Abstract
Paracoccus denitrificans strains with mutations in the genes encoding the cytochrome c550, c552, or c1 and in combinations of these genes were constructed, and their growth characteristics were determined. Each mutant was able to grow heterotrophically with succinate as the carbon and free-energy source, although their specific growth rates and maximum cell numbers fell variably behind those of the wild type. Maximum cell numbers and rates of growth were also reduced when these strains were grown with methylamine as the sole free-energy source, with the triple cytochrome c mutant failing to grow on this substrate. Under anaerobic conditions in the presence of nitrate, none of the mutant strains lacking the cytochrome bc1 complex reduced nitrite, which is cytotoxic and accumulated in the medium. The cytochrome c550-deficient mutant did denitrify provided copper was present. The cytochrome c552 mutation had no apparent effect on the denitrifying potential of the mutant cells. The studies show that the cytochromes c have multiple tasks in electron transfer. The cytochrome bc1 complex is the electron acceptor of the Q-pool and of amicyanin. It is also the electron donor to cytochromes c550 and c552 and to the cbb3-type oxidase. Cytochrome c552 is an electron acceptor both of the cytochrome bc1 complex and of amicyanin, as well as a dedicated electron donor to the aa3-type oxidase. Cytochrome c550 can accept electrons from the cytochrome bc1 complex and from amicyanin, whereas it is also the electron donor to both cytochrome c oxidases and to at least the nitrite reductase during denitrification. Deletion of the c-type cytochromes also affected the concentrations of remaining cytochromes c, suggesting that the organism is plastic in that it adjusts its infrastructure in response to signals derived from changed electron transfer routes.
Many bacteria that live in soil, sewage or sludge possess a genome that encodes a highly branched respiratory network. The encoded network comprises various types of dehydrogenase and of terminal oxidoreductase, which are capable of electron input and output flow, respectively, during oxidative phosphorylation (4). Such versatility allows these bacteria to use a variety of electron donors and terminal electron acceptors once they become available in their natural habitats. The genes or gene clusters that encode the different redox enzymes are usually tightly regulated. They are expressed only under the growth conditions at which they are required. One of the best-studied organisms in that respect is Paracoccus denitrificans. This gram-negative bacterium is able to grow heterotrophically with a variety of organic carbon and free-energy sources, as well as autotrophically, by using hydrogen, or so-called C1 substrates such as methylamine or methanol, as free-energy sources (2, 3, 17, 41, 42, 46). The electron transport chain used for aerobic heterotrophic growth possesses a full complement of proteins with counterparts in the mitochondrial respiratory chain (20, 41). Upon depletion of the organic substrates, the bacterium induces hydrogenase and methanol or methylamine dehydrogenases, as well as their dedicated electron acceptors, provided that their substrates are present (16, 49).
P. denitrificans is able to reduce molecular oxygen at a wide range of oxygen concentrations. This flexibility is the result of its potential to synthesize three types of terminal oxidase (9, 10, 32, 33, 35), which all belong to the superfamily of heme copper oxidases (15, 38, 44). The one that is most expressed at atmospheric oxygen concentrations is the aa3-type cytochrome c oxidase, which has a relatively low affinity for oxygen (31, 36). Its expression level decreases with decreasing oxygen concentrations (5). The second type of oxidase is the cbb3-type cytochrome c oxidase, which has a relatively high affinity for oxygen and which is increasingly synthesized at decreasing oxygen concentrations (10, 31). The third type of oxidase is a ba3-type quinol oxidase, which is the counterpart of the bo3-type quinol oxidase found in Escherichia coli (8, 35). The ba3-type oxidase receives electrons from ubiquinol, is expressed and increasingly active under conditions that give rise to high reduction levels of the Q-pool, and apparently serves to prevent that by rapidly transferring electrons from ubiquinol to oxygen (27).
P. denitrificans is also able to synthesize four types of N-oxide oxidoreductase, which catalyze the sequential reduction of nitrate to dinitrogen gas via the intermediates nitrite, nitric oxide, and nitrous oxide. This denitrification process takes place at nearly anoxic conditions, during which nitrate substitutes for oxygen as a terminal electron acceptor. Nitrate, nitrite, nitric oxide, and nitrous oxide reductases are expressed when nitrate is present and oxygen is virtually absent (2, 3, 14, 42).
Much of our knowledge on how electrons are transferred from the dehydrogenases to the terminal oxidoreductases stems from the analyses of mutants with mutations in genes encoding electron carriers that make up the respiratory network (9, 11, 50, 51, 52, 53). However, it has thus far been difficult to understand the exact role of the different types of cytochrome c in situ since mutations in their corresponding genes did not give clear phenotypes if at all. Apparently, cytochromes c or other small redox carriers can substitute for each other. The present study sought to elucidate the roles of the cytochromes c550 and c552 and the cytochrome bc1 complex in electron transport in P. denitrificans. The approach was to isolate P. denitrificans strains with mutations in the genes encoding these three types of cytochrome c and in combinations of these genes in order to study the resultant effects of these mutations on their growth characteristics.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. E. coli strains were cultivated in yeast-tryptone medium at 37°C. P. denitrificans strains were grown at 34°C in batch cultures either aerobically, i.e., in 2-liter flasks filled with 300 ml of medium and shaken vigorously; semiaerobically, i.e., in 1-liter flasks filled with 500 ml of medium and slowly shaken; or anaerobically, i.e., in flasks almost completely filled with medium and not shaken. The P. denitrificans strains were grown on mineral batch medium containing Lawford trace solution and methylamine (100 mM), succinate (25 mM), or butyrate (25 mM) as carbon and free-energy sources at pH 7.0 (7, 24). Anaerobic cultures were supplemented with 100 mM potassium nitrate as an electron acceptor. Inhibition of the cytochrome bc1 complex was achieved by addition of 5 μM myxothiazol to the cultures (24). Cells were harvested in the mid-exponential phase of growth when the turbidity at 660 nm was ca. 1.0 cm−1. When appropriate, antibiotics were added, i.e., rifampin (40 mg liter−1), streptomycin (50 mg liter−1), kanamycin (50 mg liter−1), or ampicillin (100 mg liter−1). Residual nitrite in denitrifying cultures was determined by using nitrite strips from Boehringer.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant genotypea | Source or reference |
|---|---|---|
| Bacteria | ||
| E. coli | ||
| TG1 | supE hsdD5 thi D(lac-proAB) F′ (traD36 proAB lacIqlacZΔM15) | 37 |
| S17-1 | Smrpro r− m+ RP4-2 integrated (Tc::Mu) (Kmr::Tn7) | 39 |
| HB101 | F−hsdS20 (rB− mB−) lacY1 proA2 recA13 | 6 |
| P. denitrificans | ||
| Pd1222 | DSM413 derivative, Rifr, enhanced conjugation frequencies, m− | 13 |
| Pd21.21 | Pd1222 derivative, cycA::Kmr | 11 |
| Pd21.31 | Pd21.21 derivative, ΔcycA | 11 |
| Pd28.22 | Pd1222 derivative, cycM::Kmr | This study |
| Pd24.21 | Pd1222 derivative, fbcC::Kmr | 11 |
| Pd24.31 | Pd24.21 derivative, ΔfbcC | This study |
| Pd92.06 | Pd21.31 derivative, ΔcycA fbcC::Kmr | 11 |
| Pd92.07 | Pd92.06 derivative, ΔcycA ΔfbcC | This study |
| Pd92.26 | Pd21.31 derivative, ΔcycA cycM::Kmr | This study |
| Pd92.28 | Pd24.31 derivative, ΔfbcC cycM::Kmr | This study |
| Pd93.14 | Pd92.07 derivative, ΔcycA ΔfbcC cycM::Kmr | This study |
| Pd93.12 | Pd1222 derivative, ΔctaDI ΔctaDII ccoN::Kmr | 10 |
| Pd93.13 | Pd1222 derivative, ΔcycA ΔctaDII cycM::Kmr | This study |
| Plasmids | ||
| pGRPd1 | oriV (colE1) AmproriT Smr (Tn1831) | 51 |
| pRVS1 | pGRPd1 derivative, PTn5 lacZ | 52 |
| pMP190 | pRK2 derivative, mcs, promoterless lacZ | 40 |
| pBL250 | pBR322 derivative, fbcC | 22 |
| PRVS2 | pBSII (KS) derivative, oriT (RK2), Smr | 48 |
| pAT3 | pBSII (KS) derivative, cycM | 43 |
| pAT8 | pAT3 derivative, cycM::Kmr | 43 |
| pRTd28.22 | pRVS2 derivative, cycM::Kmr | This study |
| pRTd24.31 | pRVS1 derivative, ΔfbcC | This study |
P, promoter. Smr, streptomycin resistance.
Isolation of intact cells and membrane fractions.
Cells were washed twice in 0.1 M of Tris (Tris hydroxymethyl aminomethaan)-HCl at pH 7.5 at 4°C and then suspended in the same buffer to a turbidity of 50 at 660 nm. After the addition of MnCl2 to 10 mM and DNase I to 2 mg ml−1, cell extracts were obtained by using a French pressure cell (American Instrument Company, Silver Spring, Md.) at 10,000 lb/in2. Cell debris was spun down for 5 min at 17,000 × g, and the resulting supernatant was centrifuged at 438,000 × g for 30 min at 4°C in order to separate the soluble and membrane fractions. The pellet containing the membrane faction was resuspended in 0.1 M Tris-HCl at pH 7.5. Protein concentrations were determined by using the BCA Kit (Pierce) with bovine serum albumin as a standard. Samples (at 10 mg of protein ml−1) were routinely stored at −80°C.
PAGE and heme staining.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out by using the Bio-Rad Mini-Protean II Gel System with 13% slab gels (23). The samples (100 μg of protein each) were diluted in sample buffer (60 mM Tris-HCl, pH 6.8; 25% glycerol [vol/vol]; 2% SDS; 50 mM 2-mercaptoethanol; 50 mM dithiothreitol) and incubated at room temperature for 10 min. The heme-binding proteins were detected by using chemiluminescent substrate “Lumi-Light Plus” (Boehringer Mannheim) on a high-performance chemiluminescence film (Amersham) (29). The program NIH-Image/ppc 1.56b61 was used for densitometric analyses.
Spectral analyses.
Spectra of dithionite (5 mg/ml) reduced cell suspensions (turbidity at 660 nm of 50) were recorded in the dual-wavelength mode of an automated Aminco/SLM DW-2 UV/Vis spectrophotometer with the reference set at 578 nm. During kinetic measurements, the reduction level of cytochromes c as a function of time was recorded at 552 nm with the reference set at 578 nm. Cuvettes were thermostated at 20°C. Oxygen was released from 20 μl of 0.3% hydrogen peroxide by intracellular catalase activity. Dithionite was added to a final concentration of 5 mg/ml.
Oxygen consumption.
Oxygen consumption rates of cells were measured polarographically with a Clark-type oxygen electrode (Yellow Springs, Ohio) in 1 ml of Tris-HCl (pH 7.5) at 25°C. Succinate (10 mM) was added as an electron donor. Where indicated, the electron flow via cytochrome bc1 was inhibited by adding antimycin A (6 μM) and myxothiazol (6 μM).
DNA manipulations.
Routine cloning procedures were performed according to standard protocols (1). The construction of marked and unmarked mutations in chromosomal genes is described in Results and was performed as described earlier (52). E. coli TG1 was used routinely for cloning procedures. E. coli HB101(pRK2020) was used as the helper strain in the conjugative transfer of plasmids via triparental mating.
Construction of cytochrome c single and multiple mutant strains.
The following strains were constructed in an earlier study (11): Pd21.21 (cycA::Kmr) with an interrupted cytochrome c550 encoding gene, Pd21.31 (ΔcycA) with an unmarked mutation in the cytochrome c550 encoding gene, Pd24.21 (fbcC::Kmr) with an interrupted cytochrome c1 encoding gene, and the double mutant strain Pd92.06 (ΔcycA fbcC::Kmr). In this study, an internal part of the chromosomal fbcC gene that contained the gene cartridge encoding kanamycin resistance (Kmr) in strains Pd24.21 and Pd92.06 was removed in order to make the fbcC mutations in these strains unmarked. For this, plasmid pBL250, which is a pBR322 derivative containing the fbcC gene and flanking regions (22), was cut with XhoI and SalI and then religated. As a result, a 1,060-bp fragment within the fbcC gene was removed. The fragment with the mutated gene was then cloned in suicide vector pRVS1, yielding plasmid pRTd24.31. This plasmid was used in the gene exchange technique described earlier (52) in order to isolate strains Pd24.31 (ΔfbcC) and Pd92.07 (ΔcycA ΔfbcC), which have unmarked mutations in their chromosomal fbcC genes. In this gene exchange technique, mutants with unmarked mutations are selected by using the E. coli lacZ gene as a suicide gene (52). Plasmid pAT8 harbors the cytochrome c552 encoding cycM gene in which the gene cartridge encoding kanamycin resistance was inserted in the internal StuI site (43). A 4.6-kb NotI fragment of this plasmid, which contains the mutated cycM gene, was cloned in suicide vector pRVS2, yielding vector pRTd28.22. This plasmid was used in the gene exchange technique described earlier (51) in order to isolate strains Pd28.22 (cycM::Kmr), Pd92.26 (ΔcycA cycM::Kmr), Pd92.28 (ΔfbcC cycM::Kmr), and Pd93.14 (ΔcycA ΔfbcC cycM::Kmr), which all have marked mutations in their chromosomal cycM genes. The correctnesses of the mutations were confirmed by Southern analyses of their chromosomal DNA. As a result of these constructions, we had mutant strains with mutations in one, two, or three of the genes encoding cytochromes c550, c552, and c1.
RESULTS
Spectral analyses of the cytochrome c single and multiple mutant strains.
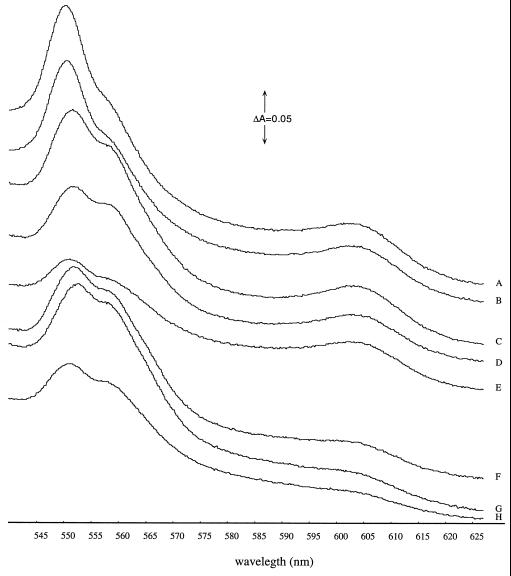
P. denitrificans wild-type and mutants with single and multiple mutations in the cycA, cycM, and fbcC genes were cultured aerobically in mineral medium containing succinate as the sole carbon and free-energy source. Cell suspensions from these cultures exhibited composite α-bands of ca. 555 nm at which cytochromes b (at ca. 560 nm) and c (at ca. 550 nm) have their maximum absorbance (Fig. 1). It should be noted that the absorbance at that wavelength reflects not only all three cytochromes c that are important here, i.e., cytochromes c550, c552, and c1, but also further contributions from other cytochromes c as well. Both the shape and the height of that composite band differed between the strains. Only the cytochrome c552-deficient mutant displayed a spectrum very similar to that of the wild-type strain. Paradoxically, strains Pd24.31 (c1 deficient) and Pd92.28 (c552/c1 deficient) had a higher cytochrome c peak than the wild-type strain, whereas that peak is lower in strains Pd21.31 and Pd92.26, which are cytochrome c550 and cytochrome c550/c552-deficient mutants, respectively. A sharp decrease in the height of that band is observed in the spectra of strains Pd92.07 and Pd93.14, which are cytochrome c550/c1- and cytochrome c550/c552/c1-deficient mutants. Another remarkable observation is that the peak at ca. 605 nm where hemes a of cytochrome aa3 have their maximum absorbance is decreased to ca. 20% of the wild-type signal in the spectra of strains Pd21.31 (cytochrome c550 deficient), Pd92.26 (cytochrome c550/c552 deficient), and Pd93.14 (cytochrome c550/c552/c1 deficient).
FIG. 1.
Dithionite-reduced absorption spectra at room temperature of cell suspensions from the wild-type strain (Pd1222 [C]) and from mutant strains deficient in cytochrome c1 (Pd24.31 [A]), cytochromes c552/c1 (Pd92.28 [B]), cytochrome c552 (Pd28.22 [D]), cytochromes c550/c1 (Pd92.07 [E]), cytochrome c550 (Pd21.31 [F]), cytochromes c550/c552 (Pd92.26 [G]), or cytochromes c550/c552/c1 (Pd93.14 [H]). The cells had been grown aerobically to their mid-exponential phase of growth (optical density at 660 nm [OD660] = ∼1.0 cm−1) in batch cultures with succinate as the carbon and free-energy source.
In order to study the effect of the oxygen concentration on the spectral properties of the mutant cells, the spectra of cells grown semiaerobically were also recorded (results not shown). These spectra show that the cytochrome c peak of strain Pd24.31 (cytochrome c1 deficient) is ca. four to five times higher than that of the wild-type strain and that the cytochrome b peak is also slightly increased. The cytochrome c550/c1-deficient double-mutant strain, in contrast, showed a decrease in both the cytochrome c and cytochrome b absorption bands. Apparently, the cytochrome c mutations, as well as the oxygen concentration, affect the concentrations of cytochromes c other than the ones that were mutated. The consequences of oxygen deprivation, as well as of the cytochrome c mutations, include reduced electron flow through the respiratory system, resulting in reduction of its constituent components. Perhaps P. denitrificans possesses a signal transduction mechanism that governs the expression of cytochrome c genes in response to that altered redox state. In order to test that hypothesis, P. denitrificans wild-type cells were cultured aerobically under different conditions, between which we expected the reduction levels of the redox enzymes to differ. In one approach, P. denitrificans wild-type cells were grown aerobically in mineral medium with either succinate or butyrate as the sole carbon and energy source. Since butyrate is a more reduced substrate than succinate, one would expect a rise in the electron-input rates and thus higher reduction states of the redox enzymes in butyrate-grown cells compared to succinate-grown cells. The cytochrome c peak of butyrate-grown cells was almost five times higher than that of the succinate-grown cells (results not shown). In another approach, P. denitrificans wild-type cells were cultured aerobically in mineral medium with succinate as the sole carbon and free-energy source and in the presence of increasing concentrations of myxothiazol, which is an inhibitor of the cytochrome bc1 complex. The growth rates of these cultures did not differ significantly from one another, indicating that the myxothiazol-treated cells had sufficiently adapted their mode of respiration in response to the presence of the inhibitor. Spectral analyses of these cultures showed an increase in the cytochrome c peak with increasing concentrations of myxothiazol ultimately resulting in a more than fivefold increase of the cytochrome c peak at a concentration of 5 μM myxothiazol (results not shown).
PAGE and heme staining of cytochrome c single and multiple mutant strains.
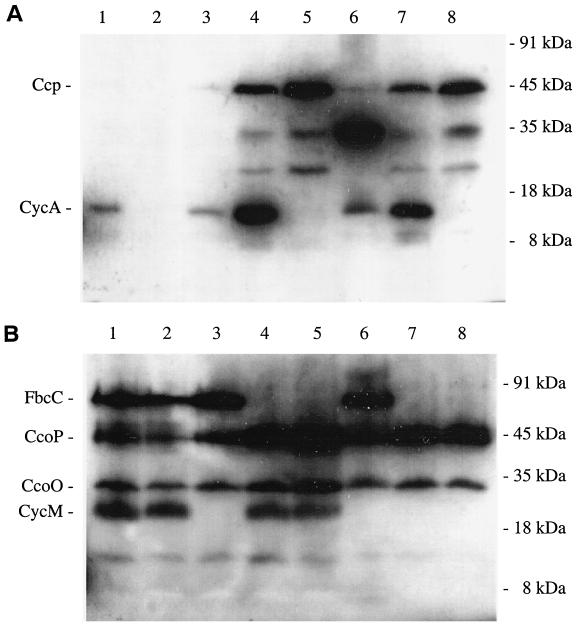
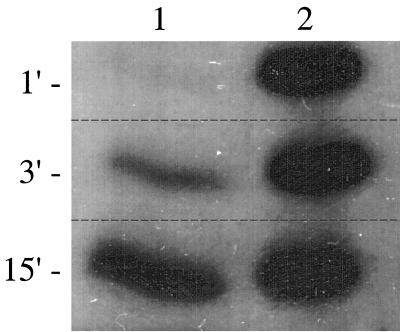
In order to study the cytochrome c composition of the mutants in more detail, cells from cultures grown aerobically on succinate were broken and centrifuged to yield soluble and membrane fractions. For each fraction, the proteins were separated by PAGE, after which the cytochromes c were visualized by a heme staining approach. Two important conclusions can be drawn from the results (Fig. 2). First, they confirm that the mutations in the cycA, cycM, and fbcC genes of the mutants had resulted in the inability to express the corresponding cytochromes c as judged by the absence of heme-stained proteins of 14 kDa (cytochrome c550) in the periplasmic fractions (Fig. 2A) and of 22 kDa (cytochrome c552) and ca. 60 kDa (cytochrome c1) in the membrane fractions (Fig. 2B) of the corresponding mutants. The faint band at ca. 14 kDa observed in the lane of the cytochrome c550/c552-deficient double mutant in Fig. 2A probably originates from a cytochrome c different from cytochrome c550 and which is specifically expressed in this mutant. This band is not present in the lane of its parent strain Pd21.31 (Fig. 2A, lane 2). We monitored the staining of the cytochromes c present in that part of the gel over time (Fig. 3) and noticed that cytochrome c550 stained much faster than the unknown one (maximally stained after 1 min and after 15 min, respectively). The second conclusion is that the knockout of cytochromes c550, c552, and bc1 singly and in various combinations had resulted in the increased expression of other types of cytochrome c. The periplasmic fractions of the wild-type strain and the cytochrome c552-deficient strain contained moderate levels of cytochrome c550, but its concentration was more than 10-fold increased in the cytochrome c1- and cytochrome c552/c1-deficient mutant strains. In addition, all of the strains that lacked the cytochrome bc1 complex induced high levels of cytochrome c peroxidase (45 kDa), as well as two cytochromes c of ca. 25 and 35 kDa. The 35-kDa cytochrome c was abundant in the cytochrome c550/c552-deficient mutant strain. Since this abundance correlated with the appearance of the unknown cytochrome c of 14 kDa, the latter might be a heme containing breakdown product of the former. In contrast to the cytochrome bc1 mutants, the cytochrome c550/c552-deficient mutant strain did not show a significantly increased induction of cytochrome c peroxidase nor of the 25-kDa cytochrome c. This correlation could indicate that the 25-kDa cytochrome c was a heme-containing breakdown product of cytochrome c peroxidase. Inspection of the cytochromes c in the membrane fraction indicated that the concentrations of the cytochrome bc1 complex and of cytochrome c552 in the mutants still able to express them were not significantly different from those of the wild-type strain. In contrast, the intensities of the bands containing the cytochrome c containing subunits of the cbb3-type oxidase, CcoO and, to a lesser extent, CcoP, were up to five times higher in the cytochrome bc1 mutants compared to the wild-type strain.
FIG. 2.
SDS-PAGE and subsequent heme staining of cytochromes c in the soluble fractions (A) and membrane fractions (B) of P. denitrificans wild-type (lanes 1) and mutant strains deficient in cytochrome c550 (lanes 2), cytochrome c552 (lanes 3), cytochrome c1 (lanes 4), cytochromes c550/c1 (lanes 5), cytochromes c550/c552 (lanes 6), cytochromes c552/c1 (lanes 7), and cytochromes c550/c552/c1 (lanes 8). Growth and harvesting conditions were as shown in the legend of Fig. 1. The fractions, containing 100 μg of protein each, were obtained from one set of experiments and analyzed in parallel. The relative concentrations of heme c-containing proteins within each gel were determined by densitometric analyses. The c-type cytochromes indicated are cytochromes c550 and c552 (CycA and CycM), cytochrome c peroxidase (Ccp), subunits of the cbb3-type oxidase (CcoO and CcoP), and cytochrome c1 (FbcC). Sizes of the molecular mass markers are indicated on the right of the gels in kilodaltons.
FIG. 3.
Time-dependent staining of soluble cytochromes c run into the gel to the position of the 14-kDa marker. Lane 1, the cytochrome c550/c552-deficient mutant; lane 2, the cytochrome c1-deficient mutant. The exposure times of the film are indicated at the left in minutes.
Growth characteristics of the cytochrome c mutants.
P. denitrificans wild-type strain and the cytochrome c550, c552, and bc1 single and multiple mutants were grown in mineral medium under three different growth conditions. The increase in cell density with time was determined, as well as their final turbidity at 660 nm. The growth conditions were (i) aerobic with succinate as the sole electron donor, (ii) aerobic with methylamine as the sole electron donor, and (iii) anaerobic with succinate as the electron donor and nitrate as the electron acceptor. The data were used to estimate the maximum specific growth rates and the maximum cell numbers (as determined from turbidity measurements) of the strains. These values are presented in Table 2.
TABLE 2.
Growth characteristics of P. denitrificans wild-type and cytochrome c mutant strains
| Strain | Characteristic(s) | Growth conditiona
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Methanol, + O2 | Methylamine + O2
|
Succinate + O2
|
Succinate + NO3, with copper
|
Succinate + NO3, without copper
|
||||||||
| μ (h−1) | OD | μ (h−1) | OD | μ (h−1) | OD | Nitrite | μ (h−1) | OD | Nitrite | |||
| Pd1222 | Wild type | G | 0.15 | 2.0 | 0.55 | 2.7 | 0.33 | 2.2 | − | 0.33 | 2.2 | − |
| Pd21.31 | CycA− | G | 0.12 | 1.8 | 0.53 | 2.5 | 0.33 | 2.2 | − | DG | 0.5 | + |
| Pd28.22 | CycM− | G | 0.14 | 2.0 | 0.53 | 2.7 | 0.33 | 2.2 | − | 0.33 | 2.2 | − |
| Pd24.21 | FbcC− | G | 0.09 | 1.7 | 0.53 | 2.4 | DG | 0.5 | + | DG | 0.5 | + |
| Pd92.06 | CycA.FbcC− | G | 0.07 | 1.6 | 0.55 | 2.5 | DG | 0.5 | + | DG | 0.5 | + |
| Pd92.26 | CycA.CycM− | G | 0.09 | 1.8 | 0.53 | 2.5 | 0.33 | 2.2 | − | DG | 0.5 | + |
| Pd92.28 | FbcC.CycM− | G | 0.08 | 1.6 | 0.55 | 2.4 | DG | 0.5 | + | DG | 0.5 | + |
| Pd93.14 | CycA.FbcC.CycM− | NG | NG | NG | 0.53 | 2.4 | DG | 0.5 | + | DG | 0.5 | + |
Growth in batch cultures with mineral medium and the electron donors and acceptors indicated. G, growth; NG, no growth; DG, disturbed growth, presumably as a consequence of nitrite accumulation, which is cytotoxic (assayed with nitur strips: −, no nitrite; +, nitrite at >80 mg/liter). Growth on methanol was studied by plating on mineral medium plates with methanol. μ, Maximum specific growth rate (per hour). The means of at least three independent experiments are shown, with standard error of the mean values of <5%. OD, the OD660 at the end of growth. The means of at least three independent experiments are shown, with standard error of the mean values of <5%.
The maximum specific growth rates of succinate-grown cells did not differ significantly between the strains. The final turbidities of the mutant cultures, however, were all ca. 10% less compared to that of the wild type. The only exception was the cytochrome c552-deficient mutant, which had wild-type growth characteristics.
More-drastic differences between the strains were observed when they were cultured in medium with methylamine as the sole free-energy source. Except for the cytochrome c552-deficient mutant, all of the strains had decreased maximum specific growth rates and final turbidities compared to the wild-type strain. The most profound decrease in growth rate was observed for the cytochrome c1-, cytochrome c550/c1-, and cytochrome c550/c552-deficient mutant strains, which grew up to 50% more slowly than the wild-type cells. The cytochrome c550/c552/c1-deficient triple mutant was unable to grow on methylamine. Apparently, a possible electron transfer route from amicyanin, the dedicated electron acceptor of methylamine dehydrogenase (53), to any of the cytochrome c oxidases cannot support growth of the mutant strain under this condition. Since all of the cytochrome c double mutants (cytochrome c550/c552-, cytochrome c550/c1-, and cytochrome c552/c1-deficient mutant strains) were able to grow on methylamine, cytochrome c550, cytochrome c552, or the cytochrome bc1 complex all suffice for electron flow from amicyanin to oxygen.
Electrons that are passed on via amicyanin to the cytochrome bc1 complex may either be directed to oxygen via the Q-pool and the ba3-type quinol oxidase in a reversed electron flow mechanism (45) or via cytochrome c1 to any of the cytochrome c oxidases. In order to study the fate of electrons in a cytochrome c550/c552-deficient mutant, this strain was grown with methylamine as an electron donor in the presence of 10 μM antimycin A and myxothiazol in order to inactivate the heme b-containing subunit of cytochrome bc1. As a control, we also cultured strain Pd93.11, a strain that lacks the cbb3- and aa3-type oxidases. This strain has been shown to be able to grow on methylamine by redirecting the electrons from cytochromes c via the cytochrome bc1 complex and the Q-pool to the ba3-type quinol oxidase (45). Both the wild-type strain and the cytochrome c550/c552-deficient mutant were still able to grow under this condition, indicating that the inhibition of the heme b-containing subunit of the cytochrome bc1 complex did not prevent electron transport in these strains. The double cytochrome c oxidase mutant, however, was unable to grow under this condition, indicating that the reversed electron transfer route from methylamine to oxygen in this strain was completely blocked. The experiments described above imply that, in the absence of cytochromes c550 and c552, electrons can flow from the cytochrome bc1 complex directly to one or both of the cytochrome c oxidases, most probably to the cbb3-type oxidase since the aa3-type oxidase is hardly present in the cytochrome c550/c552-deficient mutant. In order to test this hypothesis, we constructed a triple-mutant strain, which lacks cytochromes c550 and c552, as well as the aa3-type oxidase. This strain grew as fast on methylamine as the cytochrome c550/c552-deficient strain, indicating that electrons from the cytochrome bc1 complex can indeed be directly transferred to the cbb3-type oxidase.
All mutant strains were also able to grow on plates supplemented with methanol as the sole source of free energy, except for the triple mutant lacking cytochromes c550 and c552 and the cytochrome bc1 complex. Quantitative data are not included since the cells grew poorly in batch cultures with methanol as sole source of carbon and free energy.
In the course of our studies on the role of the cytochromes c in electron transport under denitrifying conditions, we noticed that cytochrome c550-deficient mutants that were cultured in brain heart infusion (BHI) broth were unable to reduce nitrite and grew to low turbidities. This may well have been due to the cytotoxicity of nitrite, which accumulated in these cultures. Earlier studies with that mutant, however, had indicated that its growth characteristics during denitrification were similar to those of the wild-type strain when they were raised in mineral medium supplemented with succinate and nitrate (51). Apparently, a component essential for denitrifying growth in the latter medium was absent from the BHI broth. We therefore added the individual ingredients of the mineral medium to BHI broth and studied the resultant effects on denitrifying growth. We found that the cultures of the cytochrome c550-deficient mutant that had copper added to it were able to denitrify again. In order to study the importance of that metal on the denitrifying growth of the mutant strains, we determined their characteristics of growth in media with or without added copper. The results presented in Table 2 show that the cytochrome bc1 mutant strains had disturbed growth, accumulated nitrite, and reached final turbidities of ca. 25% of the wild-type value. All other mutants showed characteristics comparable to those of the wild-type when they were cultured in medium that contained copper. In the absence of copper, however, both the cytochrome c550 and cytochrome c550/c552-deficient mutant strains displayed characteristics similar to those of the cytochrome bc1 mutants. They showed disturbed growth as a consequence of nitrite accumulation and did not produce gas in the culture medium, which would be diagnostic for the operation of a fully competent denitrification pathway. Apparently, electron transfer to at least the nitrite reductase requires a copper-containing component, which compensates for the loss of cytochrome c550. We also noticed that the supernatants of the cytochrome c550 and cytochrome c550/c552-deficient mutants, which were raised under denitrifying growth conditions in mineral medium without copper, were yellow, whereas the supernatants of the other strains were not. The component responsible for this yellow color could not be precipitated with trichloroacetic acid, indicating that it was not a protein. The spectrum of the yellow component displayed an absorption band with a maximum at ca. 360 nm.
Kinetics of electron transport in cytochrome c mutants.
P. denitrificans wild-type strain and cytochrome c mutants were grown under aerobic conditions with succinate as the sole carbon and free-energy source. Cells grown to their mid-exponential phase of growth were harvested, washed, and resuspended. The oxygen consumption rates of these suspensions were determined with succinate as the electron donor (Table 3). The data show that the oxygen consumption rates of the strains that still had the potential to synthesize the cytochrome bc1 complex were comparable. Those that lacked the complex had rates ca. 15% higher than that of the wild-type strain. In the presence of antimycin A and myxothiazol, inhibitors of the cytochrome bc1 complex, the oxygen consumption rates of the wild-type and cytochrome c552-deficient mutant were almost halved compared to the uninhibited condition, whereas they were ca. 25% decreased in cytochrome c550- and cytochrome c550/c552-deficient mutants. The respiratory inhibitor hardly if at all affected the oxygen consumption rates of the cytochrome c1-deficient strains.
TABLE 3.
Oxygen consumption rates of whole-cell suspensions of P. denitrificans strains
| Strain | Characteristics | O2 uptake ratea (nmol/min/mg of protein):
|
|
|---|---|---|---|
| Without AA-MYXO | With AA-MYXO | ||
| Pd1222 | Wild-type | 132 | 65 |
| Pd21.31 | ΔcycA | 135 | 86 |
| Pd28.22 | cycM::Kmr | 130 | 60 |
| Pd24.31 | ΔfbcC | 152 | 148 |
| Pd92.07 | ΔcycA ΔfbcC | 150 | 146 |
| Pd92.26 | ΔcycA cycM::Kmr | 136 | 80 |
| Pd92.28 | ΔfbcC cycM::Kmr | 148 | 140 |
| Pd93.14 | ΔcycA ΔfbcC cycM::Kmr | 155 | 150 |
Rates were determined after the addition of succinate as an electron donor and either without or with of 5 μM antimycin A and 5 μM myxothiazol (AA-MYXO).
We also studied the kinetics of oxidation and reduction of cytochromes c present in the wild-type and the cytochrome c1-deficient mutant strains. The suspensions were incubated with succinate as reductant to the cytochromes, the reduction level of which was recorded spectrophotometrically in time (results not shown). Oxygen was supplied to these suspensions at a concentration of 1 mM by adding hydrogen peroxide, which is rapidly converted into oxygen and water by intracellular catalase activity (12). The traces show that the cytochromes c became more oxidized upon the addition of oxygen and became reduced again after oxygen had been fully consumed. The wild-type strain consumed the supplied oxygen at a rate of ca. 130 nmol of oxygen min−1 mg of protein−1 (results not shown). Cytochrome bc1-deficient strains did so between 10 to 15% faster. The subsequent reduction of the cytochromes c in the latter strains occurred at an at least 40-fold-lower rate than that of the wild-type strain (results not shown).
DISCUSSION
The studies presented in this study have shed more light on the roles of cytochromes c550, c552, and c1 in controlling the rates of electron transfer in P. denitrificans grown at various growth conditions. The data also indicate that cytochrome c deficiencies by the mutations are in many cases accompanied by changed levels of expression of other respiratory enzymes.
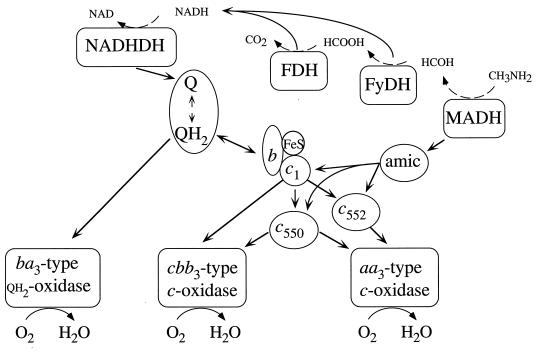
Much of our knowledge on the makeup and function of the P. denitrificans respiratory system at various growth conditions has been described in earlier studies (reviewed in references 2, 3, and 46). The roles of cytochromes c550, c552, and c1 in that electron transport have now been defined by careful examination of the growth characteristics of mutant strains lacking one or more of these types of cytochrome. A scheme of the flexible respiratory network of methylamine grown cells as based on these studies is presented in Fig. 4. It shows that cytochrome c550 is a potential electron donor to any of the cytochrome c oxidases. Cytochrome c552 is a dedicated donor to only the aa3-type oxidase, in agreement with the observation that cytochrome c552 made part of a purified supercomplex containing the cytochrome bc1-complex and the aa3-type oxidase, which was functional in electron transport (4). Our results further show that cytochrome c552 is not a donor to the cbb3-type oxidase in agreement with an earlier observation that a mutant strain lacking cytochromes c1 and c550, as well as the aa3-type oxidase, was unable to grow with methylamine as the sole free-energy source, whereas it could still express cytochrome c552 and the cbb3-type oxidase (11).
FIG. 4.
Scheme of the respiratory network of P. denitrificans during growth with methylamine as sole source of free energy. The sequential oxidation of methylamine to carbon dioxide (dashed arrows) is mediated by methylamine dehydrogenase (MADH), formaldehyde dehydrogenase (FyDH), and formate dehydrogenase (FDH). Physiologically important electron transfer routes (straight arrows) from NADH dehydrogenase (NADHDH) and MADH to the terminal oxidases were deduced from the studies in this study. amic, amicyanin.
The studies described here present a qualitative view on all possible electron transfer pathways during methylamine oxidation as they operate in the various mutant strains. Our observation do not prove that all of these pathways operate to the same extent in the wild-type cells as they do in these mutants. The deletions of cytochromes c1 and c550 resulted in decreased maximum specific growth rates of the mutant strains by 40 and 20%, respectively. These cytochromes are apparently essential for optimal performance of the methylamine oxidizing pathways since they have multiple tasks in connecting quinol and amicyanin oxidation to cytochrome c reduction. However, it is not possible to deduce from these numbers the relative electron fluxes through the parallel electron transfer pathways in the wild-type cells. This is because every mutation in these mutants gives rise to changes in the kinetics of the remaining electron transfer reactions as a consequence of (i) changes in the expression levels of the respiratory components (28) and (ii) changes in the reduction levels of them (27).
The respiratory network of methanol-grown cells is quite similar to that of methylamine-grown cells. The difference is that methylamine dehydrogenase and its dedicated electron acceptor amicyanin (19, 53) are replaced by a methanol dehydrogenase and its dedicated electron acceptor cytochrome c551i (18, 50). Just like amicyanin, this cytochrome requires another cytochrome c as electron acceptor and is unable to donate to any of the cytochrome c oxidases.
When the cytochrome c-deficient strains were grown with succinate as the carbon and free-energy source, their maximum specific growth rates were quite comparable to that of the wild-type strain, indicating that these mutant cells adapt completely in that respect to compensate for the loss of any of these cytochromes c. That adaptation involves higher electron transfer fluxes through the ba3-type quinol oxidases since these mutants display lower maximum cell numbers (since this pathway is less efficient with respect to free-energy transduction) and higher maximum oxygen uptake activity via the ba3-type oxidase. This result is in agreement with earlier studies on cytochrome bc1-deficient mutants (27).
The analyses of the mutants grown under denitrifying conditions indicated that cytochrome c1 is essential for the reduction of nitrite to dinitrogen gass. This result is in agreement with the view that the cytochrome bc1 complex is a compulsory intermediate in electron transfer from the Q-pool to the nitrite, nitric oxide, and nitrous oxide reductases (reviewed in reference 34). At least up to and including nitrite reductase, the pathway also involves cytochrome c550, which appeared to be an essential electron donor to this enzyme during copper limitation. In the presence of copper, however, the mutation in the gene encoding cytochrome c550 had hardly any effect on the denitrifying potential of that mutant strain, indicating that a copper-dependent electron carrier substituted for cytochrome c550 (51). A likely candidate is pseudoazurin, which is a type I blue copper protein and able to mediate electron transfer between the cytochrome bc1 complex and the nitrite, nitric oxide, and nitrous oxide reductases in vitro (21, 25, 26). Indeed, it has been shown that a mutant strain deficient in cytochrome c550 and pseudoazurin failed to reduce nitrite (30; Stuart Ferguson, unpublished data). Cytochrome c552 is apparently redundant in the denitrifying pathways since we observed virtually no effect of its absence on the denitrifying potential of the mutant cells and since it was unable to compensate for the loss of cytochrome c550 under copper limitation.
In the course of our studies, we noticed that the expression levels of certain types of cytochrome c was increased in aerobically grown mutant strains, especially in those that lacked the cytochrome bc1 complex. Apparently, the mutations in the cytochrome c-deficient mutants cause metabolic changes that result in changes in transcription activation of the genes encoding these cytochromes c. One of these metabolic changes affects the activity of the FnrP protein, which is a transcription activator regulating the expression of genes in response to oxygen limitation (47). This conclusion is based on the observation that the concentrations of the cytochrome c-containing subunits of the cbb3-type oxidase and cytochrome c peroxidase, the genes of which are regulated by FnrP (47), are increased in the mutant strains. Some of the mutants, i.e., the cytochrome c550 single mutant, the cytochrome c550/c552 double mutant, and the cytochrome c550/c552/c1 triple mutant, also displayed lower levels of the aa3-type oxidase as judged by spectroscopic analyses. That phenomenon has been described earlier for the cytochrome c550-deficient mutant (51). The reason for the observed decrease is not clear at present but cannot be merely ascribed to the cycA mutation alone since the cytochrome c550/c1-deficient double mutant has wild-type levels of the aa3-type oxidase.
Taken together, these observations indicate that the performance of the respiratory network is continuously monitored and that its makeup adapted in response to changes in that performance apparently in order to maintain an optimal electron flux at each given growth condition.
ACKNOWLEDGMENTS
This work was supported by the Chemical Sciences Foundation (CW), with additonal financial aid from The Netherlands Organization for Scientific Research (NWO).
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1993. [Google Scholar]
- 2.Baker S C, Ferguson S J, Ludwig B, Page M D, Richter O M H, Van Spanning R J M. Molecular genetics of the genus Paracoccus: metabolically versatile bacteria with bioenergetic flexibility. Microbiol Mol Biol Rev. 1998;62:1046–1078. doi: 10.1128/mmbr.62.4.1046-1078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berks B C, Ferguson S J, Moir J W B, Richardson D J. Enzymes and associated electron transport systems that catalyse the respiratory reduction of nitrogen oxides and oxyanions. Bba-Bioenergetics. 1995;1232:97–173. doi: 10.1016/0005-2728(95)00092-5. [DOI] [PubMed] [Google Scholar]
- 4.Berry E A, Trumpower B L. Isolation of ubiquinol oxidase from Paracoccus denitrificans and resolution into cytochrome bc1 and cytochrome c-aa3 complexes. J Biol Chem. 1985;260:2458–2467. [PubMed] [Google Scholar]
- 5.Bosma G, Braster M, Stouthamer A H, Van Verseveld H W. Isolation and characterization of ubiquinol oxidase complexes from Paracoccus denitrificans cells cultured under various limiting conditions in the chemostat. Eur J Biochem. 1987;165:657–663. doi: 10.1111/j.1432-1033.1987.tb11491.x. [DOI] [PubMed] [Google Scholar]
- 6.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 7.Chang J P, Morris J G. Studies on the utilization of nitrate by Micrococcus denitrificans. J Gen Microbiol. 1962;29:301–310. doi: 10.1099/00221287-29-2-301. [DOI] [PubMed] [Google Scholar]
- 8.De Gier J W. The terminal oxidases of Paracoccus denitrificans. Ph.D. thesis. Amsterdam, The Netherlands: Free University; 1995. [Google Scholar]
- 9.De Gier J W L, Lubben M, Reijnders W N M, Tipker C A, Slotboom D J, Van Spanning R J M, Stouthamer A H, Van der Oost J. The terminal oxidases of Paracoccus denitrificans. Mol Microbiol. 1994;13:183–196. doi: 10.1111/j.1365-2958.1994.tb00414.x. [DOI] [PubMed] [Google Scholar]
- 10.De Gier J W L, Schepper M, Reijnders W N M, Van Dyck S J, Slotboom D J, Warne A, Saraste M, Krab K, Finel M, Stouthamer A H, Van Spanning R J M, Van der Oost J. Structural and functional analysis of aa3-type and cbb3-type cytochrome c oxidases of Paracoccus denitrificans reveals significant differences in proton-pump design. Mol Microbiol. 1996;20:1247–1260. doi: 10.1111/j.1365-2958.1996.tb02644.x. [DOI] [PubMed] [Google Scholar]
- 11.De Gier J W L, Van der Oost J, Harms N, Stouthamer A H, Van Spanning R J M. The oxidation of methylamine in Paracoccus denitrificans. Eur J Biochem. 1995;229:148–154. doi: 10.1111/j.1432-1033.1995.0148l.x. [DOI] [PubMed] [Google Scholar]
- 12.De Gier J W L, Van Spanning R J M, Oltmann L F, Stouthamer A H. Oxidation of methylamine by a Paracoccus denitrificans mutant impaired in the synthesis of the bc1 complex and the aa3-type oxidase. Evidence for the existence of an alternative cytochrome c oxidase in this bacterium. FEBS Lett. 1992;306:23–26. doi: 10.1016/0014-5793(92)80829-6. [DOI] [PubMed] [Google Scholar]
- 13.De Vries G E, Harms N, Hoogendijk J, Stouthamer A H. Isolation and characterization of Paracoccus denitrificans mutants with increased conjugation frequencies and pleiotropic loss of a n(GATC)n DNA-modifying property. Arch Microbiol. 1989;152:52–57. [Google Scholar]
- 14.Ferguson S J. Denitrification and its control. Anton Leeuwenhoek Int J Gen M. 1994;66:89–110. doi: 10.1007/BF00871634. [DOI] [PubMed] [Google Scholar]
- 15.Garciahorsman J A, Barquera B, Rumbley J, Ma J X, Gennis R B. The superfamily of heme-copper respiratory oxidases. J Bacteriol. 1994;176:5587–5600. doi: 10.1128/jb.176.18.5587-5600.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harms N, Ras J, Koning S, Reijnders W N M, Stouthamer A H, Van Spanning R J M. Genetics of C1 metabolism regulation in Paracoccus denitrificans. In: Lidstrom M E, Tabita F R, editors. Microbial growth on C1 compounds. Amsterdam, The Netherlands: Kluwer Academic Publishers; 1996. pp. 126–132. [Google Scholar]
- 17.Harms N, Van Spanning R J M. C1 metabolism in Paracoccus denitrificans: genetics of Paracoccus denitrificans. J Bioenerg Biomembr. 1991;23:187–210. doi: 10.1007/BF00762217. [DOI] [PubMed] [Google Scholar]
- 18.Husain M, Davidson V L. Characterization of two inducible c-type cytochromes from Paracoccus denitrificans. J Biol Chem. 1986;261:8577–8580. [PubMed] [Google Scholar]
- 19.Husain M, Davidson V L. An inducible periplasmic blue copper protein from Paracoccus denitrificans: purification, properties, and physiological role. J Biol Chem. 1985;260:14626–14629. [PubMed] [Google Scholar]
- 20.John P, Whatley F R. Paracoccus denitrificans and the evolutionary origin of the mitochondrion. Nature. 1975;254:495–498. doi: 10.1038/254495a0. [DOI] [PubMed] [Google Scholar]
- 21.Koutny M, Kucera I, Tesarik R, Turanek J, Van Spanning R J M. Pseudoazurin mediates periplasmic electron flow in a mutant strain of Paracoccus denitrificans lacking cytochrome c550. FEBS Lett. 1999;448:157–159. doi: 10.1016/s0014-5793(99)00345-2. [DOI] [PubMed] [Google Scholar]
- 22.Kurowski B, Ludwig B. The genes of the Paracoccus denitrificans bc1 complex. Nucleotide sequence and homologies between bacterial and mitochondrial subunits. J Biol Chem. 1987;262:13805–13811. [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Lawford H G, Cox J C, Garland P B, Hadock B A. Electron transport in aerobically grown Paracoccus denitrificans: kinetic characterization of the membrane-bound cytochromes and the stoichiometry of respiratory driven-proton translocation. FEBS Lett. 1976;64:369–374. doi: 10.1016/0014-5793(76)80330-4. [DOI] [PubMed] [Google Scholar]
- 25.Moir J W B, Baratta D, Richardson D J, Ferguson S J. The purification of a cd1-type nitrite reductase from, and the absence of a copper-type nitrite reductase from, the aerobic denitrifier Thiosphaera pantotropha: the role of pseudoazurin as an electron donor. Eur J Biochem. 1993;212:377–385. doi: 10.1111/j.1432-1033.1993.tb17672.x. [DOI] [PubMed] [Google Scholar]
- 26.Moir J W B, Ferguson S J. Properties of a Paracoccus denitrificans mutant deleted in cytochrome c550 indicate that a copper protein can substitute for this cytochrome in electron transport to nitrite, nitric oxide and nitrous oxide. Microbiology. 1994;140:389–397. [Google Scholar]
- 27.Otten M F, Reijnders W N, Bedaux J J, Westerhoff H V, Krab K, Van Spanning R J. The reduction state of the Q-pool regulates the electron flux through the branched respiratory network of Paracoccus denitrificans. Eur J Biochem. 1999;261:767–774. doi: 10.1046/j.1432-1327.1999.00334.x. [DOI] [PubMed] [Google Scholar]
- 28.Otten M F, Stork D M, Reijnders W N, Westerhoff H V, Van Spanning R J. Regulation of expression of terminal oxidases in Paracoccus denitrificans. Eur J Biochem. 2001;268:2486–2497. doi: 10.1046/j.1432-1327.2001.02131.x. [DOI] [PubMed] [Google Scholar]
- 29.Otto B R, van Dooren S J, Nuijens J H, Luirink J, Oudega B. Characterization of a hemoglobin protease secreted by the pathogenic Escherichia coli strain EB1. J Exp Med. 1998;188:1091–1103. doi: 10.1084/jem.188.6.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pearson I. Cloning, mutagenesis and expression studies of the pazS gene encoding pseudoazurin from Paracoccus denitrificans. Ph.D. thesis. Oxford, England: Oxford University; 2000. [Google Scholar]
- 31.Preisig O, Zufferey R, Thonymeyer L, Appleby C A, Hennecke H. A high-affinity cbb3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum. J Bacteriol. 1996;178:1532–1538. doi: 10.1128/jb.178.6.1532-1538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raitio M, Jalli T, Saraste M. Isolation and analysis of the genes for cytochrome c oxidase in Paracoccus denitrificans. EMBO J. 1987;6:2825–2833. doi: 10.1002/j.1460-2075.1987.tb02579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raitio M, Pispa J M, Metso T, Saraste M. Are there isoenzymes of cytochrome c oxidase in Paracoccus denitrificans? FEBS Lett. 1990;261:431–435. doi: 10.1016/0014-5793(90)80609-m. [DOI] [PubMed] [Google Scholar]
- 34.Richardson D J. Bacterial respiration: a flexible process for a changing environment. Microbiology. 2000;146:551–571. doi: 10.1099/00221287-146-3-551. [DOI] [PubMed] [Google Scholar]
- 35.Richter O M H, Tao J S, Turba A, Ludwig B. A cytochrome ba3 functions as a quinol oxidase in Paracoccus denitrificans—Purification, cloning, and sequence comparison. J Biol Chem. 1994;269:23079–23086. [PubMed] [Google Scholar]
- 36.Riistama S, Puustinen A, Verkhovsky M I, Morgan J E, Wikstrom M. Binding of O2 and its reduction are both retarded by replacement of valine 279 by isoleucine in cytochrome c oxidase from Paracoccus denitrificans. Biochemistry. 2000;39:6365–6372. doi: 10.1021/bi000123w. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Saraste M, Castresana J. Cytochrome oxidase evolved by tinkering with denitrification enzymes. FEBS Lett. 1994;341:1–4. doi: 10.1016/0014-5793(94)80228-9. [DOI] [PubMed] [Google Scholar]
- 39.Simon R, Priefer U, Pühler A. Vector plasmids for in vivo and in vitro manipulations of gram-negative bacteria. In: Pühler A, editor. Molecular genetics of the bacteria-plant interactions. Berlin, Germany: Springer-Verlag KG; 1983. pp. 98–106. [Google Scholar]
- 40.Spaink H P, Okker R J H, Wijffelman C A, Pees E, Lugtenberg B J J. Promoters in the nodulation region of Rhizobium leguminosarum Sym plasmid pRL1J1. Plant Mol Biol. 1987;9:27–39. doi: 10.1007/BF00017984. [DOI] [PubMed] [Google Scholar]
- 41.Stouthamer A H. Metabolic pathways in Paracoccus denitrificans and closely related bacteria in relation to the phylogeny of prokaryotes. Antonie Leeuwenhoek Int J Gen M. 1992;61:1–33. doi: 10.1007/BF00572119. [DOI] [PubMed] [Google Scholar]
- 42.Stouthamer A H. Metabolic regulation including anaerobic metabolism in Paracoccus denitrificans. J Bioenerg Biomembr. 1991;23:163–185. doi: 10.1007/BF00762216. [DOI] [PubMed] [Google Scholar]
- 43.Turba A, Jetzek M, Ludwig B. Purification of Paracoccus denitrificans cytochrome c552 and sequence analysis of the gene. Eur J Biochem. 1995;231:259–265. [PubMed] [Google Scholar]
- 44.Van der Oost J, Deboer A P N, Degier J W L, Zumft W G, Stouthamer A H, Van Spanning R J M. The heme-copper oxidase family consists of three distinct types of terminal oxidases and is related to nitric oxide reductase. FEMS Microbiol Lett. 1994;121:1–9. doi: 10.1111/j.1574-6968.1994.tb07067.x. [DOI] [PubMed] [Google Scholar]
- 45.Van der Oost J, Schepper M, Stouthamer A H, Westerhoff H V, Van Spanning R J M, De Gier J W L. Reversed electron transfer through the bc1 complex enables a cytochrome c oxidase mutant (Δaa3/cbb3) of Paracoccus denitrificans to grow on methylamine. FEBS Lett. 1995;371:267–270. doi: 10.1016/0014-5793(95)00900-t. [DOI] [PubMed] [Google Scholar]
- 46.Van Spanning R J M, De Boer A P N, Reijnders W N M, De Gier J W L, Delorme C O, Stouthamer A H, Westerhoff H V, Harms N, Van der Oost J. Regulation of oxidative phosphorylation: The flexible respiratory network of Paracoccus denitrificans. J Bioenerg Biomembr. 1995;27:499–512. doi: 10.1007/BF02110190. [DOI] [PubMed] [Google Scholar]
- 47.Van Spanning R J M, De Boer A P N, Reijnders W N M, Westerhoff H V, Stouthamer A H, Van der Oost J. FnrP and NNR of Paracoccus denitrificans are both members of the FNR family of transcriptional activators but have distinct roles in respiratory adaptation in response to oxygen limitation. Mol Microbiol. 1997;23:893–907. doi: 10.1046/j.1365-2958.1997.2801638.x. [DOI] [PubMed] [Google Scholar]
- 48.Van Spanning R J M, De Boer A P N, Slotboom D-J, Reijnders W N M, Stouthamer A H. Isolation and characterization of a novel insertion sequence element, IS1248, in Paracoccus denitrificans. Plasmid. 1995;34:11–21. doi: 10.1006/plas.1995.1029. [DOI] [PubMed] [Google Scholar]
- 49.Van Spanning R J M, De Vries S, Harms N. Coping with formaldehyde during C1 metabolism of Paracoccus denitrificans. J Mol Catal B Enzymatic. 2000;8:37–50. [Google Scholar]
- 50.Van Spanning R J M, Wansell C W, De Boer T, Hazelaar M J, Anazawa H, Harms N, Oltmann L F, Stouthamer A H. Isolation and characterization of the moxJ, moxG, moxI, and moxR genes of Paracoccus denitrificans: inactivation of moxJ, moxG, and moxR and the resultant effect on methylotrophic growth. J Bacteriol. 1991;173:6948–6961. doi: 10.1128/jb.173.21.6948-6961.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Spanning R J M, Wansell C W, Harms N, Oltmann L F, Stouthamer A H. Mutagenesis of the gene encoding cytochrome c550 of Paracoccus denitrificans and analysis of the resultant physiological effects. J Bacteriol. 1990;172:986–996. doi: 10.1128/jb.172.2.986-996.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Spanning R J M, Wansell C W, Reijnders W N M, Harms N, Ras J, Oltmann L F, Stouthamer A H. A method for introduction of unmarked mutations in the genome of Paracoccus denitrificans: construction of strains with multiple mutations in the genes encoding periplasmic cytochromes c550, c551i, and c553i. J Bacteriol. 1991;173:6962–6970. doi: 10.1128/jb.173.21.6962-6970.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Spanning R J M, Wansell C W, Reijnders W N M, Oltmann L F, Stouthamer A H. Mutagenesis of the gene encoding amicyanin of Paracoccus denitrificans and the resultant effect on methylamine oxidation. FEBS Lett. 1990;275:217–220. doi: 10.1016/0014-5793(90)81475-4. [DOI] [PubMed] [Google Scholar]