Abstract
Despite abundant research demonstrating that platelets can promote tumor cell metastasis, whether primary tumors affect platelet-producing megakaryocytes remains understudied. In this study, we used a spontaneous murine model of breast cancer to show that tumor burden reduced megakaryocyte number and size and disrupted polyploidization. Single-cell RNA sequencing demonstrated that megakaryocytes from tumor-bearing mice exhibit a pro-inflammatory phenotype, epitomized by increased Ctsg, Lcn2, S100a8, and S100a9 transcripts. Protein S100A8/A9 and lipocalin-2 levels were also increased in platelets, suggesting that tumor-induced alterations to megakaryocytes are passed on to their platelet progeny, which promoted in vitro tumor cell invasion and tumor cell lung colonization to a greater extent than platelets from wild-type animals. Our study is the first to demonstrate breast cancer–induced alterations in megakaryocytes, leading to qualitative changes in platelet content that may feedback to promote tumor metastasis.
Breast cancer creates pro-inflammatory megakaryocytes, producing “super-charged” platelets that exacerbate tumor cell metastasis.
INTRODUCTION
Blood platelets are small, anucleate cells produced by megakaryocytes (MKs) to act as essential mediators of primary hemostasis. While most platelets remain quiescent within circulation, activated platelets release bioactive compounds from specialized granules and establish adhesive connections between neighboring platelets and damaged blood vessels to preserve vascular integrity (1). However, inappropriate platelet activation can contribute to nonhemostatic processes, particularly cancer progression (2–6). Extensive studies have characterized how platelets mediate several hallmarks of cancer, including evasion of the immune system (7–9), tumor-supportive inflammation (10), and neoangiogenesis (11, 12).
Despite an intense focus on platelet-tumor interactions (13), it remains an understudied topic of research whether and how primary tumors affect platelet-producing MKs and thrombopoiesis. Tumor-derived interleukin-6 up-regulates hepatic thrombopoietin (TPO) production, leading to paraneoplastic thrombocytosis in ovarian cancer patients (14). In preclinical murine models, Lewis lung carcinoma increased levels of anti-angiogenic thrombospondin 1 (Thbs1) in both MKs and platelets (12), and hyperglycemia, which is an established risk factor for tumor metastasis, was shown to alter MK metabolism, leading to the production of platelets with increased in vitro adherence to melanoma cells (15). However, despite its prevalence (16) and ability to systemically manipulate the bone marrow (BM) microenvironment (17), the impact of breast cancer on MKs has not been investigated.
In this work, we characterized MK development, maturation, gene expression, and protein levels in several murine models of breast cancer. BM MKs from tumor-bearing mice displayed abnormal ploidy and a pro-inflammatory phenotype, with increased expression of various genes associated with tumor growth and metastasis, most notably Ctsg, Lcn2, S100a8, and S100a9 (18–23). Changes in MK gene expression were reflected in the protein content of circulating platelets and likely contribute to the platelet-tumor interactions that promote tumor cell invasion and metastasis. Both murine models and breast cancer patients had elevated serum but not plasma S100A8/A9, which was not observed in cancer-free controls, suggesting that activated platelets contribute to circulating S100A8/A9 in breast cancer. However, serum levels between breast cancer patients and cancer-free controls were comparable. Our findings highlight a tumor-induced generation of pro-inflammatory platelets that occur via alterations to BM MKs.
RESULTS
Morphological differences in MKs from tumor-bearing mice
To assess megakaryopoiesis in breast cancer, we used several preclinical breast cancer models, predominantly MMTV-PyMT transgenic mice, which form palpable autochthonous tumors in mammary fat pads from 8 weeks of age and were sacrificed at 13 weeks of age (Fig. 1A). We also used transplantable Met-1 and AT-3 cell lines, which were initially isolated from PyMT-driven tumors in FVB/NJ and C57BL/6J mice, respectively (24, 25). Mice were sacrificed 21 or 33 days after implantation of Met-1 or AT-3 cells, respectively (Fig. 1, B and C).
Fig. 1. Morphological characterization of MKs from mice with breast cancer.
Spontaneous tumor growth within the mammary fat pads of (A) MMTV-PyMT (PyMT) mice and tumor growth in mice orthotopically inoculated with either Met-1 (B) or AT-3 (C) cells. Arrows depict age of PyMT animals or days post-inoculation upon sacrifice. n = 5 to 10 mice per condition. (D to F) Femurs from PyMT and wild-type (WT) mice, or orthotopically injected animals were immunofluorescently stained for MKs (CD41, cyan), nuclei (DAPI, gray), and blood vessels (laminin, magenta). Phosphate-buffered saline (PBS)–injected mice were used as orthotopic tumor-naïve controls. Scale bars, 50 μm; n = 4 to 5 mice per condition. P values were determined by two-sample t tests. (G to I) Polyploidization of native MKs from either PyMT and WT mice or orthotopic Met-1 and AT-3 models. n = 4 to 5 mice per condition. *P < 0.05 and **P < 0.01, as determined by one-way ANOVA.
Femurs from 13-week-old PyMT mice had significantly fewer (P = 0.0074) and smaller (P = 0.0280) MKs compared to their PyMT-null [wild type (WT)] littermate controls (Fig. 1D), whereas the number and size of MKs from the Met-1 or AT-3 models were comparable to vehicle control [phosphate-buffered saline (PBS)]–injected animals (Fig. 1, E and F). Plasma levels of TPO showed a trending increase in PyMT mice (1.60-fold, P = 0.072) but were unaffected in either Met-1 or AT-3 models (fig. S1, A to C). MKs from PyMT mice appeared to have otherwise normal ultrastructure as determined by electron microscopy (fig. S2A). MKs from the BM of PyMT mice displayed abnormal ploidy, with increased numbers of 2N, 4N, and 32N MKs at the expense of 8N and 16N MKs (Fig. 1G). Despite increased 32N MKs in Met-1–implanted mice, no other abnormalities in MK polyploidization were observed in either the Met-1 or AT-3 orthotopic models (Fig. 1, H and I).
Given that MK size and polyploidization were altered in PyMT mice, we next assessed whether breast cancer affected progenitor differentiation during the early stages of megakaryopoiesis. Clear decreases were observed in early MK-erythroid progenitors but not terminally differentiated MK progenitors (fig. S3, A and B) and were associated with down-regulated red blood cells (RBCs; fig. S4A). We also detected myeloid expansion in the BM of both PyMT and Met-1 tumor–bearing mice, highlighted by increased myeloid-biased multipotent progenitors (MPP3; fig. S3, A and B) and white blood cells (WBCs; fig. S4, B and C). These findings suggest that despite alterations in MK polyploidization and size, there were no apparent abnormalities to early megakaryopoiesis in murine breast cancer.
Extramedullary MKs also reside outside of the BM niche (26), which prompted us to quantify MKs in the spleens of 13-week-old PyMT mice. PyMT mice had clear (WT = 0.11 ± 0.01 g; PyMT = 0.21 ± 0.05 g) splenomegaly (fig. S5, A and B) likely due to granulocytic hyperplasia (27), and absolute numbers of MKs were increased (fig. S5, C and D). However, when the spleen area was normalized to account for splenomegaly, PyMT mice had unaltered numbers of splenic MKs (fig. S5D). There was also no change in the number of CD41+ cells in the lungs of PyMT mice (fig. S6, A and B), suggesting organ-selective effects of this model on BM-resident MKs.
Differential gene expression in MKs from tumor-bearing mice
Several studies have identified altered RNA and protein content in platelets of cancer patients (28–31). Since it is unknown whether these changes arise from altered platelet production or a modified uptake from the circulation, we next assessed RNA and protein expression in murine MKs. Single-cell RNA sequencing (scRNA-seq) of fluorescently sorted (CD41+, DAPIlow) BM cells from 13-week-old PyMT and WT mice revealed three MK subpopulations (MK1, MK2, and MK3) based on a predetermined panel of MK-associated genes (Fig. 2A and fig. S7, C to E). Two MK progenitor populations (G1 and G2/M) were also identified on the basis of transcripts associated with early MK maturation (Gata1, Runx1, Etv6, Itga2b, Pf4, and Vwf) (fig. S7, C and E) and trajectory analysis (fig. S3, D and E). Dimensionality reduction using Uniform Manifold Approximation and Projection (UMAP) segregated G1, G2/M, MK1, MK2, and MK3 clusters from contaminating lymphocyte, monocyte, and neutrophil progenitors that expressed low levels of CD41 (Fig. 2A and figs. S7A and S8). Projecting UMAP coordinates of cells from PyMT (n = 3817) and control mice (n = 5213) suggested that cell clusters from both cohorts were evenly represented (Fig. 2B). Differential gene expression of MKs from PyMT mice was most prominent within the MK2 and MK3 subpopulations (Fig. 2C), with significant alterations in gene ontologies associated with hematological function, cell development, and inflammatory responses (Fig. 2D). Genes with increased expression were predominantly related to the innate immune response (MK2: Ctsg, 5.96-fold; Lcn2, 4.91-fold; Elane, 4.13-fold; Camp, 2.72-fold; S100a8, 3.21-fold; S100a9, 2.48-fold). Four genes associated with inflammatory responses were selected for subsequent investigation, including S100a8 and S100a9, whose expression was increased across all MK subpopulations (Fig. 2E), Ctsg, and Thbs1, the latter of which has previously been shown to increase in MKs of mice with Lewis lung carcinoma (12).
Fig. 2. Transcriptomic profiling of MKs from a model of murine breast cancer.
CD41+ DAPIlow cells were isolated from the BM of a 13-week-old PyMT mouse and a WT littermate for scRNA-seq. (A) Dimensionality reduction using Uniform Manifold Approximation and Projection (UMAP) of pooled PyMT and WT samples. Cells of MK lineage (orange) were separated into five clusters (G1 = 1699, G2/M = 1395, MK1 = 871, MK2 = 537, and MK3 = 344 cells) based on differential gene expression. Gray cells from non-MK clusters (n = 4184 cells) were excluded. (B) Overlay of PyMT (red, n = 3817) and WT (blue, n = 5213) cells. (C) Heatmaps clustered according to average log fold change depicting the top 50 differentially expressed genes between PyMT and WT cells in MK1, MK2, and MK3. Scatterplots depict gene expression of all recorded transcripts. Red lines model no change in gene expression between WT and PyMT samples. (D) Diseases and functions associated with differentially expressed genes in MKs from PyMT mice. (E) Expression levels of thrombospondin 1 (Thbs1), cathepsin G (Ctsg), S100a8, and S100a9 mRNA transcripts among MK subclusters.
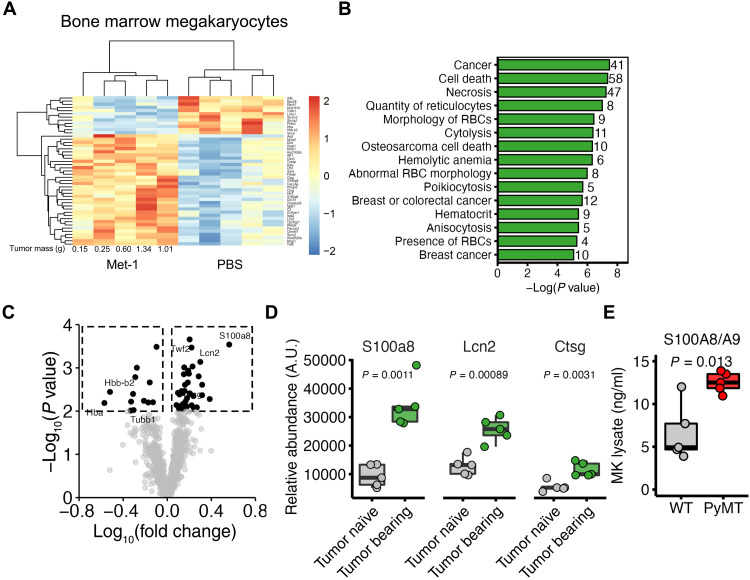
To assess whether the abnormal transcriptional profile of breast cancer MKs resulted in altered protein content, BM MKs were subjected to bulk proteomics 3 weeks after orthotopic implantation of Met-1 cells (tumor mass = 0.68 ± 0.22 g). Blinded hierarchal clustering segregated samples from Met-1 tumor–bearing and vehicle control (PBS)–implanted cohorts (Fig. 3A). We identified statistically significant differences for 49 of 1046 identified proteins (36 up-regulated and 13 down-regulated, P ≤ 0.01), which were predominantly associated with cancer and abnormal hematopoiesis (Fig. 3, B and C). In line with our scRNA-seq experiments on PyMT-derived MKs, S100A8, lipocalin-2 (Lcn-2), and cathepsin G (Ctsg), protein levels were increased in BM MKs from Met-1 tumor–bearing mice (Fig. 3D). To validate these findings in our PyMT model, we quantified S100A8/A9 levels in MKs from 13-week-old PyMT and WT mice by enzyme-linked immunosorbent assay (ELISA) and found that PyMT-derived MKs expressed higher levels of S100A8/A9 (P = 0.013). These findings collectively demonstrate that breast cancer induces pro-inflammatory gene expression in BM MKs.
Fig. 3. MK protein expression from mice with mammary tumors.
(A) Heatmap of differentially expressed (P < 0.01) proteins from native BM MKs of mice bearing Met-1 tumors for 21 days or tumor-naïve controls (PBS); n = 5 mice per condition. (B) Diseases and functions of differentially expressed proteins from Met-1 and PBS-only injected mice. The number of proteins associated with each disease or function is shown to the right of the plot. (C) Volcano plot highlighting differentially expressed proteins. Factors of interest are annotated. (D) Protein expression of candidate proteins S100A8 (S100a8), lipocalin-2 (Lcn2), and cathepsin G (Ctsg). n = 5 mice per condition. P values were determined by two-sample t tests. (E) S100A8/A9 levels in MKs isolated from 13-week-old PyMT and WT mice as determined by ELISA. n = 5 mice per condition. P value determined by two-sample t test.
Increased platelet size, RNA, and protein expression in platelets from PyMT mice
To determine whether abnormal megakaryopoiesis translates into altered platelet production, we assessed the number, size, life span, and activity of platelets from PyMT and WT mice. Despite no discernible change in platelet count, platelets from PyMT mice had increased mean platelet volume (MPV), platelet distribution width (PDW), and an elevated immature platelet fraction (IPF) (Fig. 4A). These findings suggested an increase in the proportion of large and reticulated platelets, which was confirmed by thiazole orange staining of platelet RNA (Fig. 4B). In vivo labeling of circulating platelets with biotin determined that platelet clearance was unaltered (Fig. 4C), suggesting that platelets from PyMT mice have elevated RNA content throughout their life span. While platelet count, PDW, MPV, and IPF were unchanged in Met-1 tumor–bearing mice (fig. S9, A to D), we observed a significant (P = 0.005) increase in platelet counts in AT-3 tumor–bearing mice (fig. S9E). Plasma platelet factor 4 (PF4) levels were unchanged in 13-week-old PyMT mice when compared to littermate PyMT-null controls (Fig. 4D), and there were no differences in surface P-selectin (CD62p) expression upon thrombin stimulation (Fig. 4E), implying that basal degranulation and sensitivity to platelet agonists were unchanged. There was also no difference in the proportion of platelet-leukocyte aggregates (WT = 2.58 ± 0.14%; PyMT = 1.95 ± 0.28%) in the blood of 13-week-old WT and PyMT mice (fig. S10).
Fig. 4. Platelet characteristics and protein expression in PyMT mice.
(A) Platelet counts, mean platelet volume (MPV), platelet distribution width (PDW), and immature platelet fraction in 7- to 13-week-old PyMT and WT mice. n = 10 mice per condition. *P < 0.05 and **P < 0.01, as determined by one-way ANOVA. (B) Thiazole orange (TO) staining of platelets from PyMT and WT mice. n = 6 to 7 mice per condition. P values were determined by two-sample t tests. (C) Proportion of biotin+ platelets isolated from PyMT and WT mice following in vivo labeling with biotin (0 hours). Dashed line depicts basal signal from unlabeled platelets. n = 5 mice per condition. Error bars depict SEM. (D) Plasma levels of platelet factor 4 (PF4) from PyMT and WT mice. n = 9 to 11 mice per condition. P values were determined by two-sample t tests. (E) Surface expression of P-selectin (CD62p) on the surface of platelets isolated from PyMT or WT mice following thrombin stimulation. n = 4 to 5 mice per condition. Error bars depict SEM. (F) S100A8 and S100A8/A9 levels in platelets from PyMT and WT mice. n = 9 to 19 mice per condition. P values were determined by two-sample t tests. (G) S100A8/A9 from the plasma of PyMT and WT mice. n = 4 to 7 mice per condition. (H) Lcn-2 levels of platelets from 13-week-old PyMT and WT mice. n = 5 mice per condition. P values were determined by two-sample t tests. (I) Lcn-2 levels from the plasma of PyMT and WT mice. n = 3 mice per condition. P values were determined by two-sample t tests.
We next sought to determine whether the same factors that were elevated in MKs were also increased in circulating platelets, which would imply their transfer from MKs during thrombopoiesis. S100A8 and the S100A8/A9 heterodimer were elevated in platelets of 13-week-old PyMT mice (Fig. 4, F and H), whereas Ctsg and Thbs1 were unchanged (fig. S11). Serum but not plasma levels of S100A8/A9 were also increased in PyMT mice (Fig. 4G), implying that S100A8/A9 secreted by platelets during clot formation can contribute to circulating levels in this breast cancer model. Lcn-2 levels were also increased in both platelets (Fig. 4H) and plasma (Fig. 4G), demonstrating that several pro-inflammatory regulators are up-regulated in circulating platelets.
Platelets from tumor-bearing mice potentiate tumor cell invasion and lung colonization
We and others have previously demonstrated that the releasate of activated platelets promotes cancer cell invasion (3, 32). On the basis of our findings of enhanced inflammatory protein signatures in platelets of tumor-bearing mice, we next tested whether the releasate of activated platelets from tumor-bearing mice promotes tumor cell migration and invasion to a higher extent than that of WT controls. Time-lapse imaging of fluorescently labeled tumor cells showed no difference in their migratory capacity between PyMT and WT releasates but revealed a statistically significant (P = 0.015) increase in tumor cell invasion upon incubation of tumor cells with PyMT releasates (Fig. 5E and fig. S12). These results demonstrate that platelets produced in tumor-bearing mice increase the invasive capacity of tumor cells. Pretreating the releasate of PyMT-derived platelets with paquinimod (a direct S100A8/A9 inhibitor) partially reduced the number of migrating tumor cells (fig. S12). However, cell migration was only marginally inhibited, suggesting that other platelet cytokines contribute toward tumor cell chemotaxis. To elucidate other cytokines in the PyMT platelet releasate that may promote tumor cell invasion, we conducted cytokine arrays on platelet releasates (Fig. 5, E and F) and found increased levels of Lcn-2, which was also elevated in the platelets (Fig. 4H) and BM MKs (Fig. 3D) of tumor-bearing mice.
Fig. 5. Tumor cell migration, invasion, and lung colonization in response to platelets and activated platelet releasate.
Time-lapse quantification of (A) migrating or (C) invading Met-1 cells toward the releasate of activated platelets. n = 4 mice per condition for migration and n = 8 mice per condition for invasion. Error = 95% confidence interval of the difference. Control indicates serum-free medium as the attractant. Endpoint quantification of the number of (B) migratory or (D) invaded cells per well. n = 4 mice per condition for invasion and n = 8 mice per condition for invasion. P values were determined by unpaired two-sample t tests. (E and F) Cytokine profiling of pooled (n = 5) activated releasate from the platelets of either 13-week-old PyMT or WT mice. (G) Experimental design for AT-3 tumor cell lung colonization following the transfusion of either PyMT- or WT-derived platelets into Mpl−/− recipients. Schematic created using BioRender.com. (H) Representative gating strategy to quantify platelets (CD41+, FSClow) from blood sampled before and after platelet transfusion. FSC, forward scatter. (I) Fold change in platelet count 2 hours after transfusion. P values were determined by unpaired two-sample t tests. (J) Selection criteria and (K) quantification of CMFDA+ AT-3 cells in the lungs of Mpl−/− mice 48 hours after intravenous injection. P values were determined by unpaired two-sample t tests; n = 5 recipient mice per condition.
In addition to pro-invasive signals from cytokines, platelets can directly bind tumor cells in blood circulation, which may impair immunosurveillance (8). We, therefore, tested whether platelets generated in the setting of cancer bind more readily to tumor cells by coincubating green fluorescent protein (GFP)–expressing Met-1 cells with blood from either WT or PyMT mice. There was no statistically significant (P = 0.12) difference in platelet expression on tumor cells incubated with blood from WT or PyMT animals (fig. S13), suggesting that platelets of tumor-bearing mice have an increased propensity for influencing tumor cells through cytokine release and not direct cell-cell interactions.
To test whether platelets generated in the setting of cancer promote metastasis more than those created under normal conditions, platelets from PyMT or WT mice were transfused into recipient mice lacking the TPO receptor Mpl (Mpl−/−), which have approximately 10% the number of circulating platelets (fig. S14). CellTracker™ Green CMFDA-labeled AT-3 cells were injected 2 hours after platelet transfusion, and experimental lung metastasis was quantified after 48 hours (Fig. 5G). Transfusion of either PyMT-derived or WT platelets similarly increased circulating platelet counts in recipient mice (Fig. 5, H to I), and platelet counts did not correlate with the number of tumor cells colonizing the lung (fig. S15), implying that any change in tumor cell number was instead driven by platelet content or function. There was a statistically significant (P = 0.0078) increase in the number of tumor cells colonizing the lungs of mice that received PyMT-derived platelets (Fig. 5, J and K), suggesting that platelets generated in the setting of cancer potentiate tumor cell metastasis.
Enhanced S100A8/A9 secretion from platelets of early-stage breast cancer patients
Given the pro-inflammatory protein expression of platelets from PyMT mice (Fig. 4), we next investigated whether similar changes occurred in platelets isolated from 10 newly diagnosed and untreated breast cancer patients. Platelet counts and MPV were within normal ranges (Fig. 6A), and plasma PF4 was unchanged when compared to age-matched cancer-free controls (Fig. 6B), indicating no change to basal platelet activation. Platelet levels of S100A8/A9 were also unchanged in relation to cancer-free controls, although platelet S100A8/A9 may increase in later-stage (II to III) patients (Fig. 6C). As with PyMT mice, serum levels of S100A8/A9 were higher than that of plasma, but only in breast cancer patients (Fig. 6D), suggesting that activated platelets selectively contributed to circulating S100A8/A9 in breast cancer patients. However, the trending but not statistically significant increase (P = 0.13) in S100A8/A9 serum levels of cancer patients compared to cancer-free controls (Fig. 6D) leads us to conclude that further studies are required to definitively determine platelet and serum levels of S100A8/A9 in breast cancer patients.
Fig. 6. Enhanced release of S100A8/A9 from platelets of early-stage breast cancer patients.
(A) Median age, platelet count, and MPV from recently diagnosed and untreated patients with early-stage (I to III) breast cancer. (B) Plasma levels of PF4 from breast cancer patients and age-matched cancer-free female controls. n = 6 control subjects and 9 patients per condition. (C) Levels of S100A8/A9 in platelets isolated from cancer-free subjects and early-stage breast cancer patients. n = 6 to 10 subjects/patients per condition. (D) S100A8/A9 levels in the plasma and serum of cancer-free subjects and early-stage breast cancer patients. n = 5 to 10 subjects/patients per condition. N/A in cancer patients indicates no assigned stage due to the lack of lymph node examination. P values were determined by one-way ANOVA.
DISCUSSION
Platelets influence several hallmarks of cancer through diverse mechanisms (33); however, few studies have examined whether primary tumors affect platelet-producing MKs within the BM niche. In this study, we used a genetically engineered murine model of breast cancer, PyMT, that recapitulates human breast cancer progression (34, 35). PyMT mice had fewer and smaller BM MKs with abnormal polyploidization (Fig. 1, D and G), while no changes were observed in WT mice implanted with cell lines derived from PyMT tumors (Fig. 1, E and F and H and I). Such discrepancies suggest that tumor-induced changes to MKs predominantly occur in spontaneous and chronic models, which closely reflect human breast cancer progression (34, 35). Simultaneously, acute tumor generation following orthoptic implantation of tumor cells may not provide adequate time to influence megakaryopoiesis and platelet production. Recent findings from Sun et al. (36) suggest that polyploidization correlates to nonconventional functions of MKs within the BM, with 2N, 4-8N, and 16-32N cells linked to inflammatory responses, active cell cycling, and hematopoietic stem cell niche-support, respectively. In line with our data, they also observed that 2N MKs were smaller than other MK subpopulations. Therefore, the significant increase in 2N MKs and decrease of 16N MKs in PyMT mice (Fig. 1G) suggest that breast cancer enhances the proportion of inflammatory response MKs while lowering those involved in niche regulation. In line with this correlation, we also observed a shift toward an inflammatory and cancer gene signature in PyMT BM MKs (Figs. 2 and 3). Beyond the BM, lung MKs, which are predominantly 2N and smaller than BM MKs, have been associated with immunomodulatory roles (37). Given the well-established effects of breast cancer on the lung microenvironment (38, 39), we examined CD41+ cells in the lungs of 13-week-old WT and PyMT mice and found no difference in cell number (fig. S6B). Similar observations were also made in the spleen (fig. S5, C and D), suggesting that MK reprogramming in cancer predominantly occurs within the BM niche.
In line with previous reports (40), we did not detect differences in platelet counts between WT and PyMT mice (Fig. 4A). However, platelets from PyMT mice had increased size (Fig. 4A) and RNA content (Fig. 4B), indicating qualitative changes to platelet production. Typically, platelets with high levels of RNA are referred to as immature (41), are hyperactive (42), and have been implicated in several diseases including non–small cell lung carcinoma (43). However, the platelets of PyMT mice in our study displayed unaltered longevity and reactivity upon thrombin stimulation (Fig. 4, C and D), leading us to conclude that this subpopulation is distinct from reticulated platelets.
Advances in scRNA-seq allow for a more stringent examination of rare and fragile cell populations (44). Such approaches have recently been adopted to study the MK lineage (45), MK heterogeneity (26, 36, 37, 46), and the effects of age-associated inflammation or myelofibrosis (46, 47). In this study, we used scRNA-seq to identify tumor-induced alterations in MK gene expression. This approach allowed us to separate viable and legitimate MKs from contaminating or nonviable cells, which are a common concern when isolating rare cell populations for bulk analysis (fig. S7, A and B). Gene ontologies from MKs of PyMT mice were predominantly associated with hematological development and inflammatory responses (Fig. 2D). Increased expression of transcripts associated with the innate immune response (e.g., Ctsg, Lcn2, Elane, Camp, S100a8, and S100a9) (Fig. 2, C to E) suggests a tumor-driven pro-inflammatory MK phenotype.
Of our differentially expressed genes, we found that S100a8 and S100a9 were markedly increased in the MKs (Figs. 2E and 3D) and platelets (Fig. 4F) of tumor-bearing mice. In cancer, myeloid-derived S100A8/A9 increases the migration and invasion of colon and lung carcinoma cells (22). In the clinic, breast cancer patients with elevated tumor S100A8 had a shorter disease-free and overall survival (48), implying that S100A8/A9 within mammary tumors holds clinical importance. Although neutrophils are the archetypal reservoir for S100A8/A9 (49) and can transfer S100A8 to activated platelets when cocultured in vitro (50), increased MK S100a8 and S100a9 mRNA in our in vivo models (Fig. 2E) imply that MKs directly synthesize these proteins. Recently, platelets from coronavirus disease 2019 (COVID-19) patients were shown to have increased S100a8 and S100a9 transcripts, possibly due to MK infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (51). Platelet S100A8/A9 is also elevated in patients with systemic lupus erythematosus (52) and early-stage pancreatic or lung cancer (31), collectively suggesting the role of platelet S100A8/A9 to be augmented under certain pathologies.
Platelets store S100A8/A9 in granules, leading to its release upon platelet activation and during clot formation (52, 53), which likely contributes to elevated serum levels in cancer patients (54–56). In our study, S100A8/A9 was increased in platelets from 13-week-old PyMT mice (Fig. 4F) but not in early-stage breast cancer patients (Fig. 6, C and D). This discrepancy may result from a small patient cohort, differing breast cancer subtypes, or reduced tumor burden in early-stage patients compared to our murine model, where 13-week-old PyMT mice have a high incidence of lung metastasis (35). Serum levels of S100A8/A9 exceeded plasma levels from both PyMT mice (Fig. 4G) and breast cancer patients (Fig. 6D) but were unchanged in cancer-free controls, suggesting that activated platelets selectively contribute to circulating S100A8/A9 in breast cancer. In addition to S100A8/A9, Lcn-2 was also increased in the MKs (Fig. 3D) and platelets (Fig. 4H) of tumor-bearing mice. Lcn-2 is a pro-inflammatory protein highly associated with estrogen receptor–negative breast tumors (57, 58) and promotes the invasion and metastasis of breast cancer cells in some but not all (59) preclinical models by up-regulating epithelial to mesenchymal transition (60). Although Lcn-2 has previously been detected in the human platelet proteome, its functional roles and possible contribution to inflammatory or metastatic processes have not been elucidated.
The roles of platelets in promoting tumor cell invasion and metastasis are well established (3, 61), which prompted us to investigate whether platelets produced by pro-inflammatory MKs further potentiate tumor cell invasion and metastasis. In line with this hypothesis, our findings show that increased tumor cell invasion in response to the releasate of activated platelets from PyMT mice (Fig. 5, D to F, and fig. S12) and Mpl−/− donor mice transfused with PyMT-derived platelets had increased tumor cell lung colonization compared to mice transfused with WT platelets (Fig. 5, G to K). Collectively, these findings lead us to hypothesize that platelets produced in the setting of cancer potentiate both early (i.e., invasion and intravasation) and late (i.e., extravasation and colonization) stages of tumor cell metastasis. This hypothesis is supported by previous work from Labelle et al. (3), who demonstrated that platelets induce epithelial to mesenchymal transition in tumor cells, which enabled tumor cell lung colonization. Ongoing studies aim to delineate the role of platelets in the formation of a premetastatic microenvironment and the distinct contributions of platelet-derived S100A8/A9 or Lcn-2 therein.
Despite the findings presented in this study, it remains largely undetermined which tumor-derived factors affect megakaryopoiesis during breast cancer progression. Tumor interleukin-6 increases platelet counts in ovarian cancer through increased TPO production (14). However, TPO levels (fig. S1A) and platelet counts (Fig. 4A) were not significantly increased in PyMT mice, suggesting an alternative mechanism of action. Chemokine ligand 5 is an inflammatory cytokine up-regulated in the plasma of breast cancer patients (62) and has previously been shown to affect MK maturation in vitro (63). However, given that both interleukin-6 and chemokine ligand 5 are increased in other chronic inflammatory conditions (64, 65), future experiments should determine whether cancer directly affects MKs through tumor-derived signals or by indiscriminately up-regulating inflammatory responses.
To summarize, we demonstrate that MKs from a progressive breast cancer model are smaller, have abnormal polyploidization, and display an inflammatory gene expression profile. Proinflammatory genes associated with tumor progression were also expressed at the protein level in circulating platelets, which likely contribute to the enhanced pro-metastatic properties of platelets generated in models of malignancy.
MATERIALS AND METHODS
Experimental design
The objective of this study was to assess alterations to MKs and platelets in the setting of breast cancer. To achieve this, our experimental design used several preclinical murine models of breast cancer, outlined below.
Animals
For orthotopic tumor generation, 8-week-old female FVB/NJ or C57BL/6J mice were purchased from The Jackson Laboratory (stock nos. 001800 and 000664). For autochthonous tumor progression, FVB/N-Tg 634Mul/J (MMTV-PyMT) transgenic mice were purchased from The Jackson Laboratory (stock no. 002374) and then bred inhouse, with nontransgenic littermates (WT) used as cancer-free controls. Mpl−/− mice on a C57BL/6J background were a gift from A. Mullally. All animal experiments were conducted in accordance with the regulations of the Brigham and Women’s Hospital (protocol no. 2019N000011) and Institutional Animal Care and Use Committee guidelines.
Cell culture
Met-1 murine cells (24) were a gift from J. Joyce (with permission from A. Borowsky). AT-3 mammary carcinoma cells were purchased from Sigma-Aldrich (catalog no. SCC178).
Tumor generation models
For orthotopic tumor generation, 2.5 × 105 Met-1 or AT-3 cells were prepared in PBS and injected into the fourth left mammary fat pad of 9-week-old FVB/NJ or C57BL/6J mice, respectively. Tumors were measured every 2 to 3 days using digital calipers, and tumor volume was calculated as (width2 × length) ÷ 2. Mice were sacrificed either 21 days (Met-1) or 33 days (AT-3) after mammary fat pad injections. For spontaneous tumor generation, MMTV-PyMT transgenic mice and PyMT-null WT controls were housed from birth to 13 weeks of age, with the collective tumor volume measured from week 7.
Immunofluorescent staining of MKs in situ
Femurs were fixed in 4% paraformaldehyde overnight at 4°C, and a sucrose gradient was performed over three consecutive days. Femora were cut at 10 μm and transferred onto slides using a tape-transfer system (Kawamoto) at a cryostat (Leica). Sections were blocked using 3% (v/v) goat serum and stained using antibodies against laminin (Sigma-Aldrich, L9393) and CD41 (BioLegend, 133902). Secondary antibodies were purchased from Invitrogen, and nuclei were visualized using 4′,6-diamidino-2-phenylindole (DAPI). Image acquisition was performed using either a confocal microscope (Zeiss LSM880, 20×) or a Lionheart FX automated microscope. MK numbers were quantified using Gen5 software (BioTek, version 3.10).
Electron microscopy
Femurs from 13-week-old PyMT mice or littermate controls were fixed with 1.25% paraformaldehyde, 0.03% picric acid, and 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) for at least 1 hour. Samples were then processed by the Harvard Medical School Electron Microscopy core facility. Femurs were fixed with 1% osmium tetroxide, dehydrated through a series of alcohols, infiltrated with propylene oxide, and embedded in epoxy resin. Sections were stained and examined with a Tecnai G2 Spirit BioTwin electron microscope (Hillsboro, OR) at an accelerating voltage of 80 kV. Images were recorded with an Advanced Microscopy Techniques (AMT) 2-K charge-coupled device camera, using AMT digital acquisition and analysis software (AMT, Danvers, MA).
Isolation of BM cells and MKs
Tibias and femurs were dissected from euthanized mice and underwent centrifugation (2500g, 1 min) to isolate BM cells (BMCs). BMCs were passed through 100-μm strainers (Corning, 352360) and washed in PBS, before RBC lysis (BD Pharm Lyze, 555899). For MK enrichment before ELISAs, BMCs were stained with CD61 MicroBeads (Miltenyi Biotec, 130-109-678) and passed through large cell separation columns (Miltenyi Biotec, 130-042-202). MKs were further enriched by layering CD61+ cells onto a 1.5 to 3.0% bovine serum albumin gradient, and the bottom layer was isolated following a density-based cell separation (66, 67).
MK polyploidization
BMCs were fixed and permeabilized with ice-cold ethanol and then stained with Alexa Fluor 488–conjugated anti-CD41/61 antibodies (Emfret, M021-1). Propidium iodide (Thermo Fisher Scientific, P1304MP) and ribonuclease A (Sigma-Aldrich, R6513) were used to stain DNA. Samples were analyzed using a Cytek Aurora spectral cytometer.
Analysis of lung and splenic MKs
Murine lungs were perfused with PBS before extraction, minced, and then digested in digestion buffer [1% (w/v) collagenase VI (Thermo Fisher Scientific, 17104019), 10% (v/v) fetal bovine serum in Dulbecco’s Modified Eagle Medium (DMEM)] for 30 min at 37°C. Digested tissue was passed through 100-μm cell strainers, followed by RBC lysis (BioLegend, 420301). For spleens, organs were extracted and pressed through 100-μm cell strainers, followed by RBC lysis. Single-cell suspensions were stained with DAPI and anti-CD41 fluorescein isothiocyanate (FITC) antibodies (BioLegend, 133903) and assessed using a Cytek Aurora spectral cytometer.
Single-cell RNA sequencing
BMCs from one 13-week-old PyMT mouse and one WT control were stained with anti-CD41 phycoerythrin (PE)–conjugated antibodies (BioLegend, 303705), before fluorescence-activated cell sorting, for PE+, DAPIlow single cells. Samples were processed using the Brigham and Women’s Hospital Single Cell Genomics Core. Cells (10,000) were loaded onto a single lane (chromium chip, 10× Genomics) followed by encapsulation in lipid droplets (Single Cell 3′ kit V2, 10× Genomics) and library generation per the manufacturer’s protocol. Complementary DNA (cDNA) libraries were sequenced to an average of 50,000 reads per cell using Illumina NextSeq 500. scRNA-seq reads were processed with Cell Ranger v2.1 (10× Genomics). In-house data analysis was performed using the Seurat package within R (68). Cells exhibiting high proportions of mitochondrial gene expression (>10%) were excluded from downstream analysis.
Proteomics
CD41+ BMCs were processed at the Mass Spectrometry and Proteomics Research Laboratory at the Harvard University. Proteins were extracted using a Covaris Pre-Analytical Sample Preparation System, before their precipitation and digestion in solution. Samples were labeled after digestion using a TMT10plex Isobaric Label Reagent Set (Thermo Fisher Scientific, 90110) and fractionated to 20 fractions, which were run on the Orbitrap Fusion Lumos Tribrid Mass Spectrometer. All data were searched on Proteome Discoverer 2.3.
Murine platelet, plasma, and serum isolation
For nonterminal murine blood collection, lateral incisions were made across a tail vein and 15 μl was collected using a pipette with an EDTA-prewetted tip. Blood was diluted 1:10 in EDTA (5 mM), and full blood counts were assessed using a Sysmex Hematology XE-5000 analyzer. For terminal procedures, blood was collected via cardiac puncture into syringes containing EDTA (5 mM) and transferred to EDTA-coated microtainer capillary blood collection tubes (BD, 365974). Blood was spun to isolate the platelet-rich plasma (PRP) and WBCs (117g, 8 min), and PRP/WBCs were spun (200g, 3 min) to isolate PRP. For platelet isolation, prostaglandin E1 (1 μM, Sigma-Aldrich, P5515) was added to PRP, spun (1000g, 5 min), and resuspended in platelet resuspension buffer (10 mM Hepes, 140 mM NaCl, 3 mM KCl, 0.5 mM MgCl2, NaHCO3, and 10 mM glucose; pH 7.4). Plasma was obtained by centrifugation of platelet-poor plasma (PPP) at 10,000g for 5 min. For serum, whole blood was collected without anticoagulant and left at 37°C for 30 min. Serum was separated from blood clots and cells by centrifugation (10,000g, 5 min). Activated platelet releasate was generated by incubating platelets (2 × 108/ml) for 30 min in DMEM at 37°C.
Human platelet, plasma, and serum isolation
Blood collection from newly diagnosed untreated breast cancer patients and age-matched cancer-free volunteers was performed as previously described in accordance with the Declaration of Helsinki and ethics regulations with Institutional Review Board approval from the Brigham and Women’s Hospital and the Dana-Farber Cancer Institute (32). Citrated whole blood was spun (177g, 20 min) to isolate PRP. Platelets were separated from PRP using the same protocol as murine platelets, and platelet counts were adjusted using a Cytek Aurora spectral cytometer. For serum, whole blood was recalcified (2 mM CaCl2) and left at 37°C for 30 min. Serum was isolated by centrifugation (10,000g, 5 min).
Platelet activation
Murine platelets were activated with thrombin (Sigma-Aldrich, T4648) for 20 min at 37°C. Samples were then stained for 20 min with anti-mouse CD62p allophycocyanin (APC) (BioLegend, 148304) to quantify alpha granule secretion. Platelets were fixed in 2% (v/v) paraformaldehyde (Thermo Fisher Scientific, AAJ19943K2), and fluorescence intensity was assessed using a Cytek Aurora spectral cytometer.
Protein arrays and ELISAs
Protein concentrations in platelet lysates, plasma, and serum were determined using DuoSet ELISA Kits from R&D Systems for S100A8 (DY3059), S100A8/9 (DY8596), and Lcn-2 (DY1857) or Aviva Systems Biology ELISA kits for Thbs1 (OKEH00550) and Ctsg (OKEH06828). Quantikine Colorimetric Sandwich ELISA Kits from R&D Systems were used for plasma TPO (MTP00). Proteome Profiler Mouse XL Cytokine Arrays from R&D Systems (ARY028) were used to characterize platelet releasate.
Platelet interactions with leukocytes or tumor cells
Met-1 cells (5000) expressing GFP were incubated in 5 μl of anticoagulated blood for 1 hour at 37°C. Samples were then diluted 1:20 in Hanks’ balanced salt solution (HBSS) containing CD41 PE (BioLegend, 133905) and CD45 BV605 (BioLegend, 103139) for 20 min at room temperature. Samples were further diluted 1:4 in HBSS before immediate analysis of platelet binding (PE+) within tumor cell (GFP+) and leukocyte (CD45+) populations using a Cytek Aurora spectral cytometer.
Tumor cell invasion and migration
To assess tumor cell migration and invasion, Met-1 tumor cells were labeled with CellTracker Deep Red (Thermo Fisher Scientific, C34565), and 15,000 cells were added to semipermeable (8 μm pore) transwell inserts of a Falcon Fluoroblok 96-well plate (Fisher Scientific, 08-771-006) that were either left uncoated or coated with Matrigel (250 μg/ml; Corning, 356231), respectively. In some cases, tumor cells and platelet releasate were pretreated with the S100A8/A9 inhibitor paquinimod (Sigma-Aldrich, SML2883) for 30 min before the addition of tumor cells to transwells. Tumor cells that passed through transmembrane inserts were quantified every hour for 24 hours using a Lionheart FX automated microscope. All conditions were performed in triplicate, and the average was used for subsequent analysis.
Platelet transfusion and experimental metastasis
Platelets isolated from either 13-week-old WT and PyMT mice were pooled, and 2 × 108 cells were injected via the left lateral tail vein into recipient Mpl−/− mice. Platelet counts were assessed before and 2 hours after transfusion by staining whole blood with anti-mouse CD41 PE (BioLegend, 148304). Two hours after platelet transfusion, mice were injected via the right lateral tail vein with 2.5 × 105 CellTracker™ Green CMFDA-labeled (Invitrogen, C2925) AT-3 tumor cells. Mice were sacrificed 48 hours after tumor cell injection, and tumor colonization was assessed by digesting lung tissue in collagenase/hyaluronidase (STEMCELL Technologies, 07912) and quantifying the number of CMFDA+ cells using a Cytek Aurora spectral cytometer.
Statistical analysis
Statistical analyses and graphics were generated using R (v4.0.4, The R Foundation for Statistical Computing, Vienna, Austria). Biological replicates (all at or exceeding 3), sources of error (SD, SEM, or 95% confidence intervals), and P values are all reported in figures and/or figure legends. Statistical tests used include unpaired and two-tailed t tests, one-way analyses of variance (ANOVAs), with post hoc Tukey’s, multiple comparisons testing, with values <0.05 considered statistically significant.
Acknowledgments
We thank P. van Galen for support and contributions to scRNA-seq data analysis. We also thank K. Santos and M. Hughes for providing deidentified patient information.
Funding: This work was supported by NIH grant 5R01CA200748 (E.M.B. and J.E.I.) and American Cancer Society grant RSG-17-161-01 (E.M.B.).
Author contributions: Conceptualization: H.G.R., J.E.I., and E.M.B. Methodology: H.G.R., G.J.G., Q.G., and I.C.B. Investigation: H.G.R., M.W.M., G.J.G., Q.G., and I.C.B. Data curation: H.G.R. Visualization: H.G.R. Supervision: J.E.I., S.S.M., E.A.M., and E.M.B. Resources: E.A.M. and S.S.M. Funding acquisition: E.M.B. and J.E.I. Writing—original draft: H.G.R. Writing—review and editing: H.G.R., G.J.G., I.C.B., and E.M.B.
Competing interests: J.E.I. has financial interest in and is a founder of PlateletBio, a company that aims to produce donor-independent human platelets from human-induced pluripotent stem cells at scale. The interests of J.E.I. were reviewed and are managed by the Boston Children’s Hospital in accordance with their conflict of interest policies. E.A.M. reports compensated service on scientific advisory boards for AstraZeneca, Exact Sciences (formerly Genomic Health), Merck, and Roche/Genentech; uncompensated service on steering committees for Bristol Myers Squibb, Lilly, and Roche/Genentech; and institutional research support from Roche/Genentech (via SU2C grant) and Gilead. She receives research funding from Susan Komen for the Cure for which she serves as a Scientific Advisor. She also reports uncompensated participation as a member of the American Society of Clinical Oncology Board of Directors. The authors declare no other competing interests.
Data and materials availability: Data from scRNA-seq and proteomics experiments can be found at https://dataverse.harvard.edu/dataset.xhtml?persistentId=doi:10.7910/DVN/KSEDXJ on the Harvard Dataverse with the digital object identifier doi:10.7910/DVN/KSEDXJ. Upon publication, manuscript data will be uploaded to the Harvard Dataverse as an .xlsx file. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S15
Tables S1 and S2
REFERENCES AND NOTES
- 1.Bye A. P., Unsworth A. J., Gibbins J. M., Platelet signaling: A complex interplay between inhibitory and activatory networks. J. Thromb. Haemost. 14, 918–930 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gay L. J., Felding-Habermann B., Contribution of platelets to tumour metastasis. Nat. Rev. Cancer 11, 123–134 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Labelle M., Begum S., Hynes R. O., Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer Cell 20, 576–590 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olsson A. K., Cedervall J., The pro-inflammatory role of platelets in cancer. Platelets 29, 569–573 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Plantureux L., Crescence L., Dignat-George F., Panicot-Dubois L., Dubois C., Effects of platelets on cancer progression. Thromb. Res. 164 ( Suppl 1), S40–S47 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Xu X. R., Yousef G. M., Ni H., Cancer and platelet crosstalk: Opportunities and challenges for aspirin and other antiplatelet agents. Blood 131, 1777–1789 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Cluxton C. D., Spillane C., O’Toole S. A., Sheils O., Gardiner C. M., O’Leary J. J., Suppression of natural killer cell NKG2D and CD226 anti-tumour cascades by platelet cloaked cancer cells: Implications for the metastatic cascade. PLOS ONE 14, e0211538 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kopp H. G., Placke T., Salih H. R., Platelet-derived transforming growth factor-beta down-regulates NKG2D thereby inhibiting natural killer cell antitumor reactivity. Cancer Res. 69, 7775–7783 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Metelli A., Wu B. X., Riesenberg B., Guglietta S., Huck J. D., Mills C., Li A., Rachidi S., Krieg C., Rubinstein M. P., Gewirth D. T., Sun S., Lilly M. B., Wahlquist A. H., Carbone D. P., Yang Y., Liu B., Li Z., Thrombin contributes to cancer immune evasion via proteolysis of platelet-bound GARP to activate LTGF-β. Sci. Transl. Med. 12, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Labelle M., Begum S., Hynes R. O., Platelets guide the formation of early metastatic niches. Proc. Natl. Acad. Sci. U.S.A. 111, E3053–E3061 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klement G. L., Yip T. T., Cassiola F., Kikuchi L., Cervi D., Podust V., Italiano J. E., Wheatley E., Abou-Slaybi A., Bender E., Almog N., Kieran M. W., Folkman J., Platelets actively sequester angiogenesis regulators. Blood 113, 2835–2842 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaslavsky A., Baek K. H., Lynch R. C., Short S., Grillo J., Folkman J., Italiano J. E. Jr., Ryeom S., Platelet-derived thrombospondin-1 is a critical negative regulator and potential biomarker of angiogenesis. Blood 115, 4605–4613 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlesinger M., Role of platelets and platelet receptors in cancer metastasis. J. Hematol. Oncol. 11, 125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stone R. L., Nick A. M., McNeish I. A., Balkwill F., Han H. D., Bottsford-Miller J., Rupairmoole R., Armaiz-Pena G. N., Pecot C. V., Coward J., Deavers M. T., Vasquez H. G., Urbauer D., Landen C. N., Hu W., Gershenson H., Matsuo K., Shahzad M. M., King E. R., Tekedereli I., Ozpolat B., Ahn E. H., Bond V. K., Wang R., Drew A. F., Gushiken F., Lamkin D., Collins K., DeGeest K., Lutgendorf S. K., Chiu W., Lopez-Berestein G., Afshar-Kharghan V., Sood A. K., Paraneoplastic thrombocytosis in ovarian cancer. N. Engl. J. Med. 366, 610–618 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu B., Ye Y., Xie S., Li Y., Sun X., Lv M., Yang L., Cui N., Chen Q., Jensen L. D., Cui D., Huang G., Zuo J., Zhang S., Liu W., Yang Y., Megakaryocytes mediate hyperglycemia-induced tumor metastasis. Cancer Res. 81, 5506–5522 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Siegel R. L., Miller K. D., Jemal A., Cancer statistics, 2020. CA Cancer J. Clin. 70, 7–30 (2020). [DOI] [PubMed] [Google Scholar]
- 17.McAllister S. S., Weinberg R. A., The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat. Cell Biol. 16, 717–727 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Acharyya S., Oskarsson T., Vanharanta S., Malladi S., Kim J., Morris P. G., Manova-Todorova K., Leversha M., Hogg N., Seshan V. E., Norton L., Brogi E., Massague J., A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell 150, 165–178 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen B., Miller A. L., Rebelatto M., Brewah Y., Rowe D. C., Clarke L., Czapiga M., Rosenthal K., Imamichi T., Chen Y., Chang C. S., Chowdhury P. S., Naiman B., Wang Y., Yang D., Humbles A. A., Herbst R., Sims G. P., S100A9 induced inflammatory responses are mediated by distinct damage associated molecular patterns (DAMP) receptors in vitro and in vivo. PLOS ONE 10, e0115828 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiratsuka S., Watanabe A., Sakurai Y., Akashi-Takamura S., Ishibashi S., Miyake K., Shibuya M., Akira S., Aburatani H., Maru Y., The S100A8-serum amyloid A3-TLR4 paracrine cascade establishes a pre-metastatic phase. Nat. Cell Biol. 10, 1349–1355 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Jo S. H., Heo W. H., Son H. Y., Quan M., Hong B. S., Kim J. H., Lee H. B., Han W., Park Y., Lee D. S., Kwon N. H., Park M. C., Chae J., Kim J. I., Noh D. Y., Moon H. G., S100A8/A9 mediate the reprograming of normal mammary epithelial cells induced by dynamic cell-cell interactions with adjacent breast cancer cells. Sci. Rep. 11, 1337 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim S. Y., Yuzhalin A. E., Gordon-Weeks A. N., Muschel R. J., Tumor-infiltrating monocytes/macrophages promote tumor invasion and migration by upregulating S100A8 and S100A9 expression in cancer cells. Oncogene 35, 5735–5745 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shabani F., Farasat A., Mahdavi M., Gheibi N., Calprotectin (S100A8/S100A9): A key protein between inflammation and cancer. Inflamm. Res. 67, 801–812 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Borowsky A. D., Namba R., Young L. J., Hunter K. W., Hodgson J. G., Tepper C. G., McGoldrick E. T., Muller W. J., Cardiff R. D., Gregg J. P., Syngeneic mouse mammary carcinoma cell lines: Two closely related cell lines with divergent metastatic behavior. Clin. Exp. Metastasis 22, 47–59 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Guy C. T., Cardiff R. D., Muller W. J., Induction of mammary tumors by expression of polyomavirus middle T oncogene: A transgenic mouse model for metastatic disease. Mol. Cell. Biol. 12, 954–961 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeung A. K., Villacorta-Martin C., Hon S., Rock J. R., Murphy G. J., Lung megakaryocytes display distinct transcriptional and phenotypic properties. Blood Adv. 4, 6204–6217 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DuPre S. A., Hunter K. W. Jr., Murine mammary carcinoma 4T1 induces a leukemoid reaction with splenomegaly: Association with tumor-derived growth factors. Exp. Mol. Pathol. 82, 12–24 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Best M. G., Sol N., Kooi I., Tannous J., Westerman B. A., Rustenburg F., Schellen P., Verschueren H., Post E., Koster J., Ylstra B., Ameziane N., Dorsman J., Smit E. F., Verheul H. M., Noske D. P., Reijneveld J. C., Nilsson R. J. A., Tannous B. A., Wesseling P., Wurdinger T., RNA-seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell 28, 666–676 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calverley D. C., Phang T. L., Choudhury Q. G., Gao B., Oton A. B., Weyant M. J., Geraci M. W., Significant downregulation of platelet gene expression in metastatic lung cancer. Clin. Transl. Sci. 3, 227–232 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roweth H. G., Battinelli E. M., Lessons to learn from tumor-educated platelets. Blood 137, 3174–3180 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabrkhany S., Kuijpers M. J. E., Knol J. C., Olde Damink S. W. M., Dingemans A. C., Verheul H. M., Piersma S. R., Pham T. V., Griffioen A. W., Oude Egbrink M. G. A., Jimenez C. R., Exploration of the platelet proteome in patients with early-stage cancer. J. Proteomics 177, 65–74 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Johnson K. E., Ceglowski J. R., Roweth H. G., Forward J. A., Tippy M. D., El-Husayni S., Kulenthirarajan R., Malloy M. W., Machlus K. R., Chen W. Y., Italiano J. E. Jr., Battinelli E. M., Aspirin inhibits platelets from reprogramming breast tumor cells and promoting metastasis. Blood Adv. 3, 198–211 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braun A., Anders H. J., Gudermann T., Mammadova-Bach E., Platelet-cancer interplay: Molecular mechanisms and new therapeutic avenues. Front. Oncol. 11, 665534 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Attalla S., Taifour T., Bui T., Muller W., Insights from transgenic mouse models of PyMT-induced breast cancer: Recapitulating human breast cancer progression in vivo. Oncogene 40, 475–491 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin E. Y., Jones J. G., Li P., Zhu L., Whitney K. D., Muller W. J., Pollard J. W., Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am. J. Pathol. 163, 2113–2126 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun S., Jin C., Si J., Lei Y., Chen K., Cui Y., Liu Z., Liu J., Zhao M., Zhang X., Tang F., Rondina M. T., Li Y., Wang Q. F., Single-cell analysis of ploidy and the transcriptome reveals functional and spatial divergency in murine megakaryopoiesis. Blood 138, 1211–1224 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pariser D. N., Hilt Z. T., Ture S. K., Blick-Nitko S. K., Looney M. R., Cleary S. J., Roman-Pagan E., Saunders J. II, Georas S. N., Veazey J., Madere F., Santos L. T., Arne A., Huynh N. P., Livada A. C., Guerrero-Martin S. M., Lyons C., Metcalf-Pate K. A., McGrath K. E., Palis J., Morrell C. N., Lung megakaryocytes are immune modulatory cells. J. Clin. Invest. 131, e137377 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y., Cao X., Characteristics and significance of the pre-metastatic niche. Cancer Cell 30, 668–681 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Wculek S. K., Malanchi I., Neutrophils support lung colonization of metastasis-initiating breast cancer cells. Nature 528, 413–417 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shirai T., Revenko A. S., Tibbitts J., Ngo A. T. P., Mitrugno A., Healy L. D., Johnson J., Tucker E. I., Hinds M. T., Coussens L. M., McCarty O. J. T., Monia B. P., Gruber A., Hepatic thrombopoietin gene silencing reduces platelet count and breast cancer progression in transgenic MMTV-PyMT mice. Blood Adv. 3, 3080–3091 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffmann J. J., Reticulated platelets: Analytical aspects and clinical utility. Clin. Chem. Lab. Med. 52, 1107–1117 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Allan H. E., Hayman M. A., Marcone S., Chan M. V., Edin M. L., Maffucci T., Joshi A., Menke L., Crescente M., Mayr M., Zeldin D. C., Armstrong P. C., Warner T. D., Proteome and functional decline as platelets age in the circulation. J. Thromb. Haemost. 19, 3095–3112 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Best M. G., Sol N., In ’t Veld S., Vancura A., Muller M., Niemeijer A. N., Fejes A. V., Tjon Kon Fat L. A., Huis In ’t Veld A. E., Leurs C., Le Large T. Y., Meijer L. L., Kooi I. E., Rustenburg F., Schellen P., Verschueren H., Post E., Wedekind L. E., Bracht J., Esenkbrink M., Wils L., Favaro F., Schoonhoven J. D., Tannous J., Meijers-Heijboer H., Kazemier G., Giovannetti E., Reijneveld J. C., Idema S., Killestein J., Heger M., de Jager S. C., Urbanus R. T., Hoefer I. E., Pasterkamp G., Mannhalter C., Gomez-Arroyo J., Bogaard H. J., Noske D. P., Vandertop W. P., van den Broek D., Ylstra B., Nilsson R. J. A., Wesseling P., Karachaliou N., Rosell R., Lee-Lewandrowski E., Lewandrowski K. B., Tannous B. A., de Langen A. J., Smit E. F., van den Heuvel M. M., Wurdinger T., Swarm intelligence-enhanced detection of non-small-cell lung cancer using tumor-educated platelets. Cancer Cell 32, 238–252.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nguyen A., Khoo W. H., Moran I., Croucher P. I., Phan T. G., Single cell RNA sequencing of rare immune cell populations. Front. Immunol. 9, 1553 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang H., He J., Xu C., Chen X., Yang H., Shi S., Liu C., Zeng Y., Wu D., Bai Z., Wang M., Wen Y., Su P., Xia M., Huang B., Ma C., Bian L., Lan Y., Cheng T., Shi L., Liu B., Zhou J., Decoding human megakaryocyte development. Cell Stem Cell 28, 535–549.e8 (2021). [DOI] [PubMed] [Google Scholar]
- 46.Davizon-Castillo P., McMahon B., Aguila S., Bark D., Ashworth K., Allawzi A., Campbell R. A., Montenont E., Nemkov T., D’Alessandro A., Clendenen N., Shih L., Sanders N. A., Higa K., Cox A., Padilla-Romo Z., Hernandez G., Wartchow E., Trahan G. D., Nozik-Grayck E., Jones K., Pietras E. M., DeGregori J., Rondina M. T., Di Paola J., TNF-α–driven inflammation and mitochondrial dysfunction define the platelet hyperreactivity of aging. Blood 134, 727–740 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Psaila B., Wang G., Rodriguez-Meira A., Li R., Heuston E. F., Murphy L., Yee D., Hitchcock I. S., Sousos N., O’Sullivan J., Anderson S., Senis Y. A., Weinberg O. K., Calicchio M. L.; NIH Intramural Sequencing Center, Iskander D., Royston D., Milojkovic D., Roberts I., Bodine D. M., Thongjuea S., Mead A. J., Single-cell analyses reveal megakaryocyte-biased hematopoiesis in myelofibrosis and identify mutant clone-specific targets. Mol. Cell 78, 477–492.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang D., Liu G., Wu B., Chen L., Zeng L., Pan Y., Clinical significance of elevated S100A8 expression in breast cancer patients. Front. Oncol. 8, 496 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hessian P. A., Edgeworth J., Hogg N., MRP-8 and MRP-14, two abundant Ca(2+)-binding proteins of neutrophils and monocytes. J. Leukoc. Biol. 53, 197–204 (1993). [PubMed] [Google Scholar]
- 50.Joshi A., Schmidt L. E., Burnap S. A., Lu R., Chan M. V., Armstrong P. C., Baig F., Gutmann C., Willeit P., Santer P., Barwari T., Theofilatos K., Kiechl S., Willeit J., Warner T. D., Mathur A., Mayr M., Neutrophil-derived protein S100A8/A9 alters the platelet proteome in acute myocardial infarction and is associated with changes in platelet reactivity. Arterioscler. Thromb. Vasc. Biol. 42, 49–62 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barrett T. J., Cornwell M., Myndzar K., Rolling C. C., Xia Y., Drenkova K., Biebuyck A., Fields A. T., Tawil M., Luttrell-Williams E., Yuriditsky E., Smith G., Cotzia P., Neal M. D., Kornblith L. Z., Pittaluga S., Rapkiewicz A. V., Burgess H. M., Mohr I., Stapleford K. A., Voora D., Ruggles K., Hochman J., Berger J. S., Platelets amplify endotheliopathy in COVID-19. Sci. Adv. 7, eabh2434 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lood C., Tyden H., Gullstrand B., Jonsen A., Kallberg E., Morgelin M., Kahn R., Gunnarsson I., Leanderson T., Ivars F., Svenungsson E., Bengtsson A. A., Platelet-derived S100A8/A9 and cardiovascular disease in systemic lupus erythematosus. Arthritis Rheumatol. 68, 1970–1980 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Piersma S. R., Broxterman H. J., Kapci M., de Haas R. R., Hoekman K., Verheul H. M., Jimenez C. R., Proteomics of the TRAP-induced platelet releasate. J. Proteomics 72, 91–109 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Khorrami S., Tavakoli M., Safari E., Clinical value of serum S100A8/A9 and CA15-3 in the diagnosis of breast cancer. Iran J. Pathol. 14, 104–112 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim H. J., Kang H. J., Lee H., Lee S. T., Yu M. H., Kim H., Lee C., Identification of S100A8 and S100A9 as serological markers for colorectal cancer. J. Proteome Res. 8, 1368–1379 (2009). [DOI] [PubMed] [Google Scholar]
- 56.Popescu I. D., Codrici E., Albulescu L., Mihai S., Enciu A. M., Albulescu R., Tanase C. P., Potential serum biomarkers for glioblastoma diagnostic assessed by proteomic approaches. Proteome Sci. 12, 47 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gruvberger S., Ringner M., Chen Y., Panavally S., Saal L. H., Borg A., Ferno M., Peterson C., Meltzer P. S., Estrogen receptor status in breast cancer is associated with remarkably distinct gene expression patterns. Cancer Res. 61, 5979–5984 (2001). [PubMed] [Google Scholar]
- 58.Stoesz S. P., Friedl A., Haag J. D., Lindstrom M. J., Clark G. M., Gould M. N., Heterogeneous expression of the lipocalin NGAL in primary breast cancers. Int. J. Cancer 79, 565–572 (1998). [DOI] [PubMed] [Google Scholar]
- 59.Cramer E. P., Glenthoj A., Hager M., Juncker-Jensen A., Engelholm L. H., Santoni-Rugiu E., Lund L. R., Laerum O. D., Cowland J. B., Borregaard N., No effect of NGAL/lipocalin-2 on aggressiveness of cancer in the MMTV-PyMT/FVB/N mouse model for breast cancer. PLOS ONE 7, e39646 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang J., Bielenberg D. R., Rodig S. J., Doiron R., Clifton M. C., Kung A. L., Strong R. K., Zurakowski D., Moses M. A., Lipocalin 2 promotes breast cancer progression. Proc. Natl. Acad. Sci. U.S.A. 106, 3913–3918 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haemmerle M., Stone R. L., Menter D. G., Afshar-Kharghan V., Sood A. K., The platelet lifeline to cancer: Challenges and opportunities. Cancer Cell 33, 965–983 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dehqanzada Z. A., Storrer C. E., Hueman M. T., Foley R. J., Harris K. A., Jama Y. H., Shriver C. D., Ponniah S., Peoples G. E., Assessing serum cytokine profiles in breast cancer patients receiving a HER2/neu vaccine using Luminex technology. Oncol. Rep. 17, 687–694 (2007). [PubMed] [Google Scholar]
- 63.Machlus K. R., Johnson K. E., Kulenthirarajan R., Forward J. A., Tippy M. D., Soussou T. S., El-Husayni S. H., Wu S. K., Wang S., Watnick R. S., Italiano J. E. Jr., Battinelli E. M., CCL5 derived from platelets increases megakaryocyte proplatelet formation. Blood 127, 921–926 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Falasca K., Ucciferri C., Dalessandro M., Zingariello P., Mancino P., Petrarca C., Pizzigallo E., Conti P., Vecchiet J., Cytokine patterns correlate with liver damage in patients with chronic hepatitis B and C. Ann. Clin. Lab. Sci. 36, 144–150 (2006). [PubMed] [Google Scholar]
- 65.Marques R. E., Guabiraba R., Russo R. C., Teixeira M. M., Targeting CCL5 in inflammation. Expert Opin. Ther. Targets 17, 1439–1460 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Becker I. C., Nagy Z., Manukjan G., Haffner-Luntzer M., Englert M., Heib T., Vogtle T., Gross C., Bharti R., Dietrich S., Mott K., Heck J., Stegmaier S., Baranowsky A., Schinke T., Schlegel N., Heckel T., Stegner D., Pleines I., Ignatius A., Schulze H., Nieswandt B., G6b-B regulates an essential step in megakaryocyte maturation. Blood Adv. 6, 3155–3161 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vijey P., Posorske B., Machlus K. R., In vitro culture of murine megakaryocytes from fetal liver-derived hematopoietic stem cells. Platelets 29, 583–588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Satija R., Farrell J. A., Gennert D., Schier A. F., Regev A., Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S15
Tables S1 and S2