Abstract
The emergence and global spread of the SARS-CoV-2 Omicron variants, which carry an unprecedented number of mutations, raise serious concerns due to the reduced efficacy of current vaccines and resistance to therapeutic antibodies. Here, we report the generation and characterization of two potent human monoclonal antibodies, NA8 and NE12, against the receptor-binding domain of the SARS-CoV-2 spike protein. NA8 interacts with a highly conserved region and has a breadth of neutralization with picomolar potency against the Beta variant and the Omicron BA.1 and BA.2 sublineages and nanomolar potency against BA.2.12.1 and BA.4. Combination of NA8 and NE12 retains potent neutralizing activity against the major SARS-CoV-2 variants of concern. Cryo-EM analysis provides the structural basis for the broad and complementary neutralizing activity of these two antibodies. We confirm the in vivo protective and therapeutic efficacies of NA8 and NE12 in the hamster model. These results show that broad and potent human antibodies can overcome the continuous immune escape of evolving SARS-CoV-2 variants.
Keywords: SARS-CoV-2; immune escape; variants of concern; Omicron sublineages BA.1., BA.2, BA.2.12.1, and BA.4; neutralization; therapeutic antibodies; hamster model
Graphical abstract

The emergence of new SARS-CoV-2 variants that escape neutralization can jeopardize the efficacy of vaccines and therapeutic antibodies. Chen et al. report the isolation of potent human antibodies that neutralize emerging variants of concern, including various Omicron sublineages. These antibodies show prophylactic and therapeutic efficacy in the hamster model.
Introduction
The ongoing coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has triggered a devastating global health, social, and economic crisis, with more than 1 million deaths in the United States and over 6.5 million worldwide (https://coronavirus.jhu.edu; The Johns Hopkins Coronavirus Resource Center Home Page, 2022). Effective vaccines against SARS-CoV-2 have been developed and deployed at an unprecedented pace, but hesitancy in vaccination and a limited supply in developing countries have made the fight against SARS-CoV-2 particularly challenging. The RNA nature and broad circulation of this virus enable the accumulation of mutations (Telenti et al., 2021; Yewdell, 2021), leading to the continuous emergence of variants with increased transmissibility or pathogenicity as well as resistance to monoclonal antibodies (mAbs) and vaccine-elicited antibodies (Corti et al., 2021; Davies et al., 2021; Wang et al., 2021; Wibmer et al., 2021), highlighting the need for effective therapeutic and preventive measures with a broad spectrum of action.
As a result of viral evolution, the initial SARS-CoV-2 lineages identified early during the pandemic in Wuhan, China (Zhou et al., 2020), have progressively been replaced by several variants of concern, such as B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), B.1.617.2 (Delta) and, most recently, B.1.1.529 (Omicron). In particular, the latter is raising major concern worldwide because it has an unprecedented number of mutations, and it has rapidly spread across the globe (Bowen et al., 2022a; Bruel et al., 2022; Greaney et al., 2021; Iketani et al., 2022; Lusvarghi et al., 2021; Piccoli et al., 2020; Takashita et al., 2022; Yamasoba et al., 2022; Yu et al., 2022; Zhou et al., 2022). It comprises several distinct sublineages (World Health Organization, https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19-1-february-2022). The original Omicron variant, BA.1, was first documented in South Africa in November 2021 and became in a short time the predominant variant worldwide (World Health Organization, Classification of Omicron [B.1.1.529]: SARS-CoV-2 Variant of Concern [2021]). This variant has more than 30 mutations in the spike (S) protein (Elbe and Buckland-Merrett, 2017), 15 of which are located within the receptor-binding domain (RBD), the main target of neutralizing antibodies (Greaney et al., 2021; Piccoli et al., 2020). Most of these mutations are unique to this variant. Consistent with this high degree of genetic heterogeneity, the Omicron sublineage BA.1 has reduced or abrogated sensitivity to neutralization by most mAbs, convalescent sera, and vaccine-elicited antibodies (Aggarwal et al., 2021; Cameroni et al., 2022; Cao et al., 2022a; Cele et al., 2022; Dejnirattisai et al., 2021; Doria-Rose et al., 2021; Lusvarghi et al., 2021; Mannar et al., 2022; Planas et al., 2022; Wilhelm et al., 2021). As of March 2022, however, the BA.2 sublineage is rapidly replacing BA.1 to become the dominant variant in several areas of the world, including most European countries, India, Pakistan, the Philippines, New Zealand, and South Africa (CoVariants, Overview of Variants/Mutations, March 25, 2022; https://covariants.org). BA.2 shares 21 mutations with BA.1, but these two sublineages contain 8 and 13 unique mutations, respectively, in the spike protein. BA.2 has a significantly reduced sensitivity to neutralization by sera from convalescent patients or vaccinated individuals to a degree comparable to that reported for BA.1 (Bowen et al., 2022b; Iketani et al., 2022; Yamasoba et al., 2022; Yu et al., 2022). In addition, BA.2 has shown marked resistance to several mAbs tested, including sotrovimab, which was shown to retain activity against BA.1 (Bruel et al., 2022; Iketani et al., 2022; Takashita et al., 2022; Zhou et al., 2022). Thus, albeit related, these two sublineages exhibit different sensitivity to mAbs and vaccine-elicited antibodies. More recently, three additional Omicron sublineages have emerged, BA.2.12.1, BA.4, and BA.5, and they have rapidly replaced BA.1 and BA.2 worldwide, accounting for the vast majority of new infections globally (World Health Organization, https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19-20-july-2022). The BA.4 and BA.5 lineages have an identical spike protein and differ only outside the spike region (Tegally, 2022). As seen with the initial Omicron variants, these newly emerged sublineages show even stronger escape from neutralizing antibodies elicited by both vaccination and natural infection (Bowen et al., 2022b; Cao et al., 2022b; Hachmann et al., 2022; Qu et al., 2022; Shrestha et al., 2022). The reduced effectiveness of current therapeutic antibodies and vaccines against Omicron subvariants, the rapid spread of these variants worldwide, and the limited rate of vaccination in developing countries highlight the urgent global need for the discovery of broadly neutralizing antibodies against SARS-CoV-2.
In this study, we took advantage of our extensive experience in generating mAbs using combinatorial phage-display library technology (Chen et al., 2006, 2018) to derive mAbs from convalescent COVID-19 donors with high neutralization titers. We report the isolation of ultrapotent mAbs that neutralize diverse and highly transmissible SARS-CoV-2 variants of concern, including the difficult-to-neutralize Beta and Omicron sublineages. To further validate our data, we compared side by side the neutralizing activity of our most potent mAbs with that of seven clinically approved mAbs, which confirmed the breadth and potency of our mAbs. Cryo-electron microscopy provided the structural basis for their broad reactivity and potency. Finally, we demonstrated the protective and therapeutic activity of our mAbs in a hamster preclinical model. Overall, this study uncovers potent mAbs, which may be utilized against both present and future SARS-CoV-2 variants of concern that will continue to emerge.
Results
Generation and characterization of anti-SARS-CoV-2 monoclonal antibodies using phage-display libraries from COVID-19 convalescent patients
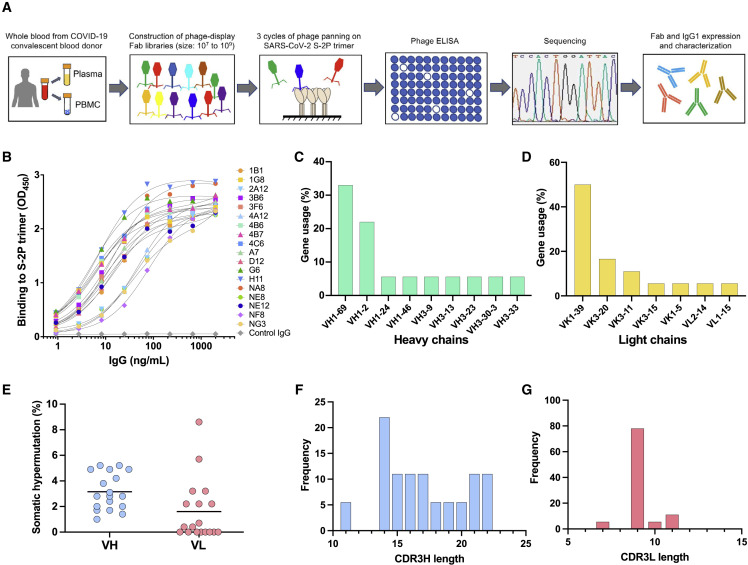
To generate mAbs against the SARS-CoV-2 spike protein by using combinatorial phage-display libraries, peripheral blood samples were collected from 12 convalescent COVID-19 plasma donors with high neutralizing serum antibody titers against SARS-CoV-2 (De Giorgi et al., 2021) (Table S1). Total RNA was extracted from peripheral blood mononuclear cells (PBMC) and used for the construction of phage-display Fab libraries (Figure 1A). A total of four phage-display Fab libraries, derived from either a single donor or multiple donors combined, were constructed, each consisting of 107 to 109 individual clones (Figure S1). Three sequential cycles of panning were carried out to enrich for specific clones using a stabilized trimeric spike protein (S-2P) derived from the original SARS-CoV-2 strain, Wuhan-Hu-1 (GenBank: MN908947; S protein sequence identical to that of the of USA/WA-1/2020 [WA-1]; GenBank: MN985325) (Wrapp et al., 2020). Subsequent screening of 672 individual clones by ELISA resulted in the identification of 538 clones that specifically bound to the S-2P protein. DNA sequencing identified 18 unique clones with distinct sequences. These clones were subcloned and expressed both as soluble Fabs and as complete IgG1 antibodies. The binding specificity of the cloned IgGs was confirmed by ELISA (Figure 1B). The binding affinity of the 18 Fabs for the soluble S-2P trimer was further assessed by surface plasmon resonance (SPR). Six Fabs showed either poor or no binding by SPR, while the remaining 12 exhibited high-affinity binding with equilibrium dissociation constants (K D) in the picomolar range for 10 and in the low nanomolar range for 2 (Figure S2; Table S2).
Figure 1.
Study design and characterization of monoclonal antibodies against the SARS-CoV-2 spike protein
(A) Schematic design of the different phases of combinatorial phage-display library production and screening leading to the development of SARS-CoV-2 neutralizing antibodies (mAbs).
(B) Normalized binding of 18 mAbs to a stabilized SARS-CoV-2 spike protein trimer, S-2P, in ELISA; OD denotes optical density.
(C and D) V-gene usage of (C) heavy chains and (D) light chains of 18 mAbs against the SARS-CoV-2 spike protein.
(E) Rate of somatic hypermutation of V genes of heavy chains (VH) and light chains (VL) of 18 mAbs.
(F and G) Amino acid length of the CDR3 loop of the (F) heavy (CDR3H) and (G) light chain (CDR3L) of 18 mAbs.
Genetic analysis of V genes of all 18 mAbs indicated that the clones primarily used the variable heavy chain (VH) genes VH1-2 and VH1-69 and the variable light chain (VK) gene VK1-39 (Figures 1C and 1D; Table S3). The dominant use of these VH genes for SARS-CoV-2 S-specific antibodies is consistent with previous studies (Andreano et al., 2021; Yuan et al., 2021). In comparison, the frequency of VH1-2 and VK1-39 gene usage only ranks 23 and 6, respectively, in the B cell repertoire of healthy individuals (Boyd et al., 2010; Prabakaran et al., 2012), suggesting that certain germline genes are naturally favored for binding to the S protein of SARS-CoV-2. Notably, the selected antibodies had very limited somatic hypermutation (SHM) with a median frequency of 3.2% in VH and 1.6% in VL (Figure 1E). These findings are consistent with previous studies, which reported the isolation of anti-SARS-CoV-2 antibodies in nearly germline configuration (Kreye et al., 2020; Liu et al., 2020; Zost et al., 2020b). The length distribution of the complementarity-determining region 3 (CDR3) in both the heavy and light chains was in line with previous observations (Figures 1F and 1G), although we found an unusual bimodal distribution of CDRH3 length as opposed to the typical bell-shaped distribution in CDRL3 (Figures 1F and 1G). No correlation was found between SHM frequency in VH and binding affinity, further supporting the notion that certain germline antibody genes are naturally fit for targeting the S protein.
Potent neutralization of diverse SARS-CoV-2 variants of concern
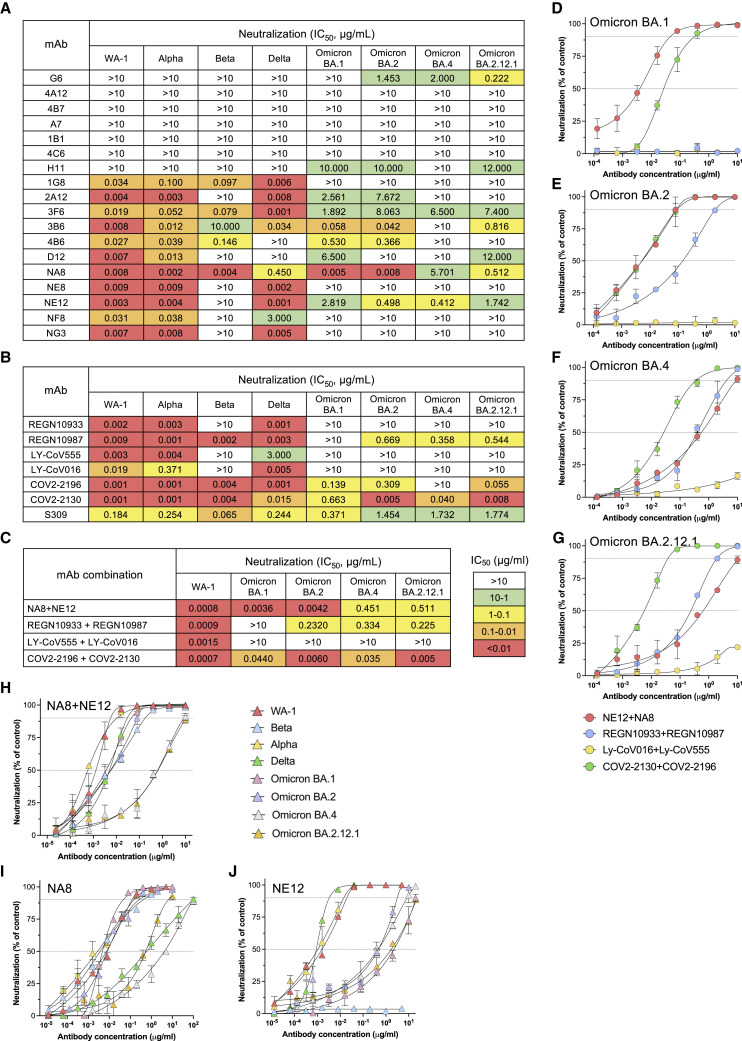
The neutralizing activity of 18 mAbs was evaluated using a pseudotype virus neutralization assay (Corbett et al., 2020a). Eleven mAbs showed potent neutralizing activity against the original SARS-CoV-2 strain (WA-1), seven with half-maximal inhibitory concentration (IC50) values in the picomolar range, below 10 ng/mL (Figure 2A), which is comparable to the potency of the best-in-class mAbs (Baum et al., 2020b; Hansen et al., 2020; Jones et al., 2021). Competition ELISA showed that all the 11 neutralizing mAbs were specific for the RBD domain (Figure S3). Cross-competition ELISA using Fab fragments and complete IgG showed high levels of cross-competition among the 11 neutralizing mAbs with the exception of two, 1G8 and 3F6, which cross-competed only between each other (Figure S4). Next, we tested the neutralizing activity of the 18 mAbs against the original North American founder strain, WA-1, as well as major variants of concern of global relevance, namely, B.1.1.7/Alpha, B.1.351/Beta, B.1.617.2/Delta, and Omicron BA.1, BA.2, BA.4, and BA.2.12.1 sublineages. All the 11 mAbs that potently neutralized the WA-1 strain retained potent activity against the Alpha variant; seven of them also neutralized the Delta variant in the picomolar range (Figure 2A). In contrast, most of the mAbs were ineffective against the Beta variant with only four retaining high neutralizing activity. Of note, one of them, NA8, neutralized Beta at picomolar concentration. Notably, NA8 also potently neutralized both Omicron BA.1 and BA.2 sublineages, which contain an unprecedented number of mutations within the S protein and are completely or partially resistant to most neutralizing mAbs as well as to serum from convalescent or vaccinated subjects (Aggarwal et al., 2021; Cameroni et al., 2022; Cao et al., 2022b; Cele et al., 2022; Dejnirattisai et al., 2022; Doria-Rose et al., 2021; Mannar et al., 2022; Planas et al., 2022; Wilhelm et al., 2021), with IC50 values in the picomolar range (Figure 2A). In addition, NA8 also neutralized at nanomolar concentrations the recently emerged BA.2.12.1 and BA.4 sublineages. Six additional mAbs neutralized different Omicron sublineages, although only one (3B6) was active in the picomolar range against BA.1 and BA.2 (Figure 2A).
Figure 2.
Neutralizing activity of 18 monoclonal antibodies derived from convalescent COVID-19 patients and seven clinically approved monoclonal antibodies against different SARS-CoV-2 variants
(A) Heatmap of pseudovirus neutralization activity of 18 mAbs derived from convalescent COVID-19 patients. IC50 values (μg/mL) are shown for neutralization of SARS-CoV-2 pseudoviruses bearing spike proteins from the USA/WA1/2020 isolate (WA-1, lineage A), the Alpha variant, Beta variant, Delta variant, and Omicron BA.1, BA.2, BA.4, and BA.2.12.1 sublineages. The color-coded legend of IC50 ranges is indicated in (C).
(B) Neutralizing activity of seven clinically approved mAbs, including REGN-10933, REGN-10987, LY-CoV555, LY-CoV016, COV2-2130, COV2-2196, and S309 (sotrovimab), against the same SARS-CoV-2 variants as in (A).
(C) Neutralizing activity of mAbs NA8 and NE12 and six clinically approved mAbs used in paired combinations against the SARS-CoV-2 WA-1 strain and the Omicron BA.1, BA.2, BA.4, and BA.2.12.1 sublineages.
(D) Neutralization curves of mAbs NE12 and NA8 and six clinically approved mAbs used in paired combinations against the Omicron BA.1 sublineage.
(E) Neutralization curves of mAbs NE12 and NA8 and six clinically approved mAbs used in paired combinations against the Omicron BA.2 sublineage.
(F) Neutralization curves of mAbs NE12 and NA8 and six clinically approved mAbs used in paired combinations against the Omicron BA.4 sublineage.
(G) Neutralization curves of mAbs NE12 and NA8 and six clinically approved mAbs used in paired combinations against the Omicron BA.2.12.1 sublineage.
(H) Neutralization curves of mAbs NE12 and NA8 used in combination against all major SARS-CoV-2 variants of concern.
(I) Neutralization curves of mAb NA8 against the major SARS-CoV-2 variants of concern.
(J) Neutralization curves of mAb NE12 against the major SARS-CoV-2 variants of concern. Results are the average of two or three independent experiments performed in duplicate using a pseudovirus assay. Values represent mean ± SD. The dotted horizontal lines indicate the IC90 and IC50.
Next, we compared our antibodies with a panel of mAbs approved for clinical use. Among seven clinical mAbs evaluated, we found that four, namely, REGN-10933 (casirivimab), REGN-10987 (imdevimab) (Baum et al., 2020b; Hansen et al., 2020), LY-CoV555 (bamlanivimab) (Jones et al., 2021), and LY-CoV016 (etesevimab) (Shi et al., 2020), had no detectable neutralizing activity against Omicron BA.1, while three, namely, S309 (sotrovimab) (Cameroni et al., 2022; Cao et al., 2022a; Cathcart et al., 2021), AZD8895/COV2-2196 (tixagevimab), and AZD1061/COV2-2130 (cilagavimab) (Zost et al., 2020a), showed neutralizing activity but were remarkably less potent than NA8 (93-, 166-, and 35-fold, respectively) (Figure 2B). Against the BA.2, BA.2.12.1, and BA.4 Omicron sublineages, only one of the approved antibodies, COV2-2130, retained potent neutralizing activity (Figure 2B).
Clinically approved mAbs are generally used in combination to ensure coverage against multiple variants (Du et al., 2021; Hammarstrom et al., 2021; Miguez-Rey et al., 2022). Thus, we compared side by side the neutralization potency of the combination of NA8 and NE12, used at equimolar concentrations (1:1), with that of six clinically approved antibodies used in paired combinations against the WA-1 strain and the Omicron sublineages BA.1 and BA.2 (Figure 2C). In agreement with previous reports (Cameroni et al., 2022; Cao et al., 2022b; Dejnirattisai et al., 2022; Planas et al., 2022; Wilhelm et al., 2021), all paired combinations were highly effective against WA-1, while only the combination of NA8 with NE12 retained high neutralization potency against Omicron BA.1. In contrast, no neutralization of Omicron BA.1 was detected with REGN-10933 in combination with REGN-10987, or with LY-CoV555 in combination with LY-CoV016, whereas the combination of COV2-2196 and COV2-2130 showed neutralizing activity that was more than 10-fold lower compared with that of the NA8-NE12 combination. Against the BA.2 sublineage, the NA8-NE12 combination was again the most potent, but a strong neutralization was also observed with the COV2-2130/COV2-2196 combination. Against the most recent variants, BA.2.12.1 and BA.4, the NA8-NE12 combination retained neutralizing activity at low nanomolar concentrations, as did the REGN-10933/REGN-10987 combination, while COV2-2130/COV2-2196 was the most potent combination and LY-CoV555/LY-CoV016 was ineffective (Figure 2C). Neutralization curves for the four paired combinations against the four Omicron sublineages are shown in Figures 2D–2G. In addition, we found that the combination of NA8 with NE12 was extremely potent against the original North American founder strain WA-1, the Alpha, Beta, and Delta variants, with IC50 values between 0.5 and 5 ng/mL (Figure 2H). These results suggest that NA8 and NE12, when used in combination, are one of the most broad and potent antibody pairs with picomolar or low nanomolar neutralizing activity against the major SARS-CoV-2 variants of concern. Based on these results, NA8 and NE12 were selected for further characterization and preclinical testing. Complete neutralization curves for each of these two mAbs against all the viral variants tested are shown in Figures 2I and 2J. The neutralizing activity of our mAbs was also tested using a live virus neutralization assay against the early WA-1 isolate and the Beta variant (Figure S5A). This assay was less sensitive than the pseudovirus assay, a finding already reported by other authors (Zost et al., 2020a). However, the neutralization profile was similar, as shown by a strong correlation between the results obtained in the two assays (Figure S5B).
Structural analysis of antibodies NE12 and NA8
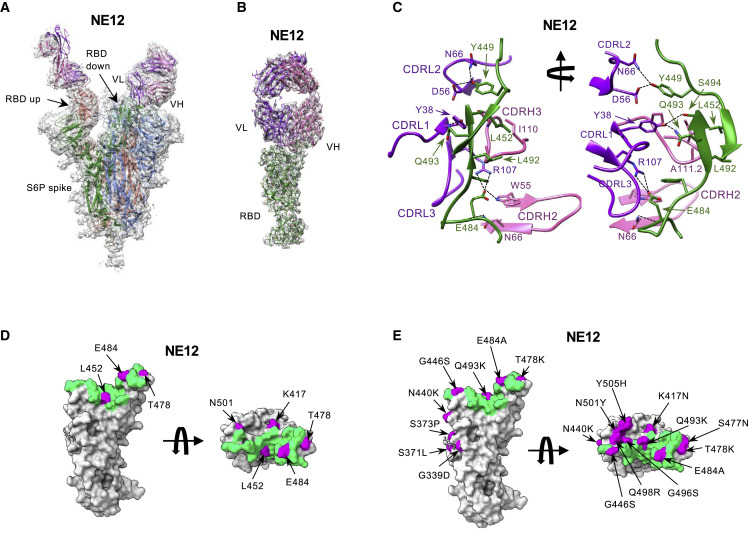
To gain structural insight into the interactions of antibodies NE12 and NA8 with the SARS-CoV-2 spike glycoprotein, we obtained cryo-EM reconstructions of Fab NE12 and Fab NA8 bound to a stabilized spike trimer (S-6P) at nominal resolutions of 3.1 Å and 2.9 Å, respectively (Table S4; Figures S6–S9). In both structures, Fabs were observed bound to each of the three RBDs of the spike irrespectively of whether it assumed the up or down position (Figures 3A and 4A ). The Fab-RBD interface was poorly resolved in the two consensus maps due to conformational variability of the S1 subunit of the spike (Figures S7 and S9). Therefore, we performed signal subtraction and focused refinement of the region that included the RBD in the down position with the bound Fab and the proximal N-terminal domain. This improved the local resolution to 3.4 Å (NE12 complex) and 3.5 Å (NA8 complex), allowing detailed structural analysis of the Fab-RBD interactions (Figures 3B, 3C, 4B, 4C, S10, and S11).
Figure 3.
Cryo-EM structures of the Fab fragment of neutralizing monoclonal antibody NE12 in complex with a stabilized SARS-CoV-2 spike protein trimer
(A) Cryo-EM map of Fab NE12 in complex with the SARS-CoV-2 S-6P spike trimer with docked spike and Fab atomic models. The heavy and light chains of the mAb are colored in pink and purple, respectively. Spike protomers are colored individually. The Fab fragment binds the receptor-binding domain (RBD) in both the up and down positions.
(B) Local refinement of a region containing one RBD with bound Fab resolved atomic details of the RBD-NE12 interactions.
(C) Ribbon diagram of the RBD/NE12 contact interface. Coloring is as in panel (B). Only the complementarity-determining regions (CDRs) and RBD fragments participating in the interaction are shown for clarity. Key residues are shown in stick representation. Dashed lines represent salt bridges and hydrogen bonds.
(D and E) Surface representation of the RBD with the epitope of NE12 colored in green. Residues mutated in B.1.1.7/Alpha, B.1.617.2/Delta, and B.1.351/Beta (D) or Omicron BA.1 sublineage (E) variants of concern are colored in magenta. The antibody residues are numbered according to the IMGT (http://www.imgt.org) (Giudicelli et al., 2005).
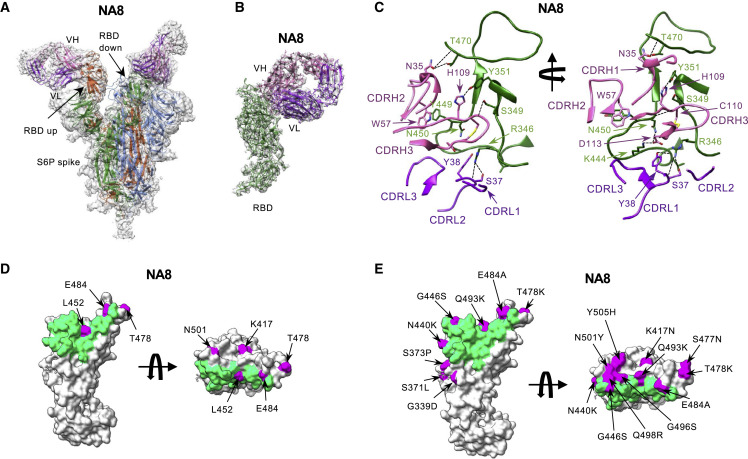
Figure 4.
Cryo-EM structures of the Fab fragment of neutralizing monoclonal antibody NA8 in complex with a stabilized SARS-CoV-2 spike protein trimer
(A) Cryo-EM map of Fab NA8 in complex with the SARS-CoV-2 S-6P spike trimer with docked spike and Fab atomic models. The heavy and light chains of the mAb are colored in pink and purple, respectively. Spike protomers are colored individually. The Fab fragment binds the receptor-binding domain (RBD) in both the up and down positions.
(B) Local refinement of a region containing one RBD with bound Fab resolved atomic details of the RBD-NA8 interactions.
(C) Ribbon diagram of the RBD/NA8 contact interface. Coloring is as in (B). Only the complementarity-determining regions (CDRs) and RBD fragment participating in the interaction are shown for clarity. Key residues are shown in stick representation. Dashed lines represent salt bridges and hydrogen bonds.
(D and E) Surface representation of the RBD with residues mutated in the B.1.1.7/Alpha, B.1.617.2/Delta, and B.1.351/Beta (D) or Omicron BA.1 sublineage (E) variants of concern are colored in magenta.
The antibody residues are numbered according to the IMGT (http://www.imgt.org) (Giudicelli et al., 2005).
Fab NE12 is positioned nearly parallel to the longest dimension of the RBD and interacts with the face of the receptor-binding ridge (Figures 3B and 3C). The interaction buries 874 Å2 of the Fab surface area, split nearly equally between the heavy (465 Å2) and light (409 Å2) chains, and 828 Å2 of the RBD surface area. The 17-residue-long CDR3 of the heavy chain (CDRH3) and the CDR1 of the light chain (CDRL1) straddle the receptor-binding ridge, while the CDRL3 and CDRH2 interact with the tip of the RBD (Figure 3C). The CDRH1 does not contribute to the paratope and is disordered, while the CDRL2 interacts with the region of the receptor-binding ridge distant from the tip. The location of the epitope, the ability to bind the RBD both in the up and down positions, and the long CDRH3 allowed the categorization of NE12 as a class 2 antibody as defined by Barnes et al. (Barnes et al., 2020). Based on the alternative classification system of Hastie et al. (Hastie et al., 2021), NE12 belongs to group RBD-2b.2. Its epitope overlaps with those of mAbs LY-CoV555 (Jones et al., 2021) and REGN-10933 (Hansen et al., 2020), as well as with the ACE2 receptor footprint (Shang et al., 2020), indicating that NE12 directly blocks the spike-receptor interaction (Figure S12). This mechanism was confirmed by binding competition studies on ACE2-expressing cells (Figure S13A). Residues mutated in the B.1.1.7/Alpha and B.1.617.2/Delta variants are located outside or at the periphery of the NE12 epitope, thus allowing for a potent neutralization of these variants (Figure 3D). In contrast, the E484K mutation of B.1.351/Beta results in clashes with CDRH2 and CDRL3 residues, compromising the neutralizing activity. Likewise, multiple replacements in the Omicron BA.1 sublineage RBD receptor-binding ridge can affect NE12-spike interactions (Figure 3E).
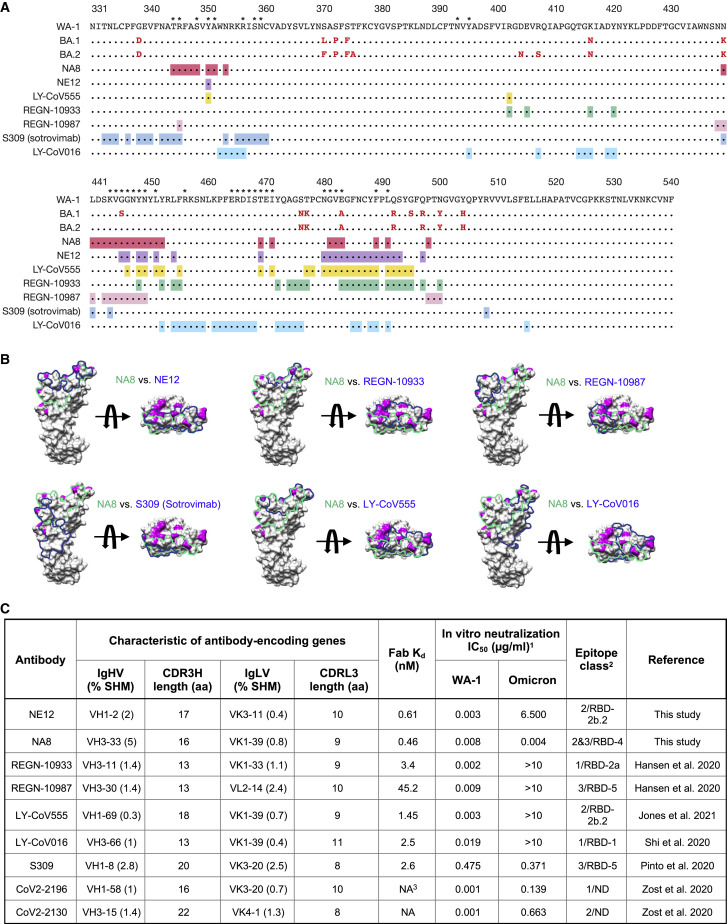
Similar to Fab NE12, Fab NA8 shows extensive contacts with the receptor-binding ridge, but this antibody binds to a distinct epitope along the outer side of the RBD distal from the tip (Figures 4B and 4C). While NA8 interacts with the periphery of the receptor-binding ridge (typical of class 2 antibodies), its epitope is shifted toward the glycosylated Asn343 (typical of class 3 antibodies), although it does not include this residue or the glycan. Thus, NA8 displays characteristics of both Barnes class 2 and class 3 antibodies (Barnes et al., 2020), while the Hastie classification system (Hastie et al., 2021) categorizes it as an RBD-4 mAb. Analysis of contact surfaces reveals a moderate overlap between the NA8 epitope and the ACE2-binding interface of the RBD (Figure S13B). Accordingly, binding competition studies on ACE2-expressing cells provided evidence that NA8 reduced the interaction between the spike trimer and ACE2 by 90% (Figure S13A). The interaction buries 981 Å2 of the Fab surface area, with the heavy chain responsible for most contacts (642 Å2) and 990 Å2 of the RBD surface area. The CDRH3 forms a long loop positioned along the side of the RBD, whereas the CDRH1 and CDRH2 contact the central region of the receptor-binding ridge. The CDRL1 and CDRL2 interact with the side of the RBD, while the CDRL3 contacts the loop formed by RBD residues 444–447. Of the residues mutated in the variants of concern, only L452 is part of the NA8 epitope (Figure 4D), which can explain the reduced neutralization of B.1.617.2/Delta. Noteworthy, all the B.1.351/Beta and Omicron BA.1 mutations fall outside the NA8 epitope or are located at its periphery (Figures 4E and 5A), which is consistent with the high neutralization potency of NA8 against these variants. Comparison of the NA8 footprint with those of NE12 and several clinically approved mAbs (Figure 5B) shows that the NA8 epitope is partially overlapping but distinct from the epitopes of REGN-10933, REGN-10987, S309 (sotrovimab), LY-CoV555, and LY-CoV016. Of note, BA.1 and BA.2 share the same mutations in the regions targeted by NA8 except for Ser446 of BA.1, which is reverted to a glycine in the BA-2 sublineage (Figure 5A); this explains the ability of NA8 to efficiently neutralize both sublineages.
Figure 5.
Comparison of binding epitopes, genetic characteristics, and neutralizing activity of NA8 and NE12 with clinically approved monoclonal antibodies
(A) Amino acid sequences of the receptor-binding domain (RBD) of Omicron sublineages BA.1 and BA.2 with mutations colored in red compared with the sequence of the original North American founder strain WA-1 (GenBank: MN985325); the asterisks indicate residues that contact the ACE receptor. The binding epitopes of different mAbs are presented below and highlighted in different colors.
(B) Surface representation of the RBD with the epitopes of NA8 (green line) and NE12 or clinically approved monoclonal antibodies (blue line) highlighted. Surface areas corresponding to the RBD residues mutated in the Omicron BA.1 sublineage are colored in magenta.
(C) Characteristics of NA8 and NE12 in comparison with clinically approved mAbs against SARS-CoV2.
In summary, based on their binding affinity, neutralization potency, and breadth, NE12 and NA8 provide a pair of potent complementary mAbs for potential clinical use (Figure 5C).
Antibodies NE12 and NA8 protect hamsters from SARS-CoV-2 infection
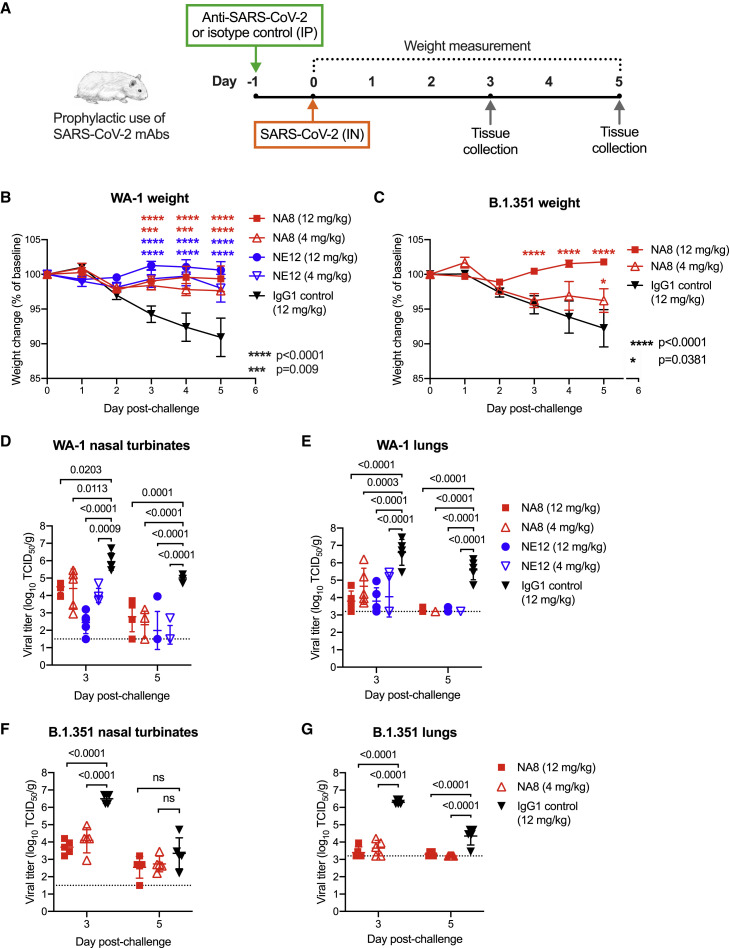
Next, we tested the in vivo prophylactic efficacy of the two selected mAbs, NE12 and NA8, in the golden Syrian hamster model, which closely mimics the severity of the disease in humans (Baum et al., 2020a; Imai et al., 2020). A total of 80 male hamsters were used for these experiments; each study group included 10 hamsters. The animals were inoculated with mAbs at two concentrations (12 and 4 mg/kg) via intraperitoneal injection 24 h before intranasal challenge with 4.5 log10 TCID50 of SARS-CoV-2 WA-1 or B.1.351/Beta (Figure 6A). An isotype-matched (IgG1) irrelevant mAb, the anti-HIV-1 VRC01, was included as a control at 12 mg/kg. Based on the in vitro neutralization results, both NA8 and NE12 were tested against the WA-1 strain (Figures 6B, 6D, and 6E), while only NA8 was tested against the B.1.351/Beta variant (Figures 6C, 6F, and 6G). Animals treated with the control IgG1 lost, on average, 9% and 8% of body weight by day 5 after challenge with WA-1 and B.1.351, respectively (Figures 6B and 6C). In contrast, treatment with NE12 or NA8 significantly protected hamsters from weight loss induced by the two viral variants, although NA8 at 4 mg/kg against the B.1.351 virus showed a lesser degree of protection (Figure 6C). The protective effect of NE12 and NA8 was confirmed by viral load measurements in nasal turbinate and lung tissues on days 3 and 5 after inoculation (n = 5 per time point per group). In animals treated with NE12 or NA8 at either 12 or 4 mg/kg, the viral titers at day 3 post-infection with the WA-1 virus were significantly reduced in both tissues compared with controls; in the lungs, the virus became undetectable at day 5 post-infection (Figures 6D and 6E). Likewise, in animals infected with the B.1.351 variant, NA8 at both concentrations significantly reduced the viral titers in nasal turbinate and lung tissues at day 3 post-infection and completely suppressed viral replication in the lungs at day 5 (Figures 6F and 6G). Overall, these data demonstrate that NA8 exerted strong prophylactic protection in vivo from two antigenically distinct SARS-CoV-2 variants, and NE12 exerted strong prophylactic protection against SARS-CoV-2 WA-1 in a susceptible animal model, despite the relatively low antibody doses used.
Figure 6.
Prophylactic efficacy of two neutralizing monoclonal antibodies, NE12 and NA8, against infection with SARS-CoV-2 in the golden Syrian hamster model
(A) Design of the study. MAbs NE12 or NA8 were administered intraperitoneally (IP) at the dose of 12 or 4 mg/kg. Control animals received an anti-HIV-1 IgG1 (VRC01) at 12 mg/kg. One day later, each group of 10 animals was challenged with 104.5 TCID50 of SARS-CoV-2 WA-1 or B.1.351 instilled intranasally (IN). The animal weight was monitored daily (days 0–3: n = 10 per group; days 4, 5: n = 5 per group) as an indicator of disease progression. Tissues were collected for virus quantification at 3 and 5 days after challenge (n = 5 per group per time point).
(B and C) Body weight changes (means ± SE) from baseline in hamsters that received neutralizing mAbs at different doses or isotype control (IgG) after challenge with (B) WA-1 or (C) B.1.351 SARS-CoV-2 strains.
(D–G) Viral titers in (D) nasal turbinate and (E) lung tissues at days 3 and 5 post-infection with (F) WA-1 or (G) the B.1.351 variant, as determined using an assay to quantify the TCID50 of infectious virus. Individual titers and means ± SD are shown for each group. Dashed lines indicate the limit of detection of the assay. Statistical comparisons of weight changes between treatment groups were performed using a repeated-measures mixed-effects model with Tukey’s multiple-comparison test (B and C). Comparisons of the viral titers between groups were performed using two-way analysis of variance (ANOVA) with Tukey’s multiple-comparison test. p values between groups treated with neutralizing mAbs and isotype control are indicated. p > 0.05, not significant (ns); ∗p = 0.0381; ∗∗∗p = 0.0009; ∗∗∗∗p < 0.0001.
Antibodies NE12 and NA8 have therapeutic activity against SARS-CoV-2 in hamsters
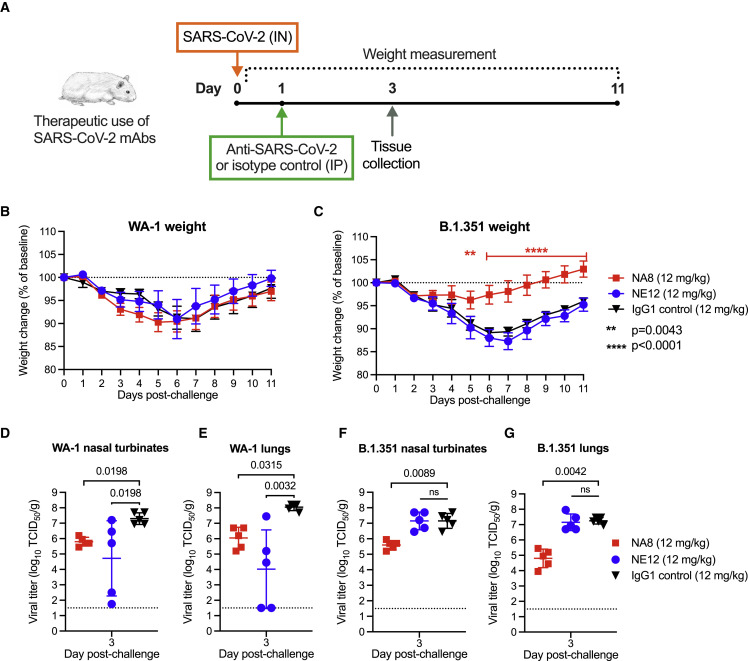
To investigate the therapeutic activity of NE12 and NA8 against SARS-CoV-2, a total of 60 male hamsters, divided in six groups of 10 animals each, were utilized for a therapeutic study. Three groups of animals were infected intranasally with the original strain, WA-1, and three groups with the B.1.351/Beta variant (4.5 log10 TCID50 per animal). The therapeutic efficacy of NE12 or NA8 (each at 12 mg/kg) was evaluated by administering the antibodies via the intraperitoneal route 24 h after intranasal challenge (Figure 7A). This is a very stringent model given the extremely rapid kinetics of replication of SARS-CoV-2. The IgG1 isotype control mAb VRC01 (12 mg/kg) was tested in parallel as a control. Five hamsters per group were sacrificed at day 3 post-infection for tissue viral load measurements, while the remaining 5 animals were weighed daily until day 11. No significant differences in weight loss were observed among the three groups of hamsters infected with WA-1 (Figure 7B). In contrast, NA8 significantly reduced the weight loss caused by infection with the B.1.351/Beta variant, which remained below 5% of the initial body weight throughout the observation period in contrast with the marked loss (nearly 15%) seen in the control group (Figure 7C). Analysis of viral titers in nasal turbinate and lung tissues showed a significant reduction in both NE12- and NA8-treated animals infected with WA-1, despite a wide data distribution in animals treated with NE12 (Figures 7D and 7E). The serum concentrations of NA8 and NE12 were measured on day 3 after infection. The three animals in the NE12/WA-1 group that showed high viral titers in nasal turbinates and lungs had low to undetectable antibody levels in serum (Figure S14), which may reflect a failure in intraperitoneal injection, as reported by other authors (Starr et al., 2021). In hamsters infected with B.1.351, treatment with NA8 induced a significant reduction in viral titers in both nasal turbinate and lung tissues, while, as expected, NE12 was ineffective (Figures 7F and 7G). Altogether, these results demonstrate that both NE12 and NA8 exerted strong therapeutic effects against sensitive SARS-CoV-2 strains in a suitable preclinical model.
Figure 7.
Therapeutic efficacy of two neutralizing mAbs, NE12 and NA8, against SARS-CoV-2 in the golden Syrian hamster model
(A) Design of the study. Each group of 10 animals was first challenged with 104.5 TCID50 of SARS-CoV-2 WA-1 or B.1.351/Beta, instilled intranasally (IN). Twenty-four hours later, the animals were injected intraperitoneally (IP) with mAbs NE12 or NA8 at the dose of 12 mg/kg. Control animals received an anti-HIV-1 IgG1 (VRC01) at 12 mg/kg. The animal weight was monitored daily as an indicator of disease progression (days 0–2: n = 10 per group; days 3–11: n = 5 per group, except for the WA-1/NE12 group (days 6–11: n = 3 due to a technical issue). Tissues were collected at day 3 after challenge for virus quantification (n = 5 per group).
(B and C) Body weight changes (means ± SE) from baseline in hamsters that were challenged with SARS-CoV-2 (B) WA-1 or (C) B.1.351/Beta.
(D–G) Quantification of viral titers at day 3 post-challenge in (D) nasal turbinate and (E) lung tissues of animals infected with WA-1 or in (F) nasal turbinate and (G) lung tissues of animals infected with the B.1.351/Beta variant, as determined using an assay to quantify the TCID50 of infectious virus. Individual titers and means ± SD are shown for each group. Dashed lines indicate the limit of detection of the assay. Statistical comparisons of weight changes between treatment groups were performed using a repeated-measures mixed-effects model with Tukey’s multiple-comparison test. Comparisons of viral titers between the treatment groups were performed using the non-parametric Kruskal-Wallis test with a Benjamini, Krieger, and Yekutieli false discovery test. p values between groups treated with neutralizing mAbs and isotype control are indicated. p > 0.05, not significant (ns); ∗∗p = 0.0043; ∗∗∗∗p < 0.0001.
Discussion
In this study, we report the generation of a panel of potent human mAbs capable of neutralizing highly diverse and transmissible SARS-CoV-2 variants of concern. Within this panel, we selected two mAbs, NA8 and NE12, as promising candidates for clinical use. The most important observation from this work is that, despite the substantial mutation of this virus, the immune system has the potential to develop RBD-directed antibodies that are capable of potent neutralization and yet retain a broad spectrum of action to overcome the continuous immune escape of evolving SARS-CoV-2 variants of concern. NA8 and NE12 have distinctive and complementary properties. NA8 features a very broad spectrum of action with ultrapotent activity, at low picomolar concentrations, against difficult-to-neutralize variants of concern, namely, Beta and Omicron sublineages BA.1 and BA.2, and at nanomolar concentrations against the most recently emerged BA.2.12.1 and BA.4. The Beta variant is particularly resistant to neutralization, and indeed very few effective mAbs are currently available (Wang et al., 2021; Wibmer et al., 2021). Likewise, the most widely clinically used mAbs, including sotrovimab (Cameroni et al., 2022; Cao et al., 2022b; Cathcart et al., 2021), have reduced or no neutralizing activity against the recently emerged Omicron variants. The Omicron BA.2 sublineage, which rapidly replaced the previously dominant BA.1 in many countries, was found to be resistant to 17 out of 19 neutralizing mAbs tested, including sotrovimab, which had retained some activity against BA.1, indicating that mAbs react in a different manner against the Omicron sublineages (Iketani et al., 2022). Among the mAbs currently approved for clinical use that were evaluated in our study, only AZD1061/COV2-2130 (cilagavimab) showed strong neutralizing activity against the BA.2 and BA.2.12.1 sublineages, but its efficacy against the BA.1 sublineage was markedly reduced. Very recently, FDA granted emergency use authorization for a new mAb, LY-CoV1404 (bebtelovimab), which has an in vitro potency against Omicron comparable to that of NA8 (Westendorf et al., 2022), but this antibody was not available for side-by-side comparison with our mAbs.
The structural basis for the breadth of neutralization of NA8, which has mixed features of both Barnes class 2 and class 3 antibodies, lies in the high conservation of its binding epitope, as revealed by cryo-EM analysis. The NA8 epitope is distinct from those of other mAbs hitherto reported in that, despite including a portion of the receptor-binding ridge, it is shifted toward the outer side of the RBD, allowing the antibody to “sidestep” the residues that are mutated in the Beta and Omicron variants. The second mAb described herein, NE12, has features of a Barnes class 2 antibody and binds to an epitope that largely overlaps with the ACE2 receptor footprint on the RBD.
NA8 and NE12 have a complementary neutralization spectrum, and indeed, when used in combination, they showed potent activity against all the major SARS-CoV-2 variants tested. Among the clinically approved mAbs tested, only the combination of COV2-2130 and COV2-2196 showed a comparable breadth. Although NA8 and NE12 bind to partially overlapping epitopes, as is the case for LY-CoV555 and LY-CoV016, their neutralization potency was not affected when used in combination. Although antibodies targeting overlapping epitopes can displace one another, particularly when their association rate is similar, it has been shown that the displacement is reversible if sufficient time is allowed to reach a state of equilibrium (Abdiche et al., 2017). This model is consistent with our experimental observations whereby an equimolar combination of NA8 and NE12 neutralized all viral variants with the same potency as that of the most effective of the two antibodies.
Considering the reduced efficacy of current vaccines and the complete loss or reduced neutralizing activity of most clinically approved antibodies to the Omicron variants (Aggarwal et al., 2021; Bowen et al., 2022a; Bruel et al., 2022; Cameroni et al., 2022; Cao et al., 2022a; Cao Y. et al., 2022b; Cathcart et al., 2021; Cele et al., 2022; Dejnirattisai et al., 2022; Doria-Rose et al., 2021; Hachmann et al., 2022; Iketani et al., 2022; Mannar et al., 2022; Planas et al., 2022; Qu et al., 2022; Shrestha et al., 2022; Takashita et al., 2022; Wilhelm et al., 2021; Zhou et al., 2022), the isolation of new antibodies capable of blocking a broad spectrum of emergent SARS-CoV-2 variants of concern is important for future therapeutic and preventive strategies. Due to the high degree of conservation of its binding epitope, our mAb NA8 has the potential to neutralize new variants that will continue to emerge over time, especially from areas of the world with low vaccination rates. Corroborating the potential clinical usefulness of our antibodies, the in vitro neutralization potency of NE12 and NA8 was confirmed in a hamster model in vivo, in which both mAbs exhibited remarkable prophylactic and therapeutic efficacy against the original WA-1 strain and the Beta variant at relatively low doses.
In summary, the identification of potent, broadly neutralizing human antibodies against SARS-CoV-2 variants of concern that resist neutralization by most mAbs, such as the Beta and the Omicron sublineages BA.1 and BA.2, is critical for creating an arsenal of therapeutic antibodies with high potency against both present and future variants of concern that will continue to emerge because of the sustained worldwide spread of this virus, leading to escape from current prophylactic and therapeutic interventions.
Limitations of the study
Although we documented the in vivo protective and therapeutic efficacies of our mAbs, NA8 and NE12, in the hamster model against two different SARS-CoV-2 strains (WA-1 and Beta variant), we did not test them against other variants of concern of global relevance such as the various Omicron sublineages. However, based on their in vitro neutralization activity, we could infer that our antibodies would be effective against these variants in the preclinical model, although physicochemical properties of our antibodies that were not addressed in this study can have a major influence on in vivo efficacy (Jain et al., 2017). Another limitation of this study is that most of the neutralization results were obtained using a pseudovirus-based assay that employs ACE2-transfected HEK293 cells as targets. However, this assay is widely used in the field and was shown to predict the in vivo neutralizing activity of mAbs, as also confirmed in this study.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| AffiniPure Goat Anti-Human IgG, Fcγ fragment specific | Jackson ImmunoResearch | 109-005-008 |
| Peroxidase AffiniPure Goat Anti-Human IgG, Fcγ fragment specific | Jackson ImmunoResearch | 109-035-190 |
| Peroxidase AffiniPure Goat Anti-Human IgG, F(ab')₂ fragment specific | Jackson ImmunoResearch | 109-035-097 |
| HRP/Anti-M13 Monoclonal Conjugate | Cytiva | 27-9421-01 |
| SARS-CoV-2 nucleoprotein-specific rabbit primary antibody | SinoBiological | 40143-R001 |
| Alexa 593-conjugated secondary antibody | Life technologies | A11037 |
| 1B1 | This manuscript | N/A |
| 3B6 | This manuscript | N/A |
| 2A12 | This manuscript | N/A |
| 1G8 | This manuscript | N/A |
| 3F6 | This manuscript | N/A |
| 4A12 | This manuscript | N/A |
| 4B6 | This manuscript | N/A |
| 4B7 | This manuscript | N/A |
| 4C6 | This manuscript | N/A |
| A7 | This manuscript | N/A |
| D12 | This manuscript | N/A |
| G6 | This manuscript | N/A |
| H11 | This manuscript | N/A |
| NA8 | This manuscript | N/A |
| NE8 | This manuscript | N/A |
| NF8 | This manuscript | N/A |
| NE12 | This manuscript | N/A |
| NG3 | This manuscript | N/A |
| REGN-10933 | VRC, NIH | N/A |
| REGN-10987 | VRC, NIH | N/A |
| LY-CoV555 | VRC, NIH | N/A |
| LY-CoV016 | VRC, NIH | N/A |
| S309 | VRC, NIH | N/A |
| COV2-2196 | VRC, NIH | N/A |
| COV2-2130 | VRC, NIH | N/A |
| Anti-polio A12 IgG | Chen et al. (2011) | N/A |
| Bacterial and virus strains | ||
| SARS-CoV-2 wild-type | CDC | GenBank: MN985325 |
| USA-WA-1/2020 | GISAID: EPI_ISL_404895 | |
| SARS-CoV-2 lineage B.1.351/Beta | Johns Hopkins University | GISAID: EPI_ISL_890360 |
| E. coli, TG1 strain | Lucigen | 60502-1 |
| M13KO7 helper phage | New England Biolabs | N0315S |
| E. coli, Top 10GF′ | Lucigen | 60061-2 |
| 5-alpha Competent E. coli | New England Biolabs | C2987H |
| Pseudovirus with SARS-CoV-2 S protein of Wuhan-1 | This manuscript | GenBank: MN908947.3 |
| Pseudovirus with SARS-CoV-2 S protein of B.1.351 | This manuscript | N/A |
| Pseudovirus with SARS-CoV-2 S protein of B.1.1.7 | This manuscript | N/A |
| Pseudovirus with SARS-CoV-2 S protein of B.1.617.2 | This manuscript | N/A |
| Pseudovirus with SARS-CoV-2 S protein of B.1.1.529 | This manuscript | N/A |
| Biological samples | ||
| Serum samples from convalescent COVID-19 patients | Blood bank of NIH | N/A |
| Whole blood samples from convalescent COVID-19 patients | Blood bank of NIH | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| SARS-CoV-2 (2019-nCoV) Spike RBD | SinoBiological | 40592-V08H |
| Stabilized SARS-CoV-2 spike protein trimer (S-2P) | This manuscript | N/A |
| Stabilized SARS-CoV-2 spike protein trimer (S-6P) | This manuscript | N/A |
| Ficoll-Paque PLUS | Cytiva | 17144002 |
| 293fectin transfection reagent | ThermoFisher Scientific | 12347019 |
| Turbo293 transfection reagent | Speed BioSystems | PXX1002 |
| HisPur Ni-NTA resin | ThermoFisher Scientific | 88221 |
| Superdex 200 16/60 | Cytiva | 28-9893-35 |
| Glucose | Sigma-Aldrich | G8270-5KG |
| PEG8000 (polyethylene glycol) | Sigma-Aldrich | 89510-1KG-F |
| NaCl | Sigma-Aldrich | S9888-1KG |
| 2xYT medium | IPM Scientific | 11006-055 |
| LB medium | IPM Scientific | 11006-004 |
| IPTG (isopropy1β-D-thiogalactoside) | Sigma-Aldrich | I6758-10G |
| Ampicillin | ThermoFisher Scientific | J63807.09 |
| Kanamycin sulfate | ThermoFisher Scientific | J17924_06 |
| HiTrap SP HP | Cytiva | 17115201 |
| HiTrap Mab Select | Cytiva | 28-4082-56 |
| HisTrap HP | Cytiva | 17-5248-02 |
| Sensor chip CM5 | Cytiva | BR-1000-2 |
| ExpiFectamine 293 Transfection kit | ThermoFisher Scientific | A14524 |
| Expi293 expression medium | ThermoFisher Scientific | A1435101 |
| SureBlue™ TMB 1-Component Microwell Peroxidase Substrate | SeraCare | 5120-0077 |
| DMEM medium | ThermoFisher Scientific | 11965092 |
| Fetal bovine serum | Sigma-Aldrich | F4135 |
| Penicillin-streptomycin | ThermoFisher Scientific | 15140122 |
| Puromycin | ThermoFisher Scientific | A1113803 |
| Bright-Glo Luciferase assay substrate | Promega | E2620 |
| XhoI | New England BioLabs | R0146M |
| SpeI-HF | New England BioLabs | R3133M |
| SacI-HF | New England BioLabs | R3156M |
| XbaI | New England BioLabs | R0145M |
| NheI-HF | New England BioLabs | R3131S |
| NotI | New England BioLabs | R3189S |
| ApaI | New England BioLabs | R0114S |
| AgeI | New England BioLabs | R3552S |
| T4 DNA ligase | New England BioLabs | M0202M |
| Bovine Serum Albumine (BSA) | Sigma-Aldrich | 3059 |
| Tween 20 | Sigma-Aldrich | 93773 |
| DMSO | Sigma-Aldrich | D2650 |
| Critical commercial assays | ||
| RNeasy mini kit | Qiagen | Cat#74106 |
| First-strand cDNA synthesis kit | Cytiva | Cat#27926101 |
| HotStarTaq DNA polymerase | Qiagen | Cat# 203205 |
| Deposited data | ||
| Antibody VH and VL sequences | This manuscript | GenBank: OM179962-OM179997 |
| Ensemble cryo-EM map of the NE12/spike trimer complex | This manuscript | EMDB: EMD-26401 |
| Local cryo-EM map of the NE12/spike trimer complex | This manuscript | EMDB: EMD-26402 Protein DataBank:7U9O |
| Ensemble cryo-EM maps of the NA8/spike trimer complex | This manuscript | EMDB: EMD-26403 |
| Local cryo-EM maps of the NA8/spike trimer complex | This manuscript | EMDB: EMD-26404 Protein DataBank:7U9P |
| Experimental models: Cell lines | ||
| Vero E6 cells | ATCC | CRL-1586 |
| 293 Freestyle cells | ThermoFisher Scientific | R79007 |
| HEK293T/17 cells | ATCC | CRL-11268 |
| ACE-2-expressing 293T cells (293T-hACE2.MF) | This manuscript | NA |
| Expi293F cells | ThermoFisher Scientific | A14527 |
| Experimental models: Organisms/strains | ||
| Golden Syrian hamster | Envigo Laboratories | N/A |
| Oligonucleotides | ||
| Primers for PCR amplification of human γ1 heavy chain Fd | Glamann et al., (1998) | N/A |
| Primers for PCR amplification of human κ chain | Glamann et al., (1998) | N/A |
| Primers for PCR amplification of human λ chain | Kang et al. (1991) | N/A |
| Recombinant DNA | ||
| pComb3H | Barbas et al. (1991) | N/A |
| IgG expression vector | This manuscript | N/A |
| Software and algorithms | ||
| Prism version 9 | GraphPad Software | N/A |
| Biacore T200 Evaluation software 3.1 | Biaeval | N/A |
| EVILFIT | Zhao et al. (2017) | N/A |
| CTFFind4 | Rohou and Grigorieff, 2015 | N/A |
| MotionCorr2 | Zheng et al., 2017 | N/A |
| RELION | Scheres, 2012 | N/A |
| UCSF Chimera | Pettersen et al., 2004 | N/A |
| ResMap | Kucukelbir et al., 2014 | N/A |
| Coot | Emsley and Cowtan, 2004 | N/A |
| Phenix | Liebschner et al., 2019 | N/A |
| Other | ||
| Operetta high content imaging system | Perkin Elmer | N/A |
| Biacore T200 | Cytiva | N/A |
| SpectraMax M5 plate reader | Molecular Devices | N/A |
| ÅKTA go | Cytiva | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Patrizia Farci (pfarci@niaid.nih.gov).
Materials availability
Aliquots of antibodies will be made available by the lead contact upon request under a Material Transfer Agreement (MTA) for non-commercial usage.
Experimental model and subject details
Convalescent COVID-19 plasma donor selection, human samples, and lymphocyte isolation
Convalescent COVID-19 plasma donors were prospectively enrolled onto an institutional review board-approved protocol (Clinical Trials Registration, NCT04360278) (De Giorgi et al., 2021) and provided written informed consent for the study. Among them, 12 convalescent plasma donors with high titer neutralizing antibodies against SARS-CoV-2 were selected, and 20-40 ml of blood was collected from each of them. Six of the 12 donors were females and 6 were males; their mean age was 54.6 years (range 35 to 66 years). Eligibility criteria included molecular or serologic evidence of past COVID-19 infection, and complete recovery from COVID-19, with no symptoms for ≥28 days or ≥14 days with a negative molecular test after recovery. PBMCs were prepared using density centrifugation on a Ficoll-Paque gradient.
Method details
Live virus neutralization assay used to test COVID-19 convalescent patients
Neutralization of live virus by patient plasmas was tested using a fluorescence reduction neutralization assay (FRNA) with SARS-CoV-2 [2019-nCoV/USA-WA1-A12/2020 (WA-1) from the US Centers for Disease Control and Prevention, Atlanta, GA, USA] at the NIH-NIAID Integrated Research Facility at Fort Detrick, MD, USA, as previously reported (Bennett et al., 2021). In these experiments, a fixed volume of diluted virus was incubated with an equivalent volume of test plasma for 1 hour at 37°C prior to adding the suspension to Vero E6 cells (ATCC, Manassas, VA, USA, #CRL-1586). The virus was allowed to propagate for 24 hours prior to fixing the cells. Following fixation, the cells were permeabilized and probed with a SARS-CoV-2 nucleoprotein-specific rabbit primary antibody (Sino Biological, Wayne, PA, USA, #40143-R001) followed by an Alexa594-conjugated secondary antibody (Life Technologies, San Diego, CA, USA, #A11037). The total number of infected cells in four fields per well with each field containing at least 1000 cells was quantified using an Operetta high content imaging system (Perkin Elmer, Waltham, MA, USA). Plasma was tested using two-fold serial dilutions from 1:40 to 1:1280 with four replicates per dilution. Results were calculated as the highest dilution of plasma leading to at least 50% reduction of SARS-CoV-2 titers. Each assay was controlled with internal addition of an S-specific neutralizing polyclonal antibodies. If a 1:40 dilution did not lead to at least 50% reduction of viral titer, results were reported as <1:40 even though some inhibition of virus propagation may have been present. To enable analysis of nAb kinetics over time, a value of 1:20 was used as a surrogate for results <1:40 since this value was the next serial 2-fold titer below the lowest positive result.
Human Fab antibody library construction
Total RNA was extracted from PBMCs using RNeasy mini kit (Qiagen) and 1st strand cDNA was reverse transcribed with oligo(dT) as a primer with a kit from Cytiva and used as a template for antibody Fab coding fragment amplification. Briefly, the γ1 heavy chain Fd (variable and first constant region) was amplified with nine human heavy chain–specific 5′ primers and a human γ1–specific 3′ primer. The κ chain was amplified by PCR with seven human κ chain–specific 5′ primers and a 3′ primer matching the end of the constant region (Glamann et al., 1998). The λ chain was amplified by PCR with six human λ chain–specific 5′ primers and a 3′ primer matching the end of the constant region (Kang et al., 1991). PCR was performed for 30 cycles at 95°C for 1 min, 52°C for 1 min, and 72°C for 1 min with HotStar Taq DNA polymerase (Qiagen). Amplified κ and λ chain DNA fragments were pooled, purified, digested with SacI and XbaI. The restriction enzyme digested light chain fragments from one donor or pooled from 5 donors were then cloned into the pComb 3H vector by electroporation (Barbas et al., 1991). The recombinant plasmid DNA was introduced into Escherichia coli Top 10 cells (Lucigen) by electroporation, yielding 1 × 107 individual clones. The plasmid DNA containing light chain sequences was digested with XhoI and SpeI, ligated with γ1 Fd DNA cut with the same enzymes. The plasmid DNAs containing light and heavy chains were transformed into E. coli Top 10 cells by electroporation. Four phage display Fab libraries with average size of 5 × 108 individual clones for each library were generated.
Expression and purification of recombinant SARS-CoV-2 spike protein trimers
The stabilized SARS-Cov-2 spike protein trimer (S-2P) was produced by transient transfection of 293 Freestyle cells as previously described (Wrapp et al., 2020). One liter of 293 Freestyle cells at concentration of about 0.9 million cells per ml was transfected with 1 mg of S-2P plasmid, pre-mixed with 1 ml of 293fectin™ Transfection Reagent. The cells were allowed to grow for 6-7 days at 125 rpm, 37°C, and 8 % CO2; then the supernatant was harvested by centrifugation and filtered through Millipore Sigma 0.22 μm PES filter. The cleared supernatant was incubated with 5 mL of HisPur™ Ni-NTA Resin ThermoFisher for three hours, then the resin was collected and washed with PBS with 25 mM imidazole, pH 7.4. The captured SARS-CoV-2 S-2P was eluted by PBS with 250 mM imidazole, pH 7.4. After elution, the trimeric protein was collected, concentrated, and applied to a Superdex 200 16/60 gel filtration column equilibrated with PBS. Peak fractions were pooled and concentrated to 1 mg/mL. The SARS-CoV-2 S-6P trimer was expressed and purified as previously described (Hsieh et al., 2020). Briefly, 1 mg of DNA encoding the S-6P spike was transfected into 293 Freestyle cells using Turbo293 transfection reagent (Speed BioSystems). The cells were grown at 37°C for 6 days, after which the supernatant was harvested and applied to His-Pure affinity resin. The resin was washed with 20 mM imidazole in PBS, and the protein eluted with 20 mM HEPES, pH 7.5, 200 mM NaCl and 300 mM imidazole. The trimeric protein was further purified on a Superdex S-200 gel filtration column equilibrated in PBS and concentrated to 1 mg/ml and flash frozen in liquid nitrogen for storage at −80°C.
Selection of specific Fab clones
For phage production, 25 ml of logarithmic bacteria culture (OD600 = 0.6) in 2×YT supplemented with 100 μg/mL ampicillin and 2% glucose (2×YT-Amp-Glu) were infected with M13KO7 helper phage (New England Biolabs, USA) at 7×109 plaque forming unit (PFU) per mL (∼1:20 multiplicity of infection) by incubating at 37°C for 30 min without shaking, followed by 30 min at 120 rpm. Infected cells were harvested by centrifugation (5000 g for 5 min) and resuspended in 100 ml 2×YT supplemented with 100 μg/mL ampicillin and 50 μg/mL kanamycin. After overnight growth at 30°C at 250 rpm, the cells were removed by centrifugation (18000 g at 4°C for 20 min). The culture supernatant containing the phages was filtered through a 0.45-μm filter and then precipitated with 1/5 volume of 20% PEG8000 (polyethylene glycol) in a 2.5 M NaCl solution, for 1 h on ice. Phage particles were pelleted by centrifugation (9000 g at 4°C for 20 min) and re-dissolved in 1 mL 1×PBS.
The phage library was panned by affinity binding of Fab-expressing phages on S-2P coated on the wells of an ELISA plate. Blocking of plates and phages was conducted for 60 min using 3% skimmed milk in 1×PBS. All washing steps were performed using PBS containing 0.05% Tween20 (PBST). For each panning cycle, 1 μg/mL antigen was used to coat the polystyrene plate and after an overnight incubation, plates were washed and blocked. For the first panning cycle, approximately 1×1011 phages were incubated with the antigen coated plates for 2 h, followed by a total of 20 washes with PBST. The eluted phages were amplified in E. coli Top 10 cells. Bacterial culture was plated on 2×YT Amp-Glu agar and incubated overnight at 30°C. Clones were harvested into 5 ml 2×YT-Amp-Glu and phage production for the next round of panning, which was conducted in 40 ml medium, as described above. Two additional panning cycles were repeated using the same protocol. Following three cycles of panning, the selected phages were used for infection of E. coli. Single colonies were randomly picked from the third cycle output and screening of specific binders was performed, using phage ELISA against S-2P, with a BSA coated-plate as a negative control.
Sequence analysis of S-2P-positive Fab clones
We sequenced the heavy-chain variable region (VH-DH-JH genes) of the ELISA-positive SARS-CoV-2 S-2P- specific clones obtained from each library using the Applied BioSystems model 3730 automated DNA sequencer with a modified Sanger method. The closest human germline V(D)J gene segments for each unique sequence were determined using DNAPLOT with IMGT sequence database (http://www.imgt.org/IMGT) (Giudicelli et al., 2005). The framework and CDRs were assigned according to IMGT nomenclature. The somatic mutations were identified by comparison of V-genes (from framework 1 to 3) with the closest germline counterpart. The first 28 nucleotides corresponding to the primer sequence were not included in the analysis. The distribution of VH and JH gene usage and HCDR3 length of anti-SARS-CoV-2 was analyzed.
Expression and purification of Fab and IgG
The phagemid containing light chain and heavy chain was cleaved with NheI and SpeI and re-circularized after removal of the phage gene III DNA fragment from the vector to encode soluble Fab. Bacteria containing circularized DNA without phage gene III were cultured in 2×TY medium containing 2% glucose, 100 μg/mL ampicillin, and 15 μg/mL tetracycline at 30°C until the OD600 reached 0.5–1. The culture was diluted 5-fold in 2×YT medium without glucose and containing 0.2 mM isopropyl β-D-thiogalactoside (IPTG), and culture was continued at 27°C for 20 h for expression of soluble Fab. Because the Fab was tagged at the C terminus with (His)6, the expressed proteins were affinity-purified on a nickel-charged column and were further purified through a cation-exchange SP column (Cytiva).
For IgG production, Expi293F™ cells (Thermo Fisher) were transiently transfected with plasmids carrying the antibody heavy chain and light chains. Cells were grown for six days at 37°C with 8% CO2 shaking at 125 rpm according to the manufacturer’s protocol (Thermo Fisher). Cell cultures were harvested six days after transfection and centrifuged at 20,000 g for 30 min at 4°C. The supernatants were collected and filtrated through a 0.22 μm filter. The expressed IgGs were purified by affinity chromatography on an immobilized protein G column (Cytiva). The purity of the Fab and IgG was evaluated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the protein concentrations were determined by optical density (OD) measurements at 280 nm, with an A280 of 1.5 corresponding to 1.0 mg/mL.
ELISA assays
For conventional antigen-binding ELISA, 96-well plates were coated with 100 μl/well containing 1 μg/mLprotein in 1×PBS (pH 7.4) and incubated at room temperature overnight. Serial dilutions of soluble Fab, IgG, or phage were added to the wells, and plates were incubated for 2 h at room temperature. The plates were washed, and the secondary conjugated antibody (anti-human Fab-HRP, anti-human IgG Fc-HRP, or anti-M13-HRP) was added and incubated for 1 h at room temperature. The plates were washed, and the color was developed by adding tetramethylbenzidine reagent (KPL; Gaithersburg, MD), and the development was stopped with H2SO4 after 10 min. The plates were read at OD450 in an ELISA plate reader. The data were plotted, and the dose-response curves were generated with Prism software (Graphpad Software, Inc., San Diego, CA).
For RBD-competition ELISA, 96-well plates were coated with 100 μl/well containing 1 μg/mL of recombinant S-2P trimer in 1×PBS (pH 7.4), and the plates were incubated at room temperature overnight. Each Fab was 3-fold serially diluted from a starting concentration of 3 μg/mL in 3% milk/PBS or in 3% milk/PBS containing recombinant RBD protein (Sino Biological) at fixed concentration (5 μg/mL). Following incubation for 2 h at room temperature, the plate was washed, and a secondary anti-human Fab-HRP antibody (Jackson ImmunoResearch) was added for 1 h at room temperature. The plate was then washed, and the reaction was developed by adding tetramethylbenzidine substrate (KPL; Gaithersburg, MD). The reaction was stopped after 10 min by addition of sulfuric acid. The plates were read at OD450 in an ELISA plate reader. The data were plotted, and dose-response curves with and without inhibition with RBD were generated using Prism software (Graphpad Software, Inc., San Diego, CA).
For antibody cross-competition ELISA, 96-well ELISA plates were coated with 100 μl/well containing 1 μg/mL of recombinant S-2P trimer in 1×PBS (pH 7.4) and the plates were incubated at room temperature overnight. To test cross-competition among the mAbs, the IgG version of each mAb was incubated at 0.1 μg/mL with each of 11 Fabs at 1 μg/ml. An in house-produced anti-poliovirus IgG antibody was used as a negative control (Chen et al., 2011). Following incubation for 2 h at room temperature, the plates were washed, and a secondary anti-human IgG Fc-HRP antibody (Jackson ImmunoResearch) was added and incubated for 1 h at room temperature. The plates were then washed, and the reaction was developed by adding tetramethylbenzidine substrate (KPL; Gaithersburg, MD). The reaction was stopped after 10 min by addition of sulfuric acid. The plates were read at OD450 in an ELISA plate reader. The average value from three wells was used to calculate the percent competition using the following formula: [1- (average OD from wells containing test IgG with Fab – average OD from control wells)/(average OD from wells containing test IgG without Fab – average OD from control wells)] x100%.
Surface plasmon resonance (SPR)
The SPR experiments were performed on a Biacore™ T200 (Cytiva) at 25°C in 10 mM HEPES pH 7.2, 150 mM NaCl, 3 mM EDTA, 0.05% Tween-20. The recombinant SARS-CoV-2 spike 2p (S-2P) protein was immobilized on a Series S-CM5 sensor chip (Cytiva) by amine coupling (NHS/EDC) to flow cells. A mock coupled surface was used for background subtraction. Binding and kinetics studies were performed multiple times for each Fab. Increasing amounts of each Fab were injected over the surface at a flow rate of 30 μl/min with an association time of 120 sec and dissociation time of 180 sec, followed by a regeneration with 100mM CAPS, pH 11.5 for 90 sec. Binding data were analyzed using Biacore™ T200 Evaluation Software 3.1 (Biaeval) with fits to the Langmuir binding equation for a 1:1 interaction model. In addition, data were analyzed by surface site affinity distribution analysis by EVILFIT (Zhao et al., 2017). Values obtained with the two methods were consistent.
Pseudotype neutralization assay
The SARS-Cov-2 pseudovirus neutralization assays were performed as previously reported (Corbett et al., 2020b). Briefly, single-round luciferase-expressing pseudoviruses were generated by co-transfection of plasmids encoding the full-length SARS-CoV-2 S protein (Wuhan-1, GenBank accession number, MN908947.3; B.1.351/Beta; B.1.1.7/Alpha; B.1.617.2./Delta; Omicron BA.1 and BA.2 sublineages; luciferase (pHR′ CMV Luc), a lentivirus backbone (pCMV ΔR8.2), and human transmembrane protease serine 2 (TMPRSS2) at a ratio of 1:20:20:0.3 into HEK293T/17 cells (ATCC) using the transfection reagent LiFect293™. The pseudoviruses were harvested after 72 hours. The supernatants were collected by centrifugation at 1500 rpm for 10 minutes, then filtered through a 0.45 μm filter, aliquoted and titrated before the neutralization assay. To test antibody-mediated neutralization, an 8-point, 5-fold dilution series was prepared for each antibody in culture medium (DMEM medium supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin and 3 μg/mL puromycin). Each antibody dilution (50 μL) was mixed with 50 μL of diluted pseudovirus in 96-well plates and incubated for 30 min at 37°C. The mixture was then incubated with 104 ACE-2-expressing 293T cells (293T-hACE2.MF) in a final volume of 200 μL. Seventy-two hours later, the supernatant was carefully removed, the cells were lysed with Bright-Glo™ Luciferase Assay substrate (Promega), and the luciferase activity (relative light units, RLU) was measured. Percent neutralization was normalized considering uninfected cells as 100% neutralization and cells infected in the absence of antibodies as 0% neutralization. IC50 titers were determined using a log (agonist) vs. normalized response (variable slope) nonlinear function in Prism v8 (GraphPad).
Live virus neutralization assay
SARS-CoV-2 neutralization using live virus was determined in the BSL3 laboratory using SARS-CoV-2 USA-WA1/2020 [WA-1; GenBank MN985325; GISAID: EPI_ISL_404895; obtained from Dr. Natalie Thornburg, Centers for Disease Control and Prevention (CDC)] and USA/MD-HP01542/2021 (lineage B.1.351/Beta variant; GISAID: EPI_ISL_890360; obtained from Dr. Andrew Pekosz, Johns Hopkins University). WA-1 was passaged twice on Vero E6 cells. The USA/MD-HP01542/2021 was passaged on TMPRSS2-expressing Vero E6 cells. The SARS-CoV-2 stocks were titrated in Vero E6 cells by determination of the 50% tissue culture infectious dose (TCID50) as previously described (Subbarao et al., 2004). The mAbs were serially diluted in Opti-MEM and mixed with an equal volume of SARS-CoV-2 (100 TCID50) and then incubated at 37°C for 1 h. Mixtures were added to quadruplicate wells of Vero E6 cells in 96-well plates and incubated for four days. The 50% neutralizing dose (ND50) was defined as the highest dilution of serum that completely prevented cytopathic effect in 50% of the wells and was expressed as a log10 reciprocal value (Liu et al., 2021). The dilution of antibodies that completely prevented cytopathic effect in 50% of the wells (ND50) was calculated by the Reed and Muench formula (Reed and Muench, 1938).
Cryo-EM specimen preparation and data collection
SARS-CoV-2 S-6P spike at a concentration of 0.5 mg/mL in PBS was mixed with Fab NA8 or Fab NE12 using a Fab-to-RBD molar ratio of 1:1, and the complex was used for specimen preparation after a brief (5-10 min) incubation at 4°C. To prepare cryo-EM specimens, Quantifoil R 2/2 gold grids were glow-discharged using a PELCO easiGlow device (air pressure: 0.39 mBar, current: 20 mA, duration: 30 s) immediately before vitrification using an FEI Vitrobot Mark IV plunger. The Vitrobot chamber was kept at 4°C and 95% humidity, and the drop volume was 2.7 μL. Datasets were collected at the National CryoEM Facility (NCEF), National Cancer Institute, using a Thermo Scientific Titan Krios G3 electron microscope equipped with a K3 direct electron detector and Gatan Quantum GIF energy filter (slit width: 20 eV) (Table S4).
Single particle analysis of cryo-EM data
Single particle analysis was performed using the Frederick Research Computing Environment (FRCE) computing cluster. MotionCorr2 was used for patch-based movie frame alignment (Zheng et al., 2017). Contrast transfer function parameters were estimated with ctffind 4.1 (Rohou and Grigorieff, 2015). The following steps were performed using Relion 3.1.0 (Scheres, 2012), unless otherwise stated. Particle picking was performed using the template-free Laplacian-Gaussian filter-based approach. Particles were first extracted with 4x binning and subjected to 2D and 3D classification, with the low-pass filtered cryo-EM map of individual SARS-CoV-2 S-2P spike at pH 7.4 serving as the initial 3D model. Particles contributing to high-quality 3D classes were then re-extracted with 2x binning, and the above procedure was repeated. Finally, the best particles were re-extracted without binning, and the 3D map was iteratively improved with rounds of 3D auto-refinement, CTF refinement, and Bayesian polishing. For local refinement, a soft mask encompassing one RBD in the down position with the bound Fab and the neighboring NTD was created after segmenting the refined ensemble map in UCSF Chimera (Pettersen et al., 2004) and used for auto-refinement with limited angular search (-sigma_ang = 5). Resolution was calculated using the gold-standard approach (Henderson et al., 2012) at the FSC curve threshold of 0.143. Local resolution was determined with ResMap 1.1.4 (Kucukelbir et al., 2014).
Atomic model generation
An RBD from the cryo-EM structure of the S-2P spike at pH 4.0 (PDB ID 6xlu) was docked into the locally refined cryo-EM map, along with an NTD from the same structure and a homology model of the corresponding Fab prepared using the SWISS-MODEL server (Waterhouse et al., 2018). The atomic model was refined by alternating rounds of model building in Coot (Emsley and Cowtan, 2004) and real-space refinement in Phenix (Liebschner et al., 2019). Structure validation was performed with Molprobity (Davis et al., 2004). Map-model correlations were evaluated with phenix.mtriage (Afonine et al., 2018). Molecular graphics and analyses were performed with UCSF Chimera (Pettersen et al., 2004).
Prophylactic and therapeutic studies in the hamster model
The hamster studies were approved by the NIAID Animal Care and Use Committee. All the animal experiments were carried out following the Guide for the Care and Use of Laboratory Animals by the NIH. Golden Syrian hamsters (Mesocricetus auratus, Envigo Laboratories, Frederick, MD) were used in experiments conducted in BSL2 and BSL3 facilities approved by the USDA and CDC. In the prophylactic study, 80 male Syrian hamsters, aged 8-9 weeks, were randomly divided into groups of 10 animals each, bled for baseline serology and inoculated intraperitoneally with 0.5 mL of NE12, NA8, or IgG1 control at 12 mg/kg or 4 mg/kg, diluted to the concentration of 2.4 mg/mL or 0.8 mg/mL in PBS. Twenty-four hours later, the animals were challenged intranasally with 4.5 log10 TCID50 of SARS-CoV-2 WA-1 or B.1.351 in 100 μL volumes of L-15 medium (ThermoFisher) per animal. Intranasal instillations were performed under light isoflurane anesthesia. Animals were euthanized by CO2 inhalation prior to necropsy. Body weights and clinical symptoms were monitored daily from day −3 before challenge through day 5 after challenge. On days 3 and 5 post-challenge, 5 animals per group were necropsied; nasal turbinate and lung tissues were collected. Tissues were weighed, mixed with L-15 medium (10 mL per gram of tissue), homogenized, and clarified by centrifugation. Aliquots were snap-frozen and stored at −80°C. The presence of the challenge virus in clarified tissue homogenates was evaluated by limiting-dilution titrations on Vero E6 cells. The titration of SARS-CoV-2 was performed by determination of the TCID50 in Vero E6 cells and expressed as TCID50 per gram of tissue, as previously described (Subbarao et al., 2004).
In the therapeutic study, 60 male Syrian hamsters, aged 9-10 weeks, were randomly divided into three groups of 10 animals each, bled for serology and inoculated intranasally with 4.5 log10 TCID50 of SARS-CoV-2 WA-1 or B.1.351 in 100 μL volumes per animal. Twenty-four hours later, the animals were inoculated intraperitoneally with 0.5 mL of NE12, NA8, or IgG1 control at 12 mg/kg, diluted to the concentration of 2.4 mg/mL in PBS. Body weights and clinical symptoms were monitored daily from day −4 before virus inoculation through day 11 after inoculation. On day 3 post-inoculation, five animals per group were necropsied; nasal turbinates and lungs were collected and processed as above. Virus titers were determined by limiting-dilution titrations on Vero E6 cells as described above (Subbarao et al., 2004).
Quantification and statistical analysis
Statistical analysis
Data sets were assessed for significance using one-way ANOVA with Tukey’s multiple comparison test, or, if indicated, by non-parametric Kruskal-Wallis test with a Benjamini, Krieger and Yekutieli false-discovery test. Statistical comparisons of weight changes between treatment groups were performed using a repeated-measures mixed-effects model with Tukey’s multiple-comparison test. Data were only considered significant at p 0.05. Analyses were performed using Prism 9 (GraphPad Software).
Acknowledgments
We wish to thank John R. Mascola, Kizzmekia S. Corbett, Nicole Doria-Rose, Lingshu Wang, Kevin Carlton, and the Vaccine Production Program of the Vaccine Research Center (VRC), NIAID, NIH, for providing mAbs REGN-10933, REGN-10987, LY-CoV555, LY-CoV016, S309, and VRC01, plasmids to express the SARS-CoV-2 stabilized S-2P and S-6P spike trimers, membrane-bound spike proteins from various SARS-CoV-2 strains, the lentivirus backbone pCMV DR8.2, and human TMPRSS2; Michael Farzan for providing 293-ACE2 cells; Craig Martens for sequencing the SARS-CoV-2 stocks; and the staff of the NIAID Comparative Medicine Branch for animal study support. We also wish to thank Dr. Anthony S. Fauci, Dr. Steven Holland, and Dr. Jeffrey I. Cohen for critically reading the manuscript and for their helpful suggestions. This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), and by the Intramural Research Program of the VRC, NIAID, NIH. This work was also supported by Federal funds from the NIAID, NIH, under Contract No. HHSN272201800013C, by Federal funds from the Frederick National Laboratory for Cancer Research (FNLCR), NIH, under Contract HHSN261200800001 (Y.T., T.S.). Cryo-EM datasets were collected at the National CryoEM Facility (NCEF) of the National Cancer Institute (NCI) with the support of the NCI National Cryo-EM Facility at the FNLCR under contract HSSN261200800001E. Chimera was developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH P41-GM103311. The Frederick Research Computing Environment (FRCE) high-performance computing cluster was used for processing cryo-EM datasets. R.G. and M.R.H. performed this work as employees of Laulima Government Solutions, LLC, while E.P performed this work as an employee of Tunnell Government Services. The content of this publication does not necessarily reflect the views or policies of the US Department of Health and Human Services (DHHS) or of the institutions and companies affiliated with the authors.
Author contributions
Z.C. and P.F conceived the research and designed the study, and Z.C., P.Z., Y.M., Y.T., D.H.M., P.D.K., H.J.A., U.J.B., P.L., and P.F designed experiments, and K.W., V.D.G., and H.J.A contributed to donor recruitment and blood sample collection, and Z.C. generated Fab libraries and cloned monoclonal antibodies, and H.N. and A.P. contributed to plasmid DNA preparation and tissue culture for IgG production, and B.Z. produced antibodies, and P.Z. and P.L. produced pseudoviruses and performed pseudovirus neutralization assays, and E.P., R.G., M.R.H., Y.M., and C.S performed live virus neutralization assays, and L.F.B. and D.H.M performed binding assays and data analysis, and Y.M. and C.S. performed animal studies, and Y.M. and U.J.B. analyzed results, and N.L.G. and R.F.J. generated and characterized the SARS-CoV-2 stocks for the animal studies, and A.S.O. expressed, purified, and provided S-6P spike for cryo-EM structural analysis, and T.S. carried out specimen preparation for cryo-EM experiments, and Y.T. performed cryo-EM experiments and with P.D.K. performed data analysis and interpretation of the cryo-EM structures, and P.F. with Z.C., Y.T., U.J.B., and P.L. wrote the manuscript with all authors providing revisions and comments, and P.F. provided supervision for the entire project.
Declaration of interests
Z.C., P.F., K.W., P.Z., P.L., U.J.B., and Y.M. are named on an unpublished patent application filed by the National Institutes of Health related to the work described herein.
Published: September 30, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111528.
Supplemental information
Data and code availability
-
-
The antibody sequences reported in this paper have been deposited in the GenBank database (accession nos. OM179962-OM179997).
-
-
The ensemble cryo-EM maps of the NE12/spike trimer and NA8/spike trimer complexes were deposited to the Electron Microscopy Data Bank (EMDB) with accession numbers EMD-26401 and EMD-26403, respectively. The locally refined maps of the NE12/spike trimer and NA8/spike trimer complexes were deposited to EMDB with accession numbers EMD-26402 and EMD-26404, respectively, and the corresponding coordinates were deposited to the Protein Data Bank with accession numbers 7U9O and 7U9P.
-
-
Any additional information required to reanalyze the data reported is available from the lead contact upon request.
References
- Abdiche Y.N., Yeung A.Y., Ni I., Stone D., Miles A., Morishige W., Rossi A., Strop P. Antibodies targeting closely adjacent or minimally overlapping epitopes can displace one another. PLoS One. 2017;12:e0169535. doi: 10.1371/journal.pone.0169535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine P.V., Klaholz B.P., Moriarty N.W., Poon B.K., Sobolev O.V., Terwilliger T.C., Adams P.D., Urzhumtsev A. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D Struct. Biol. 2018;74:814–840. doi: 10.1107/S2059798318009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal A., Stella A.O., Walker G., Akerman A., Milogiannakis V., Brilot F., Amatayakul-Chantler S., Roth N., Coppola G., Schofield P., et al. SARS-CoV-2 Omicron: evasion of potent humoral responses and resistance to clinical immunotherapeutics relative to viral variants of concern. medRxiv. 2021 doi: 10.1101/2021.12.14.21267772. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano E., Nicastri E., Paciello I., Pileri P., Manganaro N., Piccini G., Manenti A., Pantano E., Kabanova A., Troisi M., et al. Extremely potent human monoclonal antibodies from COVID-19 convalescent patients. Cell. 2021;184:1821–1835.e16. doi: 10.1016/j.cell.2021.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbas C.F., 3rd, Kang A.S., Lerner R.A., Benkovic S.J. Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc. Natl. Acad. Sci. USA. 1991;88:7978–7982. doi: 10.1073/pnas.88.18.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.M.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Ajithdoss D., Copin R., Zhou A., Lanza K., Negron N., Ni M., Wei Y., Mohammadi K., Musser B., et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science. 2020;370:1110–1115. doi: 10.1126/science.abe2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett R.S., Postnikova E.N., Liang J., Gross R., Mazur S., Dixit S., Kocher G., Yu S., Georgia-Clark S., Gerhardt D., et al. Scalable, micro-neutralization assay for assessment of SARS-CoV-2 (COVID-19) virus-neutralizing antibodies in human clinical samples. Viruses. 2021;13 doi: 10.3390/v13050893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen J.E., Sprouse K.R., Walls A.C., Mazzitelli I.G., Logue J.K., Franko N.M., Ahmed K., Shariq A., Cameroni E., Gori A., et al. Omicron BA.1 and BA.2 neutralizing activity elicited by a comprehensive panel of human vaccines. bioRxiv. 2022 doi: 10.1101/2022.03.15.484542. Preprint at. [DOI] [Google Scholar]
- Bowen J.E., Addetia A., Dang H., Stewart C., Brown J.T., Sharkey W.K., Sprouse K.R., Walls A.C., Mazzitelli I.G., Logue J.K., et al. Science. 2022:eabq0203. doi: 10.1126/science.abq0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd S.D., Gaëta B.A., Jackson K.J., Fire A.Z., Marshall E.L., Merker J.D., Maniar J.M., Zhang L.N., Sahaf B., Jones C.D., et al. Individual variation in the germline Ig gene repertoire inferred from variable region gene rearrangements. J. Immunol. 2010;184:6986–6992. doi: 10.4049/jimmunol.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruel T., Hadjadj J., Maes P., Planas D., Seve A., Staropoli I., Guivel-Benhassine F., Porrot F., Bolland W.H., Nguyen Y., et al. Serum neutralization of SARS-CoV-2 Omicron sublineages BA.1 and BA.2 in patients receiving monoclonal antibodies. Nat. Med. 2022;28:1297–1302. doi: 10.1038/s41591-022-01792-5. [DOI] [PubMed] [Google Scholar]
- Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., Pinto D., VanBlargan L.A., De Marco A., di Iulio J., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602:664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Yisimayi A., Jian F., Song W., Xiao T., Wang L., Du S., Wang J., Li Q., Chen X., et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022;608:593–602. doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cathcart L.A., Havenar-Daughton C., Lempp F.A., Ma D., Schmid M.A., Agostini M.L., Guarino B., Di iulio J., Rosen L.E., Tucker H., et al. The dual function monoclonal antibodies VIR-7831 and VIR-7832 demonstrate potent in vitro and in vivo activity against SARS-CoV-2. bioRxiv. 2021 doi: 10.1101/2021.03.09.434607. Preprint at. [DOI] [Google Scholar]
- Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H., San J.E., Cromer D., Scheepers C., Amoako D.G., et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Chumakov K., Dragunsky E., Kouiavskaia D., Makiya M., Neverov A., Rezapkin G., Sebrell A., Purcell R. Chimpanzee-human monoclonal antibodies for treatment of chronic poliovirus excretors and emergency postexposure prophylaxis. J. Virol. 2011;85:4354–4362. doi: 10.1128/JVI.02553-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Diaz G., Pollicino T., Zhao H., Engle R.E., Schuck P., Shen C.H., Zamboni F., Long Z., Kabat J., et al. Role of humoral immunity against hepatitis B virus core antigen in the pathogenesis of acute liver failure. Proc. Natl. Acad. Sci. USA. 2018;115:E11369–E11378. doi: 10.1073/pnas.1809028115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Moayeri M., Zhou Y.H., Leppla S., Emerson S., Sebrell A., Yu F., Svitel J., Schuck P., St Claire M., Purcell R. Efficient neutralization of anthrax toxin by chimpanzee monoclonal antibodies against protective antigen. J. Infect. Dis. 2006;193:625–633. doi: 10.1086/500148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schäfer A., Ziwawo C.T., DiPiazza A.T., et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O'Connell S., Bock K.W., Minai M., et al. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N. Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D., Purcell L.A., Snell G., Veesler D. Tackling COVID-19 with neutralizing monoclonal antibodies. Cell. 2021;184:3086–3108. doi: 10.1016/j.cell.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., Pearson C.A.B., Russell T.W., Tully D.C., Washburne A.D., et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372 doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis I.W., Murray L.W., Richardson J.S., Richardson D.C. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Giorgi V., West K.A., Henning A.N., Chen L.N., Holbrook M.R., Gross R., Liang J., Postnikova E., Trenbeath J., Pogue S., et al. Naturally acquired SARS-CoV-2 immunity persists for up to 11 Months following infection. J. Infect. Dis. 2021;224:1294–1304. doi: 10.1093/infdis/jiab295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Huo J., Zhou D., Zahradnik J., Supasa P., Liu C., Duyvesteyn H.M.E., Ginn H.M., Mentzer A.J., Tuekprakhon A., et al. Omicron-B.1.1.529 Leads to Widespread Escape from Neutralizing Antibody Responses. 2021. [DOI] [PMC free article] [PubMed]
- Dejnirattisai W., Huo J., Zhou D., Zahradník J., Supasa P., Liu C., Duyvesteyn H.M.E., Ginn H.M., Mentzer A.J., Tuekprakhon A., et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185:467–484.e15. doi: 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose N.A., Shen X., Schmidt S.D., O'Dell S., McDanal C., Feng W., Tong J., Eaton A., Maglinao M., Tang H., et al. Booster of mRNA-1273 strengthens SARS-CoV-2 omicron neutralization. medRxiv. 2021 doi: 10.1101/2021.12.15.21267805. Preprint at. [DOI] [Google Scholar]
- Du L., Yang Y., Zhang X. Neutralizing antibodies for the prevention and treatment of COVID-19. Cell. Mol. Immunol. 2021;18:2293–2306. doi: 10.1038/s41423-021-00752-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Global Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Giudicelli V., Chaume D., Lefranc M.P. IMGT/GENE-DB: a comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res. 2005;33:D256–D261. doi: 10.1093/nar/gki010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glamann J., Burton D.R., Parren P.W., Ditzel H.J., Kent K.A., Arnold C., Montefiori D., Hirsch V.M. Simian immunodeficiency virus (SIV) envelope-specific Fabs with high-level homologous neutralizing activity: recovery from a long-term-nonprogressor SIV-infected macaque. J. Virol. 1998;72:585–592. doi: 10.1128/JVI.72.1.585-592.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D., et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29:44–57.e9. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarström L., Marcotte H., Piralla A., Baldanti F., Pan-Hammarström Q. Antibody therapy for COVID-19. Curr. Opin. Allergy Clin. Immunol. 2021;21:553–558. doi: 10.1097/ACI.0000000000000787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachmann N.P., Miller J., Collier A.R.Y., Ventura J.D., Yu J., Rowe M., Bondzie E.A., Powers O., Surve N., Hall K., Barouch D.H. Neutralization escape by SARS-CoV-2 omicron subvariants BA.2.12.1, BA.4, and BA.5. N. Engl. J. Med. 2022;387:86–88. doi: 10.1056/NEJMc2206576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie K.M., Li H., Bedinger D., Schendel S.L., Dennison S.M., Li K., Rayaprolu V., Yu X., Mann C., Zandonatti M., et al. Defining variant-resistant epitopes targeted by SARS-CoV-2 antibodies: a global consortium study. Science. 2021;374:472–478. doi: 10.1126/science.abh2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R., Sali A., Baker M.L., Carragher B., Devkota B., Downing K.H., Egelman E.H., Feng Z., Frank J., Grigorieff N., et al. Outcome of the first electron microscopy validation task force meeting. Structure. 2012;20:205–214. doi: 10.1016/j.str.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.L., Goldsmith J.A., Schaub J.M., DiVenere A.M., Kuo H.C., Javanmardi K., Le K.C., Wrapp D., Lee A.G., Liu Y., et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369:1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iketani S., Liu L., Guo Y., Liu L., Chan J.F.W., Huang Y., Wang M., Luo Y., Yu J., Chu H., et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature. 2022;604:553–556. doi: 10.1038/s41586-022-04594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P.J., Nakajima N., Watanabe T., Ujie M., Takahashi K., Ito M., et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. USA. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain T., Sun T., Durand S., Hall A., Houston N.R., Nett J.H., Sharkey B., Bobrowicz B., Caffry I., Yu Y., et al. Biophysical properties of the clinical-stage antibody landscape. Proc. Natl. Acad. Sci. USA. 2017;114:944–949. doi: 10.1073/pnas.1616408114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B.E., Brown-Augsburger P.L., Corbett K.S., Westendorf K., Davies J., Cujec T.P., Wiethoff C.M., Blackbourne J.L., Heinz B.A., Foster D., et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med. 2021;13:eabf1906. doi: 10.1126/scitranslmed.abf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang A.S., Barbas C.F., Janda K.D., Benkovic S.J., Lerner R.A. Linkage of recognition and replication functions by assembling combinatorial antibody Fab libraries along phage surfaces. Proc. Natl. Acad. Sci. USA. 1991;88:4363–4366. doi: 10.1073/pnas.88.10.4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreye J., Reincke S.M., Kornau H.C., Sánchez-Sendin E., Corman V.M., Liu H., Yuan M., Wu N.C., Zhu X., Lee C.C.D., et al. A therapeutic non-self-reactive SARS-CoV-2 antibody protects from lung pathology in a COVID-19 hamster model. Cell. 2020;183:1058–1069.e19. doi: 10.1016/j.cell.2020.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A., Sigworth F.J., Tagare H.D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D., Afonine P.V., Baker M.L., Bunkóczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J., et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F.W., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Liu X., Luongo C., Matsuoka Y., Park H.S., Santos C., Yang L., Moore I.N., Afroz S., Johnson R.F., Lafont B.A.P., et al. A single intranasal dose of a live-attenuated parainfluenza virus-vectored SARS-CoV-2 vaccine is protective in hamsters. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2109744118. e2109744118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusvarghi S., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B.E., de Oliveira T., Vermeulen M., van der Berg K., et al. SARS-CoV-2 Omicron neutralization by therapeutic antibodies, convalescent sera, and post-mRNA vaccine booster. bioRxiv. 2021 doi: 10.1101/2021.12.22.473880. Preprint at. [DOI] [Google Scholar]
- Mannar D., Saville J.W., Zhu X., Srivastava S.S., Berezuk A.M., Tuttle K.S., Marquez A.C., Sekirov I., Subramaniam S. SARS-CoV-2 Omicron variant: antibody evasion and cryo-EM structure of spike protein-ACE2 complex. Science. 2022;375:760–764. doi: 10.1126/science.abn7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miguez-Rey E., Choi D., Kim S., Yoon S., Săndulescu O. Monoclonal antibody therapies in the management of SARS-CoV-2 infection. Expert Opin. Invest. Drugs. 2022;31:41–58. doi: 10.1080/13543784.2022.2030310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., Bolland W.H., Porrot F., Staropoli I., Lemoine F., et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2022;602:671–675. doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- Prabakaran P., Chen W., Singarayan M.G., Stewart C.C., Streaker E., Feng Y., Dimitrov D.S. Expressed antibody repertoires in human cord blood cells: 454 sequencing and IMGT/HighV-QUEST analysis of germline gene usage, junctional diversity, and somatic mutations. Immunogenetics. 2012;64:337–350. doi: 10.1007/s00251-011-0595-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu P., Faraone J., Evans J.P., Zou X., Zheng Y.M., Carlin C., Bednash J.S., Lozanski G., Mallampalli R.K., Saif L.J., et al. Neutralization of the SARS-CoV-2 omicron BA.4/5 and BA.2.12.1 subvariants. N. Engl. J. Med. 2022;386:2526–2528. doi: 10.1056/nejmc2206725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938;27:493–497. [Google Scholar]
- Rohou A., Grigorieff N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H.W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Shrestha L.B., Foster C., Rawlinson W., Tedla N., Bull R.A. Evolution of the SARS-CoV-2 omicron variants BA.1 to BA.5: implications for immune escape and transmission. Rev. Med. Virol. 2022;32:e2381. doi: 10.1002/rmv.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Czudnochowski N., Liu Z., Zatta F., Park Y.J., Addetia A., Pinto D., Beltramello M., Hernandez P., Greaney A.J., et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature. 2021;597:97–102. doi: 10.1038/s41586-021-03807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao K., McAuliffe J., Vogel L., Fahle G., Fischer S., Tatti K., Packard M., Shieh W.J., Zaki S., Murphy B. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J. Virol. 2004;78:3572–3577. doi: 10.1128/jvi.78.7.3572-3577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashita E., Kinoshita N., Yamayoshi S., Sakai-Tagawa Y., Fujisaki S., Ito M., Iwatsuki-Horimoto K., Chiba S., Halfmann P., Nagai H., et al. Efficacy of antibodies and antiviral drugs against covid-19 omicron variant. N. Engl. J. Med. 2022;386:995–998. doi: 10.1056/NEJMc2119407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., et al. Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa. Nature Meicine. 2022;28:1785–1790. doi: 10.1038/s41591-022-01911-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telenti A., Arvin A., Corey L., Corti D., Diamond M.S., García-Sastre A., Garry R.F., Holmes E.C., Pang P.S., Virgin H.W. After the pandemic: perspectives on the future trajectory of COVID-19. Nature. 2021;596:495–504. doi: 10.1038/s41586-021-03792-w. [DOI] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westendorf K., Žentelis S., Wang L., Foster D., Vaillancourt P., Wiggin M., Lovett E., van der Lee R., Hendle J., Pustilnik A., et al. LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. bioRxiv. 2022 doi: 10.1101/2021.04.30.442182. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B.E., de Oliveira T., Vermeulen M., van der Berg K., et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- Wilhelm A., Widera M., Grikscheit K., Toptan T., Schenk B., Pallas C., Metzler M., Kohmer N., Hoehl S., Helfritz F.A., et al. Reduced neutralization of SARS-CoV-2 omicron variant by vaccine sera and monoclonal antibodies. medRxiv. 2021 doi: 10.1101/2021.12.07.21267432. Preprint at. [DOI] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasoba D., Kimura I., Nasser H., Morioka Y., Nao N., Ito J., Uriu K., Tsuda M., Zahradnik J., Shirakawa K., et al. Virological characteristics of SARS-CoV-2 BA.2 variant. bioRxiv. 2022 doi: 10.1101/2022.02.14.480335. Preprint at. [DOI] [Google Scholar]
- Yewdell J.W. Antigenic drift: understanding COVID-19. Immunity. 2021;54:2681–2687. doi: 10.1016/j.immuni.2021.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Collier A.Y., Rowe M., Mardas F., Ventura J.D., Wan H., Miller J., Powers O., Chung B., Siamatu M., et al. Comparable neutralization of the SARS-CoV-2 omicron BA.1 and BA.2 variants. medRxiv. 2022 doi: 10.1101/2022.02.06.22270533. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Liu H., Wu N.C., Wilson I.A. Recognition of the SARS-CoV-2 receptor binding domain by neutralizing antibodies. Biochem. Biophys. Res. Commun. 2021;538:192–203. doi: 10.1016/j.bbrc.2020.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Boyd L.F., Schuck P. Measuring protein interactions by optical biosensors. Curr. Protoc. Protein Sci. 2017;88:20.2.1–20.2.25. doi: 10.1002/cpps.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Tada T., Dcosta B.M., Landau N.R. Neutralization of SARS-CoV-2 omicron BA.2 by therapeutic monoclonal antibodies. bioRxiv. 2022 doi: 10.1101/2022.02.15.480166. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Chen R.E., Case J.B., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Chen E.C., et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 2020;26:1422–1427. doi: 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
-
The antibody sequences reported in this paper have been deposited in the GenBank database (accession nos. OM179962-OM179997).
-
-
The ensemble cryo-EM maps of the NE12/spike trimer and NA8/spike trimer complexes were deposited to the Electron Microscopy Data Bank (EMDB) with accession numbers EMD-26401 and EMD-26403, respectively. The locally refined maps of the NE12/spike trimer and NA8/spike trimer complexes were deposited to EMDB with accession numbers EMD-26402 and EMD-26404, respectively, and the corresponding coordinates were deposited to the Protein Data Bank with accession numbers 7U9O and 7U9P.
-
-
Any additional information required to reanalyze the data reported is available from the lead contact upon request.