Abstract
The cydAB genes from Mycobacterium smegmatis have been cloned and characterized. The cydA and cydB genes encode the two subunits of a cytochrome bd oxidase belonging to the widely distributed family of quinol oxidases found in prokaryotes. The cydD and cydC genes located immediately downstream of cydB encode a putative ATP-binding cassette-type transporter. At room temperature, reduced minus oxidized difference spectra of membranes purified from wild-type M. smegmatis displayed spectral features that are characteristic of the γ-proteobacterial type cytochrome bd oxidase. Inactivation of cydA or cydB by insertion of a kanamycin resistance marker resulted in loss of d-heme absorbance at 631 nm. The d-heme could be restored by transformation of the M. smegmatis cyd mutants with a replicating plasmid carrying the highly homologous cydABDC gene cluster from Mycobacterium tuberculosis. Inactivation of cydA had no effect on the ability of M. smegmatis to exit from stationary phase at 37 or 42°C. The growth rate of the cydA mutant was tested under oxystatic conditions. Although no discernible growth defect was observed under moderately aerobic conditions (9.2 to 37.5 × 102 Pa of pO2 or 5 to 21% air saturation), the mutant displayed a significant growth disadvantage when cocultured with the wild type under extreme microaerophilia (0.8 to 1.7 × 102 Pa of pO2 or 0.5 to 1% air saturation). These observations were in accordance with the two- to threefold increase in cydAB gene expression observed upon reduction of the pO2 of the growth medium from 21 to 0.5% air saturation and with the concomitant increase in d-heme absorbance in spectra of membranes isolated from wild-type M. smegmatis cultured at 1% air saturation. Finally, the cydA mutant displayed a competitive growth disadvantage in the presence of the terminal oxidase inhibitor, cyanide, when cocultured with wild type at 21% air saturation in an oxystat. In conjunction with these findings, our results suggest that cytochrome bd is an important terminal oxidase in M. smegmatis.
The major human pathogen, Mycobacterium tuberculosis is responsible for 3 million deaths and 8 million new cases of tuberculosis per annum (12). Most cases of tuberculosis arise from reactivation of an infection acquired years before the onset of symptoms, reflecting the emergence of actively replicating organisms from a latent or dormant state (4, 34, 55). Even after prolonged adequate therapy, M. tuberculosis DNA can be detected in the sputum of patients, and the organism may reactivate to cause disease following immunosuppression (15, 21). It is widely believed that oxygen limitation, amino acid starvation, and carbon source restriction are involved in establishing and maintaining dormancy in M. tuberculosis (10, 24, 49). Several in vitro models for studying the physiology of the “dormant” state of M. tuberculosis have been developed on the basis of these assumptions (42, 59). The Wayne model of mycobacterial dormancy is based on interrupting the aeration of logarithmic-phase cultures. After cessation of growth and autolysis of most of the bacilli, a small proportion of survivors are able to resume growth at a reduced rate (59). The evidence suggests that mycobacteria, once thought to be obligately aerobic, can adapt and perhaps thrive under reduced oxygen conditions (59–62). Significantly, Segal found evidence that the virulence of mycobacteria, and of M. tuberculosis specifically, is related to the ability to grow under reduced concentrations of O2 (48).
Aerobic and facultative aerobic bacteria are able to respond to altered levels of oxygen in the environment by utilizing alternative respiratory pathways (41). As such, many bacterial species possess more than one terminal oxidase, which catalyzes the oxidation of either cytochrome c or quinol. The cytochrome bd oxidase is a widely distributed prokaryotic quinol oxidase, which performs a variety of physiological functions in vivo (17, 27, 33, 41). It is involved in energy-transducing respiration in Escherichia coli (27) and Bacillus species (45). Cytochrome bd also plays a role in aerotolerant nitrogen fixation and protection against metal toxicity and oxidative stress in Azotobacter vinelandii (13, 28, 30). Significantly, Way et al. recently demonstrated a positive correlation between the virulence of the bacterial pathogen, Shigella flexneri, and the level of cytochrome bd expression (58), suggesting that cytochrome bd may be particularly important for the growth and survival of pathogens that encounter environments in which O2 is progressively limited. Sequence analysis of the M. tuberculosis genome (7) revealed the presence of a cydABDC gene cluster, the cydAB genes of which encode a putative cytochrome bd oxidase that is highly homologous to its counterpart in E. coli. By analogy with the physiological function of the E. coli oxidase, we speculated that the mycobacterial bd oxidase might play a role in adaptation to conditions of reduced oxygen. To test this hypothesis, we investigated the function of the cydAB genes in Mycobacterium smegmatis, which is a fast-growing nonpathogenic saprophyte that demonstrates oxygen depletion-induced dormancy similar to that observed with M. tuberculosis (11) and is regarded as an important model for studying mycobacterial physiology (35, 52, 66). In this study, we report the gene cloning, allelic disruption, spectral analysis, expression analysis, and phenotypic characterization of cyd-inactivated mutant strains, and we demonstrate that this oxidase plays an important respiratory role in M. smegmatis.
MATERIALS AND METHODS
Materials, bacterial strains, plasmids, and growth conditions.
Enzymes were from Boehringer Mannheim or Amersham, radiochemicals were from ICN, and all other chemicals were from Sigma. The bacterial strains and plasmids used in this study are listed in Table 1. E. coli DH5α was used for plasmid manipulations and JM101 was used for M13 cloning (47). For E. coli, ampicillin, kanamycin (KAN), and hygromycin (HYG) were used at 100, 50 and 200 μg/ml, respectively. For M. smegmatis, KAN was used at 10 (solid medium) or 25 (liquid medium) μg/ml, and HYG was used at 50 μg/ml. E. coli strains were grown in Luria-Bertani broth (LB) or agar (LA). M. smegmatis strains were grown in LB for targeted knockout mutagenesis or in MADC-Tw (Middlebrook 7H9 broth [Difco] supplemented with 0.085% NaCl, 0.2% glucose, 0.2% glycerol, and 0.05% Tween 80) and on LA as the solid medium.
TABLE 1.
Strains and plasmids used in this study
| M. smegmatis strain or plasmid | Characteristicsa | Source or reference |
|---|---|---|
| M. smegmatis | ||
| mc2155 | High-frequency transformation mutant of M. smegmatis ATCC 607 | 53 |
| mc2155 (pHINT) | mc2155 with pHINT integrated at the attP locus; Hygr | This work |
| mc2155(pOTBCYD) | mc2155 carrying M. tuberculosis cydABDC gene cluster on pOTBCYD; Hygr | This work |
| cydA::aph | cydA knockout mutant of mc2155; Kmr | This work |
| cydA::aph(pOTBCYD) | cydA::aph carrying M. tuberculosis cydABDC gene cluster on pOTBCYD; Kmr Hygr | This work |
| cydB::aph | cydB knockout mutant of mc2155; Kmr | This work |
| cydB::aph(pOTBCYD) | cydB::aph carrying M. tuberculosis cydABDC gene cluster on pOTBCYD; Kmr Hygr | This work |
| mc2155::pBK4 | mc2155 carrying pBK4 integrated at the cyd locus; Kmr | This work |
| Plasmids | ||
| pGEM32F(+) | E. coli cloning vector; Apr | Promega |
| pOLYG | Multicopy E. coli-Mycobacterium shuttle vector; Hygr | 37 |
| p6.11 | pOLYG carrying hsp60-sacB cassette; Hygr | S. Quan |
| pHINT | Integrative shuttle vector that integrates at the Mycobacterium attP locus; Hygr Apr | 37 |
| p2NIL | Suicide plasmid used for cloning homologous recombination substrates | 40 |
| pOTBCYD | pOLYG carrying the M. tuberculosis cydABDC genes carried on an 11kb EcoRI fragment; Hygr | This work |
| pSMT3 | Derivative of pOLYG carrying M. bovis BCG hsp60 promoter; Hygr | 37 |
| pSlacZ | Derivative of pSMT3 carrying hsp60-lacZ cassette; Hygr | This work |
| p45C10 | pBluescript SK(+) construct containing partial M. smegmatis cydAB region | M. Everett |
| p16N11 | pBluescript SK(+) construct containing partial M. smegmatis cydA and upstream promoter region | M. Everett |
| pBK1 | pGEM32F(+) carrying 2.4-kb KpnI fragment from M. smegmatis containing part of cydA and cydB genes | This work |
| pBK2 | pGEM32F(+) carrying 10-kb BglII fragment from M. smegmatis containing entire cydABDC gene cluster | This work |
| pCYDAKO | pBK1 carrying cydA::aph, hsp60-sacB, and hsp60-lacZ; used for targeted knockout of M. smegmatis cydA; Kmr | This work |
| pCYDBKO | pBK1 carrying cydB::aph, hsp60-sacB, and hsp60-lacZ; used for targeted knockout of M. smegmatis cydB; Kmr | This work |
| pATB12 | Multicopy E. coli-Mycobacterium shuttle vector; lacZ, Hygr | M. Everett |
| pBK3 | pATB12 carrying 2,634-bp PstI fragment from p16N11 containing M. smegmatis cyd promoter region, cloned immediately upstream of lacZ; Hygr | This work |
| pBK4 | p2NIL containing the cydA′::lacZ cassette from pBK3; Kmr | This work |
Apr, ampicillin resistance; Hygr, HYG resistance.
Cloning and sequencing of the M. smegmatis cyd gene cluster.
A cosmid carrying the M. tuberculosis cydABDC genes was isolated from a pYUB328::H37Rv cosmid library (3) by using a probe derived from the closely linked polA gene (22). A 2.5-kb SalI fragment carrying the cydAB genes was isolated from the cosmid and used to probe a gridded plasmid library of M. smegmatis (kindly provided by M. Everettt, Glaxo Wellcome, Stevenage, United Kingdom). A positive clone, p45C10, was found to contain an insert showing a high degree of homology to cydA and cydB from M. tuberculosis. This 1-kb BamHI-HindIII fragment was then used to probe 2- to 3-kb KpnI and 9- to 11-kb BglII size-fractionated libraries of M. smegmatis genomic DNA cloned in pGEM32F(+), from which pBK1and pBK2 were identified. The complete sequence of the cydABDC cluster was determined by sequencing 0.5- to 1.2-kb inserts of pBK1 and pBK2 subcloned in M13mp18/19. All clones were sequenced on both strands by using a combination of universal and internal primers. DNA sequencing was carried out with an ABI Prism 377 automated DNA sequencer, and analysis was performed by using the Lasergene suite of programs (DNASTAR, Madison, Wis.) and the BLAST server located at the National Centre for Biotechnology Information (Bethesda, Md.; http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST). Preliminary sequence data for M. smegmatis mc2155 was obtained from The Institute for Genomic Research website (http://www.tigr.org).
Construction of cydA::aph and cydB::aph knockout mutants of M. smegmatis
The suicide plasmid, pCYDAKO, carrying a cydA::aph allele, was constructed from pBK1 for use in allelic exchange by cloning the Tn903 KAN resistance (Kmr) (aph) cassette from pY6002 (23) into the PstI site in cydA. The hsp60-lacZ cassette from pSlacZ and the hsp60-sacB cassette from p6.11 were then sequentially cloned into the SalI and XbaI sites of the vector to form pCYDAKO. The plasmid pCYDBKO was constructed by cloning the aph gene into the SphI site located in the cydB gene, followed by sequential cloning of the selection markers, as described for pCYDAKO. These plasmids conferred sucrose sensitivity on E. coli and resulted in the development of blue color when plated on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)-containing indicator medium. M. smegmatis was transformed with pCYDAKO and pCYDBKO, as previously described (6), and recombinants were selected by plating on medium containing KAN and X-Gal. Blue, Kmr colonies were picked, diluted, and plated on medium containing KAN, X-Gal, and 5% sucrose. Sucrose-resistant colonies that remained white after patching on fresh indicator medium (3 of 200 in both cases) were selected as candidate allelic exchange mutants and were genotyped by Southern blot analysis by previously described methods (6, 20).
Construction of cydA′::lacZ reporter strain.
A 2,634-bp PstI fragment from p16N11 (isolated from the gridded plasmid library of M. smegmatis), containing the first 959 bp of cydA flanked by 1,675 bp of upstream sequence was used to make the reporter construct. The 1,675-bp region consists of a 95-bp intergenic spacer immediately upstream of cydA (probably containing the cyd promoter), preceded by an open reading frame that is highly homologous to the Rv1624c gene from M. tuberculosis (7), preceded by a further 962 bp of upstream sequence. The PstI fragment was cloned into in the ScaI site of pATB12, which is located immediately upstream of the lacZ reporter, to form pBK3. The cydA′::lacZ cassette was excised from pBK3 as a NotI-PstI fragment and cloned in NotI-PstI-digested p2NIL to produce pBK4. M. smegmatis was transformed with pBK4, as previously described (6), and recombinants were selected by plating on media containing KAN and X-Gal. Southern blot analysis was used to confirm that the blue colonies contained pBK4 integrated site specifically at the cyd locus.
Temperature sensitivity assays.
To determine whether the cyd mutants were temperature sensitive for growth and/or survival, overnight cultures of the wild-type and cydA::aph strains were diluted to an optical density at 600 nm (OD600) of 0.03 to 0.04 and grown at 37°C with shaking (100 rpm) to late stationary phase. Aliquots were removed during the late exponential and stationary phases of growth, and CFU were enumerated by plating at least three serial dilutions in duplicate and incubating them at 37 or 42°C.
Growth of M. smegmatis under oxystatic conditions.
Growth of M. smegmatis under defined oxygen tensions (pO2) was carried out in a Braun BIOSTAT B fermentor in batch cultures. The water-jacketed, 3-liter fermentor vessel had a 2-liter working volume and was equipped with baffles, two six-bladed impellers, a temperature probe, a ring sparger, a sampling port, and a Mettler Toledo dissolved-O2 probe. The probes and water jacket were linked to a fermentor control unit that monitored and adjusted all of the variables in the system, except for the airflow rate. For air supply, compressed air was passed into the ring sparger through a rotometer and a 0.22-μm (pore-size) sterile filter. The rotometer allowed for control of the air supply from 0 to 400 ml/min, and this parameter was set manually, depending on the air saturation level required. The temperature was maintained by connecting the vessel water jacket to a circulatory water-cooling unit. Stirring was accomplished by using an overhead motor to turn the impeller shaft. The fermentor control unit was also equipped with a dissolved-O2 cascade control, which linked the stirring rate to the dissolved-O2 level. Agitation speeds ranged from 100 to 600 rpm. All experiments conducted in this system were carried out at the research facility of the Process Biotechnology Division of the Council for Scientific and Industrial Research in Modderfontein, Johannesburg, South Africa, which is located 1,800 m above sea level, at an atmospheric pressure of 84.2 kPa.
The fermentor vessel was first sterilized with 1.5 liter of Middlebrook 7H9 plus 3 ml of glycerol. As the vessel cooled down, Tween 80, NaCl, and glucose were added, and the final volume was made up to 1.6 liter. The vessel was allowed to equilibrate overnight. Thereafter, the dissolved-O2 probe was calibrated to 0% air saturation by sparging the medium with 99.99% nitrogen and to 100% air saturation by sparging with 100% compressed air (21% O2; 176.8 Pa). Precultures were prepared by inoculating 100 ml of MADC plus Tw medium with 4 ml of a freezer stock of the culture and growing the mixture for 16 h on a rotary shaker (100 rpm), at which time the OD600 of the culture had reached 2 (ca. 108 CFU/ml). For competitive growth experiments with the mc2155::pHINT (wild type) and cydA::aph (mutant) strains, the precultures were mixed and added to the 1.6 liter of medium contained in the vessel to yield a starting culture containing a total of ca. 107 CFU/ml. An 800-μl aliquot of sterile antifoam (Durapol; AECI Bioproducts) was added to the culture, the stirrer was set to 100 rpm, air inflow was shut off, and the bacteria were allowed to consume the oxygen present in the medium. Once the air saturation had dropped to the desired level, the rotometer was set at a flow rate of 60 ml/min (for 21% air saturation), 40 ml/min (for 5% air saturation), or 20 ml/min (for 1 and 0.5% air saturation), and the stirrer was set to cascade control. Under these conditions, the pO2 could be maintained within ±10% of the desired level over the duration of the experiment (24 h). Samples were withdrawn at 3-h intervals for the first 12 h of the experiment, and a final sample was taken 12 h later. Growth was monitored by plating on LA containing either KAN (to select for cydA::aph) or HYG (to select for mc2155::pHINT).
Oxystatic growth of the mc2155::pBK4 reporter strain was carried out by inoculating 0.6 liter of culture medium (MADC plus Tw containing KAN [10 μg/ml]) with 200 ml of a preculture to give a starting cell density of ca. 7 × 107 CFU/ml. After the addition of 400 μl of antifoam, the culture was grown for 6 h, with air saturation being controlled as described above. Duplicate samples (1 to 2 ml) were withdrawn at 2-h intervals, and the experiment was carried out in triplicate at each level of air saturation (0.5, 1, 5, and 21%).
Inhibition by cyanide under oxystatic conditions.
A New Brunswick Scientific Bioflow 3000 fermentor system (University of Pennsylvania) was used for the cyanide inhibition studies. The mechanical organization of this system, the pO2 probe calibration, vessel preparation, and other relevant methods were similar to those of the Braun fermentor system decribed above. The strains used in this study were mc2155 and cydA::aph, which were grown in MADC medium, but without Tween and antifoam additives. Instead, clumps were disrupted by vortexing samples with 1/8-in. glass beads (3 ml of beads in 4 ml of culture, vortexed for 90 s in a 22-by-70-mm vial). For the competitive growth experiments, the final working volume was 3 liters, to which was added an inoculum of 5 ml of a frozen preculture of each strain (ca. 106 CFU of each/ml) to yield a starting coculture containing ca. 102 CFU of each strain/ml. KCN (1 mM) was added 30 min after inoculation, and CFU were enumerated by plating in duplicate on 7H9 agar in the presence (to score mutant CFU) or absence (to score total CFU) of KAN.
β-Galactosidase assays.
Assays were performed on cell extracts, as previously described (18), with the following modifications. Cells were lysed in Z-buffer in a Savant BIO101 Fastprep cell disruptor according to the manufacturer's instructions. Assays were carried out in Z-buffer (without β-mercaptoethanol), with 50 to 100 μl of soluble cell extract, in a final volume of 1 ml by using 2-nitrophenyl-β-d-galactopyranoside (ONPG) at 0.8 mg/ml as the substrate. Protein concentrations in the cell extracts were determined by using a Bradford-based Bio-Rad kit and following the manufacturer's instructions. For a given experiment, the specific activities determined for the four duplicate samples taken over the 6-h time course were averaged to obtain the mean for a given air saturation value. Overall means and standard deviations were obtained from the data generated from three independent growth experiments.
Spectral analysis of membrane fractions.
Unless otherwise indicated, cultures of M. smegmatis were grown aerobically in MADC plus Tw for 3 days at 37°C, with shaking at 100 rpm. Cultures were then transferred to 1-liter Schott bottles, the headspaces were purged with argon, and the bottles were sealed and maintained at 37°C for a further 5 to 6 days. During this period, the bacteria settled slowly to the bottom of the vessel. The settled bacteria were then harvested by centrifugation. Cells (8 g [wet weight]) were washed with 5 ml of Tris-cholate (TC) buffer (10 mM Tris-HCl [pH 7.4], 16 mM cholate) and resuspended in the same buffer. The bacteria were lysed by passage through a French press at 14,000 kPa, and the lysate was centrifuged at 12,000 × g for 15 min to remove cellular debris. The supernatant was further clarified by centrifugation at 50,000 × g for 50 min. The ultracentrifuged supernatant was used as the membrane-containing fraction for spectral studies. Further centrifugation at 150,000 × g for 90 min to isolate membrane particles did not improve the quality of the spectra. Protein concentrations were determined by bicinchoninic acid assay (Pierce) by using bovine serum albumin as the standard. Reduced minus oxidized spectra were recorded at room temperature on a Cary 4E dual-beam spectrophotometer, by using a few grains of fresh solid sodium dithionite as the reductant and 100 μM potassium ferricyanide as the oxidant.
Quantitation of cytochrome bd oxidase.
The amount of cytochrome bd oxidase was determined as described by Jünemann and Wrigglesworth (26) by using an extinction coefficient of 27 mM−1cm−1 for the 629- to 650-nm wavelength pair.
RESULTS
Identification and characterization of the M. smegmatis cyd gene cluster.
The high degree of homology between the cydAB genes of M. tuberculosis and their counterparts from M. smegmatis allowed the entire M. smegmatis cyd gene cluster to be cloned by using a cydAB probe derived from M. tuberculosis (Fig. 1A). The cydABDC organization in M. smegmatis parallels that observed in M. tuberculosis (7), B. subtilis (63), and Streptomyces coelicolor. The spacing of 13 bp between cydA and cydB suggests that these genes, like their counterparts from E. coli and B. subtilis, are polycistronic. However, the intergenic spacing of 90 bp between cydB and cydD suggests that the cydDC genes may be expressed independently from cydAB. As expected, the M. smegmatis cyd genes encode polypeptides showing the greatest homology to their counterparts from M. tuberculosis (78, 67, 65, and 64% identity for CydA, CydB, CydC, and CydD, respectively). In the γ-proteobacterial CydA prototype from E. coli, His19, His186, and Met393 provide three of the four axial ligands to the irons of the hemes in the cytochrome bd complex (14, 29). These three residues are conserved in the CydA polypeptides from B. subtilis, M. smegmatis, and M. tuberculosis. The counterparts of the Glu99, Glu107 residues of E. coli CydA, which have been proposed to form part of a proton channel leading from the cytoplasmic side of the membrane to the oxygen reactive site (38) are also conserved in M. smegmatis and M. tuberculosis CydA. Furthermore, the GRQPW motif, which is implicated in the binding of the quinol or heme-d in E. coli CydA (38), is also conserved in the mycobacterial CydA homologues, but the C-terminal half of the Q-loop of E. coli CydA is deleted. The mycobacterial cytochrome bd oxidases therefore belong to the more widely distributed and evolutionarily older class of cytochrome bd oxidases found in gram-positive, cyanobacterial and archaeal organisms (46).
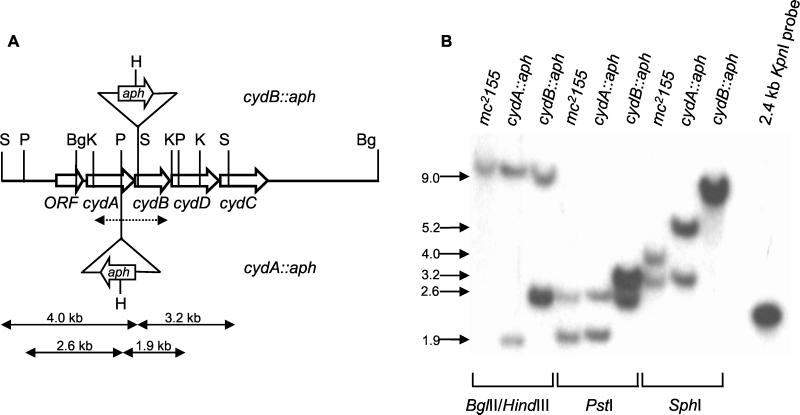
FIG. 1.
Construction and genotypic characterization of M. smegmatis cyd mutants. (A) Restriction map of the cyd locus of M. smegmatis showing the insertion sites of the aph cassette in the cyd knockout mutants. The 2.4-kb KpnI fragment from pBK1, used as the probe for Southern blot analysis (see panel B), is denoted by a dashed, double-headed arrow. Restriction sites: Bg, BglII; H, HindIII; K, KpnI; P, PstI; S, SphI. The sequence extending from the PstI site upstream of cydA to a SalI site downstream of cydC has been deposited in GenBank under accession no. AF196488. A hypothetical open reading frame that showed high homology to Rv1624c from M. tuberculosis was found upstream of cydA, confirming that the genetic organization at this locus is conserved between M. smegmatis and M. tuberculosis. (B) Southern blot analysis of cyd mutants. Genomic DNA from the various strains was digested with the indicated enzyme(s) and probed with the 2.4-kb KpnI fragment from pBK1. In all cases, the sizes of the hybridizing fragments derived from the mutant strains were consistent with site-specific, allelic exchange.
Construction and characterization of cyd knockout mutants of M. smegmatis
To investigate the in vivo function of the cydAB-encoded cytochrome bd oxidase in M. smegmatis, we constructed cydA::aph and cydB::aph mutants by homologous recombination. The mutant alleles were delivered on suicide vectors carrying lacZ (39) and sacB markers (42). Southern blot analysis confirmed that the white, sucrose-resistant clones recovered from the mutagenesis procedure with pCYDAKO and pCYDBKO were site-specific, allelic exchange mutants (Fig. 1B). Inactivation of cydA and/or cydB genes had no discernible effect on the rate of growth of M. smegmatis during the exponential phase, when the organism was grown in shaking flasks (data not shown). To investigate whether cyd gene inactivation conferred a stationary-phase exit defect analogous to that observed in E. coli cyd mutants (19, 51), the M. smegmatis cydA::aph mutant was tested for temperature sensitivity for growth and survival. The mutant displayed no defect during the exponential phase of growth when cultured at 42°C. Moreover, comparison of the plating efficiencies at 37 and 42°C of cultures sampled 6 to 14 h after the cessation of growth revealed that the wild-type and mutant strains were indistinguishable (data not shown).
Spectral analysis of cytochromes in membranes of wild-type and cyd mutant strains of M. smegmatis
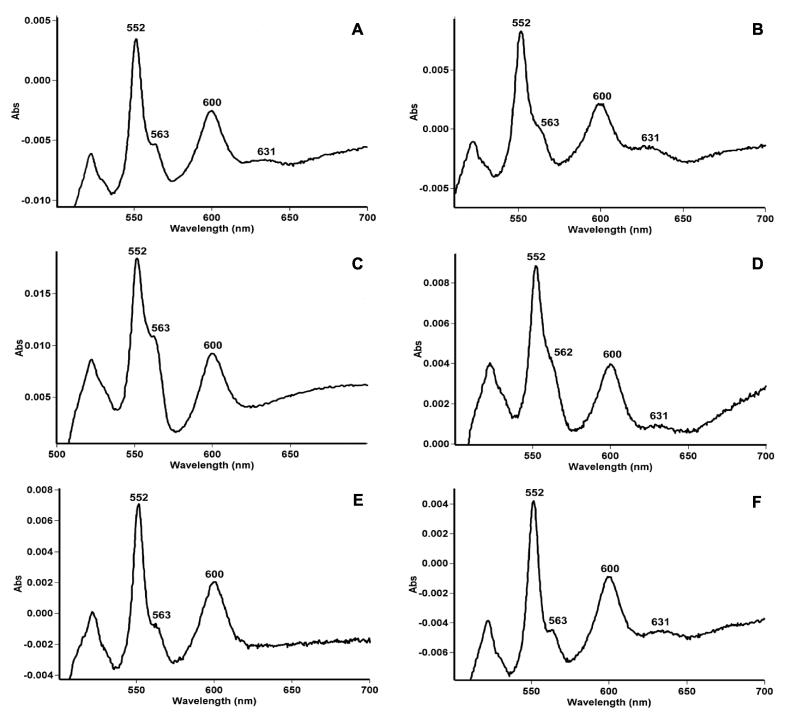
Reduced minus oxidized difference spectroscopy at room temperature was used to analyze the cytochrome content of M. smegmatis (Fig. 2A). Major absorbance peaks observed at 600, 563, and 552 nm were characteristic of a-, b-, and c-type cytochromes, respectively (25). A discrete peak at 631 nm denoted the presence of heme-d, which is characteristic of the cytochrome bd oxidase in E. coli (36) and B. subtilis (63). The spectra of the cydA::aph and cydB::aph mutants lacked the features of heme-d but retained all other absorbance peaks (Fig. 2C and E), a finding consistent with a specific disruption of cytochrome bd. The persistence of the major peaks at 558 and 595 nm in the mutant is indicative of other respiratory heme molecules that can absorb in this area, e.g., the prominent peak at 600 nm could be due to the presence of a cytochrome aa3-type respiratory oxidase in this organism, which is substantiated by the preliminary genome sequence, as described below. The various strains were transformed with a plasmid carrying the entire cydABDC gene cluster from M. tuberculosis. The presence of pOTBCYD in the cydA::aph and cydB::aph strains restored the heme-d631 absorbance peak (Fig. 2D and F), confirming that the M. tuberculosis cydABDC genes were able to functionally complement the cytochrome bd defect of the M. smegmatis mutants.
FIG. 2.
Sodium dithionite-reduced minus potassium ferricyanide-oxidized difference spectra on wild-type and cyd mutant membranes. (A) mc2155, (B) mc2155(pOTBCYD), (C) cydA::aph, (D) cydA::aph(pOTBCYD), (E) cydB::aph, or (F) cydB::aph(pOTBCYD). In all cases, samples were suspended in 1 ml of TC buffer (pH 7.4) at a protein concentration of 2.6 mg/ml.
Comparative growth rates under oxystatic conditions.
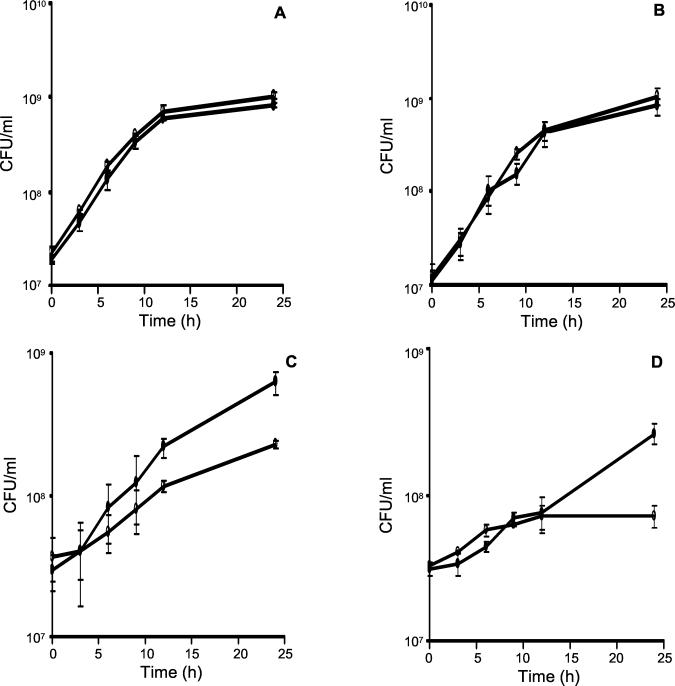
To test whether the bd oxidase of M. smegmatis serves a respiratory function under microaerophilic conditions, we monitored the growth dependence of the wild-type and cydA::aph mutant strains on the O2 tension (pO2) of the growth medium over a wide pO2 range by using an oxystat to maintain the pO2 at the desired level. The growth of cocultures inoculated with approximately equal numbers of wild-type (mc2155::pHINT) and mutant (cydA::aph) organisms was monitored at each pO2 level by scoring the time-dependent increase of CFU from each strain by plating on medium containing HYG or KAN. Inactivation of cydA had no discernible effect on the rate of growth of M. smegmatis at 21 or 5% air saturation (Fig. 3A and B). However, the, growth rate of the wild-type strain was considerably lower at 0.5 and 1% air saturation compared to that observed at higher aeration, indicating that limited availability of O2 restricts the growth of M. smegmatis, in accordance with the observations of Dick et al. (11). Importantly, a growth phenotype was revealed under these conditions. At 1% air saturation, the cyd mutant was significantly impaired for growth and entered stationary phase at a lower cell density than the wild type (Fig. 3C). The growth defect was exacerbated at 0.5% air saturation, with little growth of the mutant being observed over the time course of the experiment (Fig. 3D).
FIG. 3.
Growth of cocultures of wild-type and cyd mutant strains at various air saturation levels. The growth curves of mixed cultures of mc2155::pHINT (●) and cydA::aph (○) are shown for 21% (A), 5% (B), 1% (C), or 0.5% (D) air saturation for 24 h (see Materials and Methods for more details). The data are shown as the averages and standard deviations from three independent experiments. Statistical analysis of the data by using an analysis of variance test revealed that the cell counts of the mutant after 24 h of growth were significantly lower than for wild type at the lower aerations (0.5 and 1% air saturation; P < 0.05).
Dependence of cyd gene expression on pO2.
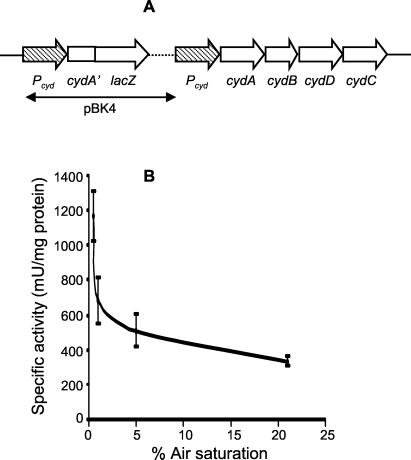
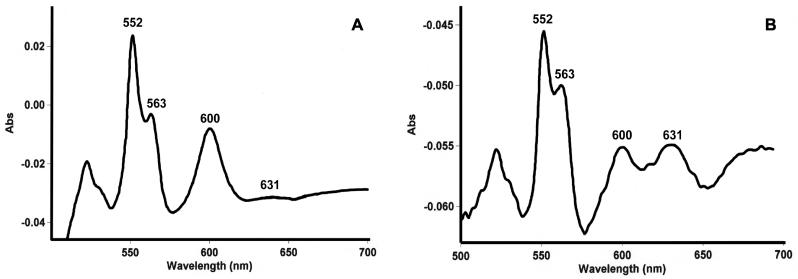
To investigate the dependence of cyd expression on pO2, a partial merodiploid reporter strain carrying a cydA′::lacZ transcriptional fusion integrated at the cyd locus of M. smegmatis was constructed by homologous recombination with the suicide plasmid pBK4 to create mc2155::pBK4 (Fig. 4A). In this strain, lacZ expression is under the control of the cyd regulatory region, and the cyd gene cluster is flanked by 1,675 bp of upstream sequence. Since this region had been shown to contain a functional promoter (data not shown), we concluded that the cyd gene cluster would be functionally expressed in the reporter strain. The dependence of lacZ expression during exponential-phase growth on the pO2 of the growth medium was monitored over a 0.5 to 21% air saturation range. The expression profile of the cydAB genes (Fig. 4B) revealed that cyd expression increased two- to threefold at between 5 and 0.5% air saturation. This increase in expression was corroborated by the relative spectral increase in absorbance of the d-heme peak (631 nm) in the membrane fraction of wild-type M. smegmatis grown in an oxystat at 1% air saturation (Fig. 5B) relative to a 21% control (Fig. 5A). The cytochrome bd oxidase content in membrane fractions isolated from bacteria grown at 1% air saturation was 1.26 pmol of bd oxidase/mg of oxidase versus a content of 0.295 pmol of bd oxidase/mg of oxidase in membrane fractions from bacteria grown at 21% air saturation.
FIG. 4.
Expression analysis of the M. smegmatis cyd gene cluster. (A) Genetic organization at the cyd locus of the reporter strain, mc2155::pBK4. The hatched arrow denotes the putative cyd promoter, Pcyd. The genotype of this site-specific single recombinant was confirmed by Southern blotting (not shown). (B) Dependence of β-galactosidase reporter activity on pO2. The specific activity of a reporter gene product (β-galactosidase) was studied at 21, 5, 1, and 0.5% air saturation as described in Materials and Methods. The data shown are an average of three independent experiments for each air saturation level. A statistical pairwise comparison showed that the data followed a significant second-order relationship, as represented in the figure (P < 0.05).
FIG. 5.
Spectral characterization of membrane fractions isolated from M. smegmatis grown in an oxystat. The spectral profiles of purified membrane fractions from M. smegmatis grown at 21% (A) and 1% (B) air saturation are shown. Reduction was accomplished by the addition of sodium dithionite, and oxidation was accomplished by the addition of potassium ferricyanide.
Sensitivity of wild-type and cyd mutant strains to cyanide.
The preliminary M. smegmatis genome sequence data from The Institute for Genomic Research that became available during the course of this study suggested the presence of a putative aa3-type cytochrome c oxidase in this organism. On the basis of these observations, we compared the sensitivity of the wild-type and cydA mutant strains to the terminal oxidase inhibitor, cyanide, which is expected to preferentially inhibit the cytochrome c oxidase branch of the respiratory chain, since some of the bd oxidases have been shown to be relatively insensitive to cyanide (9, 31). In preliminary experiments carried out in shaking culture tubes, the mutant was found to display an enhanced sensitivity to cyanide, as assessed by CFU enumeration of cultures containing KCN at concentrations of 10 to 300 μM (data not shown). The cyanide hypersensitivity of the mutant was then confirmed in a mixed culture growth experiment in the presence of 1 mM KCN, carried out at 21% air saturation in an oxystat, in which the inoculum contained approximately equal CFU of wild type and mutant. Under these conditions, the wild type showed a competitive growth advantage over the mutant, as evidenced by the 10-fold difference in CFU after 140 h of growth (Fig. 6).
FIG. 6.
Cyanide inhibition of wild-type and cydA::aph strains under oxystatic conditions. The growth of mixed cultures of mc2155 (●) and cydA::aph (○) in the presence of 1 mM KCN are shown. KCN was added 30 min postinoculation, and the growth of each strain was monitored by differential plating as described in Materials and Methods. The data are shown as the averages and standard errors from two independent experiments.
DISCUSSION
In this study, we used targeted gene knockout to investigate the function of the cydAB-encoded cytochrome bd oxidase of M. smegmatis. While the presence of cytochromes in mycobacteria has been known for more than 50 years (56), detailed work failed to recognize heme-d-containing cytochromes in these organisms (1, 32). However, the spectroscopic evidence provided herein confirms the presence of a heme-d-containing cytochrome in M. smegmatis, which was lost by insertional inactivation of the cydA or cydB genes. In this and other respects, the cydAB-encoded cytochrome bd oxidase of M. smegmatis displayed spectral features analogous to the well-characterized prototypical γ-proteobacterial-type enzyme of E. coli (31).
DNA sequence analysis suggested that the M. smegmatis cydA and cydB genes are arranged on an operon that may extend to also include the cydD and cydC genes. The phenotypic consequences of cydA inactivation may therefore be due to loss of the cytochrome bd oxidase itself and/or loss of the putative ATP-binding cassette-type transporter as a result of polar effects of the cydA::aph mutation on the downstream genes. Unlike the stationary-phase viability defect observed in cyd mutants of E. coli (50) and A. vinelandi (13), the M. smegmatis cydA mutant displayed no loss of viability in the stationary phase.
In other systems, the levels of cytochrome bd oxidase have been shown to vary in response to the pO2 of the growth medium. The regulatory mechanisms governing cyd gene expression have been elucidated in E. coli. In this organism, arcAB-dependent upregulation of the cydAB genes results in maximal expression at 7% air saturation (57), but expression is attenuated at lower pO2 levels by fnr-mediated repression (8). Studies in B. subtilis have revealed that expression of its cydABDC gene cluster also increased under reduced pO2 (63) but, unlike E. coli, induction of cyd persists as the pO2 is further reduced, as evidenced by the high level of induction observed under anaerobiosis (65). Our data suggest that the M. smegmatis cyd gene cluster is also transcriptionally regulated in response to the pO2 of the growth medium; however, induction of expression occurs under conditions different from those previously observed for other organisms. The threshold pO2 for cyd induction in M. smegmatis (ca. 1% air saturation) is significantly lower than that of E. coli (10% air saturation), suggesting that, as in B. subtilis (65), the cytochrome bd oxidase is induced as the transition to anaerobiosis is approached.
A phenotype for the M. smegmatis cydA mutant was revealed in competitive growth experiments with the parental wild type under oxystatic conditions. The competitive growth defect of the cydA mutant was only apparent at or below 1% air saturation, which corresponds to the threshold pO2 level for induction of the bd oxidase. An analogous concordance between expression and in vivo function has been observed in E. coli, where the threshold for attenuation of growth of E. coli cyd mutants matches that for induction (10% air saturation [5]). In conjunction, these results suggest that the cytochrome bd oxidase of M. smegmatis has a different functional or kinetic range with respect to oxygen availability compared to that of E. coli. The mycobacterial bd oxidase is required specifically for growth under extreme microaerophilia, whereas the E. coli enzyme bridges the gap between aerobic respiration and fermentation over the pO2 range in which both pathways are poorly expressed (5).
The M. smegmatis cydA mutant was also significantly growth impaired relative to its parental wild type when the two strains were cocultured in the presence of 1 mM cyanide at 21% air saturation. This observation suggests that, in addition to the relatively cyanide-insensitive bd oxidase (9, 31), M. smegmatis contains a cyanide-sensitive terminal(s) that plays a role in aerobic respiration. The fact that the mutant displayed a competitive growth disadvantage compared to the wild type under moderate aeration (21% air saturation) in the presence of cyanide may be indicative of a respiratory role for the bd oxidase even under conditions of high O2, particularly since cyd gene expression was detectable at this pO2. However, we cannot exclude the possibility that bd oxidase function under these conditions is a compensatory response occurring only when other terminals are inhibited by cyanide.
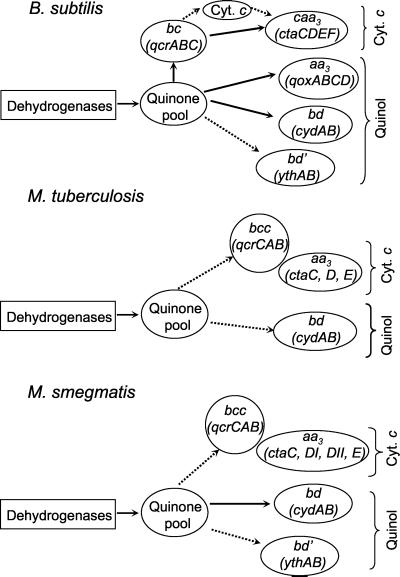
To place our results on the bd oxidase of M. smegmatis in the context of aerobic respiratory branches in mycobacteria, we analyzed the preliminary genome sequence of this organism for its repertoire of respiratory genes and compared it to those of M. tuberculosis and B. subtilis. A schematic representation of the branched electron transport chains in these organisms is shown in Fig. 7. The well-studied, aerobic respiratory chain of B. subtilis is the most complex, terminating with cytochromes bd (cydABDC), cytochrome caa3 (ctaCDEF), cytochrome aa3 (qoxABCD), or cytochrome bd′ (ythAB) (41, 64). There is also evidence for a bb′-type terminal oxidase in this organism (2), which appears to be bd-type, with the heme-d substituted by high spin heme-b. Therefore, terminal branches of the respiratory chain cannot be inferred solely from the genome sequence, since macromolecular reorganization of terminal oxidase subunits and prosthetic groups could give rise to alternate oxidases. In B. subtilis at least one quinol oxidase, cytochrome bd or cytochrome aa3, is required for aerobic growth, the ythAB genes encoding a putative quinol oxidase are involved in sporulation, and the cytochrome c oxidase branch is not expressed until the stationary phase (64).
FIG. 7.
Proposed aerobic respiratory pathways in M. smegmatis and related organisms. The pathways in B. subtilis were adapted from references 41 and 64. The M. tuberculosis pathways were deduced from its genome sequence (7), and those of M. smegmatis are based on this study and analysis of the preliminary genome sequence data obtained from The Institute for Genomic Research website (http://www.tigr.org). Solid arrows denote confirmed pathways, and dashed arrows denote putative pathways. The mycobacterial cytochrome reductase complex (QcrCAB) is denoted as “bcc” in accordance with the designation of QcrC as cytochrome cc (54).
Other than the early studies of Todd (56), which confirmed that mycobacteria contained cytochromes and the subsequent demonstration of a complex containing cytochromes aa3 and o in Mycobacterium phlei (43), the respiratory chains of mycobacteria are poorly understood. The aerobic respiratory chain of M. tuberculosis deduced from its genome sequence (7) is far simpler than that of B. subtilis, terminating in only two main branches, namely, cytochrome bd and an aa3-type cytochrome c oxidase belonging to the heme-copper respiratory oxidase superfamily (Fig. 7). Assignment of this aa3-type oxidase as a cytochrome c oxidase rather than a quinol oxidase is based on the presence in subunit II (CtaC) of a CuA binding site (16). The M. smegmatis respiratory chain is of intermediate complexity, possessing the same two branches as M. tuberculosis and a third possible branch terminating in the YthAB quinol oxidase. The other notable difference is the presence of two distinct ctaD alleles in M. smegmatis versus one in M. tuberculosis. We therefore speculate that alternate isoforms of cytochrome c oxidase may exist in M. smegmatis, as has been proposed for Paracoccus denitrificans, which also contains ctaDI and ctaDII alleles (44).
The cytochrome c branch of mycobacteria has some unusual features that are shared by other high-G+C-content gram-positive bacteria (54). The only identifiable c-type cytochrome in M. smegmatis and M. tuberculosis is the di-heme c-type cytochrome QcrC, which, like its counterpart from Corynebacterium glutamicum, is a tandem product of two class I cytochromes c (54). This cytochrome has been proposed to accept electrons from the Rieske Fe-S cluster and to donate them to a terminal oxidase (54), suggesting that the quinol cytochrome c reductase (QcrCAB) may form a complex with cytochrome c oxidase (CtaCDE) in mycobacteria and related organisms (Fig. 7). Interestingly, the qcrCAB operon in M. tuberculosis and M. smegmatis is located immediately downstream of ctaE, suggesting that these genes may be coordinately regulated.
In summary, we have shown that the cydAB-encoded quinol oxidase plays a particularly important respiratory role in M. smegmatis under conditions of extreme microaerophilia. Although the bd oxidase might also operate under moderately aerobic conditions, we propose that the major oxidase used under these conditions is the cytochrome c oxidase. Experiments aimed at testing this hypothesis and further elucidating the respiratory chains of M. smegmatis are currently under way in our laboratories. In addition, the confirmed respiratory role for the M. smegmatis bd oxidase in microaerophilic growth has obvious implications for studies of respiration in M. tuberculosis. In this respect, in vivo phenotyping of recently constructed cyd mutant strains of M. tuberculosis (S. S. Dawes and V. Mizrahi, unpublished data) should reveal whether loss of the bd oxidase affects the virulence of this organism.
ACKNOWLEDGMENTS
This work was supported by grant AI-43420 (to H.R.) from the National Institutes of Health and by grants from the Medical Research Council of South Africa, the DACST-MRC Lead Programme for Tuberculosis, the National Research Foundation of South Africa, and the South African Institute for Medical Research (to V.M.). V.M was also supported by an International Scholar's grant from the Howard Hughes Medical Institute, and B.K. was supported by an NRF Prestigious bursary.
We thank the Council for Scientific and Industrial Research for the use of the Braun fermentor and are indebted to Paul Axelsen for allowing us the use of his Cary spectrophotometer. We also thank Brian James, as well as Bhavna Gordhan and other members of the Mizrahi lab, for advice and assistance; Selwyn Quan for providing p6.11; Peadar O'Gaora for pOLYG and pSMT3; and Martin Everett for providing the gridded plasmid library and for recovering clones. We thank Piet Becker of the South African MRC Biostatistics Unit for assistance with the statistical analysis of our data. Preliminary sequence data was obtained from The Institute for Genomic Research website at http://www.tigr.org.
REFERENCES
- 1.Asano A, Brodie A F. Oxidative phosphorylation in fractionated bacterial systems. 18. Phosphorylation coupled to different segments of the respiratory chains of Mycobacterium phlei. J Biol Chem. 1965;240:4002–4010. [PubMed] [Google Scholar]
- 2.Azarkina N, Siletsky S, Borisov V, Von Wachenfeldt C, Hederstedt L, Konstantinov A. A cytochrome bb′-type quinol oxidase in Bacillus subtilis strain 168. J Biol Chem. 1999;274:32810–32817. doi: 10.1074/jbc.274.46.32810. [DOI] [PubMed] [Google Scholar]
- 3.Balasubramanian V, Pavelka M S, Jr, Bardarov S S, Martin J, Weisbrod T R, McAdam R A, Bloom B R, Jacobs W R., Jr Allelic exchange in Mycobacterium tuberculosis with long linear recombination substrates. J Bacteriol. 1996;178:273–279. doi: 10.1128/jb.178.1.273-279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes P F, el-Hajj H, Preston-Martin S, Cave M D, Jones B E, Otaya M, Pogoda J, Eisenach K D. Transmission of tuberculosis among the urban homeless. JAMA. 1996;275:305–307. [PubMed] [Google Scholar]
- 5.Becker S, Vlad D, Schuster S, Pfeiffer P, Unden G. Regulatory O2 tensions for the synthesis of fermentation products in Escherichia coli and relation to aerobic respiration. Arch Microbiol. 1997;168:290–296. doi: 10.1007/s002030050501. [DOI] [PubMed] [Google Scholar]
- 6.Boshoff H I M, Mizrahi V. Purification, gene cloning, targeted knockout, overexpression, and biochemical characterization of the major pyrazinamidase from Mycobacterium smegmatis. J Bacteriol. 1998;180:5809–5814. doi: 10.1128/jb.180.22.5809-5814.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail M A, Rajandream M A, Rogers J, Rutter S, Seeger K, Skelton J, Squares S, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 8.Cotter P A, Melville S B, Albrecht J A, Gunsalus R P. Aerobic regulation of cytochrome d oxidase (cydAB) operon expression in Escherichia coli: roles of Fnr and ArcA in repression and activation. Mol Microbiol. 1997;25:605–615. doi: 10.1046/j.1365-2958.1997.5031860.x. [DOI] [PubMed] [Google Scholar]
- 9.Cunningham L, Pitt M, Williams H D. The cioAB genes from Pseudomonas aeruginosa code for a novel cyanide-insensitive terminal oxidase related to the cytochrome bd quinol oxidases. Mol Microbiol. 1997;24:579–591. doi: 10.1046/j.1365-2958.1997.3561728.x. [DOI] [PubMed] [Google Scholar]
- 10.Cunningham A F, Spreadbury C L. Mycobacterial stationary phase induced by low oxygen tension: cell wall thickening and localization of the 16-kilodalton alpha-crystallin homolog. J Bacteriol. 1998;180:801–808. doi: 10.1128/jb.180.4.801-808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dick T, Lee B H, Murugasu-Oei B. Oxygen depletion induced dormancy in Mycobacterium smegmatis. FEMS Microbiol Lett. 1998;163:159–164. doi: 10.1111/j.1574-6968.1998.tb13040.x. [DOI] [PubMed] [Google Scholar]
- 12.Dye C, Scheele S, Dolin P, Pathania V, Raviglione M C. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. W.H.O. Global Surveillance and Monitoring Project. JAMA. 1999;282:677–686. doi: 10.1001/jama.282.7.677. [DOI] [PubMed] [Google Scholar]
- 13.Edwards S E, Loder C S, Wu G, Corker H, Bainbridge B W, Hill S, Poole R K. Mutation of cytochrome bd quinol oxidase results in reduced stationary phase survival, iron deprivation, metal toxicity and oxidative stress in Azotobacter vinelandii. FEMS Microbiol Lett. 2000;185:71–77. doi: 10.1111/j.1574-6968.2000.tb09042.x. [DOI] [PubMed] [Google Scholar]
- 14.Fang H, Lin R J, Gennis R B. Location of heme axial ligands in the cytochrome d terminal oxidase complex of Escherichia coli determined by site-directed mutagenesis. J Biol Chem. 1989;264:8026–8032. [PubMed] [Google Scholar]
- 15.Gangadharam P R J. Mycobacterial dormancy. Tuber Lung Dis. 1995;76:477–479. doi: 10.1016/0962-8479(95)90521-9. [DOI] [PubMed] [Google Scholar]
- 16.García-Horsman J A, Barquera B, Rumbley J, Ma J, Gennis R B. The superfamily of heme-copper respiratory oxidases. J Bacteriol. 1994;176:5587–5600. doi: 10.1128/jb.176.18.5587-5600.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gennis R B, Stewart V. Respiration. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 217–261. [Google Scholar]
- 18.Ghanekar K, McBride A, Dellagostin O, Thorne S, Mooney R, McFadden J. Stimulation of transposition of the Mycobacterium tuberculosis insertion sequence IS6110 by exposure to microaerobic environment. Mol Microbiol. 1999;33:982–993. doi: 10.1046/j.1365-2958.1999.01539.x. [DOI] [PubMed] [Google Scholar]
- 19.Goldman B S, Gabbert K K, Kranz R G. The temperature-sensitive growth and survival phenotypes of Escherichia coli cydDC and cydAB strains are due to deficiencies in cytochrome bd and are corrected by exogenous catalase and reducing agents. J Bacteriol. 1996;178:6348–6351. doi: 10.1128/jb.178.21.6348-6351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gordhan B G, Andersen S J, De Meyer A R, Mizrahi V. Construction by homologous recombination and phenotypic characterization of a polA mutant of Mycobacterium smegmatis. Gene. 1996;178:125–130. doi: 10.1016/0378-1119(96)00350-2. [DOI] [PubMed] [Google Scholar]
- 21.Grange J M. The mystery of the mycobacterial persistor. Tuberc Lung Dis. 1992;73:249–251. doi: 10.1016/0962-8479(92)90128-7. [DOI] [PubMed] [Google Scholar]
- 22.Huberts P, Mizrahi V. Cloning and sequence analysis of the gene encoding the DNA polymerase I from Mycobacterium tuberculosis. Gene. 1995;164:133–136. doi: 10.1016/0378-1119(95)00453-d. [DOI] [PubMed] [Google Scholar]
- 23.Husson R N, James B E, Young R A. Gene replacement and expression of foreign DNA in mycobacteria. J Bacteriol. 1990;172:519–524. doi: 10.1128/jb.172.2.519-524.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imboden P, Schoolnik G K. Construction and characterization of a partial Mycobacterium tuberculosis cDNA library of genes expressed at reduced oxygen tension. Gene. 1998;213:107–117. doi: 10.1016/s0378-1119(98)00192-9. [DOI] [PubMed] [Google Scholar]
- 25.Jones C W, Poole R K. The analysis of cytochromes. In: Gottschalk G, editor. Methods in microbiology. London, England: Academy Press; 1985. pp. 285–328. [Google Scholar]
- 26.Jünemann S, Wrigglesworth J M. Cytochrome bd oxidase from Azotobacter vinelandii. J Biol Chem. 1995;270:16213–16220. doi: 10.1074/jbc.270.27.16213. [DOI] [PubMed] [Google Scholar]
- 27.Jünemann S. Cytochrome bd terminal oxidase. Biochim Biophys Acta. 1997;1321:107–127. doi: 10.1016/s0005-2728(97)00046-7. [DOI] [PubMed] [Google Scholar]
- 28.Juty N S, Moshiri F, Merrick M, Anthony C, Hill S. The Klebsiella pneumoniae cytochrome bd terminal oxidase complex and its role in microaerobic nitrogen fixation. Microbiology. 1997;143:2673–2683. doi: 10.1099/00221287-143-8-2673. [DOI] [PubMed] [Google Scholar]
- 29.Kaysser T M, Ghaim J B, Georgiou C, Gennis R B. Methionine-393 is an axial ligand of the heme b558 component of the cytochrome bd ubiquinol oxidase from Escherichia coli. Biochemistry. 1995;34:13491–13501. doi: 10.1021/bi00041a029. [DOI] [PubMed] [Google Scholar]
- 30.Kelly M J S, Poole R K, Yates M G, Kennedy C. Cloning and mutagenesis of genes encoding the cytochrome bd terminal oxidase complex in Azotobacter vinelandii: mutants deficient in the cytochrome d complex are unable to fix nitrogen in air. J Bacteriol. 1990;172:6010–6019. doi: 10.1128/jb.172.10.6010-6019.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kita K, Konishi K, Anraku Y. Purification and properties of two terminal oxidase complexes of Escherichia coli aerobic respiratory chain. Methods Enzymol. 1986;126:94–113. doi: 10.1016/s0076-6879(86)26012-7. [DOI] [PubMed] [Google Scholar]
- 32.Kusaka T, Sato R, Shoji K. Comparison of cytochromes in mycobacteria grown in vitro and in vivo. J Bacteriol. 1964;87:1383–1388. doi: 10.1128/jb.87.6.1383-1388.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kusumoto K, Sakiyama M, Sakamoto J, Noguchi S, Sone N. Menaquinol oxidase activity and primary structure of cytochrome bd from the amino-acid fermenting bacterium Corynebacterium glutamicum. Arch Microbiol. 2000;173:390–397. doi: 10.1007/s002030000161. [DOI] [PubMed] [Google Scholar]
- 34.Le H Q, Davidson P T. Reactivation and exogenous infection: their relative roles in the pathogenesis of tuberculosis. Curr Clin Top Infect Dis. 1996;16:260–276. [PubMed] [Google Scholar]
- 35.Lee B H, Murugasu-Oei B, Dick T. Upregulation of a histone-like protein in dormant Mycobacterium smegmatis. Mol Gen Genet. 1998;260:475–479. doi: 10.1007/s004380050919. [DOI] [PubMed] [Google Scholar]
- 36.Lorence R M, Koland J G, Gennis R B. Coulometric and spectroscopic analysis of the purified cytochrome d complex of Escherichia coli: evidence for the identification of “cytochrome a1” as cytochrome b595. Biochemistry. 1986;25:2314–2321. doi: 10.1021/bi00357a003. [DOI] [PubMed] [Google Scholar]
- 37.O'Gaora P, Barnini S, Hayward C, Filley E, Rook G, Young D, Thole J. Mycobacteria as immunogens: development of expression vectors for use in multiple mycobacterial species. Med Princ Pract. 1997;6:91–96. [Google Scholar]
- 38.Osborne J P, Gennis R B. Sequence analysis of cytochrome bd oxidase suggests a revised topology for subunit I. Biochim Biophys Acta. 1999;1410:32–50. doi: 10.1016/s0005-2728(98)00171-6. [DOI] [PubMed] [Google Scholar]
- 39.Parish T A, Gordhan B G, McAdam R A, Duncan K, Mizrahi V, Stoker N G. Production of mutants in amino acid biosynthesis genes of Mycobacterium tuberculosis by homologous recombination. Microbiology. 1999;145:3497–3503. doi: 10.1099/00221287-145-12-3497. [DOI] [PubMed] [Google Scholar]
- 40.Parish T, Stoker N G. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology. 2000;146:1969–1975. doi: 10.1099/00221287-146-8-1969. [DOI] [PubMed] [Google Scholar]
- 41.Poole R K, Cook G. Redundancy of aerobic respiratory chains in Bacteria? Routes, reasons, and regulation. Adv Microb Physiol. 2000;43:165–224. doi: 10.1016/s0065-2911(00)43005-5. [DOI] [PubMed] [Google Scholar]
- 42.Primm T P, Andersen S J, Mizrahi V, Avarbock D, Rubin H, Barry C E., III The stringent response of Mycobacterium tuberculosis is required for long-term survival. J Bacteriol. 2000;182:4889–4898. doi: 10.1128/jb.182.17.4889-4898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Revsin B, Marquez E D, Brodie A F. Cytochromes from Mycobacterium phlei: isolation and spectral properties of a mixture of cytochromes (a + a3) (o) Arch Biochem Biophys. 1970;139:114–120. doi: 10.1016/0003-9861(70)90052-4. [DOI] [PubMed] [Google Scholar]
- 44.Raitio M, Pispa J M, Metso T, Saraste M. Are there isoenzymes of cytochrome c oxidase in Paracoccus denitrificans? FEBS Lett. 1990;261:431–435. doi: 10.1016/0014-5793(90)80609-m. [DOI] [PubMed] [Google Scholar]
- 45.Sakamoto J, Matsumoto A, Oobuchi K, Sone N. Cytochrome bd-type quinol oxidase in a mutant of Bacillus stearothermophilus deficient in caa3-type cytochrome c oxidase. FEMS Microbiol Lett. 1996;143:151–158. doi: 10.1111/j.1574-6968.1996.tb08474.x. [DOI] [PubMed] [Google Scholar]
- 46.Sakamoto J, Koga E, Mizuta T, Sato C, Noguchi S, Sone N. Gene structure and quinol oxidase activity of a cytochrome bd-type oxidase from Bacillus stearothermophilus. Biochim Biophys Acta. 1999;1411:147–158. doi: 10.1016/s0005-2728(99)00012-2. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 48.Segal W. Growth dynamics of in vivo and in vitro grown mycobacterial pathogens. In: Kubica G P, Wayne L G, editors. The Mycobacteria—a sourcebook. New York, N.Y: Marcel Dekker; 1984. pp. 547–573. [Google Scholar]
- 49.Sherman D R, Voskuil M, Schnappinger D, Liao R, Harrell M I, Schoolnik G K. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding α-crystallin. Proc Natl Acad Sci USA. 2001;98:7534–7539. doi: 10.1073/pnas.121172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siegele D A, Kolter R. Isolation and characterization of an Escherichia coli mutant defective in resuming growth after starvation. Genes Dev. 1993;7:2629–2640. doi: 10.1101/gad.7.12b.2629. [DOI] [PubMed] [Google Scholar]
- 51.Siegele D A, Imlay K R C, Imlay J A. The stationary-phase-exit defect of cydC (surB) mutants is due to the lack of a functional terminal cytochrome oxidase. J Bacteriol. 1996;178:6091–6096. doi: 10.1128/jb.178.21.6091-6096.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smeulders M J, Keer J, Speight R A, Williams H D. Adaptation of Mycobacterium smegmatis to stationary phase. J Bacteriol. 1999;181:270–283. doi: 10.1128/jb.181.1.270-283.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Snapper S B, Melton R E, Mustafa S, Keiser T, Jacobs W R., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 54.Sone N, Nagata K, Kojima H, Tajima J, Kodera Y, Kanamaru T, Noguchi S, Sakamoto J. A novel hydrophobic diheme c-type cytochrome: purification from Corynebacterium glutamicum and analysis of the qcrCBA operon encoding three subunit proteins of a putative cytochrome reductase complex. Biochim Biophys Acta. 2001;1503:279–290. doi: 10.1016/s0005-2728(00)00205-x. [DOI] [PubMed] [Google Scholar]
- 55.Starke J R. Tuberculosis in children. Curr Opin Pediatr. 1995;7:268–277. doi: 10.1097/00008480-199506000-00005. [DOI] [PubMed] [Google Scholar]
- 56.Todd C M. Occurrence of cytochrome and coproporphyrin in mycobacteria. Biochem J. 1949;45:386–390. doi: 10.1042/bj0450386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tseng C P, Albrecht J, Gunsalus R P. Effect of microaerophilic cell growth conditions on expression of the aerobic (cyoABCDE and cydAB) and anaerobic (narGHJI, frdABCD, and dmsABC) respiratory pathway genes in Escherichia coli. J Bacteriol. 1996;178:1094–1098. doi: 10.1128/jb.178.4.1094-1098.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Way S S, Sallustio S, Magliozzo R S, Goldberg M B. Impact of either elevated or decreased levels of cytochrome bd expression on Shigella flexneri virulence. J Bacteriol. 1999;181:1229–1237. doi: 10.1128/jb.181.4.1229-1237.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wayne L G, Diaz G A. Autolysis and secondary growth of Mycobacterium tuberculosis in submerged culture. J Bacteriol. 1967;93:1374–1381. doi: 10.1128/jb.93.4.1374-1381.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wayne L G, Lin K Y. Glyoxylate metabolism and adaptation of Mycobacterium tuberculosis to survival under anaerobic conditions. Infect Immun. 1982;37:1042–1049. doi: 10.1128/iai.37.3.1042-1049.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wayne L G. Dormancy of Mycobacterium tuberculosis and latency of disease. Eur J Clin Microbiol Infect Dis. 1994;13:908–914. doi: 10.1007/BF02111491. [DOI] [PubMed] [Google Scholar]
- 62.Wayne L G, Hayes L G. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64:2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winstedt L, Yoshida K, Fujita Y, von Wachenfeldt C. Cytochrome bd biosynthesis in Bacillus subtilis: characterization of the cydABCD operon. J Bacteriol. 1998;180:6571–6580. doi: 10.1128/jb.180.24.6571-6580.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Winstedt L, von Wachenfeldt C. Terminal oxidases of Bacillus subtilis strain 168: one quinol oxidase, cytochrome aa3 or cytochrome bd, is required for aerobic growth. J Bacteriol. 2000;182:6557–6564. doi: 10.1128/jb.182.23.6557-6564.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ye R W, Tao W, Bedzyk L, Young T, Chen M, Li L. Global gene expression profiles of Bacillus subtilis grown under anaerobic conditions. J Bacteriol. 2000;182:4458–4465. doi: 10.1128/jb.182.16.4458-4465.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Young D B, Duncan K. Prospects for new interventions in the treatment and prevention of mycobacterial disease. Annu Rev Microbiol. 1995;49:641–673. doi: 10.1146/annurev.mi.49.100195.003233. [DOI] [PubMed] [Google Scholar]