ABSTRACT
Hepatitis delta virus (HDV) is a defective satellite virus that uses hepatitis B virus (HBV) envelope proteins to form its virions and infect hepatocytes via the HBV receptors. Concomitant HDV/HBV infection continues to be a major health problem, with at least 25 million people chronically infected worldwide. N6-methyladenine (m6A) modification of cellular and viral RNAs is the most prevalent internal modification that occurs cotranscriptionally, and this modification regulates various biological processes. We have previously described a wider range of functional roles of m6A methylation of HBV RNAs, including its imminent regulatory role in the encapsidation of pregenomic RNA. In this study, we present evidence that m6A methylation also plays an important role in the HDV life cycle. Using the methylated RNA immunoprecipitation (MeRIP) assay, we identified that the intracellular HDV genome and antigenome are m6A methylated in HDV- and HBV-coinfected primary human hepatocytes and HepG2 cell expressing sodium taurocholate cotransporting polypeptide (NTCP), while the extracellular HDV genome is not m6A methylated. We observed that HDV genome and delta antigen levels are significantly decreased in the absence of METTL3/14, while the extracellular HDV genome levels are increased by depletion of METTL3/14. Importantly, YTHDF1, an m6A reader protein, interacts with the m6A-methylated HDV genome and inhibits the interaction between the HDV genome and antigens. Thus, m6A of the HDV genome negatively regulates virion production by inhibiting the interaction of the HDV genome with delta antigens through the recruitment of YTHDF1. This is the first study that provides insight into the functional roles of m6A in the HDV life cycle.
IMPORTANCE The functional roles of N6-methyladenine (m6A) modifications in the HBV life cycle have been recently highlighted. Here, we investigated the functional role of m6A modification in the HDV life cycle. HDV is a subviral agent of HBV, as it uses HBV envelope proteins to form its virions. We found that m6A methylation also occurs in the intracellular HDV genome and antigenome but not in the extracellular HDV genome. The m6A modification of the HDV genome recruits m6A reader protein (YTHDF1) onto the viral genome. The association of YTHDF1 with the HDV genome abrogates the interaction of delta antigens with the HDV genome and inhibits virion assembly. This study describes the unique effects of m6A on regulation of the HDV life cycle.
KEYWORDS: hepatitis delta virus, N6-methyladenine modification, virion assembly, HDV life cycle
INTRODUCTION
Hepatitis delta virus is a significant human pathogen and a natural subviral agent of the hepatitis B virus (HBV) (1 and 2). At least 25 million people are estimated to be chronically coinfected with both HBV and HDV (3). Superinfection with HDV in individuals chronically infected with HBV inflicts additional liver damage associated with accelerated liver disease, cirrhosis, liver failure, and hepatocellular carcinoma (4). Currently, there are no effective antiviral treatments or therapeutic vaccines for chronic HDV infection (5). A better understanding of the HDV life cycle may facilitate the identification of novel promising drug targets to interfere with the infectious process.
HDV virions contain a ribonucleoprotein (RNP) complex composed of a circular negative-sense single-stranded circular RNA (HDV genome) of almost 1.7 kb and the only viral proteins, called small and large delta antigens (S-HDAg and L-HDAg) (6–8). Because HDV virions are coated with HBV envelope proteins, they enter the hepatocytes via the HBV receptor sodium taurocholate cotransporting polypeptide (NTCP) (9). After infection, HDV replicates via RNA-directed RNA synthesis through an RNA intermediate called the antigenome, which is present in the nucleus but not in the HDV virions (10, 11). The HDV antigenome is fully complementary to the HDV genome; it bears the open reading frames (ORFs) for delta antigens but does not serve for their translation (12). HDV replicates by host RNA polymerase II (RNA Pol II) via a rolling-circle mechanism, in which the circular genomic RNA serves as the template for the synthesis of the antigenome and mRNA and the circular antigenome serves as the template for the synthesis of the genome (13–15). Small self-cleaving RNA sequences (ribozymes) identified in the genome and antigenome are responsible for the cleavage of multimeric RNA molecules that arise during the rolling-circle amplification and represent the only enzymatic activity reported for HDV (16). The HDV genome forms an extended quasi-double-stranded RNA (quasi-dsRNA) rod-like structure containing numerous internal loops and bulges (6). HDAg binds to the unbranched quasi-double-stranded HDV RNA as multimers, and these RNP complexes protect the genome and antigenome RNAs from host nuclease activity (17). Its association with HDAg would depend more on its secondary structure than on its primary sequence (18). Although S-HDAg and L-HDAg bind to the HDV genome and antigenome and differ in a very short sequence at the C-terminus, they have very distinct roles in the viral life cycles (19–26). S-HDAg is required for viral genome replication and mRNA transcription (19–22), while L-HDAg is essential for virus particle assembly but negatively inhibits viral genome replication (23–26). Recently, it was shown that myrcludex B, a myristoylated peptide of 47 amino acids derived from the preS1-domain of the HBV large envelope protein, efficiently blocks the entry of HBV and HDV in cell lines and a liver-humanized mice model; this drug is in clinical trials (27).
Chemical modifications of RNAs have been previously described and intensely characterized for tRNAs and rRNAs (28). Of all the chemical modifications of mRNAs, the N6-methyladenine (m6A) modification is the most abundant and well characterized, functionally implicated in a wide range of biological processes, including differentiation, sex determination, metabolic disorders, cancer, and viral infections (29–34). The consensus sequence for m6A is 5′-DRA*CH-3′ (where D = A, G, or U; R = G or A; A = methylated adenosine; C = C; and H = A, C, or U) (35). The m6A modification is a dynamic cotranscriptional process that is reversibly catalyzed by m6A “writers,” including the complex of methyltransferase-like protein 3 (METTL3), METTL14, and Wilms’ tumor 1-associating protein (WTAP), and “erasers,” such as fat mass- and obesity-associated protein (FTO) and ALKBH5 (33, 34, 36–38). The m6A-modified mRNA directly interacts with YT521-B homology (YTH) domain family proteins (YTHDF1 to YTHDF3) (33, 34, 39, 40). These proteins regulate the RNA stability and translation of m6A-methylated RNA. Functionally, YTHDF1 enhances m6A-modified mRNA translation, while YTHDF2 promotes target mRNA degradation. YTHDF3 regulates the degradation and translation of m6A-modified mRNA through cooperation with YTHDF1 and YTHDF2. In particular, m6A modification of RNA viral genomes and transcripts of DNA viruses has been characterized (41–49). m6A modification of viral RNAs has been shown to affect various aspects of the viral life cycle and associated pathogenesis. m6A methyltransferases (METTL3/14) and m6A reader proteins (YTHDFs) play important roles in regulating the life cycles of both DNA and RNA viruses (42, 46–49). While m6A modification of viral transcripts has been identified, its effects on viral replication and translation continue to be characterized.
Previously, we extensively studied the role of m6A modification in the HBV life cycle and found that m6A methylation plays diverse roles of regulation (44, 46–53). In this study, we investigated the functional role of m6A methylation of HDV RNAs in their life cycle. We observed that m6A methylation occurs in the HDV genome and antigenome during HBV/HDV coinfection as well as HDV transfection. However, virion-associated (extracellular) HDV genomes are not m6A methylated. The silencing of cellular m6A methyltransferases (METTL3/METTL14) and YTHDF1 reduces the intracellular HDV genome levels but increases HDV virion production. Important, m6A methylation of the HDV genome abrogates the interaction between HDV genome and delta antigens by recruiting YTHDF1 and negatively regulates virion production. This study reveals novel functional roles of m6A methylation in the HDV life cycle.
RESULTS
m6A methylation occurs in the intracellular HDV genome and antigenome.
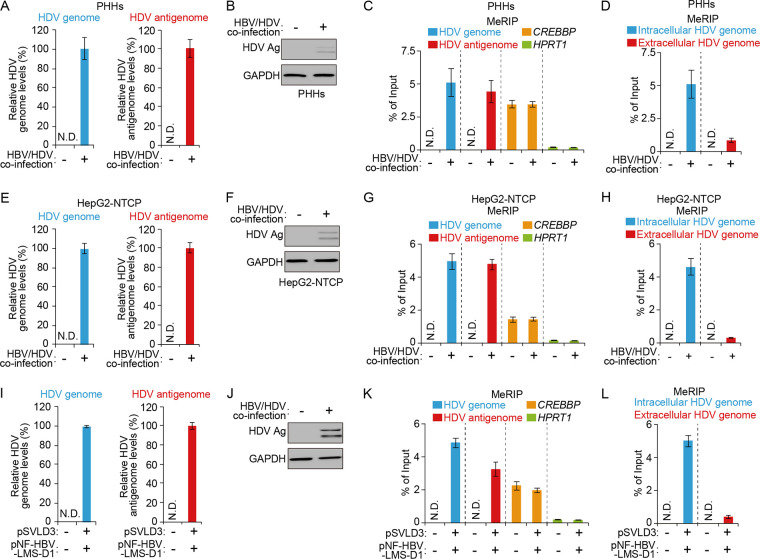
m6A modification has been identified in several viral RNAs, including HBV, but its function on HDV RNAs has not been studied. We first determined whether m6A methylation occurs in HDV RNAs using a methylated RNA immunoprecipitation (MeRIP) assay with an m6A-specific antibody, as described previously in HBV and HDV coinfection systems (42, 47). Primary human hepatocytes (PHHs) were coinfected with HBV and HDV infectious particles, and HDV replication was evaluated using a strand-specific real-time reverse-transcription quantitative PCR (RT-qPCR) assay (22) and Western blot analysis (Fig. 1A and B). For the MeRIP assay, total RNA was extracted and immunoprecipitated using an m6A-specific antibody. Immunoprecipitated RNA was then analyzed using RT-qPCR with primers to specific HDV RNAs. Interestingly, we found that the intracellular HDV genome and antigenome RNAs contained m6A modifications (Fig. 1C). In the MeRIP-RT-qPCR assay (Fig. 1C), cellular CREBBP mRNA was used as the positive m6A control and HPRT1 mRNA was used as a nonmethylated RNA control (39). Importantly, however, the m6A methylation level of the extracellular HDV genome was dramatically lower than that of the intracellular HDV genome (Fig. 1D). We next validated these results using HepG2-NTCP cells coinfected with HBV and HDV infectious particles and found similar results: we observed that m6A methylation occurs in the intracellular HDV genome and antigenome in this system but not in the extracellular HDV genome (Fig. 1E to H). Since HBV infection is essential for HDV replication and affects the m6A methylation profile of cellular RNAs, the m6A methylation of HDV RNAs may be affected by HBV replication. To confirm this possibility, we conducted a MeRIP assay using total RNA from HepG2 cells cotransfected with plasmid pSVLD3 (which initiates HDV genome replication) and plasmid pNF-HBV-LMS-D1 (which expresses HBV envelope proteins) (54). In the transfection system, the intracellular HDV genome and antigenome also contained m6A modification, but the HDV genome from virions did not contain m6A methylation, suggesting that m6A modification occurs in the HDV genome and antigenome without HBV expression. These results clearly show that the intracellular HDV genome and antigenome contain m6A methylation, while the extracellular HDV genome is not m6A methylated.
FIG 1.
Intracellular HDV genome and antigenome contain m6A methylation. (Top) PHHs were HBV/HDV coinfected and analyses conducted to determine (A) HDV genome (left) and antigenome (right) levels, (B) protein expression levels, (C) intracellular m6A-methylated RNA levels (CREBBP and HPRT1 were analyzed as the positive and negative controls, respectively), and (D) m6A methylation levels of the extracellular HDV genome. (Middle) HepG2-NTCP cells were HBV/HDV coinfected and analyses conducted to assess (E) HDV genome (left) and antigenome (right) levels, (F) protein expression levels, and (G and H) m6A-methylated RNA levels. (Bottom) HepG2 cells were cotransfected with plasmids pSVLD3 and pNF-HBV-LMSD1 and analyses conducted to determine (I) HDV genome (left) and antigenome (right) levels, (J) protein expression levels, and (K and L) m6A-methylated RNA levels. In all panels, data are mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (unpaired one-tailed Student’s t test). N.D., not detected; Ag, antigen.
m6A-methylated HDV genome is bound by YTHDF1 protein.
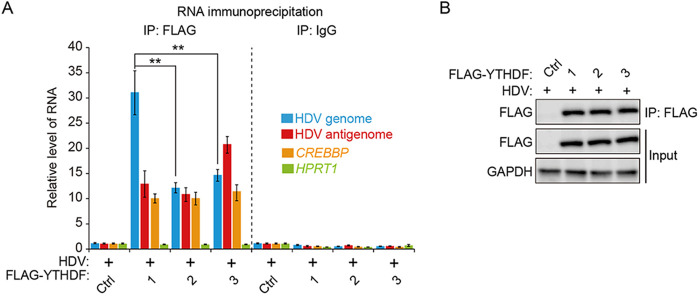
We next analyzed whether HDV RNAs are recognized by the YTHDF proteins, because the m6A reader proteins (YTHDF1 to YTHDF3) are cellular m6A RNA-binding proteins that regulate the RNA stability and translation activity of m6A-methylated RNAs (33, 34, 39, 40). We transfected HBV-/HDV-coinfected HepG2-NTCP cells with each plasmid encoding FLAG-YTHDF1, FLAG-YTHDF2, or FLAG-YTHDF3 protein, respectively. These cells were irradiated with UV to capture the RNA-protein complexes (55), and cellular lysates were extracted and subjected to immunoprecipitation using anti-FLAG-antibodies. Immunoprecipitated RNA was purified from RNA-protein complexes and analyzed using RT-qPCR assay. The results of the RT-qPCR analysis showed that both the HDV genome and antigenome were enriched in all YTHDF immunoprecipitates relative to the IgG control, suggesting that the HDV genome and antigenome are bound by the YTHDF1, YTHDF2, and YTHDF3 proteins (Fig. 2) Cellular CREBBP and HPRT1 RNAs were used for m6A-positive and -negative controls, respectively (Fig. 2A) (39). Importantly, the HDV genome was more strongly recognized by YTHDF1 than by YTHDF2 and YTHDF3, suggesting that YTHDF1 may play an important role in the HDV life cycle. Taken together, these results reveal that m6A methylation occurs in the intracellular HDV genome and antigenome and binds cellular m6A reader proteins (YTHDF1, YTHDF2, and YTHDF3).
FIG 2.
HDV genome and antigenome bind to m6A reader proteins. HepG2-NTCP cells were HBV/HDV coinfected and transfected with FLAG-YTHDF (1, 2, and 3) plasmids. Cellular lysates were subjected to immunoprecipitation with an anti-FLAG antibody. Immunoprecipitated RNA was quantified by RT-qPCR as fold enrichment relative to the control (A). Proteins were analyzed by Western blotting (B). In all panels, data are mean ± SD. **, P < 0.01 by unpaired one-tailed Student’s t test. IP, immunoprecipitation; ctrl, control.
m6A RNA methylation modulates the HDV life cycle.
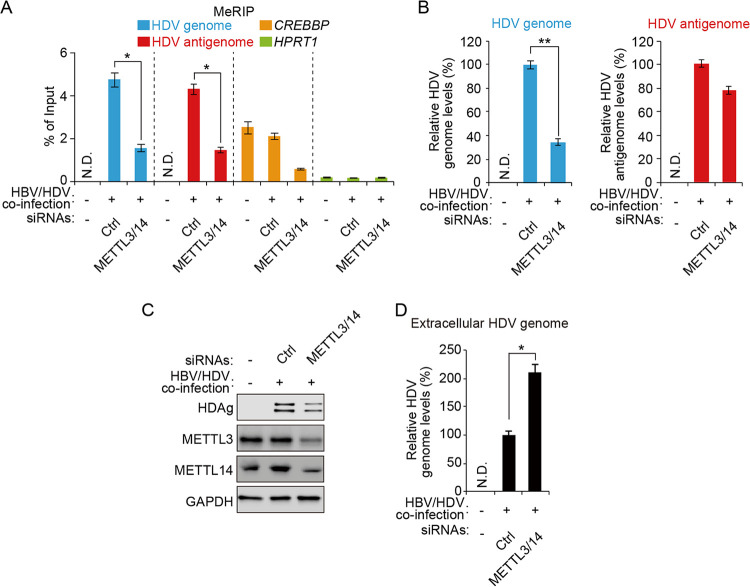
m6A methylation is installed onto RNA by a complex of m6A methyltransferases (METLL3 and METTL14) (36). To determine whether the cellular m6A methyltransferases regulate the HDV life cycle by placing m6A methylation on HDV RNAs, we depleted both METTL3 and METTL14 in HBV- and HDV-coinfected HepG2-NTCP cells using small interfering RNAs (siRNAs) and observed that the m6A modification levels of HDV RNAs were decreased by the silencing of METTL3/METTL14, suggesting that cellular m6A methyltransferases (METTL3 and METTL14) catalyze m6A modification in the HDV genome and antigenome (Fig. 3A). We then analyzed the HDV RNA and protein levels in METTL3- and METTL14-depleted cells and found that the absence of these proteins resulted in decreased accumulation of the intracellular HDV genome and antigen compared with control siRNA-treated cells, as revealed by RT-qPCR and immunoblotting (Fig. 3B and C). However, the HDV antigenome levels were slightly decreased in METTL3- and METTL14-depleted cells (Fig. 3B), suggesting that m6A methylation may not regulate HDV antigenome levels. We next analyzed the effect of silencing METTL3 and METTL14 on the extracellular HDV genome levels, because the unmethylated HDV genome accumulated inside the virions (Fig. 1D). The results showed that depletion of METTL3 and METTL14 significantly increased the accumulation of the extracellular HDV genome (Fig. 3D). Taken together, these results suggest that m6A methylation of HDV RNAs positively regulates the intracellular HDV genome and antigen expression but negatively affects HDV virion production.
FIG 3.
Cellular m6A methyltransferases regulate the HDV life cycle. HepG2-NTCP cells were HBV/HDV coinfected and transfected with control or METTL3/14 siRNAs. After 3 days, total RNA and cellular lysates were extracted to (A) conduct MeRIP-RT-qPCR and (B) assess HDV genome (left) and antigenome (right) levels and (C) protein expression levels. (D) Extracellular HDV genome was extracted and analyzed by RT-qPCR. In all panels, data are mean ± SD. *, P < 0.05; **, P < 0.01 (unpaired one-tailed Student’s t test). N.D., not detected.
YTHDF1 protein affects the HDV life cycle.
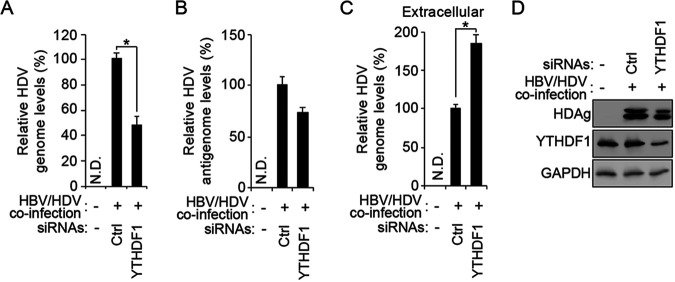
After determining that m6A methyltransferases positively regulate HDV genome and antigen accumulation but negatively affect the extracellular HDV genome levels, we next tested whether m6A reader proteins could similarly affect the HDV life cycle. In this study, we focused on the effect of YTHDF1 in the regulation of the m6A-mediated HDV life cycle, because the HDV genome was strongly recognized by YTHDF1, as shown in Fig. 2, and YTHDF1 promotes the translation of m6A-methylated RNAs (33, 34). We depleted YTHDF1 protein in HBV- and HDV-coinfected HepG2 cells using siRNAs. We found that the depletion of YTHDF1 significantly reduced the intracellular HDV genome and antigen levels but only slightly decreased the antigenome levels (Fig. 4), suggesting that YTHDF1 protein also positively regulates HDV genome and antigen accumulation, similar to m6A methyltransferases. Importantly, the depletion of YTHDF1 also significantly increased the extracellular HDV genome levels (Fig. 4C), indicating that YTHDF1 protein affects the HDV life cycle. Together, these results suggest that the m6A methylation of the HDV genome regulates viral replication and assembly by recruiting YTHDF1 protein.
FIG 4.
YTHDF1, m6A reader protein, regulates the HDV life cycle. HepG2-NTCP cells were HBV/HDV coinfected and transfected with control or YTHDF1 siRNAs. Using RT-qPCR, levels of HDV genome (A), antigenome (B), and extracellular genome (C) were quantified; proteins were analyzed by Western blotting (D). In all panels, data are mean ± SD. *, P < 0.05 by unpaired one-tailed Student’s t test. N.D., not detected.
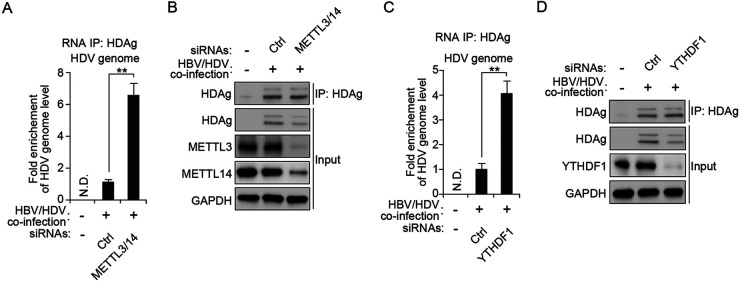
m6A RNA methylation of the HDV genome affects it’s interaction with HDV antigen.
Both S-HDAg and L-HDAg interact with the HDV genome, and these interactions are important in viral RNA replication and assembly (19–26). In particular, the interaction of L-HDAg with the HDV genome promotes the assembly of the HDV RNP complex with HBV envelope proteins in order to form virions and to facilitate their egress (23). In this context, we tested whether m6A methylation of the HDV genome affects its interactions with HDV antigens to regulate the viral life cycle. We depleted METTL3 and METTL14 in HBV- and HDV-coinfected HepG2-NTCP cells using siRNAs and irradiated these cells with UV to capture the RNA-protein complex (55). Cellular lysates were extracted and immunoprecipitated using an antidelta antigen-antibody. The interaction between HDV RNA and antigen was evaluated using RT-qPCR analysis. Interestingly, we found that the silencing of METTL3 and METTL14 significantly increased the interactions between the HDV genome and delta antigens (Fig. 5A and B), suggesting that m6A modification of the HDV genome inhibits the interaction of delta antigens to regulate viral RNA replication and virion assembly. We further validated whether YTHDF1 could similarly affect the interaction of the HDV genome with delta antigens and found that depletion of YTHDF1 also increased the interaction between HDV RNAs and antigens (Fig. 5C and D). Therefore, these results reveal that m6A methylation of the HDV genome decreases the interaction of HDV antigens by recruiting YTHDF1 protein. The reduced occupancy of delta antigens on the HDV genome affects the HDV genome and virion production levels.
FIG 5.
m6A methylation of the HDV genome inhibits the interaction with HDV antigen by recruiting YTHDF1. (A and B) HepG2-NTCP cells were HBV/HDV coinfected and transfected with control or METTL3/14 siRNAs. Cellular lysates were extracted and immunoprecipitated with an antidelta antigen-antibody. The immunoprecipitated HDV genome was quantified using RT-qPCR (A). Proteins were analyzed by Western blotting (B). (C and D) HepG2-NTCP cells were HBV/HDV coinfected and transfected with control or YTHDF1 siRNAs. Cellular lysates were extracted and immunoprecipitated with an anti-HDV antigen-antibody. The immunoprecipitated HDV genome was quantified using RT-qPCR (C). Proteins were analyzed by Western blotting (D). In all panels, data are mean ± SD. **, P < 0.01 by unpaired one-tailed Student’s t test. N.D., not detected.
DISCUSSION
m6A RNA modification has been shown to regulate the function of viral RNAs (41–49). Here, we investigated whether HDV RNAs in infected cells were m6A modified. We found that the HDV genome and antigenome, isolated from cells coinfected with HBV and HDV, as well as from cells only expressing HDV in culture, contain m6A methylation (Fig. 1). The silencing of the cellular m6A machinery and YTHDF1 protein increases the extracellular HDV genome accumulation but decreases intracellular HDV genome levels (Fig. 3 and 4). Importantly, the loss of m6A methylation of HDV RNA enhanced the interaction of delta antigens; depletion of YTHDF1 also resulted in a similar interaction between the HDV genome and delta antigens (Fig. 5). Based on these results, we conclude that m6A methylation of the HDV genome regulates the interaction of delta antigens by recruiting YTHDF1 and thereby negatively regulating viral assembly. Since m6A methylation is reversibly catalyzed by cellular m6A methyltransferases or demethylases (33, 34, 36–38), m6A methylation cannot occur in all HDV genomes. Thus, m6A-methylated and -unmethylated HDV genomes can coexist in HDV-infected cells. In this respect, only the m6A-methylated HDV genome is sequestered by YTHDF1 from delta antigens, resulting in the accumulation of unmethylated RNA genomes in the HDV particles. Therefore, this study points to a novel role of m6A modification in the HDV virion assembly.
m6A modification regulates both cellular and viral RNA biology by diverse pathways, and these effects of m6A methylation depend on the recognition of different m6A reader proteins (29–34, 41–49). For instance, m6A methylation in the 3′ untranslated regions (UTR) of mRNAs reduces the RNA stability mediated by interaction with YTHDF2 protein, while YTHDF1 proteins can protect their target RNA from degradation and promote the translation of m6A-methylated mRNA. In this study, although we focused on the effect of YTHDF1 mediated by m6A methylation in HDV assembly, we observed that the HDV genome also interacts with YTHDF2 and YTHDF3 (Fig. 2). In this respect, YTHDF2 and YTHDF3 proteins should also be able to affect the HDV life cycle. If this is true, m6A can regulate the HDV life cycle in more than one way. In addition, m6A methylation can alter the RNA secondary structure to regulate the binding between the RNA and protein (56). In this context, m6A methylation of the HDV genome could reduce the binding affinity with HDV antigens by altering the RNA structure. Further studies are needed to better understand the role of m6A methylation in HDV replication. Identifying the exact m6A sites within the HDV genome is fundamental to sorting out the various functions of m6A RNA methylation in the HDV life cycle.
HDV uses the HBV surface antigen (HBsAg) proteins to form its virions and to enter hepatocytes via HBV receptors (9). HBsAg is synthesized in three forms of envelope proteins; small (S), middle (M), and large (L) proteins (57). HDV virion assembly occurs through interactions between the HDV RNP complex and all three forms of HBs antigens (6). Although HBs proteins are required for HDV virion production, HDV infection temporarily or permanently suppresses HBV replication (58). Thus, beyond interacting with HBsAg, HDV infection may regulate the HBV life cycle by other mechanisms. In this respect, m6A modification may play an important role in the interplay between HDV and HBV, because m6A methylation occurs in both HBV transcripts and HDV RNAs, and this methylation regulates the HBV and HDV life cycles by diverse pathways (47–53, 59). Understanding the m6A methylation-mediated interplay between HDV and HBV could provide a clearer understanding of the mechanisms of this interaction between these two viruses.
S-HDAg and L-HDAg bind to unbranched quasi-double-stranded HDV RNA to regulate viral genome and antigenome replications and viral mRNA transcription (19–26). In particular, S-HDAg binds to HDV RNA as an octamer and interacts with the linker histone H1e, RNA Pol II, and transcription initiation factors (60). These interactions positively regulate the synthesis of HDV genome and mRNAs but not HDV antigenome synthesis (19–22). A smaller amount of L-HDAg inhibits HDV genome synthesis, whereas the synthesis of HDV antigenome and mRNA is inhibited only when the L-HDAg content vastly exceeds that of S-HDAg (61). L-HDAg disrupts the positive effect of S-HDAg on HDV RNA synthesis by interaction with the homo-oligomeric S-HDAg multimer bound to the HDV antigenome (26). Generally, m6A methylation is installed onto RNAs during transcription (33, 34). Thus, delta antigens may contribute to m6A methylation in HDV RNAs by the interaction with cellular m6A machinery during HDV genome and antigenome replications and thus regulate m6A-mediated HDV replication. We previously showed that HBV X (HBx) protein, which regulates HBV transcription and replication, directly binds and recruits m6A methyltransferases onto the HBV covalently closed circular DNA (cccDNA) genome to affect cotranscriptional m6A modification of viral RNAs (50). Whether HBx contributes to m6A methylation of the HDV RNAs by the recruitment of m6A methyltransferases onto HDV RNA templates should be investigated.
Our work opens a new avenue for studying the functional role of m6A methylation in the HDV life cycle. Novel roles of m6A methylation are continuously being uncovered in various biological processes, viral infections, metabolic diseases, psychiatric disorders, cancers, etc. (29–34). Dysregulation of m6A modification has been implicated in cancer development and progression, and an inhibitor of m6A methyltransferases has been identified as a novel anticancer drug target (62, 63). In this context, our work may lead to the design of new therapeutic opportunities against HDV/HBV coinfections.
MATERIALS AND METHODS
Plasmids, antibodies, and reagents.
The pSVLD3 construct was used to start HDV genome replication, and plasmid pNF-HBV-LMS-D1 was used to express the HBV envelope proteins of one of the variants of the genotype D of HBV, as previously described (54). Antibodies were obtained as follows: anti-FLAG (number 14793) and anti-YTHDF1 (number 86463) antibodies from Cell Signaling Technology (Danvers, MA, USA), antiglyceraldehyde-3-phosphate dehydrogenase (GAPDH; number SC-47724) antibodies from Santa Cruz Biotechnology (Santa Cruz, CA, USA), anti-METTL3 (number 15073-1-AP) antibody from Proteintech Group (Rosemont, IL, USA), and anti-METTL14 (number HPA038002) antibody from Sigma-Aldrich (San Jose, CA, USA). Polyclonal rabbit antidelta antigen antibodies were generated against recombinant small delta antigen (54). The antibodies were used at a 1:1,000 ratio in a 5% bovine serum albumin (BSA) buffer for immunoblotting. The ON-TARGET plus siRNAs of METTL3 (number L-005170-02-0005), METTL14 (number L-014169-02-0005), and YTHDF1 (number L-018095-02-0005) were obtained from Dharmacon (Lafayette, CO, USA).
Cell culture and transfection.
Primary human hepatocytes (PHHs) were obtained from Gibco and cultured according to the manufacturer’s protocol. HepG2 cells were obtained from ATCC. HepG2-NTCp cells were provided by Wenhui Li (National Institute of Biological Sciences, Beijing, China) (64). HepG2 and HepG2-NTCP cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). HepAD38 cells were provided by Christoph Seeger (Fox Chase Cancer Center, Philadelphia, PA, USA). The HepAD38 cells were maintained in Roswell Park Memorial Institute (RPMI) 1640 medium with 20% FBS. HepAD38 and HepG2-NTCP cells were cultured in the presence of 400 μg/mL G418 during maintenance and passage. The medium was supplemented with 2 mM l-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin, and 0.1 mM nonessential amino acid under standard culture conditions (5% CO2, 37°C). HepG2 and HepG2-NTCP cells were transfected with plasmids using Mirus TransIT-LT1 according to the manufacturer’s protocol. siRNAs were transfected into cells using Lipofectamine RNAiMAX (Invitrogen), according to the manufacturer’s instructions.
Virus production and cell infection.
HDV particles were harvested from the supernatants of HepG2 cells cotransfected with plasmids pSVLD3 and pNF-HBV-LMS-D1 at a 1:1 ratio. HBV particles were concentrated from the supernatants of HepAD38 cells. The culture medium was centrifuged at 4°C and 10,000 × g for 15 min and concentrated about 100-fold with 5% polyethylene glycol (PEG) 8000 overnight at 4°C. The HBV and HDV titers were quantified using RT-qPCR (65) and diluted in a serum-free medium with 4% PEG 8000 and 2% dimethyl sulfoxide (DMSO) for infection. PHHs and HepG2-NTCP cells were infected with HBV (100 viral genome equivalents [vge]/cell). After 24 h, the cells were washed three times with phosphate-buffered saline (PBS). For HDV infection, HBV-infected cells were then incubated with infectious HDV particles (100 vge/cell) for 24 h (66, 67). After the incubation with HDV particles, the PHHs and HepG2-NTCP cells were washed with a culture medium and further cultured for 9 days.
Real-time reverse-transcription quantitative PCR.
Total RNA was extracted using the RNeasy minikit (Qiagen) and treated with DNase prior to reverse transcription. The extracellular RNA was 10-fold concentrated using the Amicon Ultra centrifugal filter (Millipore) and extracted using the PureLink viral RNA kit (Invitrogen). RNA was reverse-transcribed using iScript reverse transcription supermix (Bio-Rad). The quantitative PCR was assessed with SsoAdvanced Universal SYBR green supermix (Bio-Rad). For strand-specific detection of HDV genome and antigenome, the total RNA was reverse transcribed using the iScript Select cDNA synthesis kit (Bio-Rad) with either Tag1 HDV genome RT primer (5′-GATGCCTACGACTGAAGCCTAGTGAATAAAGCGGGTTTCCACTCACAG-3′) or Tag2 HDV antigenome RT primer (5′-GTAGGCTACGACTGAAGCCTAGTGTCTCTCTCGAGTTCCTCTAACTTCTTTCTTCC-3′), respectively (22). HDV genome and antigenome levels were quantified using SsoAdvanced universal probes supermix (Bio-Rad) with either Tag1 HDV genome F (5′-GATGCCTACGACTGAAGCCTAGT-3′) and HDV genome R (5′-GTGGCTCTCCCTTAGCCATC-3′) for the HDV genome or Tag2 HDV antigenome F (5′-GTAGGCTACGACTGAAGCCTAGT-3′) and HDV antigenome R (5′-GTCCAGCAGTCTCCTCTTTACAG-3′) for the HDV antigenome. Viral RNA and cellular mRNA levels, normalized to GAPDH, were analyzed using the ΔΔ threshold cycle (ΔΔCT) method.
Methylated RNA immunoprecipitation RT-qPCR.
Total RNA was isolated using the RNeasy minikit (Qiagen) and treated with DNase. Isolated total RNA was incubated with an anti-m6A antibody (Thermo Fisher Scientific) conjugated to protein G Dynabeads (Thermo Fisher Scientific) in MeRIP buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, and 0.1% NP-40) with RNase inhibitor overnight at 4°C. The beads were then washed with MeRIP buffer 5 times, and m6A-modified RNA was eluted in with MeRIP buffer containing 5 mM m6A 5′-monophosphate sodium salt (Sigma-Aldrich). The eluted RNA was concentrated using the RNeasy minikit (Qiagen) and reverse transcribed into cDNA. HDV genome and antigenome were quantified using a strand-specific RT-qPCR.
RNA immunoprecipitation.
Cells were cross-linked with 245 nm UV for 250 mJ/cm and lysed in SDS lysis buffer (0.5% SDS, 50 mM Tris-HCl [pH 6.8], 1 mM EDTA, 1 mM dithiothreitol [DTT], 150 mM NaCl) supplemented with a protease inhibitor and RNase inhibitor (Thermo Fisher Scientific). The cellular lysates were incubated with antidelta antigen or anti-FLAG antibody conjugated to protein G Dynabeads (Thermo Fisher Scientific) overnight at 4°C in a rotating wheel. The beads were washed with lysis buffer 5 times. Half of the beads were incubated in elution buffer (20 mM Tris-HCl [pH 7.5], 5 mM EDTA, 50 mM NaCl, 1% SDS, proteinase K 50 μg/mL) for 2 h at 58°C, and then RNA was extracted using the RNeasy minikit (Qiagen) (3). The other half of the beads were analyzed by Western blot assay.
Western blotting.
Cells were lysed in a radioimmunoprecipitation assay (RIPA) lysis buffer (Thermo Fisher Scientific) supplemented with a protease inhibitor (Thermo Fisher Scientific) for 15 min at 4°C, and the cellular lysates were centrifuged for 20 min at 14,000 rpm at 4°C. The clarified cellular lysates were resolved by SDS-PAGE gel and transferred to nitrocellulose membranes (Bio-Rad). The membranes were incubated with various primary antibodies, and the chemiluminescence signals were detected using the ChemiDoc MP imaging system (Bio-Rad).
Statistical analysis.
All results are representative of three independent experiments. For each result, error bars represent the standard deviations (SD) from at least three independent experiments. The P value was calculated using a one-tailed unpaired Student’s t test.
Data availability.
All study data are included in the article.
ACKNOWLEDGMENTS
This study was supported in part by NIH grant AI139234 to A.S.; S.O.G. was supported by NIH grant R21 AI144923.
We thank David Durantel and Julie Lucifora (Lyon, France) and Maura Dandri (Hamburg, Germany) for helpful discussions.
We declare that we have no conflicts of interest.
Contributor Information
Geon-Woo Kim, Email: gekim@health.ucsd.edu.
J.-H. James Ou, University of Southern California.
REFERENCES
- 1.Rizzetto M, Canese MG, Arico S, Crivelli O, Trepo C, Bonino F, Verme G. 1977. Immunofluorescence detection of new antigen-antibody system (delta/anti-delta) associated to hepatitis B virus in liver and in serum of HBsAg carriers. Gut 18:997–1003. 10.1136/gut.18.12.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rizzetto M, Canese MG, Gerin JL, London WT, Sly DL, Purcell RH. 1980. Transmission of the hepatitis B virus-associated delta antigen to chimpanzees. J Infect Dis 141:590–602. 10.1093/infdis/141.5.590. [DOI] [PubMed] [Google Scholar]
- 3.Wedemeyer H, Negro F. 2019. Devil hepatitis D: an orphan disease or largely underdiagnosed? Gut 68:381–382. 10.1136/gutjnl-2018-317403. [DOI] [PubMed] [Google Scholar]
- 4.Buti M, Homs M, Rodriguez-Frias F, Funalleras G, Jardi R, Sauleda S, Tabernero D, Schaper M, Esteban R. 2011. Clinical outcome of acute and chronic hepatitis delta over time: a long-term follow-up study. J Viral Hepat 18:434–442. 10.1111/j.1365-2893.2010.01324.x. [DOI] [PubMed] [Google Scholar]
- 5.Lutgehetmann M, Mancke LV, Volz T, Helbig M, Allweiss L, Bornscheuer T, Pollok JM, Lohse AW, Petersen J, Urban S, Dandri M. 2012. Humanized chimeric uPA mouse model for the study of hepatitis B and D virus interactions and preclinical drug evaluation. Hepatology 55:685–694. 10.1002/hep.24758. [DOI] [PubMed] [Google Scholar]
- 6.Wang KS, Choo QL, Weiner AJ, Ou JH, Najarian RC, Thayer RM, Mullenbach GT, Denniston KJ, Gerin JL, Houghton M. 1986. Structure, sequence and expression of the hepatitis delta (delta) viral genome. Nature 323:508–514. 10.1038/323508a0. [DOI] [PubMed] [Google Scholar]
- 7.Luo GX, Chao M, Hsieh SY, Sureau C, Nishikura K, Taylor J. 1990. A specific base transition occurs on replicating hepatitis delta virus RNA. J Virol 64:1021–1027. 10.1128/JVI.64.3.1021-1027.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mandell GL, Douglas RG, Bennett JE, Dolin R. 2005. Mandell, Douglas, and Bennett's principles and practice of infectious diseases. Elsevier/Churchill Livingstone; 9th ed. Book review (E-STREAMS). https://www.elsevier.com/books/mandell-douglas-and-bennetts-principles-and-practice-of-infectious-diseases/bennett/978-0-323-48255-4. Accessed 8 August 2019. [Google Scholar]
- 9.Urban S, Bartenschlager R, Kubitz R, Zoulim F. 2014. Strategies to inhibit entry of HBV and HDV into hepatocytes. Gastroenterology 147:48–64. 10.1053/j.gastro.2014.04.030. [DOI] [PubMed] [Google Scholar]
- 10.Kuo MY, Sharmeen L, Dinter-Gottlieb G, Taylor J. 1988. Characterization of self-cleaving RNA sequences on the genome and antigenome of human hepatitis delta virus. J Virol 62:4439–4444. 10.1128/JVI.62.12.4439-4444.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeng KS, Daniel A, Lai MM. 1996. A pseudoknot ribozyme structure is active in vivo and required for hepatitis delta virus RNA replication. J Virol 70:2403–2410. 10.1128/JVI.70.4.2403-2410.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen PJ, Kalpana G, Goldberg J, Mason W, Werner B, Gerin J, Taylor J. 1986. Structure and replication of the genome of the hepatitis delta virus. Proc Natl Acad Sci USA 83:8774–8778. 10.1073/pnas.83.22.8774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang J, Nie X, Chang HE, Han Z, Taylor J. 2008. Transcription of hepatitis delta virus RNA by RNA polymerase II. J Virol 82:1118–1127. 10.1128/JVI.01758-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macnaughton TB, Shi ST, Modahl LE, Lai MMC. 2002. Rolling circle replication of hepatitis delta virus RNA is carried out by two different cellular RNA polymerases. J Virol 76:3920–3927. 10.1128/jvi.76.8.3920-3927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu TB, Taylor J. 1993. The RNAs of hepatitis delta virus are copied by RNA polymerase II in nuclear homogenates. J Virol 67:6965–6972. 10.1128/JVI.67.12.6965-6972.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharmeen L, Kuo MY, Dinter-Gottlieb G, Taylor J. 1988. Antigenomic RNA of human hepatitis delta virus can undergo self-cleavage. J Virol 62:2674–2679. 10.1128/JVI.62.8.2674-2679.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Defenbaugh DA, Johnson M, Chen R, Zheng YY, Casey JL. 2009. Hepatitis delta antigen requires a minimum length of the hepatitis delta virus unbranched rod RNA structure for binding. J Virol 83:4548–4556. 10.1128/JVI.02467-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffin BL, Chasovskikh S, Dritschilo A, Casey JL. 2014. Hepatitis delta antigen requires a flexible quasi-double-stranded RNA structure to bind and condense hepatitis delta virus RNA in a ribonucleoprotein complex. J Virol 88:7402–7411. 10.1128/JVI.00443-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuo MY, Chao M, Taylor J. 1989. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J Virol 63:1945–1950. 10.1128/JVI.63.5.1945-1950.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glenn JS, Taylor JM, White JM. 1990. In vitro-synthesized hepatitis delta virus RNA initiates genome replication in cultured cells. J Virol 64:3104–3107. 10.1128/JVI.64.6.3104-3107.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glenn JS, White JM. 1991. trans-dominant inhibition of human hepatitis delta virus genome replication. J Virol 65:2357–2361. 10.1128/JVI.65.5.2357-2361.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harichandran K, Shen Y, Stephenson Tsoris S, Lee SC, Casey JL. 2019. Hepatitis delta antigen regulates mRNA and antigenome RNA levels during hepatitis delta virus replication. J Virol 93:e01989-18. 10.1128/JVI.01989-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang FL, Chen PJ, Tu SJ, Wang CJ, Chen DS. 1991. The large form of hepatitis delta antigen is crucial for assembly of hepatitis delta virus. Proc Natl Acad Sci USA 88:8490–8494. 10.1073/pnas.88.19.8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee CH, Chang SC, Wu CH, Chang MF. 2001. A novel chromosome region maintenance 1-independent nuclear export signal of the large form of hepatitis delta antigen that is required for the viral assembly. J Biol Chem 276:8142–8148. 10.1074/jbc.M004477200. [DOI] [PubMed] [Google Scholar]
- 25.Chao M, Hsieh SY, Taylor J. 1990. Role of two forms of hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J Virol 64:5066–5069. 10.1128/JVI.64.10.5066-5069.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia YP, Lai MM. 1992. Oligomerization of hepatitis delta antigen is required for both the trans-activating and trans-dominant inhibitory activities of the delta antigen. J Virol 66:6641–6648. 10.1128/JVI.66.11.6641-6648.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bogomolov P, Alexandrov A, Voronkova N, Macievich M, Kokina K, Petrachenkova M, Lehr T, Lempp FA, Wedemeyer H, Haag M, Schwab M, Haefeli WE, Blank A, Urban S. 2016. Treatment of chronic hepatitis D with the entry inhibitor myrcludex B: first results of a phase Ib/IIa study. J Hepatol 65:490–498. 10.1016/j.jhep.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 28.Roundtree IA, Evans ME, Pan T, He C. 2017. Dynamic RNA modifications in gene expression regulation. Cell 169:1187–1200. 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yue Y, Liu J, He C. 2015. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev 29:1343–1355. 10.1101/gad.262766.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li S, Mason CE. 2014. The pivotal regulatory landscape of RNA modifications. Annu Rev Genomics Hum Genet 15:127–150. 10.1146/annurev-genom-090413-025405. [DOI] [PubMed] [Google Scholar]
- 31.Saletore Y, Meyer K, Korlach J, Vilfan ID, Jaffrey S, Mason CE. 2012. The birth of the epitranscriptome: deciphering the function of RNA modifications. Genome Biol 13:175. 10.1186/gb-2012-13-10-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. 2015. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 526:591–594. 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi H, Wei J, He C. 2019. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell 74:640–650. 10.1016/j.molcel.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyer KD, Jaffrey SR. 2017. Rethinking m6A readers, writers, and erasers. Annu Rev Cell Dev Biol 33:319–342. 10.1146/annurev-cellbio-100616-060758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. 2015. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods 12:767–772. 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, Dai Q, Chen W, He C. 2014. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 10:93–95. 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, He C. 2011. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 7:885–887. 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang C-M, Li CJ, Vagbo CB, Shi Y, Wang W-L, Song S-H, Lu Z, Bosmans RPG, Dai Q, Hao Y-J, Yang X, Zhao W-M, Tong W-M, Wang X-J, Bogdan F, Furu K, Fu Y, Jia G, Zhao X, Liu J, Krokan HE, Klungland A, Yang Y-G, He C. 2013. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 49:18–29. 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C. 2014. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505:117–120. 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, He C. 2014. Reading RNA methylation codes through methyl-specific binding proteins. RNA Biol 11:669–672. 10.4161/rna.28829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gonzales-van Horn SR, Sarnow P. 2017. Making the mark: the role of adenosine modifications in the life cycle of RNA viruses. Cell Host Microbe 21:661–669. 10.1016/j.chom.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gokhale NS, McIntyre ABR, McFadden MJ, Roder AE, Kennedy EM, Gandara JA, Hopcraft SE, Quicke KM, Vazquez C, Willer J, Ilkayeva OR, Law BA, Holley CL, Garcia-Blanco MA, Evans MJ, Suthar MS, Bradrick SS, Mason CE, Horner SM. 2016. N6-methyladenosine in Flaviviridae viral RNA genomes regulates infection. Cell Host Microbe 20:654–665. 10.1016/j.chom.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lichinchi G, Gao S, Saletore Y, Gonzalez GM, Bansal V, Wang Y, Mason CE, Rana TM. 2016. Dynamics of the human and viral m6A RNA methylomes during HIV-1 infection of T cells. Nat Microbiol 1:16011. 10.1038/nmicrobiol.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Imam H, Kim G-W, Siddiqui A. 2020. Epitranscriptomic (N6-methyladenosine) modification of viral RNA and virus-host interactions. Front Cell Infect Microbiol 10:584283. 10.3389/fcimb.2020.584283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim G-W, Siddiqui A. 2021. The role of N6-methyladenosine modification in the life cycle and disease pathogenesis of hepatitis B and C viruses. Exp Mol Med 53:339–345. 10.1038/s12276-021-00581-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim G-W, Siddiqui A. 2021. N6-methyladenosine modification of HCV RNA genome regulates cap-independent IRES-mediated translation via YTHDC2 recognition. Proc Natl Acad Sci USA 118:e2022024118. 10.1073/pnas.2022024118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Imam H, Khan M, Gokhale NS, McIntyre ABR, Kim G-W, Jang JY, Kim S-J, Mason CE, Horner SM, Siddiqui A. 2018. N6-methyladenosine modification of hepatitis B virus RNA differentially regulates the viral life cycle. Proc Natl Acad Sci USA 115:8829–8834. 10.1073/pnas.1808319115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim G-W, Imam H, Siddiqui A. 2021. The RNA binding proteins YTHDC1 and FMRP regulate the nuclear export of N(6)-methyladenosine-modified hepatitis B virus transcripts and affect the viral life cycle. J Virol 95:e0009721. 10.1128/JVI.00097-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim G-W, Siddiqui A. 2022. Hepatitis B virus X protein expression is tightly regulated by N6-methyladenosine modification of its mRNA. J Virol 96:e0165521. 10.1128/JVI.01655-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim G-W, Siddiqui A. 2021. Hepatitis B virus X protein recruits methyltransferases to affect cotranscriptional N6-methyladenosine modification of viral/host RNAs. Proc Natl Acad Sci USA 118:e2019455118. 10.1073/pnas.2019455118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim G-W, Imam H, Khan M, Mir SA, Kim S-J, Yoon SK, Hur W, Siddiqui A. 2021. HBV-induced increased N6 methyladenosine modification of PTEN RNA affects innate immunity and contributes to HCC. Hepatology 73:533–547. 10.1002/hep.31313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim G-W, Moon J-S, Siddiqui A. 2022. N6-methyladenosine modification of the 5′ epsilon structure of the HBV pregenome RNA regulates its encapsidation by the viral core protein. Proc Natl Acad Sci USA 119:e2120485119. 10.1073/pnas.2120485119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Imam H, Kim G-W, Mir SA, Khan M, Siddiqui A. 2020. Interferon-stimulated gene 20 (ISG20) selectively degrades N6-methyladenosine modified hepatitis B virus transcripts. PLoS Pathog 16:e1008338. 10.1371/journal.ppat.1008338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Freitas N, Cunha C, Menne S, Gudima SO. 2014. Envelope proteins derived from naturally integrated hepatitis B virus DNA support assembly and release of infectious hepatitis delta virus particles. J Virol 88:5742–5754. 10.1128/JVI.00430-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poria DK, Ray PS. 2017. RNA-protein UV-crosslinking assay. Bio Protoc 7:e2193. 10.21769/BioProtoc.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. 2017. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res 45:6051–6063. 10.1093/nar/gkx141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seeger C, Mason WS. 2015. Molecular biology of hepatitis B virus infection. Virology 479–480:672–686. 10.1016/j.virol.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jardi R, Rodriguez F, Buti M, Costa X, Cotrina M, Galimany R, Esteban R, Guardia J. 2001. Role of hepatitis B, C, and D viruses in dual and triple infection: influence of viral genotypes and hepatitis B precore and basal core promoter mutations on viral replicative interference. Hepatology 34:404–410. 10.1053/jhep.2001.26511. [DOI] [PubMed] [Google Scholar]
- 59.Kim G-W, Imam H, Khan M, Siddiqui A. 2020. N6-methyladenosine modification of hepatitis B and C viral RNAs attenuates host innate immunity via RIG-I signaling. J Biol Chem 295:13123–13133. 10.1074/jbc.RA120.014260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cao D, Haussecker D, Huang Y, Kay MA. 2009. Combined proteomic-RNAi screen for host factors involved in human hepatitis delta virus replication. RNA 15:1971–1979. 10.1261/rna.1782209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Macnaughton TB, Lai MM. 2002. Large hepatitis delta antigen is not a suppressor of hepatitis delta virus RNA synthesis once RNA replication is established. J Virol 76:9910–9919. 10.1128/jvi.76.19.9910-9919.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yankova E, Blackaby W, Albertella M, Rak J, De Braekeleer E, Tsagkogeorga G, Pilka ES, Aspris D, Leggate D, Hendrick AG, Webster NA, Andrews B, Fosbeary R, Guest P, Irigoyen N, Eleftheriou M, Gozdecka M, Dias JML, Bannister AJ, Vick B, Jeremias I, Vassiliou GS, Rausch O, Tzelepis K, Kouzarides T. 2021. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 593:597–601. 10.1038/s41586-021-03536-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Su R, Dong L, Li Y, Gao M, Han L, Wunderlich M, Deng X, Li H, Huang Y, Gao L, Li C, Zhao Z, Robinson S, Tan B, Qing Y, Qin X, Prince E, Xie J, Qin H, Li W, Shen C, Sun J, Kulkarni P, Weng H, Huang H, Chen Z, Zhang B, Wu X, Olsen MJ, Muschen M, Marcucci G, Salgia R, Li L, Fathi AT, Li Z, Mulloy JC, Wei M, Horne D, Chen J. 2020. Targeting FTO suppresses cancer stem cell maintenance and immune evasion. Cancer Cell 38:79–96.e11. 10.1016/j.ccell.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, Fu L, Song M, Chen P, Gao W, Ren B, Sun Y, Cai T, Feng X, Sui J, Li W. 2012. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife 1:e00049. 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ferns RB, Nastouli E, Garson JA. 2012. Quantitation of hepatitis delta virus using a single-step internally controlled real-time RT-qPCR and a full-length genomic RNA calibration standard. J Virol Methods 179:189–194. 10.1016/j.jviromet.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 66.Tham CYL, Kah J, Tan AT, Volz T, Chia A, Giersch K, Ladiges Y, Loglio A, Borghi M, Sureau C, Lampertico P, Lutgehetmann M, Dandri M, Bertoletti A. 2020. Hepatitis delta virus acts as an immunogenic adjuvant in hepatitis B virus-infected hepatocytes. Cell Rep Med 1:100060. 10.1016/j.xcrm.2020.100060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alfaiate D, Lucifora J, Abeywickrama-Samarakoon N, Michelet M, Testoni B, Cortay JC, Sureau C, Zoulim F, Deny P, Durantel D. 2016. HDV RNA replication is associated with HBV repression and interferon-stimulated genes induction in super-infected hepatocytes. Antiviral Res 136:19–31. 10.1016/j.antiviral.2016.10.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All study data are included in the article.