Abstract
An enzyme exhibiting NADH oxidase (diaphorase) activity was isolated from the hyperthermophilic sulfate-reducing anaerobe Archaeoglobus fulgidus. N-terminal sequence of the protein indicates that it is coded for by open reading frame AF0395 in the A. fulgidus genome. The gene AF0395 was cloned and its product was purified from Escherichia coli. Like the native NADH oxidase (NoxA2), the recombinant NoxA2 (rNoxA2) has an apparent molecular mass of 47 kDa, requires flavin adenine dinucleotide for activity, has NADH-specific activity, and is thermostable. Hydrogen peroxide is the product of bivalent oxygen reduction by rNoxA2 with NADH. The rNoxA2 is an oxidase with diaphorase activity in the presence of electron acceptors such as tetrazolium and cytochrome c. During purification NoxA2 remains associated with the enzyme responsible for d-lactate oxidation, the d-lactate dehydrogenase (Dld), and the genes encoding NoxA2 and Dld are in the same transcription unit. Together these results suggest that NADH oxidase may be involved in electron transfer reactions resulting in sulfate respiration.
The sulfur oxide sulfate is the most favored electron acceptor in anoxic environments where sulfate predominates. Dissimilatory sulfate reducers use sulfate as an electron sink in the generation of energy. The evolved product of sulfate reduction, H2S, can be used as a substrate for growth by other organisms or is released into the environment, where it may be involved in the detoxification or precipitation and removal of compounds such as iron in a sulfide complex (FeS).
Sulfate reducers play an integral part in the complex redox cycle for sulfur. Archaeoglobus fulgidus is a hyperthermophilic anaerobe that can use d- or l-lactate, pyruvate, or hydrogen as an energy source (34). Members of this genus are the only known sulfate reducers in the domain Archaea. As a dissimilatory sulfate reducer, A. fulgidus transfers electrons through intermediary carriers to ATP-activated sulfate to obtain energy. The enzymes from sulfate-reducing organisms involved in the later stages of sulfate respiration (ATP sulfurylase, adenylylsulfate reductase, and sulfite reductase) have been characterized (7, 32). However the mechanisms by which respiratory enzymes such as lactate dehydrogenases and NADH dehydrogenase interact with electron carriers such as cytochromes and quinones during sulfate reduction are less well known (4, 10).
To better understand the physiology involved in lactate catabolism and the transfer of electrons during anaerobic sulfate respiration in the archaeon Archaeoglobus, we previously characterized the d-lactate dehydrogenase (Dld) from A. fulgidus (28). During purification of the Dld, an enzyme with NADH oxidase (Nox) activity remained closely associated with the fractions having Dld activity. The role for NADH oxidase in A. fulgidus, a strict anaerobe, is enigmatic because these enzymes catalyze the oxidation of NADH, with subsequent electron transfer to oxygen (oxidase activity). NADH oxidases carry out the bivalent reduction of oxygen to peroxide or the tetravalent reduction of oxygen to water, and they may also transfer electrons univalently to oxygen to form superoxides (20). A subset of NADH oxidases have diaphorase activity and can donate electrons to an acceptor like cytochrome c.
NADH oxidase (EC 1.6.99.3) can be classified as an oxidoreductase, dehydrogenase, diaphorase, or cytochrome c reductase. This type of Nox acts on NADH and transfers electrons to an acceptor. In addition, NAD(P)H dehydrogenase (quinone) (EC 1.6.99.2), mitochondrial NADH oxidase (ubiquinone) (EC 1.6.5.3), and NADH peroxidase (EC 1.11.1.1) are sometimes referred to as NADH oxidases. NADH oxidase (Nox) has been characterized from the eukaryotic mitochondria and eubacteria and more recently in archaea (23). Nox enzymes purified thus far come in a wide range of molecular weights (Mr 29,000 to 215,000) as monomeric, dimeric, or hexameric forms and are often associated with flavin cofactors and iron-sulfur complexes.
NADH oxidase (Nox) activity in anaerobes may contribute to antioxidant activities. For example, during aeration the catalytic activity of Nox in Lactobacillus results in a significant accumulation of H2O2, yet Nox has a very minimal energetic role in the cell (22). This suggests that the role of Nox in this organism is to remove oxygen (22). In Streptococcus mutans growing anaerobically, expression of Nox is induced with increased oxygen levels, probably in response to oxygen toxicity (12).
Evidence exists for NADH oxidoreductase involvement in the electron transfer reactions of respiration. A 47-kDa Nox from Escherichia coli has a noncovalently bound flavin adenine dinucleotide (FAD) that catalyzes the reduction of a quinone, probably ubiquinone-8, in vivo (27). Additionally, a defect in Nox in E. coli can be complemented by the respiratory d-lactate dehydrogenase, hinting at a common role for these enzymes in respiration (37). The sequence of the hmc operon in the sulfate reducer Desulfovibrio includes a cytochrome c reductase gene, believed to be part of a protein complex involved in hydrogen oxidation and sulfate reduction (30). Desulfovibrio's flavin mononucleotide (FMN)-containing Nox can transfer electrons from NADH directly to the adenylyl phosphosulfate (APS) reductase, suggesting a role for Nox in sulfate reduction (5). Although some data indicate that the NADH oxidase in sulfate-reducing bacteria can reduce menaquinones, NADH is not generally used as the carrier of the reducing equivalents from the substrate to the electron transport chain of sulfate reducers (9, 10).
To begin to understand the type of NADH oxidase (dehydrogenase) produced by A. fulgidus, the sequence of the N-terminal end of Nox that copurifies with d-lactate dehydrogenase was obtained. Sequence showed Nox to be coded for by a gene adjacent to the gene that codes for the Dld. Together these data suggested that Dld and NoxA2 might have related roles in electron transfer reactions or that Nox acts as a shield to protect components involved in lactate metabolism from O2. Here we describe the expression of open reading frame (ORF) AF0395 (noxA2) in E. coli and biochemical characterization of NoxA2 enzyme.
MATERIALS AND METHODS
Strains, vectors, and reagents.
Archaeoglobus fulgidus VC-16 (DSM4303) was obtained from Karl Stetter (Lehrstuhl für Mikrobiologie, Universität Regensburg). E. coli strains BL21(DE3) and JM107 were used for cloning DNA and expression of recombinant protein (26, 36). E. coli strains carrying plasmids were grown in Luria-Bertani (LB) medium supplemented with kanamycin sulfate (40 μg/ml). All antibiotics and chemicals were obtained from Aldrich, Fisher, and Sigma. Restriction enzymes obtained from New England Biolabs (NEB; Beverly, Mass.) were used as recommended by the manufacturer. Vector pET24b DNA was obtained from Novagen (Madison, Wis.). PCR product was purified using the Gibco PCR purification resins (Rockville, Md.). Plasmid DNA was purified over Qiagen columns (Valencia, Calif.). Talon affinity resin for recombinant protein purification was obtained from Clonetech (Palo Alto, Calif.). Fast protein liquid chromatography (FPLC) columns and column resins were purchased from Amersham-Pharmacia (Piscataway, N.J.). Ultrafiltration cartridges and membranes were obtained from Millipore-Amicon (Beverly, Mass.).
Growth of Archaeoglobus.
To obtain protein and DNA from A. fulgidus, cells were grown anaerobically at 83°C in glass carboys. Modified sulfate-thiosulfate-lactate (STL) medium 3, growth (1), and harvest conditions are described elsewhere (11, 28). The STL buffer was modified by using 20 mM Tris and 20 mM maleic acid, adjusted to a pH of 6.1 with NaOH. The medium contained 22 mM dl-sodium lactate, 0.5 g of yeast extract per liter, basal salts, and trace minerals. The medium was reduced with 1 mM Na2S and 1 mM Na2S2O3, and 0.2% resazurin was added as a reduction indicator. A 40-liter carboy was inoculated with 500 ml of logarithmic-phase A. fulgidus cells. Growth was monitored with a Milton-Roy Spectronic 21D spectrophotometer. Following harvest of cells by ultrafiltration with an S3Y100 spiral cartridge and centrifugation, cell mass was stored under O2-free N2 gas at −70°C.
Purification of NADH oxidase.
To purify the NADH oxidase, NoxA2, from A. fulgidus, frozen cell paste was thawed and cells were washed in salt solution (50 mM Tris [pH 7.8], 5 mM KCl, 300 mM NaCl, 15 mM MgCl2, 6 mM Na2S2O3, 1 mM dl-dithiothreitol [DTT]). Cells were centrifuged under anoxic conditions at 21,000 × g for 10 min at 4°C and suspended in one-third volume of suspension buffer (5 mM DTT, 50 mM Tris, pH 7.8). Cells were lysed by passage through a cold French pressure cell at 20,000 lb/in2 three times and collected under a stream of N2 gas. Microscopic analysis showed that lysis of A. fulgidus cells was complete after passage through the French pressure cell. After centrifugation at 14,000 × g for 30 min to remove insoluble salts and debris, the supernatant was collected for subsequent protein purification.
Ammonium sulfate and streptomycin sulfate dissolved in buffer A (25 mM sodium pyruvate, 100 mM Tris, pH 8.0) were added under anoxic conditions to the supernatant to final concentrations of 35% and 5%, respectively. The mixture was incubated in an anaerobic chamber with stirring for 1 h at room temperature before ultracentrifugation at 300,000 × g for 1 h at 4°C in a Sorvall S100AT5 rotor. Proteins were precipitated from the supernatant by addition of solid (NH4)2SO4 (65% saturation) and centrifugation at 20,000 × g for 30 min at 4°C. The protein pellet was suspended in 50 mM Tris (pH 7.8) and dialyzed at 25°C for 12 h against three changes of anoxic buffer B (200 mM NaCl, 20 mM Tris, pH 8.0), each at a volume 60 times the sample volume. Dialyzed sample was passed through a 0.45-μm filter and applied under oxic conditions to a fast protein liquid chromatography (FPLC) Q Sepharose Fast Flow ion-exchange column that had been equilibrated with buffer B.
The sample was eluted stepwise from the ion exchange column with NaCl (255 to 285 mM). Ammonium acetate was added to the eluted fractions to 1 M final concentration (fc) in buffer C (50 mM Tris, pH 7.7) and applied to a phenyl-Sepharose 6 Fast Flow column. The fractions containing d-lactate dehydrogenase and Nox oxidase/diaphorase activities were eluted stepwise in 600 to 450 mM ammonium sulfate in buffer C. Proteins were concentrated by ultrafiltration using a Microcon YM10 membrane and dialyzed against three changes of buffer C, each at 400× volumes. Samples were sparged with O2-free N2 gas and stored at 4°C.
Gel analysis.
To analyze protein purity, samples were heated at 95°C for 5 min in sample buffer (18), then separated in 10% sodium dodecyl sulfate (SDS)-Tricine-polyacrylamide gels (31) and stained with Coomassie GelCode blue staining reagent according to the manufacturer's specifications (Pierce, Rockford, Ill.). Protein concentration was determined by the Bradford method (3). Bovine serum albumin (BSA) and γ-globulin were used as standards.
To detect enzyme activity, protein fractions were assayed in nondenaturing gels. Loading buffer (500 mM Tris [pH 8.8]), 10% glycerol, 2% 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate ([CHAPS], 0.001% bromophenol blue dye) was added to fractions containing protein, and samples were separated by 10% Tris-glycine-polyacrylamide gel electrophoresis (PAGE) at 25°C. Electrophoresis and subsequent enzyme assays were carried out in an anaerobic chamber (Coy).
To identify Dld and Nox activities after phenyl-Sepharose fractionation, samples were denatured in sample buffer and separated by 10% Tricine-SDS-PAGE. Enzymes were allowed to renature in the gel after three 20-min incubations at 25°C in 20 mM Tris (pH 7.5) and assayed first for Dld and then for NoxA2 activity.
Dld activity was detected in gels by the formation of blue formazan precipitate after incubation at 50°C for 60 min in 20 mM Tris (pH 7.8 at 50°C)–10 mM MgSO4–5 μM FAD–65 μM phenazine methosulfate (PMS)–240 μM 3(4,5-dimethylthiazol-2,yl)-2,5-diphenyl tetrazolium (MTT)–2 mM d-lactate.
NoxA2 enzyme activity was assayed in gels at 50°C by a slight modification of a published procedure (21). Proteins separated under nondenaturing PAGE conditions were assayed using 200 mM Tris (pH 6.8 at 50°C), 2 mM 2-methyl-1,4-naphthoquinone (menadione; dissolved in absolute ethanol), 150 μM nitro blue tetrazolium (NBT), and 470 μM NADH. Enzyme activity was detected after 30 min by the appearance of dark formazan bands. To assay the activity of Nox after separation by SDS-PAGE, protein in the gel was allowed to renature in buffer lacking SDS and then incubated in 50 mM sodium phosphate (NaPi; pH 6.6 at 50°C), 10 μM FAD, 500 μM menadione, 150 μM NBT or 240 μM MTT, and 200 μM NADH. Although menadione was typically added to the gel assay, it was not required for diaphorase activity.
N-terminal sequence.
To sequence partially purified protein from A. fulgidus exhibiting Nox activity, samples were heated to 50°C for 10 min in SDS sample buffer and separated by 10% Tricine-SDS-PAGE at 4°C. The gel was allowed to polymerize for 36 h prior to electrophoresis to eliminate compounds that might artificially modify the N terminus of proteins and inhibit sequencing. The gel was rinsed in transfer buffer (10 mM 3-[cyclohexylamino]-1-propanesulfonic acid [CAPS, pH 11.0], 10% methanol) for 20 min, and proteins were transferred by tank blotting to polyvinylidene difluoride (PVDF) nylon membrane (Immobilon; psq) in transfer buffer. The membrane was rinsed with water for 5 min, stained with 0.2% Ponceau S dye in 3% trichloroacetic acid, and destained with water for 5 min. Protein bands in the membrane were excised, and the N terminus of the protein was sequenced with an Applied Biosystems 475A protein sequencer.
A. fulgidus DNA purification and cloning of ORF AF0395.
Chromosomal DNA was prepared by suspending harvested A. fulgidus cells in 25% sucrose and 10 mM Tris (pH 7.5) and then treating the cells with one-half volume of lysis solution (5% SDS, 0.125 M EDTA, 0.5 M Tris, pH 9.4) for 1 h at 50°C (28). Lysis was complete following an overnight incubation at 37°C with pronase E (2 mg/ml). Following the addition of 0.5 M potassium acetate for 10 min at 37°C and then 1 h at 4°C, protein and cell debris were precipitated by a 15-min centrifugation (10,000 × g). DNA was spooled from the supernatant with a glass rod following the addition of 2 volumes of absolute ethanol. DNA was washed (70% ethanol, 10 mM Tris, 10 mM MgCl2, 1 mM EDTA, pH 8.0), dried, and dissolved in TE (10 mM Tris, 1 mM EDTA, pH 8.0).
A. fulgidus genomic DNA was used as the template to amplify ORF AF0395 by PCR. The oligonucleotide primers 5′GGAATTCCATATGAGGGTAGTTGTTATCG and 5′CCGCTCGAGGAATTTAAGAATTCTTGCCG included NdeI and XhoI sites (underlined) designed to generated an in-frame fusion between AF0395 and the six-histidine tag in vector pET24b. PCR amplification was performed by incubating samples containing 1× polymerase buffer, 2 mM MgCl2, 200 μM each of the four deoxynucleoside triphosphates (dNTPs), 200 nM primer (each), 2 U of Taq Gold polymerase (Perkin-Elmer), and 0.3 μg of A. fulgidus chromosomal DNA for 30 cycles (30 s at 95°C, 30 s at 55°C, and 3 min at 72°C). A single 1.3-kb PCR product was obtained. Purified PCR product and plasmid pET24b were each digested with NdeI and XhoI, extracted with phenol-chloroform, ethanol precipitated, resuspended in TE, and ligated overnight. Recombinant plasmids carrying AF0395 were recovered in E. coli JM107 after electroporation, selection on LB agar with kanamycin, and restriction analysis. The recombinant plasmid named pDR8 was used for subsequent analysis.
Induction and expression of NoxA2 protein in E. coli.
Plasmid pDR8 was transformed into E. coli strain BL21, a λDE3 lysogen containing a copy of the inducible gene for T7 RNA polymerase, to express the recombinant form of NoxA2. To determine if the NoxA2 C-terminal His6-tagged protein was produced in E. coli, strain BL21 containing pDR8 was induced with isopropyl-β-d-thiogalactopyranoside (IPTG), and cell extracts were analyzed by SDS-PAGE for the appearance of a 48-kDa protein.
To maximize expression of the fusion protein, an isolated colony of BL21 carrying pDR8 was incubated overnight at 37°C in LB-kanamycin, diluted 1:1,000 in LB-kanamycin, and grown at 37°C in a flask with shaking at 190 rpm until the optical density at 600 nm reached 0.45 to 0.5 (about 4 h). IPTG was added to a final concentration of 1 mM, incubated for 3 h at 37°C, and harvested by centrifugation at 3,000 × g for 15 min, and the cell pellet was suspended in 20 mM Tris (pH 7.5) and stored at −70°C.
Purification of recombinant NoxA2.
The 48-kDa recombinant NoxA2-His6 protein (rNoxA2) was purified from E. coli using the Talon-immobilized metal affinity chromatography resin. Frozen cell suspensions in Tris buffer were quickly thawed, an equal volume of cold lysis buffer (100 mM NaCl, 20 mM Tris, pH 8.0) was added, and cells were lysed by three passages through a French pressure cell. Cell debris was removed by ultracentrifugation at 300,000 × g for 15 min at 4°C, and resin in lysis buffer (12.5 volumes) was added to the supernatant. The sample was gently agitated on a rocker for 20 min at room temperature and centrifuged at 700 × g for 2 min. Twelve bed volumes of lysis buffer were added to the rNoxA2 protein-bound resin, and the sample was agitated for 10 min at 25°C and centrifuged at 700 × g for 5 min. This step was repeated. The resin was suspended in 8 bed volumes of lysis buffer, transferred to a gravity flow column, and washed two times with 6 bed volumes of lysis buffer, and the recombinant protein was eluted with 100 mM NaCl, 50 mM imidazole, and 20 mM Tris (pH 8.0). The eluate was concentrated (to 1.5 to 2 mg/ml) by ultrafiltration with a Microcon YM10 membrane and washed in 25 mM NaPO4 (pH 6.0) with 50 mM NaCl, and the rNoxA2 histidine-tagged protein was stored in the same buffer at 4°C or in 25% glycerol at −70°C.
Spectrophotometric enzyme assays.
Enzyme activity was assayed in quartz cuvettes (Starna) on a dual-beam Perkin Elmer Lambda 12 UV/VIS spectrophotometer equipped with a PTP-6 temperature block and supported by a Dell Optiplex XMT590 work station with UV-Winlab software. Absorption spectra were taken from 200 to 900 nm.
To measure the oxidation of NADH to NAD+ by rNoxA2, samples (0.5 ml) were monitored at 340 nm for a decrease in absorbance of NADH. The molar amount of NADH oxidized was determined by using the molar extinction coefficient ɛ340 = 6220 M−1 cm−1. Aerobic assay buffer contained 50 mM NaPi (pH 7.6, measured at 55°C), 0.2% CHAPS, 5 μM FAD, 100 μM NADH, and 0.25 to 1 μg of protein. The 7-mg/ml (10 mM) NADH stock solution was prepared fresh in 10 mM Tris (pH 7.5), kept on ice, and protected from light.
To identify potential electron acceptors for rNoxA2, spectrophotometric assays were performed under anoxic conditions. Reagents prepared in O2-free water were added to cuvettes in an anaerobic chamber, stoppered, and removed from the chamber. Anoxic NADH was added to the reaction mix with a gas-tight syringe and immediately placed in the preheated spectrophotometer to initiate the reaction. Each sample contained 1 μg of NoxA2 in 50 mM NaPi (pH 7.0)–200 μM NADH–5 μM FAD–0.5% CHAPS–100 μM each of the potential electron acceptors. The ability of rNoxA2 to reduce the acceptors was monitored at the following wavelengths: MTT, 578 nm; dimethylnaphthoquinone (DMN), 270 nm (24); dichlorophenolindophenol (DCIP), 600 nm; menadione, 340 nm; K3Fe(CN)6, 420 nm; and cytochrome c, 550 nm. The ability of rNoxA2 to oxidize and reduce H2O2, NaNO2, NaNO3, and Na2SO4 was determined by observing the oxidation of NADH at 340 nm.
Cofactor analysis.
Purified rNoxA2 was examined spectrophotometrically to identify potential cofactors. Cofactor was released from the protein and identified by ascending paper chromatography analysis (14). Cofactor was extracted from rNoxA2 (7 to 60 μg) using 10% trichloroacetic acid (TCA) (17), heat, or hot methanol (33). Extracts containing the cofactor and controls (200 μM FAD, 200 μM FMN, and 300 μM riboflavin) were spotted on Whatman no. 1 paper or Silica Gel 60 plates and allowed to dry. Samples were chromatographed in the dark with 5% Na2HPO4, n-butanol–acetic acid–H2O (4:5:1) or n-butanol–acetic acid–H2O (12:3:5), and analyzed with long-UV (365 nm).
To determine the molar ratio of FAD associated with rNoxA2, 530 μg of rNoxA2-His6 (estimated molecular mass, 48.6 kDa) was incubated with excess FAD (320 μM) in buffer D (50 mM NaCl, 25 mM NaPi, pH 6.0) for 18 h at 4°C. Unassociated FAD was removed by dialysis against buffer D (five changes of 200 volumes each). The absorption of a control without enzyme was subtracted from the absorption of the sample as measured in a spectrophotometer.
To determine the specificity and requirement for flavin, NoxA2 was denatured at 95°C for 4 min, and cofactor was separated from apoprotein following electrophoresis in 10% Tricine-SDS-PAGE. Enzyme was allowed to renature in the gel as described above and assayed in the gel at 55°C alone and in the presence of 30 μM each of the flavins FAD, FMN, riboflavin, and deazaflavin F420.
NADH oxidase, H2O2, and cytochrome c assays.
To determine if oxygen is a terminal electron acceptor for the oxidation of NADH by NoxA2, rNoxA2 (190 μg) was incubated with FAD and then dialyzed to remove excess FAD (see above). The sample was flushed with a stream of O2-free N2 gas for 1 h to remove oxygen from the sample, the volume was adjusted for loss of water due to evaporation, and the absorbance from 250 to 550 nm was measured in an anaerobic cuvette at 25°C. Anoxic NADH (400 μM) was added using a gas-tight syringe, and the temperature was increased to 55°C. After monitoring the absorbance for 30 min, the stopper was removed and the sample was stirred to introduce O2. After 10 min, spectral readings were taken.
To determine if hydrogen peroxide is a product in the NoxA2 oxidation reaction (O2 → H2O2), NADH (50 μM) was oxidized at 55°C by rNoxA2 (10 μg) in 50 mM NaPi buffer (pH 6.6) with 0.5% CHAPS and 20 μM FAD. Following oxidation of NADH, 2.5 U of horseradish peroxidase (EC 1.11.1.7), dissolved in 50 mM NaPi (pH 7.0), was added with 50 mM (fc) sodium acetate (pH 5.0) and 500 μM (fc) 3′,3′,5′,5′-tetramethylbenzidine (TMBZ; dissolved in methanol) (21). The mixture was incubated at 25°C and monitored at 650 nm for peroxidase activity (H2O2 → H2O) by the formation of the oxidized blue-green dye. We chose 650 nm because oxidized FAD and NAD+ do not interfere with the measurements at this wavelength. To show that TMBZ did not serve as a direct electron acceptor for rNoxA2 oxidation, it was included in a control assay without horseradish peroxidase.
Cytochrome c was tested as a terminal electron acceptor for NADH oxidation by rNoxA2. Reagents were prepared under anaerobic conditions, and the reaction proceeded as described above. Cytochrome c from horse heart and Saccharomyces cerevisiae was added to the rNoxA2 (1 μg) solution, and cytochrome c reduction was monitored as an increase in absorption at 550 nm.
Yeast two-hybrid analysis.
S. cerevisiae strain PJ69-4A (MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ) is Ura−, Met−, and Lys+ but His+, Ade+, and LacZ+ in the presence of functional GAL4 (15). Plasmids pDR6 and pDR11 were engineered to express d-lactate dehydrogenase as a fusion to the GAL4 binding domain (BD) and NoxA2 as a fusion to the GAL4 activation domain (AD), respectively. To confirm the protein-protein interaction, reciprocal two-hybrid tests were performed. The dld and noxA2 fragments in the pDR6 and pDR11 vectors were removed after digestion with BglII and EcoRI purified from an agarose gel, and subcloned into pGAD and pGBD vectors, respectively, digested with the same enzymes to produce pDR9 (AD-Dld fusion) and pDR10 (BD-Nox fusion).
Plasmids were transformed into PJ69-4A using the method of Gietz (8) and maintained in SC medium lacking leucine, tryptophan, and histidine (SC-Leu-Trp-His) (Technical Tips Online, http://tto.trends.com). Colonies that grew on SC-Leu-Trp-His were transferred to SC-Leu-Trp-Ade to screen for GAL2-ADE2 reporter activity. White colonies that grew on the SC medium without Leu, Trp, and Ade were tested for β-galactosidase activity using the flash-freezing filter assay.
The yeast strains that were His+, Ade+, and LacZ+ were grown in 5 ml of SC without Leu, Trp, and His at 30°C. Plasmid DNA was purified by the method of Rose et al. (29) and used to transform a naive PJ69-4A to His+ Ade+ to confirm the interaction between the BD fusion and the AD fusion.
RESULTS
Archaeoglobus species, in the domain Archaea, are the only hyperthermophiles that can use sulfate as the terminal electron acceptor to generate energy. To better understand the processes involved in the initial stages of sulfate respiration, enzymes involved in electron capture have been studied. The d-lactate dehydrogenase (Dld) coded for by AF0394 (dld) is the enzyme responsible for d-lactate catabolism leading to the eventual transfer of electrons to sulfate. Analysis of the A. fulgidus genome indicates that dld is a third gene of five that likely comprise an operon. AF0395, the gene adjacent to the gene encoding Dld, is predicted to encode a 47-kDa NADH oxidase (NoxA2).
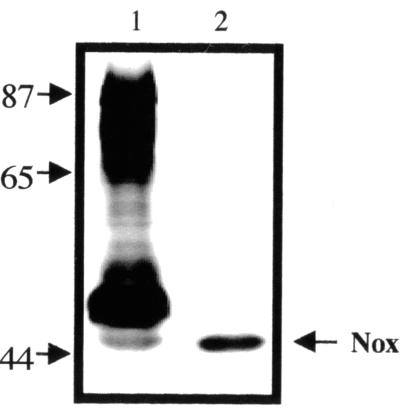
To determine if A. fulgidus encodes an NADH oxidase, cell extracts were separated by nondenaturing gel electrophoresis and assayed for activity with NADH as the electron donor and NBT as the artificial electron acceptor. Multiple proteins, ranging in size from about 45 to 90 kDa, had NADH oxidase activity (Fig. 1, lane 1). This is consistent with the fact that the A. fulgidus genome includes at least eight genes that code for proteins having identity with NADH oxidases. A least four nox genes (noxA-1, noxA-2, noxA-4, and noxA-5) code for proteins of about 48 kDa, noxA-3 codes for a 60-kDa protein, and noxB-1 and noxB-2 code for 68-kDa proteins. The noxC gene codes for a 19-kDa protein, which was not detected in our gel assay. Because dld and noxA2 are transcribed together (V. Pagala and P. Hartzell, unpublished results), NoxA2 and Dld might function in the same energy-yielding pathway. Indeed, a 47-kDa protein with Nox activity was present in samples of partially purified Dld from A. fulgidus after salt precipitation and two chromatographic steps (Fig. 1, lane 2).
FIG. 1.
Purified NoxA2 activity. Samples were separated by nondenaturing polyacrylamide and assayed for NADH oxidase (diaphorase) activity with Tris, menadione, NBT, and NADH. Activity was detected by the appearance of dark formazan precipitate. Lanes: 1, French-pressed cells (20 μg); 2, phenyl-Sepharose-purified fraction (0.16 μg). Sizes are shown in kilodaltons.
N-terminal sequence of NADH oxidase.
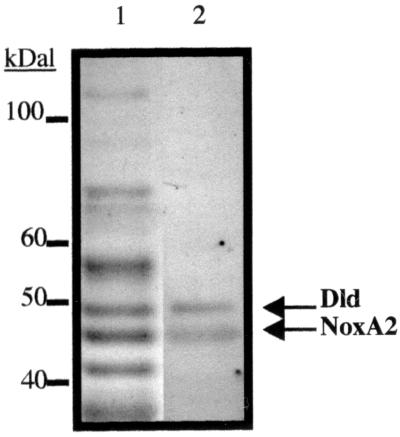
Column fractions containing Nox were identified after gel electrophoresis and treatment with NBT or MTT to assay for activity. Polyacrylamide gel strips stained by Coomassie dye or with enzyme assay reagents were treated for the same time period to preserve the position of each protein. Activities for Dld and Nox correlated precisely with two distinct protein bands in the Coomassie-stained gel lane (Fig. 2). The 47-kDa band with Nox activity was targeted for sequencing.
FIG. 2.
Purification and comigration of Dld and NoxA2 enzyme activities. Lane 1, partially purified sample from A. fulgidus obtained after phenyl-Sepharose fractionation (3.3 μg) was separated by SDS–10% PAGE and stained with Coomassie dye. Lane 2, the same phenyl-Sepharose fraction was incubated in Tris (pH 7.5) and then assayed with Tris, MgSO4, PMS, MTT, and D-lactate to detect Dld activity and with NaPi, menadione, NBT, and NADH to detect NoxA2 activity.
To identify the gene coding for the 47-kDa Nox, partially purified Nox from the phenyl-Sepharose column was separated in a Tricine gel and transferred to a PVDF membrane. The protein band on the membrane was excised, and the sequence of amino acids at the amino terminus was determined. Comparison of the sequence (MxVVVIxGGAA) against the A. fulgidus genome indicated that only two ORFs, AF0395 and AF0400, both coding for putative NADH oxidases, fit the N-terminal sequence profile. The 436-amino-acid AF0395 is predicted to yield a 48-kDa protein, whereas the 551-amino-acid AF0400 is predicted to yield a 60-kDa protein. Because the estimated mass of the partially purified Nox is about 47 kDa, we propose that the Nox protein that copurifies with Dld is the product of AF0395.
Expression of AF0395 and purification of NoxA2 from E. coli.
To confirm that AF0395 codes for a protein with Nox activity, the gene was expressed in E. coli. The 1,300-bp AF0395 gene was amplified by PCR using chromosomal DNA from A. fulgidus as the template and cloned into pET24b to generate plasmid pDR8. This construct is expected to generate a 48-kDa protein (NoxA2-His6) that includes six His residues at the carboxyl end of the protein.
The NoxA2-His6 protein was expressed in E. coli strain BL21 following induction with IPTG and separated by SDS-PAGE. A protein with an apparent mass of 48 kDa was clearly evident in the induced sample compared to the noninduced sample. When the E. coli growth medium was augmented with 6.3 μM riboflavin, trace minerals required for A. fulgidus growth (see above), and additional elements [33 μM each of ZnCl2, MgSO4, CuCl3, Fe(NH4)2(SO4)2, BeSO4, and CaCl2], the 48-kDa Coomassie-stained protein band in the polyacrylamide gel was significantly more intense. We attribute this increase to mean that standard LB medium may be limiting in a cofactor or element that is needed for production of large amounts of Nox.
The 48-kDa NoxA2-His6 protein was purified to homogeneity by Talon resin affinity chromatography. To determine if the recombinant protein had NADH oxidase activity, samples were electrophoresed under nondenaturing conditions and assayed at 55°C with NADH and NBT. The appearance of a single intense band of activity indicates that AF0395 codes for a protein that has NADH oxidase (diaphorase) activity similar to the purified Nox from A. fulgidus.
NoxA2 requires FAD for activity.
Some NADH oxidases are flavin-containing enzymes, including the NoxA2-related Enterococcus faecalis 10C1 FAD- and Desulfovibrio vulgaris FMN-containing NADH oxidases (5, 20). Purified rNoxA2 has absorption maxima at 373 and 451 nm with a shoulder at 480 nm, spectral features which are characteristic of proteins with bound flavin cofactors.
The flavin cofactor associated with rNoxA2 was identified by thin-layer chromatography. The rNoxA2 cofactor was extracted from the protein by boiling, hot methanol, or TCA treatment and compared with standards by ascending paper or silica gel chromatography. As shown in Table 1, the R value of the NoxA2 cofactor was most similar to that of FAD, which shows that the active form of rNoxA2 contains an FAD cofactor that is associated noncovalently with the protein.
TABLE 1.
Rf values of flavins by ascending thin-layer chromatographya
| Solvent system |
Rf value
|
|||
|---|---|---|---|---|
| Flavin standard
|
rNoxA2 cofactor | |||
| FMN | Riboflavin | FAD | ||
| A | 0.56 | 0.29 | 0.39 | 0.41 |
| B | 0.29 | 0.48 | 0.11 | 0.13 |
| C | 0.40 | 0.63 | 0.25 | 0.23 |
Samples were separated on Whatman no. 1 paper for solvents A and B and on Silica Gel 60 plates for solvent C. Cofactor was released by TCA extraction for rNoxA2 (30 μg) for solvent A, heat extraction from rNoxA2 (60 μg) for solvent B, and hot methanol extraction from rNoxA2 (7 μg) for solvent C. Solvent system A was 5% disodium hydrogen phosphate in H2O. Solvent system B was n-butanol–acetic acid–H2O (4:1:5). Solvent system C was n-butanol–acetic acid–H2O (12:3:5).
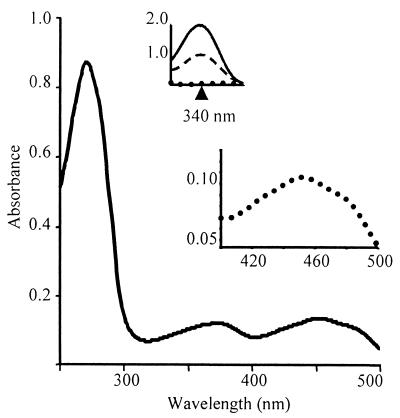
Initial stoichiometric measurements showed that rNoxA2 had about 0.35 mol of FAD per mol of protein, based on ɛ450 = 11,300 M−1 cm−1 for FAD. Because the stoichiometry is expected to be ≥1, either the flavin was not fully oxidized or a portion of the sample contained rNoxA2 apoprotein. To determine if rNoxA2 could bind more FAD, purified protein was incubated aerobically with excess FAD and then dialyzed to remove unbound FAD. A sample of FAD without protein was dialyzed in parallel to establish a baseline. When the concentration of FAD was determined for the treated rNoxA2, the stoichiometry was calculated to be 1.1 mol of FAD per mol of NoxA2 (Fig. 3).
FIG. 3.
Absorption spectra of rNADH oxidase A2. Purified rNoxA2 (190 μg) was incubated with 25 mM NaPi (pH 6.0) and 50 mM NaCl with 320 μM FAD and then dialyzed in the same buffer to remove unbound FAD. The spectrum shows peaks characteristic of FAD at 373 and 451 nm and a shoulder at 480 nm. The ratio of FAD to rNoxA2 was 1.1:1. (Insets) Redox peaks of FAD (lower) at 451 nm and NADH (upper) at 340 nm. Dotted line, absorption spectrum of enzyme (190 μg) following flushing with nitrogen gas; solid line, spectrum of rNoxA2 following addition of NADH (400 μM); dashed line, spectrum of rNoxA2 following the mixing of oxygen with the enzyme.
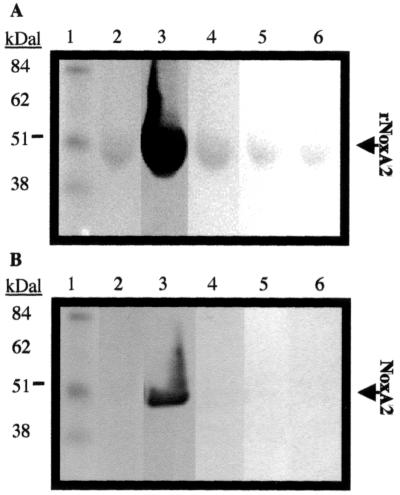
To determine if FAD is required for the function of rNoxA2 and if FAD is the only flavin required for activity of NoxA2 in A. fulgidus, apoprotein was assayed in the presence and absence of different flavins. Purified rNoxA2 and partially purified NoxA2 from A. fulgidus were denatured with heat and SDS to remove bound cofactor and separated by PAGE under denaturing conditions. Following electrophoresis, the proteins in the gel were allowed to renature with buffer or buffer plus deazaflavin F420, FMN, riboflavin, or FAD and assayed for activity (Fig. 4). Both NoxA2 and rNoxA2 were active only after renaturation in the presence of FAD.
FIG. 4.
Reconstitution of active NADH oxidase after extraction of cofactor. (A) rNoxA2 (40 μg) and (B) NoxA2 (13 μg, total protein) were electrophoresed under denaturing conditions, renatured, and then assayed in the presence of cofactors for Nox activity. Activity was observed by the formation of a blue formazan precipitate from MTT. Lanes: 1, benchmark prestained protein standards; 2, no cofactor; 3, FAD; 4, F420; 5, FMN; 6, riboflavin.
rNoxA2 is an FAD-dependent NADH oxidase.
To show that rNoxA2 is an NADH oxidase using oxygen as a terminal electron acceptor, an O2-free NoxA2 sample was assayed at 55°C for FAD reduction at 450 nm and NADH oxidation at 340 nm before and after addition of saturating NADH. As indicated in Fig. 3 (insets), the addition of NADH resulted in rapid reduction of the FAD cofactor, which shows that electrons are transferred from NADH to FAD in the absence of oxygen. Following the introduction of O2 to the sample and regeneration of FAD, the absorbance at 340 nm decreased due to the oxidation of NADH, which was present in excess. These results show that the FAD moiety of NoxA2 can accept electrons from NADH and transfer them to O2.
In early experiments, MTT was used as the terminal electron acceptor to confirm that FAD is required for enzymatic activity of both rNoxA2 and NoxA2. To show that the FAD moiety of NoxA2 is required when O2 is the terminal electron acceptor, we monitored electron flow from NADH to O2 using apoNoxA2 and NoxA2 with FAD, FMN, or riboflavin cofactors. Assays showed that although FMN and riboflavin increased oxidase activity slightly above that in the no-flavin sample, only FAD significantly restored the oxidase activity to apoNoxA2.
H2O2 is the product of NADH oxidation.
To identify the end product of NADH oxidation, NoxA2 and NADH and control reactions without enzyme or NADH were incubated at 55°C in the presence of O2 and then assayed with horseradish peroxidase. If NADH-dependent reduction of O2 by NoxA2 yields H2O2, the chromogenic substrate TMBZ will be oxidized upon the addition of horseradish peroxidase at 25°C. The change in absorbance of TMBZ at 650 nm showed that H2O2 is an end product of rNoxA2 under aerobic conditions. An assay of rNoxA2 using TMBZ as the sole electron acceptor failed to show a color change, showing that rNoxA2 did not transfer electrons directly to TMBZ.
rNoxA2 has diaphorase activity, including affinity to cytochrome c.
Like the native NoxA2 from A. fulgidus, the recombinant NoxA2 has diaphorase activity. Under anaerobic conditions, NBT, MTT, DCIP, potassium ferricyanide [K3Fe(CN)6], menadione, and cytochrome c each served as a terminal electron acceptor during oxidation of NADH. MTT and K3Fe(CN)6 served as electron acceptors even in the presence of oxygen, suggesting that some factors may serve as better electron acceptors than oxygen. The addition of H2O2 (50 μM) to the diaphorase reaction did not affect the rate of reduction for MTT and K3Fe(CN)6, implying that these acceptors receive electrons directly from the enzyme cofactor.
A. fulgidus produces a single, membrane-associated c-type cytochrome (D. Reed, V. Reddy Pagala, K. Kashefi, and P. Hartzell, submitted for publication). To determine if the A. fulgidus cytochrome c might serve as an electron acceptor during oxidation of NADH, commercially available c-type cytochromes were tested. The S. cerevisiae cytochrome c served as an electron acceptor only in the absence of oxygen, and it was necessary to remove all traces of oxygen from the sample. However, horse heart cytochrome c was reduced even in the presence of O2, albeit at a slightly lower rate than in the absence of O2. These results suggest that in vivo, cytochrome c may serve as an electron acceptor for NoxA2. NADH oxidation by rNoxA2 was not detected for the alternative potential electron acceptors DMN, F420, H2O2, NaNO3, NaNO2, and Na2SO4.
Kinetic analysis of rNoxA2.
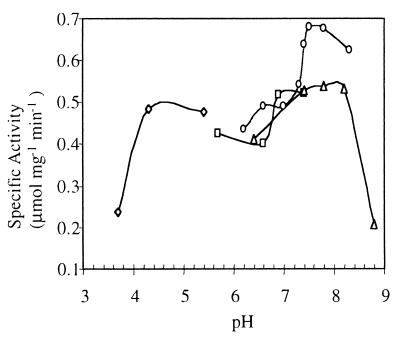
NADH (NADPH) oxidation by rNoxA2 at 55°C was monitored at 340 nm. Using oxygen as an electron acceptor, β-NADH but not β-NADPH served as an electron donor in A. fulgidus. The maximum catalytic activity of the enzyme was observed in NaPi buffer at pH 7.6 (Fig. 5). Increasing the ionic strength of phosphate from 10 to 100 mM increased rNoxA2 activity only slightly. The anionic detergent SDS (0.5%) completely inhibited the reaction, whereas sodium deoxycholate (0.5%) slightly decreased activity. The nonionic detergents Tween 20 (1.25%), Triton X-100 (0.1 to 0.5%), and n-dodecyl-β-d-maltoside (0.5%) and the zwitterionic detergent CHAPS (0.2 to 0.5%) each increased activity about twofold.
FIG. 5.
Effect of pH and buffers on rNoxA2 activity. rNoxA2 (1 μg) was assayed spectrophotometrically at 340 nm with 50 mM buffer, 0.5% CHAPS, 5 μM FAD, and 100 μM NADH at 55°C. The pH of each buffer was measured at 55°C. Buffers tested included sodium acetate (◊), Tris maleate (□), NaPi (○), and Tris-HCl (▵).
To identify ions that might stimulate NoxA2 activity, rNoxA2 was incubated with 10 mM CuCl3, K3Fe(CN)6, CaCl2, ZnCl2, CoCl2, MnCl2, Na2B4O7, Na2MoO4, AlCl3, NiCl2, Na2WO4, MgCl2, KCl, NaCl, CsCl2, BeSO4, Cd(C2H3O2)2, NaSeO4, EGTA, or EDTA for 10 min at 25°C and then diluted to 100 μM (fc) in 50 mM Tris (pH 7.8). NADH oxidation was stimulated three- to fourfold when CuCl3 or K3Fe(CN)6, which can serve as an electron acceptor for diaphorases, was added to the sample. EGTA, CaCl2, ZnCl2, CoCl2, MnCl2, and Cd(C2H3O2)2 each slightly increased activity. EDTA did not appear to inhibit activity, suggesting that NoxA2 does not require a metal cofactor for activity.
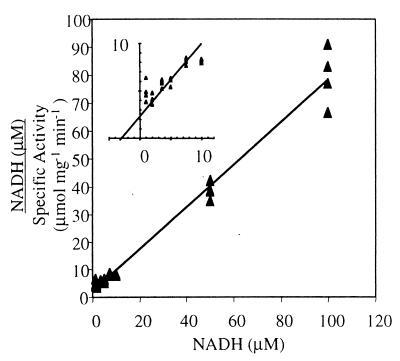
To characterize the catalytic properties of NoxA2, Km and Vmax values were measured. The Km was determined by plotting initial rates against the substrate concentration using the linear Hanes-Woolf plot [S]/v versus [S] (Fig. 6). The Km of rNoxA2 for NADH was 3.1 μM at 55°C, and the Vmax was estimated to be 1.3 μmol mg−1 min−1.
FIG. 6.
Hanes-Woolf plot of rNoxA2-catalyzed oxidation of NADH shows Michaelis-Menten kinetics. rNoxA2 (0.2 μg) was assayed spectrophotometrically at 340 nm with 50 mM NaPi (pH 7.6), 0.2% CHAPS, 5 μM FAD, and 0 to 500 μM NADH at 55°C. A Km of 3 μM for NADH was determined from the plot at 98% confidence. NADH approached saturation for rNoxA2 near 10 μM. (Inset) Expanded plot near the y intercept. x intercept = Km (3.1 μM); slope = Vmax−1 (1.3 μmol mg−1 min−1).
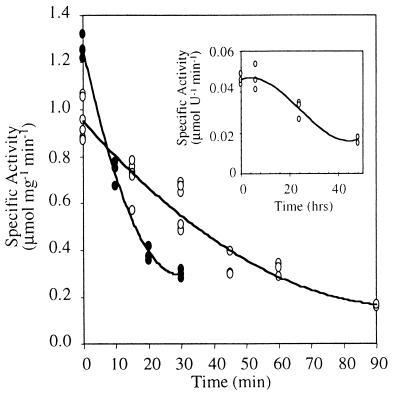
Because A. fulgidus grows at 83°C, the recombinant NoxA2 was incubated at this temperature over time to determine its half-life. rNoxA2 retained activity after a 40-min incubation in phosphate buffer at pH 7.6, but when incubated in Tris buffer at pH 7.8, the half-life of the recombinant enzyme was only 12 min (Fig. 7). In contrast, the partially purified native NoxA2 had a half-life of 35 h (Fig. 7, inset). Attempts to stabilize rNoxA2 with FAD, NADH, glycerol, CHAPS, or salt [(NH4)2SO4 and NaCl] were unsuccessful.
FIG. 7.
Thermal stability of the recombinant form of NoxA2. rNoxA2 was incubated at 83°C in 50 mM NaPi (pH 7.6, as determined at 55°C) (open circles) or 50 mM Tris (pH 7.8, as determined at 55°C) (solid circles). Aliquots of rNoxA2 (0.5 μg) were removed and monitored at 340 nm for NADH oxidation in 50 mM NaPi (pH 7.6), 0.2% CHAPS, 20 μM FAD, and 100 μM NADH at 55°C. Half-life of rNox following incubation in buffer was estimated to be 40 min in NaPi and 12 min in Tris. (Inset) NoxA2 (phenyl-Sepharose fraction) from A. fulgidus was incubated at 83°C in NaPi prior to assaying as explained above. One unit equals 1 μg of total protein; the half-life of NoxA2 averaged 35 h.
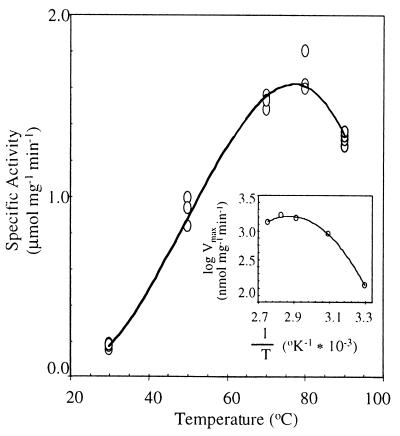
The temperature for maximum catalytic activity of rNoxA2 was near 80°C, the optimal temperature of growth for A. fulgidus (Fig. 8). The enzyme is active from room temperature to 90°C. At 50°C, the activity of rNoxA2 was half that at 80°C, with the activation energy increasing 19.2 kJ/mol in that range.
FIG. 8.
Effect of temperature on rNoxA2 activity. The activity of rNoxA2 (1 μg) was monitored at 340 nm for NADH oxidation at the temperatures indicated. The enzyme was assayed in 50 mM NaPi (pH 7.6, as determined at 55°C), 0.2% CHAPS, 20 μM FAD, and saturating 200 μM NADH. (Inset) An average of maximum velocities determined by the method of Arrhenius.
Interaction between NADH oxidase and d-lactate dehydrogenase.
The association of NoxA2 with Dld during the purification of Dld hints that these proteins interact in vivo. To test this idea independently, the yeast two-hybrid system was used to check for protein-protein interactions. The yeast strain PJ-694A, constructed by James et al. (15), has been designed to eliminate the risk of false-positive tests by placing the HIS, ADE, and lacZ genes under control of a GAL4-dependent promoter. Transformants that are able to grow in medium lacking His and Ade likely carry fusion proteins that interact to produce active GAL4 protein and can be tested further for β-galactosidase activity. When pDR6 and pDR11 were introduced into PJ-694A, the transformants were able to grow on medium lacking His and Ade. Similarly, pDR10 and pDR9 also passed this initial screen. Controls, pDR6 plus pGAD and pDR11 plus pGBD, failed to grow on medium without His and Ade. Yeast carrying pDR6 and pDR11 produced pale blue colonies on yeast medium with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). The positive controls, pGAD-Fos and pGBD-Jun, produced strong blue colonies on X-Gal-containing medium. These results show that Dld and NoxA2 interact, albeit weakly. Yeast carrying pDR6 (GBD-Dld) and pDR9 (GAD-Dld) also showed a weak interaction, which suggests that Dld interacts with itself. This is consistent with earlier observations that some Dld migrates as a dimer on a Superose gel filtration column. Attempts to show an interaction between NoxA2 and the A. fulgidus c-type cytochrome (the product of ORF AF503) in the yeast two-hybrid system were unsuccessful.
A series of coupled enzyme assays were done to determine if the interaction between Dld and NoxA2 has a biochemical basis. Addition of rNoxA2 to a reaction designed to measure the reduction of NBT by Dld in the presence of d-lactate stimulated the rate of NBT reduction twofold. Stimulation was independent of NAD(H), which suggests that stimulation has a physical rather than a biochemical basis. When Dld alone or with d-lactate or pyruvate was added to a NoxA2 assay, no change in the activity for Nox was detected.
DISCUSSION
During dissimilatory sulfate respiration, electrons are removed from reduced substrates such as d- and l-lactate and eventually are passed to oxidized sulfur compounds, such as sulfate, to generate a proton motive force. The d-lactate dehydrogenase (Dld) transfers electrons from d-lactate to its FAD cofactor to other intermediate carriers. To begin to identify proteins that might interact with Dld during electron transfer, we examined a protein that remains associated with Dld during purification. N-terminal sequence of this protein matches two proteins, both putative NADH oxidases, encoded by AF395 and AF400 on the A. fulgidus genome. The protein that copurifies with Dld is about 48 kDa. AF0395 codes for the expected size (47 kDa) of the protein, whereas AF0400 codes for a much larger protein (60 kDa).
AF0395 (noxA2) is the gene immediately upstream of dld within a cluster of five genes that appear to form an operon (V. Pagala and P. Hartzell, unpublished results). NoxA2 has homology with NADH oxidases from organisms such as E. (Streptococcus) faecalis and Deinococcus radiodurans. Like other NADH oxidases, the predicted product of noxA2 has an N-terminal binding site for the ADP moiety of FAD, a binding site for the flavin moiety of FAD, a putative internal NAD+ binding site, and a conserved cysteine (Cys42) that has been shown to be critical for redox chemistry of Nox from E. faecalis (Fig. 9). Consistent with this, we found that both partially purified and recombinant NoxA2 proteins contain a tightly bound FAD cofactor and can oxidize NADH in the presence of various electron acceptors.
FIG. 9.
Primary sequence of noxA2 derived from A. fulgidus AF0395. Conserved ADP binding residues are in bold; conserved redox-active cysteine (Cys42) is starred; the flavin ribityl binding site is underlined (conserved amino acids are italicized).
By the classic definition, NADH oxidases donate electrons to molecular oxygen and regenerate critical reserves of NAD+. NoxA2 has oxidase activity because in the presence of NADH, O2 can serve as an electron acceptor. In the presence of oxygen, NoxA2 catabolism of NADH results in the production of H2O2. NoxA2 also has diaphorase activity, because it can donate electrons to acceptors such as NBT, MTT, DCIP, potassium ferricyanide, menadione, and cytochrome c. These activities are similar to the NADH oxidase activities of thermophilic prokaryotes that transfer electrons to O2 and artificial electron carriers (25, 35).
The FAD cofactor is required for oxidase and diaphorase activities of both recombinant and native NoxA2. NoxA2 has a Km of about 3 μM for NADH, similar to the Km determined for NADH oxidase from Thermus thermophilus (25) and a Vmax near 1.3 μmol mg−1 min−1 was similar to the Vmax of 0.21 μmol mg−1 min−1 reported for Desulfovibrio vulgaris (5). Like the partially purified enzyme from A. fulgidus, rNoxA2 was stable in the presence of O2 and was active over a range of temperatures from 25 to 85°C. rNoxA2 oxidase and diaphorase activities were optimal at temperatures near 80°C, indicating that AF0395 codes for an enzyme that folds properly when expressed at 37°C under aerobic conditions in E. coli. Although the half-life of the recombinant enzyme ranged from 12 to 40 min at 83°C, preparations of the partially purified form from A. fulgidus had a half-life of about 35 h. Hence, the native NoxA2 may be protected from denaturation by additional proteins or a heat-stable conformation only attainable in A. fulgidus.
The role of Nox in vivo and the need for so many forms of Nox, which appear to be produced constitutively under anaerobic conditions, is mysterious. Because A. fulgidus is a strict anaerobe, it is unlikely that Nox acts in the classic sense to transfer electrons to O2 for the purpose of regenerating NAD+. Nox enzymes may instead play a protective role and/or interact with other electron transfer components in novel ways. In E. faecalis and Streptococcus mutans, the NADH oxidase (peroxidase) is involved in defense against oxygen toxicity by direct reduction of O2 to water (6, 12). Typically these enzymes are found in organisms that are unable to synthesize heme, which is needed to produce catalase. The NADH oxidase in A. fulgidus may similarly be involved in protecting the cell from oxidative stress. In this case NoxA2 may reduce oxygen toxicity by the formation of H2O2. Although H2O2 can potentially cause cellular damage, A. fulgidus has a gene, AF2233, which is predicted to encode a peroxidase to detoxify H2O2. Indeed, although A. fulgidus can only grow under anaerobic conditions, cells are not killed upon exposure to oxygen, but rather form a protective biofilm (19). Cells remain viable for up to 24 h in medium contaminated with oxygen, particularly if the temperature is below 80°C.
The finding that NoxA2 specifically copurifies and interacts with the d-lactate dehydrogenase hints that Nox may play a role in energy-yielding reactions. NoxA2 may behave like the respiratory NADH dehydrogenase in E. coli that has oxidase activity and is involved in electron transfer reactions (27). The bacterial sulfate reducer D. vulgaris produces an H2O2-forming NADH oxidase that transfers electrons to APS reductase (4). Because this NADH oxidase is functionally related to NoxA2, it is exciting to speculate that NoxA2 might interact with APS reductase in A. fulgidus. In D. vulgaris, NADH oxidase and cytochrome c reductase are postulated to form part of a membrane complex that, together with ubiquinol, catalyzes the reduction of sulfate (30). Likewise, the A. fulgidus Nox may interact directly with cytochrome c or indirectly with an intermediate, such as the A. fulgidus 7-menaquinone, to catalyze sulfate reduction. This is consistent with the finding that rNoxA2 can donate electrons to the quinoline-like compound menadione.
The ability of NoxA2 to reduce yeast and horse heart cytochrome c is of interest because cytochrome c is a potential electron carrier in vivo. Cytochrome c in A. fulgidus is located in the “periplasm” (between the membrane and the proteinaceous S-layer), where it associates with the membrane. If NoxA2 interacts with a c-type cytochrome in vivo, then NoxA2 must be located in the periplasm. Indeed, we found that NoxA2 is located almost exclusively in the periplasmic space in A. fulgidus cells (V. Pagala and P. Hartzell, unpublished results).
Although the topology of other membrane-bound NADH oxidases (2, 13, 16) is unclear, they may have periplasmic domains that facilitate interaction with periplasmic electron carriers. If cytochrome c is an electron acceptor of NADH oxidation by NoxA2 in A. fulgidus, then NoxA2 may have a respiratory role under anaerobic conditions. We have been unable to detect a change in absorbance of cytochrome using partially purified NoxA2 and crude A. fulgidus membrane preparations. This assay may not be sensitive enough if the level of cytochrome is low or if other factors interfere in a crude preparation. Alternatively, the c-type cytochrome may not interact with NoxA2 under these or any other conditions.
ACKNOWLEDGMENT
This work was supported by grant MCB 9906433 from the Naitonal Science Foundation to P.L.H.
REFERENCES
- 1.Balch W E, Fox G E, Magrum L J, Woese C R, Wolfe R S. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979;43:260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borneleit P, Kleber H P. Purification and properties of the membrane-bound NADH dehydrogenase from hydrocarbon-grown Acinetobacter calcoaceticus. Biochim Biophys Acta. 1983;722:94–101. [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Castresana J, Moreira D. Respiratory chains in the last common ancestor of living organisms. J Mol Evol. 1999;49:453–460. doi: 10.1007/pl00006568. [DOI] [PubMed] [Google Scholar]
- 5.Chen L, Legall J, Xavier A V. Purification, characterization and properties of an NADH oxidase from Desulfovibrio vulgaris (Hildenborough) and its coupling to adenylyl phosphosulfate reductase. Biochem Biophys Res Commun. 1994;203:839–844. doi: 10.1006/bbrc.1994.2259. [DOI] [PubMed] [Google Scholar]
- 6.Claiborne A, Ross R P, Parsonage D. Flavin-linked peroxide reductatses protein-sulfeneic acids and the oxidative stress response. Trends Biochem Sci. 1992;17:183–186. doi: 10.1016/0968-0004(92)90263-9. [DOI] [PubMed] [Google Scholar]
- 7.Dahl C, Koch H, Keuken O, Trüper H G. Purification and characterization of ATP sulfurylase from the extremely thermophilic archaebacterial sulphate-reducer, Archaeoglobus fulgidus. FEMS Microbiol Lett. 1990;67:27–32. [Google Scholar]
- 8.Gietz R D, Triggs-Raine B, Robbins A, Graham K C, Woods R A. Identification of proteins that interact with a protein of interest: applications of the yeast two-hybrid system. Mol Cell Biochem. 1997;172:67–79. [PubMed] [Google Scholar]
- 9.Hansen T A. Physiology of sulphate-reducing bacteria. Microbiol Sci. 1988;5:81–84. [PubMed] [Google Scholar]
- 10.Hansen T A. Metabolism of sulfate-reducing prokaryotes. Antonie Van Leeuwenhoek. 1994;66:165–185. doi: 10.1007/BF00871638. [DOI] [PubMed] [Google Scholar]
- 11.Hartzell P L, Millstein J, LaPaglia C L. Biofilm formation in a hyperthermophilic archaeon. Methods Enzymol. 1999;310:335–349. doi: 10.1016/s0076-6879(99)10027-2. [DOI] [PubMed] [Google Scholar]
- 12.Higuchi M. Reduced nicotinamide adenine-dinucleotide oxidase involvement in defense against oxygen-toxicity of Streptococcus mutans. Oral Microbiol Immun of. 1992;7:309–314. doi: 10.1111/j.1399-302x.1992.tb00594.x. [DOI] [PubMed] [Google Scholar]
- 13.Hochstei L I, Dalton B P. Studies of a halophilic NADH dehydrogenase 1. Purification and properties of enzyme. Biochim Biophys Acta. 1973;302:216–228. doi: 10.1016/0005-2744(73)90150-2. [DOI] [PubMed] [Google Scholar]
- 14.Huennekens F M, Felton S P. Preparation and enzymatic assay of FAD and FMN. Methods Enzymol. 1957;3:950–959. [Google Scholar]
- 15.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaworowski A, Mayo G, Shaw D C, Campbell H D, Young I G. Characterization of the respiratory NADH dehydrogenase of Escherichia coli and reconstitution of NADH oxidase in ndh mutant membrane-vesicles. Biochemistry. 1981;20:3621–3628. doi: 10.1021/bi00515a049. [DOI] [PubMed] [Google Scholar]
- 17.Kohn L D, Kaback H R. Mechanisms of active transport in isolated bacterial membrane vesicles. XV. Purification and properties of the membrane-bound d-lactate dehydrogenase from Escherichia coli. J Biol Chem. 1973;248:7012–7017. [PubMed] [Google Scholar]
- 18.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.LaPaglia C, Hartzell P L. Stress-induced production of biofilm in the hyperthermophile Archaeoglobus fulgidus. Appl Environ Microbiol. 1997;63:3158–3163. doi: 10.1128/aem.63.8.3158-3163.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mallett T C, Claiborne A. Oxygen reactivity of an NADH oxidase C42S mutant: evidence for a C(4a)-peroxyflavin intermediate and a rate-limiting conformational change. Biochemistry. 1998;37:8790–8802. doi: 10.1021/bi9803630. [DOI] [PubMed] [Google Scholar]
- 21.Manchenko G P. Handbook of detection of enzymes on electrophoretic gels. Boca Raton, Fla: CRC Press; 1994. [Google Scholar]
- 22.Marty-Teysset C, de la Torre F, Garel J R. Increased production of hydrogen peroxide by Lactobacillus delbrueckii subsp. bulgaricus upon aeration: involvement of an NADH oxidase in oxidative stress. Appl Environ Microbiol. 2000;66:262–267. doi: 10.1128/aem.66.1.262-267.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masullo M, Raimo G, DelloRusso A, Bocchini V, Bannister J V. Purification and characterization of NADH oxidase from the archaea Sulfolobus acidocaldarius and Sulfolobus solfataricus. Biotechnol Appl Biochem. 1996;23:47–54. [PubMed] [Google Scholar]
- 24.Moller-Zinkhan D, Borner G, Thauer R K. Function of methanofuran, tetrahydromethanopterin, and coenzyme F420 in Archaeoglobus fulgidus. Arch Microbiol. 1989;152:362–368. [Google Scholar]
- 25.Park H J, Reiser C O A, Kondruweit S, Erdmann H, Schmid R D, Sprinzl M. Purification and characterization of a NADH oxidase from the thermophile Thermus thermophilus HB8. Eur J Biochem. 1992;205:881–885. doi: 10.1111/j.1432-1033.1992.tb16853.x. [DOI] [PubMed] [Google Scholar]
- 26.Phillips T A, Van Bogelen R A, Neidhardt F C. lon gene product of Escherichia coli is a heat shock protein. J Bacteriol. 1984;159:283–287. doi: 10.1128/jb.159.1.283-287.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poulis M I, Shaw D C, Campbell H D, Young I G. In vitro synthesis of the respiratory NADH dehydrogenase of Escherichia coli — role of UUG as initiation codon. Biochemistry. 1981;20:4178–4185. doi: 10.1021/bi00517a035. [DOI] [PubMed] [Google Scholar]
- 28.Reed D W, Hartzell P L. The Archaeoglobus fulgidusd-lactate dehydrogenase is a Zn2+ flavoprotein. J Bacteriol. 1999;181:7580–7587. doi: 10.1128/jb.181.24.7580-7587.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rose M D, Winston F, Hieter P, editors. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. Assay of β-galactosidase in yeast; pp. 155–159. [Google Scholar]
- 30.Rossi M, Pollock W B R, Reij M W, Keon R G, Fu R D, Voordouw G. The hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough encodes a potential transmembrane redox protein complex. J Bacteriol. 1993;175:4699–4711. doi: 10.1128/jb.175.15.4699-4711.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:386–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 32.Speich N, Dahl C, Heisig P, Klein A, Friedrich L, Stetter K O, Trüper H G. Adenylylsulphate reductase from the sulphate-reducing archaeon Archaeoglobus fulgidus: cloning and characterization of the genes and comparison of the enzyme with other iron-sulphur flavoproteins. Microbiology. 1994;140:1273–1284. doi: 10.1099/00221287-140-6-1273. [DOI] [PubMed] [Google Scholar]
- 33.Stanton T B, Jensen N S. Purification and characterization of NADH oxidase from Serpulina (Treponema) hyodysenteriae. J Bacteriol. 1993;175:2980–2987. doi: 10.1128/jb.175.10.2980-2987.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stetter K O, Lauerer G, Thomm M, Neuner A. Isolation of extremely thermophilic sulfate reducers: evidence for a novel branch of archaebacteria. Science. 1987;236:822–824. doi: 10.1126/science.236.4803.822. [DOI] [PubMed] [Google Scholar]
- 35.Toomey D, Mayhew S G. Purification and characterisation of NADH oxidase from Thermus aquaticus YT-1 and evidence that it functions in a peroxide-reduction system. Eur J Biochem. 1998;251:935–945. doi: 10.1046/j.1432-1327.1998.2510935.x. [DOI] [PubMed] [Google Scholar]
- 36.Yanisch-Perron C, Viera J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 37.Young I G, Jaworowski A, Poulis M. Cloning of the gene for the respiratory d-lactate dehydrogenase of Escherichia coli. Biochemistry. 1982;21:2092–2095. doi: 10.1021/bi00538a017. [DOI] [PubMed] [Google Scholar]