Abstract
CRISPR/Cas9 is a revolutionary genome editing technology with the tremendous advantages such as precisely targeting/shearing ability, low cost and convenient operation, becoming an efficient and indispensable tool in biological research. As a disruptive technique, CRISPR/Cas9 genome editing has a great potential to realize a future breakthrough in the clinical bone and cartilage repairing as well. This review highlights the research status of CRISPR/Cas9 system in bone and cartilage repair, illustrates its mechanism for promoting osteogenesis and chondrogenesis, and explores the development tendency of CRISPR/Cas9 in bone and cartilage repair to overcome the current limitations.
Keywords: CRISPR/Cas9, Genome editing, Bone repair, Cartilage repair
Graphical abstract
Highlights
-
•
Presenting a comprehensive discuss of the technical principles of the CRISPR/Cas9 system.
-
•
Highlighting the applications of the CRISPR/Cas9 system for promoting osteogenesis and chondrogenesis.
-
•
Summarizing the challenges of CRISPR/Cas9 systems in bone and cartilage repair.
-
•
Discussing the prospect and future direction of CRISPR/Cas9 application in orthopaedics.
1. Introduction
For the regeneration of bone and cartilage, it is crucial to promote the proliferation or differentiation of osteoblasts and chondroblasts. In the ordinary bone or cartilage defect repair, fixation and suture repair are preferred methods in clinic. While large bone defects always need autologous bone or allogeneic bone transplantation, which has numerous side effects like chronic pain, nerve injury, infection, high risk of disease transmission and immune rejection [1]. Furthermore, implantation of orthopaedic materials is another important strategy in current clinic treatment. However, long-term biocompatible problems of some widely used non-bioabsorbable biomaterials are reported frequently, such as poly-ether-ether-ketone (PEEK) and polymethyl methacrylate (PMMA), may causing some unavoidable drawbacks, such as osteolysis, secondary surgery requirement and increasing risk of postoperative complications (Table 1) [2].
Table 1.
Comparison of mainstream bone and cartilage repair methods.
| Repair methods | Example | Advantages | Disadvantages |
|---|---|---|---|
| Autogenous and Allogeneic bone | / | No immunological rejection | Chronic pain, nerve injury |
| Non-bioabsorbable materials | PMMA, PEEK | 1) Rapid Curing 2) No magnetism |
Secondary surgery |
| Bioabsorbable metal materials | Magnesium, Zinc | 1) Reduce stress shielding 2) High biocompatibility |
Low mechanical strength and corrosion |
| Bioabsorbable polymer materials | Collagen, Chitosan | 1) Can be cross-linked or blended 2) Promote cell adhesion |
Poor stability and mechanical properties |
| Bioabsorbable inorganic materials | Hydroxyapatite, Bio-glass | Poor stability and mechanical properties | Low toughness and high brittleness |
| Bioabsorbable compound materials | Mineralized collagen | Better osteogenic capacity | Process complexity |
Gene editing is regarded as a monumental technology in life science. The early techniques relied on site-specific identification of DNA loci based on nucleases, including Zinc Finger nucleases (ZFNs) and Transcription Activator-Like Effector nucleases (TALENs) [3]. However, the difficulties in protein design and synthesis were limited the widespread use of these nucleases [4,5]. In 1980s, a defense mechanism of some prokaryotes against the virus was found based on a sequence called Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) to identify of exogenous pathogenic DNA [6]. CRISPR is guided by RNA, and is epoch-making in the current application of gene editing while thanks to the specificity and codability of RNA sequences [7]. The gene in CRISPR locus nearby was named as CRISPR-associated (Cas). After years of research, the mature CRISPR system was simplified with two parts: single guide RNA (sgRNA) and CRISPR-associated protein 9 (Cas9) nuclease. Since then, CRISPR/Cas9 technology has brought a huge impact to the life science field, and researches was recognized by the Nobel Prize in Chemistry in 2020.
In bone and cartilage repair field, CRISPR/Cas9 system as a third-generation genome editing technology can overcome many shortcomings of traditional strategies. It can provide powerful ability to regulate genome sequence and gene expression, which could change or correct gene function and defects for long-term to achieve therapeutic effects at the genetic level. Usually, bone and cartilage repair are regulated by several growth factors, such as transforming growth factor (TGF-β), insulin-like growth factor-1 (IGF-1) and basic fibroblast growth factor (bFGF) [8], a), b), [9], a), b). Besides, some osteogenesis-related proteins such as alkaline phosphatase (ALP), bone morphogenetic protein (BMP) and osteocalcin (OCN) are also involved in the synthesis and mineralization of bone matrix and the value-added and differentiation of osteoblasts [10], a), b), [11]. Gene therapy for the repair of bone and cartilage defects usually involves the introduction of target gene fragments into the organism so that the relative expression products can be sustainably expressed in the subsequent repair with the aim of promoting osteogenesis. Unlike direct regulation of osteogenic-related factors, the CRISPR/Cas9 system can reprogram pluripotent stem cells and induce their differentiation towards bone or cartilage [12]. By exploiting the multifunctional differentiation properties of bone marrow mesenchymal stem cells (BMSCs), BMSCs are widely used in the treatment and repair of bone and soft tissues and are shown to be a viable approach to mediate bone regeneration [13]. Compared with the traditional gene therapies, the CRISPR/Cas9 system enables precise target gene insertion, knockout and editing. It can not only circumvent the systemic side effects of traditional surgery and conventional drugs, but also induce large amounts of phenotypic proteins at once to complete the treatment in a targeted manner, providing a promising new pathway for bone and cartilage repair (Fig. 1).
Fig. 1.
Schematic diagram of CRISPR/Cas9 genome editing for bone and cartilage repair.
In recent years, rapid advances in molecular biology and genomics greatly promote the possibility of gene editing techniques in the treatment of bone and cartilage diseases. Here, CRISPR/Cas9 system, as the most powerful genome engineering tool, benefits from programmable RNA that can rapidly generate specific sequences and easily accomplish applications [14]. In this review, we presented a comprehensive discuss of the molecular mechanisms and technical principles of the CRISPR/Cas9 system and detailly described the classification and research progress of delivery vectors. We highlighted the applications and challenges of the CRISPR/Cas9 system for promoting osteogenesis and chondrogenesis. Finally, we summarized the off-target effects and editing efficiency of CRISPR/Cas9 systems in bone and cartilage repair and discuss the direction of their subsequent development.
2. Mechanism of CRISPR/Cas9 gene editing technology
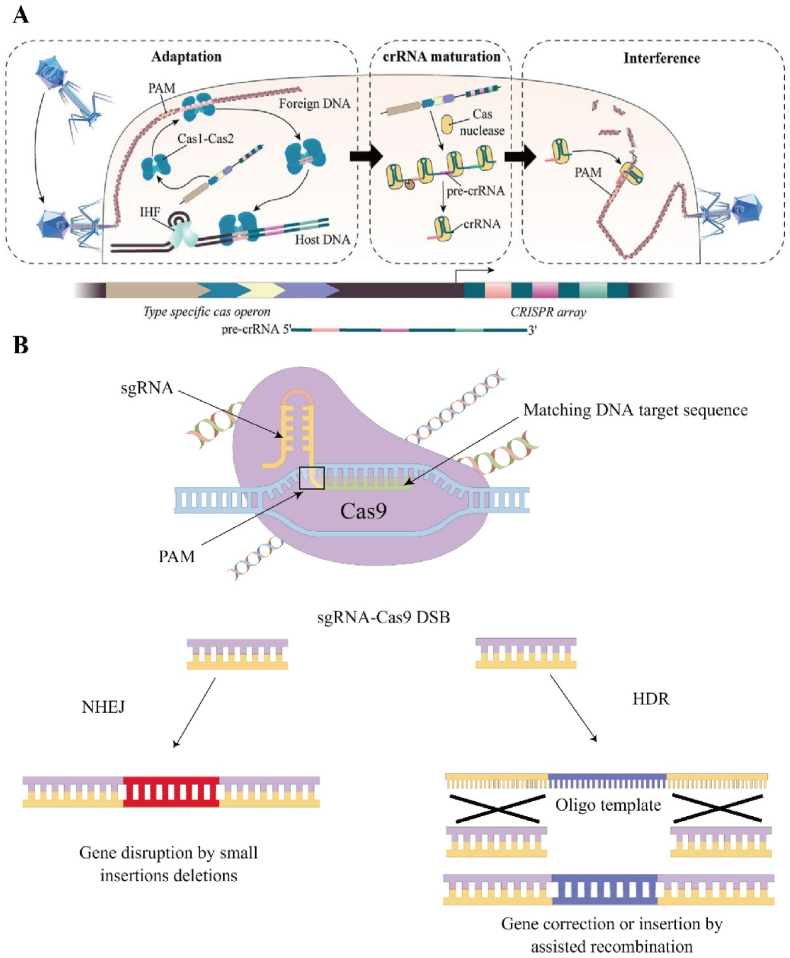
Generally, CRISPR consists of a leader (promoter), repeats and genome-targeting sequences (spacers) [15]. When an exogenous gene is invaded into a prokaryotic organism, the original spacer in its genome is recognized by the Cas proteins through the protospacer adjacent motif (PAM) sequence downstream of the original spacer and then cut (Fig. 2A). Cas proteins insert the clipped original spacer into the middle of the leader and the repeat to form a new spacer [16]. The A-T-rich leader contains a promoter, which is used to initiate transcription of repeat and spacer sequences. The transcribed long-stranded RNA is called precursor transcript CRISPR RNA (pre-crRNA), which is then processed into mature CRISPR RNA (crRNA) by related enzymes [17]. Cas proteins can form new Cas protein complexes together with crRNA and trans-activating crRNA (tracrRNA). The small guide RNA (sgRNA) formed by the fusion of crRNA and tracrRNA complementarily pairs with the gene in the exogenous gene and directs the Cas protein to shear the exogenous gene fragment [18].
Fig. 2.
A) Schematic diagram of the CRISPR/Cas9 system. Starting with the Cas1-Cas2 complex, the target sequence is recognized by PAM and later integrated into the host sequence during the adaptation phase. During the crRNA maturation phase, the transcribed long-stranded RNA is called precursor transcript CRISPR RNA (pre-crRNA), which is then processed into mature CRISPR RNA (crRNA) by related enzymes. In the final interference phase, crRNA induces Cas protein to shear the exogenous gene fragment. Reproduced with permission [151]. Copyright 2020, The Authors. B) Cas9 can cleave exogenous DNA into double DSB DNA. There are two repair methods: the first is NHEJ and the other is HDR.
The recognition of target DNA by the CRISPR/Cas9 system differs from traditional protein-guided gene editing techniques in that it uses small molecules of RNA sequences, which not only avoids the cumbersome protein engineering associated with DNA recognition, but also greatly improves its applicability for high-throughput genomic manipulation or screening [19]. As mentioned previously, Cas9 needs to assemble with crRNA and tracrRNA to form a new complex in order to achieve recognition and cleavage of the target DNA. sgRNA of the Cas9 complex matches the target DNA according to the principle of Watson-Crick base pairing, while localizing at a specific site [20]. PAM sequence which near this specific site is a short sequence associated with a specific Cas protein and necessary for complementary pairing [21]. Different Cas proteins often match different short sequences, for example Cas9 from Streptococcus pyogenes (SpCas9) matches 5′-NGG-3’ [22]. The relatively simple PAM sequence has a higher probability of occurring in the genome and is more conducive to the widespread use of Cas9. The sequence from staphylococcus aureus is 5′-NNGRRT-3′, and the longer PAM sequence increases the specificity of its recognition and helps prevent off-target effects at the same time [23]. Upon binding to the target gene, the complementary single strand (complementary paired with crRNA) and non-complementary single strand of the target gene are recognized and cleaved by the HNH and RuvC structural domains of Cas9 respectively, producing a double stranded break (DSB) at the flat end of the target gene [24].
There are three different Cas systems using different mechanisms. Type I and Type III require a large protein complex composed of different Cas proteins when cutting the target gene, while Type II requires only one Cas protein such as Cas9 [25]. Cas9 in the Type II CRISPR system can cleave exogenous DNA into double DSB DNA. There are two types of repair methods when a DNA double strand is cut (Fig. 2B). The first one is nonhomologous end joining (NHEJ), which urgently links the broken DNA double strands like spontaneous “SOS” (“save our souls” signal) repair in living organisms. However, this repair method is stochastic and prone to insertion or deletion mutations that can damage the target gene [26]. The other one is homology-directed repair (HDR), which is a small fragment of DNA with the same sequence at both ends and the broken sequence can be homologously recombined with the broken gene, thus completing the exact recombination of the gene [27]. The most commonly used synthetic CRISPR system is the Type II CRISPR system which uses a single cut of Cas9 protein. Its convenience and speed have given it a potential that cannot be underestimated in medical and scientific fields.
3. Delivery methods of CRISPR/Cas9 system in bone and cartilage repairing
CRISPR/Cas9 changes the current status quo using a single conventional treatment in bone and cartilage tissue diseases. As a powerful gene editing tool, CRISPR/Cas9 systems are essential to be delivered to bone tissue targets such as the synovial membrane of the joint cavity or cartilage tissue, thereby allowing the long-term stable expression of a specific gene for the ultimate therapeutic purpose. Many common physical delivery methods are limited by in vivo applications while highly efficient in delivery. So far, nanotechnology-based non-viral vectors or conventional viral vectors are gaining more and more attention. Herein, delivery/transfer vectors of CRISPR/Cas9 system should be focused (Fig. 3). In order to assess the potential of conventional delivery methods for application, it is particularly important to consider the advantages and disadvantages of various methods in an integrated manner (Table 2).
Fig. 3.
Classification of CRISPR/Cas9 delivery methods.
Table 2.
Methods of CRISPR/Cas9 transfer.
| Transfer methods | Principle | Advantages | Disadvantages |
|---|---|---|---|
| Electroporation | Enhance membrane permeability by short electric impulses | Non-immunogenicity and efficient | Lower cellular viability |
| Magnetofection | Directed to cells by using the magnetic field | Without damaging the cell membranes | High cost |
| Microfluidics | Exploiting the deformability of cell membranes | Integration | Inability to complete delivery in vivo |
| Ultrasonic | Induced pore formation in cell membranes | Non-invasiveness | Low controllability |
| Microinjection | With the use of microneedles | High transfection efficiency | Need high proficiency requirements |
| Lentivirus | Interaction with target cell surface receptors | Sustained expression | Recombination occurs in vivo |
| Adenovirus | Interaction with target cell surface receptors | High security | Momentary expression |
| Dendrimers | Electrostatic interaction | Precise and controllable physical and chemical properties | Cytotoxicity |
| Liposomes | Electrostatic interaction | High transfection efficiency | Cytotoxicity |
| Micelles | Electrostatic interaction | Low cytotoxicity | Low transfection efficiency |
3.1. Physical delivery methods
Physical delivery methods include electroporation, magnetofection, microfluidics, ultrasonic, and microinjection [[28], [29], [30], [31], [32]].
3.1.1. Electroporation
Previously, electroporation has been widely used as a non-virus delivery method for cell transfection. Electroporation also called electrotransfection. Electrotransfection transiently increases the permeability of the cell membrane by applying a high-intensity electric field to the cell, thereby allowing the entry of exogenous genes or drugs into the cell [33]. And the high intensity of the electric field can reduce complications associated with targeting and immunogenicity [34]. Beyond that, electroporation has the ideal property of transfecting non-dividing cells such as nerves, cartilage or bone. It has extraordinary advantages in the field of tissue engineering repair [[35], [36], [37]]. Despite its many advantages, electroporation still has some limitations. Cells often die or have increased toxicity due to high voltages and lower cell viability is one of the main disadvantages of electroporation [38]. If in vivo transfection therapy is performed, different transmitter devices are required with high costs and varying degrees of tissue damage resulting from invasive operations [39]. Tröder et al. used a modified electroporation method to act on fertilized eggs of C57BL/6 mice to obtain CRISPR/cas9-mediated specific mutant mice [40]. Not only was the specific mutation successfully introduced without affecting embryonic development, but the live birth rate of embryos was also significantly increased. Miao et al. targeted the germ cell-specific gene nanos2 by pre-assembling sgRNA/Cas9 RNPs and electroporated fertilized eggs from mice, cattle and pigs, which stably produced mutant progeny [41]. Xu et al. reported a tubular electroporation technique and experimental results showed that the new technique could enable efficient genome editing in mammalian cells especially for human-derived stem cells that are difficult to transfect. It may be a new strategy for delivering CRISPR/Cas9 [42] (Fig. 4A).
Fig. 4.
A) Gene editing by CRISPR/Cas9 in human stem and primary cells using tube electroporation. (i) Devices for tube electroporation. (ii) Transfection of human iPSC with Cas9/gRNA RNP-targeted APP gene, without ssODN template. (iii) Different types of cells were transfected with pCMV-GFP plasmid, and transfection efficiency was detected by fluorescence microscopy and flow cytometry after 48 h. From left to right are the cell bright field plots, fluorescence plots and flow cytometry dot plots, where red is the transfected cells and black is the mock-transfected control cells, respectively. Reproduced with permission [42]. Copyright 2018, The Authors. B) Magnetic induction is the process of delivering nucleic acids under the influence of a magnetic field acting on a nucleic acid carrier associated with a magnetic nanoparticle. (i) Schematic diagram of the magnetically guided nanoparticles targeting method. (ii) Applied magnetic field in mice experiments. (iii) Magnetic regimes of magnetite and maghemite as a function of their size. Reproduced with permission [43]. Copyright 2015, The Authors. C) Microfluidic transfection device. (i) Plasmids encoding sgRNA and Cas9 proteins are mixed with cells and flow through the chip. (ii) Flow chart of delivery mechanism. (iii) Scanning electron microscope (SEM) images of the device structure. (iv) Diagrammatic representation of cellular stress gradient formation across the cell membrane. D) Characterization of microfluidic chip. i) Delivery of FITC-labeled ssDNA to HEK293T cells via two different chips. ii) Delivery efficiency. iii) Cell viability. iv) Western blotting for PC-3 cells after 48 h of delivery with three different siRNA oligonucleotides targeting Akt1.l v) Cell Counting. vi) Transfer efficiency of different cell lines. Reproduced with permission [49]. Copyright 2015, The Authors.
3.1.2. Magnetofection
Magnetofection technology uses the surface activity of magnetic nanoparticles like iron oxide to combine with target genes to form gene-loaded magnetic microspheres [43] (Fig. 4B). Although the magnetofection technique does not directly improve transfection efficiency, the powerful magnetic attraction capacity allows the number and speed of nucleic acid molecules to enter the cell, maximizing the use of nucleic acids [44], a), b). Different from electroporation, the magnetofection technique does not require disruption of the cell membrane and uses the endocytosis mechanism to complete uptake which effectively avoids the low activity state of cells after transfection [45]. Mykhaylyk et al. successfully constructed magnetically responsive siRNA complexes that accomplished intracellular translocation and effectively silenced the target gene [46]. Hryhorowicz et al. used polyethyleneimine-coated magnetic iron oxide nanoparticles to construct magnetic plasmid DNA complexes (PEI-Mag2) to facilitate CRISPR/Cas9 plasmid transfection of porcine fetal fibroblasts (PFFs). The transfection efficiency can be improved using PEI-Mag2, which may be due to its magnetic properties that accelerate the deposition of nucleic acids on the cell surface [47].
3.1.3. Microfluidics
The microfluidic membrane deformation method exploits the deformation capacity of the cell membrane, which undergoes rapid deformation under mechanical action to produce temporary pores through which nucleic acid or protein molecules enter the cytoplasm [48]. The integrated property of microfluidics itself allows selective screening of untransfected cells or repeated transfection again by microarray to improve the gene editing activity. Han et al. first applied microfluidic membrane deformation to CRISPR/Cas9 delivery and successfully completed RNA-guided gene editing [49] (Fig. 4C and D). This rapid and high-throughput platform can provide a promising strategy for CRISPR/Cas9-mediated gene editing and gene analysis. Microfluidic membrane deformation does not depend on exogenous material, endocytosis or chemical modification of the target molecule. Transfection can be done reproducibly with only microfluidic chips [50]. Because of the limitation of microfluidic chip, it cannot be delivered in vivo which limits its clinical application potential.
3.1.4. Ultrasonic
Lipid bilayers are capable of directly converting acoustic energy into mechanical stress and strain at the subcellular and cellular levels [51], a), b). High-energy ultrasonic produces local shear in the extracellular fluid, inducing the formation of pores in the cell membrane and increasing the permeability to DNA or RNA [52], a), b). Ultrasonic methods can be safely used in vivo due to their non-invasive nature and are currently used in clinical cases [53]. However, its low control leads to low transfection efficiency, low success rate and high equipment requirements [54]. Ryu et al. constructed microbubble nano-liposome particles as Cas9/sgRNA nucleoprotein complex carriers and successfully transferred the protein complex into dermal papilla cells of hair follicles in male bald animals under ultrasonic activation The research validated the inhibitory effect of CRISPR/Cas9 on SRD5A2 in vivo and in vitro which finally restored hair growth [55]. To deliver CRISPR plasmids, Dong et al. developed a dual ultrasonic/magnetic responsive microdrop that can effectively deliver plasmids to cancer cells, which could be a potential strategy for clinical application of ultrasonic methods for cancer treatment [56].
3.1.5. Microinjection
Microinjection is a simple and direct non-virus transfection method that allows the introduction of DNA or RNA into the cytosol using only micron-sized pipettes. Its transfection efficiency tends to be high because the injection volume and location can be precisely controlled [57], a), b). However, due to its huge optical instrumentation requirements and the results are related to the proficiency of the technician, microinjection is only used in specific settings [58]. Ai et al. successfully completed the knockout of cotton bollworm genes using the CRISPR/Cas9 system by microinjection method, which improved the efficiency of gene editing [59]. Li et al. explored the effect of microinjection time on the embryonic developmental capacity of the CRISPR/Cas9 system and showed that microinjection time had different degrees of effect on embryonic development and gene editing rate of in vitro fertilized porcine embryos. Proper optimization of sgRNA or Cas9 concentration and precise determination of injection time could better accomplish gene editing [60] (Fig. 5A–C).
Fig. 5.
A) Schematic of the embryo injection strategy to determine the optimal timing of CRISPR/Cas9 microinjection. In vitro fertilization (IVF) and somatic cell nuclear transfer (SCNT)-derived embryos were microinjected with sgRNA and Cas9 mRNA. B) The priming RNA was designed to target exon 5 of the interleukin 2 receptor gamma (IL2RG) locus in the porcine genome (Sscrofa11.1). Nucleotides in blue text indicate designed gRNAs, and letters in red text indicate PAM sequences (NGG). C) The expression of Cas9 protein was assessed by immunofluorescence staining 6 h after microinjection. Reproduced with permission [60]. Copyright 2021, The Authors. D) Different approaches to in vivo and in vitro gene therapy. E) Current frequency of use of different viral vectors. F) Third generation lentiviral vector constructed from four plasmids. G) Schematic diagram of the wild-type adenovirus type 5 (Ad5) genome and genetic modifications of common Ad5-based vectors. Reproduced with permission [69]. Copyright 2021, The Authors.
3.2. Virus vector delivery
Virus vectors are the vectors of choice for gene editing and gene therapy. They are widely used not only in vitro but also in clinical settings. However, the safety of vectors is often a concern due to the specificity of viruses. To avoid recombination of lentiviral vectors in vivo, their genome is first split into multiple different structures. Secondly, the promoter or enhancer sequences in their terminal repeat sequences are deleted to avoid activation of related genes. Alternatively, the glycoprotein is wrapped around the surface of the virus vector and modified to limit the host range [61]. All these safety measures can generate inactivated vectors for safe transfer.
3.2.1. Lentivirus vector
Lentivirus vectors are retrovirus vectors based on HIV-1 [62]. The entry of lentivirus vectors into target cells is mediated by the interaction between glycoproteins immobilized on the outer membrane and receptors on the surface of the target cells. Lentivirus vectors have a large capacity of 9 kb, which is sufficient to package the various components of CRISPR [63]. As an integrative vector, lentivirus vectors can also carry multiple genomic fractions which may be a key element in the treatment of certain diseases. In addition, lentivirus vectors can transfect cells in stationary phase whereas ordinary retrovirus vectors can only transfect cells in division. It increases the scope of application and indirectly enhancing the gene therapy effect [64]. Lentiviruses, as carriers of CRISPR/Cas9, not only have the ability to gene edit, but also take full advantage of the advantages that lentivirus vectors themselves possess [65]. Lentivirus vectors can efficiently integrate exogenous genes into the chromosomes of target cells, thus expressing target genes in a sustained manner. Lentivirus vectors are preferred for cells that are more difficult to transfect, such as primary cells or stem cells. It can significantly improve the transduction efficiency of target genes and achieve long-term and stable expression of target genes conveniently and quickly. Joung et al. selected the lentivirus as a vector for CRISPR/Cas9 by considering the insertion capacity and cell type. And gene editing was successfully completed proving that lentivirus vectors are a powerful tool for gene transduction [66].
3.2.2. Adenovirus vector
Adenovirus vectors are double-stranded DNA viruses that enter the target cells through receptor-mediated endocytosis and can transduce different cell types without being restricted by the division phase. Likewise, adenovirus vectors do not integrate into the host cell genome, staying outside the chromosome and achieving only instantaneous expression with a high safety profile [67]. Adenovirus vectors have many unique advantages over other viral vectors. Firstly, most human cells can express adenovirus receptors, which makes adenovirus vectors have a wide range of application and high transduction efficiency. Secondly, modified adenovirus vectors can easily escape the body's intrinsic immune system defenses. Adenovirus vectors are also becoming the most commonly used vectors in clinical trials worldwide [68]. Adenovirus vectors have not only been used for vaccine development and tumor therapy, but also have potential for gene editing [69] (Fig. 5D–G). Tsukamoto et al. applied adenovirus vector-loaded AsCpf1 gene to primary culture of humanized mouse hepatocytes and showed that the adenovirus vector-mediated CRISPR/AsCpf1 system provides a useful tool for genome editing of human hepatocytes [70].
3.3. Nanocarriers
The development of nanocarriers as an alternative to traditional physical transfection methods and viral delivery methods has been very rapid. Non-viral vectors are more secure and easier to mass-produce than viral vectors. Both dendrimers and liposomes offer extraordinary advantages in terms of targeting, high encapsulation and low immunogenicity. More and more nanocarriers are taking the stage for bone or cartilage gene therapy.
3.3.1. Dendrimers
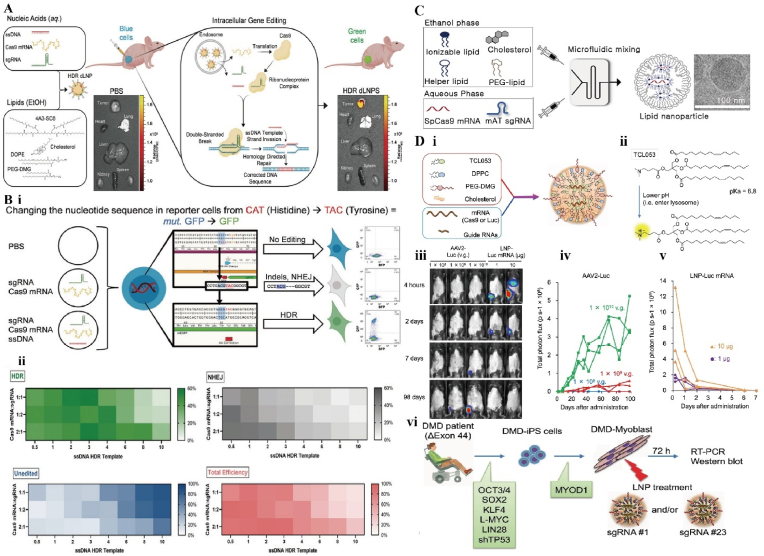
Dendrimers are macromolecules with a dendritic structure, consisting of oligomers repeatedly and linearly linked by dendritic units. Due to the controllability of its monomer, the physicochemical properties of dendrimers can be precisely controlled and personalized preparation can be accomplished. Meanwhile, the dendrimers contain a cavity structure inside, forming a dense three-dimensional sphere structure that can be perfectly used for gene encapsulation. Also, many functional groups are exposed on its surface which can be modified to accomplish better performance optimization thanks to the spherical structure [71] (Fig. 6A and B). Farbiak et al. developed a dendrimer-based lipid nanoparticle in order to accomplish CRISPR/Cas9-mediated HDR while avoiding the NHEJ error-prone mechanism. The team encapsulated Cas9 mRNA, sgRNA and donor ssDNA (single-stranded DNA) in nanoparticles with low cytotoxicity and no charge at neutral pH. At the appropriate ratio, the nanoparticles are able to complete HDR. This may be related to the simultaneous delivery of three nucleic acids [72]. Liu et al. reported a boric acid-containing dendrimer that can deliver Cas9 protein with high efficiency. Not only that, it can also be used for protein delivery with different isoelectric points or sizes without chemical modification, overcoming the problem of difficulty in forming stable complexes with proteins [73].
Fig. 6.
A) Schematic diagram of HDR gene editing mediated by dendrimers lipid nanoparticles. B) Nanoparticles successfully complete HDR in HEK293 cells containing Y66H mutant GFP. (i) Fluorescence disappears when NHEJ is performed and can return to normal levels when HDR is performed. (ii) Trends in HDR, NHEJ, total efficiency and unedited cell groups were observed in three different ratios of Cas9 mRNA: sgRNA. Reproduced with permission [71]. Copyright 2021, Wiley. C) Schematic diagram of liposome synthesis using microfluidic system. Reproduced with permission [76]. Copyright 2022, The Authors. D) Characterization of liposome-CRISPR complexes. (i) Composition of complexes. (ii) Chemical formula of liposome. (iii) In vivo imaging of mice after intramuscular injection. (iv-v) Fluorescence signal analysis of different nanoparticles after injection. (vi) Gene therapy for duchenne muscular dystrophy (DMD). Reproduced with permission [77]. Copyright 2021, The Authors.
3.3.2. Liposomes
Liposomes are bilayer vesicles that mimic cell membranes and show promise in the delivery field because of their efficient delivery capacity and good biocompatibility [74]. Cationic lipids are a key component of liposomes and play an important role in nucleic acid encapsulation and cellular delivery. Pre-liposomes as siRNA delivery vectors have difficulty in encapsulating large molecules of Cas9 protein and sgRNA. To overcome this obstacle, Rosenblum et al. developed a liposome system for CRISPR/Cas9 gene editing [75]. The results showed that the system successfully delivered Cas9 mRNA and sgRNA into tumor cells and led to apoptosis of tumor cells and improved survival. Han et al. prepared a series of liposomes containing Cas9 mRNA and modified sgRNA using microfluidic technology. The end-modified sgRNA could achieve 85% encapsulation rate, and this liposome-mediated gene editing became a new safe therapeutic method [76] (Fig. 6C). Unlike viral vectors for in vivo delivery, Kenjo et al. reported a liposome that can be repeatedly injected into skeletal muscle tissue [77] (Fig. 6D). It was injected into the gastrocnemius muscle of mice and maintained stable levels in vivo for close to 100 days. Its low immunogenicity and repeatable administration are important features for the future use of liposomal carriers for bone or cartilage repair.
3.3.3. Micelles
Micelles are ordered aggregates of molecules that begin to form in large numbers after the surfactant concentration reaches a certain value in an aqueous solution [78]. The micelles rely mainly on hydrophobic and electrostatic interactions for nucleic acid loading [79]. Abbasi et al. explored polymorphic micelles (PMs) prepared from polyethylene glycol (PEG)-poly(cationic block copolymers) for delivery of CRISPR/Cas9 components. It was found that loading Cas9 mRNA and sgRNA together into 1 p.m. significantly improved the stability of sgRNA compared to loading sgRNA alone. The team reported the successful co-encapsulation of Cas9 mRNA and sgRNA in PM and the successful completion of gene editing [80]. Self-assembled micelles consisting of quaternary ammonium terminated poly(propylene oxide) (PPO–NMe3) and amphiphilic Pluronic F127 were designed. Lao et al. optimized micelle performance to target the HPV18-E7 oncogene. While assessing gene silencing capacity, the micelles demonstrated strong protection and delivery capabilities [[81], a)].
So far, although there are numerous methods of gene delivery, none of them is perfect. We should consider the scope of applicability and the advantages and disadvantages of various delivery vectors or methods, and reasonably assess their potential for introduction into clinical applications. Some elements that may determine the efficiency or biosafety of gene transfer such as carrying capacity [81b], immunogenicity [81c] and economic cost [81c], are the key conditions that we should examine comprehensively.
4. CRISPR/Cas9 for bone repair
The basis of bone repair is the formation of bone scabs by osteoblasts and osteoclasts in response to various cytokines. Gene editing technique based on CRISPR/Cas9 demonstrates a great potential to be translated into clinical applications for bone repairing [82]. Currently, long bone fractures are self-healing with strong internal fixation, but healing is often less optimistic due to many factors such as osteoporosis or advanced age. In the face of segmental defects with a defect length of more than twice the diameter of the diaphysis, non-healing of the broken end often occurs [83]. Conventional autologous bone grafts, allogeneic bone grafts and material implants are more or less subject to adverse reactions such as pain, infection or immune rejection. Nowadays, the concept of introducing one or more osteogenic genes into a non-healing patient with the expectation of modifying the healing pattern of the organism through gene editing has become very attractive (Table 3). There are two main strategies of gene editing for the treatment of bone defects. First, the gene-loaded vector is applied directly to the defect site, or it can be combined with a scaffold before implantation. Second, suitable tissues such as bone or muscle tissue are collected in vivo, edited and modified in vitro and then reimplanted in vivo [84]. In either strategy, the ultimate goal is to promote osteogenesis and healing, using gene editing to complete the treatment of bone defect diseases.
Table 3.
CRISPR/Cas9 for bone repair.
| Target gene | Repair Mechanism | Research progress | Literature |
|---|---|---|---|
| ELMO1 | ELMO1 promotes enhanced osteoclast activity and increases bone resorption activity. Knock-out. | Deleted the ELMO1 gene in Hoxb8 macrophages (osteoclast precursor cells) via CRISPR/Cas9 and sgRNA. Then the transfected macrophages developed functional defects. | Arandjelovic et al., [86] 2021 |
| Noggin | High expression of BMP2 upregulates the expression of osteogenic genes and induces differentiation of stem cells into osteoblasts. Noggin can bind BMP2 and prevent its docking with stem cell surface receptors. Knock-down. | Gene editing mediated by a hybrid baculovirus system that prolongs BMP2 expression and reduces Noggin expression. | Hsu et al., [89] 2020 |
| BMP9 | BMP9 induces differentiation of stem cells to osteoblasts by activating Smad-dependent signaling pathways. Upregulation. | MSCs were genetically edited to overexpress BMP9 using the CRISPR/Cas9 system. | Freitas et al., [92] 2021 |
| Sox9 and PPAR-γ | Sox9 and PPAR-γ act as major transcription factors for cartilage and adipose formation respectively, and PPAR-γ inhibits the action of Sox9. Activation of Sox9 while inhibiting PPAR-γ promotes bone healing. Knock-down. | The CRISPRai system was constructed using the large capacity of the baculovirus vector. CRISPRai could be combined to stimulate tissue regeneration for bidirectional regulation. | Truong et al., [93] 2019 |
| USP11 | USP11 overexpression enhances the expression of osteogenic factors in BMSCs. MSX1 balances the level of protein degradation during normal osteogenesis. Upregulation. | Used the CRISPR/Cas9 system to screen for deubiquitinating enzymes (DUBs) that regulate MSX1 protein. | Kaushal et al., [95] 2022 |
| Wnt16 | The Wnt pathway plays an important role in the osteogenic differentiation of BMSCs. Wnt16 is a ligand that affects stem cell proliferation, differentiation and migration through the Wnt pathway. Knock-out. | Generated stable Wnt16 −/− mutant zebrafish lines using CRISPR/Cas9 technology to study their effects on bone tissue using tissue mineral density (TMD) as an observation. | McGowan et al., [98] 2021 |
| Satb2 | Deletion of Satb2 may result in incomplete expression of osteogenic genes or poor skeletal development. Mutation. | Used the CRISPR/Cas9 system to induce mutations in the Satb2 gene in MC3T3-E1 cells. When Satb2 expression is reduced, the growth rate of osteoblasts is slowed down. | Dowrey et al., [100] 2019 |
Bone repair and reconstruction consists of resorption of old bone by osteoclasts and formation of new bone by osteoblasts [85]. If osteoclasts are deficient, the fracture ends are atrophied on both sides, the bone marrow cavity is closed or sclerosis of the bone ends occurs, often resulting in non-healing consequences. ELMO1 promotes enhanced osteoclast activity and increases bone resorption activity. Arandjelovic et al. deleted the ELMO1 gene in Hoxb8 macrophages via CRISPR/Cas9 and sgRNA. Then the transfected macrophages developed functional defects [86] (Fig. 7A and B). To further explore whether ELMO1 can control other signaling markers in osteoblasts, the group continued to target proteins that may play a role such as athepsin G (Ctsg) and myeloperoxidase (Mpo). The results showed that this macrophage reduced the degradation function after knocking down the mRNA of this protein using CRISPR/Cas9, indicating that ELMO1 is a key part of the functional network regulating bone degradation in osteoclasts. However, targeting the resorptive activity of osteoclasts is often more beneficial than the quantity. A rational selection of target genes could help find new breakthroughs in bone repair.
Fig. 7.
A) Elmo1-deficient mice exhibit reduced bone erosion in two models of arthritis. (i) Schematic diagram of collagen-induced arthritis (CIA) induction in Elmo−/− DBA/1J mice. (ii) RT-PCR analysis of the expression of the indicated genes. (iii) MicroCT images of the ankle joint in mice. (iv) Quantification of bone erosion in the heel bone region of mice. (v) Schematic representation of K/BxN serum transfer-mediated induction of arthritis in Elmo−/− C57BL/6 mice. (vi) On day 10 after K/BxN serum injection, the expression of the indicated genes. (vii) On day 10 after K/BxN serum injection, H & E staining of histological sections of mouse ankle. B) ELMO1 is a signaling hub that regulates osteoclast function. (i) Genes are sorted from left to right according to the magnitude of change. (ii) TRAP staining was performed 7 days after differentiation of different Hoxb8 cells into osteoblasts. Hoxb8 osteoblasts tested for resorptive function on OsteoAssay plates. (iii) (iv) Osteoclast surface and total ELMO1 protein interaction group. Reproduced with permission [86]. Copyright 2021, The Authors. C) BMP2 binds to surface receptors, activates downstream signaling and promotes osteogenesis, leading to upregulation of Nog, antagonism of BMP2 and inhibition of osteogenesis (left). Baculovirus (BV) provides CRISPRi to inhibit Nog and reduce the antagonistic effect of Nog on BMP2, thereby promoting osteogenesis (right). D) Concepts of BV design. E) Alizarin red staining after CRISPRi-mediated Nog inhibition. F) qRT-PCR analysis of OSX and OCN. Reproduced with permission [89]. Copyright 2021, Elsevier.
BMP2 is known as a regulator of bone and cartilage formation and is a potent osteoinductive growth factor, but its clinical application is limited due to its high cost because of the high dosage required for its efficacy [87]. High expression of BMP2 upregulates the expression of osteogenic genes and induces differentiation of stem cells into osteoblasts. However, bone precursor cells express a Noggin protein in response to BMP2 stimulation to antagonize the biological activity of BMP2 [88]. Noggin can bind BMP2 and prevent its docking with stem cell surface receptors, thereby inhibiting stem osteogenic differentiation. Therefore, inhibition of Noggin expression may reduce the antagonistic effect of Noggin on BMP2 and indirectly promote osteogenesis. Hsu et al. fused Cas9 with transcriptional repressors such as VPR to generate dCas9-VPR that binds to specific sgRNA, and performed in vitro gene editing using baculovirus as a vector targeting Noggin. The results showed that Noggin gene was knocked out and BMP2 was overexpressed, which could effectively promote osteogenic differentiation of adipose stem cells and promote bone healing [89] (Fig. 7C–F).
BMP9 induces differentiation of stem cells to osteoblasts by activating Smad-dependent signaling pathways and has a higher osteogenic potential compared to BMP2 [90,91]. To evaluate the role of BMP9 in promoting bone defect repair in vitro and in vivo, MSCs were genetically edited to overexpress BMP9 using the CRISPR/Cas9 system [92]. Osteogenic markers, such as Runx2, Sp7, ALP and Oc, were increased to varying degrees. In vivo experiments showed accelerated new bone formation and increased bone density in rats injected with gene edited stem cells. This is the first demonstration that CRISPR-edited MSCs overexpressing BMP9 can successfully promote bone formation, providing a new option for the application of gene therapy in the field of bone defects.
Until now, healing of large segmental bone defects remains difficult. Truong et al. attempted to improve it by stimulating cartilage formation through implantation of stem cells [93]. The group constructed a Cas9, sgRNA-based CRISPR activation/repression (CRISPRai) system and verified that Cas9 synergizes with sgRNAa (activator) to activate the mCherry activator and with sgRNAi (inhibitor) to activate the d2EGFP repressor. Mesenchymal stem cells from rats introduced via baculovirus were simultaneously assayed for the blocking effect of Sox9 and PPAR-γ. Sox9 and PPAR-γ act as major transcription factors for cartilage and adipose formation respectively, and PPAR-γ inhibits the action of Sox9 [94]. Therefore, activation of Sox9 while inhibiting PPAR-γ promotes bone healing. The CRISPRai system was constructed using the large capacity of the baculovirus vector. It was demonstrated that CRISPRai could be combined to stimulate tissue regeneration for bidirectional regulation for the first time and provided a more flexible tool.
The osteogenic differentiation capacity of BMSCs needs to be tightly regulated. Too little bone formation may not complete repair, but too much bone formation may lead to ectopic ossification or osteosclerosis. MSX1 balances the level of protein degradation during normal osteogenesis. In the present study, Kaushal et al. used the CRISPR/Cas9 system to screen for deubiquitinating enzymes (DUBs) that regulate MSX1 protein and identified the Ubiquitin carboxyl-terminal hydrolase 11 (USP11) as a regulator of MSX1 [95]. USP11 overexpression enhances the expression of osteogenic factors in BMSCs. Also, it affects calcification and ALP activity if USP11 is lacking. The group selected 50 sgRNAs of USP family genes and validated the selected USP11. There are no functional reports on the interaction of USP11 with MSX1 in BMSCs, and the group demonstrated a novel role of USP11 in the osteogenic differentiation process.
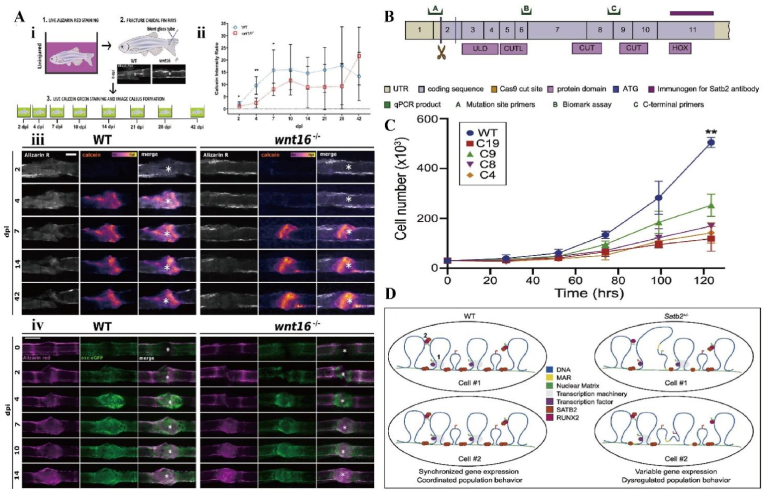
The Wnt signaling pathway is a central regulator of bone development and repair. And the Wnt pathway is an attractive therapeutic target that plays an important role in the osteogenic differentiation of BMSCs [96]. Wnt16 is a ligand that affects stem cell proliferation, differentiation and migration through the Wnt pathway [97]. McGowan et al. generated stable Wnt16 −/− mutant zebrafish lines using CRISPR/Cas9 technology to study their effects on bone tissue using tissue mineral density (TMD) as an observation [98] (Fig. 8A). Bone defects were subsequently induced in the caudal fin of Wnt16−/− zebrafish. Compared to wild-type zebrafish, Wnt16 mutants exhibit delayed bone mineralization during bone repair. Osteoblast recruitment was also significantly delayed in Wnt16 mutants after bone defect. This study effectively demonstrates that Wnt16 may regulate Wnt activity through Runx2a to promote osteoblast differentiation and bone matrix deposition. Appropriate treatment targeting Wnt16 may prevent fractures and promote bone repair.
Fig. 8.
A) Delayed osteoblast recruitment and bone mineralization. (i) Marking of old and new bone. (ii) Reduced callus formation in the Wnt mutant group 2–7 days after injury. (iii) Alizarin red (gray) labeling of old bone and callus labeled by calcein. White asterisks are fracture centers. (iv) Calcified bone (alizarin red) and osteoblasts (osx:GFP). Reproduced with permission [98]. Copyright 2021, The Authors. B) Location map of Satb2 protein and Cas9 cleavage site. C) Satb2 gene mutation reduces osteoblast proliferation rate. D) Satb2-mediated models of molecular and cellular outcome variation. Reproduced with permission [100]. Copyright 2019, Elsevier.
The function of some osteogenic transcription factors can be enhanced by the specific AT-rich sequence binding protein 2 (Satb2). Deletion of Satb2 may result in incomplete expression of osteogenic genes or poor skeletal development [99]. To further explore the molecular mechanism of Satb2-mediated osteogenic function, Dowrey et al. used the CRISPR/Cas9 system to induce mutations in the Satb2 gene in MC3T3-E1 cells [100] (Fig. 8B–D). When Satb2 expression is reduced, the growth rate of osteoblasts is slowed down. In addition, Satb2 mutations lead to nuclear abnormalitie. And the osteoblast value-added process, in which Satb2 is involved, can help to demonstrate the genetic background for the repair of bone defects.
5. CRISPR/Cas9 for cartilage repair
Cartilage has no nerves or blood vessels and therefore lacks the ability to heal itself. The repair of cartilage defects remains a great challenge at this time. Traditional methods include microfractures, cartilage grafts or scaffold implants, but these methods do not fully restore the natural cartilage tissue. Current research is exploring the potential of MSCs in cartilage repair, and CRISPR/Cas9 is an excellent tool for this purpose. As the preferred tool for gene editing, how to promote cartilage regeneration is an important direction for its development (Table 4).
Table 4.
CRISPR/Cas9 for cartilage repair.
| Target gene | Repair Mechanism | Research progress | Literature |
|---|---|---|---|
| DANCR | Activation of DANCR significantly promotes the differentiation of adipose stem cells to chondrocytes and enhances cartilage formation in vitro. Upregulation | Packaged dCas9-VPR and its corresponding gRNA into a baculovirus for gene transfection and compared four dCas9-VPRs derived from different bacteria, showing that SadCas9-VPR derived from Staphylococcus aureus successfully induced DANCR activation in rat adipose stem cells. | Nguyen et al., [102] 2021 |
| MMP13 | Reduction of total MMP13 secretion by CRISPR/Cas9 indirectly reduces degradation of the extracellular matrix. Knock-down. | Reconstructed human articular chondrocyte populations using a CRISPR/Cas9-mediated gene editing strategy that stably reduced MMP13 expression in cartilage. | Seidl et al., [103] 2019 |
| ACAN and Col2 | As a major component of the ECM, collagen not only provides mechanical support but also controls the growth and differentiation of cells. Upregulation. | Used the dCas-VPR CRISPR gene activation system to upregulate aggrecan (ACAN) and Col2. | Farhang et al., [104] 2020 |
| TRPV4 | A mutation in TRPV 4 is a functional mutation that can lead to an increase in intracellular calcium ion levels. Repair template. | In the presence of TRPV 4-specific agonists, the mutant group showed significantly accelerated cartilage differentiation at early stages and upregulated Sox9 mRNA expression. | Nonaka et al., [106] 2019 |
| Cx43 | High levels of transmembrane protein Cx43 expression in osteoarthritic cartilage. Cx43 also upregulated p16INK4a and NF-κB to cause senescence and apoptosis in chondrocytes. Knock-down. | Used CRSIPR/Cas9 to downregulate Cx43 expression and successfully slowed cartilage degeneration. | Varela-Eirín et al., [109] 2018 |
| COL2A1-GFP | Purification by identifying surface markers would greatly improve chondrogenic efficiency. Knock-in. | The CD146+/CD166+/PDGFRβ+/CD45- subpopulation of chondrogenic progenitor cells possessed more powerful chondrogenic ability. | Dicks et al., [117] 2020 |
LncRNA DANCR was reported to induce differentiation of human synovial-derived stem cells and synovial-derived MSCs toward cartilage [101]. Nguyen et al. packaged dCas9-VPR and its corresponding gRNA into a baculovirus for gene transfection and compared four dCas9-VPRs derived from different bacteria, showing that SadCas9-VPR derived from Staphylococcus aureus successfully induced DANCR activation in rat adipose stem cells [
2] (Fig. 9A–C). Activation of DANCR significantly promotes the differentiation of adipose stem cells to chondrocytes and enhances cartilage formation in vitro. Activation of DANCR with CRISPR/Cas9 can dramatically upregulate the expression of Smad3 and enhance bone defect repair, which is expected to be a novel therapeutic target for future cartilage repair.
Fig. 9.
A) CRISPRa activates DANCR to promote skull formation. B) Evaluation of different dCas9-VPR direct homologs on DANCR activation. C) Stimulatory effect of DANCR activation on rASC cartilage formation. (i-iii) Expression of chondrogenic markers (Sox9, Col2a1, Acan) at 7 dpt measured by qRT-PCR. (iv) Effects of DANCR activation at 1 and 14 dpt on cell morphology during differentiation of chondrogenic cells. (v) Alcian blue staining at 14 dpt. Reproduced with permission [102]. Copyright 2021, Elsevier. D) Cas9 editing strategy for human chondrocytes. E) Immunofluorescence of COL2A1, comparing unedited and edited spheres. Scale bars = 100 μm. N, unedited; E, MMP13 edited. F) Western blotting for COL2A1. Reproduced with permission [103]. Copyright 2021, Elsevier.
Seidl et al. reconstructed human articular chondrocyte populations using a CRISPR/Cas9-mediated gene editing strategy that stably reduced MMP13 expression in cartilage [103] (Fig. 9D–F). Reduction of total MMP13 secretion by CRISPR/Cas9 indirectly reduces degradation of the extracellular matrix. Firstly, a 3D tissue model was established to simulate the natural environment of cells and tissues. Next, the level of collagen type 2 (Col2) was detected to determine whether the secretion and activity of MMP13 was successfully inhibited. Tissues like cartilage which lack vascularization and have low self-proliferative activity are good candidates for gene editing and are well protected from migration of other accidental edits from cartilage tissue. This approach would greatly improve the efficacy of current cell-based cartilage repair.
Similar to the previous approaches, stem cell therapy has been demonstrating a central role in clinical regenerative therapy. Whether it is exogenous implantation of stem cells or re-induction of differentiation using growth factors, etc., the aim is to precisely control protein expression and enhance the activity of target cells. To address the uncontrollable in vivo environment, Farhang et al. used the dCas-VPR CRISPR gene activation system to upregulate aggrecan (ACAN) and Col2 [104]. By RNA-seq analysis, Col upregulation was found to mediate broader cartilage gene expression. Besides, dual overexpression of ACAN and Col2 resulted in deposition of sGAG and Col2. As a major component of the ECM, collagen not only provides mechanical support but also controls the growth and differentiation of cells [105]. In conclusion, the dCas-VPR CRISPR system serves as a method to up-regulate endogenous ECM proteins which can well regulate the cell phenotype.
Metatropic dysplasia caused by mutations in the TRPV4 (transient receptor potential vanilloid 4) gene is a form of congenital skeletal dysplasia. Nonaka et al. repaired single base mutations in TRPV4 using CRSIPR/Cas9. The results showed that in the presence of TRPV 4-specific agonists, the mutant group showed significantly accelerated cartilage differentiation at early stages and upregulated Sox9 mRNA expression [106]. In brief, a mutation in TRPV 4 is a functional mutation that can lead to an increase in intracellular calcium ion levels. Currently, most clinical cartilage repair involves upregulation of a gene or correction of a mutation. This study provides a new direction for prompting a mutation in a gene to treat cartilage injury.
Osteoarthritis is a degenerative disease of cartilage, often resulting from disruption of cartilage integrity or subchondral bone plate lesions [107]. Previous studies have found high levels of transmembrane protein connexin43 (Cx43) expression in osteoarthritic cartilage [108]. Varela-Eirín et al. found that Cx43 maintains the presence of various immature cells present in cartilage by increasing the expression of Twist-1 and MMPs [109] (Fig. 10A). Next, Cx43 also upregulated p16INK4a and NF-κB to cause senescence and apoptosis in cells such as chondrocytes. In the present study, the investigators used CRSIPR/Cas9 to downregulate Cx43 expression and successfully slowed cartilage degeneration. Cx43 is a specific mechanism for chondrocytes towards senescence. Controlling chondrocyte plasticity and apoptosis through targeted treatment of Cx43 is a new approach to treat cartilage diseases. This could also be a potential candidate for promoting cartilage repair in regenerative medicine.
Fig. 10.
A) Down-regulation of Cx43 in osteoarthritis promotes cartilage repair. Reproduced with permission [109]. Copyright 2018, The Authors. B) Differentiation of RVR cell lines carrying the COL2A1-GFP reporter gene into chondroprogenitor cells. C) Higher population of triple positive cells. D) Chondrogenic progenitor cells were identified by RNA sequencing to contain at least 9 populations. There are 3 major groups: neurogenic cells (blue dashed circles), chondrogenic cells (green dashed circles) and mesenchymal (brown dashed circles). E) Characteristic genes of chondrogenic cells. F) Cells expressing these desired markers were sorted from wild-type chondroprogenitor cells. Reproduced with permission [117]. Copyright 2020, The Authors.
Liu et al. designed a therapeutic protocol using modified mesenchymal stem cells (MSC) as implants for cartilage repair [110]. MSC not only contribute structurally to cartilage repair, but also have potent immunomodulatory activity. It can interact with macrophages to coordinate tissue repair in rheumatoid arthritis [111]. The research team selected human synovial-derived MSC to further improve the chondrogenic ability of MSC by modifying the target gene via CRISPR/Cas9. The target gene modified MSC would then become a novel therapeutic option for chronic diseases of cartilage damage like rheumatoid arthritis.
Although pluripotent stem cells currently have multiple chondrogenic differentiation options, incomplete differentiation and cellular heterogeneity remain major barriers to cartilage formation [112,113]. MSCs expressing CD105, CD166 and CD146 have been reported to have a higher chondrogenic potential [[114], [115], [116]]. Dicks et al. used CRISPR-Cas9-edited COL2A1-GFP knock-in human pluripotent stem cells [117] (Fig. 10B–F). The aim was to identify cell surface markers of true chondroprogenitor cell populations. Single cell RNA sequencing was then used to analyze the different subpopulations. The results showed that the CD146+/CD166+/PDGFRβ+/CD45- subpopulation of chondrogenic progenitor cells possessed more powerful chondrogenic ability. Or purification by identifying surface markers would greatly improve chondrogenic efficiency.
6. Establishment of bone disease model with CRISPR/Cas9
The establishment of several bone disease models will help to further explore new models of the role of CRISPR/Cas9 in bone repair. Conventional mechanisms of bone repair such as osteoclast-osteoblast homeostasis, matrix mineralization, and scab plasticity are represented in common bone disease models. Osteogenesis imperfecta (OI) is an autosomal dominant disorder that often causes fragility fractures and recurrent fractures [118]. OI is a rare genetic disease and various treatment modalities have failed to achieve good outcomes [119]. With the development of induced pluripotent stem cell (iPSC) strategies, gene editing techniques are gradually being applied in the study of OI. CRISPR/Cas9 can correct patient genes, correct genetic mutations, and repair pathophysiological changes in a comprehensive manner across all processes.
The main pathogenic factor of OI originates from impaired type I collagen synthesis. Mutations in COL1A1 and COL1A2 are the direct cause of impaired type I collagen α-chain synthesis [120]. As an important component of the bone matrix, collagen causes impaired bone mineralization and osteoporosis. Jung et al. isolated iPSCs from patients with COL1A1 mutation, and osteogenic induced differentiation resulted in lower than normal collagen type I levels as expected [121]. Increased osteoblast differentiation potential and improved collagen levels after correction of the mutant gene COL1A1 by CRISPR/Cas9.
Rauch et al. successfully generated a type V OI mouse model using CRISPR/Cas9 [122]. Type V OI is mainly caused by the MALEP-BRIL mutation in the chromosome-producing IFITM5 gene. The modeling results showed reduced cranial mineralization, curved and shortened long bones and increased rib fragility. The histological findings also showed no formation of primary ossification centers and less cortical bone. The expression of relevant osteogenic markers and angiogenic factors was reduced by monitoring the genetic level of the model. Low level of MALEP-BRIL expression may affect the induced differentiation of osteoblasts.
7. CRISPR/Cas9 technology improvements
7.1. CRISPR/Cas9 off-target effect
CRISPR/Cas9 is a powerful gene editing tool with low cost and high efficiency. It has been widely used in biomedical fields and opened a new avenue for regenerative medicine research. However, off-target effects remain one of the non-negligible drawbacks of the CRISPR/Cas9 system [123]. The first step in editing by CRISPR/Cas9 system is to select a target region thus determine the sequence of the target sgRNA. As mentioned earlier, Cas9 cuts the DNA and then completes further modification of the site after the sgRNA recognizes the PAM of the target genome. However, many Cas9 complexes can also bind to non-target regions and accomplish unexpected gene modifications called off-target effects [124]. Off-target effects often result in unwanted sequence mutations or deletions and can even activate oncogenes or cause cell death [125]. This potential side effect of CRISPR/Cas9 greatly limits its application in in vivo translational medicine research and poses a high risk to clinical treatment. If stem cells are edited using CRISPR/Cas9, off-target effects can cause irreversible apoptosis or transdifferentiation of stem cells. The original osteoblasts or chondrogenic cells may become osteoclasts or fibroblasts, which is detrimental to bone or cartilage repair. How to increase the efficiency of precision editing and reduce the off-target rate has become another challenge for researchers.
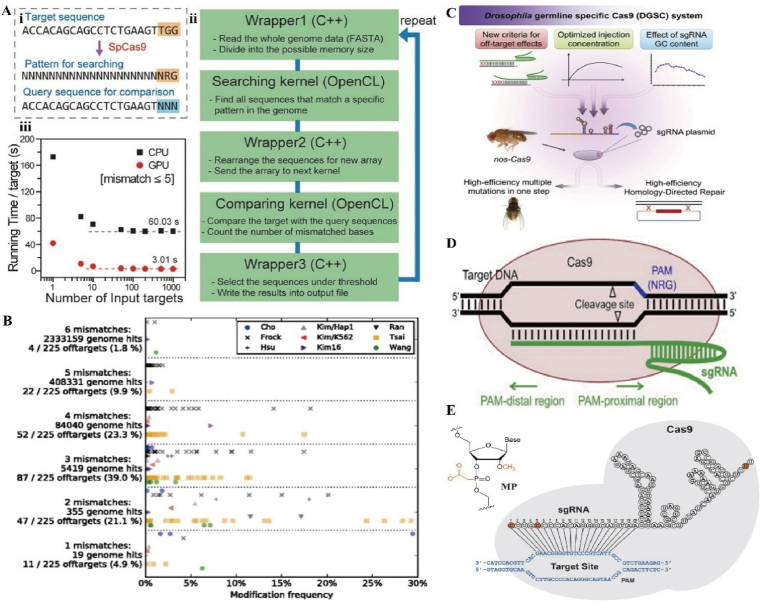
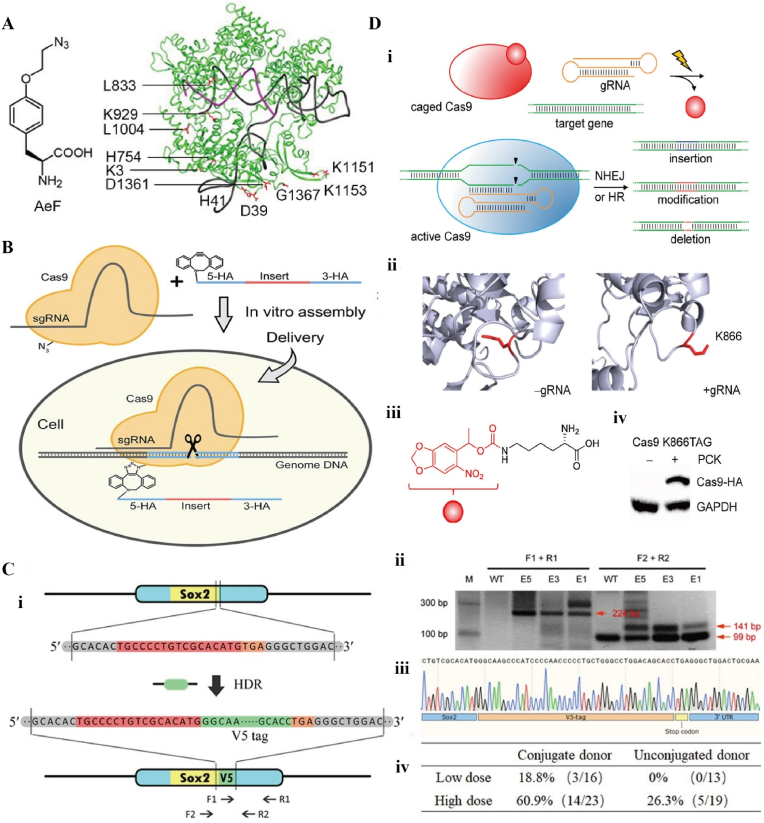
It is well known that the editing efficiency of CRISPR/Cas9 is influenced by the recognition efficiency of sgRNA and PAM, and more than three mismatches between the target sequence and sgRNA may lead to off-target effects [126]. The human genome tends to be many times smaller than the bacterial genome. CRISPR/Cas9 derived from the natural immune system of bacteria tends to have poor specificity in humans and a higher probability of off-target effects than in bacteria [127]. The efficiency of gene editing would be improved if the off-target effects could be detected or quantified by some method in vitro or in vivo. First, the design of sgRNA is crucial. Scientists have developed algorithms based on computer simulation prediction models to detect off-target effects. A computerized prediction program called Cas-OFFinder is not limited by the number of mismatches and allows PAM changes to find potential off-target sites. It is now freely accessible [128] (Fig. 11A). Similar to Cas-OFFinder, CRISPOR (http://crispor.org) is an off-target site scoring tool that allows for better off-target prediction and saves time for researchers' screening [129] (Fig. 11B).
Fig. 11.
A) Basic structure of Case-OFFinder. (i) Diagram of Case-OFFinder. (ii) Workflow of Case-OFFinder. (iii) Running time per target site as a function of the number of target sites input via CPU and GPU. Reproduced with permission [128]. Copyright 2014, Oxford University Press. B) 225 off-target modification frequencies of 26 sgRNAs separated by mismatch number. Reproduced with permission [129]. Copyright 2016, The Authors. C) Optimizing sgRNA parameters to improve the specificity and efficiency of the drosophila CRISPR/Cas9 system. D) Schematic diagram of the Cas9/sgRNA system. Reproduced with permission [133]. Copyright 2014, Elsevier. E) Chemically modified sgRNA. Reproduced with permission [135]. Copyright 2017, Oxford University Press.
The sgRNA is considered to be a key factor for target specificity. Designing a low off-target sgRNA is a challenging task [130]. To reduce off-target effects, scientists have used strategies such as adjusting GC content and sgRNA length. sgRNAs with GC content between 40% and 60% possess a low off-target rate because higher GC content stabilizes the DNA/RNA duplex and reduces binding to non-target regions [131]. Not only that, repetitive bases (a continuous stretch of identical bases) are also associated with new DNA synthesis. Four adjacent guanines are most strongly correlated with low CRISPR activity, which can lead to sgRNAs that do not bind easily to the target sequence [132]. Ren et al. found that the sgRNA GC content of six PAM proximal nucleotides (PAMPNs) was positively correlated with editing efficiency which may provide a more optimal modification [133] (Fig. 11C and D). Interestingly, the length of sgRNAs also affects off-targeting. Fu et al. found that sgRNAs with 17 or 18 nucleotide fragments usually function more specifically than longer sgRNAs [134]. Appropriate shortening of the length of sgRNA might allow for a more efficient CRISPR/Cas9 system. The remaining modifications such as the incorporation of 2ʹ-O-methyl-3ʹ-phosphonoacetate in sgRNA [135] (Fig. 11E), modification of the 5ʹ-end hairpin structure [136], improvement of non-virus delivery methods [137] or selection of Cas variants [138] can reduce the off-target effect to some extent. Rational improvement of some of the factors affecting off-target can help to complete bone or cartilage repair safely and efficiently.
7.2. CRISPR/Cas9 editing efficiency
CRISPR/Cas9 editing efficiency often refers to the percentage of target genes that are inserted, replaced or deleted. A higher editing efficiency not only costs less but also accomplishe the desired effect better. Appropriate improvement of editing efficiency is crucial for the application of CRISPR/Cas9 in bone and cartilage repair. Bone and cartilage repair is often accompanied by a longer healing period. Lower editing efficiency may require repeated implantation and testing, or increase cytotoxicity thereby reducing efficacy and prolonging recovery time. Currently, the editing efficiency is mainly improved by optimizing CRISPR/Cas9 sequences, improving delivery systems or changing gene repair strategies.
Ma et al. improved gene editing efficiency by optimizing plant codons [139]. However, gene editing in plants and animals is slightly different. The same methods may not be reproducible, but it points to a potential direction. Not only that, the team also suggested that the editing efficiency of CRISPR/Cas9 is also related to the Cas9 expression level, the composition of the target sequence (GC content) and the secondary structure of the sgRNA. This matches the factors associated with the efficiency of animal editing. Similarly, Farboud et al. designed an sgRNA with a GC sequence at the 3’ end. This simple design induced efficient editing [140]. The order of bases determines the specificity of codons and even genes [141]. During the sequence modification or optimization of sgRNA, there are some tricks that may help to improve the editing efficiency. Firstly, guanine will perform better at positions −1 and −2 [142]. Secondly, thymine is not preferred at the four positions close to the PAM. Finally, the −3 position is preferred for cytosine and −5 to −12 for adenine [143]. Consistent with the previous findings, nucleotides downstream of the PAM influence the efficiency of sgRNA more than nucleotide sequences upstream [144].
For CRISPR/Cas9, the mechanisms of NHEJ- and HDR-mediated repair are very different and the editing efficiency varies greatly from cell to cell [145]. In the current study, HDR repair is predominant which acts preferentially in the S or G2 phase and is slightly slower [146]. In contrast, mutations induced by NHEJ are more frequent which acts throughout the cell cycle. This competing repair relationship inspired scientists. Ma et al. mixed Scr7 (DNA ligase IV inhibitor) into the Cas9 protein complex to inhibit NHEJ [147]. The HDR-mediated precision modification was enhanced by inhibiting NHEJ to improve the efficiency of gene editing. Conversely, focusing solely on HDR strategies may also lead to inefficiencies. He et al. integrated the promoterless ires-eGFP fragment into the GAPDH locus and were able to highly express GFP in somatic cells [148]. NHEJ-based editing may be more effective than HDR. For bone and cartilage repair, the differentiation ability of pluripotent stem cells determines the timing and efficiency of repair. Rapid completion of the corresponding editing and significant reduction of healing time are what we need to consider. These results provide a valuable pathway for human stem cell editing.
Currently, there is an increasing number of methods to chemically modify Cas9 proteins to improve editing efficiency. Modification of Cas9 proteins by azide-containing noncanonical amino acids (ncAA) has also been reported [149] (Fig. 12A–C). Such a modification allows the recruitment of donor DNA templates to the Cas9 complex. Hemphill et al. designed a light-controlled Cas9, aiming to achieve precise spatiotemporal control by light-regulated Cas9 function [150] (Fig. 12D). The caged Cas9 protein has a specific site for incorporation into photocaged lysine, which is inactivated prior to UV irradiation. It can be restored to normal levels by irradiation at 365 nm for 120 s. This light-activated CRISPR/Cas9 system can edit genes with high precision and also artificially reduce the toxicity of mutations that occur at certain time points. These methods greatly improve the efficiency of HDR-mediated editing and show great potential in the treatment of bone or cartilage damage.
Fig. 12.
A) Chemical structure of ncAA AeF (4-(2-azidoethoxy)-l-phenylalanine) and the crystal structure of SpyCas9 with gRNA. Aef: labeled in red. B) Bio-orthogonal strain-promoted alkyne-azide cycloaddition of azide-modified Cas9 with DBCO (dibenzylcyclooctyne)-modified single-stranded oligodeoxynucleotide. C) Precise genome editing of chemically modified Cas9-adaptor in mouse fertilized eggs. (i) Schematic representation of targeted Sox2 gene editing with insertion of V5 tag. (ii) Validation of V5 gene insertion. (iii) DNA sequencing results confirmed the integration of the V5 tag at the end of the Sox2 gene. (iv) HDR efficiency was determined by genotyping PCR. Reproduced with permission [149]. Copyright 2020, The Authors. D) CRISPR/Cas9 optical control system. (i) The caged Cas9 protein contains a specific site and is incorporated into the photocaged lysine. (ii) Conformation of gRNA before (left) and after (right) binding to Cas9. K866: labeled in red. (iii) Photocaged lysine (PCK). (iv) Western blotting of PCK-dependent Cas9 K866TAG expression. Reproduced with permission [150]. Copyright 2015, American Chemical Society.
8. Summary, challenges and outlook
Since CRISPR/Cas9 technique emerged, it has been perfected by many scientists and been playing a major role in several fields of life sciences, agriculture, and bioengineering. Undoubtedly, orthopaedic researchers also endeavored to apply this transgenerational tool in their relevant area, solving the challenges of treating severe bone or cartilage defects, along with bringing new treatment models and promising cures to clinicians.
Compared to traditional gene editing techniques that rely on protein guidance, the CRISPR/Cas9 system relies mainly on sgRNA for the recognition of target DNA. This particular mechanism grants the CRISPR/Cas9 system the possibility of high-throughput operation and higher matching precision, and avoids the tedious and costly upfront protein construction engineering. Not only that, Type II CRISPR/Cas9 can accomplish more precise HDR after cutting exogenous DNA. This has a non-underestimated application potential in the balance of human NHEJ-based repair mechanism.
The ideal CRISPR/Cas9 delivery methods should be characterized by high efficiency and low toxicity. The commonly used physical or biological delivery methods have been extensively investigated, but physical delivery methods mainly electroporation are limited in clinical applications. Generally, CRISPR/Cas9 for bone or cartilage gene therapy is applied in three main routes. First, the CRISPR/Cas9 gene editing is performed in vitro on osteoblasts or chondroblasts, and then the edited cells are reimplanted in vivo for treatment. Second, gene editing is performed on the reproductive or embryonic cells, by this means to obtain healthier offspring for patients with genetic diseases. However, gene editing for reproductive purposes is the most controversial route which is still an absolute research no-go area. Third, editing happens in vivo after delivery of CRISPR/Cas9 systems to the body via virus or non-viral carriers to complete relevant repairs in vivo. Since all CRISPR/Cas9 clinical trials conducted at this stage are mainly in vitro experiments, the cells need to be isolated from the patients' bodies, edited and then infused back into the patients. This makes CRISPR/Cas9 delivery heavily dependent on viral carriers or nanocarriers. The long-term stability of viral carriers and the modifiability of nanocarriers have demonstrated powerful bone or cartilage repair capabilities in the field of smart delivery.
As of now, the gene editing produced by CRISPR/Cas9 is similar to natural mutation in that it does not produce foreign genes, but only involves modifications based on the original genes. In addition, the gene editing tools represented by CRISPR/Cas9 are able to converge more precisely to a purpose that benefits the organism. In the face of many common orthopaedic diseases such as fractures, osteoarthritis, cartilage injuries and bone tumors, relevant vectors were used rationally to knock out disease-causing genes or overexpress antagonistic genes to accomplish fundamental cures from the transcriptional level. Degenerative diseases such as osteoarthritis (OA) are often accompanied by many pathological features involving the upregulation of many genes in the joint tissue. Changes in MMPs or some inflammatory cytokines play an important role in the pathophysiological process of OA. Blockade of certain cytokines by CRISPR/Cas9 will provide new ideas for safe and effective treatment of OA. Also for rheumatoid arthritis (RA) or osteoporosis, CRISPR/Cas9 can target specific sites for editing. Gene therapy can significantly improve the efficacy of inflammatory or immune diseases, and obtaining specific phenotypes by knockdown warrants further attempted studies. Although orthopaedic diseases are influenced by many factors, the relevant target factors are relatively mature and stable. While known loci are studied, relevant effects due to mutations continue to be explored.
For orthopaedic diseases, stem cell editing, osteogenic targeting or chondrogenic targeting editing has become a new therapeutic direction. Theoretically, the edited stem cells can be implanted into the body to play a corresponding osteogenic or chondrogenic role, otherwise, the CRISPR/Cas9 components also can be delivered to osteoblasts or chondrogenic cells via vectors. At present, CRISPR/Cas9 technology in orthopaedics is still applied in a single direction, stagnating in the overexpression or silencing of the corresponding mature targets. In the future, we should focus on other cells such as macrophages or fibroblasts, and pay attention to the dynamic process of osteogenesis-osteolysis, and deeply investigate the mechanism of extracellular matrix mineralization. Rational use of CRISPR/Cas9 tools at different entry points to accomplish better orthopaedic applications. Significant breakthroughs have been made in CRISPR/Cas9 technology, but there are still some issues worth discussing in the future.
-
1.
Off-target effects remain the biggest limitation for the development of CRISPR/Cas9 systems at present. Mutations caused by off-targeting may reduce the repair effect. In severe cases, they may alter the phenotype and even trigger inflammatory storms or death. It is not only a matter of editing success rate, but also a matter of safety. Altering the original normal genes or triggering unnecessary mutations can cause irreversible and permanent damage to humans. While improving the way of detecting mutations, the research on the causes of off-targeting should be strengthened. Optimize the conditions of various parameters to ensure safety as much as possible.
-
2.
The efficiency of CRISPR/Cas9 editing can have a direct impact on bone or cartilage therapeutic outcomes. Whether it is improving delivery vectors or modifying sgRNA sequences, these efforts stop at the level of a single cell or a single disease. In the face of frequent bone or cartilage defects, how to maintain the high efficiency editing level is the future direction of development. It is hoped that single cell sequencing technology, microfluidic technology, and microarray screening technology will all be involved in the application of CRISPR/Cas9 editing efficiency enhancement.
-
3.
Faster advancement of human clinical trials based on completion at the cellular and animal levels. Although CRISPR/Cas9 is still quite some time away from being used in the clinic, we cannot stop moving forward. As mentioned earlier, there are very few clinical trials completed and they are mostly oncology related. In the future, more therapeutic modalities such as stem cell editing, mutation gene knockout, etc. are expected in the field of orthopaedics. The challenge of bone or cartilage defects can be faced in a shorter time and with better rehabilitation results.
-
4.
We need to pay attention not only to the efficacy of CRISPR/Cas9 applications, but also to the ethical issues of CRISPR/Cas9-based gene editing therapies. Some side effects of CRISPR/Cas9 such as off-target effects may cause more serious harm to patients. On the other hand, the emergence of gene editing may permanently change the sequence of the human genome and cause irreversible reproductive catastrophe. We need to consider a combination of potential risks, cultural, ethical, regulatory, policy, and public outreach issues. The rational use of this powerful tool can benefit humanity. But how to avoid the risks and maximize the research and development of CRISPR/Cas9 is a question that deserves our consideration.
Ethics approval and consent to participate
I confirm that I have obtained all consents required by applicable law for the publication of any personal details or images of patients, research subjects or other individuals that are used in the materials submitted to KeAi. I have retained a written copy of all such consents and I agree to provide KeAi with copies of the consents or evidence that such consents have been obtained if requested by KeAi.
Statement of originality
This material has not been published and is not currently under consideration with another journal.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
The authors thank Figdraw.com for providing some of the elements in Fig. 1, Fig. 2, Fig. 3. This work was supported by the National Natural Science Foundation of China (91949203, 22105127), Non-profit Central Research Institute Fund of the Chinese Academy of Medical Sciences (2019PT320001), Shanghai Pujiang Program (21PJD045) and Clinical Research Project of Health Industry of Shanghai (202140128)
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Contributor Information
Wenguo Cui, Email: wgcui80@hotmail.com.
Wei Chen, Email: surgeonchenwei@126.com.
References
- 1.de Girolamo L., Ragni E., Cucchiarini M., van Bergen C.J.A., Hunziker E.B., Chubinskaya S. Cells, soluble factors and matrix harmonically play the concert of allograft integration. Knee Surg. Sports Traumatol. Arthrosc. 2019;27:1717–1725. doi: 10.1007/s00167-018-5182-1. [DOI] [PubMed] [Google Scholar]
- 2.Li C., Lv H., Du Y., Zhu W., Yang W., Wang X., Wang J., Chen W. Biologically modified implantation as therapeutic bioabsorbable materials for bone defect repair. Regenerative therapy. 2021;19:9–23. doi: 10.1016/j.reth.2021.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rudin N., Sugarman E., Haber J.E. Genetic and physical analysis of double-strand break repair and recombination in Saccharomyces cerevisiae. Genetics. 1989;122:519–534. doi: 10.1093/genetics/122.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhakta M.S., Henry I.M., Ousterout D.G., Das K.T., Lockwood S.H., Meckler J.F., Wallen M.C., Zykovich A., Yu Y., Leo H., Xu L., Gersbach C.A., Segal D.J. Highly active zinc-finger nucleases by extended modular assembly. Genome Res. 2013;23:530–538. doi: 10.1101/gr.143693.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., Kay S., Lahaye T., Nickstadt A., Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 6.Jansen R., Embden J.D., Gaastra W., Schouls L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 7.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Jaklenec A., Hinckfuss A., Bilgen B., Ciombor D.M., Aaron R., Mathiowitz E. Sequential release of bioactive IGF-I and TGF-beta 1 from PLGA microsphere-based scaffolds. Biomaterials. 2008;29:1518–1525. doi: 10.1016/j.biomaterials.2007.12.004. [DOI] [PubMed] [Google Scholar]; b) Wu G., Xiao M., Xiao J., Guo L., Ke Y., Li H., Fang L., Deng C., Liao H. Elastic polyurethane bearing pendant TGF-β1 affinity peptide for potential tissue engineering applications. Mater Sci Eng C Mater Biol Appl. 2018;83:67–77. doi: 10.1016/j.msec.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 9.a) Schmal H., Zwingmann J., Fehrenbach M., Finkenzeller G., Stark G.B., Südkamp N.P., Hartl D., Mehlhorn A.T. bFGF influences human articular chondrocyte differentiation. Cytotherapy. 2007;9:184–193. doi: 10.1080/14653240601182846. [DOI] [PubMed] [Google Scholar]; b) Shen X., Zhu T., Xue J., Zhang Y., Lu Y., Yang H., Yu Z., Zhu Y., Zhu X. Influence of bFGF on in vitro expansion and chondrogenic construction of articular cartilage-derived progenitor cells. Ann. Transl. Med. 2022;10:36. doi: 10.21037/atm-21-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Wang X., Oyane A., Tsurushima H., Sogo Y., Li X., Ito A. BMP-2 and ALP gene expression induced by a BMP-2 gene-fibronectin-apatite composite layer. Biomed. Mater. 2011;6 doi: 10.1088/1748-6041/6/4/045004. [DOI] [PubMed] [Google Scholar]; b) Tenkumo T., Vanegas Sáenz J.R., Nakamura K., Shimizu Y., Sokolova V., Epple M., Kamano Y., Egusa H., Sugaya T., Sasaki K. Prolonged release of bone morphogenetic protein-2 in vivo by gene transfection with DNA-functionalized calcium phosphate nanoparticle-loaded collagen scaffolds. Mater Sci Eng C Mater Biol Appl. 2018;92:172–183. doi: 10.1016/j.msec.2018.06.047. [DOI] [PubMed] [Google Scholar]
- 11.Komori T. Functions of osteocalcin in bone, pancreas, testis, and muscle. Int. J. Mol. Sci. 2020;21:7513. doi: 10.3390/ijms21207513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu M.N., Chang Y.H., Truong V.A., Lai P.L., Nguyen T.K.N., Hu Y.C. CRISPR technologies for stem cell engineering and regenerative medicine. Biotechnol. Adv. 2019;37 doi: 10.1016/j.biotechadv.2019.107447. [DOI] [PubMed] [Google Scholar]
- 13.Arthur A., Gronthos S. Clinical application of bone marrow mesenchymal stem/stromal cells to repair skeletal tissue. Int. J. Mol. Sci. 2020;21:9759. doi: 10.3390/ijms21249759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S.W., Gao C., Zheng Y.M., Yi L., Lu J.C., Huang X.Y., Cai J.B., Zhang P.F., Cui Y.H., Ke A.W. Current applications and future perspective of CRISPR/Cas9 gene editing in cancer. Mol. Cancer. 2022;21:57. doi: 10.1186/s12943-022-01518-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang F., Wen Y., Guo X. CRISPR/Cas9 for genome editing: progress, implications and challenges. Hum. Mol. Genet. 2014;23:R40–R46. doi: 10.1093/hmg/ddu125. [DOI] [PubMed] [Google Scholar]
- 16.Lino C.A., Harper J.C., Carney J.P., Timlin J.A. Delivering CRISPR: a review of the challenges and approaches. Drug Deliv. 2018;25:1234–1257. doi: 10.1080/10717544.2018.1474964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savić N., Schwank G. Advances in therapeutic CRISPR/Cas9 genome editing. Transl. Res. 2016;168:15–21. doi: 10.1016/j.trsl.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 18.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346 doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 19.Pavlovic K., Tristán-Manzano M., Maldonado-Pérez N., Cortijo-Gutierrez M., Sánchez-Hernández S., Justicia-Lirio P., Carmona M.D., Herrera C., Martin F., Benabdellah K. Using gene editing approaches to fine-tune the immune system. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.570672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi J., Chen W., Suiter C.C., Lee C., Chardon F.M., Yang W., Leith A., Daza R.M., Martin B., Shendure J. Precise genomic deletions using paired prime editing. Nat. Biotechnol. 2022;40:218–226. doi: 10.1038/s41587-021-01025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corsi G.I., Qu K., Alkan F., Pan X., Luo Y., Gorodkin J. CRISPR/Cas9 gRNA activity depends on free energy changes and on the target PAM context. Nat. Commun. 2022;13:3006. doi: 10.1038/s41467-022-30515-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S., Koonin E.V., Sharp P.A., Zhang F. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishimasu H., Cong L., Yan W.X., Ran F.A., Zetsche B., Li Y., Kurabayashi A., Ishitani R., Zhang F., Nureki O. Crystal structure of Staphylococcus aureus Cas9. Cell. 2015;162:1113–1126. doi: 10.1016/j.cell.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang F., Doudna J.A. CRISPR-Cas9 structures and mechanisms. Annu. Rev. Biophys. 2017;46:505–529. doi: 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- 25.Makarova K.S., Wolf Y.I., Shmakov S.A., Liu Y., Li M., Koonin E.V. Unprecedented diversity of unique CRISPR-cas-related systems and Cas1 homologs in asgard archaea. CRISPR J. 2020;3:156–163. doi: 10.1089/crispr.2020.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romsdahl J., Schultzhaus Z., Chen A., Liu J., Ewing A., Hervey J., Wang Z. Adaptive evolution of a melanized fungus reveals robust augmentation of radiation resistance by abrogating non-homologous end-joining. Environ. Microbiol. 2021;23:3627–3645. doi: 10.1111/1462-2920.15285. [DOI] [PubMed] [Google Scholar]
- 27.Scully R., Panday A., Elango R., Willis N.A. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 2019;20:698–714. doi: 10.1038/s41580-019-0152-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi J., Ma Y., Zhu J., Chen Y., Sun Y., Yao Y., Yang Z., Xie J. A review on electroporation-based intracellular delivery. Molecules. 2018;23:3044. doi: 10.3390/molecules23113044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bassetto M., Sen M., Poulhes F., Arango-Gonzalez B., Bonvin E., Sapet C., Ueffing M., Zelphati O. New method for efficient siRNA delivery in retina explants: reverse magnetofection. Bioconjugate Chem. 2021;32:1078–1093. doi: 10.1021/acs.bioconjchem.1c00132. [DOI] [PubMed] [Google Scholar]
- 30.DiTommaso T., Cole J.M., Cassereau L., Buggé J.A., Hanson J., Bridgen D.T., Stokes B.D., Loughhead S.M., Beutel B.A., Gilbert J.B., Nussbaum K., Sorrentino A., Toggweiler J., Schmidt T., Gyuelveszi G., Bernstein H., Sharei A. Cell engineering with microfluidic squeezing preserves functionality of primary immune cells in vivo. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E10907–E10914. doi: 10.1073/pnas.1809671115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shapiro G., Wong A.W., Bez M., Yang F., Tam S., Even L., Sheyn D., Ben-David S., Tawackoli W., Pelled G., Ferrara K.W., Gazit D. Multiparameter evaluation of in vivo gene delivery using ultrasound-guided, microbubble-enhanced sonoporation. J. Contr. Release. 2016;223:157–164. doi: 10.1016/j.jconrel.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu W. Microinjection and micromanipulation: a historical perspective. Methods Mol. Biol. 2019;1874:1–16. doi: 10.1007/978-1-4939-8831-0_1. [DOI] [PubMed] [Google Scholar]
- 33.Cervia L.D., Chang C.C., Wang L., Mao M., Yuan F. Enhancing electrotransfection efficiency through improvement in nuclear entry of plasmid DNA. Mol. Ther. Nucleic Acids. 2018;11:263–271. doi: 10.1016/j.omtn.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mellott A.J., Forrest M.L., Detamore M.S. Physical non-viral gene delivery methods for tissue engineering. Ann. Biomed. Eng. 2013;41:446–468. doi: 10.1007/s10439-012-0678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yockell-Lelièvre J., Riendeau V., Gagnon S.N., Garenc C., Audette M. Efficient transfection of endothelial cells by a double-pulse electroporation method. DNA Cell Biol. 2009;28:561–566. doi: 10.1089/dna.2009.0915. [DOI] [PubMed] [Google Scholar]
- 36.Saraf A., Mikos A.G. Gene delivery strategies for cartilage tissue engineering. Adv. Drug Deliv. Rev. 2006;58:592–603. doi: 10.1016/j.addr.2006.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saijilafu Hur EM., Zhou F.Q. Genetic dissection of axon regeneration via in vivo electroporation of adult mouse sensory neurons. Nat. Commun. 2011;2:543. doi: 10.1038/ncomms1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ozyigit Gene transfer to plants by electroporation: methods and applications. Mol. Biol. Rep. 2020;47:3195–3210. doi: 10.1007/s11033-020-05343-4. [DOI] [PubMed] [Google Scholar]
- 39.Cervia L.D., Yuan F. Current progress in electrotransfection as a nonviral method for gene delivery. Mol. Pharm. 2018;15:3617–3624. doi: 10.1021/acs.molpharmaceut.8b00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tröder S.E., Ebert L.K., Butt L., Assenmacher S., Schermer B., Zevnik B. An optimized electroporation approach for efficient CRISPR/Cas9 genome editing in murine zygotes. PLoS One. 2018;13 doi: 10.1371/journal.pone.0196891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miao D., Giassetti M.I., Ciccarelli M., Lopez-Biladeau B., Oatley J.M. Simplified pipelines for genetic engineering of mammalian embryos by CRISPR-Cas9 electroporation. Biol. Reprod. 2019;101:177–187. doi: 10.1093/biolre/ioz075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu X., Gao D., Wang P., Chen J., Ruan J., Xu J., Xia X. Efficient homology-directed gene editing by CRISPR/Cas9 in human stem and primary cells using tube electroporation. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-30227-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Estelrich J., Escribano E., Queralt J., Busquets M.A. Iron oxide nanoparticles for magnetically-guided and magnetically-responsive drug delivery. Int. J. Mol. Sci. 2015;16:8070–8101. doi: 10.3390/ijms16048070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.a) Plank C., Schillinger U., Scherer F., Bergemann C., Rémy J.S., Krötz F., Anton M., Lausier J., Rosenecker J. The magnetofection method: using magnetic force to enhance gene delivery. Biol. Chem. 2003;384:737–747. doi: 10.1515/BC.2003.082. [DOI] [PubMed] [Google Scholar]; b) Sizikov A.A., Kharlamova M.V., Nikitin M.P., Nikitin P.I., Kolychev E.L. Nonviral locally injected magnetic vectors for in vivo gene delivery: a review of studies on magnetofection. Nanomaterials (Basel) 2021;11:1078. doi: 10.3390/nano11051078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laurentt N., Sapet C., Le Gourrierec L., Bertosio E., Zelphati O. Nucleic acid delivery using magnetic nanoparticles: the Magnetofection technology. Ther. Deliv. 2011;2:471–482. doi: 10.4155/tde.11.12. [DOI] [PubMed] [Google Scholar]
- 46.Mykhaylyk O., Zelphati O., Hammerschmid E., Anton M., Rosenecker J., Plank C. Recent advances in magnetofection and its potential to deliver siRNAs in vitro. Methods Mol. Biol. 2009;487:111–146. doi: 10.1007/978-1-60327-547-7_6. [DOI] [PubMed] [Google Scholar]
- 47.Hryhorowicz M., Grześkowiak B., Mazurkiewicz N., Śledziński P., Lipiński D., Słomski R. Improved delivery of CRISPR/Cas9 system using magnetic nanoparticles into porcine fibroblast. Mol. Biotechnol. 2019;61:173–180. doi: 10.1007/s12033-018-0145-9. [DOI] [PubMed] [Google Scholar]
- 48.Han X., Liu Z., Ma Y., Zhang K., Qin L. Cas9 ribonucleoprotein delivery via microfluidic cell-deformation chip for human T-cell genome editing and immunotherapy. Adv Biosyst. 2017;1 doi: 10.1002/adbi.201600007. [DOI] [PubMed] [Google Scholar]
- 49.Han X., Liu Z., Jo M.C., Zhang K., Li Y., Zeng Z., Li N., Zu Y., Qin L. CRISPR-Cas9 delivery to hard-to-transfect cells via membrane deformation. Sci. Adv. 2015;1 doi: 10.1126/sciadv.1500454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sharei A., Zoldan J., Adamo A., Sim W.Y., Cho N., Jackson E., Mao S., Schneider S., Han M.J., Lytton-Jean A., Basto P.A., Jhunjhunwala S., Lee J., Heller D.A., Kang J.W., Hartoularos G.C., Kim K.S., Anderson D.G., Langer R., Jensen K.F. A vector-free microfluidic platform for intracellular delivery. Proc. Natl. Acad. Sci. U. S. A. 2013;110:2082–2087. doi: 10.1073/pnas.1218705110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.a) Krasovitski B., Frenkel V., Shoham S., Kimmel E. Intramembrane cavitation as a unifying mechanism for ultrasound-induced bioeffects. Proc. Natl. Acad. Sci. U.S.A. 2011;108:3258–3263. doi: 10.1073/pnas.1015771108. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Vasan A., Orosco J., Magaram U., Duque M., Weiss C., Tufail Y., Chalasani S.H., Friend J. Ultrasound mediated cellular deflection results in cellular depolarization. Adv Sci (Weinh). 2022;9 doi: 10.1002/advs.202101950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.a) O'Neill B.E., Li K.C. Augmentation of targeted delivery with pulsed high intensity focused ultrasound. Int. J. Hyperther. 2008;24:506–520. doi: 10.1080/02656730802093661. [DOI] [PubMed] [Google Scholar]; b) Sitta J., Howard C.M. Applications of ultrasound-mediated drug delivery and gene therapy. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms222111491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sum C.H., Shortall S.M., Wong S., Wettig S.D. Non-viral Gene Delivery. 2018;110:3–68. doi: 10.1007/978-3-319-78259-1_2. [DOI] [PubMed] [Google Scholar]
- 54.Böhmer M.R., Klibanov A.L., Tiemann K., Hall C.S., Gruell H., Steinbach O.C. Ultrasound triggered image-guided drug delivery. Eur. J. Radiol. 2009;70:242–253. doi: 10.1016/j.ejrad.2009.01.051. [DOI] [PubMed] [Google Scholar]
- 55.Ryu J.Y., Won E.J., Lee H., Kim J.H., Hui E., Kim H.P., Yoon T.J. Ultrasound-activated particles as CRISPR/Cas9 delivery system for androgenic alopecia therapy. Biomaterials. 2020;232 doi: 10.1016/j.biomaterials.2019.119736. [DOI] [PubMed] [Google Scholar]
- 56.Dong W., Huang A., Huang J., Wu P., Guo S., Liu H., Qin M., Yang X., Zhang B., Wan M., Zong Y. Plasmid-loadable magnetic/ultrasound-responsive nanodroplets with a SPIO-NP dispersed perfluoropentane core and lipid shell for tumor-targeted intracellular plasmid delivery. Biomater. Sci. 2020;8:5329–5345. doi: 10.1039/d0bm00699h. [DOI] [PubMed] [Google Scholar]
- 57.a) Kucherlapati R., Skoultchi A.I. Introduction of purified genes into mammalian cells. CRC Crit. Rev. Biochem. 1984;16:349–379. doi: 10.3109/10409238409108719. [DOI] [PubMed] [Google Scholar]; b) Xu W. Microinjection and micromanipulation: a historical perspective. Methods Mol. Biol. 2019;18741:1–16. doi: 10.1007/978-1-4939-8831-0_1. [DOI] [PubMed] [Google Scholar]
- 58.Gurumurthy C.B., O'Brien A.R., Quadros R.M., Adams J., Jr., Alcaide P., Ayabe S., Ballard J., Batra S.K., Beauchamp M.C., Becker K.A., Bernas G., Brough D., Carrillo-Salinas F., Chan W., Chen H., Dawson R., DeMambro V., D'Hont J., Dibb K.M., Eudy J.D.…Burgio G. Reproducibility of CRISPR-Cas9 methods for generation of conditional mouse alleles: a multi-center evaluation. Genome Biol. 2019;20:171. doi: 10.1186/s13059-019-1776-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ai D., Wang B., Fan Z., Fu Y., Yu C., Wang G. Embryo microinjection and knockout mutant identification of CRISPR/Cas9 genome-edited helicoverpa armigera (hübner) JoVE. 2021:173. doi: 10.3791/62068. [DOI] [PubMed] [Google Scholar]
- 60.Li Y., Adur M.K., Wang W., Schultz R.B., Hale B., Wierson W., Charley S.E., McGrail M., Essner J., Tuggle C.K., Ross J.W. Effect of ARTEMIS (DCLRE1C) deficiency and microinjection timing on editing efficiency during somatic cell nuclear transfer and in vitro fertilization using the CRISPR/Cas9 system. Theriogenology. 2021;170:107–116. doi: 10.1016/j.theriogenology.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schambach A., Zychlinski D., Ehrnstroem B., Baum C. Biosafety features of lentiviral vectors. Hum. Gene Ther. 2013;24:132–142. doi: 10.1089/hum.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gutierrez-Guerrero A., Cosset F.L., Verhoeyen E. Lentiviral vector pseudotypes: precious tools to improve gene modification of hematopoietic cells for research and gene therapy. Viruses. 2020;12:1016. doi: 10.3390/v12091016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Naldini L., Blömer U., Gallay P., Ory D., Mulligan R., Gage F.H., Verma I.M., Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 64.Bruner K.M., Wang Z., Simonetti F.R., Bender A.M., Kwon K.J., Sengupta S., Fray E.J., Beg S.A., Antar A., Jenike K.M., Bertagnolli L.N., Capoferri A.A., Kufera J.T., Timmons A., Nobles C., Gregg J., Wada N., Ho Y.C., Zhang H., Margolick J.B.…Siliciano R.F. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature. 2019;566:120–125. doi: 10.1038/s41586-019-0898-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen Y., Liu X., Zhang Y., Wang H., Ying H., Liu M., Li D., Lui K.O., Ding Q. A self-restricted CRISPR system to reduce off-target effects. Mol. Ther. 2016;24:1508–1510. doi: 10.1038/mt.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Joung J., Konermann S., Gootenberg J.S., Abudayyeh O.O., Platt R.J., Brigham M.D., Sanjana N.E., Zhang F. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc. 2017;12:828–863. doi: 10.1038/nprot.2017.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chang J. Adenovirus vectors: excellent tools for vaccine development. Immune Netw. 2021;21:e6. doi: 10.4110/in.2021.21.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee C.S., Bishop E.S., Zhang R., Yu X., Farina E.M., Yan S., Zhao C., Zheng Z., Shu Y., Wu X., Lei J., Li Y., Zhang W., Yang C., Wu K., Wu Y., Ho S., Athiviraham A., Lee M.J., Wolf J.M.…He T.C. Adenovirus-mediated gene delivery: potential applications for gene and cell-based therapies in the new era of personalized medicine. Genes & diseases. 2017;4:43–63. doi: 10.1016/j.gendis.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bulcha J.T., Wang Y., Ma H., Tai P.W.L., Gao G. Viral vector platforms within the gene therapy landscape. Signal Transduct. Targeted Ther. 2021;6:53. doi: 10.1038/s41392-021-00487-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsukamoto T., Sakai E., Iizuka S., Taracena-Gándara M., Sakurai F., Mizuguchi H. Generation of the adenovirus vector-mediated CRISPR/Cpf1 system and the application for primary human hepatocytes prepared from humanized mice with chimeric liver. Biol. Pharm. Bull. 2018;41:1089–1095. doi: 10.1248/bpb.b18-00222. [DOI] [PubMed] [Google Scholar]
- 71.Svenson S., Tomalia D.A. Dendrimers in biomedical applications--reflections on the field. Adv. Drug Deliv. Rev. 2005;57:2106–2129. doi: 10.1016/j.addr.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 72.Farbiak L., Cheng Q., Wei T., Álvarez-Benedicto E., Johnson L.T., Lee S., Siegwart D.J. All-in-one dendrimer-based lipid nanoparticles enable precise HDR-mediated gene editing in vivo. Adv. Mater. 2021;33 doi: 10.1002/adma.202006619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu C., Wan T., Wang H., Zhang S., Ping Y., Cheng Y. A boronic acid-rich dendrimer with robust and unprecedented efficiency for cytosolic protein delivery and CRISPR-Cas9 gene editing. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aaw8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zahednezhad F., Saadat M., Valizadeh H., Zakeri-Milani P., Baradaran B. Liposome and immune system interplay: challenges and potentials. J. Contr. Release. 2019;305:194–209. doi: 10.1016/j.jconrel.2019.05.030. [DOI] [PubMed] [Google Scholar]
- 75.Rosenblum D., Gutkin A., Kedmi R., Ramishetti S., Veiga N., Jacobi A.M., Schubert M.S., Friedmann-Morvinski D., Cohen Z.R., Behlke M.A., Lieberman J., Peer D. CRISPR-Cas9 genome editing using targeted lipid nanoparticles for cancer therapy. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abc9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Han J.P., Kim M., Choi B.S., Lee J.H., Lee G.S., Jeong M., Lee Y., Kim E.A., Oh H.K., Go N., Lee H., Lee K.J., Kim U.G., Lee J.Y., Kim S., Chang J., Lee H., Song D.W., Yeom S.C. In vivo delivery of CRISPR-Cas9 using lipid nanoparticles enables antithrombin gene editing for sustainable hemophilia A and B therapy. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abj6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kenjo E., Hozumi H., Makita Y., Iwabuchi K.A., Fujimoto N., Matsumoto S., Kimura M., Amano Y., Ifuku M., Naoe Y., Inukai N., Hotta A. Low immunogenicity of LNP allows repeated administrations of CRISPR-Cas9 mRNA into skeletal muscle in mice. Nat. Commun. 2021;12:7101. doi: 10.1038/s41467-021-26714-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guerrero-Hernández L., Meléndez-Ortiz H.I., Cortez-Mazatan G.Y., Vaillant-Sánchez S., Peralta-Rodríguez R.D. Gemini and bicephalous surfactants: a review on their synthesis, micelle formation, and uses. Int. J. Mol. Sci. 2022;23:1798. doi: 10.3390/ijms23031798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Navarro G., Pan J., Torchilin V.P. Micelle-like nanoparticles as carriers for DNA and siRNA. Mol. Pharm. 2015;12:301–313. doi: 10.1021/mp5007213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Abbasi S., Uchida S., Toh K., Tockary T.A., Dirisala A., Hayashi K., Fukushima S., Kataoka K. Co-encapsulation of Cas9 mRNA and guide RNA in polyplex micelles enables genome editing in mouse brain. J. Contr. Release. 2021;332:260–268. doi: 10.1016/j.jconrel.2021.02.026. [DOI] [PubMed] [Google Scholar]
- 81.a) Lao Y.H., Li M., Gao M.A., Shao D., Chi C.W., Huang D., Chakraborty S., Ho T.C., Jiang W., Wang H.X., Wang S., Leong K.W. HPV oncogene manipulation using nonvirally delivered CRISPR/Cas9 or natronobacterium gregoryi argonaute. Adv. Sci. 2018;5 doi: 10.1002/advs.201700540. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sun J., Chen Y., Xu J., Song X., Wan Z., Du Y., Ma W., Li X., Zhang L., Li S. High loading of hydrophobic and hydrophilic agents via small immunostimulatory carrier for enhanced tumor penetration and combinational therapy. Theranostics. 2020;10:1136–1150. doi: 10.7150/thno.38287. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zhao Y., Li Z., Zhu X., Cao Y., Chen X. Improving immunogenicity and safety of flagellin as vaccine carrier by high-density display on virus-like particle surface. Biomaterials. 2020;249 doi: 10.1016/j.biomaterials.2020.120030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Betz V.M., Betz O.B., Harris M.B., Vrahas M.S., Evans C.H. Bone tissue engineering and repair by gene therapy. Front. Biosci. 2008;13:833–841. doi: 10.2741/2724. [DOI] [PubMed] [Google Scholar]
- 83.Schemitsch E.H. Size matters: defining critical in bone defect size. J. Orthop. Trauma. 2017;31:S20–S22. doi: 10.1097/BOT.0000000000000978. [DOI] [PubMed] [Google Scholar]
- 84.De la Vega R.E., Atasoy-Zeybek A., Panos J.A., Van Griensven M., Evans C.H., Balmayor E.R. Gene therapy for bone healing: lessons learned and new approaches. Transl. Res. 2021;236:1–16. doi: 10.1016/j.trsl.2021.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kylmaoja E., Nakamura M., Tuukkanen J. Osteoclasts and remodeling based bone formation. Curr. Stem Cell Res. Ther. 2016;11:626–633. doi: 10.2174/1574888x10666151019115724. [DOI] [PubMed] [Google Scholar]
- 86.Arandjelovic S., Perry J., Zhou M., Ceroi A., Smirnov I., Walk S.F., Shankman L.S., Cambré I., Onengut-Gumuscu S., Elewaut D., Conrads T.P., Ravichandran K.S. ELMO1 signaling is a promoter of osteoclast function and bone loss. Nat. Commun. 2021;12:4974. doi: 10.1038/s41467-021-25239-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jones A.L., Bucholz R.W., Bosse M.J., Mirza S.K., Lyon T.R., Webb L.X., Pollak A.N., Golden J.D., Valentin-Opran A. BMP-2 Evaluation in Surgery for Tibial Trauma-Allgraft (BESTT-ALL) Study Group, Recombinant human BMP-2 and allograft compared with autogenous bone graft for reconstruction of diaphyseal tibial fractures with cortical defects. A randomized, controlled trial. J. Bone Joint Surg. 2006;88:1431–1441. doi: 10.2106/JBJS.E.00381. [DOI] [PubMed] [Google Scholar]
- 88.Wu X.B., Li Y., Schneider A., Yu W., Rajendren G., Iqbal J., Yamamoto M., Alam M., Brunet L.J., Blair H.C., Zaidi M., Abe E. Impaired osteoblastic differentiation, reduced bone formation, and severe osteoporosis in noggin-overexpressing mice. J. Clin. Investig. 2003;112:924–934. doi: 10.1172/JCI15543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hsu M.N., Yu F.J., Chang Y.H., Huang K.L., Pham N.N., Truong V.A., Lin M.W., Kieu Nguyen N.T., Hwang S.M., Hu Y.C. CRISPR interference-mediated noggin knockdown promotes BMP2-induced osteogenesis and calvarial bone healing. Biomaterials. 2020;252 doi: 10.1016/j.biomaterials.2020.120094. [DOI] [PubMed] [Google Scholar]
- 90.Beederman M., Lamplot J.D., Nan G., Wang J., Liu X., Yin L., Li R., Shui W., Zhang H., Kim S.H., Zhang W., Zhang J., Kong Y., Denduluri S., Rogers M.R., Pratt A., Haydon R.C., Luu H.H., Angeles J., Shi L.L.…He T.C. BMP signaling in mesenchymal stem cell differentiation and bone formation. J. Biomed. Sci. Eng. 2013;6:32–52. doi: 10.4236/jbise.2013.68A1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lauzon M.A., Drevelle O., Daviau A., Faucheux N. Effects of BMP-9 and BMP-2 on the PI3K/Akt pathway in mc3t3-E1 preosteoblasts. Tissue Eng. 2016;22:1075–1085. doi: 10.1089/ten.TEA.2016.0151. [DOI] [PubMed] [Google Scholar]
- 92.Freitas G.P., Lopes H.B., Souza A., Gomes M., Quiles G.K., Gordon J., Tye C., Stein J.L., Stein G.S., Lian J.B., Beloti M.M., Rosa A.L. Mesenchymal stem cells overexpressing BMP-9 by CRISPR-Cas9 present high in vitro osteogenic potential and enhance in vivo bone formation. Gene Ther. 2021;28:748–759. doi: 10.1038/s41434-021-00248-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Truong V.A., Hsu M.N., Kieu Nguyen N.T., Lin M.W., Shen C.C., Lin C.Y., Hu Y.C. CRISPRai for simultaneous gene activation and inhibition to promote stem cell chondrogenesis and calvarial bone regeneration. Nucleic Acids Res. 2019;47:e74. doi: 10.1093/nar/gkz267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ren X., Zheng D., Guo F., Liu J., Zhang B., Li H., Tian W. PPARγ suppressed Wnt/β-catenin signaling pathway and its downstream effector SOX9 expression in gastric cancer cells. Medical oncology (Northwood, London, England) 2015;32:91. doi: 10.1007/s12032-015-0536-8. [DOI] [PubMed] [Google Scholar]
- 95.Kaushal K., Tyagi A., Karapurkar J.K., Kim E.J., Tanguturi P., Kim K.S., Jung H.S., Ramakrishna S. Genome-wide CRISPR/Cas9-Based screening for deubiquitinase subfamily identifies ubiquitin-specific protease 11 as a novel regulator of osteogenic differentiation. Int. J. Mol. Sci. 2022;23:856. doi: 10.3390/ijms23020856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Houschyar K.S., Tapking C., Borrelli M.R., Popp D., Duscher D., Maan Z.N., Chelliah M.P., Li J., Harati K., Wallner C., Rein S., Pförringer D., Reumuth G., Grieb G., Mouraret S., Dadras M., Wagner J.M., Cha J.Y., Siemers F., Lehnhardt M.…Behr B. Wnt pathway in bone repair and regeneration - what do we know so far. Front. Cell Dev. Biol. 2019;6:170. doi: 10.3389/fcell.2018.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nalesso G., Thomas B.L., Sherwood J.C., Yu J., Addimanda O., Eldridge S.E., Thorup A.S., Dale L., Schett G., Zwerina J., Eltawil N., Pitzalis C., Dell'Accio F. WNT16 antagonises excessive canonical WNT activation and protects cartilage in osteoarthritis. Ann. Rheum. Dis. 2017;76:218–226. doi: 10.1136/annrheumdis-2015-208577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McGowan L.M., Kague E., Vorster A., Newham E., Cross S., Hammond C.L. Wnt16 elicits a protective effect against fractures and supports bone repair in zebrafish. JBMR Plus. 2021;5 doi: 10.1002/jbm4.10461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dobreva G., Chahrour M., Dautzenberg M., Chirivella L., Kanzler B., Fariñas I., Karsenty G., Grosschedl R. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell. 2006;125:971–986. doi: 10.1016/j.cell.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 100.Dowrey T., Schwager E.E., Duong J., Merkuri F., Zarate Y.A., Fish J.L. Satb2 regulates proliferation and nuclear integrity of pre-osteoblasts. Bone. 2019;127:488–498. doi: 10.1016/j.bone.2019.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fang P., Zhang L.X., Hu Y., Zhang L., Zhou L.W. Long non-coding RNA DANCR induces chondrogenesis by regulating the miR-1275/MMP-13 axis in synovial fluid-derived mesenchymal stem cells. Eur. Rev. Med. Pharmacol. Sci. 2019;23:10459–10469. doi: 10.26355/eurrev_201912_19685. [DOI] [PubMed] [Google Scholar]
- 102.Nguyen N., Chang Y.H., Truong V.A., Hsu M.N., Pham N.N., Chang C.W., Wu Y.H., Chang Y.H., Li H., Hu Y.C. CRISPR activation of long non-coding RNA DANCR promotes bone regeneration. Biomaterials. 2021;275 doi: 10.1016/j.biomaterials.2021.120965. [DOI] [PubMed] [Google Scholar]
- 103.Seidl C.I., Fulga T.A., Murphy C.L. CRISPR-Cas9 targeting of MMP13 in human chondrocytes leads to significantly reduced levels of the metalloproteinase and enhanced type II collagen accumulation. Osteoarthritis Cartilage. 2019;27:140–147. doi: 10.1016/j.joca.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 104.Farhang N., Davis B., Weston J., Ginley-Hidinger M., Gertz J., Bowles R.D. Synergistic CRISPRa-regulated chondrogenic extracellular matrix deposition without exogenous growth factors. Tissue Eng. 2020;26:1169–1179. doi: 10.1089/ten.tea.2020.0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yue K., Trujillo-de Santiago G., Alvarez M.M., Tamayol A., Annabi N., Khademhosseini A. Synthesis, properties, and biomedical applications of gelatin methacryloyl (GelMA) hydrogels. Biomaterials. 2015;73:254–271. doi: 10.1016/j.biomaterials.2015.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nonaka K., Han X., Kato H., Sato H., Yamaza H., Hirofuji Y., Masuda K. Novel gain-of-function mutation of TRPV4 associated with accelerated chondrogenic differentiation of dental pulp stem cells derived from a patient with metatropic dysplasia. Biochem. Biophy. Rep. 2019;19 doi: 10.1016/j.bbrep.2019.100648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Burr D.B., Gallant M.A. Bone remodelling in osteoarthritis. Nat. Rev. Rheumatol. 2012;8:665–673. doi: 10.1038/nrrheum.2012.130. [DOI] [PubMed] [Google Scholar]
- 108.Mayan M.D., Carpintero-Fernandez P., Gago-Fuentes R., Martinez-de-Ilarduya O., Wang H.Z., Valiunas V., Brink P., Blanco F.J. Human articular chondrocytes express multiple gap junction proteins: differential expression of connexins in normal and osteoarthritic cartilage. Am. J. Pathol. 2013;182:1337–1346. doi: 10.1016/j.ajpath.2012.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Varela-Eirín M., Varela-Vázquez A., Guitián-Caamaño A., Paíno C.L., Mato V., Largo R., Aasen T., Tabernero A., Fonseca E., Kandouz M., Caeiro J.R., Blanco A., Mayán M.D. Targeting of chondrocyte plasticity via connexin43 modulation attenuates cellular senescence and fosters a pro-regenerative environment in osteoarthritis. Cell Death Dis. 2018;9:1166. doi: 10.1038/s41419-018-1225-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu S. Mesenchymal stem cell engineering. Methods Mol. Biol. 2018;1868:145–150. doi: 10.1007/978-1-4939-8802-0_15. [DOI] [PubMed] [Google Scholar]
- 111.Luz-Crawford P., Jorgensen C., Djouad F. Mesenchymal stem cells direct the immunological fate of macrophages. Results Probl. Cell Differ. 2017;62:61–72. doi: 10.1007/978-3-319-54090-0_4. [DOI] [PubMed] [Google Scholar]
- 112.Yoshida Y., Yamanaka S. Recent stem cell advances: induced pluripotent stem cells for disease modeling and stem cell-based regeneration. Circulation. 2010;122:80–87. doi: 10.1161/CIRCULATIONAHA.109.881433. [DOI] [PubMed] [Google Scholar]
- 113.Cahan P., Daley G.Q. Origins and implications of pluripotent stem cell variability and heterogeneity. Nat. Rev. Mol. Cell Biol. 2013;14:357–368. doi: 10.1038/nrm3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vinod E., Boopalan P.R.J.V.C., Sathishkumar S. Reserve or resident progenitors in cartilage? Comparative analysis of chondrocytes versus chondroprogenitors and their role in cartilage repair. Cartilage. 2018;9:171–182. doi: 10.1177/1947603517736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Su X., Zuo W., Wu Z., Chen J., Wu N., Ma P., Xia Z., Jiang C., Ye Z., Liu S., Liu J., Zhou G., Wan C., Qiu G. CD146 as a new marker for an increased chondroprogenitor cell sub-population in the later stages of osteoarthritis. J. Orthop. Res. 2015;33:84–91. doi: 10.1002/jor.22731. [DOI] [PubMed] [Google Scholar]
- 116.Yang Y.K., Ogando C.R., Wang See C., Chang T.Y., Barabino G.A. Changes in phenotype and differentiation potential of human mesenchymal stem cells aging in vitro. Stem Cell Res. Ther. 2018;9:131. doi: 10.1186/s13287-018-0876-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dicks A., Wu C.L., Steward N., Adkar S.S., Gersbach C.A., Guilak F. Prospective isolation of chondroprogenitors from human iPSCs based on cell surface markers identified using a CRISPR-Cas9-generated reporter. Stem Cell Res. Ther. 2020;11:66. doi: 10.1186/s13287-020-01597-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bardai G., Moffatt P., Glorieux F.H., Rauch F. DNA sequence analysis in 598 individuals with a clinical diagnosis of osteogenesis imperfecta: diagnostic yield and mutation spectrum. Osteoporos. Int. 2016;27:3607–3613. doi: 10.1007/s00198-016-3709-1. [DOI] [PubMed] [Google Scholar]
- 119.Marini J.C., Forlino A., Bächinger H.P., Bishop N.J., Byers P.H., Paepe A., Fassier F., Fratzl-Zelman N., Kozloff K.M., Krakow D., Montpetit K., Semler O. Osteogenesis imperfecta. Nat. Rev. Dis. Prim. 2017;3 doi: 10.1038/nrdp.2017.52. [DOI] [PubMed] [Google Scholar]
- 120.Prockop D.J. Seminars in medicine of the Beth Israel Hospital, Boston. Mutations in collagen genes as a cause of connective-tissue diseases. N. Engl. J. Med. 1992;326:540–546. doi: 10.1056/NEJM199202203260807. [DOI] [PubMed] [Google Scholar]
- 121.Jung H., Rim Y.A., Park N., Nam Y., Ju J.H. Restoration of osteogenesis by CRISPR/Cas9 genome editing of the mutated COL1A1 gene in osteogenesis imperfecta. J. Clin. Med. 2021;10:3141. doi: 10.3390/jcm10143141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rauch F., Geng Y., Lamplugh L., Hekmatnejad B., Gaumond M.H., Penney J., Yamanaka Y., Moffatt P. Crispr-Cas9 engineered osteogenesis imperfecta type V leads to severe skeletal deformities and perinatal lethality in mice. Bone. 2018;107:131–142. doi: 10.1016/j.bone.2017.11.013. [DOI] [PubMed] [Google Scholar]
- 123.Choudhury A., Fankhauser R.G., Freed E.F., Oh E.J., Morgenthaler A.B., Bassalo M.C., Copley S.D., Kaar J.L., Gill R.T. Determinants for efficient editing with cas9-mediated recombineering in Escherichia coli. ACS Synth. Biol. 2020;9:1083–1099. doi: 10.1021/acssynbio.9b00440. [DOI] [PubMed] [Google Scholar]
- 124.Alkan F., Wenzel A., Anthon C., Havgaard J.H., Gorodkin J. CRISPR-Cas9 off-targeting assessment with nucleic acid duplex energy parameters. Genome Biol. 2018;19:177. doi: 10.1186/s13059-018-1534-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chen S., Yao Y., Zhang Y., Fan G. CRISPR system: discovery, development and off-target detection. Cell. Signal. 2020;70 doi: 10.1016/j.cellsig.2020.109577. [DOI] [PubMed] [Google Scholar]
- 12.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pattanayak V., Lin S., Guilinger J.P., Ma E., Doudna J.A., Liu D.R. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bae S., Park J., Kim J.S., Cas-Offinder A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Haeussler M., Schönig K., Eckert H., Eschstruth A., Mianné J., Renaud J.B., Schneider-Maunoury S., Shkumatava A., Teboul L., Kent J., Joly J.S., Concordet J.P. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17:148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O., Cradick T.J., Marraffini L.A., Bao G., Zhang F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang Y., Cheng X., Shan Q., Zhang Y., Liu J., Gao C., Qiu J.L. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 2014;32:947–951. doi: 10.1038/nbt.2969. [DOI] [PubMed] [Google Scholar]
- 132.Wong N., Liu W., Wang X., Wu-Crispr Characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol. 2015;16:218. doi: 10.1186/s13059-015-0784-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ren X., Yang Z., Xu J., Sun J., Mao D., Hu Y., Yang S.J., Qiao H.H., Wang X., Hu Q., Deng P., Liu L.P., Ji J.Y., Li J.B., Ni J.Q. Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell Rep. 2014;9:1151–1162. doi: 10.1016/j.celrep.2014.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ryan D.E., Taussig D., Steinfeld I., Phadnis S.M., Lunstad B.D., Singh M., Vuong X., Okochi K.D., McCaffrey R., Olesiak M., Roy S., Yung C.W., Curry B., Sampson J.R., Bruhn L., Dellinger D.J. Improving CRISPR-Cas specificity with chemical modifications in single-guide RNAs. Nucleic Acids Res. 2018;46:792–803. doi: 10.1093/nar/gkx1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kocak D.D., Josephs E.A., Bhandarkar V., Adkar S.S., Kwon J.B., Gersbach C.A. Increasing the specificity of CRISPR systems with engineered RNA secondary structures. Nat. Biotechnol. 2019;37:657–666. doi: 10.1038/s41587-019-0095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Vakulskas C.A., Behlke M.A. Evaluation and reduction of CRISPR off-target cleavage events. Nucleic Acid Therapeut. 2019;29:167–174. doi: 10.1089/nat.2019.0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z., Gonzales A.P., Li Z., Peterson R.T., Yeh J.R., Aryee M.J., Joung J.K. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ma X., Zhang Q., Zhu Q., Liu W., Chen Y., Qiu R., Wang B., Yang Z., Li H., Lin Y., Xie Y., Shen R., Chen S., Wang Z., Chen Y., Guo J., Chen L., Zhao X., Dong Z., Liu Y.G. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant. 2015;8:1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 140.Farboud B., Meyer B.J. Dramatic enhancement of genome editing by CRISPR/Cas9 through improved guide RNA design. Genetics. 2015;199:959–971. doi: 10.1534/genetics.115.175166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Goffeau A., Barrell B.G., Bussey H., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Louis E.J., Mewes H.W., Murakami Y., Philippsen P., Tettelin H., Oliver S.G. Life with 6000 genes. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 142.Wang T., Wei J.J., Sabatini D.M., Lander E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Xu H., Xiao T., Chen C.H., Li W., Meyer C.A., Wu Q., Wu D., Cong L., Zhang F., Liu J.S., Brown M., Liu X.S. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 2015;25:1147–1157. doi: 10.1101/gr.191452.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Doench J.G., Hartenian E., Graham D.B., Tothova Z., Hegde M., Smith I., Sullender M., Ebert B.L., Xavier R.J., Root D.E. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chen M., Mao A., Xu M., Weng Q., Mao J., Ji J. CRISPR-Cas9 for cancer therapy: opportunities and challenges. Cancer Lett. 2019;447:48–55. doi: 10.1016/j.canlet.2019.01.017. [DOI] [PubMed] [Google Scholar]
- 146.Deriano L., Roth D.B. Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu. Rev. Genet. 2013;47:433–455. doi: 10.1146/annurev-genet-110711-155540. [DOI] [PubMed] [Google Scholar]
- 147.Ma Y., Chen W., Zhang X., Yu L., Dong W., Pan S., Gao S., Huang X., Zhang L. Increasing the efficiency of CRISPR/Cas9-mediated precise genome editing in rats by inhibiting NHEJ and using Cas9 protein. RNA Biol. 2016;13:605–612. doi: 10.1080/15476286.2016.1185591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.He X., Tan C., Wang F., Wang Y., Zhou R., Cui D., You W., Zhao H., Ren J., Feng B. Knock-in of large reporter genes in human cells via CRISPR/Cas9-induced homology-dependent and independent DNA repair. Nucleic Acids Res. 2016;44:e85. doi: 10.1093/nar/gkw064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ling X., Xie B., Gao X., Chang L., Zheng W., Chen H., Huang Y., Tan L., Li M., Liu T. Improving the efficiency of precise genome editing with site-specific Cas9-oligonucleotide conjugates. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aaz0051. eaaz0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hemphill J., Borchardt E.K., Brown K., Asokan A., Deiters A. Optical control of CRISPR/Cas9 gene editing. J. Am. Chem. Soc. 2015;137:5642–5645. doi: 10.1021/ja512664v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang Z., Cui W. CRISPR-Cas system for biomedical diagnostic platforms. View. 2020;1 [Google Scholar]