ABSTRACT
Members of the mosquito-borne flavivirus genus such as dengue (DENV), West Nile (WNV), and Zika (ZIKV) viruses cause distinct diseases and affect different tissues. We previously found that the secreted flaviviral nonstructural protein 1 (NS1) interacts with endothelial cells and disrupts endothelial barrier function in a tissue-specific manner consistent with the disease tropism of the respective viruses. However, the underlying molecular mechanism of this tissue-specific NS1-endothelial cell interaction is not well understood. To elucidate the distinct role(s) that the wing and β-ladder domains of NS1 play in NS1 interactions with endothelial cells, we constructed flavivirus NS1 chimeras that exchanged the wing and β-ladder domains in a pairwise manner between DENV, WNV, and ZIKV NS1. We found that both the NS1 wing and β-ladder domains conferred NS1 tissue-specific endothelial dysfunction, with the wing conferring cell binding and the β-ladder involved in inducing endothelial hyperpermeability as measured by transendothelial electrical resistance. To narrow down the amino acids dictating cell binding specificity, we utilized the DENV-WNV NS1 chimera and identified residues 91 to 93 (GDI) of DENV NS1 as a molecular motif determining binding specificity. Further, using an in vivo mouse model of localized leak, we found that the GDI motif of the wing domain was essential for triggering DENV NS1-induced vascular leak in mouse dermis. Taken together, we identify molecular determinants of flavivirus NS1 that confer NS1 binding and vascular leak and highlight the importance of the NS1 wing domain for flavivirus pathogenesis.
IMPORTANCE Flavivirus NS1 is secreted into the bloodstream from infected cells during a viral infection. Dengue virus NS1 contributes to severe dengue pathology such as endothelial dysfunction and vascular leak independently of the virus. We have shown that multiple flavivirus NS1 proteins result in endothelial dysfunction in a tissue-specific manner consistent with their respective viral tropism. Here, we aimed to identify the molecular determinants that make some, but not other, flavivirus NS1 proteins bind to select endothelial cells in vitro and cause vascular leak in a mouse model. We identified the wing domain of NS1 as a primary determinant conferring differential endothelial dysfunction and vascular leak and narrowed the contributing amino acid residues to a three-residue motif within the wing domain. The insights from this study pave the way for future studies on the effects of flavivirus NS1 on viral dissemination and pathogenesis and offer potential new avenues for antiviral therapies.
KEYWORDS: flavivirus, NS1, dengue virus, endothelial dysfunction, vascular leak, tissue specificity
INTRODUCTION
Dengue virus (DENV) is a mosquito-borne positive-stranded RNA virus of the Flavivirus genus, consisting of four serotypes (DENV1-4). DENV causes ~100 million symptomatic infections per year globally, ranging from classic dengue fever to the more severe dengue haemorrhagic fever and dengue shock syndrome (1, to ,3). The severe forms of dengue are characterized by clinical presentation of vascular leak as a result of endothelial dysfunction (4). Traditionally, endothelial dysfunction has been attributed to a hyperactivated immune response as a result of uncontrolled viral infection and immune cell activation (5, 6). Recently, we and others have demonstrated that secreted nonstructural protein 1 (NS1) directly contributes to endothelial dysfunction and vascular leak through interactions with endothelial and immune cells that result in the breakdown of endothelial barriers such as the endothelial glycocalyx and intercellular junctions (7, to ,14). In addition to DENV, West Nile (WNV) and Zika (ZIKV) viruses are also members of the Flavivirus genus that cause human diseases of public health significance. Like DENV NS1, the NS1 proteins of WNV and ZIKV have been shown to modulate endothelial barriers. WNV NS1 has also been shown to contribute to WNV infection in the brain (15), while ZIKV NS1 has been shown to modulate the barrier integrity of placental explants (10) as well as the testis, contributing to ZIKV infection of Sertoli cells (9, 16).
NS1 is an ~55-kDa glycoprotein that is highly conserved across the flaviviruses (17). It dimerizes in the endoplasmic reticulum and is found as a component of the viral replication complex. The dimers can also trimerize to form hexamers that are secreted by DENV-infected cells into the bloodstream as a soluble lipoprotein containing lipid cargo (18, to ,20). In severe dengue patients, the levels of detectable NS1 in the blood are generally reported as 1 to 10 μg/mL (21, 22). Secreted NS1 has been shown to associate with components of the innate immune system such as toll-like receptors (13, 23) and complement proteins (24, to 26), as well as components of the blood-clotting cascade such as platelets (27, to ,29), which results in both host immune evasion and modulation of barrier integrity of endothelial cells. Separately, NS1 can also directly disrupt endothelial cell-barrier integrity by promoting the degradation of endothelial glycocalyx components that line the surface of endothelial cells (7, 12, 30, 31) and disrupting junctional protein complexes that mediate cell–cell interactions (8, 32, 33). In mouse models, NS1 vaccination has been shown to be protective against lethal DENV challenge. Conversely, addition of NS1 to sublethal DENV infection was shown to exacerbate pathology, resulting in a lethal infection, suggesting the potential of NS1 as a vaccine component and therapeutic target (12).
NS1 proteins from multiple flaviviruses have been shown to interact with endothelial cells of distinct tissue origin in a manner consistent with the tropism of their respective flaviviruses (34). For example, DENV NS1 interacts with multiple cell lines, including human pulmonary microvascular endothelial cells (HPMEC), which correlates with the systemic nature of DENV infection that can be observed in the lungs. In contrast, WNV NS1 interacts well with brain endothelial cells but minimally with HPMEC, as WNV causes neurological pathologies, such as meningitis and encephalitis, but not pulmonary pathology. Similarly, ZIKV NS1 interacts well with both brain and umbilical vein endothelial cells but minimally with HPMEC, as ZIKV causes neurological and congenital but not pulmonary pathologies. However, how these tissue-specific NS1 interactions are modulated remains unclear.
Flavivirus NS1 contains three domains that are highly conserved: the β-roll (residues 1 to 29), wing (residues 30 to 180), and β-ladder (residues 181 to 352) (35). The NS1 wing domain contains many hydrophobic residues that are predicted to interact with the cell’s plasma membrane as well as lipid cargo in its hexameric form (36, 37). Interestingly, these surface-exposed residues are conserved among DENV serotypes but divergent across the Flavivirus genus (e.g., between DENV and WNV), suggesting their possible roles in mediating NS1 tissue-specific interactions. Additionally, prior studies have suggested that the three domains of NS1 may have distinct functions. DENV NS1 wing domain has been shown to be immunodominant in both humans and mice (12, 38, 39), where conserved, hydrophobic residues in the flexible loop (residues 108 to 129) within the wing domain (Trp-115, Trp-118, Gly-119) have been identified to partially contribute to DENV NS1 binding to HPMEC (36, 40). In contrast, select residues in the β-ladder domain (such as residues N207, A303, E326, and D327) have been implicated as important for NS1-induced endothelial hyperpermeability, while being dispensable for binding endothelial cells (32, 40). While these data offer clues about NS1 molecular determinants of tissue-specific interactions with endothelial cells, a systematic investigation of these different domains in distinct flavivirus NS1 proteins has not been undertaken.
In this study, we provide new insights on the tissue-specific interactions between flavivirus NS1 and endothelial cells using NS1 chimeras that exchange the wing and β-ladder domains of DENV, WNV, and ZIKV in a pairwise manner with one another. We found that both the wing and β-ladder domains confer tissue-specific barrier dysfunction, with the wing influencing initial attachment to endothelial cells and the β-ladder involved in inducing endothelial hyperpermeability in vitro. We further identified a variable 3-amino-acid (aa) motif in the wing domain as a molecular determinant for tissue specificity. Finally, we identify the wing domain and the 3-aa motif as a driver of DENV-triggered vascular leak in vivo. Taken together, our results provide insights into the tissue-specific interactions of flavivirus NS1.
RESULTS
The wing domain of flavivirus NS1 confers tissue-specific binding to endothelial cells.
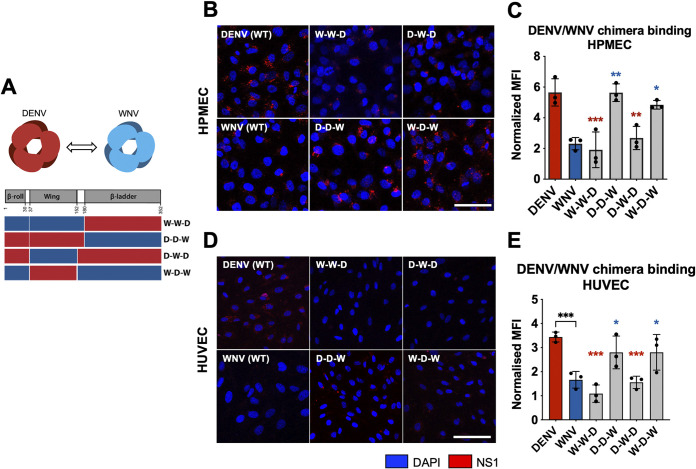
In previous work, we showed that DENV NS1 binds to human lung endothelial cells (HPMEC) at higher levels than WNV and ZIKV NS1 (34), correlating with the capacity of DENV NS1 but not WNV or ZIKV NS1 to cause endothelial hyperpermeability of HPMEC and pleural effusion in humans. Similarly, DENV and ZIKV NS1 exhibited higher binding than WNV NS1 to human umbilical vein microvascular endothelial cells (HUVEC) and caused endothelial hyperpermeability of HUVEC (34). To uncover the molecular determinants of flavivirus NS1 that dictate this tissue-specific endothelial cell tropism, we generated chimeric NS1 proteins that exchanged either the wing or the β-ladder domains between DENV and WNV NS1 (Fig. 1A). We cloned and expressed the C-terminally His-tagged NS1 proteins in HEK-293 cells, which were successfully secreted, and then purified NS1 oligomers using cobalt affinity chromatography (Fig. 2). Chimeric proteins are annotated by their 3 domains from the respective flavivirus NS1 proteins in sequence “β-roll–wing–β-ladder” (e.g., D-D-W designating DENV β-roll – DENV wing – WNV β-ladder). We treated HPMEC with either wild-type (WT) or DENV-WNV chimeric NS1 proteins and measured the levels of NS1 binding to HPMEC using an immunofluorescence microscopy assay (IFA). We found that the chimeric NS1 constructs containing DENV NS1 wing domain (D-D-W and W-D-W) bound to HPMEC at comparable levels to WT DENV NS1, whereas the constructs containing WNV NS1 wing (W-W-D and D-W-D) exhibited significantly lower binding than WT DENV NS1, but at comparable levels to WT WNV NS1 (Fig. 1B and C). A similar pattern was observed when we treated HUVEC with the same DENV-WNV chimeric NS1 proteins, where constructs containing the DENV NS1 wing domain drove binding to HUVEC at comparable levels as WT DENV NS1 (Fig. 1D and E).
FIG 1.
The wing domain of DENV NS1 confers tissue-specific binding to endothelial cells. (A) Schematic representation of chimeric NS1 proteins that exchange wing or β-ladder domains between DENV and WNV NS1. Red box represents DENV NS1, and blue box represents WNV NS1. (B) Recombinant WT or chimeric NS1 (10 μg/mL) was added to HPMEC and incubated at 37°C for 1 h. NS1 binding was assessed by immunofluorescence microscopy, and representative images from three experiments are shown. (C) Quantification of (B), normalized to untreated controls. (D) NS1 binding as in (B), but on HUVEC. (E) Quantification of (D), normalized to untreated controls. MFI, mean fluorescence intensity. Images represent 3 independent experiments. Scale bars, 100 μm. Data plotted as mean ± standard deviation (SD). *, P < 0.05; **, P < 0.01; ***, P < 0.005; ****, P < 0.001 by one-way ANOVA with multiple comparisons. Star colors indicate the respective control to which each construct was compared.
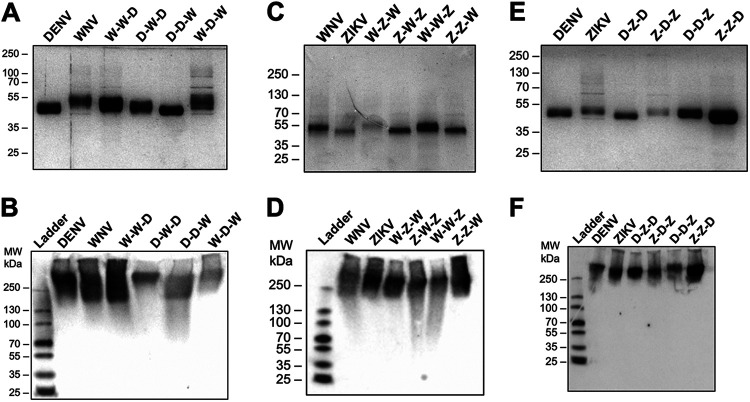
FIG 2.
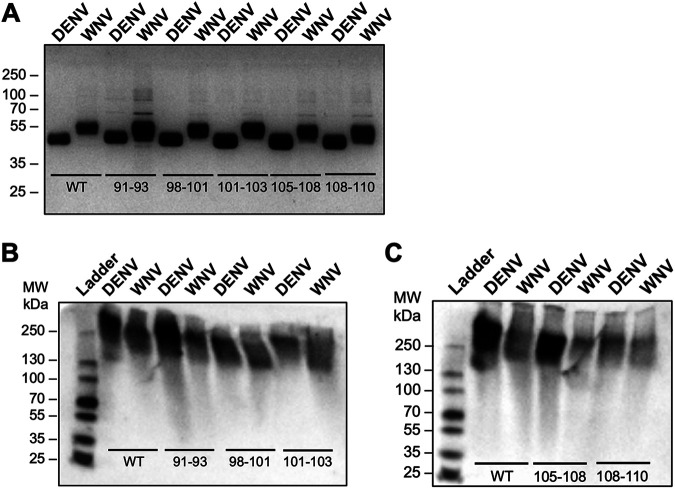
Production and quality control of flavivirus NS1 chimeric constructs. Proteins were evaluated by silver stain following SDS-PAGE (A, C, E) and native PAGE (B, D, F). Two micrograms of each NS1 construct were used. For native gels, proteins were detected using anti-His MAb. (A) Silver stain of DENV-WNV NS1 chimeras. (B) Native PAGE of DENV-WNV NS1 chimeras. (C) Silver stain of WNV-ZIKV NS1 chimeras. (D) Native PAGE of WNV-ZIKV NS1 chimeras. (E) Silver stain of DENV-ZIKV NS1 chimeras. (F) Native PAGE of DENV-ZIKV NS1 chimeras.
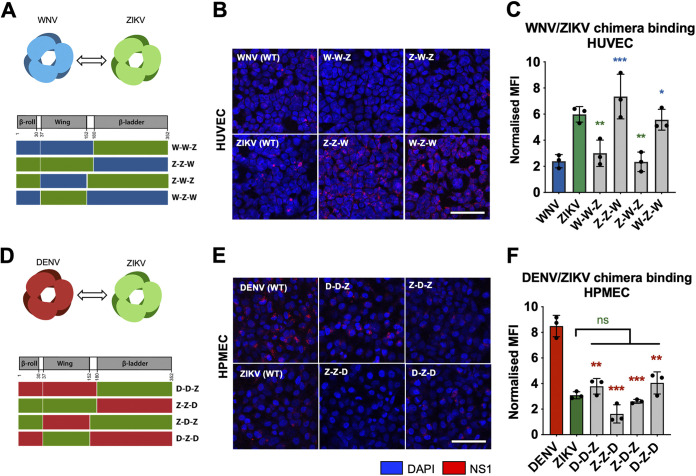
To investigate whether the wing domain of other flaviviruses conferred NS1 binding to endothelial cells, we constructed similar chimeric NS1 proteins that exchanged the wing and β-ladder domains between WNV and ZIKV NS1 and DENV and ZIKV NS1, respectively (Fig. 3A and D). We treated HUVEC with WNV-ZIKV chimeric NS1 and found that constructs containing the ZIKV NS1 wing domain (Z-Z-W and W-Z-W) bound to HUVEC at comparable levels to WT ZIKV NS1, whereas the constructs containing the WNV NS1 wing (W-W-Z and Z-W-Z) bound to HUVEC at lower levels, consistent with WT WNV NS1 (Fig. 3B and C). We subsequently treated HPMEC with DENV-ZIKV chimeric NS1 and found that while constructs with ZIKV NS1 wing (Z-Z-D and D-Z-D) exhibited significantly diminished binding to HPMEC, the constructs with DENV NS1 wing (D-D-Z and Z-D-Z) did not gain binding to HPMEC comparably to WT DENV NS1 (Fig. 3E and F), possibly due to differential effects of interdomain interactions between flaviviruses. Overall, these results indicate that the tissue-specific patterns of flavivirus NS1 binding to endothelial cells are driven by the wing domain of NS1.
FIG 3.
The wing domain of flavivirus NS1 confers tissue-specific binding to endothelial cells. (A, D) Schematic representation of chimeric NS1 proteins produced with WNV and ZIKV NS1 (A) or DENV and ZIKV NS1 (D). Red box represents DENV NS1, blue box represents WNV NS1, and green box represents ZIKV NS1. (B) Recombinant WT or WNV-ZIKV chimeric NS1 (10 μg/mL) was added to HUVEC and incubated at 37°C for 1 h. NS1 binding was assessed by immunofluorescence microscopy, and representative images from three experiments are shown. (C) Quantification of (B), normalized to untreated controls. (E) NS1 binding as in (B), but with DENV-ZIKV chimeric NS1 to HPMEC. (F) Quantification of (E), normalized to untreated controls. MFI, mean fluorescence intensity. Images represent 3 independent experiments. Scale bars, 100 μm. Data plotted as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.005; ****, P < 0.001 by one-way ANOVA with multiple comparisons. Star colors indicate the respective control to which each construct was compared.
The wing and β-ladder domains of flavivirus NS1 proteins are necessary for inducing endothelial hyperpermeability.
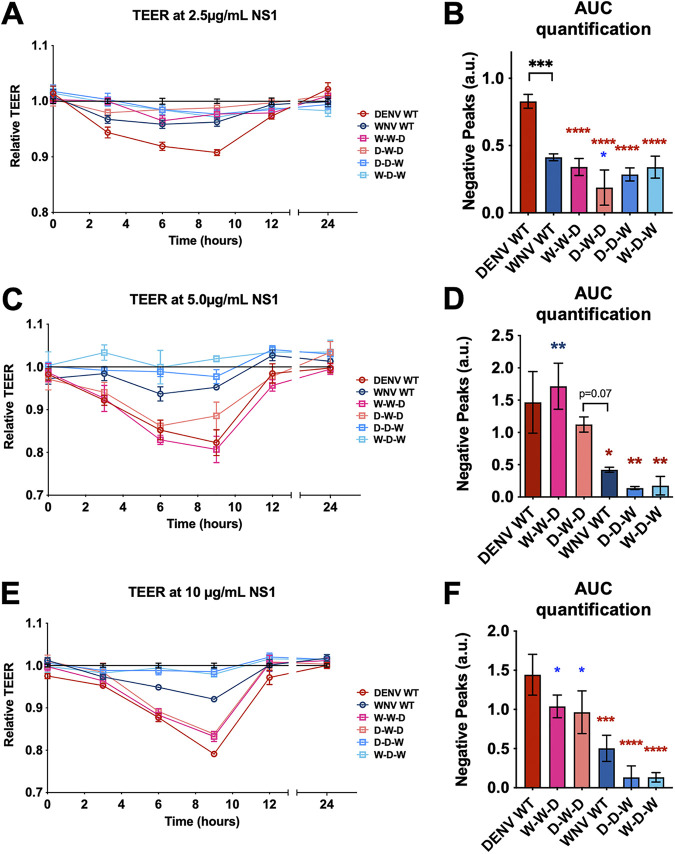
Following binding to endothelial cells, DENV NS1 is taken up by cells via clathrin-mediated endocytosis, traffics to endosomes, and activates enzymes that degrade the endothelial glycocalyx and disrupt intercellular junctional complexes (7, 10, 14, 31, 32). This results in disruption of endothelial barrier integrity, making endothelial cells hyperpermeable to solutes and fluids. We have previously shown that DENV NS1 can cause endothelial barrier dysfunction by inducing endothelial hyperpermeability independently of the virus (7, 12, 31). Given this finding, we next explored whether the NS1 wing domain that conferred DENV NS1 binding to HPMEC was also responsible for causing HPMEC hyperpermeability. To investigate how different flavivirus NS1 domains influence the capacity of NS1 to trigger endothelial hyperpermeability, we utilized the DENV-WNV NS1 chimeras in a transendothelial electrical resistance (TEER) assay. We treated HPMEC seeded in the apical chamber of a Transwell with either WT or chimeric NS1 (2.5 μg/mL) and measured the TEER values between the apical and the basolateral chambers of the Transwell (Fig. 4A and B). Consistent with previous observations (34), WT DENV NS1 resulted in significantly greater TEER reduction of HPMEC than WNV NS1, indicating greater endothelial barrier hyperpermeability. Interestingly, the constructs containing either the DENV wing or β-ladder domain (D-D-W, W-D-W, W-W-D, D-W-D) were unable to cause TEER reduction to the same extent as WT DENV NS1, indicating that while the wing domain confers cell binding, both the DENV wing and the DENV β-ladder domains are required for NS1 to trigger endothelial hyperpermeability of HPMEC (Fig. 4A and B).
FIG 4.
The wing and β-ladder domains of flavivirus NS1 are necessary for inducing endothelial cell hyperpermeability. (A) HPMEC monolayer was seeded in the apical chambers of 24-well Transwell inserts and treated with either WT or chimeric NS1 proteins at 2.5 μg/mL. The transendothelial electrical resistance (TEER) between the apical and basolateral chamber was measured over time and normalized to the untreated controls at each respective time point. (B) Quantification of the area between the curve and Y = 1.0 in (A) (“area under curve”), correlating to a decrease in electrical resistance and an increase in permeability. (C, E) Endothelial hyperpermeability (TEER) assay as in (A), but with the indicated NS1 protein at (C) 5 and (E) 10 μg/mL, respectively. (D, F) Area of the curve of (C) and (E), respectively. AUC, area under the curve. a.u., arbitrary units. Data represent at least n = 3 biological replicates, plotted as mean ± standard error of the mean (SEM). *, P < 0.05; **, P < 0.01; ***, P < 0.005; ****, P < 0.001 by one-way ANOVA with multiple comparisons. Star colors indicate the respective control to which each construct was compared.
Since we have previously observed that NS1 triggers endothelial hyperpermeability in a dose-dependent manner (7, 8), we tested whether our chimeric NS1 proteins would trigger endothelial hyperpermeability when cells were treated with a higher concentration. We repeated the TEER experiments using 5 and 10 μg/mL of our NS1 proteins (Fig. 4C to F) and observed that at both concentrations, the constructs containing the DENV NS1 β-ladder (W-W-D and D-W-D) caused a decrease in TEER on HPMEC comparably to WT DENV NS1, whereas the constructs containing the WNV β-ladder domain (D-D-W and W-D-W) did not cause endothelial hyperpermeability, similar to WT WNV NS1. This implicates a role for the DENV β-ladder in inducing endothelial hyperpermeability as measured by TEER, consistent with our previous findings (32, 40).
A 3-amino-acid motif in the wing domain of DENV NS1 confers endothelial cell binding specificity between DENV and WNV NS1.
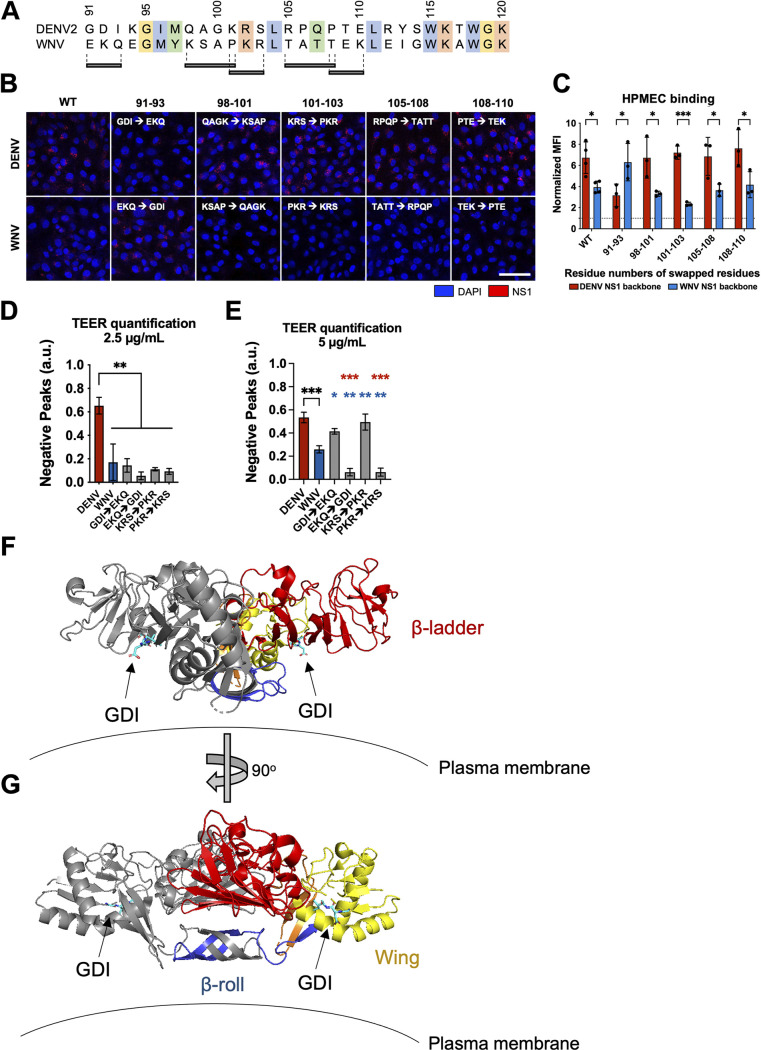
While NS1 is highly conserved across the Flavivirus genus (41), regions of variability can be found within the wing domain, specifically between amino acid (aa) residues 90 and 120 (Fig. 5A). We have previously identified the same stretch of residues within the NS1 wing domain to be immunodominant, eliciting robust antibody responses in DENV2-infected mice and natural human infections (38). Multiple structural studies have reported a flexible loop structure located near residues 90 to 120 that is thought to be critical for endothelial cell binding, given its exposure on the surface of dimeric NS1 and its predicted proximity to cell membranes (36, 42). This suggests a functional importance that could explain the tissue-specificity of flavivirus NS1. As such, we hypothesized that amino acids within residues 90 to 120 of the wing domain confer tissue-specific endothelial binding.
FIG 5.
Residues 91 to 93 of DENV NS1 drive DENV NS1–endothelial cell interactions. (A) Sequence alignment of DENV and WNV NS1 from amino acid residue 91 to 120, with black bars indicating the 3 to 4 residue motifs swapped in the site-directed mutants between DENV and WNV NS1. Colors of the residues indicate their similarities based on biochemical properties. (B) NS1 binding assay where 10 μg/mL of WT or mutant NS1 as indicated were added to HPMEC and imaged using immunofluorescence microscopy. (C) Quantification of (B). (D) HPMEC monolayer was seeded in the apical chamber of a Transwell and treated with either WT or site-directed mutant NS1 proteins at 2.5 μg/mL. Transendothelial electrical resistance (TEER) was measured over time, normalized to the untreated controls of respective time points. Area-under-the-curve (AUC) quantification of TEER curves is shown. (E) Same as (D) but treating with NS1 proteins at 5.0 μg/mL. AUC quantification of TEER curves is shown. (F, G) Dimeric DENV2 NS1 structure at 2.89 Å (PDB 7K93 [40]) was annotated to denote the location of the 91 to 93 GDI motif within the wing domain in spatial and structural reference to the rest of NS1. One monomer is colored gray while the other monomer is colored as follows: blue for β-roll, yellow for wing, red for β-ladder, and orange for interdomain connecting regions. Arrows point toward GDI motif on both monomers, colored in cyan. Plasma membrane is shown to indicate the position in which NS1 is proposed to interact. (F) and (G) are rotated 90° along the y axis from each other. All data are from at least 3 biological replicates, plotted as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.005; ****, P < 0.001 by one-way ANOVA with multiple comparisons. a.u., arbitrary units. Star colors indicate the respective control to which each construct has been compared.
To test this hypothesis, we aligned the DENV and WNV NS1 sequences to identify differentially conserved residues that were predicted to be surface-exposed, reasoning that these divergent residues may dictate tissue specificity of NS1 (36, 42). We identified five sites of 3 to 4 aa between residues 90 and 120 that contained divergent residues across flaviviruses but were conserved within each flavivirus (Fig. 5A). To test the involvement of these residues in the differential DENV-WNV NS1 cell-binding phenotype, we generated 5 pairs of site-specific mutants that exchanged these motifs between DENV and WNV NS1 (Fig. 5 and 6). We then treated HPMEC with either WT NS1 or site-specific NS1 mutants and measured the levels of NS1 binding to HPMEC by IFA (Fig. 5B). Interestingly, we found that while 4 pairs retained the binding pattern of their parental NS1 protein, one pair exhibited the opposite pattern. WNV NS1 containing DENV2 residues 91 to 93 (WNV-GDI) gained the capacity to bind to HPMEC at comparable levels to WT DENV NS1, whereas the DENV NS1 containing residues 91 to 93 from WNV NS1 (DENV-EKQ) had reduced capacity to bind to HPMEC, exhibiting levels similar to WT WNV NS1 (Fig. 5B and C). These results suggest that residues 91 to 93 of DENV NS1 form a motif that contributes to the DENV-WNV tissue-specific endothelial cell binding pattern we observe.
FIG 6.
Production and quality control of flavivirus NS1 site-directed mutants. Proteins were evaluated by silver stain following SDS-PAGE gel (A) and native PAGE (B, C). Two micrograms of each NS1 construct were used. For native gels, proteins were detected using anti-His MAb. Top-row labels indicate the flavivirus NS1 backbone. Numbering under bands refers to the residues that were swapped between DENV and WNV NS1. (A) Silver stain of site-directed NS1 mutants between DENV and WNV NS1. (B, C) Native PAGE of site-directed NS1 mutants between DENV and WNV NS1, divided into two gels each with respective WT DENV and WNV NS1 controls. WT, wild type.
To determine if residues 91 to 93 conferring NS1 cell binding specificity between DENV and WNV also dictate the capacity of DENV-WNV NS1 to trigger endothelial hyperpermeability, we conducted a TEER assay on HPMEC with NS1 mutants swapped at residues 91 to 93 and 101 to 103 for comparison (Fig. 5D). Similar to our observations above, at low NS1 levels (2.5 μg/mL), only WT DENV NS1 was able to induce endothelial hyperpermeability, in contrast to WT WNV and mutants from the DENV-WNV 91 to 93 and 101 to 103 swaps that did not noticeably alter permeability, suggesting that while the residues in the wing domain are important for inducing endothelial hyperpermeability, they alone are not sufficient.
We next repeated the TEER experiments at 5 μg/mL to confirm if the wing deficiency phenotype of DENV NS1 could be overcome by utilization of a higher dose of NS1 (Fig. 5E). WT DENV NS1 and NS1 constructs containing WNV motifs of either residues 91 to 93 and 101 to 103 (both containing DENV β-ladder) were able to trigger endothelial hyperpermeability, whereas WT WNV NS1 caused less TEER decrease, and WNV NS1 constructs containing DENV NS1 motifs of both residues 91 to 93 and 101 to 103 (both containing WNV β-ladder) remained unable to cause TEER decrease. Together, these results are consistent with the previous TEER data involving NS1 domain chimeras (Fig. 4), where at the higher NS1 concentrations of 5 and 10 μg/mL, only the constructs containing DENV β-ladder triggered endothelial hyperpermeability. Using a published DENV2 NS1 structure at 2.89 Å resolution (40), we highlight the location and orientation of the GDI motif (Fig. 5F and G), indicating its predicted interaction with the plasma membrane.
The wing domain of DENV NS1 confers NS1-induced vascular leak in vivo.
To explore whether the tissue-specific NS1-endothelial cell interactions we observed in vitro could be recapitulated in vivo, we next asked which NS1 domains were required for tissue-specific vascular leak in vivo. We used a mouse model of localized dermal leak, in which we have previously shown that DENV NS1 causes significantly higher leak than WNV NS1 (31). We shaved the hair from the back (dorsal side) of wild-type C57BL/6 mice and administered injections intradermally (ID): PBS as baseline vehicle control, WT parental NS1, and DENV-WNV NS1 chimeras. Immediately following ID injections, we retro-orbitally administered dextran conjugated to Alexa-680. Two hours post-NS1 treatment, we excised the dorsal dermis and used a fluorescent scanner to measure the extent of vascular leak as indicated by dextran-associated fluorescence, since dextran is a small molecule that can extravasate into tissues (Fig. 7A and B).
FIG 7.
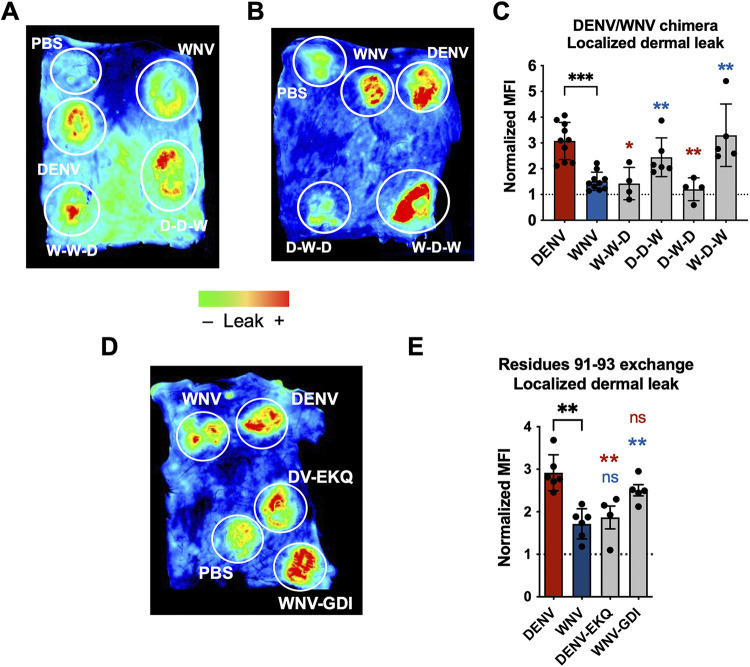
The wing domain of DENV NS1 and residues 91 to 93 within the wing domain confer NS1-induced localized vascular leak in vivo. (A, B) Wild-type C57BL/6J mice were shaved by removing hair from their backs 3 days before the experiment. On the day of experiment, PBS, WT, and chimeric NS1 proteins (15 μg) were administered intradermally into discrete spots on the shaved dorsal dermis. Immediately following NS1 injections, 25 μg of Dextran conjugated to Alexa Fluor 680 was administered retro-orbitally. Two hours postinjection, the dermis of each mouse was collected and visualized using a fluorescent scanner. Representative images of the dorsal dermis from at least 4 mice are shown. Chimeric proteins were analyzed in different mice with each mouse containing both positive and negative controls (WT DENV and WNV NS1, respectively). (C) Mean fluorescent intensity (MFI) quantification of (A) and (B), normalized to PBS injection. (D) Same as (A) and (B), but using site-directed NS1 mutants that exchanged residues 91 to 93 between DENV and WNV NS1. Representative image of the dorsal dermis from at least 4 mice backs are shown. (E) MFI quantification of (D), normalized to PBS injection. Data plotted as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.005; ****, P < 0.001 by unpaired Mann-Whitney U test. Star colors indicate the respective control to which each construct was compared.
Using this model, we found that the NS1 chimeras containing DENV wing domain (D-D-W and W-D-W) caused comparable levels of leak as WT DENV NS1, which is significantly higher leak than the leak caused by WT WNV NS1 (Fig. 7C). Conversely, the NS1 chimeras containing WNV wing domain—and DENV β-ladder (W-W-D and D-W-D)—caused leak comparable to WT WNV NS1, and significantly less than WT DENV NS1. It should be noted that while WT WNV NS1 caused visible vascular leak, the leak was not statistically significant compared to PBS. These data suggest that the DENV NS1 wing is driving NS1-induced vascular leak in the dorsal dermis of mice in vivo.
Since we identified residues 91 to 93 of DENV as a determinant of wing-mediated endothelial hyperpermeability in vitro, we asked whether this motif was also a determinant for wing-mediated dermal leak in vivo. Using the same dermal leak model as above, we intradermally injected PBS, WT parental NS1 proteins, and DENV NS1 with WNV NS1 residues 91 to 93 and vice versa, followed by retro-orbital administration of Dextran-A680 (Fig. 7D). We found that WNV NS1 with DENV residues 91 to 93 gained the ability to trigger vascular leak at comparable levels as WT DENV NS1, while DENV NS1 with WNV residues 91 to 93 caused significantly less leak, comparable to WT WNV NS1 (Fig. 7E). Taken together, these results suggest that the DENV NS1 wing domain, along with residues 91 to 93 (GDI), are required to trigger localized vascular leak in this in vivo model.
DISCUSSION
In this study, we determined that both the wing and β-ladder domains of flavivirus NS1 proteins drive differential interactions with human endothelial cells, with the wing influencing binding to endothelial cells and the β-ladder involved in inducing endothelial hyperpermeability in vitro. We further identified residues 91 to 93 (GDI) within the flexible loop of the DENV NS1 wing domain as a critical molecular determinant for tissue-specific NS1-endothelial cell binding. Finally, we showed that the wing domain, along with the GDI motif, also dictates the capacity of NS1 to trigger vascular leak in vivo. Our finding that the NS1 wing domain contributes to endothelial binding and hyperpermeability is consistent with our previous observations (40) and with studies reporting that in DENV NS1-vaccinated mice, anti-wing antibodies were highly protective against lethal DENV infection (39). While in its cell-bound dimeric form, residues within the 90 to 120 region of the wing domain are expected to interact with the plasma membrane (35, to ,37, 42), which our findings support. In addition, the hydrophobic residues in the DENV NS1 wing domain have also been implicated to be involved in membrane remodeling (43).
It could be expected that flavivirus NS1 contains both conserved core residues mediating interactions with endothelial cells, as well as divergent residues that confer tissue specificity. In agreement with this hypothesis, previous work from our group uncovered NS1 residues involved in endothelial cell binding that are conserved across the Flavivirus genus (W115-W118-G119); these residues are located in the flexible loop within the wing domain (40). In that study, we mutated the W-W-G residues to alanines; the DENV NS1-WWG mutant bound to HPMEC at lower levels than WT DENV NS1 and had reduced capacity to cause endothelial hyperpermeability in HPMEC. This finding suggests that the highly conserved WWG residues may mediate a baseline level of NS1-endothelial cell binding across all flavivirus NS1 proteins, while the tissue specificity is likely mediated by nonconserved molecular determinants. The conserved WWG residues contrast with the DENV-GDI motif we identified in the present study, which is highly variable among flaviviruses.
One interesting observation in our data was how the DENV-WNV NS1 chimeras that bound weakly at 2.5 μg/mL to HPMEC (W-W-D and D-W-D, containing WNV NS1 wing domain and DENV NS1 β-ladder), at much lower levels than WT DENV NS1, could induce HPMEC hyperpermeability at comparable levels to WT DENV NS1 at the higher NS1 concentrations of 5 and 10 μg/mL (Fig. 4). We observed the same trend again when we tested the capacity of the site-directed mutants to cause endothelial hyperpermeability, where at higher concentrations, the DENV-EKQ mutant NS1 bound at a low level to HPMEC while remaining able to cause HPMEC hyperpermeability, whereas WNV-GDI NS1 bound HPMEC but could not cause HPMEC hyperpermeability. This implicates the β-ladder in conferring endothelial cell hyperpermeability and is consistent with prior studies that identified residues in the β-ladder as important for endothelial hyperpermeability at a step following endothelial cell binding (32, 40). Specifically, a glutamine point mutant of the glycosylated residue N207 (32) and targeted mutations at residues T301, A303, E326, and D327 (40), which are in the β-ladder domain, were able to bind to HPMEC but were deficient in causing hyperpermeability. While the present study tested the relative contributions of the wing and β-ladder domains to binding and hyperpermeability, the contribution of the β-roll domain cannot be ruled out.
It is important to note that in the TEER model used to measure endothelial hyperpermeability in vitro, cells are treated with NS1 and left undisturbed over time under static conditions, in contrast to DENV infection in vivo, where NS1 travels through the bloodstream with constant blood flow. As a result, when the DENV-WNV NS1 chimeras (containing WNV wing but DENV β-ladder) that bound poorly to HPMEC were left undisturbed on HPMEC in TEER assays, we hypothesized that their low-level binding was then sufficient to induce hyperpermeability, which was driven by the DENV β-ladder domain. In the in vivo environment, we observed that the vascular leak phenotype was conferred by the wing domain, which is consistent with the observation of wing-driven endothelial binding in vitro. This suggests that in the fluid condition of the in vivo environment, it is critical for NS1 to have strong interactions with host factors on the cell surface such that NS1 binding, which is mediated by the wing domain, can occur despite blood flow.
Further, the complexity of the in vivo condition likely accounts for the discrepancy we observe between the in vitro endothelial permeability system and the in vivo murine dermal leak model, where constructs containing DENV wing and WNV β-ladder could cause vascular leak in vivo despite not causing endothelial hyperpermeability in vitro. The in vitro TEER assay contains only endothelial cells, whereas in the in vivo mouse model, other non-endothelial intrinsic factors may be at play, which could be mediated by the wing domain of NS1. In addition, the kinetics differed in our in vivo model of localized leak versus in vitro TEER assay. Finally, the NS1 proteins might have different effects on the dermal cells in the in vivo system compared to pulmonary cells in the in vitro system. Future studies are needed to fully characterize the relative contribution of non-endothelial factors to NS1-mediated vascular leak.
A critical question to address next is the mechanism by which the residues 91 to 93 of the wing domain mediate tissue-specific endothelial cell binding. We propose two models to explain the manner in which residues 91 to 93 may mediate tissue-specific interactions. One possibility is that the motif directly interacts with specific host factors on the surface of endothelial cells such as glycans or proteins. In particular, the carboxylate side chain of D92 points directly toward the cell surface; it may interact with host factors, distinct from the WWG motif, which may facilitate association with the plasma membrane (Fig. 5F and G). A second possibility is that the aa 91 to 93 motif modulates the flexibility of the flexible loop containing the WWG motif (40), which is ultimately the site predicted to interact with endothelial cells.
On the other side of NS1 tissue-specific interactions lies the host endothelial cell. The endothelial cell surface is made up of the endothelial glycocalyx, which is a network of luminal membrane-bound glycoproteins and proteoglycans with both short, branched carbohydrates and long, unbranched glycosaminoglycan side chains (44, 45). Glycocalyx components include sialic acid, heparan sulfate, chondroitin sulfate, and hyaluronan (44). The composition and ratio of the components that make up the glycocalyx can vary significantly depending on the tissue of origin of endothelial cells, which in turn can result in differential binding of viral proteins to distinct tissues. As such, different residues in both the aa 91 to 93 motif and the broader wing domain may influence the interactions with specific types of glycans on different endothelial cells.
Our study begins to uncover the molecular determinants for flavivirus NS1 binding and tissue tropism. We establish the flavivirus NS1 wing domain to be important for binding endothelial cells and causing vascular leak, and in the case of DENV NS1, identify residues 91 to 93 within the wing domain as key determinants driving such interactions. This new molecular insight into what determines flavivirus NS1 tissue specificity is crucial for understanding pan-flaviviral pathogenesis and offers new approaches for antiviral therapies.
MATERIALS AND METHODS
Cell lines.
FreeStyle 293F suspension cells (Thermo Fisher Scientific) were used for production of recombinant NS1 proteins. 293F cells were cultured in FreeStyle 293 Expression medium (Thermo Fisher Scientific) containing 1% penicillin/streptomycin (P/S) and grown in a CO2 incubator at 37°C with 8% CO2 and maintained on a cell shaker at ~130 rpm. HPMEC (HPMEC-ST1.6r) were kindly donated by J.C. Kirkpatrick at Johannes Gutenberg University, Germany, and were used for NS1 cell binding and TEER assays. HUVEC were kindly gifted from Melissa Lodoen at the University of California, Irvine. HUVEC are primary endothelial cells obtained from a single female donor (Lonza). Both HPMEC and HUVEC cell lines were propagated (passages 5 to 10) and maintained in endothelial growth medium 2 (EGM-2) using the EGM-2 bullet kit from Lonza following the manufacturer’s specifications and grown in a CO2 incubator at 37°C with 5% CO2.
NS1 mutagenesis and cloning chimeric NS1 proteins.
Chimeric NS1 proteins were produced by amplifying fragments of β-roll, wing, and β-ladder domains from the WT DENV2 NS1 (Thailand/16681), WNV NS1 (NY99), or ZIKV NS1 (Uganda MR766), using primers listed in Table 1. The N-terminus of β-roll and C-terminus of β-ladder primer sequences were flanked with nucleotide bases complementary to the protein expression vector plasmid “pMAB.” The pMAB vector encodes a N-terminal CD33 signal sequence and C-terminal 6×His tag, a kind gift from Michael Diamond, Washington University at St. Louis. The domain fragments and pMAB vector were fused together using overlap extension PCR. Site-directed NS1 mutants were produced using a site-directed mutagenesis kit (QuikChange XL site-directed mutagenesis kit, Agilent) following the manufacturer’s instructions, with primers listed in Table 1. All mutant NS1 constructs were sequence-verified with 5′ and 3′ primers that recognize the pMAB vector beyond the mutagenesis insertion region.
TABLE 1.
Primers used for mutagenesis in this study
| Construct | NS1 backbone | Mutation/insertion | Primer direction | Sequence |
|---|---|---|---|---|
| Plasmid pMAB | -a | Forward | CTCTAGACCGGTACGCGTGCCGCCACC | |
| Plasmid pMAB | Reverse | GGATCCCTGCAGCTCGAGTCAGTGATGGTGATGGTGATG | ||
| W-W-D | WNV | DENV β-ladder | Forward | AACACAACTGAATGCGACTCAAAACTCATGTCAGCG |
| WNV | DENV β-ladder | Reverse | TGACATGAGTTTTGAGTCGCATTCAGTTGTGTTGC | |
| D-D-W | DENV | WNV β-ladder | Forward | AGGATGTATTCTGCGACTCGAAGATCATTGG |
| DENV | WNV β-ladder | Reverse | AATGATCTTCGAGTCGCAGAATACATCCTG | |
| D-W-D | DENV | DENV β-roll + WNV wing | Reverse | TAGGCCTTGTGGCGTTTCTGGTTGGAAC |
| DENV | DENV β-roll + WNV wing | Forward | AAGTTCCAACCAGAAACGCCACAAGG | |
| DENV | WNV wing + DENV β-ladder | Reverse | AACTTCCAACGAATTCCAAGCGCG | |
| DENV | WNV wing + DENV β-ladder | Forward | ATCGCGCTTGGAATTCGTTGGAAGTTG | |
| W-D-W | WNV | WNV β-roll + DENV wing | Reverse | GTTTTGAAGGGGATTCAGGGTAATACTTGTAC |
| WNV | WNV β-roll + DENV wing | Forward | GTACAAGTATTACCCTGAATCCCCTTCAAAAC | |
| WNV | DENV wing + WNV β-ladder | Reverse | TCCACTTCTAAGCTATTCCAAGCTC | |
| WNV | DENV wing + WNV β-ladder | Forward | TAGAGCTTGGAATAGCTTAGAAGTGG | |
| W-Z-W | WNV | WNV β-roll + ZIKV wing | Reverse | TCTACGGGGGGATTCAGGGTAATAC |
| WNV | WNV β-roll + ZIKV wing | Forward | TATTACCCTGAATCCCCCCGTAGATTGG | |
| WNV | ZIKV wing + WNV β-ladder | Reverse | TCCACTTCTAAGCTGTTCCATGCTCTATG | |
| WNV | ZIKV wing + WNV β-ladder | Forward | ATAGAGCATGGAACAGCTTAGAAGTGGAGG | |
| Z-W-Z | ZIKV | ZIKV β-roll + WNV wing | Reverse | TAGGCCTTGTGGCGTGTCAGGATGGTAC |
| ZIKV | ZIKV β-roll + WNV wing | Forward | TACCATCCTGACACGCCACAAGG | |
| ZIKV | WNV wing + ΖΙΚV β-ladder | Reverse | ACAAGAAAGCTATTCCAAGC | |
| ZIKV | WNV wing + ΖΙΚV β-ladder | Forward | TTGGAATAGCTTTCTTGTGG | |
| W-W-Z | WNV | ZIKV β-ladder | Reverse | AATAACGGCTGGGTCGCATTCAGTTG |
| WNV | ZIKV β-ladder | Forward | AACTGAATGCGACCCAGCCGTTATTGG | |
| Z-Z-W | ZIKV | WNV β-ladder | Reverse | TGATCTTCGAATCACACTC |
| ZIKV | WNV β-ladder | Forward | AGAGTGTGATTCGAAGATCATTGG | |
| D-Z-D | DENV | DENV β-roll + ZIKV wing | Reverse | ATCTACGGGGGGATTCTGGTTGGAAC |
| DENV | DENV β-roll + ZIKV wing | Forward | AGTTCCAACCAGAATCCCCCCGTAGATTGG | |
| DENV | ZIKV wing + DENV β-ladder | Reverse | TCAACTTCCAACGAGTTCCATGCTCTATG | |
| DENV | ZIKV wing + DENV β-ladder | Forward | TAGAGCATGGAACTCGTTGGAAGTTG | |
| Z-D-Z | ZIKV | ZIKV β-roll + DENV wing | Reverse | AGTTTTGAAGGGGAGTCAGGATGGTAC |
| ZIKV | ZIKV β-roll + DENV wing | Forward | TACCATCCTGACTCCCCTTCAAAAC | |
| ZIKV | DENV wing + ZIKV β-ladder | Reverse | ACAAGAAAGCTATTCCAAGCTCT | |
| ZIKV | DENV wing + ZIKV β-ladder | Forward | TAGAGCTTGGAATAGCTTTCTTGTGG | |
| D-D-Z | DENV | ZIKV β-ladder | Reverse | ATAACGGCTGGGTCGCAGAATAC |
| DENV | ZIKV β-ladder | Forward | TATTCTGCGACCCAGCCGTTATTG | |
| Z-Z-D | ZIKV | DENV β-ladder | Reverse | ACATGAGTTTTGAATCACACTCTAATGA |
| ZIKV | DENV β-ladder | Forward | AGAGTGTGATTCAAAACTCATG | |
| Site-directed mutants | DENV | 91 to 93 EKQ (WNV sequence) | Forward | AACTATTATGACAGAGAAACAGAAAGGAATCATGCAGG |
| DENV | 91 to 93 EKQ (WNV sequence) | Reverse | TGCATGATTCCTTTCTGTTTCTCTGTCATAATAGTTAAC | |
| WNV | 91 to 93 GDI (DENV sequence) | Forward | TAGTGTCGTGGTTGGAGACATCGAGGGAATGTACAAG | |
| WNV | 91 to 93 GDI (DENV sequence) | Reverse | TTGTACATTCCCTCGATGTCTCCAACCACGACACTAAGG | |
| DENV | 98 to 101 KSAP (WNV sequence) | Forward | ACATCAAAGGAATCATGAAGTCAGCACCTCGATCTCTGCGGCCTCAGC | |
| DENV | 98 to 101 KSAP (WNV sequence) | Reverse | TGAGGCCGCAGAGATCGAGGTGCTGACTTCATGATTCCTTTGATGTC | |
| WNV | 98 to 101 QAGK (DENV sequence) | Forward | AACAGGAGGGAATGTACCAGGCAGGAAAAAAACGCCTCACCGCC | |
| WNV | 98 to 101 QAGK (DENV sequence) | Reverse | TGGCGGTGAGGCGTTTTTTTCCTGCCTGGTACATTCCCTCCTGTTTC | |
| DENV | 101 to 103 PKR (WNV) | Forward | TGCAGGCAGGACCTAAACGCCTGCGGCCTCAGC | |
| DENV | 101 to 103 PKR (WNV) | Reverse | TGAGGCCGCAGGCGTTTAGGTCCTGCCTGCATG | |
| WNV | 101 to 103 KRS (DV) | Forward | TACAAGTCAGCAAAACGATCTCTCACCGCCACC | |
| WNV | 101 to 103 KRS (DV) | Reverse | TGGCGGTGAGAGATCGTTTTGCTGACTTGTAC | |
| DENV | 105 to 108 TATT (WNV) | Forward | AAAACGATCTCTGACCGCCACCACGACTGAGCTGAAG | |
| DENV | 105 to 108 TATT (WNV) | Reverse | TTCAGCTCAGTCGTGGTGGCGGTCAGAGATCGTTTTCC | |
| WNV | 105 to 108 RPQP (DV) | Forward | ACCTAAACGCCTCCGGCCTCAGCCCGAAAAATTGGAA | |
| WNV | 105 to 108 RPQP (DV) | Reverse | TTCCAATTTTTCGGGCTGAGGCCGGAGGCGTTTAGGTG | |
| DENV | 108 to 110 TEK (WNV) | Forward | TCTGCGGCCTCAGACGGAAAAACTGAAGTATTCATG | |
| DENV | 108 to 110 TEK (WNV) | Reverse | ATGAATACTTCAGTTTTTCCGTCTGAGGCCGCAG | |
| WNV | 108 to 110 PTE (DV) | Forward | TCACCGCCACCCCCACTGAGTTGGAAATTGGC | |
| WNV | 108 to 110 PTE (DV) | Reverse | AGCCAATTTCCAACTCAGTGGGGGTGGCGGTGAG |
-, not applicable.
NS1 protein production and purification.
Plasmids containing WT or mutant NS1 sequence were transfected into FreeStyle 293F cells using polyethylenimine (PEI) (40K) (Sigma) according to the manufacturer’s instructions. Forty-eight to 72 h posttransfection, NS1-containing supernatants were collected, filtered through a 0.45-μm cellulose acetate membrane to remove cell debris, and stored at −80° prior to protein purification. The NS1-containing supernatants were thawed, mixed 1:1 with binding buffer (20 mM sodium phosphate, 500 mM sodium chloride, 20 mM imidazole, pH 7.4), and bound to HisPur cobalt resin (Thermo Fisher Scientific) with shaking for 2 h at room temperature. The NS1-resin mixture was then transferred to a column and washed 5 times in wash buffer (20 mM sodium phosphate, 500 mM sodium chloride, 25 mM imidazole, pH 7.4). NS1 was then eluted from the HisPur cobalt resin with elution buffer (20 mM sodium phosphate, 500 mM sodium chloride, 200 mM imidazole, pH 7.4) over 5 fractions. The purified NS1 stocks were then subjected to dialysis against 1× PBS for 48 h at 4°C and concentrated using Amicon filters with 10,000 molecular weight cutoff (Millipore). The Pierce BCA protein quantitation kit (Thermo Fisher Scientific) was used to quantify the purified recombinant proteins according to manufacturer’s instructions. These proteins were used for all experiments within this study.
SDS-PAGE and Western blot.
Recombinant proteins were collected in protein sample buffer (0.1 M Tris pH 6.8, 4% SDS, 4 mM EDTA, 286 mM 2-mercaptoethanol, 3.2 M glycerol, 0.05% bromophenol blue) and then resolved by SDS-PAGE. For native gels, the same protocol was followed except that the sample buffer contained no SDS and 2-mercaptoethanol. Proteins were then transferred onto nitrocellulose membranes and probed with primary antibodies diluted in Tris-buffered saline with 0.1% Tween20 (TBST) containing 5% nonfat dry milk. Membranes and antibodies were incubated overnight rocking at 4°C. The next day, membranes were washed three times with TBST before being probed with anti-mouse IgG conjugated to horseradish peroxidase (HRP) secondary antibodies diluted in 5% milk in TBST at a dilution of 1:5,000 at room temperature for 1 h. Afterwards, membranes were washed with TBST three more times before being developed with enhanced chemiluminescence (ECL) reagents and imaged on a ChemiDoc system with Image Lab software (Bio-Rad) The following antibodies were used: mouse anti-His (MA1-21315, Thermo Scientific), goat anti-mouse HRP (405306, Biolegend).
NS1 cell binding assays.
To measure binding of WT and mutant NS1 proteins to HPMEC and HUVEC, 1 × 105 cells were seeded on glass coverslips in 24-well plates. Cells were allowed to form a fully confluent monolayer for 3 days, with medium change every other day. On the day of experiment, 10 μg/mL (3 μg in 300 μL) of NS1 proteins were prepared in 10 μL medium, then added to the cells. Untreated wells were used as negative controls. NS1 and cells were incubated for 1 h at 37°C. Mouse anti-6×His antibody conjugated to Alexa Fluor 647 (Novus Biologicals) was then added at a dilution of 1:200, together with Hoechst 33342 (Immunochemistry) at a 1:2,000 dilution for staining of nuclei, for 30 min at 37°C. Cells were then washed twice in 1× PBS followed by fixation in 4% formaldehyde diluted in 1× PBS (Thermo Fisher Scientific). Coverslips were mounted onto microscope slides on a drop of ProLong Gold (Thermo Fisher Scientific) and imaged using a Zeiss LSM 710 inverted confocal microscope (CRL Molecular Imaging Center, UC Berkeley). Images were processed using ImageJ software.
Transendothelial electrical resistance (TEER).
The transendothelial electrical resistance assay was used to measure the functional effect of NS1 on endothelial barrier function in HPMEC, as previously described (34). Briefly, 1 × 105 cells (HPMEC) were seeded in 300 μL of medium on the polycarbonate membrane insert of a transwell (Transwell permeable support, 0.4 μm, 6.5 mm insert; Corning Inc.). The transwell were placed in a well on a 24-well plate, becoming the apical (upper) chamber. Then, 1.5 mL of medium was added to the basolateral (lower) chamber. Cells were allowed to form a monolayer for 3 days with media changes in both apical and basolateral chambers every day, until the interchamber electrical resistance reached about 60 Ω difference between transwells seeded with (~150Ω) and without cells (~90Ω). On the day of experiment, 2.5, 5, or 10 μg/mL of indicated NS1 proteins (0.75, 1.5, or 3 μg proteins, respectively) were mixed with media up to 10 μL and added to the apical chambers of the transwells. Electrical resistance between the apical and basolateral chambers was measured in ohms using an Epithelial Volt Ohm Meter (EVOM) with an electrode pair (World Precision Instruments), at the times indicated in the figures. Transwells containing no cells and untreated transwells containing only cells were used as negative controls to calculate the baseline electrical resistance at each time point. Relative TEER was calculated as a ratio of resistance values (). Area under the curve (AUC) analyses were calculated using the area between TEER curves and Y = 1.
Mouse model of localized vascular leak.
Five- to 8-week-old WT C57BL/6 male mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and maintained under specific pathogen-free conditions at the University of California, Berkeley, Animal Facility. Mice were housed in a controlled temperature environment on a 12-h light/dark cycle, with food and water provided ad libitum. All experimental procedures involving animals were preapproved by the Animal Care and Use Committee (ACUC) of the University of California, Berkeley. Three to 4 days prior to experiment, the dorsal dermises of 6- to 10-week-old WT C57BL/6 female mice (Jackson Laboratory) were shaved using hair clippers, and residual hair removed using Nair (Church & Dwight). On the day of experiment, 15 μg of WT or mutant NS1 was mixed with PBS in a total volume 50 μL each. NS1 mixtures and PBS were then injected intradermally (ID) into discrete spots in the shaved mouse dermis. Immediately following ID injections, 25 μg of 10-kDa dextran conjugated to Alexa Fluor 680 (1 mg/mL; Sigma) was delivered intravenously (IV) through the retro-orbital route. Two hours postinjection, mice were euthanized, and the dorsal dermis was removed and placed in petri dishes. The dermis was then placed on a fluorescent scanner (LI-COR Odyssey CLx Imaging System) to visualize the fluorescence signal accumulation at a wavelength of 700 nm. Vascular leak at the ID injection sites was quantified using Image Studio software (LI-COR Biosciences) as described previously (31, 32).
Statistics.
All quantitative analyses were conducted, and all data were plotted, using GraphPad Prism 9 software. Experiments were repeated at least 3 times, to ensure reproducibility. All experiments were designed and performed with both positive and negative controls (indicated in the figures), which were used for inclusion/exclusion determination. For immunofluorescence microscopy experiments, images of random fields were captured. For all experiments with quantitative analysis, data are displayed as mean ± standard deviation (SD) or mean ± standard error of the mean (SEM), as indicated in the figure legends. All cell-binding and TEER quantitative data were analyzed using a one-way ANOVA analysis with Tukey’s multiple-comparison test. For the localized dermal leak experiments, a nonparametric, unpaired Mann-Whitney U test was used to determine statistical significance between groups. The resulting P values from the above statistical tests were displayed as n.s., not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.005; ****, P < 0.001.
ACKNOWLEDGMENTS
We thank Janet Smith at the University of Michigan for her helpful discussions about NS1 structure-function relationships. We also thank Elias Duarte, Evan Juan, Bryan Castillo-Rojas, and Richard Ruan of the Harris laboratory for their technical assistance. Confocal imaging experiments were conducted on a Zeiss LSM 710 microscope at the CRL Molecular Imaging Center at UC Berkeley, which is supported by the Gordon and Betty Moore Foundation. This study was supported by NIH grant R01 AI124493 to E.H. S.B.B was partially supported as an Open Philanthropy Awardee of the Life Science Research Foundation.
N.T.N.L., S.B.B., and E.H. conceived the study. N.T.N.L., S.Z.R., and N.T.N.L. cloned chimeric NS1 constructs. N.T.N.L. and S.Z.R. produced and purified the recombinant NS1 proteins. N.T.N.L. performed the experiments and data analysis in this study. S.Z.R. assisted with imaging and image analysis. M.P.W. conducted experiments during the peer review process. S.B.B. and E.H. provided project guidance. E.H. acquired funding and provided resources. N.T.N.L. wrote the initial manuscript draft. N.T.N.L., S.B.B., and E.H. reviewed and edited the manuscript, and all authors provided editorial comments.
We declare no competing interests.
Contributor Information
Scott B. Biering, Email: sbiering@berkeley.edu.
Eva Harris, Email: eharris@berkeley.edu.
Rebecca Ellis Dutch, University of Kentucky College of Medicine.
REFERENCES
- 1.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, William WG, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stanaway JD, Shepard DS, Undurraga EA, Halasa YA, Coffeng LE, Brady OJ, Hay SI, Bedi N, Bensenor IM, Castañeda-Orjuela CA, Chuang T-W, Gibney KB, Memish ZA, Rafay A, Ukwaja KN, Yonemoto N, Murray CJL. 2016. The global burden of dengue: an analysis from the Global Burden of Disease Study 2013. Lancet Infect Dis 16:712–723. 10.1016/S1473-3099(16)00026-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pierson TC, Diamond MS. 2020. The continued threat of emerging flaviviruses. Nat Microbiol 5:796–812. 10.1038/s41564-020-0714-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glasner DR, Puerta-Guardo H, Beatty PR, Harris E. 2018. The good, the bad, and the shocking: the multiple roles of dengue virus nonstructural protein 1 in protection and pathogenesis. Annu Rev Virol 5:227–253. 10.1146/annurev-virology-101416-041848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Srikiatkhachorn A, Mathew A, Rothman AL. 2017. Immune-mediated cytokine storm and its role in severe dengue. Semin Immunopathol 39:563–574. 10.1007/s00281-017-0625-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malavige GN, Jeewandara C, Ogg GS. 2020. Dysfunctional innate immune responses and severe dengue. Front Cell Infect Microbiol 10:590004. 10.3389/fcimb.2020.590004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puerta-Guardo H, Glasner DR, Harris E. 2016. Dengue virus NS1 disrupts the endothelial glycocalyx, leading to hyperpermeability. PLoS Pathog 12:e1005738. 10.1371/journal.ppat.1005738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rastogi M, Singh SK. 2020. Zika virus NS1 affects the junctional integrity of human brain microvascular endothelial cells. Biochimie 176:52–61. 10.1016/j.biochi.2020.06.011. [DOI] [PubMed] [Google Scholar]
- 9.Hui L, Nie Y, Li S, Guo M, Yang W, Huang R, Chen J, Liu Y, Lu X, Chen Z, Yang Q, Wu Y. 2020. Matrix metalloproteinase 9 facilitates Zika virus invasion of the testis by modulating the integrity of the blood-testis barrier. PLoS Pathog 16:e1008509. 10.1371/journal.ppat.1008509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puerta-Guardo H, Tabata T, Petitt M, Dimitrova M, Glasner DR, Pereira L, Harris E. 2020. Zika virus nonstructural protein 1 disrupts glycosaminoglycans and causes permeability in developing human placentas. J Infect Dis 221:313–324. 10.1093/infdis/jiz331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh S, Anupriya MG, Modak A, Sreekumar E. 2018. Dengue virus or NS1 protein induces trans-endothelial cell permeability associated with VE-Cadherin and RhoA phosphorylation in HMEC-1 cells preventable by Angiopoietin-1. J Gen Virol 99:1658–1670. 10.1099/jgv.0.001163. [DOI] [PubMed] [Google Scholar]
- 12.Beatty PR, Puerta-Guardo H, Killingbeck SS, Glasner DR, Hopkins K, Harris E. 2015. Dengue virus NS1 triggers endothelial permeability and vascular leak that is prevented by NS1 vaccination. Sci Transl Med 7:304ra141. 10.1126/scitranslmed.aaa3787. [DOI] [PubMed] [Google Scholar]
- 13.Modhiran N, Watterson D, Muller DA, Panetta AK, Sester DP, Liu L, Hume DA, Stacey KJ, Young PR. 2015. Dengue virus NS1 protein activates cells via Toll-like receptor 4 and disrupts endothelial cell monolayer integrity. Sci Transl Med 7:304ra142. 10.1126/scitranslmed.aaa3863. [DOI] [PubMed] [Google Scholar]
- 14.Puerta-Guardo H, Biering SB, de Sousa FTG, Shu J, Glasner DR, Li J, Blanc SF, Beatty PR, Harris E. 2022. Flavivirus NS1 triggers tissue-specific disassembly of intercellular junctions leading to barrier dysfunction and vascular leak in a GSK-3β-dependent manner. Pathogens 11:615. 10.3390/pathogens11060615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wessel AW, Dowd KA, Biering SB, Zhang P, Edeling MA, Nelson CA, Funk KE, DeMaso CR, Klein RS, Smith JL, Cao TM, Kuhn RJ, Fremont DH, Harris E, Pierson TC, Diamond MS. 2021. Levels of circulating NS1 impact West Nile virus spread to the brain. J Virol 95:e00844-21. 10.1128/JVI.00844-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siemann D, Strange D, Maharaj P, Shi PY, Verma S. 2017. Zika virus infects human Sertoli cells and modulates the integrity of the in vitro blood-testis barrier model. J Virol 91:e00623-17. 10.1128/JVI.00623-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flamand M, Megret F, Mathieu M, Lepault J, Rey FA, Deubel V. 1999. Dengue virus type 1 nonstructural glycoprotein NS1 is secreted from mammalian cells as a soluble hexamer in a glycosylation-dependent fashion. J Virol 73:6104–6110. 10.1128/JVI.73.7.6104-6110.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutsche I, Coulibaly F, Voss JE, Salmon J, d'Alayer J, Ermonval M, Larquet E, Charneau P, Krey T, Mégret F, Guittet E, Rey FA, Flamand M. 2011. Secreted dengue virus nonstructural protein NS1 is an atypical barrel-shaped high-density lipoprotein. Proc Natl Acad Sci USA 108:8003–8008. 10.1073/pnas.1017338108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller DA, Young PR. 2013. The flavivirus NS1 protein: molecular and structural biology, immunology, role in pathogenesis and application as a diagnostic biomarker. Antiviral Res 98:192–208. 10.1016/j.antiviral.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 20.Edeling MA, Diamond MS, Fremont DH. 2014. Structural basis of Flavivirus NS1 assembly and antibody recognition. Proc Natl Acad Sci USA 111:4285–4290. 10.1073/pnas.1322036111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young PR, Hilditch PA, Bletchly C, Halloran W. 2000. An antigen capture enzyme-linked immunosorbent assay reveals high levels of the dengue virus protein NS1 in the sera of infected patients. J Clin Microbiol 38:1053–1057. 10.1128/JCM.38.3.1053-1057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Libraty DH, Young PR, Pickering D, Endy TP, Kalayanarooj S, Green S, Vaughn DW, Nisalak A, Ennis FA, Rothman AL. 2002. High circulating levels of the dengue virus nonstructural protein NS1 early in dengue illness correlate with the development of dengue hemorrhagic fever. J Infect Dis 186:1165–1168. 10.1086/343813. [DOI] [PubMed] [Google Scholar]
- 23.Modhiran N, Watterson D, Blumenthal A, Baxter AG, Young PR, Stacey KJ. 2017. Dengue virus NS1 protein activates immune cells via TLR4 but not TLR2 or TLR6. Immunol Cell Biol 95:491–495. 10.1038/icb.2017.5. [DOI] [PubMed] [Google Scholar]
- 24.Avirutnan P, Fuchs A, Hauhart RE, Somnuke P, Youn S, Diamond MS, Atkinson JP. 2010. Antagonism of the complement component C4 by flavivirus nonstructural protein NS1. J Exp Med 207:793–806. 10.1084/jem.20092545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Avirutnan P, Hauhart RE, Somnuke P, Blom AM, Diamond MS, Atkinson JP. 2011. Binding of flavivirus nonstructural protein NS1 to C4b binding protein modulates complement activation. J Immunol 187:424–433. 10.4049/jimmunol.1100750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chung KM, Liszewski MK, Nybakken G, Davis AE, Townsend RR, Fremont DH, Atkinson JP, Diamond MS. 2006. West Nile virus nonstructural protein NS1 inhibits complement activation by binding the regulatory protein factor H. Proc Natl Acad Sci USA 103:19111–19116. 10.1073/pnas.0605668103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malavige GN, Ogg GS. 2017. Pathogenesis of vascular leak in dengue virus infection. Immunology 151:261–269. 10.1111/imm.12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riswari SF, Tunjungputri RN, Kullaya V, Garishah FM, Utari GSR, Farhanah N, Overheul GJ, Alisjahbana B, Gasem MH, Urbanus RT, de Groot PG, Lefeber DJ, van Rij RP, van der Ven A, de Mast Q. 2019. Desialylation of platelets induced by Von Willebrand Factor is a novel mechanism of platelet clearance in dengue. PLoS Pathog 15:e1007500. 10.1371/journal.ppat.1007500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chao C-H, Wu W-C, Lai Y-C, Tsai P-J, Perng G-C, Lin Y-S, Yeh T-M. 2019. Dengue virus nonstructural protein 1 activates platelets via Toll-like receptor 4, leading to thrombocytopenia and hemorrhage. PLoS Pathog 15:e1007625. 10.1371/journal.ppat.1007625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Avirutnan P, Zhang L, Punyadee N, Manuyakorn A, Puttikhunt C, Kasinrerk W, Malasit P, Atkinson JP, Diamond MS. 2007. Secreted NS1 of dengue virus attaches to the surface of cells via interactions with heparan sulfate and chondroitin sulfate E. PLoS Pathog 3:e183. 10.1371/journal.ppat.0030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glasner DR, Ratnasiri K, Puerta-Guardo H, Espinosa DA, Beatty PR, Harris E. 2017. Dengue virus NS1 cytokine-independent vascular leak is dependent on endothelial glycocalyx components. PLoS Pathog 13:e1006673. 10.1371/journal.ppat.1006673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang C, Puerta-Guardo H, Biering SB, Glasner DR, Tran EB, Patana M, Gomberg TA, Malvar C, Lo NTN, Espinosa DA, Harris E. 2019. Endocytosis of flavivirus NS1 is required for NS1-mediated endothelial hyperpermeability and is abolished by a single N-glycosylation site mutation. PLoS Pathog 15:e1007938. 10.1371/journal.ppat.1007938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barbachano-Guerrero A, Endy TP, King CA. 2020. Dengue virus non-structural protein 1 activates the p38 MAPK pathway to decrease barrier integrity in primary human endothelial cells. J Gen Virol 101:484–496. 10.1099/jgv.0.001401. [DOI] [PubMed] [Google Scholar]
- 34.Puerta-Guardo H, Glasner DR, Espinosa DA, Biering SB, Patana M, Ratnasiri K, Wang C, Beatty PR, Harris E. 2019. Flavivirus NS1 triggers tissue-specific vascular endothelial dysfunction reflecting disease tropism. Cell Rep 26:1598–1613.e8. 10.1016/j.celrep.2019.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akey DL, Brown WC, Dutta S, Konwerski J, Jose J, Jurkiw TJ, DelProposto J, Ogata CM, Skiniotis G, Kuhn RJ, Smith JL. 2014. Flavivirus NS1 structures reveal surfaces for associations with membranes and the immune system. Science 343:881–885. 10.1126/science.1247749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown WC, Akey DL, Konwerski JR, Tarrasch JT, Skiniotis G, Kuhn RJ, Smith JL. 2016. Extended surface for membrane association in Zika virus NS1 structure. Nat Struct Mol Biol 23:865–867. 10.1038/nsmb.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu X, Song H, Qi J, Liu Y, Wang H, Su C, Shi Y, Gao GF. 2016. Contribution of intertwined loop to membrane association revealed by Zika virus full-length NS1 structure. EMBO J 35:2170–2178. 10.15252/embj.201695290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hertz T, Beatty PR, MacMillen Z, Killingbeck SS, Wang C, Harris E. 2017. Antibody epitopes identified in critical regions of dengue virus nonstructural 1 protein in mouse vaccination and natural human infections. J Immunol 198:4025–4035. 10.4049/jimmunol.1700029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lai YC, Chuang YC, Liu CC, Ho TS, Lin YS, Anderson R, Yeh TM. 2017. Antibodies against modified NS1 wing domain peptide protect against dengue virus infection. Sci Rep 7:69–75. 10.1038/s41598-017-07308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biering SB, Akey DL, Wong MP, Brown WC, Lo NTN, Puerta-Guardo H, Tramontini Gomes de Sousa F, Wang C, Konwerski JR, Espinosa DA, Bockhaus NJ, Glasner DR, Li J, Blanc SF, Juan EY, Elledge SJ, Mina MJ, Beatty PR, Smith JL, Harris E. 2021. Structural basis for antibody inhibition of flavivirus NS1-triggered endothelial dysfunction. Science 371:194–200. 10.1126/science.abc0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rastogi M, Sharma N, Singh SK. 2016. Flavivirus NS1: a multifaceted enigmatic viral protein. Virol J 13:131. 10.1186/s12985-016-0590-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akey DL, Brown WC, Jose J, Kuhn RJ, Smith JL. 2015. Structure-guided insights on the role of NS1 in flavivirus infection. Bioessays 37:489–494. 10.1002/bies.201400182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ci Y, Liu ZY, Zhang NN, Niu Y, Yang Y, Xu C, Yang W, Qin CF, Shi L. 2020. Zika NS1-induced ER remodeling is essential for viral replication. J Cell Biol 219:e201903062. 10.1083/jcb.201903062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinbaum S, Tarbell JM, Damiano ER. 2007. The structure and function of the endothelial glycocalyx layer. Annu Rev Biomed Eng 9:121–167. 10.1146/annurev.bioeng.9.060906.151959. [DOI] [PubMed] [Google Scholar]
- 45.Srikiatkhachorn A, Kelley JF. 2014. Endothelial cells in dengue hemorrhagic fever. Antiviral Res 109:160–170. 10.1016/j.antiviral.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]