Abstract
A powerful medical decision support system for classifying skin lesions from dermoscopic images is an important tool to prognosis of skin cancer. In the recent years, Deep Convolutional Neural Network (DCNN) have made a significant advancement in detecting skin cancer types from dermoscopic images, in-spite of its fine grained variability in its appearance. The main objective of this research work is to develop a DCNN based model to automatically classify skin cancer types into melanoma and non-melanoma with high accuracy. The datasets used in this work were obtained from the popular challenges ISIC-2019 and ISIC-2020, which have different image resolutions and class imbalance problems. To address these two problems and to achieve high performance in classification we have used EfficientNet architecture based on transfer learning techniques, which learns more complex and fine grained patterns from lesion images by automatically scaling depth, width and resolution of the network. We have augmented our dataset to overcome the class imbalance problem and also used metadata information to improve the classification results. Further to improve the efficiency of the EfficientNet we have used ranger optimizer which considerably reduces the hyper parameter tuning, which is required to achieve state-of-the-art results. We have conducted several experiments using different transferring models and our results proved that EfficientNet variants outperformed in the skin lesion classification tasks when compared with other architectures. The performance of the proposed system was evaluated using Area under the ROC curve (AUC - ROC) and obtained the score of 0.9681 by optimal fine tuning of EfficientNet-B6 with ranger optimizer.
Keywords: EfficientNet, Deep convolutional neural network (DCNN), Melanoma classification, Dermoscopic images
Introduction
The most common and dangerous type of cancer in humans is skin cancer. Skin cancer can be broadly classified as melanoma and non-melanoma, where non-melanoma includes basal cell carcinoma and squamous cell carcinoma. Malignant melanoma is considered to be one of the most deadly forms of skin cancer and is exponentially increasing throughout the world. To reduce the mortality rate of skin cancer early diagnosis is very much crucial, since it bestows an elevated cure rate when detected and treated at an early stage.
Skin cancer is normally screened by clinicians through visual examination which is time consuming, error prone and is more subjective. Dermoscopy is a noninvasive imaging technique that eliminates the surface reflection of the skin which is able to capture illuminated and magnified images of skin lesions to increase the clarity of the spots. However, the detection of melanoma from dermoscopic images by the dermatologist achieved less than 80% accuracy in routine clinical settings [40]. To improve the efficiency and efficacy of skin cancer detection an automated diagnosis system is necessary to assist clinicians in order to enhance the decision making. For developing automated diagnostic tools, traditional Machine Learning (ML) algorithms are used to classify melanoma and non-melanoma. But it is very hard to achieve high diagnostic performance since ML algorithms require hand crafted features and also dermoscopic images have high intra-class and low inter-class variations [43].
Unbiased diagnosis is very important for any early detection and treatment of skin cancer diseases. During the recent years many researchers are focusing on Convolutional Neural Networks (CNNs) based methods since it provides significant improvement in prediction accuracy [15]. Many researchers are focusing on skin cancer classification using Deep Learning (DL) based methods because of its automatic feature engineering and self-learning capabilities. With deep neural networks high performance can be obtained at the cost of expanding the CNN wider, deeper and increasing resolution which leads the architecture to have additional parameters resulting in high computing power for training and testing. One of the most dynamic CNN architecture is EfficientNet which can be used to achieve high accuracy by exploiting compound scaling method. By compound scaling method the architecture can be enlarged by expanding depth, width, and resolution to obtain better accuracy with fewer computational resources than other models [38]. In general, classification of skin cancer is challenging, because of the presence of artifacts, image resolution disparity and less discriminating features between different types of cancer. Thus to alleviate these problems, EfficientNet architecture can be considered as an appropriate model, due to its compound scaling property, for skin cancer classification to strengthen the accuracy.
In this research work, we have proposed an automatic classification system for skin cancer using different deep neural networks based on transfer learning techniques. We have done extensive studies to identify the best classifier suitable for dermoscopic image classification using different deep neural networks such as Google Exception, DenseNet and different variants of EfficientNet models. ISIC-2019 and 2020 image datasets along with metadata information were used to train the DNN models and achieved best accuracy using EfficientNet. The main contribution of this work are as follows:
A systematic method was investigated to classify skin cancer as melanoma and non-melanoma using various DCNN architectures.
- The issues pertaining to skin classification such as, different image resolutions and class imbalance problems observed in the dataset were addressed.
- EfficientNet architecture based on transfer learning was proposed which determines the hyper-parameters automatically resulting in improved accuracy.
- An ensemble architecture was also proposed to improve classification accuracy by combining both metadata and image features.
- The effectiveness of the proposed method was boosted using ranger optimizer which reduced the problem of hyper-tuning.
The rest of this paper is organized as follows, in Section 2 papers related to this research work are discussed, Section 3 describes the methodology, dataset and various DCNN architectures analysed in this study. Experimental results and analysis are outlined in Section 4 and finally conclusion and future work are presented in Section 5.
Related works
The extension of ML algorithms during recent years empowered researchers to develop many applications in the field of medical image analysis. Moldovanu et al. [30] segmented input images using threshold method and extracted a set of Gabor features and trained using a multilevel neural network. A framework for melanoma skin cancer detection using SVM model was proposed in [6] and trained with the help of optimized HOG features extracted from dermoscopic images. Dermoscopic score calculation with the help of ABCD (Asymmetry, Border, Color, and Diameter) features of the images used to separate benign lesions from melanoma was introduced by Kasmi et al. [20].
SVM classifier was used in [11] to train the model using combined global features and local patterns. Global features such as color and lesion shape were extracted using traditional image processing techniques whereas local patterns were extracted using DNN. Moura et al. [31] proposed a binary skin classification method using multiLayer perceptron by training the model using hybrid descriptors which combines the ABCD rule and the features from various pre-trained Convolutional Neural Networks (CNNs). A method for skin cancer localization and recognition using multilayered feed forward neural network was proposed in [22] after extracting the features using DenseNet201 and selecting most discriminating features by iteration-controlled Newton-Raphson (IcNR) method. Feature fusion approach using mutual information metric was proposed in [4], by extracting handcrafted features from ABCD rule and deep learning features from transfer learning.
Naeem et al. [33] presented a deep learning techniques for melanoma diagnosis using CNN and provide a systematic review for the challenges on the basis of similarities and differences. They also discussed about the recent research trends, challenges and opportunities in the field of melanoma diagnosis and investigated the existing solutions. An end to end and pixel-wise learning using DCNN was proposed in [1]. The issues of multi-size, multi-scale, multi-resolution and low contrast images were handled by employing local binary convolution on U-net architecture was presented in [35]. A two stage segmentation method followed by classification was proposed in [19] which uses region-based Convolution Neural Network technique to crop the ROI and binary classification was done using ResNet152 architecture. A pre-trained model using transfer learning technique leveraged with AlexNet was presented in [2]. The training of deep learning models with best set-up of hyperparamaters outperform the ensemble model of classification for different pre-trained models of ImageNet namely Xception, InceptionV3, InceptionResNetV2, NASNetLarge, ResNet101 were discussed in [7]. An optimized pipeline for seven different skin classes was proposed in [24] which learns pixel-wise features for segmentation with modified U-Net and classified using the features extracted from the DenseNet encoder. A methodology to improve the classification accuracy by combining the architectures based on the weighted output of the CNNs was studied in [13]. Fusing the classification output of the trained SVM after training the SVM with deep features from various pre-trained CNNs was proposed in [28]. The accuracy of the lower performing models has been improved by adding the metadata information along with the image features was proposed by Gessert et al. [12]. In general the accuracy of any DL model depends directly on hyper parameter tunning which is done manually by trial and error methods. Hence identifying appropriate parameters is crucial for any model to achieve high accuracy. So in this work we have proposed a method to automatically fine tune the models based on the hyper parameters obtained through compound scaling method introduced in EfficientNet model by Mingxing and Quoc [38].
EfficientNet model uses compounding-scaling method that expands in three dimensions such as scale, width and height of deep network with the help of large number of hyperparameters to obtain better accuracy. A deep learning model for the classification of remote sensing scenes using EfficientNet combined with an attention mechanism was proposed in [3]. The network learns to emphasize important regions of the scene and suppress the irrelevant regions. Deep learning networks such as EfficientNet and MixNet have been used for automated recognition of fruits in [10]. A transfer learning based approach using a pre-trained EfficientNet model was constructed to perform accurate classification of cucumber diseases with improved optimizer namely ranger which helps to obtain high accuracy was proposed in [45]. A bidirectional long short-term memory module which integrates the attention mechanism helps in accurate recognition of cow’s motion behaviour [42]. The EfficientNet architecture was proposed in [29] to achieve binary and multiclass classification for Covid-19 diseases. A deep neural network based on Efficient-B5 using ISIC 2020 Challenge Dataset was discussed in [18]. EfficientNet deep learning architecture was used in [5] based on the transfer learning approach for plant leaf disease classification. Tuberculosis detection from X-ray chest images was proposed in [32] and was done based on transfer learning using pre-trained ResNet and EfficientNet models after enhancing the images using Unsharp Masking (UM) and High-Frequency Emphasis Filtering (HEF). An ensemble model for multi-label classification was proposed in [41] to detect the retinal fundus diseases by extracting features using Efficient net and the classification was achieved using neural network. Mahbod [27] proposed an algorithm that ensembles deep features from multiple pre-trained and fine-tuned DNNs and fused the obtained prediction values of different models. In the recent research it is evident that the EfficientNet models have been used in classification to improve the accuracy but it is noticed that very few researchers applied EfficientNet for skin lesion classification. Thus, we have proposed a new model for classifying skin cancer from dermoscopic images using EfficientNet model with ranger optimizer to achieve high accuracy.
Materials and methods
The motivation for carrying out this research is to investigate the performance of transfer learning techniques using different Deep Learning (DL) architectures for skin lesion classification from dermoscopic images.
Transfer learning
During the past decade the research community has made remarkable progress in the field of medical image analysis by utilizing DL architectures. The DL models achieve incredible accuracy as they try to learn the features automatically from the data in an incremental manner. The Convolutional Neural Networks (CNN) achieves high accuracy in the field of image classification. But training a CNN from scratch is not so easy since the accuracy of the classifier depends on the hyper-parameter tuning such as initial weights, number of epochs, learning rate, dropout, optimizers and also it requires high computational power and an extensive amount of labelled training data set. These problems can be leveraged using a transfer learning techniques. In transfer learning, the training time is reduced by the weights obtained from the pre-trained model which can be utilized as the initial weights to train the new similar problems. This method of reusing the pre-trained weights results in low generalization error. This is true in the case of general images since all transfer learning models have been built based on the general images where as this may not be true for specific images such as medical images.
To overcome these problems, researchers keep increasing the depth of the layers which results in vanishing gradient problem or increasing the layers breadth wise suffers from global optimization problem. So in this paper, we propose to explore an automatic method to classify two different types of skin cancer namely melanoma and non-melanoma from dermoscopic images using different transfer learning models. To investigate an efficient method for classification of skin cancer, using ISIC dataset we have used the state-of-the-art DCNN models such as DenseNet121, ResNet50, InceptionResNetV2 and variants of EfficientNet. We have analysed the results of all transfer learning models for classification in terms of both fine tuning and feature extraction and found EfficientNet performs better since it uses compound scaling method that expands the layers in all directions.
Dataset
In this work, we have used ISIC 2019 [8, 9, 39] and ISIC 2020 [34] dataset. ISIC 2020 dataset contains 33,126 dermoscopic images collected from multiple sites for training and the images are provided in both DICOM and JPEG format along with the metadata. The metadata contains patient_id, sex, age, and general anatomic site with its target value. The training set contains two classes of images, melanoma and non-melanoma with their ground truth while the test set contains 10,982 images with the contextual information. ISIC 2019 dataset contains 25,331 dermoscopic images provided in a JPEG format. ISIC 2019 dataset includes BCN_20000 dataset [9], HAM10000 [8] dataset and MSK Dataset [39]. The test set for ISIC 2019 dataset includes 8,238 images with embedded contextual data.
DenseNet model
The Densenet architecture is popular because the DenseNet model attenuates the vanishing-gradient problem, improves feature propagation, motivates feature reuse and reduces the number of parameters. In a dense convolutional neural network each layer is connected to every other layer as a feed forward pattern. Each layer in DenseNet accepts the feature maps of all preceding layers as additional input and passes on its own feature maps to all subsequent layers. Thus the nth layer has n inputs of all preceding layers.
Generally, CNN tries to change the size of the feature map by down sampling layers. But DenseNet facilitates both down-sampling and feature concatenation by dividing the network into multiple densely connected dense blocks. The feature map size inside the blocks remains the same. Outside the dense blocks convolution and pooling operations are performed to down sample and inside the dense block the size of the feature maps are same which helps to carry out concatenation.
We have used DenseNet121 which comprises 4 dense blocks with (6,12,24,16) layers in each. Each dense layer consists of 2 convolutional layers 1*1 and 3*3. 1*1 layer is used to extract the features and the other layer used to bring down the feature depth. At the end of each dense layer a transition layer or block is added. The transition layer consists of a batch-norm layer, a 1x1 convolution followed by a 2x2 average pooling layer. The transition layer is used for changing the size of the feature maps. The last dense block is followed by a classifier at the end. Thus the DenseNet consists of 117 conv, 3 transition and 1-classification making the size of layers as 121.
ResNet50
ResNet50 supports residual learning which consists of a 50 layer residual network [16]. The ResNet architecture introduced skip connections, also called residual connections, which enables to train very deep networks and can boost the performance of the model by overcoming the vanishing gradient problem. The ResNet architecture is mainly composed of residual networks in which intermediate layers of a block learn a residual function with reference to the block input. The architecture of ResNet50 has 4 stages. The network takes the input in the multiples of 32 with 3 as channel width. Every ResNet architecture performs the initial convolution and max-pooling using 7x7 and 3x3 kernel sizes. Each stage has 3 residual blocks containing 3 layers, at each stage the size of the input is reduced to half in terms of height and width but the channel width will be doubled. There are three layers 1x1, 3x3, 1x1 convolutions in each residual block. The 1x1 convolution layers are responsible for reducing and then restoring the dimensions. The 3x3 layer is left as a bottleneck with smaller input/output dimensions. Finally, the network has an average pooling layer followed by a fully connected layer.
InceptionResNet V2
InceptionResNet V2 [37] is a variant of Inception V3 model which takes some ideas from ResNet architectures [14, 17]. So the architecture is just the combination of the Inception V3 model and ResNet model. Residual blocks are added in the inception model to replace the filter concatenation stage. This makes this model produce all the benefits of the residual approach while maintaining the same computational efficiency. The architecture contains 164 layers with input image size of 299x299. In this model every Inception block is followed by a filter expansion layer (1 x 1 convolution without activation) which is used for scaling up the dimension of the filter bank before the addition to match the depth of the input. In the case of InceptionResNet, batch normalization is used only on top of the traditional layers, but not on top of the summations. The InceptionResNet V2 architecture is more accurate than previous state of the art models.
EfficientNet model
EfficientNet has been proposed to improve accuracy and efficiency of CNN by applying a uniform scaling method to all dimensions i.e., depth, width and resolution of the network yet scaling down the model. In general, convNets scales down or up by adjusting either network depth, width or resolution. However, scaling the individual dimension of the network enhances accuracy but for bigger models the accuracy declines. So to achieve better accuracy and efficiency, it is important to scale all dimensions of network width, depth, and resolution uniformly.
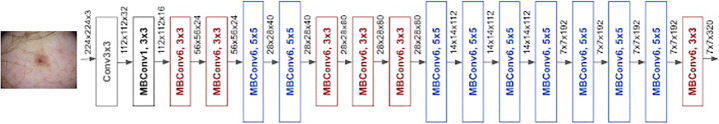
The compound scaling method proposed in [38] applies grid search to find the relationship between the different scaling dimensions of the baseline network under a fixed resource constraint. With this method, an acceptable scaling factor for depth, width and resolution dimensions is determined. These coefficients are then applied to scale the baseline network to the desired target network. This type of architecture is called EfficientNet models and it has eight different variants between EfficientNet B0 to EfficientNet B7. The accuracy of the EfficientNet increases evidently as the model number increases. They are called EfficientNets because they achieve much better accuracy and efficiency when compared to the previous Convolutional Neural Networks. The architecture of EfficientNet-B0 is shown in Fig. 1.
Fig. 1.
Architecture of EfficientNet-B0
The main building block of the EfficientNet family is the inverted bottleneck MBConv, which was derived from MobileNetV2 proposed in [36]. MBconv block is an inverted residual block that contains layers which first widens and then expands the channels, so direct connections are used between bottlenecks that connect a lesser number of channels than expansion layers. Also this architecture utilizes the in-depth separable convolutions that bring down calculation by almost k2 factor when compared to normal layers where k represents kernel size. In order to work with inverted residual connections and to improve the performance, a new activation function called Swish activation was used instead of ReLu activation function. The basic architecture of EfficientNet B0 model is depicted in Fig. 1.
The efficacy of model scaling heavily depends on the baseline network. So the new baseline network is built using the automatic machine learning (AutoML) framework which automatically searches for a CNN model that optimizes both precision and efficiency (in FLOPS). Next, the compound scaling factor was applied to the baseline network to uniformly scale depth, width, and resolution. To build the variants of the baseline model different values were substituted for the compound coefficient psi. The equation for scaling depth, width, and resolution is shown below.
| 1 |
| 2 |
| 3 |
| 4 |
where, α, β, γ are constants fixed using grid search. ψ is a user-defined coefficient that controls how many resources are available for model scaling, while α, β, γ determine how to allocate these extra resources to network width, depth, and resolution respectively. Beginning with the baseline EfficientNet-B0, compound scaling method is applied to scale up using the following two steps [38]
Determine the best values for α, β, γ by fixing ψ = 1 using grid search with the assumption that there are twice as many resources available.
Using the determined α, β, γ values as constant, scale up the baseline network to create EfficientNet-B1 to B7 with different ψ values.
We have analysed and compared the classification results obtained using different transfer learning techniques. From the results obtained it is apparent that EfficientNet produced better results when compared to other methods.
System architecture and methodology
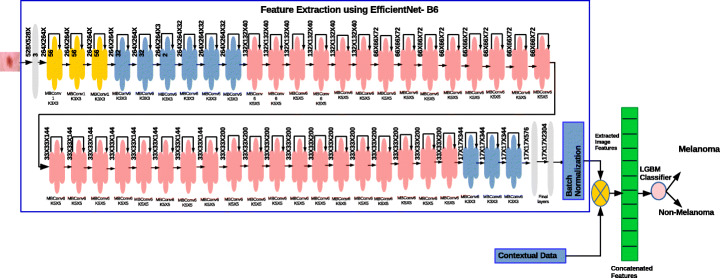
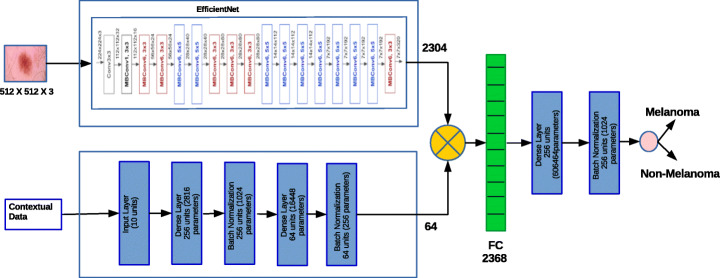
We perform different experiments using different deep learning techniques and analyze the performance. Besides using different architectures we have done extensive experiments using variants of EfficientNet. There are two major transfer learning technique such as fine tuning and feature extraction. We have analysed both the methods of transfer learning techniques with different deep learning architecture with various optimizers and found that EfficientNet-B6 along with ranger optimizer produced better results. The proposed architecture using feature extraction is depicted in Fig. 2 and fine tuning the EfficientNet is shown in Fig. 3.
Fig. 2.
Proposed feature extraction model using EfficientNet-B6
Fig. 3.
Proposed fine tuning method using EfficientNet-B6
Experimental results and analysis
This research work utilizes dermoscopic skin images and classifies them into two different skin cancers namely melanoma and non-melanoma based on the transfer learning techniques. We have used different networks such as DesneNet121, ResNet50, InceptionResnet V2 and variants of EfficientNet and analyzed their performance using classification accuracy in terms of Area Under the Receiver operating characteristic curve (AUC-ROC) using ISIC-2019 and 2020 dataset.
The Receiver Operator Characteristic (ROC) is a probability curve that plots the True Positive Rate (TPR) against False Positive Rate (FPR) for various threshold values. The Area Under the Curve (AUC) measures the entire two-dimensional area underneath the entire ROC curve. AUC is used to project an aggregate measure of performance over all possible classification thresholds.
Experimental setup
The main objective of this study is to build an automatic system to categorise two types of cancer from dermoscopic images. We have used transfer learning techniques with different neural networks as a backbone and ascertain the performance of the classification models using different experiments as follows:
The same CNN architectures are used as feature extractor with Light Gradient Boosting Machine (LGBM) Classifier.
- Fine Tuning the CNN architectures such as DenseNet121, ResNet50, InceptionResNet V2 and EfficientNet for training.
- Analysed the efficacy of fine tuning method using optimizers, Adam and Ranger.
Feature extraction using CNN architectures
To perform feature extraction on each dermoscopic image different CNN models were trained using pre-trained weights and feature representation vectors were obtained. Normally the deep learning architecture tries to extract generic features in initial layers while the later layers capture task specific features. So in this work we have extracted the features from the later layers. The extracted feature maps obtained from CNN are in high-dimensional space so we have down-sampled the feature maps by adding a global average pooling layer. Followed by a pooling layer, a drop out layer with 20% have been added to exclude the neurons during training which ensures the effectiveness of the data and prevents over-fitting. The extracted features from the images and contextual features are combined and trained using Light Gradient Boosting Machine (LGBM) classifier [21]. The LGBM classifier is a gradient boosting framework which works based on a decision tree algorithm. LGBM can handle the large size of data, takes lower memory to run and is called ‘Light’ because of its high speed. This algorithm results in better accuracy than any other boosting algorithms since this algorithm splits the tree leaf wise with best fit. The architecture of the proposed feature extraction model is presented in Fig. 2.
To train our models we have used 33,126 dermoscopic images with a learning rate of 0.001. The LGBM classifier is trained with the maximum tree depth of 6 and the number of leaves as 64. Also we have used stratified K-fold cross validation since the folds are selected in such a way that the mean response value is approximately equal in all the folds. This ensures that each fold contains approximately the same amount of the two types of class labels. Finally, the trained model is tested using 25331 images from the dataset. The results of the ISIC-2020 dataset is presented in Table 1.
Table 1.
Feature Extraction Technique for different architecture using ISIC 2020 Dataset
| Model Name | Image Size | No. of features∗ | Training AUC | Valida tion AUC | Testing AUC |
|---|---|---|---|---|---|
| Dense Net121 | 256X256X3 | 1024 + 3 | 0.9578 | 0.8512 | 0.8629 |
| Res Net50 | 224X224X3 | 2048 + 3 | 0.9577 | 0.8279 | 0.8381 |
| Inception ResNet V2 | 299X299X3 | 1536 + 3 | 0.9633 | 0.8359 | 0.8825 |
| Efficient Net-B6 | 512X512X3 | 2304 + 3 | 0.9462 | 0.9100 | 0.9174 |
* Image features + contextual features
From Table 1 it is evident that EfficientNet B6 produced 91% of AUC because the number of features extracted using EfficientNet is more when compared to other models. Hence EfficientNet have learnt more number of global discriminating features which helps to classify the images more precisely.
Fine tuning the CNN architectures
Fine tuning is the method which unfreezes few of the top layers of the trained model and adds new classifier layers. The base model is then retrained along with the newly added layers which allows to fine tune the global features that is relevant to the specific task. To achieve better accuracy we have used both contextual and image information provided in the dataset. So we have developed two different architectures one for contextual data and other one for image data and concatenated the layers of the architectures. To train the contextual information we have used a simple neural architecture which consists of a dense layer followed by batch normalization and ReLu activation function. Next to train the image information a neural network architecture such as EfficientNet B0 to B7, DenseNet, InceptionResNet V2 and ResNet50 were used by unfreezing the top layers. The last layers of both the models were concatenated and fed into the dense layers with 384 neurons with ReLu activation function. Next a batch normalization layer is added to normalize the values and we have used a dropout of 20% which was chosen randomly to avoid over fitting. Finally a dense layer with softmax activation function consisting of a single neuron for binary classification is added at the top of the model. The architecture diagram of the proposed fine tuning method is presented in Fig. 3.
To analyse the efficacy of the proposed method we have implemented different architectures with Adam optimizer for ISIC 2020 dataset. The results of different architecture using Adam optimizer for ISIC-2020 dataset is depicted in Table 2. From the results it is evident that EfficientNet produced good accuracy when compared to other models. The EfficientNet architecture produced a promising results because the network architecture automatically scales the depth, width and resolution of the network and hence we have obtained a better AUC for this model. The neural network architectures presented in the table are trained with learning rate as 0.01, optimizer as Adam and trained the model for 100 epochs.
Table 2.
Fine Tuning method for different architecture with Adam optimizer using ISIC 2020
| Model Name | Image Size | Training AUC | Validation AUC | Testing AUC |
|---|---|---|---|---|
| Dense Net121 | 256X256X3 | 0.6472 | 0.7637 | 0.7414 |
| ResNet50 | 224X224X3 | 0.66875 | 0.7627 | 0.7499 |
| Inception ResNet V2 | 299X299X3 | 0.67913 | 0.7423 | 0.7423 |
| EfficientNet-B6 | 512X512X3 | 0.7052 | 0.7369 | 0.7765 |
From the previous studies it is apparent that EfficientNet models produces high accuracy with few parameters and less training time. To study the performance of the proposed methodology, we have also conducted a few experiments with different size of input images such as 256 × 256,384 × 384,512 × 512. The obtained results for EfficientNet with different images size for ISIC-2020 and combined data set of ISIC-2019 & 2020 datasets are presented in the Tables 3 and 4 respectively. From the table it is shown that EfficientNet produced good results when compared with other models and also the increase in images size gradually increases the accuracy. We have obtained 0.7765 and 0.9486 as AUC for ISIC-2020 and ISIC-2019 & 2020 datasets respectively, using EfficientNet-B6 with learning rate as 0.00001, optimizer as Adam and for 100 epochs.
Table 3.
Results of fine tuning the EfficinetNet models using ISIC-2020 dataset
| Model Name | AUC for Image Size 256X256 | AUC for Image Size 384X384 | AUC for Image Size 512X512 |
|---|---|---|---|
| EfficeientNet-B0 | 0.6622 | 0.7306 | 0.7732 |
| EfficeientNet-B1 | 0.6507 | 0.7265 | 0.7055 |
| EfficeientNet-B2 | 0.6873 | 0.6615 | 0.7064 |
| EfficeientNet-B3 | 0.6234 | 0.677 | 0.7663 |
| EfficeientNet-B4 | 0.6608 | 0.7655 | 0.7003 |
| EfficeientNet-B5 | 0.732 | 0.7266 | 0.73 |
| EfficeientNet-B6 | 0.7122 | 0.7208 | 0.7765 |
| EfficeientNet-B7 | 0.6615 | 0.7178 | 0.7642 |
Table 4.
Fine Tuning results for EfficientNetB0 - B7 with different image size using ISIC-2019 & ISIC-2020 dataset
| Model Name | AUC for Image Size 256X256 | AUC for Image Size 384X384 | AUC for Image Size 512X512 |
|---|---|---|---|
| EfficeientNet B0 | 0.9221 | 0.9313 | 0.9199 |
| EfficeientNet B1 | 0.9254 | 0.9368 | 0.9202 |
| EfficeientNet B2 | 0.9276 | 0.9313 | 0.9283 |
| EfficeientNet B3 | 0.9296 | 0.9355 | 0.9386 |
| EfficeientNet B4 | 0.9342 | 0.9324 | 0.9368 |
| EfficeientNet B5 | 0.9302 | 0.9374 | 0.9382 |
| EfficeientNet B6 | 0.9445 | 0.9483 | 0.9486 |
| EfficeientNet B7 | 0.9336 | 0.9375 | 0.9465 |
Table 4 shows the AUC values of EfficientNet trained using the combined datasets. It shows that the EfficeientNet-B6 and B7 achieved good results, but there are no much difference in the results though EfficientNet-B7 takes more training time than EfficientNet-B6. Hence we conclude that EfficientNet-B6 is the most suitable classifier for classifying melanoma and non-melanoma.
We have also estimated and analysed the execution time for the proposed methods using Tensorflow processing Unit (TPUs) for 100 epochs and results are presented in Tables 5 and 6. From these tables it is obvious that increase in image size and complexity of the architecture increases the execution time. We believe that the setback of this work is that, the execution time is very high and this is because of the complexity of the EfficientNet architecture.
Table 5.
Execution Time for EfficientNetB0 - B7 with different image size using ISIC-2019
| Model Name | Execution Time for 256X256 | Execution Time for 384X384 | Execution Time for 512X512 |
|---|---|---|---|
| EfficeientNet-B0 | 2392.2 | 4456.9s | 7276.6s |
| EfficeientNet-B1 | 2618.5 | 5762.3s | 7710.3s |
| EfficeientNet-B2 | 2825.6 | 6256.4s | 8274.3s |
| EfficeientNet-B3 | 3137.8s | 6713.6s | 9101.0s |
| EfficeientNet-B4 | 3702.0s | 7136.4s | 10761.8s |
| EfficeientNet-B5 | 4646.7s | 7955.2s | 12954.4s |
| EfficeientNet-B6 | 5056.1s | 8655.2s | 13213.5 |
| EfficeientNet-B7 | 6972.0s | 9621.6s | 14895.2 |
Table 6.
Execution Time for EfficientNetB0 - B7 with different image size using ISIC-2020 & ISIC-2020 dataset
| Model Name | Execution Time for 256X256 | Execution Time for 384X384 | Execution Time for 512X512 |
|---|---|---|---|
| EfficeientNet-B0 | 4192.1s | 7463.6s | 11385.1s |
| EfficeientNet-B1 | 4348.5s | 9352.2s | 17045.1s |
| EfficeientNet-B2 | 4523.0s | 9623.4s | 17389.7s |
| EfficeientNet-B3 | 5012.3s | 10856.6s | 19991.5s |
| EfficeientNet-B4 | 5864.6s | 12148.5s | 22040.1s |
| EfficeientNet-B5 | 6772.0s | 14096.2s | 26146.2s |
| EfficeientNet-B6 | 8034.7s | 17644.1s | 30256.5s |
| EfficeientNet-B7 | 9795.2s | 21946.7s | 35178.4s |
Choosing an appropriate optimizer and effectively tuning its hyper parameter controls the training speed and efficiency of the learned model. To choose a suitable optimizer for training the EfficientNet we have run some experiments by training EfficientNet-B6 with different optimizers and the results are discussed in Table 7. The results of Adam [23] and Rectified Adam (RAdam) [26] optimizers are somewhat close, but using RAdam optimizer leads to more stable training and converges quickly with good generalization. Also Adam produced lower loss than Rectified Adam which implies that Rectified Adam is generalizing better and at the same time because of lower loss, Adam may lead to over-fitting. From Table 7 it is evident that Ranger [25] optimizer produced higher accuracy than other optimizers because Ranger optimizer combines RAdam and lookahead [44] into a single optimizer. RAdam uses a dynamic rectifier to adjust Adam’s adaptive momentum while lookahead reduces the task of hyper-parameter tuning and achieves faster convergence with minimal computational overhead. Therefore combining RAdam and lookahead provide advantages in different aspects of deep learning optimization and provide best results. From these experiments we find that using EfficientNet-B6 along with ranger optimizer produced better results by fine tuning very few hyper-parameters and less training time.
Table 7.
Results of Fine Tuning EfficientNet-B6 architecture with various optimizers
| Model Name | Image Size | Training AUC | Validation AUC | Testing AUC |
|---|---|---|---|---|
| EfficientNet-B6+SGD | 384X384X3 | 0.8289 | 0.8865 | 0.8775 |
| EfficientNet-B6+RMSprop | 384X384X3 | 0.6112 | 0.5451 | 0.5370 |
| EfficientNet-B6+Adam | 384X384X3 | 0.9445 | 0.9483 | 0.9486 |
| EfficientNet-B6+RAdam | 384X384X3 | 0.8805 | 0.9202 | 0.9475 |
| EfficientNet-B6+Ranger | 384X384X3 | 0.90984 | 0.9475 | 0.9681 |
We have also tested the effect of using ranger optimizer with other architectures and the results are shown in Table 8. From Table 8 it is clear that ranger optimizer produced improved results when compared with other optimizers.
Table 8.
Obtained AUC by Fine Tuning different architectures using ranger optimizer
| Model Name | Image Size | Training AUC | Validation AUC | Testing AUC |
|---|---|---|---|---|
| DenseNet121 | 256X256X3 | 0.6470 | 0.7788 | 0.7449 |
| ResNet50 | 224X224X3 | 0.63687 | 0.7656 | 0.7334 |
| Inception ResNet V2 | 299X299X3 | 0.65785 | 0.6611 | 0.7502 |
Comparison with the existing work
We have also compared the classification performance of our proposed work with other CNN models published recently. We observed that our proposed network achieved better AUC score of 0.9681 which is approximately 6% higher than the existing work presented in [18].
Conclusion and future work
This work aims to encourage the development of digital diagnosis of skin lesion and prospect the pertinence of a new CNN architecture, EfficientNet. We have proposed automated system for skin lesion classification which uses the dermoscopic images and meta data of patient information. The performance of the proposed method was evaluated using the datasets released by the popular challenges ISIC-2019 and ISIC-2020. The dataset contains different resolution images and also the dataset is highly imbalanced which may affect the final results. To address these issues we have performed several experiments using two different transfer learning techniques such as feature extractor and fine tuning. In feature extractor technique the features from the later layers and the contextual data were combined and trained using LGBM classifiers. With this model we have obtained AUC score of 0.9174 for EfficientNet B6. To improve the results further, we have applied fine tuning method which concatenates the last layers of the pre-trained architecture and a simple neural network that takes contextual data as input. With this method we achieved better AUC score of 0.9681 for Efficient B6 with ranger optimizer which reduces the problem of hyper-parameter tuning. We have also compared our results with the existing work and obtained a better score.
Declarations
Competing interests
There are no relevant financial or non-financial competing interests to report.
Footnotes
Supported by Vellore Institute of Technology, Chennai, India, and Sri Sivasubramaniya Nadar College of Engineering, Chennai, India
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jaisakthi S M, Email: jaisakthi.murugaiyan@vit.ac.in.
Mirunalini P, Email: miruna@ssn.edu.in.
Chandrabose Aravindan, Email: aravindanc@ssn.edu.in.
Rajagopal Appavu, Email: rajagopal@usf.edu.
References
- 1.A A, S V (2020) Deep convolutional network-based framework for melanoma lesion detection and segmentation. In: Blanc-Talon J, Delmas P, Philips W, Popescu D, Scheunders P (eds) advanced concepts for intelligent vision systems, pp 1294–1298
- 2.Ameri A. A deep learning approach to skin cancer detection in dermoscopy images. J Biomed Phys Eng. 2020;10:801–806. doi: 10.31661/jbpe.v0i0.2004-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alhichri H, Alsuwayed A, Bazi Y, Ammour N, Alajlan N. Classification of remote sensing images using efficientnet-b3 cnn model with attention. IEEE Access. 2021;PP:1–1. [Google Scholar]
- 4.Almaraz-Damian JA, Ponomaryov V, Sadovnychiy S, Castillejos-Fernandez H (2020) Melanoma and nevus skin lesion classification using handcraft and deep learning feature fusion via mutual information measures. Entropy 22(4) [DOI] [PMC free article] [PubMed]
- 5.Atila U, Uçar M, Akyol K, Uçar E. Plant leaf disease classification using efficientnet deep learning model. Ecol Inform. 2021;61:101182. doi: 10.1016/j.ecoinf.2020.101182. [DOI] [Google Scholar]
- 6.Bakheet S (2017) An svm framework for malignant melanoma detection based on optimized hog features. Computation 5(1):4. https://www.mdpi.com/2079-3197/5/1/4
- 7.Chaturvedi S, Tembhurne J, Diwan T. A multi-class skin cancer classification using deep convolutional neural networks. Multimed Tools Appl. 2020;79:28477–28498. doi: 10.1007/s11042-020-09388-2. [DOI] [Google Scholar]
- 8.Codella NCF, Gutman D, Celebi ME, Helba B, Marchetti MA, Dusza S, Kalloo A, Liopyris K, Mishra N, Kittler H, Halpern A (2018) Skin lesion analysis toward melanoma detection: a challenge at the 2017 international symposium on biomedical imaging (isbi) hosted by the international skin imaging collaboration (isic)
- 9.Combalia M, Codella NCF, Rotemberg V, Helba B, Vilaplana V, Reiter O, Carrera C, Barreiro A, Halpern A, Puig S, Malvehy J (2019) Bcn20000: Dermoscopic lesions in the wild [DOI] [PMC free article] [PubMed]
- 10.Duong LT, Nguyen PT, Di Sipio C, Di Ruscio D. Automated fruit recognition using efficientnet and mixnet. Comput Electron Agric. 2020;171:105326. doi: 10.1016/j.compag.2020.105326. [DOI] [Google Scholar]
- 11.Ge Z, Demyanov S, Bozorgtabar B, Abedini M, Chakravorty R, Bowling A, Garnavi R (2017) Exploiting local and generic features for accurate skin lesions classification using clinical and dermoscopy imaging. In: 2017 IEEE 14th international symposium on biomedical imaging (ISBI 2017). 10.1109/ISBI.2017.7950681, pp 986–990
- 12.Gessert N, Nielsen M, Shaikh M, Werner R, Schlaefer A. Skin lesion classification using ensembles of multi-resolution efficientnets with meta data. MethodsX. 2020;7:100864. doi: 10.1016/j.mex.2020.100864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harangi B. Skin lesion classification with ensembles of deep convolutional neural networks. J Biomed Inform. 2018;86:25–32. doi: 10.1016/j.jbi.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 14.He K, Zhang X, Ren S, Sun J (2015) Deep residual learning for image recognition. arXiv:1512.03385
- 15.He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
- 16.He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: 2016 IEEE conference on computer vision and pattern recognition (CVPR). 10.1109/CVPR.2016.9010.1109/CVPR.2016.90, pp 770–778
- 17.He K, Zhang X, Ren S, Sun J (2016) Identity mappings in deep residual networks. arXiv:1603.05027
- 18.Jiahao W, Xingguang J, Yuan W, Luo Z, Yu Z (2021) Deep neural network for melanoma classification in dermoscopic images. In: 2021 IEEE international conference on consumer electronics and computer engineering (ICCECE). 10.1109/ICCECE51280.2021.9342158, pp 666–669
- 19.Jojoa Acosta M, Caballero Tovar L, Garcia-Zapirain M. Melanoma diagnosis using deep learning techniques on dermascopic images. BMC Med Imaging. 2021;6:1471–2342. doi: 10.1186/s12880-020-00534-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasmi R, Mokrani K. Classification of malignant melanoma and benign skin lesions: implementation of automatic abcd rule. IET Image Process. 2016;10(6):448–455. doi: 10.1049/iet-ipr.2015.0385. [DOI] [Google Scholar]
- 21.Ke G, Meng Q, Finley T, Wang T, Chen W, Ma W, Ye Q, Liu TY (2017) Lightgbm: A highly efficient gradient boosting decision tree. In: Guyon I, Luxburg UV, Bengio S, Wallach H, Fergus R, Vishwanathan S, Garnett R (eds) Advances in neural information processing systems, vol 30. Curran Associates, Inc
- 22.Khan MA, Sharif M, Akram T, Bukhari SAC, Nayak RS. Developed newton-raphson based deep features selection framework for skin lesion recognition. Pattern Recogn Lett. 2020;129:293–303. doi: 10.1016/j.patrec.2019.11.034. [DOI] [Google Scholar]
- 23.Kingma DP, Ba J (2015) Adam: A method for stochastic optimization. In: Bengio Y, LeCun Y (eds) 3rd international conference on learning representations, ICLR 2015, San Diego, CA, USA, May 7-9, 2015, Conference Track Proceedings
- 24.Lee YC, Jung S, Won H (2018) Wonderm: Skin lesion classification with fine-tuned neural networks. arXiv:1808.03426
- 25.Li S, Anees A, Zhong Y, Yang Z, Liu Y, Goh RSM, Liu EX (2019) Learning to reconstruct crack profiles for eddy current nondestructive testing
- 26.Liu L, Jiang H, He P, Chen W, Liu X, Gao J, Han J (2019) On the variance of the adaptive learning rate and beyond. arXiv:1908.03265
- 27.Mahbod A, Schaefer G, Ellinger I, Ecker R, Pitiot A, Wang C. Fusing fine-tuned deep features for skin lesion classification. Comput Med Imaging Graph. 2019;71:19–29. doi: 10.1016/j.compmedimag.2018.10.007. [DOI] [PubMed] [Google Scholar]
- 28.Mahbod A, Schaefer G, Wang C, Ecker R, Ellinge I (2019) Skin lesion classification using hybrid deep neural networks. In: ICASSP 2019 - 2019 IEEE international conference on acoustics, speech and signal processing (ICASSP). 10.1109/ICASSP.2019.8683352, pp 1229–1233
- 29.Marques G, Agarwal D, de la Torre Díez I. Automated medical diagnosis of covid-19 through efficientnet convolutional neural network. Appl Soft Comput. 2020;96:106691. doi: 10.1016/j.asoc.2020.106691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moldovanu S, Obreja CD, Biswas K, Moraru L. Towards accurate diagnosis of skin lesions using feedforward back propagation neural networks. Diagnostics. 2021;11:936. doi: 10.3390/diagnostics11060936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moura N, Veras R, Aires K, Machado V, Silva R, Araújo F, Claro M. Abcd rule and pre-trained cnns for melanoma diagnosis. Multimedia Tools Appl. 2019;78(6):6869–6888. doi: 10.1007/s11042-018-6404-8. [DOI] [Google Scholar]
- 32.Munadi K, Muchtar K, Maulina N, Pradhan B. Image enhancement for tuberculosis detection using deep learning. IEEE Access. 2020;8:217897–217907. doi: 10.1109/ACCESS.2020.3041867. [DOI] [Google Scholar]
- 33.Naeem A, Farooq MS, Khelifi A, Abid A. Malignant melanoma classification using deep learning: Datasets, performance measurements, challenges and opportunities. IEEE Access. 2020;8:110575–110597. doi: 10.1109/ACCESS.2020.3001507. [DOI] [Google Scholar]
- 34.Rotemberg V, Kurtansky N, Betz-Stablein B, Caffery L, Chousakos E, Codella N, Combalia M, Dusza S, Guitera P, Gutman D, Halpern A, Kittler H, Kose K, Langer S, Lioprys K, Malvehy J, Musthaq S, Nanda J, Reiter O, Shih G, Stratigos A, Tschandl P, Weber J, Soyer HP (2020) A patient-centric dataset of images and metadata for identifying melanomas using clinical context
- 35.Salih O, Viriri S (2020) Skin lesion segmentation using local binary convolution-deconvolution architecture. Image Anal Stereology 39(3)
- 36.Sandler M, Howard A, Zhu M, Zhmoginov A, Chen LC (2018) Mobilenetv2: Inverted residuals and linear bottlenecks. In: proceedings of the IEEE conference on computer vision and pattern recognition, pp 4510–4520
- 37.Szegedy C, Ioffe S, Vanhoucke V (2016) Inception-v4, inception-resnet and the impact of residual connections on learning. arXiv:1602.07261
- 38.Tan M, Le QV (2019) Efficientnet: Rethinking model scaling for convolutional neural networks. arXiv:1905.11946
- 39.Tschandl P, Rosendahl C, Kittler H (2018) The ham10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci Data 5(1) [DOI] [PMC free article] [PubMed]
- 40.Vestergaard M, Macaskill P, Holt P, Menzies S. Dermoscopy compared with naked eye examination for the diagnosis of primary melanoma: a meta-analysis of studies performed in a clinical setting. Br J Dermatol. 2008;159(3):669–676. doi: 10.1111/j.1365-2133.2008.08713.x. [DOI] [PubMed] [Google Scholar]
- 41.Wang J, Yang L, Huo Z, He W, Luo J. Multi-label classification of fundus images with efficientnet. IEEE Access. 2020;8:212499–212508. doi: 10.1109/ACCESS.2020.3040275. [DOI] [Google Scholar]
- 42.Yin X, Wu D, Shang Y, Jiang B, Song H. Using an efficientnet-lstm for the recognition of single cow’s motion behaviours in a complicated environment. Comput Electron Agric. 2020;177:105707. doi: 10.1016/j.compag.2020.105707. [DOI] [Google Scholar]
- 43.Yu L, Chen H, Dou Q, Qin J, Heng P. Automated melanoma recognition in dermoscopy images via very deep residual networks. IEEE Trans Med Imaging. 2017;36(4):994–1004. doi: 10.1109/TMI.2016.2642839. [DOI] [PubMed] [Google Scholar]
- 44.Zhang MR, Lucas J, Hinton GE, Ba J (2019) Lookahead optimizer: k steps forward, 1 step back. arXiv:1907.08610
- 45.Zhang P, Yang L, Li D. Efficientnet-b4-ranger: A novel method for greenhouse cucumber disease recognition under natural complex environment. Comput Electron Agric. 2020;176:105652. doi: 10.1016/j.compag.2020.105652. [DOI] [Google Scholar]