Abstract
Membrane proteins are currently investigated after detergent extraction from native cellular membranes and reconstitution into artificial liposomes or nanodiscs, thereby removing them from their physiological environment. However, to truly understand the biophysical properties of membrane proteins in a physiological environment, they must be investigated within living cells. Here, we used a spin-labeled nanobody to interrogate the conformational cycle of the ABC transporter MsbA by double electron-electron resonance. Unexpectedly, the wide inward-open conformation of MsbA, commonly considered a nonphysiological state, was found to be prominently populated in Escherichia coli cells. Molecular dynamics simulations revealed that extensive lateral portal opening is essential to provide access of its large natural substrate core lipid A to the binding cavity. Our work paves the way to investigate the conformational landscape of membrane proteins in cells.
Spin-labeled nanobodies were used to address the conformational landscape of the lipid A transporter MsbA in cells.
INTRODUCTION
MsbA of Escherichia coli is a homodimeric adenosine 5′-triphosphate (ATP)–binding cassette (ABC) exporter that flips core lipid A, the precursor of lipopolysaccharide (LPS), across the cytoplasmic membrane (1, 2); it is a type IV transporter according to the newest nomenclature (3) (type I exporter in the old nomenclature), which is formed through the dimerization of two identical half-transporters, each composed of a transmembrane domain (TMD) and a nucleotide-binding domain (NBD) (4, 5). ATP binding and subsequent hydrolysis at the NBDs lead to the dimerization and dissociation of the NBD dimer, respectively (Fig. 1) (6). These large-scale conformational changes are coupled to the TMDs and result in the reorientation of a large cavity either oriented toward the cytoplasm [inward-facing (IF) conformation with separated NBDs] or toward the periplasm [outward-facing (OF) conformation with fully dimerized NBDs carrying two ATP molecules sandwiched at the dimer interface] (7, 8). An outward-occluded conformation (OFoccluded) with fully dimerized NBDs and a closed periplasmic gate at the TMDs has been described as well (9–11).
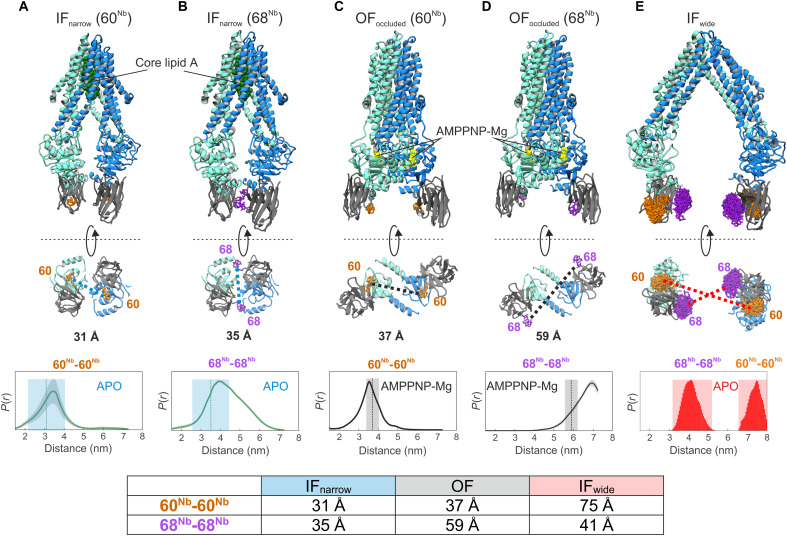
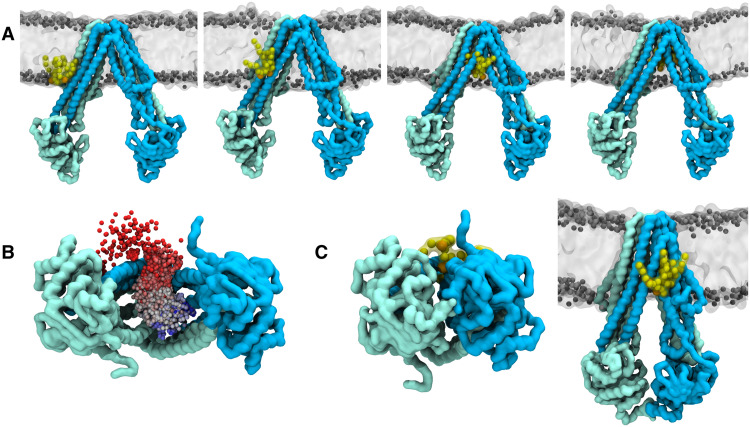
Fig. 1. Structure-based identification and experimental validation of spin-labeling sites.
(A to D) Cryo-EM structures of nanodisc-reconstituted MsbA (cyan/blue) in complex with the nanobody (gray) containing Gd-DOTA either at position 60Nb (A and C) or 68Nb (B and D), obtained in the apo conformation (IFnarrow) or in the presence of AMPPNP-Mg (OFoccluded). Gd-Gd distances are shown as dotted lines in the bottom view of the structures. The bottom of (A) to (D) shows experimentally determined DEER distance distributions using the same samples used for cryo-EM structure determination. Uncertainties estimated from the neural network analysis are shown as gray contour of the distributions (39). Dotted vertical lines denote Gd-Gd distances measured in the respective cryo-EM structures. The blue and gray shaded areas are error margins that correspond to the resolution of the structures and the degree of conformational flexibility determined from multibody refinement as described in Methods. (E) Nanobody docked onto the crystal structure of IFwide MsbA. The in silico predicted rotamers of the Gd-maleimide-DOTA spin label (39) are displayed as orange and purple sticks at nanobody positions 60Nb and 68Nb, respectively. The interspin distances are highlighted with dotted lines. The bottom panel shows simulated distance distributions for the two spin-label pairs of IFwide. Throughout the manuscript, we use the red shaded areas encompassing the full range of simulated distances to schematically represent the distances in the IFwide conformation. A table at the bottom summarizes the mean Gd-Gd distances for the respective conformation.
A hallmark of MsbA is the highly disputed wide inward-open conformation with large separation of the NBDs (IFwide). This peculiar conformation was first observed in crystal structures obtained in detergent in the absence of nucleotides (4) and later confirmed by double electron-electron resonance (DEER) analyses with detergent-solubilized proteins and in proteoliposomes (12, 13). Molecular dynamics (MD) simulations suggested that MsbA is flexible in the absence of nucleotides and substrates and that the free energy landscape is shallow. Hence, conformations covering a broad range of NBD separation distances, including IFwide, are thermally accessible (14). The coexistence of multiple IF conformations with varying degrees of NBD separation was also observed in negative-stain electron microscopy (EM) using detergent-purified MsbA (15). The IFwide conformation with its widely opened lateral portals was suggested to grant access of the large substrate core lipid A to the substrate-binding cavity (16, 17).
In contrast, high-resolution cryo-EM studies of MsbA reconstituted in nanodiscs or peptidiscs showed a much closer distance between the NBDs under apo conditions (IFnarrow conformation) (9, 18). In light of other ABC exporter apo structures exhibiting close inter-NBD distances (19–21), these more recently determined MsbA structures led to the prevailing opinion in the field that the IFwide conformation is a detergent and/or crystallization artifact of limited physiological relevance, in particular in the cellular context where ATP and adenosine 5′-diphosphate (ADP) are present at millimolar concentrations (22–25). Single-molecule fluorescence resonance energy transfer (FRET) (26) as well as small-angle neutron scattering and x-ray scattering (27) studies indeed confirmed that only the IFnarrow conformation of E. coli MsbA is populated in nanodiscs. However, a very recent cryo-EM study on the MsbA of Acinetobacter baumannii revealed the IFwide conformation in nanodiscs, suggesting that major differences exist between different MsbA homologs (28).
The only way to address the controversy regarding the relevance of the IFwide conformation is to investigate MsbA in its native environment, namely, the cytoplasmic membrane of E. coli. Although DEER, single-molecule FRET, and luminescence resonance energy transfer have been previously used to study purified and/or reconstituted MsbA (12, 13, 26, 29), these methods cannot be applied to carry out in vivo experiments with MsbA, because they would require the site-specific and near-complete labeling of engineered cysteines using thiol-reactive reporter dyes in the context of the cell’s proteome. In pioneering studies, in-cell DEER was successfully applied to spin-labeled cytosolic proteins upon electroporation into cells (30, 31), spin-labeled RNA injected into oocytes or cells (32), and bacterial outer membrane proteins directly spin-labeled in the extracellular regions (33). Recently, we used spin-labeled nanobodies to study membrane transporters in cell-derived membranes (34). Nanobodies are the variable domains of heavy chain–only antibodies that can be obtained by immunizing camelids (35) or from synthetic libraries (36). Their robustness, high binding affinity, and excellent specificity predestine them as ideal tools for DEER applications to investigate wild-type (WT) membrane proteins in their native, cellular environment.
RESULTS
We generated a nanobody called Nb_MsbA#1 and solved its crystal structure in complex with the isolated NBD of MsbA (fig. S1 and table S1). Nb_MsbA#1 binds at the bottom of the NBD and establishes contacts with around 15 NBD residues located close to the C terminus. Grating-coupled interferometry (GCI) using full-length detergent-solubilized MsbA as ligand revealed a dissociation constant (Kd) of 9 nM for the nanobody (fig. S2). Basal adenosine triphosphatase (ATPase) activity of MsbA was only moderately influenced in the presence of Nb_MsbA#1 when analyzed in detergent, nanodiscs, or proteoliposomes (fig. S3).
Taking known structures of MsbA into consideration (Fig. 1), we identified the amino acid positions A60 and T68 on the nanobody (called 60Nb and 68Nb) as ideal spin-labeling sites with high distance contrast between different conformations of MsbA. To enable in-cell DEER, the chosen sites were mutated to cysteines and modified with maleimide-gadolinium(III)-DOTA (in the following named Gd-DOTA), which is stable under the reducing environment of the cytoplasm (30). Gd-DOTA labeling did not weaken the affinity of the nanobody for MsbA (fig. S2).
To benchmark the ability of spin-labeled nanobodies to report inter-NBD distances with high fidelity, we determined cryo-EM structures of nanodisc-reconstituted MsbA in complex with Gd-DOTA–labeled nanobodies in the presence and absence of the nonhydrolyzable ATP analog AMPPNP-Mg (Fig. 1, A to D, fig. S4, and table S2). In parallel, DEER experiments on the same protein preparations were performed. In agreement with a previous cryo-EM study in nanodiscs (9), apo MsbA with bound nanobodies predominantly assumes the IFnarrow conformation with bound core lipid A in the IF cavity, whereas incubation with AMPPNP-Mg resulted in the OFoccluded conformation with fully dimerized NBDs and no density for lipid A. Global resolutions for the IFnarrow conformation range from 3.7 to 4.1 Å, while both OFoccluded maps reached 2.8-Å resolution. Hence, the structures reported here are considerably improved over previously published cryo-EM structures of E. coli MsbA (9). The nanobodies are tightly bound to the NBDs, which is in line with MD simulations performed on the crystal structure of the isolated NBD-nanobody complex (fig. S5 and table S3). A cryo-EM structure of apo MsbA determined with the same nanodisc preparation, but without the addition of nanobody, demonstrated that the nanobodies have only a small impact on the inter-NBD distance; therefore, they do not affect the stability of the IFnarrow conformation (fig. S6). This agrees with DEER control experiments performed in different environments on the spin-labeled transporter in the presence and absence of the nanobody (fig. S7).
Densities for the Gd-DOTA labels at position 60Nb are well resolved in experimental cryo-EM maps of both IFnarrow and OFoccluded. This allowed for the accurate placement of the Gd ions and measurement of interspin distances (Fig. 1, A and C, and fig. S8), which were in very close agreement with DEER measurements performed in parallel (Fig. 1, A and C, bottom). The Gd-DOTA densities at position 68Nb are less resolved, presumably because of a higher flexibility of the nanobody loop where the label was located, but still allowed for placing Gd ions (Fig. 1, B and D). Increased motional freedom is also reflected by the broader distance distribution in DEER for the 68Nb-68Nb pair compared to the 60Nb-60Nb pair (Fig. 1, B and D).
Although IFnarrow displays a continuous movement of the NBDs, as observed in multibody refinement (movie S1), the average Gd-Gd distance for the 60Nb-60Nb pair is 3.1 nm, and the largest distance does not exceed 3.6 nm. Hence, the IFwide conformation described for the apo MsbA crystal structure (4) is not populated in nanodiscs. The expected Gd-Gd distances of the IFwide conformation were therefore predicted on the basis of the apo MsbA crystal structure using a rotamer library approach. For the 60Nb position, the simulated Gd-Gd distance in the IFwide is about 7 nm and thus close to the DEER detection limit, while it is short (4 nm) and can be measured with high confidence for the 68Nb position (Fig. 1E).
Our nanobody-assisted DEER measurements cannot differentiate between the OFoccluded conformation present in nanodiscs (Fig. 1, C and D) and the OFopen conformation with the open periplasmic gate observed in crystal structures (4), because both conformations have the NBDs in close contact. We will thus simply refer to the OF conformation in the context of our DEER analyses.
Note that, owing to NBD rotation as part of the IF-OF transition (37), changes of interspin distances in response to conformational changes of MsbA strongly depend on the nanobody labeling position (Fig. 1). For 60Nb, the interspin distance is long (7 nm) for IFwide and becomes shorter (3 to 4 nm) for IFnarrow and OF. The picture is inversed for 68Nb where the interspin distance is short for IFwide and IFnarrow (around 4 nm) but long (6 to 7 nm) for OF. Hence, by performing DEER experiments with the same nanobody, either spin-labeled at position 60Nb or 68Nb, all relevant conformations can be distinguished.
Gd-DOTA–labeled nanobodies were used to probe the conformational space of WT MsbA either purified in detergent or reconstituted into proteoliposomes (Fig. 2 and fig. S9 for primary DEER data). In addition, we also probed overexpressed MsbA in inside-out membrane vesicles (ISOVs) derived from E. coli. Excellent specificity of the nanobody toward MsbA in ISOVs was demonstrated by control experiments with ISOVs containing the overexpressed ABC exporter TM287/288 (fig. S10) (38). DEER measurements were performed in the absence of nucleotides (Fig. 2, red curves) or upon addition of AMPPNP-Mg (Fig. 2, black curves). The experimentally determined interspin distances (Fig. 2) were compared with the predicted Gd-Gd distances based on IFwide (red shaded area from Fig. 1E) or with the experimental Gd-Gd distances measured in the corresponding cryo-EM structures of IFnarrow (blue shaded area from Fig. 1, A and B) and OFoccluded (dark gray shaded area from Fig. 1, C and D). Data analysis was performed using a neural network–based approach (39).
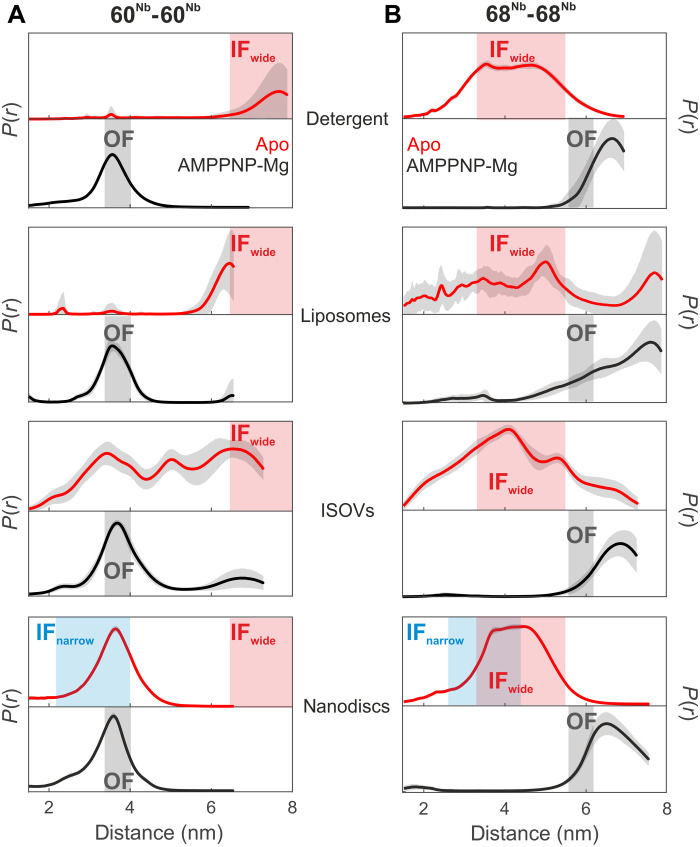
Fig. 2. Nanobody-assisted DEER analysis of MsbA embedded in different environments.
Internanobody distance distributions between pairs 60Nb-60Nb (A) and 68Nb-68Nb (B) measured in the indicated environment in the absence of nucleotides (apo state, red curves) or in the presence of AMPPNP-Mg (black curves). Uncertainties estimated from the neural network analysis are shown as gray contour of the distributions. The blue and gray shaded areas are interspin distances including error margins obtained from the cryo-EM structures of IFnarrow and OFoccluded, respectively (from Fig. 1, A to D). The red shaded areas represent the range of expected interspin distances predicted using a rotamer approach for IFwide (PDB: 3B5W; see Fig. 1E). The distances in nanodiscs correspond to a biological repeat different from that shown in Fig. 1. Primary DEER data are shown in fig. S9.
In detergent-solubilized apo MsbA, the 60Nb-60Nb pair (red curve, Fig. 2A) showed distances larger than 6 nm, which is a characteristic feature of the IFwide conformation (see the red shaded area in Fig. 2). The existence of the IFwide conformation is further corroborated by the appearance of the 4-nm distance peak in the 68Nb-68Nb pair (compare the red curve with the red shaded area in Fig. 2B). Upon addition of AMPPNP-Mg, DEER measurements reported the distance characteristic of the OF conformation, namely, 3.5 nm for the 60Nb-60Nb pair and about 6 nm for the 68Nb-68Nb pair (black curves in Fig. 2, A and B). The experimental distance for the 68Nb-68Nb pair in the OF state (Fig. 2B) was found to be longer and more broadly distributed than expected on the basis of the corresponding cryo-EM structure (Fig. 1D). This is mainly due to the larger uncertainty in determining distances >6 nm from traces with limited dipolar evolution time (fig. S9).
The DEER data of MsbA in proteoliposomes largely agree with the measurements obtained in detergent micelles (Fig. 2, A and B) but show an overall increased conformational heterogeneity in the absence of nucleotides (red curves). In ISOVs, IFwide is clearly populated in the absence of nucleotides, but other conformations corresponding to IFnarrow and/or OF were detected as well. This indicates a larger heterogeneity in the relative displacement of the NBDs within the native lipid bilayer, which may originate from residual traces of nucleotides being still present in the preparations or a broad distribution of vesicle sizes resulting in heterogeneous membrane curvatures. Addition of AMPPNP-Mg resulted in a complete transition to the OF conformation in ISOVs.
In stark contrast to all other environments, nanodisc-reconstituted MsbA predominantly adopts the IFnarrow conformation in the absence of nucleotides. This is clearly visible in the 60Nb-60Nb pair, which reports a defined distance distribution centered at 3.6 nm (Fig. 2A), which nicely agrees with the 3.1-nm mean distance extracted from the cryo-EM structure of the MsbA-nanobody complex in nanodiscs (Fig. 1A).
In contrast to the 60Nb-60Nb pair, the 68Nb-68Nb pair cannot nicely differentiate the IFwide (4.1 nm) from the IFnarrow (3.5 nm) conformation (Fig. 1). Therefore, in nanodiscs, the distance distribution looks similar to the one measured in detergent (Fig. 2B). Upon addition of AMPPNP-Mg, DEER measurements reported the distance characteristic of the OF conformation for both pairs, namely, 3.5 nm for the 60Nb-60Nb pair, which is almost invariant with respect to the distance detected in the apo state in nanodiscs, and >6 nm for the 68Nb-68Nb pair (black curves in Fig. 2, A and B), which is distinguishable from the peak centered at around 4 nm in the apo state.
Hence, the comparative analysis performed with spin-labeled nanobodies supports the notion that for apo MsbA, the NBDs are in closer proximity to each other in nanodiscs (IFnarrow) than in detergent, proteoliposomes, and ISOVs, where the IFwide conformation is substantially populated. The nanodisc-specific IFnarrow conformation was additionally confirmed with nitroxide-labeled MsbA (fig. S11) and with nitroxide-labeled nanobodies (fig. S12). The almost invariant 3.5-nm distance detected in the apo state and in the presence of AMPPNP-Mg in nanodiscs for the 60Nb-60Nb pair persists also in the presence of ADP-Mg (fig. S13). Therefore, if IFnarrow was the physiologically relevant IF conformation in the cellular environment, we would detect an invariant 3.5-nm distance peak for the 60Nb-60Nb pair under ATP turnover conditions. Therefore, the 60Nb-60Nb pair is an excellent tool to test the hypothesis whether IFwide is populated under physiologically relevant conditions.
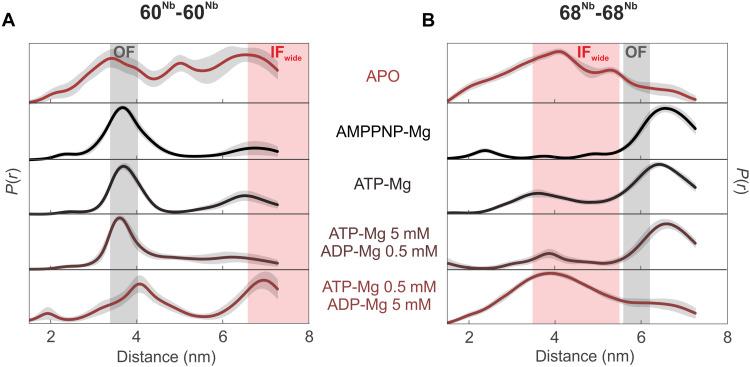
The IFwide conformation was shown to be populated in ISOVs in the absence of nucleotides (Fig. 2). To gain further insights into MsbA’s conformational spectrum in a near-physiological setting, we used the spin-labeled nanobodies to probe the overexpressed transporter in ISOVs under ATP turnover conditions (see Fig. 3 and fig. S14 for primary data and comparative analysis with Tikhonov regularization or model-based Gaussian fit). DEER analyses of the 60Nb-60Nb pair revealed that in the presence of 5 mM ATP-Mg or 5 mM ATP-Mg/0.5 mM ADP-Mg, MsbA predominantly exhibits closed NBDs, indicating a major fraction of OF conformation. Nevertheless, as clearly shown by the 68Nb-68Nb pair, a small fraction of IFwide is also present under these conditions (Fig. 3B and fig. S14). Upon inversion of the nucleotide mixture toward ADP (0.5 mM ATP-Mg/5 mM ADP-Mg), the conformational equilibrium of MsbA shifted toward the IFwide conformation (Fig. 3). Hence, DEER measurements in native membranes clearly demonstrate that the IFwide conformation is populated not only in the absence of nucleotides but also in the presence of physiological nucleotide concentrations reflecting a low-energy metabolic state. Notably, biological repeats demonstrate that the distance corresponding to the IFwide conformation is consistently present, whereas we noted some variability in the fraction of short to intermediate distances in the apo state (fig. S15A).
Fig. 3. DEER analysis of MsbA under ATP turnover conditions in inside-out vesicles from E. coli.
Distance distributions of the inter-nanobody distances for the 60Nb-60Nb pair (A) and 68Nb-68Nb pair (B) in ISOVs overexpressing WT MsbA under apo, AMPPNP-Mg, or ATP turnover conditions. The gray shaded areas represent the distance range of the OFoccluded conformation, estimated on the basis of the cryo-EM structures, and the red shaded areas correspond to the distance range predicted for the IFwide conformation using a rotamer approach (see Fig. 1). The apo and AMPPNP-Mg data are the same as shown in Fig. 2.
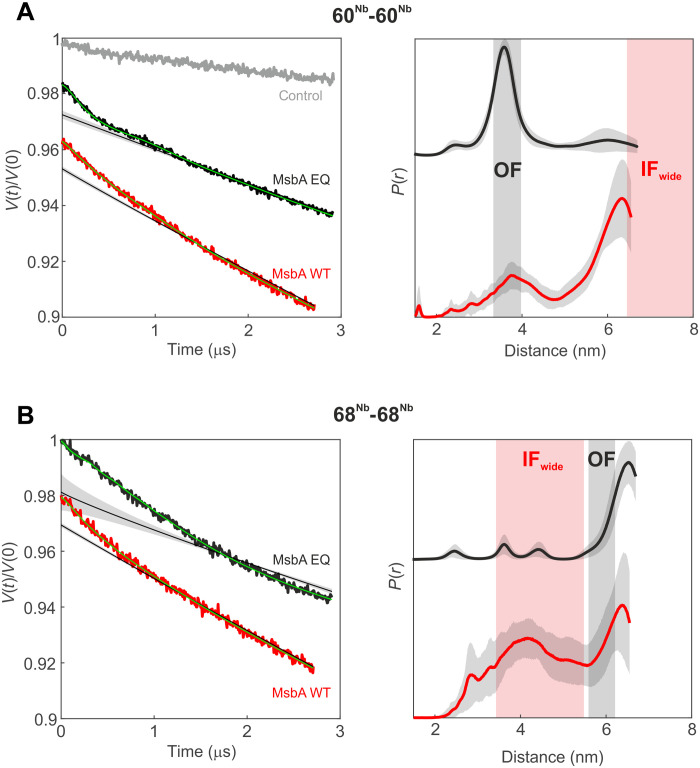
Last, we established an electroporation protocol to deliver the Gd-DOTA-labeled nanobodies into the cytosol of E. coli overexpressing MsbA (fig. S16) to directly monitor the conformational ensemble of this transporter in its physiological environment. We first used an MsbA mutant with the Walker B glutamate substituted by glutamine (EQ mutation), which is trapped in the OF conformation due to the capture of ATP at the NBDs (40). Control experiments with ISOVs indeed confirmed that MsbA mainly adopts the OF conformation even in the absence of externally added ATP (fig. S17), indicating efficient trapping of cell-derived ATP by the MsbA_EQ mutant as previously observed for an analogous EQ mutant of the ABC transporter TM287/288 (41). Supporting this notion, ATP hydrolysis induced by prolonged incubation of the MsbA_EQ-containing ISOVs in the presence of Mg2+ shifted the population toward the IFwide conformation (fig. S17). On the basis of this control experiment, the 60Nb-60Nb pair is expected to report a short distance (3.5 nm) in cells expressing MsbA_EQ. As expected, we retrieved the short 60Nb-60Nb fingerprint distance that is characteristic for the OF conformation (Fig. 4A) in E. coli cells overexpressing MsbA_EQ and electroporated with the Gd-DOTA-labeled nanobodies (Fig. 4A). The same experiment performed with the 68Nb-68Nb pair yielded a distance distribution lacking short distances, confirming that MsbA_EQ was completely trapped in the OF conformation within cells (Fig. 4B). As a control, cells containing the overexpressed ABC transporter TM287/288 were electroporated under the same conditions. These control cells did not show dipolar modulation (Fig. 4A) despite the fact that echo-detected field-sweep (EDFS) measurements demonstrated the presence of trapped gadolinium ion within the cells (fig. S18A).
Fig. 4. DEER analysis of MsbA in E. coli cells.
Primary DEER data (left) and corresponding distance distributions (right) of the 60Nb-60Nb pair (A) and the 68Nb-68Nb pair (B) in cells overexpressing mutant (EQ, black) or WT (red) MsbA. A control sample [gray in (A)] with the Gd-labeled nanobody (60Nb) electroporated in cells overexpressing TM287/288 did not show dipolar modulation. The gray shaded areas represent the distance range of the OFoccluded conformation, estimated on the basis of the cryo-EM structures, and the red shaded areas correspond to the distance range predicted for the IFwide conformation using a rotamer approach (see Fig. 1). The obtained distance distributions are further validated by analyses performed with Tikhonov regularization and model-based Gaussian fit (fig. S20).
Protein production levels were found to be slightly higher for WT MsbA than for MsbA_EQ (fig. S18B). Accordingly, we electroporated the Gd-DOTA-labeled nanobodies into E. coli overexpressing WT MsbA under comparable experimental conditions, increasing the concentration range of the electroporated Gd-DOTA-labeled nanobodies from 15 to 25 μM for MsbA_EQ to 20 to 30 μM for WT MsbA (fig. S19). The in-cell distances obtained with the nanobodies labeled at position 60Nb were distinct from those detected in the MsbA_EQ mutant and showed the concomitant presence of short and long (>6 nm) distances (see Fig. 4A, biological repeats in fig. S19, and analysis with Tikhonov regularization and model-based Gaussian fit in fig. S20). This suggests that WT MsbA under physiological conditions adopts a conformational continuum with highly varying inter-NBD distances representing all known conformations, including IFwide. Notably, the 60Nb-60Nb pair is perfectly suited to monitor the existence of IFwide in cells, because if IFnarrow had been the physiological IF conformation, only short distances would have appeared in cells expressing WT MsbA, akin to the data obtained in nanodiscs with the 60Nb-60Nb pair (Fig. 2). The corresponding DEER data on the 68Nb-68Nb pair further corroborated the notion that all conformations of MsbA coexist within cells (see Fig. 4 and comparative analysis with Tikhonov regularization and model-based Gaussian fit in fig. S20). Notably, the fraction of IFwide in cells cannot be inferred from the data presented. Although the cells electroporated with the nanobodies and recovered for 20 min were found to be intact and impermeable, as judged on the basis of fluorescence microscopy (fig. S16B) and external addition of ATP-Mg before freezing the DEER sample (fig. S21A), they exhibit high ADP/ATP ratios and thus are in a low-energy metabolic state (fig. S21B). Therefore, the fraction of IFwide in the ensemble was relatively high, as is the case in ISOVs at high ADP/ATP ratios (Fig. 3). A prolonged recovery time (4 hours) was found to be sufficient to restore ADP/ATP ratios as present in metabolically active cells, but it was not suitable for reliable detection of long distances because of the poorer Gd signal in the samples. Nevertheless, short DEER traces recorded at 20 min or 4 hours of recovery time did not show relevant changes in the dipolar decay for the 68Nb-68Nb pair, suggesting that, within the sensitivity of our method, we can conclude that a fraction of the IFwide still appears to be present in fully recovered cells (fig. S21C).
To compare IFnarrow and IFwide in terms of their distinct capability to bind lipid A, multi-microsecond coarse-grained MD simulations were carried out on MsbA embedded in a mixed lipid bilayer with 10 mole percent of core lipid A in the cytoplasmic leaflet. In the simulations, a lipid A molecule spontaneously entered from the membrane via a lateral portal into the TMD cavity of IFwide (Fig. 5, A and B). Lipid A binding was not observed in the simulations of IFnarrow, where lipid A attaches from the outside to the lateral portals but does not enter the TMD cavity (Fig. 5C). The MD simulations thus show that the presence of the IFwide conformation plays a key role for lowering the kinetic barrier for lipid A binding to the MsbA cavity, which is a prerequisite for transport.
Fig. 5. Binding of core lipid A inside the TMD cavity of IFwide.
(A) Shown are snapshots from a representative coarse-grained MD simulation of the IFwide conformation. The MsbA chains are colored as in Fig. 1. The lipid A molecule that binds inside MsbA is shown in yellow with the phosphate beads in orange. The bilayer lipids are shown in gray; solvent molecules are not shown for clarity. (B) Binding pathway of lipid A to the MsbA TMD cavity viewed from the cytosol. The dots represent the position of the phosphate groups in the lipid A headgroup and are colored from red to blue as a function of the simulation time to show the entrance pathway. Similar lipid A entry pathways were found in four independent repeat simulations (each 50 μs long), whereas lipid A did not enter into the TMD cavity in the corresponding simulations of IFnarrow. (C) Left: Shown is the last snapshot from one of the simulations of the IFnarrow conformation viewed from the cytosol with a lipid A molecule located next to one of the lateral portals. Right: The same snapshot viewed from the side.
DISCUSSION
Using nanobody-assisted DEER, we demonstrated that the IFwide conformation of MsbA is populated in E. coli cells and in cell-derived membranes in the presence of nucleotides added at the millimolar concentration range. MD simulations in addition show that the large degree of opening of the lateral portal as present in IFwide is essential for lipid A binding. Owing to the fact that wide inward-open structures have been described for a large number of ABC exporters, including P-glycoprotein (42–45), it is plausible to assume that extensive lateral portal opening is a common mechanism by which ABC transporters capture substrates from the membrane. Intriguingly, the conformational equilibrium of MsbA in ISOVs is shifted in response to changes in physiologically relevant ADP/ATP ratios, suggesting that the transporter’s structural ensemble and, therefore, the substrate turnover are modulated in response to the availability of ATP. The higher abundance of the wide inward-open conformation at low ATP and high ADP concentrations could represent a mechanism to slow down lipid A flipping (and thus LPS incorporation and cellular expansion) under low-energy conditions, which would enable a faster physiological response than the shutdown of lipid A biosynthesis. Our unexpected experimental findings stand in seeming contradiction to cryo-EM structures determined in nanodiscs and rebut the prevailing opinion that the IFwide conformation is a detergent and crystallization artifact. Instead, our data indicate nanodisc-mediated funneling of the IFnarrow conformation. Since apo MsbA has a shallow energy landscape (14), allowing for the coexistence of both IFwide and IFnarrow conformations (15), small perturbations, such as differences in the membrane environment in nanodiscs versus ISOVs, or substrate availability, can have a major effect on the relative distribution of distinct conformations. It is therefore important to be cautious when interpreting transporter conformations observed in nanodiscs. Our work paves the way to investigate membrane transporters in situ with precise experimental tools to shed light on their molecular physiology in the context of the native cell membrane.
METHODS
Molecular biology
The generation of Cys-less MsbA and mutant N191C was described in a previous study (13). MsbA NBDs were amplified from pBAD24_MsbA using primers EcMsbA_NBD_FX_for1 (5′-ATA TGC TCT TCT AGT GGC GAC GTG GAA TTC CGC) and EcMsbA_NBD_FX_rev1 (5′-TAT AGC TCT TCA TGC TTG GCC AAA CTG CAT TTT GTG) and cloned into pETM11-small ubiquitin-related modifier (SUMO) (46) or pBXNH3CA vector using FX cloning (47). These vectors are called pETM11-SUMO-MsbA-NBD and pBXNH3CA-MsbA-NBD hereafter. Cysteine mutants of Nb_MsbA#1 were generated by QuikChange site-directed mutagenesis. To introduce a cysteine at position A60 or position T68, primers Nb_MsbA#1_A60C_fw (5′-GGA CAA GCT ATT GCG ACT CCG TG) and Nb_MsbA#1_A60C_rv (5′-CAC GGA GTC GCA ATA GCT TGT CC) or Nb_MsbA#1_T68C_fw (5′-GGG CCG ATT CTG CAT CTC CAA AG) and Nb_MsbA#1_T68C_rv (5′-CTT TGG AGA TGC AGA ATC GGC CC) were used.
Purification of MsbA-NBD
Vectors pETM11-SUMO-MsbA-NBD and pBXNH3CA-MsbA-NBD were used to transform E. coli C43 (DE3) or E. coli MC1061, respectively. A single colony was used to inoculate an overnight culture in terrific broth (TB) medium, supplemented with kanamycin (50 μg/ml; pETM11-SUMO construct) or ampicillin (100 μg/ml; pBXNH3CA construct). Fresh TB medium containing antibiotics was inoculated with 1:100 of overnight culture and grown for 1.5 hours at 37°C followed by 1.5 hours at 30°C to reach an optical density at 600 nm (OD600) of 1.0 to 1.5. Protein expression was induced by adding 0.5 mM isopropyl-β-d-thiogalactopyranoside (for tag-less MsbA-NBD from pETM11-SUMO-MsbA-NBD) or 0.01% l-arabinose (for Avi-tagged MsbA-NBD from pBXNH3CA-MsbA-NBD), and protein production proceeded for 3 hours at 30°C. Cells were collected at 5000g for 10 min and resuspended in 20 mM tris-HCl (pH 7.5) and 150 mM NaCl supplemented with deoxyribonuclease (DNase; Sigma-Aldrich). Cells were lysed by passing three times through an M-110P Microfluidizer (Microfluidics) at 25,000 psi. Unbroken cells were removed by centrifugation at 8000g for 30 min at 4°C. The supernatant was supplemented with 30 mM imidazole and loaded onto Ni-NTA (nitrilotriacetic acid) gravity flow columns. After washing with 20 column volumes with 50 mM imidazole (pH 7.5), 200 mM NaCl, and 10% glycerol, the NBDs were eluted with 5 column volumes of 200 mM imidazole (pH 7.5), 200 mM NaCl, and 10% glycerol. SUMO fusion was cleaved using SenP2 protease (50 μg/ml) while dialyzing against 20 mM tris-HCl (pH 7.5) and 150 mM NaCl for 16 hours. Avi-tagged MsbA-NBD was first concentrated to 360 μl using Amicon Ultra-4 concentrator units with a 10-kDa molecular weight cutoff (MWCO), and 3C cleavage and enzymatic biotinylation were performed simultaneously in a total reaction mixture of 4 ml containing 3C protease (0.2 mg/ml), 330 nM BirA (0.016 mg/ml), 20 mM imidazole (pH 7.5), 10 mM magnesium acetate, 200 mM ATP, 200 mM NaCl, 10% glycerol (v/v), and a 1.2-fold molar excess of biotin. Using reverse immobilized metal affinity chromatography (IMAC), tags were removed by loading cleaved NBDs onto Ni-NTA gravity flow columns and directly collecting flow-through as well as three additional column volumes of 20 mM tris-HCl (pH 7.5) and 150 mM NaCl. NBDs were concentrated using Amicon Ultra-4 concentrator units with 10-kDa MWCO and finalized by size exclusion chromatography (Superdex-200 10/300 GL) in 20 mM tris-HCl (pH 7.5) and 150 mM NaCl.
Purification of MsbA and MsbA_E506Q and preparation of spin-labeled MsbA
E. coli MsbA, MsbA_E506Q, and cysteine mutant MsbA_N191C were expressed from pBAD24 vector with an N-terminal His tag, 3C-cleavage site, and a linker (D-E-A-E-K-L-F-N-Q) (13). The Avi-tagged version of MsbA was expressed from vector pBXNH3CA (46). TB medium containing ampicillin (100 μg/ml) was inoculated 1:100 (w/v) with an overnight culture of freshly transformed plasmid in E. coli MC1061 and grown at 37°C for 1.5 hours followed by incubation at 30°C until an OD600 of 1.2 to 1.5 was reached. Protein expression was induced by the addition of 0.01% (w/v) l-arabinose, and cells were harvested after 4 hours. Cell pellets were resuspended with 20 mM tris-HCl (pH 7.5) and 150 mM NaCl supplemented with DNase (Sigma-Aldrich) and disrupted with an M-110P Microfluidizer (Microfluidics). Intact cells and cell debris were removed by centrifugation at 8000g for 30 min, and membranes were collected by centrifugation of the supernatant at 170,000g for 1 hour. Membranes were resuspended in 20 mM tris-HCl (pH 7.5), 150 mM NaCl, and 10% glycerol; snap-frozen in liquid nitrogen; and stored at −80°C until further use. MsbA was solubilized with 1% (w/v) β-DDM (n-dodecyl-b-d-maltoside) for 3 hours at 4°C, and the insoluble fraction was removed by centrifugation at 170,000g for 1 hour. The cleared lysate was supplemented with 30 mM imidazole (pH 7.5) and loaded onto Ni-NTA gravity flow columns. After washing with 20 column volumes of 50 mM imidazole (pH 7.5), 200 mM NaCl, and 0.03% (w/v) β-DDM, MsbA was eluted with 4 column volumes of 200 mM imidazole (pH 7.5), 200 mM NaCl, and 0.03% (w/v) β-DDM. The eluate was concentrated to 2.5 ml using Amicon Ultra-4 concentrator units with a 50-kDa MWCO and loaded onto a PD10 desalting column (GE Healthcare) equilibrated with 20 mM tris-HCl (pH 7.5), 150 mM NaCl, and 0.03% (w/v) β-DDM. MsbA was eluted with 3.5 ml of 20 mM tris-HCl (pH 7.5), 150 mM NaCl, and 0.03% (w/v) β-DDM and incubated with 400 μg of 3C protease overnight at 4°C. Reverse IMAC was performed to remove His-tag and 3C protease by loading cleaved MsbA onto Ni-NTA gravity flow columns and directly collecting flow-through as well as three additional column volumes of 20 mM tris-HCl (pH 7.5), 150 mM NaCl, and 0.03% (w/v) β-DDM. The sample was again concentrated and separated by size exclusion chromatography (Superdex-200 10/300 GL) in 20 mM tris-HCl (pH 7.5), 150 mM NaCl, and 0.03% (w/v) β-DDM. For mutant MsbA_N191C, 2 mM dithiothreitol was included in the solubilization and IMAC buffer, and purification proceeded as described for MsbA. After IMAC, the protein was concentrated to 2.5 ml, loaded onto a PD10 desalting column, and eluted with 3.2 ml of 20 mM tris-HCl (pH 7.5), 150 mM NaCl, and 0.03% (w/v) β-DDM. Immediately after, a fivefold excess of MTSSL [(1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl) methanethiosulfonate] was added, and the samples were incubated at 4°C overnight. Excess of spin label was removed by size exclusion chromatography on a Superdex-200 10/300 GL column with 20 mM tris-HCl (pH 7.5), 150 mM NaCl, and 0.03% (w/v) β-DDM. For Avi-tagged MsbA, enzymatic biotinylation was performed during 3C cleavage using 3C protease (0.2 mg) and 330 nM BirA (0.016 mg/ml) in a buffer containing 20 mM imidazole (pH 7.5), 10 mM magnesium acetate, 200 mM ATP, 200 mM NaCl, 10% glycerol (v/v), 0.03% (w/v) β-DDM, and a 1.2-fold molar excess of biotin.
Nanobody selection
To obtain nanobodies against MsbA, an alpaca was immunized four times in biweekly intervals with purified MsbA in 20 mM tris-HCl (pH 7.5), 150 mM NaCl, and 0.03% (w/v) β-DDM, and phage libraries were generated as described previously (41, 48). Immunizations of alpacas were approved by the Cantonal Veterinary Office in Zürich, Switzerland (animal experiment license no. 188/2011). Two rounds of phage display were performed against detergent-solubilized, Avi-tagged MsbA (13), and nanobody enrichment was determined by quantitative polymerase chain reaction (605-fold enrichment after the second and final round of phage display) (36). The enriched library was then subcloned into the pSb_init vector by FX cloning and subsequently used for screening with enzyme-linked immunosorbent assay (ELISA) using biotinylated full-length MsbA and MsbA-NBD as targets (36). Ninety-four single clones were screened, and 86 gave ELISA signals against the full-length MsbA, out of which 11 were also positive against MsbA-NBD. Sanger sequencing revealed three distinct families based on the CDR (complementarity determining region) composition. From each family, one candidate was subcloned into vector pBXNPHM3 (Addgene #110099) and attempted to crystallize, of which one nanobody called Nb_MsbA#1 yielded diffracting crystals.
Purification and spin labeling of Nb_MsbA#1
Nb_MsbA#1 was expressed in E. coli MC1061 from vector pBXNPHM3 (Addgene #110099) for the production of tag-free binder for crystallization (48). Cysteine mutants Nb_MsbA#1_A60C and Nb_MsbA_T68C were expressed from the same vector, purified under reducing conditions, and labeled with a 3.6 molar excess of Gd-DOTA as previously described (34).
Crystallization and structure determination
MsbA-NBD at 10 mg/ml was mixed with a 1.1 molar excess of Nb_MsbA#1 and incubated for 5 min. Sitting drop vapor diffusion was used to obtain crystals after 3 days. Crystals were grown at 20°C in 100 mM Na-acetate (pH 5.5), 0.15 M KSCN, and 15% (w/v) polyethylene glycol 6000. Crystals were cryoprotected in reservoir solution containing 30% (v/v) ethylenglycol and flash-frozen in liquid nitrogen. Data were collected at beamline X06DA of the Swiss Light Source (SLS; Villigen, Switzerland), setting the wavelength to 1 Å at 100 K. XDS was used to process diffraction data, and molecular replacement was performed in Phaser2 using MsbA-NBD [Protein Data Bank (PDB): 3B5W] and a nanobody backbone as search models. Models were refined using PHENIX (49), and Coot (50) was used for model building (Ramachandran favored/outliers: 97.14%/0.29%).
Affinity determination by GCI
Affinity of nanobodies was determined using GCI on the WAVEsystem (Creoptix AG, Switzerland), a label-free biosensor. Avi-tagged full-length MsbA was captured onto a streptavidin PCP-STA WAVEchip (polycarboxylate quasi-planar surface; Creoptix AG) to a density of 2564 pg/mm2. Binding kinetics were measured at 25°C with increasing concentration ranging from 1 to 243 nM (threefold serial dilution, six concentrations) in 20 mM tris (pH 7.5), 150 mM NaCl, and 0.03% (w/v) β-DDM. Nanobodies were injected at a flow rate of 50 μl/min for 500 s, and dissociation was continued for 1500 s. Data were analyzed on the WAVEcontrol (Creoptix AG) software. After double-referencing by subtracting the signals from blank injections and from the reference channel, a Langmuir 1:1 model was used for data fitting.
ATPase activity assays
ATPase activity of MsbA in detergent, nanodiscs, and proteoliposomes was determined with or without 200 nM Nb_MsbA#1 and labeled variants thereof. Nanobodies were dialyzed three times against 20 mM tris-HCl (pH 7.5) and 150 mM NaCl to remove the remaining traces of phosphate before use in the assay. ATPase assays were performed in 150 mM NaCl and 10 mM MgSO4, containing, in addition, 0.03% (w/v) β-DDM in case the detergent-purified MsbA was analyzed. ATPase activities were measured using 8 nM reconstituted MsbA (proteoliposomes or nanodiscs) or 20 nM detergent-purified MsbA at 30°C for 15 min with 2 mM ATP. Liberated phosphate was detected using the molybdate/malachite green method (38). To this end, 90 μl of the reaction mixture was mixed with filtered malachite green solution [150 μl, ammonium molybdate (10.5 mg/ml), 0.5 M H2SO4, malachite green (0.34 mg/ml), and 0.1% Triton X-100], and absorption was measured at 650 nm.
Reconstitution of MsbA into nanodiscs
Membrane scaffold protein MSP1E3D1 was produced as previously described (41). E. coli polar lipids (Avanti, 100600C) were mixed 3:1 (w/w) with l-α-phosphatidylcholine from egg yolk (Sigma-Aldrich, P3556), and chloroform was removed by evaporation. The optimal ratio of 1:36 (MSP1E3D1 to lipids) was determined by reconstituting empty nanodiscs and assessing the size exclusion profiles using a Superdex-200 10/300 GL column. Detergent-purified MsbA was reconstituted into nanodiscs using MSP1E3D1 and E. coli polar lipids (Avanti Polar Lipids) at a ratio of 1:7:250 MsbA monomer/MSP/lipids in Na-Hepes (pH 8.0) with 30 mM cholate in a total volume of 800 μl. The reconstitution mixture was incubated at room temperature (RT) for 30 min. Two hundred milligrams of bio-beads was added and incubated overnight at 4°C while rocking at 650 rpm. After removing the bio-beads, reconstituted MsbA was mixed with a threefold molar excess of Gd-labeled nanobodies, and the complex was further purified by size exclusion chromatography using a Superdex-200 10/300 GL column equilibrated with 20 mM tris-HCl (pH 7.5) and 150 mM NaCl. MsbA-nanodiscs were concentrated using Amicon Ultra-4 concentrator units with a 100-kDa MWCO, split into aliquots, flash-frozen in liquid nitrogen, and stored at −80°C. These aliquots stemming from the exact same reconstitution were then used for cryo-EM structure determination, DEER measurements, and ATPase activity assays.
Cryo-EM structure determination
Three microliters of E. coli MsbA in complex with Gd-DOTA–labeled nanobody Nb_MsbA#1 (or without, in the case of Apo control) was applied onto freshly glow-discharged C-flat holey carbon grids (CF1.2/1.3) at a concentration of 2.6 to 2.9 mg/ml and plunge-frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific) with the environmental chamber set to 100% humidity and 4°C. To obtain structures in OF conformation, MsbA-nanobody complexes were incubated with 1 mM AMPPNP-Mg for 1 min before plunge-freezing. Micrographs (fig. S4) were recorded automatically with EPU, using a Titan Krios microscope (Thermo Fisher Scientific) operated at 300 kV and equipped with a BioQuantum energy filter and a K3 camera (Gatan), at a nominal magnification of ×105,000, corresponding to a pixel size of 0.837 Å. Dose-fractionated movies, composed of 50 frames, were acquired at an electron flux of 15 e−/pixel per second over 2.3 s, resulting in a total electron dose of approximately 50 e−/Å2. Images were recorded in the defocus range from −0.5 to −2.5 μm. Data collection quality was monitored in CryoSPARC Live. Frame-based motion correction was performed in RELION-3.1 (51), using its own implementation of MotionCor2 (52), and the contrast transfer function was estimated, using CTFFIND-4.1. For the first dataset of Msba-60Nb apo, the particles were picked, using template picker in RELION-3.1. For all other datasets, the particle picks were obtained with Topaz (53), using the model pretrained on the subset of selected particles from CryoSPARC Live. CryoSPARC Live was also used to generate the initial models. Picked particles were extracted at a box size of 64 pixels with 4.185 Å/pixel and directly subjected to a single round of multimodel three-dimensional (3D) classification in RELION-3.1 to eliminate bad picks. The selected particles were reextracted at full pixel size and refined using C2 symmetry. The particles were then polished and ctf-refined as implemented in RELION-3.1. To further sort out conformational variability, refined particles were subjected to the next round of 3D classification with no alignment. The resulting best classes were then refined, and an additional round of polishing and CTF refinement was performed. The polished particles were transferred to CryoSPARC to yield the final maps, using nonuniform refinement (54). CryoSPARC was also used for local resolution estimation. All datasets were processed in a similar manner, but because of the larger degree of flexibility, Apo datasets required an additional round of 3D classification. Msba-60Nb apo particles were additionally subjected to multibody refinement (55) in RELION-3.1, treating each MsbA “leg” as an individual body. To aid model building, density modification with phenix.resolve_cryo_em (56) was carried out, using the final half-maps, obtained in CryoSPARC. The previous cryo-EM structures in nanodiscs (PDB: 5TV4 and 5TTP for the IFnarrow and OFoccluded conformations, respectively) were used as initial templates together with the crystal structure of Nb_MsbA#1, solved in this study. All structures were manually edited in COOT and refined using phenix.real_space_refine (57) in combination with rigid-body refinement and several rounds of rebuilding in COOT. The quality check for each structure was performed with MolProbity (49). Refinement and validation statistics are summarized in table S2. All structural figures were generated, using UCSF Chimera or ChimeraX (58). The uncertainties in the Gd-Gd distances shown in Figs. 1 to 4 were extracted from the four cryo-EM structures and represent the ±σ interval. For the OFoccluded structures determined in the presence of AMPPNP-Mg, σ corresponds to the nominal resolution of the respective cryo-EM structures. In case of the IFnarrow conformations, where multibody refinement revealed continuous motion of the NBDs (movie S1), the largest and smallest distances, extracted from the multicomponent analysis, plus the resolution of the structure were used to estimate σ.
Reconstitution of MsbA into proteoliposomes
E. coli polar lipids and l-α-phosphatidylcholine were mixed at a ratio of 3:1 and dissolved in chloroform (20 mg/ml). Chloroform was evaporated, and the dried lipid film was dissolved in 50 mM K-Hepes (pH 7) to a final concentration of 20 mg/ml. Lipids were sonicated in six cycles at an intensity of 4 μm on ice to obtain small unilamellar vesicles (SUVs), which were then flash-frozen in liquid nitrogen and stored at −80°C until further use. Three freeze-thaw cycles converted SUVs into large multilamellar vesicles, which were extruded 11 times through a 400-nm polycarbonate filter prewashed with methanol, ethanol, and 50 mM K-Hepes (pH 7) to obtain large unilamellar vesicles (LUVs) at a final concentration of 4 mg/ml. LUVs were destabilized with 2.5 mM Triton X-100. Detergent-purified MsbA or MsbA_N191C-MTSL was added to 5 ml of destabilized LUVs at a protein/lipid ratio of 1:50 and incubated at 25°C for 30 min. Two hundred milligrams of bio-beads was added, and the mixture was incubated for 30 min at 25°C. Bio-beads were replaced three times with 200 mg of fresh beads and incubated at 4°C for 1 hour, 16 hours, and 1 hour in between. Proteoliposomes were separated from bio-beads and harvested by ultracentrifugation at 120,000g for 40 min. Proteoliposomes were resuspended to 20 mg/ml in 50 mM K-Hepes (pH 7), and the protein concentration was determined by SYPRO Ruby Protein Gel Stain (Bio-Rad, #1703126) using a dilution series of detergent-purified MsbA as a standard curve. Labeled nanobodies were added at a threefold molar excess to MsbA reconstituted in proteoliposomes and incubated on ice for 3 min. Proteoliposomes were washed using two rounds of centrifugation (170,000g, 30 min, 4°C) and resuspension [2 ml of 50 mM K-Hepes (pH 7)] to remove unbound nanobodies. The last resuspension was done with 90 μl of 50 mM K-Hepes (pH 7) (prepared with deuterium oxide) and 20% glycerol-d8 (Sigma-Aldrich), and 40 μl was added to electron paramagnetic resonance (EPR) tubes and snap-frozen in liquid N2.
Preparation of inside-out vesicles containing overexpressed MsbA, MsbA_E506Q, or TM287/288
Plasmids pBAD24_MsbA, pBAD24_MsbA_E506Q (13), and pBXNH3L_TM287/288 (19) were freshly transformed into chemically competent E. coli MC1061 cells and streaked onto agar plates containing ampicillin (100 μg/ml). From a single colony, overnight cultures were prepared in TB medium containing ampicillin (100 μg/ml), from which 10 ml was used to inoculate 1 liter of TB containing ampicillin (100 μg/ml). The culture was grown at 37°C for 1.5 hours followed by incubation at 30°C until an OD600 of 1.2 to 1.5 was reached. Protein expression was induced by the addition of l-arabinose [0.01% (w/v) for MsbA and 0.0017% (w/v) for TM287/288], and protein was produced for 4 hours at 30°C. Cells were harvested and diluted in 20 mM tris-HCl (pH 7.5) and 150 mM NaCl supplemented with DNase (Sigma-Aldrich). The cell solution was passed three times through an M-110P Microfluidizer (Microfluidics) at 25,000 lb. in−2 while cooling to 4°C. Intact cells and cell debris were removed by centrifugation at 8000g for 30 min, and vesicles were harvested by ultracentrifugation at 170,000g for 1 hour and resuspended with 4 ml of 20 mM tris-HCl (pH 7.5) and 150 mM NaCl per liter of expression culture. ISOVs were then aliquoted, flash-frozen in liquid nitrogen, and stored at −80°C until further use or used directly. To 100 μl of ISOVs, 18 μM Gd-labeled nanobodies was added and incubated for 3 min on ice. After two rounds of washing by centrifugation (170,000g, 1 hour, 4°C) and resuspension in phosphate-buffered saline (PBS), the ISOVs containing bound nanobody were resuspended in 90 μl of PBS (prepared with deuterium oxide) and 20% glycerol-d8 (Sigma-Aldrich). Nucleotides (ATP, ADP, or AMPPNP) were added at a concentration of 5 or 0.5 mM (as indicated in Results), and the mixtures were filled into EPR tubes, incubated for 3 min at 37°C, and directly snap-frozen in liquid N2.
Preparation of E. coli cells loaded with Gd-labeled nanobodies
To generate electrocompetent cells, 600 ml of TB medium supplemented with ampicillin (100 μg/ml) was inoculated 1:100 with overnight cultures of E. coli MC1061 containing either plasmid pBAD_MsbA, pBAD_MsbA_E506Q, or pBXNH3L_TM287/288 (control) and grown for 1.5 hours at 37°C and 1.5 hours at 30°C until OD600 reached 1.2 to 1.5. Expression was induced by adding l-arabinose [0.01% (w/v) for MsbA and 0.0017% (w/v) for TM287/288] and proceeded for 2.5 to 3 hours at 30°C. Cells were harvested at RT at 3300g for 10 min. Cells were washed twice with sterile ddH2O at RT and resuspended with 0.5 ml of sterile ddH2O. Gd-labeled nanobodies spin-labeled either at position 60 or 68 and purified in 20 mM tris-HCl (pH 7.5) and 150 mM NaCl were concentrated to 0.5 to 1 mM and dialyzed against 2 mM tris-HCl (pH 7.5) and 15 mM NaCl. Two hundred microliters of competent cells in ddH2O was then mixed with stock solutions of Gd-labeled nanobodies to reach final concentrations of 20, 25, or 30 μM (15-20-25 for the MsbA_E506Q mutant) (fig. S16). The mixture was transferred to cooled 2-mm electroporation cuvettes (Bio-Rad, 1652082) and electroporated at 2.5 kV, 200 ohms, and 25 μF. After electroporation, 1 ml of cooled LB was added immediately to the cuvettes, and the mixture was transferred to 100 ml of LB. Cells were recovered at 37°C for 20 min and then harvested by centrifugation at 3300g for 10 min. To remove noninternalized nanobody, the recovered cells were washed at 4°C by centrifugation at 3300g (fig. S16). The first washing step was performed with 5 ml of PBS, 0.005% Triton X-100, and 100 mM NaCl, followed by a washing step with 5 ml of PBS. The cell pellet was lastly resuspended with 500 μl of PBS, loaded to two 0.45-μm spin filters (Ultrafree_CL, Millipore) prewashed with PBS, and centrifuged for 3 min at 800g. The cells were washed two more times by resuspending them with 500 μl of PBS, applying them to the same spin filters and centrifugation at 800g for 3 min. Washed cells were then collected with 1 ml of PBS per spin filter and pooled into a 15-ml falcon tube. Washed cells were pelleted at 3300g for 10 min, and the pellet was 1:1 (v/v) diluted and resuspended with PBS (prepared with deuterium oxide) and 20% glycerol-d8 (Sigma-Aldrich). Forty microliters of the suspension was filled into EPR tubes and flash-frozen in liquid nitrogen. For control experiments, cells were incubated for 20 min at 4°C with 500 μM nonlabeled Nb_MsbA#1, pelleted at 3300g for 10 min, and the pellet was 1:1 (v/v) diluted and resuspended with PBS (prepared with deuterium oxide) and 20% glycerol-d8 (Sigma-Aldrich).
Test experiments to assess the electroporation protocol using fluorescently labeled nanobodies
Nb_MsbA#1_T68C was labeled with a 3.6 molar excess of Alexa Fluor 594 maleimide (Thermo Fisher Scientific) following the same protocol used to attach Gd-DOTA labels. Electroporation of E. coli cells with fluorescently labeled nanobody was carried out as described above, using 25 μM nonlabeled nanobody during electroporation. Fluorescence was assessed in a microplate reader (Cytation 5, BioTek) with the following settings: excitation at 575 nm and emission at 620 nm. For fluorescent micrographs of E. coli cells loaded with fluorescently labeled nanobody, cells were fixed in an eight-well μ-Slide (ibidi). One microliter of bacterial suspension in PBS was added. To a corner of the well, 50 μl of dissolved and warm 1% agarose in PBS was added and allowed to solidify at RT. Microscopic images were observed at ×63 magnification using a Leica HC PL APO CS2 63×/1.40 objective with Leica Type F immersion oil on a Leica SP8 X CLSM microscope. Fluorescence of Alexa Fluor 594 was detected with a red light filter (excitation at 598 nm and emission at 603 to 766 nm), and images were processed with Leica LAS X software. Image overlaying was done by combining the consecutive images with and without a red light fluorescence filter. Images were processed with ImageJ image processing software.
Determination of ADP/ATP ratios in E. coli cells
Cells (1.5 ml) at an OD600 of 1 were collected by centrifugation (3 min, 3300g), and the pellet was resuspended with 50 μl of ddH2O. Cell suspensions were added to 50 μl of ice-cold 10% trichloroacetic acid (w/v) supplemented with 10 mM EDTA, vortexed for 10 s, and incubated on ice for 20 min. After centrifugation (5 min, 20,000g, 4°C), the supernatant was aliquoted and stored at −20°C for up to 5 days. Sample (10 μl) was neutralized with 40 μl of 0.5 M Na-Hepes (pH 8.5) and the ADP/ATP ratio was determined with the ADP/ATP Ratio Assay Kit (Sigma-Aldrich, MAK135).
MD simulations
All-atom MD simulations of NBD-nanobody complexes were performed with GROMACS version 2020.1 (59) using the AMBER ff99SB-disp (60) protein force field with the corresponding modified TIP4P-D water model (61). The sizes and compositions of all the simulated systems are listed in table S3. From the x-ray crystal structure of the NBD-nanobody tetramer complex, two dimeric simulation systems were generated, each composed of one NBD chain (residues 340 to 580 and 340 to 579 for chains A and B, respectively) in complex with one nanobody chain (residues 1 to 113). Crystal water molecules were kept upon setting up the simulations. The dimers were solvated with ca. 45,000 water molecules in periodic cubic boxes with an edge length of ca. 11.2 nm; ions were added to neutralize the systems (see table S3). After energy minimization with steepest descent (for about 1000 steps), the systems were equilibrated for 20 ns with harmonic position restraints on all protein heavy atoms, followed by subsequent 30 ns with only the protein backbone atoms restrained, using force constants of 1000 kJ/mol per square nanometer. The initial equilibrations were performed under NPT conditions at 300 K and 1 bar, using the v-rescale thermostat (62) (coupling time constant τt = 0.5 ps) and the Berendsen barostat (63) (coupling time constant τp = 2.0 ps and a compressibility of 4.5 × 10−5/bar), respectively. Nonbonded Coulomb and Lennard-Jones interactions were treated up to a cutoff of 1 nm. Long-range electrostatics were treated with the particle-mesh Ewald method (64) with default settings. The LINCS algorithm was used to constrain bond lengths (65), allowing the integration of the equations of motion with 2-fs time steps. For each NDB-nanobody dimer complex, two unrestrained production simulations of 800 ns were carried out, starting from different initial atomic velocities and with the same parameters as described above, except that the pressure was controlled by the Parrinello-Rahman barostat (66). Coordinates were saved to the disk every 50 ps. All analyses were carried out on the trajectories from the production runs. The GROMACS analysis tools gmx rms, gmx rmsf, and gmx distance were used to calculate the root mean square deviation (RMSD), root mean square fluctuation (RMSF), and distances, respectively. For the RMSF calculation, the first 200 ns of the production run was discarded. Molecular graphics were generated with VMD 1.9.4 (www.ks.uiuc.edu/Research/vmd/). Data were plotted using Grace (http://plasma-gate.weizmann.ac.il/Grace/).
Coarse-grained MD simulations of full-length MsbA were carried out with GROMACS version 2021.1 with the Martini 2.2 force field (67). For modeling the IFwide conformation of full-length MsbA, the E. coli MsbA protein sequence (UniProt ID: P60752) was aligned to the Salmonella typhimurium MsbA (UniProt ID: P63359) using Clustal Omega (68). The 2.80-Å-resolution x-ray crystal structure from S. typhimurium (PDB: 6BL6) (16) was used as a template. Target and template share 96% of sequence identity, and the structural superposition over the Cα atoms of the target 6BL6 and the E. coli IFwide Cα structure PDB: 3B5W (4) yielded an RMSD of 1.29 Å. The homology modeling was carried out using the MODELLER software (69), and after 300 iterations, the best MsbA E. coli structural model was selected according to the discrete optimized protein energy (DOPE) score. For modeling the IFnarrow conformation of full-length MsbA, the starting coordinates were taken from the MsbA E. coli cryo-EM structure PDB: 6UZ2 (4.20-Å resolution). Missing C-terminal residues and side chains in the NBD domains were added with UCSF Chimera, and the coordinates of the first 10 N-terminal residues were taken from the IFwide model described above. The E. coli MsbA atomistic models described above were converted to the coarse-grained resolution using the martinize.py script. An elastic network with a cutoff distance of 12 Å was used (eu 1.2 and ef 1000 settings in martinize.py) to maintain the structural integrity of the protein. The protein structure was initially energy-minimized and inserted into a bilayer resembling the E. coli lipid composition using an in-house modified version of the insane.py tool (70) to include E. coli lipid A (see below). The composition of the outer leaflet was 67% 1-palmitoyl-2-oleyl-phosphtidylethanolamine (POPE), 23% 1-palmitoyl-2-oleyl-phosphtidylglycerol (POPG), and 10% cardiolipin. The composition of the inner leaflet was 62% POPE, 18% POPG, 10% cardiolipin, and 10% lipid A. The total number of lipids in the outer and inner bilayer leaflets was 291 and 270, respectively. The Martini topology for the E. coli lipid A was adapted from that of the Campylobacter jejuni lipid A (71) by removing the beads C4C2 and C3D2 from tails C and D, respectively. The solvated systems were neutralized with a 150 mM concentration of NaCl and subsequently energy-minimized with steepest descent until machine precision. The minimized systems were equilibrated by 100 ns of MD simulation with the protein backbone beads restrained (force constants of 2000 kJ/mol per square nanometer) to allow the bilayer to equilibrate around MsbA. The integration time step for the coarse-grained simulations was 20 fs. The “new-RF” simulation parameters were used, as suggested by de Jong et al. (72). Equilibration was performed at 330 K, with protein, lipids, and solvent separately coupled to an external bath using the v-rescale thermostat (62) with coupling time constant τT = 1.0 ps. Pressure was maintained at 1 bar using the stochastic cell rescaling (c-rescale) barostat (73) with semi-isotropic conditions (coupling time constant τp = 12.0 ps and compressibility = 3.0 × 10−4/bar). For both IFwide and IFnarrow, four independent production simulations (each 50 μs in length) were carried out using different random seeds for the initial velocities.
Distance distribution simulations and EPR experiments
Interspin distances between two Nb_MsbA#1 nanobodies spin-labeled at position 60 or 68 were performed with a rotamer-based approach in MMM 2018.2 (39, 74). Simulations were carried out using the available ambient temperature rotamer libraries for MTSL and Gd-DOTA. The nanobody-NDB crystal structure was superimposed to the available crystal or cryo-EM structures by aligning the NBDs in PyMOL (75).
The labeling efficiency for the MTSL-labeled nanobody was determined via CW-EPR spectroscopy by comparison of the double spectral integrals with a nitroxide standard at a known concentration, resulting in values of 90% for A60C and 100% for T68C. Labeling efficiencies of the Gd-DOTA-labeled nanobodies were determined by mass spectrometry as previously described (fig. S2) (34).
The final concentration of the WT MsbA used for DEER measurements was 20 μM for the detergent-purified protein and 10 and 20 μM for the reconstituted transporter in proteoliposomes and nanodiscs, respectively. The nanobody was added to the transporter in a 1:2 transporter/nanobody stoichiometric ratio. All AMPPNP-Mg samples contained 5 mM AMPPNP and 5 mM MgCl2 and were incubated at 298 K for 8 min. Glycerol-d8 (10%, v/v) was added as a cryoprotectant to all samples immediately before shock-freezing in liquid nitrogen. Forty microliters of the sample was loaded in quartz tubes (Aachener Quarzglas Heinrich) with an outer diameter of 3 mm. The preparation of ISOVs and cells for DEER has been described in the previous dedicated sections.
Biological repeats were performed for samples in each environment. In particular, two batches of MsbA WT reconstituted in nanodiscs (fig. S22), two batches of MsbA reconstituted in proteoliposomes (see fig. S23 for the data on the second batch), four batches of ISOVs overexpressing MsbA WT [see fig. S15 (B and C) for comparison of AMPPNP-Mg states on different batches], and several batches of E. coli cells overexpressing MsbA WT or EQ (fig. S19) were prepared. Moreover, all conditions (apart from in-cell data) were tested with gadolinium- and nitroxide-labeled nanobodies to corroborate the conclusions.
EDFS (echo-detected field sweep) spectra (see fig. S18 for some examples) were recorded using a 16- to 32-ns sequence with Gaussian pulse shapes with an interpulse delay of 400 ns, and the echo was integrated over 32 ns. The field axis of the spectra was frequency-normalized in the figures for better comparison. The Hahn echo decays were recorded with a 16–τ–32 ns sequence with Gaussian pulses, with τ incremented in steps of 32 ns starting from 180 ns. The echo intensity (integrated over 32 ns) was plotted versus 2τ. The optimal microwave power for the pulses was determined via nutation experiments recorded at the maximum of the spectra. An example of Hahn echo decay for each investigated environment is shown in fig. S24.
DEER measurements were performed at 50 K (nitroxide-nitroxide) or 10 K (Gd-Gd) on a Bruker ELEXSYS E580Q-AWG (arbitrary waveform generator) dedicated pulse Q-band spectrometer equipped with a 150-W TWT amplifier and a homemade resonator. A four-pulse DEER sequence with a Gaussian, nonselective observer and pump pulses of 32 ns length (corresponding to 13.6 ns full width at half maximum) with 100-MHz frequency separation was used (76). The pump position was always chosen as the maximum of the gadolinium or nitroxide signal.
DEER experiments were performed using the dead-time free four-pulse DEER sequence (π/2)obs-(d1)-(π)obs-(d1 + T)-(π)pump-(d2-T)-(π)obs-(d2)-(echo) with a 16-step phase cycling. For the Gd-Gd measurements, we found that a small zero-time artifact due to pulse overlap could be removed by using 10 dB of attenuation in the main attenuator. The initial value of the interpulse delay τ1 was set to 400 ns for nitroxide DEER and 800 ns for gadolinium DEER. Deuterium ESEEM was suppressed incrementing τ1 eight times by 16 ns and summing the individual traces (nuclear modulation averaging). The dipolar evolution time τ2 ranged from 2000 to 5000 ns in the different samples, according to the dipolar frequency contained. The shot repetition time was set to 1 ms for nitroxide DEER and 1 or 2 ms for gadolinium DEER.
Data were analyzed using DEERNet (39, 77, 78), a neural network–based approach recently implemented in Spinach (79) that enables to extract distance distributions from DEER traces with an error estimation. Moreover, this analysis removes the user bias in the choice of the background function. In its latest version, the program provides estimation on the background function and fitting of the primary trace, with relative uncertainties. The output distributions, which are normalized by the modulation depth, have been intensity-normalized for better visualization, with the modulation depth being slightly different in different environments because of mismatch in the MsbA-nanobody stoichiometric ratio, which can be caused by the difficulty in determining protein concentration in certain environments. The distance distributions obtained by DEERNet for ISOVs and cells were further validated by analysis with Tikhonov and a model-based Gaussian fit from DeerAnalysis (80) to provide an independent data analysis. Details of the fitting and of the validations are reported in the corresponding legends in the Supplementary Materials.
DEER data processing in DEERNet
DEER data processing was performed using the DEERNet module (39) of Spinach 2.6 (79). Each DEER trace was resampled to 512 time points and fed into a set of fully connected neural networks with the following topology: vector input–(fully connected–batch normalization–softplus)5–probability normalization–vector regression. The weight matrices in the fully connected layers had dimensions of 512 × 512; the probability normalization layer divides the vector by its sum to ensure that the statistical requirement for the probabilities to add up to 100% is fulfilled.
Output confidence interval estimation was done by running statistics over a set of normalized distance distributions {pk(r)} obtained from 32 networks trained from different random initial guesses against different databases. To obtain background and modulation depth confidence intervals, distance distributions were converted back into form factors {fk(t)} using the analytical DEER kernel γ(r, t) (39). The form factors were mixed with the background signals
| (S1) |
using modulation depth μ and a stretched exponential background model with an effective dimension N and a decay rate constant k. The resulting function was fitted to the experimental DEER trace to extract a set of backgrounds and modulation depths. Confidence intervals were obtained by running statistics over the resulting set of 32 background signals and 32 modulation depths. The annotated MATLAB source code for the procedures described in this section is available as part of the Spinach library version 2.6 (http://spindynamics.org).
Acknowledgments
We acknowledge B. Blattmann and C. Stutz-Ducommun of the Protein Crystallization Center UZH for performing the crystallization screening and the staff of the SLS beamline X06DA for the support during data collection. We thank S. Štefanić of the Nanobody Service Facility (NSF), University of Zürich, for alpaca immunization. We thank C. von Ballmoos for giving technical advice on how to measure ATP/ADP ratios. We are grateful to W. Kühlbrandt and staff for access to the cryo-EM facility at the Max Planck Institute of Biophysics.
Funding: Research in the laboratory of I.K. was supported by grants from Leverhulme Trust (RPG-2019-048) and MathWorks and used NVIDIA Tesla A100 GPUs through NVIDIA Academic Grants Programme. Research in the laboratory of A.M. was funded by DFG through CRC944. Research in the laboratory of E.B. and L.V.S. was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy-EXC 2033-390677874–RESOLV and grant SCHA 1574/6-1. Research in the laboratory of M.A.S. was funded by the European Research Council (ERC) (consolidator grant no. 772190), an SNSF Professorship of the Swiss National Science Foundation (PP00P3_144823), and a project grant of the Swiss National Science Foundation (310030_188817).
Author contributions: E.B. and M.A.S. conceived the project. G.M. generated the nanobody and solved its crystal structure with the help of M.A.S. L.G. suggested the positions to be labeled. G.M. purified and spin-labeled all proteins for nanobody selection, DEER measurements, and cryo-EM analyses. G.M. carried out all reconstitutions in nanodiscs and proteoliposomes and prepared ISOVs. G.M. performed affinity measurements and ATPase activity assays and established the nanobody electroporation protocol with the support of B.S. and H.H. L.G. and G.M. identified optimal experimental conditions for the electroporation protocols. L.G. prepared DEER samples, performed and analyzed all EPR measurements, and interpreted the results together with E.B. D.J. prepared grids, recorded and processed cryo-EM data, and interpreted the structures together with A.M. D.J. and K.P. built and validated cryo-EM structures. D.V. built the MsbA atomistic models, performed MD simulations and discussed the results with L.V.S. I.K. optimized the neural network analysis with inputs of L.G. and E.B. All authors discussed and analyzed the results. L.G. prepared all EPR figures, G.M. prepared the supplementary figures related to the nanobody characterization and electroporation, D.J. and L.G. prepared Fig. 1, D.J. prepared the cryo-EM–related figures, and D.V. prepared the MD simulation figures. E.B., M.A.S., L.G., G.M., D.J., A.M., and L.V.S. wrote the paper and all authors edited the paper.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The expression plasmid encoding for Nb_MsbA#1 was made available via Addgene (#186428). The crystal structure of Nb_MsbA#1 in complex with the MsbA-NBD was deposited under the accession number 7NDF. The four 3D cryo-EM density maps of nanodisc-reconstituted MsbA in complex with spin-labeled nanobody Nb_MsbA#1 were deposited to the Electron Microscopy Data Bank under the following accession numbers: EMD-13404 for MsbA-60Nb IFnarrow, EMD-13405 for MsbA-60Nb OFoccluded, EMD-13409 for MsbA-68Nb IFnarrow, and EMD-13406 for MsbA-68Nb OFoccluded. The corresponding atomic models have been deposited to the PDB under accession numbers 7PH2, 7PH3, 7PH7, and 7PH4, accordingly (table S2). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The primary DEER traces of the main figures are available at https://doi.org/10.26037/yareta:o3t4cjzskrbxlpnz5rehlcatuu.
Supplementary Materials
This PDF file includes:
Figs. S1 to S24
Tables S1 to S3
Other Supplementary Material for this manuscript includes the following:
Movie S1
REFERENCES AND NOTES
- 1.Polissi A., Georgopoulos C., Mutational analysis and properties of the msbA gene of Escherichia coli, coding for an essential ABC family transporter. Mol. Microbiol. 20, 1221–1233 (1996). [DOI] [PubMed] [Google Scholar]
- 2.Zhou Z. M., White K. A., Polissi A., Georgopoulos C., Raetz C. R. H., Function of Escherichia coli MsbA, an essential ABC family transporter, in lipid A and phospholipid biosynthesis. J. Biol. Chem. 273, 12466–12475 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Thomas C., Aller S. G., Beis K., Carpenter E. P., Chang G., Chen L., Dassa E., Dean M., Duong van Hoa F., Ekiert D., Ford R., Gaudet R., Gong X., Holland I. B., Huang Y., Kahne D. K., Kato H., Koronakis V., Koth C. M., Lee Y., Lewinson O., Lill R., Martinoia E., Murakami S., Pinkett H. W., Poolman B., Rosenbaum D., Sarkadi B., Schmitt L., Schneider E., Shi Y., Shyng S. L., Slotboom D. J., Tajkhorshid E., Tieleman D. P., Ueda K., Váradi A., Wen P. C., Yan N., Zhang P., Zheng H., Zimmer J., Tampé R., Structural and functional diversity calls for a new classification of ABC transporters. FEBS Lett. 594, 3767–3775 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ward A., Reyes C. L., Yu J., Roth C. B., Chang G., Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc. Natl. Acad. Sci. U.S.A. 104, 19005–19010 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woebking B., Reuter G., Shilling R. A., Velamakanni S., Shahi S., Venter H., Balakrishnan L., van Veen H. W., Drug-lipid A interactions on the Escherichia coli ABC transporter MsbA. J. Bacteriol. 187, 6363–6369 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Higgins C. F., Linton K. J., The ATP switch model for ABC transporters. Nat. Struct. Mol. Biol. 11, 918–926 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Dawson R. J., Locher K. P., Structure of a bacterial multidrug ABC transporter. Nature 443, 180–185 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Lacabanne D., Orelle C., Lecoq L., Kunert B., Chuilon C., Wiegand T., Ravaud S., Jault J. M., Meier B. H., Böckmann A., Flexible-to-rigid transition is central for substrate transport in the ABC transporter BmrA from Bacillus subtilis. Commun. Biol. 2, 149 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mi W., Li Y., Yoon S. H., Ernst R. K., Walz T., Liao M., Structural basis of MsbA-mediated lipopolysaccharide transport. Nature 549, 233–237 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choudhury H. G., Tong Z., Mathavan I., Li Y., Iwata S., Zirah S., Rebuffat S., van Veen H. W., Beis K., Structure of an antibacterial peptide ATP-binding cassette transporter in a novel outward occluded state. Proc. Natl. Acad. Sci. U.S.A. 111, 9145–9150 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hofmann S., Januliene D., Mehdipour A. R., Thomas C., Stefan E., Brüchert S., Kuhn B. T., Geertsma E. R., Hummer G., Tampé R., Moeller A., Conformation space of a heterodimeric ABC exporter under turnover conditions. Nature 571, 580–583 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou P., Bortolus M., McHaourab H. S., Conformational cycle of the ABC transporter MsbA in liposomes: Detailed analysis using double electron-electron resonance spectroscopy. J. Mol. Biol. 393, 586–597 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mittal A., Böhm S., Grütter M. G., Bordignon E., Seeger M. A., Asymmetry in the homodimeric ABC transporter MsbA recognized by a DARPin. J. Biol. Chem. 287, 20395–20406 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moradi M., Tajkhorshid E., Mechanistic picture for conformational transition of a membrane transporter at atomic resolution. Proc. Natl. Acad. Sci. U.S.A. 110, 18916–18921 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moeller A., Lee S. C., Tao H., Speir J. A., Chang G., Urbatsch I. L., Potter C. S., Carragher B., Zhang Q., Distinct conformational spectrum of homologous multidrug ABC transporters. Structure 23, 450–460 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Padayatti P. S., Lee S. C., Stanfield R. L., Wen P.-C., Tajkhorshid E., Wilson I. A., Zhang Q., Structural insights into the lipid a transport pathway in MsbA. Structure 27, 1114–1123.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho H., Miu A., Alexander M. K., Garcia N. K., Oh A., Zilberleyb I., Reichelt M., Austin C. D., Tam C., Shriver S., Hu H., Labadie S. S., Liang J., Wang L., Wang J., Lu Y., Purkey H. E., Quinn J., Franke Y., Clark K., Beresini M. H., Tan M. W., Sellers B. D., Maurer T., Koehler M. F. T., Wecksler A. T., Kiefer J. R., Verma V., Xu Y., Nishiyama M., Payandeh J., Koth C. M., Structural basis for dual-mode inhibition of the ABC transporter MsbA. Nature 557, 196–201 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Angiulli G., Dhupar H. S., Suzuki H., Wason I. S., van Hoa F. D., Walz T., New approach for membrane protein reconstitution into peptidiscs and basis for their adaptability to different proteins. eLife 9, e53530 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hohl M., Briand C., Grütter M. G., Seeger M. A., Crystal structure of a heterodimeric ABC transporter in its inward-facing conformation. Nat. Struct. Mol. Biol. 19, 395–402 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Lee J. Y., Yang J. G., Zhitnitsky D., Lewinson O., Rees D. C., Structural basis for heavy metal detoxification by an Atm1-type ABC exporter. Science 343, 1133–1136 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shintre C. A., Pike A. C. W., Li Q., Kim J. I., Barr A. J., Goubin S., Shrestha L., Yang J., Berridge G., Ross J., Stansfeld P. J., Sansom M. S. P., Edwards A. M., Bountra C., Marsden B. D., von Delft F., Bullock A. N., Gileadi O., Burgess-Brown N. A., Carpenter E. P., Structures of ABCB10, a human ATP-binding cassette transporter in apo- and nucleotide-bound states. Proc. Natl. Acad. Sci. U.S.A. 110, 9710–9715 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Locher K. P., Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 23, 487–493 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Wright J., Muench S. P., Goldman A., Baker A., Substrate polyspecificity and conformational relevance in ABC transporters: New insights from structural studies. Biochem. Soc. Trans. 46, 1475–1484 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Loo T. W., Bartlett M. C., Clarke D. M., Human P-glycoprotein is active when the two halves are clamped together in the closed conformation. Biochem. Biophys. Res. Common. 395, 436–440 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Perez C., Gerber S., Boilevin J., Bucher M., Darbre T., Aebi M., Reymond J. L., Locher K. P., Structure and mechanism of an active lipid-linked oligosaccharide flippase. Nature 524, 433–438 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Liu Y., Liu Y., He L., Zhao Y., Zhang X. C., Single-molecule fluorescence studies on the conformational change of the ABC transporter MsbA. Biophys. Rep. 4, 153–165 (2018). [Google Scholar]
- 27.Josts I., Nitsche J., Maric S., Mertens H. D., Moulin M., Haertlein M., Prevost S., Svergun D. I., Busch S., Forsyth V. T., Tidow H., Conformational states of ABC transporter MsbA in a lipid environment investigated by small-angle scattering using stealth carrier nanodiscs. Structure 26, 1072–1079.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Thélot F. A., Zhang W., Song K. K., Xu C., Huang J., Liao M., Distinct allosteric mechanisms of first-generation MsbA inhibitors. Science 374, 580–585 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooper R. S., Altenberg G. A., Association/dissociation of the nucleotide-binding domains of the ATP-binding cassette protein MsbA measured during continuous hydrolysis. J. Biol. Chem. 288, 20785–20796 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Theillet F. X., Binolfi A., Bekei B., Martorana A., Rose H. M., Stuiver M., Verzini S., Lorenz D., van Rossum M., Goldfarb D., Selenko P., Structural disorder of monomeric α-synuclein persists in mammalian cells. Nature 530, 45–50 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Yang Y., Chen S. N., Yang F., Li X. Y., Feintuch A., Su X. C., Goldfarb D., In-cell destabilization of a homodimeric protein complex detected by DEER spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 117, 20566–20575 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collauto A., Bülow S., Gophane D. B., Saha S., Stelzl L. S., Hummer G., Sigurdsson S. T., Prisner T. F., Compaction of RNA duplexes in the cell*. Angew. Chem. Int. Ed. Engl. 59, 23025–23029 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joseph B., Jaumann E. A., Sikora A., Barth K., Prisner T. F., Cafiso D. S., In situ observation of conformational dynamics and protein ligand-substrate interactions in outer-membrane proteins with DEER/PELDOR spectroscopy. Nat. Protoc. 14, 2344–2369 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galazzo L., Meier G., Timachi M. H., Hutter C. A. J., Seeger M. A., Bordignon E., Spin-labeled nanobodies as protein conformational reporters for electron paramagnetic resonance in cellular membranes. Proc. Natl. Acad. Sci. U.S.A. 117, 2441–2448 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pardon E., Laeremans T., Triest S., Rasmussen S. G. F., Wohlkönig A., Ruf A., Muyldermans S., Hol W. G. J., Kobilka B. K., Steyaert J., A general protocol for the generation of nanobodies for structural biology. Nat. Protoc. 9, 674–693 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zimmermann I., Egloff P., Hutter C. A. J., Arnold F. M., Stohler P., Bocquet N., Hug M. N., Huber S., Siegrist M., Hetemann L., Gera J., Gmür S., Spies P., Gygax D., Geertsma E. R., Dawson R. J. P., Seeger M. A., Synthetic single domain antibodies for the conformational trapping of membrane proteins. eLife 7, e34317 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Göddeke H., Timachi M. H., Hutter C. A. J., Galazzo L., Seeger M. A., Karttunen M., Bordignon E., Schäfer L. V., Atomistic mechanism of large-scale conformational transition in a heterodimeric ABC exporter. J. Am. Chem. Soc. 140, 4543–4551 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Hohl M., Hürlimann L. M., Böhm S., Schöppe J., Grütter M. G., Bordignon E., Seeger M. A., Structural basis for allosteric cross-talk between the asymmetric nucleotide binding sites of a heterodimeric ABC exporter. Proc. Natl. Acad. Sci. U.S.A. 111, 11025–11030 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Worswick S. G., Spencer J. A., Jeschke G., Kuprov I., Deep neural network processing of DEER data. Sci. Adv. 4, eaat5218 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schultz K. M., Merten J. A., Klug C. S., Characterization of the E506Q and H537A dysfunctional mutants in the E. coli ABC transporter MsbA. Biochemistry 50, 3599–3608 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hutter C. A. J., Timachi M. H., Hürlimann L. M., Zimmermann I., Egloff P., Göddeke H., Kucher S., Štefanić S., Karttunen M., Schäfer L. V., Bordignon E., Seeger M. A., The extracellular gate shapes the energy profile of an ABC exporter. Nat. Commun. 10, 2260 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ward A. B., Szewczyk P., Grimard V., Lee C. W., Martinez L., Doshi R., Caya A., Villaluz M., Pardon E., Cregger C., Swartz D. J., Falson P. G., Urbatsch I. L., Govaerts C., Steyaert J., Chang G., Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain. Proc. Natl. Acad. Sci. U.S.A. 110, 13386–13391 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Esser L., Zhou F., Pluchino K. M., Shiloach J., Ma J., Tang W. K., Gutierrez C., Zhang A., Shukla S., Madigan J. P., Zhou T., Kwong P. D., Ambudkar S. V., Gottesman M. M., Xia D., Structures of the multidrug transporter P-glycoprotein reveal asymmetric ATP binding and the mechanism of polyspecificity. J. Biol. Chem. 292, 446–461 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin M. S., Oldham M. L., Zhang Q., Chen J., Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans. Nature 490, 566–569 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Futamata R., Ogasawara F., Ichikawa T., Kodan A., Kimura Y., Kioka N., Ueda K., In vivo FRET analyses reveal a role of ATP hydrolysis-associated conformational changes in human P-glycoprotein. J. Biol. Chem. 295, 5002–5011 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bukowska M. A., Hohl M., Geertsma E. R., Hürlimann L. M., Grütter M. G., Seeger M. A., A transporter motor taken apart: Flexibility in the nucleotide binding domains of a heterodimeric ABC exporter. Biochemistry 54, 3086–3099 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Geertsma E. R., Dutzler R., A versatile and efficient high-throughput cloning tool for structural biology. Biochemistry 50, 3272–3278 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Zimmermann I., Egloff P., Hutter C. A. J., Kuhn B. T., Bräuer P., Newstead S., Dawson R. J. P., Geertsma E. R., Seeger M. A., Generation of synthetic nanobodies against delicate proteins. Nat. Protoc. 15, 1707–1741 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zivanov J., Nakane T., Forsberg B. O., Kimanius D., Hagen W. J. H., Lindahl E., Scheres S. H. W., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng S. Q., Palovcak E., Armache J. P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bepler T., Morin A., Rapp M., Brasch J., Shapiro L., Noble A. J., Berger B., Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Punjani A., Zhang H., Fleet D. J., Non-uniform refinement: Adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Nakane T., Kimanius D., Lindahl E., Scheres S. H., Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. eLife 7, e36861 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Terwilliger T. C., Ludtke S. J., Read R. J., Adams P. D., Afonine P. V., Improvement of cryo-EM maps by density modification. Nat. Methods 17, 923–927 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Afonine P. V., Poon B. K., Read R. J., Sobolev O. V., Terwilliger T. C., Urzhumtsev A., Adams P. D., Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goddard T. D., Huang C. C., Meng E. C., Pettersen E. F., Couch G. S., Morris J. H., Ferrin T. E., UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abraham M. J., Murtola T., Schulz R., Páll S., Smith J. C., Hess B., Lindahl E., GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2, 19–25 (2015). [Google Scholar]
- 60.Robustelli P., Piana S., Shaw D. E., Developing a molecular dynamics force field for both folded and disordered protein states. Proc. Natl. Acad. Sci. U.S.A. 115, E4758–E4766 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Piana S., Donchev A. G., Robustelli P., Shaw D. E., Water dispersion interactions strongly influence simulated structural properties of disordered protein states. J. Phys. Chem. B 119, 5113–5123 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Bussi G., Donadio D., Parrinello M., Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F., DiNola A., Haak J. R., Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 (1984). [Google Scholar]
- 64.Darden T., York D., Pedersen L., Particle mesh Ewald: An N.Log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993). [Google Scholar]
- 65.Hess B., Bekker H., Berendsen H. J. C., Fraaije J. G. E. M., LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 (1997). [Google Scholar]
- 66.Parrinello M., Rahman A., Polymorphic transitions in single-crystals—A new molecular-dynamics method. J. Appl. Phys. 52, 7182–7190 (1981). [Google Scholar]
- 67.de Jong D. H., Singh G., Bennett W. F. D., Arnarez C., Wassenaar T. A., Schäfer L. V., Periole X., Tieleman D. P., Marrink S. J., Improved parameters for the martini coarse-grained protein force field. J. Chem. Theory Comput. 9, 687–697 (2013). [DOI] [PubMed] [Google Scholar]
- 68.Sievers F., Higgins D. G., Clustal omega for making accurate alignments of many protein sequences. Protein Sci. 27, 135–145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Webb B., Sali A., Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. 86, 2.9.1–2.9.37 (2016). [DOI] [PubMed] [Google Scholar]
- 70.Wassenaar T. A., Ingolfsson H. I., Bockmann R. A., Tieleman D. P., Marrink S. J., Computational lipidomics with insane: A versatile tool for generating custom membranes for molecular simulations. J. Chem. Theory Comput. 11, 2144–2155 (2015). [DOI] [PubMed] [Google Scholar]
- 71.Ma H., Cummins D. D., Edelstein N. B., Gomez J., Khan A., Llewellyn M. D., Picudella T., Willsey S. R., Nangia S., Modeling diversity in structures of bacterial outer membrane lipids. J. Chem. Theory Comput. 13, 811–824 (2017). [DOI] [PubMed] [Google Scholar]
- 72.de Jong D. H., Baoukina S., Ingolfsson H. I., Marrink S. J., Martini straight: Boosting performance using a shorter cutoff and GPUs. Comput. Phys. Commun. 199, 1–7 (2016). [Google Scholar]
- 73.Bernetti M., Bussi G., Pressure control using stochastic cell rescaling. J. Chem. Phys. 153, 114107 (2020). [DOI] [PubMed] [Google Scholar]
- 74.Polyhach Y., Bordignon E., Jeschke G., Rotamer libraries of spin labelled cysteines for protein studies. Phys. Chem. Chem. Phys. 13, 2356–2366 (2011). [DOI] [PubMed] [Google Scholar]
- 75.The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.
- 76.Teucher M., Bordignon E., Improved signal fidelity in 4-pulse DEER with Gaussian pulses. J. Magn. Reson. 296, 103–111 (2018). [DOI] [PubMed] [Google Scholar]
- 77.Amey J. L., Keeley J., Choudhury T., Kuprov I., Neural network interpretation using descrambler groups. Proc. Natl. Acad. Sci. U.S.A. 118, e2016917118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Keeley J., Choudhury T., Galazzo L., Bordignon E., Feintuch A., Goldfarb D., Russell H., Taylor M. J., Lovett J. E., Eggeling A., Fábregas Ibáñez L., Keller K., Yulikov M., Jeschke G., Kuprov I., Neural networks in pulsed dipolar spectroscopy: A practical guide. J. Magn. Reson. 338, 107186 (2022). [DOI] [PubMed] [Google Scholar]
- 79.Hogben H. J., Krzystyniak M., Charnock G. T., Hore P. J., Kuprov I., Spinach—A software library for simulation of spin dynamics in large spin systems. J. Magn. Reson. 208, 179–194 (2011). [DOI] [PubMed] [Google Scholar]
- 80.Jeschke G., Chechik V., Ionita P., Godt A., Zimmermann H., Banham J., Timmel C. R., Hilger D., Jung H., DeerAnalysis2006—A comprehensive software package for analyzing pulsed ELDOR data. Appl. Magn. Reson. 30, 473–498 (2006). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S24
Tables S1 to S3
Movie S1