Abstract
Salmonella enterica serovar Typhimurium requires Mn2+, but only a few Mn2+-dependent enzymes have been identified from bacteria. To characterize Mn2+-dependent enzymes from serovar Typhimurium, two putative PPP-family protein phosphatase genes were cloned from serovar Typhimurium and named prpA and prpB. Their DNA-derived amino acid sequences showed 61% identity to the corresponding Escherichia coli proteins and 41% identity to each other. Each phosphatase was expressed in E. coli and purified to near electrophoretic homogeneity. Both PrpA and PrpB absolutely required a divalent metal for activity. As with other phosphatases of this class, Mn2+ had the highest affinity and stimulated the greatest activity. The apparent Ka of PrpA for Mn2+ of 65 μM was comparable to that for other bacterial phosphatases, but PrpB had a much higher affinity for Mn2+ (1.3 μM). The pH optima were pH 6.5 for PrpA and pH 8 for PrpB, while the optimal temperatures were 45 to 55°C for PrpA and 30 to 37°C for PrpB. Each phosphatase could hydrolyze phosphorylated serine, threonine, or tyrosine residues, but their relative specific activities varied with the specific substrate tested. These differences suggest that each phosphatase is used by serovar Typhimurium under different growth or environmental conditions such as temperature or acidity.
MntH, the Salmonella enterica serovar Typhimurium ortholog of the eukaryotic NRAMP proteins (natural resistance-associated macrophage protein), is a highly selective H+-stimulated Mn2+ transporter (6). Subsequently, the mammalian NRAMP1 protein, expressed in monocytes and macrophages, was shown to mediate marked changes in intracellular Mn2+ in macrophages (5). Transcription of mntH in serovar Typhimurium is induced by hydrogen peroxide and cation starvation, and mutation of mntH makes serovar Typhimurium cells more sensitive to killing by peroxide. These data suggest that Mn2+ is required for maintenance of some physiological functions and that Mn2+ could play a role in virulence, especially in resistance to reactive oxygen species (6). Any role for Mn2+ is presumably mediated by its use as an enzyme cofactor. Surprisingly, very few known enzymes are known to be Mn2+ dependent. In an effort to understand the role of Mn2+ in cell function and pathogenesis, we have begun to characterize each of the few known Mn2+-cofactored enzymes. One such family is the PPP class of protein phosphatases (2, 8, 17).
PPP phosphatases are metalloenzymes that include the catalytic subunits of mammalian PP1, PP2A, PP2B, and related protein phosphatases. Two divalent metal ions in their active sites catalyze a single-step dephosphorylation reaction. In eukaryotes, PPP phosphatases are involved in regulating multiple cellular functions such as cell division, glycogen metabolism, and muscle contraction. Homologs of PPP phosphatases can be discerned in many bacterial and archaeal genomes (for reviews, see references 8 and 17). All bacterial PPP phosphatases characterized to date require Mn2+ for maximal activation (7, 10, 12–16, 18–20). To investigate the role of Mn2+ further, we have cloned, purified, and characterized the two PPP phosphatase homologs evident in the extant genome of serovar Typhimurium. As predicted each is a Mn2+-dependent protein phosphatase; however, the two enzymes have markedly different properties suggestive of distinct physiological roles.
MATERIALS AND METHODS
Standard procedures.
Protein concentrations were measured with the bicinchoninic acid protein assay kit from Pierce (Rockford, Ill.). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described by Laemmli (9), and the gels were stained as described by Fairbanks et al. (4).
Cloning and sequencing prpA and prpB from serovar Typhimurium.
The prpA and prpB genes were cloned from MM1255 (wild-type serovar Typhimurium LT2) by PCR. Primers (Genosys, The Woodlands, Tex.) were based on available serovar Typhimurium genomic sequence fragments (http://genome.wustl.edu /bacterial.salmonella/shtml). The upstream and downstream primers for prpA were 5′-GCTTACTAAGCAAATGCATCTGCACACGC-3′ and 5′-CCTGCGCAGCAGGATCCCGATTTTTAACCC-3′, respectively. The upstream and downstream primers for prpB were 5′-GGCCTACTTTTTATGCATAATACCAGACTC-3′ and 5′-GAAGGCTATATCGGATCCCATAATGCAGGG-3′, respectively. Restriction enzyme digestion of PCR products gave clones for each gene that began with an NsiI site 30 to 40 bp 5′ of the presumed start codon and ended with a BamHI site 30 to 40 bp 3′ of the presumed stop codon. These were ligated into the NsiI and BamHI sites of pDGK101 to give pDGK235 (containing prpA) and pDGK236 (containing prpB). Each plasmid contained 2,916 bp of pBluescriptII SK(+) from the BamHI site through the SalI site in the polylinker (Stratagene, La Jolla, Calif.) followed by 505 bp of the serovar Typhimurium corA promoter on a SalI-NsiI fragment, followed by the prpA or prpB insert.
Inserts in pDGK235 and pDGK236 were sequenced using the above oligonucleotides as primers at the Howard Hughes Medical Institute Biopolymer/Keck Foundation Biotechnology Resource Laboratory, Yale University. Both sequences agree with the serovar Typhimurium LT2 sequence available at the above website. prpA and prpB map to approximately 40.2 and 62.8 min on the serovar Typhimurium chromosome, comparable to E. coli prpA and prpB at 41 and 61.5 min, respectively (3).
Expression of ST-PrpA and ST-PrpB.
The open reading frames for prpA and prpB were amplified by PCR with Pfu DNA polymerase (Stratagene). The forward primers were 5′-TGGGGATCCATGAACGACAGGAAAAACATGATG-3′ for prpA and 5′-TGGGAATCCATGGAATTAATTCGTTATGCTFAC-3′ for prpB, while the reverse primers were 5′-TGGAAGCTTGCTATTGTATCCGCGCTAACG-3′ for prpA and 5′-TGGAAGCTTTTACTTTATTTTAAAAAAAGACAGATTG-3′ for prpB. To facilitate cloning, a BamHI restriction was introduced at the 5′ end of the forward primers and a HindIII restriction site was introduced at the 5′ end of the reverse primers. The resulting PCR products were ligated into the pRSET-A expression vector (Invitrogen, Carlsbad, Calif.) and transformed into E. coli DH5α cells (Gibco BRL, Rockville, Md.). After plasmid purification, both strands of the insert were sequenced to verify the fidelity of the PCR amplification. Competent E. coli BL21(DE3) cells (Promega, Madison, Wis.) were transformed with plasmid, grown to an optical density at 600 nm of 0.6 to 1, induced with isopropyl-β-d-thiogalactopyranoside (IPTG) at a final concentration of 0.1 mM for 4 h at 37°C, and harvested by centrifugation. The pellet was resuspended in 40 ml of buffer A (20 mM Tris [pH 7], 500 mM NaCl, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM imidazole-HCl) with 2 mg of lysozyme per ml, 100 μg of DNase I per ml, and 100 μg of RNase per ml and incubated on ice for 30 min. The cells were lysed by being passed through a French press three times at 8,000 lb/in2; lysates were centrifuged at 17,000 × g at 4°C for 30 min. The supernatant was loaded on a 3-by 5-cm column of Ni-nitrilotriacetic acid agarose (Qiagen, Valencia, Calif.) charged with NiCl2 and equilibrated with buffer A. The column was washed with 100 ml of buffer A and then with 100 ml of buffer B (20 mM Tris [pH 7], 500 mM NaCl, 1 mM PMSF, 40 mM imidazole-HCl). The fusion proteins were then eluted from the column with 150 ml of buffer C (20 mM Tris [pH 7], 500 mM NaCl, 1 mM PMSF, 80 mM imidazole-HCl), 10-ml fractions were collected, and 10 μl of each fraction removed and assayed for phosphatase activities, and peak fractions were pooled. Samples containing ST-PrpA were dialyzed against 3 liters of buffer D (10 mM Tris-HCl [pH 7], 1 mM dithiothreitol, 1 mM PMSF, 1 mM EDTA) plus 10 mM NaCl overnight, and loaded on a 3- by 5-cm column of Macro-Prep High S Support (Bio-Rad, Hercules, Calif.) that had been equilibrated with buffer D plus 10 mM NaCl. The column was washed with 100 ml of buffer D plus 50 mM NaCl and eluted with a salt gradient of 50 to 400 mM NaCl in buffer D. Fractions (5 ml) were collected and assayed for phosphatase activity. Active fractions were pooled, dialyzed with 1 liter of buffer E (50 mM Tris-HCl [pH 7], 1 mM dithiothreitol, 1 mM PMSF, 1 mM EDTA, 10% [vol/vol] glycerol) overnight, and stored at −20°C.
ST-PrpB samples were concentrated with a Centriprep-10 column and loaded on a 2- by 65-cm column of Superose 6 (Amersham Pharmacia Biotech, Piscataway, N.J.) equilibrated with buffer F (10 mM ammonium formate [pH 6], 1 mM PMSF, 1 mM EDTA). The column was eluted with 180 ml of buffer F; 1-ml fractions were collected and assayed for phosphatase activity. Peak fractions were pooled, concentrated with a Centriprep-10 column, resuspended with buffer E, and stored at −20°C. All purification procedures were carried out at 4°C except gel filtration chromatography, which was performed at room temperature. Expression and purification of ST-PrpA and ST-PrpB were monitored by Western hybridization with anti-Xpress antibody (Invitrogen, Carlsbad, Calif.).
Preparation of phosphorylated MBP.
Serine/threonine- or tyrosine-phosphorylated myelin basic protein (MBP) was prepared with [γ-33P]ATP as specified by the manufacturer (New England BioLabs, Beverly, Mass.). Peptide substrates were purchased from Promega.
Phosphatase assay.
Purified phosphatase protein (200 ng) was incubated at 37°C in 30 μl of solution containing 50 mM imidazole (pH 6.5 for ST-PrpA and pH 8 for ST-PrpB), 1 mM MnCl2, 10 μg of bovine serum albumin per ml, BSA, and 2 μM [33P]phospho-MyBP. The initial rate of the reaction was linear for at least 30 min for all substrates. The reaction was terminated by the addition of 100 μl of 20% (wt/vol) trichloroacetic acid, mixed, and centrifuged for 3 min in a microcentrifuge at maximum speed. 33P radioactivity was determined using 50 μl of supernatant liquid. A modified molybdic acid extraction method (7) was used to verify the reaction product as inorganic phosphate. The peptide substrates were assayed as specified by the manufacturer (Promega).
pNPP phosphatase activities were assayed at 37°C in 200 μl of solution containing 50 mM imidazole (pH 6.5 for ST-PrpA and pH 8 for ST-PrpB) 1 mM MnCl2, and 1 mM pNPP. ST-PrpA or ST-PrpB was added to start the reaction. After 30 min, the reaction was stopped with 800 μl of 0.5 M sodium borate (pH 9). Released p-nitrophenol was measured at 410 nm on a Beckman DU-64 spectrophotometer. Most phosphatase assays were performed on proteins carrying the His6 tag and linking sequence. However, removal of the fusion domains of ST-PrpA and ST-PrpB with enterokinase did not change their affinity for Mn2+ or other properties tested. For pH optimum experiments, controls indicated that the composition of the buffer had no significant influence on phosphatase activity. Final concentrations of inhibitors used were 10 mM EDTA, 10 mM sodium pyrophosphate, 50 mM NaF, 10 mM sodium potassium tartrate, 1 mM tetramisole, 1 mM sodium orthovanadate, 150 μM trifluoperazine, 10 μM microcystin-LR, 5 μM okadaic acid, and 1 mM AppppA (diadenosine tetraphosphate).
Nucleotide sequence accession numbers.
The prpA and prpB sequences have been deposited with GenBank under accession no. AY149950 and AY049951, respectively.
RESULTS
Cloning of prpA and prpB.
Genes for two PPP family protein phosphatases identified in extant serovar Typhimurium genomic sequence by a BLAST (1) search were cloned by PCR amplification from serovar Typhimurium genomic DNA. Derived amino acid sequences were each 61% identical to their respective homologs in E. coli (Fig. 1); therefore, the serovar Typhimurium genes were designated prpA and prpB. The predicted protein product of prpA, ST-PrpA, consisted of 222 amino acids with a predicted molecular mass of 25,879 Da and an isoelectric point of 8.8. ST-PrpB was predicted to have 218 amino acids, a molecular mass of 25,024 Da, and a pI of 4.6. The serovar Typhimurium proteins were each 61% identical to their respective E. coli homologs and 41% identical to each other (Fig. 1).
FIG. 1.
Sequence alignment of serovar Typhimurium and E. coli PPP phosphatases. The predicted amino acid sequences of ST-PrpA, ST-PrpB, and the E. coli phosphatases were aligned using CLUSTAL W and modified slightly by eye to optimize alignment at the termini. Dashes indicate sequence gaps introduced to optimize the alignment of conserved regions; asterisks indicate identical residues in all four phosphatases; colons indicate either three of four residues identical or all four residues conserved.
Expression and purification ST-PrpA and ST-PrpB.
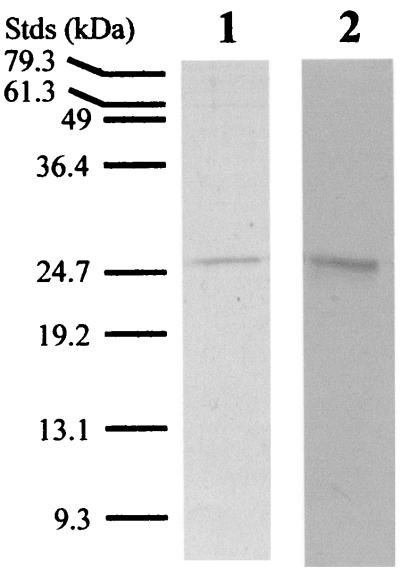
To facilitate purification by metal-chelate affinity chromatography, an amino-terminal His6 sequence from the pRSET-A vector was introduced into the recombinant ST-PrpA and ST-PrpB carried in pRSRT-A and expressed in E. coli after induction with IPTG. After metal-chelate affinity chromatography, ST-PrpB was concentrated and further purified by gel filtration chromatography. ST-PrpA, however, became aggregated when concentrated. Each was further purified by cation-exchange chromatography. Both ST-PrpA and ST-PrpB were purified to near electrophoretic homogeneity (Fig. 2), and their identities were confirmed by Western blot analysis with anti-Xpress antibody (data not shown). The yield from 1 liter of cells was 1 mg of ST-PrpA or 3 mg of ST-PrpB.
FIG. 2.
Analysis of purified ST-PrpA and ST-PrpB by SDS-PAGE. ST-PrpA and ST-PrpB were expressed in E. coli and purified by metal-chelate affinity chromatography and ion-exchange or gel filtration chromatography as described in Materials and Methods. Portions of 1 μg of ST-PrpA and 2 μg of ST-PrpB were analyzed by SDS-PAGE on a 15% (wt/vol) acrylamide gel. Lanes: 1, ST-PrpA; 2, ST-PrpB. The positions of protein standard (Stds) are indicated at the left.
Properties of ST-PrpA and ST-PrpB.
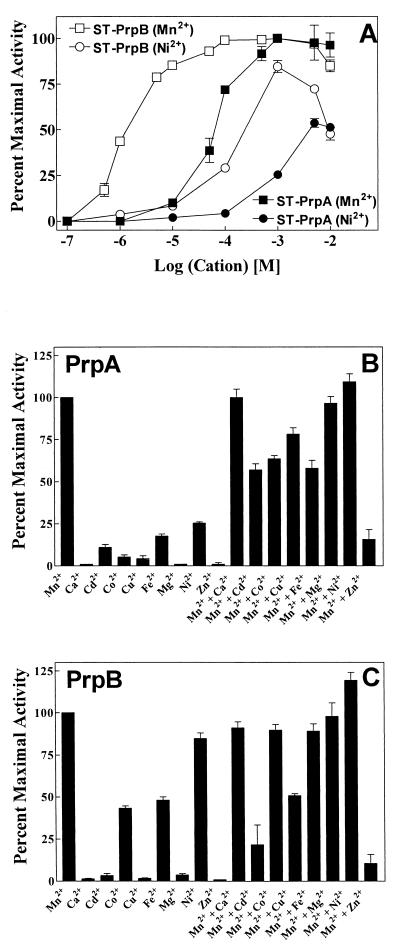
Both ST-PrpA and ST-PrpB showed divalent metal ion-dependent protein phosphatase activity toward [33P]phosphoseryl MBP (Fig. 3). Although a few other cations supported some phosphatase activity, Mn2+ clearly produced the greatest stimulation. Surprisingly, ST-PrpA and ST-PrpB displayed different affinities for Mn2+ (Fig. 3A). The Mn2+ Ka of ST-PrpB was 1.3 μM, over 50 times lower than the Mn2+ Ka of ST-PrpA at 65 μM (Table 1). Ni2+, Co2+, and Fe2+ could support the phosphatase activity of ST-PrpB (Fig. 3C) but with Ka values in the mM range (Fig. 3A; Table 1). Ni2+ was also a weak activator of PrpA activity. Neither Mg2+ nor Ca2+ had stimulatory effects on either phosphatase, while Cd2+, Cu2+, and Zn2+ inhibited both phosphatases when added concomitantly with Mn2+. Fe2+ and Co2+ slightly inhibited Mn2+-stimulated ST-PrpA protein phosphatase activity (Fig. 3B).
FIG. 3.
Effects of divalent metal ions on the catalytic activity of ST-PrpA and ST-PrpB. (A) The activity of 200 ng each of purified ST-PrpA and ST-PrpB was assayed under standard conditions (see Materials and Methods) using [33P]phosphoseryl MBP as substrate with the indicated concentrations of Mn2+ or Ni2+. Results are given as the percent activity observed with 1 mM of Mn2+ and the standard error of the mean (n = 4). For points with no error bar, the error was smaller than the size of the symbol. For ST-PrpA, 100% activity was equal to 0.0302 pmol of 33Pi released per min, while the corresponding value for ST-PrpB was 0.0304 pmol/min. (B and C) The activity of purified ST-PrpA (B) and ST-PrpB (C) was assayed under the conditions described for panel A, except that, where indicated, the compounds listed were substituted for the activating divalent metal ion, Mn2+. All compounds were present at a final concentration of 1 mM. Iron was maintained in the ferrous state by the addition of ascorbic acid to a final concentration of 1 mM. All results are reported as the percentage of activity relative to that observed with Mn2+ and standard error of the mean (n = 4).
TABLE 1.
Ka of ST-PrpA or ST-PrpB for cations
| Phosphatase |
Ka (mM) for:
|
|||
|---|---|---|---|---|
| Mn2+ | Ni2+ | Fe2+ | Co2+ | |
| ST-PrpA | 0.065 | 1 | NDa | ND |
| ST-PrpB | 0.0013 | 0.14 | 1.3 | 5.2 |
ND, not determined.
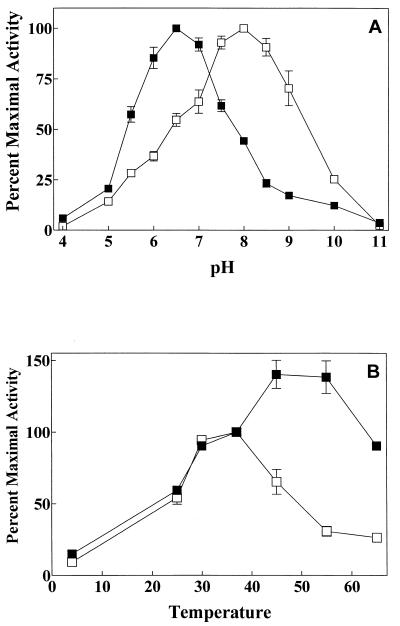
Both ST-PrpA and ST-PrpB were active toward pNPP over a pH spectrum from 5 to 10. However, optimal ST-PrpA activity occurred at pH 6 to 7, while the optimal pH for ST-PrpB activity was pH 7.5 to 8.5 (Fig. 4A). Each was active over a temperature range from 0 to 65°C. Maximal activity was seen between 45 and 55°C for ST-PrpA but 30 to 37°C for ST-PrpB. ST-PrpA but not ST-PrpB was heat stable, with PrpA maintaining substantial activity at 65°C (Fig. 4B). ST-PrpA and ST-PrpB demonstrated a similar specific activity toward [33P]-phosphoseryl MBP, but the activity of ST-PrpA toward [33P]-phosphotyrosyl MBP was less than 10% that of ST-PrpB. In contrast, activity toward synthetic phosphopeptide substrates, whether phosphotyrosyl or phosphothreonyl, was much greater with ST-PrpA than with ST-PrpB (Table 2).
FIG. 4.
pH and temperature dependence of ST-PrpA and ST-PrpB. (A) The activity of 2 μg of purified ST-PrpA and ST-PrpB was assayed for phosphatase activity against pNPP under standard conditions (see Materials and Methods), except that the pH was varied as indicated using different buffers. Shown is the relative protein phosphatase activity detected as a function of pH and the standard error of the mean (n = 3). 100% activity was defined as 0.84 nmol of p-nitrophenol released per min for ST-PrpA at pH 6.5 and 0.92 nmol/min for ST-PrpB at pH 8. Buffer salts (50 mM) were sodium acetate (pH 4 to 5), imidazole (pH 5 to 8), Tris-HCl (pH 8 to 9), and glycine (pH 9 to 11). (B) The activity of purified ST-PrpA and ST-PrpB was assayed for pNPP phosphatase activity under standard conditions (see Materials and Methods) while varying the temperature. All results are reported as the percent activity relative to that observed at 37°C and the standard error of the mean (n = 3). For points with no error bar, the error was smaller than the size of the symbol.
TABLE 2.
Protein phosphatase activity of ST-PrpA and ST-PrpB
| Substrate | Enzyme activity (pmol min−1 · mg−1 [n = 3])
|
|
|---|---|---|
| ST-PrpA | ST-PrpB | |
| Phosphoseryl/threonyl MBP | 151 ± 6 | 152 ± 6 |
| Phosphotyrosyl MBP | 60 ± 4 | 733 ± 37 |
| RRA(pT)VA | 18,200 ± 870 | 700 ± 170 |
| END(pY)INADSL | 39,800 ± 8,000 | 2,240 ± 120 |
| DADE(pY)LIPQQG | 8,660 ± 880 | 1,960 ± 140 |
The effect of protein phosphatase and other phosphomonoesterase inhibitors (11, 14, 16) on the activity of ST-PrpA and ST-PrpB was also examined (data not shown). At the concentrations tested, pyrophosphate, the chelating agent EDTA, the nonspecific phosphatase inhibitor NaF, and the tyrosine phosphatase inhibitor vanadate inhibited >90% of activity from either enzyme with [33P]-phosphoseryl MBP as substrate. Diadenosine tetraphosphate (AppppA) inhibited the activity of both enzymes by 20 to 30%, but the acid phosphatase inhibitor tartrate, the alkaline phosphatase inhibitor tetramisole, the protein phosphatase 2B inhibitor trifluoperazine, and the protein phosphatase 1 and 2A inhibitors microcystin-LR and okadaic acid did not inhibit ST-PrpA and ST-PrpB activity.
DISCUSSION
Little is known about the physiological functions of Mn2+. Mn2+ is often a functional substitute for Mg2+. Intuitively, one might thus expect to find a fairly large number of Mn2+-dependent enzymes. However, only a handful of Mn2+-dependent metalloenzymes are known. Because of our interest in Mn2+ transport and its possible role in pathogenesis (6), we are characterizing several of these Mn2+-dependent enzymes, including the two Mn2+-dependent protein phosphatases of serovar Typhimurium described in this report.
prpA and prpB of serovar Typhimurium are clear homologs of the corresponding genes in E. coli (13), but their sequences diverge much more than many homologs between such closely related organisms, at only 61% identity between species for both PrpA and PrpB. In contrast, the percent identity between serovar Typhimurium and E. coli for other Mn2+-dependent enzymes is 91 to 97% for SodA (Mn2+-superoxide dismutase), GpmM (phosphoglyceromutase), SpoT (AppppA synthase/hydrolase), and PepP (Aminopeptidase P). This could be indicative of a different origin for the phosphatases compared to other Mn2+-dependent enzymes.
The Ka of ST-PrpA for Mn2+ (65 μM) is within a factor of 3 of the Ka for Mn2+ for the homologous phosphatases PP1-arch1 from the archaeon Sulfolobus solfataricus and PP1-cyano1 and PP1-cyano2 from Microcystis sp. (7, 12, 16). In contrast, the Mn2+ Ka of 1.3 μM for ST-PrpB is much lower than that for other prokaryotic PPP phosphatases previously characterized. Although Ni2+, Co2+, and Fe2+ can support some phosphatase activity, their relative Ka values compared to that of Mn2+ indicate that Mn2+ is the physiologically relevant cation.
PrpA and PrpB of E. coli have also been purified and partially characterized (13). Unfortunately, divalent cation selectivity cannot be compared since this was not determined for the E. coli enzymes. The effects of several phosphatase inhibitors on ST-PrpA and ST-PrpB were generally similar not only to each other but also to the effects of the same inhibitors on the homologous phosphatases PP1-cyano1 from Microcystis aeruginosa PCC 7820 and PP1-cyano2 from M. aeruginosa UTEX 2063 (16). In contrast, although fluoride ion could completely block phosphatase activity in the serovar Typhimurium and M. aeruginosa enzymes, it had little effect on the two E. coli phosphatases. Vanadate inhibited both phosphatases from serovar Typhimurium but only PrpA from E. coli (13). Finally, substrate specificity may differ between serovar Typhimurium and E. coli. In both species, PrpA and PrpB have similar activity toward phosphoseryl MBP, but the activities of the E. coli phosphatases toward phosphotyrosyl MBP were equivalent whereas ST-PrpB had 10-fold greater activity than ST-PrpA toward phosphotyrosyl MBP. In contrast, ST-PrpA had high activity against phosphotyrosyl peptide substrates. Therefore, until physiological substrates are determined, substrate specificity cannot be determined. Our preliminary data (L. Shi and M. E. Maguire, unpublished results) indicate that mutation of prpA or prpB markedly alters the peroxide and temperature sensitivity. Proteins whose expression is altered with a change in temperature or exposure to reactive oxygen species might therefore be candidates for substrates for these phosphatases.
Not all bacterial genomes have PPP-class phosphatases. For those that have PPP phosphatases, almost all have only a single gene (17). Serovar Typhimurium and E. coli are the only bacteria so far studied to have two PPP phosphatase genes. The E. coli phosphatases are also the only bacterial PPP phosphatases for which a function can be assigned. They regulate the transcription of htrA via the CpxR-CpxA two-component signal transduction pathway that is responsive to protein misfolding in the periplasm (13). htrA encodes a protease that degrades misfolded proteins, thus avoiding potential cell toxicity. The similarities between the PrpA phosphatase in E. coli and serovar Typhimurium suggest that they probably have a similar function in serovar Typhimurium. In contrast, the rather different properties of the PrpB enzymes from the two species suggest the possibility of overlapping but not identical physiological roles.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Barford D. Molecular mechanisms of the protein serine/threonine phosphatases. Trends Biochem Sci. 1996;21:407–412. doi: 10.1016/s0968-0004(96)10060-8. [DOI] [PubMed] [Google Scholar]
- 3.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 4.Fairbanks G, Steck T L, Wallach D F. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry. 1971;10:2606–2617. doi: 10.1021/bi00789a030. [DOI] [PubMed] [Google Scholar]
- 5.Jabado N, Jankowski A, Dougaparsad S, Picard V, Grinstein S, Gros P. Natural resistance to intracellular infections. Natural resistance-associated macrophage protein 1 (NRAMP1) functions as a pH-dependent manganese transporter at the phagosomal membrane. J Exp Med. 2000;192:1237–1248. doi: 10.1084/jem.192.9.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kehres D G, Zaharik M L, Finlay B B, Maguire M E. The NRAMP proteins of Salmonella typhimurium and Escherichia coli are selective manganese transporters involved in the response to reactive oxygen. Mol Microbiol. 2000;36:1085–1100. doi: 10.1046/j.1365-2958.2000.01922.x. [DOI] [PubMed] [Google Scholar]
- 7.Kennelly P J, Oxenrider K A, Leng J, Cantwell J S, Zhao N. Identification of a serine/threonine-specific protein phosphatase from the archaebacterium Sulfolobus solfataricus. J Biol Chem. 1993;268:6505–6510. [PubMed] [Google Scholar]
- 8.Kennelly P J, Potts M. Life among the primitives: protein O-phosphatases in prokaryotes. Front Biosci. 1999;4:D372–D385. doi: 10.2741/kennelly. [DOI] [PubMed] [Google Scholar]
- 9.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 10.Leng J, Cameron A J, Buckel S, Kennelly P J. Isolation and cloning of a protein-serine/threonine phosphatase from an archaeon. J Bacteriol. 1995;177:6510–6517. doi: 10.1128/jb.177.22.6510-6517.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MacKintosh C, MacKintosh R W. Inhibitors of protein kinases and phosphatases. Trends Biochem Sci. 1994;19:444–448. doi: 10.1016/0968-0004(94)90127-9. [DOI] [PubMed] [Google Scholar]
- 12.Mai B, Frey G, Swanson R V, Mathur E J, Stetter K O. Molecular cloning and functional expression of a protein- serine/threonine phosphatase from the hyperthermophilic archaeon Pyrodictium abyssi TAG11. J Bacteriol. 1998;180:4030–4035. doi: 10.1128/jb.180.16.4030-4035.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Missiakas D, Raina S. Signal transduction pathways in response to protein misfolding in the extracytoplasmic compartments of E. coli: role of two new phosphoprotein phosphatases PrpA and PrpB. EMBO J. 1997;16:1670–1685. doi: 10.1093/emboj/16.7.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oxenrider K A, Rasche M E, Thorsteinsson M V, Kennelly P J. Inhibition of an archaeal protein phosphatase activity by okadaic acid, microcystin-LR, or calyculin A. FEBS Lett. 1993;331:291–295. doi: 10.1016/0014-5793(93)80355-x. [DOI] [PubMed] [Google Scholar]
- 15.Rusnak F, Yu L, Todorovic S, Mertz P. Interaction of bacteriophage lambda protein phosphatase with Mn(II): evidence for the formation of a [Mn(II)]2 cluster. Biochemistry. 1999;38:6943–6952. doi: 10.1021/bi982606u. [DOI] [PubMed] [Google Scholar]
- 16.Shi L, Carmichael W W, Kennelly P J. Cyanobacterial PPP family protein phosphatases possess multifunctional capabilities and are resistant to microcystin-LR. J Biol Chem. 1999;274:10039–10046. doi: 10.1074/jbc.274.15.10039. [DOI] [PubMed] [Google Scholar]
- 17.Shi L, Potts M, Kennelly P J. The serine, threonine, and/or tyrosine-specific protein kinases and protein phosphatases of prokaryotic organisms: a family portrait. FEMS Microbiol Rev. 1998;22:229–253. doi: 10.1111/j.1574-6976.1998.tb00369.x. [DOI] [PubMed] [Google Scholar]
- 18.Solow B, Young J C, Kennelly P J. Gene cloning and expression and characterization of a toxin-sensitive protein phosphatase from the methanogenic archaeon Methanosarcina thermophila TM-1. J Bacteriol. 1997;179:5072–5075. doi: 10.1128/jb.179.16.5072-5075.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voegtli W C, White D J, Reiter N J, Rusnak F, Rosenzweig A C. Structure of the bacteriophage lambda Ser/Thr protein phosphatase with sulfate ion bound in two coordination modes. Biochemistry. 2000;39:15365–15374. doi: 10.1021/bi0021030. [DOI] [PubMed] [Google Scholar]
- 20.Zhuo S, Clemens J C, Hakes D J, Barford D, Dixon J E. Expression, purification, crystallization, and biochemical characterization of a recombinant protein phosphatase. J Biol Chem. 1993;268:17754–17761. [PubMed] [Google Scholar]