Abstract
Hepatic steatosis is a major etiological factor in hepatocellular carcinoma (HCC), but factors causing lipid accumulation leading to HCC are not understood. We identify BNIP3 (a mitochondrial cargo receptor) as an HCC suppressor that mitigates against lipid accumulation to attenuate tumor cell growth. Targeted deletion of Bnip3 decreased tumor latency and increased tumor burden in a mouse model of HCC. This was associated with increased lipid in bnip3−/− HCC at early stages of disease, while lipid did not accumulate until later in tumorigenesis in wild-type mice, as Bnip3 expression was attenuated. Low BNIP3 expression in human HCC similarly correlated with increased lipid content and worse prognosis than HCC expressing high BNIP3. BNIP3 suppressed HCC cell growth by promoting lipid droplet turnover at the lysosome in a manner dependent on BNIP3 binding LC3. We have termed this process “mitolipophagy” because it involves the coordinated autophagic degradation of lipid droplets with mitochondria.
Lipid droplet turnover with mitochondria in “mitolipophagy” requires BNIP3 and is disrupted in hepatocellular carcinoma.
INTRODUCTION
Obesity-associated fatty liver disease, including nonalcoholic fatty liver disease (NAFLD) and nonalcoholic steatohepatitis (NASH), has emerged as the major etiological factor contributing to hepatocellular carcinoma (HCC) in western societies (1, 2). The relative risk of mortality from HCC is elevated (1.5- to 4-fold) among obese patients compared to nonobese individuals, more than for any other human cancer (3, 4). Given the continuing increase in obesity rates in the U.S. population (1, 3), efforts to understand how fatty liver promotes HCC are likely to be highly impactful in terms of identifying biomarkers predictive of disease onset or progression, in addition to finding novel targets for therapy.
Altered mitochondrial function has been reported in human NAFLD (5, 6) and may contribute to altered lipid metabolism in a variety of ways, such as changes in rates of fatty acid oxidation (FAO), export of citrate to the cytosol for lipid synthesis, and production of metabolic intermediates and mitochondrial reactive oxygen species that affect lipid species produced (7). Independent of lipid metabolism, dysfunctional mitochondria also modulate production of proinflammatory signals and the type and extent of cell death that contributes to tumor progression (8).
Mitochondrial quality control relies on the turnover of dysfunctional and/or excess mitochondria at the autolysosome by mitophagy (9). Bcl2/E1B19K-interacting protein 3 (BNIP3) is a mitochondrial cargo receptor that is induced in the liver by nutrient stress (10) and inserts into the outer mitochondrial membrane (OMM) where it interacts directly with processed LC3 at the autophagosome to target mitochondria for degradation at the autolysosome, thereby contributing to metabolic zonation in the liver (11). Here, we examined a previously unknown role for BNIP3 in liver tumorigenesis where we show that targeted deletion of Bnip3 in a mouse model of HCC caused reduced tumor latency and increased tumor growth rate that was associated with increased lipid content in tumors at earlier stages of disease. Low BNIP3 expression in human HCC also correlated with high lipid content and worse overall survival. BNIP3 decreased HCC growth rates by increasing rates of lipid droplet (LD) turnover at the lysosome in a process that we have termed “mitolipophagy” in which LDs get turned over in association with BNIP3-expressing mitochondria.
RESULTS
Loss of Bnip3 reduces HCC tumor latency and promotes HCC tumor growth
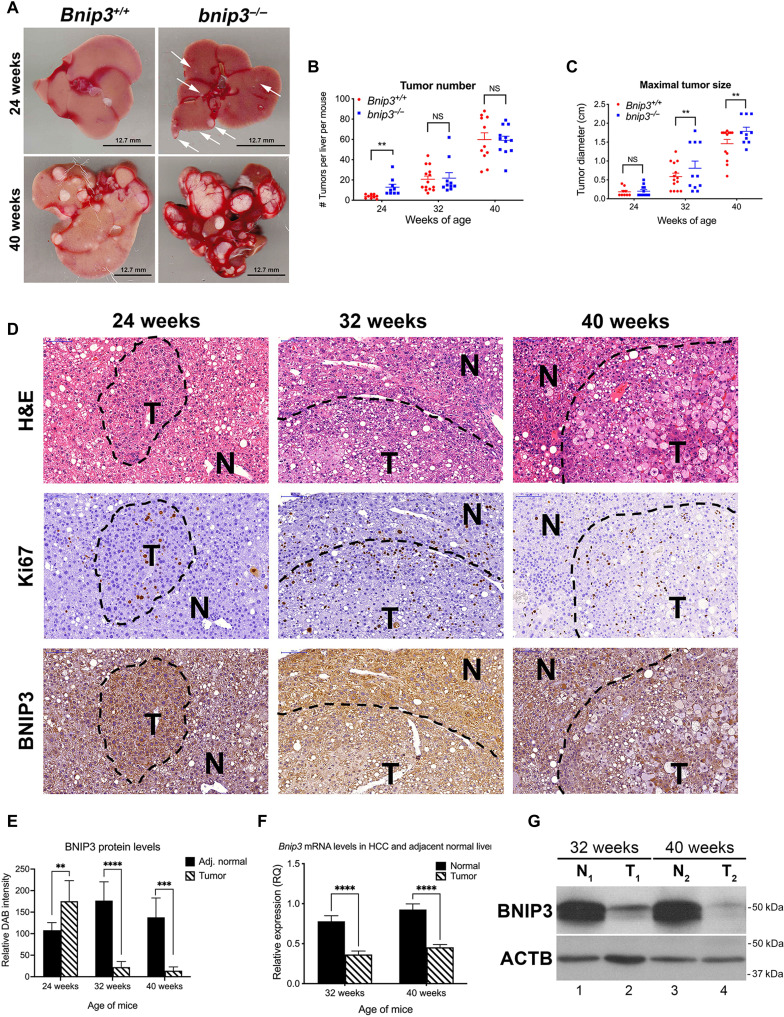
To examine the role of BNIP3 in HCC, we injected 15-day-old Bnip3+/+ and bnip3−/− mice with the chemical carcinogen diethylnitrosamine (DEN) that is known to induce HCC with predicted latency (12, 13). We harvested mice at 24, 32, and 40 weeks of age to assess the effect of Bnip3 loss on latency and growth of HCC. At 24 weeks of age, macroscopic liver lesions were apparent on the surface of the bnip3−/− liver (Fig. 1A, top right, arrows) but not on Bnip3+/+ liver (Fig. 1A, top left). By 40 weeks of age, large tumors were obvious in both the Bnip3+/+ and bnip3−/− livers, although the lesions on bnip3−/− liver were visibly larger than those on Bnip3+/+ liver (Fig. 1A). Quantification of tumor number (total per liver on serial sections through each liver) and tumor size (diameter) supported visual assessment that tumors form earlier and grow faster in DEN-treated bnip3−/− mice than wild-type (WT) mice. Specifically, by 24 weeks of age, there were significantly more tumors detected in the DEN-treated bnip3−/− mice than in DEN-treated WT control mice (Fig. 1B), and by 32 and 40 weeks, the bnip3−/− tumors were significantly larger than those forming in WT mice (Fig. 1C). In summary, loss of Bnip3 reduces tumor latency and increases tumor growth rate of DEN-induced HCC in mice.
Fig. 1. Loss of BNIP3 promotes HCC initiation and tumor growth.
(A) Representative images of HCC tumors forming in the liver of Bnip3+/+ and bnip3−/− mice at 24 and 40 weeks of age following DEN injection. Scale bars, 12.7 mm. (B) Graph of tumor number forming in the liver of Bnip3+/+ (red) and bnip3−/− (blue) mice at 24 weeks (n = 9 per genotype), 32 weeks (n = 14 per genotype), and 40 weeks (n = 13 per genotype) of age following DEN injection. (NS, not significant; **P < 0.01). (C) Graph of tumor size in the liver of Bnip3+/+ (red) and bnip3−/− (blue) mice at 24 weeks (n = 9 per genotype), 32 weeks (n = 14 per genotype), and 40 weeks (n = 13 per genotype) of age following DEN injection (NS, not significant; ** P < 0.01). Scale bars, 100 μm (top left). (D) Liver sections from Bnip3+/+ mice at 24, 32, and 40 weeks of age following DEN injection, stained with H&E (top row), Ki67 (middle row), and BNIP3 (bottom row). (E) Quantification of immunohistochemical staining for BNIP3 on liver sections from Bnip3+/+ mice at 24 weeks (n = 5), 32 weeks (n = 7), and 40 weeks (n = 5) of age following DEN injection (**P < 0.05, ***P < 0.005, and ****P < 0.0005). (F) qPCR for Bnip3 mRNA isolated from HCC lesions and adjacent normal liver in Bnip3+/+ mice at 32 weeks (n = 12) and 40 weeks (n = 12) of age following DEN injection (****P < 0.0001). (G) Western blot for BNIP3 in protein lysates isolated from HCC tumor lesions (T) and adjacent normal (N) liver in Bnip3+/+ mice at 32 and 40 weeks of age following DEN injection.
In WT mice, BNIP3 expression was elevated in HCC tumors at 24 weeks compared to adjacent normal liver (Fig. 1, D and E). By contrast, BNIP3 levels were lower in HCC tumors compared to adjacent normal liver at 32 and 40 weeks (Fig. 1, D and E). The up-regulation of BNIP3 in HCC tumors at 24 weeks compared to adjacent normal liver is likely mediated by elevated nuclear Hif1α expression, which is also elevated in HCC tumors at 24 weeks (fig. S1A). Quantitative polymerase chain reaction (qPCR) showed that Bnip3 mRNA was down-regulated in macroscopic tumors at 32 and 40 weeks of age compared to adjacent normal tissue (Fig. 1F), consistent with gene silencing. Similarly, Western blot confirmed reduced BNIP3 protein in HCC tumor in WT mice compared to adjacent normal at both 32 and 40 weeks (Fig. 1G). Such analyses were not performed on tumors at 24 weeks due to the absence of macroscopic tumors in WT liver at this time point (Fig. 1A), and thus, analysis of BNIP3 expression at 24 weeks relied on immunohistochemical analysis (Fig. 1, D and E). These results show that while BNIP3 is expressed at higher levels in early-stage HCC lesions than in adjacent normal liver, there is selection for loss of endogenous Bnip3 expression at 32 and 40 weeks, as WT tumors progress to later stages of hepatocellular tumorigenesis.
Loss of Bnip3 is associated with increased lipid accumulation in HCC
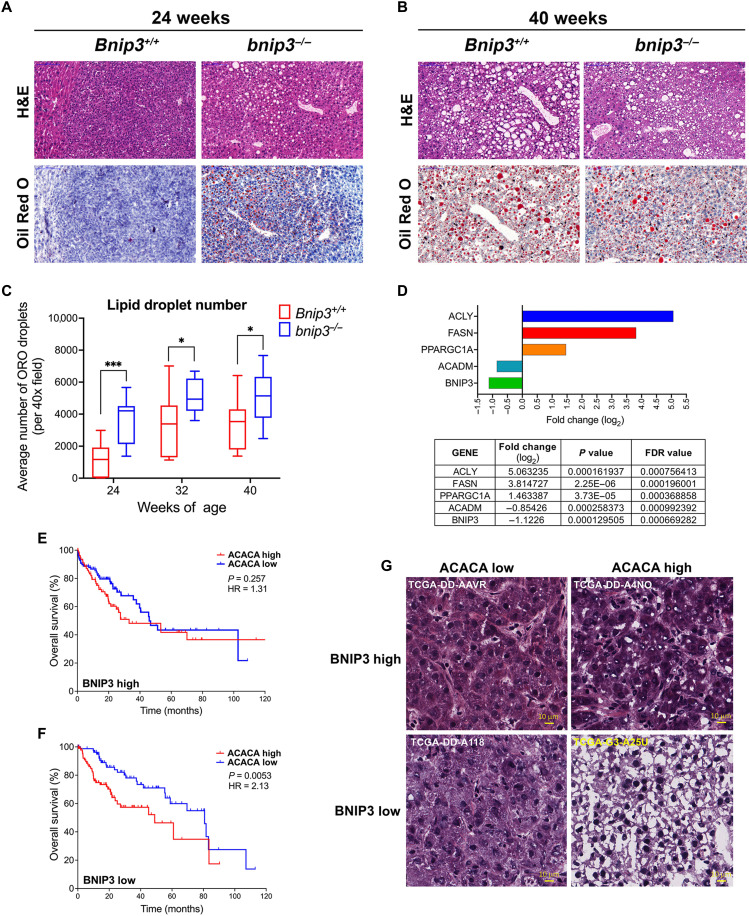
In addition to loss of BNIP3 expression in WT tumors at 32 to 40 weeks of age, there was also a change in the appearance of HCC tumors forming in WT mice. At 24 weeks of age, WT HCCs contain small, tightly packed tumor cells (Figs. 1D and 2A, left), whereas by 32 and 40 weeks of age, HCC tumors contained larger tumor cells with more “bubbles” (Figs. 1D and 2B, left), suggesting increased lipid accumulation in WT HCC at later stages of progression. Consistent with this pattern detected by hematoxylin and eosin (H&E) staining, we observed by Oil Red O staining that WT tumors at 24 weeks (Fig. 2A, left, and fig. S1B, left) contained less lipid compared to WT tumors at 40 weeks (Fig. 2B, left, and fig. S1C, left). By contrast, HCC tumors forming in bnip3−/− livers at 24 weeks (Fig. 2A, right, and fig. S1B, right) contained considerably more lipid than tumors in WT mice at 24 weeks (Fig. 2A, left, and fig. S1B, left). However, tumors growing in 40-week-old WT mouse liver that had lost Bnip3 expression (Fig. 1, D to G) exhibited as high lipid content (Fig. 2B, left, and fig. S1C, left) as that detected in bnip3−/− tumors at 40 weeks (Fig. 2B, right, and fig. S1C, right). Quantification of Oi Red O staining confirmed these observations (Fig. 2C). At 24 weeks, there was increased lipid content in bnip3−/− tumors compared to WT (Fig. 2, A and C), but these differences in lipid content diminished over time at 32 to 40 weeks, as WT tumors lost Bnip3 expression (Fig. 1, D to G) and simultaneously accumulated more lipid (Fig. 2, B and C). In summary, our findings show that loss of Bnip3 either by genetic deletion in bnip3−/− mice or via silencing at late stages of tumorigenesis in WT mice is associated with increased lipid accumulation in HCC.
Fig. 2. Low BNIP3 expression correlates with high lipid levels and poor survival in mouse and human HCC.
(A and B) Liver tumor sections from Bnip3+/+ and bnip3−/− mice at 24 weeks (A) and 40 weeks (B) of age following DEN injection, stained with H&E (top) or Oil Red O (bottom) at ×200 magnification. Scale bars, 100 μm (top left). (C) Graph of Oil Red O (ORO)–positive LD number in the liver of Bnip3+/+ (red) and bnip3−/− (blue) mice at 24 weeks (n = 12 per genotype), 32 weeks (n = 16 per genotype), and 40 weeks (n = 17 per genotype) of age following DEN injection (NS, not significant; *P < 0.05 and ***P < 0.001). (D) Comparison of relative expression of ACLY, FASN, PPARGC1A, ACADM, and BNIP3 in healthy human liver compared to HCC (14), showing fold change (log2), statistical significance (P value) of fold change, and false discovery rates (FDR) for each gene. (E and F) Overall HCC survival data obtained from the TCGA database for tumors showing BNIP3HIGH expression (above the mean, E) versus BNIP3LOW expression (below the mean, F) with either ACACALOW or ACACAHIGH expression. (G) H&E-stained sections of human BNIP3HIGH-expressing HCC (top) or BNIP3LOW-expressing HCC (bottom), with either ACACALOW (left)– or ACACAHIGH (right)–expressing HCC. Scale bars, 10 μm.
Low BNIP3 expression in human HCC correlates with worse overall survival
We examined publicly available RNA-sequencing data (14) and found increased expression of genes involved in lipid metabolism in human HCC compared to healthy liver (fig. S2, A to C), in line with previously published data (15–17). We also observed that BNIP3 expression was significantly decreased in HCC where lipid synthesis genes like fatty acid synthase (FASN) and adenosine triphosphate (ATP) citrate lyase (ACLY) were increased (Fig. 2D). Peroxisome proliferator-activated receptor-γ coactivator-1-α (PPARGC1A) that promotes mitochondrial biogenesis (a process that counters mitophagy) was increased in HCC compared to healthy liver, while medium-chain acyl-CoA dehydrogenase (ACADM) that promotes FAO was decreased (Fig. 2D). Linear regression analyses showed a highly significant inverse correlation between FASN, ACLY, and PPARGC1A levels compared to BNIP3 (fig. S2, D to F) and a direct correlation between levels of ACADM and BNIP3 (fig. S2G). These data indicate that in human HCC where lipogenic programs are increased, BNIP3 is significantly decreased.
We further explored The Cancer Genome Atlas (TCGA) data looking at how expression levels of BNIP3 affect overall survival of patients with HCC and identified a correlation between levels of BNIP3, acetyl CoA carboxylase-1 (ACACA), and patient survival. The ACACA gene encodes an enzyme that catalyzes conversion of acetyl coenzyme A (CoA) to malonyl CoA to promote lipid synthesis and is frequently up-regulated in human cancers, including HCC (18, 19). We found that high BNIP3 expression levels in combination with high ACACA expression portended a highly significant increase in overall survival rates (Fig. 2E) compared to the combination of high ACACA expression with low BNIP3 expression that had the worst prognosis for overall survival (Fig. 2F). Moreover, when we examined the histology of liver sections from these HCC patients, we observed that low BNIP3 expression in combination with high ACACA (Fig. 2G, bottom right) was associated with a markedly different histology than that associated with high BNIP3 expression and high ACACA expression (Fig. 2G, top right). Human HCC tumor with low BNIP3 in combination with high ACACA expression (Fig. 2G, bottom right) showed extensive cell ballooning, was more translucent, and exhibited less overall H&E staining than HCC sections with high BNIP3 expression and high ACACA expression (Fig. 2G, top right). This indicates that high BNIP3 mitigated against lipid accumulation linked to high ACACA expression (Fig. 2G, top right), in contrast to low BNIP3 where human HCC sections appeared to contain more lipid when associated with high ACACA expression (Fig. 2G, bottom right). Consistent with human data, we also observed elevated ACACA expression in mouse HCC relative to adjacent normal tissue, which was more marked in bnip3−/− HCC (fig. S2H). Overall, these results indicate that high BNIP3 expression is associated with decreased lipid accumulation, less aggressive HCC, and increased patient survival (Fig. 2, E and G) when linked to high ACACA expression, while conversely low BNIP3 predicts increased lipid accumulation and worse patient outcome (Fig. 2, F and G).
BNIP3 promotes mitophagy and inhibits HCC cell growth
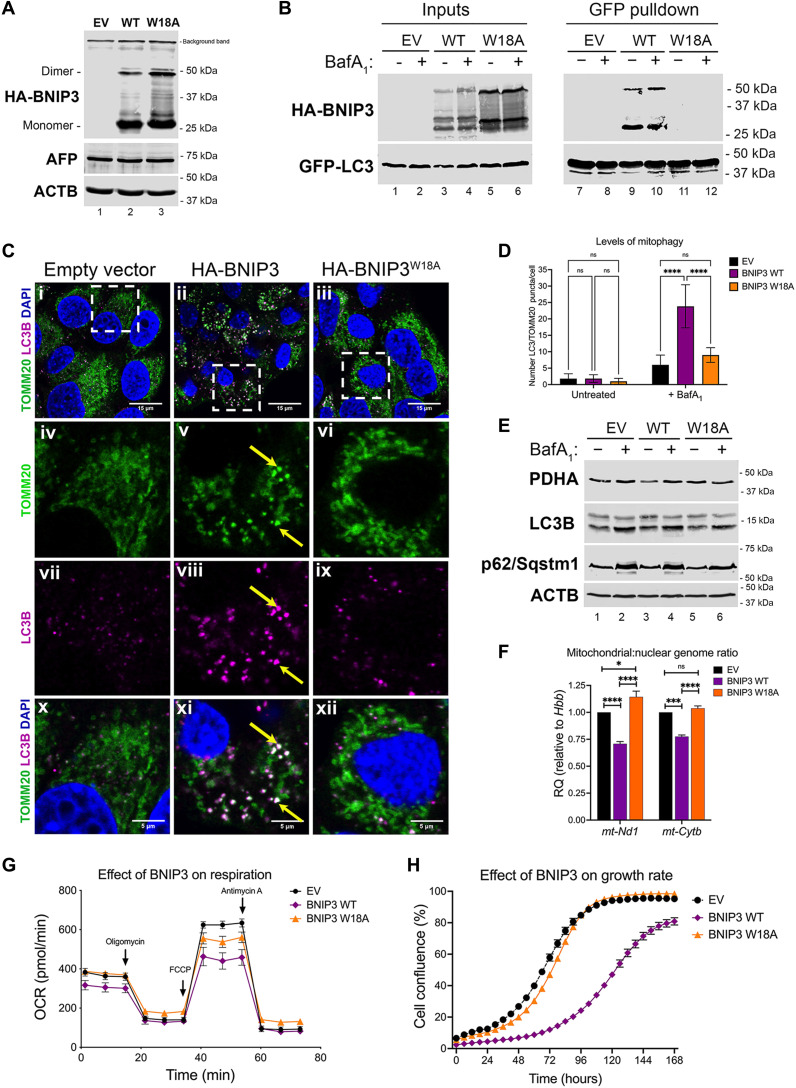
To gain mechanistic insight into how BNIP3 modulates lipid content in HCC, we established primary cell lines from bnip3−/− tumors engineered to stably express either empty vector (EV), hemagglutinin (HA)-tagged WT BNIP3 (HA-BNIP3), or HA-tagged BNIP3 mutated at W18 to inhibit LC3 interaction and block mitophagy (HA-BNIP3W18A) (11). We validated expression of EV, HA-BNIP3, or HA-BNIP3W18A in bnip3−/− lines (Fig. 3A) and confirmed expression of α-fetoprotein (AFP), an oncofetal protein expressed during liver development and reexpressed in HCC tumors (Fig. 3A). We also confirmed by pulldown assay in this HCC cell system that while HA-BNIP3 interacted with green fluorescent protein (GFP)–LC3 (Fig. 3B, lanes 9 and 10), HA-BNIP3W18A did not (Fig. 3B, lanes 11 and 12). Immunostaining for overlap between processed LC3B (an autophagosome marker) and TOMM20 (a mitochondrial marker) in the presence of bafilomycin A1 further confirmed that expression of HA-BNIP3, but not EV or HA-BNIP3W18A, promoted mitophagy (Fig. 3C). Exogenous HA-BNIP3 expression decreased staining for TOMM20 (Fig. 3Cv) and induced significant overlap in LC3B-TOMM20 staining (Fig. 3Cxi), whereas EV and HA-BNIP3W18A did not (Fig. 3, C, x and xii, and D). The overlap between LC3 and TOMM20 could only be detected in the presence of bafilomycin A1 (Fig. 3, Cxi and D, and fig. S3A), indicating that there was increased mitophagic flux when HA-BNIP3 was expressed but not with EV or HA-BNIP3W18A expression. We also noted that bafilomycin A1 caused primarily fragmented mitochondria to accumulate in HCC cells expressing HA-BNIP3 (Fig. 3Cv) compared to the same cells grown in the absence of bafilomycin A1 (fig. S3A) or compared to expression of EV or HA-BNIP3W18A (Fig. 3C, iv or vi)). These fragmented mitochondria also appeared to be the ones that overlapped with LC3B staining (Fig. 3Cxi), indicating that these are mitophagosomes (mitochondria engulfed by autophagosomes). Together, these results indicate that BNIP3 promotes mitophagy of fragmented mitochondria at the autolysosome, which is dependent on the interaction of BNIP3 with LC3.
Fig. 3. BNIP3-dependent mitophagy suppresses mitochondrial mass, glucose oxidation, and HCC cell growth.
(A) Western blot for α-HA and α-AFP in bnip3−/− HCC cells reconstituted with EV, HA-BNIP3WT, or HA-BNIP3W18A. (B) Pulldown for GFP-LC3 in bnip3−/− HCC cells reconstituted with EV, HA-BNIP3WT, or HA-BNIP3W18A in the presence (+)/absence (−) of 100 nM bafilomycin A1 and blotted for GFP (LC3) or HA (BNIP3) in input (left) or pulldown lysates (right). (C) Immunofluorescent staining for TOMM20 and LC3 in bnip3−/− HCC cells reconstituted with EV, HA-BNIP3WT, or HA-BNIP3W18A in the presence of 100 nM bafilomycin A1. Yellow arrows in (v), (viii), and (xi) point to representative fragmented mitochondria staining positive for TOMM20 and LC3B. (D) Quantification of LC3-positive puncta (magenta) overlapping (white) with TOMM20 staining (green) per cell in bnip3−/− HCC cells reconstituted with EV, HA-BNIP3WT, or HA-BNIP3W18A in the presence (+) or absence (−) of 100 nM bafilomycin A1 (NS, not significant; ****P < 0.0001; at least five cells per field, each data point is one field, n = 10 fields per condition). (E) Western blot analysis for PDHA, LC3B, and p62/Sqstm1 in bnip3−/− HCC cells reconstituted with EV, HA-BNIP3WT, or HA-BNIP3W18A in the presence (+)/absence (−) of 100 nM bafilomycin A1. (F) qPCR for mt:nuc DNA ratio in bnip3−/− HCC cells reconstituted with EV, HA-BNIP3WT, or HA-BNIP3W18A (NS, not significant; *** P < 0.001 and ****P < 0.0001). Experiments performed 12 times per condition, n = 2 experiments. (G) OCR (25 mM d-glucose and 2 mM l-glutamine) of bnip3−/− HCC cells reconstituted with EV, HA-BNIP3WT, or HA-BNIP3W18A. Cells were plated 12 times per plate/time point/genotype/experiment, n = 3 experiments. (H) Rate of cell growth determined by IncuCyte of bnip3−/− HCC cells reconstituted with EV, HA-BNIP3WT, or HA-BNIP3W18A, n = 3 experiments.
Western blot analysis showed that while HA-BNIP3 (but not EV or HA-BNIP3W18A) decreased the levels of mitochondrial matrix protein pyruvate dehydrogenase A (PDHA), it did not alter levels of processed LC3B-II or cause p62/Sqstm1 to accumulate (Fig. 3E, lane 3), indicating that general autophagy was not overtly affected by BNIP3. Treatment of cells (EV, HA-BNIP3, and HA-BNIP3W18A) with bafilomycin A1 increased levels of LC3B-II and p62/Sqstm1 (Fig. 3E, lanes 2, 4, and 6 compared to lanes 1, 3, and 5), also indicating that autophagy is broadly functional in these HCC cells independent of BNIP3 status. However, levels of PDHA only increased with bafilomycin A1 treatment in cells expressing HA-BNIP3, and not when EV or HA-BNIP3W18A was expressed, supporting the conclusion that BNIP3 is primarily affecting mitophagy downstream of LC3 and is not affecting the general autophagy machinery.
Mitochondrial genome copy number (amplifying for Nd1 and Cytb) relative to nuclear DNA (amplifying β-hemoglobin/Hbb) was also shown to be decreased in HCC cells expressing HA-BNIP3 but not EV or HA-BNIP3W18A (Fig. 3F), consistent with increased mitophagy induced by HA-BNIP3. As expected, if BNIP3-dependent mitophagy is decreasing mitochondrial mass, we also observed decreased oxygen consumption rate (OCR) by HCC cells expressing HA-BNIP3 but not EV or HA-BNIP3W18A (Fig. 3G). Exogenous HA-BNIP3 also decreased extracellular acidification of growth media following a glycolysis stress test (fig. S3B) and suppressed glucose uptake by HCC cells in culture (fig. S3C). Conversely, loss of Bnip3 increased glucose uptake into HCCs in bnip3−/− liver compared to Bnip3+/+ liver as measured by fluorodeoxyglucose–positron emission tomography (FDG-PET) analysis of live mice at 32 weeks of age (fig. S3, D and E). Overall, these results show that exogenous HA-BNIP3, but not EV or HA-BNIP3W18A, promotes mitophagy, resulting in decreased mitochondrial mass in HCC cells. In addition, BNIP3 reduces reliance of HCC tumor cells on glucose to fuel either oxidative phosphorylation or glycolysis.
Last, we observed that exogenous HA-BNIP3 also markedly slowed the growth rate of bnip3−/− HCC cells in contrast to HCC cells expressing either EV or HA-BNIP3W18A (Fig. 3H). The failure of HA-BNIP3W18A to suppress HCC cell growth suggests that the ability of BNIP3 to inhibit tumor growth is dependent on binding to LC3, although it cannot be ruled out that the W18A mutation affects other functions of BNIP3. In summary, BNIP3 induced mitophagy, decreased mitochondrial mass, decreased dependence on glucose as a fuel source, and decreased the growth rate of HCC cells.
BNIP3 reverses lipid accumulation in HCC cells by promoting LD turnover
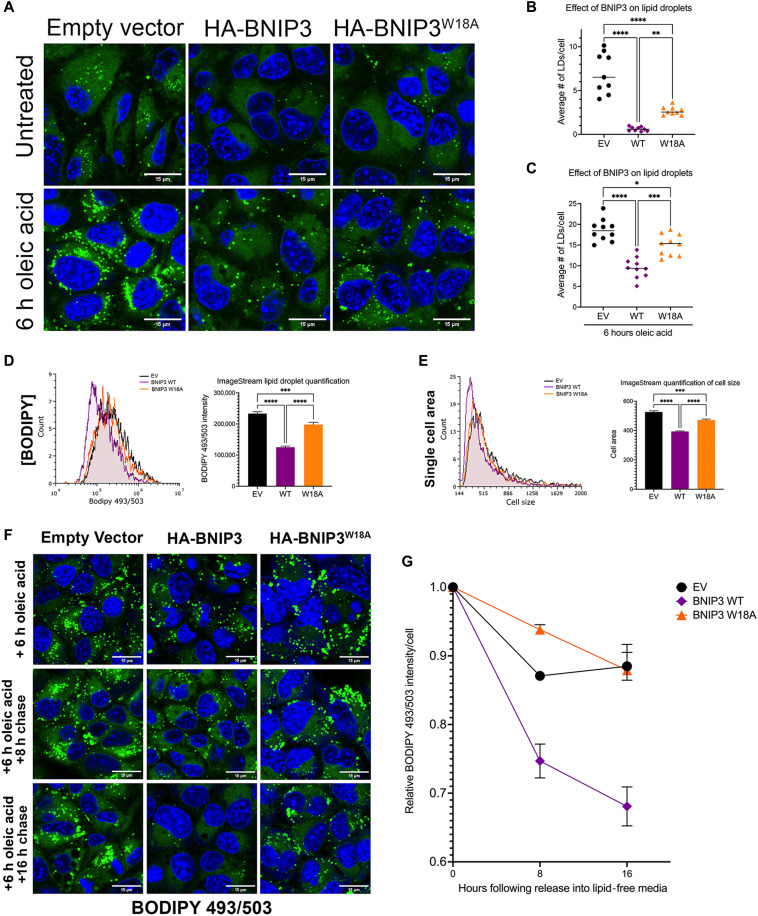
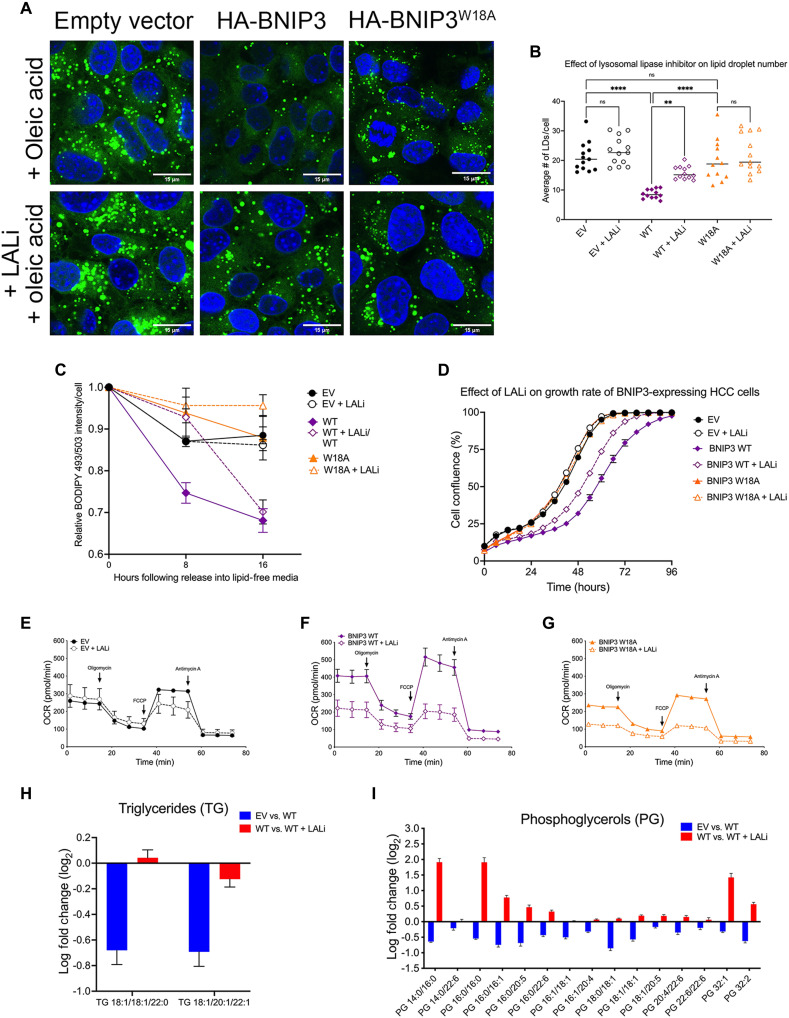
Given the steatotic phenotype in vivo in HCC tumors growing in the absence of BNIP3, we examined lipid content of HCC cells in culture expressing EV, HA-BNIP3, or HA-BNIP3W18A by staining cells with the lipophilic dye BODIPY 493/503. We observed that expressing exogenous HA-BNIP3 decreased the number of BODIPY-positive LDs in HCC cells compared to EV-expressing HCC cells (Fig. 4, A and B, and fig. S4A). Although HA-BNIP3W18A was unable to promote mitophagy or suppress cell growth (Fig. 3, C, D, and H), we observed that this mutant form of BNIP3 retained partial ability to decrease LD number compared to EV, although not as effectively as HA-BNIP3 (Fig. 4, A and B). When cells were challenged with oleic acid to further increase cellular lipid content, HA-BNIP3 was significantly more effective at decreasing LD number than either EV or HA-BNIP3W18A (Fig. 4, A and C, and fig. S4A). Analysis of BODIPY staining of HCC cells by ImageStream flow cytometric analysis confirmed results obtained by immunofluorescence microscopy in showing decreased LD numbers when HA-BNIP3 was expressed but not EV, and less so with HA-BNIP3W18A (Fig. 4D). ImageStream flow cytometric analysis also showed HCC cells expressing HA-BNIP3 to be smaller in size than HCC cells expressing EV or HA-BNIP3W18A (Fig. 4E). Previous work has linked lower mitochondrial mass and function to decreased overall cell size (20, 21).
Fig. 4. BNIP3 suppresses LD accumulation in HCC cells.
(A) LD staining (BODIPY 493/503) in bnip3−/− HCC cells reconstituted with EV, HA-BNIP3WT, or HA-BNIP3W18A ± 100 μM oleic acid. (B) Graph of BODIPY 493/503 staining in bnip3−/− HCC cells reconstituted with EV, HA-BNIP3WT, or HA-BNIP3W18A (***P < 0.001; ****P < 0.0001; at least 20 cells per field, each data point is one field, n ≥ 10 fields per condition). (C) Graph of BODIPY 493/503 staining in bnip3−/− HCC cells reconstituted with EV, HA-BNIP3WT, or HA-BNIP3W18A + oleic acid (***P < 0.001; ****P < 0.0001; at least 20 cells per field, each data point is one field, n ≥ 10 fields per condition). (D) Quantification of BODIPY 493/503 ImageStream flow cytometric analysis of bnip3−/− HCC cells reconstituted with EV, HA-BNIP3WT, or HA-BNIP3W18A (***P < 0.001 and ****P < 0.0001; at least 10,000 cells per condition were quantified for n = 2 experiments). (E) Quantification of cell size generated by ImageStream flow cytometric analysis of bnip3−/− HCC cells reconstituted with EV, HA-BNIP3WT, or HA-BNIP3W18A (***P < 0.001 and ****P < 0.0001; at least 10,000 cells per condition were quantified for n = 2 experiments). (F) Lipid droplet staining (BODIPY 493/503) in bnip3−/− HCC cells reconstituted with EV, HA-BNIP3WT, or HA-BNIP3W18A grown for 6 hours in 100 μM oleic acid and then released into lipid-free media for 0, 8, and 16 hours. (G) ImageStream flow cytometric quantification of relative numbers of LDs in bnip3−/− HCC cells reconstituted with EV, HA-BNIP3WT, or HA-BNIP3W18A grown for 6 hours in 100 μM oleic acid and then released into lipid free–media for 0, 8, and 16 hours (at least 10,000 cells per condition were quantified for n = 2 experiments).
To assess whether the ability of BNIP3 to suppress cellular lipid content was due to decreased uptake of lipid from the culture media or to a more specific role in LD turnover, we performed pulse-chase experiments in which HCC cells were grown in the presence of oleic acid for 6 hours and then released into fatty acid–free media and numbers of LDs were quantified at 8 and 16 hours following release (Fig. 4, F and G). HCC cells expressing HA-BNIP3 showed significantly lower numbers of LDs immediately after growth for 6 hours in oleic acid (Fig. 4F and fig. S4B) and showed a further marked decrease in LD numbers at 8 hours after release from oleic acid (Fig. 4, F and G, and fig. S4B). ImageStream flow cytometric analysis revealed that the rate at which LD numbers decreased in HA-BNIP3–expressing HCC cells (Fig. 4G, purple) was significantly greater than in EV-expressing (Fig. 4G, black) or HA-BNIP3W18A–expressing (Fig. 4G, orange) HCC cells. HCC cells expressing EV or HA-BNIP3W18A were barely able to clear LDs at all and had similar numbers of LDs at 16 hours as they had at 0 hours (Fig. 4G, black and orange). By contrast, HA-BNIP3–expressing HCC cells see a significant drop in LD number by 8 hours, which continues to decrease after that (Fig. 4G, purple). In summary, these findings show that BNIP3 promotes clearance of LDs from HCC cells in a manner that is at least partially dependent on its ability to promote mitophagy and is not affected by the ability to uptake lipid from the microenvironment.
BNIP3 promotes FAO, but this does not explain how BNIP3 decreases lipid levels or inhibits tumor cell growth
Lipid accumulation in cells can arise for various reasons, including FAO (22), and because FAO takes place in the mitochondrial matrix, we postulated that BNIP3 may be promoting lipid turnover by increasing FAO. To examine this, we measured OCR of HCC cells expressing EV, HA-BNIP3, or HA-BNIP3W18A using palmitate as a substrate for oxidation (fig. S5, A to C). While HA-BNIP3 lowered OCR of cells using glucose as their primary carbon source (Fig. 3G), we were surprised to observe that HA-BNIP3 markedly increased OCR when palmitate was supplied for oxidation (fig. S5B) compared to cells expressing EV or HA-BNIP3W18A (fig. S5, A and C). The OCR of HA-BNIP3–expressing HCC cells was also more sensitive to etomoxir (ETO), which suppresses carnitine palmitoyl transferase-1 (CPT1) and blocks fatty acid uptake into the mitochondria to suppress FAO (fig. S5B). ETO collapsed the OCR of HA-BNIP3–expressing HCC cells grown in palmitate down to levels seen in HCC cells expressing EV or HA-BNIP3W18A (fig. S5B). By contrast, ETO had no effect on OCR of HCC cells expressing EV or HA-BNIP3W18A (fig. S5, A and C). These results suggest that HA-BNIP3 is promoting FAO, and thus, we were interested to determine whether this explained how HA-BNIP3 was able to promote lipid turnover in HCC cells.
Unexpectedly, HA-BNIP3 retained the ability to decrease LD content in HCC cells, even when FAO was inhibited (fig. S5, D and E). This ability remained evident in HCC cells fed oleic acid in the presence of ETO (fig. S5, D and F), suggesting that while HA-BNIP3 does promote FAO, HA-BNIP3 has an additional activity that regulates LD content in HCC cells.
In support of BNIP3 eliciting its effect on LD numbers independent of its ability to increase FAO, treatment of HA-BNIP3–expressing HCC cells with ETO had no effect on cell growth such that HA-BNIP3, but not EV or HA-BNIP3W18A, was equally able to suppress HCC cell growth in the presence or absence of ETO (fig. S5G). These results show that while BNIP3 promotes FAO in bnip3−/− HCC cells, this does not explain how BNIP3 decreases lipid content or how BNIP3 suppresses HCC cell growth.
BNIP3 does not decrease lipid levels or HCC cell growth via effects on lipogenesis
To assess whether BNIP3 was decreasing lipid content by inhibiting de novo lipogenesis (DNL), we examined the effect of treating cells with an active site inhibitor of FASN called TVB-3664 that is active on mouse cells (23). Treatment of cells for 24 hours with TVB-3664 markedly increased expression of fatty acid synthesis genes (Fasn, Acaca, and Acly) irrespective of genotype (fig. S6A), as expected due to transcriptional feedback effects when lipid synthesis is repressed (24). Inhibiting FASN is also expected to increase cellular levels of malonyl CoA that are depleted through use as a substrate for FASN. Only cells expressing HA-BNIP3, not EV or HA-BNIP3W18A, showed increased malonyl CoA when treated with the TVB-3664 FASN inhibitor (fig. S6B), indicating that FASN is more active in HA-BNIP3–expressing HCC cells than in cells expressing either EV or HA-BNIP3W18A, as might be expected due to their lower levels of lipid. TVB-3664 eliminated lipid content independent of BNIP3 because it decreased lipid content to undetectable levels in all cell lines including EV-expressing cells (fig. S6, C and D). However, by increasing the amount of lipid in cells by feeding them with oleic acid, we observed that HA-BNIP3, but not EV or HA-BNIP3W18A, remained able to decrease lipid content even when FASN was inhibited (fig. S6, D and E), arguing that the ability of BNIP3 to decrease lipid content in HCC cells was independent of effects on DNL. Furthermore, the repressive effect of FASN inhibition on growth of HA-BNIP3–expressing cells was synergistic with the effect of exogenous HA-BNIP3 (fig. S6F), suggesting that BNIP3 is suppressing tumor cell growth in a manner independent of effects on lipid synthesis. In addition, TVB-3664 inhibition of FASN was also able to inhibit growth of EV- and HA-BNIP3W18A–expressing HCC cells (fig. S6F), consistent with the growth suppressive effects of TVB-3664 being BNIP3 independent. In summary, while TVB-3664–mediated inhibition of FASN reduces HCC cell growth, this is independent of BNIP3, and conversely, BNIP3 suppresses HCC cell growth independently of TVB-3664 and FASN activity.
We measured fatty acid uptake and detected decreased fatty acid uptake by BNIP3-expressing cells compared to either EV or BNIP3W18A control cells (fig. S6G). However, interpretation of this data is confounded by the fact that BNIP3-expressing cells are smaller than either EV- or BNIP3W18A-expressing cells (Fig. 4E). Moreover, pulse-chase data in which we monitored the effects of BNIP3 on decreasing LD number following release into fatty acid–free media confirmed a role for BNIP3 in limiting lipid content independent of fatty acids in the media (Fig. 4, F and G). Thus, while we cannot at this time exclude an effect of BNIP3 on fatty acid uptake, BNIP3 has additional functions that limit lipid content and cell growth that are not explained by effects on FAO, DNL, or fatty acid uptake.
BNIP3 limits lipid content and growth by promoting LD turnover at the lysosome
Another mechanism by which cells regulate lipid content is via turnover of LDs that involves both lipolysis and lipophagy (25, 26). LDs play important roles in fatty acid trafficking within the cell and in storing lipids to buffer the cell against lipotoxicity (25, 26). LD turnover by lipophagy relies on their engulfment by autophagosomes and the activity of acid lipases at the autolysosome (26–28), although direct uptake of lipid by the lysosome via microlipophagy has also been described (29, 30). LDs are known to associate with mitochondria (31–33), and given the role of BNIP3 in mitophagy, we examined a potential role for BNIP3 in promoting LD turnover via a mechanism that integrates mitophagy with lipophagy. When we inhibited lysosomal lipases with Lalistat2 (LALi) (28), we observed increased numbers of LDs accumulating in HCC cells when HA-BNIP3 was expressed but less with EV or HA-BNIP3W18A (fig. S7, A and B), suggesting that BNIP3 relied on lysosomal lipases to elicit at least part of its effect in decreasing LD numbers. By contrast, inhibition of the cytosolic lipase adipose triglyceride lipase (ATGL) failed to significantly alter the effect of BNIP3 on LD numbers (fig. S7C). ImageStream flow cytometric analysis confirmed that LALi only increased BODIPY LD number significantly when HA-BNIP3 was expressed and less so when HA-BNIP3W18A or EV was expressed (fig. S7, D and E).
LALi also decreased the ability of HA-BNIP3 to clear LDs when cells were cultured in oleic acid for 6 hours (Fig. 5, A and B) but did not have any significant effect on LD numbers in bnip3−/− HCC cells expressing EV or HA-BNIP3W18A (Fig. 5, A and B). In pulse-chase experiments, we observed that when cells were grown in oleic acid for 6 hours and then switched to fatty acid–free media, LALi inhibited the marked effect of HA-BNIP3 on the rate of LD turnover over time (Fig. 5C, purple solid line compared to purple dashed line). Although HA-BNIP3–expressing cells always exhibit fewer LDs, when treated with LALi, the rate at which they were able to clear LDs between 0 and 8 hours was now similar to that detected in EV- and HA-BNIP3W18A–expressing cells that were unable to clear LDs and markedly less affected by LALi (Fig. 5C, black and orange dashed lines compared to solid lines). In summary, these results show that BNIP3 decreases lipid content in HCC cells, at least in part, by promoting LD turnover by acid lipases at the lysosome.
Fig. 5. BNIP3 promotes lipophagy, and inhibiting lysosomal lipase limits HCC cell growth.
(A) Fluorescent microscopy imaging of BODIPY 493/503 staining in bnip3−/− HCC cells reconstituted with EV, HA-BNIP3WT, or HA-BNIP3W18A ± 50 μM LALi + 100 μM oleic acid. (B) Graph of BODIPY 493/503 staining in bnip3−/− HCC cells reconstituted with EV, HA-BNIP3WT, or HA-BNIP3W18A ± 50 μM LALi + 100 μM oleic acid (ns, not significant; **P < 0.01 and ****P < 0.0001; at least 20 cells per field, each data point is one field, n ≥ 10 fields per condition). (C) ImageStream flow cytometric quantification of LDs in bnip3−/− HCC cells reconstituted with EV, HA-BNIP3WT, or HA-BNIP3W18A grown for 24 hours in 50 μM LALi + 6 hours in 100 μM oleic acid and then released into lipid free–media for 0, 8, and 16 hours. At least 10,000 cells per condition were quantified for n = 2 experiments. (D) Rate of cell growth determined by IncuCyte of bnip3−/− HCC cells reconstituted with EV, HA-BNIP3WT, or HA-BNIP3W18A ± 50 μM LALi in n = 3 experiments for each condition. (E to G) OCR (166 μM palmitate and 2 mM d-glucose) in bnip3−/− HCC cells reconstituted with EV (E), HA-BNIP3WT (F), or HA-BNIP3W18A. Cells were plated 12 times per plate for each time point per genotype per experiment for n = 3 experiments. (G) ±50 μM LALi. (H and I) LC-MS analysis of triglycerides and phosphoglycerols decreased (fold change > 2.0 and P > 0.05) in bnip3−/− HCC cells expressing BNIP3 WT compared to EV (blue bars) and increased by 24-hour treatment with 50 μM LALi (red bars). Lipidomics were performed on five biological replicates per condition.
Treatment of cells with LALi significantly increased the growth rate of HCC cells expressing HA-BNIP3 (Fig. 5D, purple dashed line compared to solid line) but had no effect on the growth of cells expressing EV or HA-BNIP3W18A that expanded more rapidly independent of any effect of LALi (Fig. 5D, black and orange lines), suggesting that BNIP3 suppresses HCC cell growth partly by promoting lysosomal turnover of LDs in a mitophagy-dependent manner. LALi treatment also markedly decreased palmitate oxidation in cells expressing HA-BNIP3, suggesting that fatty acids liberated by lysosomal lipases are fueling FAO in HCC cells (Fig. 5F). The effect of LALi on palmitate oxidation was markedly stronger in HCC cells expressing HA-BNIP3 (Fig. 5F) compared to either control EV–expressing cells (Fig. 6E) or cells expressing HA-BNIP3W18A (Fig. 5G). These findings indicate that BNIP3 promotes FAO and suppresses HCC cell growth by promoting LD turnover.
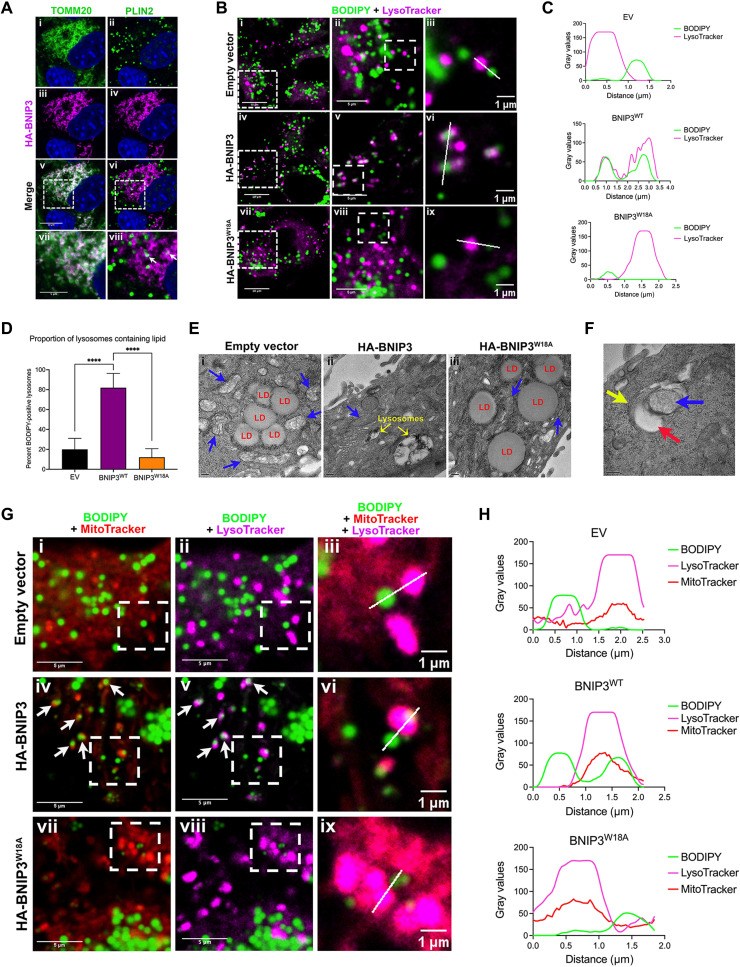
Fig. 6. BNIP3 promotes LD-lysosome interactions at mitochondria.
(A) Fluorescent microscopy of bnip3−/− HCC cells expressing HA-BNIP3WT, stained for HA-BNIP3 (magenta) and TOMM20 or PLIN2 (green) to assess overlap (white, arrows in viii). White dashed square indicates area magnified below. (B) Fluorescent microscopy of bnip3−/− HCC cells expressing EV, HA-BNIP3WT, or HA-BNIP3W18A grown in 50 μM LALi + 100 μM oleic acid, stained with BODIPY 493/503 and LysoTracker to visualize overlap (white) between LDs (green) and lysosomes (magenta). (C) Linescan analysis colocalizing LDs with lysosomes from (B, iii, vi, and ix) from bnip3−/− HCC cells expressing EV, HA-BNIP3WT, or HA-BNIP3W18A, showing only BNIP3WT-promoted LD-lysosome overlap. (D) Quantification of percent BODIPY-positive lysosomes/cell for bnip3−/− HCC cells expressing EV, HA-BNIP3WT, or HA-BNIP3W18A [****P < 0.0001; >2 regions of interest (ROIs)/field, each data point is one ROI, n ≥ 10 fields/condition]. (E) TEM on bnip3−/− HCC cells expressing EV, HA-BNIP3WT, or HA-BNIP3W18A + 50 μM LALi + 100 μM oleic acid (red: LDs; blue: mitochondria; yellow: lysosomes). Scale bars, 200 nm (bottom left). (F) TEM on bnip3−/− HCC cells expressing HA-BNIP3WT + 50 μM LALi + 100 μM oleic acid (red: LDs; blue: mitochondria; yellow: lysosomes). Scale bar, 200 nm (bottom left). (G) Fluorescent microscopy of bnip3−/− HCC cells expressing EV, HA-BNIP3WT, or HA-BNIP3W18A grown in 50 μM LALi + 100 μM oleic acid, stained with BODIPY 493/503 (green), MitoTracker (red), and LysoTracker (magenta). Arrows point to areas of overlap between mitochondria and LDs (iv) and lysosomes with LDs (v), consistent with colocalization of LDs, mitochondria, and lysosomes. (H) Linescan analysis colocalizing LDs, mitochondria, and lysosomes in bnip3−/− HCC cells expressing EV, HA-BNIP3WT, or HA-BNIP3W18A in (F, iii, vi, ix), showing that only BNIP3WT promoted LD-mitochondria-lysosome overlap.
To further assess the effect of HA-BNIP3 on lipid content in HCC cells, we performed unbiased liquid chromatography–mass spectrometry (LC-MS)–based lipidomics (34). Consistent with BNIP3 promoting mitophagy, we observed increased turnover of mitochondrial cardiolipins in bnip3−/− HCC cells expressing HA-BNIP3 (WT) compared to EV (fig. S8A, blue bars). Treatment of HA-BNIP3–expressing bnip3−/− HCC cells (WT) with LALi (WT + LALi) reversed the effect of HA-BNIP3 on cardiolipin levels (fig. S8A, red bars). Similarly, we observed decreased levels of Bis(monoacylglycero)phosphate (BMP) phospholipids in bnip3−/− HCC cells expressing HA-BNIP3 (WT) compared to EV (fig. S8B, blue bars). BMPs are enriched at late endosomes and the lysosome, and their decreased levels with HA-BNIP3 (WT) that are reversed by LALi treatment (fig. S8B, red bars) are consistent with BNIP3 promoting enhanced lysosomal activity. LDs store triglycerides (TGs), and both TG species detected by lipidomic analysis were decreased in levels in HA-BNIP3– expressing HCC cells (WT) compared to control EV–expressing cells (Fig. 5H, blue bars), an effect that was reversed by treatment of WT cells with LALi (Fig. 5H, red bars). Other classes of lipids that were decreased by HA-BNIP3 (WT) in a manner dependent on lysosomal lipase activity were phosphoglycerols (Fig. 5I), phosphoethanolamines (fig. S8C), phosphoserines (fig. S8D), phosphocholines (fig. S8E), and phosphoinositols (fig. S8F). These phospholipids make up the lipid monolayer of LDs and other organelles in the cell, and decreased overall levels of these specific lipid species are supportive of our finding that BNIP3 promotes LD turnover at the lysosome. A class of lipids that showed the opposite response to HA-BNIP3 expression and LALi were ceramides that were increased by HA-BNIP3 (WT) expression compared to control EV–expressing cells (fig. S8, G and H, blue bars) and decreased by LALi (fig. S8, G and H, red bars). Ceramides are hydrolyzed at the mitochondria, and by eliminating mitochondria, BNIP3 may be promoting ceramide synthesis at the expense of hydrolysis. In summary, BNIP3 promotes the turnover of lipids that make up LDs and other organelles, including TGs and phospholipids, possibly explaining the smaller size of BNIP3-expressing cells (Fig. 4E) if phospholipids are limiting for membrane expansion.
BNIP3 promotes the accumulation of LDs with mitochondria at the lysosome
Given the effect of HA-BNIP3 in promoting LD turnover at the lysosome (Fig. 5), we examined whether BNIP3 localized to LDs or played a role in promoting LD turnover through its canonical localization at the OMM. Staining HA-BNIP3–expressing HCC cells with antibodies to HA (BNIP3), TOMM20 (mitochondria), and PLN2 (LDs) (Fig. 6A), we observed extensive overlap in staining for BNIP3 and TOMM20, consistent with the primary function of BNIP3 in mitophagy at the OMM. All detectable BNIP3 staining (Fig. 6Aiii, magenta) overlapped (Fig. 6Av, white) with TOMM20 (Fig. 6Ai, green), suggesting that most if not all BNIP3 is at the mitochondria. When we looked for overlap in staining between BNIP3 (magenta) and PLIN2 (green) (Fig. 6Avi), we observed that most LDs (Fig. 6Aii, green) did not show overlapping staining (Fig. 6A, vi and vii, white) with BNIP3 (Fig. 6Avi, magenta). A few PLIN2-positive LDs (green) did show overlapping staining (white) with BNIP3 (Fig. 6A, vi and viii, magenta arrows). Similarly, overlap between TOMM20 (green) and PLIN2 (magenta) was restricted to a small proportion of LDs (fig. S9Aiii), indicating that while some LDs associate with mitochondria, most do not. In addition, the overlap between BNIP3 and PLIN2 (Fig. 6A, vi and viii, arrows) was identical to that of TOMM20 and PLIN2 (fig. S9Aiv, arrows), indicating that BNIP3 overlap with PLN2-positive LDs was likely secondary to the interaction of LDs with TOMM20-positive mitochondria. In summary, these data suggest that BNIP3 is not at the LD but is restricted to the OMM and that its association with LDs is limited to regions of the mitochondrial network that interact with LDs.
To further assess the overlap between LDs and lysosomes, we costained cells with BODIPY 493/503 (green) and LysoTracker (magenta). Experiments were performed in the presence of oleic acid and LALi to allow LDs to accumulate in lysosomes. While a few overlapping LD/lysosomes were detected in HCC cells expressing EV (Fig. 6B, top) or HA-BNIP3W18A (Fig. 6B, bottom), there was a marked increase in overlap (white) between BODIPY (green) and LysoTracker (magenta) in cells expressing HA-BNIP3 (Fig. 6B, middle). Higher-magnification images demonstrated that while HA-BNIP3 promoted extensive overlap between LDs and lysosomes (Fig. 6B, iv to vi), EV and HA-BNIP3W18A did not (Fig. 6B, i to iii and vii to ix). Linescan analysis of the overlap of LDs with lysosomes shown in Fig. 6B (EV, iii; BNIP3, vi; and BNIP3W18A, ix) is shown in Fig. 6C and illustrates how BODIPY-positive LDs only significantly overlap with LysoTracker-positive lysosomes in bnip3−/− HCC cells when HA-BNIP3 is expressed and not when either EV or HA-BNIP3W18A is expressed (Fig. 6C). Quantification of the number of lysosomes that have BODIPY accumulation in these images shows that while expressing HA-BNIP3 in bnip3−/− HCC cells increases the percentage of BODIPY-positive lysosomes, expressing EV and HA-BNIP3W18A does not (Fig. 6D). These results show that HA-BNIP3, but not HA-BNIP3W18A, promotes LD uptake by lysosomes.
When we examined the ultrastructure of HCC cells expressing EV, HA-BNIP3, or HA-BNIP3W18A by transmission electron microscopy (TEM) (Fig. 6E), we observed more LDs in both EV- and HA-BNIP3W18A–expressing cells (Fig. 6E, i and iii) but fewer LDs in the HA-BNIP3–expressing HCC cells (Fig. 6Eii), consistent with HA-BNIP3 decreasing LD number. Lysosomes containing lipid-like material (low electron density) were also more evident in HA-BNIP3–expressing cells (Fig. 6Eii, yellow arrows) than in control or HA-BNIP3W18A–expressing cells (Fig. 6E, i and iii). We also observed LDs (Fig. 6F, red arrow) associated with membrane-bound structures that resembled fragmented mitochondria (Fig. 6F, blue arrow) surrounded by a double membrane (Fig. 6F, yellow arrow) that resembled the overlapping structures detected by imaging of BODIPY/LysoTracker-stained cells (Fig. 6Bvi).
To test more definitively whether LDs were colocalizing with both mitochondria and lysosomes, we stained cells for MitoTrackerDR (red, mitochondria), BODIPY 493/503 (green, LDs), and LysoTracker (magenta, lysosomes) (Fig. 6G and fig. S9B). EV- and BNIP3W18A-expressing cells appeared to have more MitoTrackerDR staining (fig. S9B, i and viii) than HA-BNIP3–expressing HCC cells (fig. S9Biv), which is consistent with HA-BNIP3 promoting mitophagy more effectively than EV or HA-BNIP3W18A, although this was not quantified. BODIPY-positive LDs were abundant in all cells due in part to growth in oleic acid and to inhibition of lysosomal lipases with LALi, but overlap between LDs (BODIPY) and mitochondria (MitoTrackerDR) was only readily detected in the cells expressing HA-BNIP3 (Fig. 6Giv) and not in cells expressing either EV (Fig. 6Gi) or HA-BNIP3W18A (Fig. 6Gvii). Areas of overlap (yellow) between BODIPY-positive LDs (green) and MitoTracker-positive (red) mitochondria (Fig. 6Giv) indicated that smaller LDs interacted closely with mitochondria (Fig. 6Gv, arrows) when HA-BNIP3 was expressed but not EV or HA-BNIP3W18A (Fig. 6G, ii and viii). These findings indicate that BNIP3, but not BNIP3W18A, promotes the interaction of small LDs with mitochondria. Notably, overlap between LDs and lysosomes occurred at the exact same sites as overlap in staining for LDs and mitochondria (Fig. 6G, v compared to iv, arrows). The colocalization of LDs with lysosomes at mitochondria was only seen in cells expressing HA-BNIP3 and was not detected in cells expressing EV or the HA-BNIP3W18A mutant that cannot bind LC3 (Fig. 6G, ii compared to i and viii compared to vii). To assess the extent of overlap between staining for BODIPY, MitoTracker, and LysoTracker, we performed linescan analysis on representative structures (Fig. 6H). Linescan plots illustrate how only in HA-BNIP3–expressing bnip3−/− HCC cells (Fig. 6H, middle) do we observe coincident staining for lipid (green), mitochondria (red), and lysosomes (magenta). These results indicated that BNIP3 promotes the colocalization of LDs with lysosomes at the mitochondria.
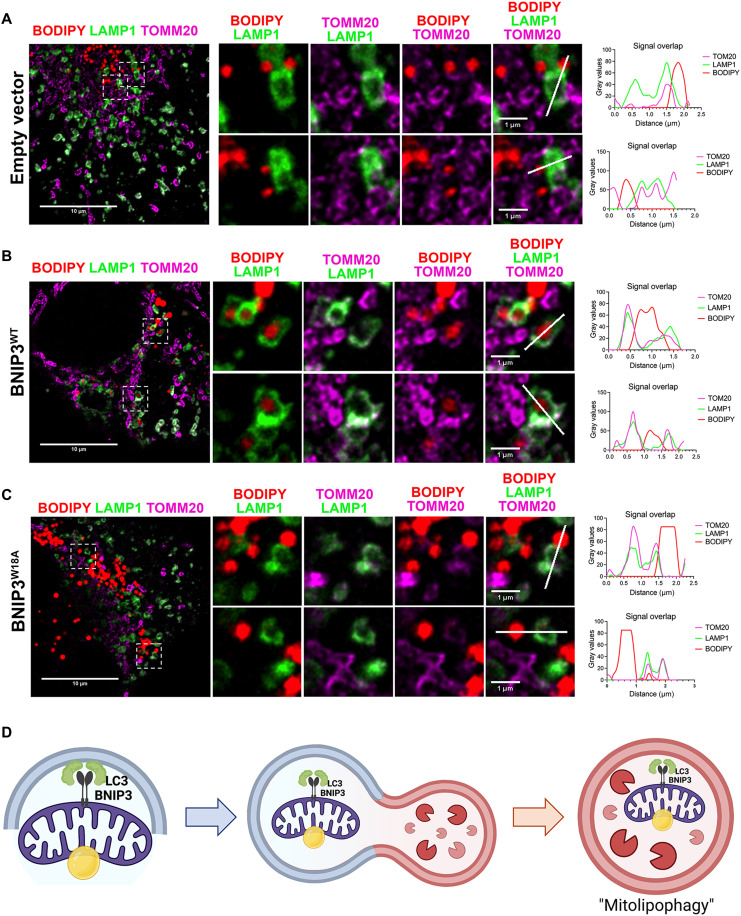
To validate these results using standard fixed immunofluorescence techniques rather than live-cell dyes, we stained bnip3−/− HCC cells expressing EV, HA-BNIP3, or HA-BNIP3W18A for TOMM20 (magenta) and LAMP1 (green) expression in combination with BODIPY (red) and again showed that only HA-BNIP3WT (fig. S10A, middle, white arrows) and not EV or HA-BNIP3W18A (fig. S10A, top and bottom) increased BODIPY-positive lysosomes found in cells treated with oleic acid and LALi. Representative high-magnification images and linescan analysis of these interactions are shown in fig. S10B and illustrate how only cells expressing HA-BNIP3WT (middle, yellow arrow) and not EV or HA-BNIP3W18A exhibit overlap in BODIPY-positive LDs (red) with LAMP1-positive lysosomes (green) and TOMM20-positive mitochondria (magenta). The red BODIPY signal was found inside the green lysosome when HA-BNIP3WT was expressed, and that was also coincident with magenta TOMM20 signal indicating that mitochondria colocalized with lipid-positive lysosomes (fig. S10B, middle). This was not observed with either EV-expressing (fig. S10B, top) or HA-BNIP3W18A–expressing (fig. S10B, bottom) cells.
In a complementary approach, we repeated staining of bnip3−/− HCC cells expressing EV, HA-BNIP3, or HA-BNIP3W18A for TOMM20 (magenta) and LAMP1 (green) expression in combination with BODIPY staining (red), but this time made use of superresolution microscopy, specifically Leica’s STED (stimulated emission depletion) modality that permits an even higher degree of resolution to visualize subdiffraction organelle interactions (Fig. 7, A to C). Consistent with data presented in Fig. 6G and fig. S10, we show that expression of HA-BNIP3WT (Fig. 7B) promotes colocalization of LDs (red), mitochondria (magenta), and lysosomes (green). Red LDs can be clearly visualized encircled by green lysosomes (Fig. 7B) while also surrounded by magenta-colored mitochondria that in turn overlapped with green lysosomes (Fig. 7B, white indicates magenta-green overlap). Linescan analysis of two different representative LD-mitochondria-lysosome structures shown suggests that mitochondria surrounding LDs are enveloped by lysosomes for degradation when HA-BNIP3WT is expressed (Fig. 7B). By contrast, this LD-mitochondria-lysosome coincidence is not seen in bnip3−/− HCC cells expressing either EV (Fig. 7A) or HA-BNIP3W18A (Fig. 7C), where LDs were seen outside or alongside lysosomes but rarely inside the lysosome. These data support our observations from Fig. 6G and fig. S10 (A to C) that BNIP3 promotes autophagic engulfment of LDs in association with mitochondria (Fig. 7D).
Fig. 7. BNIP3 promotes colocalization of LDs, mitochondria, and lysosomes in mitolipophagy.
(A to C) Superresolution immunofluorescence microscopy of bnip3−/− HCC cells reconstituted with EV (A), HA-BNIP3WT (B), or HA-BNIP3W18A (C) grown for 24 hours in 50 μM LALi + 6 hours in 100 μM oleic acid and then stained with BODIPY 493/503 (red), TOMM20 (magenta), and LAMP1 (green) to visualize overlap between LDs (green), mitochondria (magenta), and lysosomes (magenta). Two representative high-magnification images (highlighted by white dashed square, left) are shown for each in (A), (B), and (C). Linescan analysis of colocalization of LDs (red) with mitochondria (magenta) and lysosomes (green) is shown for each of the two representative high-magnification images from bnip3−/− HCC cells reconstituted with EV (A), HA-BNIP3WT (B), or HA-BNIP3W18A (C), illustrating that only BNIP3WT promoted LD-lysosome overlap. (D) Graphical representation of model proposed.
In summary, our data demonstrate that the role of BNIP3 in promoting LD turnover at the lysosome (lipophagy) depends on the interaction of BNIP3 with LC3, and we propose that BNIP3 is promoting lipophagy indirectly by eliminating LDs that are associated with mitochondria via its primary role in mitophagy. We refer to this as mitolipophagy wherein LDs are passengers turned over with mitochondria at the autolysosome in a BNIP3-dependent manner (Fig. 7D).
DISCUSSION
The role of obesity in causing cancer is perhaps best defined in HCC, where the increasing incidence of HCC has been attributed to the ever-growing prevalence of NAFLD and NASH in the U.S. population (1, 35–38). Excess lipid in hepatocytes, lipotoxicity, cycles of cell death, and regenerative proliferation with associated inflammation and ensuing fibrosis contribute to the progression of fatty liver disease to HCC (1, 8). Cell death due to lipid overload (lipotoxicity) is avoided in part via storage of excess lipids in LDs (25, 26), while functional mitochondria modulate the balance between FAO and DNL to also determine lipid levels and cell viability (19, 39–41).
Defects in LD management have been previously implicated in human liver disease (42). In particular, PNPLA3 mutations are a major human genetic risk factor for NAFLD that contribute to race-dependent differences in hepatic fat content and susceptibility to NAFLD (43). Mutant PNPLA3 accumulates at LDs altering ATGL lipase activity by interfering with CGI-58, a critical cofactor for ATGL (44–46). Genomic analyses also identified the CIDEB gene, which is involved in LD metabolism, as being enriched for somatic mutations linked to fatty liver disease relative to healthy control (47), while deletion of LAMP2A in mouse liver caused hepatic steatosis due to defective lipophagy (48). Thus, given the importance of LD biology in the etiology of liver disease, more needs to be understood about the mechanisms controlling LD accumulation and turnover (49, 50).
Our work identifies a previously unknown role for the BNIP3 mitophagy adaptor in mitochondrial homeostasis and LD turnover that prevents lipid accumulating by promoting mitolipophagy in which LDs are turned over in concert with mitochondria at the lysosome (Fig. 7D). This activity of BNIP3 contributes to its ability to attenuate HCC cell growth, and decreased expression of BNIP3 was associated with elevated lipid and decreased overall survival in human HCC patients. Deletion of Parkin in mice also leads to hepatic steatosis (51, 52), suggesting that mitophagy may promote lipid homeostasis in the liver more generally. Recently, defective mitophagy was also shown to contribute to Niemann-Pick type C neurodegenerative disease as a result of cholesterol accumulation in lysosomes (53), again emphasizing the importance of mitophagy for lipid homeostasis, this time in the nervous system.
The original report of lipophagy proposed engulfment of LDs by phagophore membranes in a classical macroautophagy process in hepatocytes that required LC3 (27). The role of LC3 in control of LD numbers in the cell is complex, with LC3 also implicated in LD formation at the endoplasmic reticulum (ER) (54, 55). In addition, LC3 is required for ATGL-mediated lipolysis via direct interaction with ATGL, suggesting a mechanism by which lipolysis and lipophagy may be coordinated (56), although ATGL inhibitors did not inhibit the ability of BNIP3 to promote LD turnover in our studies. Hepatocytes also use a form of microlipophagy in which LDs associate directly with the lysosome and extrude lipid directly into the lysosome for lipolysis, in the absence of an LC3-dependent phagophore (29, 30). Recently, PLIN3 was identified as promoting lipophagy in the liver in a manner dependent on its phosphorylation by mammalian target of rapamycin (mTOR), suggesting a way in which LD turnover is coordinated with growth control more generally (57). Our work contributes to this field by showing a role for BNIP3 in bringing LDs and lysosomes together specifically at mitochondria (Fig. 6, B, D, and G) and implicating LC3 in this process because the W18A mutant form of BNIP3 was significantly impaired in its ability to promote these interactions.
We were unable to detect BNIP3 at LDs, and it remains to be determined how BNIP3 promotes the interaction of LDs with lysosomes, except that it happens at the mitochondria. Generally, much remains to be found about how LDs interact with mitochondria and lysosomes. Various studies have implicated the Rab7 and Rab10 small guanosine triphosphatases (GTPases) in promoting the interaction of LDs with lysosomes during lipophagy (58, 59) via LC3 recruitment (59) or by acting as a “lipophagic synapse” that tethers the LD to the lysosome via as yet unknown interactions (58). Rab GTPases are also implicated in mitophagy where Rab5 and Rab7 have been shown to be involved in Parkin-mediated mitophagy via an endosomal pathway to degradation (60–62), while Rab7 promotes mitochondria-lysosome interactions (63) and BNIP3 may rely on Rab5 for mitochondrial clearance (64). However, how this could contribute to the role we report here for mitochondrial BNIP3 in LD turnover at the lysosome remains to be determined.
Various mechanisms have also been proposed to explain how LDs interact with mitochondria, which can occur independent of the LD-lysosome interaction (32). Such mechanisms include LD-mitochondria tethering via PLIN5 at the LD (65, 66) and MIGA2 at mitochondria (31, 32). In brown adipose tissue, LD size is partly determined by surface area of contact with peri-droplet mitochondria (66) such that LD size increased in proportion to the size of the mitochondria with which it is associated (66). However, as for LD-lysosome interactions, much remains to be determined about how LDs interact with mitochondria, including how these mechanisms differ between tissues and also between normal cells and tumor cells. Our work reported here is carried out using HCC tumor cells, while most of the work in the field of LD-lysosome interactions and lipophagy has been performed on primary hepatocytes (27, 29, 58). Rapidly growing small tumor cells are less amenable to imaging analyses of such interactions, and our ongoing work is pursuing the mechanistic basis by which BNIP3 modulates interactions of LDs with the lysosome at the mitochondria in primary hepatocytes, where our previous work showed a critical role for BNIP3 in lipid handling (67). This raises the question of whether mitolipophagy is unique to HCC cells or also occurs in normal hepatocytes, and if mitolipophagy is a subversion of normal processes by tumor cells. Hepatocytes have relatively high mitochondrial mass compared to other cell types, and the liver is also a major site of lipid catabolism in the body, particularly under fasting conditions (68), so it is possible that mitolipophagy is more prevalent in the liver than other tissues, which remains to be determined.
There is also the link between the ER, LDs, and mitochondria that has not been examined here but is clearly important to be looked at in future studies. LDs are generated at the ER and relocation of ER-resident enzymes involved in lipid synthesis promotes LD growth (69), while ER-located DGAT1 funnels fatty acids released by autophagy to LDs that are specifically associated with mitochondria (70). The ER wraps around mitochondria to modulate lipid flux requiring ER-resident Rrbp1 for ER-mitochondria interactions, specifically at MAMs (mitochondrial-associated ER membranes) such that silencing of Rrbp1 in liver gave rise to steatosis (71). Rrbp1 also stimulates mitophagy in skeletal muscle by promoting LC3 lipidation (72), suggesting a broader role for Rrbp1 in lipid homeostasis through interactions at the mitochondria and the ER. While we have not detected BNIP3 at other subcellular locations other than mitochondria, future work will examine whether BNIP3 is enriched at sites of mitochondrial contact with ER (MAMs) and mitochondrial-LD contacts.
Mitochondrial fission is known to precede mitophagy (73, 74), and BNIP3 promotes mitochondrial fragmentation as a prelude to turnover of smaller fragmented mitochondria by mitophagy (75) that in turn shifts the balance of mitochondria in the cell to a more fused state. Previous work has indicated that mitochondrial fission is required for Ras-driven transformation (76–78), although it is not understood why. Mitochondrial fission likely slows FAO by limiting exchange of reducing agents and metabolic intermediates, such as occurs within the matrix of more fused mitochondria (79). This in turn may switch the reliance of tumor cells from oxidizing lipid to oxidizing other carbon sources, such as glucose, and we certainly see BNIP3 suppressing glucose oxidation and promoting oxidation of lipid (Fig. 3G and fig. S5B), consistent with BNIP3 promoting an increased proportion of fused mitochondria in cells. The interaction of LDs with mitochondria is dependent on mitochondrial fusion that, like BNIP3-dependent mitophagy (10, 11), is increased by starvation (26, 80). Thus, BNIP3 could be indirectly promoting increased LD association with mitochondria (Figs. 6G and 7B) by pushing cellular mitochondria to a more fused state. The W18A mutant retains the ability to promote mitochondrial fragmentation but is unable to promote mitophagy, causing smaller fragmented mitochondria to accumulate, which may explain why the W18A mutant cannot promote the association of LDs with lysosomes, if this only takes place at fused mitochondria.
Selective autophagy implies that only the selected cargo gets turned over and certainly up-regulation of specific cargo receptors like BNIP3 promotes increased mitophagy preferentially over other selective forms of autophagy. However, analysis of electron micrographs indicates that while mitochondria make up the bulk of the cargo during mitophagy, other organelles, such as associated ER and ribosomes, also get turned over by association with targeted mitochondria (81). On the basis of the data presented, including work showing BNIP3 to be located at the mitochondria and not at LDs (Fig. 6A), we have proposed that BNIP3 promotes LD turnover through mitolipophagy in which LDs get turned over as passengers with associated mitochondria (Fig. 7D). We further propose that the ability of BNIP3 to promote LD turnover inhibits HCC growth by limiting the cellular pool of available phospholipids for cellular membranes (Fig. 5I and fig. S8). This BNIP3-dependent tumor-suppressive mechanism may be uniquely relevant to liver where accumulating lipid promotes both NAFLD and NASH, which are precursor disease states to HCC.
MATERIALS AND METHODS
Mice
All mice (WT and bnip3−/− mice) were maintained on a pure C57Bl/6J genetic background. Mice were maintained in an environmentally controlled specific pathogen–free barrier facility and provided ad libitum with water and chow. Tumors were induced in mice by intraperitoneal injection of 15-day-old male mice with DEN (25 mg/kg).
Cells
Primary HCC cell lines were established from bnip3−/− mice using a standard two-step liver perfusion technique, as described previously by our laboratory (11). Macroscopic tumors evident in the excised and collagenase perfused liver were dissected away from surrounding nontumor liver parenchyma in a culture dish containing isolation media [Dulbecco’s modified Eagle’s medium (DMEM)/glucose (4.5 g/liter), 1 mM lactate, 2 mM l-glutamine, 15 mM Hepes, 100 nM dexamethasone, 10% defined fetal bovine serum (HyClone), and penicillin (100 U/ml)/streptomycin (0.1 mg/ml)]. Tumors were disaggregated by pipetting, filtered through a 75-μm filter, and washed three times in isolation media. The HCC cell pellet was resuspended in defined HCC growth media [DMEM/F12, 10% defined fetal bovine serum (HyClone), 100 nM dexamethasone, epidermal growth factor (20 μg/liter), 1× insulin-transferrin-selenium (Gibco, catalog no. 41400045), and penicillin (100 U/ml)/streptomycin (0.1 mg/ml)]. HCC cell lines were expanded and infected overnight in the presence of polybrene with lentivirus (8 μg/ml) (pLVX) expressing either EV sequences, HA-BNIP3 or HA-BNIP3W18A, and selection for expressing lines was performed in hygromycin (200 μg/ml). Cell lines were validated for exogenous BNIP3 expression by Western blot with α-HA and α-AFP and by immunofluorescence for HA (see below). Experiments were conducted in HCC-defined media (no hygromycin) in the presence or absence of 100 μM bovine serum albumin (BSA)–conjugated oleic acid (Sigma-Aldrich, catalog no. O3008). Drug treatments included 100 nM bafilomycin A1 (Enzo, catalog no. BML-CM110) for 4 hours, 10 μM ETO (Sigma-Aldrich, catalog no. 509455) for 24 hours, 12 nM TVB-3664 (3-V Biosciences) for 24 hours, 20 μM Atglistatin (ATGLi) (Cayman Chemical, catalog no. 15284) for 24 hours, or 50 μM LALi (Cayman Chemical, catalog no. 25347) for 24 hours.
Analysis of OCRs
HCC cells were seeded in Seahorse XF96 microplates at a density of 0.75 × 104 cells per well. The next day, the cellular mitochondrial stress test was performed according to the manufacturer’s protocol [1 μM oligomycin, 0.75 μM carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP), and 5 μM antimycin A], using the Seahorse XF96 analyzer in the Biophysics Core at the University of Chicago. Briefly, 2× DMEM base medium was used to make 1× DMEM supplemented with 25 mM glucose, 2 mM glutamine, and 1 mM sodium pyruvate, with a pH adjusted to 7.35. Cells were rinsed with phosphate-buffered saline (PBS) before addition of 175 μl of 1× DMEM, and the plate was incubated in the absence of CO2 for approximately 1 hour. Alternatively, cells were incubated in substrate-limited growth media (no glutamine, 2 mM glucose, no sodium pyruvate, and 0.5 mM l-carnitine) for 24 hours, and then 166 μM palmitate was added as the main carbon source according to the manufacturer’s protocol in a palmitate oxidation stress kit from Agilent Plc (catalog no. 103693-100). Data were normalized by cell number using Hoechst 33342 nuclear counterstain and fluorescence quantification using a microplate reader. Normalized OCR data were then analyzed using Agilent Seahorse Wave software.
Measurement of cell growth rate
HCCs that stably express HA-BNIP3 or HA-BNIP3W18A or EV as control were seeded into 12 wells each per condition (104 cells per well) on 96-well plates. Cells were treated or not with different drugs (12 nM TVB-3664, 10 μM ETO, or 50 μM LALi), and confluence was measured over time using the IncuCyte Live-Cell Analysis System (Sartorius) over a 7-day period and normalized to day 0 using IncuCyte custom software. Photographs of cells were taken with a 10× objective every 6 hours from four separate regions per well, and values from four regions of each well were pooled and averaged across 24 replicates.
RNA extraction
Cells in a six-well plate were washed twice with 2 ml of Dulbecco’s PBS (DPBS), followed by addition of 1 ml of TRIzol. Wells were incubated for 5 min at room temperature (RT) and collected in Eppendorf tubes. At this step, samples could be frozen at −80°C or immediately extracted for RNA. For extraction, 200 μl of chloroform was added to each sample, followed by vigorous shaking for 15 s and incubation for 3 min at RT. Tubes were centrifuged at 12,000g, 15 min, at 4°C. The aqueous upper phase (~400 μl) was transferred into a fresh tube, followed by addition of 1 volume of 70% EtOH (~400 μl) and vigorous shaking. Samples were incubated for 5 min and then applied to RNeasy columns. The remainder of the extraction was performed according to the RNeasy Mini kit protocol (Qiagen) and included on-column deoxyribonuclease I digestion. RNA was eluted in 50 μl of ribonuclease (RNase)–free water, concentrations were measured using the NanoDrop Spectrophotometer, and samples were stored at −80°C.
Quantitative PCR
To make complementary DNA (cDNA), 1 to 2 μg of RNA were reverse-transcribed using the High Capacity RNA-to-cDNA Kit (Applied Biosystems). The concentration of cDNA was measured by NanoDrop, and samples were stored at −20°C. For gene expression analysis, we performed quantitative real-time PCR on 250 ng of cDNA per sample using TaqMan gene-specific fluorogenic probes (Applied Biosystems/Thermo Fisher Scientific). Primers used for qPCR included in this article are as follows: genomic copy number (Mt:Nuc gDNA): Cyba, Mm00241140_cn; Ndufa1, Mm00526370_cn; and Hbb-bh1, Mm00216612_cn; gene expression: Bnip3, Mm01275601_g1; Fasn, Mm01253292_m1; Acaca, Mm01304277_m1; Acly, Mm00652520_m1; Scd1, Mm00772290_m1; Cpt1a, Mm00550438_m1; Cpt2, Mm00487205_m1; Acadm, Mm01323360_g1; Acadl, Mm00599660_m1; and Rps12, Mm03030276_g1.
Immunohistochemistry and Oil Red O staining
Immunohistochemistry on mouse liver sections was carried out as described previously (11) using heat denaturation in citrate buffer (pH 6.0) to expose the epitope. Stained slides were digitized using an Allied Vision Technologies Stingray F146C color slide scanner and quantified using the Spectrum Plus Image analysis software (Aperio). Antibodies were used in immunohistochemistry as follows: Ki67 (1:100; LabVision, catalog no. RM9106), α-BNIP3 (1:100; Sigma Prestige, HPA003015), α-FASN (1:100; Cell Signaling Technology, catalog no. 3180), and α-ACACA (1:200; Cell Signaling Technology, catalog no. 3676). Oil Red O staining was performed on frozen liver sections that were warmed to RT, fixed for 10 min in cold 10% neutral buffered formalin, and allowed to air dry. Slides were incubated in propylene glycol for 3 min and then in Oil Red O/propylene glycol solution for 10 min, followed by 3 min in 85% propylene glycol and washing three times in water. Oil Red O reagents were obtained from Newcomer Supply (catalog no. 9119A). Oil Red O–stained sections were then counterstained in hematoxylin and mounted using Vectashield (Vector Laboratories). Oil Red O droplets were quantified in the red channel following deconvolution and thresholding, using ImageJ (NIH).
Protein extraction
For harvesting of cells, plates were washed in ice-cold DPBS, followed by scraping in 1 ml of DPBS containing protease inhibitors [0.5 mM phenylmethylsulfonyl fluoride, aprotinin (1 μg/ml), leupeptin (1 μg/ml), and 1 mM Na3VO4]. Cells were pelleted at 3000g for 3 min at 4°C and resuspended in radioimmunoprecipitation assay lysis buffer [10 mM tris-HCl (pH 8.0), 150 mM NaCl, 1% sodium deoxycholate, 0.1% SDS, and 1% Triton X-100] containing protease and phosphatase inhibitors (Roche PhosSTOP inhibitor cocktail tablet). Samples were incubated on ice for 15 min with vortexing every 5 min and centrifuged at full speed for 15 min at 4°C. The supernatant was transferred to prechilled Eppendorf tubes, and protein concentration was measured on a NanoDrop spectrophotometer and stored frozen at −80°C.
Western blot and IP assays
Protein samples were denatured by boiling for 5 min with SDS-reducing sample buffer [400 mM tris (pH 6.8), 10% SDS, and 500 mM β-mercaptoethanol] and sample loading dye [60% glycerol and bromophenol blue (BPB)]. The amount of protein loaded per sample varied depending on the proteins being probed, but typically, 75 μg was loaded onto SDS–polyacrylamide gel electrophoresis (SDS-PAGE) gels, followed by transfer to nitrocellulose (0.2-μm pore, GE Healthcare) or polyvinylidene difluoride (0.45-μm pore, GE Healthcare) membranes. Membranes were blocked in 5% nonfat milk in tris-buffered saline (TBS)/0.05% Tween (TBST) for 30 min at RT with shaking and incubated with primary antibodies overnight at 4°C on a rocker, in 5% BSA/TBST for antibodies from Cell Signaling Technology, and in 5% nonfat milk/TBST for all others. The next day, membranes were washed three times with TBST and incubated with horseradish peroxidase–conjugated secondary antibody (Dako) in 5% nonfat milk/TBST for 2 hours at RT on a shaker. Membranes were washed three times in TBST, and proteins were visualized by chemiluminescence and exposure on x-ray film. Antibodies used for Western blots are as follows: α-HA (1:2000; Cell Signaling Technology, 3724), α-BNIP3 (1:500; Cell Signaling Technology, #3769), α-AFP (1:500; Santa Cruz Biotechnology, sc-15375), α-LC3B (1:1000; Cell Signaling Technology, 2775), α-p62/Sqstm1 (1:1000; Cell Signaling Technology, 5114), α-PDHA (1:1000; Abcam, ab 168379), and α-GFP (1:1000; Santa Cruz Biotechnology, sc-9996). For GFP-LC3 pulldown assays, HCC cell lines were infected overnight in the presence of pLVX (8 μg/ml) expressing GFP-LC3 and cultured for 48 hours, when cells were treated or not with 100 nM bafilomycin A1 (Enzo, catalog no. BML-CM110) for the last 4 hours of the experiment. Following experimental treatments, plates were scraped in 1 ml of NP-40 immunoprecipitation (IP) lysis buffer [50 mM tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% IGEPAL, and 0.01% β-mercaptoethanol] containing protease and phosphatase inhibitors and sonicated at 10% power for 30 s using a Fisher Sonic Dismembrator (model 500). GFP-tagged proteins were immunoprecipitated using GFP-Trap magnetic beads (Chromotek). Lysates were incubated on beads for 1 hour at 4°C on a rotator and washed three times with NP-40 IP lysis buffer for 10 min, and finally resuspended in 2× sample loading buffer [1:2:2, 10× SDS:5× BPB:ddH2O]. Immunoprecipitates were resolved on SDS-PAGE and subjected to indicated Western blotting as described above.
LD and live-cell dye imaging and quantification
Cells were seeded onto eight-well chambered coverslips (Ibidi, #80826) and grown in HCC growth media. For MitoTracker and LysoTracker experiments, live cells were pretreated with 50 nM LysoTracker (Invitrogen, catalog no. L7528) and/or 50 nM MitoTracker Deep Red (Invitrogen, catalog no. M22426) 3 hours before experimental end point. At experimental end point, media were aspirated and wells were washed in DPBS followed by fixation in 4% paraformaldehyde (PFA) (Alfa Aesar, catalog no. J61899AP) for 10 min at RT. Cells were then incubated with BODIPY 493/503 (0.5 mg/ml; Invitrogen catalog no. D3922) for 20 min, washed 1× with DPBS, and mounted with Ibidi mounting medium with 4′,6-diamidino-2-phenylindole (DAPI) (Ibidi, catalog no. 50011). All images were collected using a 63× oil-immersion objective on a Leica TCS SP8 laser scanning confocal microscope. Ten to 15 representative images were obtained for each well. Quantification of BODIPY-positive LDs was performed using ImageJ software [National Institutes of Health (NIH)]. An ImageJ macro was written to autothreshold and quantify LDs/nuclei for all images.
Immunofluorescence and confocal microscopy
For exclusively immunofluorescence staining, cells were seeded onto eight-well chambered coverslips and grown in HCC growth media. At experimental end point, media were aspirated and wells were washed in DPBS followed by fixation in 4% PFA for 10 min at RT, followed by permeabilization in ice-cold 100% methanol for 10 min at −20°C. Wells were blocked in 10% goat serum in 0.5% TBST for 30 min. Wells were incubated with primary antibodies [anti-TOMM20 (Abcam, ab56783; 1:200), anti-LC3B (Cell Signaling, 3868S; 1:200), anti-HA (Bethyl Laboratories, A190-106A; 1:100), anti-PLN2 (Abcam, ab52356; 1:400), and anti-LAMP1 (Abcam, ab25245; 1:200)] in 10% goat serum in 0.5% TBST overnight at 4°C. The next day, wells were washed in 0.5% TBST for 3 × 5 min, followed by incubation in appropriate fluorescent secondary antibodies in 10% goat serum/TBST for 1 hour at RT. Wells were washed in 0.5% TBST for 3 × 5 min and mounted with Ibidi mounting medium with DAPI (Ibidi, catalog no. 50011). All images were collected using a 63× oil-immersion objective on a Leica TCS SP8 laser scanning confocal microscope. Ten to 15 representative images were obtained for each condition.
For immunofluorescence staining that included BODIPY staining, cells were seeded onto eight-well chambered coverslips and grown in HCC growth media. At experimental end point, media were aspirated and wells were washed in DPBS followed by fixation in 4% PFA for 10 min at RT. Wells were blocked in 10% goat serum in 0.1% saponin for 30 min. Wells were incubated with primary antibodies [anti-TOMM20 (Abcam, ab56783; 1:200) and anti-LAMP1 (Abcam, ab25245; 1:200)] in 10% goat serum in 0.1% saponin overnight at 4°C. The next day, wells were washed in 0.1% saponin for 3 × 5 min, followed by incubation in appropriate fluorescent secondary antibodies in 10% goat serum/saponin for 1 hour at RT. Secondary antibodies used were specific Alexa Fluor (Invitrogen Molecular Probes). Wells were washed in 0.1% saponin for 3 × 5 min and then incubated with BODIPY 493/503 (0.5 mg/ml) for 20 min, washed ×1 with DPBS, and mounted with Ibidi mounting medium with DAPI. Images were collected using a 63× oil-immersion objective on a Leica TCS SP8 laser scanning confocal microscope, and Leica’s Lightning Adaptive Image Deconvolution software was applied to all images to achieve subdiffraction resolution. Ten to 15 representative images were obtained for each condition. Image analysis and all linescans were generated using ImageJ software (NIH).
Immunofluorescence and superresolution microscopy
Cells were seeded onto eight-well chambered coverslips and grown in HCC growth media. At experimental end point, media were aspirated and wells were washed in DPBS followed by fixation in 4% PFA for 10 min at RT. Wells were blocked in 10% goat serum in 0.1% saponin for 30 min. Wells were incubated with primary antibodies [anti-TOMM20 (Abcam, ab56783; 1:200) and anti-LAMP1 (Abcam, ab25245; 1:200)] in 10% goat serum in 0.1% saponin overnight at 4°C. The next day, wells were washed in 0.1% saponin for 3 × 5 min, followed by incubation in appropriate fluorescent secondary antibodies in 10% goat serum/saponin for 1 hour at RT. Secondary antibodies used for STED microscopy were STAR Red, STAR Orange, and STAR 580 (Abberior STAR dyes). Wells were washed in 0.1% saponin for 3 × 5 min and then incubated with BODIPY 493/503 (0.5 mg/ml) for 20 min, washed once with DPBS, and mounted with Ibidi mounting medium without DAPI (Ibidi, catalog no. 50001). Images were collected using Leica’s STED modality on the TCS SP8 laser scanning confocal microscope platform with a 100× oil-immersion objective. Leica’s Lightning Adaptive Image Deconvolution software was applied to all images to achieve higher resolution. Ten to 15 representative images were obtained for each condition. Image analysis and all linescans were generated using ImageJ software (NIH).
Measurement of malonyl CoA
Cells were collected from confluent 15-cm plates, scraped in 1 ml of DPBS, and immediately frozen at −20°C. Malonyl CoA levels were measured using a mouse malonyl coenzyme A enzyme-linked immunosorbent assay (ELISA) kit (MyBiosource, catalog no. MBS705127) according to the manufacturer’s instructions. Values were normalized by total protein levels measured using a Bradford assay protein quantification kit (Bio-Rad, catalog no. 5000201).
Glucose uptake and fatty acid uptake
In vitro glucose uptake by HCC tumor cells was measured in cell lysates according to the manufacturer’s instructions using the luciferase-based Glucose Uptake-Glo assay from Promega (catalog no. J1341). Fatty acid uptake into HCC cell in vitro was quantified using a fluorescent dodecanoic acid substrate, and uptake was measured by plate reader at 515 nm according to the manufacturer’s instructions (Sigma-Aldrich, catalog no. MAK156). In vivo glucose uptake by liver tumors was determined by performing micro-PET/computed tomography (CT) imaging using a FLEX Triumph micro-PET/single photon emission computerized tomography (SPECT)/computerized tomography (CT) system (Trifoil Imaging, Northridge, CA). Animals were fasted overnight, and 100 μCi of 18F-FDG was injected into the tail vein. Micro-CT images for anatomical reference were first acquired (60 kV, 140 lA), and micro-PET acquisition was started 30 min after 18F-FDG administration. Static image acquisition was performed for 30 min per animal.
Electron microscopy
Cells were fixed in situ in 2% glutaraldehyde/4% PFA for 1 hour at RT and then gently scraped and pelleted at 900g. The cell pellet was processed for sectioning and electron microscopy by the Electron Microscopy Core Facility at the University of Chicago.
ImageStream flow cytometric analysis
Data collection and analysis of LD content and cell size were collected using the Amnis ImageStreamX Mk II and IDEAS Software (Luminex, version 6.3) and FCS Express (FCS Express, version 7.08.0018 4). Briefly, cells were fixed for 10 min at RT and then cells were stained with BODIPY 493/503 (Invitrogen, catalog no.D3922) at a 1:1000 dilution for 20 min at RT. Cells were washed three times with DPBS and then resuspended in DPBS before flow cytometry analysis. Acquisition data analysis was performed using IDEAS software using an 8-μm core with Speed Beads, which are 1-μm polystyrene beads that allow calibration of focus and flow. Fluorochrome was excited with a 488-nm laser, and single-focused cells were gated using Gradient RMS (focus). A total of 5000 events were collected for each sample. The cell size and BODIPY signal of individual cells were measured by generating an area plot and intensity plot, respectively. The measurements shown are given as means for different cell populations.
Extraction and LC-MS analysis of crude lipid extracts
Crude lipid extracts from cell pellets (100 μg of protein equivalent) were prepared via liquid-liquid extraction using the Folch method. SPLASH II LIPIDOMIX Mass Spec Standard (Avanti) was added as an internal standard before lipid extraction. The lipid extracts were dried and resuspended in 100 μl of methanol:chloroform (9:1, v/v) before the use of LC-MS analyzer (Agilent 6546 Q-ToF LC-MS system controlled by the Agilent Mass Hunter acquisition software). The mass spectrometer was operated in 2-GHz extended dynamic range in both negative and positive ion modes [negative ion mode, mass/charge ratio (m/z) 112.9855 and 980.0163; positive ion mode, m/z 121.0509 and 922.0098]. Mobile phase A was 90:10 H2O:methanol (v/v) with 10 mM ammonium acetate plus 0.5 mM ammonium fluoride, and B was (5:3:2) isopropanol:methanol:acetonitrile (v/v/v) with 10 mM ammonium acetate plus 0.5 mM ammonium fluoride. Then, the resuspended lipid extracts (2 μl) were loaded onto Agilent Poroshell C18, 2.7-μm column (Agilent Technologies Inc., Santa Clara, CA, USA), and separation was performed using the Agilent 1290 ultra-performance liquid chromatography (UPLC) system (gradient B: 70% at 0 to 1 min, 86% at 3.5 to 10 min, and 100% at 11 to 17 min at a flow rate of 400 μl/min) with the source parameters described here: gas temperature (200°C), drying gas (11 liters/min), nebulizer (35 psi), sheath gas temperature (350°C), sheath gas flow (12 liters/min), VCap (3000 V), and fragmentor (145 V). Data were collected, and relative quantification was performed using a scan speed of 4 mass spectrometry (MS) spectra per second.
Bioinformatic analysis
Expression data from dataset GSE84073 were downloaded from the Gene Expression Omnibus (GEO) database, and BioJupies web application (82) was used to perform bioinformatic analysis of BNIP3 expression and lipid-mitochondrial genes between healthy liver samples (GSM2653819 and GSM2653820) versus HCC samples (GSM2653823 and GSM2653824). Linear regression correlation analysis between BNIP3 and FASN, ACLY, PPARGC1A, and ACADM was performed using BioVinci (Bioturing, San Diego, CA, USA).
Liver hepatocellular carcinoma (LIHC) patient’s survival analysis was performed using cBioPortal (83, 84) and data from the TCGA PanCancer Atlas (n = 372 patients). LIHC patients were classified into different groups according to the BNIP3 and ACACA median expression value. By Kaplan-Meier method, we obtained the overall survival and comparison between curves was made using the log-rank test. Data obtained from cBioPortal database do not require ethical approval.
Statistics
All statistical analyses were carried out using GraphPad Prism of raw data. The data were analyzed using one-way or two-way analysis of variance (ANOVA) with Tukey’s posttest with a 95% confidence interval for datasets involving single parameters or single groups of data. Other datasets involving comparisons among multiple groups used Wilcoxon rank sum analyses with a 95% confidence interval. Data are shown as the mean ± SEM. Values of P ≤ 0.05 are considered significant. *P ≤ 0.05; **P ≤ 0.01; *** P ≤ 0.001; ****P ≤ 0.0001.
Study approval
All work was approved by the University of Chicago Institutional Animal Care and Use Committee under protocols 71155 and 72056.
Acknowledgments
We thank S. DeVries at the Northwestern University for supplying the anti-rat STAR 580 secondary antibody for STED imaging purposes; G. Kemble and others at 3-V Biosciences for supplying TVB-3664; L. Leoni and B. Roman for assistance in performing in vivo FDG-PET imaging; S. Cologna, M. Nguyen, and H. Chen at UIC for advice on analyzing lipidomics data; and C. Labno and V. Bindokas for advice on superresolution microscopy using STED.
Funding: This work was supported by funding from NIH R01 CA200310 (K.F.M.) and NIH T32 CA009594 (K.F.M. for A.B.-H.). This work made use of the Human Tissue Resource Center, the Integrated Small Animal Imaging Facility, the Advanced Electron Microscopy Facility, and the Integrated Light Microscope Facility that are supported by the University of Chicago Comprehensive Cancer Center Support Grant (P30 CA 014599).
Author contributions: Maintenance of mouse model: A.R.T., L.E.D., and G.B. Tumor analysis and characterization: A.R.T., L.E.D., and K.F.M. Cell line generation and characterization: D.E.B., L.E.D., and K.F.M. qPCR: D.E.B. and A.R.T. Western blots: D.E.B. and L.E.D. Fluorescence microscopy: A.B.-H. Seahorse analyses: D.E.B. Flow cytometry: D.E.B. IncuCyte growth assays: D.E.B. ELISA assays: A.B.-H. Electron microscopy: A.B.-H. and K.F.M. Human TCGA/bioinformatics analyses: D.E.B. Lipidomics analysis: A.R.T. and K.F.M. Study conceptualization: D.E.B., A.B.-H., and K.F.M. Manuscript preparation: D.E.B., A.B.-H., and K.F.M. Manuscript review: All authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
REFERENCES AND NOTES
- 1.Loomba R., Friedman S. L., Shulman G. I., Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell 184, 2537–2564 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang D. Q., El-Serag H. B., Loomba R., Global epidemiology of NAFLD-related HCC: Trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 18, 223–238 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calle E. E., Rodriguez C., Walker-Thurmond K., Thun M. J., Overweight, obesity and mortality from cancer in a prospectively studied cohort of U.S. adults. N. Engl. J. Med. 348, 1625–1638 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Larsson S. C., Wolk A., Overweight, obesity and risk of liver cancer: A meta-analysis of cohort studies. Br. J. Cancer 97, 1005–1008 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunt E. M., Nonalcoholic steatohepatitis. Semin. Liver Dis. 24, 3–20 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Begriche K., Igoudjil A., Pessayre D., Fromenty B., Mitochondrial dysfunction in NASH: Causes, consequences and possible means to prevent it. Mitochondrion 6, 1–28 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Mansouri A., Gattolliat C. H., Asselah T., Mitochondrial dysfunction and signaling in chronic liver diseases. Gastroenterology 155, 629–647 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Font-Burgada J., Sun B., Karin M., Obesity and cancer: The oil that feeds the flame. Cell Metab. 23, 48–62 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Macleod K. F., Mitophagy and mitochondrial dysfunction in cancer. Annu. Rev. Cancer Biol. 4, 41–60 (2020). [Google Scholar]
- 10.Lee J. M., Wagner M., Xiao R., Kim K. H., Feng D., Lazar M. A., Moore D. D., Nutrient-sensing nuclear receptors coordinate autophagy. Nature 516, 112–115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Springer M. Z., Poole L. P., Drake L. E., Bock-Hughes A., Boland M. L., Smith A. G., Hart J., Chourasia A. H., Liu I., Bozek G., Macleod K. F., BNIP3-dependent mitophagy promotes cytosolic localization of LC3B and metabolic homeostasis in the liver. Autophagy 17, 3530–3546 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verna L., Whysner J., Williams G. M., N-nitrosodiethylamine mechanistic data and risk assessment: Bioactivation, DNA-adduct formation, mutagenicity, and tumor initiation. Pharmacol. Ther. 71, 57–81 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Maeda S., Kamata H., Luo J. L., Leffert H., Karin M., IKKβ couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell 121, 977–990 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Broutier L., Mastrogiovanni G., Verstegen M. M. A., Francies H. E., Gavarró L. M., Bradshaw C. R., Allen G. E., Arnes-Benito R., Sidorova O., Gaspersz M. P., Georgakopoulos N., Koo B. K., Dietmann S., Davies S. E., Praseedom R. K., Lieshout R., IJzermans J. N. M., Wigmore S. J., Saeb-Parsy K., Garnett M. J., van der Laan L. J. W., Huch M., Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat. Med. 23, 1424–1435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yahagi N., Shimano H., Hasegawa K., Ohashi K., Matsuzaka T., Najima Y., Sekiya M., Tomita S., Okazaki H., Tamura Y., Iizuka Y., Ohashi K., Nagai R., Ishibashi S., Kadowaki T., Makuuchi M., Ohnishi S., Osuga J. I., Yamada N., Co-ordinate activation of lipogenic enzymes in hepatocellular carcinoma. Eur. J. Cancer 41, 1316–1322 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Yamashita T., Honda M., Takatori H., Nishino R., Minato H., Takamura H., Ohta T., Kaneko S., Activation of lipogenic pathway correlates with cell proliferation and poor prognosis in hepatocellular carcinoma. J. Hepatol. 50, 100–110 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Calvisi D. F., Wang C., Ho C., Ladu S., Lee S. A., Mattu S., Destefanis G., Delogu S., Zimmermann A., Ericsson J., Brozzetti S., Staniscia T., Chen X., Dombrowski F., Evert M., Increased lipogenesis, induced by Akt-mTORC1-RPS6 signaling, promotes development of human hepatocellular carcinoma. Gastroenterology 140, 1071–1083.e5 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Svensson R. U., Parker S. J., Eichner L. J., Kolar M. J., Wallace M., Brun S. N., Lombardo P. S., van Nostrand J. L., Hutchins A., Vera L., Gerken L., Greenwood J., Bhat S., Harriman G., Westlin W. F., Harwood H. J. Jr., Saghatelian A., Kapeller R., Metallo C. M., Shaw R. J., Inhibition of acetyl-CoA carboxylase suppresses fatty acid synthesis and tumor growth of non–small cell lung cancer in preclinical models. Nat. Med. 22, 1108–1119 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lally J. S. V., Ghoshal S., DePeralta D. K., Moaven O., Wei L., Masia R., Erstad D. J., Fujiwara N., Leong V., Houde V. P., Anagnostopoulos A. E., Wang A., Broadfield L. A., Ford R. J., Foster R. A., Bates J., Sun H., Wang T., Liu H., Ray A. S., Saha A. K., Greenwood J., Bhat S., Harriman G., Miao W., Rocnik J. L., Westlin W. F., Muti P., Tsakiridis T., James Harwood H. Jr., Kapeller R., Hoshida Y., Tanabe K. K., Steinberg G. R., Fuchs B. C., Inhibition of acetyl-CoA carboxylase by phosphorylation or the inhibitor ND-654 suppresses lipogenesis and hepatocellular carcinoma. Cell Metab. 29, 174–182.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitami T., Logan D. J., Negri J., Hasaka T., Tolliday N. J., Carpenter A. E., Spiegelman B. M., Mootha V. K., A chemical screen probing the relationship between mitochondrial content and cell size. PLOS ONE 7, e33755 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miettinen T. P., Björklund M., Cellular allometry of mitochondrial functionality establishes the optimal cell size. Dev. Cell 39, 370–382 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snaebjornsson M. T., Janaki-Raman S., Schulze A., Greasing the wheels of the cancer machine: The role of lipid metabolism in cancer. Cell Metab. 31, 62–76 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Benjamin D. I., Li D. S., Lowe W., Heuer T., Kemble G., Nomura D. K., Diacylglycerol metabolism and signaling is a driving force underlying FASN inhibitor sensitivity in cancer cells. ACS Chem. Biol. 10, 1616–1623 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimano H., Sato R., SREBP-regulated lipid metabolism: Convergent physiology-divergent pathophysiology. Nat. Rev. Endocrinol. 13, 710–730 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Zechner R., Madeo F., Kratky D., Cytosolic lipolysis and lipophagy: Two sides of the same coin. Nat. Rev. Mol. Cell Biol. 18, 671–684 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Olzmann J. A., Carvalho P., Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 20, 137–155 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A. M., Czaja M. J., Autophagy regulates lipid metabolism. Nature 458, 1131–1135 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schott M. B., Weller S. G., Schulze R. J., Krueger E. W., Drizyte-Miller K., Casey C. A., McNiven M. A., Lipid droplet size directs lipolysis and lipophagy catabolism in hepatocytes. J. Cell Biol. 218, 3320–3335 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schulze R. J., Krueger E. W., Weller S. G., Johnson K. M., Casey C. A., Schott M. B., McNiven M. A., Direct lysosome-based autophagy of lipid droplets in hepatocytes. Proc. Natl. Acad. Sci. U.S.A. 117, 32443–32452 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodman J. M., The importance of microlipophagy in liver. Proc. Natl. Acad. Sci. U.S.A. 118, e2024058118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freyre C. A. C., Rauher P. C., Ejsing C. S., Klemm R. W., MIGA2 Links mitochondria, the ER, and lipid droplets and promotes de novo lipogenesis in adipocytes. Mol. Cell 76, 811–825.e14 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Veliova M., Petcherski A., Liesa M., Shirihai O. S., The biology of lipid droplet-bound mitochondria. Semin. Cell Dev. Biol. 108, 55–64 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benador I. Y., Veliova M., Liesa M., Shirihai O. S., Mitochondria bound to lipid droplets: Where mitochondrial dynamics regulate lipid storage and utilization. Cell Metab. 29, 827–835 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pergande M. R., Serna-Perez F., Mohsin S. B., Hanek J., Cologna S. M., Lipidomic analysis reveals altered fatty acid metabolism in the liver of the symptomatic niemann-pick, type C1 mouse model. Proteomics 19, e1800285 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Baffy G., Brunt E. M., Caldwell S. H., Hepatocellular carcinoma in non-alcoholic fatty liver disease: An emerging menace. J. Hepatol. 56, 1384–1391 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Younossi Z. M., Stepanova M., Younossi Y., Golabi P., Mishra A., Rafiq N., Henry L., Epidemiology of chronic liver diseases in the USA in the past three decades. Gut 69, 564–568 (2020). [DOI] [PubMed] [Google Scholar]
- 37.McGlynn K. A., Petrick J. L., El-Serag H. B., Epidemiology of hepatocellular carcinoma. Hepatology 73( Suppl. 1), 4–13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X., Ramadori P., Pfister D., Seehawer M., Zender L., Heikenwalder M., The immunological and metabolic landscape in primary and metastatic liver cancer. Nat. Rev. Cancer 21, 541–557 (2021). [DOI] [PubMed] [Google Scholar]
- 39.Schafer Z. T., Grassian A. R., Song L., Jiang Z., Gerhart-Hines Z., Irie H. Y., Gao S., Puigserver P., Brugge J. S., Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature 461, 109–113 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zaugg K., Yao Y., Reilly P. T., Kannan K., Kiarash R., Mason J., Huang P., Sawyer S. K., Fuerth B., Faubert B., Kalliomäki T., Elia A., Luo X., Nadeem V., Bungard D., Yalavarthi S., Growney J. D., Wakeham A., Moolani Y., Silvester J., ten A. Y., Bakker W., Tsuchihara K., Berger S. L., Hill R. P., Jones R. G., Tsao M., Robinson M. O., Thompson C. B., Pan G., Mak T. W., Carnitine palmitoyltransferase 1C promotes cell survival and tumor growth under conditions of metabolic stress. Genes Dev. 25, 1041–1051 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Svensson R. U., Shaw R. J., Lipid synthesis is a metabolic liability of non–small cell lung cancer. Cold Spring Harb. Symp. Quant. Biol. 81, 93–103 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Filali-Mouncef Y., Hunter C., Roccio F., Zagkou S., Dupont N., Primard C., Proikas-Cezanne T., Reggiori F., The ménage à trois of autophagy, lipid droplets and liver disease. Autophagy 18, 50–72 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romeo S., Kozlitina J., Xing C., Pertsemlidis A., Cox D., Pennacchio L. A., Boerwinkle E., Cohen J. C., Hobbs H. H., Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 40, 1461–1465 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smagris E., BasuRay S., Li J., Huang Y., Lai K. M. V., Gromada J., Cohen J. C., Hobbs H. H., Pnpla3I148M knockin mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology 61, 108–118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y., Kory N., BasuRay S., Cohen J. C., Hobbs H. H., PNPLA3, CGI-58, and inhibition of hepatic triglyceride hydrolysis in mice. Hepatology 69, 2427–2441 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.BasuRay S., Wang Y., Smagris E., Cohen J. C., Hobbs H. H., Accumulation of PNPLA3 on lipid droplets is the basis of associated hepatic steatosis. Proc. Natl. Acad. Sci. U.S.A. 116, 9521–9526 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ng S. W. K., Rouhani F. J., Brunner S. F., Brzozowska N., Aitken S. J., Yang M., Abascal F., Moore L., Nikitopoulou E., Chappell L., Leongamornlert D., Ivovic A., Robinson P., Butler T., Sanders M. A., Williams N., Coorens T. H. H., Teague J., Raine K., Butler A. P., Hooks Y., Wilson B., Birtchnell N., Naylor H., Davies S. E., Stratton M. R., Martincorena I., Rahbari R., Frezza C., Hoare M., Campbell P. J., Convergent somatic mutations in metabolism genes in chronic liver disease. Nature 598, 473–478 (2021). [DOI] [PubMed] [Google Scholar]
- 48.Kaushik S., Cuervo A. M., Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat. Cell Biol. 17, 759–770 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mejhert N., Gabriel K. R., Frendo-Cumbo S., Krahmer N., Song J., Kuruvilla L., Chitraju C., Boland S., Jang D.-K., von Grotthuss M., Costanzo M. C., Rydén M., Olzmann J. A., Flannick J., Burtt N. P., Farese R. V. Jr., Walther T. C., The lipid droplet knowledge portal: A resource for systematic analyses of lipid droplet biology. Dev. Cell 57, 387–397.e4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schott M. B., Rozeveld C. N., Weller S. G., McNiven M. A., Lipophagy at a glance. J. Cell Sci. 135, jcs259402 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williams J. A., Ni H. M., Ding Y., Ding W. X., Parkin regulates mitophagy and mitochondrial function to protect against alcohol-induced liver injury and steatosis in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 309, G324–G340 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edmunds L. R., Xie B., Mills A. M., Huckestein B. R., Undamatla R., Murali A., Pangburn M. M., Martin J., Sipula I., Kaufman B. A., Scott I., Jurczak M. J., Liver-specific Prkn knockout mice are more susceptible to diet-induced hepatic steatosis and insulin resistance. Mol. Metab. 41, 101051 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davis O. B., Shin H. R., Lim C.-Y., Wu E. Y., Kukurugya M., Maher C. F., Perera R. M., Ordonez M. P., Zoncu R., NPC1-mTORC1 signaling couples cholesterol sensing to organelle homeostasis and is a targetable pathway in niemann-pick type C. Dev. Cell 56, 260–276.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shibata M., Yoshimura K., Furuya N., Koike M., Ueno T., Komatsu M., Arai H., Tanaka K., Kominami E., Uchiyama Y., The MAP1-LC3 conjugation system is involved in lipid droplet formation. Biochem. Biophys. Res. Commun. 382, 419–423 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Shibata M., Yoshimura K., Tamura H., Ueno T., Nishimura T., Inoue T., Sasaki M., Koike M., Arai H., Kominami E., Uchiyama Y., LC3, a microtubule-associated protein1A/B light chain3, is involved in cytoplasmic LD formation. Biochem. Biophys. Res. Commun. 393, 274–279 (2010). [DOI] [PubMed] [Google Scholar]
- 56.Martinez-Lopez N., Garcia-Macia M., Sahu S., Athonvarangkul D., Liebling E., Merlo P., Cecconi F., Schwartz G. J., Singh R., Autophagy in the CNS and periphery coordinate lipophagy and lipolysis in the brown adipose tissue and liver. Cell Metab. 23, 113–127 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garcia-Macia M., Santos-Ledo A., Leslie J., Paish H. L., Collins A. L., Scott R. S., Watson A., Burgoyne R. A., White S., French J., Hammond J., Borthwick L. A., Mann J., Bolaños J. P., Korolchuk V. I., Oakley F., Mann D. A., A mammalian target of rapamycin-perilipin 3 (mTORC1-Plin3) pathway is essential to activate lipophagy and protects against hepatosteatosis. Hepatology 74, 3441–3459 (2021). [DOI] [PubMed] [Google Scholar]
- 58.Schroeder B., Schulze R. J., Weller S. G., Sletten A. C., Casey C. A., McNiven M. A., The small GTPase Rab7 as a central regulator of hepatocellular lipophagy. Hepatology 61, 1896–1907 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Z., Schulze R. J., Weller S. G., Krueger E. W., Schott M. B., Zhang X., Casey C. A., Liu J., Stöckli J., James D. E., McNiven M. A., A novel Rab10-EHBP1-EHD2 complex essential for the autophagic engulfment of lipid droplets. Sci. Adv. 2, e1601470 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamano K., Fogel A. I., Wang C., van der Bliek A. M., Youle R. J., Mitochondrial Rab GAPs govern autophagosome biogenesis during mitophagy. eLife 3, e01612 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hammerling B. C., Najor R. H., Cortez M. Q., Shires S. E., Leon L. J., Gonzalez E. R., Boassa D., Phan S., Thor A., Jimenez R. E., Li H., Kitsis R. N., Dorn G. W. 2nd, Sadoshima J., Ellisman M. H., Gustafsson Å. B., A Rab5 endosomal pathway mediates Parkin-dependent mitochondrial clearance. Nat. Commun. 8, 14050 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamano K., Wang C., Sarraf S. A., Münch C., Kikuchi R., Noda N. N., Hizukuri Y., Kanemaki M. T., Harper W., Tanaka K., Matsuda N., Youle R. J., Endosomal Rab cycles regulate Parkin-mediated mitophagy. eLife 7, e31326 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wong Y. C., Ysselstein D., Krainc D., Mitochondria-lysosome contacts regulate mitochondrial fission via RAB7 GTP hydrolysis. Nature 554, 382–386 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hammerling B. C., Shires S. E., Leon L. J., Cortez M. Q., Gustafsson Å. B., Isolation of Rab5-positive endosomes reveals a new mitochondrial degradation pathway utilized by BNIP3 and Parkin. Small GTPases 11, 69–76 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang H., Sreenivasan U., Hu H., Saladino A., Polster B. M., Lund L. M., Gong D. W., Stanley W. C., Sztalryd C., Perilipin 5, a lipid droplet-associated protein, provides physical and metabolic linkage to mitochondria. J. Lipid Res. 52, 2159–2168 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Benador I. Y., Veliova M., Mahdaviani K., Petcherski A., Wikstrom J. D., Assali E. A., Acín-Pérez R., Shum M., Oliveira M. F., Cinti S., Sztalryd C., Barshop W. D., Wohlschlegel J. A., Corkey B. E., Liesa M., Shirihai O. S., Mitochondria bound to lipid droplets have unique bioenergetics, composition, and dynamics that support lipid droplet expansion. Cell Metab. 27, 869–885.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Glick D., Zhang W., Beaton M., Marsboom G., Gruber M., Simon M. C., Hart J., Dorn G. W. II, Brady M. J., Macleod K. F., BNip3 regulates mitochondrial function and lipid metabolism in the liver. Mol. Cell. Biol. 32, 2570–2584 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin H. V., Accili D., Hormonal regulation of hepatic glucose production in health and disease. Cell Metab. 14, 9–19 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilfling F., Wang H., Haas J. T., Krahmer N., Gould T. J., Uchida A., Cheng J. X., Graham M., Christiano R., Fröhlich F., Liu X., Buhman K. K., Coleman R. A., Bewersdorf J., Farese R. V. Jr., Walther T. C., Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell 24, 384–399 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nguyen T. B., Louie S. M., Daniele J. R., Tran Q., Dillin A., Zoncu R., Nomura D. K., Olzmann J. A., DGAT1-dependent LD biogenesis protects mitochondrial function during starvation-induced autophagy. Dev. Cell 42, 9–21.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Anastasia I., Ilacqua N., Raimondi A., Lemieux P., Ghandehari-Alavijeh R., Faure G., Mekhedov S. L., Williams K. J., Caicci F., Valle G., Giacomello M., Quiroga A. D., Lehner R., Miksis M. J., Toth K., de Aguiar Vallim T. Q., Koonin E. V., Scorrano L., Pellegrini L., Mitochondria-rough-ER contacts in the liver regulate systemic lipid homeostasis. Cell Rep. 34, 108873 (2021). [DOI] [PubMed] [Google Scholar]
- 72.Killackey S. A., Bi Y., Soares F., Hammi I., Winsor N. J., Abdul-Sater A. A., Philpott D. J., Arnoult D., Girardin S. E., Mitochondrial protein import stress regulates the LC3 lipidation step of mitophagy through NLRX1 and RRBP1. Mol. Cell 82, 2815–2831.e5 (2022). [DOI] [PubMed] [Google Scholar]
- 73.Gomes L. C., Di Benedetto G., Scorrano L., During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 13, 589–598 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rambold A. S., Kostelecky B., Elia N., Lippincott-Schwartz J., Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl. Acad. Sci. U.S.A. 108, 10190–10195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee Y. K., Lee H.-Y., Hanna R. A., Gustafsson A. B., Mitochondrial autophagy by Bnip3 involves Drp1-mediated mitochondrial fission and recruitment of Parkin in cardiac myocytes. Am. J. Physiol. Heart Circ. Physiol. 301, H1924–H1931 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Serasinghe M. N., Wieder S. Y., Renault T. T., Elkholi R., Asciolla J. J., Yao J. L., Jabado O., Hoehn K., Kageyama Y., Sesaki H., Chipuk J. E., Mitochondrial division is requisite to RAS-induced transformation and targeted by oncogenic MAPK pathway inhibitors. Mol. Cell 57, 521–536 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kashatus J. A., Nascimento A., Myers L. J., Sher A., Byrne F. L., Hoehn K. L., Counter C. M., Kashatus D. F., Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol. Cell 57, 537–551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nagdas S., Kashatus J. A., Nascimento A., Hussain S. S., Trainor R. E., Pollock S. R., Adair S. J., Michaels A. D., Sesaki H., Stelow E. B., Bauer T. W., Kashatus D. F., Drp1 promotes KRas-driven metabolic changes to drive pancreatic tumor growth. Cell Rep. 28, 1845–1859.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rossignol R., Gilkerson R., Aggeler R., Yamagata K., Remington S. J., Capaldi R. A., Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 64, 985–993 (2004). [DOI] [PubMed] [Google Scholar]
- 80.Rambold A. S., Cohen S., Lippincott-Schwartz J., Fatty acid trafficking in starved cells: Regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev. Cell 32, 678–692 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mizushima N., Levine B., Autophagy in mammalian development and differentiation. Nat. Cell Biol. 12, 823–830 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Torre D., Lachmann A., Ma’ayan A., BioJupies: Automated generation of interactive notebooks for RNA-Seq data analysis in the cloud. Cell Syst. 7, 556–561.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., Jacobsen A., Byrne C. J., Heuer M. L., Larsson E., Antipin Y., Reva B., Goldberg A. P., Sander C., Schultz N., The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gao J., Aksoy B. A., Dogrusoz U., Dresdner G., Gross B., Sumer S. O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., Schultz N., Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10