Abstract
The biosynthesis of [NiFe] hydrogenases is a complex process that requires the function of the Hyp proteins HypA, HypB, HypC, HypD, HypE, HypF, and HypX for assembly of the H2-activating [NiFe] site. In this study we examined the maturation of the regulatory hydrogenase (RH) of Ralstonia eutropha. The RH is a H2-sensing [NiFe] hydrogenase and is required as a constituent of a signal transduction chain for the expression of two energy-linked [NiFe] hydrogenases. Here we demonstrate that the RH regulatory activity was barely affected by mutations in hypA, hypB, hypC, and hypX and was not substantially diminished in hypD- and hypE-deficient strains. The lack of HypF, however, resulted in a 90% decrease of the RH regulatory activity. Fourier transform infrared spectroscopy and the incorporation of 63Ni into the RH from overproducing cells revealed that the assembly of the [NiFe] active site is dependent on all Hyp functions, with the exception of HypX. We conclude that the entire Hyp apparatus (HypA, HypB, HypC, HypD, HypE, and HypF) is involved in an efficient incorporation of the [NiFe] center into the RH.
Hydrogen plays a major role in bacterial energy metabolism. Many microorganisms can generate reducing power by hydrogen oxidation, while others can release excess reducing equivalents in the form of dihydrogen. Both reactions are catalyzed by enzymes called hydrogenases. The family of [NiFe] hydrogenases is most widespread in nature (for a review, see reference 1). Crystallographic and spectroscopic analyses of hydrogenases from sulfate-reducing bacteria revealed a structure consisting of a large catalytic site-containing subunit and a small three iron sulfur cluster-containing electron-transferring subunit. The H2-activating site is a bimetallic center carrying a nickel and an iron atom. The two metals are coordinated by thiolate groups provided by four cysteine residues, and the iron bears three nonprotein ligands: one CO and two CN−'s (17, 30, 44).
The assembly of the [NiFe] active site is a complex process that requires at least six accessory gene products, the HypA, HypB, HypC, HypD, HypE, and HypF proteins (for a review, see reference 7). HypB is able to bind Ni2+ ions (16, 35) and displays GTPase activity, which is required for nickel incorporation (26). HypC is considered a chaperone assisting metal center assembly (24). Recent studies showed that HypF is involved in the incorporation of CO and/or CN− and that carbamoylphosphate serves as the source of these diatomic ligands (29). The precise roles of HypA, HypD, and HypE are not yet defined. Recently, it was demonstrated that HypE and HypF of Helicobacter pylori interact in the yeast two-hybrid assay (33). A few organisms contain an additional open reading frame, HypX, that is necessary to obtain high level of hydrogenase activity (6, 11, 34). The last step in the maturation of [NiFe] hydrogenases is catalyzed by a specific endopeptidase which cleaves off a short peptide from the C terminus of the large subunit prior to oligomerization of the polypeptides (15, 40).
The facultative lithoautotrophic proteobacterium Ralstonia eutropha H16 possesses two energy-linked [NiFe] hydrogenases, a membrane-bound hydrogenase (MBH) coupled to the respiratory chain via a b-type cytochrome (3, 36) and a cytoplasmic hydrogenase (SH) that displays NAD+-reducing activity (37, 42). The SH and MBH structural genes are clustered on megaplasmid pHG1 of R. eutropha in two distinct operons, together with MBH- and SH-specific accessory genes (38). A complete set of hyp genes (hypA1B1F1CDEX) is associated with the MBH operon (10). Three of the hyp genes form a second copy (hypA2B2F2) downstream of the SH genes (45). Mutations in any of the hyp genes have a pleiotropic effect on the SH and MBH, leading to a substantial decrease or a complete loss of enzymatic activity due to a failure to assemble the [NiFe] active site (6, 10, 45). The duplicated hyp gene products compensate for each other physiologically.
Hydrogenase gene expression in a number of R. eutropha strains depends on the availability of molecular hydrogen. H2 is recognized by the cells via an intracytoplasmic protein complex consisting of a regulatory hydrogenase (RH) and the histidine protein kinase HoxJ. The signal is transmitted on the DNA level by the response regulator HoxA (21). The hydrogen-sensing RH shows typical features of a subclass of [NiFe] hydrogenases (18). Counterparts of this protein are present in Rhodobacter capsulatus (13) and Bradyrhizobium japonicum (5). Studies with soluble extracts (31) and purified RH from R. eutropha (2) showed a [NiFe] active site with electron paramagnetic resonance and Fourier transform infrared (FTIR) spectral properties resembling those of standard [NiFe] hydrogenases. The redox properties and the activity, however, dramatically differed. Although RH-like proteins show enzymatic activity in assays, such as the H2-dependent reduction of redox dyes or the D2/H+ exchange, the absolute activity is ca. 2 orders of magnitude lower than that of energy-linked [NiFe] hydrogenases (2, 43). Furthermore, unlike standard cases, the RH-type protein lacks a C-terminal extension in the large subunit; therefore, it is conceivable to exclude a proteolytic step in the maturation of this protein (18). This observation raises the question as to whether the Hyp protein-assisted metal center assembly process participates in RH activation.
We show here that HypF is almost indispensable for the synthesis of active RH. Mutations in the remaining hyp genes affect the regulatory capacity of the RH to a lesser extent but clearly decrease its H2-oxidizing activity if the RH is expressed at an elevated level.
MATERIALS AND METHODS
Strains and plasmids.
The strains and plasmids used in this study are listed in Table 1. Strains with the initials HF were derived from wild-type R. eutropha H16. Escherichia coli JM109 (46) was used for standard cloning procedures, and E. coli S17-1 (39) was used for conjugative plasmid transfer to R. eutropha strains.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| R. eutropha | ||
| H16 | Wild type, SH+ MBH+ RH+ HoxJ− (hoxJg1264a) | DSM428, ATCC 17699 |
| HF470 | SH− (hoxHΔ) MBH− (hoxGΔ) RH+ | This study |
| HF510 | SH− (hoxHΔ) MBH− (hoxGΔ) RH− (hoxCΔ) | This study |
| HF410 | hypA1Δ hypA2Δ HoxJ− | 45 |
| HF417 | hypB1Δ hypB2Δ HoxJ− | 45 |
| HF441 | hypF1Δ hypF2Δ HoxJ− | 45 |
| HF340 | hypCΔ HoxJ− | 10 |
| HF338 | hypDΔ HoxJ− | 10 |
| HF339 | hypEΔ HoxJ− | 10 |
| HF469 | hypXΔ HoxJ− | 6 |
| HF503 | hypA1Δ hypA2Δ SH− (hoxHΔ) MBH− (hoxGΔ) | This study |
| HF504 | hypB1Δ hypB2Δ SH− (hoxHΔ) MBH− (hoxGΔ) | This study |
| HF505 | hypF1Δ hypF2Δ SH− (hoxHΔ) MBH− (hoxGΔ) | This study |
| HF506 | hypCΔ SH− (hoxHΔ) MBH− (hoxGΔ) | This study |
| HF507 | hypDΔ SH− (hoxHΔ) MBH− (hoxGΔ) | This study |
| HF508 | hypEΔ SH− (hoxHΔ) MBH− (hoxGΔ) | This study |
| HF509 | hypXΔ SH− (hoxHΔ) MBH− (hoxGΔ) | This study |
| HF573 | SH− (hoxHΔ) MBH− (hoxGΔ) nor(R2A2B2)Δ::Φ(hoxK′-lacZ) | This study |
| HF439 | hyp1Δ (hyp[A1B1F1CDEX]Δ) | This study |
| HF575 | hyp1Δ hyp2Δ (hyp[A1B1F1CDEX]Δ) (hyp[A2B2F2]Δ) | This study |
| HF581 | hyp1Δ hyp2Δ SH− (hoxHΔ) MBH− (hoxGΔ) RH+nor(R2A2B2)Δ::Φ(hoxK′-lacZ) | This study |
| HF582 | hyp1Δ hyp2Δ SH− (hoxHΔ) MBH− (hoxGΔ) RH− (hoxCΔ) nor(R2A2B2)Δ::Φ(hoxK′-lacZ) | This study |
| E. coli | ||
| JM109 | F′ traD36 lacIq Δ(lacZ)M15 proA+B+/e14− (McrA−) Δ(lac-proAB) thi gyrA96 (Nalr) endA1 hsdR17(rK− mK+) relA1 supE44 recA1 | 46 |
| S17-1 | Tra+recA pro thi hsdR chr::RP4-2 | 39 |
| Plasmids | ||
| pBluescript SK− | Apr, lacZ′, T7 gene 10 promoter, f1 ori | Stratagene Cloning Systems |
| LITMUS 28 | Apr, lacZ′, ColE1 ori | New England Biolabs |
| pLO1/pLO2 | Kmr, sacB, RP4 oriT, ColE1 ori | 22 |
| pEDY309 | RK2 ori, Tcr, Mob+, promoterless lacZ gene | 18 |
| pCH297 | 3.5-kb KpnI fragment containing ′hypDEXhoxA′ in pBluescript SK− | 22 |
| pCH412 | 2.1-kb SmaI-PvuII fragment containing hypA1Δ in pLO1 (PmeI cut) | 9 |
| pCH547 | 2.7-kb fragment containing a 6.3-kb in-frame deletion in hypA1B1F1CDEX in pLO1 | This study |
| pCH371 | 2.7-kb KpnI-XbaI fragment containing hypB1F1 in pTZ18R | J. Dernedde and B. Friedrich |
| pCH424 | pLO1 with a 2.2-kb SalI-SmaI fragment containing hoxGΔ | 4 |
| pCH455 | 15-kb HindIII fragment of pGE15 containing the SH gene region in pBluescript KS+ | 27 |
| pCH474 | pLO1 with a 976-bp SacI fragment containing hoxHΔ | 27 |
| pCH615 | 0.8-kb Eco47III fragment containing ′hoxJ into PmeI-digested pLO3 | 21 |
| pCH644 | pLO1 with a 2.45-kb PstI fragment containing hoxCΔ | 21 |
| pCH857 | 5.1-kb Ecl136II-BglII fragment of pCH455 cloned into LITMUS 28 (EcoRV-BglII cut) | This study |
| pCH858 | 4.1-kb fragment of a PvuII partial digest of pCH857, religated | This study |
| pCH859 | 1.4-kb BglII fragment (Klenow treated) of pCH858 cloned into the PmeI site of pLO2 | This study |
| pCH872 | nor(R2A2B2)Δ::Φ(hoxK′-lacZ) in pLO1 | O. Lenz and B. Friedrich |
| pGE6 | 8.6-kb EcoRI fragment containing the hyp1 region in pVK101 | 12 |
| pGE15 | 15.0-kb HindIII fragment of pHG1 in pVK101 | 42 |
| pGE151 | Derivative of pRK404 | 19 |
| pGE301 | Φ(hoxK′-lacZ), Tcr | 22 |
| pGE378 | pEDY309 with a 2.8-kb HindIII-XbaI fragment containing PSH-hoxB-hoxC and with a 2.2-kb PvuII-Ecl136II fragment containing Plac-hoxA | 18 |
| pGE457 | 1.4-kb PstI-EcoRV fragment of pCH371 containing hypF1 cloned into PstI-Ecl136II-cut pGE151 | This study |
Nalr, nalidixic acid resistant; Tcr, tetracycline resistant; Apr, ampicillin resistant; Kmr, kanamycin resistant.
A 1.4-kb SspI-Ecl136II fragment of pCH297 was cloned into the Ecl136II site of pCH412. The resulting plasmid pCH547 harbors a 6,303-bp in-frame deletion in the hyp1 region of the MBH operon (hyp[A1B1F1CDEX]Δ; hyp1Δ). For construction of a deletion in the hyp2 region of the SH operon, a pCH455-derived 5.1-kb Ecl136II-BglII fragment was subcloned into the EcoRV-BglII-cut LITMUS 28. The resulting plasmid, pCH857, was partially digested with PvuII, and a 4.1-kb fragment was religated to give pCH858, which contains a 3,780-bp deletion in the hyp2 region (hyp[A2B2F2]Δ; hyp2Δ). Finally, hyp2Δ was inserted as a 1.4-kb Klenow-treated BglII fragment into the PmeI site of pLO2, yielding plasmid pCH859.
For complementation studies, hypF1 was cloned as a 1.4-kb PstI-EcoRV fragment derived from pCH371 into PstI-Ecl136II-cut pGE151 to give pGE457.
Media and growth conditions.
E. coli strains were grown in Luria broth (LB). R. eutropha strains were grown in modified LB medium containing 0.25% (wt/vol) sodium chloride (LSLB) or in mineral salts medium (38) containing 0.4% fructose (FN) or a mixture of fructose and glycerol (0.2% [wt/vol] each; FGN) as the carbon sources. Sucrose-resistant segregants of sacB-harboring strains were selected on LSLB plates containing 15% (wt/vol) sucrose (22). Solid media contained 1.5% (wt/vol) agar. Antibiotics were used at the following concentrations: 350 μg of kanamycin ml−1 and 15 μg of tetracycline ml−1 for R. eutropha and 25 μg of kanamycin ml−1, 15 μg of tetracycline ml−1, and 100 μg of ampicillin ml−1 for E. coli.
Conjugative plasmid transfer and gene replacement.
Mobilizable plasmids were transferred from E. coli to R. eutropha by using a spot mating technique (39). Gene replacement in R. eutropha was achieved by using an allelic exchange procedure based on the conditionally lethal sacB gene (22). The resulting isolates were screened for the presence of the desired mutation by PCR amplification of the respective target site (4). Deletion-carrying isolates were identified on the basis of the altered electrophoretic mobility of the amplification products. Suicide plasmids pCH424 (hoxGΔ), pCH474 (hoxHΔ), and pCH644 (hoxCΔ) were used for the deletion of the genes for the large subunits of the MBH, SH, and RH, respectively. The hoxJa1264g exchange was achieved by using pCH615. The hyp1 region of the MBH operon was deleted in R. eutropha H16 by using plasmid pCH547, yielding HF439 (hyp[A1B1F1CDEX]Δ; hyp1Δ). Subsequently, the hyp2 region of the SH region was deleted in HF439 by using plasmid pCH859 to generate HF575 (hyp[A1B1F1CDEX]Δhyp[A2B2F2]Δ; hyp1Δ hyp2Δ). Plasmid pCH872 was used for the introduction of the Φ(hoxK′-lacZ) gene fusion into the chromosomal norR2A2B2 gene region of R. eutropha strains.
Cell fractionation and immunoblot analysis.
R. eutropha cells were disrupted by two passages through a chilled French pressure cell (Amicon) at 900 lb/in2. Cell debris and membranes were separated from the soluble fraction by ultracentrifugation (90,000 × g). Soluble proteins of R. eutropha extracts were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 12% gels and subsequently transferred to Protran BA85 nitrocellulose membranes (Schleicher & Schuell) according to a standard protocol (41). The RH subunits HoxC and HoxB were detected by using anti-HoxC serum (diluted 1:1,000) and anti-HoxB serum (diluted 1:10,000), respectively, and alkaline phosphatase-labeled goat anti-rabbit immunoglobulin G (Dianova, Hamburg, Germany).
In-gel activity staining.
Soluble proteins of R. eutropha extracts were separated by native PAGE (4 to 15%). Subsequently, the gel was incubated in H2-saturated 50 mM potassium phosphate buffer (pH 7.0) containing 0.09 mM phenazine methosulfate (PMS) and 0.06 mM nitroblue tetrazolium (NBT) under an atmosphere of 100% H2. Purple bands occured upon incubation at 30°C in the dark, indicating PMS-mediated reduction of NBT.
63Ni labeling.
Cells were grown in the presence of 120 nM 63NiCl2 (6.38 mCi/ml; Amersham). Soluble extracts were prepared and subjected to native PAGE. Gels were dried and autoradiographed by using an SI 550 storage PhosphorImager (Molecular Dynamics) as described earlier (18).
FTIR spectroscopy.
FTIR spectra were obtained with a Bio-Rad FTS 60A spectrometer equipped with a mercury cadmium telluride (MCT) detector. Spectra were recorded at room temperature with a resolution of 2 cm−1. Typically, averages of 1,524 spectra were determined against proper blanks. Samples of soluble extracts (10 μl) were loaded into a gastight transmission cell (CaF2, 56-μm pathlength). Samples of whole cells were prepared by drying 100-μl aliquots of a cell suspension on a CaF2 window, and the dried placard was measured. The spectra were corrected for the baseline by using a spline function provided by the Bio-Rad software.
Assays.
H2-oxidizing activity was quantified by an amperometric H2 uptake assay as described previously by using an H2 electrode with methylene blue as the electron acceptor (31). β-Galactosidase activity was determined as described previously (47), and the activities were calculated according to the Miller method (28), except that the cell density was measured at 436 nm. The proteins of the soluble extracts were determined according to the protocol of Lowry (23).
RESULTS
Regulatory properties of mutants with deletions in individual hyp genes.
For H2-responding strains of R. eutropha, such as HF470 (Table 1), a functional RH is absolutely necessary to express the genes for the SH and MBH. Mutants with impaired RH fail to grow on H2 as an energy source (21). To test whether mutations in the various hyp genes affect the regulatory activity of the RH, we examined two different sets of hyp mutants (Table 1). The first group of mutants carried single site in-frame deletions in hypC (HF340), hypD (HF338), hypE (HF339), and hypX (HF469), and the second group of mutants was characterized by deletions in both copies of the respective hyp genes, i.e., hypA1A2 (HF410), hypB1B2 (HF417), and hypF1F2 (HF441). We knocked out the SH and MBH in these strains by deletions of the corresponding structural genes hoxH and hoxG, respectively, in order to avoid interferences with their dominant activities in the enzyme assays. Furthermore, since the hyp mutants were originally constructed from the non-H2-responding strain R. eutropha H16 (Table 1), the activity of the histidine kinase HoxJ was restored in the hyp mutants by site-directed mutagenesis as described previously (21). A codon conversion in hoxJ replaced serine at position 422 by a glyine residue.
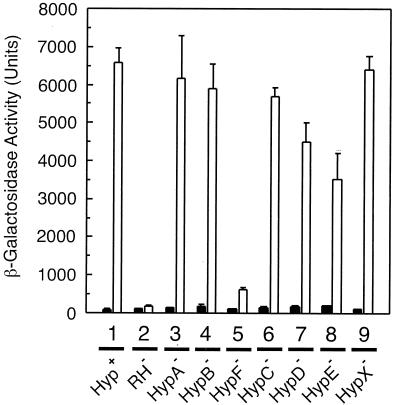
Since the MBH and SH genes are regulated coordinately, transcription was monitored by using the plasmid-borne MBH gene fusion Φ(hoxK′-lacZ) as a representative parameter. The hoxK gene encodes the MBH small subunit and is the first gene of the MBH operon (19). As expected, the reference strain HF470 (Fig. 1, lane 1) showed low β-galactosidase reporter activity in the absence of H2 and high activity in the presence of H2. The RH-negative strain failed to activate the MBH promoter under both conditions (Fig. 1, lane 2). The loss of HypA (lane 3), HypB (lane 4), HypC (lane 6), and HypX (lane 9) scarcely affected the MBH promoter activity, whereas mutations in hypD (lane 7) and hypE (lane 8) led to a moderate decrease of β-galactosidase to a level of 50 to 70%. A dramatic downregulation occurred by mutation of hypF (lane 5). Mutant HF505 retained only 10% of the MBH promoter activity. These results showed that HypF is a major component for the H2-sensing function of the RH, whereas the other hyp gene products seem to play a subordinate role in the synthesis of active RH.
FIG. 1.
RH regulatory activity in the hyp deletion strains. R. eutropha strains harboring the plasmid-based Φ(hoxK′-lacZ) fusion were grown in FGN medium in the absence (black bars) or in the presence (white bars) of hydrogen. Cells were harvested at an optical density at 436 nm (OD436) of 8.0 ± 0.3, and the β-galactosidase activity was determined according to the protocol of Miller (28). Lane 1, HF470; lane 2, HF510; lane 3, HF503; lane 4, HF504; lane 5, HF505; lane 6, HF506; lane 7, HF507; lane 8, HF508; lane 9, HF509.
Effects of hyp mutations on the biochemical characteristics of the RH.
To explore whether the regulatory properties of the hyp mutants correlate with the enzymatic activity of the RH, the mutants were cultivated in fructose-glycerol minimal medium supplied with H2. Soluble extracts were prepared and H2-oxidizing activity was determined amperometrically by using methylene blue as the electron acceptor. The data are summarized in Table 2. In a regular RH-producing background (column 1), the RH− and HypF− mutants were the only strains that were severely affected in their enzymatic activity. The level of activity obtained with the HypD− and HypE− strains correlated well with the diminished MBH promoter activity (Fig. 1). The wild-type-like activity profile of the remaining Hyp− mutants was in line with the regulatory data.
TABLE 2.
RH-mediated H2-oxidizing activities in the hyp deletion strains
| Straina | Relevant characteristic | Sp actb (mU/mg of protein)
|
63Ni incorporationc (RHoverpre) | |
|---|---|---|---|---|
| RHwtd | RHoverpre | |||
| HF470 | Hyp+ RH+ | 0.34 | 58.61 | ++ |
| HF510 | Hyp+ RH− | <0.05 | <0.05f | −f |
| HF503 | hypA1ΔA2Δ | 0.28 | 3.73 | + |
| HF504 | hypB1ΔB2Δ | 0.34 | 1.74 | + |
| HF505 | hypF1ΔF2Δ | <0.05 | <0.05 | − |
| HF506 | hypCΔ | 0.27 | 0.88 | + |
| HF507 | hypDΔ | 0.19 | 0.20 | − |
| HF508 | hypEΔ | 0.23 | 0.42 | − |
| HF509 | hypXΔ | 0.29 | 49.42 | ++ |
All strains are SH− MBH− HoxJ+.
Cells were grown in FGN medium in the presence of H2. The specific activities were determined amperometrically with methylene blue as an electron acceptor. The values are the mean of two independent experiments.
Cells were grown in the presence of 63NiCl2. Proteins of soluble extracts were separated by native PAGE, and the gel was subjected to autoradiography. ++, strong signal; +, weak signal; −, no signal.
RHwt, RH expressed from megaplasmid pHG1.
RHoverpr, overproduced RH expressed from plasmid pGE378.
Strain HF510 containing control vector pEDY309 instead of pGE378.
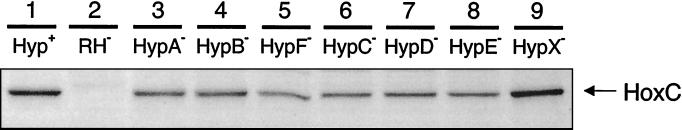
The pattern changed substantially in strains that produced the RH at an elevated level caused by the introduction of the hoxBC-harboring plasmid pGE378 (Table 2). With the exception of the hypX mutant, all of the hyp-deficient strains showed a dramatic decrease of hydrogenase activity, which directly correlated with low 63Ni incorporation by the hyp strains (Table 2). Western blot analysis, conducted with an antibody raised against the large HoxC subunit of the RH, confirmed the expression of RH protein in the hyp mutants (Fig. 2). Nevertheless, with the exception of the hypX mutant, the rest of the hyp strains exhibited a decreased band intensity pointing to less-stable RH protein.
FIG. 2.
RH protein stability in the hyp deletion strains. R. eutropha strains harboring plasmid pGE378 for RH overproduction were grown in FGN medium under hydrogenase derepressing conditions. The presence of the RH large subunit HoxC in soluble extracts was analyzed by the immunoblot technique. A total of 20 μg of protein was applied to each lane. Lane 1, HF470; lane 2, HF510 (containing control vector pEDY309 instead of pGE378); lane 3, HF503; lane 4, HF504; lane 5, HF505; lane 6, HF506; lane 7, HF507; lane 8, HF508; lane 9, HF509.
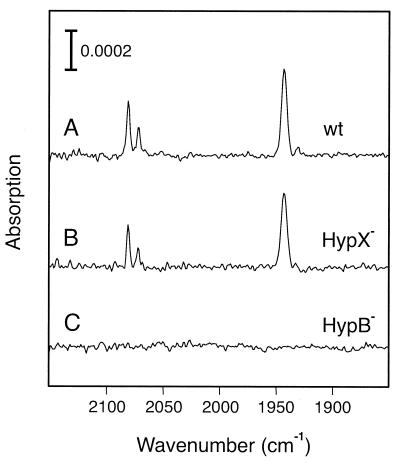
Evidence for changes in the structure of the active site of RH mutant proteins was also obtained by FTIR spectroscopy. This method can be applied only to extracts from RH-overproducing strains due to sensitivity limits (31). Thus, extracts prepared from the pGE378-containing hyp mutants were analyzed for the presence of infrared bands from metal-bound CO and CN− (Fig. 3). As expected, the Hyp+ control (trace A) showed a strong absorption at 1,943 cm−1, which corresponds to one CO ligand and the two bands at 2,072 cm−1 and 2,081 cm−1 are indicative for the presence of two CN− as reported previously (2, 31). A similar spectrum was obtained with extracts of the hypX mutant (Fig. 3, trace B), a finding which is in good agreement with its wild-type-like phenotype (Fig. 1 and Table 2). No FTIR bands in the 2,150 to 1,850 cm−1 spectral region could be detected in the spectra of the rest of the hyp mutants, even in those derivatives which showed residual promoter and hydrogenase activities. As an example, the spectrum of the hypB1Δ hypB2Δ mutant extract is shown (Fig. 3, trace C). Obviously, the concentration of intact RH molecules in the mutants extracts was below the detection limit of the instrument. The use of intact mutant cells for FTIR analysis to circumvent the possibility of the destruction of labile RH maturation intermediates during the extract preparation yielded the same results.
FIG. 3.
FTIR spectra of soluble extracts containing the overproduced RH. Soluble extracts were prepared from R. eutropha strains grown in FGN medium under hydrogenase-derepressing conditions. The spectra were recorded by using the as-isolated, concentrated extracts containing the oxidized RH. Trace A, HF470(pGE378); trace B, HF509(pGE378); trace C, HF504(pGE378).
Complete deletion of the two megaplasmid-borne hyp DNA regions.
The previous data indicate a graded significance of the various hyp gene products in the RH synthesis. If the Hyp proteins are instrumental as chaperones in a series of concerted steps, the loss of one of the seven proteins by mutation may be phenotypically suppressed and less apparent. Therefore, we completely deleted all known hyp genes in R. eutropha and raised the question of whether and to what extent introduction of the individudal hyp genes restored the loss of the hyp gene regions.
Large in-frame deletions in both the hyp1 and hyp2 regions yielded mutant HF575. As described above, the SH and MBH activities were also blocked by mutations in the subunit genes hoxH and hoxG, and the kinase HoxJ was reactivated by a Ser/Gly replacement. To prepare the strain for subsequent plasmid-based complementation, the Φ(hoxK′-lacZ) gene fusion was inserted into the NO reductase gene region norR2A2B2 on the chromosome, a locus which is dispensable for hydrogen metabolism (32).
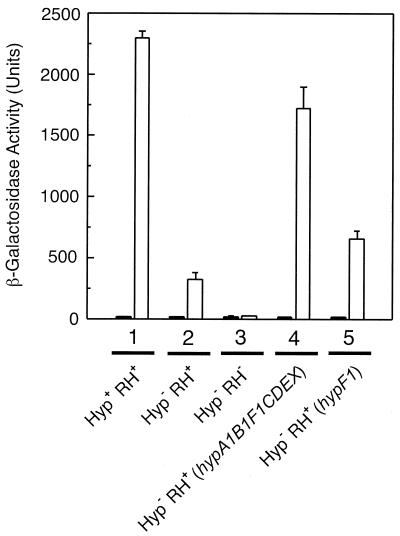
The resulting mutant HF581 was cultivated in fructose-glycerol minimal medium with or without H2 supplementation and tested for β-galactosidase activity. Surprisingly, the mutant still exhibited 10% of the wild-type activity (Fig. 4, lane 2), which corresponds to the level of activity observed before with the HypF− strain (Fig. 1, lane 5). A knockout of the RH gene hoxC on the other hand completely abolished the MBH promoter activity (Fig. 4, lane 3). This result indicates that the residual expression of β-galactosidase in the hyp-negative strain HF581 is mediated by a small population of NiFe-containing RH molecules.
FIG. 4.
RH regulatory activity in the hyp-negative strain HF581. R. eutropha strains harboring the Φ(hoxK′-lacZ) fusion integrated into the norR2A2B2 gene region of the chromosome were grown in FGN medium in the absence (black bars) or in the presence (white bars) of hydrogen. Cells were harvested at an optical density at 436 nm of 8.0 ± 0.3 and the β-galactosidase activity was determined according to the protocol of Miller (28). Lane 1, HF573(pGE151); lane 2, HF581(pGE151); lane 3, HF582(pGE151); lane 4, HF581(pGE6); lane 5, HF581(pGE457).
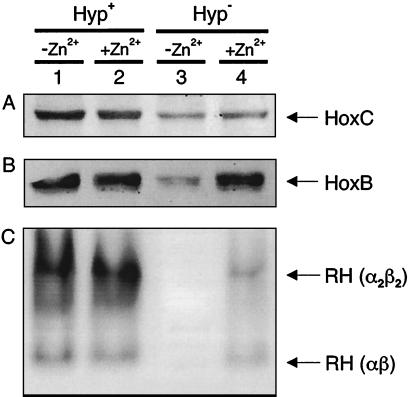
The amperometric assay was not sensitive enough to detect a low level of hydrogenase activity in the hyp-negative strain HF581. The in-gel hydrogenase assay with PMS as the electron acceptor is more appropriate for detecting even traces of enzymatic activity. This method initially also failed to demonstrate H2-oxidizing activity in extracts of the the hyp-negative strain HF581 (Fig. 5A, lane 3). In the course of characterizing mutants with alterations in the SH protein, it was observed that addition of Zn2+ to the growth medium had a stabilizing effect on the structure of the SH (C. Massanz and B. Friedrich, unpublished results). Therefore, we grew cells of the hyp-negative strain HF581 in minimal medium supplemented with 1 μM ZnCl2. The resulting extract clearly developed hydrogenase activity (Fig. 5 A, lane 4). Immunoblot analysis showed that addition of Zn2+ had a particularly stabilizing effect on the small subunit HoxB of the RH (Fig. 5 B, lane 4). Stabilization of the RH protein was not observed by supplementing the minimal medium with Co2+, Cu2+, or Mn2+ ions (data not shown).
FIG. 5.
RH protein stability and RH-mediated H2-oxidizing activity in the hyp-negative strain HF581. R. eutropha strains were grown in FGN medium supplemented with or without 1 μM ZnCl2 as indicated above the figures. (A) Immunoblot against the RH large subunit HoxC, with 20 μg of soluble protein in each lane. (B) Immunoblot against the RH small subunit HoxB, with 20 μg of soluble protein in each lane. (C) In-gel activity assay. A total of 500 μg of soluble proteins were separated by native PAGE. Dark-colored bands indicate H2-oxidizing activity of the RH by the PMS-mediated reduction of NBT. The fact that the RH forms an α2β2 oligomer was previously described (2). Lanes 1 and 2, HF573; lanes 3 and 4, HF581.
Complementation of RH activity.
It was reported before that the activity of the two energy-linked hydrogenases was completely restored in mutants devoid of hyp gene products by introducing the respective hyp gene on a plasmid (10, 45). An analogous complementation experiment was conducted by using the hyp-negative strain HF581 as the recipient. Plasmid pGE6 harboring the complete hyp1 region (hypA1B1F1CDEX) was introduced into HF581. The resulting transconjugants were able to activate the MBH promoter in the presence of H2 up to 80% of the wild-type level (Fig. 4, lane 4). If the product of hypF is the major player in the RH cofactor insertion, introduction of hypF1 on plasmid pGE457 should substantially complement the MBH promoter activity. In fact 30% of β-galactosidase activity were recovered in the transconjugants (Fig. 4, lane 5). This level was not enhanced by introducing the alternative copy hypF2. Moreover, plasmids harboring one of the other hyp genes had no complementation capacity at all (data not shown).
DISCUSSION
The H2-sensing hydrogenase of R. eutropha belongs to a new subclass of [NiFe] hydrogenases which exhibits some unique structural and biochemical features (2, 18). Although its active site has the spectral properties of a normal [NiFe] site, the active site can exist in only two redox states and does not react with O2 or CO. In addition, RH has a very low H2-oxidizing activity with artificial electron acceptors. The lack of a C-terminal extension in the large subunit indicates the absence of a proteolytic step in RH maturation and raised the question whether metal-center assembly requires auxiliary proteins as demonstrated for [NiFe] hydrogenases involved in energy metabolism (7, 25). In addition to the H2-sensing hydrogenases (18), the CO-induced hydrogenase in Rhodospirillum rubrum (14) and the Ech hydrogenase from Methanosarcina barkeri (20), which are physiologically quite diverse, are devoid of a C-terminal extension in the large subunit. This observation suggests that the final proteolysis is not an obligate step in [NiFe] center assembly.
To examine whether Hyp proteins are involved in metal center assembly of the RH, a collection of hyp mutants of R. eutropha was analyzed for its regulatory capacity, for its ability to oxidize H2 with redox dyes and for some structural features. It has been reported that the product of hypD is necessary for the synthesis of active HupUV protein in R. capsulatus (43) and that HypF participates in the regulation of hydrogenase synthesis through maturation of HupUV (8). In the present study we show that complete deletion of the known hyp genes in R. eutropha HF581 dramatically affects both the H2-sensing and H2-oxidizing activity of the RH. Only 10% of β-galactosidase activity, expressed from a Φ(hoxK′-lacZ) gene fusion, was recovered and trace amounts of hydrogenase activity were identified in a native gel after stabilization of the RH by the addition of ZnCl2. This result clearly argues for a requirement of the Hyp proteins in the maturation of the H2-sensing hydrogenase. The fact that mutants devoid of RH have completely lost the regulatory and enzymatic activities confirms the notion that the residual activity in the hyp-negative strains derived from some NiFe-containing RH molecules. A strict correlation between both the RH regulatory and enzymatic activity and the availability of nickel in the medium has been reported previously (18).
Our results showed that of the seven hyp gene products in R. eutropha HypX had barely any effect on the regulatory and enzymatic activity of the RH under all conditions tested. Even the CO- and CN−-related infrared absorptions were not affected in extracts of the hypX-deficient strain. Therefore, the function of HypX which, under certain conditions, may participate in the delivery of the C1 compounds (34) appears to be restricted to the maturation of the SH and MBH (6). From the phenotypic behavior of the corresponding mutants, it is inferred that the functional significance of the remaining hyp gene products follow a graded pattern. HypA, HypB, and HypC mutants showed a decrease of maximal 10% in MBH promoter activity which correlated with a slightly altered enzymatic activity. These mutations, however, had a severe effect on the level of enzymatic RH activity if the hoxBC genes were expressed from a multiple-copy plasmid. Obviously, the cells need these Hyp proteins when hydrogenase synthesis proceeds at a high level. Mutants with deletions of hypD, hypE and, in particular, hypF had low if any regulatory and enzymatic activity. Provided HypF of R. eutropha has a similar function as postulated for E. coli (29), incorporation of the nonprotein ligands CO and CN− is also a crucial reaction for metal-center assembly into the RH that can hardly be accomplished without the function of HypF. If HypF incorporates the Fe(CO)(CN)2 moiety, then this reaction is essential for the H2-sensing function of the RH. We previously showed that both the regulatory and enzymatic activity of the RH are dependent on Ni (18). Taken together, these two observations indicate that signal transduction requires an H2 sensor with an intact [NiFe] active site. The question whether a simple binding of H2 or further electron transfer processes are required for H2-signaling remains open.
ACKNOWLEDGMENTS
This work was funded by the Deutsche Forschungsgemeinschaft and by the Fonds der Chemischen Industrie.
We thank J. Dernedde for the construction of R. eutropha HF439 and O. Lenz for providing plasmid pCH872.
REFERENCES
- 1.Albracht S P. Nickel hydrogenases: in search of the active site. Biochim Biophys Acta. 1994;1188:167–204. doi: 10.1016/0005-2728(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 2.Bernhard M, Buhrke T, Bleijlevens B, De Lacey A L, Fernandez V M, Albracht S P, Friedrich B. The H2 sensor of Ralstonia eutropha: biochemical characteristics, spectroscopic properties, and its interaction with a histidine protein kinase. J Biol Chem. 2001;276:15592–15597. doi: 10.1074/jbc.M009802200. [DOI] [PubMed] [Google Scholar]
- 3.Bernhard M, Benelli B, Hochkoeppler A, Zannoni D, Friedrich B. Functional and structural role of the cytochrome b subunit of the membrane-bound hydrogenase complex of Alcaligenes eutrophus H16. Eur J Biochem. 1997;248:179–186. doi: 10.1111/j.1432-1033.1997.00179.x. [DOI] [PubMed] [Google Scholar]
- 4.Bernhard M, Schwartz E, Rietdorf J, Friedrich B. The Alcaligenes eutrophus membrane-bound hydrogenase gene locus encodes functions involved in maturation and electron transport coupling. J Bacteriol. 1996;178:4522–4529. doi: 10.1128/jb.178.15.4522-4529.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black L K, Fu C, Maier R J. Sequence and characterization of hupU and hupV genes of Bradyrhizobium japonicum encoding a possible nickel-sensing complex involved in hydrogenase expression. J Bacteriol. 1994;176:7102–7106. doi: 10.1128/jb.176.22.7102-7106.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buhrke T, Friedrich B. hoxX (hypX) is a functional member of the Alcaligenes eutrophus hyp gene cluster. Arch Microbiol. 1998;170:460–463. doi: 10.1007/s002030050667. [DOI] [PubMed] [Google Scholar]
- 7.Casalot L, Rousset M. Maturation of the [NiFe] hydrogenases. Trends Microbiol. 2001;9:228–237. doi: 10.1016/s0966-842x(01)02009-1. [DOI] [PubMed] [Google Scholar]
- 8.Colbeau A, Elsen S, Tomiyama M, Zorin N A, Dimon B, Vignais P M. Rhodobacter capsulatus HypF is involved in regulation of hydrogenase synthesis through the HupUV proteins. Eur J Biochem. 1998;251:65–71. doi: 10.1046/j.1432-1327.1998.2510065.x. [DOI] [PubMed] [Google Scholar]
- 9.Dernedde J. Ph.D. thesis. Berlin, Germany: Freie Universität zu Berlin; 1996. [Google Scholar]
- 10.Dernedde J, Eitinger T, Patenge N, Friedrich B. hyp gene products in Alcaligenes eutrophus are part of a hydrogenase maturation system. Eur J Biochem. 1996;235:351–358. doi: 10.1111/j.1432-1033.1996.00351.x. [DOI] [PubMed] [Google Scholar]
- 11.Durmowicz M C, Maier R J. Roles of HoxX and HoxA in biosynthesis of hydrogenase in Bradyrhizobium japonicum. J Bacteriol. 1997;179:3676–3682. doi: 10.1128/jb.179.11.3676-3682.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eberz G, Hogrefe C, Kortlüke C, Kamienski A, Friedrich B. Molecular cloning of structural and regulatory hydrogenase genes (hox) of Alcaligenes eutrophus H16. J Bacteriol. 1986;168:636–641. doi: 10.1128/jb.168.2.636-641.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elsen S, Colbeau A, Chabert J, Vignais P M. The hupTUV operon is involved in negative control of hydrogenase synthesis in Rhodobacter capsulatus. J Bacteriol. 1996;178:5174–5181. doi: 10.1128/jb.178.17.5174-5181.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fox J D, Kerby R L, Roberts G P, Ludden P W. Characterization of the CO-induced, CO-tolerant hydrogenase from Rhodospirillum rubrum and the gene encoding the large subunit of the enzyme. J Bacteriol. 1996;178:1515–1524. doi: 10.1128/jb.178.6.1515-1524.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fritsche E, Paschos A, Beisel H G, Böck A, Huber R. Crystal structure of the hydrogenase maturating endopeptidase HYBD from Escherichia coli. J Mol Biol. 1999;288:989–998. doi: 10.1006/jmbi.1999.2719. [DOI] [PubMed] [Google Scholar]
- 16.Fu C, Olson J W, Maier R J. HypB protein of Bradyrhizobium japonicum is a metal-binding GTPase capable of binding 18 divalent nickel ions per dimer. Proc Natl Acad Sci USA. 1995;92:2333–2337. doi: 10.1073/pnas.92.6.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Happe R P, Roseboom W, Pierik A J, Albracht S P, Bagley K A. Biological activation of hydrogen. Nature. 1997;385:126. doi: 10.1038/385126a0. [DOI] [PubMed] [Google Scholar]
- 18.Kleihues L, Lenz O, Bernhard M, Buhrke T, Friedrich B. The H2 sensor of Ralstonia eutropha is a member of the subclass of regulatory [NiFe] hydrogenases. J Bacteriol. 2000;182:2716–2724. doi: 10.1128/jb.182.10.2716-2724.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kortlüke C, Horstmann K, Schwartz E, Rohde M, Binsack R, Friedrich B. A gene complex coding for the membrane-bound hydrogenase of Alcaligenes eutrophus H16. J Bacteriol. 1992;174:6277–6289. doi: 10.1128/jb.174.19.6277-6289.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Künkel A, Vorholt J A, Thauer R K, Hedderich R. An Escherichia coli hydrogenase-3-type hydrogenase in methanogenic archaea. Eur J Biochem. 1998;252:467–476. doi: 10.1046/j.1432-1327.1998.2520467.x. [DOI] [PubMed] [Google Scholar]
- 21.Lenz O, Friedrich B. A novel multicomponent regulatory system mediates H2 sensing in Alcaligenes eutrophus. Proc Natl Acad Sci USA. 1998;95:12474–12479. doi: 10.1073/pnas.95.21.12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lenz O, Schwartz E, Dernedde J, Eitinger M, Friedrich B. The Alcaligenes eutrophus H16 hoxX gene participates in hydrogenase regulation. J Bacteriol. 1994;176:4385–4393. doi: 10.1128/jb.176.14.4385-4393.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 24.Magalon A, Böck A. Analysis of the HypC-HycE complex, a key intermediate in the assembly of the metal center of the Escherichia coli hydrogenase 3. J Biol Chem. 2000;275:21114–21120. doi: 10.1074/jbc.M000987200. [DOI] [PubMed] [Google Scholar]
- 25.Maier T, Böck A. Nickel incorporation into hydrogenases. In: Hausinger R P, Eichhorn G L, Marzilli L G, editors. Advances in inorganic biochemistry: mechanisms of metallocenter assembly. New York, N.Y: VHC Publishers, Inc.; 1996. pp. 173–192. [Google Scholar]
- 26.Maier T, Lottspeich F, Böck A. GTP hydrolysis by HypB is essential for nickel insertion into hydrogenases of Escherichia coli. Eur J Biochem. 1995;230:133–138. [PubMed] [Google Scholar]
- 27.Massanz C, Schmidt S, Friedrich B. Subforms and in vitro reconstitution of the NAD-reducing hydrogenase of Alcaligenes eutrophus. J Bacteriol. 1998;180:1023–1029. doi: 10.1128/jb.180.5.1023-1029.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 29.Paschos A, Glass R S, Böck A. Carbamoylphosphate requirement for synthesis of the active center of [NiFe]-hydrogenases. FEBS Lett. 2001;488:9–12. doi: 10.1016/s0014-5793(00)02408-x. [DOI] [PubMed] [Google Scholar]
- 30.Pierik A J, Roseboom W, Happe R P, Bagley K A, Albracht S P. Carbon monoxide and cyanide as intrinsic ligands to iron in the active site of [NiFe]-hydrogenases. NiFe(CN)2CO: biology's way to activate H2. J Biol Chem. 1999;274:3331–3337. doi: 10.1074/jbc.274.6.3331. [DOI] [PubMed] [Google Scholar]
- 31.Pierik A J, Schmelz M, Lenz O, Friedrich B, Albracht S P J. Characterization of the active site of a hydrogen sensor from Alcaligenes eutrophus. FEBS Lett. 1998;438:231–235. doi: 10.1016/s0014-5793(98)01306-4. [DOI] [PubMed] [Google Scholar]
- 32.Pohlmann A, Cramm R, Schmelz K, Friedrich B. A novel NO-responding regulator controls the reduction of nitric oxide in Ralstonia eutropha. Mol Microbiol. 2000;38:626–638. doi: 10.1046/j.1365-2958.2000.02157.x. [DOI] [PubMed] [Google Scholar]
- 33.Rain J-C, Selig L, De Reuse H, Battaglia V, Reverdy C, Simon S, Lenzen G, Petel F, Wojcik J, Schächter V, Chemama Y, Labigne A, Legrain P. The protein-protein interaction map of Helicobacter pylori. Nature. 2001;409:211–215. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- 34.Rey L, Fernandez D, Brito B, Hernando Y, Palacios J M, Imperial J, Ruiz-Argueso T. The hydrogenase gene cluster of Rhizobium leguminosarum bv. viciae contains an additional gene (hypX), which encodes a protein with sequence similarity to the N10-formyltetrahydrofolate-dependent enzyme family and is required for nickel-dependent hydrogenase processing and activity. Mol Gen Genet. 1996;252:237–248. doi: 10.1007/BF02173769. [DOI] [PubMed] [Google Scholar]
- 35.Rey L, Imperial J, Palacios J M, Ruiz-Argueso T. Purification of Rhizobium leguminosarum HypB, a nickel-binding protein required for hydrogenase synthesis. J Bacteriol. 1994;176:6066–6073. doi: 10.1128/jb.176.19.6066-6073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schink B, Schlegel H G. The membrane-bound hydrogenase of Alcaligenes eutrophus. I. Solubilization, purification, and biochemical properties. Biochem Biophys Acta. 1979;567:315–324. doi: 10.1016/0005-2744(79)90117-7. [DOI] [PubMed] [Google Scholar]
- 37.Schneider K, Schlegel H G. Purification and properties of the soluble hydrogenase from Alcaligenes eutrophus H16. Biochim Biophys Acta. 1976;452:66–80. doi: 10.1016/0005-2744(76)90058-9. [DOI] [PubMed] [Google Scholar]
- 38.Schwartz E, Gerischer U, Friedrich B. Transcriptional regulation of Alcaligenes eutrophus hydrogenase genes. J Bacteriol. 1998;180:3197–3204. doi: 10.1128/jb.180.12.3197-3204.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:717–743. [Google Scholar]
- 40.Thiemermann S, Dernedde J, Bernhard M, Schröder W, Massanz C, Friedrich B. Carboxy-terminal processing of the soluble, NAD-reducing hydrogenase of Alcaligenes eutrophus requires the hoxW gene product. J Bacteriol. 1996;178:2368–2374. doi: 10.1128/jb.178.8.2368-2374.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of protein from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4357. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tran-Betcke A, Warnecke U, Böcker C, Zaborosch C, Friedrich B. Cloning and nucleotide sequences of the genes for the subunits of NAD-reducing hydrogenase of Alcaligenes eutrophus H16. J Bacteriol. 1990;172:2920–2929. doi: 10.1128/jb.172.6.2920-2929.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vignais P M, Dimon B, Zorin N A, Tomiyama M, Colbeau A. Characterization of the hydrogen-deuterium exchange activities of the energy-transducing HupSL hydrogenase and H2-signaling HupUV hydrogenase in Rhodobacter capsulatus. J Bacteriol. 2000;182:5997–6004. doi: 10.1128/jb.182.21.5997-6004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Volbeda A, Charon M-H, Piras C, Hatchikan E C, Frey M, Fontecilla-Camps J C. Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas. Nature. 1995;373:580–587. doi: 10.1038/373580a0. [DOI] [PubMed] [Google Scholar]
- 45.Wolf I, Buhrke T, Dernedde J, Pohlmann A, Friedrich B. Duplication of hyp genes involved in maturation of [NiFe] hydrogenases in Alcaligenes eutrophus H16. Arch Microbiol. 1998;170:451–459. doi: 10.1007/s002030050666. [DOI] [PubMed] [Google Scholar]
- 46.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 47.Zimmer D, Schwartz E, Tran-Betcke A, Gewinner P, Friedrich B. Temperature tolerance of hydrogenase expression in Alcaligenes eutrophus is conferred by a single amino acid exchange in the transcriptional activator HoxA. J Bacteriol. 1995;177:2373–2380. doi: 10.1128/jb.177.9.2373-2380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]