Abstract
Background
Preventing Salmonella infection and colonization in young birds is key to improving poultry gut health and reducing Salmonella contamination of poultry products and decreasing salmonellosis for human consumption (poultry meat and eggs). Probiotics can improve poultry health. The present study was conducted to investigate the impact of a probiotics, Enterococcus faecium NCIMB 11181 (E. faecium NCIMB 11181) on the intestinal mucosal immune responses, microbiome and barrier function in the presence or absence of Salmonella Typhimurium (S. Typhimurium, ST) infection.
Methods
Two hundred and forty 1-day-old Salmonella-free male broiler chickens (Arbor Acres AA+) were randomly allocated to four groups with 6 replicate cages of 10 birds each. The four experimental groups were follows: (1) negative control (NC), (2) S. Typhimurium, challenged positive control (PC), (3) the E. faecium NCIMB 11181-treated group (EF), (4) the E. faecium NCIMB 11181-treated and S. Typhimurium-challenged group (PEF).
Results
Results indicated that, although continuous feeding E. faecium NCIMB 11181 did not obviously alleviate growth depression caused by S. Typhimurium challenge (P > 0.05), E. faecium NCIMB 11181 addition significantly blocked Salmonella intestinal colonization and translocation (P < 0.05). Moreover, supplemental E. faecium NCIMB 11181 to the infected chickens remarkably attenuated gut morphological structure damage and intestinal cell apoptosis induced by S. Typhimurium infection, as evidenced by increasing gut villous height and reducing intestinal TUNEL-positive cell numbers (P < 0.05). Also, E. faecium NCIMB 11181 administration notably promoting the production of anti-Salmonella antibodies in intestinal mucosa and serum of the infected birds (P < 0.05). Additionally, 16S rRNA sequencing analysis revealed that E. faecium NCIMB 11181 supplementation ameliorated S. Typhimurium infection-induced gut microbial dysbiosis by enriching Lachnospiracease and Alistipes levels, and suppressing Barnesiella abundance. Predicted function analysis indicated that the functional genes of cecal microbiome involved in C5-branched dibasic acid metabolism; valine, leucine and isoleucine biosynthesis; glycerolipid metabolism and lysine biosynthesis were enriched in the infected chickens given E. faecium NCIMB 11181. While alanine, asparate and glutamate metabolism; MAPK signal pathway-yeast; ubiquine and other terpenoid-quinore biosynthesis, protein processing in endoplasmic reticulum; as well as glutathione metabolism were suppressed by E. faecium NCIMB 11181 addition.
Conclusion
Collectively, our data suggested that dietary E. faecium NCIBM 11181 supplementation could ameliorate S. Typhimurium infection-induced gut injury in broiler chickens. Our findings also suggest that E. faecium NCIMB 11181 may serve as an effective non-antibiotic feed additive for improving gut health and controlling Salmonella infection in broiler chickens.
Keywords: Broiler chickens, Enterococcus faecium, Gut health, Salmonella Typhimurium
Background
Salmonella enterica var. Typhimurium (S. Typhimurium, ST), a rod-shaped, flagellated, aerobic and Gram-negative intracellular pathogen, which is one of the most prevalent serotypes of Salmonella in broiler chickens, and cause gastroenteritis in the human by entering the human food chain through animal products, particularly raw poultry products [1, 2]. S. Typhimurium infection in chicks younger than 2 weeks old results in poor growth rate, severe enteric and systemic disease with a high mortality rate along with persistent Salmonella infection in tolerant chickens. While S. Typhimurium infection in older chickens results in asymptomatic cecal colonization and persistent shedding of the organisms in feces, resulting in Salmonella contamination of poultry products, vertical transmission to offspring along with a cumulative economic loss [3–6]. Prolonged persistent infection with S. Typhimurium in the gut of chickens throughout their lifespan could not only alter the development of gut microbiota and have detrimental effect on the overall gut health of the chicken host, especially exposed to stress stimulation or other pathogens challenge. Therefore, reducing S. Typhimurium in the intestinal tract of chickens not only could reduce morbidity and mortality caused by Salmonella infection in young chicks, but also reduce Salmonella prevalence in poultry along with decrease contamination of poultry products and salmonellosis in humans.
Various preventive strategies, which include cleaning up the breeding herbs, general strict hygiene and biosecurity measures in the farm, vaccination, genetic selection of chicken lines with improved immunity, antibiotics, as well as supplementation with feed additives, such as organic acids, essential oils, bacteriophages, prebiotics, probiotics and etc., have been made by farmers to prevent Salmonella infection in poultry [5, 7]. Among above these measures, probiotics were considered as one of the most safest and effective measures employed in controlling Salmonella infections in poultry, because plenty of studies have demonstrated that probiotics could confer the health benefit on the host when administered in adequate amounts in chickens [8–11].
Enterococcus faecium (E. faecium) is a lactic acid bacterium and normal inhabitant in the gut. Although some strains of Enterococcus are pathogenic to human or animals, other strains such as Enterococcus faecium EF1, NCIMB 11181, NCIMB 10415, E. faecium SF68 and E. faecium M-74 are nonpathogenic and often used as commercial probiotics in medicine, food and animal feed, because of their resistance to low pH and bile salts, and encountered in digestion and produced enterococins [12–14]. Previous studies have demonstrated that feed or drinking water supplementation with E. faecium strain facilitates systemic and intestinal local mucosal immune responses [15–18], enhances disease resistance to pathogenic infection, partially prevents or treats diarrhea in pigs [19–23]. Furthermore, the addition of E. faecium to pig directly or indirectly modifies intestinal bacterial communities by increasing the prevalence of beneficial bacteria and reducing pathogenic bacteria load and/or increases growth performances [24–27]. Results from poultry experiments have revealed that supplementation of the diet with E. faecium strain improves growth performance, immune function, eggshell quality and modulates intestinal microflora composition [28–30]. Moreover, probiotics E. faecium supplementation has been reported to regulate intestinal mucosal immune responses and enhance chicken resistance to intestinal pathogen infection, such as Salmonella [31–34], Escherichia coli (E. coli) [35]. Indeed, probiotic strains differ regarding the properties and clinical effects that they elicit even if the strains belong to the same bacterial species. E. faecium strain NCIMB 11181 is currently authorized by the EFSA Panel on additives and products or substances used in animal feed as a supplement for fattening and improving the performance of animals. This strain has been shown to effectively increase daily weight gain, improve feed conversion and gut microbiota composition, together with enhance gut health in pigs [36]. In addition, our previous study had demonstrated that dietary E. faecium NCIMB 11181 addition could improve growth performance, and enhance cellular and humoral immunity of broiler chickens reared under non-challenged conditions [37]. We also found that pre-administration of E. faecium NCIMB 11181 could ameliorate necrotic enteritis-induced intestinal barrier injury in broilers [38], improve growth and reduced the death rate together with maintaining the intestinal integrity in Escherichia coli O78-challenged broiler chickens [39]. Some probiotics have been shown to be useful as antibiotics alternative for the control of some subclinical infections in poultry. Nevertheless, it is unknown whether dietary E. faecium NCIMB 11181 addition could be helpful for protecting intestinal health in broiler chickens infected with S. Typhimurium. Therefore, this study was conducted to investigate the effects of E. faecium NCIMB 11181 addition on Salmonella colonization and invasion, development of intestinal pathological lesions, intestinal immune response, together with intestinal barrier function in broiler chickens challenged with S. Typhimurium. In addition, we further assess the shifts in intestinal microbial community structure induced by dietary treatment and/or ST challenge to explain the possible protective effects of E. faecium NCIMB 11181 addition on broilers infected with ST.
Materials and methodsf
Animal ethics statement
Animal experiment was reviewed and approved by the Animal Care and Use Committee of China Agricultural University (Beijing, P. R. China).
Experimental design, birds, diets and animal management
Two hundred and forty (n = 240) 1-day-old Salmonella-free male broiler chickens (Arbor Acres AA+) were purchased from a local supplier (Beijing Arbor Acres Poultry Breeding Company, Beijing, China). Birds were used to evaluate the protective efficacy of E. faecium NCIMB 11181 feed supplementation against ST infection. Meconium from each individual chicken was collected and checked it for Salmonella negativity using the plating method. Samples were pre-enriched with tetrathionate broth (CM 203–01, Beijing Land Bridge Technology Co., Ltd., Beijing, China) at 37 °C for 24 h, and then streaked on bismuth sulfite agar (CM 207, Beijing Land Bridge Technology Co., Ltd., Beijing, China) to confirm that the chicks were free of Salmonella. Subsequently the chicks were randomly divided into four experimental groups. These 240 two-day-old Salmonella-negative chickens was randomly assigned into four groups including: no additive and no challenge with S. Typhimurium (negative control, NC); no additive but challenged with S. Typhimurium (positive control, PC); E. faecium-supplemented but uninfected (EF); E. faecium-supplemented and infected with S. Typhimurium (PEF). Each group contained six replicate pens with 10 birds per pen and fed a balanced, un-medicated corn and soybean meal-based pelleted diet that contained either 0 or 200 mg/kg E. faecium NCIMB 11181, (viable count ≥ 2 × 109 CFU/g; manufactured by Probiotics International Ltd. Co., UK). To avoid cross-contamination, all uninfected birds were reared in one clean separate room, whereas all infected birds were housed in another room under the same environmental conditions. Antibiotic-free and coccidiostat-free corn-soybean meal-based pelleted diets were formulated to meet or exceed National Research Council (1994) requirements [40] and tested for the presence of Salmonella. The composition and nutrient levels of the basal diet is presented in Table 1. The experimental diet was formulated by mixing the basal diet with 200 g of E. faecium (2 × 109 CFU/g of the product) to reach 4 × 108 CFU/kg of diet. The feed samples were taken and the E. faecium number was counted by Enterococcus faecium agar (bile aesculin azide agar, HB0133-3) to ensure the probiotic dosages were performed correctly.
Table 1.
Composition and nutrient levels of the experimental basal diet (as-fed basis unless stated otherwise, %)
| Items | Content | Items | Content |
|---|---|---|---|
| Ingredients, % | Calculated Nutrient levelsc | ||
| Corn (CP 8.0%) | 61.00 | Metabolizable energy, kcal/kg | 2.9072 |
| Soybean meal (CP 43.0%) | 33.00 | Crude protein, % | 19.24 |
| Soybean oil | 2.00 | Total calcium, % | 0.97 |
| Limestone-calcium carbonate | 1.21 | Available phosphorus, % | 0.43 |
| Calcium hydrogen phosphate | 1.83 | Lysine, % | 1.03 |
| Sodium chloride | 0.30 | Methionine, % | 0.41 |
| DL-Methionine (98%) | 0.10 | ||
| L-Lysine HCL (78%) | 0.10 | ||
| Vitamin premixa | 0.03 | ||
| Mineral premixb | 0.20 | ||
| Choline chloride (50%) | 0.20 | ||
| Ethoxyquin (33%) | 0.03 | ||
| Total, % | 100.00 |
aVitamin premix provided the following per kg of diets: vitamin A (retinylacetate), 12,500 IU; vitamin D3 (cholecalciferol), 2500 IU; vitamin E (DL-a-tocopherol acetate), 18.75 mg; vitamin K3 (menadione sodium bisulfate), 2.65 mg; VB1, 2 mg; VB2, 6 mg; VB6, 6 mg; vitamin B12 (cyanocobalamin), 0.025 mg; biotin, 0.0325 mg; folic acid, 1.25 mg; pantothenic acid, 1.25 mg; nicotinic acid, 50 mg
bMineral premix provided per kilogram of complete diet: Cu (as copper sulfate) 8 mg, Zn (as zinc sulfate) 75 mg, Fe (as ferrous sulfate) 80 mg, Mn (as manganese sulfate) 100 mg, Se (as sodium selenite) 0.15 mg, I (as potassium iodide) 0.35 mg
cCalculated value based on the analyzed data of experimental diets
The chicks were reared on net floor cages in a closed and ventilated house. Each pen had a floor space of 7200 (120 × 60) cm2 and was equipped with a separate feeding trough. Water was supplied through nipple drinkers. Water and feed were provided ad libitum. In accordance with the AA+ Broiler Management Guide, all chicks received continuous light for the first 24 h, and were then maintained under a 23-h light/1-h dark cycle for the remainder of the study. The room temperature was maintained at 33–34 °C on the first 3 days, and then gradually decreased by 2 °C/week until a final room temperature of 22–24 °C of reached. The relative humidity was kept at 60%–70% during the first week and then 50%–60% thereafter.
Salmonella Typhimurium challenge
The Salmonella enterica serovar Typhimurium CVCC 2232 was obtained from the China Veterinary Culture Collection Center (Beijing, China). The frozen culture was recovered by using sterile buffered peptone water (CM201, BPW, Beijing Land Bridge Technology Co., Ltd, Beijing, China). ST pre-culture was transferred to 100 mL of tryptone soy broth (CM201, TSB, Beijing Land Bridge Technology Co., Ltd, Beijing, China) and incubated at 37 °C with orbital shaking for 16 to 18 h. The concentration of viable ST in the culture was counted on bismuth sulfite agar (CM207, BS, Beijing Land Bridge Technology Co., Ltd, Beijing, China) at 37 °C for 24 h and the stock culture was adjusted to a final concentration of 1 × 109 CFU/mL ST. At 10 and 11 days of age, birds in the ST-challenged groups were inoculated with 1 mL of bacterial suspension containing approximately 1 × 109 colony forming units (CFU) of ST suspension by gavage. Unchallenged groups received 1.0 mL of PBS without ST on the same date. Feed was withdrawn from all birds 10 h before challenge.
Measurement of growth performance
Body weight of broiler in each replicate was measured individually at 1, 10 and 18 days of bird age. Average body weight (ABW) and average body weight gain (BWG) were calculated during different periods (during d 1 to 10, and d 11 to 18).
Samples collection
On d 7 after the S. Typhimurium challenge, all birds from each group were euthanized via cervical dislocation, and livers, spleen and left cecal contents of each bird were aseptically harvested and assessed for ST content as soon as possible. At the same time, blood, ileum and cecal contents from only 8 chickens in each treatment group were collected for subsequent analysis. Details are as follows, blood were collected for serum anti-Salmonella specific IgG analysis; the right cecal contents were aseptically collected, snap-frozen in liquid nitrogen and then stored at –80 °C for intestinal microbial 16S rDNA-based analysis. Proximal ileum segments were flushed with 0.05 mol/L PBS, pH 7.2 and fixed in 4% (w/v) polyoxymethylene solution for histological and immunohistochemistry examination. Distal ileum parts were collected, washed for 2 times with ice-cold PBS, and then snap-frozen in liquid nitrogen for mRNA determination. Ileal mucosa from each bird was collected and homogenized in ice-cold PBS (pH 7.2), and centrifuged, then the supernatant was collected and stored at –20 °C for anti-Salmonella specific IgA analysis.
Detection of Salmonella in cecal contents and internal organs
Salmonella numbers in cecal contents and internal organ were determined as described previously [5]. Briefly, samples of liver, spleen and cecal contents were weighed, homogenized in BPW (10% w/v suspensions) for 1 min using a Stomacher respectively and serially diluted tenfold (1:10) with sterile PBS to appropriate levels for Salmonella numeration on xylose lysine tergitol 4 agar (CM219-07, XLT4, Beijing Land Bridge Technology Co., Ltd, Beijing, China) plates containing 100 μg/mL nalidixic acid. The number of black bacterial colonies was determined counted on XLT4 agar plates after incubation for 24 h at 37 °C and expressed as mean ± standard error of the mean log10 CFU/g feces or tissues. Samples that were positive only after enrichment with tetrathionate broth (CM203, TTB, Beijing Land Bridge Technology Co., Ltd, Beijing, China) and then streaked into XLT4 agar plated containing 100 μg/mL nalidixic acid solution were counted as 1 CFU/g, and samples that yielded no Salmonella growth after enrichment were counted as 0 CFU/g. A Salmonella-positive bird was defined based on recovery of Salmonella from any of the internal organs (liver, spleen) studied in an assay. The percent efficacy of protection for a particular group was calculated based on the number of Salmonella-positive birds out of the total number of birds in a group.
Intestinal histology and immunohistochemical staining analysis
Ileal samples were collected and fixed in 4% paraformaldehyde after postmortem examination, and then processed, trimmed, and embedded in paraffin by routine methods. The serial paraffin Sections (5 μm) were prepared and stained with hematoxylin–eosin (HE) for histological (Villous height (VH) and crypt depth (CD), magnification × 40) [5]. In addition, HE-stained 5-µm-thick sections was determined intestinal inflammation or pathological scores as previously described [4] using a light microscope (Leica model DMi8, Leica, Wetzlar, Germany) at magnification of × 200. All scores were obtained in a blinded fashion by two independent investigators. The terminal-deoxynucleoitidyl transferase mediated nick end labeling (TUNEL) assay of ileum-tissue sections was performed by using immunostaining following the same procedure as described in our previous study [39]. The integral optical density (IOD) of TUNEL-positive cells in the ileum was assessed by a digital microscope and camera system (Nikon DS-Ri1, Japan).
Quantitative real-time PCR
Total RNA isolation, reverse transcription, and real-time PCR were carried out as previously described [39]. The primers for real-time PCR are listed in Table 2. The efficiency of all tested genes was between 90% and 110%. All the tissue samples for the cDNA synthesis and in the following PCR amplifications were run in triplicate. Gene expression for immune-related genes (TLR4, MyD88, NF-κB, IFN-γ, IL-1β, IL-6, IL-8, TNF-SF15, TGF-β4, PIgR, A20, Tollip and PI3K), tight junction proteins-related genes (Claudin-1, Occludin, ZO-1, ZO-2 and MLCK) was analyzed using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an endogenous control. The method of 2−ΔΔCt was used to analyze the real-time PCR data [41] and results were expressed as the fold change relative to the average value of the negative control group (the non-treated and non-challenged control).
Table 2.
Sequences of the oligonucleotide primers used for quantitative real-time PCR for immune-related genes expreesiona
| Genes | Forward primer sequence (5′ → 3′) | Reverse primer sequence (5′ → 3′) | GenBank accession No | PCR size, bp |
|---|---|---|---|---|
| TLR4 | F:CCACTATTCGGTTGGTGGAC | R:ACAGCTTCTCAGCAGGCAAT | NM_001030693.1 | 120 |
| MyD88 | F:TGCAAGACCATGAAGAACGA | R:TCACGGCAGCAAGAGAGATT | NM_001030962.3 | 123 |
| NF-kB | F:TGGAGAAGGCTATGCAGCTT | R:CATCCTGGACAGCAGTGAGA | NM_205134.1 | 117 |
| IL-1β | F:TCATCTTCTACCGCCTGGAC | R:GTAGGTGGCGATGTTGACCT | NM_204524.1 | 149 |
| IL-6 | F:GATCCGGCAGATGGTGATAA | R:AGGATGAGGTGCATGGTGAT | NM_204628.1 | 126 |
| IL-8 | F-GGCTTGCTAGGGGAAATGA | R-AGCTGACTCTGACTAGGAAACTGT | NM_205498.1 | 200 |
| TNF-SF15 | F-CCCCTACCCTGTCCCACAA | R-TGAGTACTGCGGAGGGTTCAT | NM_204267.1 | 67 |
| IFN-γ | F:CTTCCTGATGGCGTGAAGA | R:GAGGATCCACCAGCTTCTGT | NM_205149.1 | 127 |
| TGF-β4 | F:AGAGCATTGCCAAGAAGCAC | R:GCAGTAGTCGGTGTCGAGGT | NM_001318456.1 | 119 |
| A20 | F:GAGAACGCAGAGCCTACACC | R:CCAACCTTCTTCCTGCACAT | NM_001277522.1 | 95 |
| Tollip | F:CATGGTACCTGTGGCAATACC | R:GCACTGAGCGGATTACTTCC | NM_001006471 | 122 |
| PI3K | F:AACATCTGGCAAAACCAAGG | R:CTGCAATGCTCCCTTTAAGC | NM001004410 | 150 |
| PIgR | F:ATGAAGCAGAGCCAGGAGAC | R:GAGTAGGCGAGGTCAGCATC | NM001044644.1 | 128 |
| MLCK | F:TTGACATGGAGGTTGTGGAA | R:GAAGTGACGGGACTCCTTGA | NM_001322361.1 | 119 |
| Claudin1 | F:AAGTGCATGGAGGATGACCA | R:GCCACTCTGTTGCCATACCA | NM_001013611.2 | 119 |
| Occludin | F:AGTTCGACACCGACCTGAAG | R:TCCTGGTATTGAGGGCTGTC | NM_205128.1 | 124 |
| ZO-1 | F:ACAGCTCATCACAGCCTCCT | R:TGAAGGGCTTACAGGAATGG | XM_015278981.1 | 125 |
| ZO-2 | F:CACCACCACCTGTTTCTGTG | R:TTCACTCCCTTCCTCTTCCA | NM_204918.1 | 119 |
aPrimers were designed and synthesized by Sango Biotech (Shanghai, China) Co., Ltd. F: forward, R: reverse
TLR Toll-like receptor, MyD88 myeloid differential protein-88, TRAF-6 TNF receptor-associated factor 6, NF-κB nuclear factor kappa-light-chain-enhancer of activated B cells, TNFSF15 tumor necrosis factor superfamily member 15, IL interleukin, IFN-γ interferon γ, Tollip Toll-interacting protein, PI3K phosphatidylinositol 3-kinase, A20 protein A20, SOCS suppressor of cytokine signaling, ZO-1 zonula occludens-1, EGFR epidermal growth factor receptor, GLP-2 lucagon-like peptide-2, IGF-2 insulin-like growth factor-2, TGF- β3 transforming growth factor beta 3
Measurement of anti-Salmonella specific antibody in the intestine and serum
Briefly, ST CVCC 2232 (108 CFU/mL) cells were washed 3 times and lysed by an ultrasonic processor 250 (USA) at 85 W and 30-s intervals on ice for 5 min. The lysed cells were centrifuged at 10,000 × g for 10 min, and the resultant supernatant was collected and stored at –70 °C until use. Flat-bottomed 96-well ELISA microplates (Corning, NY, USA) were coated with 100 μL of 20 μg/mL of the antigen diluted in 0.1 mol/L carbonate-bicarbonate buffer (15 mmol/L Na2CO3, 35 mmol/L NaHCO3, 0.3 mmol/L NaN3) for the measurement of anti–ST specific IgG in the serum and specific IgA in intestinal mucosa homogenate, respectively, using an indirect enzyme-linked immunosorbent assay (ELISA) as described previously [42]. Serum samples were diluted 1:100 and intestinal wash samples were diluted 1:5 in PBST with 1% bovine serum albumin (BSA). Absorbance values (optical density, OD) were read at 450 nm using an automatic ELISA reader (Bio-Tek EL311sx autoreader, Bio-Tek, USA). Each serum sample or intestinal sample was tested in duplicate.
Microbial DNA extraction, 16S rRNA amplification, sequencing and bioinformatic analysis
Microbial DNA was extracted from cecum contents of broilers using QIAamp DNA Stool Mini Kits protocol (Qiagen Inc, Germany) according to the manufacturers’ protocol. The quality and quantity of DNA samples were determined using a Nanodrop ND-1000, and agarose gel electrophoresis was used to confirm the absence of degradation. The bacterial 16S rRNA gene V3-V4 region was amplified using the KAPA HiFi Hotstart ReadyMix PCR kit (Kapa Biosystems, USA) and primers F341 and R806 (F341: 5′-ACTCCTACGGGRSGCAGCAG-3′, R806: 5′-GGACTACVVGGGTATCTAATC-3′). 16S rRNA gene sequencing was performed using the Illumina HiSeq PE250 sequencing platform (Illumina, San Diego, United States) at Biomarker Technology Co., Ltd. (Beijing, China). FLASH (FLASH: fast length adjustment of short reads to improve genome assemblies) was applied for the assembly of resulting 300-bp paired-end reads [43]. Additional sequence read processing, which included quality filtering based on a quality score > 25 and removal of mismatched barcodes and sequences below length thresholds, was performed within QIIME (version 1.9.1) [44]. USEARCH (version 7, 64-bit) was further utilized for denoising and chimera detection [45]. All of the effective reads from each sample were clustered into operational taxonomic units (OTUs) based on a 97% sequence similarity identified by UCLUST in QIIME (version 1.9.1 [44]. Taxonomic classification at different taxonomic levels of OUT sequences were performed by comparing sequences to the GreenGene v13.8 database [46]. Shannon and Simpson indices, Chao1 and ACE estimators were included in α-diversity analysis by using the MOTHUR v1.31.2 [47]. The principal coordinates analysis (PCoA) and partial least squares discriminant analysis (PLS-DA) plots based on weighted and unweighted Unifrac distance matrices were used to estimate pairwise distances among samples and to establish β-diversity. Analysis of similarities (ANOSIM) with 999 permutations was used to detect statistical significances between microbial communities in different groups. This test measures a value of R, normally scaled from 0 to 1, which is based on the average rank similarity among groups and replicates within each group [48]. R = 0 indicates that two groups are similar, whereas R = 1 shows a perfect separation between groups. Linear discriminant analysis (LDA) combined effect size measurements (LEfSe) and non-parametric t-test (with Metastats software) were further employed to identify the biological differences in the microbial composition among groups [49]. LDA was performed from the phylum to genus level, and LDA scores ≥ 4.0, and P-values < 0.05 were selected for plotting and further analysis.
Functional analysis of the gut microbiota
Metagenome functional content from high-quality 16S rDNA was predicted using the phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) software [50], based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology database. Significance analysis was performed by two-way ANOVA using GraphPad Prism 5 (GraphPad Software, San Diego, CA, USA). Data were then analyzed with Statistical Analysis of Taxonomic and Functional Profiles (STAMP) version 2.1.3 [51]. Differentially represented functional pathways (level 2 in hierarchy, representing KEGG pathways) between the two conditions (presented in extended error bar plots) were analyzed with two-sided Welch’s t-test on every pair of means where P < 0.05 was considered significant. Confidence intervals of 95% were obtained by inverting the Welch’s tests.
Statistical analysis
The growth performance data, intestinal structure data, intestinal apoptosis index, intestinal and internal organs salmonella numbers, antibody levels and gene expression data were subjected to two-way ANOVA by using the GLM procedure of the SPSS, version 18.0 (SPSS Inc., Chicago, IL, USA). The model included the main effect of probiotics treatments, Salmonella challenge and their interaction. The relative abundance of microorganisms obtained from 16S rRNA sequencing was analyzed using the Kruskal–Wallis rank sum test to compare the difference between the comparison groups. Significance was set at P < 0.05, and a trend towards significance at P < 0.10 was seen. Data in the tables were expressed as means and pooled SEM.
Results
Growth performance
Data on growth performance including body weight (BW), body weight gain (BWG) of different phase are shown in Table 3. S. Typhimurium challenge resulted in a significant reduce in BW (at d 21) and BWG (during d 12 to 21) compared with the non-challenged birds (P < 0.05), while the addition of E. faecium NCIMB 11181 in feed had no remarkable influence on growth performance (BW and BWG) in broiler chickens irrespective of ST challenge (P > 0.05). No significant difference for mortality rate was observed in all groups during the experimental period (P > 0.05).
Table 3.
Effect of dietary Enterococcus faecium NCIMB 11181 supplementation on broiler chicken performance challenged with Salmonella Typhimurium
| Items | ST1 | Body weight, g/bird | Body weight gain, g/bird | ||
|---|---|---|---|---|---|
| d 10 | d 18 | d 1–10 | d 11–18 | ||
| Dosage, mg/kg | |||||
| 0 | – | 182 | 500 | 144 | 318 |
| 200 | – | 193 | 530 | 154 | 337 |
| 0 | + | 183 | 482 | 144 | 298 |
| 200 | + | 176 | 458 | 138 | 281 |
| SEM2 | 2.78 | 9.87 | 2.73 | 7.60 | |
| Main factors | |||||
| 0 | 183 | 491 | 144 | 308 | |
| 200 | 185 | 494 | 146 | 309 | |
| Non-challenged | 188 | 515a | 149 | 327a | |
| Challenged | 180 | 470b | 141 | 290b | |
| Main factors and Interaction (P value)3 | |||||
| Enterococcus faecium | 0.697 | 0.819 | 0.694 | 0.905 | |
| Challenged | 0.161 | 0.011 | 0.176 | 0.006 | |
| Enterococcus faecium × Challenged | 0.108 | 0.082 | 0.136 | 0.106 | |
1Challenged with Salmonella Typhimurium; –, without ST challenged; +, with ST challenged
2SEM, standard error of the mean
3P-value represent the main effect of the diet, the main effect of ST challenged, and the interaction between the dietary treatments and ST challenged
a,bMeans in the same column without common superscripts differ significantly (P < 0.05)
Intestinal histopathological scores and intestinal cell apoptosis index
A small number of inflammatory cells were found only in the ileum of the ST group, indicating a mild inflammatory reaction at 7 days post S. Typhimurium infection (dpi), while no obvious inflammatory cells infiltration were found in the other three groups (Fig. 1). Moreover, S. Typhimurium strongly increased the inflammation score, and TUNEL-positive cell numbers, significantly decreased villous height, and the ratio of villous height to crypt depth (V/C) (P < 0.05) as compared to the non-infected groups. E. faecium NCIBM 11181 pretreatment significantly decreased the inflammation score and TUNEL-positive cells content, moreover, promoted the growth of villous height irrespective of ST infection (P < 0.05), but the crypt depth and V/C was not affected by E. faecium NCIBM 11181 (P > 0.05) (Table 4; Fig. 2). However, no significant interaction effect was observed in intestinal histopathological scores, cell apoptosis rates at 7 dpi in the ileum among the four treatment groups (P > 0.05).
Fig. 1.

Histopathological changes in the ileum of the NC group (a), the PC group (b), the EF group (c), and the PEF group (d). The histological structure of chicken ileum was observed by H&E staining. Magnification was 40 × , and the scale bar was 20 μm. Red arrow, lymphocytes infiltration; blue arrow, hemorrhage; black arrow, defects of epithelium at the tip of villus. NC non-infected and untreated negative control, PC ST-infected positive control without probiotics addition, EF E. faecium-treated group without ST infection, PEF both E. faecium-treated and ST-infected group. ST Salmonella Typhimurium
Table 4.
Effect of dietary Enterococcus faecium NCIMB 11181 supplementation on intestinal morphology, histopathological scores and intestinal cells apoptosis index (Tunel) of broiler chickens challenged with Salmonella Typhimurium
| Items | ST1 | Villous height, μm | Crypt depth, μm | V/C | HE scores | Tunel |
|---|---|---|---|---|---|---|
| Dosage, mg/kg | ||||||
| 0 | – | 899.2 | 134.5 | 6.81 | 1.46 | 7767 |
| 200 | – | 957.7 | 145.4 | 6.59 | 1.00 | 3514 |
| 0 | + | 743.4 | 133.2 | 5.58 | 4.00 | 7538 |
| 200 | + | 832.2 | 142.6 | 5.85 | 3.40 | 5391 |
| SEM2 | 43.27 | 5.92 | 0.293 | 0.334 | 1275.7 | |
| Main factors | ||||||
| 0 | 821.7a | 133.9 | 6.18 | 2.81a | 7652a | |
| 200 | 895.3b | 144.0 | 6.22 | 2.09b | 4452b | |
| Non-challenged | 928.3a | 140.5 | 6.70a | 1.24b | 5641b | |
| Challenged | 788.7b | 137.8 | 5.71b | 3.70a | 6464a | |
| Main factors and Interaction (P-value)3 | ||||||
| Enterococcus faecium | 0.042 | 0.498 | 0.537 | 0.047 | 0.016 | |
| Challenged | 0.001 | 0.251 | 0.048 | 0.000 | 0.037 | |
| Enterococcus faecium × Challenged | 0.298 | 0.497 | 0.264 | 0.392 | 0.057 | |
1Challenged with Salmonella Typhimurium, –, without ST challenged, + , with ST challenged
2SEM, standard error of the mean
3P-value represent the main effect of the diet, the main effect of ST challenged, and the interaction between the dietary treatments and ST challenged
a,bMeans in the same column without common superscripts differ significantly (P < 0.05)
Fig. 2.

A TUNEL assay in the ileum sections after 7 days of Salmonella Typhimurium (ST) infection in broiler chickens from (a) the NC group, (b) the PC group, (c) the EF group, and (d) the PEF group. The blue color represents the live cells in the jejunal villus, and the brown color represents the apoptotic cells. The red arrow points out a typical TUNEL-positive cell. Magnification = 200 × . NC non-infected and untreated negative control, PC ST-infected positive control without probiotics addition, EF E. faecium-treated group without ST infection, PEF both E. faecium-treated and ST-infected group. ST Salmonella Typhimurium
Salmonella numbers in cecal contents and internal organs
Bacterial translocation occurs when the barrier function of the intestine is impaired. Therefore, we checked the number of Salmonella colonies in the intestines and internal organs. All samples that were taken from uninfected control chicks were negative for S. Typhimurium. The efficacy of E. faecium NCIMB 11181 supplementation in reducing Salmonella colonization and invasion was evaluated by bacterial counting of the ST challenge strain in liver, spleen and cecal content of broiler chickens (Table 5). Table 5 shows that at 7 days post-challenge, Salmonella was detected in the liver, spleen and cecal content only after enrichment in all challenged groups (PC and PEF) with significant differences in Salmonella burden in the liver and cecum compared with un-challenged groups (P < 0.05). Challenged-birds fed diets with E. faecium NCIMB 11181 resulted in significant reduction in cecal Salmonella counts in comparison with the challenged but un-supplemented birds. Salmonella load in the liver of the E. faecium NCIMB 11181-treated birds tended to be lower than that of the single ST-challenged group, though not statistically significantly. After direct enrichment cultures, the number of Salmonella -positive birds (liver) was significantly lower in the E. faecium NCIMB 11181-treated birds (6/29) than in the single challenged control (14/29) at d 7 post-challenge, whereas no significant difference was found in the spleen. The results showed that compared with the ST group, the number of the liver and cecum in the ST-infected birds fed E. faecium NCIMB 11181 group was significantly lower in both liver tissue and cecum content, indicating that E. faecium NCIMB 11181 can reduce bacterial colonization and prevent bacterial translocation.
Table 5.
Effect of dietary Enterococcus faecium NCIMB 11181 supplementation on the number of Salmonella in the liver, spleen and cecal contents (CFU/g) and the percentage of Salmonella-positive bird (liver, spleen) in Salmonella-challenged broilers chickens
| Items | ST1 | Salmonella recovery5 | ||||
|---|---|---|---|---|---|---|
| Dosage, mg/kg | Liver | Spleen | Cecum | |||
| CFU/g4 | No. positive6 | CFU/g4 | No. positive6 | |||
| 0 | – | 0.00 | 0/30 | 0.00 | 0/30 | 0.00c |
| 200 | – | 0.00 | 0/30 | 0.00 | 0/30 | 0.00c |
| 0 | + | 0.74 | 14/29 | 0.29 | 7/29 | 6.85a |
| 200 | + | 0.29 | 6/29 | 0.22 | 5/29 | 5.32b |
| SEM2 | 0.125 | 0.089 | 0.146 | |||
| Main factors | ||||||
| 0 | 0.37 | 0.14 | 3.43 | |||
| 200 | 0.14 | 0.11 | 2.66 | |||
| Non-challenged | 0.00b | 0.00 | 0.00 | |||
| Challenged | 0.52a | 0.25 | 6.09 | |||
| Main factors and Interaction (P-value)3 | ||||||
| Enterococcus faecium | 0.350 | 0.842 | 0.042 | |||
| Challenged | 0.038 | 0.171 | < 0.001 | |||
| Enterococcus faecium × Challenged | 0.350 | 0.842 | 0.031 | |||
1Challenged with Salmonella Typhimurium, –, without ST challenged, + , with ST challenged
2SEM, standard error of the mean
3P-value represent the main effect of the diet, the main effect of ST challenged, and the interaction between the dietary treatments and ST challenged
4CFU/g recovered from tissues expressed in log10 values
5Challenge strain recovery by direct and enrichment cultures of liver, spleen, and cecum of birds
6Number of positive samples per total number of samples after bacterial recovery
a,bMean ± SD in the same column without common superscripts differ significantly (P < 0.05)
Ileum immune-related genes expression
To explain the anti-inflammatory action of E. faecium NCIMB 11181, the mRNA levels of TLR-mediated signal pathway molecules, i.e. TLR4, MyD88, NF-κB, IL-1β, IL-6, IL-8, TNF-α, TGF-β4, IFN-γ, pIgR and negative regulators A20, Tollip and PI3K in the chicken ileum mucosa were measured at 7 days post ST infection. Results showed that the expression levels of MyD88, NF-κB, IFN-γ, pIgR and negative regulators Tollip and PI3K in the chickens of the infected group were significantly upregulated (P < 0.05) than those of the non-challenged control groups (Table 6). ST infection also showed an increased trend for TLR4 (P = 0.099) and TNF-α (P = 0.096) mRNA levels. While only pIgR and Tollip genes expression are remarkably up regulated by the addition of E. faecium NCIMB 11181 to the diets. Significant upregulation of pIgR and Tollip mRNA expression level in the ileum was observed in the E. faecium NCIMB 11181-treated groups compared with the non-supplemented birds. However, there was no significant cooperative effects on immune-related molecules expression between ST challenge and E. faecium NCIMB 11181 addition.
Table 6.
Effect of dietary Enterococcus faecium NCIMB 11181 supplementation on immune-related gene expression in the ileum of broilers challenged with Salmonella Typhimurium
| Items | ST1 | TLR4 | MyD88 | NF-κB | IL-1β | IL-6 | IL-8 | TNF-SF15 | IFN-γ | TGF-β4 | PlgR | A20 | Tollip | PI3K |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dosage, mg/kg | ||||||||||||||
| 0 | – | 1.03 | 1.07 | 1.02 | 1.07 | 1.12 | 1.08 | 1.02 | 1.02 | 1.02 | 1.04 | 1.01 | 1.03 | 1.02 |
| 200 | – | 1.00 | 0.94 | 1.09 | 1.38 | 0.75 | 1.93 | 1.01 | 1.52 | 0.80 | 1.28 | 1.08 | 1.21 | 1.16 |
| 0 | + | 1.33 | 1.23 | 1.19 | 1.85 | 1.14 | 1.96 | 1.13 | 2.61 | 0.77 | 1.60 | 1.11 | 1.26 | 1.56 |
| 200 | + | 1.31 | 1.47 | 1.33 | 2.03 | 0.82 | 1.88 | 1.50 | 2.20 | 0.76 | 2.39 | 1.05 | 1.65 | 1.44 |
| SEM | 0.086 | 0.074 | 0.047 | 0.214 | 0.149 | 0.287 | 0.090 | 0.184 | 0.084 | 0.125 | 0.032 | 0.070 | 0.083 | |
| Main factors2 | ||||||||||||||
| 0 | 1.15 | 1.15 | 1.11 | 1.46 | 1.13 | 1.52 | 1.08 | 1.82 | 0.90 | 1.23b | 1.06 | 1.15b | 1.29 | |
| 200 | 1.16 | 1.20 | 1.21 | 1.71 | 0.78 | 1.91 | 1.26 | 1.83 | 0.78 | 1.71a | 1.06 | 1.43a | 1.30 | |
| Non-challenged | 1.02 | 1.01b | 1.05b | 1.23 | 0.94 | 1.50 | 1.02 | 1.27b | 0.92 | 1.16b | 1.04 | 1.12b | 1.09b | |
| Challenged | 1.32 | 1.35a | 1.26a | 1.94 | 0.98 | 1.92 | 1.32 | 2.43a | 0.77 | 1.99a | 1.08 | 1.46a | 1.50a | |
| Main factors and Interaction (P-value)3 | ||||||||||||||
| Enterococcus faecium | 0.910 | 0.699 | 0.237 | 0.573 | 0.279 | 0.523 | 0.296 | 0.883 | 0.516 | 0.049 | 0.954 | 0.019 | 0.950 | |
| Challenged | 0.099 | 0.017 | 0.024 | 0.110 | 0.898 | 0.493 | 0.096 | 0.001 | 0.417 | 0.004 | 0.572 | 0.006 | 0.014 | |
| Enterococcus faecium × Challenged | 0.975 | 0.170 | 0.714 | 0.882 | 0.940 | 0.447 | 0.282 | 0.116 | 0.552 | 0.520 | 0.320 | 0.339 | 0.426 | |
1Challenged with Salmonella Typhimurium, –, without ST challenged, + , with ST challenged
2SEM, standard error of the mean
3P-value represent the main effect of the diet, the main effect of ST challenged, and the interaction between the dietary treatments and ST challenged
a,bMeans in the same column without common superscripts differ significantly (P < 0.05)
Gene expressions of intestinal tight junction
In order to investigate why the intestinal integrity was changed by S. Typhimurium and E. faecium NCIMB 11181, expression of selected MLCK and tight junction genes was measured by RT-PCR. As showed in Table 7, in comparison with the non-challenged group, Salmonella infection significantly decreased mRNA levels of tight junction Claudin-1, Occludin, ZO-1 and ZO-2 while increased the mRNA level of MLCK (P < 0.05). No significant difference in MLCK, Claudin-1, Occludin, ZO-1 and ZO-2 at the mRNA level was observed in the E. faecium NCIMB 11181 group compared with the un-supplemented group regardless of ST infection (P > 0.05). Moreover, there was no significant interactive effects on MLCK and tight junction expression at 7 d post-infection between ST challenge and E. faecium NCIMB 11181 supplementation.
Table 7.
Effect of dietary Enterococcus faecium NCIMB 11181 supplementation on mRNA abundance of tight junction in the ileum of broiler chickens challenged with Salmonella Typhimurium
| Items | ST1 | MLCK | Claudin-1 | Occludin | ZO-1 | ZO-2 |
|---|---|---|---|---|---|---|
| Dosage, mg/kg | ||||||
| 0 | – | 1.03 | 1.05 | 1.06 | 1.02 | 1.01 |
| 200 | – | 0.85 | 0.86 | 0.74 | 0.98 | 1.04 |
| 0 | + | 1.61 | 0.48 | 0.47 | 0.69 | 0.68 |
| 200 | + | 2.03 | 0.58 | 0.59 | 0.74 | 0.66 |
| SEM2 | 0.153 | 0.070 | 0.074 | 0.052 | 0.054 | |
| Main factors | ||||||
| 0 | 1.30 | 0.79 | 0.79 | 0.87 | 0.86 | |
| 200 | 1.44 | 0.73 | 0.68 | 0.86 | 0.85 | |
| Non-challenged | 0.94b | 0.95a | 0.90a | 1.00a | 1.02a | |
| Challenged | 1.84a | 0.53b | 0.52b | 0.72b | 0.67b | |
| Main factors and Interaction (P -value)3 | ||||||
| Enterococcus faecium | 0.638 | 0.682 | 0.419 | 0.981 | 0.971 | |
| Challenged | 0.002 | 0.001 | 0.007 | 0.005 | 0.000 | |
| Enterococcus faecium × Challenged | 0.233 | 0.197 | 0.082 | 0.660 | 0.763 | |
1Challenged with Salmonella Typhimurium, –, without ST challenged, + , with ST challenged
2SEM, standard error of the mean
3P-value represent the main effect of the diet, the main effect of ST challenged, and the interaction between the dietary treatments and ST challenged
a,bMeans in the same column without common superscripts differ significantly (P < 0.05)
Humoral immune response
The induction of humoral immune responses against the S. Typhimurium-specific antigen was monitored during the weeks after ST infection to evaluate the immune-regulatory capacity of the E. faecium NCIMB 11181. As illustrated in Table 8, intestinal mucosa anti-Salmonella IgA and serum anti-ST specific IgG levels were significantly elevated (P < 0.05) at 7 days following ST infection in broiler chickens. E. faecium NCIMB 11181 addition remarkably promoted intestinal mucosa anti-Salmonella specific antibody production compared with those in the non-supplemented groups (P < 0.05). Moreover, there was significant cooperative effects on intestinal mucosa anti-Salmonella IgA titers between ST challenge and E. faecium NCIMB 11181 supplementation. Infected birds given E. faecium NCIMB 11181 displayed the highest anti-Salmonella IgA content compared with the other three groups (P < 0.05), infected birds alone showed higher IgA content as compared to that of the non-infected groups.
Table 8.
Effect of dietary Enterococcus faecium NCIMB 11181 supplementation on anti-Salmonella specific IgA (OD450nm) of ileum mucosa and serum anti-Salmonella IgG (OD450nm) of broiler chicken challenged with Salmonella Typhimurium
| Items | ST1 | IgA | IgG |
|---|---|---|---|
| Dosage, mg/kg | |||
| 0 | – | 0.07c | 0.45 |
| 200 | – | 0.12c | 0.49 |
| 0 | + | 0.33b | 1.05 |
| 200 | + | 0.48a | 0.84 |
| SEM2 | 0.015 | 0.066 | |
| Main factors | |||
| 0 | 0.20 | 0.75 | |
| 200 | 0.30 | 0.66 | |
| Non-challenged | 0.10 | 0.47b | |
| Challenged | 0.41 | 0.94a | |
| Main factors and Interaction (P-value)3 | |||
| Enterococcus faecium | 0.047 | 0.340 | |
| Challenged | 0.009 | < 0.001 | |
| Enterococcus faecium × Challenged | 0.039 | 0.170 | |
1Challenged with Salmonella Typhimurium, –, without ST challenged, + , with ST challenged
2SEM, standard error of the mean
3P-value represent the main effect of the diet, the main effect of ST challenged, and the interaction between the dietary treatments and ST challenged
a,bMeans in the same column without common superscripts differ significantly (P < 0.05)
Cecal microbiome
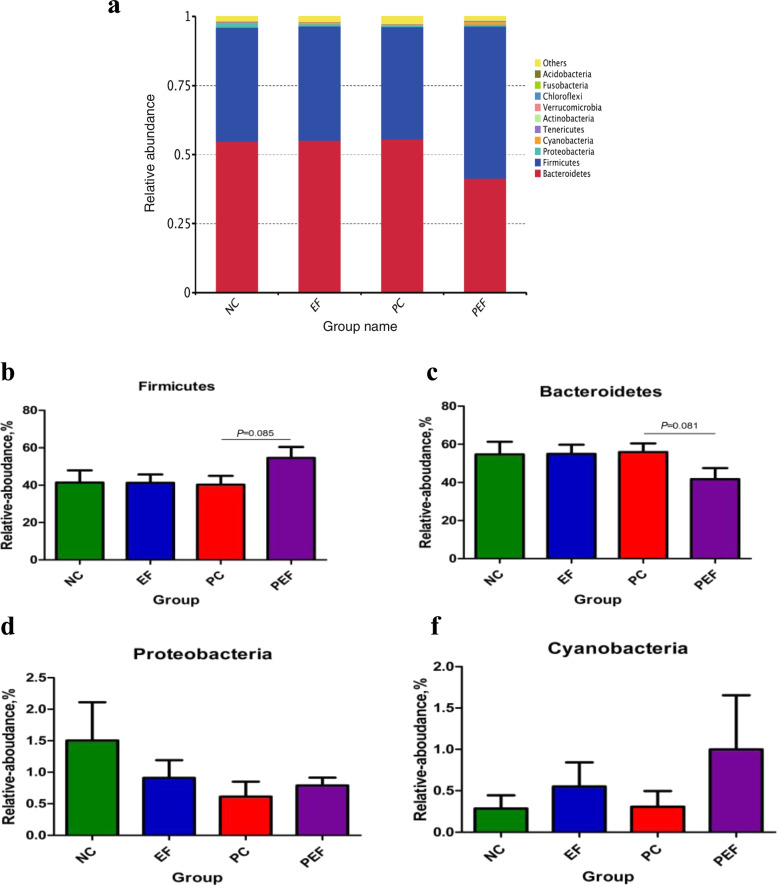
As shown in Table 9, a total of 542,151 high-quality sequences were obtained from 4 groups. All of OTUs were defined at 97% species similarity level, 25,765 OTUs were obtained from cecal contents samples, with an average of 1074 OTUs per sample. The species richness (observed OTUs, Chao, good-coverage) and the community diversity (Shannon, Simpson) was not influenced by ST challenge, E. faecium NCIMB 11181 supplementation or the interaction between E. faecium NCIMB 11181 and ST challenge (Fig. 3a–e). Venn diagram (Fig. 3f) indicated 4786 common core OTUs were shared among all groups, while 17,014, 21,904, 16,981, and 17,776 OTUs were unique to groups NC, PC, EF and PEF, respectively. PCA and PCoA was to be visualized β-diversity (Fig. 4a–b). Results showed that there was no obvious clustering tree associated with dietary treatments or S. Typhimurium infection. Conversely, the infected birds fed E. faecium NCIMB 11181 form a unique cluster separated from all other three groups, especially separated from the single ST-infected birds. The single ST-infected birds displayed little similarity with the other three groups (0 < R = 0.3704 < 1; P = 0.011). However, no distinct separation for cecal microbiota was found between the EF and NC groups. Differences in microbial community abundance at the phylum level between all groups were shown in Fig. 5. At phylum level, the Top 5 dominant phylum includes Firmicute, Bacteriodetes, Cyanobacteria, Proteobacteria and Tenericutes. Higher abundance of Firmicutes (P = 0.085) and lower abundance of Bacteriodetes (P = 0.061) were detected in PEF compared to the PC control. The top 10 microbes at the genus level (Fig. 6) were Alistipes, Barnesiella, Bacterioides, Lachonclostridum, Faecalibacterium, Ruminococcaceae-UCG-014, Ruminiclostridium-5, Ruminococcaceae-UCG-005, Phascolarctobacterium and Subdoligranulum. The abundance of genus Alistipes at 7 days after ST infection was notably decreased, while remarkably was increased by E. faecium NCIMB 11181 addition compared with the PC control (P < 0.05). The infected birds treated with E. faecium NCIMB 11181 showed an increased trend for Lachnoclostridum, while reduced the percentage of the genus Barnesiella (P < 0.05) and display a reduced trend in the abundance of Bacterioides (0.05 < P < 0.1). LEfSe analysis (Fig. 7) highlighted that the infected birds fed E. faecium NCIMB 11181 showed higher abundance in the genus Lachnospiracease, genus Alistipes, Rikenellaceae family and Lachonclostridum, which was similar to the changes of intestinal microbial communities of the non-infected control compared with the single ST-infected control. Under non-infected conditions, E. faecium NCIMB 11181 addition enriched cecal Anaerotruncus and Flavonifractor abundance as compared to the NC group.
Table 9.
Effect of dietary Enterococcus faecium NCIMB 11181 supplementation on α-diversity of cecal microbiota of broiler chickens challenged with Salmonella Typhimurium
| Items | ST1 | Clean reads/tags | Effective reads | OTUs | Chao1 | Goods-coverage | Shannon | Simpson |
|---|---|---|---|---|---|---|---|---|
| Dosage, mg/kg | ||||||||
| 0 | – | 141,844 | 130,059 | 5982 | 5894.15 | 0.98 | 6.41 | 0.92 |
| 200 | – | 157,548 | 139,395 | 7020 | 7289.19 | 0.98 | 6.76 | 0.93 |
| 0 | + | 139,543 | 125,817 | 6338 | 6173.16 | 0.98 | 6.74 | 0.92 |
| 200 | + | 159,097 | 146,880 | 6425 | 6524.50 | 0.98 | 6.76 | 0.94 |
| SEM2 | 20,771 | 17,075 | 1864 | 429.555 | 0.001 | 0.121 | 0.006 | |
| Main factors | ||||||||
| 0 | 140,694 | 127,938 | 6160 | 6033.65 | 0.98 | 6.57 | 0.92 | |
| 200 | 158,318 | 143,138 | 6723 | 6906.84 | 0.98 | 6.76 | 0.94 | |
| Non-challenged | 149,696 | 134,727 | 6501 | 6591.67 | 0.98 | 6.58 | 0.93 | |
| Challenged | 149,320 | 136,349 | 6382 | 6348.83 | 0.98 | 6.75 | 0.93 | |
| Main factors and Interaction (P-value)3 | ||||||||
| Enterococcus faecium | 0.044 | 0.030 | 0.482 | 0.339 | 0.234 | 0.472 | 0.178 | |
| Challenged | 0.964 | 0.806 | 0.892 | 0.788 | 0.713 | 0.509 | 0.942 | |
| Enterococcus faecium × Challenged | 0.816 | 0.379 | 0.550 | 0.565 | 0.898 | 0.514 | 0.775 | |
1Challenged with Salmonella Typhimurium; –, without ST challenged; + , with ST challenged
2SEM, standard error of the mean
3P-value represent the main effect of the diet, the main effect of ST challenged, and the interaction between the dietary treatments and ST challenged
Fig. 3.
Alpha-diversity analysis of cecal microbiota communities among groups at 7 days post ST infection. a Goods-coverage, b Observed species, c Chao 1, d Shannon index, e Simpson index, f Venn diagram showing the shared OTUs by groups. NC non-infected and untreated negative control, PC ST-infected positive control without probiotics addition, EF E. faecium-treated group without ST infection, PEF both E. faecium-treated and ST-infected group. ST Salmonella Typhimurium
Fig. 4.
β-diversity analysis of cecal microbiota communities of broiler challenged with Salmonella Typhimurium (ST) at 7 days post ST infection. (a) Principal component analysis of the caecal microbiota based on weighted Unifrac distance (PCA plot), (b) Principal co-ordinates analysis (PCoA) plot based on unweighted UniFrac distance. NC non-infected and untreated negative control, PC ST-infected positive control without probiotics addition, EF E. faecium-treated group without ST infection, PEF both E. faecium-treated and ST-infected group. ST Salmonella Typhimurium
Fig. 5.
Composition of cecal microbiota of broiler chickens at the phylum level (a) and comparison of relative abundances of the dominant phylum within different groups (b, c, d, e). Values are presented as mean ± SEM. NC non-infected and untreated negative control, PC ST-infected positive control without probiotics addition, EF E. faecium-treated group without ST infection, PEF both E. faecium-treated and ST-infected group. ST Salmonella Typhimurium
Fig. 6.
Composition of cecal microbiota of broiler chickens at the genus level (a) and comparison of relative abundances of the selected dominant bacterial genus in the four groups (b, c, d, e). Values are presented as mean ± SEM. Asterisks (∗P < 0.05, ∗∗P < 0.01) indicate statistical differences between the treatment group and the pc group. NC non-infected and untreated negative control, PC ST-infected positive control without probiotics addition, EF E. faecium-treated group without ST infection, PEF both E. faecium-treated and ST-infected group. ST Salmonella Typhimurium
Fig. 7.

Histogram of the Linear Discriminant Analysis (LDA) score computed for differentially abundant taxa with cut-off LDA score > 2.0. NC non-infected and untreated negative control, PC ST-infected positive control without probiotics addition, EF E. faecium-treated group without ST infection, PEF both E. faecium-treated and ST-infected group. ST Salmonella Typhimurium
Predicting the function of intestinal bacteria based on 16S rDNA data
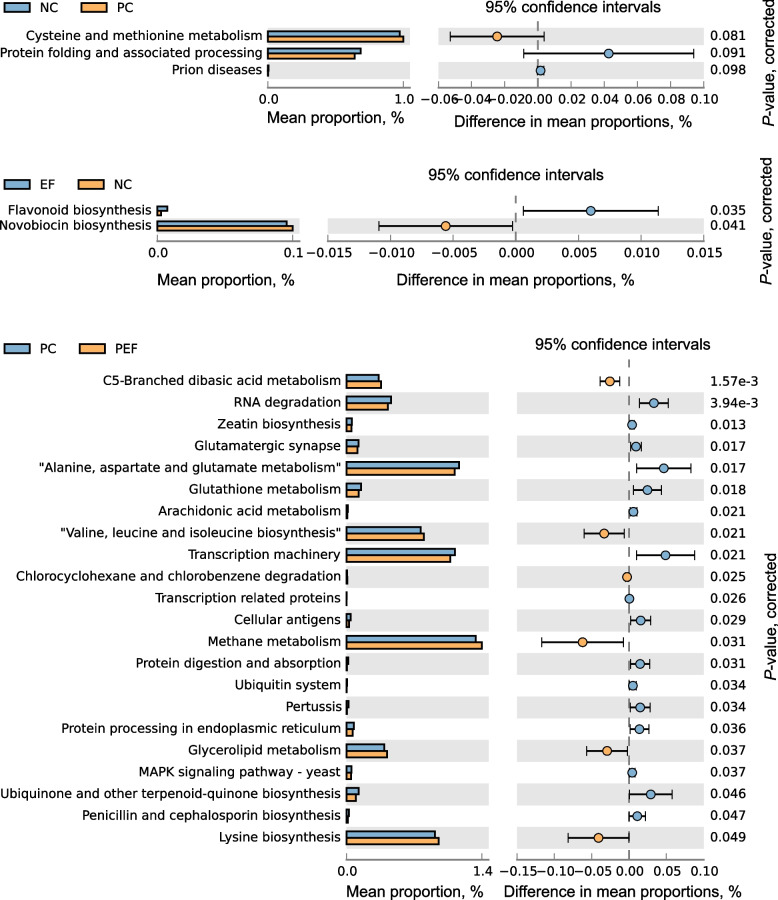
As presented in Fig. 8, STAMP analysis revealed that pretreatment with E. faecium NCIMB 11181 enriched the abundance of functional genes related to flavanoid biosynthesis pathway (P = 0.035) but suppressed novobiocin pathway (P = 0.041) of cecal microbiota of the uninfected birds (at KEGG level 2). Compared with the infected chickens, infected birds given E. faecium NCIMB 11181 had greater numbers of functional genes involved in C5-branched dibasic acid metabolism; valine, leucine and isoleucine biosynthesis; methane metabolism; and glycerolipid metabolism and lysine biosynthesis (P < 0.05). While alanine, asparate and glutamate metabolism; RNA degradation, transcription machinery; MAPK signal pathway-yeast; ubiquine and other terpenoid-quinore biosynthesis; protein processing in endoplasmic reticulum; as well as glutathione metabolism (P < 0.05) of the cecal microbiota were suppressed in the infected birds received E. faecium NCIMB 11181. Thus, dietary supplementation with E. faecium NCIMB 11181 affected important predicted functions of the intestinal microbiota of the S. Typhimurium-infected birds.
Fig. 8.
PICRUSt metagenome inference analysis (level 2) based on 16S rRNA dataset. Mean proportion of functional pathways is illustrated with bar plots and dot plots indicate the differences in mean proportions between two groups based on P-values obtained from two-sided Welch’s t-test. NC non-infected and untreated negative control, PC ST-infected positive control without probiotics addition, EF E. faecium-treated group without ST infection, PEF both E. faecium-treated and ST-infected group. ST Salmonella Typhimurium
Discussion
The current study investigated whether dietary E. faecium NCIMB 11181 addition could be helpful for controlling S. Typhimurium infection and protecting intestinal health in broiler chickens. Our results showed that S. Typhimurium challenge caused a significant negative effect on broiler growth performance, which was similar to previous findings [5, 52, 53]. However, feeding E. faecium NCIMB 11181 had no remarkable growth-improving influences on chicken growth performance regardless of S. Typhimurium challenge. In contrast to our findings, some previous studies have showed that appropriate dose of probiotic Enterococcus faecium supplementation has positively affected broiler performance by increased weight gain or decreased FCR under non-challenged rearing environments [28, 54–57]. Similarly, our previous results demonstrated that dietary supplementation of low dose of E. faecium NCIMB 11181 (50 mg/kg) remarkably improved growth performance, and high dose of E. faecium NCIMB 11181 (200 mg/kg) notably enhanced immunity of broiler chickens under non-challenged conditions [37]. Additionally, Cao et al. [35] reported that E. coli K88-infected broiler chickens fed Enterococcus faecium showed increased growth performance, improved intestinal morphology and cecal microflora. Furthermore, our research group had reported that pretreatment with E. faecium NCIMB 11181 could alleviate the growth suppression caused by Eimeria spp./Clostridium perfringens co-challenge in broilers [38] and E.coli O78-challenged birds [39]. The inconsistent results in growth performance was possible attribute to the difference in biological features and properties of probiotics strain Enterococcus faecium, additive amount of E. faecium, feeding schedules, age of broiler chickens, challenged or not, as well as type of pathogens used for infection. In view of results of growth performance in this study, we suggested that dietary E. faecium NCIMB 11181 addition (200 mg/kg) did not mitigate the adverse effects of Salmonella Typhimurium on broiler growth performance.
Intestinal morphology (villus height, crypt depth and the V/C ratio), lesion scores, histopathological grades, bacterial colonization and translocation together with intestinal cell proliferative and apoptosis indices are important indicators of intestinal health, mucosal barrier function, integrity, permeability and recovery [58]. Additionally, intestinal epithelial cells apical junctional proteins including Claudins, Occludins, ZOs, junctional adhesion molecules and E-cadherins, also play a vital role in regulating intestinal permeability and maintaining gut barrier integrity, defense pathogens infection and inflammation response [59]. In this study, S. Typhimurium infection caused intestinal inflammation and gut wall impairment, as evidenced by the infiltration of inflammatory cells, shorter villus height, reduced V/C ratio, and increased TUNEL-positive cell numbers in the ileum. Also, S. Typhimurium infection damaged intestinal barrier function, as indicated by higher Salmonella load in cecal content and liver, together with downregulated tight junction Claudin-1, Occludin, and ZO-1 mRNA levels and upregulated MLCK mRNA level in the ileum. These observation was partially in consistent with the results of previous studies [5, 52, 53, 60] in chickens, suggesting that S. Typhimurium infections induced the damage of intestinal morphology, promoted villus cells apoptosis, compromised the intestinal barrier integrity, and increased gut permeability of broiler chickens, thereby bacterial translocation. However, these changes caused by S. Typhimurium were partially alleviated by dietary inclusion of E. faecium NCIMB 11181, indicating that E. faecium NCIMB 11181 addition seem to mildly mitigate gut barrier injury induced by S. Typhimurium infection through improving gut morphological structure, decreasing intestinal cells apoptosis and inflammatory cells infiltration. Consistent with our findings, Enterococcus faecium addition not only mitigated gut injury caused by Clostridium perfringens [38] and Escherichia. coli O78 infection [35, 39], but also attenuated intestinal inflammation or gut barrier injury in chicks challenged with Salmonella Enteritidis [31–34] and Salmonella Typhimurium [60], resulting in preventing Salmonella infection in chickens. Based on our findings, we suggested that the addition of the probiotic product E. faecium NCIMB 11181 might be moderate in controlling Salmonella Typhimurium infection in broiler chickens. This protective and anti-Salmonella action induced by probiotics Enterococcus faecium may be attributed to its producing antimicrobial substances such as organic acids, bacteriocin and hydrogen peroxide, and adhesion inhibitors [9, 10, 61].
Intestinal inflammation will lead to destruction of intestinal tight junction and epithelial integrity. Decreased expression of tight junction proteins will aggravate intestinal inflammation. To elucidate why dietary probiotic E. faecium NCIMB 11181 addition could attenuate intestinal barrier impairment induced by S. Typhimurium, we further evaluated the changes in the intestinal mucosal immune and humoral immune responses in ST-infected broiler chickens. TLR-mediated signaling pathways are involved in regulating intestinal mucosal immune defense and epithelial barrier integrity as well as maintaining maintain mucosal and commensal homeostasis [62]. Pro-inflammatory cytokines were reported to increase intestinal permeability and tissue damage via the dysregulation of tight junction proteins [63, 64], while anti-inflammatory cytokines (TGF-β, IL-4 and IL-10), growth factors (EGF, GLP-2 and IGF-2) have been demonstrated to protect intestinal barrier function by regulating tight junction expression and facilitating the repair of damaged gut tissue [65]. In the current study, infection with S. Typhimurium upregulating ileal TLR4, MyD88, NF-κB, IFN-γ, TNF-α, pIgR, and the negative regulators (Tollip and PI3K) mRNA levels at the early stage of infection; which was in similar with observations of previous studies in chickens infected with S. Typhimurium [4, 5, 53, 66–71]. Meanwhile, intestinal mucosa anti-ST IgA and serum anti-ST specific IgG levels were also significantly elevated following ST infection in broiler chickens, which was in consistent with previous results [3, 5, 69, 70]. These observations showed that S. Typhimurium infection triggered intestinal local inflammation, thereby causing the disruption of intestinal barrier and the increase of gut permeability, resulting in Salmonella translocation and systemic inflammatory response. Interestingly, the present study found that E. faecium NCIMB 11181 addition only upregulated pIgR and Tollip mRNA expression level, but did not alter other genes expression profiles in the ileum of TLR-related signal pathway regardless of S. Typhimurium infection. On the contrary, results from previous studies have demonstrated that inclusion of E. faecium in the diet remarkably altered the genes expression profiles of intestinal TLR-mediated signal pathway when subjected to S. Enteritidis challenge in chickens [31–33]. The discrepancy of these findings might be associated with strains and administration dose of probiotics E. faecium; strains type and virulence of challenged Salmonella, sampling time-point and sampled tissues. Tollip is a negative modulator which can suppress activation of TLR-related signal pathway. Increased Tollip expression in the ileum of the chickens treated with E. faecium NCIMB 11181 indicated that E. faecium NCIMB 11181 seem to have the capability to inhibit the over-activation of TLR signal pathway. In addition, administration of E. faecium NCIMB 11181 promoted Salmonella-specific IgA production in intestinal mucosa of Salmonella-infected broiler chickens. Secretory IgA (sIgA) and its transcytosis receptor, polymeric immunoglobulin receptor (pIgR), along with mucus form the first lines of intestinal mucosal defenses, mainly defensing or neutralizing pathogenic bacteria and enteric toxins [72]. In this study, increased intestinal pIgR expression here may mean more mucosal secretory IgA antibody production in the gut, which help in reducing cecal Salmonella load and facilitating Salmonella elimination from the gut lumen during the recovery phase of infection. Higher levels of ileal sIgA together with lower Salmonella burden in the intestine and liver of the infected chickens fed E. faecium NCIMB 11181, showing that feeding E. faecium NCIMB 11181 had ability to provide protection against Salmonella infection through enhancing specific sIgA production. The findings further showed that pretreatment with probiotic E. faecium NCIMB 11181 could mildly alleviate S. Typhimurium-induced intestinal injury in chickens, possibly associated with promoting the production of sIgA in the gut.
The chicken gastrointestinal tract is colonized by trillions of microorganisms, constituting a dynamic ecosystem with significant impacts on host metabolism, productivity, immune responses and health status including gut health. Consequently, modulation of the gut microbiota and modification of the intestinal microenvironment could assist in preventing animal colonization by the pathogen [73]. More importantly, changes in gut microbe populations may be closely related to the degree of intestinal inflammation and disease resistance, which is one of the characteristics of S. Typhimurium infection [1, 73]. In the current study, our results revealed that neither S. Typhimurium infection nor dietary probiotics E. faecium NCIMB 11181 treatments significantly altered α-diversity of chicken caecal microbiota, indicated that the caecal microbiota diversity remained relatively stable. Nevertheless, S. Typhimurium infection significantly modified the indigenous microbiota composition and relative abundance of some bacterial species in the cecum of chickens, as showed by increasing relative abundance of Barnesiella, whereas decreasing Alistipes abundances, which was similar to results of Azcarate-Peril et al. [1]. In similar to our findings, S. Enteritidis infection also disturbed microbial composition of gut microbiota of chickens, as evidenced by expanding relative abundance of potential harmful bacteria such as Enterobacteriaceae, whereas decreasing potential beneficial bacteria including (i.e., butyrate-producing bacterira Lachnospiracease, Bifidobacterium and Lactobacillus) abundances [74]. Thus, our findings showed that S. Typhimurium infection disrupted microbial composition of gut microbiota of chickens, besides impairment in intestinal barrier structure. Additionally, taxonomic analysis showed that E. faecium NCIMB 11181 addition enriched the relative abundance of the phylum Firmicutes and the genera Alistipes while suppressed the relative population of the genera Barnesiella of the cecal microbiota of the infected birds when comparing with the single S. Typhimurium infected control. LEFsE analysis also indicated that the infected birds received E. faecium NCIMB 11181 showed higher abundance in the genus Lachnospiracease, Alistipes, Rikenellaceae family and Lachonclostridum, which was similar to the changes of intestinal microbial communities of the non-infected control compared with the single ST-infected control. Our study demonstrated that E. faecium NCIBM 11181 administration modified the structure of the gut microbiome of the Salmonella-infected chickens. Alistipes, a sub-branch genus of the Bacteroidetes phylum, exhibited protective effects against some diseases, including liver fibrosis, inflammatory colitis, cancer immunotherapy, and cardiovascular disease [75]. Butyrate-producing Lachnospiraceae positively correlated with good FCR performance and gut health [76], and an increase in its abundance has shown to limit expansion of aerobic enteric pathogens, reduce inflammatory diseases and prevent gut barrier dysfunction in systemic chronic lower-grade inflammation mice [77]. The genus Barnesiella also was reported to negatively be linked with anti-inflammatory responses but strongly correlated with proinflammatory responses in chickens [78]. Thus, higher proportion of Lachnospiracease, Alistipes and lower abundance of Barnesiella, accompanied by reduced Salmonella carrier in the cecum of Salmoenlla-challenged broiler chickens following E. faecium NCIMB 11181 administration, suggesting that pretreatment with probiotics E. faecium NCIMB 11181 could control Salmonella infection via improving gut microbiome. These data also indicated that the barrier-protecting effects of E. faecium NCIMB 11181 is possibly associated with the improvement of intestinal microbiome.
PICRUSt analysis revealed that functional genes related to C5-branched dibasic acid metabolism; valine, leucine and isoleucine biosynthesis; methane metabolism; glycerolipid metabolism and lysine biosynthesis were enriched; whereas functional genes involved in alanine, asparate and glutamate metabolism; RNA degradation, transcription machinery; MAPK signal pathway-yeast; ubiquine and other terpenoid-quinore biosynthesis, protein processing in endoplasmic reticulum; as well as glutathione metabolism were depleted in the cecum of the Salmonella-infected chickens given E. faecium NCIMB 11181. The findings indicated that probiotics E. faecium NCIMB 11181 administration altered functional changes of intestinal microbiota induced by S. Typhimurium infection. Amino acids supply was associated with energy supply, immune regulation and damage repair of gut cells, especially under challenge conditions [79]. Salmonella infection induced up-regulation of glycolytic process and the catabolism of amino acids at the middle and later of infection, resulting in exhaustion of energy and amino acids in chickens [80]. Such increase in amino acids biosynthesis and C5-branched dibasic acid metabolism might suggest that feeding probiotics E. faecium NCIMB 11181 to Salmonella-infected chickens potentially promoted amino acids biosynthesis processes of intestinal microbe, thereby contributing to energy supply of gut cells and repair of gut barrier impairment as well as dampening of Salmonella-induced intestinal inflammatory responses in broiler chickens. Asparate and glutamate metabolism, glutathione metabolism and ubiquinone biosynthesis, protein processing in endoplasmic reticulum, and other terpenoid-quinore biosynthesis reported to be involved in host nucleotide synthesis, energy metabolism of mitochondria, and redox status. Over-activation of these pathways meant that host was being exposed to stress stimulus and in a state of the imbalance of redox, resulting in oxidative stress. Suppressed pathways of functional genes of gut microbiota obtained in infected chickens after feeding E. faecium NCIMB 11181 indicated that E. faecium NCIMB 11181 pretreatment could lighten oxidative stress induced by Salmonella infection. We further identified that inhibition of MAPK signal pathway-yeast in the infected birds after feeding E. faecium NCIMB 11181, meant that E. faecium NCIMB 11181 could prevent Salmonella-induced intestinal inflammation. Hence, the increased amino acids biosynthesis, and the decreased MAPK signal pathway and redox pathway suggested that E. faecium NCIBM 11181 administration might play a role in alleviating Salmonella-induced intestinal inflammation by regulating gut microbiome, which in turn affects amino acids biosynthesis, redox pathway metabolism and MAPK signal pathway. Further experiments would be essential to confirm this possibility.
Conclusion
In summary, Salmonella Typhimurium infection disrupted the balance of gut microbiota, induced intestinal inflammation and downregulated tight junction proteins genes expression, resulting in gut barrier injury and bacterial translocation in broiler chickens. Nevertheless, continuous feeding Enterococcus faecium NCIMB 11181 appear to alleviate Salmonella Typhimurium-induced gut injury mildly through modulating gut microbiota composition, promoting intestinal specific anti-Salmonella IgA production, along with inhibiting intestinal cells apoptosis. The results provide new information on the critical role played by dietary Enterococcus faecium NCIBM 11181 in controlling Salmonella infection in broiler chickens.
Abbreviations
- A20
Protein A20
- BW
Body weight
- BWG
Body weight gain
- CD
Crypt depth
- CFU
Colony-forming unit
- E. coli
Escherichia coli
- E. faecium
Enterococcus faecium
- EGF
Epidermal growth factor
- FCR
Feed conversion ratio
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- GLP-2
Glucagon-like peptide-2
- HE
Hematoxylin–eosin
- HFD
High fat diet
- IFN-γ
Interferon- γ
- IGF-2
Insulin-like growth factor-2
- IL
Interleukin
- IOD
Integral optical density
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LDA
Linear discriminant analysis (LDA)
- LEfSe
Linear discriminant analysis combined effect size measurements (LEfSe)
- MAPK
Mitogen-activated protein kinase
- MLCK
Myosin light chain kinase
- MyD88
Myeloid differential protein-88
- NF-κB
Nuclear factor kappalight-chain-enhancer of activated B cells
- NRC
National Research Council
- PCoA
Principal coordinate analysis
- PCR
Polymerase chain reaction
- PIgR
Polymeric immunoglobulin receptor
- PI3K
Phosphatidylinositol 3-kinase
- PLS-DA
Partial least squares discriminant analysis
- SIgA
Secretory immunoglobulin A
- TGF-β4
Transforming growth factor beta 3
- TLR
Toll-like receptor
- TNFSF15
Tumor necrosis factor superfamily 15
- Tollip
Toll-interacting protein
- TUNEL
Terminal-deoxynucleoitidyl transferase mediated nick end labeling
- VH
Villous height
- VH:CD ratio
The ratio of villus height to crypt depth
- ZO-1
Zonula occludens-1
Authors’ contributions
ZW conceptualized the project and designed the study. WRZ, FSG and YJS contributed to performing animal experiments, analyzing samples, doing statistical analysis, and preparing the original manuscript draft. YMG and ZW made insightful edits for the manuscript. All authors reviewed the manuscript and approved the final submission.
Funding
This work was supported by the grant from Talent Plan of Zaozhuang City (2022), Shandong, China. The company had no role in conducting the research, generating the data, interpreting the results, or writing the manuscript.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Experimental procedures and animal use were approved by the China Agricultural University, Animal Care and Use Committee, Beijing, P. R. China.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Azcarate-Peril MA, Butz N, Cadenas MB, Koci M, Ballou A, Mendoza M, et al. An attenuated Salmonella enterica serovar typhimurium strain and galacto-oligosaccharides accelerate clearance of Salmonella infections in poultry through modifications to the gut microbiome. Appl Environ Microb. 2018;84(5):e02526–2543. doi: 10.1128/AEM.02526-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shah DH, Paul NC, Sischo WC, Crespo R, Guard J. Population dynamics and antimicrobial resistance of the most prevalent poultry-associated Salmonella serotypes. Poult Sci. 2017;96(3):687–702. doi: 10.3382/ps/pew342. [DOI] [PubMed] [Google Scholar]
- 3.Beal RK, Wigley P, Powers C, Hulme SD, Barrow PA, Smith AL. Age at primary infection with Salmonella enterica serovar typhimurium in the chicken influences persistence of infection and subsequent immunity to re-challenge. Vet Immunol Immunopathol. 2004;100(3–4):151–164. doi: 10.1016/j.vetimm.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Withanage GS, Kaiser P, Wigley P, Powers C, Mastroeni P, Brooks H, et al. Rapid expression of chemokines and proinflammatory cytokines in newly hatched chickens infected with Salmonella enterica serovar typhimurium. Infect Immun. 2004;72(4):2152–2159. doi: 10.1128/IAI.72.4.2152-2159.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shao Y, Guo Y, Wang Z. Beta-1,3/1,6-glucan alleviated intestinal mucosal barrier impairment of broiler chickens challenged with Salmonella enterica serovar typhimurium. Poult Sci. 2013;92(7):1764–1773. doi: 10.3382/ps.2013-03029. [DOI] [PubMed] [Google Scholar]
- 6.Dar MA, Ahmad SM, Bhat SA, Ahmed R, Urwat U, Mumtaz PT, et al. Salmonella Typhimurium in poultry: A review. World’s Poult Sci J. 2019;73(2):345–54. 10.1017/s0043933917000204.
- 7.Vandeplas S, Dubois Dauphin R, Beckers Y, Thonart P, Thewis A. Salmonella in chicken: Current and developing strategies to reduce contamination at farm level. J Food Prot. 2010;73(4):774–785. doi: 10.4315/0362-028x-73.4.774. [DOI] [PubMed] [Google Scholar]
- 8.Chen CY, Tsen HY, Lin CL, Yu B, Chen CS. Oral administration of a combination of select lactic acid bacteria strains to reduce the Salmonella invasion and inflammation of broiler chicks. Poult Sci. 2012;91(9):2139–2147. doi: 10.3382/ps.2012-02237. [DOI] [PubMed] [Google Scholar]
- 9.Mathipa MG, Thantsha MS. Probiotic engineering: Towards development of robust probiotic strains with enhanced functional properties and for targeted control of enteric pathogens. Gut Pathog. 2017;9:28. 10.1186/s13099-017-0178-9. [DOI] [PMC free article] [PubMed]
- 10.Nava GM, Bielke LR, Callaway TR, Castaneda MP. Probiotic alternatives to reduce gastrointestinal infections: The poultry experience. Anim Health Res Rev. 2005;6(1):105–118. doi: 10.1079/ahr2005103. [DOI] [PubMed] [Google Scholar]
- 11.Tellez G, Pixley C, Wolfenden RE, Layton SL, Hargis BM. Probiotics/direct fed microbials for Salmonella control in poultry. Food Res Int. 2012;45(2):628–633. doi: 10.1016/j.foodres.2011.03.047. [DOI] [Google Scholar]
- 12.Franz CM, Huch M, Abriouel H, Holzapfel W, Galvez A. Enterococci as probiotics and their implications in food safety. Int J Food Microbiol. 2011;151(2):125–140. doi: 10.1016/j.ijfoodmicro.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 13.Zommiti M, Cambronel M, Maillot O, Barreau M, Sebei K, Feuilloley M, et al. Evaluation of probiotic properties and safety of Enterococcus faecium isolated from artisanal tunisian meat “dried ossban.” Front Microbiol. 2018;9:1685. 10.3389/fmicb.2018.01685. [DOI] [PMC free article] [PubMed]
- 14.Hanchi H, Mottawea W, Sebei K, Hammami R. The genus Enterococcus: Between probiotic potential and safety concerns-an update. Front Microbiol. 2018;9:1791. doi: 10.3389/fmicb.2018.01791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scharek L, Guth J, Reiter K, Weyrauch KD, Taras D, Schwerk P, et al. Influence of a probiotic Enterococcus faecium strain on development of the immune system of sows and piglets. Vet Immunol Immunopathol. 2005;105(1–2):151–161. doi: 10.1016/j.vetimm.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 16.Scharek-Tedin L, Filter M, Taras D, Wrede P, Schmidt MF. Influence of an Enterococcus faecium probiotic on the development of peyer’s patches b cells in piglets. Arch Anim Nutr. 2009;63(5):343–55. 10.1080/17450390903052771. [DOI] [PubMed]
- 17.Siepert B, Reinhardt N, Kreuzer S, Bondzio A, Twardziok S, Brockmann G, et al. Enterococcus faecium NCIMB 10415 supplementation affects intestinal immune-associated gene expression in post-weaning piglets. Vet Immunol Immunopathol. 2014;157:65–77. [DOI] [PubMed]
- 18.Lan R, Kim I. Enterococcus faecium supplementation in sows during gestation and lactation improves the performance of sucking piglets. Vet Med Sci. 2020;6:92–99. doi: 10.1002/vms3.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bednorz C, Guenther S, Oelgeschlager K, Kinnemann B, Pieper R, Hartmann S, et al. Feeding the probiotic Enterococcus faecium strain NCIMB 10415 to piglets specifically reduces the number of Escherichia coli pathotypes that adhere to the gut mucosa. Appl Environ Microbiol. 2013;79(24):7896–7904. doi: 10.1128/AEM.03138-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szabó I, Wieler LH, Tedin K, Tedin LS, Taras D, Hensel A, et al. Influence of a probiotic strain of Enterococcus faecium on Salmonella enterica serovar Typhimurium DE104 infection in a porcine animal infection model. Appl Environ Microbiol. 2009;75:2621–8. [DOI] [PMC free article] [PubMed]
- 21.Peng X, Wang R, Hu L, Zhou Q, Liu Y, Yang M, et al. Enterococcus faecium NCIMB 10415 administration improves the intestinal health and immunity in neonatal piglets infected by enterotoxigenic Escherichia coli k88. J Anim Sci Biotechnol. 2019;10:72. doi: 10.1186/s40104-019-0376-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeyner A, Boldt E. Effects of a probiotic Enterococcus faecium strain supplemented from birth to weaning on diarrhoea patterns and performance of piglets. J Anim Physiol Anim Nutr (Berl) 2006;90(1–2):25–31. doi: 10.1111/j.1439-0396.2005.00615.x. [DOI] [PubMed] [Google Scholar]
- 23.Buesing K, Zeyner A. Effects of oral Enterococcus faecium strain DSM 10663 NCIMB 10415 on diarrhoea patterns and performance of sucking piglets. Beneficial Microbes. 2015;6(1):41–44. doi: 10.3920/bm2014.0008. [DOI] [PubMed] [Google Scholar]
- 24.Broom LJ, Miller HM, Kerr KG, Knapp JS. Effects of zinc oxide and Enterococcus faecium SF68 dietary supplementation on the performance, intestinal microbiota and immune status of weaned piglets. Res Vet Sci. 2006;80(1):45–54. doi: 10.1016/j.rvsc.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 25.Mallo JJ, Rioperez J, Honrubia P. The addition of Enterococcus faecium to diet improves piglet’s intestinal microbiota and performance. Livest Sci. 2010;133(1–3):176–8. 10.1016/j.livsci.2010.06.057.
- 26.Wang YB, Du W, Fu AK, Zhang XP, Huang Y, Lee KH, et al. Intestinal microbiota and oral administration of Enterococcus faecium associated with the growth performance of new-born piglets. Benef Microbes. 2016;7(4):529–538. doi: 10.3920/BM2015.0099. [DOI] [PubMed] [Google Scholar]
- 27.Xie YH, Zhang CY, Wang LX, Shang QH, Zhang GG, Yang WR. Effects of dietary supplementation of Enterococcus faecium on growth performance, intestinal morphology, and selected microbial populations of piglets. Livest Sci. 2018;210:111–117. doi: 10.1016/j.livsci.2018.02.010. [DOI] [Google Scholar]
- 28.Lan RX, Lee SI, Kim IH. Effects of Enterococcus faecium SIB 120 on growth performance, blood parameters, relative organ weight, breast muscle meat quality, excreta microbiota shedding, and noxious gas emission in broilers. Poult Sci. 2017;96(9):3246–3253. doi: 10.3382/ps/pex101. [DOI] [PubMed] [Google Scholar]
- 29.Gheisar MM, Hosseindoust A, Kim IH. Effects of dietary Enterococcus faecium on growth performance, carcass characteristics, faecal microbiota, and blood profile in broilers. Vet Med Czech. 2016;61:28–34. doi: 10.17221/8680-VETMED. [DOI] [Google Scholar]
- 30.Park JW, Jeong JS, Lee SI, Kim IH. Effect of dietary supplementation with a probiotic (Enterococcus faecium) on production performance, excreta microflora, ammonia emission, and nutrient utilization in ISA brown laying hens. Poult Sci. 2016;95(12):2829–2835. doi: 10.3382/ps/pew241. [DOI] [PubMed] [Google Scholar]
- 31.Kuritza LN, Loureco MC, Miglino L, Pickler L, Kraieski AL, Santin E. Effects of Enterococcus faecium on diet in the dynamics of CD4+ and CD8+ cell infiltration in the intestinal mucosa of broilers challenged with Salmonella Minnesota. Int J Poult Sci. 2013;12:523–528. doi: 10.3923/ijps.2013.523.528. [DOI] [Google Scholar]
- 32.Karaffova V, Bobikova K, Husakova E, Levkut M, Herich R, Revajova V, et al. Interaction of TGF-β4 and IL-17 with IgA secretion in the intestine of chickens fed with E. faecium AL41 and challenged with S. Enteritidis. Res Vet Sci. 2015;100:75–9. doi: 10.1016/j.rvsc.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 33.Levkut M, Revajova V, Laukova A, Sevcikova Z, Spisakova V, Faixova Z, et al. Leukocytic responses and intestinal mucin dynamics of broilers protected with Enterococcus faecium EF55 and challenged with Salmonella Enteritidis. Res Vet Sci. 2012;93:195–201. [DOI] [PubMed]
- 34.Beirao BCB, Ingberman M, Favaro C, Jr, Mesa D, Bittencourt LC, Fascina VB, et al. Effect of an Enterococcus faecium probiotic on specific IgA following live Salmonella Enteritidis vaccination of layer chickens. Avian Pathol. 2018;47(3):325–333. doi: 10.1080/03079457.2018.1450487. [DOI] [PubMed] [Google Scholar]
- 35.Cao GT, Zeng XF, Chen AG, Zhou L, Zhang L, Xiao YP, et al. Effects of a probiotic, Enterococcus faecium, on growth performance, intestinal morphology, immune response, and cecal microflora in broiler chickens challenged with Escherichia coli k88. Poult Sci. 2013;92(11):2949–2955. doi: 10.3382/ps.2013-03366. [DOI] [PubMed] [Google Scholar]
- 36.Chae JP, Pajarillo EAB, Oh JK, Kim H, Kang DK. Revealing the combined effects of lactulose and probiotic enterococci on the swine faecal microbiota using 454 pyrosequencing. Microb Biotechnol. 2016;9:486–495. doi: 10.1111/1751-7915.12370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu Y, Zhen W, Geng Y, Wang Z, Guo Y. Effects of dietary Enterococcus faecium NCIMB 11181 supplementation on growth performance and cellular and humoral immune responses in broiler chickens. Poult Sci. 2019;98(1):150–163. doi: 10.3382/ps/pey368. [DOI] [PubMed] [Google Scholar]
- 38.Wu YY, Zhen WR, Geng YQ, Wang Z, Guo YM. Pretreatment with probiotic Enterococcus faecium NCIMB 11181 ameliorates necrotic enteritis-induced intestinal barrier injury in broiler chickens. Sci Rep. 2019;9:10256. doi: 10.1038/s41598-019-46578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang L, Luo L, Zhang Y, Wang Z, Xia Z. Effects of the dietary probiotic, Enterococcus faecium NCIMB 11181, on the intestinal barrier and system immune status in Escherichia coli O78-challenged broiler chickens. Probiotics Antimicrob Proteins. 2019;11(3):946–956. doi: 10.1007/s12602-018-9434-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.NRC. Nutrient requirements of poultry. 9th rev. Washington, DC: National Academy Press; 1994.
- 41.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta c(t)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 42.Shao Y, Wang Z, Tian X, Guo Y, Zhang H. Yeast beta-d-glucans induced antimicrobial peptide expressions against Salmonella infection in broiler chickens. Int J Biol Macromol. 2016;85:573–584. doi: 10.1016/j.ijbiomac.2016.01.031. [DOI] [PubMed] [Google Scholar]
- 43.Magoc T, Salzberg SL. Flash: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. Qiime allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. Uchime improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clarke KR. Non-parametric multivariate analysis of changes in community structure. Aust J Ecol. 1993;18:117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x. [DOI] [Google Scholar]
- 49.White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol. 2009;5(4):e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31(9):814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. Stamp: Statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30(21):3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fasina YO, Hoerr FJ, McKee SR, Conner DE. Influence of Salmonella enterica serovar typhimurium infection on intestinal goblet cells and villous morphology in broiler chicks. Avian Dis. 2010;54(2):841–847. doi: 10.1637/9055-090809-Reg.1. [DOI] [PubMed] [Google Scholar]
- 53.Marcq C, Cox E, Szalo IM, Thewis A, Beckers Y. Salmonella Typhimurium oral challenge model in mature broilers: Bacteriological, immunological, and growth performance aspects. Poult Sci. 2011;90(1):59–67. doi: 10.3382/ps.2010-01017. [DOI] [PubMed] [Google Scholar]
- 54.Luo J, Zheng A, Meng K, Chang W, Bai Y, Li K, et al. Proteome changes in the intestinal mucosa of broiler (gallus gallus) activated by probiotic Enterococcus faecium. J Proteomics. 2013;91:226–241. doi: 10.1016/j.jprot.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 55.Samli HE, Dezcan S, Koc F, Ozduven ML, Okur AA, Senkoylu N. Effects of Enterococcus faecium supplementation and floor type on performance, morphology of erythrocytes and intestinal microbiota in broiler chickens. Br Poult Sci. 2010;51(4):564–568. doi: 10.1080/00071668.2010.507241. [DOI] [PubMed] [Google Scholar]
- 56.Samli HE, Senkoylu N, Koc F, Kanter M, Agma A. Effects of Enterococcus faecium and dried whey on broiler performance, gut histomorphology and intestinal microbiota. Arch Anim Nutr. 2007;61(1):42–49. doi: 10.1080/17450390601106655. [DOI] [PubMed] [Google Scholar]
- 57.Mountzouris KC, Tsitrsikos P, Palamidi I, Arvaniti A, Mohnl M, Schatzmayr G, et al. Effects of probiotic inclusion levels in broiler nutrition on growth performance, nutrient digestibility, plasma immunoglobulins, and cecal microflora composition. Poult Sci. 2010;89(1):58–67. doi: 10.3382/ps.2009-00308. [DOI] [PubMed] [Google Scholar]
- 58.Watson AJ, Chu S, Sieck L, Gerasimenko O, Bullen T, Campbell F, et al. Epithelial barrier function in vivo is sustained despite gaps in epithelial layers. Gastroenterology. 2005;129(3):902–912. doi: 10.1053/j.gastro.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 59.Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9(11):799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 60.Zhang H, Wang M, Jia J, Zhao J, Radebe SM, Yu Q. The protective effect of E.faecium on S. Typhimurium infection induced damage to intestinal mucosa. Front Vet Sci. 2021;8:740424.10.3389/fvets.2021.740424. [DOI] [PMC free article] [PubMed]
- 61.Levkut M, Pistl J, Laukova A, Revajova V, Herich R, Sevcikova Z, et al. Antimicrobial activity of Enterococcus faecium EF 55 against Salmonella Enteritidis in chicks. Acta Vet Hung. 2009;57(1):13–24. doi: 10.1556/AVet.57.2009.1.2. [DOI] [PubMed] [Google Scholar]
- 62.Kawai T, Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int Immunol. 2009;21(4):317–337. doi: 10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Al-Sadi R, Boivin M, Ma T. Mechanism of cytokine modulation of epithelial tight junction barrier. Front Biosci (Landmark Ed) 2009;14(7):2765–2778. doi: 10.2741/3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Edelblum KL, Turner JR. The tight junction in inflammatory disease: Communication breakdown. Curr Opin Pharmacol. 2009;9(6):715–720. doi: 10.1016/j.coph.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee SH. Intestinal permeability regulation by tight junction: Implication on inflammatory bowel diseases. Intest Res. 2015;13(1):11–18. doi: 10.5217/ir.2015.13.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rychlik I, Elsheimer-Matulova M, Kyrova K. Gene expression in the chicken caecum in response to infections with non-typhoid Salmonella. Vet Res. 2014;45:119. doi: 10.1186/s13567-014-0119-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fasina YO, Holt PS, Moran ET, Moore RW, Conner DE, McKee SR. Intestinal cytokine response of commercial source broiler chicks to Salmonella Typhimurium infection. Poult Sci. 2008;87(7):1335–1346. doi: 10.3382/ps.2007-00526. [DOI] [PubMed] [Google Scholar]
- 68.Khan S, Chousalkar KK. Transcriptome profiling analysis of caeca in chicks challenged with Salmonella Typhimurium reveals differential expression of genes involved in host mucosal immune response. Appl Microbiol Biotechnol. 2020;104(21):9327–9342. doi: 10.1007/s00253-020-10887-3. [DOI] [PubMed] [Google Scholar]
- 69.Beal RK, Powers C, Wigley P, Barrow PA, Smith AL. Temporal dynamics of the cellular, humoral and cytokine responses in chickens during primary and secondary infection with Salmonella enterica serovar Typhimurium. Avian Pathol. 2004;33(1):25–33. doi: 10.1080/03079450310001636282. [DOI] [PubMed] [Google Scholar]
- 70.Dar MA, Urwat U, Ahmad SM, Ahmad R, Kashoo ZA, Dar TA, et al. Gene expression and antibody response in chicken against Salmonella Typhimurium challenge. Poult Sci. 2019;98(5):2008–2013. doi: 10.3382/ps/pey560. [DOI] [PubMed] [Google Scholar]
- 71.Withanage GS, Wigley P, Kaiser P, Mastroeni P, Brooks H, Powers C, et al. Cytokine and chemokine responses associated with clearance of a primary Salmonella enterica serovar Typhimurium infection in the chicken and in protective immunity to rechallenge. Infect Immun. 2005;73(8):5173–5182. doi: 10.1128/IAI.73.8.5173-5182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Johansen FE, Kaetzel CS. Regulation of the polymeric immunoglobulin receptor and IgA transport: New advances in environmental factors that stimulate pIgR expression and its role in mucosal immunity. Mucosal Immunol. 2011;4(6):598–602. doi: 10.1038/mi.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stanley D, Hughes RJ, Moore RJ. Microbiota of the chicken gastrointestinal tract: Influence on health, productivity and disease. Appl Microbiol Biotechnol. 2014;98(10):4301–4310. doi: 10.1007/s00253-014-5646-2. [DOI] [PubMed] [Google Scholar]
- 74.Videnska P, Sisak F, Havlickova H, Faldynova M, Rychlik I. Influence of Salmonella enterica serovar Enteritidis infection on the composition of chicken cecal microbiota. BMC Vet Res. 2013;9:140. doi: 10.1186/1746-6148-9-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Parker BJ, Wearsch PA, Veloo ACM, Rodriguez-Palacios A. The genus alistipes: Gut bacteria with emerging implications to inflammation, cancer, and mental health. Front Immunol. 2020;11:906. doi: 10.3389/fimmu.2020.00906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stanley D, Hughes RJ, Geier MS, Moore RJ. Bacteria within the gastrointestinal tract microbiota correlated with improved growth and feed conversion: Challenges presented for the identification of performance enhancing probiotic bacteria. Front Microbiol. 2016;7:187. doi: 10.3389/fmicb.2016.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rivera-Chavez F, Zhang LF, Faber F, Lopez CA, Byndloss MX, Olsan EE, et al. Depletion of butyrate-producing Clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe. 2016;19(4):443–454. doi: 10.1016/j.chom.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu Y, Li Q, Liu J, Liu Y, Xu Y, Zhang R, et al. Integrating serum metabolome and gut microbiome to evaluate the benefits of lauric acid on lipopolysaccharide challenged broilers. Front Immunol. 2021;12:759323. 10.3389/fimmu.2021.759323. [DOI] [PMC free article] [PubMed]
- 79.Gaggini M, Carli F, Rosso C, Buzzigoli E, Marietti M, Della Latta V, et al. Altered amino acid concentrations in NAFLD: Impact of obesity and insulin resistance. Hepatology. 2018;67(1):145–158. doi: 10.1002/hep.29465. [DOI] [PubMed] [Google Scholar]
- 80.Kogut MH, Arsenault RJ. Immunometabolic phenotype alterations associated with the induction of disease tolerance and persistent asymptomatic infection of Salmonella in the chicken intestine. Front Immunol. 2017;8:372. doi: 10.3389/fimmu.2017.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.