Abstract
Silver nanoparticles (AgNPs) are potent antimicrobial agents, but their utility is limited due to their relatively high cytotoxicity. In this work, we used trehalose as the ligand to reduce the cytotoxicity of AgNPs without affecting their antimicrobial activities. Trehalose is a disaccharide that is unique to mycobacteria. We showed that trehalose-functionalized AgNPs, AgNP-Tre, drastically increased the viability of A549 cells, especially at high concentrations, for example, from 4% for AgNPs to 67% for AgNP-Tre at 64 μg/mL. The trehalose ligand slowed down the release of silver, and the amount of silver released from AgNP-Tre was less than half of that from AgNPs in the culture medium. Intriguingly, while the maltose (Mal) or tri(ethylene glycol) (TEG) ligand reduced the antibacterial activity of AgNPs against M. smegmatis (minimal inhibitory concentration (MIC) of AgNP-Mal and AgNP-TEG: 4 μg/mL for 7 nm AgNPs), the activity of AgNP-Tre was similar to that of AgNPs (MIC of AgNP-Tre: 1 μg/mL for 7 nm AgNPs). Uptake experiments revealed that the intracellular concentration of AgNP-Tre was 87 and 114% higher than those of AuNP-Mal and AgNP-TEG, respectively. The increased uptake was attributed to the enhanced interactions of AgNP-Tre with mycobacteria promoted by the trehalose ligand.
Keywords: silver nanoparticles, trehalose, glyconanoparticles, mycobacteria, uptake, cytotoxicity
Graphical Abstract
INTRODUCTION
The medicinal use of silver as an antimicrobial agent dates back to ancient times, including uses in water and food preservation and for skin and eye infections.1–3 The antibacterial action of silver has been attributed to silver ions; for example, silver metal is antibacterial by releasing Ag+ ions at the metal surface.4 Silver nanoparticles (AgNPs), owing to their high specific surface areas, release Ag+ ions at a faster rate and provide better contact during interactions with bacteria.5,6 As such, enhanced antimicrobial activities have been observed for smaller AgNPs.7–11 Several mechanisms have been suggested to account for the antimicrobial activity of AgNPs, including inhibition of DNA replication through the interaction of Ag+ with the sulfur and phosphorus of DNA, inhibition of protein synthesis by denaturing ribosomes, disruption to ATP production, generation of reactive oxygen species, and damage to the cell wall.12,13
A major drawback of AgNPs is that the released silver ions also cause cytotoxicity.14,15 One way to counter this is to lower the cytotoxicity while at the same time maintain the antimicrobial activity. Various methods have been developed to reduce the cytotoxicity of AgNPs. One approach is through shape control. For examples, Liu and co-workers demonstrated that silver nanocubes had lower toxicity to plant cells compared to spherical nanoparticles while maintaining high toxicity to bacteria.16 Another method to reduce cytotoxicity of AgNPs is through surface chemistry. It has been reported that negatively charged AgNPs had lower cytotoxicity while preserving their antibacterial activity.17 Cysteine ligand has also shown to lower the cytotoxicity of AgNPs toward human cells.18 Carbohydrates as the surface ligand bring about several advantages, such as exceptional water solubility and biocompatibility.19–22 Goswami and co-workers reported that polysaccharide-capped AgNPs had reduced cytotoxicity but still had the ability to inhibit biofilm formation by multidrug-resistant bacteria.23 β-Cyclodextrin-functionalized AgNPs were found to have lower cytotoxicity and enhanced biofilm inhibition.24
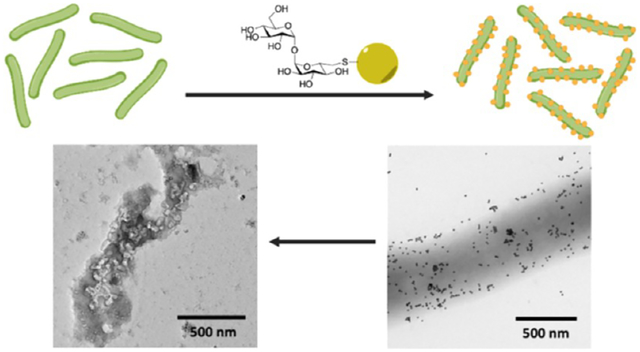
In this study, we report the use of a bacterium-specific carbohydrate, trehalose, to reduce the cytotoxicity while maintaining the antibacterial activity of AgNPs against mycobacteria. Trehalose is a disaccharide of d-glucose linked by an α-1,1-glycosidic bond. It is essential for the survival and pathogenicity of Mycobacterium tuberculosis, which causes tuberculosis (TB), but is absent in mammalian biology.25 In the cytosol, trehalose is used by both M. smegmatis and M. tuberculosis as an osmoprotectant.26,27 Trehalose is also produced in the periplasm during the cell wall synthesis when the mycolyl transferase, the antigen 85 complex (Ag85), transfers the mycolyl group in trehalose monomycolate (TMM) to arabinogalactan or to another TMM to give trehalose dimycolate (TDM).27,28 We have shown that trehalose facilitated the interactions of silica nanoparticles (42 nm) with M. smegmatis as well as the uptake of trehalose-functionalized iron oxide nanoparticles (6 nm) by M. Smegmatis.29 Based on this observation, we conjugated trehalose to drug delivery vehicles like mesoporous silica nanoparticles and showed that trehalose increased the killing efficiency of the TB drug isoniazid.30,31 Most intriguingly, no interactions were observed between trehalose-functionalized nanoparticles and the mammalian cells.29 In one experiment, we treated A549 cells with mycobacteria and then with trehalose-functionalized nanoparticles. The trehalose-functionalized nanoparticles were only seen on mycobacteria and not on A549 cells. The Prussian blue staining on A549 cells treated with trehalose-functionalized iron oxide nanoparticles showed minimal uptake of the nanoparticles by A549 cells.29 We therefore hypothesize that trehalose as the surface ligand would reduce the cytotoxicity, while it will not have a negative effect on the antimicrobial activity of AgNPs against mycobacteria. In this work, we functionalized AgNPs with trehalose and showed that trehalose as the ligand indeed decreased the toxicity of AgNPs toward A549 cells. While other ligands like maltose would lower the antimicrobial activity of AgNPs, trehalose-functionalized AgNPs, AgNP-Tre, were as active as AgNPs against mycobacteria. The antimicrobial activity of AgNP-Tre was attributed to the enhanced interactions of AgNP-Tre with mycobacteria and the increased uptake of AgNP-Tre by the bacteria (Scheme 1).
Scheme 1.
Trehalose-Functionalized AgNPs are Active against Mycobacteria through Enhanced Interactions and Uptake
RESULTS AND DISCUSSION
Synthesis and Functionalization of Gold Nanoparticles (AuNPs) and AgNPs.
Since AgNPs are toxic to bacteria, AuNPs of similar size and ligand composition were used to study the interactions with bacteria. The ~5 nm AuNP or AgNP were prepared by treating HAuCl4 or AgNO3 with sodium citrate in the presence of tannic acid, which acts as both a reducing agent and a capping agent to control the shape and size distribution of the nanoparticles.32,33 The nanoparticles dispersed well and were relatively uniform in size, at 4.5 ± 0.4 and 7.2 ± 1.9 nm for AuNPs and AgNPs, respectively (Figure 1a,b). The ~15 nm nanoparticles were synthesized by adding HAuCl4 or AgNO3 to a boiling solution of sodium citrate without or with tannic acid in the case of AuNPs34 and AgNPs35 to give 14.5 ± 2.0 nm AuNPs and 16.2 ± 2.0 nm AgNPs, respectively (Figure 1c,d).
Figure 1.
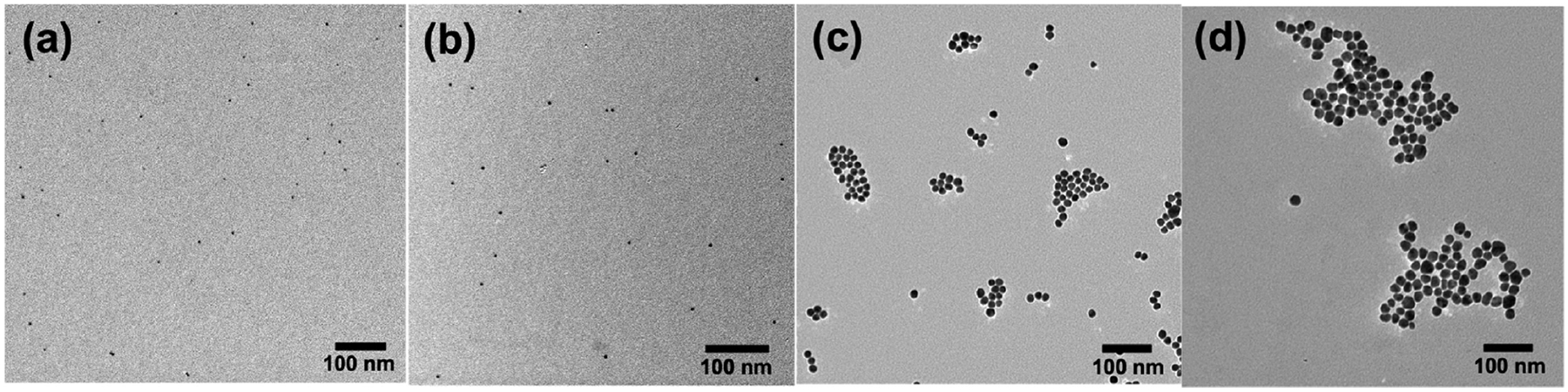
TEM images of (a) 4.5 ± 0.4 nm AuNPs, (b) 7.2 ± 1.9 nm AgNPs, (c) 14.5 ± 2.0 nm AuNPs, and (d) 16.2 ± 2.0 nm AgNPs. Particle diameters were measured from about 100 nanoparticles.
The larger nanoparticles with sizes of ≥15 nm were synthesized following the seeded-growth method by sequential addition of sodium citrate and HAuCl4 or AgNO3 to ~5 nm AuNP or ~7 nm AgNP seeds until the desired particle size was obtained.35,36 All samples showed the characteristic localized surface plasmon resonance (LSPR) absorptions associated with metal nanoparticles, and the absorption maxima red-shifted with increasing particle size after each successive growth step (Figure S1). The particles were relatively uniform as shown by both TEM (Figures S2 and S3) and dynamic light scattering (DLS) (Table S1).
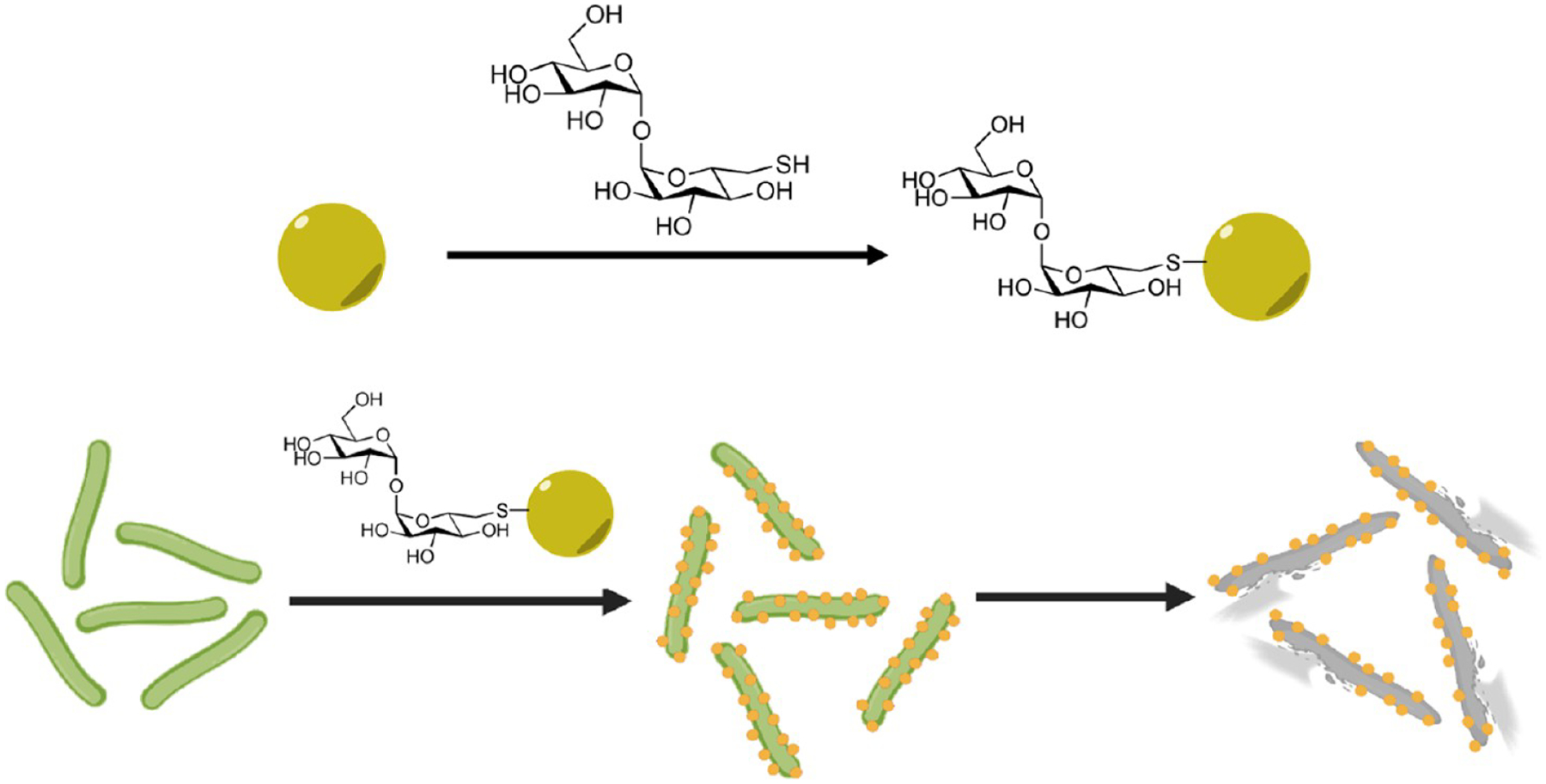
AuNPs and AgNPs were then functionalized with a thiolderivatized trehalose 5, synthesized following a reported protocol (Scheme 2A).37 Maltose (Mal), a disaccharide that consists of two glucose units like trehalose but has an α-1,4 instead of α-1,1 glycosidic linkage, and tri(ethylene glycol) (TEG) were used for comparison. Functionalization was carried out using a thiolated Mal (10, Scheme 2B) or disulfide-derivatized TEG (13, Scheme 2C) following the same protocol as thiolated trehalose 5. After functionalization with the ligand, the LSPR peaks of both AuNPs and AgNPs red-shifted, from 520 nm to 525 and 524 nm for 15 nm AuNP-Tre (Figure S4a) and 15 nm AuNP-Mal (Figure S4c), respectively. Similarly, the LSPR peak of 16 nm AgNPs at 389 nm red-shifted to 396 nm for 16 nm AgNP-Tre (Figure S4b) and 393 nm for 16 nm AgNP-Mal (Figure S4d), respectively. These results are consistent with findings from previous reports that the conjugation of carbohydrates on metal nanoparticles caused a red shift in the LSPR peak.38,39
Scheme 2.
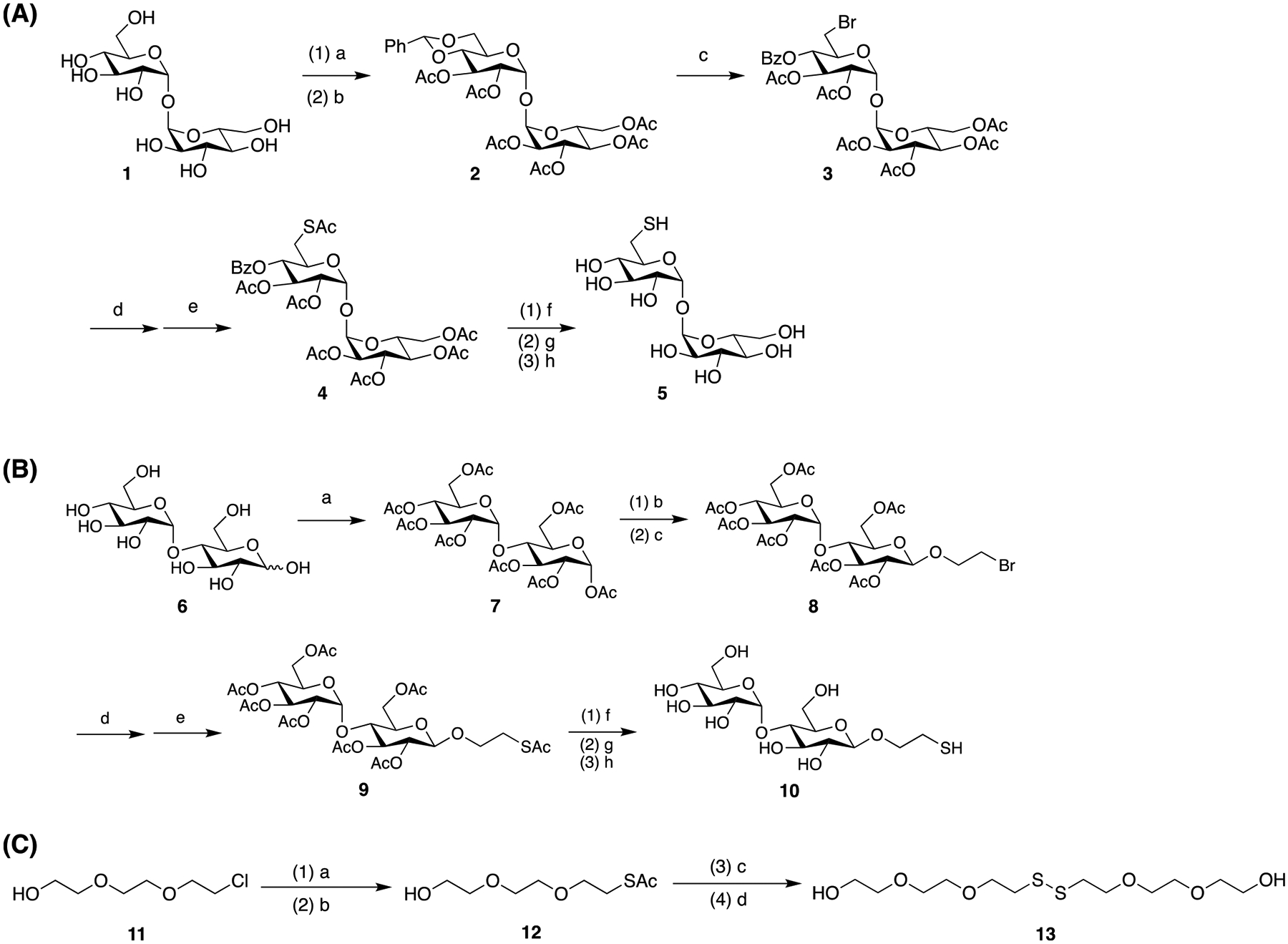
(A) Synthesis of Thiol-Derivatized Trehalose 537,37a; (B) Synthesis of Thiol-Derivatized Maltose 10b; and (C) Synthesis of TEG-Disulfidec
a(a) PhCH(OCH3)2, PTS, DMF, 23 °C, overnight; (b) triethylamine, acetic anhydride, DMF, 23 °C, overnight (38% overall yield); (c) NBS, CaCO3, CCl4, 77 °C, 3 h (91%); (d) KI, DMF, 90 °C, 5 h; (e) KSAc, DMF, N2, 8 h (66% overall yield); (f) NaOCH3, methanol, 23 °C, 6 h; (g) Amberlight IRC-120 H+ resin, pH 7; and (h) DTT, MeOH, 23 °C, 8 h (22%). b(a) Acetic anhydride, pyridine, 23 °C, 36 h (80%); (b) BF3·Et2O, dichloromethane, 2-bromoethanol, −40 °C, 1 h; (c) 23 °C, 22 h; (d) KI, DMF, 70 °C; (e) KSAc, DMF, 23 °C, 8 h; (f) NaOCH3, methanol, 23 °C, 6 h; (g) pH 7; and (h) DTT, 23 °C, 8 h. c(a) NaI, DMF, 90 °C, 2 h; (b) KSAc, DMF, 80 °C, 6 h; (c) K2CO3, methanol, 3 h; and (d) titration with saturated I2/ethanol.
To further confirm the successful conjugation of trehalose and maltose, the nanoparticles were treated with concanavalin A (Con A), a lectin consisting of four subunits that bind to carbohydrates having α-d-mannopyranosyl or α-d-glucopyranosyl structures including trehalose and maltose.40–42 Since both carbohydrate-functionalized nanoparticles and Con A are multivalent, interactions of trehalose or maltose nanoparticles with Con A would result in the formation of cross-linked Con A–nanoparticle complexes, which would precipitate out from the solution.38,43 Indeed, precipitates were observed when AuNP-Mal, AuNP-Tre, AgNP-Mal, or AgNP-Tre were treated with Con A. Bovine serum albumin (BSA), a protein that does not have Tre or Mal receptors, did not produce any precipitates when treated with these nanoparticles. No precipitation was observed when nanoparticles without Tre or Mal (AuNP, AuNP-TEG, AgNP, AgNP-TEG) were treated with Con A or BSA. These results support the specific interaction of Tre- and Mal-functionalized nanoparticles with Con A, further supporting the successful conjugation of Tre and Mal on the nanoparticles. We attempted to measure the ligand density by thermogravimetric analysis (TGA) but were not able to obtain meaningful results due to small sample quantities. On the other hand, results on cytotoxicity (Figure 2) and silver release kinetics (Figure 6) indicated that the density of the ligand (Tre, Mal, TEG) was similar and does not vary to a large extent.
Figure 2.
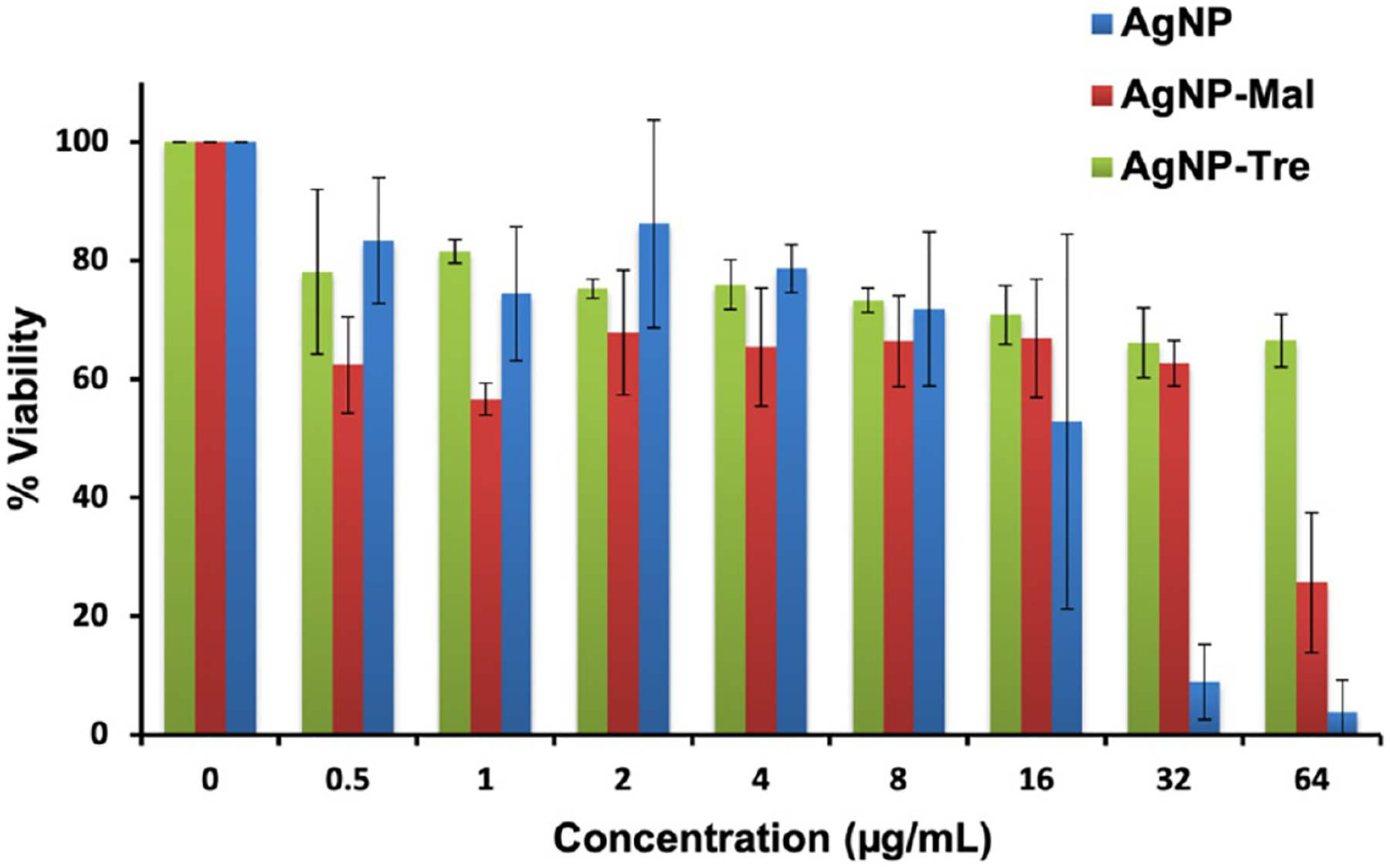
Percent viability of A549 cells treated with varying concentrations of AgNP, AgNP-Mal, or AgNP-Tre. Viability of untreated A549 cells was set to 100%. Results are averages from three independent trials.
Figure 6.
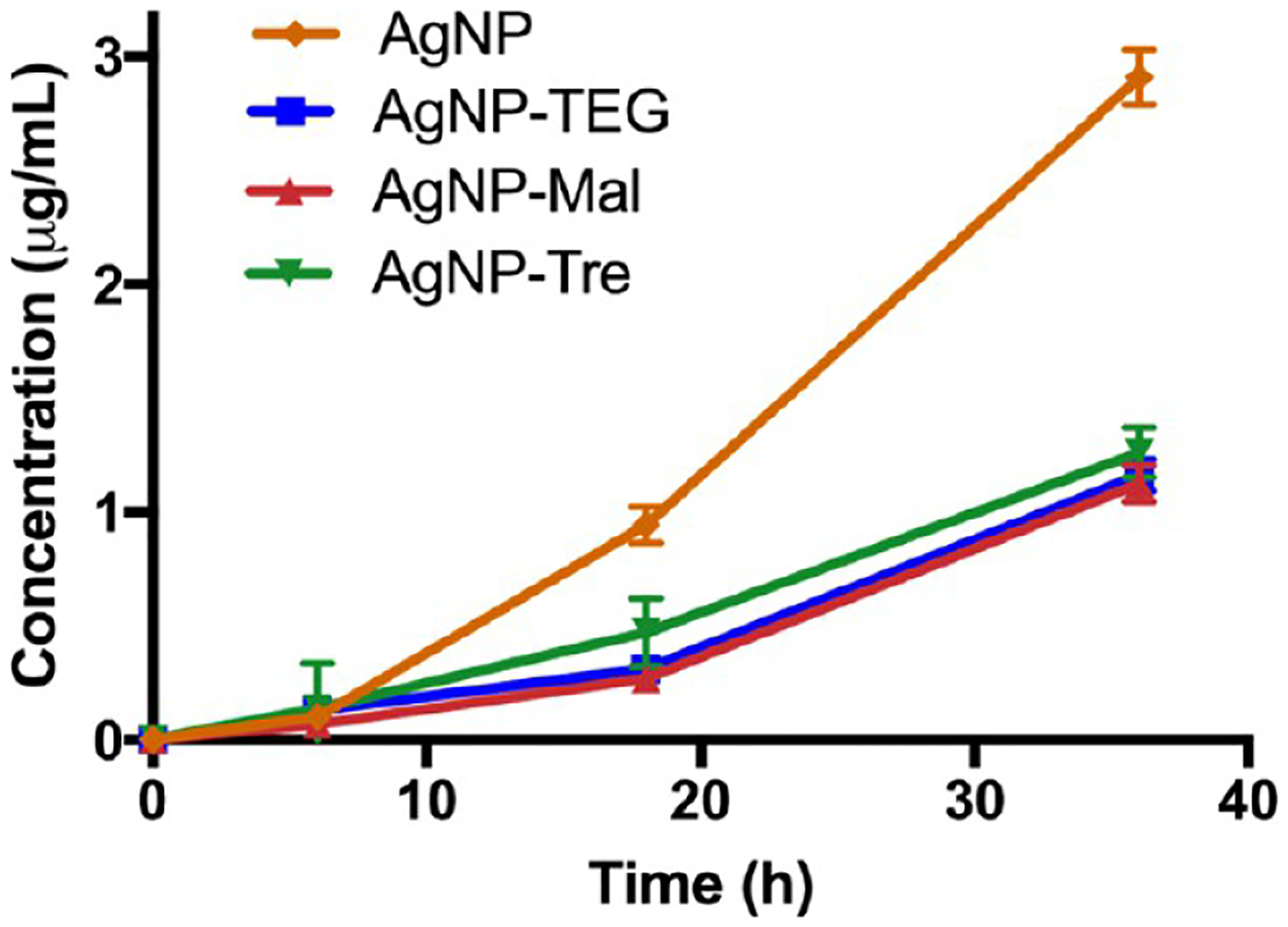
Concentrations of silver released in the culture medium (Middlebrook) from 7 nm AgNP samples (10 μg/mL) after 6, 18, and 36 h. Lines are drawn to aid visualization.
AgNP-Tre is Less Toxic to A549 Human Lung Epithelial Cells than AgNP.
We hypothesize that the conjugation of trehalose would reduce the toxicity of AgNPs to mammalian cells. On one hand, the carbohydrate ligand would slow down the release of the cytotoxic silver. More importantly, as trehalose is absent in mammalian biology, we expect minimal interactions of AgNP-Tre with mammalian cells. In a previous work, we showed that trehalose-functionalized iron oxide nanoparticles interacted specifically with mycobacteria but not with A549 cells.29 The Prussian blue staining assay on A549 cells treated with trehalose-functionalized iron oxide nanoparticles showed minimal uptake of the nanoparticles in A549 cells.
To test this, we incubated A549 human lung epithelial cells with varying concentrations of AgNP, AgNP-Mal, or AgNP-Tre (16 nm). The cell viability was measured using the MTT assay and the results are shown in Figure 2. At concentrations of 16 μg/mL or below, similar cell viability was observed for AgNP-Tre, AgNP-Mal, and AgNPs. At concentrations greater than 16 μg/mL, AgNPs showed notable inhibition on cell growth, with only 9% of cells viable at 32 μg/mL. For AgNP-Tre, however, 66% of the cells were still viable at 32 μg/mL. At 64 μg/mL, 67% of the cells treated with AgNP-Tre were viable, whereas almost no viable cells (4%) remained after treatment with AgNPs. The dose-dependent toxicity is very common in drug actions. In our case, the higher the concentration of AgNPs, the more Ag+ released and the higher the cell toxicity.
Trehalose Enhanced the Antibacterial Activity of AgNPs against Mycobacteria Compared to Maltose and TEG Ligands.
To test whether trehalose could enhance the antibacterial activity of AgNPs compared to other ligands, the minimal inhibitory concentration (MIC) was measured against M. smegmatis mc2155. The 7 nm AgNP had an MIC of 0.5 μg/mL against M. smegmatis mc2155 (entry 1, Table 1). After conjugation with Mal, the MIC increased 8 folds to 4 μg/mL for AgNP-Mal. A similar result was obtained with AgNP-TEG, which had a MIC of 4 μg/mL. Here, the surface ligand acts as a protective layer, lowering the antimicrobial activity of AgNPs.44 However, when trehalose was the ligand, the MIC of AgNP-Tre was 1 μg/mL. This is four times better than AgNP-Mal and AgNP-TEG. For larger AgNPs, the MIC increased with the particle size but a similar trend was observed (entries 2–4). The experiments were then repeated on M. smegmatis mc2651, which is an isoniazid-resistant strain.45 Similar results were obtained showing that AgNP-Tre had higher activity against M. smegmatis mc2651 than AgNP-Mal and AgNP-TEG (entries 5–8). All samples tested showed a general size-dependent activity, where smaller nanoparticles had higher antibacterial activity than the larger ones.
Table 1.
MIC (μg/mL) against M. smegmatis mc2155 and mc2651a
| entry | NP size (nm) | AgNP | AgNP-TEG | AgNP-Mal | AgNP-Tre |
|---|---|---|---|---|---|
| M. smegmatis mc2155 | |||||
| 1 | 7 | 0.5 | 4 | 4 | 1 |
| 2 | 16 | 8 | 16 | 16 | 8 |
| 3 | 42 | 2 | 4 | 4 | 4 |
| 4 | 65 | 32 | 64 | 64 | 32 |
| M. smegmatis mc2651 | |||||
| 5 | 7 | 0.25 | 0.5 | 0.5 | 0.25 |
| 6 | 16 | 1 | 4 | 4 | 1 |
| 7 | 42 | 2 | 4 | 4 | 2 |
| 8 | 65 | 4 | 8 | 8 | 4 |
All assays were done in triplicate, and the results were identical.
The morphology of mycobacteria upon exposure to AgNP-Tre was studied by transmission electron microscopy (TEM). For this, M. smegmatis mc2155 was treated with 16 nm AgNP-Tre or AgNP-TEG for 48 h at 8 μg/mL, which is the MIC of AgNP-Tre (entry 2, Table 1). Bacteria were completely disintegrated after treatment with AgNP-Tre (Figure 3a,b). For AgNP-TEG, bacterial cells were intact, although morphological changes could be seen in some bacteria (Figure 3c,d).
Figure 3.
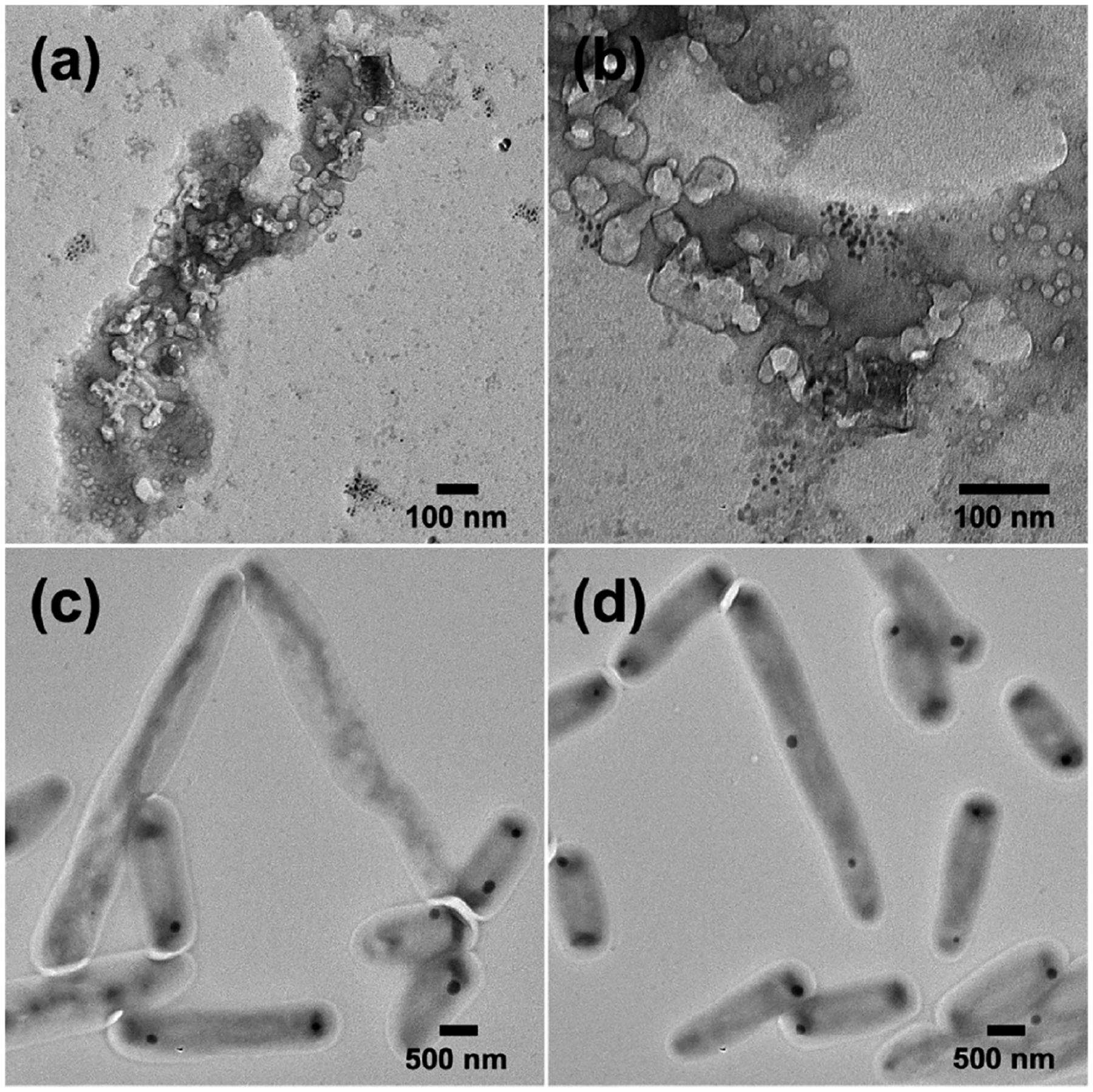
TEM images of M. smegmatis mc2155 after treatment with 16 nm (a, b) AgNP-Tre or (c, d) AgNP-TEG at the concentration of 8 μg/mL for 48 h.
Trehalose Enhances the Interactions of Nanoparticles with Mycobacteria.
We hypothesize that the higher antimicrobial activity of AgNP-Tre against mycobacteria compared to AgNP-Mal and AgNP-TEG was due to the enhanced interactions of AgNP-Tre with the mycobacterium. We used AuNPs to test the hypothesis since AgNPs are toxic to bacteria, whereas AuNPs of such size are nontoxic.46 The ~15 nm AuNPs were used as they were easier to visualize under TEM than the smaller nanoparticles and would have stronger interactions compared to the larger AuNPs based on our previous studies.47–49 After M. smegmatis mc2155 was treated with AuNP-Tre, AuNP-Mal, or AuNPs, a significantly higher number of AuNP-Tre were seen on the bacteria (Figure 4a) compared to AuNP-Mal (Figure 4b) or AuNP (Figure 4c). To test the specificity of the interactions, a Gram-negative strain E. coli ORN208 and a Gram-positive strain S. epidermidis ATCC35984 were used for comparison. Almost no AuNP-Tre was observed on E. coli (Figure 4d) or S. epidermidis (Figure 4g). These results provided strong evidence that trehalose as the ligand promoted the interactions of AgNP-Tre with M. smegmatis, and the interaction was specific for the mycobacterium. The ability of trehalose to enhance the interactions with mycobacteria is general and has been observed using silica nanoparticles, iron oxide nanoparticles, polylactide nanoparticles, and mesoporous nanoparticles of different sizes.29–31,41 AuNP-Mal appeared to preferentially adhere to E. coli (Figure 4e) and S. epidermidis (Figure 4h) than M. smegmatis (Figure 4b). Maltose is a bacterial nutrient and can be taken up by E. coli by its maltose transport system50–52 or by staphylococcus by diffusion.53,54 Previously, we have also observed that maltoheptaose enhanced the interactions of nanoparticles with E. coli.42,55 For AuNP, almost no particles were observed on all three bacteria (Figure 4c,f,i).
Figure 4.
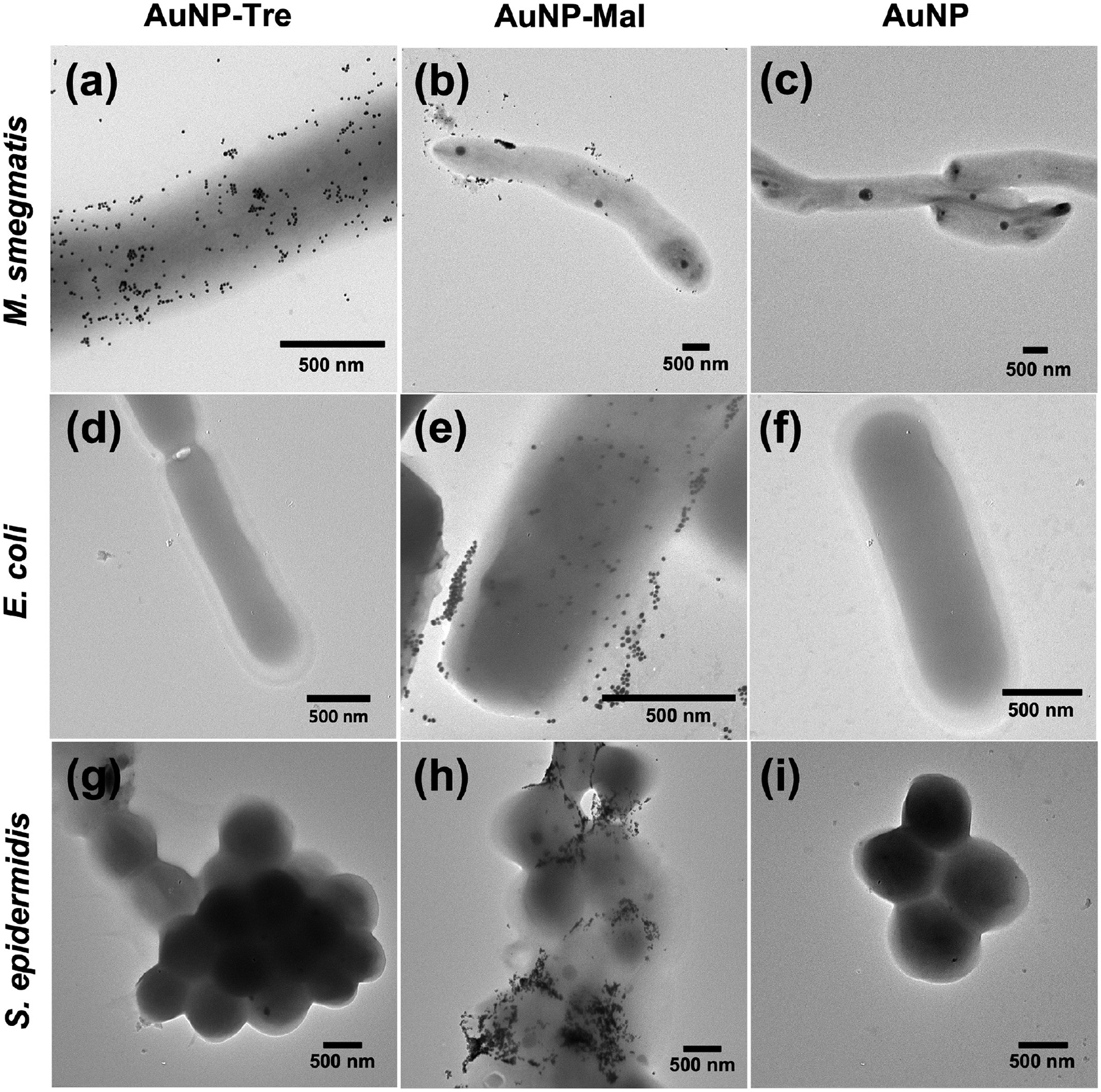
TEM images of (a–c) M. smegmatis mc2155, (d–f) E. coli ORN208, and (g–i) S. epidermidis ATCC35984 after treatment with AuNP-Tre, AuNP-Mal, or AuNP for 6 h.
Trehalose Increased the Uptake of AgNP-Tre by Mycobacteria.
We then hypothesize that the enhanced interactions between AgNP-Tre and mycobacteria would lead to increased uptake of AgNP-Tre by the bacteria and thus to higher killing efficiency. To test the hypothesis, M. smegmatis mc2155 was treated with AgNPs, AgNP-TEG, AgNP-Mal, or AgNP-Tre for 24 h. The samples were washed with pH 7.4 PBS buffer and Milli-Q water to remove excess nanoparticles and dried through a series of ethanol solutions and finally vacuum dried to obtain the dry weight. The samples were digested with concentrated nitric acid, and the concentrations of silver were determined by atomic absorption spectroscopy (AAS). Results showed that AgNP-Tre had the highest amount of Ag accumulation in mycobacteria (Figure 5). The accumulation of silver in mycobacteria was 40% higher for AgNP-Tre than AgNPs. The uptake was even greater for AgNP-Tre when compared to AgNP-Mal and AgNP-TEG, at 87 and 114% higher, respectively.
Figure 5.
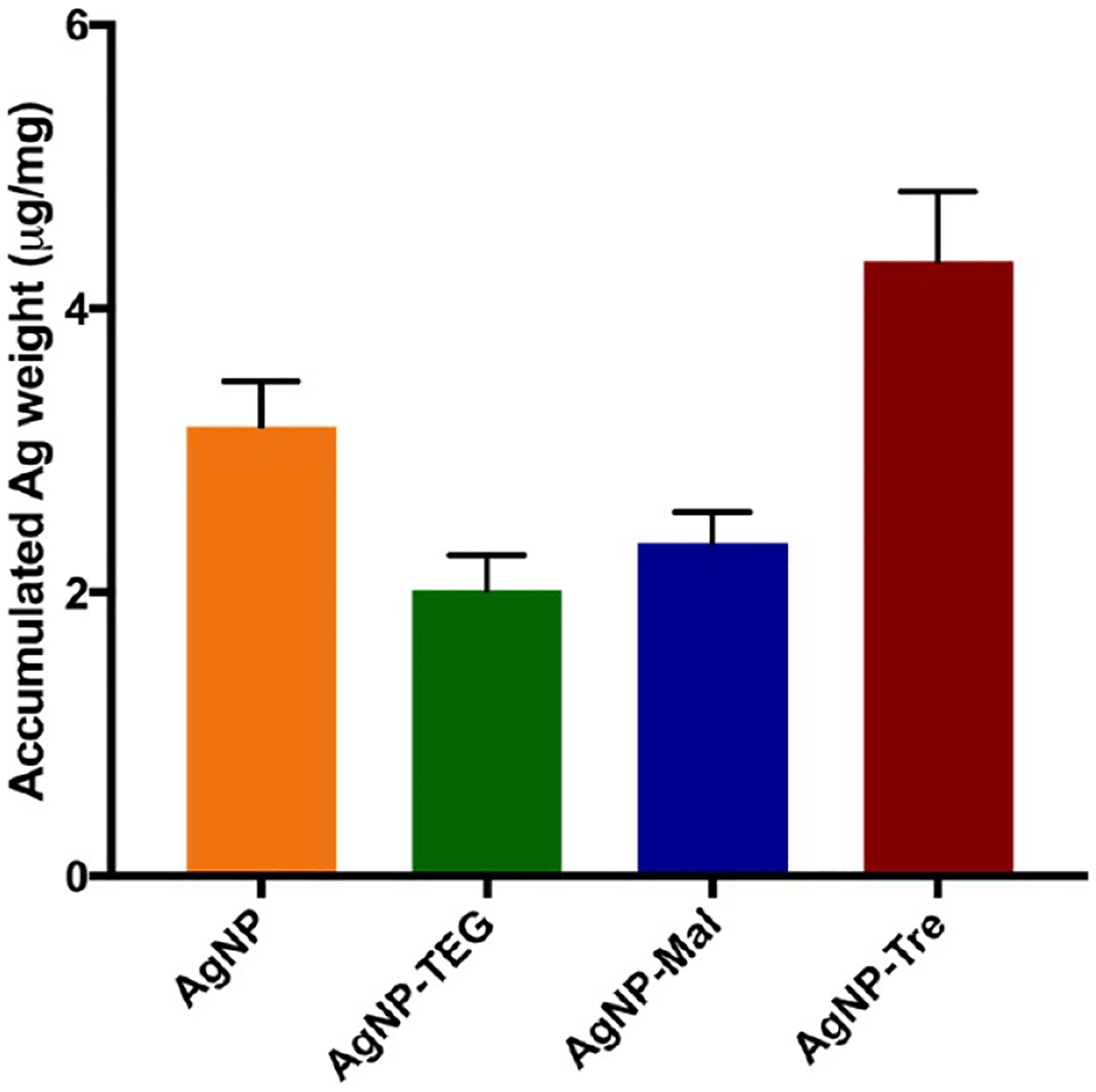
Accumulation of silver in M. smegmatis mc2155 per bacteria dry weight (μg/mg). The values are presented as μg of Ag per mg of bacteria dry weight. Each data point was the average of three independent experiments.
To exclude the possibility that the higher uptake of AgNP-Tre by mycobacteria was due to the difference in the released silver, the amount of silver released from different nanoparticle samples was measured. The 7 nm AgNP, AgNP-TEG, AgNP-Mal, or AgNP-Tre was incubated in the Middlebrook medium at 37 °C for 6, 18, and 36 h, respectively. After removing the remaining nanoparticles by centrifugation, the supernatant was analyzed by atomic absorption spectroscopy (AAS) to determine the concentration of silver released into the medium. As expected, the presence of ligands slowed down the release of silver from the nanoparticles. AgNPs had the fastest rate of release, and the amount of Ag released from AgNP-Tre was 48 and 43% of that from AgNPs at 18 and 36 h, respectively (Figure 6). AgNP-Mal and AgNP-TEG showed similar release kinetics as AgNP-Tre. The fact that all three ligand-functionalized AgNPs gave similar release kinetics and all were slower than AgNPs indicated that the density of the ligand (Tre, Mal, TEG) on AgNPs was likely very similar.
Since there was very little difference in the amount of silver released, the higher uptake of AgNP-Tre by mycobacteria than AgNP-Mal and AgNP-TEG can only be attributed to the enhanced interactions of AgNP-Tre with mycobacteria promoted by the trehalose ligand. Furthermore, although the trehalose ligand slowed down the release of silver by more than half compared to unfunctionalized AgNPs, the amount of silver accumulated in the bacteria was 40% higher. This compensated for the slower release and resulted in similar antimicrobial activities of AgNP-Tre to AgNPs.
CONCLUSIONS
Gold and silver nanoparticles having sizes ranging from ~5 to ~65 nm were successfully synthesized, and AuNPs appeared to be more uniform in size than AgNPs and both showed good stability. Trehalose, maltose, and TEG were successfully conjugated to the nanoparticles by ligand exchange reactions. The trehalose ligand lowered the toxicity of AgNPs against A549 cells, more significantly at higher concentrations. While the introduction of Mal and TEG lowered the antibacterial activity of AgNP-Mal or AgNP-TEG against M. smegmatis, trehalose did not; the MIC of AgNP-Tre was several times lower than AgNP-Mal and AgNP-TEG and was similar to AgNPs. The higher antimicrobial activity can be attributed to the increased intracellular accumulation of AgNP-Tre, which was 87 and 114% higher than those of AgNP-Mal and AgNP-TEG and 40% higher than that of AgNPs. This is in spite of decreased rate of silver release from AgNP-Tre, which was less than half of that from AgNPs. The higher uptake of AgNP-Tre was the result of enhanced interactions with mycobacteria promoted by the trehalose ligand. Using a carbohydrate ligand to target bacteria while reducing cytotoxicity of AgNPs represents a promising strategy in the clinical applications of antimicrobial AgNPs.
EXPERIMENTAL SECTION
Materials and Instrumentation.
Sodium citrate, sodium chloride, d-trehalose, d-maltose, 2-bromoethanol, sodium methoxide (MaOMe), p-toluenesulfonic acid (PTS), triethylamine (TEA), N-bromosuccinimide (NBS), d,l-dithiothreitol (DTT), potassium iodide (KI), potassium thioacetate (KSAc), and iodine were purchased from Sigma-Aldrich. Solvents were purchased from Fischer Scientific and used without purification unless otherwise noted. Deuterated solvents were obtained from Cambridge Isotope Laboratories Inc. Synthetic compounds were dried using a VWR symphony vacuum oven or a Labconco FreeZone 2.5 freeze-drier. Transmission electron microscopy (TEM) images were taken on a PHILIPS EM 200T electron microscope. Dynamic light scattering (DLS) data were obtained using a HORIBA SZ-100 nanoparticle analyzer. UV–vis spectra were recorded on a PerkinElmer Lambda 45 UV/vis spectrometer. IR spectra were recorded on a Thermo Scientific NICOLET 6700 FT-IR spectrometer. Centrifugation was done using a HERMLE, Labnet Z326 centrifuge. Atomic absorption spectroscopy (AAS) was carried out using an Agilent 200 Series AA spectrometer. Glasswares used for bacterial studies were autoclaved in a Tuttannauer EZ10 autoclave. An Epoch BioTek and TECAN infinite M200PRO plate readers were used to measure optical density (OD) and fluorescence intensity of bacteria samples.
A549 cells (CCL-185), M. smegmatis mc2155, and S. epidermidis ATCC35984 were purchased from the American Type Culture Collection (ATCC). M. smegmatis mc2651 was a gift from Prof. William Jacobs Jr. (Albert Einstein College of Medicine), and Escherichia coli ORN 208 was a gift from Prof. Paul Orndorff (North Carolina State University). Middlebrook 7H10 medium was prepared by mixing Middlebrook 7H10 Agar Base (19.47 g, Fluka Analytical) in 900 mL of distilled water and glycerol (5 mL, Acros Organics) and sterilized by autoclave. To enrich the medium, 20 mL of Middlebrook OADC growth supplement (MO678) was added.
Synthesis of 4,6-Benzylidine-2,2′,3,3′,6′-penta-O-acetyl-α,α-d-trehalose (2).
d-Trehalose dihydrate 1 (2.20 g, 5.84 mmol) was added to a solution of benzaldehyde dimethyl acetal (PhCH(OCH3)2, 1.05 mL, 7.01 mmol) in DMF (14 mL), and p-toluenesulfonic acid (223 mg, 1.17 mmol) was added. The mixture was stirred at room temperature overnight. Once thin layer chromatography (TLC) confirmed the conversion of reactants, the reaction mixture was extracted with ethyl acetate (40 mL × 3) and the solvent was evaporated. The product was dissolved in DMF (50 mL), and triethylamine (25 mL) was added followed by acetic anhydride (7.7 mL, 82 mmol). The mixture was stirred at room temperature overnight. The solvent was removed by vacuum distillation (50 °C) and was extracted with ethyl acetate (40 mL × 3). The crude product was further purified by column chromatography (2:1 v/v hexanes:ethyl acetate) to give 2 as an off-white solid (1.5 g, 38%). 1H NMR (500 MHz, CDCl3) δ (ppm): 7.34−7.24 (m, 5H), 5.58 (t, 1H), 5.49−5.46 (m, 2H), 5.33 (d, 1H), 5.23 (d, 1H), 5.03−4.96 (m, 3H), 4.23 (dd, 1H), 4.14−3.95 (4H) 3.72−3.63 (m, 2H), 1.99–2.09 (m, 18H, CH3). 13C NMR (200 MHz, CDCl3) δ (ppm) 170.76, 170.22, 170.03, 169.97, 169.78, 136.87, 129.32, 128.40, 126.33, 101.96, 93.49, 92.35, 68.27, 63.34, 61.94, 21.01, 20.83, 20.79.
Synthesis of 4-O-Benzoyl-6-bromo-2,2′,3,3′,4′,6′-penta-O-acetyl-6-deoxy-α,α-d-trehalose (3).
To a solution of 2 (1.44 g, 2.11 mmol) in CCl4 (50 mL) were added N-bromosuccinimide (413 mg, 2.32 mmol) and CaCO3 (232 mg, 2.32 mmol), and the reaction was stirred at 77 °C for 3 h. After filtering and removing the solvent, the crude product was purified by column chromatography (3:2 v/v hexanes:ethyl acetate) to give 3 as a white powder (1.43 g, 91%). 1H NMR (500 MHz, CDCl3) δ (ppm): 8.03 (d, 2H) 7.62 (t, 1H), 7.49 (t, 2H), 5.71 (t, 1H), 5.54 (t, 1H), 5.41 (d, 1H), 5.38 (d, 1H), 5.20 (m, 2H), 5.12−5.05 (m, 2H), 4.23 (t, 2H), 4.06 (t, 2H), 3.44−3.36 (two dd, 2H), 2.11−1.89 (m, 18H). 13C NMR (200 MHz, CDCl3) δ (ppm): 170.81, 170.23, 169.73, 169.65, 165.59, 134.10, 130.17, 128.91, 128.65, 92.39, 91.98, 70.40, 69.43, 68.45, 61.98, 30.75, 21.16, 20.92, 20.90, 20.81, 20.80, 20.78, 20.76.
Synthesis of 4-O-Benzoyl-6-thioethanoyl-2,2′,3,3′,4′,6′-penta-O-acetyl-6-deoxy-α,α-d-trehalose (4).
To a solution of 3 (1.00 g, 1.31 mmol) in DMF (20 mL) was added KI (1.10 g, 6.57 mmol), and the mixture was stirred at 60 °C for 5 h. The solvent was evaporated and extracted with dichloromethane (40 mL × 3). The product was dissolved in DMF (20 mL), and potassium thioacetate (450 mg, 3.9 mmol) was added and the solution was stirred at room temperature for 8 h under N2 atmosphere. The product was extracted with dichloromethane (40 mL × 3), concentrated under vacuum, and purified by column chromatography (2:1 v/v hexanes:ethyl acetate) to give 4 as an off-white powder (657 mg, 66%). 1H NMR (500 MHz, CDCl3) δ (ppm): 8.01 (d, 2H, o-benzyl C-H), 7.57 (t, 1H), 7.44 (t, 2H), 5.63 (t, 1H), 5.48 (t, 1H), 5.30 (d, 2H), 5.05 (t, 1H), 5.00−4.98 (m 3H), 4.14 (dd, 1H), 4.04 (d, 1H), 3.95−3.89 (m, 2H), 3.26 (dd, 1H), 2.87 (dd, 1H), 2.28 (s, 3H), 2.07−1.90 (m, 18H). 13C NMR (200 MHz, CDCl3) δ (ppm): 194.71, 170.07, 170.00, 169.92, 169.84, 169.77, 165.73, 133.84, 130.09, 129.02, 91.32, 91.19, 70.41, 69.66, 68.38, 62.00, 30.54, 30.34, 20.87, 20.79, 20.77.
Synthesis of 6-Mercapto-6-deoxy-α,α-d-trehalose (5).
Compound 4 (400 mg, 0.5 mmol) was dissolved in methanol (20 mL), and a solution of sodium methoxide in methanol (25.0 wt %, 133 μL, 0.582 mmol) was added slowly. The reaction was stirred at room temperature for 6 h, and the pH of the reaction was then adjusted to 7 using the Amberlight IRC-120 H+ resin. The mixture was filtered, and d,l-dithiothreitol (163 mg, 1.06 mmol) was added to the filtrate and the mixture was stirred at room temperature for 8 h. Column chromatography (1:3 v/v CHCl3:MeOH) of the crude product yielded the product 5 as an off-white solid (42 mg, 22%). 1H NMR (500 MHz, D2O) δ (ppm): 5.27 (d, 1H), 5.22 (d, 1H), 3.86−3.78 (m, 6H), 3.69 (dt, 2H), 3.46 (t, 2H), 3.02 (dd, 1H), 2.75 (dd, 1H). 13C NMR (200 MHz, D2O) δ (ppm): 93.74, 93.56, 73.06, 72.80, 72.59, 71.55, 70.11, 60.95, 49.30, 47.62, 25.52.
Synthesis of 1,2,2′,3,3′,4′,6,6′-Octata-O-acetyl-α-d-maltose (7).
Maltose 6 (5.00 g, 13.8 mmol) was suspended in acetic anhydride (40 mL), and pyridine (6.0 mL) was added to the mixture while stirring. The reaction was stirred at room temperature for 36 h until a clear solution was obtained. Water (40 mL) was added, and the mixture was stirred for another hour. The reaction mixture was then mixed with dichloromethane (50 mL) and washed with saturated NaHCO3 until no production of gas was observed. The organic layer was then washed with water followed by saturated NaCl and dried over anhydrous Na2SO4. The solvent was evaporated under vacuum to yield product 7 as a white solid (7.58 g, 80%). 1H NMR (500 MHz, CDCl3) δ (ppm) 5.72 (d, 1H), 5.38 (d, 1H), 5.33 (t, 1H), 5.27 (t, 1H), 5.06 (t, 1H), 4.96 (t, 1H), 4.84 (dd, 1H), 4.43 (dd, 1H), 4.18−4.01 (m, 4H), 3.92 (dt 1H), 3.82 (dt, 1H), 2.12−1.98 (m, 24H); 13C NMR (200 MHz, CDCl3) δ (ppm) 170.66, 170.60, 170.53, 170.16, 169.97, 169.69, 169.53, 168.90, 95.83, 91.37, 75.36, 73.10, 72.53, 71.05, 70.11, 69.41, 68.69, 68.06, 62.63, 61.56, last 4 signals for 8 carbons—20.98, 20.92, 20.79, 20.70.
Synthesis of 1-O-(2-Bromoethyl)-2,2′,3,3′,4′,6,6′-hepta-O-acetyl-β-d-maltose (8).
Compound 7 (1.00 g, 1.47 mmol) was dissolved in dichloromethane (50 mL), and 2-bromoethanol (125 μL, 1.77 mmol) was added. The reaction mixture was cooled to −40 °C (dry ice/acetonitrile) before BF3•Et2O (1.25 mL, 10 mmol) was added. The mixture was stirred for 1 h, then warmed to room temperature and stirred for another 23 h. The reaction mixture was then added to ice water (50 mL), extracted into dichloromethane (40 mL × 2), and the combined organic phase was washed with ice water, saturated NaHCO3, and ice water. The organic phase was dried over anhydrous Na2SO4, and the solvent was removed under vacuum. The crude product was purified by column chromatography (1:1 v/v hexanes:ethyl acetate) to yield 8 as a white solid (780 mg, 71%). 1H NMR (500 MHz, CDCl3) δ (ppm) 5.36 (d, 1H), 5.30 (t, 1H), 5.21 (t, 1H), 5.00 (t, 1H), 4.82−4.77 (m, 2H), 4.55 (d, 1H), 4.44 (dd, 1H), 4.19 (dt, 2H), 4.09−4.05 (m, 1H), 4.01−3.89 (m, 3H), 3.77 (dt, 1H), 3.65 (ddt, 1H), 3.44−3.36 (m, 2H) 2.09−1.95 (6s, 21H); 13C NMR (200 MHz, CDCl3) δ (ppm) 170.51, 170.48, 170.39, 170.16, 169.92, 169.65, 169.39, 100.50, 95.61, 75.21, 72.73, 72.34, 71.96, 70.05, 69.78, 69.37, 68.58, 68.11, 62.73, 61.58, 20.91, 20.84, 20.72, 20.70, 20.62, 20.59.
Synthesis of 1-O-(2-(Thioethanoyl)ethyl)-2,2′,3,3′,4′,6,6′-hepta-O-acetyl-β-d-maltose (9).
To a solution of 8 (400 mg, 0.54 mmol) in DMF (20 mL) was added KI (267 mg, 1.61 mmol). The mixture was heated to 60 °C and stirred for 5 h. After removing the solvent by distillation, the solid was extracted into dichloromethane, washed with water and brine, dried over sodium sulfate, and the solvent was removed. The residue was then dissolved in DMF (20 mL), and potassium thioacetate (184 mg, 1.61 mmol) was added. The mixture was stirred at room temperature overnight. The solvent was removed by distillation and the crude product was dissolved in dichloromethane. The organic layer was washed with water and brine, dried over sodium sulfate, and concentrated under vacuum. The product was further purified by column chromatography (2:3 v/v hexanes:ethyl acetate) to yield 9 as a white solid (325 mg, 82%). 1H NMR (500 MHz, CDCl3) δ (ppm) 5.40 (d, 1H), 5.34 (t, 1H), 5.23 (t, 1H), 5.04 (t, 1H), 4.86−4.79 (m, 2H), 4.55 (d, 1H), 4.47 (dd, 1H), 4.23 (dt, 2H), 4.05−3.90 (m, 4H) 3.68 (dt, 1H), 3.61 (dt, 1H), 3.12−3.00 (m, 2H), 2.32 (s, 3H), 2.13−1.99 (7s, 21H). 13C NMR (200 MHz, CDCl3) δ (ppm) 195.24, 170.63, 170.61, 170.54, 170.30, 170.02, 169.72, 169.52, 100.48, 95.71, 75.44, 72.91, 72.40, 72.16, 70.17, 69.51, 68.73, 68.67, 68.23, 62.92, 61.69, 30.64, 28.96, 21.02, 20.94, 20.80, 20.71, 20.69.
Synthesis of 1-(2-Mercaptoethyl) β-d-Maltose (10).
Compound 9 (288 mg, 0.39 mmol) was dissolved in methanol (20 mL), and a solution of sodium methoxide in methanol (25 wt.%, 98 μL, 0.43 mmol) was added. After stirring for 6 h, the solvent was reduced to about 10 mL, and Amberlight IRC-120 H+ resin was added until the pH reached 7. The mixture was then filtered, diluted with methanol (10 mL), and DTT (120 mg, 0.78 mmol) was added. The mixture was stirred for 8 h and concentrated under vacuum. Purification of the crude product by column chromatography (3:1 v/v dichloromethane:methanol) gave compound 10 as a white solid (139 mg, 89%). 1H NMR (500 MHz, D2O) δ (ppm) 5.42 (d, 1H), 4.52 (d, 1H), 4.03 (dt, 1H), 3.94 (dd, 1H) 3.88−3.58 (m, 10H) 3.43 (t, 1H), 3.33 (t, 1H), 2.79 (t, 2H). 13C NMR (200 MHz, D2O) δ (ppm) 102.06, 99.55, 76.71, 76.16, 74.57, 72.96, 72.82, 72.69, 71.68, 71.64, 69.31, 60.69, 60.47, 23.37.
Synthesis of TEG-Disulfide (13).
To a solution of 2-(2-(-2-chloroethoxy)ethoxy)ethanol 11 (422 mg, 2.5 mmol) in DMF (4.0 mL) was added NaI (325 mg, 5.0 mmol), and the mixture was stirred at 90 °C for 2 h. The reaction mixture was then cooled to room temperature before potassium thioacetate (5.0 mmol, 570 mg) was added and stirred at 80 °C for 6 h. After the solvent was removed, the residue was dissolved in ethyl acetate (20 mL) and washed with water (20 mL × 3). The organic layer was dried over anhydrous sodium sulfate. After the solvent was removed, the resulting solid (12) was treated with K2CO3 and MeOH for 3 h. The solvents were then evaporated, and the crude product was purified by column chromatography (4:1 v/v dichloromethane:methanol). Since this product might be a mixture of thiol and disulfide, it was dissolved in methanol (5 mL) and titrated with saturated I2 in ethanol (a few drops) until a very faint yellow color appeared. The solvent was then evaporated, and the residue was dried in a vacuum oven overnight to yield the disulfide 13 as a thick liquid (356 mg, 68%). 1H NMR (500 MHz, D2O) δ(ppm) 3.85 (t, 2H), 3.75−3.72 (m, 6H), 3.65 (t, 2H), 2.97 (t, 2H).
Synthesis of ~5 nm AuNPs.
A method reported by Puntes et al. was followed to prepare AuNPs of approximately 5 nm in size.32 To a freshly prepared aqueous solution of sodium citrate (2.2 mM, 150 mL) were added aqueous solutions of tannic acid (2.5 mM, 0.10 mL) and K2CO3 (150 mM, 1.0 mL), and the solution was heated to 70 °C under vigorous stirring. Once the temperature reached 70 °C, HAuCl4 (25 mM, 1.0 mL) was added. The solution instantly turned to blackish-gray color and then slowly to brownish-red in about 2 min. The solution was allowed to stir at the same temperature for another 5–10 min. The particles were purified by centrifugation (2000g, 25 min) three times using a centrifugal filter (MWCO 30000, Amicon). The product was stored at 4 °C.
Synthesis of ~7 nm AgNPs.
The protocol for the synthesis of the ~5 nm AuNPs above was used with a slight modification. To a freshly prepared aqueous solution of sodium citrate (2.2 mM, 150 mL) were added tannic acid (2.5 mM, 0.10 mL) and K2CO3 (150 mM, 1.0 mL), and the resulting solution was heated to 80 °C with vigorous stirring. Once the temperature reached 80 °C, AgNO3 (25 mM, 1.0 mL) was added, and the solution quickly turned into bright yellow. The solution was stirred at the same temperature for another 20 min. The particles were purified by centrifugation (2000g, 25 min) three times using a centrifugal filter (MWCO 30000, Amicon). The product was stored at 4 °C.
Synthesis of ~15 nm AuNPs.
AuNPs with an average size of around 15 nm were synthesized following a literature protocol.34 A solution of HAuCl4 (1.00 mM, 50.0 mL) was brought to boil under reflux. A solution of sodium citrate (38.8 mM, 5.00 mL) was added, and the reaction was allowed to run until a wine-red color was observed. The particles were purified by centrifugation (6000g, 20 min) three times. The product was stored at 4 °C.
Synthesis of ~15 nm AgNPs.
A literature protocol was followed for the synthesis of 15 nm AgNPs.35 An aqueous solution (100 mL) containing sodium citrate (5.0 mM) and tannic acid (0.025 mM) was heated to boiling, and a solution of AgNO3 (25 mM, 1.0 mL) was added while stirring. This resulted in a yellow solution containing silver seeds within about a minute. The temperature was then reduced to 90 °C and sodium citrate (25 mM, 100 μL), tannic acid (2.5 mM, 250 μL), and AgNO3 (50 mM, 250 μL) were added with a 1 min time delay between the addition of each reagent. This step was repeated four times, each step after 15 min from the previous. The final nanoparticles were purified by centrifugation (23 000g, 30 min) three times. The product was stored at 4 °C.
Synthesis of up to ~65 nm AuNPs.
AuNPs of sizes 15 nm or larger were synthesized using a seeded-growth method.36 To prepare ~15 nm seeds, a solution of sodium citrate (2.2 mM, 150 mL) was heated to boiling with vigorous stirring, and HAuCl4 (25 mM, 1 mL) was added. The color changed to dark gray within a minute and then turned into pink-red in about 10 min. To grow larger nanoparticles from the seeds, immediately after the seeds had been synthesized and in the same flask, the reaction temperature was lowered to 90 °C, HAuCl4 (25 mM, 1 mL) was added, and the solution was stirred at the same temperature. After 30 min, another 1.0 mL of HAuCl4 was added. Then, 55 mL of the mixture was removed, and 53 mL of distilled water was added followed by sodium citrate (60 mM, 2 mL). This process was repeated to obtain different sizes of AuNPs. The nanoparticles were purified by centrifugation (6000g, 20 min) three times and stored at 4 °C.
Synthesis of 22, 42, and 65 nm AgNPs.
A seeded-growth method was adapted to prepare ~15 nm and larger AgNPs.35 An aqueous solution (100 mL) containing sodium citrate (5.0 mM) and tannic acid (0.1 mM) was heated to boiling under vigorous stirring, and AgNO3 (25 mM, 1.0 mL) was added. The solution turned yellow, indicating the formation of seeds. To grow larger AgNPs, immediately after the formation of the seeds, 19.5 mL of the solution was taken out and 16.5 mL of Milli-Q water was added. The temperature of the solution was then adjusted to 90 °C, and sodium citrate (25 mM, 0.5 mL), tannic acid (2.5 mM, 1.5 mL), and AgNO3 (25 mM, 1.0 mL) were sequentially added with 1 min time delay between the addition of each reagent. The reaction was allowed to run for 30 min, and this process was repeated to obtain AgNPs of different sizes. AgNPs smaller than 30 nm were purified by centrifugation at 23 000g (30 min × 3), while those larger were centrifuged at 16 000g (30 min × 3). The particles were stored at 4 °C.
Functionalization of Gold and Silver Nanoparticles.
The procedure described below is for ~15 nm nanoparticles and thiolderivatized trehalose 5. An aqueous solution of 5 (5.0 mg/mL, 200 μL) was added drop-wise to a solution of as-prepared AuNPs (20.0 mL) while stirring. The solution was stirred under ambient conditions overnight. For the functionalization of AgNPs, an aqueous solution of 5 (0.5 mg/mL, 172 μL) was added to 12.0 mL of as-prepared AgNPs while stirring. The solution was stirred for 6 h. Maltose-modified nanoparticles (AuNP-Mal/AgNP-Mal) or TEG-modified nanoparticles (AuNP-TEG/AgNP-TEG) were synthesized following the same protocol using thiol-functionalized maltose 10 (5.0 mg/mL, 200 μL) or TEG-disulfide 13 (5.0 mg/mL, 184 μL). The products were purified by centrifugation at 20 000g or higher for 30 min, washing with water and repeating three times. For smaller ~5 nm particles, the products were purified by centrifugal filtration using Amicon ultra 15 centrifugal filter tubes, by adding 5.0 mL of the nanoparticle solution to the tube followed by centrifugation at 2000g for 30 min. After removing the filtrate, 5.0 mL of Milli-Q water was added and was centrifuged again. This procedure was repeated three times to obtain purified products.
Determination of Metal Concentrations in Nanoparticle Samples.
AAS was used to determine the concentration of Au or Ag in nanoparticle samples. Each type of nanoparticle solution (100 μL) was mixed with conc. HNO3 (900 μL) and Milli-Q water (9.0 mL). The concentrations of these solutions were determined by AAS against a standard calibration obtained by measuring the absorbance of a concentration series of AgNO3 or HAuCl4 (1.0, 1.5, 2.0, 2.5, and 3.0 μg/mL). The concentrations of gold or silver in the original solutions were calculated accordingly (see the SI for details).
Interactions of Nanoparticles with Con A.
A 1 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) buffer solution containing 0.1 mM Ca2+ and 0.1 mM Mn2+ was prepared. Concanavalin A (Con A) was dissolved in the HEPES buffer to give a concentration of 10 μg/mL. A 1.0 mL of ligand-functionalized nanoparticles (0.1 mg/mL) was mixed with 1.0 mL of Con A solution followed by shaking for 1 h. Two separate control experiments were carried out. In the first control, respective unmodified nanoparticles were treated with Con A in a similar manner. In the second control, ligand-functionalized nanoparticles were treated with BSA (10 μg/mL, 1.0 mL) instead of Con A following the same protocol.
Mammalian Cell Toxicity.
A549 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% fetal bovine serum and 1% each of penicillin/streptomycin, gentamicin sulfate, and amphotericin B in a humidified chamber at 37 °C and 5% CO2 until 70–90% confluency was reached. The cells were detached from the flask, and cell concentration was determined on a Countess automated cell counter to be 8 × 104 cells/mL. The cells were then seeded in a 96-well plate and incubated at 37 °C overnight to attach the cells. The fresh medium along with serially diluted solutions (80 μL) of AgNP-Tre, AgNP-Mal, or AgNP (concentrations ranging from 64–0.125 μg/mL, each in triplicate) were added, and the cells were incubated for 18 h, after which the medium was discarded, and each well was rinsed with fresh pH 7.4 PBS buffer (2 × 50 μL). Cell viability was determined by the MTT assay by incubating at 37 °C for 3 h and recording the absorbance at 575 nm on a plate reader.
Interactions of Nanoparticles with Bacteria.
M. smegmatis mc2155, E. coli ORN208, and S. epidermidis ATCC35984 were cultured in Middlebrook, Mueller Hinton, or Tryptic soy broth, respectively. Bacterial stock solutions of 108 CFU/mL having an optical density of 0.3 (at 650 nm for M. smegmatis and 600 nm for E. coli and S. epidermidis, determined by colony counting) were diluted with the growth medium to about 2 × 106 CFU/mL for subsequent experiments. AuNP samples (1.0 mL) was incubated with 1.0 mL of bacteria to obtain the final concentrations of bacteria at ~106 CFU and nanoparticles at 30 μg/mL, respectively. The mixtures were then incubated while shaking at 250 rpm and 37 °C for 6 h. The samples were centrifuged at 1700g and the supernatant was discarded. The pellet was resuspended by shaking and centrifuged in PBS buffer twice and finally in water. To prepare samples for TEM imaging, the suspension (50 μL) was dropped onto a TEM grid and dried in air for 4 h followed in a vacuum oven for 24 h.
Determination of MIC.
M. smegmatis mc2155 or M. smegmatis mc2651 were incubated under shaking at 250 rpm and 37 °C for 48 h with AgNP, AgNP-TEG, AgNP-Mal, or AgNP-Tre at serially diluted concentrations ranging from 32 to 0.0625 μg/mL with respect to each type of nanoparticle (concentration determined by AAS). The AgNP sample (100 μL) was mixed with an equal volume of the bacterial culture in a 96-well plate so that the final bacterial concentration in each well was 106 CFU/mL. The Alamar Blue assay was used to determine the MIC.56 Typically, 20 μL of Alamar Blue solution was added to each well, and the mixture was incubated in the dark at 37 °C for 2 h. The fluorescence intensity was measured at 590 nm at 560 nm excitation on a plate reader.
Determination of Silver Accumulation in Mycobacteria.
M. smegmatis mc2155 (2.0 mL, 108 CFU/mL) was mixed with AgNP-Tre, AgNP-Mal, AgNP-TEG, or AgNP (20 μg/mL, 2.0 mL) at 37 °C for 24 h. A control sample, which contained the same amount of bacteria without any nanoparticles, was also treated under the same conditions. The resulting suspensions were centrifuged (5000g × 1, 3000g × 1, 2000g × 1 in PBS followed by 2000g × 2 in water) to remove excess nanoparticles. All pellets were treated through a series of ethanol/water solutions to remove water from bacteria. The pellet was suspended in 20% ethanol and centrifuged at 2000g for 10 min, and this procedure was repeated with 30, 50, 75, 95, and 100% ethanol. Then, the pellet was dried in air for 2 h and in a vacuum oven for 48 h. The residue was weighed, from which the dry weights of the bacterial samples were obtained. The dried pellets were then dissolved in conc. HNO3 (1.0 mL) and topped to 5.0 mL with deionized water. The concentrations of Ag in the samples were determined by AAS. The amount of silver was calculated by comparing the absorbance of each sample with that of the calibration curve constructed from a concentration series of AgNO3 solutions ranging from 1.0 to 3.0 μg/mL. The results were presented as micrograms of silver per milligram of bacterial dry weight.
Determination of Silver Released in Culture Medium.
AgNP-Tre, AgNP-Mal, AgNP-TEG, or AgNP, each at the concentration of 10 μg/mL, were incubated in the Middlebrook broth medium (2.0 mL) at 37 °C for 24 h. The residual solids were removed by centrifuging at 1000g for 20 min. The supernatant was further centrifuged at 26 000g for 30 min to remove additional solids and undissociated nanoparticles. The supernatant was collected and the absorbance at 328 nm of each sample was measured by AAS.
Supplementary Material
ACKNOWLEDGMENTS
The work was supported in part by a grant from NIH (R15GM128164). The authors thank Prof. William Jacobs Jr. from Albert Einstein College of Medicine for the generous donation of M. smegmatis mc2651 and Professor Paul Orndorff (North Carolina State University) for the generous donation of E. coli ORN208. Scheme 1 and the TOC graphic were created with BioRender.com.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsanm.2c02047.
UV–vis spectra and TEM images of gold and silver nanoparticles; standard calibration curves of gold and silver; and NMR and FT-IR spectra of synthesized compounds (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acsanm.2c02047
The authors declare no competing financial interest.
Contributor Information
Samurdhi A. Wijesundera, Department of Chemistry, University of Massachusetts Lowell, Lowell, Massachusetts 01854, United States
Kalana W. Jayawardana, Department of Chemistry, University of Massachusetts Lowell, Lowell, Massachusetts 01854, United States
Mingdi Yan, Department of Chemistry, University of Massachusetts Lowell, Lowell, Massachusetts 01854, United States.
REFERENCES
- (1).Alexander JW History of the medical use of silver. Surg. Infect 2009, 10, 289–292. [DOI] [PubMed] [Google Scholar]
- (2).Mijnendonckx K; Leys N; Mahillon J; Silver S; Van Houdt R Antimicrobial silver: uses, toxicity and potential for resistance. BioMetals 2013, 26, 609–621. [DOI] [PubMed] [Google Scholar]
- (3).Sim W; Barnard RT; Barnard RT; Blaskovich MAT; Ziora ZM Antimicrobial Silver in Medicinal and Consumer Applications: A Patent Review of the Past Decade (2007–2017). Antibiotics 2018, 7, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Chernousova S; Epple M Silver as antibacterial agent: ion, nanoparticle, and metal. Angew. Chem., Int. Ed 2013, 52, 1636–1653. [DOI] [PubMed] [Google Scholar]
- (5).Rai M; Yadav A; Gade A Silver nanoparticles as a new generation of antimicrobials. Biotechnol. Adv 2009, 27, 76–83. [DOI] [PubMed] [Google Scholar]
- (6).Li WR; Xie XB; Shi QS; Zeng HY; Ou-Yang YS; Chen YB Antibacterial activity and mechanism of silver nanoparticles on Escherichia coli. Appl. Microbiol. Biotechnol 2010, 85, 1115–1122. [DOI] [PubMed] [Google Scholar]
- (7).Cui L; Chen P; Chen S; Yuan Z; Yu C; Ren B; Zhang K In situ study of the antibacterial activity and mechanism of action of silver nanoparticles by surface-enhanced Raman spectroscopy. Anal. Chem 2013, 85, 5436–5443. [DOI] [PubMed] [Google Scholar]
- (8).Lu Z; Rong K; Li J; Yang H; Chen R Size-dependent antibacterial activities of silver nanoparticles against oral anaerobic pathogenic bacteria. J. Mater. Sci.: Mater. Med 2013, 24, 1465–1471. [DOI] [PubMed] [Google Scholar]
- (9).Korshed P; Li L; Liu Z; Mironov A; Wang T Size-dependent antibacterial activity for laser-generated silver nanoparticles. J. Interdiscip. Nanomed 2019, 4, 24–33. [Google Scholar]
- (10).Szerencsés B; Igaz N; Tobias A; Prucsi Z; Ronavari A; Belteky P; Madarasz D; Papp C; Makra I; Vagvolgyi C; Konya Z; Pfeiffer I; Kiricsi M Size-dependent activity of silver nanoparticles on the morphological switch and biofilm formation of opportunistic pathogenic yeasts. BMC Microbiol. 2020, 20, 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Radniecki TS; Stankus DP; Neigh A; Nason JA; Semprini L Influence of liberated silver from silver nanoparticles on nitrification inhibition of Nitrosomonas europaea. Chemosphere 2011, 85, 43–49. [DOI] [PubMed] [Google Scholar]
- (12).Marambio-Jones C; Hoek EMV A review of the antibacterial effects of silver nanomaterials and potential implications for human health and the environment. J. Nanop. Res 2010, 12, 1531–1551. [Google Scholar]
- (13).Tang S; Zheng J Antibacterial Activity of Silver Nanoparticles: Structural Effects. Adv. Healthcare Mater 2018, 7, No. 1701503. [DOI] [PubMed] [Google Scholar]
- (14).Akter M; Sikder MT; Rahman MM; Ullah A; Hossain KFB; Banik S; Hosokawa T; Saito T; Kurasaki M A systematic review on silver nanoparticles-induced cytotoxicity: Physicochemical properties and perspectives. J. Adv. Res 2018, 9, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Sun J; Wan J; Zhai X; Wang J; Liu Z; Tian H; Xin L Silver nanoparticles: Correlating particle size and ionic Ag release with cytotoxicity, genotoxicity, and inflammatory responses in human cell lines. Toxicol. Ind. Health 2021, 37, 198–209. [DOI] [PubMed] [Google Scholar]
- (16).Gorka DE; Osterberg JS; Gwin CA; Colman BP; Meyer JN; Bernhardt ES; Gunsch CK; DiGulio RT; Liu J Reducing Environmental Toxicity of Silver Nanoparticles through Shape Control. Environ. Sci. Technol 2015, 49, 10093–10098. [DOI] [PubMed] [Google Scholar]
- (17).Salvioni L; Galbiati E; Collico V; Alessio G; Avvakumova S; Corsi F; Tortora P; Prosperi D; Colombo M Negatively charged silver nanoparticles with potent antibacterial activity and reduced toxicity for pharmaceutical preparations. Int. J. Nanomedicine 2017, Volume 12, 2517–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Oćwieja M; Barbasz A; Walas S; Roman M; Paluszkiewicz C Physicochemical properties and cytotoxicity of cysteine-functionalized silver nanoparticles. Colloids Surf., B 2017, 160, 429–437. [DOI] [PubMed] [Google Scholar]
- (19).Chen X; Ramstrom O; Yan M Glyconanomaterials: Emerging applications in biomedical research. Nano Res. 2014, 7, 1381–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Kang B; Opatz T; Landfester K; Wurm FR Carbohydrate nanocarriers in biomedical applications: functionalization and construction. Chem. Soc. Rev 2015, 44, 8301–8325. [DOI] [PubMed] [Google Scholar]
- (21).Hao N; Neranon K; Ramstrom O; Yan M Glyconanomaterials for biosensing applications. Biosens. Bioelectron 2016, 76, 113–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Ndugire W; Liyanage SH; Yan M Carbohydrate-Presenting Metal Nanoparticles: Synthesis, Characterization and Applications. Compr. Glycosci 2021, 4, 380–405. [Google Scholar]
- (23).Sanyasi S; Majhi RK; Kumar S; Mishra M; Ghosh A; Suar M; Satyam PV; Mohapatra H; Goswami C; Goswami L Polysaccharide-capped silver Nanoparticles inhibit biofilm formation and eliminate multi-drug-resistant bacteria by disrupting bacterial cytoskeleton with reduced cytotoxicity towards mammalian cells. Sci. Rep 2016, 6, No. 24929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Jaiswal S; Bhattacharya K; McHale P; Duffy B Dual effects of beta-cyclodextrin-stabilised silver nanoparticles: enhanced biofilm inhibition and reduced cytotoxicity. J. Mater. Sci.: Mater. Med 2015, 26, 5367. [DOI] [PubMed] [Google Scholar]
- (25).Yamagami H; Matsumoto T; Fujiwara N; Arakawa T; Kaneda K; Yano I; Kobayashi K Trehalose 6,6′-dimycolate (cord factor) of Mycobacterium tuberculosis induces foreign-body- and hypersensitivity-type granulomas in mice. Infect. Immun 2001, 69, 810–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Elbein AD; Mitchell M Levels of glycogen and trehalose in Mycobacterium smegmatis and the purification and properties of the glycogen synthetase. J. Bacteriol 1973, 113, 863–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Woodruff PJ; Carlson BL; Siridechadilok B; Pratt MR; Senaratne RH; Mougous JD; Riley LW; Williams SJ; Bertozzi CR Trehalose is required for growth of Mycobacterium smegmatis. J. Biol. Chem 2004, 279, 28835–28843. [DOI] [PubMed] [Google Scholar]
- (28).Hoffmann C; Leis A; Niederweis M; Plitzko JM; Engelhardt H Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc. Natl. Acad. Sci. U.S.A 2008, 105, 3963–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Jayawardana KW; Jayawardena HS; Wijesundera SA; De Zoysa T; Sundhoro M; Yan M Selective targeting of Mycobacterium smegmatis with trehalose-functionalized nanoparticles. Chem. Commun 2015, 51, 12028–12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Hao N; Chen X; Jeon S; Yan M Carbohydrate-Conjugated Hollow Oblate Mesoporous Silica Nanoparticles as Nanoantibiotics to Target Mycobacteria. Adv. Healthcare Mater 2015, 4, 2797–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Zhou J; Jayawardana KW; Kong N; Ren Y; Hao N; Yan M; Ramstrom O Trehalose-Conjugated, Photofunctionalized Mesoporous Silica Nanoparticles for Efficient Delivery of Isoniazid into Mycobacteria. ACS Biomater. Sci. Eng 2015, 1, 1250–1255. [DOI] [PubMed] [Google Scholar]
- (32).Piella J; Bastús NG; Puntes V Size-Controlled Synthesis of Sub-10-nanometer Citrate-Stabilized Gold Nanoparticles and Related Optical Properties. Chem. Mater 2016, 28, 1066–1075. [Google Scholar]
- (33).Mendis P; de Silva RM; de Silva KMN; Wijenayaka LA; Jayawardana K; Yan M Nanosilver rainbow: a rapid and facile method to tune different colours of nanosilver through the controlled synthesis of stable spherical silver nanoparticles. RSC Adv. 2016, 6, 48792–48799. [Google Scholar]
- (34).Lekeufack DD; Brioude A; Mouti A; Alauzun JG; Stadelmann P; Coleman AW; Miele P Core-shell Au@(TiO(2), SiO(2)) nanoparticles with tunable morphology. Chem. Commun 2010, 46, 4544–4546. [DOI] [PubMed] [Google Scholar]
- (35).Bastús NG; Merkoçi F; Piella J; Puntes V Synthesis of Highly Monodisperse Citrate-Stabilized Silver Nanoparticles of up to 200 nm: Kinetic Control and Catalytic Properties. Chem. Mater 2014, 26, 2836–2846. [Google Scholar]
- (36).Bastús NG; Comenge J; Puntes V Kinetically controlled seeded growth synthesis of citrate-stabilized gold nanoparticles of up to 200 nm: size focusing versus Ostwald ripening. Langmuir 2011, 27, 11098–11105. [DOI] [PubMed] [Google Scholar]
- (37).Revell DJ; Knight JR; Blyth DJ; Haines AH; Russell DA Self-Assembled Carbohydrate Monolayers: Formation and Surface Selective Molecular Recognition. Langmuir 1998, 14, 4517–4524. [Google Scholar]
- (38).Wang X; Ramstrom O; Yan M A photochemically initiated chemistry for coupling underivatized carbohydrates to gold nanoparticles. J. Mater. Chem 2009, 19, 8944–8949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Hofmann A; Schmiel P; Stein B; Graf C Controlled formation of gold nanoparticle dimers using multivalent thiol ligands. Langmuir 2011, 27, 15165–15175. [DOI] [PubMed] [Google Scholar]
- (40).Gray RD; Glew RH The kinetics of carbohydrate binding to concanavalin A. J. Biol. Chem 1973, 248, 7547–7551. [PubMed] [Google Scholar]
- (41).Chen X; Wu B; Jayawardana KW; Hao N; Jayawardena HS; Langer R; Jaklenec A; Yan M Magnetic Multivalent Trehalose Glycopolymer Nanoparticles for the Detection of Mycobacteria. Adv. Healthcare Mater 2016, 5, 2007–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Wu B; Ndugire W; Chen X; Yan M Maltoheptaose-Presenting Nanoscale Glycoliposomes for the Delivery of Rifampicin to E. coli. ACS Appl. Nano Mater 2021, 4, 7343–7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Liyanage SH; Yan M Quantification of binding affinity of glyconanomaterials with lectins. Chem. Commun 2020, 56, 13491–13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Padmos JD; Boudreau RT; Weaver DF; Zhang P Impact of protecting ligands on surface structure and antibacterial activity of silver nanoparticles. Langmuir 2015, 31, 3745–3752. [DOI] [PubMed] [Google Scholar]
- (45).Banerjee A; Dubnau E; Quemard A; Balasubramanian V; Um KS; Wilson T; Collins D; de Lisle G; Jacobs WR Jr. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 1994, 263, 227–230. [DOI] [PubMed] [Google Scholar]
- (46).Shareena Dasari TP; Zhang Y; Yu H Antibacterial Activity and Cytotoxicity of Gold (I) and (III) Ions and Gold Nanoparticles. Biochem. Pharmacol 2015, 4, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Wang X; Ramstrom O; Yan M Quantitative analysis of multivalent ligand presentation on gold glyconanoparticles and the impact on lectin binding. Anal. Chem 2010, 82, 9082–9089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Wang X; Ramstrom O; Yan M Dye-doped silica nanoparticles as efficient labels for glycans. Chem. Commun 2011, 47, 4261–4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Liu L-H; Dietsch H; Schurtenberger P; Yan M Photoinitiated Coupling of Unmodified Monosaccharides to Iron Oxide Nanoparticles for Sensing Proteins and Bacteria. Bioconjugate Chem. 2009, 20, 1349–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Davidson AL; Shuman HA; Nikaido H Mechanism of maltose transport in Escherichia coli: transmembrane signaling by periplasmic binding proteins. Proc. Natl. Acad. Sci. U.S.A 1992, 89, 2360–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Boos W; Shuman H Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev 1998, 62, 204–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Nikaido H Maltose transport system of Escherichia coli: an ABC-type transporter. FEBS Lett. 1994, 346, 55–58. [DOI] [PubMed] [Google Scholar]
- (53).Hengstenberg W; Egan JB; Morse ML Carbohydrate transport in Staphylococcus aureus. VI. The nature of the derivatives accumulated. J. Biol. Chem 1968, 243, 1881–1885. [PubMed] [Google Scholar]
- (54).Button DK; Egan JB; Hengstenberg W; Morse ML Carbohydrate transport in Staphylococcus aureus. IV. Maltose accumulation and metabolism. Biochem. Biophys. Res. Commun 1973, 52, 850–855. [DOI] [PubMed] [Google Scholar]
- (55).Jayawardena HSN; Jayawardana KW; Chen X; Yan M Maltoheptaose promotes nanoparticle internalization by Escherichia coli. Chem. Commun 2013, 49, 3034–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Franzblau SG; Witzig RS; McLaughlin JC; Torres P; Madico G; Hernandez A; Degnan MT; Cook MB; Quenzer VK; Ferguson RM; Gilman RH Rapid, Low-Technology MIC Determination with Clinical Mycobacterium tuberculosis Isolates by Using the Microplate Alamar Blue Assay. J. Clin. Microbiol 1998, 36, 362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


