Abstract
Assessment of ecological risks of chemicals in the field usually involves complex mixtures of known and unknown compounds. Herein we describe the use of pathway-based chemical and biological approaches to assess the risk of chemical mixtures in the Maumee River (OH, USA), which receives a variety of agricultural and urban inputs. Fathead minnows (Pimephales promelas) were deployed in cages for 4 d at a gradient of sites along the river and adjoining tributaries in 2012 and during two periods (April, June) in 2016, in conjunction with an automated system to collect composite water samples. More than 100 industrial chemicals, pharmaceuticals and pesticides were detected in water at some of the study sites, with the greatest number typically found near domestic wastewater treatment plants. In 2016, there was an increase in concentrations of several herbicides from April to June at upstream agricultural sites. Comparison of chemical concentrations in site water to single chemical data from vitro high-throughput screening (HTS) assays suggested potential for perturbation of multiple biological pathways, including several associated with induction or inhibition of different cytochrome P450 (CYP) isozymes. This was consistent with direct effects of water extracts in an HTS assay and induction of hepatic CYPs in caged fish. Targeted in vitro assays and measurements in the caged fish suggested minimal effects on endocrine function (e.g., estrogenicity). A nontargeted mass spectroscopy-based analysis suggested that hepatic endogenous metabolite profiles in caged fish covaried strongly with the occurrence of pesticides and pesticide degradates. These studies demonstrate application of an integrated suite of measurements to help understand the effects of complex chemical mixtures in the field.
Keywords: Mixture, Toxicity, Field, Adverse Outcome Pathways
Graphical abstract
INTRODUCTION
Assessing the potential for adverse biological effects caused by contaminant mixtures in aquatic environments is an ongoing challenge. The most common approach to mixture assessment uses instrumental analyses to measure chemicals in matrices of concern (e.g., water, sediment, tissue), followed by translation of the resultant data into possible biological effects. As analytical instrumentation has evolved, it has become possible to routinely detect hundreds of different analytes in surface water samples using high-quality targeted techniques (e.g., Bradley et al. 2017), and many hundreds more using nontargeted exploratory approaches based on high-resolution mass spectroscopy (Hollender et al. 2017). Unfortunately, even the most extensive analytical characterizations are of little utility for predicting potential biological impacts without hazard data for individual chemicals and chemical mixtures. In the past, effects data for chemicals detected in environmental samples have been largely derived from published whole-animal toxicology studies obtained through standard literature reviews or from curated databases such as the ECOTOX Knowledgebase (US Environmental Protection Agency 2020a). While this remains the most robust approach available to predict the potential effects of measured chemicals on aquatic organisms of concern, the ability to detect chemicals has far exceeded generation of the in vivo toxicological data traditionally used to assess their possible hazards. Consequently, there is a need to employ “alternative” data sources to assess the potential for chemical mixtures to elicit biological effects.
There is an ever-increasing amount of alternative data that could contribute to complex mixture assessment. For example, we recently described a chemical-gene association network modeling approach that employs linked chemical and gene/protein expression information from extensive, open-source databases, to develop hypotheses as to possible perturbation of biological pathways by complex mixtures of chemicals detected in surface waters or effluent (Martinovic-Weigelt et al. 2014; Cavallin et al. 2016; Li et al. 2017; Perkins et al. 2017; Schroeder et al. 2017; Berninger et al. 2019). One challenge associated with this chemical-gene network modeling approach is that it is based on the presence or absence of chemicals rather than concentrations in samples, so the ability to quantitively consider exposure as part of the complex mixture analysis is limited (Schroeder et al. 2016; 2017; Blackwell et al. 2017). To help address this shortcoming, we recently utilized single chemical dose-response data generated from a large-scale high-throughput screening (HTS) program supported through the USEPA and other Federal partners (ToxCast; US Environmental Protection Agency 2020b), to generate exposure-activity ratios (EARs) based on concentrations of chemicals measured in an environmental sample (Blackwell et al. 2017; Blackwell et al. 2019; Corsi et al. 2019). The ToxCast program has generated in vitro dose-response data capturing hundreds of biological pathways and processes for more than 9000 chemicals, many of which have limited or no in vivo toxicity information. As such, the ToxCast database provides a readily accessible, methodologically consistent resource enabling evaluation of potential biological effects based on chemical measurements in complex mixtures. Further, identification of specific HTS targets through the EAR calculations can provide insights as to the potential perturbation of specific biological pathways controlling higher-level apical responses (Ankley et al. 2010; Schroeder et al. 2016; Blackwell et al. 2017; Corsi et al. 2019).
Basing mixture assessments on analytical chemistry is a well-defined, widely accepted paradigm that can yield very useful information when there are data linking chemical concentrations to a relevant biological response(s). However, reliance solely on analytical chemistry for mixture assessment is problematic when contaminants of potential concern are unknown (unable to be targeted), are not identifiable from full-scan (nontargeted) analyses, cannot be detected at biologically-relevant concentrations, and/or when there may be interactions among chemicals in producing adverse impacts. To address these challenges, some regulatory/monitoring programs have complemented analytical measurements with the determination of responses in biological assays conducted with relevant environmental samples. For example, more than 30 years ago a provision for standardized biological testing was incorporated into regulatory programs focused on effluent quality in the US (Federal Register 1984). Similarly, programs focused on sediment and water quality assessment have used toxicity tests for many years to evaluate the biological effects of complex mixtures (US Environmental Protection Agency 1994; Ingersoll et al. 1995). While direct measurement of toxicity certainly can enhance mixture assessment, like targeted chemical analyses, standardized toxicity tests are limited in their coverage of the “biological universe” of possible concern, particularly in terms of reflecting perturbation of the wide range of species, pathways and associated endpoints that might be affected by components of a mixture. For example, an assay commonly used in the US effluent testing program focuses on early life-stage survival and growth in fish over 7 d (US Environmental Protection Agency 1989), which is an extremely sensitive test for some classes of toxicants but may not detect chemicals such as estrogen or androgen receptor agonists that can have profound effects on vertebrate development and reproduction.
Consequently, for complex, poorly defined mixtures, biological assessment methodologies ideally would employ not only targeted (sometimes termed supervised) tests/endpoints of concern, but also nontargeted (unsupervised) assays to detect unanticipated biological effects (Ekman et al. 2013). Nontargeted biological measurements could include characterization of global gene, protein and/or metabolite profiles in exposed organisms (commonly referred to as ‘omics; Garcia-Reyero and Perkins 2011; Martinovic-Weigelt et al. 2014; Perkins et al 2017), or evaluation of environmental samples with batteries of multi-endpoint in vitro assays, including those amenable to HTS (Escher et al. 2013; Schroeder et al. 2016; Blackwell et al. 2018). Identification of molecular or biochemical changes in vivo or in vitro using nontargeted bioassay approaches then can be used as a basis for inferring potential impacts at higher levels of biological organization through pathway-based predictions of potential adverse outcomes (Ankley et al. 2010; Schroeder et al. 2016; Blackwell et al. 2018; van der Oost et al. 2020).
Over the past several years, we have been involved in a large collaborative effort, supported through the Great Lakes Restoration Initiative (GLRI), to develop and demonstrate the use of pathway-based tools for assessing complex mixtures of aquatic contaminants (Ekman et al. 2013). As part of this, we have worked at several Great Lakes sites in the context of case studies/demonstration projects (e.g., Davis et al. 2013; 2016; Kahl et al. 2014; Cavallin et al. 2016; Li et al. 2017; Blackwell et al. 2017; 2019; Perkins et al. 2017; Mosley et al. 2018; Corsi et al. 2019). In this paper, we present data and results from studies conducted in the Maumee River (OH), which demonstrate the practical application of several different pathway-based approaches to collect and interpret analytical and biological data in the context of an integrated site assessment. In addition to conceptually demonstrating the use of pathway-based tools for complex mixture assessment, an aim of the work was to provide information that could aid local stakeholders (e.g., State of Ohio, Lake Erie Action and Management Plan managers, etc.) in designing subsequent research and monitoring efforts focused on potential impacts of contaminants in the Maumee watershed and nearshore regions of Lake Erie (US Environmental Protection Agency 2014). These studies were conducted over the course of two years (2012, 2016) using a variety of bioinformatic, in vitro and in vivo tools developed over the last decade to support GLRI activities.
MATERIALS AND METHODS
Site Selection and General Experimental Design
The Maumee River discharges into Lake Erie and is a Great Lakes Area of Concern (AOC), listed for restrictions on fish consumption, eutrophication, and fish and wildlife population degradation (US Environmental Protection Agency 2015). Due to the diversity of chemicals of potential concern, the Maumee AOC is a highly relevant setting for evaluation of pathway-based techniques for assessing potential effects of complex chemical mixtures. The Maumee River receives input from the Lucas County wastewater treatment plant (WWTP), which discharges 23 million gallons per day (MGD), as well as the Perrysburg (6 MGD) and the 60 MGD Toledo-Bay View WWTPs (Ohio Environmental Protection Agency 2013). Beyond WWTP inputs, the AOC is affected by a variety of nonpoint sources of contaminants, including input from a major tributary, Swan Creek, which is an urban stream receiving input from several combined sewer overflows (Ohio Environmental Protection Agency 2009). The Maumee River has the largest drainage area of any Great Lakes river (21,500 km2) and much of the surrounding watershed is active farmland (US Environmental Protection Agency 2015). Therefore, the region is potentially subject to seasonal fluxes of runoff containing pesticides associated with agriculture. Finally, the lower section of the river, near the mouth in Toledo, has been a major industrial port since around 1850, so there is also legacy sediment contamination (US Environmental Protection Agency 2015).
Most sites for the present work were selected to be representative of large portions of the watershed to characterize the diversity of contaminants contributing to the Maumee River while a few sites of interest focused on specific inputs potentially relevant to the AOC (e.g., WWTPs; in and near Swan Creek). A total of eight sites (Figure 1, Supplemental Information Table S1) were sampled September 18–21, 2012. In 2016, eight sites (five of which corresponded to the 2012 sites) were evaluated during two time periods spanning between April 27-May 4 and June 8–15 (Figure 1; Supplemental Information Table S1). Sampling dates in 2016 aimed to capture possible chemical mixture changes in the AOC associated with agricultural runoff relative to the planting season.
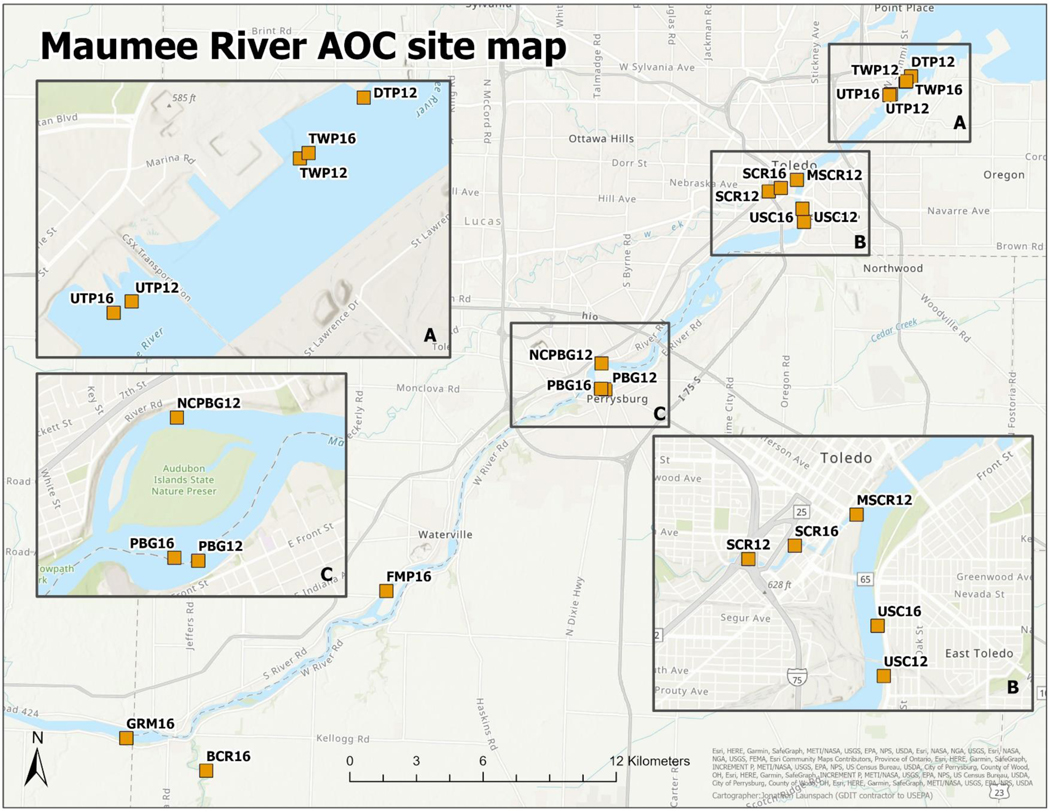
Figure 1.
A map of the study area within the Maumee River Area of Concern showing fish deployment and water collection sites. Sites are designated by alphanumeric codes where letters represent site abbreviations and two-digit number indicate the sampling year. Three magnified boxes are presented to provide finer detail for sites near the (A) Toledo-Bay View wastewater treatment plant (WWTP), (B) Swan Creek and Maumee River confluence, and (C) Perrysburg WWTP. Eight sites were sampled in 2012: three sites associated with Toledo-Bay View WWTP (upstream, UTP12; within the WWTP mixing zone, TWP12; and downstream, DTP12), two sites associated with Swan Creek, an urban draining tributary of the Maumee River, (Maumee confluence, MSCR12; 2 km upstream, SCR12), one site located in the middle of the area designated as the Maumee AOC, (USC12), and two upstream sites located near the outflow of the Perrysburg, Ohio WWTP, (PBG12; North channel of Ewing Island, NCPBG12). Five of these sites were sampled in 2016 (PBG16, USC16, UTP16, TWP16, and SCR16). Three new sites were established upstream including: a site adjacent to the Farnsworth Metro Park (FMP16), Beaver Creek, a tributary surrounded by cropland draining into the Maumee River, (BCR16), and a Maumee River site near Grand Rapids Marina, OH, (GRM16).
To facilitate presentation of data among the different sampling events and sites, we established a site-naming system to distinguish both unique and reoccuring sites assessed during 2012 and 2016. In subsequent descriptions herein, upper-case letters reflect site location abbreviations, while numeric descriptors denote the year sites were sampled (see Figure 1). Eight sites were sampled in 2012: three sites were associated with the Toledo-Bay View WWTP (upstream, UTP12; within the WWTP mixing zone, TWP12; and downstream, DTP12), two sites were associated with the Swan Creek tributary (Maumee confluence, MSCR12; 2 km upstream Swan Creek, SCR12), one site was located in the middle of the area designated as the Maumee AOC (USC12), and two upstream sites were located near the outflow of the Perrysburg WWTP (South channel of Perrysburg, PBG12; North channel of Ewing Island at Perrysburg, NCPBG12).
Five of these eight sites were sampled again during the two sampling periods in 2016 (PBG16, USC16, UTP16, TWP16, and SCR16), with sites at the same location typically within <100 m of one another between years. Three new sites were established upstream, including: a site adjacent to the Farnsworth Metro Park (FMP16), a site in the lower section of Beaver Creek, a tributary surrounded primarily by cropland draining into the Maumee River (BCR16), and a Maumee River site near Grand Rapids Marina (GRM16). These upstream sites were sampled in 2016 to optimize detection of possible seasonal fluctuations in contaminants associated with agricultural runoff.
The same basic experimental design was used at each of the study sites during all three sampling periods. This involved 4-d deployments of caged-fish (fathead minnows; Pimephales promelas) from which tissue samples were collected for both targeted and nontargeted (‘omics) analyses. Kahl et al. (2014) describe in detail aspects of the caged-fish deployment systems, including the materials and dimensions. Time-integrated composite water samples were obtained in conjunction with the caged fish studies using a novel collection system also described by Kahl et al. (2014). The water samples were used for instrumental analysis of chemicals in the water, as well as targeted and nontargeted measures of in vitro bioactivity.
Caged Fish Studies and Water Sample Collection
For the caged fish studies, we used adult male and female fathead minnows, aged 6–7 months, from the on-site culture facility at the US Environmental Protection Agency, Great Lakes Toxicology and Ecology Division (GLTED) in Duluth, MN. Field and laboratory procedures involving the fish were reviewed, and approved by an Animal Care and Use Committee, in accordance with the Animal Welfare Act and Interagency Research Animal Committee guidelines.
At GLTED, the fish were placed, in groups of six males and six females, in plastic bags filled with oxygen-fortified Lake Superior water and shipped in a cooler on ice using an over-night delivery service. Prior to field deployment fish were gradually acclimated to the temperature of site water (while in the shipping bags). When the temperature was within 2°C of site water (typically within 15 min), fish were transferred from the bags into cages held in site water. Cages were closed and encased in a fine mesh bag to capture any fish that might escape from the primary cage. This technique has been shown to result in >95% recovery of fish over the course of 4-d deployments (Kahl et al. 2014). Two cages, each containing 12 fathead minnows (six males and six females), were attached to a buoy-cable system at mid-water depth at each site. Concurrently, an automated composite water sampler (Kahl et al. 2014) was attached to the buoy cable, with a water intake hose at the level of the caged fish. The autosamplers were programmed to collect water aliquots at 10 min intervals for the entirety of the 4-d fish deployments. In 2016, a composite field blank (HPLC grade water) was collected as above for both the April and June sampling periods to account for any potential contamination from the auto-sampler collection process. A corresponding field blank was not collected in 2012.
Water depths at the deployment sites ranged from 0.7 to 4.9 m (Supplemental Information Table S1). Flow rates were not measured, but visible current was present at each site such that there was constant water exchange through the fish cages, but not so great that fish were unable to swim freely. Flow did not move the buoys at any of the sites during the deployment.
To accommodate the logistics for fish processing, sample preservation and shipment, cage deployments were staggered, so that two sites were evaluated each day. In 2012, extra fish from the daily shipping coolers were transported to a staging lab at the University of Toledo for sample collection. These fish were considered day 0 shipping controls for a given deployment. Due to a lack of observed variability in several of the endpoints (e.g., steroids, gene expression) in shipping versus the GLTED lab controls described below (data not shown), day 0 shipping controls were not employed in 2016. After a 4-d field exposure, fish were removed from cages, placed into buckets of site water with secure lids, and transported to the University of Toledo for processing. In both 2012 and 2016, fish from the same cohort as shipped to Toledo were held in a lab at the GLTED facility in a continuous flow of UV-treated, filtered Lake Superior water for 4 d and fed brine shrimp daily to satiation. For these lab controls, general water quality characteristics (mean ± standard deviation [SD]) measured over the course of the deployments were temperature, 20±1ºC in 2012, and in April and June, 2016, respectively, 15.3 ± 0.17 and 20.2 ± 0.41ºC; dissolved oxygen, 8.79 ± 0.98 and 7.37 ± 0.68 mg/L; and pH, 7.03 ± 0.58 and 7.27 ± 0.09. The lab fish were sampled in the same manner as those from the field. The GLTED lab controls were used as the basis for statistical comparisons for measurements made in the caged fish. One exception to this was the spectral data processing for the hepatic metabolomics work with samples from the four upstream sites in 2016 (BCR16, GRM16, FMP16, PBG16) where liver samples from a field reference treatment were used to help discern exogenous from endogenous chemicals (see Caged Fish Metabolomics section below for analysis details). These field reference samples were collected from fish that had been held in outdoor tanks at the Toledo Yacht Club Marina in a water bath with the temperature controlled by incoming Maumee River water. The field reference exposures corresponded with the cage deployments and had the same number of fish (six males and six females). The tanks received a 50% daily renewal of Lake Superior water (shipped from GLTED) and were fed brine shrimp once daily. Otherwise the field reference fish were treated and sampled in the same manner as the caged fish.
Corresponding with both deployment and retrieval of the caged fish, pH, dissolved oxygen, conductivity, temperature, alkalinity and hardness were measured at each site (Supplemental Information Table 1).
Fish Tissue Collection and Processing
Fish were processed, one cage at a time, within 3 h of retrieval from the field. They were anesthetized with buffered tricaine methanesulfonate (MS-222; Finquel; Argent, Redmond, WA), and wet weights were recorded. Blood was collected from the caudal peduncle with heparinized micro-hematocrit tubes and centrifuged to obtain plasma. Plasma samples were snap frozen on dry ice and stored at −80°C until used to measure vitellogenin (egg yolk precursor protein; VTG) and sex steroids. Liver samples were collected, placed in liquid nitrogen and stored at −80°C for subsequent transcriptomic or metabolomic analyses or RNA extraction and evaluation of target gene expression using quantitative real-time polymerase chain reaction (qPCR). Total gonad weights were recorded prior to subdivision to determine the gonadosomatic index. In 2012, an approximately10 mg subsample of fresh gonad was used immediately for an ex vivo steroidogenesis assay (described below), and the remainder was snap frozen for gene expression assays using qPCR or microarrays. The steroidogenesis assays were not conducted in 2016, so entire gonads were frozen for gene expression analyses. During sample processing, RNA degradation and sample cross-contamination on dissection tools was minimized by cleaning equipment with RNaseZap (Ambion, Austin, TX, USA) between samples. With the staggered field deployments, fish were processed over the course of multiple days, and collected tissues were stored in a freezer at −80°C until being shipped overnight or transported by car to the GLTED on dry ice, where they were held at −80°C until further processing.
Water Collection and Handling
The final volume of the 4-d composite water samples corresponding with the caged fish exposure typically was about 10 L. Upon retrieval, samples (in the polyethylene [2012] or Teflon [2016] collection chambers) were placed in a cooler on ice for transport to the University of Toledo for processing. One liter of the composite sample was partitioned into eight 12 mL aliquots and stored in 15 mL polypropylene centrifuge tubes at −20°C for in vitro bioassays. Four additional 1 L samples were transferred into pre-cleaned amber glass bottles (Lee et al. 2012; 2015) and shipped overnight on ice to the USGS National Water Quality Laboratory (NWQL, Denver, CO) for analyses of organic contaminants. The remainder of the composite sample was frozen at −20°C and stored at the GLTED for possible additional analyses.
Analytical Chemistry and Spatial Occurrence Analysis
Analytical measurements were usually conducted using composite water samples for all sampling events. When a composite sample was not successfully collected, grab samples were used. In 2016 additional analytical replicates were collected from the composite sampler bags at USC16 in April and FMP16 in June. Resultant chemical concentrations were averaged to create an individual value for these two instances. Samples were assessed by the NWQL using different analytical schedules employing mass spectrometry (MS). In both 2012 and 2016, schedules were used that focused specifically on pharmaceuticals (Schedule 8244 in 2012 and Schedule 2440 in 2016) and wastewater indicators (Schedule 4433, which includes several pesticides and polycyclic aromatic hydrocarbons [PAHs]). In 2012, an analytical schedule that focused specifically on steroid hormones/sterols (Schedule 4434) was used. In 2016, a pesticide schedule (Schedule 2437) was employed, which included more than 200 target analytes, many of which could be associated with agricultural activities in the AOC watershed (Shoda et al. 2018). In addition to the broad pesticide schedule, a more specialized analysis for neonicotinoid pesticides was performed with the 2016 water samples (Hladik et al. 2018). The complete list of target analytes, corresponding protocols, and quality assurance/quality control procedures are described elsewhere (Zaugg et al. 2006; Lee et al. 2015; Foreman et al. 2012; Furlong et al. 2014; Shoda et al. 2018; Hladik et al. 2018). Concentrations reported as an estimated value were categorized as detected for the different analyses described in this paper.
As described below, analytical chemistry results were used for pathway-based inferences as to possible biological effects in the Maumee system, but the analytical data also provided a basis to examine spatial and temporal contaminant occurrence trends. To this end, cluster analysis was employed to distinguish site similarities and differences based on chemical occurrence data. Due to variation associated with chemical sampling schedules, seasons, and the inclusion of several new sites in 2016, three separate cluster analyses were performed grouping sites individually by year (2012) and season (April and June) in 2016. Data for cluster analysis were prepared by normalizing chemical concentrations through mean centering and scaling to unit variance. Afterward, hierarchical clustering was performed using the correlation between sites as the distance metric and Wards criteria for the linkage to derive site clustering (Murtagh et. al. 2014). The number of site clusters was determined using the R (R Core Team 2019) function pvclust (Suzuki and Shimodaira 2006), with an alpha level set to 0.1. An exception to this approach was made for the April 2016 data where one of the clusters determined using pvclust (UTP16, TWP16, SCR16, GRM16, and USC16) was further subdivided into three smaller clusters (UTP16 and TWP16; SCR16; GRM16 and USC16). This was done to highlight the spatial patterns observed in April 2016, with chemical cluster C3 containing chemicals that had their highest concentrations in SCR16 and chemical cluster C4 containing chemicals that had their highest concentrations at the GRM16 and USC16 sites. These chemicals were also clustered using correlation as the distance metric and Wards criteria for the linkage. The elbow method was used to determine and reduce the number of clusters (Ketchen and Shook 1996). Site clusters were visualized using the percent of the maximum concentrations detected for chemistry data projected as heatmaps using the superheat R package (Barter and Yu 2018). Since cluster analyses included relative values, the data plotted in each heatmap represent the ratio of the log concentration between the highest concentration of each chemical at a given site and that chemical’s lowest concentration within the dataset.
Inferring Biological Hazards from Chemical Occurrence Data: EARs and AOPs
Chemical occurrence data were used to calculate EARs based on publicly-available HTS results from the USEPA ToxCast and Tox21 programs (US Environmental Protection Agency 2020b). Due to the nature of the analysis (i.e., screening), the EARs were based on the maximum values of individual chemical concentrations detected during either sampling period in 2016. Calculations and associated visualizations for the analyses were carried out using the functionality of the R package ToxEval (DeCicco et al. 2018).
By themselves EAR values can be indicative of chemicals with the potential to perturb one or more biological targets (Blackwell et al. 2019), taking into consideration both concentrations and chemical potency. However, the generation of EAR values is only a first step in identifying possible specific effects of a given chemical/suite of chemicals on organsims in the field. That is, the HTS data used for the EAR calculations are from in vitro systems that measure the bioactivity of a chemical in terms of molecular or biochemical alterations. To translate these types of in vitro measurements into possible apical responses in whole organisms requires knowledge concerning the consequences of changes in molecular or biochemical endpoints with regard to what might occur at higher biological levels of organization. The adverse outcome pathway (AOP) framework provides a conceptual basis for meeting this challenge (Ankley et al. 2010). Specifically, an AOP depicts causal linkages between the initial interaction of a chemical with a biological system (termed the molecular initiating event, MIE) and subsequent changes at progressively higher biological levels of organization (intermediate key events) that culminate in an adverse outcome meaningful to risk assessment (i.e., survival, growth reproduction). The HTS data used for EAR calculations typically reflect MIEs or early key events in an AOP.
Consequently it is possible to use the EAR data in conjunction with established AOPs to provide insights as to possible apical impacts caused by a given exposure associated with a complex environmental mixture (Corsi et al. 2019). In the current study, we queried an open-access AOP Wiki (Society for the Advancement of Adverse Outcome Pathways 2016), with respect to assays or gene targets associated with elevated EARs to identify AOPs and apical responses of potential concern relative to possible effects on fish in the Maumee AOC.
Pathway-Based Measurements of Potential Toxicity: Targeted Techniques
Due to well-established connections between WWTP effluents and impacts on components of the hypothalamus-pituitary-gonadal (HPG) axis in fish (Purdom et al. 1994; Hotchkiss et al. 2008), several measurements were made to assess different aspects of reproductive endocrine function. Targeted in vitro bioassays were conducted to determine total estrogenic and androgenic activity of water samples. In addition, measures of HPG axis status were determined in the caged fish, including VTG and sex steroid concentrations in plasma, ex vivo steroid synthesis by gonad tissue, and qPCR determinations in gonads and liver of transcripts of genes known to be important in endocrine function (Ankley et al. 2009). Collectively many of these in vivo and in vitro measurements can be associated with chemical impacts on fish reproduction through consideration of established AOPs (Society for Advancement of Adverse Outcome Pathways 2016). Transcripts of several genes related to HPG axis function were measured in fish from all sites in 2012. However, given the generally low responses observed in 2012, endocrine-associated measurements in 2016 were more focused. Specifically, in 2016 only hepatic vtg expression in males was measured at all sites (as a measure of estrogenicity), and ovarian expression of a suite of HPG-related genes was determined at a subset of sites with elevated herbicide concentrations (see Results).
In vitro bioassays
The T47D-KBluc and MDA-kb2 cell lines were used to estimate, respectively, total estrogenic and androgenic activity in water samples. These cell lines are stably transfected with human estrogen receptor alpha- or androgen receptor-regulated luciferase reporter gene constructs (Wilson et al. 2002; 2004). For the assays, site water samples were prepared with powdered media, which was used to directly dose the cells, as previously described (Wehmas et al. 2011; Cavallin et al. 2014). Estrogenic and androgenic activities, respectively, were quantified based on simultaneously generated duplicate 17α-ethinylestradiol (EE2) or 17β-trenbolone (TRB) standard curves. All samples were analyzed in triplicate within each plate. The background-adjusted percent maximum response of each sample replicate was interpolated against the standard curve concentrations using nonlinear regression (log agonist concentration versus response-variable intensity). The resulting EE2-equivalents (EQs) and TRB-EQs were adjusted for sample dilution in the assay. A sample was considered significantly estrogenic/androgenic if its activity exceeded three standard deviations above the mean assay media control.
In vivo measurements
Gonadal production of 17β-estradiol (E2) and testosterone (T) was determined in the 2012 caged fish using basic procedures described elsewhere (McMaster et al. 1995; Ankley et al. 2007). To help minimize variability, the assay solutions were prepared at the GLTED as a single batch, and frozen in aliquots of an appropriate volume for each sampling event. After 12-h of incubation at 25°C, gonad tissues were removed from the assay system and the remaining solution stored at −80°C until measurement of T and E2 were made using radioimmunoassay (Jensen et al. 2001).
Plasma concentrations of T and E2 in caged fish (both years) were also determined using radioimmunoassay procedures optimized for small volumes (Jensen et al. 2001). Concentrations of plasma VTG protein were measured in males from 2012 using enzyme-linked immunosorbent assays with purified fathead minnow VTG standard and a polyclonal antibody (Parks et al. 1999; Korte et al. 2000).
Due to its specificity and sensitivity for indicating exposure to exogenous estrogens (Ankley et al. 2009), vtg expression in the livers of males from all sampling sites/times was measured by qPCR. A set of ovarian gene transcripts for qPCR analysis was identified based on known roles in HPG function and previously documented chemical effects on their expression (Ankley et al. 2009; Villeneuve et al. 2007; Richter et al. 2016). Transcripts measured in ovary RNA samples from 2012 included cytochrome P450 cholesterol side-chain cleavage (cyp11a), cytochrome P450 17α hydroxylase, 17, 20 lyase (cyp17) and cytochrome P450 aromatase A (cyp19a1a). In 2016 ovarian transcripts measured in females from an upstream subset of the sampling sites included follicle stimulating hormone receptor (fshr), cyp19a1a, zona pellucida phosphoprotein (zp3), 3β-hydroxysteroid dehydrogenase (3βhsd) and steroidogenic acute regulatory protein (star).
Additional transcripts we measured in livers of the caged fish code for enzymes involved in xenobiotic metabolism, some of which can be induced by exposure to a variety of chemicals found in WWTP discharges such as pharmaceuticals and PAHs (Whyte et al. 2000; You 2004; Cavallin et al. 2016). These included two cytochrome P450s, cy1a1, cyp3a, a glutathione S transferase (gst), and a sulfotransferase (sult2).
Specific procedures for the hepatic and ovarian qPCR measurements, including kits, primers, and probe sequences used for each gene are described in detail elsewhere (Villeneuve et al. 2007; Cavallin et al. 2014; Supplemental Information Methods and Supplemental Information Table S2). The large number of qPCR measurements for this work necessitated conducting the analyses across several different assays. To help reduce variability introduced by combining data from mutiple assay plates, gene expression data were normalized to measurements made using a common set of GLTED control samples from each plate. This normalization helped to correct for normal plate-to-plate variations in qPCR efficiency.
Statistical analysis of targeted in vivo data
Data were analyzed using GraphPad Prism version 7.04 for Windows (GraphPad Software, La Jolla, CA). When plasma VTG and steroid concentrations and gene expression (qPCR) data were normally distributed and variance was homogenous, one-way analysis of variance (ANOVA) followed by Dunnett’s Test was used to test for differences between field sites and the GLTED control. When data were not normally distributed, they were either transformed (e.g., log10) and then analyzed by one-way ANOVA, or if still not normally distributed after transformation, analyzed using a nonparametric Kruskall–Wallis test followed by Dunn’s post-hoc analysis. When plasma VTG protein concentrations were below the detection limit, values of one-half the detection limit were used. Differences were considered significant at p < 0.05).
Pathway-Based Measurements of Potential Toxicity: Nontargeted Techniques
Liver and gonad samples from caged fish were used for gene expression (microarray) and metabolomics measurements. Given the size and complexity of the transcriptomics dataset and associated analyses, those results will be described elsewhere. In the current paper we describe hepatic metabolomic analyses for the caged fish, as well as results from an in vitro nontargeted approach with a commercially-available HTS platform used to measure different bioactivities in water samples (generally composites) collected in conjunction with the fish deployments.
Caged Fish Metabolomics
Metabolomics is the study of the complete set of small-molecule endogenous metabolites, or the metabolome, in an organism’s cells, organs, biofluids or tissues. The metabolome consists of semi-polar and polar compounds, such as amino acids and sugars, as well as non-polar compounds such as phospholipids. The relative proportions of these many endogenous metabolites can serve as a fingerprint of an organism’s physiological state. Exposures to potentially toxic chemicals and chemical mixtures result in changes in these fingerprints, which can often be interpreted in terms of adverse impacts in the context of different AOPs (Collette et al. 2010; Davis et al. 2017).
We focused the metabolomic analysis on the polar and semi-polar metabolites from male fathead minnow liver samples from the 2016 field season. We divided the samples into two batches that were subjected separately to the processing and analysis steps described below. One batch, hereafter referred to as the “upstream batch”, included samples (April, June) from PBG16 along with three other sites potentially influenced by agricultural activities (GRM16, BCR16, and FMP16). Corresponding samples from the field reference fish were also included in the upstream batch. The other batch (i.e., the “downstream batch”) included April and June samples from USC16, SCR16, UTP16, and TWP16, which are potentially more heavily influenced by urban activity such as combined sewer overflow and WWTP effluent. The GLTED lab control fish were also included in the downstream batch.
Male fathead minnow liver samples were processed randomly in a 96-well plate format using previously published protocols for a dual phase extraction (Davis et al. 2013). Aliquots of ultrapure water were extracted alongside study samples to serve as process blanks within each batch. The polar extracts were vacuum dried overnight and stored at −80°C. Prior to analysis, the dried extracts were reconstituted in 200 μL of water:acetonitrile (19:1,v/v), after which the samples were allowed to sit for 10 min and then vortexed at 500 rpm for 10 min. Samples were allowed to sit again for 10 min and then vortexed a final time for 10 min (500 rpm). Equal 25 μL aliquots were then collected from the reconstituted extract for each sample, pooled, and split into multiple quality control (QC) samples for high-resolution MS/MS analysis. To remove particulates, the samples were transferred to a prewetted 96-well filter plate (0.2 μm pore size) and centrifuged at 1000×g for 5 min (20°C). The filtrate was then transferred to a 96-well plate with 300 μL glass inserts and briefly centrifuged to remove air bubbles (1 min at 1000×g). Samples were stored in a refrigerated tray (4°C) during the liquid chromatography (LC)-MS/MS analysis. Corresponding QC samples were analyzed at the beginning, end, and after every tenth sample. Process blanks and liver extract samples were analyzed in a randomized fashion.
A full description of the LC and MS parameters is included in the Supplemental Information. Briefly, polar liver extracts were profiled using an LC coupled to a Q-Exactive orbitrap mass spectrometer (Thermo Fisher Scientific). Spectral peaks from the collected data were de-convoluted and aligned separately for the two batches with the XCMS package in R, an open source framework for processing and visualizing chromatographically separated mass spectral data (Smith et al. 2006; Tautenhahn et al. 2008). Isotopic features and ion adducts were annotated with the CAMERA package in R (Kuhl et al. 2012). Parameters used to extract and annotate mass spectral features are provided in Supplemental Information Table S3. Following visual inspection, features belonging to the same metabolite were combined into a single integrated intensity value per metabolite per sample using custom R scripts. Prior to statistical analyses, background components were removed, and the processed dataset was reduced to only those metabolites that were detected in the batch-relevant control/reference samples—these were retained for analysis and are hereafter referred to as “endogenous metabolites”. Other detected metabolites (not likewise found in control/reference samples) were denoted as exogenous and were not used in any of the analyses. This process was conducted to prevent site-specific detections in the dataset (for example, from xenobiotics) from heavily influencing subsequent analyses. A full description of the methods used to screen for endogenous metabolites is presented in Supplemental Information. We identified and removed duplicate records in datasets collected using the +ESI and −ESI modes. The data from these two ion modes were then combined and normalized to unit total integrated intensity and used for all subsequent analyses.
Principal component analysis (PCA) was performed on the normalized spectral data using SIMCA-P+ (Version 13.0, Umetrics) after mean-centering and scaling to unit variance. Batch-specific two component PCA score plots were used separately to assess the overall data structure and to identify potential outliers based on location outside the Hotelling’s T2 line. Using this method, three fish (out of 259 across the two batches) were identified as outliers and removed from the datasets prior to further analyses.
We used partial least-squares (PLS) regression (SIMCA-13.0, Umetrics Inc.) to compare changes in endogenous metabolite profiles with concentrations of detected organic contaminants, and to screen out those contaminants whose concentrations did not significantly covary with metabolite changes. We have previously used this modeling approach to identify environmental stressors that have (or do not have) a significant biological impact on endogenous metabolites, and full modeling details can be found in these earlier papers (Davis et al. 2016; Collette et al. 2019). Briefly, for the upstream batch, we first constructed a “global” PLS model that included all 87 organic contaminants detected in at least one of the samples from the upper-river sites. The optimal number of model components was determined by cross-validation, and model fit was assessed using the fraction of variation that could be predicted by the model components (Q2; Eriksson et al. 2006). We then used contaminant-specific analysis of variance testing of cross-validated predictive residuals (CV-ANOVA; Eriksson et al. 2008) to identify individual contaminants whose measured values were not significantly predicted by the global model. Using an iterative backward-elimination process, we excluded those organic contaminants that had the highest CV-ANOVA values, then rebuilt and revalidated a series of subsequent PLS regression models. We repeated this process until all contaminants remaining in the “final” model exhibited a CV-ANOVA value <0.10. Then, we used these CV-ANOVA values to assess the relative strength of the relationship with endogenous metabolite profiles for each organic contaminant that remained in the final PLS model. Subsequently, we repeated this entire process for the downstream batch, beginning with a global PLS regression model that included all 119 organic contaminants detected in at least one of the samples from the four downstream sites.
High-Throughput Assays
Nontargeted HTS of the Maumee samples was conducted with the Attagene platform (Attagene, Inc. Morrisville, NC), which was one of several employed for data generation in the USEPA ToxCast program. Two individual assays were used for bioactivity screening of surface water extracts, the cis-FACTORIAL and trans-FACTORIAL assays (Romanov et al. 2008). Both are HepG2 cell-based assays multiplexed for the simultaneous assessment of transcription factors (cis-FACTORIAL) or transfected human nuclear receptors (trans-FACTORIAL), together covering nearly 70 individual endpoints across several biological pathways. such as metabolism, endocrine signaling, and cell stress (Romanov et al. 2008).
Surface waters screened using the HTS assays were processed and screened in one (2012) or both (2016) HTS assays as previously described in detail by Blackwell et al. (2019). In 2012, composite water samples from four sites (DTP12, TWP12, UTP12, NCPBG12) were evaluated, while in 2016, samples from all sites and both sampling periods, including the June GLTED control (17 samples total), were used for the HTS bioactivity determinations. Briefly, whole water was filtered prior to solid phase extraction using Oasis HLB cartridges (Waters, Inc. Milford, MA). Final extracts were evaporated to dryness and reconstituted in dimethyl sulfoxide (DMSO) at a relative enrichment factor of 1000-fold. Extracts were screened in duplicate at a relative enrichment of 10-fold in both assays. Bioassay responses were expressed as fold-change relative to DMSO control by dividing sample response by that of corresponding DMSO treated controls. Assay response was batch-normalized by dividing sample response by the extraction blanks run with each assay set. A cutoff of 1.5-fold above blank response was used to identify active endpoints (Blackwell et al. 2019).
RESULTS
Sample Collection Overview
Fish for the cages were shipped, received and deployed without incident during all field seasons. After the 4-d exposure period, >99% of the fish were successfully retrieved from the cages, alive and with no noticeable physical injury (e.g., cuts, abrasions, lesions). Composite water samples were successfully collected at all sites/sampling times, except for SCR12 and GRM16 (June), for which grab samples were obtained upon retrieval of the cages (see Supplemental Information Tables S4 and S6 for a summary of the water sample types employed for the chemical and in vitro analyses). Across 21 composite samples successfully collected over all deployments in 2012 and 2016, the mean (±standard deviation) volume was 10.5 (±3.04) L over the 4-d deployment period. Basic water chemistry data (e.g., pH, conductivity, hardness, alkalinity, dissolved oxygen, total suspended solids, etc.) can be found in Supplemental Information Table S1.
Analytical Chemistry Data and Spatial Analysis-2012
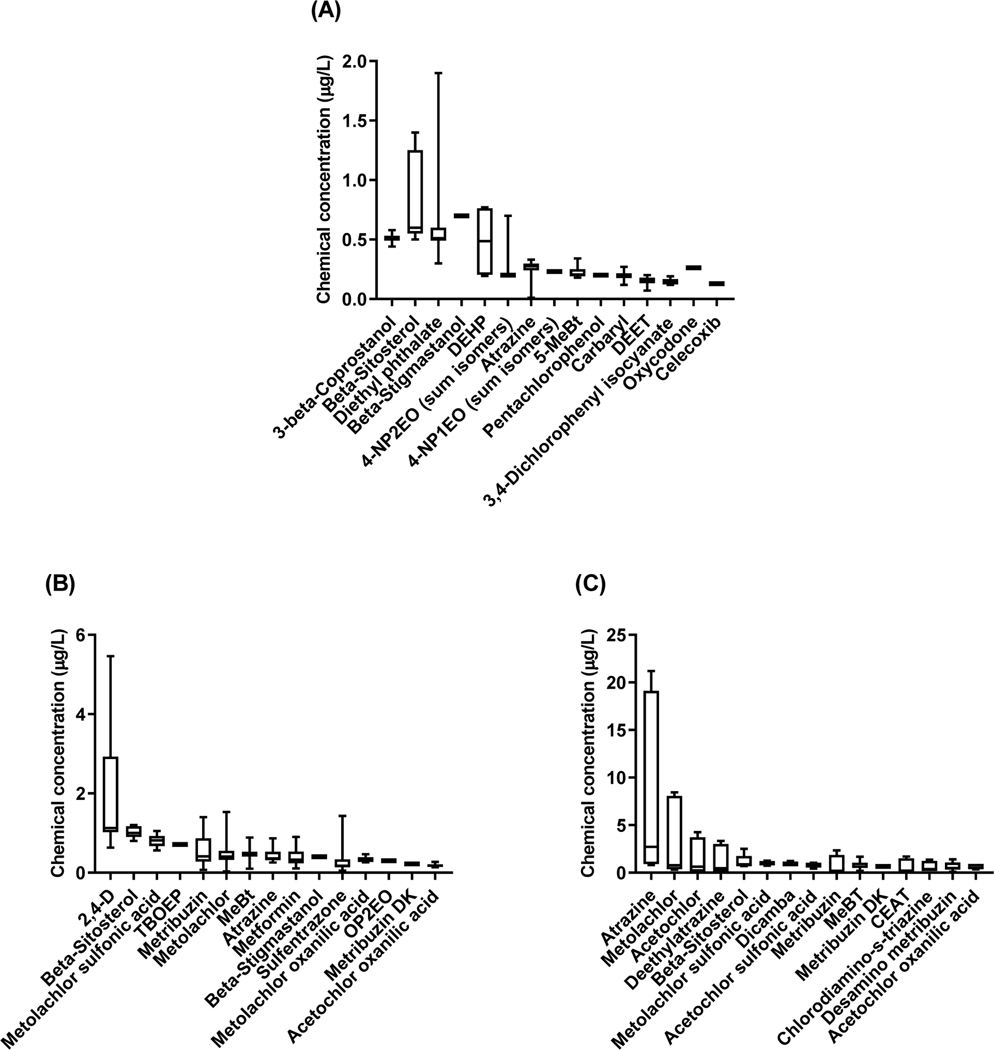
A total of 63 chemicals were detected across all sites in 2012, with members of the wastewater indicators schedule the most frequently observed, particularly at WWTP effluent-influenced sites (NCPG12, PBG12, TWP12, DTP12; Supplemental Information Table S4). Chemical detections included two of the 19 targeted steroid hormones/sterols, 18 of the 48 targeted pharmaceuticals, and 43 of the 67 chemicals in the wastewater indicator schedule. At individual sites total detections ranged from 26 to 49 chemicals. The site closest to the Toledo-Bay Field WWTP mixing zone (TWP12) had the most detectable chemicals, with average concentrations that were frequently higher than at other locations. Overall, the highest absolute concentrations observed were for phytosterols, plasticizers, analgesics, and pesticides. Of all the chemicals detected, cholesterol concentrations were by far the greatest at the various sites, for example ranging from 5.7–7.14 μg/L at UTP12, PBG12, and NCPBG12. Figure 2A depicts mean cross-site concentrations of the 15 chemicals present at the highest concentrations in 2012 (for scaling purposes cholesterol was excluded from the figure).
Figure 2.
Highest 15 average chemical concentrations (μg/L) detected across all sampling sites in the Maumee AOC during (A) September 2012, (B) April 2016, and (C) June 2016. Chemical concentrations were averaged across sites and presented as boxplots with whiskers displaying 10 th to 90 th percentiles. NOTE: Cholesterol was consistently present at relatively high concentrations at all sampling times, so was excluded from the figure to enhance resolution of concentrations of the other analytes. Abbreviations used for figure labelling clarity: 2-Chloro-6ethylamino-4-amino-s-triazine, (CEAT); 2-Chloro-4-isopropylamino-6-amino-s-triazine, (Deethylatrazine); Di(2-ethylhexyl) phthalate, (DEHP); Methyl-1H-benzotriazole, (MeBt); 5-Methyl-1H-benzotriazole, (5-MeBt); 4-Nonylphenol monoethoxylate sum of all isomers, (4-NP1EO); 4-Nonylphenol diethoxylate sum of all isomers, 4-NP2EO; 4-tert-octylphenol diethoxylate, (OP2EO); and Tris(2-butoxyethyl) phosphate, (TBOEP).
Specific chemicals detected varied greatly across the sites in the Maumee AOC (Supplemental Information Table S4). Four chemicals (cholesterol, 5-methyl-1H-benzotriazole (5-MeBt), atrazine, and DEET) were detected at all locations sampled. Beta-sitosterol was found at most sites, with other phytosterols including 3-beta-coprostanol and beta-stigmastanol detected at sites near WWTPs. Diethyl phthalate was detected at all locations except SCR12, while di(2-ethylhexyl) phthalate (DEHP) was detected predominantly in upstream locations (UTP12, PBG12, NCPBG12). The presence of pesticides throughout the entire AOC, particularly atrazine and metolachlor, likely reflects the general influence of agriculture within the watershed (USEPA 2015). This interpretation is consistent with the relatively low concentrations of pesticides at the SCR12 site, as Swan Creek drains primarily urban areas. Polycyclic aromatic hydrocarbons were also present throughout the AOC, with a different suite of PAHs observed at the upstream (PBG12, NCPG12, USC12) versus downstream (MSCR12, UTP12, TWP12, DTP12) sites, suggesting different source contributions.
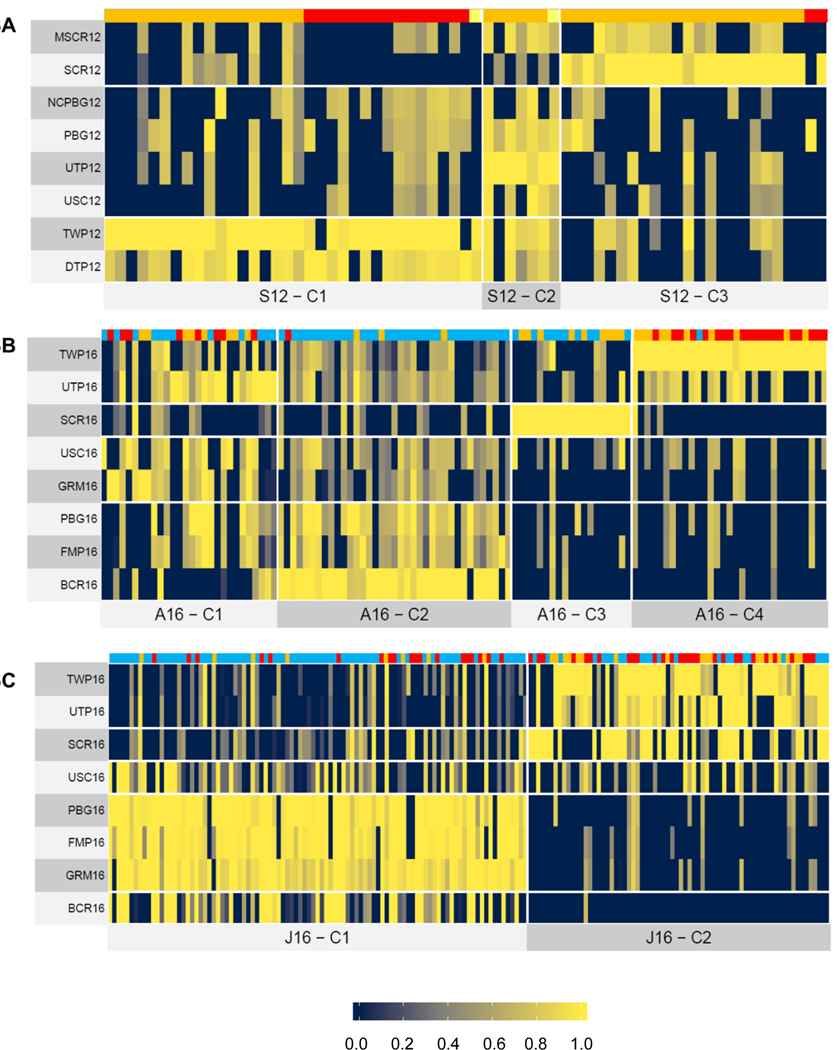
We used cluster analysis to more formally evaluate differences and commonalities among the test sites relative to the chemical occurrence data. In 2012, three distinct site clusters were noted (Figure 3A). The two sites near the Toledo-Bay Field WWTP were in cluster S12-C1 (DTP12 and TWP12), with both having a wide range of pharmaceuticals and wastewater indicators including plasticizers, PAHs, sterols, pain relievers, and antidepressants (Figure 3A; Supplemental Information Table S5). Cluster S12-C2 had four sites comprised of two sub-clusters (PBG12 and NCPBG12; USC12 and UTP12). Chemicals dominating these sites included herbicides (atrazine, metolachlor), plasticizers (diethyl-phthalate, DEHP) and cholesterol (Figure 3A; Supplemental Information Table S5). All four locations were located upstream of the Toledo-Bay View WWTP, with PBG12 and NCPBG12 located furthest upstream, USC12 mid-reach, and UTP12 just above the WWTP. Lastly, cluster S12-C3 contained the two Swan Creek sites (SCR12, MSCR12), which were predominantly characterized by wastewater indicators including PAHs, pesticides, and industrial intermediates (Figure 3A; Supplemental Information Table S5). Five chemicals were largely unique to the Swan Creek sites, and included two organophosphate pesticides (carbaryl, dichlorovos) and a non-crop herbicide (bromacil). Again, the distinct composition of the Swan Creek contaminant profile likely is due to it receiving urban runoff with little in the way of WWTP input (Ohio Environmental Protection Agency 2009).
Figure 3.
Heatmaps of Maumee AOC chemical occurrence presented as site clusters during (A) September 2012, (B) April 2016, and (C) June 2016 sampling. Rows signify sampling locations, with each column distinguishing an individual chemical. Individual chemicals are expressed as proportions of the maximum detected concentration across all locations ranging from low (dark blue, 0) to high (yellow, 1) occurrence. Chemical groups are denoted by varying color within the top row of each panel. For 2012, colors represent the following: (A) sterols/hormones (USGS Schedule 4434; Yellow), pharmaceuticals (USGS Schedule 8244; Red), and wastewater indicators (USGS Schedule 4433; Orange). For 2016 (B and C), colors represent the following: pesticides (USGS Schedule: 2437, Light Blue), pharmaceuticals (USGS Schedule 2440, Red), and wastewater indicators (USGS Schedule 4433, Orange).
Analytical Chemistry Data and Spatial Analysis-2016
The USGS NWQL pesticide and pharmaceutical schedules (2437, 2440, respectively) used in 2016 had a greater number of target analytes than the schedules used in 2012, so the overall chemical detection incidence was higher. The total number of chemicals detected across all sites in 2016 was similar between the two sampling periods with 111 in April and 110 in June (Supplemental Information Table S6). A slightly lower incidence of pesticide detections (58 vs 63) was observed in April compared to June, with somewhat fewer pharmaceutical (28 vs 25) and wastewater indicator (25 vs 22) detections in June vs April. Across the individual sites in April, the number of chemical detections varied from 36–47 for pesticides, 4–23 for pharmaceuticals, and 3–14 for wastewater indicators. Similar ranges were seen in June with detections ranging from 39–51 for pesticides, 0–21 for pharmaceuticals, and 2–18 for wastewater indicators. The most prominent changes across individual sites were observed during June, when pesticide detections and concentrations increased compared to April at all locations except BCR16 (Figure 2B,C; Supplemental Information Table S6). The largest increases in the number of pesticides detected occurred at the most upstream sites (GRM16, FMP16). In April 2016 many of the chemicals present at elevated concentrations were the same as those observed in 2012 (e.g., cholesterol, beta-sitosterol, MeBT). In contrast with 2012, the pre-emergent herbicide 2,4-D was one of the 15 chemicals detected at the highest concentrations in April, particularly at BCR16 and PBG16 (Figure 2B). Many of the analytes detected in April were also observed in June including cholesterol, beta-sitosterol, metolachlor, metribuzin, MeBT and metolachlor sulfonic acid. In contrast, maximum concentrations for the three herbicides, atrazine, metolachlor and acetochlor, were higher on average during June (Figure 2C). For example, average (SD) concentrations of atrazine across all sites increased from 0.43 (0.2) μg/L in April to 8.2 (9.2) μg/L in June. Notably, three upstream Maumee River sites (GRM16, FMP16 and PBG16) had increases in atrazine concentrations of >90% compared to April, with values in June ranging from 14.8 to 21.2 μg/L at these sites (Supplemental Information Table S6).
Cluster analysis of chemical occurrence data for April 2016 yielded four distinct site clusters. Despite differences associated with varying chemical schedules, the April 2016 clusters (Figure 3B; Supplemental Information Table S5) were often comparable to profiles of corresponding sites observed in 2012. For example, Cluster A16-C1 grouped sites USC16 and GRM16 based on occurrence of pesticides, sterols, pharmaceuticals, and PAHs, a composition similar to USC12 (cluster S12-C2). The second cluster (A16-C2) was characterized largely by agricultural land use resulting in pesticides and associated degradates comprising 89% of the chemicals at BCR16, FMP16, and PBG16. Notably, the highest absolute concentrations of herbicides (e.g., 2,4-D, metribuzin, metolachlor, atrazine) in the AOC typically were observed at these same sites. Cluster A16-C3 indicated that the Swan Creek tributary (SCR16) was largely characterized by PAHs (fluoranthene, pyrene) and pesticides (carbaryl, bromacil, diuron), similar to the pattern seen in 2012 at the two Swan Creek sites (SCR12, MSCR12). The remaining April cluster (A16-C4) consisted of pharmaceuticals and wastewater indicators detected near the WWTP sites (TWP16, UTP16), the two sites with the highest total number of chemicals detected during April 2016.
Site clusters were less defined in June 2016, with only two identified across the AOC (Figure 3C; Supplemental Information Table S5). The first cluster (J16-C1) contained two sub-clusters comprised of (a) PBG16, FMP16, and GRM16, and (b) BCR16. These sites are all upstream of the Toledo-Bay View WWTP and were mainly influenced by pesticides (79% of the cluster contribution), reminiscent of trends seen in April. The second cluster (J16-C2) included USC16, SCR16, UTP16, and TWP16, all of which are in the downstream region of the AOC. Chemical contributions to this cluster were spread across the pharmaceutical (41%), pesticide (33%), and wastewater indicator (26%) schedules. It was possible to discern several sub-clusters of J16-C2: (a) USC16, (b) SCR16, and (c) UTP16 and TWP16. As in April, UTP16 and TWP16 sites were largely dominated by pharmaceuticals and wastewater indicators.
Inferring Biological Hazards from Chemical Occurrence Data: EARs and AOPs
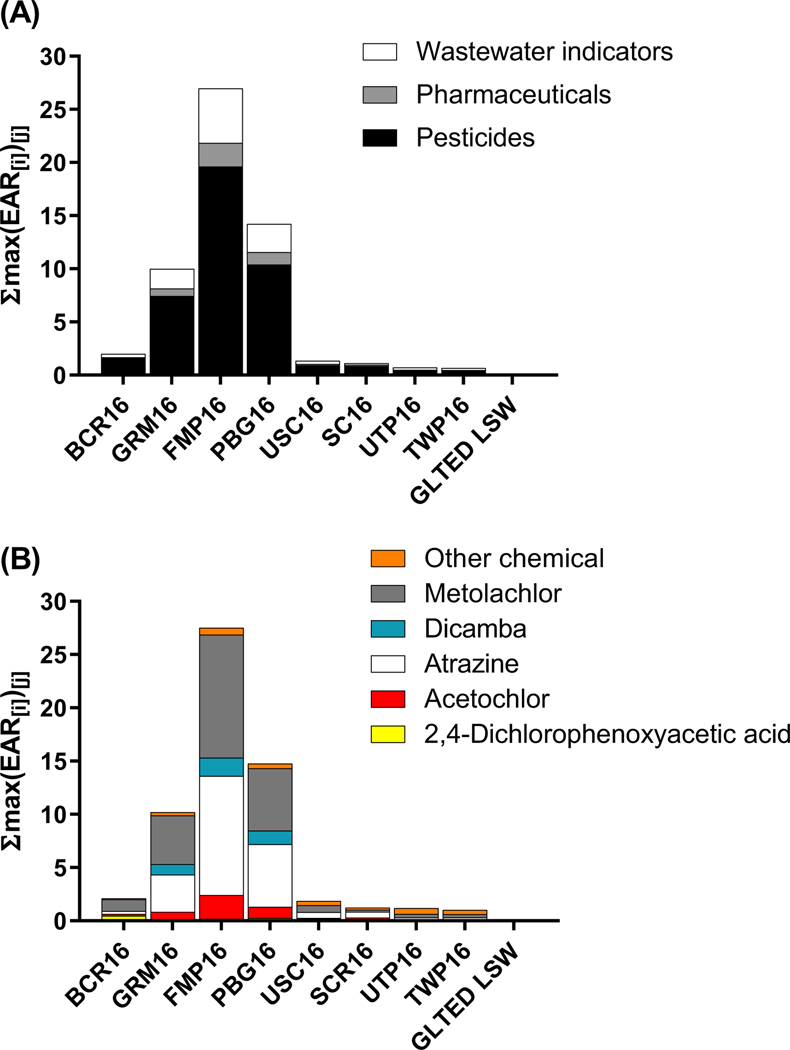
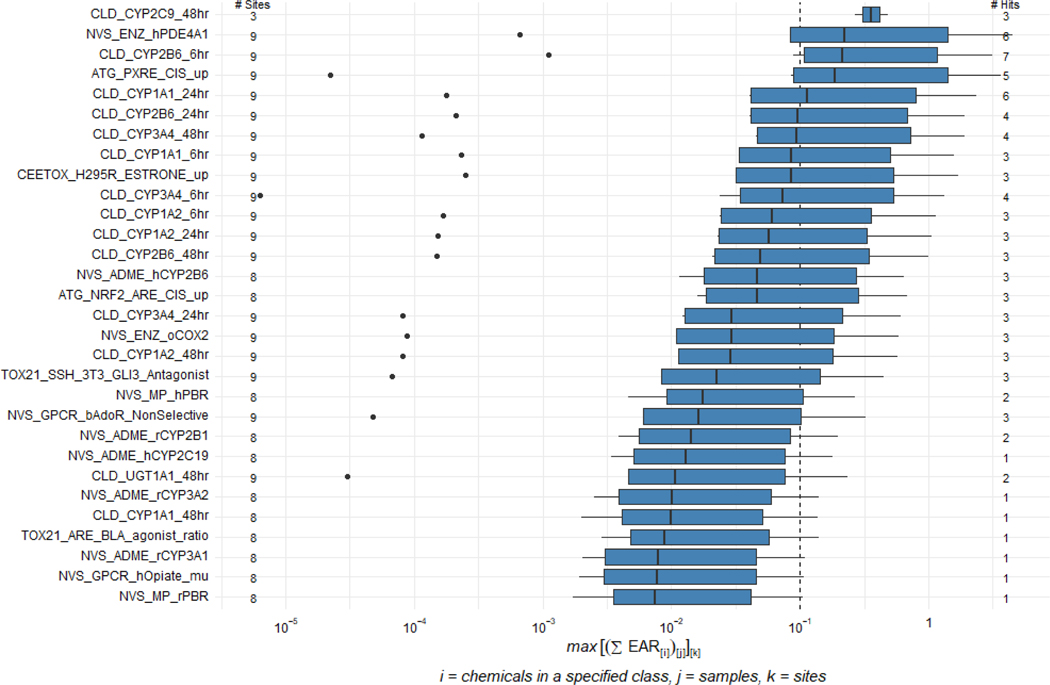
Analysis of the 2016 data using ToxEval revealed that pesticides were the predominant chemical class contributing to elevated EAR values (Figure 4A). Based on summed EAR values derived from maximum chemical concentrations measured during either sampling event in 2016, three sites were identified as having the greatest potential impact (FMP16, PBG16, GRM16), with FMP16 having the highest total EAR among all sites. The three locations are in the main channel of the Maumee River upstream of Toledo-Bay View WWTP, where the primary land use is agricultural. Overall, five pesticides (atrazine, metolachlor, acetochlor, dicamba, 2,4-D) were the main contributors to the cumulative EAR values (Figure 4B). Figure 5 depicts the specific assays/targets from the ToxCast suite that resulted in EAR values greater than a cutoff of 0.1 (see Blackwell et al. 2019 for rationale for this cutoff). It is noteworthy that several of the molecular targets/activities in the assays resulting in elevated EAR values are related to xenobiotic metabolism pathways (e.g., CYP2C9, CYP2B6, PXR, UGT1A1).
Figure 4.
Cumulative maximum exposure-activity ratio (EAR) values for 2016 water samples collected from the Maumee AOC sites expressed as a function of (A) chemical class and (B) individual chemical distributions of pesticides. The values within each bar represent the maximum EAR sum (∑max(EAR[i])[j], where i= chemicals within in a specific class, j= number of samples) within a chemical class for each site. The GLTED LSW (Great Lakes Toxicology and Ecology Division-Lake Superior Water) sample is a lab control outside of the Maumee system.
Figure 5.
ToxCast assays resulting in elevated exposure-activity ratios (EARs) across the 2016 Maumee AOC sites for chemicals within the (A) pesticide, (B) wastewater indicator, and (C) pharmaceutical classes as defined by USGS Schedules 2437, 2440 and 4433, respectively. Figures show the maximum EAR sum (max[∑ (EAR[i])[j]] [k], where i= chemicals within in a specific class, j= number of samples, k= number of sites). Boxes represent 25th to 75th percentiles with the median presented as a dark line. Whiskers represent data occurring within 1.5x of the interquartile range (IQR) and distinct points represented as circles occurring beyond the IQR interval. Number of hits refers to the number of values exceeding the 0.1 threshold.
While the derived EAR values are useful for highlighting those chemicals present in complex mixtures at concentrations high enough to produce a biological response in vitro, this alone provides limited information as to potential biological consequences in organisms exposed in the field. Adverse outcome pathways provide one avenue through which to forecast possible apical effects associated with molecular or biochemical perturbations. In the present study ToxCast assays corresponding to maximum summed EARs greater than 0.1 (Figure 5) represented 19 unique gene targets, half of which were various cytochrome P450 isoforms. Considering the type of responses of each target (e.g., agonism versus antagonism; activation versus inhibition), effects on those 19 gene targets mapped to eight key events described in the AOP-Wiki (aopwiki.org; accessed March 31, 2020; Supplemental Information Table S7). Because key events can be shared by more than one AOP, these key events were associated with a total of 14 AOPs (Supplemental Information Table S7). Six of these were a network of AOPs linking cyclooxygenase-2 inhibition to reproductive dysfunction. Another five linked aryl hydrocarbon receptor (AhR) activation to early life stage mortality, porphyria, and hepatic steatosis. The remainder of the AOPs identified various liver pathologies as potential apical hazards. It should be noted that exceedance of an EAR threshold of 0.1 does not necessarily indicate that concentrations of the active chemicals in the environment are high enough or present for durations long enough, to produce the hazards outlined in the AOPs. Rather, these are outcomes that could be expected if exposures (in terms of dose and duration) were sufficient to perturb a given pathway.
Pathway-Based Measurements of Potential Toxicity: Targeted Techniques
In vitro assays
There was no significant androgenic activity detected in the MDA-kb2 assay at any site during 2012 (data not shown), so this assay was not employed in 2016. No significant estrogenic activity occurred in the T47D-KBluc assay with composite water samples from sites across the AOC in 2012 (Supplemental Information Figure 1A). In 2012, estrogenic activity was also measured in a series of grab samples collected at the beginning of the caged-fish deployments. In these samples there was slight estrogenic activity at TWP12 (Supplemental Information Figure 1B). Estrogenic activity in composite water samples from 2016, during both the April and June sampling periods, also was uniformly low across the different sampling sites (Supplemental Information Figure 2).
Gonadal steroid production and plasma VTG and steroid concentrations
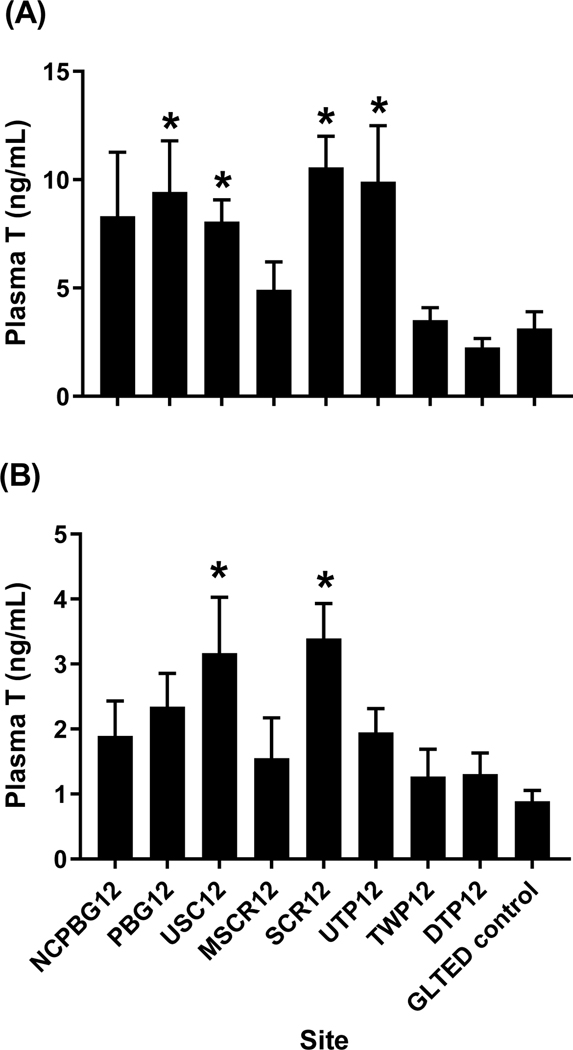
Changes in gonadal steroid (E2, T) production or plasma concentrations of T, E2 and VTG also can indicate the presence of endocrine-active chemicals, including estrogens. There was little effect on most of these endpoints in caged fish from the 2012 study. Plasma concentrations of VTG in males were not significantly affected in fish from any of the sites (Supplemental Information Figure 3). Ex vivo ovarian production of E2 and T, and plasma E2 concentrations in caged females from the eight field sites did not differ from lab controls (Supplemental Information Figure 4A-C)). Similarly, neither testicular T production nor plasma E2 concentrations were affected in males from the field in 2012 (Supplemental Information Figure 5B,C). Production of E2 by testis of males from all the field sites was significantly lower than that of control fish (Supplemental Information Figure 5A), but this measurement is near the limits of detection of the assay so is of questionable biological significance. One endocrine-associated endpoint that showed relatively consistent changes in the caged fish was plasma T concentrations. In males, plasma T was elevated at PBG12, USC12, SCR12 and UTP12, while T was significantly higher than control values in females from USC12 and SCR12 (Figure 6A,B).
Figure 6.
Plasma concentration of testosterone (T) from adult (A) male, and (B) female fathead minnows caged for 4-d at sites within the Maumee AOC in 2012. The GLTED (Great Lakes Toxicology and Ecology Division) control fish were maintained in Lake Superior water at the GLTED facility. Bars represent mean (± standard error; n = 12–14 males per site except PGB12, n = 5; NCPBG12, n = 4 and UTP12, n = 8; n = 7–10 females per site). Asterisks indicate significant difference from the GLTED control (p≤0.05).
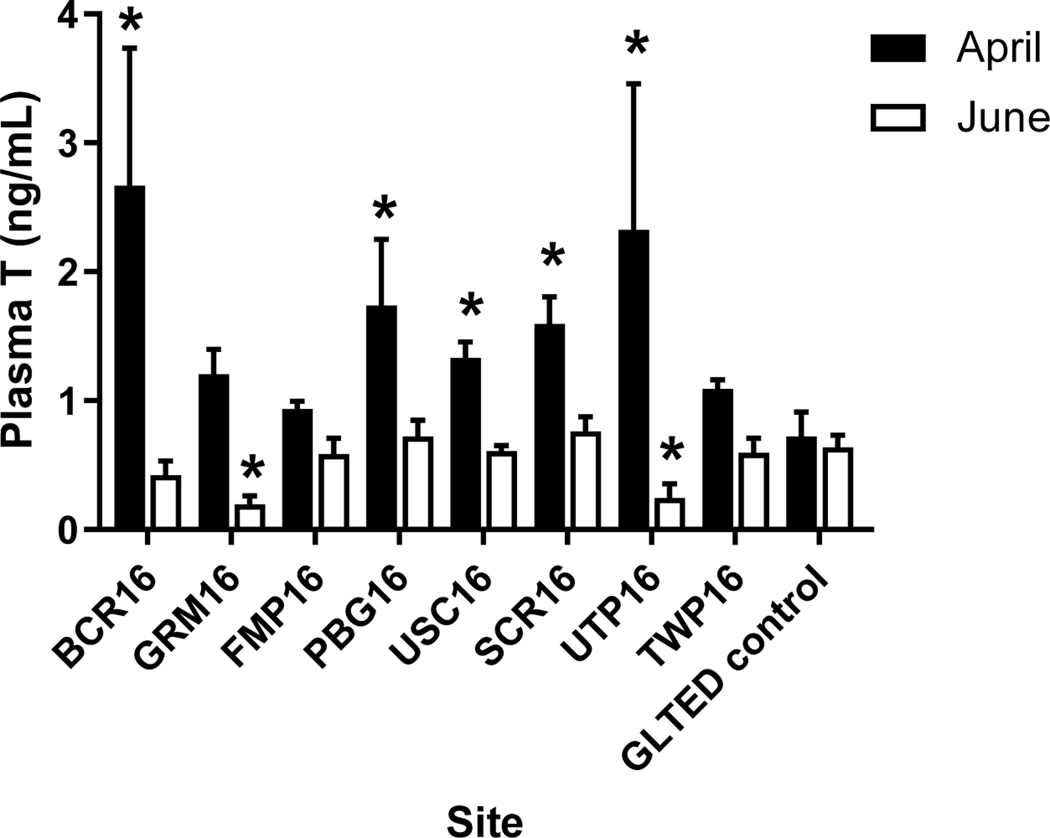
Given the overall lack of significant changes in 2012, we did not measure plasma VTG nor gonadal synthesis of T or E2 in 2016. Neither plasma T nor E2 concentrations were consistently affected in caged females in April or June, 2016 (Supplemental Information Figure 6A,B). Plasma E2 in males was not affected during either sampling period in 2016 (data not shown); however, plasma T was different in caged fish than in controls. In April, T was significantly increased in males from five of the eight study sites, with four of these corresponding to locations where T was elevated in males in 2012 (PBG, USC, SCR, UTP; Figure 7). However, T was not elevated in caged males from June 2016 and, in fact, was slightly decreased at two of the study sites (GRM16, UTP16; Figure 7).
Figure 7.
Plasma concentration of testosterone (T) in adult male fathead minnows caged for 4-d at sites within the Maumee AOC in April (black bars) and June (white bars) 2016. The GLTED (Great Lakes Toxicology and Ecology Division) control fish were maintained in Lake Superior water at the GLTED facility. Bars represent mean (± standard error; n = 8–12 per site). Asterisks indicate significant difference from the GLTED control (p≤0.05).
Gene expression
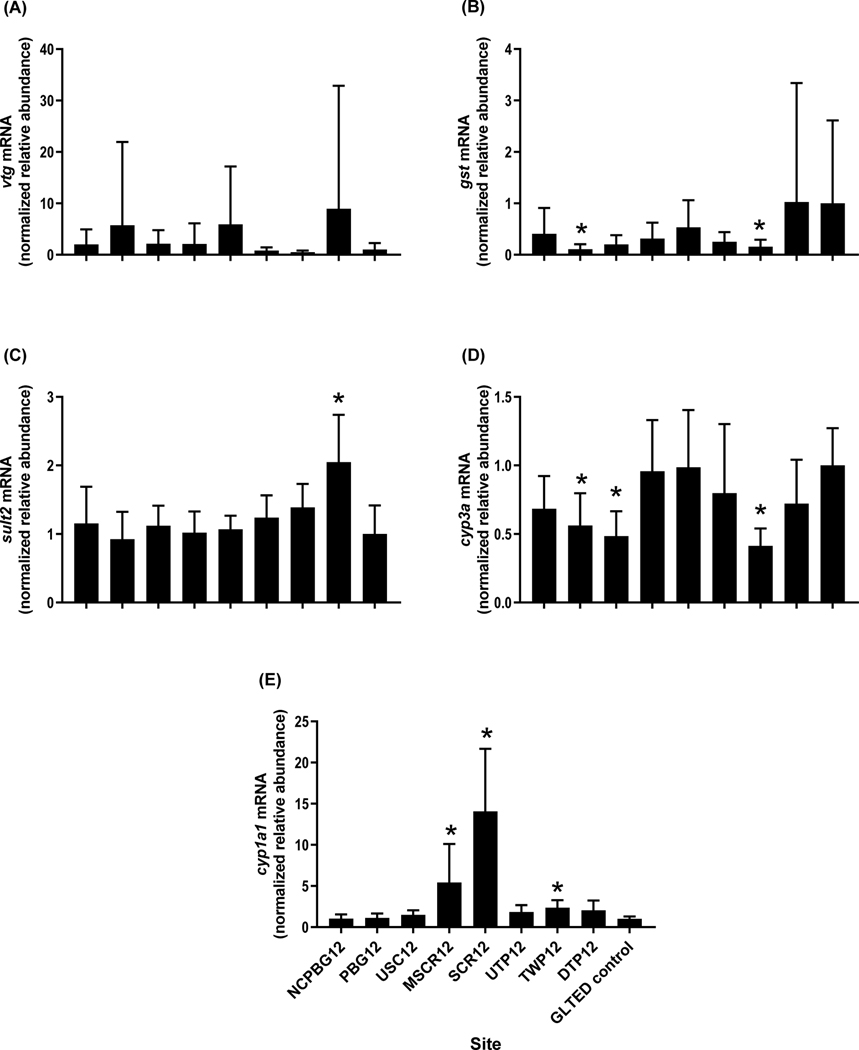
In 2012, hepatic vtg seemed to be slightly elevated in male fish from three sites, most notably DTP12, but the expression data were variable and not statistically different (Figure 8A). Transcripts of hepatic gst were lower in males from seven of the eight sites than in controls, significantly so for PBG12 and TWP12 (Figure 8B). Sulfotransferase 2 transcripts were elevated in the livers of males from DTP12 (Figure 8C). Expression of cyp3a was significantly decreased in caged males from three sites, PBG12, USC12 and TWP12 (Figure 8D). Arguably the most inducible of the hepatic CYPs, cyp1a1, was higher in the field males than in the controls, significantly so at MSCR12, SCR12 and TWP12 (Figure 8E).
Figure 8.
Relative transcript abundance of (A) vitellogenin (vtg), (B) glutathione S-transferase (gst), (C) sulfotransferase (sult2), (D) cytochrome P4503A (cyp3a) and (E) cytochrome P450 1A1 (cyp1a1) in hepatic tissue from adult male fathead minnows caged for 4-d at sites within the Maumee AOC in 2012. The GLTED (Great Lakes Toxicology and Ecology Division) control fish were maintained in Lake Superior water at the GLTED facility. Bars represent mean (± standard error; n = 10–13 per site except GLTED control, n = 8). Asterisks indicate significant difference from the GLTED control (p≤0.05).
Ovarian expression of three genes coding for steroidogenic enzymes, cyp19a1a, cyp17 and cyp11a, did not differ greatly between fish from the lab versus the field in 2012, except for a slight decrease in cyp17 at NCPBG12 and PBG12 (Supplemental Information Figure 7A-C).
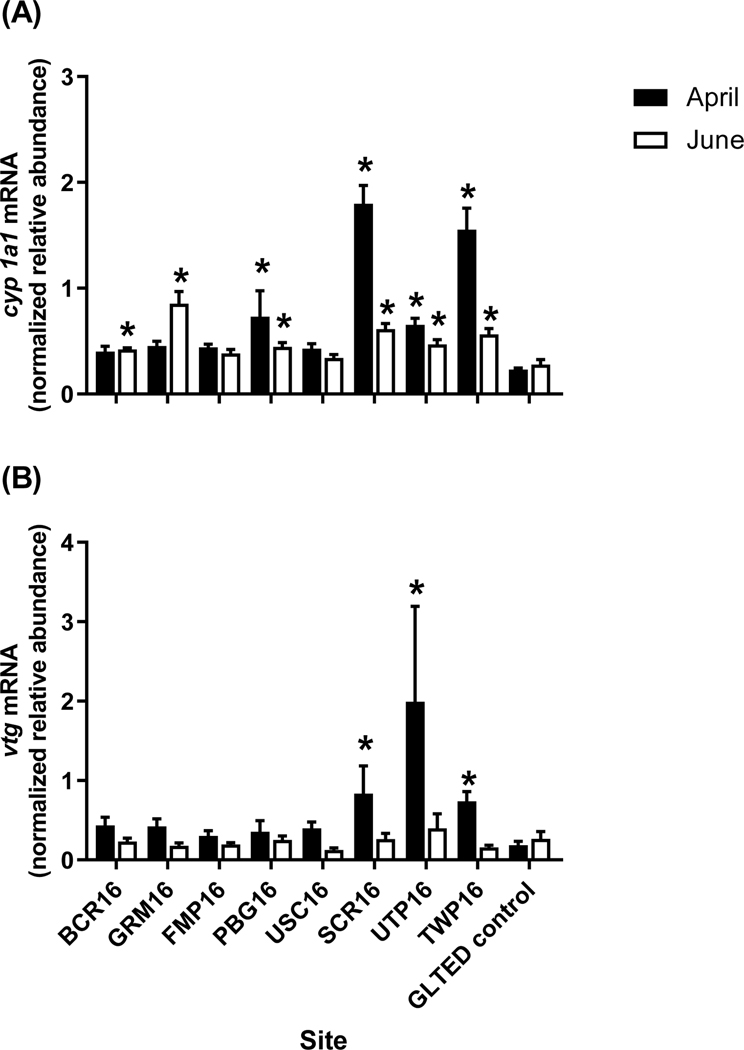
In 2016, hepatic cyp1a1 was elevated in males from half of the sites (PBG16, SCR16, UTP16 and TWP16) in April, and at all four of these sites plus GRM16 and BCR16 in June (Figure 9A). Hepatic vtg in caged male fish was increased at SCR16, UTP16 and TWP16 in April, but not at any of the deployment sites in June (Figure 9B).
Figure 9.
Relative transcript abundance of (A) cytochrome P450 1A1 (cyp1a1) and (B) vitellogenin (vtg) in hepatic tissue from adult male fathead minnows caged for 4-d at sites within the Maumee AOC in April (black bars) and June (white bars) 2016. The GLTED (Great Lakes Toxicology and Ecology Division) control fish were maintained in Lake Superior water at the GLTED facility. Bars represent the mean (± standard error; n = 10–12 per site). Asterisks indicate significant difference from the GLTED control (p≤0.05).
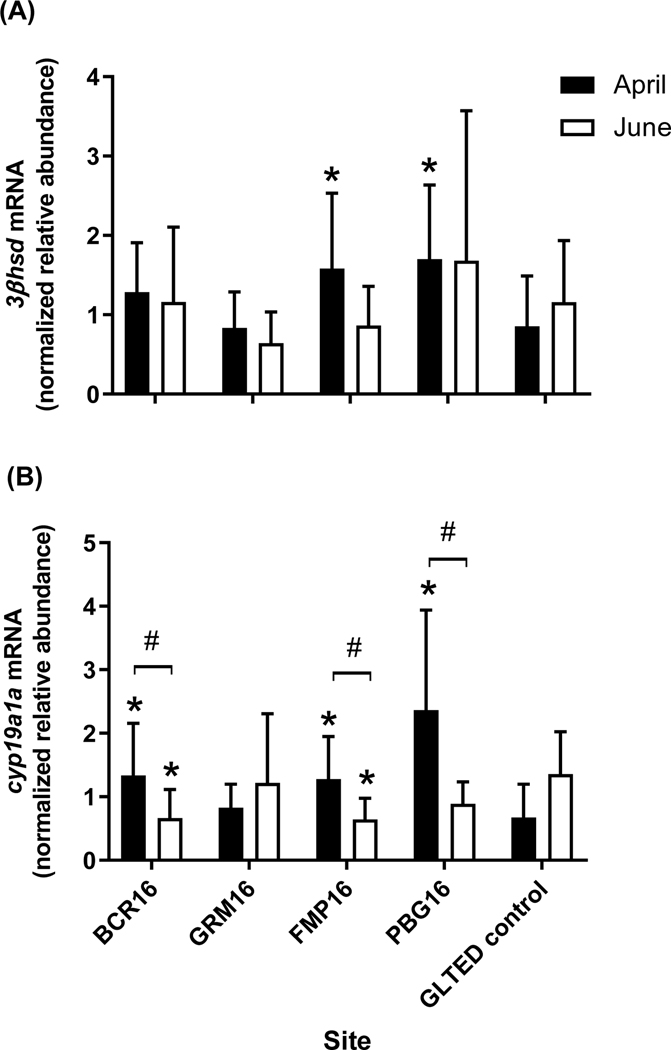
In 2012 we measured expression of several CYP-based steroidogenic genes at all sites. In contrast in 2016 we focused on a functionally broader set of HPG-related ovarian genes at four upstream sites (BCR16, GRM16, PBG16, FMP16) where there were marked seasonal changes in measured concentrations of several herbicides. In evaluating the 2016 expression data we compared females from the field to lab controls from a given sampling period and evaluated responses of the fish between April and June at the four sites using t-tests. Expression of fshr, star and zp3 did not differ between females from the field versus the lab, nor was expression different between the two sampling periods at the field sites (Supplemental Information Figure 8A-C). Transcripts for 3βhsd were higher in females from PBG16 and FMP16 than controls in April but did not differ between April and June at any site (Figure 10A). Compared to lab controls, ovarian cyp19a1a was significantly elevated at BCR16, PBG16 and FMP16 in April and decreased at BCR16 and FMP16 in June, and there were significant differences between the sampling times at BCR16, PBG16 and FMP16 (Figure 10B).
Figure 10.
Relative transcript abundance of (A) 3β-hydroxysteroid dehydrogenase (3βhsd), and (B) cytochrome P450 aromatase A (cyp19a1a) in ovarian tissue from adult female fathead minnows caged for 4-d at sites within the Maumee AOC during April (black bars) and June (white bars) 2016. The GLTED (Great Lakes Toxicology and Ecology Division) control fish were maintained in Lake Superior water at the GLTED facility. Bars represent the mean (± standard error n = 11–12 per site). Asterisks indicate a significant difference from the GLTED control and # indicates significant differences between April and June within a site (p≤0.05).
Pathway-Based Measurements of Potential Toxicity: Nontargeted Techniques
Hepatic Metabolomics
For the April and June 2016 upstream samples (sites BCR16, GRM16, PBG16 and FMP16), the global PLS regression model resulted in a Q2 value of 0.54 when comparing the concentrations of the 87 detected organic contaminants with the LC-MS/MS metabolomic data in the polar extracts of male fathead minnow liver samples. After three iterations of backward elimination, nine contaminants were omitted due to lack of covariance with the metabolite profiles, which resulted in a final model with 78 organic contaminants, with a Q2 value of 0.59 (Supplemental Information Tables S8, S9). Thus, elimination of these relatively few contaminants resulted in a final model with only marginally greater predictive power (i.e., slightly higher Q2 value) than the initial global model.
For the downstream batch (USC16, SCR16, UTP16, and TWP16), the global PLS regression model exhibited a Q2 value of 0.26 when comparing the concentrations of the 119 detected organics with the metabolomic data. After five iterations of backward elimination, 26 contaminants were omitted, leaving a final model with 93 organic contaminants with a Q2 value of 0.34 (Supplemental Information Tables S10, S11).
Note that it is possible to assess the strength of the covariance between the polar hepatic metabolite profiles and individual organic contaminants in the final models by their associated CV-ANOVA values. However, small changes in these values are usually not very meaningful. For this reason, we chose to focus on a coarse categorization of these contaminants by dividing them into groups of 25 members. The groups we hereafter refer to as the “top-25” contain those contaminants that most strongly covaried with endogenous metabolite profile changes (i.e., exhibiting the lowest CV-ANOVA values) for each of the two batches. The top 25 chemicals at the upstream sites all were pesticide or pesticide degradates, while the top 25 chemicals at the downstream sites included pesticides/pesticide degradates, pharmaceuticals, industrial chemicals and wastewater indicators (Table 1).
Table 1.
Top 25 contaminants detected in composite water samples from the Maumee AOC in 2016 that were found to most strongly covary with endogenous metabolite profile changes found with polar extracts of liver tissue collected from associated caged male fathead minnows. The analysis was conducted in two different batches comprised of upstream (BCR16, GRM16, FMP16, PBG16) and downstream sites (USC16, SCR16, UTP16, TWP16). An X in the column headed Upstream or Downstream indicates the presence of the contaminant in the top 25 for the given batch. (See text for analysis details).
| Chemical | Class | Upstream | Downstream |
|---|---|---|---|
| Imidacloprid | Insecticide | X | X |
| Metalaxyl | Fungicide | X | |
| Tebuconazole | Fungicide | X | |
| Acetochlor | Herbicide | X | |
| Ametryn | Herbicide | X | |
| Atrazine | Herbicide | X | X |
| Dicamba | Herbicide | X | |
| Flumetsulam | Herbicide | X | |
| Imazethapyr | Herbicide | X | |
| Metolachlor | Herbicide | X | |
| Prometon | Herbicide | X | |
| Propazine | Herbicide | X | X |
| Triclopyr | Herbicide | X | X |
| 2-Chloro-4-isopropylamino-6-amino-s-triazine | Herbicide (degradate) | X | |
| 2-Chloro-6-ethylamino-4-amino-s-triazine | Herbicide (degradate) | X | X |
| 2-Chloro-N-(2-ethyl-6-methylphenyl) acetamide | Herbicide (degradate) | X | |
| 2-Hydroxy-4-isopropylamino-6-amino-s-triazine | Herbicide (degradate) | X | |
| 2-Hydroxy-4-isopropylamino-6-ethylamino-s-triazine | Herbicide (degradate) | X | X |
| Acetochlor oxanilic acid | Herbicide (degradate) | X | X |
| Acetochlor sulfinylacetic acid | Herbicide (degradate) | X | |
| Acetochlor sulfonic acid | Herbicide (degradate) | X | |
| Chlorodiamino-s-triazine | Herbicide (degradate) | X | |
| Dechlorometolachlor | Herbicide (degradate) | X | |
| Diketonitrile-isoxaflutole | Herbicide (degradate) | X | X |
| Hydroxyacetochlor | Herbicide (degradate) | X | X |
| Hydroxymetolachlor | Herbicide (degradate) | X | |
| Hydroxysimazine | Herbicide (degradate) | X | |
| Isoxaflutole acid metabolite RPA 203328 | Herbicide (degradate) | X | |
| Metolachlor oxanilic acid | Herbicide (degradate) | X | X |
| N-(3,4-Dichlorophenyl)-N’-methylurea | Herbicide (degradate) | X | |
| 3,4-Dichlorophenyl isocyanate | Industrial | X | |
| Tris(dichloroisopropyl) phosphate | Industrial | X | |
| Venlafaxine | Pharmaceutical | X | |
| Carbamazepine | Pharmaceutical | X | |
| Lidocaine | Pharmaceutical | X | |
| Menthol | Pharmaceutical | X | |
| Triamterene | Pharmaceutical | X | |
| Methyl-1H-benzotriazole | Pharmaceutical precursor | X | |
| Caffeine | Wastewater indicator | X | |
| N,N-Diethyl-m-toluamide (DEET) | Wastewater indicator | X |
High-Throughput Assays
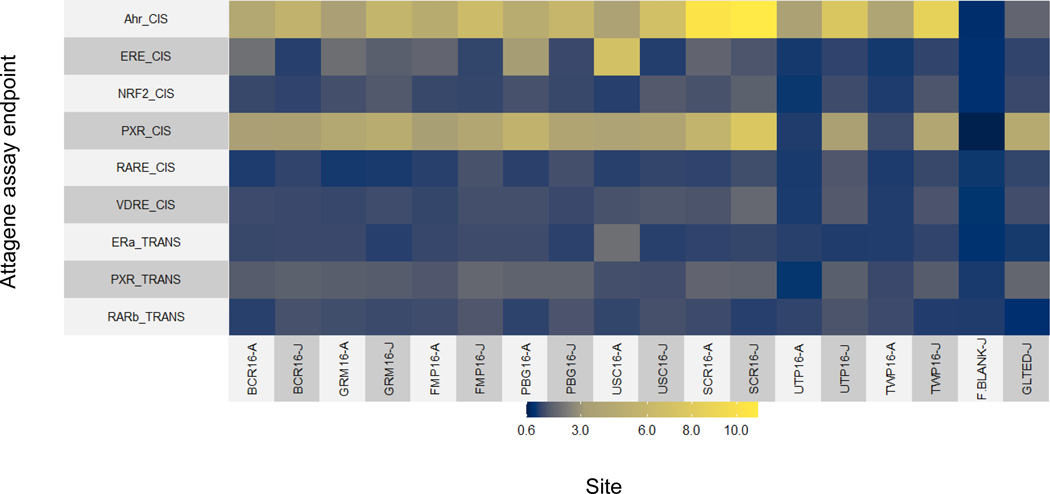
Of 69 endpoints analyzed using both Attagene assays, 15 responded (activity ≥1.5 fold) to one or more site-derived mixtures during either 2012 or 2016 (Supplemental Information Table S12). In general, bioactivity patterns were similar across 2012 and 2016 (Supplemental Information Figures 9, 10), so below we focus specifically on the more extensive 2016 dataset. Overall, nine bioactivities were affected during either the April or June sampling (Figure 11). Specific molecular targets activated included the AhR, estrogen receptor, pregnane X receptor (PXR), retinoic acid receptor, and NRF2, an indicator of oxidative stress. The most frequently elevated bioactivity was AhR (AHR_CIS), which was observed at all sites, including the laboratory control; however, AhR activity in the field samples was much higher than in the lab controls (Figure 11). Site SCR16, for both April and June samples, induced the greatest AhR activity of all the 2016 samples, indicating the occurrence of AhR agonists such as PAHs. Pregnane X receptor (PXR_CIS) was the second most frequent elevated bioactivity, present in all the 2016 samples except UTP16 and TWP16 in April (Figure 11). In general, PXR activity was elevated to a greater extent during the June sampling. Estrogen receptor (ERE_CIS) activity was the other bioactivity commonly observed, at six of eight sites from April (all except UTP16 and TWP16), and at two sites (GRM16, SCR16) in June. Changes in other biological activities observed using the two HTS assays were minimal, with three or fewer samples inducing activity above baseline.
Figure 11.
Heatmap of blank normalized responses of water from the sites in the Maumee AOC from April (site name-A) and June (site name-J) 2016 (x-axis), across select Attagene assay endpoints (y-axis). Plotted values represent the mean of duplicate assay measurements. Only endpoints with mean response ≥1.5-fold at one or more sites are included. See Supplemental Information Table 12 for details on each individual endpoint.
DISCUSSION
The goal of the present study was to provide a practical demonstration of the application of pathway-based targeted and nontargeted techniques for the identification of occurrence and potential effects of complex chemical mixtures in a Great Lakes AOC. Throughout the Great Lakes, human activities past and present have contributed to the presence of complex mixtures of legacy contaminants, current use pesticides, pharmaceuticals, flame retardants, plasticizers, and many other contaminants of emerging concern (Baldwin et al. 2016; Davis et al. 2016; Elliot et al. 2017; Venier et al. 2014). The Maumee River is the largest tributary of the Great Lakes by watershed area, significantly contributing to chemical and sediment loading to Lake Erie (Baker et al. 2014). The system is impacted by multiple point and non-point source contaminant inputs, including WWTPs and urban and agricultural runoff (Arini et al. 2016; Baker et al. 2014; Corsi et al. 2019). As such, the Maumee AOC provided an appropriate setting in which to evaluate an integrated suite of tools and associated databases designed to enhance assessment of the ecological effects of mixtures.
Inferring Biological Hazards from Chemical Occurrence Data
Analytical characterization of water samples from multiple sites in the Maumee AOC in 2012 and 2016 revealed detectable concentrations of more than 100 chemicals broadly characterized as pesticides, pharmaceuticals and wastewater indicators. Hierarchal cluster analysis of the chemical occurrence data suggested associations with different inputs/land use patterns across the AOC, an observation consistent with a more limited investigation on the Maumee River conducted by Baldwin et al. (2016). Specifically, we found distinct agricultural chemical profiles in addition to patterns indicative of urban and WWTP inputs. For example, upstream locations predominantly associated with agriculture had the greatest number of detections (and concentrations) of crop-use pesticides. In contrast, sites proximate to WWTPs (particularly the Toledo-Bay View facility) exhibited higher frequencies and concentrations of pharmaceuticals and chemicals typically associated with wastewater effluent. Other waterways entering the AOC including urban (Swan Creek) and agricultural (Beaver Creek) tributaries had unique chemical profiles that were particularly apparent during the April 2016 sampling. This suggests that potential biological impacts of chemical mixtures in the Maumee AOC are likely to vary spatially.
One of the more notable attributes of the analytical chemistry dataset was a marked increase in concentrations of several herbicides going from the April to June sampling period in 2016. The magnitude of changes in metalochlor and atrazine levels were most pronounced, with concentrations of the latter approaching and occasionally exceeding 20 μg/L at some of the upstream, more agriculturally influenced sites (GRM16, FMP16, PBG16). Seasonal variations in surface water atrazine levels in Great Lakes tributaries have been reported before (Baldwin et al. 2016), although concentrations of the herbicide were typically lower than those in the present study. Specifically, in the Baldwin et al. (2016) dataset, of 709 samples from 57 sites, there were only seven instances with atrazine concentrations greater than 5 μg/L, although five of those samples also were from the Maumee River where a maximum concentration of 9.4 μg/L was observed. Another assessment of concentrations of atrazine in several Midwestern US streams during 2013 reported an occasional occurrence of atrazine concentrations greater than 20 μg/L, with one sample as high as 120 μg/L (Mahler et al. 2017). Atrazine has been extensively evaluated in toxicity tests with a variety of terrestrial and aquatic animals and plants (for a recent compilation of test data see US Environmental Protection Agency 2016). Given its herbicidal mode of action, derivation of environmental levels of concern (LOC) for atrazine in freshwater aquatic systems often has been based on potential impacts on plant communities (Moore et al. 2017). Recently a 60-d LOC for atrazine of 15 μg/L was proposed (US Environmental Protection Agency 2019). While atrazine concentrations from multiple sites in the present study exceed this value, they were based on 4-d composite samples so may not be directly germane to evaluating longer-term plant community risks. There are considerably fewer toxicity data for metolachlor than for atrazine but, again, the greatest concern for its possible effects in aquatic systems involves changes in plant community structure, which perhaps would be exacerbated by the simultaneous occurrence of elevated concentrations of multiple herbicides (e.g., Powell et al. 2017; Nowell et al. 2018). While we cannot conclude that aquatic plants in upper reaches of the Maumee River are impacted by herbicides, data suggest that additional testing or monitoring in the system should perhaps be focused on this component of the aquatic community.
Comparing or benchmarking measured chemical concentrations with relevant in vivo toxicological information provides a relatively direct approach for predicting the possible effects of compounds detected in environmental samples. However, very few chemicals have been tested as extensively as, for example, atrazine. There may be no toxicity data available for some detected chemicals and, in the case of others, prior testing might have been limited to just a few biological responses (e.g., lethality). One method to help address limitations in toxicity data is to employ pathway-based in vitro information for benchmarking. Publicly accessible data from the USEPA ToxCast program are well-suited for this application, as the results are from a suite of assays covering a wide variety of biological targets/pathways. The assays have been conducted with several thousand chemicals in a standardized manner relative to generating dose-response relationships suitable for the calculation of risk quotients such as EAR values. In the present study we used an EAR cutoff of 0.1 or greater to “flag” detected chemical classes of potential biological concern in composite water samples from the Maumee AOC. Given in vitro to in vivo extrapolation assumptions underlying interpretation of derived EAR values, the 0.1 threshold may be regarded as somewhat arbitrary in terms of effects prediction, but the value seems justifiable based on the screening-level nature of the analysis (Blackwell et al. 2018). In the present study, individual chemicals with EARs exceeding the 0.1 threshold included dicamba, acetochlor, 2,4-D, metolachlor and atrazine (Figure 4B). Some of these same chemicals (metolachlor, atrazine, bisphenol-A) also had elevated ToxCast-based EARs in other Great Lakes tributaries (Corsi et al. 2019), but it should be noted that the earlier work used a lower cutoff value as a threshold trigger (0.001) than that of the current study. Corsi et al. (2019) also reported that EAR values for several PAHs (anthracene, benzo (a) pyrene, fluoranthene, phenanthrene, pyrene) were elevated in their analysis. Even at an EAR cutoff of 0.001 only the benzo (a) pyrene EAR would have been considered elevated in the current Maumee AOC dataset; however, several of the tributaries included in the more spatially extensive Corsi et al. (2019) analysis would be regarded as relatively industrialized (e.g., Clinton River, Rouge River, Cuyahoga River) and hence potentially more prone to PAH contamination. Also, we cannot discount the possible effects of hydrologic influences on the occurrence of chemicals like PAHs that can have a strong association with particulate matter (Baldwin et al. 2020).
Generation of EAR values using the pathway based ToxCast assays provides the ability to link chemicals or groups of chemicals to potential apical effects using the AOP framework (Corsi et al. 2019). Most of the ToxCast assays associated with maximum sum EARs greater than 0.1 were related to pathways directly relevant to hepatic metabolism of xenobiotics. For example, activation of PXR and AhR is known to induce expression of hepatic biotransformation enzymes, most notably different CYPs. While there are several AOPs linking these responses to potential liver toxicity (e.g., fatty liver) or tumor formation through the formation of reactive oxygen species, it should be noted that AOPs are developed under the assumption that exposures are of adequate concentration and duration to drive the system to an adverse apical effect. It is also possible that the chemicals in the mixtures may simply activate the biotransformation system that an exposed animal could use to cope with the exposures. The quantitative boundaries that distinguish a beneficial adaptive response from one expected to result in chronic liver disease and associated reductions in fitness are unclear. Nonetheless, the current complement of AOPs linked to the activation of biotransformation pathways suggest that monitoring field collected organisms for signs of liver toxicity may be warranted.
Two additional clusters of AOPs suggest some potential for adverse outcomes related to early development or reproduction in vertebrates. Three AOPs provide evidence linking AhR activation to early life stage mortality in fish (Supplemental Information Table S7; AOPs 21, 150, 310). The AOPs outline a number of events at the molecular, and biochemical level that could be used as potential biological indicators of progression toward an adverse early life stage effect, including increased cyclooxygenase-2 gene expression (AOP 21), reduced VEGF expression (AOP 150), or potential methylation of the gonadotropin releasing hormone receptor promoter region (AOP 310). A final cluster of AOPs derived from the elevated EAR values link inhibition of cyclooxygenase 2 to potential reproductive impairment (Supplemental Information Table S7). Examination of those AOPs suggests measurement of plasma prostaglandin concentrations as one of the more “field deployable” measures associated with this network of AOPs. Unfortunately, other events associated with cyclooxygenase inhibition, like pheromone release and/or proper spindle formation and prophase to metaphase transitions in germ cell meiosis would be challenging to measure outside of a controlled laboratory setting.
Pathway-Based Measures of Potential Toxicity: Targeted Techniques
Prediction of CYP-related responses based on the EAR calculations were consistent with observations of increased expression of cyp1a1 in the livers of caged male fish. A variety of environmental contaminants can induce expression and activity of CYP1A1 in fish; this provides the basis for an assortment of mRNA and protein biomarkers of the enzyme that have been used in environmental toxicology for many years (see Whyte et al. 2000 for a review). In the present study, hepatic cyp1a1 in caged males was significantly induced at three sites in 2012, four sites in April 2016, and six sites June 2016. There were two sites where cyp1a1 was elevated on all three occasions: SCR, which had elevated concentrations of PAHs, known inducers of hepatic CYPs, and TWP, a source of multiple contaminants, including pharmaceuticals, associated with CYP induction. Interestingly, in caged males from 2012, there were decreases in hepatic transcripts for two other xenobiotic-metabolizing enzymes, CYP3A and GST, that can be induced by many of the same chemicals that increase cyp1a1.
Given concern for the possible effects of contaminants on endocrine function, several of our targeted measurements focused on different components of the HPG axis, including those indicative of estrogens, which have been of interest for many years due to early observations of feminized fish from the vicinity of WWTPs (Purdom et al. 1994). Overall, there was no evidence of substantial estrogenic activity observed in the Maumee AOC based either on in vitro or in vivo endpoints. One water sample in 2012 would have been identified as marginally estrogenic using the T47D-KBluc assay, but the measured activity was much lower than observed at other surface water samples, including from other Great Lakes sites using the same assay (e.g., Wehmas et al. 2011; Cavallin et al. 2014; 2016). Similarly, induction of VTG (measured either as plasma protein or hepatic mRNA), a sensitive and unambiguous indicator of exposure of male fathead minnows to exogenous estrogens (Korte et al. 2000; Ankley and Jensen 2014), was not consistently seen in caged fish from the AOC. There was a slight elevation of hepatic vtg in fish from three sites during the April 2016 sampling, but no evidence of induction from these same sites in June. Overall, our findings of little estrogenicity in fish exposed to surface waters of the Maumee River are consistent with the results of Cipoletti et al. (2019), who exposed fathead minnows to water from the system under controlled conditions for 21 d and found no obvious estrogenic effects.
Changes in gonadal production and plasma concentrations of T and E2 can be indicative of chemical interactions with other components of the fish HPG axis, including androgen receptor agonists and various enzymes involved in steroid synthesis, several of which are cytochrome P450 isozymes (Ankley et al. 2009). With one exception (plasma T), there was little in terms of effects on sex steroid status in the caged fish. In 2012, there were no effects on gonadal production of T or E2 in females or T in males, and no effects on plasma concentrations of E2 in either sex. Concentrations of plasma T in females were elevated at two sites (USC12, SCR12), and plasma T concentrations in males were elevated at these same sites, as well as at PBG12 and UTP12. In 2016, ex vivo steroid production was not measured, but plasma T and E2 concentrations were not affected in females nor was plasma E2 changed in males, either in April or June. Plasma T concentrations in males were elevated during April at the same four sites where T was increased in males in 2012. However, T concentrations were not elevated in males during the June sampling and, in fact, were slightly decreased at two sites. Irrespective of the June data, the elevated plasma T in males from the same sites in 2012 and April 2016 suggests that the results were not spurious. However, possible causes of the elevated T are uncertain; the response would not appear to be a direct effect of chemicals on steroidogenesis, as there were no changes in ex vivo gonadal synthesis of steroids. Further, in past studies with a variety of chemicals affecting different targets in the fathead minnow HPG axis, elevated plasma T concentrations in the absence of other changes have not been seen (Ankley et al. 2009; Ankley and Jensen 2014). It is possible, however, that there could have been chemical effects, not on synthesis, but degradation of T. Specifically, steroids are degraded by various hepatic CYPs whose activity both can be induced (e.g., cyp1a1) and inhibited by different environmental contaminants. Interestingly, in 2012, expression of transcripts for hepatic cyp3a, which metabolizes several substrates, including T in fish (Baldwin et al. 2005; Kashiwada et al. 2005), was reduced in males at two of the same sites where elevated plasma T was observed (PBG12, USC12). Further investigation of this hypothesis, as well as the physiological significance of elevated T in males may be warranted.
A final set of targeted measurements related to endocrine function in caged fathead minnows were a suite of ovarian transcripts coding for proteins involved in steroid synthesis and homeostatic control of the HPG axis. Changes in expression of several of the genes studied in the caged females can be affected by chemicals both via direct and indirect (compensatory) mechanisms (Ankley and Villeneuve 2015). In 2012, there were no consistent differences in expression of three steroidogenic CYPs (cyp11a, cyp17, cyp19a1a) among female fathead minnows from the eight field sites and lab controls. In 2016, we took a slightly different approach, evaluating a broader functionality of proteins within the HPG axis at the four upstream sites where there were marked increases concentrations of several herbicides between the April and June sampling periods. One reason for this focus were reported changes in several HPG axis-related genes in small fish (fathead minnow, Japanese medaka) exposed to atrazine at concentrations in the range of those measured in the Maumee River, albeit for periods of time longer than 4 d (Richter et al. 2016). In the present study, there were no differences in ovarian expression of fshr, star, zp3 or 3βhsd between fish sampled in April versus June at the upstream sites. However, cyp19a1a did differ between the two sampling times in females from three of the four sites (BCR16, FMP16, PBG16), with measured transcripts higher in April than in June. Data are limited concerning the effects of different herbicides on HPG function in fish, so it is difficult to ascertain whether our observation could have been related to increased relative concentrations of herbicides at these sites (or, perhaps, seasonal differences in some other environmental variable such as temperature). Some studies indicate that atrazine can upregulate expression/activity of aromatase in vitro, potentially resulting in estrogenic effects (e.g., Sanderson et al. 2001; 2002; Tinfo et al. 2011); however, this is opposite of the trend in cyp19a1a expression observed in the caged females. The longer-term studies of Richter et al. (2016) did not find that atrazine changed ovarian expression of cyp19a1a in the fathead minnow.
Pathway-Based Measures of Potential Toxicity: Nontargeted Techniques
The Attagene HTS platform covers about 70 endpoints associated with numerous biological pathways involved in metabolism, endocrine signaling and cell stress, including several controlled by nuclear receptors (Romanov et al. 2008). The assay is mammalian-based; however, there is good evidence for conservation of many of the molecular targets in the assay (particularly nuclear receptors) across other vertebrate species (Ankley et al. 2016; LaLone et al. 2013; 2018). Although originally developed and used for single chemical testing in the context of human health, in recent years we have used the Attagene system to screen a variety of surface water samples containing complex mixtures of chemicals that could cause adverse ecological effects (Schroeder et al. 2016; Blackwell et al. 2019). In the present study, the most prevalent bioactivities elevated in water samples from the Maumee system were associated with the AhR and PXR, two targets associated with the induction of enzymes such as CYPs responsible for the metabolism of xenobiotics. We have commonly seen elevated AhR and PXR bioactivity in Attagene studies with other surface water samples containing complex mixtures of contaminants (Blackwell et al. 2019). In the present study, there did not seem to be well-defined temporal or spatial patterns in AhR or PXR activities, which perhaps is not surprising given both receptors are relatively “promiscuous”, being affected by different classes of structurally diverse xenobiotics, including PAHs and some pesticides and pharmaceuticals (see Blackwell et al. 2019 and references therein). Notably, observation of elevated AhR and PXR activities in this study is consistent both with the EAR-based predictions of the effects of detected contaminants on various CYPs, and the observed induction of hepatic cyp1a1 in caged fathead minnows.
A third Attagene target affected on multiple occasions was the estrogen receptor, which we also have observed in other studies with surface water samples (Blackwell et al. 2019). In general, the magnitude of effects of samples from the Maumee system on estrogen-bioactivity in the HTS assay were comparatively low. However, as described above, there was little evidence of estrogenicity in the same composite water samples in the targeted T47D-KBluc assay. It is possible that the Attagene assay is more sensitive to estrogenic chemicals than the T47D-KBluc system. However, preparation of the water for testing in the two assay systems differs substantially, with the Attagene samples extracted, concentrated and transferred into DMSO before testing, while the T47D-KBluc assay employs unconcentrated whole water samples. Whatever the cause of the differing results in the two in vitro assays, it is noteworthy that there was little evidence of consistent estrogenic effects in caged fish (i.e., induction of vtg in males) from the different water collection sites.
Metabolomic profiles measured in livers of the caged fish provide a nontargeted assessment of in vivo responses, and thus offer a biological perspective more directly translatable to potential effects in resident fish than possible with in vitro data. Our research team has employed metabolomic measurements in two different manners in past studies with chemical stressors in fish. One approach involves studying changes in specific metabolites or suites of metabolites to identify biological pathways affected by a given individual test chemical (e.g., Collette et al. 2010; Ekman et al. 2012; Davis et al. 2017). The second approach employs changes in metabolite profiles to provide inferences about the nature of the complex chemical mixture associated with observed biological effects (e.g., Davis et al. 2013; 2016; Collette et al. 2019). As an illustration of this second type of application, Davis et al. (2013) described a caged-fish study in which comparative metabolite profiles enabled definition of the contribution of pulp and paper mill influent to the biological effects of a WWTP effluent by conducting measurements in fish exposed before, during and after a scheduled shutdown of the mill. To build upon this approach, we subsequently developed a more direct way to associate changes in endogenous metabolite profiles with individual contaminants in complex mixtures using partial least squares (PLS) regression (Davis et al. 2016; Collette et al. 2019). For the current study, we employed this PLS-based method to identify contaminants in the measured mixtures that covaried with hepatic metabolomic changes in order to highlight (or possibly eliminate) chemicals of potential concern. Specifically, in the April and June 2016 Maumee River studies, metabolite profiles and measured contaminant concentrations at the downstream sites covaried moderately well (Q2 = 0.34) and indicated a potential relationship between hepatic metabolome responses and a variety of pesticides, pharmaceuticals, and wastewater indicators reflective of the more urban nature of the lower Maumee River (Table 1). This contrasted somewhat with the upstream sites, which displayed greater covariance (Q2 =0.59), indicating a stronger correlation between detected contaminant concentrations and biological effects. Notably, the top 25 chemicals covarying with metabolite profiles at the upstream sites were all pesticides and pesticide degradates, several of which also displayed elevated EAR values (e.g., metolachlor, dicamba, atrazine, acetochlor), providing a tentative link between the nontargeted in vivo and in vitro responses. Of particular note is the large number of pesticide degradates in this list (Table 1). This finding illustrates the importance of monitoring and hazard testing for a wide range of potential environmental transformation products and not limiting the focus solely to parent materials, be they pesticide, pharmaceutical, or industrial chemicals. It also underscores the challenge of basing either monitoring or hazard-testing programs entirely on listed (i.e., targeted) chemicals, because it is generally not feasible to anticipate all possible environmental transformation products under varied field conditions (Fenner et al. 2013).
Summary and Conclusions
The Maumee River and associated tributaries are a prototypical example of a system influenced by a mosaic of contaminant inputs from point and nonpoint sources along a gradient of land uses. Contaminants in the upper part of the Maumee River clearly reflect agricultural practices, while downstream, the suite of chemicals present includes those from agriculture in conjunction with contaminants more indicative of a general urban setting, influenced in some areas by WWTP inputs. To assess the potential effects of contaminants on aquatic biota in a system this complex requires a combination of analytical and biological monitoring techniques to provide data that can be assembled and interpreted in an integrated manner. The aim of the current paper was to provide a demonstration of this type of approach, as well as to supply actionable information of potential utility to risk assessors and managers working at the Maumee AOC.
We feel that there are three major “take home” messages from this set of studies. First, although there are a variety of contaminants present in surface waters from the Maumee AOC, crop-use pesticides as a broad class appear to be of most potential ecological concern. From an analytical perspective, one of the more notable observations from our study was the marked increase in concentrations of several herbicides at upstream sites between the April and June caged fish deployments in 2016. Further, the chemicals resulting in elevated EAR values based on data from in vitro assays were always pesticides. Finally, the metabolomics data from the caged fish, particularly at the upstream sites, showed that pesticides and pesticide degradates were strongly correlated with biological responses in vivo. These observations suggest that additional effects-based work in the Maumee AOC focused specifically on pesticides and their degradates may be warranted. The addition of nontargeted contaminant monitoring to expand the detection of pesticide degradates and other environmental transformation products should also be considered. And, given that many of the pesticides elevated in the system were herbicides, future studies may need to include assays based not just on animals but plants as well.
A second important observation from the current study involves the potential for contaminant effects on endocrine function in fish. A variety of chemicals associated with WWTPs and agriculture have been linked to disruption of endocrine pathways involved with development and reproduction in vertebrates, and there are well-documented examples of effects of endocrine-active chemicals, especially estrogens, in fish from the field (e.g., Purdom et al. 1994). Consequently, many environmental monitoring programs specifically target contaminants and/or biological responses associated with endocrine disruption (Hotchkiss et al. 2008). However, data from contaminant analyses (and associated EAR calculations), and in vitro or in vivo tests provided no consistent indication of endocrine activity/effects at the variety of sites in the Maumee AOC influenced by agricultural or urban activities. Therefore, monitoring resources in this system may be best allocated to non-endocrine contaminant-related impacts.
Finally, our studies do indicate that contaminants almost certainly are causing effects on some biological pathways in aquatic organisms in the Maumee AOC. Specifically, EAR data (and associated AOPs), HTS measurements of bioactivity in surface water, and determination of hepatic cyp1a1 in caged fish all indicate the presence of chemicals that interact with different CYPs, either in terms of inhibition or induction. Notably, studies conducted with tree swallows in 2016 at terrestrial locations adjacent to some of sites where caged fish deployments occurred also indicated effects on CYPs (induction of activity associated with CYP1A) in livers of the birds (Custer et al. 2020). Some CYPs have critical roles in processes such as the synthesis or degradation of steroids, so inhibition of these CYPs could affect normal physiological functions. While induction of other CYPs (such as cyp1a1) can be viewed as a short-term adaptive response to exposure to different types of xenobiotics, in a longer-term setting induction could have undesirable consequences, such as increased oxidative stress (van der Oost et al. 2020). Overall, although multiple lines of evidence indicate that contaminants in the Maumee have the potential to—or are—affecting biological pathways through CYP interactions, the consequences of this in terms of adverse effects on assessment endpoints directly related to survival, growth and reproduction are uncertain. Additional monitoring work in the AOC focused on endpoints relevant to possible physiological effects related to changes in CYP expression/activity might be warranted. This work could be guided by the insights afforded through the AOP-based analyses described above.
Supplementary Material
Acknowledgement:
We thank Steve Corsi and Erin Maloney for helpful comments on an earlier draft of the paper. We also thank Johan Gottgens and colleagues at the University of Toledo for providing laboratory space for this study and Terri Jicha for providing critical technical support. Portions of the work were supported through the Great Lakes Restoration Initiative administered through the USEPA Great Lakes National Program Office (GLNPO). We thank Edwin Smith, Elizabeth Murphy and Derek Ager from GLNPO for their contributions to this work.
Footnotes
Disclaimer: Mention of trade names or commercial products does not constitute endorsement or recommendation for use. The contents of this manuscript neither constitute, nor necessarily reflect US EPA policy.
Data accessibility: All data can be accessed at DOI: 10.23719/1519036
References
- Ankley GT, Jensen KM. 2014. A novel framework for interpretation of data from the fish short term reproduction assay (FSTRA) for the detection of endocrine-disrupting chemicals. Environ. Toxicol. Chem. 33:2529–2540. [DOI] [PubMed] [Google Scholar]
- Ankley GT, Villeneuve DL. 2015. Temporal changes in biological responses and uncertainty in assessing risk of endocrine-disrupting chemicals: Insights from intensive time-course studies with fish. Toxicol. Sci. 144:259–275. [DOI] [PubMed] [Google Scholar]
- Ankley GT, Jensen KM, Kahl MD, Makynen EA, Blake LS, Greene KJ, Johnson RD, Villeneuve DL. 2007. Ketoconazole in the fathead minnow (Pimephales promelas): Reproductive toxicity and biological compensation. Environ Toxicol Chem 26:1214–1223. [DOI] [PubMed] [Google Scholar]
- Ankley GT, Bencic DC, Breen MS, Collette TW, Conolly RB, Denslow ND, Edwards SW, Ekman DR, Garcia-Reyero N, Jensen KM. 2009. Endocrine disrupting chemicals in fish: developing exposure indicators and predictive models of effects based on mechanism of action. Aquat Toxicol 92:168–178. [DOI] [PubMed] [Google Scholar]
- Ankley GT, Bennett RS, Erickson RJ, Hoff DJ, Hornung MW, Johnson RD, Mount DR, Nichols JW, Russom CL, Schmieder PK, Serrano JA, Tietge JE, Villeneuve DL. 2010. Adverse outcome pathways: a conceptual framework to support ecotoxicology research and risk assessment. Environ Toxicol Chem 29:730–741. [DOI] [PubMed] [Google Scholar]
- Ankley GT, LaLone CA, Gray LE, Villeneuve DL, Hornung M. 2016. Evaluation of the scientific underpinnings for identifying estrogenic chemicals in non-mammalian taxa using mammalian test systems. Environ Toxicol Chem 35:2806–2816. [DOI] [PubMed] [Google Scholar]
- Arini A, Cavallin JE, Berninger JP, Marfil-Vega R, Mills M, Villeneuve DL, Basu N. 2016. In vivo and In vitro neurochemical-based assessments of wastewater effluents from the Maumee River area of concern. Environ Poll 211:9–19. [DOI] [PubMed] [Google Scholar]
- Baldwin AK, Corsi SR, DeCicco LA, Lenaker PL, Lutz MA, Sullivan DJ, Richards KD. 2016. Organic contaminants in Great Lakes tributaries: Prevalence and potential aquatic toxicity. Sci Total Environ 554-555:42–52. [DOI] [PubMed] [Google Scholar]
- Baldwin AK, Corsi SR, Oliver SK, Lenaker PL, Nott MA, Mills MA, Norris GA, Paatero P. 2020. Primary source of polycyclic aromatic hydrocarbons to streambed sediment in Grate Lakes tributaries using multiple lines of evidence. Environ Toxicol Chem 39:1392–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin WS, Roling JA, Peterson S, Chapman LM. 2005. Effects of nonylphenol on hepatic testosterone metabolism and the expression of acute phase proteins in the winter flounder (Pleuronectes americanus): Comparison to the effects of Saint John’s Wort. Comp. Biochem. Physiol. 140C:87–96. [DOI] [PubMed] [Google Scholar]
- Baker DB, Ewing DE, Johnson LT, Kramer JW, Merryfield BJ, Confesor RB, Richards RP, Roerdink AA. 2014. Lagrangian analysis of the transport and processing of agricultural runoff in the lower Maumee River and Maumee Bay J. Great Lakes Res 40:479–495. [Google Scholar]
- Barter RL, and Yu B. 2018. Superheat: An R package for creating beautiful and extendable heatmaps for visualizing complex data. Journal of Computational and Graphical Statistics, 1–30. 10.1080/10618600.2018.1473780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berninger JP, DeMarini DM, Warren SH, Simmons JE, Wilson VS, Conley JM, Armstrong MD, Iwanowicz LR, Kolpin DW, Kuivila KM, Reilly TJ, Romanok KM, Villeneuve DL, Bradley PM. 2019. Predictive analysis using chemical-gene interaction networks consistent with observed endocrine activity and mutagenicity of U.S. streams. Environ Sci Technol 53:8611–8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell BR, Ankley GT, Corsi SR, DeCicco LA, Houck KA, Judson RS, Li S, Martin MT, Murphy E, Schroeder AL, Smith ER, Swintek J, Villeneuve DL. 2017. An “EAR” on environmental surveillance and monitoring: A case study on the use of exposure-activity ratios (EARs) to prioritize sites, chemicals and bioactivities of concern in the Great Lakes. Environ Sci Technol 51:8713–8724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell BR, Ankley GT, Bradley PM, Houck KA, Schroeder AL, Swintek J, Villeneuve DL. 2019. Potential toxicity of complex mixtures in surface waters from a nationwide survey of United States streams: Identifying in vitro bioactivities and causative chemicals. Environ Sci Technol 53:973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley PM, Journey CA, Romanok KM, Barber LB, Buxton HT, Foreman WT, Furlong ET, Glassmeyer ST, Hladik ML, Iwanowicz LR, Jones DK, Kolpin DW, Kuivila KM, Loftin KA; Mills MA, Meyer MT, Orlando JL, Reilly TJ, Smalling KL Villeneuve DL. 2017. Expanded target-chemical analysis reveals extensive mixed-organic-contaminant exposure in U.S. streams. Environ Sci Technol 51:4792–4802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallin JE, Durhan EJ, Evans N, Jensen KM, Kahl MD, Kolpin DW, Kolodziej EP, Foreman WT, LaLone CA, Makynen EA, Seidl SM, Thomas LM, Villeneuve DL, Weberg MA, Wilson VS, Ankley GT. 2014. Integrated assessment of runoff from livestock farming operations: Analytical chemistry, in vitro bioassays, and in vivo fish exposures. Environ Toxicol Chem 33:1849–1857. [DOI] [PubMed] [Google Scholar]
- Cavallin JE, Jensen KM, Kahl MD, Villeneuve DL, Lee K, Schroeder A, Mayasich J, Eid EP, Nelson KR, Milsk RY, Blackwell BR, Berninger JP, LaLone CA, Blanksma C, Jicha T, Elonen C, Johnson R, Ankley GT. 2016. Pathway-based approaches for assessment of real-time exposure to an estrogenic wastewater treatment plant effluent on fathead minnow reproduction. Environ Toxicol Chem 35:702–716. [DOI] [PubMed] [Google Scholar]
- Cipoletti N, Jorgenson ZG, Banda JO, Hummel SL, Kohno S, Schoenfuss HL. 2019. Land use contributions to adverse biological effects in a complex agricultural and urban watershed: A case study of the Maumee River. Environ. Toxicol. Chem. 38:1035–1052. [DOI] [PubMed] [Google Scholar]
- Collette TW, Teng Q, Jensen KM, Kahl MD, Makynen EA, Durhan EJ, Villeneuve DL, Martinovic-Weigelt D, Ankley GT, Ekman DR. 2010. Impacts of an anti-androgen and an androgen/anti-androgen mixture on the metabolite profile of male fathead minnow urine. Environ Sci Technol 44:6881–6886. [DOI] [PubMed] [Google Scholar]
- Collette TW, Ekman DR, Zhen HJ, Nguyen H, Bradley P, Teng Q. 2019. Cell-based metabolomics for untargeted screening and prioritization of vertebrate-active stressors in streams across the United States. Environ Sci Technol 53:9232–9240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsi SR, De Cicco LA, Villeneuve DL, Blackwell BR, Fay KA, Ankley GT, Baldwin AK. 2019. Prioritizing chemicals of ecological concern in Great Lakes tributaries using high-throughput screening data and adverse outcome pathways. Sci Total Environ 686:995–1009. [DOI] [PubMed] [Google Scholar]
- Custer CM, Custer TW, Dummer PM, Schultz S, Tseng CY, Karouna-Reiner N, Matson CW. 2020. Legacy and contaminants of emerging concern (CECs) in tree swallows along an agricultural to industrial gradient: Maumee River, Ohio. Environ Toxicol Chem. In Press. [DOI] [PubMed] [Google Scholar]
- Davis JM, Collette TW, Villeneuve DL, Cavallin JE, Teng Q, Jensen KM, Kahl MD, Mayasich JM, Ankley GT, Ekman DR. 2013. Field-based approach for assessing the impact of treated pulp and paper mill effluent on endogenous metabolites of fathead minnows (Pimephales promelas). Environ Sci Technol 47:10628–10636. [DOI] [PubMed] [Google Scholar]
- Davis JM, Ekman DR, Teng Q, Ankley GT, Berninger JP, Cavallin JE, Jensen KM, Kahl MD, Schroeder AL, Villeneuve DL, Jorgenson ZG, Lee KE, Collette TW. 2016. Linking field-based metabolomics and chemical analyses to prioritize contaminants of emerging concern in the Great Lakes basin. Environ Toxicol Chem 35:2493–2592. [DOI] [PubMed] [Google Scholar]
- Davis JM, Ekman DR, Skelton DM, LaLone CA, Ankley GT, Cavallin JE, Villeneuve DL, Collette TW. 2017. Metabolomics for informing adverse outcome pathways: Androgen receptor activation and the pharmaceutical spironolactone. Aquat Toxicol 184:103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cicco LA, Corsi SR, Villeneuve DL, Blackwell BR, Ankley GT, 2018. toxEval: Evaluation of measured concentration data using the ToxCast high-throughput screening database or a user-defined set of concentration benchmarks. R Package version 1.0.0 [WWW Document]. URL https://code.usgs.gov/water/toxEval, doi.org/10.5066/P906UQ5I.
- Ekman DR, Hartig PC, Cardon M, Skelton D, Teng Q, Durhan EJ, Jensen KM, Kahl MD, Villeneuve DL, Gray LE, Collette TW, Ankley GT. 2012. Metabolite profiling and a transcriptional activation assay provide direct evidence of androgen receptor antagonism by bisphenol A in fish. Environ. Sci. Technol. 46:9673–9680. [DOI] [PubMed] [Google Scholar]
- Elliott SM, Brigham ME, Lee KE, Banda JA, Choy SJ, Gefell DJ, Minarik TA, Moore JN, Jorgenson ZG. 2017. Contaminants of emerging concern in tributaries to the Laurentian Great Lakes: i. Patterns of occurrence. PLoS One, 12: e0182868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekman DR, Ankley GT, Blazer VS, Collette TW, Garcia-Reyero N, Iwanowicz LR, Jorgenson ZG, Lee KE, Mazik PM, Miller DH, Perkins EJ, Smith ET, Tietge JE, Villeneuve DL. 2013. Biological effects–based tools for monitoring impacted surface waters in the Great Lakes: A multiagency program in support of the Great Lakes Restoration Initiative. Environ Prac 15:409–426. [Google Scholar]
- Eriksson L, Johansson E, Kettaneh-Wold N, Trygg J, Wikstrom C. 2006. Multi- and Megavariate Data Analysis: Basic Principles and Applications. Umetrics: Umea, Sweden. [Google Scholar]
- Eriksson L, Trygg J, Wold S. 2008. CV-ANOVA for significance testing of PLS and OPLS (R) models. J Chemom 22:594–600. [Google Scholar]
- Escher BI, Allinson M, Altenburger R, Bain PA, Balaguer P, Busch W, Crago J, Denslow ND, Dopp E, Hilscherova K. 2013. Benchmarking organic micropollutants in wastewater, recycled water and drinking water with in vitro bioassays. Environ Sci Technol 48:1940–1956. [DOI] [PubMed] [Google Scholar]
- Federal Register. 1984. Development of Water-Quality Based Permit Limitations for Toxic Pollutants. US Federal Register; 49:9016. Washington, DC. [Google Scholar]
- Fenner K, Canonica S, Wackett LP, Elsner M. 2013. Evaluating pesticide degradation in the environment: Blind spots and emerging opportunities. Science 341:752–758. [DOI] [PubMed] [Google Scholar]
- Foreman WT, Gray JL, ReVello RC, Lindley CE, Losche SA, Barber LB. 2012. Determination of steroid hormones and related compounds in filtered and unfiltered water by solid-phase extraction, derivatization, and gas chromatography with tandem mass spectrometry. US Geological Survey Techniques and Methods, Book 5, Chap. B9, 188 pp. [Google Scholar]
- Furlong ET, Kanagy CJ, Kanagy LK, Coffey LJ, Burkhardt MR. 2014. Determination of human-use pharmaceuticals in filtered water by direct aqueous injection-high-performance liquid chromatography/tandem mass spectrometry. US Geological Survey Techniques and Methods, Book 5, Chap. B10. 300 pp. [Google Scholar]
- Garcia-Reyero N, Perkins EJ. 2011. Systems biology: Leading the revolution in ecotoxicology. Environ Toxicol Chem 30:265–273. [DOI] [PubMed] [Google Scholar]
- Hladik ML, Corsi SR, Kolpin DW, Baldwin AK, Blackwell BR, Cavallin JE. 2018. Year-round presence of neonicotinoid pesticides in tributaries to the Great Lakes, USA. Environ Poll 235:1022–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander J, Schymanski EL, Singer HP, Ferguson PL. 2017. Nontarget screening with high resolution mass spectrometry in the environment: Ready to go? Environ Sci Technol 51:11505–11512. [DOI] [PubMed] [Google Scholar]
- Hotchkiss AK, Rider CV, Blystone CR, Wilson VS, Hartig PC, Ankley GT, Foster PM, Gray CL, Gray LE. 2008. Fifteen years after “Wingspread”-environmental endocrine disrupters and human and wildlife health: Where we are today and where we need to go. Toxicol. Sci. 105:235–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingersoll CG, Brunson EL, Dwyer FJ, Ankley GT, Benoit DA, Norberg-King TJ, Burton GA, Hoke RA, Landrum PF, Winger PV. 1995. Toxicity and bioaccumulation of sediment-associated contaminants using freshwater invertebrates: A review of methods and applications. Environ. Toxicol. Chem. 14:1885–1894. [Google Scholar]
- Jensen KM, Korte JJ, Kahl MD, Pasha MS, Ankley GT. 2001. Aspects of basic reproductive biology and endocrinology in the fathead minnow (Pimephales promelas). Comp Biochem Physiol 128C:127–141. [DOI] [PubMed] [Google Scholar]
- Kahl MD, Villeneuve DL, Stevens K, Schroeder A, Makynen EA, LaLone CA, Jensen KM, Hughes M, Holmen BA, Eid E, Durhan EJ, Cavallin JE, Berninger J, Ankley GT. 2014. An inexpensive, temporally integrated system for monitoring occurrence and biological effects of aquatic contaminants in the field. Environ Toxicol Chem 33:1584–1595. [DOI] [PubMed] [Google Scholar]
- Kashiwada S, Hinton DE, Kullman SW. 2005. Functional characterization of medaka CYP3A38 and CYP3A40: Kinetics and catalysis by expression in a recombinant baculovirus system. Comp. Biochem. Physiol. 141C:338–448. [DOI] [PubMed] [Google Scholar]
- Ketchen DJ Jr, and Shook CL. 1996. The application of cluster analysis in strategic management research: An analysis and critique. Strat Manag J 17: 441–459. [Google Scholar]
- Korte JJ, Kahl MD, Jensen KM, Pasha MS, Parks LG, LeBlanc GA, Ankley GT. 2000. Fathead minnow vitellogenin: Complementary DNA sequence and messenger RNA and protein expression after 17β-estradiol treatment. Environ Toxicol Chem 19:972–981. [Google Scholar]
- Kuhl C, Tautenhahn R, Bottcher C, Larson TR, Neumann S. 2012. CAMERA: An integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Anal Chem 84:283–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaLone CA, Villeneuve DL, Burgoon LD, Russom CL, Helgen HW, Berninger JP, Tietge JE, Severson MN, Cavallin JE, Ankley GT. 2013. Molecular target sequence similarity as a basis for species extrapolation to assess the risk of chemicals with known modes of action. Aquat Toxicol 144/145:141–154. [DOI] [PubMed] [Google Scholar]
- LaLone CA, Villeneuve DL, Doering JA, Blackwell BR, Transue TR, Simmons CW, Swintek J, Degitz SJ, Williams AJ, Ankley GT. 2018. Evidence for cross-species extrapolation of mammalian-based high-throughput screening assay results. Environ. Sci. Technol. 52:13960–13971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KE, Langer SK, Menheer MA, Hansen DS, Foreman WT, Furlong ET, Jorgenson ZG, Choy SJ, Moore JN, Banda J, Gefell DJ. 2015. Chemicals of emerging concern in water and bottom sediment in the Great Lakes Basin, 2012: collection methods, analytical methods, quality assurance, and study data. Report 2327–638X. US Geological Survey, Reston, VA. [Google Scholar]
- Li S, Villeneuve DL, Berninger JP, Blackwell BR, Cavallin JE, Hughes MN, Jensen KM, Jorgenson Z, Kahl MD, Schroeder AL, Stevens KE, Thomas LM, Weberg MA, Ankley GT. 2017. An integrated approach for identifying priority contaminants in the Great Lakes basin - Investigations in the Lower Green Bay/Fox River and Milwaukee Estuary areas of concern. Sci Total Environ 578:825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahler BJ, Van Metre PC, Burley TE, Loftin KA, Meyer MT, Nowell LH. 2017. Similarities and differences in occurrence and temporal fluctuations in glyphosate and atrazine in small Midwestern streams (USA) during the 2013 growing season. Sci Total Environ 579:149–158. [DOI] [PubMed] [Google Scholar]
- Martinovic-Weigelt D, Mehinto AC, Ankley GT, Denslow ND, Barber LB, Lee KE, King RJ, Schoenfuss HL, Schroeder AL, Villeneuve DL. 2014. Transcriptomic effects-based monitoring for endocrine active chemicals: assessing relative contribution of treated wastewater to downstream pollution. Environ Sci Technol 48:2385–2394. [DOI] [PubMed] [Google Scholar]
- McMaster ME, Munkittrick KR, Jardine JJ, Robinson RD, Van Der Kraak GJ. 1995. Protocol for measuring in vitro steroid production by fish gonadal tissue. Canadian Technical Report of Fisheries and Aquatic Sciences 1961. Department of Fisheries and Oceans, Burlington, Ontario, Canada. [Google Scholar]
- Moore DR, Greer CD, Manning G, Wooding K, Beckett KJ, Brain RA, Marshall G. 2017. A weight-of-evidence approach for deriving a level of concern for atrazine that is protective of aquatic plant communities. Integr Environ Assess Manag 13:686–701. [DOI] [PubMed] [Google Scholar]
- Mosley JD, Ekman DR, Cavallin JE, Villeneuve DL, Ankley GT, Collette TW. 2018. Highresolution mass spectrometry of skin mucous for monitoring physiological impacts and contaminant biotransformation products in fathead minnows exposed to wastewater effluent. Environ Toxicol Chem 37:788–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtagh F and Legendre P. 2014. Ward’s hierarchical agglomerative clustering method: which algorithms implement Ward’s criterion? Journal of Classification, 31: 274–295. [Google Scholar]
- Nowell LH, Moran PW, Schmidt TS, Norman JE, Nakagaki N, Shoda ME, Mahler BJ, Van Metre PC, Stone WW, Sandstrom MW, Hladik ML. 2018. Complex mixtures of dissolved pesticides show potential aquatic toxicity in a synoptic study of Midwestern U.S. streams. Sci Total Environ 613-614:1469–1488. [DOI] [PubMed] [Google Scholar]
- Ohio Environmental Protection Agency (OEPA). 2009. Total Maximum Daily Loads for the Swan Creek Watershed. State of Ohio Environmental Protection Agency Division of Surface Water. Columbus, OH. Available at https://www.epa.state.oh.us/portals/35/tmdl/SwanCreekTMDL_final_oct09_wo_app.pdf. Last accessed 27 October 2020. [Google Scholar]
- Ohio Environmental Protection Agency (OEPA). 2013. Ohio EPA National Pollutant Discharge Elimination System (NPDES) Fact Sheet. Ohio Environmental Protection Agency. Columbus, OH. Available at http://www.epa.ohio.gov/dsw/permits/individuals.aspx. Last accessed 27 October 2020. [Google Scholar]
- Parks LG, Cheek AO, Denslow ND, Heppell SA, McLachlan JA, LeBlanc GA, Sullivan CV. 1999. Fathead minnow (Pimephales promelas) vitellogenin: purification, characterization and quantitative immunoassay for the detection of estrogenic compounds. Comp Biochem Physiol 123C:113–125. [DOI] [PubMed] [Google Scholar]
- Perkins EJ, Habib T, Escalon BL, Cavallin JE, Thomas L, Weberg M, Hughes MN, Jensen KM, Kahl MD, Villeneuve DL, Ankley GT, Garcia-Reyero N. 2017. Prioritization of contaminants of emerging concern in wastewater treatment plant discharges using chemical-gene interactions in caged fish. Environ Sci Technol 51:8701–8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell KW, Cope WG, LePrevost CE, Augspurger T, McCarthy AM, Shea D. 2017. A retrospective analysis of agricultural herbicides in surface water reveals risk plausibility for declines in submerged aquatic vegetation. Toxics 5(3) pii: E21. doi: 10.3390/toxics5030021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdom CE, Harriman PA, Bye VVJ, Eno NC, Tyler CR, Sumpter JP. 1994. Estrogenic effects of effluent from sewage treatment works. Chem. Ecol. 8:275–285. [Google Scholar]
- R Core Team. 2019. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/. [Google Scholar]
- Richter CA., Papoulias DM, Whyte JJ, Tillitt DE. 2016. Evaluation of possible mechanisms of atrazine-induced reproductive impairment if fathead minnow (Pimephales promelas) and Japanese medaka (Oryzias latipes). Environ Toxicol Chem 35:2230–2238. [DOI] [PubMed] [Google Scholar]
- Romanov S, Medvedev A, Gambarian M, Poltoratskaya N, Moeser M, Medvedeva L, Gambarian M, Diatchenko L, Makarov S. 2008. Homogeneous reporter system enables quantitative functional assessment of multiple transcription factors. Nat Meth 5:253–260. [DOI] [PubMed] [Google Scholar]
- Sanderson JT, Letcher RJ, Heneweer M, Giesy JP, van den Berg M. 2001. Effects of chloro-s-triazine herbicides and metabolites on aromatase activity in various human cell lines and on vitellogenin production in male carp hepatocytes. Environ. Health Perspect. 109:1027–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson JT, Boerma J, Lansbergen GWA, van den Berg M. 2002. Induction and inhibition of aromatase (CYP19) activity by various classes of pesticides in H295R human adrenocortical carcinoma cells. Toxicol. Appl. Pharmacol. 182:44–54. [DOI] [PubMed] [Google Scholar]
- Schroeder AL, Ankley GT, Houck K, Villeneuve DL. 2016. Environmental surveillance and monitoring – The next frontiers for high-throughput toxicology. Environ Toxicol Chem 35:513–525. [DOI] [PubMed] [Google Scholar]
- Schroeder AL, Martinovic-Weigelt D, Ankley GT, Lee KE, Garcia-Reyero N, Perkins EJ, Schoenfuss HL, Villeneuve DL. 2017. Prior knowledge-based approach for associating contaminants with biological effects: A case study in the St. Croix River basin, MN, WI, USA. Environ Poll 221:427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoda ME, Nowell LH, Stone WW, Sandstrom MW, and Bexfield LM. 2018. Data Analysis Considerations for Pesticides Determined by National Water Quality Laboratory Schedule 2437. USGS Scientific Investigations Report 2018–5007. 459 pp 10.3133/sir20185007. [DOI] [Google Scholar]
- Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. 2006. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem 78:779–787. [DOI] [PubMed] [Google Scholar]
- Society for the Advancement of Adverse Outcome Pathways (SAAOP). 2016. Welcome to the Collaborative Adverse Outcome Pathway Wiki (AOP-Wiki). Available from: https://aopwiki.org/. Last accessed 29 October 2020. [Google Scholar]
- Suzuki R, and Shimodaira H. 2006. Pvclust: An R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22: 1540–1542. [DOI] [PubMed] [Google Scholar]
- Tautenhahn R, Bottcher C, Neumann S. 2008. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics 9: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinfo NS, Hotchkiss MG, Buckalew AR, Zorrilla LM, Cooper RL, Laws SC. 2011. Understanding the effects of atrazine on steroidogenesis in rat granulosa and H295R adrenal cortical carcinoma cells. Repro. Toxicol. 31:184–193. [DOI] [PubMed] [Google Scholar]
- US Environmental Protection Agency (USEPA). 1989. Short-Term Methods for Estimating the Chronic Toxicity of Effluents and Receiving Waters to Freshwater Organisms. Second Edition. EPA/600/4–89/01. Washington, DC. [Google Scholar]
- US Environmental Protection Agency (USEPA). 1994. Methods for Measuring the toxicity and bioaccumulation of sediment-associated contaminants with freshwater invertebrates. US Environmental Protection Agency, Duluth, MN. [Google Scholar]
- US Environmental Protection Agency (USEPA). 2014. Great Lakes Restoration Initiative: Action Plan II. https://www.glri.us/sites/default/files/glri-action-plan-2-201409-30pp.pdf.
- US Environmental Protection Agency (USEPA). 2015. Maumee River Great Lakes Area of Concern Information. US Environmental Protection Agency. Chicago, IL. Available from: https://www.epa.gov/great-lakes-aocs/maumee-aoc. Last accessed 27 October 2020. [Google Scholar]
- US Environmental Protection Agency (USEPA). 2016. Refined Ecological Risk Assessment for Atrazine. US Environmental Protection Agency, Office of Pesticide Programs, Washington, DC. [Google Scholar]
- US Environmental Protection Agency (USEPA). 2019. Atrazine Proposed Interim Registration Review Decision. Docket Number EPA-HQ-OPP-2013–0266. US Environmental Protection Agency, Washington, DC. [Google Scholar]
- US Environmental Protection Agency (USEPA). 2020a. ECOTOX Knowledgebase. US Environmental Protection Agency, Washington, DC. Available from: https://cfpub.epa.gov/ecotox/. Last accessed 27 October 2020. [Google Scholar]
- US Environmental Protection Agency (USEPA). 2020b. Comp Tox Chemicals Dashboard. US Environmental Protection Agency, Washington, DC. Available from: https://comptox.epa.gov/dashboard/. Last accessed 27 October 2020. [Google Scholar]
- van der Oost R, McKenzie DJ, Verweij F, Satumalay, van der Molen N, Winter MJ, Chipman JK. 2020. Identifying adverse outcome pathways (AOP) for Amsterdam city fish by integrated field monitoring. Environ Toxicol Pharmacol 74: 10.1016/j.etap.2019.103301. [DOI] [PubMed] [Google Scholar]
- Venier M, Dove A, Romanak K, Backus S, Hites R. 2014. Flame Retardants and Legacy Chemicals in Great Lakes’ Water. Environ Sci Technol 48: 9563–9572. [DOI] [PubMed] [Google Scholar]
- Villeneuve DL, Blake LS, Brodin JD, Greene KJ, Knoebl I, Miracle AL, Martinovic D, Ankley GT. 2007. Transcription of key genes regulating gonadal steroidogenesis in control and ketoconazole- or vinclozolin-exposed fathead minnows. Toxicol Sci 98:395–407. [DOI] [PubMed] [Google Scholar]
- Wehmas LC, Cavallin JE, Durhan EJ, Kahl MD, Martinovic D, Mayasich J, Tuominen T, Villeneuve DL, Ankley GT. 2011. Screening complex effluents for estrogenic activity with the T47D-KBluc cell bioassay: assay optimization and comparison with in vivo responses in fish. Environ Toxicol Chem 30:439–445. [DOI] [PubMed] [Google Scholar]
- Whyte JJ, Jung RE, Schmitt CJ, Tillitt DE. 2000. Ethoxyresorufin-O-deethylase (EROD) Activity in Fish as a Biomarker of Chemical Exposure. Crit Rev Toxicol 30:347–570. [DOI] [PubMed] [Google Scholar]
- Wilson VS, Bobseine K, Lambright CR, Gray LE Jr., 2002. A novel cell line, MDA-kb2, that stably expresses an androgen- and glucocorticoid-responsive reporter for the detection of hormone receptor agonists and antagonists. Toxicol Sci 66:69–81. [DOI] [PubMed] [Google Scholar]
- Wilson VS, Bobseine K, Gray LE Jr., 2004. Development and characterization of a cell line that stably expresses an estrogen-responsive luciferase reporter for the detection of estrogen receptor agonist and antagonists. Toxicol Sci 81:69–77. [DOI] [PubMed] [Google Scholar]
- You L. 2004. Steroid hormone biotransformation and xenobiotic induction of hepatic steroid metabolizing enzymes. Chemico-Biol Interact 147:233–246. [DOI] [PubMed] [Google Scholar]
- Zaugg SD, Smith SG, Schroeder MP. 2006. Determination of wastewater compounds in whole water by continuous liquid-liquid extraction and capillary-column gas chromatography/mass spectrometry. US Geological Survey Techniques and Methods, Book 5, Chap. B4, 30 pp. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.