Background:
Coronavirus disease 2019 (COVID-19) is usually mild and self-limited in children. However, a few Severe acute respiratory syndrome coronavirus 2 (SARS-CoV2) infections in children may progress to severe disease with respiratory distress or can result in a multisystem inflammatory syndrome (MIS-C) associated with COVID-19. The immune mechanisms for these differential clinical outcomes are largely unknown.
Methods:
A prospective cohort study was performed to analyze the laboratory parameters, antibody response, immune phenotypes and cytokine profiles of 51 children with different clinical presentations of COVID-19.
Results:
We found that the absolute lymphocyte counts gradually decreased with disease severity. Furthermore, SARS-CoV-2 IgG levels in the acute phase and convalescence were not significantly different in patients with different disease severity. A decrease in CD3+, CD4+ and CD8+ T cells was observed as disease severity increased. Both CD4+ and CD8+ T cells were activated in children with COVID-19, but no difference in the percentage of HLADR+-expressing cells was detected across the severity groups. In contrast, MIS-C patients exhibited augmented exhausted effector memory CD8+ T cells. Interestingly, the cytokine profile in sera of moderate/severe and MIS-C patients revealed an increase in anti-inflammatory IL-1RA and a suppression of tumor necrosis factor-α, RANTES, eotaxin and PDGF-BB. MIS-C patients also exhibited augmented IL-1β.
Conclusions:
We report distinct immune profiles dependent on severity in pediatric COVID-19 patients. Further investigation in a larger population will help unravel the immune mechanisms underlying pediatric COVID-19.
Keywords: COVID-19, pediatric, disease severity, antibody response, T lymphocyte profile, cytokine profile
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel betacoronavirus causing coronavirus disease 2019 (COVID-19) in humans.1,2 Children generally develop milder disease than adults.3,4 Different hypotheses have been considered to explain the differential presentation of COVID-19 in children such as a qualitatively different immune response, competition with other viruses in the mucus respiratory membranes of small children and differential expression of the cell receptor angiotensin-converting enzyme 2 (ACE2).5–7 Although pediatric COVID-19 is typically mild, infection of children with SARS-CoV-2 can produce a rare post-COVID syndrome known as multisystem inflammatory syndrome (MIS-C) that usually appears after 2 weeks of SARS-CoV-2 infection.3
The immune system plays a central role in determining the clinical outcomes of SARS-CoV-2-infected patients. Understanding humoral and cellular immune responses to SARS-CoV-2 are essential to define how these responses contribute to disease severity and vaccine-induced immunity.
In this context, we aim to evaluate the laboratory parameters, antibody response and the cellular immune and cytokine profiles against SARS-CoV-2 in pediatric patients with COVID-19 and their association with disease severity.
MATERIALS AND METHODS
Study Population
A prospective cohort study was conducted at Ricardo Gutierrez Children’s Hospital (HNRG) between April and December 2020. The study population consisted of hospitalized children (0−18 years of age) with laboratory-confirmed SARS-CoV-2 infection by the real-time reverse transcription-polymerase chain reaction (RT-PCR) on nasopharyngeal swab. The informed consent was signed by parents/legal guardian and/or the assent was given by patients. Upon enrollment, parents or legal guardians of all pediatric patients were asked to provide a clinical and epidemiological history; a complete physical examination was performed and clinical evolution was followed.
This study was approved by the Institutional Review Board (IRB) and Ethical Research Committee at HNRG.
Sample Collection
On admission, patients who manifested signs and symptoms compatible with COVID-19 or who had close contact with a suspected case according to the World Health Organization (WHO) case definition8 were tested by real-time RT-PCR to amplify the nucleocapside genes 1 and 2 (N1 and N2) of SARS-CoV-29 on nasopharyngeal swabs at the Virology Laboratory of HNRG. The real-time RT-PCR technique was followed as previously described.10 Blood was collected from participants upon enrollment (acute phase – within 72 h from admission) and ≥21 days post-enrollment (convalescence) by venipuncture for hematological and biochemical laboratory tests. Patients with COVID-19-associated MIS-C were included in the study, and a blood sample was collected from these patients within 72 h of admission. Sera was obtained by centrifugation, and then aliquoted and stored at −80°C for serological and cytokine determinations.
Determination of SARS-CoV-2 S IgG levels
Serum IgG against the SARS-CoV-2 spike (S) protein [including the receptor-binding domain, (RBD)] was detected by a previously described two-step ELISA, COVIDAR IgG (Laboratorio LEMOS, Buenos Aires, Argentina), following the manufacturer’s instructions.11 Serum samples with optical density (OD) 450 nm values higher than the cut-off value +10% were considered reactive for IgG antibodies against the SARS-CoV-2 S protein, samples with OD 450 nm readings in the range of cutoff value ±10% were considered undetermined, and samples with OD 450 nm values lower than the cut-off value −10% were considered non-reactive for IgG antibodies against SARS-CoV-2 S. The cut-off was defined as the negative control average OD value + 0.150.
Subsequently, samples that showed reactive IgG antibodies against the SARS-CoV-2 S protein were further assayed to determine IgG concentrations, expressed as International Units/ml (IU/ml) after normalization with the WHO International Standard for anti-SARS-CoV-2 IgG.12 To this end, a calibration curve was constructed and the SARS-CoV-2 antibody concentration of each serum sample, expressed in IU/ml, was calculated by extrapolation of the OD 450 nm value.
Determination of Neutralizing Antibodies against SARS-CoV-2
Serum neutralizing antibodies against SARS-CoV-2 that block the interaction between the RBD of the S protein and the cell receptor ACE2 were detected by a surrogate Virus Neutralization Test (sVNT, GenScript, Piscataway, NJ). For neutralization titrations, samples that showed neutralizing antibodies against SARS-CoV-2 were subsequently serially diluted and assayed again by sVNT. Best-fit titration curves were calculated by non-linear regression to a sigmoidal function and SARS-CoV-2 RBD neutralizing antibody titer was calculated as the reciprocal of serum dilution that was inhibited by 50% RBD-ACE2 interaction.
Determination of Lymphocyte Subsets by Flow Cytometry
Immunostaining of lymphocyte subsets was performed using 50 µl of whole blood collected with EDTA with the corresponding monoclonal antibody (mAb) for CD45+ cells, T cells (CD3+, CD4+, CD8+), B cells (CD19+) and NK cells (CD56+). The following fluorochrome-labeled mAbs were used: V500-anti-CD45 (2D1), APC-anti-CD3 (SK7), V450-anti-CD4 (RPA-T4), PerCP/Cy5.5-anti-CD8 (SK1), PE-Cy7-anti-CD19 (SJ25C1) and PE-anti-CD56 (N901). Determination of T cell subsets, including central memory and effector memory T cells as well as activation and exhaustion markers, is described in Supplemental Digital Content 1 http://links.lww.com/INF/E795. Cells were acquired in a FACSCanto II flow cytometer (BD) and analyzed using the FlowJo software v10.0.7 (Treestar, Inc., Ashland, OR).
Cytokine Quantification
Serum samples were assayed for cytokine determination using a Bioplex type immunoassay (Bio-Plex Pro Human, BioRad Laboratories, Hercules, CA). Twenty-seven cytokines, chemokines and growth factors were simultaneously assayed and measurements were performed using the Bio-Plex Luminex (Bio-Rad Laboratories).
Statistical Analysis
Data were analyzed using the χ2 test for categorical variables. The nonparametric Mann−Whitney or Wilcoxon matched pairs tests were used for comparisons of continuous variables between two patient groups. The Kruskal−Wallis test and Dunn’s multiple comparison post-test were used to evaluate differences of continuous variables among multiple groups. The association between continuous variables was analyzed using Spearman’s correlation. A P value <0.05 was considered statistically significant for all the tests performed. Statistical analyses were performed using Stata 13 software (StataCorp LP, College Station, TX) and graphics were made with GraphPad Prism 8.0. Principal component analysis (PCA) was performed with R Statistical Language version 4.1.0 (R Core team, 2021).
RESULTS
Patient Demographic and Clinical Characteristics
A total of 51 pediatric patients were enrolled in the study. The median age of children was 78 (IQR 10–133) months and 29 patients (56.9%) were female. Patients were classified as having asymptomatic, mild, moderate or severe disease according to the WHO classification of COVID-19 severity.13 Twelve children (23.5%) developed COVID-19-associated MIS-C and one death was reported in this group. Among the MIS-C group, 5 patients had a positive PCR at the time of enrollment, 4 patients had a history of a positive PCR within 4 weeks before the onset of symptoms, 5 patients exhibited detectable SARS-CoV-2 IgG levels (positive serology for COVID-19) and 1 patient had a recent exposure to a confirmed COVID-19 case. Thirty-five (68.2%) patients had nonsevere disease. Initially, all children with COVID-19 clinical findings and/or asymptomatic COVID-19 were admitted to the hospital to mitigate local transmission of the disease, particularly in Buenos Aires neighborhoods with limited possibilities for isolation. The demographic and clinical characteristics of each group are described in Table 1. Median age and sex distribution were not significantly different between the severity groups. Twelve patients (23.5%) were admitted to the intensive care unit. Although 39.2% of patients had at least 1 comorbidity, the presence of comorbidities was not significantly different between the severity groups. The presence of comorbidities was not associated with the development of MIS-C (OR: 0.431; 95% CI: 0.101–1.842; P = 0.256) or severe disease (OR: 1.611; 95% CI: 0.208–12.469; P = 0.648) in our pediatric population.
TABLE 1.
Demographic and Clinical Characteristics of the Pediatric Population
| Demographic and clinical characteristics | Total (n = 51) |
Asymptomatic (n = 7) |
Mild (n = 22) |
Moderate (n = 6) |
Severe (n = 4) |
MIS-C (n = 12) |
P * |
|---|---|---|---|---|---|---|---|
| Age in months median (IQR) | 78 (10–133) | 51 (5–91) | 27 (9–130) | 114 (71–189) | 174 (126–183) | 64 (41–126) | 0.078 |
| Age in months range | 2–213 | 2–185 | 3–213 | 29–206 | 85–186 | 2–198 | – |
| Female n (%) | 29 (56.9) | 3 (42.9) | 14 (63.6) | 3 (50.0) | 3 (75.0) | 6 (50.0) | 0.763 |
| PICU admission n (%) | 12 (23.5) | 0 (0) | 0 (0) | 0 (0) | 4 (100) | 8 (66.7) | <0.0001 |
| Presence of comorbidities†n (%) | 20 (39.2) | 3 (42.9) | 9 (40.9) | 3 (50.0) | 2 (50.0) | 3 (25.0) | 0.814 |
| Deaths n (%) | 1 (1.96) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (8.3) | 0.507 |
P-values were calculated using the Kruskal-Wallis test for noncategorical variables and Chi-square test for categorical variables.
Comorbidities included chronic respiratory disease (n = 4), chronic cardiac disease (n = 1), primary or secondary immunodeficiency (n = 6), oncohematological and/or transplanted patient (n = 1), nutritional alteration (n = 2), chronic renal disease (n = 1), prematurity (n = 1), genetic disorder (n = 2), neurologic disorder (n = 2), obesity (n = 1), rheumatic disease (n = 1), myasthenia/autoimmunity (n = 1).
IQR indicates interquartile range; PICU, pediatric intensive care unit; MIS-C, multisystem inflammatory syndrome in children.
Laboratory Data
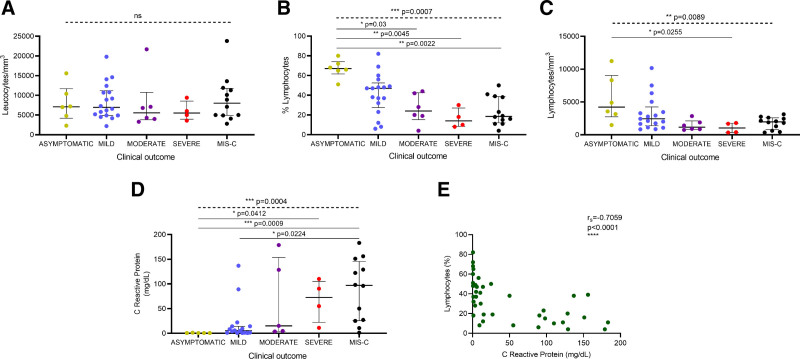
Hemogram analysis in the acute phase of the disease showed that absolute leukocyte counts did not significantly differ between the patient groups (Fig. 1A). Conversely, the percentage of lymphocytes gradually decreased with disease severity (Fig. 1B); there was a nearly 5-fold reduction in severe patients compared to asymptomatic children. Similarly, absolute lymphocyte counts also diminished with disease severity (Fig. 1C); there was a 4-fold reduction in lymphocytes in severe vs. asymptomatic infections. Likewise, patients with MIS-C exhibited a nearly 4-fold decrease in the percentage of lymphocytes and a 2-fold reduction in absolute lymphocyte counts as compared to asymptomatic children (Fig. 1B,C). Ten patients (19.6%) exhibited lymphocyte counts less than 1000/mm3; lymphopenia was more frequent in moderate/severe than in asymptomatic/mild patients (50.0% vs. 8.7%, respectively; P = 0.008). With regard to MIS-C children, 30% of patients developed lymphopenia. As expected, the C-reactive protein (CRP) increased with disease severity (Fig. 1D) and augmented levels of CRP were associated with decreased levels of %lymphocytes (Fig. 1E).
FIGURE 1.
Laboratory parameters in children with COVID-19 according to disease severity. (A) Absolute number of leucocytes, (B) percentage of lymphocytes, (C) absolute number of lymphocytes, and (D) C reactive protein levels were determined in blood samples from COVID-19 patients in acute phase of the disease. Lines represent median values with interquartile range. P values were calculated using the Kruskal-Wallis test (dashed line) and Dunn’s multiple comparison post-test (solid line). P values showing significant post-test comparisons are displayed. (E) Correlation between percentage of lymphocytes and C reactive protein levels in blood from pediatric patients in acute phase of the disease. rS, Spearman correlation coefficient. A P value <0.05 was considered significant; * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001, ns, non-significant.
Humoral Immune Response Against SARS-CoV-2
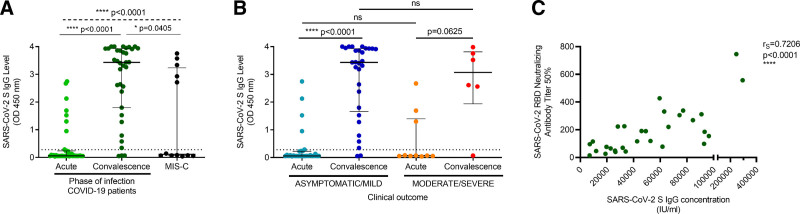
SARS-CoV-2 S IgG levels were determined in serum samples from COVID-19 pediatric patients during the acute and convalescent phases of the disease. Since convalescent serum samples could only be obtained from a subgroup of children, data were analyzed in asymptomatic/mild and moderate/severe groups, considering the clinical disease spectrum and laboratory parameter similarities of the patients. Serum SARS-CoV-2 S IgG levels significantly increased in convalescence in the overall COVID-19 pediatric population (P < 0.0001) as well as in the asymptomatic/mild group (P < 0.0001), while a trend toward increased SARS-CoV-2 IgG levels was observed in convalescent sera from moderate/severe patients (Fig. 2A,B). Thirty-one COVID-19 patients (79.5%) had undetectable antibodies at the time of sampling during the acute phase, while 3 children (7.7%) did not develop an IgG antibody response against SARS-CoV-2 in convalescence (Fig. 2A). Interestingly, SARS-CoV-2 S IgG levels in the acute and convalescent phases were not significantly different in COVID-19 patients with differing disease severity (Fig. 2B). Notably, SARS-CoV-2 S IgG levels were variable in MIS-C children; 7 MIS-C patients (58.3%) did not exhibit reactive SARS-CoV-2 S IgG antibodies (Fig. 2A).
FIGURE 2.
Humoral immune response in pediatric patients with COVID-19. Serum IgG levels against the SARS-CoV-2 S protein (measured as OD at 450 nm) (A) were determined in the COVID-19 pediatric population and in MIS-C patients and (B) were compared according to disease severity in acute and convalescent phases. (C) Correlation between SARS-CoV-2 RBD neutralizing antibody titers and SARS-CoV-2 S IgG concentrations in children in convalescence. SARS-CoV-2 RBD neutralizing antibody titers were calculated as the reciprocal of the dilution of serum that inhibited by 50% RBD-ACE2 interaction in a surrogate Virus Neutralization Test (GenScript). rS, Spearman correlation coefficient. (A,B) Lines represent median values with interquartile range. The lower dotted line indicates the cut-off value +10%, defined in the COVIDAR IgG assay. P values were calculated using (A) the Kruskal-Wallis test (dashed line) and Dunn’s multiple comparison post-test (solid line) for comparisons between multiple groups, (B) the Wilcoxon matched pairs test for comparisons between acute and convalescent phases, or the Mann−Whitney test for comparisons between severity groups. A P value <0.05 was considered significant; * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001, ns, non-significant. MIS-C, multisystem inflammatory syndrome in children; RBD, receptor-binding domain.
Subsequently, sera from pediatric patients that exhibited reactive SARS-CoV-2 S IgG antibodies were further assayed to determine IgG concentrations. Serum SARS-CoV-2 S IgG concentrations in reactive serum samples ranged from 588.7 to 75,545.0 IU/ml and 6994.3 to 293,420.7 IU/ml in the acute and convalescent phases, respectively, and SARS-CoV-2 S IgG levels positively correlated with SARS-CoV-2 S IgG concentrations (P < 0.0001; see Figure A,B, Supplemental Digital Content 2 http://links.lww.com/INF/E796). Importantly, convalescent SARS-CoV-2 RBD neutralizing antibody titers positively correlated with SARS-CoV-2 S IgG concentrations (Fig. 2C). Additionally, no associations were detected between SARS-CoV-2 S IgG concentrations in the acute and convalescent phases and patient age (see Figure C,D, Supplemental Digital Content 2 http://links.lww.com/INF/E796).
Interestingly, SARS-CoV-2 S IgG antibodies were detected up to 80 days post-symptom onset and no correlations were found between SARS-CoV-2 S IgG antibodies in convalescence and the time post-symptom onset (see Figure, Supplemental Digital Content 3 http://links.lww.com/INF/E797).
Evaluation of Lymphocyte Subset Distribution and T cell Activation Markers
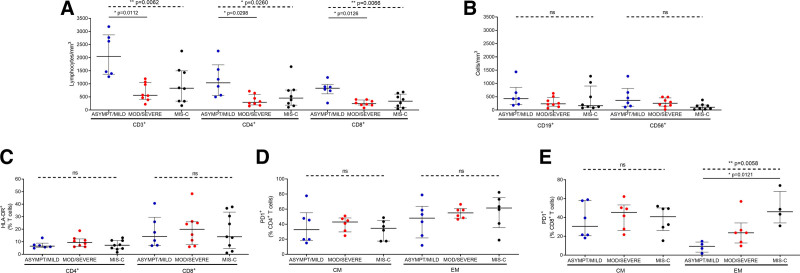
The absolute counts of CD3+, CD4+ and CD8+ T lymphocyte subsets were significantly lower in the moderate/severe vs. asymptomatic/mild groups (Fig. 3A). Conversely, CD19+ (B cells) and CD3-CD56+ (NK cells) were not significantly different between the severity groups (Fig. 3B). Both CD4+ and CD8+ T cells were activated in children with COVID-19 (% HLADR+CD4+ T cells >5% and % HLADR+CD8+ T cells >12%), but no significant difference was found in the percentage of activated cells between the severity groups (Fig. 3C). In contrast, the percentages of exhausted cells (PD1+-expressing cells) were similar between the severity groups within the CD4+T cell subsets (Fig. 3D) but augmented in effector memory CD8+ T cells in the MIS-C group (Fig. 3E).
FIGURE 3.
Cellular immune profiles in pediatric patients with COVID-19. (A) Absolute numbers of T cell populations (CD3+, CD4+ and CD8+) and (B) absolute number of B cells (CD19+) and NK cells (CD56+), in asymptomatic/mild (n = 6), moderate/severe (n = 8) and MIS-C (n = 8) groups. (C) Activated CD4+ and CD8+ T cells (HLADR+) and (D, E) exhausted cells (PD1+) within CD4+ and CD8+T cell subpopulations; central memory T cells (CM, CD45RA-CD27+) and effector memory T cells (EM, CD45RA-CD27-). Lines represent median values with interquartile range. P values were calculated using the Kruskal-Wallis test (dashed line) and Dunn’s multiple comparison post-test (solid line) for comparisons between severity groups. P values showing significant post-test comparisons are displayed. A P value <0.05 was considered significant; * P < 0.05, ** P < 0.01, ns, non-significant. ASYMPT, asymptomatic; MOD, moderate.
Cytokine Profiles Associated with Disease Severity
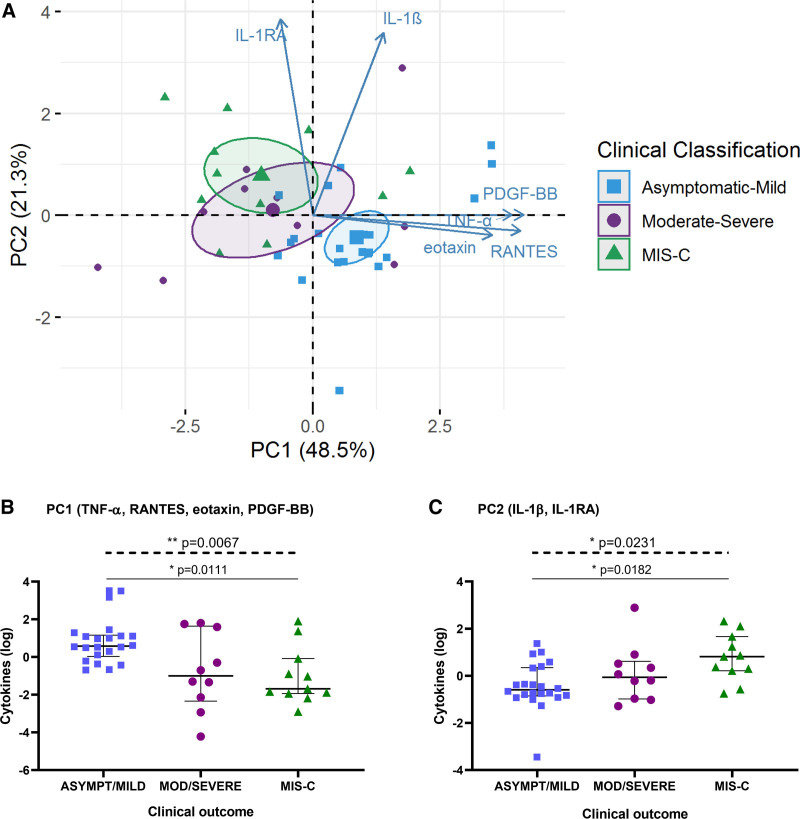
To identify cytokine profiles associated with disease severity, PCA was performed for cytokines, chemokines and growth factors that mainly contributed to interpatient variation across the severity groups during acute infection (Fig. 4; see Figure, Supplemental Digital Content 4 http://links.lww.com/INF/E798). Patients in the moderate/severe group demonstrated greater heterogeneity in the overall cytokine profile within the group, as opposed to the asymptomatic/mild and MIS-C groups that showed more homogeneity (Fig. 4A). Two distinct cytokine profiles were identified with a cumulative percentage of variance of 69.8%. Tumor necrosis factor-α (TNF-α), RANTES, eotaxin and platelet-derived growth factor BB (PDGF-BB) mainly contributed to the first principal component (PC1), whereas interleukin (IL)-1β and IL-1RA had an orthogonal contribution to the second principal component (PC2). Interestingly, these two distinct cytokine combinations were differentially associated with COVID-19 severity, with decreasing PC1 scores and augmenting PC2 scores across the severity groups (Fig. 4B,C). Patients with MIS-C exhibited lower scores for PC1 and higher scores for PC2 than patients with asymptomatic/mild disease (P = 0.0111 and P = 0.0182, respectively), while cytokine profiles from patients with moderate/severe disease were scattered between the other groups.
FIGURE 4.
Distinct cytokine profiles associated with COVID-19 clinical outcomes in children. (A) PCA of cytokines/chemokines/growth factors that most contributed to interpatient variation according to the severity groups: asymptomatic/mild, moderate/severe and MIS-C. The immune mediators were measured in serum samples from children with COVID-19 during the acute phase of the disease or during MIS-C. Ellipses represent the 95% confidence interval for each group centroid. Levels of cytokines derived from (B) PC1 and (C) PC2 for patients in each severity group. Lines represent median values with interquartile range. P values were calculated using the Kruskal-Wallis test (dashed line) and Dunn’s multiple comparison post-test (solid line). A P value <0.05 was considered significant; * P < 0.05, ** P < 0.01. P values showing significant post-test comparisons are displayed. MIS-C, multisystem inflammatory syndrome in children; PCA, principal component analysis.
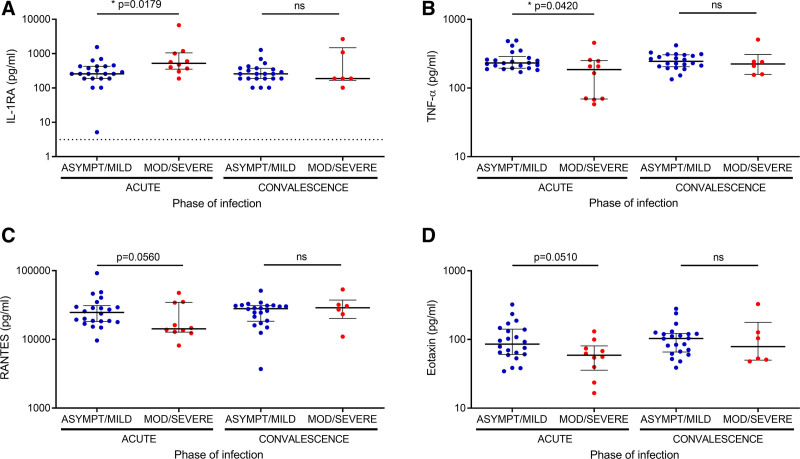
When cytokine levels were analyzed during acute and convalescent phases for COVID-19 patients, we found that acute serum levels of IL-1RA were significantly increased in patients with moderate/severe vs. asymptomatic/mild disease (Fig. 5A). Conversely, acute serum levels of TNF-α were significantly decreased in moderate/severe patients, and a trend towards diminished serum levels of RANTES and eotaxin was also observed (Fig. 5B–D). Remarkably, no differences were observed in the levels of these cytokines between the severity groups in convalescent serum, suggesting that the differential cytokine profile in each severity group was due to acute SARS-CoV-2 infection (Fig. 5A–D). Furthermore, serum levels of IL-6, a cytokine that has been associated with severe-to-critical COVID-19,14 did not differ based on the phase of infection or severity, while serum levels of IP-10 were increased in moderate/severe patients in the acute phase as compared to the convalescent phase (P = 0.0464) (see Figure, Supplemental Digital Content 5 http://links.lww.com/INF/E799).
FIGURE 5.
Cytokines and chemokines associated with distinct profiles in children with differing COVID-19 severity. (A) IL-1RA, (B) TNF-α, (C) RANTES and (E) eotaxin, were measured in serum samples from children with COVID-19 during the acute phase of the disease and during convalescence. Lines represent median values with interquartile range. Lower dotted lines indicate minimum detectable levels for each cytokine. For IL-1RA, TNF-α, RANTES, and eotaxin the minimum detectable levels were 3.16 pg/ml, 1.13 pg/ml, 3.98 pg/ml, and 0.05 pg/ml, respectively. P values were calculated using the Mann-Whitney test for comparisons between the severity groups. P values were calculated using the Wilcoxon matched pairs test for comparisons between acute and convalescent phases. A P value <0.05 was considered significant; * P < 0.05, ns, non-significant. TNF, Tumor necrosis factor.
DISCUSSION
In this study, carried out during the first wave of the COVID-19 pandemic, we found differences in laboratory parameters, host immune responses as well as cytokine profiles depending on disease severity in a pediatric population with COVID-19.
Laboratory abnormalities in children with COVID-19 included a reduction in the percentage of lymphocytes and an increase in CRP levels as disease severity augmented, while total leukocyte counts remained unaffected across the severity groups. Previous reports have described elevated CRP as a risk factor for severe COVID-19 in children.15–18 Lymphopenia has been described during SARS-CoV-2 infection in children,5 in both inpatients and outpatients,16 and has been also described in MIS-C.19 We observed that lymphopenia was more frequent in moderate, severe and MIS-C patients than in children with mild disease. Furthermore, leukopenia has been described as a laboratory abnormality in children with COVID-19 and has been associated with prolonged hospitalization,15 though this finding has not been universally replicated.17 The present study did not find significant differences in white blood cell (WBC) counts across the severity groups.
Several studies have associated the presence of comorbidities in children with severe presentations of COVID that require intensive care and prolonged hospitalization,5,15,17,20,21 though most cases of MIS-C have been observed in previously healthy individuals without comorbidities.5,19,20 In agreement with these studies, we observed that the presence of comorbidities in children was not associated with the development of MIS-C.
Previous studies have analyzed the humoral immune response in children with COVID-19. It has been reported that children principally generate IgG antibodies against the S protein18 or against accessory proteins22 and exhibit reduced neutralizing activity compared to adults with severe disease.18 In contrast, other studies reported that children with asymptomatic or mild SARS-CoV-2 infection developed robust antibody responses against SARS-CoV-2 that were detectable 2−4 months after infection23 or beyond 12 months of infection.24 Interestingly, another study showed that children with severe disease developed a weak and delayed IgG antibody response against SARS-CoV-2, and a seronegative status was observed in 25% and 12% of convalescent and MIS-C children, respectively.20 Conversely, another study found that children with MIS-C exhibited higher titers of neutralizing antibodies against the SARS-CoV-2 S protein than those from children with COVID-19 and without MIS-C.25 In our study, we observed that most children developed a robust IgG antibody response against the SARS-CoV-2 S protein that was detectable for more than 2 months post-symptom onset. In addition, we found that IgG levels against SARS-CoV-2 S were not significantly different in children with distinct disease severity and did not correlate with patient age. Interestingly, we observed that 20.5% of children had detectable IgG antibodies against the SARS-CoV-2 S protein during acute infection that were likely due to the presence of cross-reactive immune responses against common cold coronaviruses26,27 and/or an early IgG response against SARS-CoV-2.11 Furthermore, we observed that 7.7% of children remained seronegative in convalescence, which was not directly associated with the presence of comorbidities producing immune deficiencies in these patients. In our study, MIS-C patients showed variable IgG levels against SARS-CoV-2 S and 41.7% of children exhibited detectable SARS-CoV-2 S IgG antibodies.
In addition to the humoral immune response, T-lymphocytes play a key role in host defense against SARS-CoV-2 infection. In our study, we observed a reduction of CD4+ and CD8+ T lymphocytes in children with moderate/severe disease as compared to children with asymptomatic/mild disease, and no difference in the percentage of HLADR+-expressing cells was observed across the severity groups. Furthermore, increased PD1+-expressing effector memory CD8+ T cells were observed in children with MIS-C. In agreement with our study, a report showed that COVID-19 patients had a reduction in absolute numbers of CD4+ and CD8+ T lymphocytes that displayed markers related to activation or exhaustion/senescence.28 Similarly, another study reported that activation and exhaustion markers on T lymphocytes were augmented in COVID-19 patients vs. controls but comparable between severity groups.29 Another report proposed that excessive exhaustion of CD8+ T cells in severe patients may reduce the cellular immune response to SARS-CoV-2.30 It has been suggested that the virus promotes excessive immune activation at disease onset that is followed by subsequent exhaustion of CD8+ T cells.31
The characterization of the immune mediators involved in SARS-CoV-2 infection in our pediatric population revealed different cytokine profiles depending on disease severity. Children with MIS-C exhibited an immune profile similar to that from moderate/severe patients but notably different from asymptomatic/mild children. The early production of IL-1RA, an inhibitory cytokine that suppresses proinflammatory cytokines and T lymphocyte responses, was higher in patients with moderate/severe disease and MIS-C. Interleukin-1RA production could affect induction of proinflammatory and antiviral cytokines during the early phase of SARS-CoV-2 infection, which may contribute to the switch from a controlled and protective immune environment to inflammation-induced tissue damage.32 Accordingly, we also found that TNF-α, RANTES, eotaxin and PDGF-BB levels were decreased in moderate/severe and MIS-C patients. Consistent with our results, higher levels of RANTES have been reported in mild vs. severe patients during follow-up studies of COVID-19.32 Likewise, the higher production of PDGF-BB in children with asymptomatic/mild COVID-19 may contribute to more efficient tissue healing, thus limiting virus-induced injury to the lungs or other tissues.33 Furthermore, it has been reported that patients who succumbed to SARS-CoV-2 infection produced decreased TNF-α and eotaxin than patients who survived infection.34 We also found higher levels of IL-1β in children with MIS-C in our pediatric population. It has been reported that this proinflammatory cytokine is increased in autoimmune inflammatory processes.35 In our study, serum levels of IL-6 did not significantly differ between the severity groups and phases of infection, suggesting that a classical cytokine storm may not be the major cause of severe illness in COVID-19 patients in our population. These results are consistent with some studies36,37 but contrasting with other reports in children,20,38–40 highlighting the need for further investigation in this area.
The decrease in TNF-α, RANTES and eotaxin observed in patients with moderate/severe SARS-CoV-2 infection or MIS-C may be the result of an exhausted T cell immune phenotype. T cell exhaustion is a state of T cell dysfunction characterized by sustained expression of inhibitory receptors and a transcriptional state distinct from that of functional effector or memory T cells.41 The increase of exhausted CD8+T cells and IL-1RA levels may contribute to poor viral clearance and bad prognosis.
Our study has several limitations. The small sample size precluded the analysis of the immune responses in each severity group separately. Furthermore, convalescent serum samples were available in a subpopulation of patients and restricted the study of humoral immune responses and cytokine profiles in convalescence to a smaller proportion of patients.
In conclusion, we showed that most children with COVID-19 in our study exhibited a robust humoral immune response against the SARS-CoV-2 S protein that was not correlated to disease severity. Distinct immune phenotypes and cytokine profiles characterized by a reduction of CD4+ and CD8+ T cells, increased exhausted effector memory CD8+ T cells, elevation of anti-inflammatory IL-1RA and suppression of pro-inflammatory cytokines were observed in moderate/severe and MIS-C patients. These results may provide insight into understanding the immune mechanisms related to pediatric COVID-19 as well as guide future investigations in the field.
ACKNOWLEDGEMENTS
The authors thank Nicolás Fioramonti for technical assistance and guidance with the R software.
Supplementary Material
Footnotes
This work was supported by Fundación Argentina de Infectología Pediátrica (FAIP) unrestricted grant.
The authors have no conflicts of interests to disclose.
L.B.T, M.M.C. and E.L.L. designed the study. M.M.C., L.E.T. and E.W.Y. enrolled patients. L.B.T., A.T., M.P.M., M.I.G., A.C., A.B.B. and P.L.A. performed experiments. L.B.T., A.T., M.M.C., M.P.M., M.I.G., F.A.B., S.T. and E.L.L. analyzed the data. L.B.T., A.T., M.M.C. and E.L.L. wrote the manuscript. All the authors revised the final manuscript.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (www.pidj.com).
Contributor Information
Laura Beatriz Talarico, Email: ltalarico@conicet.gov.ar.
Analía Toledano, Email: anat_21@hotmail.com.
María Marta Contrini, Email: mmcontrini@gmail.com.
Lidia E. Torrado, Email: ltorrado@intramed.net.
María Paula Martínez, Email: mpaumartinez@hotmail.com.
María Isabel Gaillard, Email: migaillard@yahoo.com.
Ana Caratozzolo, Email: anacaratozzolo@hotmail.com.
Alana Brooke Byrne, Email: alanabyrne@conicet.gov.ar.
Florencia Agustina Bonnin, Email: florenciabonn616@gmail.com.
María Soledad Tineo, Email: mstineo@gmail.com.
Eduardo Walter Yfran, Email: walter10yfran@gmail.com.
Patricio Leandro Acosta, Email: patricioacosta@conicet.gov.ar.
REFERENCES
- 1.Hagmann SHF. COVID-19 in children: more than meets the eye. Travel Med Infect Dis. 2020;34:101649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.She J, Liu L, Liu W. COVID-19 epidemic: disease characteristics in children. J Med Virol. 2020;92:747–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kabeerdoss J, Pilania RK, Karkhele R, et al. Severe COVID-19, multisystem inflammatory syndrome in children, and Kawasaki disease: immunological mechanisms, clinical manifestations and management. Rheumatol Int. 2021;41:19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brodin P. Why is COVID-19 so mild in children? Acta Paediatr. 2020;109:1082–1083. [DOI] [PubMed] [Google Scholar]
- 5.Suratannon N, Dik WA, Chatchatee P, et al. COVID-19 in children: heterogeneity within the disease and hypothetical pathogenesis. Asian Pac J Allergy Immunol. 2020;38:170–177. [DOI] [PubMed] [Google Scholar]
- 6.Fialkowski A, Gernez Y, Arya P, et al. Insight into the pediatric and adult dichotomy of COVID-19: age-related differences in the immune response to SARS-CoV-2 infection. Pediatr Pulmonol. 2020;55:2556–2564. [DOI] [PubMed] [Google Scholar]
- 7.Nickbakhsh S, Mair C, Matthews L, et al. Virus-virus interactions impact the population dynamics of influenza and the common cold. Proc Natl Acad Sci USA. 2019;116:27142-27150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.WHO COVID-19 Case definition. Available at: https://apo.who.int/publications/i/item/WHO-2019-nCoV-Surveillance_Case_Definition-2020.1.
- 9.Research Use Only 2019-Novel Coronavirus (2019-nCoV) Real-time RT-PCR Primers and Probes. National Center for Immunization and Respiratory Diseases (NCIRD), Division of Viral Diseases. Available at: https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html.
- 10.Gentile A, Juarez MDV, Lucion MF, et al. COVID-19 in Children: Correlation Between Epidemiologic, Clinical Characteristics, and RT-qPCR Cycle Threshold Values. Pediatr Infect Dis J. 2022;41:666–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ojeda DS, Gonzalez Lopez Ledesma MM, Pallares HM, et al. Emergency response for evaluating SARS-CoV-2 immune status, seroprevalence and convalescent plasma in Argentina. PLoS Pathog. 2021;17:e1009161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossi AH, Ojeda DS, Varese A, et al. Sputnik V vaccine elicits seroconversion and neutralizing capacity to SARS-CoV-2 after a single dose. Cell Rep Med. 2021;2:100359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.World Health Organization 2021. Living guidance for clinical management of COVID-19. Available at: https://www.who.int/publications/i/item/WHO-2019-nCoV-clinical-2021-2.
- 14.Potere N, Batticciotto A, Vecchie A, et al. The role of IL-6 and IL-6 blockade in COVID-19. Expert Rev Clin Immunol. 2021;17:601–618. [DOI] [PubMed] [Google Scholar]
- 15.Kari JA, Shalaby MA, Albanna AS, et al. Coronavirus disease in children: a multicentre study from the Kingdom of Saudi Arabia. J Infect Public Health. 2021;14:543–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alkan G, Sert A, Emiroglu M, et al. Evaluation of hematological parameters and inflammatory markers in children with COVID-19. Ir J Med Sci. 2022;191:1725–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graff K, Smith C, Silveira L, et al. Risk factors for severe COVID-19 in children. Pediatr Infect Dis J. 2021;40:e137–e145. [DOI] [PubMed] [Google Scholar]
- 18.Weisberg SP, Connors TJ, Zhu Y, et al. Distinct antibody responses to SARS-CoV-2 in children and adults across the COVID-19 clinical spectrum. Nat Immunol. 2021;22:25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramaswamy A, Brodsky NN, Sumida TS, et al. Immune dysregulation and autoreactivity correlate with disease severity in SARS-CoV-2-associated multisystem inflammatory syndrome in children. Immunity. 2021;54:1083–1095.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sananez I, Raiden SC, Algieri SC, et al. A poor and delayed anti-SARS-CoV2 IgG response is associated to severe COVID-19 in children. EBioMedicine. 2021;72:103615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gotzinger F, Santiago-Garcia B, Noguera-Julian A, et al. COVID-19 in children and adolescents in Europe: a multinational, multicentre cohort study. Lancet Child Adolesc Health. 2020;4:653–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hachim A, Gu H, Kavian O, et al. The SARS-CoV-2 antibody landscape is lower in magnitude for structural proteins, diversified for accessory proteins and stable long-term in children. medRxiv. 2021;2021.01.03.21249180. [Google Scholar]
- 23.Garrido C, Hurst JH, Lorang CG, et al. Asymptomatic or mild symptomatic SARS-CoV-2 infection elicits durable neutralizing antibody responses in children and adolescents. JCI Insight. 2021;6:e150909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dowell AC, Butler MS, Jinks E, et al. Children develop robust and sustained cross-reactive spike-specific immune responses to SARS-CoV-2 infection. Nat Immunol. 2022;23:40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson EM, Diorio C, Goodwin EC, et al. Severe Acute Respiratory Syndrome-Coronavirus-2 (SARS-CoV-2) Antibody Responses in Children With Multisystem Inflammatory Syndrome in Children (MIS-C) and Mild and Severe Coronavirus Disease 2019 (COVID-19). J Pediatric Infect Dis Soc. 2021;10:669–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mateus J, Grifoni A, Tarke A, et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng KW, Faulkner N, Cornish GH, et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370:1339–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Biasi S, Meschiari M, Gibellini L, et al. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat Commun. 2020;11:3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christensen EE, Jorgensen MJ, Nore KG, et al. Critical COVID-19 is associated with distinct leukocyte phenotypes and transcriptome patterns. J Intern Med. 2021;290:677–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng HY, Zhang M, Yang CX, et al. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol Immunol. 2020;17:541–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azkur AK, Akdis M, Azkur D, et al. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy. 2020;75:1564–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Y, Qin L, Zhang P, et al. Longitudinal COVID-19 profiling associates IL-1RA and IL-10 with disease severity and RANTES with mild disease. JCI Insight. 2020;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan YH, Fong SW, Poh CM, et al. Asymptomatic COVID-19: disease tolerance with efficient anti-viral immunity against SARS-CoV-2. EMBO Mol Med. 2021;13:e14045:e139834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horspool AM, Kieffer T, Russ BP, et al. Interplay of antibody and cytokine production reveals CXCL13 as a potential novel biomarker of lethal SARS-CoV-2 infection. mSphere. 2021;6:e01324–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao R, Zhou H, Su SB. A critical role for interleukin-1beta in the progression of autoimmune diseases. Int Immunopharmacol. 2013;17:658–669. [DOI] [PubMed] [Google Scholar]
- 36.Remy KE, Mazer M, Striker DA, et al. Severe immunosuppression and not a cytokine storm characterizes COVID-19 infections. JCI Insight. 2020;5:e140329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson JG, Simpson LJ, Ferreira AM, et al. Cytokine profile in plasma of severe COVID-19 does not differ from ARDS and sepsis. JCI Insight. 2020;5:e140289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun D, Li H, Lu XX, et al. Clinical features of severe pediatric patients with coronavirus disease 2019 in Wuhan: a single center’s observational study. World J Pediatr. 2020;16:251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Venkataraman A, Kumar NP, Hanna LE, et al. Plasma biomarker profiling of PIMS-TS, COVID-19 and SARS-CoV2 seropositive children - a cross-sectional observational study from southern India. EBioMedicine. 2021;66:103317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu W, Yang L, Li X, et al. Early immune responses and prognostic factors in children with COVID-19: a single-center retrospective analysis. BMC Pediatr. 2021;21:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12:492–499. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.