ZL × VC2917 | qMLA2A and qPH5A | Liu et al., 2017 |
| Common Bean | Drought | AFLP (53), RAPD (2), SSRs (42), and SNPs (127) | RILs | BAT 881 × G21212 | Pv01 and 08 | Diaz et al., 2018 |
Genome-wide association studies (GWAS) for abiotic stresses in legumes
Genome wide-association mapping is another beneficial tool to mitigate the constraints which provides the benefits from the historical meiosis of the diversity panel but may also offer better precision (Breria et al., 2020). In comparison to bi-parental mapping, association mapping is much more viable and cost-effective (Narayana and von Wettberg, 2020). Experimental architecture and statistical assessments in association mapping are continually changing to reduce the effects of influencing variables, reduce false positives, and regulate minor allele consequences. Due to genetic linkage and population structure, marker-trait relationships are muddled, creating a state of instability lacking real correlations. Numerous diagnostics or efficient statistical methods were developed to reduce false positives as well as minor allele QTL influences. Investigations of developmental genes including vernalization decreased height, and photoperiod response has been conducted at the exact genomic regions that are employed as a standard of comparison to integrate phenotypic variation with markers (Plewiński et al., 2020; Gondalia et al., 2022). These identified genes, being a strong genetic basis, can adjust the legumes plants under stress by changing heading time, plant height, and maturity time. In legumes, association mapping/GWAS is gaining popularity due to its capability to improve QTL discovery precision without investing additional resources in population growth. GWAS has made it possible to access functional genetic variations for salt tolerance traits in grain legume germplasms with greater accuracy and allelic richness (Hoyos-Villegas et al., 2017). Table 5 provide comprehensive information about the application of GWAS approach for the identification of genomic regions associated with abiotic stresses in legumes.
Table 5.
Application of GWAS approach to identify the genomic regions associated with abiotic stresses in legumes.
| Crop | Abiotic stress | Markers used | Chromosomes/region/gene | References |
|---|---|---|---|---|
| Common bean | Drought stress | SNPs (8,657) and SilicoDArT (3,213) | Chr. 6, 7, 10, and 11 | Valdisser et al., 2020 |
| Drought tolerance | SNPs (3,724,159) | Chr. 01 and 06 | Wu et al., 2021 | |
| Drought tolerance | SNPs(3,832,340) | Chr. 10/PvXIP1;2 | Wu et al., 2022 | |
| Drought tolerance | SNPs (5,389) | Chr. 02, 03, 04, 06,09, 10, and 11 | Dramadri et al., 2021 | |
| Aluminum toxicity | SNPs (13,906) | Chr. 1, 4, 5, 6, 11 | Ambachew and Blair, 2021 | |
| Aluminum toxicity | SNPs (5,389) | Chr. 02, 04, 06, 07, 09, and 10 | Njobvu et al., 2020 | |
| Mung bean | Salt and drought stress | SNPs | Chr.1, 7, 9, and 11 | Breria et al., 2020 |
| Alfalfa | Salt stress | SNPs (4,653) | All chromosomes except 2 | Liu et al., 2017 |
| Phaseolus vulgaris L. | Drought stress | SNPs | Pv01 and Pv02 | Trapp et al., 2015 |
| Drought and heat stress | SNPs | Chr. 1, 2, 3, 11 | Oladzad et al., 2019 | |
| Flooding tolerance | SNP (~203 K) | Pv07 and Pv08 | Soltani et al., 2018 | |
| Heat stress | SNP (23,373) | Chr. 1–11 | Assefa et al., 2019 | |
| Glycine max (L.) | Salinity stress | SNPs | Chr. 3 | Patil et al., 2016 |
| Flooding tolerance | Multi-locus random-SNP | QTN | Yu et al., 2019 | |
| Flooding tolerance | SNPs | Chr 03, 4, 07, 13, and 19 | Wu et al., 2020 | |
| Flooding tolerance | SNPs (34,718) | Glyma.01G198000, Glyma.05G008000, and Glyma.08G348500 | Sharmin et al., 2021 | |
| Drought tolerance | SNPs (12,316) | Chr. 01–15 | Sertse et al., 2021 | |
| Cicer arietinum L. | Salinity stress | DArTseq markers (1,856) | Chr. Ca4 and Ca2 | Ahmed et al., 2021 |
| Drought tolerance | SNPs (144,777) | Chr. 01–08 | Li et al., 2018 | |
| Vicia faba L | Heat tolerances | SNPs (10,749) | Chr. 9 and Chr. 11 | Maalouf et al., 2022 |
| Macrotyloma uniflorum | Drought stress | SNPs (20,241) | – | Choudhary et al., 2022 |
| Camelina sativa | Salt tolerance | SNPs (17) | Chr. 1, 7, and 20 | Luo et al., 2020 |
| Pea | Frost tolerance | SNP (11366) | LGI, LGII, LGIII, LGV, LGVI | Beji et al., 2020 |
Genomic selection in legumes
With the development of model-based linkages with the widespread accessibility of genetic markers emerges a new concept called genomic selection which is developed to evaluate genotype pairing potential (Heffner et al., 2009). This method is used to overcome the limitations of map-based scientific studies of genetics, which only identify a few QTLs to explain the diversity in selected characteristics. Populations with much more allelic diversity of desired characteristics have a more accurate analysis of the QTL effect than a population that is strongly linked. Connection instability is regularly overstated in inter-mating individuals and it fades in subsequent meiotic activities (Nawaz and Chung, 2020). The genomic breeding potential is usually anticipated through genomic choice by altering marker-assisted selection using markers. The established model evaluated by genotypic and phenotypic facts of the research population will be applied to measure the phenotypic diversity of the selected respondent based solely on genetic makeup. This will increase yield potential in comparison to both QTL and phenotype trait selection (Shu et al., 2013; Sandhu et al., 2021). Statistical techniques are applied to create genomic selection models which depict the characteristics of many traits along with markers (Jannink et al., 2010). An optimum streamlined genome that is nonbiased assesses the genetic similarity between individuals based on molecular marker relatedness as well as evaluates the phenotypic efficiency (Nadaf et al., 2012). These models are near to the assessment of breeding value (BV) from heredity as well as physical competence of associated genetic makeup in a pedigree. According to statistical assessments, forward or mixed sort regression models can eliminate markers associated with different substantial impacts. Ridge regression also includes a penalized factor in markers design that is more than the statistically allowed number (Annicchiarico et al., 2017). The use of genomic selection (GS) in legumes began with the goal of enhancing yield and agronomic traits in soybeans. GBS was used to genotype 301 elite breeding lines, which were then phenotyped for grain yield in several localities (Jarquín et al., 2014). Several genomic selection studies have been conducted for legumes and some of them are given in Table 6.
Table 6.
Genomic selection studies in legumes.
| Legume crop | Population size | Marker type | Traits | *Genomic selection models | References |
|---|---|---|---|---|---|
| Soybean | 301 | GBS | Grain yield | G-BLUP model | Jarquín et al., 2014 |
| 5,000 | 4,236 SNPs | Yield, protein content, and height | DualCNN, deepGS, RR-BLUP | Liu et al., 2019 | |
| 249 | 23,279 SNPs | Amino acid | RR-BLUP model | Qin et al., 2019 | |
| 5,600 | 4,600 SNPs | Yield | G-BLUP | Howard and Jarquin, 2019 | |
| Alfalfa | 190 | GBS (10,000 SNPs) | Single harvest biomass Total biomass | RR-BLUP model | Chapman, 2015 |
| 278 (adapted to two different environments) | GBS | Dry matter yield | RR-BLUP model | Annicchiarico et al., 2017 | |
| Pea | 372 | 331 SNP | Date of flowering Number of seeds per plant and Thousand seed weight | LASSO PLS SPLS Bayes A, Bayes B, and G-BLUP | Burstin et al., 2015 |
| 339 | 9,824 SNPs (GenoPea 13.2 K SNP Array) | Date of flowering Number of seeds per plant Thousand seed weight | (kPLSR), LASSO, G-BLUP, Bayes A, and Bayes B | Tayeh et al., 2015 | |
| Chickpea | 320 | 3,000 DArT markers | Seed yield 100 seed weight Days to 50% flowering, Days to maturity | RR-BLUP, kinship GAUSS, Bayes Cπ, Bayes B, Bayesian LASSO, Random Forest (RF) | Roorkiwal et al., 2016 |
| 320 | 8,900 SNPs | Seed weight, harvest index and biomass | Reaction norm models | Roorkiwal et al., 2018 | |
| 132 | 144,777 SNPs | Seed number and grain yield | BL and BRR | Li et al., 2018 | |
| Groundnut | 188 | 2,356 DArT markers | Days to flowering, seed weight, and pod yield | RR-BLUP, kinship GAUSS, Bayes Cπ, Bayes B, Baysian LASSO and RF | Pandey et al., 2014 |
| 340 | 13,355 SNPs | Seed wight, yield and days to maturity | Reaction norm models | Pandey et al., 2020 | |
| 281 | 493 SNPs | Leaf let length, days to maturity and 100 seed weight | RR-BLUP | Akohoue et al., 2020 | |
| Common bean | 481 | 5,820 SNPs | Grain yield and days to maturity | G-BLUP | Keller et al., 2020 |
Transcriptome profiling for abiotic stress tolerance in legumes
Transcriptomics is a strong approach for quantifying gene expression along with the accurate image of a target cell or tissue. Transcriptomics properly identifies the gene regulatory pathway as well as candidate genes that are responsible for abiotic stress resilience in legumes and help in crop breeding (Afzal et al., 2020). Serial analysis of gene expression (SAGE) and microarrays might be used to determine extensive transcriptome information due to advanced technology like high-throughput analysis. Differential gene expression (DGEs) is usually evaluated by sequencing of nucleic acid (RNA-seq). A relatively newer invented technique known as digital gene expression (DGE) is used for the quantified gene expression measurement. RNA-seq is a low-cost, high-throughput sequencing technique for evaluating vast quantities of transcriptome data. This method has various advantages over microarray technology, including the fact that it should not need genomic data to create probe sets and can detect unique transcripts (Lowe et al., 2017). This method has been used in many studies to identify the gene regulatory process which is responsible for the tolerance to abiotic stress, particularly on pulse crops (Table 7). Using the NGS method, a transcriptome process has been generated under stressful circumstances like drought in soybean (Libault et al., 2010). The transcriptional alterations in drought-tolerant as well as drought-sensitive soybean cultivars have been documented using a comparative transcriptome process (Wang et al., 2018). Different varieties of common bean genotype which are vulnerable to biotic and abiotic conditions such as drought, aluminum toxicity, low phosphorous, and heat were evaluated for paternal polymorphism, functional and molecular genomic data association by single nucleotide polymorphisms (SNPs) markers, that was obtained from Sanger sequencing along with Illumina's GoldenGate method (Blair et al., 2013). Das et al. (2017) explained that the metabolomics monitoring of nitrogen or sugar metabolism and phytochemical metabolic activity are all important during water shortage situations in soybean. Singh et al. (2017b) identified potential genes involved in drought stress during lentil seedlings predicated on a transcriptome study, while Pandey et al. (2008) discovered dehydration-responsive proteins in chickpeas. Molina et al. (2008) detected 80,238 labels indicating 17,493 distinct transcripts in chickpea transcriptomes with drought conditions by employing SuperSAGE and deep SuperSAGE. The rapid generation of jasmonate into chickpea roots in drought situations was revealed by root transcriptome profiling of particularly oxylipin production genes. Future potential breeding initiatives may benefit from the use of RNA-seq to better comprehend the genes expressed under stress conditions (Dai et al., 2021).
Table 7.
Transcriptome profiling of legumes crops under abiotic stress using RNA-Seq.
| Legumes crops | Abiotic stress | Plant parts | Seq. platform | *Details | References |
|---|---|---|---|---|---|
| Chickpea | Drought | Roots and Shoots | Illumina HiSeq 2500 | Identification of transcription factors linked with drought tolerance | Mahdavi Mashaki et al., 2018 |
| Root | Illumina HiSeq 2500 | Identification of drought-resistant transcription factors Bhlh, C3H, NAC, AP2-EREBP, and MYB. | Shukla et al., 2006 | ||
| Leaf | Illumina HiSeq 3000 | 1,562 genes were differentially expressed in the tolerant genotype based on RNA extracted from leaf tissues. Genes that respond to drought were elevated in the tolerant genotype. | Badhan et al., 2018 | ||
| Salinity and Drought | Root apex | Roche 454 FLX | Under transcriptome profiling, miRNA-mediated post-transcriptional regulation of genes involved in lateral root development and re-patterning of root hair cells, as well as genes with a high affinity for K+ absorption. The root apex was used to dissect salt and water deprivation situations. | Khandal et al., 2017 | |
| Common bean | Drought | Leaf | Illumina GAIIx | During drought stress drought-sensitive genes were detected. | Wu et al., 2014 |
| Drought | Leaf and root | Illumina platforms (GAII and HiSeq 2000) | Data from transcriptomes showed novel genes associated with the drought stress response. | Confortin et al., 2017 | |
| Salinity | Cotyledon, hypocotyl, and radicle | Illumina HiSeq 2500 PE 150 | During the sprouting stage under salt stress, the role of zinc finger proteins (C3H) was discovered. | Zhang et al., 2020 | |
| Root | Illumina HiSeq TM 2000 | Transcriptome analysis revealed a total of 2,678 transcription factors, 441 of which were involved in salinity tolerance. | Hiz et al., 2014 | ||
| Cowpea | Drought | Leaf | Illumina deep sequencing technology | There were just drought-responsive miRNAs identified. | Barrera-Figueroa et al., 2011 |
| Drought | Leaf | For drought-responsive genes, an SSH database (http://sshdb.bi.up.ac.za/, viewed on 3 August 2021) was generated. | Coetzer et al., 2010 | ||
| Cold (Chilling) | Pods | Illumina HiSeq 2500 | Many sRNAs and miRNAs are implicated in the response to chilling, according to sRNAomic and transcriptome analyses. | Zuo et al., 2018 | |
| Leaf | Illumina HiSeq 4000 | During the vegetative and blooming stages, a total of 538 and 642 putative Transcription factors were found, respectively. | Khan et al., 2019a | ||
| Faba bean | Drought | Root | Illumina HiSeq 4000 | New DEGs with altered expression during drought were discovered. | Alghamdi et al., 2018 |
| Salinity | Cotyledons | Illumina HiSeq 4000 | A total of 1,410 salinity-responsive genes were discovered, with the salt-tolerant genotype showing considerable up-regulation of these genes. | Yang et al., 2020 | |
| Lentil | Drought | Leaf | Illumina HiSeq 2500 | Drought-tolerant genotypes had more severe upregulation of genes involved in oxidation-reduction processes, TCA cycle, organ senescence, and stomatal conductance decrease than drought-sensitive genotypes. | Razzaq et al., 2021 |
| Heat | Leaf | Illumina HiSeq 2000 | The cell wall and secondary metabolite pathways were both found to be significantly affected. | Singh et al., 2019 | |
| Mung bean | Desiccation | Seed | Illumina HiSeq 2500 with PE125 | Many transcription factors (AP2, NAC, MYB, and methyltransferase and histone genes were discovered to be differently expressed.) | Tian et al., 2016 |
*AP2-EREBP, APETALA2/Ethylene-Responsive Element Binding Protein; bHLH, beta Helix Loop Helix; bZIP, beta Leucine Zipper; DEGs, Differentially expressed genes; HSPs, Heat shock proteins; LEA, Late embryogenesis associated; MADS, minichromosome maintenance factor1, agamous, deficiens, and serum response factor; miRNA, MicroRNA; MYB, myeloblastosis; NAC, NAM/ATAF1/CUC2; RNA, Ribonucleic acid; SSH, Suppression subtractive hybridization; sRNA, small RNA; TCA, Tricarboxylic acid cycle; TFs, Transcription factors.
Metabolomics, proteomics, and ion-omics advances for abiotic stress in legumes
Abiotic stress has a significant impact on plants' metabolomes and proteomes, besides the change in genetic makeup and nucleotide sequences which are actively engaged in defense mechanisms against various stresses (Arbona et al., 2013). The proteome of an individual, which serves as a link between both the transcriptome and the metabolome more accurately represents the current level of proliferation and differentiation than Genetic markers. The transcriptome's assessment of cellular mRNA is not an exact representation of transcriptional activations (Min et al., 2019). The synthesis protein has played a role in cell signaling pathways and is implicated in stress adaption, stress repair, and also involved with other functions of the host plant. As a result, they aid the plant's recuperation from stress damage as well as make its survival much better in challenging situations (Hakeem et al., 2012). Conversely, metabolites, as opposed to mRNA or proteins, are a depiction of the transcriptional interactions that are involved in the regulation of gene expression throughout adverse conditions and have intimate linkages to the phenotype (Wienkoop et al., 2008). Metabolomics has been the most cross-functional of all systems biology and is present in many of the functions discussed (Arbona et al., 2013). Finally, detailed investigations proved that the metabolic networks participating in the growth and development of plants under a variety of environmental factors are critical for plant growth. Previously, metabolomics and proteomics were used to evaluate the molecular characteristics of legumes against abiotic stressors, as shown in Table 8. Ionomics is the science of a single tissue or the entire organism which encompasses the assessment of all elemental components in response to physiological functions or their alterations. Lahner et al. (2003) were the first who described an “ionome” as the metals, nonmetals, and metalloids present in an organism. Eventually, the terminology “ionome” was expanded to a “metallome” to relate to a collection of ecologically essential non-metals like nitrogen (N), sulfur (S), and phosphorus (P) (Salt et al., 2008). Ions play an important role in maintaining a plant's homeostasis under a variety of environmental situations. Ion transporters are also essential for the regular functioning of metabolic processes as well as managing stress. Particularly compared to various other grain crops, the ionomics technique has been widely employed in model legume plants (Lotus japonicus) and commercial legume crops including soybean (Chen et al., 2009). At the commercial level, the soybean genotypes with modified seed chemistry were found using this technique. Rapid innovations in ionomics have opened up new avenues for obtaining a precise description of the macro-and micronutrients or even the chemical compositions of legume grains in timely and cost-effective ways. Ionomics can be employed in this sense for attaining global food and nutritional security while also addressing the “hidden” starvation caused by micronutrient deficiencies (Hacisalihoglu and Settles, 2017; Ziegler et al., 2018). Finally, the ionome evidence can be used in experiments on the accessibility of micronutrients in fundamental pulses. Ionomics involves a thorough understanding of the transcriptional regulation concerned with homeostasis to assess the abiotic-stress-responsive ion transporters, genetic mutations compounds, and other constituents. Combining ionomics with other pan-omics tools, such as metabolomics and proteomics, will expand the possibilities for evaluating the impacts of abiotic stresses in legumes and their implementations helps in increasing legume productivity (Singh et al., 2021).
Table 8.
Combating abiotic stresses in legumes using proteomics and metabolomics approaches.
| Crop | Abiotic stress | Omics approach | Details | References |
|---|---|---|---|---|
| Chickpea | Drought | Proteomics | Potential resources for improving drought tolerance were identified. | Vessal et al., 2020 |
| Drought | Comparative proteomics | A total of 75 proteins were found to be differentially expressed in roots. | Gupta et al., 2020 | |
| Drought | Comparative proteomics | MALDI-TOF/TOF-MS/MS analyses revealed 24 differently expressed proteins in leaves under drought stress. | Çevik et al., 2019 | |
| Drought | Metabolomics | Effect of PGPRs under drought stress was identified using UPLC-HRMS analysis | Khan et al., 2019b | |
| Heat | Comparative proteomics | A total of 482 heat-responsive proteins were found to be engaged in heat stress tolerance. | Parankusam et al., 2017 | |
| Salinity | Comparative proteomics | Various proteins were found to be engaged in salinity tolerance. | Arefian et al., 2019 | |
| Drought | Metabolomics | Effect of PGPRs under drought stress was identified using UPLC-HRMS analysis | Khan et al., 2019b | |
| Cowpea | Drought | Metabolomics | GC-TOF-MS profiling of primary metabolites and LC-DAD profiling of secondary metabolites under drought stress. Prolonged stress irrespective of the developmental stage affected the metabolome. | Goufo et al., 2017 |
| Faba bean | Drought | Proteomics | Proteins including chitinase, Bet, and glutamate–glyoxylate aminotransferase were found to be upregulated in leaves under drought stress. | Li et al., 2018 |
| Salinity | Metabolomics | Molecules such as myo-inositol, allantoin, and glycerophosphoglycerol were found to be up-regulated in roots in response to salt stress. | Richter et al., 2019 | |
| Drought and heat | Metabolomics | Upregulation of nitrogen and metabolism under combined heat and drought stress. | Das et al., 2017 | |
| Soybean | Salinity | Comparative metabolomics | A total of 47 different metabolites were found to be responsible for salt tolerance. | Li et al., 2017 |
| Aluminum | Comparative proteomics | MALDI TOF analysis revealed differential protein expression in roots under Al stress. | Duressa et al., 2011 |
Phenomics prospective in legumes
The phenotype of an organism seems to be an observable biophysical property like appearance, behavior, and development (Pratap et al., 2019). Phenomics is a research area that involves high-throughput phenotyping analysis. Phenotype results from a complex combination of genetic capability among an organism and its environment (Deshmukh et al., 2014). Understanding any biological process needs precise phenotyping. A specific phenotype is often employed in plant and animal studies (like symptoms) to comprehend the biological condition, including disease, pest infestation, or physiological disorders. Genetic resources are employed for phenotype-based assessment of genetic markers, which arise as a result of technological advancements; known as “genetic symptoms.” The performance of genomics is determined by the consistency with which a genetic marker and phenotype are connected (Varshney et al., 2018). Plant breeding is using omics approaches to improve genetics to obtain an ideal phenotype that will provide a greater and more consistent yield under a variety of environmental situations. As a result, phenomics combined with certain other omics methods holds the most hope in plant breeding (Biswas et al., 2021). Abiotic stress tolerance seems to be a critical feature in terms of yield persistence and potential. Some traits which correlate to stress tolerance in legumes can be evaluated in a controlled condition at the time of sowing. The closing of stomata is one of the primary reasons for saline occurring in plants due to the osmotic effect of soluble compounds which are directly up taken by plant roots from groundwater. As a result, the photosynthesis process becomes slow. Therefore, an alternative measure can be exploited for stomatal responses and photosynthetic under osmotic tensity, which can also be stretched to legumes (Munns, 2010). Phenotyping concedes the imaging of characters of curiosity in real-time. NIRS (near-Infrared Spectroscopy) is one of the very dynamic techniques in advanced phenotyping study among plant breeding. For example, Jakubowski et al. (2017) chose various forage legume crops relying on their improved N2 fixation from the atmosphere by using this NIRS technique. The plants were evaluated based on the highest tissue N levels, which were proven to increase N2 fixation in the plant from the atmosphere. This depicts how phenotypic scanning would be used to assess and selected plants. The technique can be implemented to various legumes as well as more aspects.
Genome editing of legume crops against abiotic stress
Multiple candidate genes which are associated with response to abiotic stress are being explored using genome editing methods. To date, numerous biotechnological tools are present to identify these genes. Somaclonal variations, tissue culture, marker-assisted breeding, mutagenesis, wide hybridization, double haploids, and genetic transformation are the main tools of genome editing (Aasim et al., 2018). Genetic transformation (GT) is a famous biotechnological technique used to address the biotic or abiotic stresses of legume plants. Yet, no meaningful progress has been made so far anyway. Kumar and Fladung (2002) explained the genetic transformation process and revealed that the AMT (Agrobacterium Meditated Transformation) is the most useful and powerful technique applied in legumes for genome editing followed by microprojectile bombardment (MP), direct gene transfer to protoplast (DGTP), and electroporation (EP).
Genetic transformation in legumes
In the modern era, the insertion of exotic genes from wild plants to legumes and other important field crops through GT technology has become more significant (Somers et al., 2003). The reliability of transgenic germplasms for reproducing has been proven to be extremely low. Transgenic approaches aid in the production of more durable, genetically engineered germplasm with increased vigor and tolerance to different abiotic challenges, as well as enhanced crop productivity and quality (Chandra and Pental, 2003). The invention of transgenic tools has aided in the identification of mechanisms behind the transcriptional activation linked to abiotic stress resilience. The cultivation of genetically modified legume crops with better agronomic practices has been a great achievement (Aasim et al., 2013). Vadez et al. (2008) described that little progress has been made due to the complicated genetic pathways which are involved to produce crops tolerant to abiotic stress. Previously, a lot of studies revealed that genetically modified legume plants have shown more resilience to abiotic stressors such as salinity, drought, and metals (Atkinson and Urwin, 2012; Sita and Kumar, 2020).
Genome editing in legume crops
To reduce food insecurity, many strategies are being used to build a sustainable agriculture system. Genome editing (GE) for crop enhancement is one such technology that has the potential to build a climate-resilient agriculture system on a worldwide scale (Liu et al., 2013; Abdelrahman et al., 2018). Plant breeding techniques have been significantly influenced by GE technology, including novel strategies for making rapid and precise modifications in crop genomes to defend plants from diverse threats and improve crop yields (Taranto et al., 2018). Site-specific endonucleases such as zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and CRISPR-Cas9 are employed in genome editing approaches (Zhu et al., 2017). There are many studies of genome editing that have been conducted on major legume crops.
Soybeans are rich in oil and protein, and an escalating demand requires genetic development with gene-editing tools to meet rising needs and cope with fluctuating climate (Bao et al., 2020). Curtin et al. (2011) targeted the green fluorescent protein (GFP) coding region in soybean with a ZFN array developed via context-sensitive selection strategies. This approach designed two independent ZFN pairs by deleting up to 71 base pairs on the target. Curtin et al. (2016) also focused on disturbing miRNA maturation and miRNA gene expression regulation. Accordingly, two ZFN pairs were designed to target Dicer-like 1a (DCL1a) and Dicer-like 1b (DCL1b) genes in soybean. Moreover, Curtin et al. (2018) generated a suite of combinatorial mutant plants using TALENs within the G. max Dicer-like2 gene by whole-genome sequencing. In CRISPR-Cas conditions, different sites of two endogenous soybean genes (GmFEI2 and GmSHR) were targeted by a transgene (bar) and six sgRNAs, and a sgRNA was designed that resulted in mutations in the targeted DNA of hairy roots (Cai et al., 2015). Di et al. (2019) CRISPR/Cas9 system was enhanced by highly active 5 U6 promoters targeting Glyma03g36470, Glyma14g04180, and Glyma06g136900 genes. Consequently, nucleotide insertion, deletion, and substitution mutations occurred. The following year, to target 102 candidate genes Bai et al. (2020) constructed 70 CRISPR-Cas9 vectors and obtained 407 T0 sgRNAs mutant lines with 59.2% mutagenesis frequency, and 35.6% lines carrying multiplex mutations. As a result, increased nodulation of gmric1/gmric2 double mutants and gmrdn1-1/1-2/1-3 triple mutant lines with decreased nodulation were detected.
L. japonicus a model organism for legume crops with similar features to M. truncatula, organizes determinate nodules, like soybean and cowpea. In L. japonicus, CRISPR/ Cas9 can perform SNF (symbiotic nitrogen fixation)-related gene mutations with hairy root transformation (Wang et al., 2016). Cai et al. (2018a) edited cytokinin receptor Lotus histidine kinaz I-interacting protein (LjCZF1) to disclose the process of cytokinin signaling regulation of rhizobia-legume symbiosis. They found mutant lines with reduced infection threads and nodules, indicating that LjCZF1 is a better regulator of nodulation. Later, CRISPR/Cas9 technology revealed that leghemoglobin (Lbs) in L. japonicus resulted in early nodule senescence (Wang et al., 2019b).
Chickpea (C. arietinum) is a commercially important crop worldwide, and gene-editing tools can be used to eliminate the problems in its production. Badhan et al. (2021) performed a study that targeted drought tolerance-associated genes, 4- coumarate ligase (4CL) and Reveille 7 (RVE7), for CRISPR/Cas9 editing in chickpea protoplast and the knock-out of the RVE7 gene showed high-efficiency editing in vivo. These results showed that CRISPR/Cas9 DNA-free gene editing can be used for genes associated with drought tolerance in chickpea by utilizing protoplast. To date, this was the first and only study that used CRISPR/Cas9 gene editing in chickpea.
Cowpea (V. unguiculata L.) Walp., with high nutrition content and health benefits, has tolerance to low rainfall, symbiotic nitrogen fixation (SNF) capability, and low fertilization requirements that make it commercially important (Ji et al., 2019; Che et al., 2021). Ji et al. (2019) targeted the SNF genes by CRISPR/ Cas9-mediated genome editing in non-inheritable mutated hairy roots in cowpea and found restricted nodule formation in the mutants with disrupted alleles. Following them, Juranić et al. (2020) used CRISPR/Cas9 gene editing and identified three cowpea meiosis genes; REC8 (encodes meiotic recombination protein), SPO11-1 (encodes SPO11 protein; an initiator of meiotic double-stranded breaks), and OSD1 (encodes Ophiostoma scytalone dehydratase protein promoting meiotic progression) to induce cowpea asexual seed formation. They found defects in meiosis due to biallelic mutations in exon 1 and exon 3 of the SPO11-1 gene. Recently, Che et al. (2021) also used CRISPR/Cas9 and noticed mutations at the target leading to inhibition of cowpea meiosis gene VuSPO11-1.
Lentils are a type of pulse crop that is consumed all over the world, being a good source of protein, minerals, carbohydrates, vitamins, dietary fiber, and secondary metabolites like phenolic compounds. Explants from various lentil tissues, such as shoot apices, epicotyls, nodal segments, embryo axes, cotyledonary nodes, and roots, have been explored for in vitro plant regeneration for genetic modification (Mahmoudian et al., 2002; Sarker et al., 2003; Akcay et al., 2009). As the number of shoots regenerated per explant greatly affect the transformation efficiency and success of CRISPR/Cas9-based gene editing, optimization of the protocol with an appropriate combination of mineral media and hormones is required in the near future. Gene disruption to improve lentils may be made simpler, less expensive, and more accurate by gene editing.
Mung bean is a warm-season, self-pollinated and diploid (2n = 2x = 22) legume crop with a small genome size (Parida et al., 1990; Nair et al., 2013; Kang et al., 2014). One of the first legumes to have a genetic linkage map was the mung bean in the early 1990s (Fatokun et al., 1992, 1993). In the past, molecular methods with an emphasis on yield, nutrient improvement, and disease resistance have been used to improve mung beans. These methods included random amplified polymorphic DNA (RAPD), restriction fragment length polymorphism (RFLP), SNP, and SSR markers (Fatokun et al., 1993; Chankaew et al., 2011; War et al., 2017). In mung bean breeding projects, CRISPR/Cas9 gene editing has a lot of potential. A symbiosis receptor-like kinase gene was recently successfully targeted using CRISPR/Cas9 gene editing to stop symbiotic nitrogen fixation in cowpea (V. unguiculata) (Ji et al., 2019). The achievement of CRISPR/Cas9 in a Vigna system shows that gene editing can be used for other species including mung bean. Quality attributes and disease resistance would be some of the initial targets for gene editing in mung beans. Developing mung bean varieties that can withstand changing weather patterns would help in the further expansion of mung bean cultivation worldwide.
Faba bean is a diploid (2n = 2x = 12) and one of the most important cool-season grain legumes with high protein content, antioxidants, and a rich source of fiber (Ray and Georges, 2010; Sudheesh et al., 2019). A reference genome of faba bean is still lacking; however, significant progress has been made in creating genetic and genomic resources to facilitate molecular breeding. Application of CRISPR/Cas gene editing is hampered by the lack of an annotated reference genome for the intricate faba bean genome, especially when designing specific gRNA-targeted genes of interest. To date, no CRISPR/Cas9 studies have been reported for this crop.
Genome editing, particularly CRISPR/Cas9 is an effective technology for abiotic stress tolerance in legumes, which is still rarely used. Similarly, there are limited studies in the literature on the implementation of genome editing for abiotic stress tolerance in other crops (Debbarma et al., 2019). In legumes, Soybean genome editing using the CRISPR/Cas9 system has been reported to be successful (Bao et al., 2019). For example, using the CRISPR/Cas9 approach, the soybean gene GmFT2a (FT-Flower Locus T) related to flowering time was knocked out, resulting in GmFT2a mutants with delayed flowering (Cai et al., 2018b). The roles of GmHsp90A2 (Heat shock protein 90s) in soybean have recently been studied using stable transgenic lines overexpressing GmHsp90A2 as well as mutant lines produced using the CRISPR/Cas9 genome editing technique (Huang et al., 2019). Hsp90s is a stable and abundant protein chaperone involved in the protective stress response, and overexpression of GmHsp90A2 in the Arabidopsis thaliana model resulted in greater tolerance to heat stress in a prior study (Xu et al., 2013). The CRISPR/Cas9 deletion of the endogenous soybean GmHsp90A2 gene was verified up to the T1 generation in the study of Huang et al. (2019). Although there are fewer scientific studies on using accurate and targeted genome editing for abiotic stress tolerance in Fabaceae can be found, these approaches have immense potential for future use in legume molecular breeding. Here, we compiled a list of reports on the application of transgenic tools in legumes to abiotic stress resilience (Table 9).
Table 9.
Application of transgenic tools for genome editing in legumes for abiotic stresses.
| Legume crop | Performance of transgenic | Gene | Sources | Protein coded | Promoter used | References |
|---|---|---|---|---|---|---|
| Mung bean | Salt and drought stress | Coda | A. globiformis | Choline oxidase A | CaMV35S | Baloda and Madanpotra, 2017 |
| Salt stress | gly I | Brassica juncea | Glyoxalate | CaMV35S and CmYLCV | Bhomkar et al., 2008 | |
| Salinity and drought stress | ALDRXV4 | – | Osmoprotection and detoxification | CaMV35S | Singh et al., 2016 | |
| Phseolus vulgaris | Drought stress | HVA1 and bar | Hordeum vulgare | Late embryogenesis protein | CaMV35S | Nguyen and Sticklen, 2012 |
| Medicago sativa | Salt tolerance | GmDREB1 | G. max | DRE-binding protein | A. thaliana RD29A | Jin et al., 2019 |
| Drought stress | WXP1 | Medicago truncatula | AP2 domain | CaMV35S | Zhang et al., 2005 | |
| Freezing stress | Fe-SOD | Nicotiana plumbaginifolia | Fe-superoxide Dismutase | CaMV35S | McKersie et al., 2000 | |
| Aluminum toxicity stress | Malate dehydrogenase | M. sativa | Malate dehydrogenase | CaMV35S | Tesfaye et al., 2001 | |
| Freezing stress | Mn-SOD | N. plumbaginifolia | Mn-superoxide dismutas | CaMV35S | Song et al., 2019 | |
| Freezing stress | SOD | P. sativum and N. plumbaginifolia | Superoxide dismutase | CaMV35S | McKersie et al., 1993 | |
| C. Arietinum | Oxidative stress | Cod A | Arthrobacter globiformis | Choline oxidase A | CaMV35S | Sharmila et al., 2009 |
| Osmotic and drought stress | p5cs | Vigna aconitifolia | O1-pyrroline 5-carboxylate synthase | CaMV35S | Bhatnagar-Mathur et al., 2009 | |
| Arachis hypogea | Drought stress | Alfin1, PDH45, and PgHSF4 | Alfaalfa and Pea | – | CaMV35S | Ramu et al., 2016 |
| Drought stress | DREB1A | A. thaliana | DRE-binding protein | A. thaliana RD29A | Mathur et al., 2014 | |
| Drought stress | MuNAC4 | Macrotyloma uniflorum | NAC | CaMV35S | Pandurangaiah et al., 2014 | |
| Drought stress | DREB1A | A. thaliana | DRE-binding protein | A. thaliana RD29A | Vadez et al., 2013 | |
| Drought and SALINITY stress | MuWRKY3 | Macrotyloma uniflorum | WRKY | CaMV35S | Kiranmai et al., 2018 | |
| Drought and salinity stress | AtHDG11 | A. thaliana | Transcription factor | A. thaliana RD29A | Banavath et al., 2018 | |
| Drought and salinity stress | SBASR-1 | S. chiata | Abscisic acid stress | CaMV35S | Tiwari et al., 2015 | |
| Drought and salinity stress | AtNAC2 | A. thaliana | NAC | CaMV35S | Patil et al., 2014 | |
| Drought and salinity stress | AtDREB2A and AtABF3 | A. thaliana | DRE-binding protein | CaMV35S | Pruthvi et al., 2014 | |
| Drought and salinity stress | SbVPPase | Sorghum bicolor | Vacuolar proton pyrophosphatase | CaMV35S | Puli et al., 2021 | |
| Salinity stress | SbNHXLP | S. bicolor | Na1/H1 antiporterlikeprotein | CaMV35S | Kavi Kishor et al., 2018 | |
| Salt stress | SbpAPX | Salicornia brachiata | Peroxisomalascorbate peroxidase | CaMV35S | Singh et al., 2014 | |
| Salt stress | PDH45 | Pisumsativum | DNA helicase 45 | CaMV35S | Manjulatha et al., 2014 | |
| Glycine max | Drought and salinity stress | NTR1 | Bactris campestris | Jasmonic acid | CaMV35S | Xue and Zhang, 2007 |
Conclusion and future recommendations
Agriculture is dependent on the climate, and changes in climate are resulting in various stresses and causing significantl production losses each year. Legumes are multi-beneficial crops and can mitigate our food scarcity problems by providing high-quality food with low inputs. During the last two decades, significant progress has been made in utilizing state-of-the-art “OMICs” approaches to develop climate-smart legume cultivars. A huge number of legumes are conserved at the gene banks and also present in the small farmer's field and are yet to be characterized. Efforts should be done to characterize those unexplored legumes' genetic resources at both phenotypic and molecular levels to identify genetic variations that can be used for the development of improved cultivars. Nevertheless, a good number of efforts have been made to identify the genomic regions associated with abiotic stresses. However, fewer attempts have been made to validate the identified genomic region to speed up marker-assisted breeding. Therefore, it is a present-day need to develop KASP markers for speed breeding. Recent advancements in genomic resources for legumes have provided the basis to perform transformative breeding approaches like genomic selection and genome editing for crop improvement. However, these efforts are still few compared to cereals. It is very important to harness CRISPR/Cas9-based gene-editing technology for the targeted improvement of traits in legume crops.
Author contributions
MN conceived the idea and perform editing to the manuscript. MN, NB, and MA collected the literature. AA, MTA, AS, HA, and TH contributed to writing the original draft of the manuscript. AA, MTA, HA, NB, K-HB, and TK designed the tables and figures. MN, FB, MA, AS, SD, TH, and TK reviewed and edited the manuscript. MN, FB, YC, NA, and MH contributed to addressing comments raised by the reviewers. MN, FB, and YC provided resources. All authors have read and approved the final version of the manuscript.
Funding
The authors are very grateful to the Scientific Research Unit of Sivas University of Science and Technology for providing a research grant to MN through his project (Project Number: 2021-GENL-TBT-0003). Basic Science Research Program supported this research through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (2019R1A6A1A11052070).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Aasim M., Baloch F. S., Nadeem M. A., Bakhsh A., Sameeullah M., Day S. (2018). “Fenugreek (Trigonella foenum-graecum L.): an underutilized edible plant of modern world,” in Global Perspectives on Underutilized Crops (Cham: Springer; ), 381–408. [Google Scholar]
- Aasim M., Day S., Rezaei F., Hajyzadeh M. (2013). Multiple shoot regeneration of plumular apices of Chickpea. Turk J. Agric. For. 37, 33–39. 10.3906/tar-1204-38 [DOI] [Google Scholar]
- Abdelrahman M., Al-Sadi A. M., Pour-Aboughadareh A., Burritt D. J., Tran L. S. P. (2018). Genome editing using CRISPR/Cas9–targeted mutagenesis: an opportunity for yield improvements of crop plants grown under environmental stresses. Plant Physiol. Biochem. 131, 31–36. 10.1016/j.plaphy.2018.03.012 [DOI] [PubMed] [Google Scholar]
- Afzal M., Alghamdi S. S., Migdadi H. H., Khan M. A., Mirza S. B., El-Harty E. (2020). Legume genomics and transcriptomics: from classic breeding to modern technologies. Saudi J. Biol. Sci. 27, 543–555. 10.1016/j.sjbs.2019.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S. M., Alsamman A. M., Jighly A., Mubarak M. H., Al-Shamaa K., Istanbuli T., et al. (2021). Genome-wide association analysis of chickpea germplasms differing for salinity tolerance based on DArTseq markers. PLoS ONE. 16, e0260709. 10.1371/journal.pone.0260709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akcay U. C., Mahmoudian M., Kamci H., Yucel M., Oktem H. (2009). Agrobacterium tumefaciens-mediated genetic transformation of a recalcitrant grain legume, lentil (Lens culinaris Medik). Plant Cell Rep. 28, 407–417. 10.1007/s00299-008-0652-4 [DOI] [PubMed] [Google Scholar]
- Akohoue F., Achigan-Dako E. G., Sneller C., Van Deynze A., Sibiya J. (2020). Genetic diversity, SNP-trait associations and genomic selection accuracy in a west African collection of Kersting's groundnut [Macrotyloma geocarpum (Harms) Maréchal & Baudet]. PLoS ONE 15, e0234769. 10.1371/journal.pone.0234769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alghamdi S. S., Khan M. A., Ammar M. H., Sun Q., Huang L., Migdadi H. M., et al. (2018). Characterization of drought stress-responsive root transcriptome of Faba bean (Vicia faba L.) using RNA sequencing. 3 Biotech. 8, 1–19. 10.1007/s13205-018-1518-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambachew D., Blair M. W. (2021). Genome wide association mapping of root traits in the andean genepool of common bean (Phaseolus vulgaris L.) grown with and without aluminum toxicity. Front. Plant Sci. 12, 825. 10.3389/fpls.2021.628687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjum S. A., Ashraf U., Zohaib A., Tanveer M., Naeem M., Ali I., et al. (2017). Growth and development responses of crop plants under drought stress: a review. Zemdirbyste 104, 267–276. 10.13080/z-a.2017.104.034 [DOI] [Google Scholar]
- Annicchiarico P., Nazzicari N., Wei Y., Pecetti L., Brummer E. C. (2017). Genotyping-by-sequencing and its exploitation for forage and cool-season grain legume breeding. Front. Plant Sci. 8, 679. 10.3389/fpls.2017.00679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar A., Sinha A., Sharif A., Siddique M., Irshad S., Anwar W., et al. (2022). The nexus between urbanization, renewable energy consumption, financial development, and CO2 emissions: Evidence from selected Asian countries. Environ. Dev. Sustain. 24, 6556–6576. 10.1007/s10668-021-01716-2 [DOI] [Google Scholar]
- Arbona V., Manzi M., Ollas C. D., Gómez-Cadenas A. (2013). Metabolomics as a tool to investigate abiotic stress tolerance in plants. Int. J. Mol. Sci. 14, 4885–4911. 10.3390/ijms14034885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arefian M., Vessal S., Malekzadeh-Shafaroudi S., Siddique K. H., Bagheri A. (2019). Comparative proteomics and gene expression analyses revealed responsive proteins and mechanisms for salt tolerance in Chickpea genotypes. BMC Plant Biol. 19, 1–26. 10.1186/s12870-019-1793-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assefa T., Assibi Mahama A., Brown A. V., Cannon E. K., Rubyogo J. C., Rao I. M., et al. (2019). A review of breeding objectives, genomic resources, and marker-assisted methods in Common bean (Phaseolus vulgaris L.). Mol. Breed. 39, 1–23. 10.1007/s11032-018-0920-0 [DOI] [Google Scholar]
- Atkinson N. J., Urwin P. E. (2012). The interaction of plant biotic and abiotic stresses: from genes to the field. J.Exp. Bot. 63, 3523–3543. 10.1093/jxb/ers100 [DOI] [PubMed] [Google Scholar]
- Badhan S., Ball A. S., Mantri N. (2021). First report of CRISPR/Cas9 mediated DNA-free editing of 4CL and RVE7 genes in chickpea protoplasts. Int. J. Mol. Sci. 22, 396. 10.3390/ijms22010396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badhan S., Kole P., Ball A., Mantri N. (2018). RNA sequencing of leaf tissues from two contrasting chickpea genotypes reveals mechanisms for drought tolerance. Plant Physiol. Biochem. 129, 295–304. 10.1016/j.plaphy.2018.06.007 [DOI] [PubMed] [Google Scholar]
- Bai M., Yuan J., Kuang H., Gong P., Li S., Zhang Z., et al. (2020). Generation of a multiplex mutagenesis population via pooled CRISPR -Cas9 in soyabean. Plant Biotechnol. J. 18, 721–731. 10.1111/pbi.13239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloda A., Madanpotra S. (2017). Transformation of mungbean plants for salt and drought tolerance by introducing a gene for an osmoprotectant glycine betaine. J. Plant Stress Physiol. 3, 5–11. 10.19071/jpsp.2017.v3.3148 [DOI] [Google Scholar]
- Banavath J. N., Chakradhar T., Pandit V., Konduru S., Guduru K. K., Akila C. S., et al. (2018). Stress inducible overexpression of AtHDG11 leads to improved drought and salt stress tolerance in Peanut (Arachis hypogaea L.). Front. Chem. 6, 34. 10.3389/fchem.2018.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao A., Zhang C., Huang Y., Chen H., Zhou X., Cao D. (2020). Genome editing technology and application in soybean improvement. Oil Crop Sci. 5, 31–40. 10.1016/j.ocsci.2020.03.001 [DOI] [Google Scholar]
- Bao Z., Yang Z., Huang Z., Zhou Y., Cui Q., Dong D. (2019). LncRNADisease 2.0: an updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 47, D1034–D1037. 10.1093/nar/gky905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera-Figueroa B. E., Gao L., Diop N. N., Wu Z., Ehlers J. D., Roberts P. A., et al. (2011). Identification and comparative analysis of drought-associated microRNAs in two Cowpea genotypes. BMC Plant Biol. 11, 1–11. 10.1186/1471-2229-11-127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beji S., Fontaine V., Devaux R., Thomas M., Negro S. S., Bahrman N., et al. (2020). Genome-wide association study identifies favorable SNP alleles and candidate genes for frost tolerance in Pea. BMC Genom. 21, 1–21. 10.1186/s12864-020-06928-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar-Mathur P., Vadez V., Jyostna Devi M., Lavanya M., Vani G., Sharma K. K. (2009). Genetic engineering of Chickpea (Cicer arietinum L.) with the P5CSF129A gene for osmoregulation with implications on drought tolerance. Mol. Breed. 23, 591–606. 10.1007/s11032-009-9258-y [DOI] [Google Scholar]
- Bhomkar P., Upadhyay C. P., Saxena M., Muthusamy A., Shiva Prakash N., Pooggin M., et al. (2008). Salt stress alleviation in transgenic Vigna mungo (L.) Hepper (blackgram) by overexpression of the glyoxalase I gene using a novel Cestrum Yellow Leaf Curling Virus (CmYLCV) promoter. Mol. Breed. 22, 169–181. 10.1007/s11032-008-9164-8 [DOI] [Google Scholar]
- Biswas A., Andrade M. H. M. L., Acharya J. P., de Souza C. L., Lopez Y., De Assis G., et al. (2021). Phenomics-assisted selection for herbage accumulation in Alfalfa (Medicago sativa L.). Front. Plant Sci. 12, 756768–756768. 10.3389/fpls.2021.756768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair M. W., Cortés A. J., Penmetsa R. V., Farmer A., Carrasquilla-Garcia N., Cook D. R. (2013). A high-throughput SNP marker system for parental polymorphism screening, and diversity analysis in common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 126, 535–548. 10.1007/s00122-012-1999-z [DOI] [PubMed] [Google Scholar]
- Bohra A., Bharadwaj C., Radhakrishnan T., Singh N. P., Varshney R. K. (2019). Translational genomics and Mol. Breed for enhancing precision and efficiency in crop improvement programs: some examples in legumes. Indian J. Genet. Plant Breed 79, 227–240. 10.31742/IJGPB.79S.1.13 [DOI] [Google Scholar]
- Breria C. M., Hsieh C.-H., Yen T.-B., Yen J.-Y., Noble T. J., Schafleitner R. (2020). A SNP-based genome-wide association study to mine genetic loci associated to salinity tolerance in mungbean (Vigna radiata L.). Gene 11, 759. 10.3390/genes11070759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. D. H. (1989). Size and Structure of Collection: The Case for Core Collection. The Use of Plant Genetic Resources. Chichester: John Wiley & Sons, Baffins Lane, 136–156. [Google Scholar]
- Burstin J., Salloignon P., Chabert-Martinello M., Magnin-Robert J.-B., Siol M., Jacquin F., et al. (2015). Genetic diversity and trait genomic prediction in a pea diversity panel. BMC Genom. 16, 1–17. 10.1186/s12864-015-1266-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai K.K, Yin J., Chao H., Ren Y., Jin L., et al. (2018a). A C3HC4-type RING finger protein regulates rhizobial infection and nodule organogenesis in lotus japonicus. J. Integr. Plant Biol. 60, 878–896. 10.1111/jipb.12703 [DOI] [PubMed] [Google Scholar]
- Cai Y., Chen L., Liu X., Guo C., Sun S., Wu C., et al. (2018b). CRISPR/Cas9-mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean. Plant Biotechnol. J. 16, 176–185. 10.1111/pbi.12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Chen L., Liu X., Sun S., Wu C., Jiang B., et al. (2015). CRISPR/Cas9- mediated genome editing in soybean hairy roots. PLoS ONE 10, e0136064. 10.1371/journal.pone.0136064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çakir Ö., Uçarli C., Tarhan Ç., Pekmez M., Turgut-Kara N. (2019). Nutritional and health benefits of legumes and their distinctive genomic properties. Food Sci. Technol. 39, 1–12. 10.1590/fst.4211735473972 [DOI] [Google Scholar]
- Cervantes E., Saadaoui E., Tocino Á., Gómez J. J. M. (2019). “Seed shape quantification in the model legumes: Methods and applications,” in The Model Legume Medicago Truncatula (New Jersey: John Wiley & Sons; ), 92–98. [Google Scholar]
- Çevik S., Akpinar G., Yildizli A., Kasap M., Karaosmanoglu K., Ünyayar S. (2019). Comparative physiological and leaf proteome analysis between drought-tolerant chickpea Cicer reticulatum and drought-sensitive chickpea C. arietinum. J. Biosci. 44, 1–13. 10.1007/s12038-018-9836-4 [DOI] [PubMed] [Google Scholar]
- Chandra A., Pental D. (2003). Regeneration and genetic transformation of grain legumes: an overview. Curr. Sci. 84, 381–387. [Google Scholar]
- Chandrashekharaiah P., Paul V., Kushwaha S., Sanyal D., Dasgupta S. (2021). “Biotechnological approaches for enhancing stress tolerance in legumes,” in Sustainable Agriculture Reviews 51, eds P. Guleria, V. Kumar, and E. Lichtfouse (Springer), 247–293. [Google Scholar]
- Chankaew S., Isemura T., Naito K., Ogiso-Tanaka E., Tomooka N., Somta P., et al. (2014). QTL mapping for salt tolerance and domestication-related traits in Vigna marina subsp. oblonga, a halophytic species. Theor. Appl. Genet. 127, 691–702. 10.1007/s00122-013-2251-1 [DOI] [PubMed] [Google Scholar]
- Chankaew S., Somta P., Sorajjapinun W., Srinives P. (2011). Quantitative trait loci mapping of Cercospora leaf spot resistance in mungbean, Vigna radiata (L.) Wilczek. Mol. Breed. 28, 255–264. 10.1007/s11032-010-9478-1 [DOI] [Google Scholar]
- Chapman M. A. (2015). Transcriptome sequencing and marker development for four underutilized legumes. Appl. Plant Sci. 3, 1400111. 10.3732/apps.1400111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che P., Chang S., Simon M. K., Zhang Z., Shaharyar A., Ourada J., et al. (2021). Developing a rapid and highly efficient cowpea regeneration, transformation and genome editing system using embryonic axis explants. Plant J. 106, 817–830. 10.1111/tpj.15202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Watanabe T., Shinano T., Okazaki K., Osaki M. (2009). Rapid characterization of plant mutants with an altered ion-profile: a case study using Lotus japonicus. New Phytol. 181, 795–801. 10.1111/j.1469-8137.2008.02730.x [DOI] [PubMed] [Google Scholar]
- Choi H.-K. (2019). Translational genomics and multi-omics integrated approaches as a useful strategy for crop breeding. Genes Genom. 41, 133–146. 10.1007/s13258-018-0751-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary S., Isobe S., Chahota R. K. (2022). Elucidation of drought tolerance potential of horsegram (Macrotyloma uniflorum Var.) germplasm using genome wide association studies. Gene 819, 146241. 10.1016/j.gene.2022.146241 [DOI] [PubMed] [Google Scholar]
- Coetzer N., Gazendam I., Oelofse D., Berger D. K. (2010). SSHscreen and SSHdb, generic software for microarray based gene discovery: application to the stress response in Cowpea. Plant Methods 6, 1–20. 10.1186/1746-4811-6-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Confortin S. C., Meneghini V., Ono L. M., Schneider I. J., Barbosa A. R., D'orsi E. (2017). Anthropometric indicators as a screening tool for sarcopenia in older adults from Florianópolis, Santa Catarina: EpiFloripa Ageing study. Revista de Nutrição. 30, 287–296. 10.1590/1678-98652017000300002 [DOI] [Google Scholar]
- Cook D. R. (1999). Medicago truncatula-a model in the making! Curr. Opin. Plant Biol. 2, 301–304. 10.1016/S1369-5266(99)80053-3 [DOI] [PubMed] [Google Scholar]
- Curtin S. J., Michno J.-M., Campbell B. W., Gil-Humanes J., Mathioni S. M., Hammond R., et al. (2016). MicroRNA maturation and MicroRNA target gene expression regulation are severely disrupted in soybean dicer-like1 double mutants. G3 6, 423–433. 10.1534/g3.115.022137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin S. J., Xiong Y., Michno J.-M., Campbell B. W., Stec A. O., Cermák T., et al. (2018). CRISPR/Cas9 and TALENs generate heritable mutations for genes involved in small RNA processing of Glycine max and Medicago Truncatula. Plant Biotechnol. J. 16, 1125–1137. 10.1111/pbi.12857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin S. J., Zhang F., Sander J. D., Haun W. J., Starker C., Baltes N. J., et al. (2011). Targeted mutagenesis of duplicated genes in soybean with zinc-finger nucleases. Plant Physiol. 156, 466–473. 10.1104/pp.111.172981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., Zhuang Z., Boschiero C., Dong Y., Zhao P. X. (2021). LegumeIP V3: from models to crops—an integrative gene discovery platform for translational genomics in legumes. Nucleic Acids Res. 49, D1472–D1479. 10.1093/nar/gkaa976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Rushton P.J., Rohila J. S. (2017). Metabolomic profiling of soybeans (Glycine max L.) reveals the importance of sugar and nitrogen metabolism under drought and heat stress. Plants 6, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debbarma J., Sarki Y. N., Saikia B., Boruah H. P. D., Singha D. L., Chikkaputtaiah C. (2019). Ethylene response factor (ERF) family proteins in abiotic stresses and CRISPR–Cas9 genome editing of ERFs for multiple abiotic stress tolerance in crop plants: a review. Mol. Biotechnol. 61, 153–172. 10.1007/s12033-018-0144-x [DOI] [PubMed] [Google Scholar]
- Deshmukh R., Sonah H., Patil G., Chen W., Prince S., Mutava R., et al. (2014). Integrating omic approaches for abiotic stress tolerance in soybean. Front. Plant Sci. 5, 244. 10.3389/fpls.2014.00244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhaliwal S. K., Talukdar A., Gautam A., Sharma P., Sharma V., Kaushik P. (2020). Developments and prospects in imperative underexploited vegetable legumes breeding: a review. Int. J. Mol. Sci. 21, 9615. 10.3390/ijms21249615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Y. H., Sun X. J., Hu Z., Jiang Q. Y., Song G. H., Zhang B., et al. (2019). Enhancing the CRISPR/Cas9 system based on multiple GmU6 promoters in soybean. Biochem. Biophysical Res. Commun. 519, 819–823. 10.1016/j.bbrc.2019.09.074 [DOI] [PubMed] [Google Scholar]
- Diaz L. M., Ricaurte J., Tovar E., Cajiao C., Teran H., Grajales M., et al. (2018). QTL analyses for tolerance to abiotic stresses in a common bean (Phaseolus vulgaris L.) population. PLoS ONE 13, e0202342. 10.1371/journal.pone.0202342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dita M. A., Rispail N., Prats E., Rubiales D., Singh K. B. (2006). Biotechnology approaches to overcome biotic and abiotic stress constraints in legumes. Euphytica 147, 1–24. 10.1007/s10681-006-6156-9 [DOI] [Google Scholar]
- Dramadri I. O., Nkalubo S. T., Kramer D. M., Kelly J. D. (2021). Genome-wide association analysis of drought adaptive traits in common bean. Crop Sci. 61, 3232–3253. 10.1002/csc2.20484 [DOI] [Google Scholar]
- Dumont E., Fontaine V., Vuylsteker C., Sellier H., Bodèle S., Voedts N., et al. (2009). Association of sugar content QTL and PQL with physiological traits relevant to frost damage resistance in pea under field and controlled conditions. Theor. Appl. Genet. 118, 1561–1571. 10.1007/s00122-009-1004-7 [DOI] [PubMed] [Google Scholar]
- Duressa D., Soliman K., Taylor R., Senwo Z. (2011). Proteomic analysis of soybean roots under aluminum stress. Int. J. Plant Genom. 2011, 282531. 10.1155/2011/282531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatokun C. A., Danesh D., Young N. D., Stewart E. L. (1993). Molecular taxonomic relationships in the genus Vigna based on RFLP analysis. Theoret. Appl. Genet. 86, 97–104. 10.1007/BF00223813 [DOI] [PubMed] [Google Scholar]
- Fatokun C. A., Menancio-Hautea D. I., Danesh D., Young N. D. (1992). Evidence for orthologous seed weight genes in Cowpea and Mung bean based on RFLP mapping. Genetics 132, 841–846. 10.1093/genetics/132.3.841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes C., Figueira E., Tauler R., Bedia C. (2018). Exposure to chlorpyrifos induces morphometric, biochemical and lipidomic alterations in Greenbeans (Phaseolus vulgaris). Ecotoxicol. Environ. Saf. Eco. Environ. Safe 156, 25–33. 10.1016/j.ecoenv.2018.03.005 [DOI] [PubMed] [Google Scholar]
- Frankel O. H. (1984). Genetic perspectives of germplasm conservation. Genet Manipul Impact Man Soc. 161, 170. [Google Scholar]
- Gill S. S., Tuteja N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant. Physiol. Biochem. 48, 909–930. 10.1016/j.plaphy.2010.08.016 [DOI] [PubMed] [Google Scholar]
- Gondalia N., Vashi R., Barot V., Sharma F., Anishkumar P., Chatterjee M., et al. (2022). “Genomic designing for abiotic stress tolerance in pea (Pisum Sativum L.),” in Genomic Designing for Abiotic Stress Resistant Pulse Crops, ed C. Kole (Springer), 45–113. [Google Scholar]
- Goufo P., Moutinho-Pereira J. M., Jorge T. F., Correia C. M., Oliveira M. R., Rosa E. A., et al. (2017). Cowpea (Vigna unguiculata L. Walp.) metabolomics: osmoprotection as a physiological strategy for drought stress resistance and improved yield. Front. Plant Sci. 8, 586. 10.3389/fpls.2017.00586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresta F., Mercati F., Santonoceto C., Abenavoli M. R., Ceravolo G., Araniti F., et al. (2016). Morpho-agronomic and AFLP characterization to explore guar (Cyamopsis tetragonoloba L.) genotypes for the Mediterranean environment. Ind. Crops Prod. 86, 23–30. 10.1016/j.indcrop.2016.03.038 [DOI] [Google Scholar]
- Gupta S., Mishra S. K., Misra S., Pandey V., Agrawal L., Nautiyal C. S., et al. (2020). Revealing the complexity of protein abundance in chickpea root under drought-stress using a comparative proteomics approach. Plant Physiol. Biochem. 151, 88–102. 10.1016/j.plaphy.2020.03.005 [DOI] [PubMed] [Google Scholar]
- Gupta S., Pandey S. (2021). “Exploiting the potential of plant growth-promoting rhizobacteria in legume production,” in Abiotic Stress and Legumes, eds V. P. Singh, S. Singh, and D. K. Chauhan (Elsevier), 1–32. [Google Scholar]
- Gupta S., Wardhan V., Kumar A., Rathi D., Pandey A., Chakraborty S., et al. (2015). Secretome analysis of chickpea reveals dynamic extracellular remodeling and identifies a Bet v1-like protein, CaRRP1 that participates in stress response. Sci. Rep. 5, 1–14. 10.1038/srep18427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacisalihoglu G., Settles A. M. (2017). Quantification of seed ionome variation in 90 diverse soybean (Glycine max) lines. J. Plant Nutr. 40, 2808–2817. 10.1080/01904167.2017.1382528 [DOI] [Google Scholar]
- Hakeem K. R., Chandna R., Ahmad P., Iqbal M., Ozturk M. (2012). Relevance of proteomic investigations in plant abiotic stress physiology. OMICS J. Integr. Biol. 16, 621–635. 10.1089/omi.2012.0041 [DOI] [PubMed] [Google Scholar]
- Heffner E. L., Sorrells M. E., Jannink J. L. (2009). Genomic selection for crop improvement. Crop Sci. 49, 1–12. 10.2135/cropsci2008.08.0512 [DOI] [Google Scholar]
- Hiz M. C., Canher B., Niron H., Turet M. (2014). Transcriptome analysis of salt tolerant common bean (Phaseolus vulgaris L.) under saline conditions. PLoS ONE 9, e92598. 10.1371/journal.pone.0092598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard R., Jarquin D. (2019). Genomic prediction using canopy coverage image and genotypic information in soybean via a hybrid model. Evol. Bioinform. 15, 1176934319840026. 10.1177/1176934319840026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyos-Villegas V., Song Q., Kelly J. D. (2017). Genome-wide association analysis for drought tolerance and associated traits in common bean. Plant Genome 10, 0122. 10.3835/plantgenome2015.12.0122 [DOI] [PubMed] [Google Scholar]
- Huang H., Ullah F., Zhou D.-X., Yi M., Zhao Y. (2019). Mechanisms of ROS regulation of plant development and stress responses. Front. Plant Sci. 10, 800. 10.3389/fpls.2019.00800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idrissi O., Udupa S. M., De Keyser E., McGee R. J., Coyne C. J., Saha G. C., et al. (2016). Identification of quantitative trait loci controlling root and shoot traits associated with drought tolerance in a lentil (Lens culinaris Medik.) recombinant inbred line population. Front. Plant Sci. 7, 1174. 10.3389/fpls.2016.01174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias-García R., Prats E., Fondevilla S., Satovic Z., Rubiales D. (2015). Quantitative trait loci associated to drought adaptation in Pea (Pisum Sativum L.). Plant Mol. Biol. Rep. 33, 1768–1778. 10.1007/s11105-015-0872-z [DOI] [Google Scholar]
- Jakubowski A. R., Casler M. D., Jackson R. D. (2017). Legume addition to perennial warm-season grass swards increases harvested biomass. Crop Sci. 57, 3343–3351. 10.2135/cropsci2017.04.0266 [DOI] [Google Scholar]
- Jannink J.-L., Lorenz A. J., Iwata H. (2010). Genomic selection in plant breeding: from theory to practice. Brief Funct. Geno. 9, 166–177. 10.1093/bfgp/elq001 [DOI] [PubMed] [Google Scholar]
- Jarquín D., Kocak K., Posadas L., Hyma K., Jedlicka J., Graef G., et al. (2014). Genotyping by sequencing for genomic prediction in a soybean breeding population. BMC Genom. 15, 1–10. 10.1186/1471-2164-15-740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha U. C., Bohra A., Jha R., Parida S. K. (2019). Salinity stress response and ‘omics' approaches for improving salinity stress tolerance in major grain legumes. Plant Cell Rep. 38, 255–277. 10.1007/s00299-019-02374-5 [DOI] [PubMed] [Google Scholar]
- Jha U. C., Bohra A., Nayyar H. (2020). Advances in “omics” approaches to tackle drought stress in grain legumes. Plant Breed. 139, 1–27. 10.1111/pbr.12761 [DOI] [Google Scholar]
- Ji J., Zhang C., Sun Z., Wang L., Duanmu D., Fan Q. (2019). Genome editing in cowpea vigna unguiculata using CRISPR-Cas9. Int. J. Mol. Sci. 20, 2471. 10.3390/ijms20102471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X., Yin X., Ndayambaza B., Zhang Z., Min X., Lin X., et al. (2019). Genome-wide identification and expression profiling of the ERF gene family in Medicago sativa (L.) under various abiotic stresses. DNA and Cell Biol. 38, 1056–1068. 10.1089/dna.2019.4881 [DOI] [PubMed] [Google Scholar]
- Joshi P., Rao P. P. (2017). Global pulses scenario: status and outlook. Ann. N. Y. Acad. Sci. 1392, 6–17. 10.1111/nyas.13298 [DOI] [PubMed] [Google Scholar]
- Juranić M., Nagahatenna D. S. K., Salinas-Gamboa R., Hand M. L., SánchezLeón N., Leong W. H., et al. (2020). A detached leaf assay for testing transient gene expression and gene editing in Cowpea (Vigna Unguiculata [L.] Walp.). Plant Methods 16, 88. 10.1186/s13007-020-00630-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahraman A., Kusmenoglu I., Aydin N., Aydogan A., Erskine W., Muehlbauer F. J. (2004). QTL mapping of winter hardiness genes in lentil. Crop Sci. 44, 13–22. 10.2135/cropsci2004.1300 [DOI] [Google Scholar]
- Kamau E. H., Nkhata S. G., Ayua E. O. (2020). Extrusion and nixtamalization conditions influence the magnitude of change in the nutrients and bioactive components of cereals and legumes. Food Sci. Nutr. 8, 1753–1765. 10.1002/fsn3.1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y. J., Kim S. K., Kim M. Y., Lestari P., Kim K. H., Ha B. K., et al. (2014). Genome sequence of mungbean and insights into evolution within Vigna species. Nat. Commun. 5, 1–9. 10.1038/ncomms6443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavi Kishor P., Venkatesh K., Amareshwari P., Hima Kumari P., Punita D., Anil Kumar S., et al. (2018). Genetic engineering for salt and drought stress tolerance in Peanut (Arachis hypogaea L.). Indian J. Plant Physiol. 23, 647–652. 10.1007/s40502-018-0421-5 [DOI] [Google Scholar]
- Keerthi C. M., Ramesh S., Byregowda M., Vaijayanthi P. V. (2018). Simple Sequence Repeat (SSR) Marker assay-based genetic diversity among dolichos bean (Lablab purpureus L. Sweet) advanced breeding lines differing for productivity per se traits. Int. J. Curr. Microbiol. Appl. Sci. 7, 3736–3744. Available online at: https://www.ijcmas.com/7-5-2018/C.M.%20Keerthi,%20et%20al.pdf [Google Scholar]
- Keller B., Ariza-Suarez D., de la Hoz J., Aparicio J. S., Portilla-Benavides A. E., Buendia H. F., et al. (2020). Genomic prediction of agronomic traits in common bean (phaseolus vulgaris l.) under environmental stress. Front. Plant Sci. 11, 1001. 10.3389/fpls.2020.01001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. A., Alghamdi S. S., Ammar M. H., Sun Q., Teng F., Migdadi H. M., et al. (2019a). Transcriptome profiling of faba bean (Vicia faba L.) drought-tolerant variety hassawi-2 under drought stress using RNA sequencing. Electron. J. Biotechnol. 39, 15–29. 10.1016/j.ejbt.2019.02.004 [DOI] [Google Scholar]
- Khan N., Bano A., Rahman M. A., Guo J., Kang Z., Babar M. (2019b). Comparative physiological and metabolic analysis reveals a complex mechanism involved in drought tolerance in chickpea (Cicer arietinum L.) induced by PGPR and PGRs. Sci. Rep. 9, 1–19. 10.1038/s41598-019-38702-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandal H., Parween S., Roy R., Meena M. K., Chattopadhyay D. (2017). MicroRNA profiling provides insights into post-transcriptional regulation of gene expression in Chickpea root apex under salinity and water deficiency. Sci. Rep. 7, 1–14. 10.1038/s41598-017-04906-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiranmai K., Lokanadha Rao G., Pandurangaiah M., Nareshkumar A., Amaranatha Reddy V., Lokesh U., et al. (2018). A novel WRKY transcription factor, MuWRKY3 (Macrotyloma uniflorum Lam. Verdc.) enhances drought stress tolerance in transgenic Groundnut (Arachis hypogaea L.) plants. Front. Plant Sci. 9, 346. 10.3389/fpls.2018.00346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korir P. C., Zhang J., Wu K., Zhao T., Gai J. (2013). Association mapping combined with linkage analysis for aluminum tolerance among soybean cultivars released in Yellow and Changjiang River Valleys in China. Theor. Appl. Genet. 126, 1659–1675. 10.1007/s00122-013-2082-0 [DOI] [PubMed] [Google Scholar]
- Kumar J., Sen Gupta D., Djalovic I., Kumar S., Siddique K. H. (2021). Root-omics for drought tolerance in cool-season grain legumes. Physiol. Planta. 172, 629–644. 10.1111/ppl.13313 [DOI] [PubMed] [Google Scholar]
- Kumar P., Choudhary M., Halder T., Prakash N. R., Singh V., Sheoran S., et al. (2022). Salinity stress tolerance and omics approaches: revisiting the progress and achievements in major cereal crops. Heredity 1–22. 10.1038/s41437-022-00516-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Fladung M. (2002). “Gene targeting in plants,” in Molecular Techniques in Crop Improvement, eds S. Mohan Jain, D. S. Brar, and B. S. Ahloowalia (Springer), 481–499. [Google Scholar]
- Kumar S., Palve A. S., Patel S. K., Selvanayagam S., Sharma R., Rathore A. (2020). Development of genomic microsatellite markers in cluster bean using next-generation DNA sequencing and their utility in diversity analysis. Curr. Plant Biol. 21, 100134. 10.1016/j.cpb.2019.100134 [DOI] [Google Scholar]
- Kushwah A., Bhatia D., Rani U., Yadav I. S., Singh I., Bharadwaj C., et al. (2021). Molecular mapping of quantitative trait loci for ascochyta blight and botrytis grey mould resistance in an inter-specific cross in chickpea (Cicer arietinum L.) using genotyping by sequencing. Breed. Sci. 71, 229–239. 10.1270/jsbbs.20085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahner B., Gong J., Mahmoudian M., Smith E. L., Abid K. B., Rogers E. E., et al. (2003). Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nat. Biotechnol. 21, 1215–1221. 10.1038/nbt865 [DOI] [PubMed] [Google Scholar]
- Langridge P., Fleury D. (2011). Making the most of ‘omics' for crop breeding. Trends Biotechnol. 29, 33–40. 10.1016/j.tibtech.2010.09.006 [DOI] [PubMed] [Google Scholar]
- Lee G., Boerma H., Villagarcia M., Zhou X., Carter T., Li Z., et al. (2004). A major QTL conditioning salt tolerance in S-100 soybean and descendent cultivars. Theor. Appl. Genet. 109, 1610–1619. 10.1007/s00122-004-1783-9 [DOI] [PubMed] [Google Scholar]
- Leonforte A., Sudheesh S., Cogan N. O., Salisbury P. A., Nicolas M. E., Materne M., et al. (2013). SNP marker discovery, linkage map construction and identification of QTLs for enhanced salinity tolerance in field pea (Pisum Sativum L.). BMC Plant Biol. 13, 1–14. 10.1186/1471-2229-13-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Guo R., Jiao Y., Jin X., Zhang H., Shi L. (2017). Comparison of salt tolerance in Soja based on metabolomics of seedling roots. Front. Plant Sci. 8, 1101. 10.3389/fpls.2017.01101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Ruperao P., Batley J., Edwards D., Khan T., Colmer T. D., et al. (2018). Investigating drought tolerance in chickpea using genome-wide association mapping and genomic selection based on whole-genome resequencing data. Front. Plant Sci. 9, 190. 10.3389/fpls.2018.00190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M., Farmer A., Joshi T., Takahashi K., Langley R. J., Franklin L. D., et al. (2010). An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 63, 86–99. 10.1111/j.1365-313X.2010.04222.x [DOI] [PubMed] [Google Scholar]
- Liu C., Wu J., Wang L., Fan B., Cao Z., Su Q., et al. (2017). Quantitative trait locus mapping under irrigated and drought treatments based on a novel genetic linkage map in mungbean (Vigna radiata L.). Theor. Appl. Genet. 130, 2375–2393. 10.1007/s00122-017-2965-6 [DOI] [PubMed] [Google Scholar]
- Liu W., Yuan J. S., Stewart C. N., Jr. (2013). Advanced genetic tools for plant biotechnology. Nat. Rev. Genet. 14, 781–793. 10.1038/nrg3583 [DOI] [PubMed] [Google Scholar]
- Liu Y., Wang D., He F., Wang J., Joshi T., Xu D. (2019). Phenotype prediction and genome-wide association study using deep convolutional neural network of soybean. Front. Genet. 10, 1091 10.3389/fgene.2019.01091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe R., Shirley N., Bleackley M., Dolan S., Shafee T. (2017). Transcriptomics technologies. PLoS Comput. Biol. 13, e1005457. 10.1371/journal.pcbi.1005457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M. R., Ehlers J. D., Huynh B.-L., Diop N.-N., Roberts P. A., Close T. J. (2013). Markers for breeding heat-tolerant cowpea. Mol. Breed. 31, 529–536. 10.1007/s11032-012-9810-z [DOI] [Google Scholar]
- Luo H., Guo J., Yu B., Chen W., Zhang H., Zhou X., et al. (2021). Construction of ddRADseq-based high-density genetic map and identification of quantitative trait loci for trans-resveratrol content in Peanut seeds. Front. Plant Sci. 12, 438. 10.3389/fpls.2021.644402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z., Szczepanek A., Abdel-Haleem H. (2020). Genome-wide association study (GWAS) analysis of Camelina seedling germination under salt stress condition. Agronomy. 10, 1444. 10.3390/agronomy10091444 [DOI] [Google Scholar]
- Maalouf F., Abou-Khater L., Babiker Z., Jighly A., Alsamman A. M., Hu J., et al. (2022). Genetic dissection of heat stress tolerance in faba bean (Vicia faba L.) using GWAS. Plants 11, 1108. 10.3390/plants11091108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay M. C., Street K., Mitrofanova O., Zuev E., Strelchenko P., Pertziger F. (2004). “Focused identification of germplasm strategy–FIGS,” in Proceedings 11th Wheat Breeding Assembly, 20–24. [Google Scholar]
- Mahdavi Mashaki K., Garg V., Nasrollahnezhad Ghomi A. A., Kudapa H., Chitikineni A., Zaynali Nezhad K., et al. (2018). RNA-Seq analysis revealed genes associated with drought stress response in kabuli chickpea (Cicer arietinum L.). PLoS ONE 13, e0199774. 10.1371/journal.pone.0199774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud G. A.-E. (2021). “Biotic stress to legumes: fungal diseases as major biotic stress factor,” in Sustainable Agriculture Reviews 51, eds P. Guleria, V. Kumar, and E. Lichtfouse (Springer), 181–212. [Google Scholar]
- Mahmoudian M., Yücel M., Öktem H. A. (2002). Transformation of lentil (Lens culinaris M.) cotyledonary nodes by vacuum infiltration of Agrobacterium tumefaciens. Plant Mol. Biol. Repor. 20, 251–257. 10.1007/BF02782460 [DOI] [Google Scholar]
- Manjulatha M., Sreevathsa R., Kumar A. M., Sudhakar C., Prasad T., Tuteja N., et al. (2014). Overexpression of a pea DNA helicase (PDH45) in peanut (Arachis hypogaea L.) confers improvement of cellular level tolerance and productivity under drought stress. Mol. Biotechnol. 56, 111–125. 10.1007/s12033-013-9687-z [DOI] [PubMed] [Google Scholar]
- Mathur S., Agrawal D., Jajoo A. (2014). Photosynthesis: response to high temperature stress. J. Photochem. Photobiol. B Biol. 137, 116–126. 10.1016/j.jphotobiol.2014.01.010 [DOI] [PubMed] [Google Scholar]
- McKersie B. D., Chen Y., de Beus M., Bowley S. R., Bowler C., Inzé D., et al. (1993). Superoxide dismutase enhances tolerance of freezing stress in transgenic alfalfa (Medicago sativa L.). Plant Physiol. 103, 1155–1163. 10.1104/pp.103.4.1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKersie B. D., Murnaghan J., Jones K. S., Bowley S. R. (2000). Iron-superoxide dismutase expression in transgenic alfalfa increases winter survival without a detectable increase in photosynthetic oxidative stress tolerance. Plant Physiol. 122, 1427–1438. 10.1104/pp.122.4.1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefleh M., Pasqualone A., Caponio F., Faccia M. (2022). Legumes as basic ingredients in the production of dairy-free cheese alternatives: a review. J. Sci. Food Agric. 102, 8–18. 10.1002/jsfa.11502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min C. W., Gupta R., Agrawal G. K., Rakwal R., Kim S. T. (2019). Concepts and strategies of soybean seed proteomics using the shotgun proteomics approach. Expert Rev. Proteom. 16, 795–804. 10.1080/14789450.2019.1654860 [DOI] [PubMed] [Google Scholar]
- Missanga J. S., Venkataramana P. B., Ndakidemi P. A. (2021). Recent developments in Lablab purpureus genomics: a focus on drought stress tolerance and use of genomic resources to develop stress-resilient varieties. Legume Sci. 3, e99. 10.1002/leg3.99 [DOI] [Google Scholar]
- Molina C., Rotter B., Horres R., Udupa S. M., Besser B., Bellarmino L., et al. (2008). SuperSAGE: the drought stress-responsive transcriptome of chickpea roots. BMC Genom. 9, 1–28. 10.1186/1471-2164-9-553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchero W., Ehlers J. D., Close T. J., Roberts P. A. (2009). Mapping QTL for drought stress-induced premature senescence and maturity in Cowpea [Vigna unguiculata (L.) Walp.]. Theor. Appl. Genet. 118, 849–863. 10.1007/s00122-008-0944-7 [DOI] [PubMed] [Google Scholar]
- Mugabe D., Coyne C. J., Piaskowski J., Zheng P., Ma Y., Landry E., et al. (2019). Quantitative trait loci for cold tolerance in chickpea. Crop Sci. 59, 573–582. 10.2135/cropsci2018.08.050432161601 [DOI] [Google Scholar]
- Munns R. (2010). “Approaches to identifying genes for salinity tolerance and the importance of timescale,” in Plant Stress Tolerance, ed R. Sunkar (Springer), 25–38. [DOI] [PubMed] [Google Scholar]
- Nadaf J., Riggio V., Yu T.-P., Pong-Wong R. (2012). “Effect of the prior distribution of SNP effects on the estimation of total breeding value,” in BMC Proceedings (BioMed Central: ), 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair R. M., Yang R. Y., Easdown W. J., Thavarajah D., Thavarajah P., Hughes J., et al. (2013). Biofortification of mungbean (Vigna radiata) as a whole food to enhance human health. J. Sci. Food Agric. 93, 1805–1813. 10.1002/jsfa.6110 [DOI] [PubMed] [Google Scholar]
- Narayana P. K., von Wettberg E. J. (2020). “GWAS and genomic approaches in legumes, an expanding toolkit for examining responses to abiotic stresses,” in The Plant Family Fabaceae, eds M. Hasanuzzaman, S. Araújo, S. S. Gill (Springer), 161–180. [Google Scholar]
- Narayanan S., Zoong-Lwe Z. S., Gandhi N., Welti R., Fallen B., Smith J. R., et al. (2020). Comparative lipidomic analysis reveals heat stress responses of two soybean genotypes differing in temperature sensitivity. Plants 9, 457. 10.3390/plants9040457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz M. A., Chung G. (2020). Genetic improvement of cereals and grain legumes. Genes. 11, 1255. 10.3390/genes11111255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T., Sticklen M. (2012). Genetic transformation of common bean (Phaseolus vulgaris L.) with the gus color marker, the bar herbicide resistance, and the barley (Hordeum vulgare) HVA1 drought tolerance genes. Int. J. Agron. 2012, 198960. 10.1155/2012/198960 [DOI] [Google Scholar]
- Njobvu J., Hamabwe S. M., Munyinda K., Kelly J. D., Kamfwa K. (2020). Quantitative trait loci mapping of resistance to aluminum toxicity in common bean. Crop Sci. 60, 1294–1302. 10.1002/csc2.2004319436988 [DOI] [Google Scholar]
- Odong T. L., Van Heerwaarden J., Jansen J., van Hintum T. J., Van Eeuwijk F. A. (2011). Determination of genetic structure of germplasm collections: are traditional hierarchical clustering methods appropriate for molecular marker data?. Theoret. Appl. Genet. 123, 195–205. 10.1007/s00122-011-1576-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y., Takano K., Saito K. (2017). Lipidomic analysis of soybean leaves revealed tissue-dependent diference in lipid remodeling under phosphorus-limited growth conditions. Plant Biotechnol (Tokyo). 34, 57–63. 10.5511/plantbiotechnology.17.0113a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oladzad A., Porch T., Rosas J. C., Moghaddam S. M., Beaver J., Beebe S. E., et al. (2019). Single and multi-trait GWAS identify genetic factors associated with production traits in common bean under abiotic stress environments. G3 Genes Genomes Genet. 9, 1881–1892. 10.1534/g3.119.400072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk Gokce Z. N., Akbas S., Ayten S., Azimi M. H., Das R., Guven S. B., et al. (2020). “Abiotic stress tolerance in field crops: integration of Omics approaches,” in Agronomic Crops, ed M. Hasanuzzaman (Springer), 503–526. [Google Scholar]
- Pandey A., Chakraborty S., Datta A., Chakraborty N. (2008). Proteomics approach to identify dehydration responsive nuclear proteins from Chickpea (Cicer arietinum L.). Mol. Cell. Proteom. 7, 88–107. 10.1074/mcp.M700314-MCP200 [DOI] [PubMed] [Google Scholar]
- Pandey A. K., Basandrai A. K. (2021). Will Macrophomina phaseolina spread in legumes due to climate change? A critical review of current knowledge. J. Plant Dis. Prot. 128, 9–18. 10.1007/s41348-020-00374-2 [DOI] [Google Scholar]
- Pandey A. K., Rubiales D., Wang Y., Fang P., Sun T., Liu N., et al. (2021). Omics resources and omics-enabled approaches for achieving high productivity and improved quality in Pea (Pisum Sativum L.). Theor. Appl. Genet. 134, 755–776. 10.1007/s00122-020-03751-5 [DOI] [PubMed] [Google Scholar]
- Pandey M. K., Chaudhari S., Jarquin D., Janila P., Crossa J., Patil S. C., et al. (2020). Genome-based trait prediction in multi-environment breeding trials in groundnut. Theor. Appl. Genet. 133, 3101–3117. 10.1007/s00122-020-03658-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey M. K., Upadhyaya H. D., Rathore A., Vadez V., Sheshshayee M., Sriswathi M., et al. (2014). Genomewide association studies for 50 agronomic traits in peanut using the ‘reference set'comprising 300 genotypes from 48 countries of the semi-arid tropics of the world. PLoS ONE 9, e105228. 10.1371/journal.pone.0105228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R., Aretano R., Gupta A. K., Meena D., Kumar B., Alatalo J. M. (2017). Agroecology as a climate change adaptation strategy for smallholders of Tehri-Garhwal in the Indian Himalayan region. Small Scale Forest. 16, 53–63. 10.1007/s11842-016-9342-1 [DOI] [Google Scholar]
- Pandurangaiah M., Lokanadha Rao G., Sudhakarbabu O., Nareshkumar A., Kiranmai K., Lokesh U., et al. (2014). Overexpression of horsegram (Macrotyloma uniflorum Lam. Verdc.) NAC transcriptional factor (MuNAC4) in groundnut confers enhanced drought tolerance. Mol. Biotechnol. 56, 758–769. 10.1007/s12033-014-9754-0 [DOI] [PubMed] [Google Scholar]
- Parankusam S., Bhatnagar-Mathur P., Sharma K. K. (2017). Heat responsive proteome changes reveal molecular mechanisms underlying heat tolerance in Chickpea. Environ. Exp. Bot. 141, 132–144. 10.1016/j.envexpbot.2017.07.007 [DOI] [Google Scholar]
- Parida A., Raina S. N., Narayan R. K. J. (1990). Quantitative DNA variation between and within chromosome complements of Vigna species (Fabaceae). Genetica 82, 125–133. 10.1007/BF00124642 [DOI] [Google Scholar]
- Parveen S., Gupta D. B., Dass S., Kumar A., Pandey A., Chakraborty S., et al. (2016). Chickpea ferritin CaFer1 participates in oxidative stress response and promotes growth and development. Sci. Rep. 6, 1–14. 10.1038/srep31218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil G., Do T., Vuong T. D., Valliyodan B., Lee J.-D., Chaudhary J., et al. (2016). Genomic-assisted haplotype analysis and the development of high-throughput SNP markers for salinity tolerance in soybean. Sci. Rep. 6, 1–13. 10.1038/srep19199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil M., Ramu S., Jathish P., Sreevathsa R., Reddy P. C., Prasad T., et al. (2014). Overexpression of AtNAC2 (ANAC092) in groundnut (Arachis hypogaea L.) improves abiotic stress tolerance. Plant Biotechnol. Rep. 8, 161–169. 10.1007/s11816-013-0305-0 [DOI] [Google Scholar]
- Paul P. J., Samineni S., Thudi M., Sajja S. B., Rathore A., Das R. R., et al. (2018). Molecular mapping of QTLs for heat tolerance in chickpea. Int. J. Mol. Sci. 19, 2166. 10.3390/ijms19082166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plewiński P., Cwiek-Kupczyńska H., Rudy E., Bielski W., Rychel-Bielska S., Stawiński S., et al. (2020). Innovative transcriptome-based genotyping highlights environmentally responsive genes for phenology, growth and yield in a non-model grain legume. Plant Cell Environ. 43, 2680–2698. 10.1111/pce.13880 [DOI] [PubMed] [Google Scholar]
- Pratap A., Gupta S., Nair R. M., Gupta S., Schafleitner R., Basu P., et al. (2019). Using plant phenomics to exploit the gains of genomics. Agronomy 9, 126. 10.3390/agronomy9030126 [DOI] [Google Scholar]
- Pruthvi V., Narasimhan R., Nataraja K. N. (2014). Simultaneous expression of abiotic stress responsive transcription factors, AtDREB2A, AtHB7 and AtABF3 improves salinity and drought tolerance in peanut (Arachis hypogaea L.). PLoS ONE 9, e111152. 10.1371/journal.pone.0111152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puli C. O. R., Akila C. S., Pandit V., Konduru S., Kandi S. R., Chinta S. (2021). “Peanut (Arachis hypogaea L.) transgenic plants for abiotic stress tolerance,” in Genetically Modified Crops, eds P. B. K. Kishor, M. V. Rajam, and T. Pullaiah (Springer), 139–173. [Google Scholar]
- Pushpavalli R., Krishnamurthy L., Thudi M., Gaur P. M., Rao M. V., Siddique K. H., et al. (2015). Two key genomic regions harbour QTLs for salinity tolerance in ICCV 2 × JG 11 derived chickpea (Cicer arietinum L.) recombinant inbred lines. BMC Plant Biol. 15, 1–15. 10.1186/s12870-015-0491-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Shi A., Song Q., Li S., Wang F., Cao Y., et al. (2019). Genome wide association study and genomic selection of amino acid concentrations in soybean seeds. Front. Plant Sci. 10, 1445. 10.3389/fpls.2019.01445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramu V. S., Swetha T. N., Sheela S. H., Babitha C. K., Rohini S., Reddy M. K., et al. (2016). Simultaneous expression of regulatory genes associated with specific drought-adaptive traits improves drought adaptation in peanut. Plant Biotechnol. J. 14, 1008–1020. 10.1111/pbi.12461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray H., Georges F. (2010). A genomic approach to nutritional, pharmacological and genetic issues of faba bean (Vicia faba L.): Prospects for genetic modifications. GM Crops 1, 99–106. 10.4161/gmcr.1.2.11891 [DOI] [PubMed] [Google Scholar]
- Raza A., Razzaq A., Mehmood S. S., Zou X., Zhang X., Lv Y., et al. (2019). Impact of climate change on crops adaptation and strategies to tackle its outcome: a review. Plants 8, 34. 10.3390/plants8020034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzaq M. K., Aleem M., Mansoor S., Khan M. A., Rauf S., Iqbal S., et al. (2021). Omics and CRISPR-Cas9 approaches for molecular insight, functional gene analysis, and stress tolerance development in crops. Int. J. Mol. Sci. 22, 1292. 10.3390/ijms22031292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A. R., Chaitanya K. V., Vivekanandan M. (2004). Drought-induced responses of photosynthesis and antioxidant metabolism in higher plants. J. Plant Physiol. 161, 1189–1202. 10.1016/j.jplph.2004.01.013 [DOI] [PubMed] [Google Scholar]
- Rehman A., Malhotra R., Bett K., Tar'An B., Bueckert R., Warkentin T. (2011). Mapping QTL associated with traits affecting grain yield in chickpea (Cicer arietinum L.) under terminal drought stress. Crop Sci. 51, 450–463. 10.2135/cropsci2010.03.0129 [DOI] [Google Scholar]
- Richter J. A., Behr J. H., Erban A., Kopka J., Zörb C. (2019). Ion-dependent metabolic responses of Vicia faba L. to salt stress. Plant Cell Environ. 42, 295–309. 10.1111/pce.13386 [DOI] [PubMed] [Google Scholar]
- Roorkiwal M., Jain A., Kale S. M., Doddamani D., Chitikineni A., Thudi M., et al. (2018). Development and evaluation of high-density Axiom® Cicer SNP Array for high-resolution genetic mapping and breeding applications in chickpea. Plant Biotechnol. 16, 890–901. 10.1111/pbi.12836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roorkiwal M., Pandey S., Thavarajah D., Hemalatha R., Varshney R. K. (2021). Molecular mechanisms and biochemical pathways for micronutrient acquisition and storage in legumes to support biofortification for nutritional security. Front. Plant Sci. 12, 682842. 10.3389/fpls.2021.682842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roorkiwal M., Rathore A., Das R. R., Singh M. K., Jain A., Srinivasan S., et al. (2016). Genome-enabled prediction models for yield related traits in chickpea. Front. Plant Sci. 7, 1666. 10.3389/fpls.2016.01666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saade S., Maurer A., Shahid M., Oakey H., Schmöckel S. M., Negrão S., et al. (2016). Yield-related salinity tolerance traits identified in a nested association mapping (NAM) population of wild barley. Sci. Rep. 6, 1–9. 10.1038/srep32586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallam A., Arbaoui M., El-Esawi M., Abshire N., Martsch R. (2016). Identification and verification of QTL associated with frost tolerance using linkage mapping and GWAS in winter faba bean. Front. Plant Sci. 7, 1098. 10.3389/fpls.2016.01098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salt D. E., Baxter I., Lahner B. (2008). Ionomics and the study of the plant ionome. Annu. Rev. Plant Biol. 59, 709–733. 10.1146/annurev.arplant.59.032607.092942 [DOI] [PubMed] [Google Scholar]
- Salvi S., Tuberosa R. (2015). The crop QTLome comes of age. Curr. Opin. Biotechnol. 32, 179–185. 10.1016/j.copbio.2015.01.001 [DOI] [PubMed] [Google Scholar]
- Sampaio A. M., Araújo S. d.S, Rubiales D., Vaz Patto M.C. (2020). Fusarium wilt management in legume crops. Agronomy 10, 1073. 10.3390/agronomy10081073 [DOI] [Google Scholar]
- Sandhu K. S., Merrick L. F., Sankaran S., Zhang Z., Carter A. H. (2021). Prospectus of genomic selection and phenomics in cereal, legume and oilseed breeding programs. Front. Genet. 12, 829131. 10.3389/fgene.2021.829131 [DOI] [Google Scholar]
- Sarker R. H., Biswas A., Mustafa B. M., Mahbub S., Hoque M. I. (2003). Agrobacterium-mediated transformation of lentil (Lens culinaris Medik.). Plant Tissue Cult. 13, 1–12. [Google Scholar]
- Sertse D., You F. M., Ravichandran S., Soto-Cerda B. J., Duguid S., Cloutier S. (2021). Loci harboring genes with important role in drought and related abiotic stress responses in flax revealed by multiple GWAS models. Theor. Appl. Genet. 134, 191–212. 10.1007/s00122-020-03691-0 [DOI] [PubMed] [Google Scholar]
- Settele J., Bishop J., Potts S. (2016). Climate change impacts on pollination. Nat. Plants 2, 16092. 10.1038/nplants.2016.92 [DOI] [PubMed] [Google Scholar]
- Shabala S., Munns R. (2012). Salinity stress: physiological constraints and adaptive mechanisms. Plant Stress Physiol. 1, 59–93. 10.1079/9781845939953.0059 [DOI] [Google Scholar]
- Sharma A. D., Sharma H., Lightfoot D. A. (2011). The genetic control of tolerance to aluminum toxicity in the ‘Essex'by ‘Forrest'recombinant inbred line population. Theor. Appl. Genet. 122, 687–694. 10.1007/s00122-010-1478-3 [DOI] [PubMed] [Google Scholar]
- Sharmila P., Phanindra M., Anwar F., Singh K., Gupta S., Saradhi P. P. (2009). Targeting prokaryotic choline oxidase into chloroplasts enhance the potential of photosynthetic machinery of plants to withstand oxidative damage. Plant Physiol. Biochem. 47, 391–396. 10.1016/j.plaphy.2009.01.001 [DOI] [PubMed] [Google Scholar]
- Sharmin R. A., Karikari B., Chang F., Al Amin G., Bhuiyan M. R., Hina A., et al. (2021). Genome-wide association study uncovers major genetic loci associated with seed flooding tolerance in soybean. BMC Plant Biol. 21, 1–17. 10.1186/s12870-021-03268-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter A. M., Keppler B. D., Liu X., Lichtenberg J., Welch L. R. (2016). Bioinformatic identification and analysis of hydroxyproline-rich glycoproteins in Populus trichocarpa. BMC Plant Biol. 16, 1–34. 10.1186/s12870-016-0912-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., Yu D., Wang D., Bai X., Zhu Y., Guo C. (2013). Genomic selection of seed weight based on low-density SCAR markers in soybean. Genet. Mol. Res. 12, 2178–2188. 10.4238/2013.July.3.2 [DOI] [PubMed] [Google Scholar]
- Shukla R. K., Raha S., Tripathi V., Chattopadhyay D. (2006). Expression of CAP2, an APETALA2-family transcription factor from chickpea, enhances growth and tolerance to dehydration and salt stress in transgenic tobacco. Plant Physiol. 142, 113–123. 10.1104/pp.106.081752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Pratap Singh A., Ramaswamy H. S. (2016). A controlled agitation process for improving quality of canned green beans during agitation thermal processing. J. Food Sci. 81, E1399–E1411. 10.1111/1750-3841.13308 [DOI] [PubMed] [Google Scholar]
- Singh D., Chaudhary P., Taunk J., Singh C. K., Singh D., Tomar R. S. S., et al. (2021). Fab advances in fabaceae for abiotic stress resilience: from ‘omics' to artificial intelligence. Int. J. Mol. Sci.22, 10535. 10.3390/ijms221910535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D., Singh C. K., Singh Tomar R. S., Pal M. (2017a). Genetics and molecular mapping of heat tolerance for seedling survival and pod set in lentil. Crop Sci. 57, 3059–3067. 10.2135/cropsci2017.05.0284 [DOI] [Google Scholar]
- Singh D., Singh C. K., Taunk J., Jadon V., Pal M., Gaikwad K. (2019). Genome wide transcriptome analysis reveals vital role of heat responsive genes in regulatory mechanisms of lentil (Lens culinaris Medikus). Sci. Rep. 9, 1–19. 10.1038/s41598-019-49496-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D., Singh C. K., Taunk J., Tomar R. S. S., Chaturvedi A. K., Gaikwad K., et al. (2017b). Transcriptome analysis of lentil (Lens culinaris Medikus) in response to seedling drought stress. BMC Genom. 18, 1–20. 10.1186/s12864-017-3596-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N., Mishra A., Jha B. (2014). Ectopic over-expression of peroxisomal ascorbate peroxidase (SbpAPX) gene confers salt stress tolerance in transgenic peanut (Arachis hypogaea). Gene 547, 119–125. 10.1016/j.gene.2014.06.037 [DOI] [PubMed] [Google Scholar]
- Singh N., Rai V., Singh N. K. (2020). Multi-omics strategies and prospects to enhance seed quality and nutritional traits in pigeonpea. Nucleus 63, 249–256. 10.1007/s13237-020-00341-0 [DOI] [Google Scholar]
- Sita K., Kumar V. (2020). Role of Gamma Amino Butyric Acid (GABA) against abiotic stress tolerance in legumes: a review. Plant Physiol. Rep. 25, 654–663. 10.1007/s40502-020-00553-1 [DOI] [Google Scholar]
- Soltani A., MafiMoghaddam S., Oladzad-Abbasabadi A., Walter K., Kearns P. J., Vasquez-Guzman J., et al. (2018). Genetic analysis of flooding tolerance in an Andean diversity panel of Dry bean (Phaseolus vulgaris L.). Front. Plant Sci. 9, 767. 10.3389/fpls.2018.00767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers D. A., Samac D. A., Olhoft P. M. (2003). Recent advances in legume transformation. Plant Physiol. 131, 892–899. 10.1104/pp.102.017681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Lv J., Ma Z., Dong W. (2019). The mechanism of alfalfa (Medicago sativa L.) response to abiotic stress. Plant Growth Regul. 89, 239–249. 10.1007/s10725-019-00530-1 [DOI] [Google Scholar]
- Stai J. S., Yadav A., Sinou C., Bruneau A., Doyle J. J., Fernández-Baca D., et al. (2019). Cercis: a non-polyploid genomic relic within the generally polyploid legume family. Front. Plant Sci. 10, 345. 10.3389/fpls.2019.00345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke S., Sato S., Sandal N., Koyama M., Kaneko T., Tabata S., et al. (2004). Exploitation of colinear relationships between the genomes of Lotus japonicus, Pisum Sativum and Arabidopsis thaliana, for positional cloning of a legume symbiosis gene. Theor. Appl. Genet. 108, 442–449. 10.1007/s00122-003-1438-2 [DOI] [PubMed] [Google Scholar]
- Subba P., Barua P., Kumar R., Datta A., Soni K. K., Chakraborty S., et al. (2013). Phosphoproteomic dynamics of chickpea (Cicer arietinum L.) reveals shared and distinct components of dehydration response. J. Proteome Res. 12, 5025–5047. 10.1021/pr400628j [DOI] [PubMed] [Google Scholar]
- Sudheesh S., Kimber R., Braich S., Forster J., Paull J., Kaur S. (2019). Construction of an integrated genetic linkage map and detection of quantitative trait loci for Ascochyta blight resistance in faba bean (Vicia faba L.). Euphytica 215, 42. 10.1007/s10681-019-2365-x [DOI] [Google Scholar]
- Taranto F., Nicolia A., Pavan S., De Vita P., D'Agostino N. (2018). Biotechnological and digital revolution for climate-smart plant breeding. Agronomy 8, 277. 10.3390/agronomy8120277 [DOI] [Google Scholar]
- Tayeh N., Aluome C., Falque M., Jacquin F., Klein A., Chauveau A., et al. (2015). Development of two major resources for pea genomics: the GenoPea 13.2 K SNP Array and a high-density, high-resolution consensus genetic map. Plant J. 84, 1257–1273. 10.1111/tpj.13070 [DOI] [PubMed] [Google Scholar]
- Tesfaye M., Temple S. J., Allan D. L., Vance C. P., Samac D. A. (2001). Overexpression of malate dehydrogenase in transgenic alfalfa enhances organic acid synthesis and confers tolerance to aluminum. Plant Physiol. 127, 1836–1844. 10.1104/pp.010376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thudi M., Palakurthi R., Schnable J. C., Chitikineni A., Dreisigacker S., Mace E., et al. (2021). Genomic resources in plant breeding for sustainable agriculture. J. Plant Physiol. 257, 153351. 10.1016/j.jplph.2020.153351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Li S., Liu Y., Liu X. (2016). Transcriptomic profiling reveals metabolic and regulatory pathways in the desiccation tolerance of Mungbean (Vigna radiata [L.] R. Wilczek). Front. Plant Sci. 7, 1921. 10.3389/fpls.2016.01921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari V., Chaturvedi A. K., Mishra A., Jha B. (2015). Introgression of the SbASR-1 gene cloned from a halophyte Salicornia brachiata enhances salinity and drought endurance in transgenic groundnut (Arachis hypogaea) and acts as a transcription factor. PLoS ONE 10, e0131567. 10.1371/journal.pone.0131567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapp J. J., Urrea C. A., Cregan P. B., Miklas P. N. (2015). Quantitative trait loci for yield under multiple stress and drought conditions in a dry bean population. Crop Sci. 55, 1596–1607. 10.2135/cropsci2014.11.0792 [DOI] [Google Scholar]
- Upadhyaya H. D., Ortiz R. (2001). A mini core subset for capturing diversity and promoting utilization of chickpea genetic resources in crop improvement. Theoret. Appl. Genet. 102, 1292–1298. 10.1007/s00122-001-0556-y [DOI] [Google Scholar]
- Upadhyaya H. D., Wang Y. H., Gowda C. L. L., Sharma S. (2013). Association mapping of maturity and plant height using SNP markers with the sorghum mini core collection. Theoret. Appl. Genet. 126, 2003–2015. 10.1007/s00122-013-2113-x [DOI] [PubMed] [Google Scholar]
- Urva P. (2021). Detection and classification of grain crops and legumes disease: a survey. Sparklinglight Transact Artif Intell Quantum Comp. 1, 41–55. 10.55011/STAIQC.2021.1105 [DOI] [Google Scholar]
- Vadez V., Krishnamurthy L., Thudi M., Anuradha C., Colmer T. D., Turner N. C., et al. (2012). Assessment of ICCV 2 × JG 62 chickpea progenies shows sensitivity of reproduction to salt stress and reveals QTL for seed yield and yield components. Mol. Breed. 30, 9–21. 10.1007/s11032-011-9594-6 [DOI] [Google Scholar]
- Vadez V., Rao J. S., Bhatnagar-Mathur P., Sharma K. (2013). DREB1A promotes root development in deep soil layers and increases water extraction under water stress in groundnut. Plant Bio. 15, 45–52. 10.1111/j.1438-8677.2012.00588.x [DOI] [PubMed] [Google Scholar]
- Vadez V., Rao S., Kholova J., Krishnamurthy L., Kashiwagi J., Ratnakumar P., et al. (2008). Root research for drought tolerance in legumes: Quo vadis? J. Food Legumes 21, 77–85. [Google Scholar]
- Valdisser P. A. M. R., Müller B. S., de Almeida Filho J. E., Morais Junior O. P., Guimarães C. M., Borba T. C., et al. (2020). Genome-wide association studies detect multiple QTLs for productivity in Mesoamerican diversity panel of common bean under drought stress. Front. Plant Sci. 1563, 574674. 10.3389/fpls.2020.574674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney R. K., Thudi M., Pandey M. K., Tardieu F., Ojiewo C., Vadez V., et al. (2018). Accelerating genetic gains in legumes for the development of prosperous smallholder agriculture: integrating genomics, phenotyping, systems modelling and agronomy. J. Exp. Bot. 69, 3293–3312. 10.1093/jxb/ery088 [DOI] [PubMed] [Google Scholar]
- Vessal S., Arefian M., Siddique K. H. (2020). Proteomic responses to progressive dehydration stress in leaves of chickpea seedlings. BMC Genom. 21, 1–15. 10.1186/s12864-020-06930-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Q., Chen S., Shan Z., Yang Z., Chen L., Zhang C., et al. (2017). Stability evaluation of reference genes for gene expression analysis by RT-qPCR in soybean under different conditions. PLoS ONE 12, e0189405. 10.1371/journal.pone.0189405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Dong S., Liu L., Ma Y., Li S., Zu W. (2018). Transcriptome profiling reveals PEG-simulated drought, heat and combined stress response mechanisms in soybean. Comput. Biol. Chem. 77, 413–419. 10.1016/j.compbiolchem.2018.09.012 [DOI] [PubMed] [Google Scholar]
- Wang L., Rubio M. C., Xin X., Zhang B., Fan Q., Wang Q., et al. (2019b). CRISPR/Cas9 knockout of leghemoglobin genes in lotus japonicusuncovers their synergistic roles in symbiotic nitrogen fixation. New Phytol. 224, 818–832. 10.1111/nph.16077 [DOI] [PubMed] [Google Scholar]
- Wang L., Wang L., Tan Q., Fan Q., Zhu H., Hong Z., et al. (2016). Efficient inactivation of symbiotic nitrogen fixation related genes in lotus japonicus using CRISPR-Cas9. Front. Plant Sci. 7, 1333. 10.3389/fpls.2016.01333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Cheng Y., Yang C., Yang C., Mu Y., Xia Q., et al. (2019a). QTL mapping for aluminum tolerance in RIL population of soybean (Glycine max L.) by RAD sequencing. PLoS ONE 14, e0223674. 10.1371/journal.pone.0223674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- War A. R., Murugesan S., Naresh B., Ramasamy S., Nair R. (2017). Mechanism of resistance in mungbean [Vigna radiata (L.) R. Wilczek var. radiata] to bruchids, Callosobruchus spp. (Coleoptera: Bruchidae). Front. Plant Sci. 8, 1031. 10.3389/fpls.2017.01031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienkoop S., Morgenthal K., Wolschin F., Scholz M., Selbig J., Weckwerth W. (2008). Integration of metabolomic and proteomic phenotypes: analysis of data covariance dissects starch and RFO metabolism from low and high temperature compensation response in Arabidopsis thaliana. Mol. Cell. Proteom. 7, 1725–1736. 10.1074/mcp.M700273-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Mozzoni L. A., Moseley D., Hummer W., Ye H., Chen P., et al. (2020). Genome-wide association mapping of flooding tolerance in soybean. Mol. Breed. 40, 1–14. 10.1007/s11032-019-1086-0 [DOI] [Google Scholar]
- Wu J., Wang L., Li L., Wang S. (2014). De novo assembly of the common bean transcriptome using short reads for the discovery of drought-responsive genes. PLoS ONE 9, e109262. 10.1371/journal.pone.0109262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Chang Y., Wang L., Wang S., Wu J. (2021). Genome-wide association analysis of drought resistance based on seed germination vigor and germination rate at the bud stage in common bean. Agronomy 113, 2980–2990. 10.1002/agj2.20683 [DOI] [Google Scholar]
- Wu L., Chang Y., Wang L., Wang S., Wu J. (2022). The aquaporin gene PvXIP1; 2 conferring drought resistance identified by GWAS at seedling stage in common bean. Theoret. Appl. Genet. 135, 485–500. 10.1007/s00122-021-03978-w [DOI] [PubMed] [Google Scholar]
- Xu J., Xue C., Xue D., Zhao J., Gai J., Guo N., et al. (2013). Overexpression of GmHsp90s, a heat shock protein 90 (Hsp90) gene family cloning from soybean, decrease damage of abiotic stresses in Arabidopsis thaliana. PLoS ONE 8, e69810. 10.1371/journal.pone.0069810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue R., Zhang B. (2007). Increased endogenous methyl jasmonate altered leaf and root development in transgenic soybean plants. J. Genet. Genom. 34, 339–346. 10.1016/S1673-8527(07)60036-8 [DOI] [PubMed] [Google Scholar]
- Yang F., Chen H., Liu C., Li L., Liu L., Han X., et al. (2020). Transcriptome profile analysis of two Vicia faba cultivars with contrasting salinity tolerance during seed germination. Sci. Rep. 10, 1–10. 10.1038/s41598-020-64288-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H., Song L., Schapaugh W. T., Ali M. L., Sinclair T. R., Riar M. K., et al. (2020). The importance of slow canopy wilting in drought tolerance in soybean. J. Exp. Bot. 71, 642–652. 10.1093/jxb/erz150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X., Sakata K., Komatsu S. (2014). Phosphoproteomics reveals the effect of ethylene in soybean root under flooding stress. J. Proteome Res. 13, 5618–5634. 10.1021/pr500621c [DOI] [PubMed] [Google Scholar]
- Yu Z., Chang F., Lv W., Sharmin R. A., Wang Z., Kong J., et al. (2019). Identification of QTN and candidate gene for seed-flooding tolerance in soybean [Glycine max (L.) Merr.] using genome-wide association study (GWAS). Gene 10, 957. 10.3390/genes10120957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zate Z. Z. (2021). Quantitative Trait Loci mapping of agronomic traits in a Cowpea (Vigna unguiculata L.) bi-parental cross. J. Sustain. Dev. 9, 39–58. 10.20372/au.jssd.9.1.2021.0258 [DOI] [Google Scholar]
- Zhang J. Y., Broeckling C. D., Blancaflor E. B., Sledge M. K., Sumner L. W., Wang Z. Y. (2005). Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic Alfalfa (Medicago sativa). Plant J. 42, 689–707. 10.1111/j.1365-313X.2005.02405.x [DOI] [PubMed] [Google Scholar]
- Zhang Q., Zhang W.-J., Yin Z.-G., Li W.-J., Zhao H.-H., Zhang S., et al. (2020). Genome-and transcriptome-wide identification of C3Hs in common bean (Phaseolus vulgaris L.) and structural and expression-based analyses of their functions during the sprout stage under salt-stress conditions. Front. Genet. 1080, 564607. 10.3389/fgene.2020.564607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Bortesi L., Baysal C., Twyman R. M., Fischer R., Capell T., et al. (2017). Characteristics of genome editing mutations in cereal crops. Trends Plant Sci. 22, 38–52. 10.1016/j.tplants.2016.08.009 [DOI] [PubMed] [Google Scholar]
- Ziegler G., Nelson R., Granada S., Krishnan H. B., Gillman J. D., Baxter I. (2018). Genomewide association study of ionomic traits on diverse Soybean populations from germplasm collections. Plant Direct. 2, e00033. 10.1002/pld3.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J., Wang Y., Zhu B., Luo Y., Wang Q., Gao L. (2018). sRNAome and transcriptome analysis provide insight into chilling response of cowpea pods. Gene 671, 142–151. 10.1016/j.gene.2018.05.064 [DOI] [PubMed] [Google Scholar]
- Zwart R. S., Thudi M., Channale S., Manchikatla P. K., Varshney R. K., Thompson J. P. (2019). Resistance to plant-parasitic nematodes (PPNs) in Chickpea: current status and future perspectives. Front. Plant Sci. 10, 966. 10.3389/fpls.2019.00966 [DOI] [PMC free article] [PubMed] [Google Scholar]