Abstract
Emergent RNA technologies employ sequence and structural information to perform a diversity of biological functions. Synthetic RNA molecules have been developed for a wide array of applications, including genetic regulation, environmental sensing, and diagnostics devices. Recent advances in chemical synthesis and computational design of RNA have enhanced our ability to program novel functions and expand upon current biomedical applications for therapeutics and diagnostics. In this review, we highlight recent advances in synthetic RNA devices that have been engineered for biomedical systems, while addressing the current limitations and challenges of translating these engineered functional RNAs to clinical applications.
Introduction
RNA is a versatile biomolecule with significant capacity to encode genetic information, perform catalytic cleavage reactions [1], regulate mRNA stability through RNA interference (RNAi) [2], and adopt three-dimensional structures to bind ligands through hydrogen bonding networks, pi-stacking, and electrostatic interactions [3]. RNA structure is governed by Watson-Crick base pairing rules and thus RNA molecules can be programmed to perform these various functions in biomedical engineering applications, providing differential and orthogonal layers of regulation for virtually any biological target of interest [4]. Given the capacity of RNA molecules to interact with various biomolecular inputs, researchers have coupled environmental sensing with functional outputs to generate RNA-based devices with input/output control, expanding the availability of synthetic parts for programming biology [5]. Computational tools and in vitro library screening methods have enhanced the potential for RNA devices to be rapidly designed, synthesized, assayed, and explored for diagnostics and in vivo therapeutic applications, as well as aiding in biomedical discoveries and drug screening [6]. Regulatory RNAs such as microRNAs (miRNAs) and small-interfering RNAs (siRNAs) [2,7], RNA scaffolds and aptamers to bind nucleic acids, small molecules, proteins, and whole cells [8,9], ribozyme-mediated genetic control [10], or recent advances in CRISPR/Cas genome engineering [11] demonstrate the engineering potential of synthetic RNA devices for controlling a diverse array of biomedical targets. As the time and cost of nucleic acid synthesis continues to decrease [12], selection protocols for functional RNA molecules become higher throughput, and therapeutic delivery challenges are addressed, these devices will be more readily deployed in clinical applications.
In this review, we discuss recent advances in RNA device engineering for various biomedical applications, ranging from biomedical diagnostics to potential therapeutic gene circuits. Specifically, we discuss efforts to leverage antisense- and CRISPR-based RNAs for in vitro diagnostic devices. We also discuss efforts to use engineered self-cleaving ribozyme and miRNA devices to regulate natural and synthetic gene circuits for applications in cellular and gene therapies. We highlight the design and selection methodologies to generate these devices, the modularity and flexibility inherent in RNA device engineering, and efforts to expand the number of targets and ligands available for controlling biological systems. Finally, we discuss translational efforts to improve the pipeline of RNA devices for clinical applications, the challenges of delivery and methods to improve selection and design of novel functional RNA molecules. Advances in chemical modifications and viral delivery systems will promote biomedical use, while considerations around safety and efficacy are paramount to their ultimate clinical translation.
RNA Devices Enable Diagnostic Platforms with Rapid and Sensitive Detection
While Watson-Crick base pairing rules for RNA nucleotides provide a simple set of defined hydrogen bond interactions (A:U vs C:G), wobble base pairing of G:U, a flexible phosphodiester backbone, and various sugar puckers expand the possible three-dimensional folded structures [3]. SELEX (Systematic Evolution of Ligands by EXponential Enrichment) relies on the library diversity of a pool of RNA oligonucleotides to recognize any ligand of interest through conformational sampling of the given sequences and identify the tightest binders, generating RNA aptamers that can be used directly as “chemical antibodies” or coupled to other functional RNA molecules to generate environmental sensing devices [13,14]. Given their rapid and cost-effective production and selection, non-immunogenicity, and lower batch-to-batch variation compared to antibodies or other detection methods, RNA aptamers have been explored as novel biomedical diagnostics tools for sensitive and rapid detection of clinically relevant ligands, including small molecule drugs [15], bacterial virulence factors [16], proteins implicated in cancer phenotypes [17–20], and whole cells [21,22]. High-affinity aptamers have been selected for antagonistic activities, and have been employed for diagnostics or conjugated to therapeutic drug vehicles for targeted delivery in vivo [14,23].
Toehold switches are rapidly designed and deployed nucleic acid diagnostic tools
RNA technologies have been recently explored as an in vitro diagnostics platform for nucleic acids and viral RNA, offering a faster and cheaper detection paradigm for remote and resource-limited areas compared with PCR and antibody-based methods, which are employed clinically in developed nations. Researchers developed a set of RNA regulators called toehold switches that bind trans-acting trigger RNA molecules and transduce an encoded biological output in vivo, generating novel gene regulatory switches in Escherichia coli via rational design [24]. Cis-regulatory translational elements, including the Shine-Dalgarno (SD) sequence and start codon, are sequestered in a hairpin structure that is disrupted through cognate RNA binding to the toehold sequence, leading to activation of protein production. Because the binding interaction between the trigger RNA and the toehold switch relies on an antisense linear strand displacement mechanism as opposed to loop-mediated interactions, the kinetic and thermodynamic properties of toehold switches can be predicted [25]. In addition, toehold switches are computationally designed via a forward engineering approach and can be rapidly synthesized, characterized, and deployed as genetic control elements in prokaryotes.
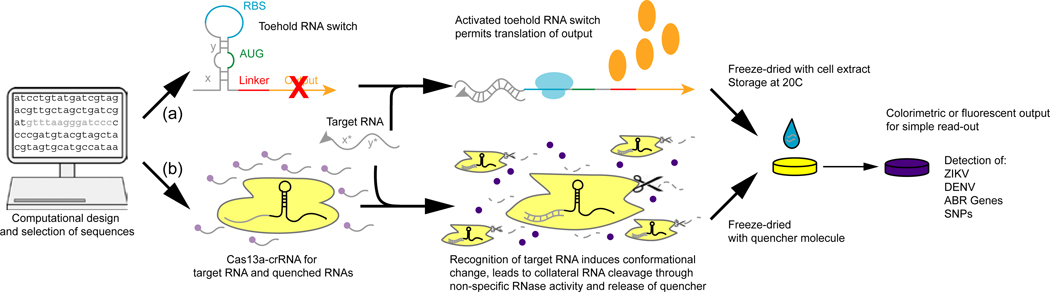
Toehold switches have been recently applied to the design of freeze-dried paper-based synthetic gene networks and diagnostic devices for Zika virus (ZIKV) and Ebola virus (EBOV) [26,27]. Toehold switches are freeze-dried with E. coli cell extract, which provides the translational machinery to produce an output indicating the absence or presence of viral RNA. By coupling the presence of ZIKV or EBOV viral RNA with in silico designed toehold switches triggering the expression of LacZ (colorimetric output) or GFP (fluorescence output), diagnostic devices based on simple visual outputs or easy-to-operate optical readers can be deployed (Figure 1). The switches distinguish between similar strains with single base pair resolution and can be combined with amplification methods to detect low femtomolar amounts of viral RNA in plasma samples. The shelf life and stability of freeze-dried components at room temperature was demonstrated for up to a year, with diagnostic results achievable in 3 hours. The paper-based diagnostics based on toehold RNA switches offer several advantages over serological diagnostics in resource-limited areas, while also providing rapid and sensitive detection when PCR-based analysis cannot be employed. The discriminatory power of RNA-RNA sequence interactions with strain-specific viruses, compared to cross-reactive antibodies, is essential for providing proper treatment and mitigating antiviral resistance to RNA viruses [28]. Continued work on increasing clinical sensitivity and expansion to other RNA viruses and parasites will dramatically improve point-of-care health outcomes for infected patients [29,30].
Figure 1: In vitro RNA devices recognize target nucleic acids to produce a diagnostic output in paper-based systems.
Genome sequences and computational design of target sequences produce nucleic acid sensors for viral genomes, SNP variants, or other pathogenic markers. Using toehold switches (a) to produce a protein output or SHERLOCK (b) to collaterally cleave fluorescently quenched RNAs upon recognition of the target viral nucleic acid, freeze-dried paper-based systems allow for simple visual or optical outputs such as colorimetric dyes or fluorescence for sub-nanomolar detection. (Figure adapted from [26,27,35])
RNA-guided CRISPR systems improve sensitivity of nucleic acid detection for diagnostics
The discovery of CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) systems and their associated RNA-guided nucleases has been explored for genome editing given the flexibility in guide RNA (gRNA) design and synthesis [31]. However, given its ability to recognize and cut a specific DNA or RNA sequence, CRISPR has been implicated as a sensitive detection system and diagnostic tool. Rapid molecular diagnostics have been achieved with the type VI CRISPR-Cas13a system to detect small amounts of RNA through its dual RNase activities for pre-crRNA processing and specific recognition of a target RNA sequence and more general non-specific RNase activity for collateral cleavage of proximal non-target nucleic acids [32]. Upon recognition of a specific target sequence by the crRNA bound to Cas13a, the enzyme adopts a catalytically active conformation that cleaves all RNAs, which potentially evolved as a programmed cell death mechanism to prevent viral replication upon infection in prokaryotes [33,34]. Based on this specific and general RNase activity, researchers developed the SHERLOCK (Specific High Sensitivity Enzymatic Reporter UnLOCKing) in vitro detection platform for rapid and attomolar-sensitive detection of ZIKV, Dengue Virus (DENV), and bacterial pathogens from clinical isolates with high strain and sequence discriminatory power [35]. Solution and paper-based systems were demonstrated with purified Leprotrichia wadei Cas13a (LwCas13a) incubated with gRNA for the target of interest and a quenched fluorescent RNA molecule that is collaterally cleaved when Cas13a recognizes its cognate RNA. The sensitivity of the SHERLOCK platform was increased to single-molecule detection through the incorporation of reverse transcription, recombinase polymerase amplification (RPA) and T7 transcription steps to amplify initial amounts of viral RNA. The SHERLOCK platform has also been applied to infer genotype and the presence of low frequency cancer mutations (~0.1% allelic fractions) using cell-free DNA samples.
Taken together, the examples highlight the potential of these nucleic acid-based detection systems for developing affordable diagnostics platforms in areas of global health, infectious diseases, and genetic testing. Assays can be developed for novel biomarkers within a few days and paper-based diagnostics can then be rapidly distributed for less than $1 per test in resource-limited locations or when serological testing is unavailable. However, the isothermal amplification techniques that many of these systems rely on are susceptible to contaminating nucleic acids, producing off-target amplicons and ultimately higher false positive rates that may pose therapeutic challenges in resource-limited regions [36,37]. In addition, the high rates of genetic drift in RNA viruses such as Zika and Ebola, may pose challenges for the design of probes and crRNAs to detect these viruses with the necessary discriminatory power. Toehold switches and SHERLOCK can tolerate some base pair mismatches, which may be useful to still detect the virus of interest, but efficacy may suffer and result in false negatives. Yet, given the ease of computationally assisted design and nucleic acid synthesis, detection platforms based on RNA devices have the advantage of being rapidly developed and deployed, expanding clinically relevant diagnostic power to developed and developing regions.
Synthetic RNA Devices Interface with the Environment to Conditionally Regulate Gene Expression
Regulating expression of endogenous genes and exogenously delivered transgenes through programmable inputs remains a significant challenge in biomedical applications and clinical translation of biotechnologies [38]. Natural RNA elements provide inspiration for engineering synthetic gene-regulatory devices by leveraging structural cues, catalytic activities, and antisense mechanisms of gene silencing with rational design and evolution to select for desired functions [39]. Based on structure and sequence information, RNA devices can be engineered to be responsive to specific molecular inputs, such as small-molecule or protein ligands or other RNA molecules, resulting in gene-regulatory devices that can regulate protein expression without accessory proteins or transcription factors [5,40]. In addition, RNA devices are typically compact (i.e., less than 200 nucleotides long) and can be readily integrated into most genetic constructs.
Ribozyme switches are a broadly deployed class of ligand-responsive gene-regulatory devices
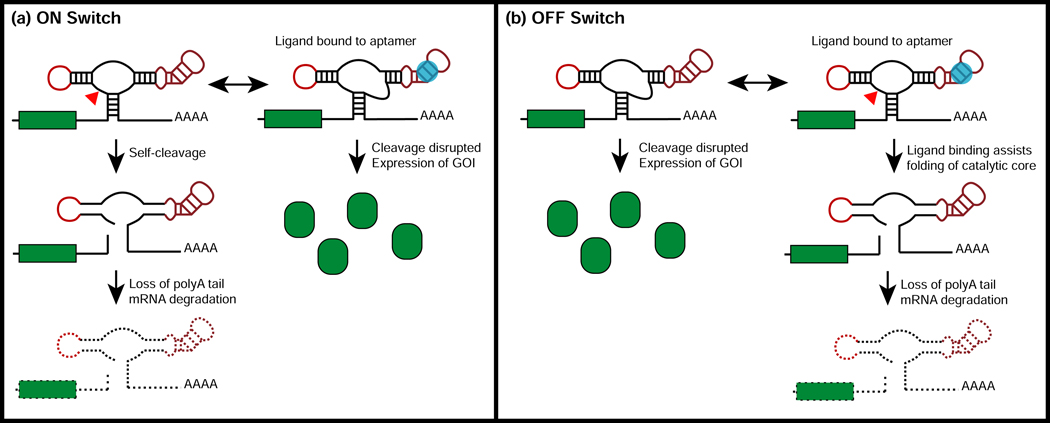
Ribozyme switches are a class of RNA-based gene-control devices that have been utilized in several engineered cell systems with biomedical relevance. The majority of ribozyme switches described to-date have been based on coupling the hammerhead ribozyme of the satellite RNA of tobacco ringspot virus (sTRSV) with an RNA aptamer in a manner that achieves ligand control over cleavage activity [5,41], although researchers have also engineered RNA switches based on the Hepatitis Delta Virus [42] and twister ribozymes [43]. In eukaryotic systems, ribozyme switches are typically integrated into the 3′ untranslated region (3′ UTR) of a gene of interest. The ligand bound state of the switch can either be associated with a ribozyme inactive conformation, in which the ribozyme structure is disrupted, resulting in increased transcript stability and protein production (i.e., a gene expression “ON” switch), or a ribozyme active conformation, resulting in transcript cleavage and subsequent rapid degradation and thus lowered protein production (i.e., a gene expression “OFF” switch) (Figure 2) [40]. Removing the inducer molecule has been shown to reverse the effects on gene expression state. Greater dynamic range in gene expression (defined as the ratio of gene expression between the ON and OFF states) has been achieved by integrating multiple copies of the ribozyme switch after the gene of interest, although this strategy generally results in a lower basal level of expression [44].
Figure 2: Ribozyme switches recognize diverse ligands to conditionally control target gene expression.
In the ON switch architecture (a), the catalytically-inactive conformation of the ribozyme is associated with ligand binding to the aptamer, preventing self-cleavage and leading to rapid degradation of the mRNA, thereby allowing for increased expression of the gene of interest. In the OFF switch architecture (b), the catalytically-active conformation of the ribozyme is associated with ligand binding to the aptamer, leading to self-cleavage and rapid degradation of the mRNA, thereby allowing for reduced expression of the gene of interest. Small molecule and protein aptamers have been integrated into the ribozyme switch architecture for conditional gene-control in mammalian systems. (Figure adapted from [60])
While the modular nature of RNA device architectures support the potential of generating tailored genetic regulators to any ligand of interest, the current set of ligands available for conditional gene expression control via RNA devices is limited [45]. Most small-molecule ribozyme switches to-date rely on the theophylline or tetracycline aptamers based on their compact structure, well-characterized binding and specificity, and subsequent studies to examine physical coupling strategies with the sTRSV ribozyme [46–49]. Although theophylline and tetracycline are FDA-approved drugs suitable for clinical translation of these ligands, the concentrations required in cell culture models are low millimolar, such that achieving these concentrations through systemic delivery would be highly toxic for any clinical applications of these switches [50]. As a result, these ligand/aptamer pairs have been more relevant for building and validating model systems that can be expanded upon with higher affinity ribozyme switches and further work to improve small-molecule ligands available for genetic control to improve the safety and efficacy of ribozyme switch-based therapeutics is necessary. Protein-responsive ribozyme switches to a diverse set of protein ligands have also been developed for conditional control in mammalian cells, including switches recognizing the MS2 coat protein [51], the E2F1 oncogene [52], beta-catenin [53], and the N-peptide from bacteriophage lambda [54].
Ribozyme switches control complex cellular growth and division phenotypes
Cellular control with ribozyme switches has been explored for regulating therapeutic circuits of ex vivo genetically modified cells. In an early example, researchers used ribozyme switches to regulate expression of the cytokines IL-2 and IL-15 in T cells to link in vivo proliferation to the presence of a drug as a persistence mechanism of transfused cells for adoptive immunotherapies [44]. In this early proof-of-concept demonstration the modularity of the sensor component was demonstrated by replacing the theophylline aptamer with the tetracycline aptamer and the modularity of the device output was demonstrated for two different cytokine transgenes. In a separate example, theophylline-responsive ribozyme switches were used to regulate the expression of herpes simplex virus-thymidine kinase and achieve suicide gene control of tumor cells in the presence of two drug inputs - ganciclovir and theophylline [50].
While these previous examples regulated the highest node of a cellular pathway to induce apoptosis or cell proliferation, ribozyme switches have also been engineered to control complex systems behaviors, such as cell cycle progression through mitosis. Researchers identified key regulatory factors in the cell cycle that arrested cells in either G0/1 phase (p27) or in G2/M phase (cyclinB1 mutant), engineered theophylline-responsive ON ribozyme switches after these transgenes, and integrated these devices into cells for stable expression [55]. Induction with theophylline arrested 77% of the population in G0/1 compared to 59% of uninduced cells, a comparable change to the constitutively expressed p27 control, with a similar result achieved with G2/M arrest - 73% versus 55%. With stable integration of these cell cycle controller devices into the genome, cells maintained cell cycle arrest in the presence of theophylline over six weeks and were generally reversible upon removal of the inducer, although leakiness of the device did affect basal fractions between the cell phases.
Strategies to increase safety and genetic control of viral vectors with ribozyme switches
Recently, viral vectors have gained more traction for biomedical applications as delivery vehicles for therapeutic genes, treatment strategies for cancer, and vaccines encoding viral antigens to raise immunity against infections [56]. Given its intrinsic cis-regulatory mechanism of self-cleavage to regulate gene expression, researchers have explored ribozyme switch-mediated control of viral vectors, as a way to circumvent challenges associated with introducing protein-based genetic regulation in a constricted packaging size [57]. In one example, conditional transgene expression was demonstrated in a replication-deficient adenovirus and replication-competent oncolytic adenovirus when theophylline-responsive ribozyme switches were integrated into the 5′- and 3′-UTRs of the transgenes [58]. The work demonstrates that ribozyme switches can serve as a powerful tool for optimal dosing or as a safety mechanism for viral therapies. In a second example, researchers incorporated theophylline-responsive ribozyme switches into specific locations of a viral genome in order to limit replication efficiency and reduce infectivity of a DNA virus, a negative-strand ssRNA measles virus [57], and in a later study a positive-strand ssRNA alphavirus [59]. By conditionally controlling viral replication with a small molecule, the authors developed a mechanism to achieve safer therapies based on engineered viruses. These safety switches rely on mechanisms different from standard antivirals, and thus would present different evolutionary pressures to resistance. One potential challenge with the approach of using RNA switches to control the infectivity and replication efficiency of RNA viruses is that the higher rates of genetic drift associated with these viruses may favor the selection of mutants that inactivate the switch and regain infectivity, thereby reducing safety and efficacy. However, small fractions of escape mutants that recover infectivity in replication-deficient or limited viruses will likely not reach adverse levels and infection can be eliminated by the immune system before significant risks arise for the patient [59]. Other considerations, such as integrating multiple ribozyme switches for replication control, tropism for specific cell types, and replication-deficient or replication-limited viruses will be essential for clinical translation.
High-throughput screening generates better ribozyme switches for genetic control
Most RNA switches described to-date are characterized by relatively small dynamic ranges, limiting their use as robust genetic controllers in certain applications. Leveraging high-throughput screening technologies such as FACS and Next-Generation Sequencing (NGS) can be used to generate novel ligand-responsive switches, improve activation ratios, and achieve faster self-cleavage kinetics, all desirable features for improving the gene-regulatory activities of this class of RNA devices. Researchers recently demonstrated a FACS-seq method for generating ribozyme switches in yeast with the above features using an in vivo FACS-seq platform and a tertiary loop-loop interaction architecture as opposed to earlier architectures based on secondary structure interactions [60]. The method integrates libraries of ribozyme switches into the 3’ UTR of a GFP reporter in a two-color plasmid system, where a mCherry reporter is used as a control to account for cell-to-cell variability in gene expression. Cells transformed with the library plasmids are grown in the presence and absence of the ligand and each cell population is sorted into discrete bins based on the ratio of GFP to mCherry expression. The library members in each bin are recovered, amplified with unique barcodes, and NGS is used to count the number of times a unique sequence occurs in each bin. From these counts, the gene-regulatory activity is reconstructed for each library member. The FACS-Seq method thus provides activity data for all members of the library, allowing for the identification of highly functional switches as well as improved insight into the overall design space for this important class of RNA devices.
Given that ribozyme switches tested and analyzed in yeast have correlative activity in mammalian cells [61], yeast can serve as a rapid prototyping model organism for mammalian RNA switches. However, selection and optimization of ribozymes switches in mammalian cells may be better suited for biomedical translation to capture the transcriptional dynamics and intracellular environment of the appropriate host cell. Researchers have also performed ribozyme switch selections in a luciferase plate-based assay in mammalian cells [62], which exhibits a much lower throughput than a FACS-based screening approach. A combination of high throughput or automated screening and selection of ribozyme switches in mammalian cells for novel ligands with less toxicity, improved dynamic range, and faster kinetics will contribute to the translation of this important class of gene-regulatory devices to biomedical applications.
miRNA-responsive switches rely on cell-specific cues to activate a synthetic RNA device
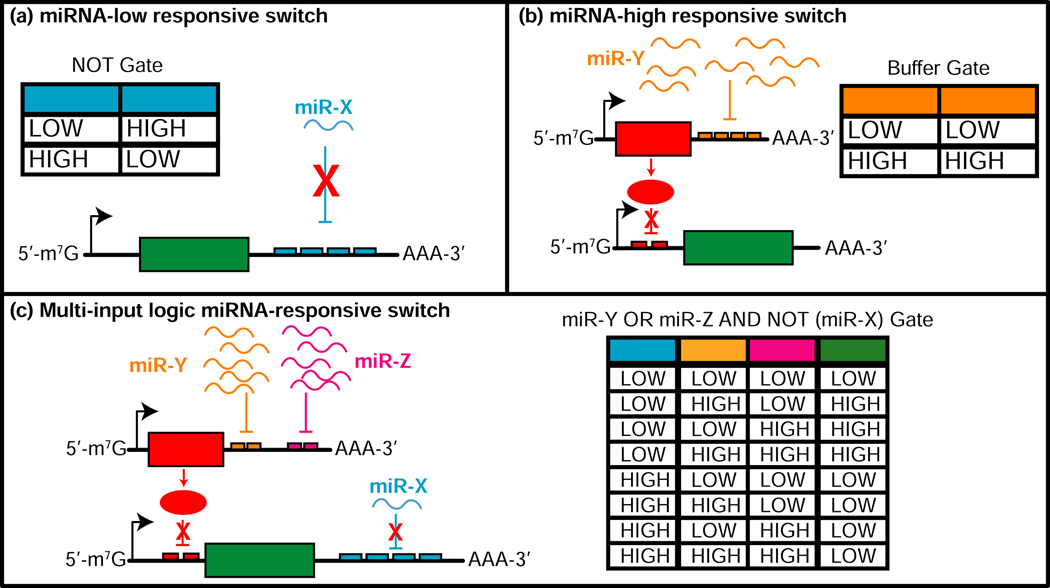
RNAi is an endogenous regulatory system that has been used to advance biomedical discoveries, target-drug screens, gain/loss-of-function studies, and new therapeutics [2,4]. Natural miRNA and siRNA silencing activity is based on sequence complementarity to a target transcript - miRNAs with partial complementarity lead to translational repression, whereas siRNAs that exhibit full complementarity direct endonucleolytic cleavage of the target transcript via RISC and AGO2. Endogenous miRNA profiles have been used to classify [63] and direct expression of target genes to specific cell populations [64]. Cell type-specific miRNA profiles and miRNA dysregulation in cancer cells provide unique signatures that when combined with endogenous RNAi machinery can be processed by RNA devices to repress or silence genes of interest in targeted cell populations. In an early example, researchers demonstrated that characteristic miRNA profiles in HeLa cells can be detected in a multi-logic gate circuit (or miRNA classifier device) such that apoptosis is induced when the anticipated miRNA profile is detected (Figure 3) [63]. Leveraging cell-specific miRNA profiles to activate a therapeutic circuit will reduce off-target effects and increase safety of these therapies for systemic delivery.
Figure 3: miRNA-responsive switches respond to high- or low-levels of endogenously expressed miRNAs.
In the miRNA-low switch (NOT gate) architecture (a), low levels of miRNA do not significantly inhibit mRNA translation, allowing for production of the protein of interest. In the miRNA-high switch (Buffer gate) architecture (b), a double-inversion module is employed, where high levels of endogenous miRNAs inhibit the production of a repressor protein, allowing mRNA translation of the gene of interest. miRNA-low and high switch designs can be multiplexed into multi-input higher-order logic gates (c), which encode protein production under specific patterns of miRNA expression levels characteristic of a specific cell type of interest. (Figure adapted from [63,65])
One recently explored application of such miRNA classifier devices has been for exogenous RNA-only circuits and mRNA therapies for non-viral delivery of post-transcriptional gene products [65]. mRNA therapies provide transient control of gene expression without random genome integration typical of conventional gene therapies. Researchers explored an RNA-only classifier circuit that induced apoptosis preferentially in HeLa cells by recognizing high levels of miR-21 and low levels of miR-141, miR-142(3p), and miR-146a in a two-part repression cascade. Production of modified RNA (modRNA) devices with a polyA tail and 7-methylguanylate cap allowed for stability and direct translation of mRNA to proteins, with miRNA targeting sites incorporated in the 3’ UTR to direct expression to the desired cell type. In a follow-up study using a similar architecture, a miRNA-responsive switch system was demonstrated to induce Cas9 gene editing in HeLa and human induced pluripotent stem cells (hIPSCs) using cell marker-specific miRNAs [66]. This additional consideration for genome editing can eliminate off-target DNA cleavage in non-target cell populations, a concern for safety of in vivo therapeutics.
A similar approach was used to purify a heterogeneous hIPSC population into differentiated cell types by transfecting a modRNA encoding an apoptotic gene and miRNA targeting sites in the 5’ UTR corresponding to highly abundant miRNAs for that specific cell type. The researchers identified miRNAs present in hIPSC-derived cardiomyocytes, hepatocytes, endothelial cells, and insulin-producing cells to include sites targeting these miRNAs in the modRNA design in order to purify differentiated cells with >95% efficiency [67]. Cardiomyocytes selected with miR-1 and miR-208a-responsive switches exhibited zero incidences of tumorigenesis when transplanted into mouse hearts, highlighting efficient purification and translation for other cell types with difficult isolation protocols. Delivering an autonomous apoptotic circuit to purify differentiated cells from a heterogeneous population reduces the need for cell sorting techniques that often kill many cells, when large numbers of viable cells are essential for regenerative cell therapies.
Conclusions and Future Directions
One of the key advantages of RNA-based devices are their ability to recognize a diverse range of ligands, spanning nucleic acids, small molecules, and proteins, to transduce a desired biological output. Rational engineering aided by computer design supports rapid prototyping and validation to generate RNA molecules responsive to new inputs, particularly other nucleic acids where Watson-Crick base pairing dictates the desired interaction. However, engineering novel ligand-binding aptamers to clinically relevant small molecule and protein inputs, that can be used in designing RNA switches with desirable properties for biomedical translation, such as low-toxicity, high-affinity, and rapid kinetics, has proven to be more challenging. Advances in high-throughput screening will aid future development efforts in order to expand the number of clinically relevant ligands that RNA devices can respond to. RNA switches typically exhibit a narrower dynamic range of gene expression when compared to highly engineered transcription factor-based systems. However, recent work has demonstrated that the application of computational folding tools [68], in vivo screening methods [62], and incorporation of genetic amplification strategies can improve the gene-regulatory performance of RNA devices [69].
The use of RNA-only devices as in vivo biosensors or therapeutics can face challenges resulting from short half-lives and potential immune stimulation [70]. While the use of modRNAs can overcome some of these limitations for mRNA transfections, they may run into challenges with long-term stability and robust expression in human therapeutics. RNA chemical modifications, including those of the 2′ hydroxyl group or the phosphodiester backbone, have been used to prevent recognition by endogenous RNases and immune stimulation and have been extensively explored for therapeutic RNAi agents [70,71] and more recently for Cas9 gRNA engineering [72,73]. Such chemical modification approaches will be important for mRNA-based therapeutics to improve the delivery and transient expression of target genes.
RNA devices possess natural functions and engineered modalities that can be harnessed in vitro and in vivo for a variety of biomedical applications. The modularity, diversity of function, and relative ease of synthesis and design of RNA devices make these functional biomolecules important tools for biomedical engineering and clinical applications.
Highlights.
RNA devices sense environmental cues to control diverse biological outcomes
RNA devices are modular, programmable, and can be integrated into complex systems
RNA can be engineered to exhibit diverse biological functions
Clinical translation of RNA devices remains a significant engineering challenge
Acknowledgements
C.M.K is financially supported by the National Institute of General Medical Sciences of the National Institutes of Health (Award Number T32GM008412) and in part by the Stanford Vice Provost for Graduate Education EDGE-STEM Fellowship. C.D.S is supported by the National Institutes of Health and the Human Frontiers Science Research Program. C.D.S. is a Chan Zuckerberg Biohub investigator. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lee K, Lee B: Structural and Biochemical Properties of Novel Self-Cleaving Ribozymes. Molecules 2017, 22:678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deng Y, Wang CC, Choy KW, Du Q, Chen J, Wang Q, Li L, Chung TKH, Tang T: Therapeutic potentials of gene silencing by RNA interference: Principles, challenges, and new strategies. Gene 2014, 538:217–227. [DOI] [PubMed] [Google Scholar]
- 3.Gelinas AD, Davies DR, Janjic N: Embracing proteins: Structural themes in aptamer-protein complexes. Curr. Opin. Struct. Biol 2016, 36:122–132. [DOI] [PubMed] [Google Scholar]
- 4.Lam JKW, Chow MYT, Zhang Y, Leung SWS: siRNA Versus miRNA as Therapeutics for Gene Silencing. Mol. Ther. Nucleic Acids 2015, 4:e252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang AL, Wolf JJ, Smolke CD: Synthetic RNA switches as a tool for temporal and spatial control over gene expression. Curr. Opin. Biotechnol 2012, 23:679–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chappell J, Watters KE, Takahashi MK, Lucks JB: A renaissance in RNA synthetic biology: new mechanisms, applications and tools for the future. Curr. Opin. Chem. Biol 2015, 28:47–56. [DOI] [PubMed] [Google Scholar]
- 7.Morris KV, Mattick JS: The rise of regulatory RNA. Nat Rev Genet 2014, 15:423–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lienert F, Lohmueller JJ, Garg A, Silver PA: Synthetic biology in mammalian cells: next generation research tools and therapeutics. Nat. Rev. Mol. Cell Biol 2014, 15:95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Myhrvold C, Silver P a: Using synthetic RNAs as scaffolds and regulators. Nat. Struct. & Mol. Biol 2015, 22:8–10. [DOI] [PubMed] [Google Scholar]
- 10.Mathur M, Xiang JS, Smolke CD: Mammalian synthetic biology for studying the cell. J. Cell Biol 2017, 216:73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katrekar D, Hu M, Mali P: Advances in CRISPR-Cas Based Genome Engineering. Curr. Opin. Biomed. Eng 2017, doi: 10.1016/j.cobme.2017.04.001. [DOI]
- 12.Hughes RA, Ellington AD: Synthetic DNA Synthesis and Assembly : Putting the Synthetic in Synthetic Biology. 2017, doi: 10.1101/CSHPERSPECT.A023812. [DOI] [PMC free article] [PubMed]
- 13.Ellington AD, Szostak JW: In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346:818–22. [DOI] [PubMed] [Google Scholar]
- 14.Zhou J, Rossi J: Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov 2017, 16:181–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKeague M, Wang YH, Smolke CD: In Vitro Screening and in Silico Modeling of RNA-Based Gene Expression Control. ACS Chem. Biol 2015, 10:2463–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakyi SA, Aboagye SY, Otchere ID, Liao AM, Caltagirone TG, Yeboah-Manu D: RNA Aptamer That Specifically Binds to Mycolactone and Serves as a Diagnostic Tool for Diagnosis of Buruli Ulcer. PLoS Negl. Trop. Dis 2016, 10:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L, Rashid F, Shah a, Awan HM, Wu M, Liu a, Wang J, Zhu T, Luo Z, Shan G: The isolation of an RNA aptamer targeting to p53 protein with single amino acid mutation. Proc Natl Acad Sci U S A 2015, 112:10002–10007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gijs M, Penner G, Blackler GB, Impens NREN, Baatout S, Luxen A, Aerts AM: Improved aptamers for the diagnosis and potential treatment of HER2-positive cancer. Pharmaceuticals 2016, 9:15–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon S, Armstrong B, Habib N, Rossi JJ: Blind SELEX Approach Identifies RNA Aptamer that Regulate EMT and Inhibit Metastasis. Mol. Cancer Res 2017, doi: 10.1158/1541-7786.MCR-16-0462. [DOI] [PMC free article] [PubMed]
- 20.Lee YJ, Lee SW: Regression of hepatocarcinoma cells using RNA aptamer specific to alpha-fetoprotein. Biochem. Biophys. Res. Commun 2012, 417:521–527. [DOI] [PubMed] [Google Scholar]
- 21.Dua P, Ren S, Lee SW, Kim J-K, Shin H, Jeong O-C, Kim S, Lee D-K: Cell-SELEX Based Identification of an RNA Aptamer for Escherichia coli and Its Use in Various Detection Formats. Mol. Cells 2016, 39:807–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen M, Yu Y, Jiang F, Zhou J, Li Y, Liang C, Dang L, Lu A, Zhang G: Development of cell-SELEX technology and its application in cancer diagnosis and therapy. Int. J. Mol. Sci 2016, 17:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jo H, Ban C: Aptamer-nanoparticle complexes as powerful diagnostic and therapeutic tools. Exp. Mol. Med 2016, 48:e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Green AA, Silver PA, Collins JJ, Yin P: Toehold switches: de-novo-designed regulators of gene expression. Cell 2014, 159:925–39. *The authors present the computational design and testing of toehold switches in prokaryotes to achieve a robust dynamic range in gene expression control. The thermodynamic parameters discussed for forward-engineering of switches is important in designing toehold switches for diagnostic devices.
- 25.Takahashi MK, Lucks JB: A modular strategy for engineering orthogonal chimeric RNA transcription regulators. Nucleic Acids Res. 2013, 41:7577–7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pardee K, Green AA, Ferrante T, Cameron DE, DaleyKeyser A, Yin P, Collins JJ: Paper-based synthetic gene networks. Cell 2014, 159:940–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pardee K, Green AA, Takahashi MK, Braff D, Lambert G, Lee JW, Ferrante T, Ma D, Donghia N, Fan M, et al. : Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell 2016, 165:1255–1266. **Given the Zika virus epidemic, the authors rapidly designed and prototyped a paper-based diagnostic test with freeze-dried cell extracts and toehold switches, with the ability to distinguish between American and African strains at low femtomolar concentrations when isothermal RNA amplification is used. The cost for the test can be as low as $1/test, and a LacZ colorimetric output provides for easy visual diagnosis of infection.
- 28.Saiz J, Martín-Acebes MA: The Race To Find Antivirals for Zika Virus. Antimicrob. Agents Chemother 2017, 61:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slomovic S, Pardee K, Collins JJ: Synthetic biology devices for in vitro and in vivo diagnostics. Proc. Natl. Acad. Sci. U. S. A 2015, 112:14429–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei TY, Cheng CM: Synthetic Biology-Based Point-of-Care Diagnostics for Infectious Disease. Cell Chem. Biol 2016, 23:1056–1066. [DOI] [PubMed] [Google Scholar]
- 31.Mali P, Esvelt KM, Church GM: Cas9 as a versatile tool for engineering biology. Nat. Methods 2013, 10:957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.East-Seletsky A, O’Connell MR, Knight SC, Burstein D, Cate JHD, Tjian R, Doudna JA: Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 2016, 538:270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DBT, Shmakov S, Makarova KS, Semenova E, Minakhin L, et al. : C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science (80-. ). 2016, 353:aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu L, Li X, Wang J, Wang M, Chen P, Yin M, Li J, Sheng G, Wang Y: Two Distant Catalytic Sites Are Responsible for C2c2 RNase Activities. Cell 2017, 168:121–134.e12. [DOI] [PubMed] [Google Scholar]
- 35. Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, et al. : Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356:438–442. *The authors develop SHERLOCK using the CRISPR-Cas13a system and RNA amplification for single-molecule detection of RNA and DNA targets, representing viral genomes, SNPs, and antibiotic resistance genes. The sensitivity and specificity of CRISPR-based approaches and their rapid prototyping motivates their use in diagnostics and detection of nucleic acids.
- 36.Zanoli LM, Spoto G: Isothermal amplification methods for the detection of nucleic acids in microfluidic devices. Biosensors 2013, 3:18–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cordray MS, Richards-Kortum RR: Review: Emerging nucleic acid-based tests for point-of-care detection of malaria. Am. J. Trop. Med. Hyg 2012, 87:223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ausländer S, Ausländer D, Fussenegger M: Synthetic Biology—The Synthesis of Biology. Angew. Chemie - Int. Ed 2017, 56:6396–6419. [DOI] [PubMed] [Google Scholar]
- 39.Liang JC, Bloom RJ, Smolke CD: Engineering Biological Systems with Synthetic RNA Molecules. Mol. Cell 2011, 43:915–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKeague M, Wong RS, Smolke CD: Opportunities in the design and application of RNA for gene expression control. Nucleic Acids Res. 2016, 44:2987–2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wittmann A, Suess B: Engineered riboswitches: Expanding researchers’ toolbox with synthetic RNA regulators. FEBS Lett. 2012, 586:2076–2083. [DOI] [PubMed] [Google Scholar]
- 42.Nomura Y, Zhou L, Miu A, Yokobayashi Y: Controlling mammalian gene expression by allosteric hepatitis delta virus ribozymes. ACS Synth. Biol 2013, 2:684–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Felletti M, Stifel J, Wurmthaler LA, Geiger S, Hartig JS: Twister ribozymes as highly versatile expression platforms for artificial riboswitches. Nat. Commun 2016, 7:12834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen YY, Jensen MC, Smolke CD: Genetic control of mammalian T-cell proliferation with synthetic RNA regulatory systems. Proc. Natl. Acad. Sci. U. S. A 2010, 107:8531–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Auslander S, Fussenegger M: Synthetic RNA-based switches for mammalian gene expression control. Curr. Opin. Biotechnol 2017, 48:54–60. [DOI] [PubMed] [Google Scholar]
- 46.Jenison RD, Gill SC, Pardi A, Polisky B: High-resolution molecular discrimination by RNA. Science 1994, 263:1425–9. [DOI] [PubMed] [Google Scholar]
- 47.Win MN, Smolke CD: A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc. Natl. Acad. Sci 2007, 104:14283–14288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beilstein K, Wittmann A, Grez M, Suess B: Conditional Control of Mammalian Gene Expression by Tetracycline-Dependent Hammerhead Ribozymes. ACS Synth. Biol 2015, 4:526–534. [DOI] [PubMed] [Google Scholar]
- 49.Zhong G, Wang H, Bailey CC, Gao G, Farzan M: Rational design of aptazyme riboswitches for efficient control of gene expression in mammalian cells. Elife 2016, 5. [DOI] [PMC free article] [PubMed]
- 50.Zhang Y, Wang J, Cheng H, Sun Y, Liu M, Wu Z, Pei R: Conditional control of suicide gene expression in tumor cells with theophylline-responsive ribozyme. Gene Ther. 2017, 24:84–91. [DOI] [PubMed] [Google Scholar]
- 51.Kennedy AB, Vowles JV, D’Espaux L, Smolke CD: Protein-responsive ribozyme switches in eukaryotic cells. Nucleic Acids Res. 2014, 42:12306–12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bloom RJ, Winkler SM, Smolke CD: Synthetic feedback control using an RNAi-based gene-regulatory device. J. Biol. Eng 2015, 9:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bloom RJ, Winkler SM, Smolke CD: A quantitative framework for the forward design of synthetic miRNA circuits. Nat Methods 2014, 11:1147–1153. [DOI] [PubMed] [Google Scholar]
- 54.Ausländer S, Stücheli P, Rehm C, Ausländer D, Hartig JS, Fussenegger M: A general design strategy for protein-responsive riboswitches in mammalian cells. Nat. Methods 2014, 11:1154–1160. [DOI] [PubMed] [Google Scholar]
- 55. Wei KY, Smolke CD: Engineering dynamic cell cycle control with synthetic small molecule-responsive RNA devices. J. Biol. Eng 2015, 9:21. *Identifying key regulatory nodes in a complex pathway, the authors integrated theophylline-responsive ribozyme switches to regulate checkpoint cell cycle genes to arrest cells in a particular phase. These regulators can be used to better study the dynamics of cell cycle and designing novel therapeutics for cell cycle dysregulation.
- 56.Schott JW, Morgan M, Galla M, Schambach A: Viral and Synthetic RNA Vector Technologies and Applications. Mol. Ther 2016, 24:1513–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ketzer P, Kaufmann JK, Engelhardt S, Bossow S, von Kalle C, Hartig JS, Ungerechts G, Nettelbeck DM: Artificial riboswitches for gene expression and replication control of DNA and RNA viruses. Proc. Natl. Acad. Sci 2014, 111:E554–E562. **The authors integrate theophylline-responsive ribozyme swtiches in their adenovirus and measles virus to control infectivity and replication, important considerations for clinical uses of viral gene delivery. Synthetic viral constructs can be engineered with these ribozymes more easily given their small size and intrinsic self-cleavage mechanism.
- 58.Ketzer P, Haas SF, Engelhardt S, Hartig JS, Nettelbeck DM: Synthetic riboswitches for external regulation of genes transferred by replication-deficient and oncolytic adenoviruses. Nucleic Acids Res. 2012, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bell CL, Yu D, Smolke CD, Geall AJ, Beard CW, Mason PW: Control of alphavirus-based gene expression using engineered riboswitches. Virology 2015, 483:302–11. [DOI] [PubMed] [Google Scholar]
- 60.Townshend B, Kennedy AB, Xiang JS, Smolke CD: High-throughput cellular RNA device engineering. Nat. Methods 2015, 12:989–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wei KY, Chen YY, Smolke CD: A yeast-based rapid prototype platform for gene control elements in mammalian cells. Biotechnol. Bioeng 2013, 110:1201–1210. [DOI] [PubMed] [Google Scholar]
- 62.Rehm C, Klauser B, Hartig JS: Engineering aptazyme switches for conditional gene expression in mammalian cells utilizing an in vivo screening approach. Methods Mol. Biol 2015, 1316:127–40. [DOI] [PubMed] [Google Scholar]
- 63.Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y: Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science 2011, 333:1307–11. [DOI] [PubMed] [Google Scholar]
- 64.Mullokandov G, Baccarini A, Ruzo A, Jayaprakash AD, Tung N, Israelow B, Evans MJ, Sachidanandam R, Brown BD: High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat. Methods 2012, 9:840–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wroblewska L, Kitada T, Endo K, Siciliano V, Stillo B, Saito H, Weiss R: Mammalian synthetic circuits with RNA binding proteins for RNA-only delivery. Nat. Biotechnol 2015, 33:839–41. *The authors incorporated miRNA-responsive switches in modRNAs for transient RNA-only delivery to regulate the circuit’s expression in a specific cell type. RNA binding proteins were also included to generate repression cascades and two-state switches, expanding the RNA device logic for cell-specific regulation of externally-delivered modRNAs.
- 66.Hirosawa M, Fujita Y, Parr CJC, Hayashi K, Kashida S, Hotta A, Woltjen K, Saito H: Cell-type-specific genome editing with a microRNA-responsive CRISPR–Cas9 switch. Nucleic Acids Res. 2017, doi: 10.1093/nar/gkx309. [DOI] [PMC free article] [PubMed]
- 67. Miki K, Endo K, Takahashi S, Funakoshi S, Takei I, Katayama S, Toyoda T, Kotaka M, Takaki T, Umeda M, et al. : Efficient Detection and Purification of Cell Populations Using Synthetic MicroRNA Switches. Cell Stem Cell 2015, 16:699–711. **From an initial iPSC pool of differentiating cells, the authors delivered modRNAs carrying apoptotic genes that purified specific cell types based on the endogenous miRNA profile. Purified cardiomyocytes selected for low miR-1 and mir-208a were transplanted in mice and showed no incidence of tumorigenesis, promoting this strategy to select for difficult-to-isolate cell types for safe and effective iPS cell-based therapeutics.
- 68.Dotu I, Garcia-Martin JA, Slinger BL, Mechery V, Meyer MM, Clote P: Complete RNA inverse folding: Computational design of functional hammerhead ribozymes. Nucleic Acids Res. 2014, 42:11752–11762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang YH, McKeague M, Hsu TM, Smolke CD: Design and Construction of Generalizable RNA-Protein Hybrid Controllers by Level-Matched Genetic Signal Amplification. Cell Syst. 2016, 3:549–562.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Burnett JC, Rossi JJ: RNA-Based Therapeutics: Current Progress and Future Prospects. Chem. Biol 2012, 19:60–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Engels JW: Gene silencing by chemically modified siRNAs. N. Biotechnol 2013, 30:302–307. [DOI] [PubMed] [Google Scholar]
- 72.Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S, Steinfeld I, Lunstad BD, Kaiser RJ, Wilkens AB, et al. : Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol 2015, 33:985–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rahdar M, McMahon MA, Prakash TP, Swayze EE, Bennett CF, Cleveland DW: Synthetic CRISPR RNA-Cas9-guided genome editing in human cells. Proc. Natl. Acad. Sci 2015, 112:E7110–7117. [DOI] [PMC free article] [PubMed] [Google Scholar]