ABSTRACT
The nucleolus is the largest structure in the nucleus, and it plays roles in mediating cellular stress responses and regulating cell proliferation, as well as in ribosome biosynthesis. The nucleolus is composed of a variety of nucleolar factors that interact with each other in a complex manner to enable its function. Many viral proteins interact with nucleolar factors as well, affecting cellular morphology and function. Here, to investigate the association between mumps virus (MuV) infection and the nucleolus, we evaluated the necessity of nucleolar factors for MuV proliferation by performing a knockdown of these factors with small interfering (si)RNAs. Our results reveal that suppressing the expression of Treacle, which is required for ribosome biosynthesis, reduced the proliferative potential of MuV. Additionally, the one-step growth kinetics results indicate that Treacle knockdown did not affect the viral RNA and protein synthesis of MuV, but it did impair the production of infectious virus particles. Viral matrix protein (M) was considered a candidate Treacle interaction partner because it functions in the process of particle formation in the viral life cycle and is partially localized in the nucleolus. Our data confirm that MuV M can interact with Treacle and colocalize with it in the nucleolus. Furthermore, we found that viral infection induces relocalization of Treacle in the nucleus. Together, these findings suggest that interaction with Treacle in the nucleolus is important for the M protein to exert its functions late in the MuV life cycle.
IMPORTANCE The nucleolus, which is the site of ribosome biosynthesis, is a target organelle for many viruses. It is increasingly evident that viruses can favor their own replication and multiplication by interacting with various nucleolar factors. In this study, we found that the nucleolar protein Treacle, known to function in the transcription and processing of pre-rRNA, is required for the efficient propagation of mumps virus (MuV). Specifically, our data indicate that Treacle is not involved in viral RNA or protein synthesis but is important in the processes leading to viral particle production in MuV infection. Additionally, we determined that MuV matrix protein (M), which functions mainly in viral particle assembly and budding, colocalized and interacted with Treacle. Furthermore, we found that Treacle is distributed throughout the nucleus in MuV-infected cells. Our research shows that the interaction between M and Treacle supports efficient viral growth in the late stage of MuV infection.
KEYWORDS: M protein, Treacle, mumps virus, nucleolus
INTRODUCTION
Viruses in the family Paramyxoviridae, within the order Mononegavirales, harbor a nonsegmented single negative-sense RNA genome. The group of paramyxoviruses includes many human and animal pathogens, such as mumps virus (MuV), measles virus (MeV), human parainfluenza virus, Nipah virus (NiV), Hendra virus (HeV), and Newcastle disease virus (NDV). These viruses cause a variety of illnesses with symptom severities ranging widely, from asymptomatic to fatal (1–3).
MuV (taxonomic name Mumps orthorubulavirus) is the causative agent of mumps, which is a common contagious childhood disease characterized by fever and painful swelling of the salivary glands (parotitis) often accompanied by serious complications, such as aseptic meningitis, deafness, pancreatitis, orchitis, and oophoritis (4, 5). The viral genome of MuV is 15,384 nucleotides in length and contains seven genes encoding eight proteins: the nucleocapsid (N), phospho- (P), V, matrix (M), fusion (F), small hydrophobic (SH), hemagglutinin-neuraminidase (HN), and large (L) proteins (6, 7).
Paramyxovirus M proteins interact with both the viral ribonucleoprotein complex, composed of genomic RNA and three viral proteins (N, P, and L), and the viral envelope proteins (F and HN), and they induce viral particle formation at the plasma membrane (8, 9). Although the paramyxovirus life cycle is completed in the cytoplasm, M protein is known to also localize in the nucleolus during many paramyxovirus infections (10). M protein has both a nuclear localization signal and a leucine-rich nuclear export signal and is considered to shuttle between the cytoplasm and the nucleus (10, 11). It has been reported that the transport of M protein from the nucleolus to the cytoplasm is dependent on the ubiquitination of lysine residues within the nuclear localization signal of the protein and that inhibition of M protein ubiquitination via drugs or gene knockdown reduces the formation of viral particles (10–13).
The nucleolus is a large nonmembranous organelle within the nucleus known primarily for its role in ribosome biogenesis. The nucleolus is divided into three regions: the fibrillar center (FC), the dense fibrillar component (DFC), and the granular component (GC). Ribosomal (r)DNA is transcribed by RNA polymerase I (Pol I) at the boundary between the FC and the DFC, and then preribosomal (pre-r)RNA is processed at the DFC and the GC. Pre-rRNA and ribosomal proteins assemble into the precursors of the 40S and 60S subunits at the GC, after which these subunits are transported to the cytoplasm where ribosome maturation occurs (14, 15).
Many of the sublocalizations of many nucleolar proteins have been reported; for example, Treacle (16, 17), NOLC1 (14), and NOP58 (18) function in the FC/DFC, whereas B23 (14), NOP2 (19, 20), and NOL12 (21) function in the DFC/GC. Treacle, encoded by the TCOF1 gene, has been predicted to be a nucleolar phosphoprotein, and a genetic mutation within TCOF1 is a known cause of Treacher Collins-Franceschetti syndrome (22). In the process of ribosome biosynthesis, Treacle is involved in the transcription and methylation of pre-rRNA (17, 23).
The nucleolus also has other functions; it plays roles in cell cycle regulation, cell proliferation, and stress sensing (24). Nucleolus functions are important in several viral infections (25–29), and some nucleolar proteins, such as B23, fibrillarin, upstream binding factor (UBF), and Treacle, have been identified as interaction partners of paramyxovirus (NiV, HeV, or NDV) M proteins. The suppression of these proteins increases or decreases the level of viral production, depending on the virus (10, 30–32). However, the relationship between MuV infection and nucleolar factors has not been reported in detail and remains poorly understood. Here, we investigated whether nucleolar proteins are involved in MuV infection and are associated with M proteins.
RESULTS
Depletion of Treacle inhibits MuV propagation.
To investigate whether nucleolus functions are involved in MuV infection, we selected six nucleolar factors for examination: Treacle, NOLC1, NOP58, B23, NOP2, and NOL12. We first confirmed that cell viability was not affected by the knockdown of any target factor at 168 h posttransfection (Fig. 1A). Viral growth was then assessed by using a recombinant MuV expressing an Aequorea coerulescens green fluorescent protein (rMuV-AcGFP) (33). As shown in Fig. 1B, a Treacle knockdown significantly reduced virus propagation, whereas knockdowns of the other target nucleolar proteins did not affect viral growth.
FIG 1.
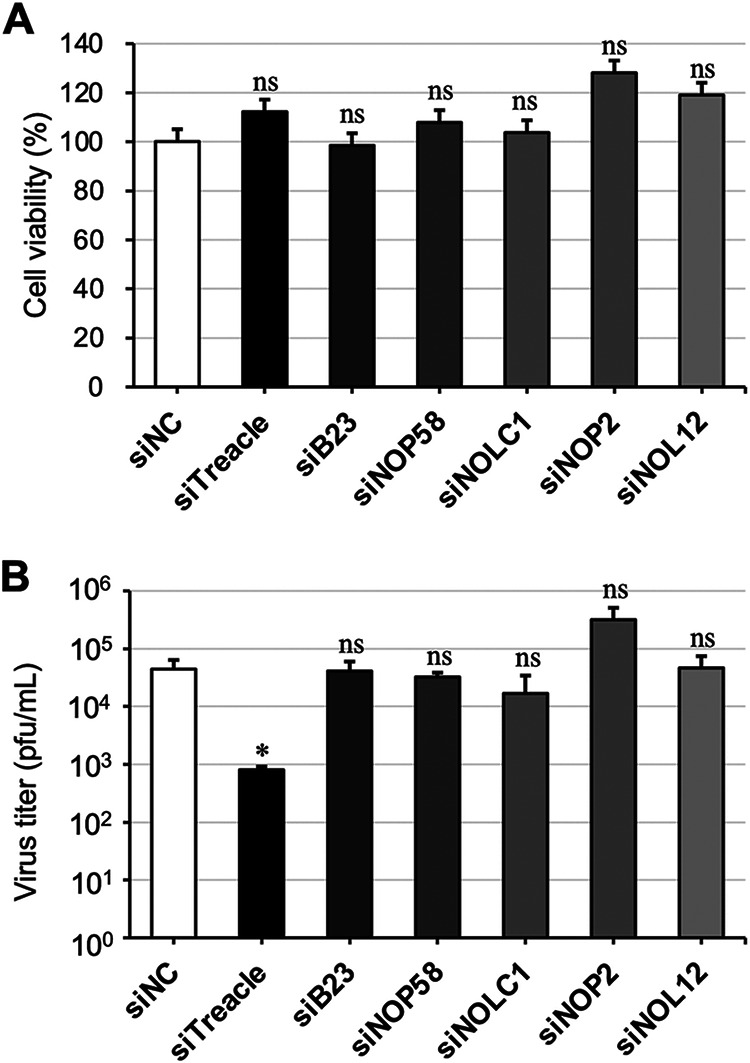
The effect of nucleolar protein knockdowns on mumps virus (MuV) infection. (A) A549 cells were transfected with siNC, siTreacle, siB23, siNOP58, siNOLC1, siNOP2, or siNOL12. At 168 h posttransfection, the cell viabilities were measured. (B) A549 cells transfected with each small interfering RNA (siRNA) were infected with a recombinant MuV expressing an Aequorea coerulescens green fluorescent protein (rMuV-AcGFP) at a multiplicity of infection (MOI) of 0.1 at 72 h posttransfection and then cultured for 96 h. Infectious viral titers were determined by conducting a plaque assay. The presented data are the means of triplicate measurements, with error bars indicating the standard deviation (SD). The significance of the difference between cells treated with siNC and cells treated with siTreacle, siB23, siNOP58, siNOP2, or siNOL12 was determined by performing a Student’s t test. *, P < 0.05; ns, not significant.
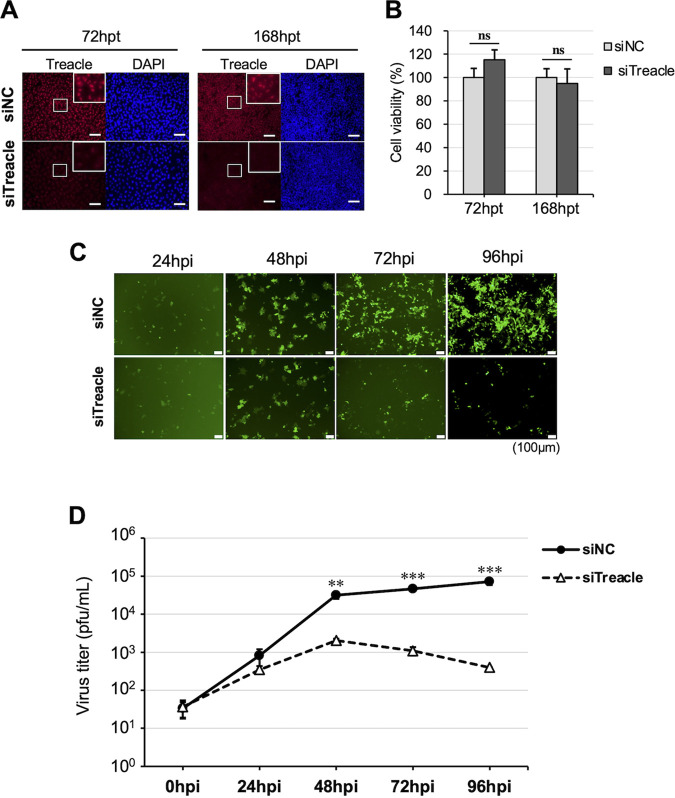
We then further assessed the viral replication kinetics by conducting a time-course analysis of MuV in Treacle-knockdown cells. The suppression of Treacle was confirmed at 72 and 168 h posttransfection by immunofluorescence assay (Fig. 2A). No cytotoxicity was observed at 72 or 168 h after treatment with Treacle-specific short interfering RNA (siTreacle) (Fig. 2B). The AcGFP signal representing virus infection increased in the cells treated with control small interfering RNA (siNC) up to 96 h postinfection (hpi), whereas the expression of AcGFP was remarkably lower in Treacle-knockdown cells at 48 to 96 hpi (Fig. 2C). Consistent with the above finding, the infectious viral titers at 48 to 96 hpi in Treacle-knockdown cells were lower than those in control cells (Fig. 2D). These results indicate that the nucleolar protein Treacle is important for MuV propagation.
FIG 2.
The effect of Treacle depletion on viral propagation. A549 cells were transfected with siNC or siTreacle. (A) At 72 or 168 h posttransfection, the cells were stained with anti-Treacle rabbit polyclonal antibody (pAb), followed by Alexa Fluor 594-conjugated goat anti-rabbit IgG. Enlarged images are shown in the white boxed areas. Bars, 20 μm. (B) The cell viabilities were measured at 72 h (left) or 168 h (right) posttransfection. (C, D) At 72 h posttransfection, the cells were infected with rMuV-AcGFP at an MOI of 0.1. These cells were observed by fluorescence microscopy at 0, 24, 48, 72, and 96 h postinfection (hpi) (C), and viral titers in the supernatant at 0, 24, 48, 72, and 96 hpi were determined by a plaque assay (D). Bars, 100 μm. The presented data are the means of triplicate measurements, with error bars indicating the SD. The significance of the difference between siNC-treated cells and siTreacle-treated cells was determined by a Student’s t test. **, P < 0.01; ***, P < 0.001; ns, not significant; DAPI, 4′,6-diamidino-2-phenylindole.
Treacle is involved in the late stages of the MuV life cycle.
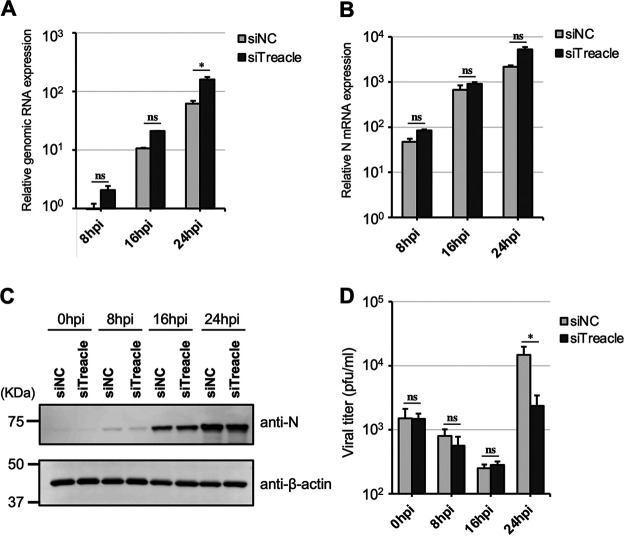
To determine the MuV life cycle stages in which Treacle is involved, we evaluated the effects of Treacle knockdown on RNA synthesis, protein synthesis, and viral production in a one-step growth assay. As shown in Fig. 3A and B, the levels of genomic RNA and mRNA in the cells treated with siTreacle were not significantly different from those in the cells treated with siNC. The only exception was that the amount of genomic RNA was higher in Treacle-knockdown cells than in control cells at 24 hpi. The expression levels of N protein in both groups of cells were comparable up to 24 hpi (Fig. 3C). Although there was no difference in viral titer between the cells treated with siNC and those treated with siTreacle at 0 to 16 hpi, the amount of infectious virus released into the supernatant of Treacle-knockdown cells at 24 hpi was significantly lower than that of control cells (Fig. 3D). These data indicate that Treacle plays important roles in the late stages of MuV infection (after the completion of viral RNA and protein synthesis).
FIG 3.
One-step growth kinetics of MuV in Treacle-knockdown cells. A549 cells were transfected with siNC or siTreacle. At 72 h posttransfection, the cells were infected with rMuV-AcGFP at a multiplicity of infection (MOI) of 3 and then cultured for 0, 8, 16, or 24 h. (A, B) The expression levels of genomic RNA (A) and mRNA (B) were determined by reverse transcription-quantitative PCR (qRT-PCR). The values were normalized to the value for the control gene HPRT1. (C) The expression of N protein was evaluated by Western blotting. (D) The viral titer in the supernatant was determined by a plaque assay. The presented data are the means of triplicate measurements, with error bars indicating the SD. The significance of the difference between siNC-treated and siTreacle-treated cells was determined by a Student’s t test. *, P < 0.05; ns, not significant.
The MuV M protein colocalizes with FC/DFC nucleolar proteins.
Because M protein is the only viral protein that has been detected in the nucleolus, we next confirmed the localization of M protein within the nucleolus by using nucleolar marker proteins. M protein partially colocalized with UBF (an FC marker) and fibrillarin (a DFC marker), as highlighted by white arrows in Fig. 4A and B. In contrast, nucleolin (a GC marker) was localized around M protein (Fig. 4C). These results suggest that the MuV M protein is localized in the FC and DFC rather than in the GC of the nucleolus.
FIG 4.
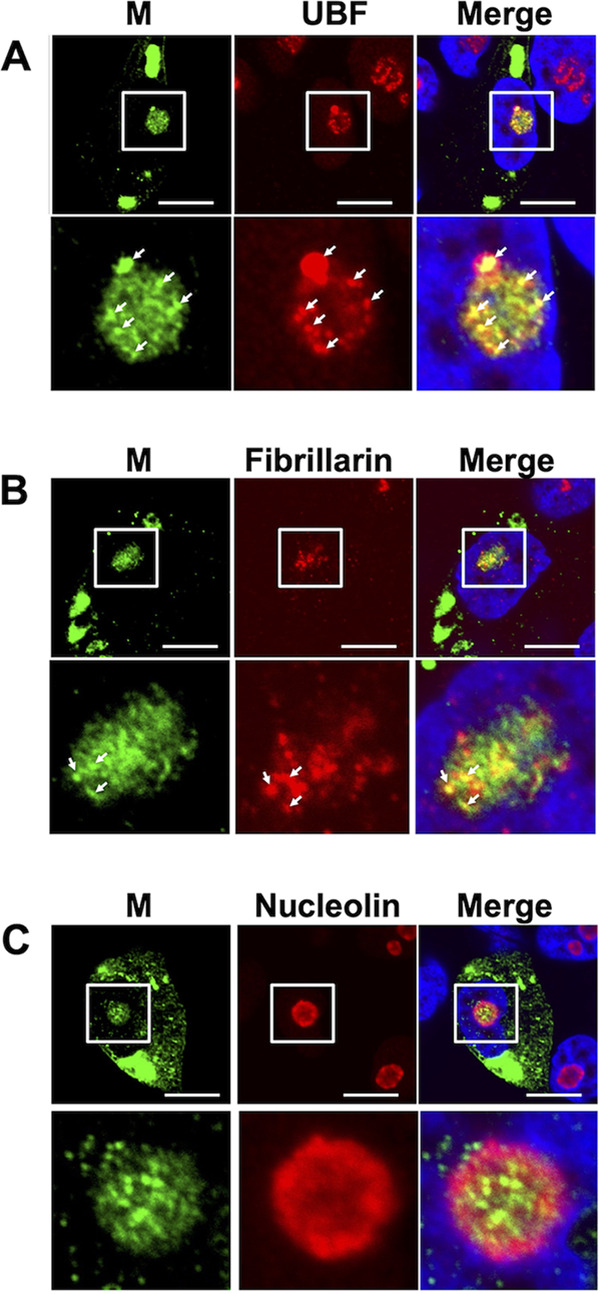
Nucleolar localization of M protein in MuV-infected cells. Vero cells were infected with MuV. At 16 hpi, the cells were incubated with anti-MuV M antibody and anti-upstream binding factor (UBF) (A), anti-fibrillarin (B), or anti-nucleolin antibodies (C), followed by Alexa Fluor 488- and Alexa Fluor 594-conjugated secondary antibodies. The enlarged images in the lower panels correspond to the white boxes in the upper panels. White arrows highlight the colocalization of M protein and UBF (A) or fibrillarin (B). Bars, 10 μm.
The M proteins of MuV and MeV colocalize with Treacle, and Treacle relocalization is induced in virus-infected cells.
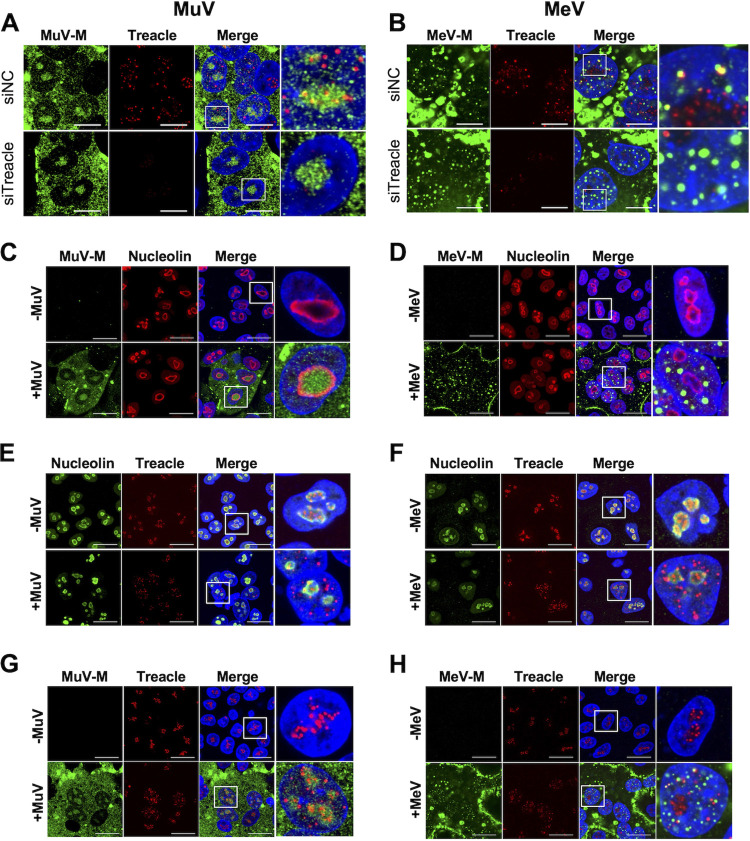
Treacle functions in the FC and the DFC of the nucleolus (17). Thus, we next examined whether the MuV M protein and Treacle colocalize in MuV-infected cells. Our data show that MuV M protein was partially colocalized with Treacle in the nucleus (Fig. 5A, upper panel). However, a Treacle knockdown did not affect the transport to the nucleus of MuV M protein (Fig. 5A, lower panels). To determine whether Treacle also colocalizes with the M protein of another paramyxovirus, we investigated MeV-infected cells. Like the MuV M protein, the MeV M protein colocalized with Treacle in the nucleus (Fig. 5B, upper panels), and its transport was not affected by a Treacle knockdown (Fig. 5B, lower panels). These data suggest that the MuV M and MeV M proteins were each transported to the nucleus independently of Treacle function but that the M proteins of both viruses colocalized with Treacle when they were translocated into the nucleus. Despite both the MuV M and MeV M proteins colocalizing with Treacle in the nucleus, the intranuclear dynamics of the MeV M protein appeared to be different from those of the MuV M protein. In virus-infected cells, the MuV M protein was concentrated inside the area of staining for nucleolin (the “nucleolin-rich compartment”) (Fig. 5C), whereas MeV M protein was concentrated on or outside the nucleolin-rich compartment (Fig. 5D). Furthermore, we found that the nuclear dynamics of Treacle are altered by viral infection. Treacle was concentrated inside the nucleolin-rich compartment in uninfected cells (Fig. 5E and F, upper panels), but it was relocated to outside the nucleolin-rich compartment and distributed throughout the nucleus in MuV- or MeV-infected cells (Fig. 5E and F, lower panels). Additionally, we revealed that although the MuV M protein colocalized with the Treacle present inside the nucleolin-rich compartment (Fig. 5G), the MeV M protein colocalized with the Treacle present outside the nucleolin-rich compartment (Fig. 5H). These results suggest that MuV and MeV infections induce a relocalization of Treacle in the nucleus.
FIG 5.
Colocalization of M protein and Treacle and relocalization of Treacle in MuV- or measles virus (MeV)-infected cells. (A, B) A549 or A549/hSLAM cells that had been transfected with siNC or siTreacle were subsequently infected with MuV (A) or MeV (B) at a multiplicity of infection (MOI) of 0.1. At 24 hpi, the cells were incubated with a combination of anti-Treacle antibody and anti-MuV M or anti-MeV M antibody, followed by appropriate secondary antibodies. (C to H) A549 cells or A549/hSLAM cells were infected with MuV (C, E, and G) or MeV (D, F, and H) at an MOI of 0.1. At 24 hpi, the cells were incubated with a combination of anti-MuV or MeV M antibody and anti-nucleolin antibody (C, D) or with a combination of anti-nucleolin and anti-Treacle antibodies (E, F) or with a combination of anti-MuV or MeV M antibody and anti-Treacle antibody (G, H), followed by appropriate secondary antibodies. The enlarged images shown in the right panels correspond to the white boxes in the panels to their left. Noninfected cells and MuV- or MeV-infected cells are shown in the upper and lower panels, respectively. Bars, 10 μm.
To investigate whether Treacle interacts with the MuV M protein, we constructed two plasmids: one for the expression of N-terminal hemagglutinin tag-fused Treacle (HA-Treacle) and one for the expression of C-terminal FLAG- and Strep tag-fused M protein (M-FOS). We confirmed that the cellular localizations of HA-Treacle and M-FOS were not different from those of wild-type Treacle and M protein, respectively (Fig. 6A and B). The results of a pulldown assay show that M-FOS interacted with HA-Treacle (Fig. 6C). Together, these findings reveal that the MuV M protein interacts with Treacle.
FIG 6.
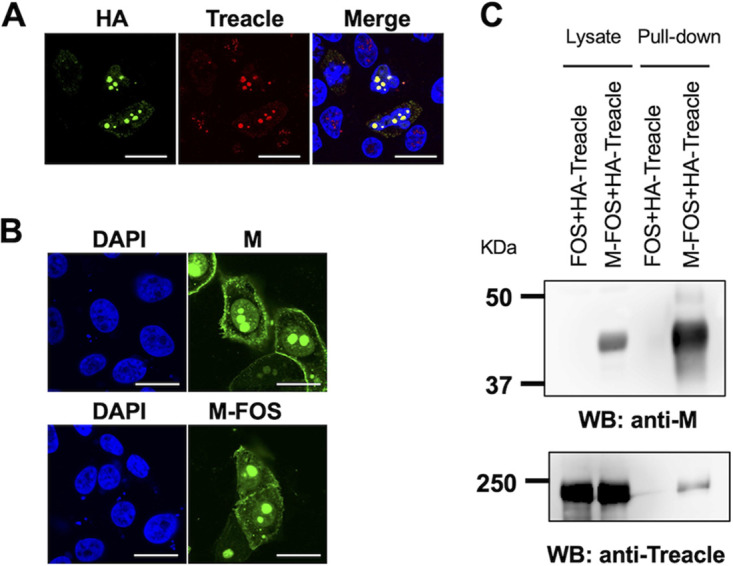
Interaction of MuV M protein with Treacle by pulldown assay. (A) A549 cells were transfected with the hemagglutinin (HA)-Treacle expression plasmid. At 48 h posttransfection, the cells were incubated with anti-HA and anti-Treacle antibodies, followed by appropriate secondary antibodies. Bars, 10 μm. (B) Vero cells were transfected with the expression plasmids for M or FLAG- and Strep tag-fused M protein (M-FOS) protein. At 48 h posttransfection, the cells were incubated with an anti-M antibody, followed by an Alexa Fluor 488-conjugated secondary antibody. Bars, 20 μm. (C) 293T cells were cotransfected with the expression plasmid for HA-Treacle and that for M-FOS protein or an empty vector. At 48 h posttransfection, the cell lysate was pulled down using Strep-Tactin beads, and the precipitants were then subjected to SDS-PAGE and Western blotting (WB) with anti-MuV M and anti-Treacle antibodies.
Treacle is involved in the propagation of both MuV and MeV.
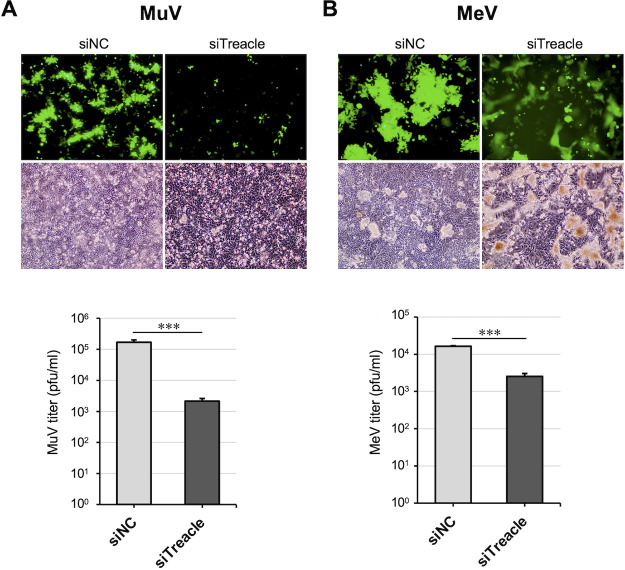
Finally, we examined whether Treacle is important for infections with other paramyxoviruses by investigating the effect of Treacle depletion on MeV propagation. In this experiment, a recombinant MeV expressing EGFP (rMeV-eGFP) (34) was used. As shown in Fig. 7A, a Treacle knockdown reduced the amount of rMuV-AcGFP growth by ~100-fold, which is similar to the results shown in Fig. 2D and 3D. Following rMeV-eGFP infection, enhanced membrane fusion and a strong cytopathic effect were observed in the Treacle-knockdown cells. The rMeV-eGFP viral titer in Treacle-knockdown cells was ~10-fold lower than that in control cells (Fig. 7B). These results suggest that Treacle is involved in MeV propagation.
FIG 7.
The effect of Treacle knockdown on MeV propagation. (A, B) A549 cells (A) or A549/hSLAM cells (B) that had been transfected with siNC or siTreacle were subsequently infected with rMuV-AcGFP (A) or a recombinant MeV expressing enhanced green fluorescent protein (rMeV-eGFP) (B) at a multiplicity of infection (MOI) of 0.05. The cells were observed by fluorescence microscopy and phase-contrast microscopy at 72 hpi (upper panels). The virus titers in the supernatant at 72 hpi were determined by a plaque assay. The presented virus titer is the mean of triplicate measurements, with error bars indicating the SD. The significance of the difference between siNC- and siTreacle-treated cells was determined by a Student’s t test. ***, P < 0.001; ns, not significant.
DISCUSSION
Although the majority of mononegaviruses complete their life cycle in the cytoplasm, various components of these viruses are localized in the nucleolus of infected cells (10, 29, 35). Several nucleolar proteins that interact with viral proteins have been identified, and they may be involved in the regulation of viral replication (25, 26, 28, 29). Thus, interaction with the nucleolus is an important event in the life cycles of many mononegaviruses, even if the virus does not use the nucleus for its replication. In paramyxoviruses, the nucleolus is thought to be a viral target because their M proteins localize in the nucleolus and interact with nucleolar factors, thereby regulating viral propagation (36). The present study demonstrated that Treacle plays an important role in MuV propagation. The primary function of Treacle is to activate RNA Pol I with UBF for the transcription of rDNA in the FC. Although synthesized pre-rRNA is methylated in the DFC by Box C/D of the snoRNP complex composed of fibrillarin, NOP56, NOP58, and NHP2L1, Treacle is involved in this methylation via NOP56 (14, 17, 37, 38). Furthermore, Treacle and its paralog NOLC1 are each monoubiquitinated by CUL3-KBTBD8 to form the Treacle-NOLC1 complex, which regulates RNA Pol I and is involved in pre-rRNA pseudouridylation (H/ACA complex) and the small-subunit (SSU) process that controls ribosomal subunit maturation and modification (39). Our data show that MuV proliferation was not affected in cells in which NOLC1 expression was suppressed, suggesting that the Treacle-NOLC1 complex may not be involved in MuV infection.
Treacle is also associated with the cellular response to DNA double-strand breaks that are induced by exposure to various stresses in the nucleolus (14, 40). Although double-strand breaks on rDNA induce the DNA damage response, i.e., DNA damage detection and the DNA damage repair system, some factors, such as NBS-1 and TOPBP1, are recruited to rDNA by Treacle, after which rDNA is quarantined from the nucleolus (41, 42). Recently, Rawlinson et al. identified Treacle as an interaction partner of the HeV M protein and found that, in HeV-infected cells, M protein interacts with Treacle to inhibit pre-rRNA synthesis by mimicking the function of NBS-1 (32). Notably, in contrast with our results, their report shows that suppressing Treacle expression increases the HeV titer (32). This difference suggests that Treacle function may differ between MuV and HeV infections. We also observed that a Treacle knockdown affected the cell-cell fusion activity in MeV-infected cells but not in MuV-infected cells. Persistent MeV infection of the brain causes a fatal progressive neurological disorder known as subacute sclerosing panencephalitis. In many patients with this condition, MeV has defective M protein expression (43) and fails to produce cell-free viruses. Additionally, previous reports showed that mutations inhibiting interaction between the M protein and the viral surface glycoproteins enhance cell-cell fusion and that an M-less MeV exhibits a hyperfusion phenotype (44). Elimination of the Treacle-M protein interaction may have promoted membrane fusion and reduced viral production.
Unexpectedly, given the reports from previous studies that B23 (30, 45–47), NOP58 (31), NOP2 (20), and NOLC1 (48) are involved in the regulation of many other viral infections, knockdowns of other nucleolar factors did not impair MuV propagation. B23 and NOP58 bind to the M protein of NDV or NiV, and knockdowns of these factors suppress NDV and NiV replication (30, 31). Thus, the findings of our study and those of other groups together indicate that, despite belonging to the same family (Paramyxoviridae), many paramyxoviruses use different nucleolar factors for viral propagation. Furthermore, the function of M protein is unlikely to be conserved among paramyxoviruses, as the M protein of NiV or HeV is sufficient to induce virus-like particles, whereas MuV particle formation requires not only M protein but also N and F proteins (10, 49–54).
This study provides new insight into the relocalization of Treacle from the nucleolus to the nucleoplasm in MuV- or MeV-infected cells. Some studies on herpes simplex virus 1 (HSV-1) indicated that UBF becomes redistributed throughout the nucleus and accumulates in viral replication components that are formed in the nucleus during HSV-1 infection (55, 56). In addition, nucleolin and B23 are also redistributed from the nucleolus to the nucleoplasm during HSV-1 infection, which has been suggested to be dependent on the expression of viral protein UL24, which localizes in the nucleoli (57, 58). Although we have not analyzed other nucleolar proteins in this study, it is likely that many components are altered by infection with MuV or MeV. It is also possible that M proteins, like the UL24 protein of HSV-1, may be involved in Treacle relocalization during viral infection. Further studies are necessary to clarify the significance of the dynamic changes in nucleolar components that occur in cells infected with MuV or MeV.
In conclusion, we demonstrated the importance of the association between the MuV M protein and Treacle during viral infection, and our results suggest that the nucleolus is closely related to the MuV life cycle. However, the detailed molecular mechanisms that promote the late stage of MuV propagation are still unknown. Nevertheless, the identification of a host nucleolar factor involved in paramyxovirus propagation is an important step toward understanding paramyxoviruses.
MATERIALS AND METHODS
Cells.
Vero (Africa green monkey kidney) cells provided by the U.S. Food and Drug Administration and 293T (human embryonic kidney) (ATCC CRL-3216) and A549 (human lung epithelial) (ATCC CCL-185) cells obtained from ATCC were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Nacalai Tesque, Kyoto, Japan) containing 100 mg/mL of streptomycin, 100 U/mL penicillin, and 8% fetal bovine serum (FBS). A549/hSLAM (59) and Vero/hSLAM (60), described previously, were each cultured in DMEM containing 0.2 mg/mL of G418 and 8% FBS.
Viruses.
We used the MuV Odate strain isolated from a patient with aseptic meningitis (61), rMuV-AcGFP (33), the MeV IC-B strain isolated from an acute measles patient (62), and rMeV-eGFP (34). For virus titration, Vero or Vero/hSLAM cells were infected with a serial dilution of MuV or MeV solution for 1 h, overlaid with DMEM containing 0.5% agarose, and cultured for 4 to 5 days at 37°C. The cells were subsequently stained with 0.01% neutral red solution (Sigma-Aldrich, St. Louis, MO, USA) for 24 h, and then plaques were counted after the agarose was removed.
Antibodies.
Anti-MuV M (79D) and anti-MeV M (E388) mouse monoclonal antibodies (MAbs) and anti-MuVs M and anti-MuV N rabbit polyclonal antibodies (pAbs) have been described previously (63, 64). Anti-TCOF1 (Treacle) rabbit pAb (HPA038237, Sigma-Aldrich), anti-UBF rabbit pAb (HPA006385, Sigma-Aldrich), anti-nucleolin (D4C7O) rabbit MAb (14574, Cell Signaling Technology, Danvers, MA, USA), anti-fibrillarin (EPR10823[B]) rabbit MAb (ab166630, Abcam, Cambridge, UK), anti-GAPDH (3E12) mouse MAb (bsm-0978M, Bioss Antibodies, MA, USA), anti-β-actin (AC-15) mouse MAb (A1978, Sigma-Aldrich), and anti-HA (Poly9023) rabbit pAb (PRB-101C, BioLegend, CA, USA) were purchased and used as primary antibodies. Alexa Fluor 488-conjugated goat anti-mouse immunoglobulin (Ig)G (H+L) and Alexa Fluor 594-conjugated goat anti-rabbit IgG (H+L) highly cross-adsorbed antibodies (Thermo Fisher Scientific, Waltham, MA, USA) were purchased and used as secondary antibodies.
Plasmids.
The construction of pCAGGS-M was described previously (64). For the construction of pCAGGS-M-FOS, the MuV M gene was amplified from pCAGGS-M by conducting PCR using the primers Odate-M-FOS-F (5′-CAAACGCGTCTCGAGATGGCAGGATCACAG-3′) and Odate-M-FOS-R (5′-ATCGAATCCAGATCTTAGGTTGCTCATTGA-3′) and then inserted into pCAG-MCS2-FOS for the expression of C-terminally FOS-tagged protein (65) using an In-Fusion HD cloning kit (Clontech, Mountain View, CA, USA) in accordance with the manufacturer’s instructions.
To construct a plasmid for the expression of N-terminally HA-tagged Treacle (HA-Treacle), total RNA was isolated from A549 cells by using an Rneasy minikit (Qiagen, Hilden, Germany), and cDNA was synthesized using a PrimeScript RT reagent kit (TaKaRa Bio, Shiga, Japan). The cDNA encoding human TCOF-1 was amplified by conducting PCR using the primers pCAGGS-nHA-Treacle_F (5′-GCTAAGCTTGGTACCGCCGAGGCCAGGAAG-3′) and pCAGGS-nHA-Treacle_R (5′-CCCTCTAGACTCGAGTCATACAGTCTGCTC-3′) and inserted into pCAGGS-HA with an In-Fusion HD cloning kit. The sequences of each plasmid were confirmed using a BigDye Terminator version 3.1 cycle sequencing kit and an ABI PRISM 3500xL genetic analyzer (Thermo Fisher Scientific).
Plasmid transfection.
The cells were seeded in a 24- or 6-well plate and then transfected with plasmids using TransIT-LTI (for Vero or A549 cells) or TransIT-293 (for 293T cells) (Mirus Bio LLC, Madison, WI, USA). Opti-MEM (Gibco, Thermo Fisher Scientific) was used for the dilution of plasmid and reagent.
Indirect fluorescence assay.
For observing samples by confocal microscopy, Vero cells, A549 cells, or A549/hSLAM were seeded in a 24-well plate containing 12-mm round cover glass (Matsunami Glass, Osaka, Japan). Cells that had been transfected with plasmids or infected with virus were washed three times in phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde in PBS for 15 min, and permeabilized with 0.2% Triton X-100 for 15 min. Each sample was incubated with 2% bovine serum albumin (BSA) in PBS for 20 min to block nonspecific antibody binding. To detect target proteins, the cells were treated with primary antibodies followed by Alexa Fluor 488/594-conjugated secondary antibodies in 2% BSA in PBS. Finally, each sample was placed on a glass slide with SlowFade Gold antifade reagent with 4′,6-diamidino-2-phenylindole (DAPI) (Thermo Fisher Scientific) and then observed on an FV3000 Confocal Microscope (Olympus, Tokyo, Japan).
Knockdown experiment.
To suppress the expression of target genes, commercially available siRNA pools targeting each protein and a control nontargeting siRNA pool (siGENOME SMARTpool) were purchased from Dharmacon (Horizon Discovery, Cambridge, UK). The siRNAs were transfected into A549 cells by using Lipofectamine RNAi-MAX (Thermo Fisher Scientific) in accordance with the manufacturer’s protocol.
Cell viability assay.
The cells were seeded in a 96-well plate and then transfected with siRNA (siNC, siTreacle, siB23, siNOP2, siNOP58, siNOL12, or siNOLC1) by using Lipofectamine RNAi-MAX. At 72 or 168 h posttransfection, 10 μL of Cell Count Reagent SF (Nacalai Tesque) was added to each well, and the cells were incubated at 37°C for 30 to 40 min. The absorbance value was measured at 450 nm on a GloMax Discover Microplate Reader (Promega, Madison, WI, USA).
Western blotting.
The cells were washed with PBS three times, lysed in NP-40/TNE buffer (10 mM Tris-HCl [pH 7.5], 200 mM NaCl, and 10 mM EDTA, with 1% NP-40) and protease inhibitor cocktail and centrifuged at 12,000 × g for 1 h at 4°C. The resulting supernatant was resolved with Tris-SDS sample buffer with 2-mercaptoethanol (2-ME) (Nacalai Tesque), boiled for 5 min at 100°C, and subjected to SDS-PAGE. The proteins were then transferred to a polyvinylidene difluoride membrane (Millipore, MA, USA). The membrane was incubated with blocking buffer (PBS containing 2% skim milk) for 1 h, treated with primary antibodies at an appropriate dilution in blocking buffer for 1 h, and finally incubated with 1/10,000-diluted secondary antibodies labeled with horseradish peroxidase for 1 h. The proteins were visualized using SuperSignal West Femto maximum sensitivity substrate (Thermo Fisher Scientific) and detected using an LAS-3000 image analyzer system (Fuji Film, Tokyo, Japan).
Pulldown assay.
The cells expressing M-FOS and HA-Treacle were rinsed with PBS three times and then lysed in NP-40/TNE buffer. StrEP-Tactin Sepharose beads (Nacalai Tesque) were added to the lysate, and the resulting mixture was incubated for 4 h at 4°C. The beads were then washed with 0.02% Tween 20 in PBS at least five times. The proteins bound to the beads were eluted in Tris-SDS sample buffer with 2-ME and then boiled for 5 min at 100°C. The resulting supernatant was subjected to SDS-PAGE and Western blotting.
Quantitative Reverse Transcription-PCR (qRT-PCR).
Total RNA was extracted using an Rneasy minikit and was then reverse transcribed into cDNA by using PrimeScript reverse transcriptase (TaKaRa Bio) and the primer 5′-ACCAAGGGGAGAAAGTAAAATCAATTTTTT-3′ for MuV genomic RNA or oligo(dT) primer for viral and cellular mRNAs, in accordance with the manufacturer’s protocol. Quantification of target cDNA was performed by using LightCycler 480 Probes Master and Universal ProbeLibrary (Roche Diagnostics, Basel, Switzerland) in accordance with the manufacturer’s protocol. The primers UPL-MuV-N-F (5′-TTCCTCCAGTTCAACAGCAA-3′) and UPL-MuV-N-R (5′-AACGTCGTCATCTGATTCCT-3′) were used for the measurement of genomic RNA and N mRNA, and the primers UPL-HPRT1-F (5′-GGGAGGCCATCACATTGTAG-3′) and UPL-HPRT1-R (5′-CACTATTTCTATTCAGTGCTTTGA-3′) were used for the measurement of hypoxanthine phosphoribosyltransferase 1 (HPRT1) mRNA (33). Thermal cycling was carried out using the LightCycler 480 system (Roche). The expression of target genomic RNA or MuV-N mRNA was normalized to that of HPRT1 mRNA.
Statistical analysis.
For all of the presented data, independent experiments were repeated at least twice. Differences between groups were evaluated using unpaired Student’s t tests. Error bars indicate the standard deviations of triplicate measurements. Statistical significance was assumed as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ns, not significant.
ACKNOWLEDGMENTS
This work was supported by grants from the Japanese Society for the Promotion of Science (JSPS) (Grant-in-Aid for Scientific Research I 19K07584) and the Japan Agency for Medical Research and Development (AMED) (Japan Program for Infectious Diseases Research and Infrastructure grant JP21wm0325024j0002 and Research Program on Emerging and Re-emerging Infectious Diseases grants JP21fk0108623j0001, JP21fk0108617j0201, and JP21fk0108087j0403).
We thank Katie Oakley, Ph.D., from Edanz for editing a draft of the manuscript.
A.W. and H.K. designed the research; A.W., F.K., and H.K. performed the research; A.W., H.K., and M.T. wrote the paper.
Contributor Information
Aika Wakata, Email: awakata@niid.go.jp.
Rebecca Ellis Dutch, University of Kentucky College of Medicine.
REFERENCES
- 1.Ganar K, Das M, Sinha S, Kumar S. 2014. Newcastle disease virus: current status and our understanding. Virus Res 184:71–81. 10.1016/j.virusres.2014.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeltina A, Bowden TA, Lee B. 2016. Emerging paramyxoviruses: receptor tropism and zoonotic potential. PLoS Pathog 12:e1005390. 10.1371/journal.ppat.1005390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Navaratnarajah CK, Generous AR, Yousaf I, Cattaneo R. 2020. Receptor-mediated cell entry of paramyxoviruses: mechanisms, and consequences for tropism and pathogenesis. J Biol Chem 295:2771–2786. 10.1074/jbc.REV119.009961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hviid A, Rubin S, Muhlemann K. 2008. Mumps. Lancet 371:932–944. 10.1016/S0140-6736(08)60419-5. [DOI] [PubMed] [Google Scholar]
- 5.Rubin S, Eckhaus M, Rennick LJ, Bamford CG, Duprex WP. 2015. Molecular biology, pathogenesis and pathology of mumps virus. J Pathol 235:242–252. 10.1002/path.4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paterson RG, Lamb RA. 1990. RNA editing by G-nucleotide insertion in mumps virus P-gene mRNA transcripts. J Virol 64:4137–4145. 10.1128/JVI.64.9.4137-4145.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rima B, Balkema-Buschmann A, Dundon WG, Duprex P, Easton A, Fouchier R, Kurath G, Lamb R, Lee B, Rota P, Wang L, Ictv Report C . 2019. ICTV virus taxonomy profile: paramyxoviridae. J Gen Virol 100:1593–1594. 10.1099/jgv.0.001328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takimoto T, Portner A. 2004. Molecular mechanism of paramyxovirus budding. Virus Res 106:133–145. 10.1016/j.virusres.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 9.Harrison MS, Sakaguchi T, Schmitt AP. 2010. Paramyxovirus assembly and budding: building particles that transmit infections. Int J Biochem Cell Biol 42:1416–1429. 10.1016/j.biocel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pentecost M, Vashisht AA, Lester T, Voros T, Beaty SM, Park A, Wang YE, Yun TE, Freiberg AN, Wohlschlegel JA, Lee B. 2015. Evidence for ubiquitin-regulated nuclear and subnuclear trafficking among Paramyxovirinae matrix proteins. PLoS Pathog 11:e1004739. 10.1371/journal.ppat.1004739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang YE, Park A, Lake M, Pentecost M, Torres B, Yun TE, Wolf MC, Holbrook MR, Freiberg AN, Lee B. 2010. Ubiquitin-regulated nuclear-cytoplasmic trafficking of the Nipah virus matrix protein is important for viral budding. PLoS Pathog 6:e1001186. 10.1371/journal.ppat.1001186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coleman NA, Peeples ME. 1993. The matrix protein of Newcastle disease virus localizes to the nucleus via a bipartite nuclear localization signal. Virology 195:596–607. 10.1006/viro.1993.1411. [DOI] [PubMed] [Google Scholar]
- 13.Ciancanelli MJ, Basler CF. 2006. Mutation of YMYL in the Nipah virus matrix protein abrogates budding and alters subcellular localization. J Virol 80:12070–12078. 10.1128/JVI.01743-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Day DH, Catalano A. 2013. Proteins of the nucleolus: regulation, translocation, & biomedical functions. Springer, New York, NY. [Google Scholar]
- 15.Lafontaine DLJ, Riback JA, Bascetin R, Brangwynne CP. 2021. The nucleolus as a multiphase liquid condensate. Nat Rev Mol Cell Biol 22:165–182. 10.1038/s41580-020-0272-6. [DOI] [PubMed] [Google Scholar]
- 16.Isaac C, Marsh KL, Paznekas WA, Dixon J, Dixon MJ, Jabs EW, Meier UT. 2000. Characterization of the nucleolar gene product, treacle, in Treacher Collins syndrome. Mol Biol Cell 11:3061–3071. 10.1091/mbc.11.9.3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valdez BC, Henning D, So RB, Dixon J, Dixon MJ. 2004. The Treacher Collins syndrome (TCOF1) gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proc Natl Acad Sci USA 101:10709–10714. 10.1073/pnas.0402492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao RW, Xu G, Wang Y, Shan L, Luan PF, Wang Y, Wu M, Yang LZ, Xing YH, Yang L, Chen LL. 2019. Nascent pre-rRNA sorting via phase separation drives the assembly of dense fibrillar components in the human nucleolus. Mol Cell 76:767–783.e11. 10.1016/j.molcel.2019.08.014. [DOI] [PubMed] [Google Scholar]
- 19.de Beus E, Brockenbrough JS, Hong B, Aris JP. 1994. Yeast NOP2 encodes an essential nucleolar protein with homology to a human proliferation marker. J Cell Biol 127:1799–1813. 10.1083/jcb.127.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong W, Biswas A, Zhou D, Fiches G, Fujinaga K, Santoso N, Zhu J. 2020. Nucleolar protein NOP2/NSUN1 suppresses HIV-1 transcription and promotes viral latency by competing with Tat for TAR binding and methylation. PLoS Pathog 16:e1008430. 10.1371/journal.ppat.1008430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scott DD, Trahan C, Zindy PJ, Aguilar LC, Delubac MY, Van Nostrand EL, Adivarahan S, Wei KE, Yeo GW, Zenklusen D, Oeffinger M. 2017. Nol12 is a multifunctional RNA binding protein at the nexus of RNA and DNA metabolism. Nucleic Acids Res 45:12509–12528. 10.1093/nar/gkx963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dixon J, Trainor P, Dixon MJ. 2007. Treacher Collins syndrome. Orthod Craniofac Res 10:88–95. 10.1111/j.1601-6343.2007.00388.x. [DOI] [PubMed] [Google Scholar]
- 23.Hayano T, Yanagida M, Yamauchi Y, Shinkawa T, Isobe T, Takahashi N. 2003. Proteomic analysis of human Nop56p-associated pre-ribosomal ribonucleoprotein complexes: possible link between Nop56p and the nucleolar protein treacle responsible for Treacher Collins syndrome. J Biol Chem 278:34309–34319. 10.1074/jbc.M304304200. [DOI] [PubMed] [Google Scholar]
- 24.Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. 2007. The multifunctional nucleolus. Nat Rev Mol Cell Biol 8:574–585. 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 25.Hiscox JA. 2002. The nucleolus—a gateway to viral infection? Arch Virol 147:1077–1089. 10.1007/s00705-001-0792-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hiscox JA. 2007. RNA viruses: hijacking the dynamic nucleolus. Nat Rev Microbiol 5:119–127. 10.1038/nrmicro1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zakaryan H, Stamminger T. 2011. Nuclear remodelling during viral infections. Cell Microbiol 13:806–813. 10.1111/j.1462-5822.2011.01596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salvetti A, Greco A. 2014. Viruses and the nucleolus: the fatal attraction. Biochim Biophys Acta 1842:840–847. 10.1016/j.bbadis.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rawlinson SM, Moseley GW. 2015. The nucleolar interface of RNA viruses. Cell Microbiol 17:1108–1120. 10.1111/cmi.12465. [DOI] [PubMed] [Google Scholar]
- 30.Duan Z, Chen J, Xu H, Zhu J, Li Q, He L, Liu H, Hu S, Liu X. 2014. The nucleolar phosphoprotein B23 targets Newcastle disease virus matrix protein to the nucleoli and facilitates viral replication. Virology 452–453:212–222. 10.1016/j.virol.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 31.Deffrasnes C, Marsh GA, Foo CH, Rootes CL, Gould CM, Grusovin J, Monaghan P, Lo MK, Tompkins SM, Adams TE, Lowenthal JW, Simpson KJ, Stewart CR, Bean AG, Wang LF. 2016. Genome-wide siRNA screening at biosafety level 4 reveals a crucial role for fibrillarin in henipavirus infection. PLoS Pathog 12:e1005478. 10.1371/journal.ppat.1005478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rawlinson SM, Zhao T, Rozario AM, Rootes CL, McMillan PJ, Purcell AW, Woon A, Marsh GA, Lieu KG, Wang LF, Netter HJ, Bell TDM, Stewart CR, Moseley GW. 2018. Viral regulation of host cell biology by hijacking of the nucleolar DNA-damage response. Nat Commun 9:3057. 10.1038/s41467-018-05354-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katoh H, Kubota T, Nakatsu Y, Tahara M, Kidokoro M, Takeda M. 2017. Heat shock protein 90 ensures efficient mumps virus replication by assisting with viral polymerase complex formation. J Virol 91:e02220-16. 10.1128/JVI.02220-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hashimoto K, Ono N, Tatsuo H, Minagawa H, Takeda M, Takeuchi K, Yanagi Y. 2002. SLAM (CD150)-independent measles virus entry as revealed by recombinant virus expressing green fluorescent protein. J Virol 76:6743–6749. 10.1128/jvi.76.13.6743-6749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gunther M, Bauer A, Muller M, Zaeck L, Finke S. 2020. Interaction of host cellular factor ANP32B with matrix proteins of different paramyxoviruses. J Gen Virol 101:44–58. 10.1099/jgv.0.001362. [DOI] [PubMed] [Google Scholar]
- 36.Watkinson RE, Lee B. 2016. Nipah virus matrix protein: expert hacker of cellular machines. FEBS Lett 590:2494–2511. 10.1002/1873-3468.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzales B, Henning D, So RB, Dixon J, Dixon MJ, Valdez BC. 2005. The Treacher Collins syndrome (TCOF1) gene product is involved in pre-rRNA methylation. Hum Mol Genet 14:2035–2043. 10.1093/hmg/ddi208. [DOI] [PubMed] [Google Scholar]
- 38.Hunziker M, Barandun J, Petfalski E, Tan D, Delan-Forino C, Molloy KR, Kim KH, Dunn-Davies H, Shi Y, Chaker-Margot M, Chait BT, Walz T, Tollervey D, Klinge S. 2016. UtpA and UtpB chaperone nascent pre-ribosomal RNA and U3 snoRNA to initiate eukaryotic ribosome assembly. Nat Commun 7:12090. 10.1038/ncomms12090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Werner A, Iwasaki S, McGourty CA, Medina-Ruiz S, Teerikorpi N, Fedrigo I, Ingolia NT, Rape M. 2015. Cell-fate determination by ubiquitin-dependent regulation of translation. Nature 525:523–527. 10.1038/nature14978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harper JW, Elledge SJ. 2007. The DNA damage response: ten years after. Mol Cell 28:739–745. 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 41.Larsen DH, Hari F, Clapperton JA, Gwerder M, Gutsche K, Altmeyer M, Jungmichel S, Toledo LI, Fink D, Rask MB, Grofte M, Lukas C, Nielsen ML, Smerdon SJ, Lukas J, Stucki M. 2014. The NBS1-Treacle complex controls ribosomal RNA transcription in response to DNA damage. Nat Cell Biol 16:792–803. 10.1038/ncb3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mooser C, Symeonidou IE, Leimbacher PA, Ribeiro A, Shorrocks AK, Jungmichel S, Larsen SC, Knechtle K, Jasrotia A, Zurbriggen D, Jeanrenaud A, Leikauf C, Fink D, Nielsen ML, Blackford AN, Stucki M. 2020. Treacle controls the nucleolar response to rDNA breaks via TOPBP1 recruitment and ATR activation. Nat Commun 11:123. 10.1038/s41467-019-13981-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cattaneo R, Schmid A, Eschle D, Baczko K, ter Meulen V, Billeter MA. 1988. Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell 55:255–265. 10.1016/0092-8674(88)90048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cathomen T, Mrkic B, Spehner D, Drillien R, Naef R, Pavlovic J, Aguzzi A, Billeter MA, Cattaneo R. 1998. A matrix-less measles virus is infectious and elicits extensive cell fusion: consequences for propagation in the brain. EMBO J 17:3899–3908. 10.1093/emboj/17.14.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okuwaki M, Iwamatsu A, Tsujimoto M, Nagata K. 2001. Identification of nucleophosmin/B23, an acidic nucleolar protein, as a stimulatory factor for in vitro replication of adenovirus DNA complexed with viral basic core proteins. J Mol Biol 311:41–55. 10.1006/jmbi.2001.4812. [DOI] [PubMed] [Google Scholar]
- 46.Tsuda Y, Mori Y, Abe T, Yamashita T, Okamoto T, Ichimura T, Moriishi K, Matsuura Y. 2006. Nucleolar protein B23 interacts with Japanese encephalitis virus core protein and participates in viral replication. Microbiol Immunol 50:225–234. 10.1111/j.1348-0421.2006.tb03789.x. [DOI] [PubMed] [Google Scholar]
- 47.Zeng Y, Ye L, Zhu S, Zheng H, Zhao P, Cai W, Su L, She Y, Wu Z. 2008. The nucleocapsid protein of SARS-associated coronavirus inhibits B23 phosphorylation. Biochem Biophys Res Commun 369:287–291. 10.1016/j.bbrc.2008.01.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu C, Zheng F, Zhu J, Liu M, Liu N, Li X, Zhang L, Deng Z, Zhao Q, Liu H. 2017. The interaction between NOLC1 and IAV NS1 protein promotes host cell apoptosis and reduces virus replication. Oncotarget 8:94519–94527. 10.18632/oncotarget.21785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coronel EC, Murti KG, Takimoto T, Portner A. 1999. Human parainfluenza virus type 1 matrix and nucleoprotein genes transiently expressed in mammalian cells induce the release of virus-like particles containing nucleocapsid-like structures. J Virol 73:7035–7038. 10.1128/JVI.73.8.7035-7038.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takimoto T, Murti KG, Bousse T, Scroggs RA, Portner A. 2001. Role of matrix and fusion proteins in budding of Sendai virus. J Virol 75:11384–11391. 10.1128/JVI.75.23.11384-11391.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pantua HD, McGinnes LW, Peeples ME, Morrison TG. 2006. Requirements for the assembly and release of Newcastle disease virus-like particles. J Virol 80:11062–11073. 10.1128/JVI.00726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patch JR, Crameri G, Wang LF, Eaton BT, Broder CC. 2007. Quantitative analysis of Nipah virus proteins released as virus-like particles reveals central role for the matrix protein. Virol J 4:1. 10.1186/1743-422X-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pohl C, Duprex WP, Krohne G, Rima BK, Schneider-Schaulies S. 2007. Measles virus M and F proteins associate with detergent-resistant membrane fractions and promote formation of virus-like particles. J Gen Virol 88:1243–1250. 10.1099/vir.0.82578-0. [DOI] [PubMed] [Google Scholar]
- 54.Li M, Schmitt PT, Li Z, McCrory TS, He B, Schmitt AP. 2009. Mumps virus matrix, fusion, and nucleocapsid proteins cooperate for efficient production of virus-like particles. J Virol 83:7261–7272. 10.1128/JVI.00421-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stow ND, Evans VC, Matthews DA. 2009. Upstream-binding factor is sequestered into herpes simplex virus type 1 replication compartments. J Gen Virol 90:69–73. 10.1099/vir.0.006353-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lymberopoulos MH, Pearson A. 2010. Relocalization of upstream binding factor to viral replication compartments is UL24 independent and follows the onset of herpes simplex virus 1 DNA synthesis. J Virol 84:4810–4815. 10.1128/JVI.02437-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lymberopoulos MH, Pearson A. 2007. Involvement of UL24 in herpes-simplex-virus-1-induced dispersal of nucleolin. Virology 363:397–409. 10.1016/j.virol.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 58.Lymberopoulos MH, Bourget A, Ben Abdeljelil N, Pearson A. 2011. Involvement of the UL24 protein in herpes simplex virus 1-induced dispersal of B23 and in nuclear egress. Virology 412:341–348. 10.1016/j.virol.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 59.Takeda M, Ohno S, Seki F, Nakatsu Y, Tahara M, Yanagi Y. 2005. Long untranslated regions of the measles virus M and F genes control virus replication and cytopathogenicity. J Virol 79:14346–14354. 10.1128/JVI.79.22.14346-14354.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ono N, Tatsuo H, Hidaka Y, Aoki T, Minagawa H, Yanagi Y. 2001. Measles viruses on throat swabs from measles patients use signaling lymphocytic activation molecule (CDw150) but not CD46 as a cellular receptor. J Virol 75:4399–4401. 10.1128/JVI.75.9.4399-4401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saito H, Takahashi Y, Harata S, Tanaka K, Sano T, Suto T, Yamada A, Yamazaki S, Morita M. 1996. Isolation and characterization of mumps virus strains in a mumps outbreak with a high incidence of aseptic meningitis. Microbiol Immunol 40:271–275. 10.1111/j.1348-0421.1996.tb03346.x. [DOI] [PubMed] [Google Scholar]
- 62.Kobune F, Sakata H, Sugiura A. 1990. Marmoset lymphoblastoid cells as a sensitive host for isolation of measles virus. J Virol 64:700–705. 10.1128/JVI.64.2.700-705.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tahara M, Takeda M, Yanagi Y. 2007. Altered interaction of the matrix protein with the cytoplasmic tail of hemagglutinin modulates measles virus growth by affecting virus assembly and cell-cell fusion. J Virol 81:6827–6836. 10.1128/JVI.00248-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Katoh H, Kubota T, Kita S, Nakatsu Y, Aoki N, Mori Y, Maenaka K, Takeda M, Kidokoro M. 2015. Heat shock protein 70 regulates degradation of the mumps virus phosphoprotein via the ubiquitin-proteasome pathway. J Virol 89:3188–3199. 10.1128/JVI.03343-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Katoh H, Okamoto T, Fukuhara T, Kambara H, Morita E, Mori Y, Kamitani W, Matsuura Y. 2013. Japanese encephalitis virus core protein inhibits stress granule formation through an interaction with Caprin-1 and facilitates viral propagation. J Virol 87:489–502. 10.1128/JVI.02186-12. [DOI] [PMC free article] [PubMed] [Google Scholar]