Abstract
The aims of the present study were (i) to develop and test a sensitive and reproducible method for the study of gene expression in staphylococci and (ii) to study the expression of five housekeeping genes which are involved in nucleic acid metabolism (gmk, guanylate kinase; the dihydrofolate reductase [DHFR] gene), glucose metabolism (tpi, triosephosphate isomerase), and protein metabolism (the 16S rRNA gene; hsp-60, heat-shock protein 60) during in vitro exponential and stationary growth. A modified method for instant mRNA isolation was combined with gene quantification via Taqman real-time quantitative PCR. The detection limit of our method was 10 copies of RNA. The average intersample variability was 16%. A 10-fold increase in the expression of the hsp-60 gene was induced by exposure to a 10°C heat shock (37 to 47°C) for 10 min. During in vitro growth, the expression of all five housekeeping genes showed rapid up-regulation after inoculation of the bacteria in brain heart infusion medum and started to decline during the mid-exponential-growth phase. Maximal gene expression was 110- to 300-fold higher than gene expression during stationary phase. This indicates that housekeeping metabolism is a very dynamic process that is extremely capable of adapting to different growth conditions. Expression of the 16S rRNA gene decreases significantly earlier than that of other housekeeping genes. This confirms earlier findings for Escherichia coli that a decline in bacterial ribosomal content (measured by 16S rRNA gene expression) precedes the decline in protein synthesis (measured by mRNA expression).
In recent years, coagulase-negative staphylococci (CNS) have emerged as major pathogens that are mainly associated with indwelling or implanted foreign body infections (22, 29, 30, 33). Their impact on public health is enormous (24, 31). It remains enigmatic why these normally innocent skin saprophytes become virulent in association with indwelling foreign bodies (17). CNS infections seem to be the result of a complex interaction between bacterium-related factors, host-related factors, and foreign body-related-factors. Genes involved in cell accumulation (13, 33, 34) and in initial adhesion (12) are presumed virulence factors in initial foreign body colonization and biofilm formation. A state of bacterial dormancy and a suppressed housekeeping metabolism are hypothesized to contribute to the persistent nature of foreign body-related CNS infections (5, 25, 26).
For study of the pathogenesis of infectious diseases, researchers have access to a rapidly growing amount of genetic information. However, the exact links between the information encoded in the genome and the final virulence and housekeeping behavior of bacteria remain unclear. Methods to unravel these links are mutagenesis and the study of gene expression. Mutagenesis is a valuable phenotypical assay (1). However, mutations in important genes may lead to only minor phenotypical changes or to lethal mutants. In these cases it is not possible to draw firm conclusions on the roles of these genes in the pathogenesis of infections. Current methods to study gene expression such as Northern hybridization, quantitative competitive PCR, and RNase protection assays are laborious, have a small dynamic range, and lack sensitivity, with a detection limit ranging from 105 to 108 mRNA copies (14, 35). The aims of the present study were (i) to develop and to test a sensitive and easy-to-perform method for the study of gene expression in staphylococci and (ii) to explore the expression of several genes involved in basic housekeeping metabolism in CNS during the exponential- and stationary-growth phases in vitro and under different conditions in order to create a reference for further gene expression studies.
Given the very short half-life of mRNA, gene expression experiments require a rapid technique of RNA isolation. Such a technique was optimized and combined with gene quantification by Taqman quantitative PCR. Taqman quantitative PCR has proven to be a very accurate and reproducible tool for gene quantification (9, 11). It is superior to both Northern hybridization (14) and RNase protection assays (35) in mRNA quantification. Results obtained by Taqman PCR correlate well with those obtained by conventional methods but have a larger dynamic range and a much higher sensitivity (14, 35).
(Parts of this investigation were presented orally [O302 and O303] at the 11th European Congress of Clinical Microbiology and Infectious Diseases, Istanbul, Turkey, 1 to 4 April 2001.)
MATERIALS AND METHODS
Bacterial strain.
For all studies a previously described clinical Stapylococcal epidermidis strain (called 10b) was used (32). This strain was isolated from a patient with a proven catheter-related bloodstream infection. It was identified as S. epidermidis using conventional laboratory techniques, the Staph-zyme kit (Rosco Diagnostica, Taastrup, Denmark), and tRNA intergenic spacer length polymorphism (20). It is a biofilm-producing strain (growth as black colonies on Congo red agar [38]).
Gene identification.
Using the partial sequences of the triosephosphate isomerase (tpi) and the guanylate kinase (gmk) genes of Staphylococcus aureus (8), we identified the complete sequences of these genes in S. aureus (http://www.sanger.ac.uk/Projects/S_aureus). On the basis of these sequences, primers were designed and used to amplify the similar genes in S. epidermidis 10b under low-stringency conditions (annealing temperature of 55°C). PCR was performed on a GeneAmp PCR System 9700 (Perkin-Elmer Applied Biosystems, Foster City, Calif.). Primers for the tpi gene were 5Cy′-GGTCATTCTGAACGTCGTGA-3′ and 5Cy′-TGATAAACGATACGTCCTGCAC-3′. Primers for the gmk gene were 5Cy′-GGATAATGAAAAAGGATTGTTAATCG-3′ and 5Cy′-GCTTCTACGCGCTCTCTTTT-3′. All primers and probes were provided by Eurogentec (Seraing, Belgium). For gene sequencing, we used the Thermo Sequenase fluorescent-labeled primer cycle sequencing kit with 7-deaza-dGTP (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, England). The sequences of the 16S rRNA gene (EMBL D83363) and the heat-shock protein 60 gene (hsp-60 [EMBL AF029245]) were retrieved from the National Center for Biotechnology Information (NCBI) GenBank. The sequence of the dihydrofolate reductase (DHFR) gene has been published elsewhere (6).
Cloning in plasmids and quantification of number of copies of the plasmid.
All genes were cloned in the pGEM-T easy vector system (Promega, Madison, Wis.) according to the manufacturer's instructions. Before cloning in the pGEM-T easy vector, the genes were amplified. Pure plasmid DNA was obtained using the High Pure Plasmid Isolation Kit (Roche Diagnostics GmbH, Mannheim, Germany). For the gmk and tpi genes, fragments of 585 and 709 bp, respectively, were amplified with the primers mentioned above. For the 16S rRNA gene a fragment of 1,443 bp was amplified with 5′-TACATGCAAGTCGAGCGAAC-3′ and 5′-AATCATTTGTCCCACCTTCG-3′; for the hsp-60 gene a fragment of 553 bp was amplified with 5′-AGCAACAGTTTTAGCACAATCAA-3′ and 5′-TGTTCCACGCATACGGTTTA-3′; and for the DHFR gene a fragment of 486 bp was amplified with 5′-TTGTCGCTCACGATAAACAAA-3′ and 5′-TCCCTTTTCTACGCACTAAATGT-3′. Gene quantification was performed with the GeneQuant RNA/DNA calculator (Amersham Pharmacia Biotech) at a wavelength of 260 nm. The number of gene copies per microliter of plasmid was 2.84 × 1010 (95% confidence interval [95% CI], 2.76 × 1010 to 2.91 × 1010) for the tpi gene, 2.66 × 1010 (95% CI, 2.59 × 1010 to 2.73 × 1010) for the gmk gene, 3.77 × 1010 (95% CI, 3.72 × 1010 to 3.82 × 1010) for the DHFR gene, 2.11 × 1010 (95% CI, 2.06 × 1010 to 2.17 × 1010) for the 16S rRNA gene, and 2.08 × 1010 (95% CI, 2.00 × 1010 to 2.17 × 1010) for the hsp-60 gene.
RNA isolation and cDNA synthesis.
All cultures were grown in brain heart infusion (BHI) (Oxoid Ltd., Basingstoke, Hampshire, England) in a shaking incubator at 37°C. These cultures, with a maximum of 109 CFU and a volume ranging from 10 to 1,000 μl, were rapidly cooled on ice. Cultures were centrifuged for 5 min at 3,600 × g at −4°C (RC 5B Plus; Sorvall, Newtown, Conn.). The initial steps of RNA isolation were performed as described by Cheung et al. (2) with some modifications (partially adapted from reference 7). The pellet was suspended in 500 μl of acidified phenol-chloroform (5:1) (pH 4.5) (Ambion, Austin, Tex.) at room temperature and added with 500 μl of NAES buffer (50 mM sodium acetate [pH 5.1], 10 mM EDTA, 1% sodium dodecyl sulfate) to a FastRNA tube-blue (Bio 101, Carlsbad, Calif.). These silica bead-containing tubes were shaken for 23 s at 6,000 rpm in a FastPrep instrument (FP 120; Bio 101, Savant, Holbrook, N.Y.). After shaking, the tubes were centrifuged for 5 min at 12,000 × g and 90% of the supernatant (450 μl) was precipitated with 520 μl of isopropyl alcohol and 35 μl of 3 M sodium acetate. The pellet was washed with 70% ethanol and resuspended in 100 μl of RNase-free water. For purposes of comparison, some RNA extractions were performed with the Trizol reagent (Gibco BRL, Grand Island, N.Y.) instead of acid phenol with NAES.
The sample was further purified with the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Samples were treated with RNase-free DNase (Qiagen) on the RNeasy columns according to the manufacturer's instructions. RNA was finally dissolved in 60 μl of RNase-free water.
For reverse transcription we used 100 U of Moloney murine leukemia virus with the supplied buffer (Promega), 20 U of RNasin (Promega), 100 μM random hexamers (Amersham Pharmacia Biotech), 1 mM each deoxynucleoside triphosphate, and 3 μl of RNA sample per 20-μl reaction volume. The final reaction volume was 20, 40, 60, or 120 μl. Reaction conditions were as follows: preheating of the RNA sample for 10 min at 72°C, addition of the reaction mixture on ice, heating for 1 h at 42°C, heating at 99°C for 2 min for enzyme denaturation, and rapid cooling to 4°C.
Taqman quantitative PCR.
Gene quantification was performed on the ABI Prism 7700 Sequence Detection System (Perkin-Elmer Applied Biosystems). Taqman primers and probes were designed using Primer Express 1.0 from Perkin-Elmer Applied Biosystems. Primers and probes are summarized in Table 1. Probes were labeled with the reporter dye 6-carboxyfluorescein (6′-FAM) at the 5′ end and with the quencher dye 6-carboxytetramethylrodamine (TAMRA) at the 3′ end. Quantitative PCR was performed with 2 μl of cDNA, 12.5 μl of 2× Taqman PCR master mix (Perkin-Elmer Applied Biosystems), 900 nmol of each primer, and a 200-nmol probe in a final volume of 25 μl. Thermal cycling conditions were as follows: 2 min at 50°C, 10 min at 95°C followed by 45 repeats of 15 s at 95°C, and 1 min at 60°C. Data collection was performed during each annealing phase. During each run, a standard dilution of the plasmid with a known quantity was included to permit gene quantification using the supplied software according to the instructions of the manufacturer. In each run a negative control (distilled water) and an RNA sample without a reverse transcriptase step (to determine genomic DNA contamination) was included. For each RNA isolation, measurements of gene expression were taken three times, and the mean of these values was used for further analysis.
TABLE 1.
Taqman primers and probes (5′→3′)
| Gene | Forward primer | Probe: labeled FAM-5′ and 3′-TAMRA | Reverse primer |
|---|---|---|---|
| gmk | AAGGTGCTAAGCAAGTAAGAAAGAAATT | ATGCGTTGTCATATTTTTAGCGCCTCCA | CAACAAGACGTTCTTTCAAGTCATCT |
| DHFR gene | GGGAAACCATTGCCAAATAGAC | CGTCGTACTCACTAACCAAGCTTCATTTCACC | CGAATAACGTTTGTCCTCCAAATA |
| tpi | CATCTGATAAACCTTCGACAGCTTT | CCAGCTTCACGTTCTTCATCAGATTCACC | TGCTATCTTCAATCACGGTATGACA |
| 16S rRNA gene | TACACACCGCCCGTCACA | CACCCGAAGCCGGTGGAGTAACC | CTTCGACGGGCTAGCTCCAAAT |
| hsp-60 | TCTTAAGAATGTTACAAGTGGTGCAA | TTGCACTGCTTTGTCAATACCTTGTCTTAAGCCT | AATCTCATGGAGCGCTTCTATAGC |
Quantification of bacteria.
Simultaneously with the RNA isolation, the bacteria were quantified. A 10-fold dilution of the inoculum was made in saline on ice. An appropriate dilution was counted in a Bürker (Marienfeld, Germany) counting chamber (average of 20 high resolution fields). At the same time at least six tryptone soy agar plates (Oxoid) were inoculated with a Spiral Plater (Spiral Systems, Cincinnati, Ohio). The number of bacteria was defined as the average of each quantitative culture. The correlation between manual cell counting and quantitative culture was high.
Relative gene expression.
It is obvious that more bacteria have the potential to produce more RNA. For this reason, the number of copies of cDNA per milliliter (as a measure of the amount of mRNA) obtained by the Taqman PCR was divided by the number of bacteria per milliliter. This quotient (number of cDNA copies per CFU) represents the amount of RNA expressed per viable bacterium.
Exponential growth, heat shock, and glucose challenge.
To obtain a culture in exponential-growth phase, 20 μl of an overnight-grown culture in BHI was inoculated in 5 ml of BHI and incubated for 45 min. Cultures were grown in a shaking incubator at 37°C. Heat shock was performed by increasing the temperature for 10 min to 47°C (versus 37°C in controls). To study the effect of glucose, glucose was added to an overnight-grown culture to a final concentration of 5%. All experiments were carried out at least in duplicate and were independently repeated.
During exponential and stationary growth, glucose concentration was measured with the Kodak Ektachem 700 Analyser C (Eastman Kodak Company, Rochester, N.Y.), and pH was measured with the PHM82 Standard pH Meter (Radiometer A/S, Copenhagen, Denmark).
Contamination with gDNA.
Genomic DNA (gDNA) was isolated from an overnight culture using the Wizard Genomic DNA Purification Kit (Promega) according to the instructions of the manufacturer. The number of copies of gDNA was quantified using the Taqman PCR with the primers, the probe, and the plasmid of the gmk gene. Bacteria were assumed to contain 1 copy of gmk gDNA per bacterium. RNA was isolated from both an early-exponential-phase culture (t = 45 min) and a late-stationary-phase culture (t = 16 h). Bacteria were counted in a Bürker counting chamber. A number of gDNA copies were added to the sample during RNA isolation, after washing of the RNA pellet with 70% ethanol (see “RNA isolation and cDNA synthesis” above), in order to obtain gDNA contamination (expressed as the number of copies of gmk gDNA) of 2, 5, 10, 50, and 100 times the number of bacteria.
Statistical methods.
To facilitate the comparison between the expressions of the different genes, the results were rescaled as a percentage with respect to the first measurement at 45 min. The results at 45 min have been given the value of 100%. To evaluate the evolution of the expression of each gene over time, only those time points with at least four independent observations were used. The 95% CIs were calculated for these differences. Since the observations within one experiment were not independent, a random-effects model was used, by which the correlation between the two subpopulations of one experiment (i.e., two different RNA extractions from the same culture at the same time) was taken into account. Weighted least squares have been used to assure that the error structure is constant over the different time values (the reciprocals of the estimated variance of the residuals of the ordinary least squares solution have been used as weights). Approximate t tests, with the degrees-of-freedom calculations detailed by Kenward and Roger (15), were used to estimate all reported pairwise differences. The expression of the 16S rRNA gene has been compared with the expressions of the other four genes, using the difference of expression between the 16S rRNA gene and the other genes at each time point as the response in the statistical analysis. The same statistical methodology has been used as for the analysis of the evolution over time within the expression of a particular gene. All analyses were performed with the PROC MIXED procedure (SAS, version 8.1).
RESULTS
gmk gene and tpi gene sequences.
Recently, the S. epidermidis sequences of the gmk gene (EMBL AF270133; bp 845 to 1468) and the tpi gene (EMBL AF269838; bp 2321 to 3082) became available at the NCBI GenBank. These sequences are 98% identical to the partial sequences of the gmk and tpi genes that we found in S. epidermidis strain 10b.
The homologies of the sequences in S. epidermidis 10b and S. aureus at the protein level were 91 and 87% for the gmk and tpi genes, respectively. This strongly suggests the same protein structure and gene function.
Dynamic range and reproducibility of RNA isolation and Taqman PCR.
RNA isolation was performed on samples containing 104 to 109 CFU. With samples containing more than 109 CFU, extraction efficacy decreased due to saturation of the Qiagen columns. Repeated RNA extractions of the same sample always gave similar results, showing good interassay reproducibility of the method (average variability, 16%; 95% CI, 13 to 18.9%; median variability, 13.5%; range, 0.8 to 39.8%). Quantification by Taqman PCR was also very stable, resulting in high intra-assay reproducibility (mean variability, 5.6%). The dynamic range of the Taqman PCR was between 0 and 107 copies. In the range of 10 copies, results were less reproducible due to the statistical variance inherent to low numbers. Changes in gene expression as small as 30% were detected and confirmed by repeated assays.
Contamination with gDNA—comparison with other methods of RNA isolation.
With the method described by Cheung et al. (2), the RNeasy kit without DNase treatment (Qiagen), or the Trizol reagent, the amount of gDNA contamination was within the same magnitude as the amount of mRNA for those genes that had the lower range of expression (the DHFR gene and gmk). For this reason these extraction methods are less useful in gene expression experiments, for which high sensitivity is needed. With the method described in this report, contamination with gDNA is between 1 and 2% of the total amount of mRNA for the gmk gene and less than 1% for the other genes during both the exponential- and stationary-growth phases.
Supplementary addition of 5, 50, and 100 times more copies of gDNA than the number of bacteria in the exponential-phase culture resulted in average residual gDNA contamination of 0.52, 1.22, and 3.76%, respectively, for the gmk gene and 0.46, 1.25, and 4.7%, respectively, for the DHFR gene. Addition of 0, 2, 5, 10, 50, and 100 times more copies of gDNA than the number of bacteria in the stationary-phase culture resulted in average residual gDNA contamination of 0.65, 7.16, 4.67, 19.88, 34.85, and 34.3%, respectively, for the gmk gene and 0.57, 6.51, 6.91, 18.02, 59.7, and 55.38%, respectively, for the DHFR gene. For the 16S rRNA gene, residual gDNA contamination was less than 1% after addition of as much as 100 times more gDNA copies than bacteria, both in the exponential-growth phase and in the stationary-growth phase.
Expression of housekeeping genes after heat shock.
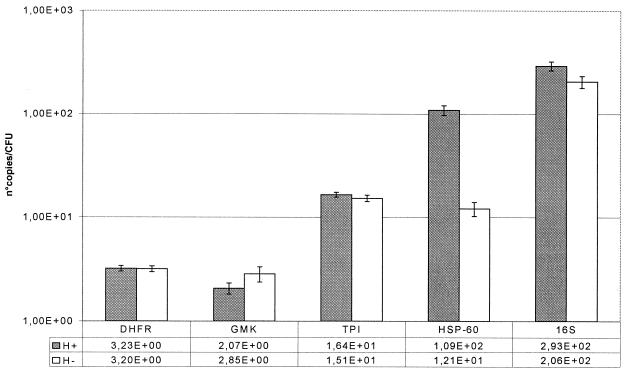
Changes in expression after heat shock during the exponential-growth phase are summarized in Fig. 1. After a heat shock of 10°C given for 10 min, the expression of the hsp-60 gene increased 9-fold (P = 1.82 × 10−13) and the expression of the 16S rRNA gene increased slightly (1.4-fold [P = 0.001]). The expression of the tpi gene and the DHFR gene did not change significantly, and the expression of the gmk gene decreased slightly (0.7-fold [P = 0.009]).
FIG. 1.
Gene expression levels after a heat shock of 10°C given for 10 min. Shaded bars, heat-shocked culture (47°C); open bars, controls (37°C). Each bar represents the number of copies of mRNA per CFU; that value is also given below the bar graph.
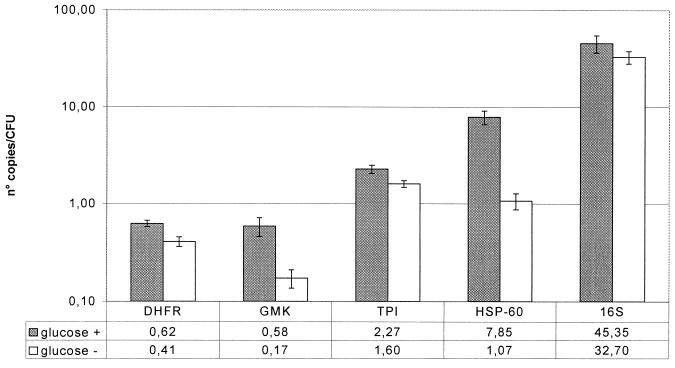
Expression of housekeeping genes after challenge with glucose.
Changes in expression after addition of glucose to a final concentration of 5% to a stationary-phase culture are summarized in Fig. 2. Ten minutes after addition of glucose, there was a general increase in the expression of all five housekeeping genes. The expression of the DHFR gene increased 1.5-fold (P = 2.41 × 10−7); that of the gmk gene increased 3.4-fold (P = 6.67 × 10−6); that of the tpi gene, 1.4-fold (P = 4.36 × 10−5); that of the hsp-60 gene, 7.3-fold (P = 3.86 × 10−9); and that of the 16S rRNA gene, 1.4-fold (P = 0.02).
FIG. 2.
Effect of glucose challenge in a stationary-phase culture. Shaded bars, gene expression in a stationary-phase culture after a glucose challenge (final concentration, 5%) for 10 min. Open bars, expression levels of the same culture without glucose challenge. Each bar represents the number of copies of mRNA per CFU; that value is also given below the bar graph.
Expression of housekeeping genes during exponential and stationary growth in vitro.
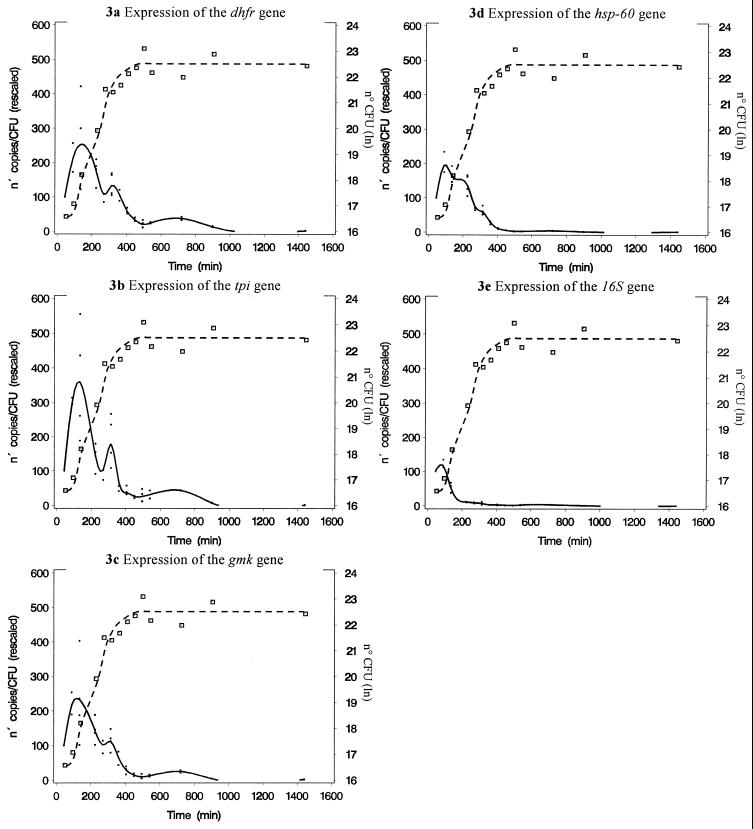
The expression of the housekeeping genes studied during the exponential- and stationary-growth phases is summarized in Fig. 3 and Table 2. The expression of all housekeeping genes increased rapidly after inoculation of the culture. Expression reached a maximum during early-exponential growth. Thereafter, a gradual decline was noticed. The decline in gene expression temporarily decelerated during the switch from the mid- to the late-exponential-growth phase (time point, 315 min). For the tpi gene, and to a lesser extent for the gmk and DHFR genes, there was a slight increase in gene expression at that time point. A rapid depletion of glucose in the culture medium and a dip in culture pH (6.5 versus the baseline of 7.1) were also observed at this time point. Maximal and minimal gene expression differed remarkably. The expression of the gmk gene declined 149-fold; that of the DHFR gene declined 112-fold; that of the tpi gene, 244-fold; that of the hsp-60 gene, 297-fold; and that of the 16S rRNA gene, 281-fold. The absolute expression (given as the number of cDNA copies per CFU) of the 16S rRNA gene (minimum, 7.7; maximum, 4.2 × 103) was higher than the expression of the hsp-60 gene (minimum, 0.4; maximum, 124) and much higher than the expression of the gmk gene (minimum, 0.05; maximum, 11.9), the DHFR gene (minimum, 0.06; maximum, 10.4), and the tpi gene (minimum, 0.06; maximum, 23.5). The expression of the 16S rRNA gene decreased faster and earlier than the expression of other housekeeping genes. This difference in expression profile was significant, as shown in Table 3.
FIG. 3.
Expression of housekeeping genes during the exponential- and stationary-growth phases in vitro. Gene expression is rescaled as a percentage with respect to the measurement at 45 min. The left y axis, dots, and solid line represent the rescaled expression of each gene. Each open square, referring to the right y axis, represents the ln of CFU at a given time after inoculation. Dashed line, growth curve.
TABLE 2.
Decrease in gene expression and P value for the evolution of gene expression during in vitro exponential and stationary growth for each gene
| Time change (min) | Decrease (P value) in expression of:
|
||||
|---|---|---|---|---|---|
| gmk | DHFR gene | tpi | hsp-60 | 16S rRNA gene | |
| 45 vs 135 | 0.47 (0.025) | 0.45 (0.003) | 0.29 (0.007) | 0.60 (<0.001) | 1.88 (<0.001) |
| 135 vs 225 | 1.54 (0.14) | 1.32 (0.15) | 2.44 (0.06) | 1.27 (0.18) | 4.65 (0.009) |
| 225 vs 315 | 1.25 (0.30) | 1.27 (0.30) | 0.76 (0.57) | 2.07 (<0.001) | 1.61 (0.57) |
| 315 vs 405 | 4.00 (<0.001) | 2.32 (0.01) | 4.39 (0.16) | 6.24 (<0.001) | 2.38 (0.56) |
| 405 vs 495 | 2.94 (0.009) | 2.73 (0.045) | 1.47 (0.86) | 4.67 (0.02) | 1.84 (0.83) |
TABLE 3.
Changes in expression of the 16S rRNA gene compared to the changes in expression in other genesa
| Gene and time change | Absolute difference in expression change (estimate) | SE | P |
|---|---|---|---|
| DHFR gene vs the 16S rRNA gene | |||
| 45–135 min | 201.69 | 38.55 | 0.0001 |
| 135–225 min | 37.31 | 45.84 | 0.4292 |
| 225–315 min | 34.98 | 34.78 | 0.3308 |
| 315–405 min | 75.27 | 25.68 | 0.0104 |
| 405–495 min | 36.33 | 10.73 | 0.0617 |
| tpi vs the 16S rRNA gene | |||
| 45–135 min | 309.50 | 58.22 | 0.0017 |
| 135–225 min | 178.84 | 76.02 | 0.074 |
| 225–315 min | 52.78 | 68.99 | 0.5058 |
| 315–405 min | 146.75 | 67.59 | 0.1391 |
| 405–495 min | 12.38 | 66.27 | 0.8676 |
| gmk vs the 16S rRNA gene | |||
| 45–135 min | 181.27 | 47.87 | 0.0006 |
| 135–225 min | 49.98 | 51.94 | 0.35 |
| 225–315 min | 23.97 | 23.97 | 0.33 |
| 315–405 min | 81.16 | 13.87 | <0.0001 |
| 405–495 min | 18.36 | 6.31 | 0.07 |
| hsp-60 vs the 16S rRNA gene | |||
| 45–135 min | 110.68 | 10.07 | 0.0006 |
| 135–225 min | 17 | 14.66 | 0.31 |
| 225–315 min | 73.56 | 14.54 | 0.007 |
| 315–405 min | 46.75 | 13.44 | 0.045 |
| 405–495 min | 6.65 | 12.88 | 0.65 |
The expression of each gene is rescaled as a percentage of the expression of that gene after 45 min. Values indicate if the change in expression in a given gene between two moments is different for the 16S rRNA gene and the comparator. The estimate indicates the absolute difference in change.
DISCUSSION
The aims of the present study were (i) to develop and to test a simple, sensitive, and reproducible method for the study of gene expression in staphylococci and (ii) to explore basic housekeeping gene expression in S. epidermidis during in vitro exponential and stationary growth and under different conditions in order to create a reference for further gene expression experiments with staphylococci. To study housekeeping gene expression, five genes were selected. The 16S rRNA is an essential part of the ribosomal complex. In prokaryotic cells the rRNA concentration is the limiting step in ribosomal synthesis, and the cellular concentration of ribosomes is proportional to total protein synthesis and thus to total cellular metabolic activity (19, 23, 28). The hsp-60 gene is involved in protein folding and assembly and in the reactivation of denaturated proteins (10, 37). The gmk gene encodes an enzyme essential for guanosine and thus nucleic acid synthesis. The tpi gene product is essential in glucose metabolism, and DHFR is essential in folic acid synthesis.
By combining an instant method for RNA isolation with gene quantification by Taqman real-time quantitative PCR, a highly sensitive and reproducible method for the study of gene expression in staphylococci in response to changing environmental conditions was developed. Rapid extraction of the mRNA with direct fixation of the RNases is necessary due to the very short half-life of mRNA. For this reason, protocols based on enzymatic digestion of the bacteria are not appropriate in gene expression experiments (2). Given the high contamination with gDNA in conventional methods for RNA extraction in staphylococci, further RNA purification and DNase treatment are indispensable. With this protocol, gDNA contamination was at least 2 log units lower than the cDNA concentration. In contrast to the conventional techniques for mRNA quantification (14, 35), Taqman quantitative PCR permits the detection of both small quantities of mRNA (as few as 10 copies in a sample) and small changes in expression. The average intersample variability for in vitro experiments was 16%.
To validate this method, the expression of the selected housekeeping genes and the hsp-60 gene after a heat shock of 10°C given for 10 min was investigated. Heat shock proteins and the heat shock response have been extensively studied in Escherichia coli (10, 37). The heat shock response in S. aureus seems to be quite similar (27). Although heat shock proteins in S. aureus and in CNS are very similar (18), very little is known about the heat shock response in CNS. In comparison with data in other studies, the ninefold increase in hsp-60 expression (P < 0.00001) after a increase in the temperature for 10 min from 37 to 47°C seems very acceptable. In E. coli the up-regulation of dnaK expression at the mRNA level reaches a maximum at about 6 min after initiation of the heat shock and then declines to a value of about 8 to 10 times the baseline expression after 10 min (37). Although an extensive study of the heat shock response in S. epidermidis is beyond the scope of our paper, we are the first to document this increase in hsp-60 expression directly after heat shock in S. epidermidis. The only study that briefly dealt with the heat shock response in S. epidermidis used less-sensitive and only semiquantitative protein assays, and found comparable changes (27). Also, the slight decrease in gmk gene expression after heat shock is logical in view of the current understanding of the bacterial heat shock response (10, 37).
When glucose was added to a stationary-phase culture after 24 h of incubation, the expression of all housekeeping genes increased 1.4- to 7.3-fold. The only previous studies to which these results could be compared are studies on starvation recovery (3, 36). In contrast to a culture starved for 1 week, in which the majority of staphylococci have died, most bacteria in a stationary-phase culture remain viable (3). In studies on starvation recovery in S. aureus, addition of glucose and amino acids to a starved culture resulted in a strong increase in mRNA synthesis (3). The relatively high increase in our experiments in hsp-60 gene expression (7.3-fold) compared to that of the other genes may suggest that recovery from stationary-phase culture is mediated not only by de novo mRNA transcription and transcription from long-standing RNA as previously described (3) but also by the refolding and reactivation of denatured proteins in the cytoplasm. This hypothesis is consistent with the key function of the chaperonin hsp-60 in protein assembling and reassembling under stress conditions (10).
Finally, the expression of these five housekeeping genes during in vitro exponential and stationary growth was explored. The amount of 16S rRNA per bacterial cell was much greater than the amount of other mRNA at every time point, which is consistent with previous findings for E. coli that indicated that during the exponential-growth phase ribosomal content may comprise as much as 40% of cell mass (23). A rapid up-regulation of all genes was followed by a marked decrease in expression varying from 297-fold for the hsp-60 gene to 112-fold for the DHFR gene. Similar large changes in total mRNA production have been observed in S. aureus after starvation recovery (3). This indicates that the regulation of housekeeping metabolic activity is a very dynamic process that is capable of an enormous increase or decrease in gene expression according to the situation. The potential for rapid adaptation is undoubtedly an advantage in bacteria that can cause infection and that have to survive and to grow in very divergent situations (4). This potential for an extensive and rapid adaptation in housekeeping metabolic activity in bacteria is probably the conditio sine qua non for bacterial virulence. In the presence of this metabolic plasticity, additional factors—the classical virulence factors—can determine the final virulence of invasive pathogens.
The expression of the 16S rRNA gene decreases significantly more rapidly and earlier than the expression of other housekeeping genes. These findings are consistent with some older studies with E. coli that demonstrated that during the exponential-growth phase in prokaryotic cells, a major increase in ribosomal content precedes the synthesis of bacterial proteins (23). The synthesis of the ribosomal complex is regulated at the level of rRNA expression (23, 28), and thus 16S rRNA content is a good marker for ribosomal content. In response to changing and more hostile environmental conditions such as starvation, the total amount of ribosomes declines rapidly in order to conserve energy for other metabolic processes (19). Initially protein synthesis remains relatively stable; it can go on for a few hours before it declines significantly (23). In this study mRNA expression was used instead of protein synthesis, and similar changes were found.
In hematologic and immunologic quantitative reverse transcriptase PCRs, a housekeeping gene such as the β-actin gene, for which a constant level of expression is supposed, is used as an internal standard (16). This internal mRNA standard serves as a marker of the number of cells in the sample. Given the rapid and exponential growth kinetics of bacteria and the marked changes in the expression of housekeeping genes during in vitro exponential and stationary growth and under varying conditions, the use of an internal RNA standard is questionable in bacteriological gene expression studies.
In conclusion, a promising method for the study of gene expression in staphylococci under various in vitro conditions was developed and validated. This method was used to explore the basic housekeeping metabolism of CNS. The expression of housekeeping genes changes considerably during in vitro cell growth. These data provide a good reference for further gene expression experiments with staphylococci. As stated elsewhere, understanding the whens and wheres of the expression of genes is fundamental for the understanding of bacterial behavior and virulence (21). The method described here offers a powerful and reproducible tool with which to study the role of presumed virulence genes during in vitro and in vivo infection. It may also be used to study the effects of antibiotics on target genes involved in bacterial replication and resistance.
ACKNOWLEDGMENTS
S. J. Vandecasteele is supported by a grant of the FWO, Belgium. W. E. Peetermans has been awarded the R. van Furth Chair in Infectious Diseases, and J. Van Eldere has been awarded the Glaxo-Wellcome Chair in Medical Microbiology, at the Catholic University of Leuven, Leuven, Belgium.
We thank Steffen Fieuws of the Biostatistical Centre, School of Public Health, University of Leuven, for statistical analysis of the results.
REFERENCES
- 1.Brückner R. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol Lett. 1997;151:1–8. doi: 10.1111/j.1574-6968.1997.tb10387.x. [DOI] [PubMed] [Google Scholar]
- 2.Cheung A L, Eberhardt K J, Fischetti V A. A method to isolate RNA from gram-positive bacteria and mycobacteria. Anal Biochem. 1994;222:511–514. doi: 10.1006/abio.1994.1528. [DOI] [PubMed] [Google Scholar]
- 3.Clements M O, Foster S J. Starvation recovery of Staphylococcus aureus 8325-4. Microbiology. 1998;144:1755–1763. doi: 10.1099/00221287-144-7-1755. [DOI] [PubMed] [Google Scholar]
- 4.Clements M O, Foster S J. Stress resistance in Staphylococcus aureus. Trends Microbiol. 1999;7:458–462. doi: 10.1016/s0966-842x(99)01607-8. [DOI] [PubMed] [Google Scholar]
- 5.Costerton J W, Stewart P S, Greenberg E P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 6.Dale G E, Broger C, Hartman P G, Langen H, Page M G, Then R L, Stüber D. Characterization of the gene for the chromosomal dihydrofolate reductase (DHFR) of Staphylococcus epidermidis ATCC 14990: the origin of the trimethoprim-resistant S1 DHFR from Staphylococcus aureus? J Bacteriol. 1995;177:2965–2970. doi: 10.1128/jb.177.11.2965-2970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Saizieu A, Certa U, Warrington J, Gray C, Keck W, Mous J. Bacterial transcript imaging by hybridization of total RNA to oligonucleotide arrays. Nat Biotechnol. 1998;16:45–48. doi: 10.1038/nbt0198-45. [DOI] [PubMed] [Google Scholar]
- 8.Enright M C, Day N P J, Davies C E Z, Peacock S J, Spratt B G. Multilocus sequence typing for characterization of methicillin-resistant and methillin-susceptible clones of Staphyloccus aureus. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibson U E, Heid C A, Williams P M. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- 10.Hartl F U. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 11.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 12.Heilmann C, Hussain M, Peters G, Götz F. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol. 1997;24:1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- 13.Hussain M, Herrmann M, von Eiff C, Perdreau-Remington F, Peters G. A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect Immun. 1997;65:519–524. doi: 10.1128/iai.65.2.519-524.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson M R, Wang K, Smith J B, Heslin M J, Diasio R B. Quantitation of dihydropyrimidine dehydrogenase expression by real-time reverse transcription polymerase chain reaction. Anal Biochem. 2000;278:175–184. doi: 10.1006/abio.1999.4461. [DOI] [PubMed] [Google Scholar]
- 15.Kenward M G, Roger J H. Small sample inference for fixed effects from restricted maximum likelihood. Biometrics. 1997;53:983–997. [PubMed] [Google Scholar]
- 16.Kidd V, Lion T. Debate round-table. Appropriate controls for RT-PCR. Leukemia. 1997;11:871–881. doi: 10.1038/sj.leu.2400711. [DOI] [PubMed] [Google Scholar]
- 17.Kloos W E. Systematics and the natural history of staphylococci. Soc Appl Bacteriol Symp Ser. 1990;19:25S–37S. doi: 10.1111/j.1365-2672.1990.tb01795.x. [DOI] [PubMed] [Google Scholar]
- 18.Kwok A Y, Su S C, Reynolds R P, Bay S J, Av-Gay Y, Dovichi N J, Chow A W. Species identification and phylogenetic relationships based on partial HSP60 gene sequences within the genus Staphylococcus. Int J Syst Bacteriol. 1999;49:1181–1192. doi: 10.1099/00207713-49-3-1181. [DOI] [PubMed] [Google Scholar]
- 19.Lamond A I, Travers A A. Stringent control of bacterial transcription. Cell. 1985;41:6–8. doi: 10.1016/0092-8674(85)90050-9. [DOI] [PubMed] [Google Scholar]
- 20.Maes N, De Gheldre Y, De Ryck R, Vaneechoutte M, Meugnier H, Etienne J, Struelens M J. Rapid and accurate identification of Staphylococcus species by tRNA intergenic spacer length polymorphism analysis. J Clin Microbiol. 1997;35:2477–2481. doi: 10.1128/jcm.35.10.2477-2481.1997. . (Erratum, 36:1468, 1998.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mekalanos J J. Environmental signals controlling expression of virulence determinants in bacteria. J Bacteriol. 1992;174:1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.National Nosocomial Infections Surveillance System. National Nosocomial Infections Surveillance (NNIS) System report: data summary from October 1986–April 1998, issued June 1998. Am J Infect Control. 1998;26:522–533. doi: 10.1016/s0196-6553(98)70026-4. [DOI] [PubMed] [Google Scholar]
- 23.Nomura M, Gourse R, Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- 24.Pittet D, Tarara D, Wenzel R P. Nosocomial bloodstream infection in critically ill patients. Excess length of stay, extra costs, and attributable mortality. JAMA. 1994;271:1598–1601. doi: 10.1001/jama.271.20.1598. [DOI] [PubMed] [Google Scholar]
- 25.Proctor R A. Microbial pathogenic factors: small colony variants. In: Bisno A L, Waldvogel F A, editors. Infections associated with indwelling medical devices. Washington, D.C.: American Society for Microbiology; 1994. pp. 79–90. [Google Scholar]
- 26.Proctor R A, Kahl B, von Eiff C, Vaudaux P E, Lew D P, Peters G. Staphylococcal small colony variants have novel mechanisms for antibiotic resistance. Clin Infect Dis. 1998;27(Suppl. 1):S68–S74. doi: 10.1086/514906. [DOI] [PubMed] [Google Scholar]
- 27.Qoronfleh M W, Streips U N, Wilkinson B J. Basic features of the staphylococcal heat shock response. Antonie Leeuwenhoek. 1990;58:79–86. doi: 10.1007/BF00422721. [DOI] [PubMed] [Google Scholar]
- 28.Reznikoff W S, Siegele D A, Cowing D W, Gross C A. The regulation of transcription initiation in bacteria. Annu Rev Genet. 1985;19:355–387. doi: 10.1146/annurev.ge.19.120185.002035. [DOI] [PubMed] [Google Scholar]
- 29.Stickler D. Prosthetic device-associated infections: what's new? Curr Opin Infect Dis. 1996;9:265–269. [Google Scholar]
- 30.Stickler D, McLean R. Biomaterials associated infections: the scale of the problem. Cell Materials. 1995;5:167–182. [Google Scholar]
- 31.Sugarman B, Young E J. Infections associated with prosthetic devices: magnitude of the problem. Infect Dis Clin N Am. 1989;3:187–198. [PubMed] [Google Scholar]
- 32.Van Wijngaerden E, Peetermans W E, Vandersmissen J, Van Lierde S, Bobbaers H, Van Eldere J. Foreign body infection: a new rat model for prophylaxis and treatment. J Antimicrob Chemother. 1999;44:669–674. doi: 10.1093/jac/44.5.669. [DOI] [PubMed] [Google Scholar]
- 33.von Eiff C, Heilmann C. Staphylococcus epidermidis: why is it so successful? Clin Microbiol Infect. 1998;4:297–299. [Google Scholar]
- 34.von Eiff C, Heilmann C, Peters G. New aspects in the molecular basis of polymer-associated infections due to staphylococci. Eur J Clin Microbiol Infect Dis. 1999;18:843–846. doi: 10.1007/s100960050417. [DOI] [PubMed] [Google Scholar]
- 35.Wang T, Brown M J. mRNA quantification by real time TaqMan polymerase chain reaction: validation and comparison with RNase protection. Anal Biochem. 1999;269:198–201. doi: 10.1006/abio.1999.4022. [DOI] [PubMed] [Google Scholar]
- 36.Watson S P, Clements M O, Foster S J. Characterization of the starvation-survival response of Staphylococcus aureus. J Bacteriol. 1998;180:1750–1758. doi: 10.1128/jb.180.7.1750-1758.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yura T, Nagai H, Mori H. Regulation of the heat-shock response in bacteria. Annu Rev Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]
- 38.Ziebuhr W, Heilmann C, Götz F, Meyer P, Wilms K, Straube E, Hacker J. Detection of the intercellular adhesion gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect Immun. 1997;65:890–896. doi: 10.1128/iai.65.3.890-896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]