Biomarkers of response to immunotherapy in lung cancer remain elusive. Valencia et al. identify DSTYK as a cancer cell–intrinsic modulator of sensitivity to CD8+ T cell activity. By regulating the mTOR pathway and stimulating protective autophagy, DSTYK blunts TNF-α–mediated killing by T cells, thereby limiting the efficacy of immunotherapy.
Abstract
Despite evidence for clinical benefit in patients suffering from lung cancer following treatment with immune checkpoint inhibitors (ICI), it is still uncertain how to predict which patients are likely to experience a significant response. In their work, Valencia et al. (2022. J. Exp. Med. https://doi.org/10.1084/jem.20220726) identify the DSTYK kinase as a cancer cell–intrinsic modulator of response to immunotherapy. Through regulation of the mTOR pathway and stimulation of protective autophagy, DSTYK blunts CD8+ T cell–mediated killing of cancer cells. Accordingly, lung cancers with increased expression of DSTYK are less responsive to ICI treatment. These observations could be useful in the clinic towards the development of predictive biomarkers and novel therapeutic strategies.
The new wave of immunomodulatory drugs is rapidly changing the field of cancer therapy. Long-lasting clinical responses have been observed in several tumor types, including cancers classified as “hard-to-treat” and for which standard treatments were generally poorly effective. In non-small cell lung cancer (NSCLC), immune checkpoints inhibitors (ICI) have already been included as first-line therapy for advanced stage disease, and evidence is building towards their efficacy also in early-stage tumors (Grant et al., 2021). Unfortunately, however, not all patients benefit from these treatments, and markers to guide personalized therapy are not satisfactory. In this context, identification of tumor (or tumor microenvironment [TME]) features that could shape response to immunotherapy is a very relevant topic of research with important clinical implications.

Insights from Roberto Ferrara and Luca Roz.
In this evolving scenario, DSTYK is described by Valencia et al. (2022) as a key cancer cell–intrinsic protecting hub which mediates tumor immune escape, inducing autophagy and oxidative stress response.
DSTYK is a dual serine/threonine and tyrosine non-receptor protein kinase previously identified by the same authors to be altered in a lung cancer cell line isolated from chemically induced murine lung squamous cell carcinoma. Data on a potential involvement of DSTYK in cancer development and progression are present in the literature, but its specific activity and the role in lung cancer were largely unknown. The manuscript presents experimental data on the involvement of DSTYK in the regulation of a “protective” form of autophagy in lung cancer cells and identifies an interesting novel role in the regulation of immune response.
In an article by Valencia et al. (2022) in this issue of JEM, mutations or copy number gains (CNG; three or more copies) of DSTYK are observed in 6.4% of lung cancer samples included in The Cancer Genome Atlas dataset, and CNG is also detected in 13.3% of 45 frozen NSCLC samples from a single institution. Importantly, CNG correlates with increased DSTYK expression and poor prognosis.
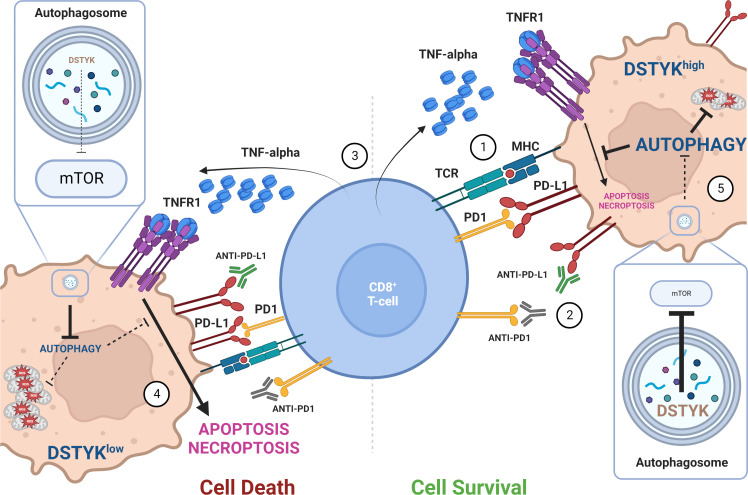
In vitro experiments show that DSTYK regulates the autophagy process in lung cancer cell lines, interacting with specific autophagy adaptors such as LC3 and P62. DSTYK also appears to be involved in the oxidative stress response, and DSTYK KO lung cancer cells are accordingly characterized by a significant increase in ROS levels and a striking accumulation of damaged mitochondria, which cannot be eliminated in the absence of DSTYK-driven cytoprotective autophagy, ultimately leading to cancer cell death. In particular, DSTYK inhibition does not increase the number of intratumoral T cells but induces TNF-α–mediated CD8+ cells’ killing of cancer cells, resulting in tumor regression (and synergy with immune checkpoint blockade) in different syngeneic mouse models of lung cancer. Correlative data are also presented on a potential relevance of DSTYK in regulating response to ICI in lung cancer patients. Altogether, these results suggest that DSTYK is a master regulator of cytoprotective cancer cell autophagy, and high (≥3) DSTYK CNG is associated with primary resistance to ICI. DSTYK genetic inhibition or autophagy pharmacological interruption (i.e., by chloroquine) drives cells towards a cellular oxidative crush characterized by high ROS levels and dysfunctional mitochondria engulfment, which sensitize cancer cells to T-lymphocytes killing (Fig. 1).
Figure 1. DSTYK protects cancer cells from oxidative stress and TNF-α–mediated CD8+ T cell killing. Tumor antigens presented on MHC complexes are recognized by CD8+ cells through interaction with the T cell receptor. T cell responses can be negatively regulated by immune checkpoints, including the PD-1/PD-L1 axis (1). Use of ICI (2) allows T cell activation and production of effector molecules including TNF-α (3). Valencia et al. (2022) show that in DSTYKlow cells (left), autophagic activity is low, resulting in oxidative stress, accumulation of damaged mitochondria, and activation of TNF-α–mediated apoptosis and necroptosis (4). When DSTYK is expressed at high levels (right), its inhibitory function on the mTOR pathway favors activation of protective autophagy, resulting in reduction of oxidative stress damage and resistance to T cell–mediated cell killing (5). Figure created with BioRender.com.
These findings could pave the way for combination treatments based on DSTYK targeting to improve efficacy of ICIs in lung cancer patients. After more than one decade of success with immunotherapy, efficacy of ICI for individual patients with lung cancer is still hard to predict. Biomarkers such as PD-1/PD-L1 and tumor mutational burden have been thoroughly investigated, but their clinical utility is far from optimal. The reason for this difficulty in validating predictive biomarkers is likely multifactorial, reflecting the numerous steps that influence recognition and killing of cancer cells by T cells. Among these factors, the presence of tumor-associated antigens (plus the correct presentation by the HLA machinery), the selection of activated T cell clones, and the repertoire of co-stimulatory and inhibitory immune molecules play a central role, but other mechanisms such as metabolic checkpoints, through which tumor cells can survive in an energetically hostile TME and blunt T cell responses, have raised the battlefield to an entirely novel and more complex dimension.
Tumors with specific mutations have indeed been shown to be poorly responsive to ICI, as exemplified by LKB1-mutated tumors (Koyama et al., 2016) and in particular KRAS-LKB1–co-mutated tumors (KL tumors; Skoulidis et al., 2018). As a promising paradigm, the identification of the subtending mechanisms, including the generation of an immunosuppressive microenvironment dominated by neutrophils, Th17-related markers, and immune checkpoints, has however opened the way for the successful evaluation in preclinical models of experimental therapies such as STAT3 inhibition in reversing LKB1-driven blockade of immune responses (Pore et al., 2021). Identification of DSTYK as a regulator of TNF-α–mediated T cell killing therefore has the potential to lead further work toward the development of novel strategies to reverse this blockade.
In this scenario, stress-induced cancer genetic vulnerabilities such as LKB1, KEAP1, and NRF2 alterations have emerged as potential mechanisms of resistance to ICI in subsets of patients with NSCLC, being reported in 20–30% (Carretero et al., 2004), 12–18%, and 3.5–15% (Nadal et al., 2019) of NSCLC respectively, with NRF2 mutations mainly occurring in patients with squamous cell lung carcinoma.
LKB1 loss induces mTOR upregulation and an anabolic switch in cancer cells, characterized by increase glucose and glutamine uptake and upregulation of fatty acids synthesis facilitating tumor growth and energy production under unfavorable metabolic conditions (Shackelford and Shaw, 2009). Autophagy inhibition in LKB1 mutant NSCLC restores HLA-mediated antigen presentation and effector T cell expansion (Deng et al., 2021). Although no data are reported in the article by Valencia et al. (2022), the increased CD8+ T cell killing of DSTYK KO cells could be explained also by an increased HLA expression upon autophagy inhibition. In this regard, HLA loss of heterozygosis (detected in up to 30% of NSCLC) has been reported to influence the impact of tumor mutational burden as a predictor of response to ICI (Shim et al., 2020); therefore, correlation between HLA expression and DSTYK CNG would probably deserve further research. Similarly, PD-L1 expression by tumor cells is also regulated by autophagy (Wang et al., 2019), and the survival benefit observed upon ICI in NSCLC patients with low-CNG DSTYK could be potentially related to higher PD-L1 expression following decreased autophagy.
Open issues remain that could pave the way for further research. One of the key points would be to define a practical and robust way to identify “DSTYK-driven” lung cancers at individual tumor level. In their report, the authors have used a genomic based cut-off (copy number variation ≥ 3), which could be difficult to implement widely and may be dependent on the techniques used for assessment. This could be one of the reasons subtending the different frequency of lung cancers with DSTYK CNG reported in The Cancer Genome Atlas dataset (4%), compared with clinical samples from different cohorts (13–60%). Alternatively, this discrepancy could be due to differences in cohort composition (e.g., early stages vs. late stages) and provide further evidence on the negative prognostic role of DSTYK alterations. Furthermore, other mechanisms of DSTYK deregulation could also be present, including epigenetic regulation: interestingly, in the setting of KL tumors it has been reported that epigenetic alterations such as loss of miR-17 can mimic a KL-like state, influencing tumor behavior and potentially guiding therapy (Borzi et al., 2021).
From a clinical perspective, stress-induced cancer vulnerabilities such as LKB1 mutations have been associated not only with primary resistance to ICI but also with a paradoxical acceleration of lung cancer tumor growth upon immune checkpoint blockade known as hyperprogressive disease (HPD; Ferrara et al., 2018). Although a common and unique mechanism of HPD has not yet been described, a highly immunosuppressive TME characterized by myeloid-derived suppressor cell tumor infiltration and neutrophil-systemic inflammation, together with low T cell tumor infiltration, as reported in LKB1-mutated NSCLC patients, could be crucial HPD players. Unfortunately, no data were available regarding the TME composition of high-CNG DSTYK cancers to suggest an involvement of this pathway in HPD. However, the overlap of progression-free survival curves of the CNG-high versus -low DSTYK arms in the first months of ICI treatment and the relatively longer median progression-free survival in high-CNG DSTYK compared to HPD NSCLC patients (4.2 vs. 1.2 mo; Ferrara et al., 2018) suggest that DSTYK CNG alone could not discriminate HPD phenomenon.
From a therapeutic point of view, high-CNG DSTYK could be the patient population more suitable for ICI and autophagy inhibitors combinations. In fact, although autophagy inhibitors such as hydroxychloroquine (HCQ) have failed to improve outcome in combination with platinum-based chemotherapy (Malhotra et al., 2019) in NSCLC patients, their potential role in inducing HLA or PD-L1 upregulation and the absence of impairment of T cell function by systemic inhibition of autophagy (Starobinets et al., 2016) are hypothesis-generating bases for further clinical investigation of non-selective (i.e., HCQ) or selective (i.e., ULK inhibitors) autophagy inhibitors in combination with PD-1/PD-L1 blockade. Similarly, the accumulation of autophagosomes due to autophagy interruption in DSTYK-expressing cells upon HCQ treatment suggests that a triple combination of autophagy inhibitors together with chemotherapy and ICI in high-CNG DSTYK patients could increase the performance of some chemotherapy drugs in inducing immunogenic cell death. In fact, after autophagy interruption due to exposure to autophagy inhibitors, autophagosomes containing multiple tumor antigens progressively accumulate, and upon chemotherapy exposure they could be released in the extracellular spaces, taken by antigen-presenting cells, and cross-presented to T cells, ultimately potentiating the activity of ICI (Ma et al., 2013).
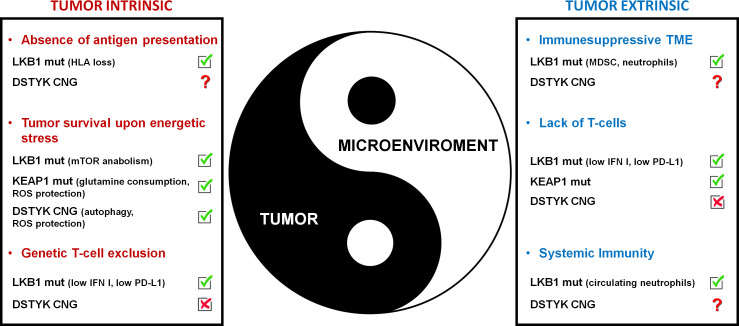
Figure 2. Tumor-intrinsic and -extrinsic mechanisms of resistance to ICI in NSCLC with cellular stress vulnerabilities. Different mechanisms have been identified as drivers of resistance to ICI therapy in NSCLC with genetically determined stress vulnerabilities, ranging from HLA downregulation to T cell exclusion. Valencia et al. (2022) provide evidence that NSCLC with high expression of DSTYK escape T cell killing through autophagy and protection from ROS-induced mitochondrial damage. On the contrary, there is apparently no effect of DSTYK in T cell recruitment, while potential regulation of antigen presentation or generation of immunosuppressive local and systemic microenvironments remains to be investigated.
Could DSTYK be a good novel target? Theoretically, the answer is yes, considering its nature as a kinase and the data supporting a potential therapeutic window obtained in non-transformed cells. Indeed, if DSTYK-driven protective autophagy will be confirmed as a specific trait of (at least a subset of) lung cancers, it would represent a promising metabolic dependency to be explored as a novel targeting strategy. Autophagy interruption is associated with increased tumor T cell killing and could be a potential exploitable pathway, paving the way to combinatorial treatment strategies including autophagy inhibitors together with ICI and chemotherapy in the context of high-CNG DSTYK NSCLC patients. In conclusion, the identification of high-CNG DSTYK as a pathway able to fuel tumor growth and survival by autophagy upregulation and insensitivity to redox imbalance expands the universe of stress-induced vulnerabilities involved in primary resistance to ICI (Fig. 2). Further research is however warranted before DSTYK could become a new player in the ICI arena.
Acknowledgments
L. Roz is supported by the Italian Ministry of Health through grant RF-2016–02362946 and the Italian Association for Cancer Research (AIRC) through Investigator Grant No. IG21431.
Disclosures: R. Ferrara has an advisory role with MSD and BeiGene. No other disclosures were reported.
References
- Borzi, C, et al. 2021. J. Thorac. Oncol. 10.1016/j.jtho.2021.04.005 [DOI] [PubMed] [Google Scholar]
- Carretero, J, et al. 2004. Oncogene. 10.1038/sj.onc.1207502 [DOI] [PubMed] [Google Scholar]
- Deng, J., et al. 2021. Nat. Cancer. 10.1038/s43018-021-00208-6 [DOI] [Google Scholar]
- Ferrara, R., et al. 2018. JAMA Oncol. 10.1001/jamaoncol.2018.3676 [DOI] [Google Scholar]
- Grant, M.J., et al. 2021. Nat. Rev. Clin. Oncol. 10.1038/s41571-021-00520-1 [DOI] [PubMed] [Google Scholar]
- Koyama, S., et al. 2016. Cancer Res. 10.1158/0008-5472.CAN-15-1439 [DOI] [Google Scholar]
- Ma, Y., et al. 2013. Immunity. 10.1016/j.immuni.2013.07.017 [DOI] [Google Scholar]
- Malhotra, J., et al. 2019. Cancer Treat. Res. Commun. 10.1016/j.ctarc.2019.100158 [DOI] [PubMed] [Google Scholar]
- Nadal, E., et al. 2019. J. Thorac. Oncol. 10.1016/j.jtho.2019.08.005 [DOI] [PubMed] [Google Scholar]
- Pore, N., et al. 2021. Cancer Discov. 10.1158/2159-8290.CD-20-1543 [DOI] [Google Scholar]
- Shackelford, D.B., and Shaw R.J.. 2009. Nat. Rev. Cancer. 10.1038/nrc2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim, J.H., et al. 2020. Ann. Oncol. 10.1016/j.annonc.2020.04.004 [DOI] [Google Scholar]
- Skoulidis, F., et al. 2018. Cancer Discov. 10.1158/2159-8290.CD-18-0099 [DOI] [Google Scholar]
- Starobinets, H., et al. 2016. J. Clin. Invest. 10.1172/JCI85705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia, K., et al. 2022. J. Exp Med. 10.1084/jem.20220726 [DOI] [Google Scholar]
- Wang, X., et al. 2019. J. Exp. Clin. Cancer Res. 10.1186/s13046-019-1148-5 [DOI] [Google Scholar]