Abstract
Expression of the Staphylococcus aureus plasmid-encoded QacA multidrug transporter is regulated by the divergently encoded QacR repressor protein. To circumvent the formation of disulfide-bonded degradation products, site-directed mutagenesis to replace the two cysteine residues in wild-type QacR was undertaken. Analysis of a resultant cysteineless QacR derivative indicated that it retained full DNA-binding activities in vivo and in vitro and continued to be fully proficient for the mediation of induction of qacA expression in response to a range of structurally dissimilar multidrug transporter substrates. The cysteineless QacR protein was used in cross-linking and dynamic light-scattering experiments to show that its native form was a dimer, whereas gel filtration indicated that four QacR molecules bound per DNA operator site. The addition of inducing compounds led to the dissociation of the four operator-bound QacR molecules from the DNA as dimers. Binding of QacR dimers to DNA was found to be dependent on the correct spacing of the operator half-sites. A revised model proposed for the regulation of qacA expression by QacR features the unusual characteristic of one dimer of the regulatory protein binding to each operator half-site by a process that does not appear to require the prior self-assembly of QacR into tetramers.
Multidrug efflux transporters are membrane proteins found in both prokaryotes and eukaryotes that confer resistance to a wide range of structurally unrelated cytotoxic compounds, typically hydrophobic cations. The mechanisms used by these transporters to bind and export such a broad range of substrates remain unknown, largely due to the difficulties posed by the structural analysis of integral membrane proteins. In the case of bacteria, a fruitful alternative approach has been the study of cytosolic multidrug-binding proteins that regulate the expression of specific multidrug transporters at the local level. Examples of both activators (1, 2) and repressors (7, 16, 17) of transcription have been described and can be typically found encoded adjacent to the gene encoding the membrane pump. Structural analysis of BmrR, a dimeric Bacillus subtilis transcriptional activator that binds a range of ligands similar to its cognate transporter, Bmr (18, 19, 35), revealed a crucial negatively charged residue buried at the base of an internal drug-binding pocket lined with hydrophobic residues (38, 39).
Although such an arrangement represents an ideal solution to the problem of binding structurally diverse, hydrophobic, cationic ligands, it remains to be seen if different regulatory proteins, and the efflux pumps themselves, employ a similar, functionally analogous mode of multidrug binding. However, it is somewhat surprising that the multidrug-binding domains of these bacterial regulatory proteins show no apparent homology even though many of them bind a number of common ligands. Thus, further analysis of distinct multidrug-binding proteins is required in order to dissect whether they employ any common themes in their interactions with ligands or instead possess discrete substrate-binding mechanisms.
For the important human pathogen Staphylococcus aureus, a number of plasmid-encoded multidrug resistance transporters have been described, including the closely related major facilitator superfamily members QacA and QacB and the small multidrug resistance protein Smr (15, 22). Expression of both qacA and qacB is regulated by a divergently encoded transcriptional repressor, QacR, a member of the TetR family of repressors (25). IR1, a large inverted repeat located immediately adjacent to and downstream from the qacA and qacB promoters, has been shown to be the site of QacR binding (7). IR1 is unusually large for an operator sequence bound by a TetR family regulator, comprising 15-bp half-sites separated by a 6-bp spacer region (7). In contrast, the DNA-bound structure of TetR indicated that a dimer of TetR, or a similar protein such as QacR, would be unlikely to span a 6-bp spacer (20). Furthermore, TetR binding is prevented by a 1-bp increase or decrease in the single-base-pair spacing between the two tet operator half-sites (37).
Addition of structurally diverse QacA substrates from a wide range of chemical classes, including the compounds benzalkonium, dequalinium, ethidium, proflavine, and rhodamine 6G, has been demonstrated to result in both derepression of the qacA promoter in vivo and dissociation of QacR from qacA promoter-operator DNA in vitro (7). In order to accommodate this range of ligands, it has been proposed that a QacR binding pocket would have to differ substantially from that of the BmrR multidrug-binding protein (13). However, further biochemical and structural investigations of the DNA- and ligand-binding properties of QacR have been seriously hampered by rapid postpurification formation of nonnative disulfide-bonded monomers and oligomeric aggregates (7).
In this paper, the significance of the two cysteine residues within QacR, C72 and C141, to DNA and ligand binding is examined, with a view to generating a fully functional cysteineless QacR derivative for further in vitro studies. Analysis of a cysteineless QacR derivative resulted in the demonstration of an intriguing oligomerization state for DNA-bound QacR, which was found to be dependent on correctly spaced operator half-sites.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli strain DH5α (26), used as the host for all the procedures described throughout this work, was cultured at 37°C in Luria-Bertani (LB) medium containing ampicillin (100 μg ml−1) to select for plasmids. The plasmid pSK5203, containing the qacA promoter (PqacA) fused to a chloramphenicol acetyltransferase (cat) reporter gene, and the related plasmid pSK5212, in which qacR is present in cis to the PqacA-cat fusion, have been described previously (7), as has pSK5210, a clone of wild-type qacR in the expression vector pTTQ18 (32). Site-directed mutagenesis reactions were done with either single-stranded template DNA and a single primer (10) or the QuickChange kit (Stratagene), employing a pair of complementary oligonucleotides and a double-stranded DNA template.
Three restriction endonuclease cleavage sites that do not occur in most commonly used plasmid vectors were inserted into a qacR-encoding DNA fragment by site-directed mutagenesis to facilitate future manipulations. A BglII site was first inserted into pSK5212 between the promoter and ribosome-binding site of qacR, at position 692 of the originally described qacA-qacR sequence (25), to create pSK5213. This plasmid was used as the template for further site-directed mutagenesis, inserting an MluI site at position 293 and a BsrGI site at position 425 to generate pSK5618. Both these changes were within the coding region of qacR but did not result in the alteration of any QacR amino acids.
Mutagenesis of the cysteine residue at position 72 (C72) in QacR was performed by replacing the BglII-BsrGI fragment of pSK5618 with a fragment generated by PCR, using the primer 5′-CATTGTACAAATAAAATTTTTCTCTATTAGTTTTAGNTTTGATTTGTTCC-3′, where N is A, C, G, or T and the BsrGI site is in italics, with the second primer being the same as that used originally to insert the BglII site. pSK5618 was also used as the template for the site-directed mutagenesis of the C141 residue of QacR, using the primer pair 5′ - T TAAATGGCGAATGGNC TAT TATGACG TCAATGC TG T TAG TAAA - 3′ and 5′-TTTACTAACAGCATTGACGTCATTAATAATAGNCCATTCGCCATTTAA-3′, with the AatII site introduced by these primers in italics. Plasmids encoding cysteineless QacR derivatives were produced by employing the unique PstI-MluI sites in these plasmids (Fig. 1) to replace the PstI-MluI fragment of the C72A single mutant with the corresponding fragments from the C141S- and C141A-encoding plasmids, generating pSK5637 (Fig. 1) and pSK5638, respectively. For overexpression of the C72A/C141S double mutant, the cysteineless QacR-encoding fragment from pSK5637 was recloned in the expression vector pTTQ18, as described previously for wild-type QacR (7), yielding the construct pSK5676.
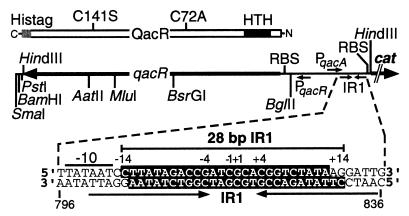
FIG. 1.
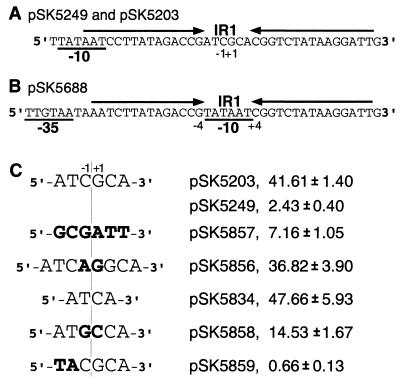
The cysteineless QacR derivative encoded by pSK5637 and relevant details of the cloned HindIII fragment in this plasmid. The QacR polypeptide encoded by pSK5637 is represented as a rectangular box showing the positions of the amino-terminal (N) HTH DNA-binding motif, the carboxy-terminal (C) hexahistidine tag (Histag), and the two mutations at cysteine residues 72 and 141 that have been introduced into this QacR derivative. Below this is a linear map of the cloned qacR-encoding fragment in plasmid pSK5637, depicting the locations of the qacR and qacA ribosome-binding sites (RBS), the qacR promoter, PqacR, and the divergent qacA promoter, PqacA, which in this construct is fused to a cat reporter gene. Also shown are the AatII, MluI, BsrGI, and BglII restriction endonuclease sites introduced during the course of this work and the position of the QacR binding site, IR1. The sequence of the PqacA −10 hexamer (overlined) is given, downstream from which the highlighted IR1 bases represent nucleotides protected in DNase I footprinting experiments (7). The sequence of the 28-bp IR1 annealed oligonucleotide duplex used throughout this work corresponds to positions −14 to +14 of the IR1 sequence. The nucleotide numbering is the same as for the originally published qacA-qacR sequence (25).
For mutagenesis of nucleotides within IR1, plasmid pSK5249 was constructed, which was essentially identical to pSK5213 except for the possession of a KpnI site located between the IR1 sequence and HindIII site. Mutations were then produced in the 6-bp spacer region separating the two IR1 half-sites by using PCR-generated fragments amplified from pSK5249 to replace the PstI-KpnI fragment of this plasmid. The primers used to achieve this were 5′-GAATTCCCGGGGATCCGTCGACCTG-3′ and 5′-TTGGGTACCAATCCTTATAGACCGXCGGTCTATAAGGATTATAATC-3′, where X is ATCA (pSK5834), ATCAGGCA (pSK5856), GCGATT (pSK5857), ATGCCA (pSK5858), or TACGCA (pSK5859). An initial mutant, pSK5688, in which the qacA promoter had been repositioned so that its −10 region, TATAAT, entirely replaced the 6-bp spacer region, was also constructed by PCR in a similar fashion.
CAT assays.
Chloramphenicol acetyltransferase (CAT) assays to ascertain the level of transcription from PqacA were performed as described previously (7), except that the level of CAT activity was adjusted for the protein concentration in the cell lysates, determined using the Coomassie protein assay reagent kit (Pierce).
Protein purification and stability.
Overexpression and purification of wild-type (encoded by pSK5210) and the C72A/C141S cysteineless QacR derivative (encoded by pSK5676) were carried out with a C-terminal His tag as described previously (7), with the following minor modifications. All the buffers were pH 7.5 and contained 20 mM 2-mercaptoethanol and 20 mM Tris-HCl or, in the case of the sonication buffer, 40 mM Tris-HCl. Exchange of proteins into alternative buffers was achieved by passage through Sephadex G50 (Amersham Pharmacia Biotech) columns previously equilibrated with the new buffer.
The in vitro stability of QacR was assessed by incubation of the indicated amounts of purified protein in 20 mM Tris-HCl (pH 7.5) in a final volume of 40 μl for 4 h at 22°C. This was followed by the addition of 0.5 volume of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer containing 30 mM EDTA, and for selected reactions dithiothreitol (DTT) was added to a final concentration of 33 mM, before heating of the samples at 100°C for 4 min and separation by SDS–15% PAGE (11), followed by silver staining.
Gel mobility shift assays were performed as described previously (7). As an indicator of the in vivo stability of QacR mutants, whole-cell lysates prepared from overnight cultures harboring QacR-encoding plasmids were separated by SDS–15% PAGE before being transferred to a Hybond-C nitrocellulose membrane (Amersham Pharmacia Biotech). QacR was then detected immunologically with an anti-His (C terminus) immunoglobulin G monoclonal antibody (Invitrogen) at a dilution of 1:250, but otherwise as described in the protocol supplied by the manufacturer.
Gel filtration.
A fast protein liquid chromatography Superose 12 HR 10/30 column (Amersham Pharmacia Biotech) with a mobile phase of 300 mM NaCl, 5% (vol/vol) glycerol, and 20 mM Tris-HCl, pH 7.5, was used in all gel filtration experiments. Blue dextran (Sigma) was used to determine the column void volume, and proteins for use as gel filtration molecular mass standards were purchased from Sigma (carbonic anhydrase and β-amylase) and Bio-Rad (myoglobin, ovalbumin, and gamma globulin). The molecular weights of the experimental samples were determined following the protocols supplied by the manufacturers. Gel filtration was also used to separate QacR from high-molecular-mass nonspecific protein aggregates before the resultant fractions were stored in dilute single-use aliquots at −70°C at a concentration of approximately 10 μg ml−1 for use in cross-linking reactions. Subsequent chromatography failed to detect the formation of any further aggregates following storage of these low-concentration samples.
Protein cross-linking.
Cross-linking reactions were performed in 1× cross-linking buffer (100 mM KCl, 15 mM Tris-HCl, pH 7.5) in a total volume of 40 μl, containing the specified amounts of C72A/C141S QacR and, where indicated, annealed 28-bp gel-purified IR1 oligonucleotide duplexes (Fig. 1). Reactions were incubated at 22°C for 15 min before the addition of gluteraldehyde to a final concentration of 0.01%, followed by a 1.5-min incubation to cross-link any QacR complexes. Alternatively, formaldehyde was used as the cross-linking agent at a final concentration of 1%, with a 10-min incubation at 22°C. Cross-linking reactions were stopped by the addition of 20 μl of 2× SDS gel loading buffer, and the samples were heated at 95°C for 3 min before separation by SDS–12.5% PAGE and transfer to a Hybond-C membrane. QacR was detected immunologically using a 1:1,500 dilution of a rabbit polyclonal antibody raised against the purified C72A/C141S QacR derivative at the Institute of Medical and Veterinary Science, South Australia. The resultant serum was used in standard Western blotting procedures with Blotto as the blocking agent (26).
DLS.
The concentration of apo-QacR used in dynamic light-scattering (DLS) experiments was 10 μM (monomer concentration) in a solution of 300 mM NaCl, 1 M imidazole, 5% glycerol, and 50 mM Tris, pH 7.5. When either the 28-bp IR1 or a 33-bp noncognate site was included, the QacR and DNA concentrations were both 40 μM (monomer concentration), and the measurements were conducted in solutions of 50 mM NaCl and 20 mM Tris, pH 7.5. Each experiment used 100-μl samples, which were microfiltered using a 0.02-μm-diameter filter. DLS was measured using a DynaPro 801 dynamic light scattering instrument (Protein Solutions). Data were adjusted for glycerol content and analyzed using the instrument control software package Dynamics 4.0 provided by the manufacturer. Up to 40 readings were recorded during each DLS experiment. Readings in which the baseline error of the data was less than 1.005 and the sum of squares error was below 5.000 (indicative of a monodisperse solution) were fit with a monomodal analysis; all other data were fit using a bimodal analysis.
RESULTS
Site-directed mutagenesis of QacR cysteine residues.
The formation of disulfide bonds during the purification and storage of proteins results from the oxidation of free thiol groups, a process catalyzed by divalent metal cations (36), as has been demonstrated for QacR (7). Relatively high levels of reducing agents and the addition of EDTA only served to partially slow the formation of the disulfide-bonded forms of QacR (7). Therefore, site-directed mutagenesis was used to alter the QacR cysteine codons, producing eight QacR single-amino-acid substitution mutants, with C72 and C141 changed separately to alanine (A), serine (S), threonine (T), or proline (P).
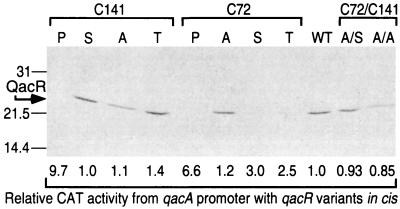
The in vivo stability of the mutant QacR proteins was then assessed by Western analysis using a C-terminal His tag-specific monoclonal antibody. Not surprisingly, both the radical replacements of cysteine with proline resulted in highly unstable proteins; no band corresponding to the 23-kDa QacR protein could be detected immunologically (Fig. 2). An unanticipated observation was the very unstable nature in vivo of mutants in which the C72 residue was replaced with either serine or threonine (Fig. 2), both representing changes that are normally considered relatively conservative substitutions. All eight QacR derivatives were also analyzed for their in vivo DNA-binding abilities. The degree to which each mutant continued to suppress transcription from PqacA closely correlated to its intracellular stability (Fig. 2). The nearly wild-type levels of repression observed for the QacR C72A and C141S derivatives clearly indicated that neither of the two cysteine residues plays a significant role in functions related to DNA binding.
FIG. 2.
In vivo stability and DNA-binding abilities of QacR variants. The cysteine residues (C141 and C72) in wild-type (WT) QacR were individually replaced with proline (P), serine (S), alanine (A), or threonine (T), in addition to the production of two cysteineless QacR derivatives, C72A/C141S and C72A/C141A. Equal amounts of whole-cell lysates, as judged by Coomassie staining, were separated by SDS–15% PAGE before being transferred to nitrocellulose, and a Western blot was performed with a C-terminal His tag-specific primary antibody to determine the in vivo stability of the QacR derivatives. The position of migration of QacR (23 kDa) relative to molecular size standards is indicated on the left-hand side. The ability of the QacR mutants to continue to bind IR1 in vivo and hence repress transcription from PqacA was assessed by CAT assays with the divergent fusion of PqacA to a cat reporter gene that is available in the plasmids encoding these proteins (Fig. 1). CAT activity is reported relative to the level of expression from PqacA in the presence of wild-type QacR, which was set at 1.
Cysteineless QacR derivative is fully functional.
CAT assays were used to determine the in vivo inducibility of the most promising cysteine substitutions in response to the presence of a monovalent or bivalent inducing compound, ethidium or dequalinium, respectively. Following the addition of these compounds, wild-type levels of induction from PqacA were observed in the presence of the divergently encoded C72A, C141A, or C141S mutant QacR proteins (data not shown). These results indicated that the cysteine residues in QacR could be individually replaced by these alternative amino acids without adversely affecting the ability of the protein to bind structurally diverse ligands. Therefore, two cysteineless QacR mutants, with C72 changed to alanine and C141 changed to alanine (pSK5638) or serine (pSK5637), were produced. For the C72A/C141S double mutant, both the in vivo protein stability (Fig. 2) and the amount of transcription it permitted from PqacA, as determined by CAT assays (0.93-fold the activity found for wild-type QacR; Fig. 2), were essentially indistinguishable from that of QacR containing two cysteine residues. However, even though the QacR C72A/C141A double mutant was slightly less stable in vivo, it exhibited enhanced repression of PqacA (0.85-fold) (Fig. 2).
Both of the QacR cysteineless mutants continued to be completely proficient for the induction of expression from PqacA in response to the presence of a wide range of monovalent and bivalent inducing compounds from a number of chemical classes (Table 1). Thus, since the C72A/C141S derivative exhibited wild-type properties for all the attributes tested, it was chosen as the fully functional cysteineless QacR variant to be the subject of further analysis. Subsequent to its recloning in the expression vector pTTQ18, the QacR C72A/C141S derivative was overexpressed and purified in tandem with wild-type QacR protein. Gel mobility shift assays demonstrated that the purified cysteineless QacR protein bound to IR1-containing DNA in vitro with equal or greater affinity than the wild-type protein (data not shown). Purified C72A/C141S QacR was also used to raise a rabbit polyclonal antibody, which proved to have significantly improved sensitivity for the immunological detection of QacR in Western blot analysis in comparison to commercially available His tag-specific monoclonal antibodies (data not shown).
TABLE 1.
Induction of wild-type and cysteineless QacR derivatives
| Inducing compound | Concn (μg/ml) | Relative CAT activitya
|
||
|---|---|---|---|---|
| Wild type | C72A/ C141S | C72A/ C141A | ||
| Benzalkonium | 10 | 7.5 | 7.0 | 7.0 |
| Dequaliniumb | 10 | 12.9 | 14.1 | 16.8 |
| Ethidium | 30 | 1.4 | 1.5 | 1.4 |
| Proflavine | 10 | 3.0 | 4.5 | 3.4 |
| Rhodamine 6G | 100 | 6.7 | 6.4 | 5.3 |
The amount of CAT activity detected from PqacA for each of the constructs in the absence of any inducing compound was set at 1.0. Expression from PqacA fused to a cat reporter gene with qacR in cis, encoding either wild-type QacR or a cysteineless derivative, was assayed as described previously (7).
Bivalent inducing compound.
Purified cysteineless QacR has greatly enhanced in vitro stability.
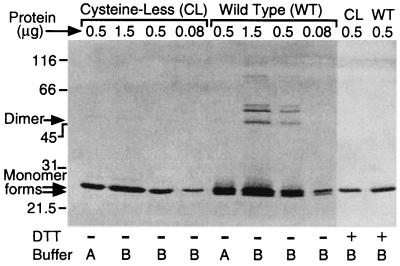
Examination by SDS-PAGE of purified C72A/C141S and wild-type QacR proteins after a 4-h incubation at room temperature indicated that the cysteineless derivative exhibited greatly enhanced in vitro stability compared to the wild-type protein (Fig. 3). The C72A/C141S QacR protein lacked the many oligomeric forms that were observed for wild-type QacR in the absence of reducing agents (Fig. 3).
FIG. 3.
In vitro stability of purified wild-type and cysteineless QacR. Following the purification of wild-type (WT) QacR or the cysteineless (CL) C72A/C141S derivative, the indicated amounts of purified protein were incubated for 4 h at 22°C before the samples were heated without (−) or with (+) 33 mM DTT. After separation by SDS–15% PAGE, the proteins were visualized by silver staining. Samples labeled A were in elution buffer containing 20 mM 2-mercaptoethanol, whereas samples labeled B had first been exchanged into a buffer devoid of reducing agents. Arrows indicate the migration positions of the two monomeric forms of QacR that were detected and also the position of the small amounts of a QacR species equivalent in size to a dimer that was detected in the first two lanes for the cysteineless derivative. The positions of migration of size standards (in kilodaltons) are shown on the right-hand side.
As described previously (7), the large number of multimeric forms presumably consisted of different combinations of inter- and intramolecular disulfide bonds (Fig. 3). These multimers have also been demonstrated to form, albeit more slowly, in the presence of reducing agents (7). Importantly, in contrast to the substantial amounts of wild-type QacR that existed as a faster-migrating disulfide-bonded monomeric form, even in the presence of 20 mM 2-mercaptoethanol, the cysteineless QacR protein preparations completely lacked this species (Fig. 3). However, silver staining did detect very small amounts of purified cysteineless QacR migrating at a position equivalent to that expected for a QacR dimer (46 kDa; Fig. 3, first two lanes), suggesting that QacR exists as dimers in solution, which can subsequently become covalently linked by degradation pathways other than the formation of disulfide bonds. This supported the proposition that the active form of QacR would most likely be a dimer, as for other TetR family members.
Detection of QacR dimers in solution.
Passage of the C72A/C141S QacR protein through a gel filtration column produced two peaks, one consistent with a monomeric form of QacR, while the second, smaller peak eluted close to the void volume of the column, indicating highly aggregated protein. Attempts to concentrate QacR beyond 5 mg ml−1, even in the presence of 300 mM NaCl, resulted in the protein precipitating out of solution. However, by addition of IR1 DNA, the solubility of purified cysteineless QacR could be markedly increased, similar to the prevention of aggregate formation that has been observed for the BmrR multidrug-binding regulatory protein (18).
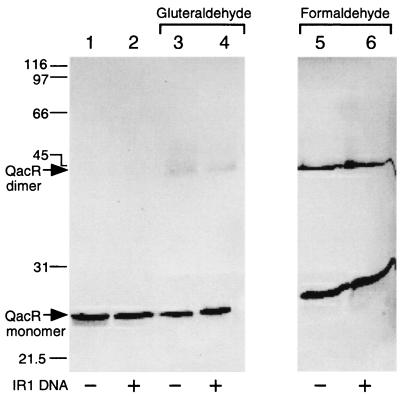
To identify the active oligomeric form of QacR, cross-linking experiments were carried out with dilute cysteineless QacR that had first been separated from any aggregated protein by gel filtration. After treatment with the cross-linking reagent gluteraldehyde or formaldehyde, separation by SDS-PAGE, and subsequent immobilization on a nitrocellulose membrane, monomeric and any covalently joined oligomeric forms of QacR were detected immunologically using the polyclonal QacR-specific antibody. Treatment with either reagent resulted in the detection of a covalently linked QacR complex of a size close to that expected for dimeric QacR, 46 kDa (Fig. 4). Formation of cross-linked QacR dimers was not significantly enhanced by the presence of IR1 DNA despite its marked effect on the solubility of the protein, nor did the addition of IR1 DNA promote the formation of any other higher-order oligomeric forms (Fig. 4). The inclusion of compounds that are inducers of qacA expression in cross-linking reactions also failed to have an impact on the outcome of these experiments (data not shown).
FIG. 4.
Detection of cross-linked QacR dimers. Purified C72A/C141S QacR protein, 80 ng (lanes 1 to 4) or 160 ng (lanes 5 and 6), was preincubated for 15 min with no (−) DNA (lanes 1, 3, and 5) or with (+) 15 ng of 28-bp IR1 DNA (lanes 2 and 4) or 30 ng of 28-bp IR1 DNA (lane 6). Samples in lanes 3 and 4 were cross-linked with 0.01% gluteraldehyde for 1.5 min and those in lanes 5 and 6 were cross-linked with 1% formaldehyde for 10 min before the reactions were stopped by the addition of one-half volume of 2× SDS-PAGE loading buffer, heated at 95°C for 3 min, and separated by SDS–12.5% PAGE. After transfer to nitrocellulose, proteins were detected with an anti-QacR polyclonal antibody. On the left-hand side, the positions of migration of monomeric and dimeric forms of QacR and molecular size markers (in kilodaltons) are indicated.
Two QacR dimers bind per IR1 operator site.
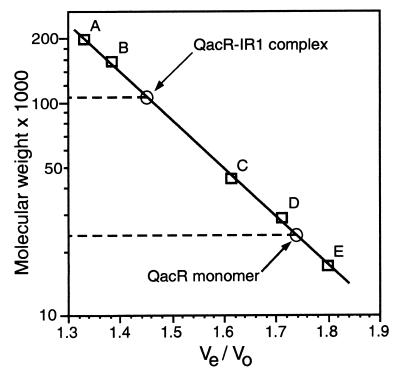
In contrast to the cross-linking results presented above, gel filtration indicated that four QacR molecules bound each IR1 DNA site. In three independent gel filtration experiments, using the C72A/C141S QacR derivative preincubated with molar ratios of between 0.25 and 0.5 of the purified, complementary, annealed 28-mer IR1 oligonucleotides (Fig. 1), an average molecular mass of 105.7 ± 3.1 kDa for a QacR-DNA complex was obtained. This value is in good agreement with the theoretical mass of 109.3 kDa for four QacR molecules bound to the 28-bp IR1 DNA fragment. The results of a representative gel filtration experiment are depicted in Fig. 5.
FIG. 5.
Gel filtration experiment demonstrating that four QacR molecules are bound per operator site. The x axis values (Ve/V0) were calculated by dividing the elution volume (Ve) of each standard or experimental sample by the void volume of the column (V0). Standards used were: A, β-amylase (Mr 200,000); B, gamma globulin (Mr 158,000); C, ovalbumin (Mr 44,000); D, carbonic anhydrase (Mr 29,000); and E, myoglobin (Mr 17,000). The experimentally determined values for purified C72A/C141S QacR and the complex it forms with the 28-bp IR1 DNA fragment are indicated.
The oligomerization state of QacR was also examined by DLS measurements in the absence of both DNA and drug (apo-QacR), the presence of the high-affinity IR1 DNA-binding site, the presence of a 33-bp noncognate DNA sequence, and the presence of known inducing compounds. DLS measurements of a 10 μM solution of apo-QacR yielded a molecular mass of 43 ± 5 kDa as fit by a bimodal analysis, which showed 98% of the species with this molecular mass and the remaining 2% as an aggregate with a mass of 980 kDa. The former molecular mass is consistent with a QacR dimer. In the presence of the 28-bp IR1 site, the QacR-DNA solution was completely monodisperse, with no indication of other molecular mass species. The single molecular mass species observed was 120 ± 3 kDa (the average of three independent experiments in which the individual molecular masses were 120, 124, and 116 kDa). This molecular mass describes a macromolecular complex in which a tetramer of QacR is bound to a duplex IR1 site.
Also investigated was the oligomerization state of QacR following the addition of the QacA monovalent substrate proflavine or the bivalent substrate dequalinium, compounds that have been shown to both induce expression of qacA in vivo and disassociate QacR from operator DNA in vitro (7). Addition of proflavine at a greater than 20-fold molar excess to the QacR-IR1 solution, followed by extensive filtering to remove particulates, reduced the molecular mass to 43 ± 8 kDa, again using a monomodal analysis. A similar result was observed when dequalinium was added to a QacR-IR1 solution (data not shown). The sole presence of the 43-kDa molecular mass species indicates that two dimers of QacR are induced off the IR1 operator site. At these concentrations, there was no evidence of monomeric or tetrameric QacR-drug complexes. In contrast to the QacR-IR1 measurements, when a noncognate DNA oligonucleotide was included with QacR, a single molecular mass species of 43 ± 6 kDa was found, which again is consistent with only a QacR dimer. This confirmed the previously demonstrated specificity of QacR for the IR1 operator (7) and furthermore established that formation of DNA-bound QacR tetramers occurs explicitly in the presence of IR1.
Binding of QacR to DNA requires correctly spaced IR1 half-sites.
A mutagenic approach was used to investigate the significance of the IR1 spacer region to the ability of QacR dimers to bind operator DNA. Due to the availability of the previously constructed plasmid pSK5688, in which PqacA had been repositioned so that the 6 bp normally separating the two IR1 half-sites had been replaced with the −10 hexamer of this promoter (Fig. 6), analysis of this alternative PqacA/IR1 arrangement provided an initial indication of the importance of the 6-bp spacer.
FIG. 6.
Relevant portion of the wild-type PqacA/IR1 sequence carried by pSK5249 and pSK5203 (A) and the alternative PqacA/IR1 arrangement in pSK5688, in which the −10 and −35 hexamers (underlined) of the qacA promoter have been repositioned so that the −10 region is located between the two half-sites of IR1 (B). (C) The sequence of the central region separating the two IR1 half-sites is given for plasmids that contain either the wild-type 6 bp (pSK5203 and pSK5249), substitutions to all 6 bp (pSK5857), a 2-bp increase (pSK5856) or decrease (pSK5834) in the spacing between the two half-sites, transversions in the −1 and +1 positions (pSK5858), or transversions in the −3 and −2 positions (pSK5859). A line delineates the center of the twofold symmetry between the IR1 half-sites. The bases are numbered as for Fig. 1, with non-wild-type IR1 nucleotides indicated in bold. The absolute CAT activities for fusions of these various PqacA/IR1 sequences to a cat reporter gene (expressed as micromoles of chloramphenicol acetylated per minute per milligram of protein) were determined for plasmids with qacR in cis, with the exception of pSK5203, which lacked the qacR gene.
CAT assays demonstrated that, compared to the level of repression observed for the wild-type promoter (pSK5249; Fig. 6A), the variant PqacA/IR1 (pSK5688; Fig. 6B) exhibited an almost 10-fold increase in the extent to which its transcription was repressed (data not shown). In contrast, in the absence of qacR, the altered PqacA/IR1 sequence directed 0.89-fold the transcription observed for the equivalent plasmid containing a wild-type promoter (pSK5203). Although the dramatic enhancement of the ability of the variant PqacA in pSK5688 to be repressed by QacR may well be due to an increased efficiency in the blocking of RNA polymerase binding, as has been demonstrated for the lac system (12), it could not be ruled out that the alteration of IR1 had instead produced a sequence which was bound more efficiently by QacR.
Hence, to more accurately investigate what significance the size and/or sequence of the 6-bp spacer region had to QacR binding, a number of further mutations more closely related to the wild-type PqacA/IR1 sequence were produced in pSK5249, and the absolute CAT activities of these fusions were determined as shown in Fig. 6C. The simultaneous alteration of all 6 bp in pSK5857 led to only a small decrease in the ability of QacR to repress expression from PqacA in comparison to the wild-type (pSK5857, 7.16, versus pSK5249, 2.43; Fig. 6C). However, a CAT activity that was almost comparable to the level of transcription that occurs in the absence of QacR (pSK5203, 41.61) resulted from a 2-bp increase in the separation of the IR1 half-sites (pSK5856, 36.82). This indicated that QacR effectively failed to bind the altered IR1 sequence in which the spacing of the half-sites had been increased by 2 bp.
Likewise, the value of 47.66 obtained for pSK5834 was indicative of a total lack of QacR binding to an IR1 sequence in which the spacing between the two half-sites had been decreased by 2 bp (Fig. 6C). In combination with the mutations that changed all 6 bp, these results indicated that the primary function of the central region of IR1 is to correctly space the half-sites. The requirement for two half-sites was verified by gel mobility shift assays, which used a DNA fragment containing a single IR1 half-site and the wild-type 6-bp spacer region. QacR did not alter the mobility of the probe DNA in these experiments (data not shown).
To further investigate the relatively small reduction in repression observed for the 6-bp mutant (pSK5857; Fig. 6C), two further mutations that introduced transversions at positions −1 and +1 (pSK5858) or −3 and −2 (pSK5859) were constructed. Surprisingly, transversions at the central 2 bp (pSK5858; Fig. 6C) produced an operator that was bound less efficiently than the one in which all 6 bp had been mutated (pSK5857; Fig. 6C), perhaps because the latter construct contained only a transversion at the −1 position, with a transition at +1. In addition to bp −1 and +1 contributing to the overall twofold symmetry of the IR1 sequence (Fig. 6A), their presence in an organism with a very low GC content further reinforces the finding that the actual sequence of the central 2 bp appears to be more important than that of the surrounding 4 bp. The increased repression of the mutant operator containing transversions at the −3 and −2 positions (pSK5859; Fig. 6C) was also a somewhat unexpected result. Interestingly, the initial “−10” mutation in pSK5688, which was strongly repressed by QacR, also possessed the same transversions at positions −3 and −2 as the pSK5859 operator, in addition to introducing transitions, and not transversions, at the central 2 bp (Fig. 6).
DISCUSSION
A number of proteins, including human fibroblast growth factor 1 (5), T4 lysozyme (23), and Bacillus α-amylases (33), have, like QacR, been observed to form disulfide-bonded multimeric degradation products following purification. Of particular concern, in the case of QacR, was the large amount of the monomeric species containing an intramolecular disulfide bond (Fig. 3), which appeared to form either during or immediately upon purification. Circumvention of this problem required the removal of the two cysteine residues, which resulted in the production of an engineered, fully functional, cysteineless QacR (C72A/C141S) derivative with greatly enhanced in vitro stability.
The cysteine residues of some regulatory proteins have been found to be essential, e.g., forming part of the DNA-binding domain in zinc finger proteins (21) or playing a fundamental role in the binding of cationic metal ligands by other regulatory proteins, such as MerR (34) and ArsR (28). Retention of full DNA-binding capabilities by the cysteineless derivative was validated both in vivo (Fig. 2) and in vitro, demonstrating that the C72 and C141 residues were not required for the binding of QacR to IR1 DNA. Likewise, the ability of cysteineless QacR to continue to mediate wild-type levels of induction from PqacA in vivo (Table 1) indicated that the two cysteine residues do not play an important role in drug binding or the communication of the ligand-binding state to the DNA-reading head.
The gel filtration data (Fig. 5), taken together with the cross-linking (Fig. 4) and DLS results, show that two separate QacR dimers bind to IR1 DNA either independently or, alternatively, with only limited dimer-dimer interactions. A revised model for the regulation of qacA expression by QacR is therefore proposed, in which induction results in the dissociation of the two dimers bound to an IR1 site, to form two separate ligand-bound dimers. DLS experiments provided proof of this event, demonstrating that the four molecules of QacR bound to operator DNA again assumed dimeric forms in the presence of inducing compounds. This result also strongly supported the postulate that QacR does not self-assemble into a tetrameric form, as did the failure of the cross-linking experiments to produce any covalently linked complexes larger in size than a dimeric form of QacR. As both formaldehyde and gluteraldehyde are generally considered to be zero-length cross-linking agents, the two QacR dimers bound to each IR1 site are therefore likely to have no or only very limited direct dimer-dimer interactions, such that no side-chains are available for cross-linking by these reagents. Formaldehyde and gluteraldehyde also did not produce any covalently linked QacR-DNA complexes that contained four protein molecules, which is consistent with the failure to obtain cross-linked protein-operator complexes for other sequence-specific DNA-binding regulatory proteins (29). The apparent anomaly of apo-QacR eluting from the Superose gel filtration column as a monomer is likely to reflect the ability of this matrix on rare occasions to temporarily monomerize native dimers, a situation previously observed for the Toxoplasma gondii uracil phosphoribosyltransferase (M. A. Schumacher and R. G. Brennan, unpublished data).
Despite all the evidence obtained in this study indicating that QacR does not self-assemble into a tetrameric form prior to DNA binding, no intermediate forms equivalent to one QacR dimer bound per IR1 site were detected in gel mobility shift assays, even at protein concentrations that did not bind all the available operator DNA (7). This result is not surprising, considering that both in vivo (7) and in vitro QacR failed to bind to DNA that contained only a single IR1 half-site. Some form of distance-dependent cooperativity in the binding of a pair of dimers to IR1 was clearly supported by the almost complete inability of QacR to bind operator sequences in which the spacing of the IR1 half-sites had been increased or decreased by 2 bp (Fig. 6C), particularly in light of the continued ability of QacR to bind operator sequences which contained substitutions to all of the central 6 bp.
A mechanism for cooperative binding of QacR dimers to IR1 that involved indirect interactions through the DNA would be consistent with all the results presented in this work. Such a scenario is not unwarranted, as monomers of the human regulatory protein RFX1 have been shown to bind DNA cooperatively, forming dimers that have no direct protein-protein interactions; instead, the cooperativity appears to occur via protein-induced deformation of the binding site (4, 6). Thus, the actual sequence of the central 6 bp of IR1 may make a contribution to the cooperative binding of QacR dimers via influencing the local structure of the operator DNA, in addition to their primary role of correctly spacing the half-sites. Alternatively, data from the DNase I footprint (Fig. 1), together with that from the mutations in the central 6 bp (Fig. 6C), could be taken to indicate that QacR makes some direct DNA contacts to the central spacer that are for the most part sequence nonspecific.
Of interest was the finding that a simple 2-bp mutation introduced at positions −3 and −2 of IR1 produced an operator that exhibited significantly improved repression (pSK5859; Fig. 6C). This observation provides some corroboration for an earlier proposal that the natural QacR/IR1 system is designed to provide a significant basal level of qacA expression in order to protect the cell against compounds that are substrates of the QacA multidrug pump but not ligands of the QacR transcriptional repressor (7).
The demonstration that two QacR dimers bind to IR1 was unexpected, considering that the other TetR family members for which the DNA-binding stoichiometry is known, bind as one dimer per operator, e.g., TetR (9) and CamR (3). However, in contrast to the above proteins, QacR binds to an exceptionally large, 36-bp inverted repeat (Fig. 1). The DNase I footprint (7), in combination with the spacer region mutations (Fig. 6), suggests that QacR interacts directly with a region of DNA consisting of 20 to 28 bp, or at least 10 bp per half-site, which is in stark contrast to the 6-bp DNA-binding capacity of a typical helix-turn-helix (HTH) motif (8, 31). Therefore, in order to bind a single IR1 half-site, the DNA-reading heads of both molecules in one QacR dimer appear to be required, which would account for the observed QacR DNA-binding stoichiometry (Fig. 5). Although the DNA “recognition” helix of TetR compensates for its abnormally short length by making an exceptionally large number of contacts with tet operator DNA (20), QacR may have acquired an alternative solution to ensure that an adequate number of DNA contacts are made by employing a dimer to bind to each operator half-site.
Although MetJ family regulatory proteins also employ one dimer to bind each of their operator half-sites, unlike QacR, these proteins have been shown to form tetramers that involve substantial dimer-dimer interactions, in addition to binding DNA with an antiparallel β-sheet motif and not an HTH (30). The lack of any apparent internal symmetry within an IR1 half-site (Fig. 1) suggests that the contacts a QacR dimer makes to DNA may be largely nonsymmetrical, as has been found for a MetJ family member, the bacteriophage P22 repressor Arc (24). Further prokaryotic regulatory proteins that bind DNA as higher-order oligomers have been shown to self-assemble from dimers into tetramers, as in the case of LacR (14), and from dimers into octamers, in the case of the lambda repressor (27). In both of the above examples, the dimeric units making up the higher-order oligomers are used to bind distinct operators.
Analysis of the region of DNA encoding the qacA and qacR genes failed to detect any additional sequences exhibiting similarity to IR1, and it has been established that QacR does not interact with the dissimilar IR2 inverted repeat located in the vicinity of the qacR promoter (7). This further supports a model in which separate QacR dimers are used to bind the two IR1 operator half-sites by a cooperative process that does not require the prior self-assembly of QacR into tetramers.
ACKNOWLEDGMENTS
The excellent technical assistance provided by Kate Hardie is acknowledged.
This work was supported in part by Project Grant 153818 from the National Health and Medical Research Council (Australia) and grant AI48593 from the National Institutes of Health (U.S.A.). M.A.S. is a Burroughs Wellcome Career Awardee. S.G. was the recipient of an Australian Postgraduate Award.
REFERENCES
- 1.Ahmed M, Borsch C M, Taylor S S, Vázquez-Laslop N, Neyfakh A A. A protein that activates expression of a multidrug efflux transporter upon binding the transporter substrates. J Biol Chem. 1994;269:28506–28513. [PubMed] [Google Scholar]
- 2.Ahmed M, Lyass L, Markham P N, Taylor S S, Vázquez-Laslop N, Neyfakh A A. Two highly similar multidrug transporters of Bacillus subtilis whose expression is differentially regulated. J Bacteriol. 1995;177:3904–3910. doi: 10.1128/jb.177.14.3904-3910.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aramaki H, Sagara Y, Kabata H, Shimamoto N, Horiuchi T. Purification and characterization of a cam repressor (CamR) for the cytochrome P-450cam hydroxylase operon on the Pseudomonas putida CAM plasmid. J Bacteriol. 1995;177:3120–3127. doi: 10.1128/jb.177.11.3120-3127.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornille F, Emery P, Schuler W, Lenoir C, Mach B, Roques B P, Reith W. DNA binding properties of a chemically synthesized DNA binding domain of hRFX1. Nucleic Acids Res. 1998;26:2143–2149. doi: 10.1093/nar/26.9.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engleka K A, Maciag T. Inactivation of human fibroblast growth factor-1 (FGF-1) activity by interaction with copper ions involves FGF-1 dimer formation induced by copper-catalyzed oxidation. J Biol Chem. 1992;267:11307–11315. [PubMed] [Google Scholar]
- 6.Gajiwala K S, Burley S K. Winged helix proteins. Curr Opin Struct Biol. 2000;10:110–116. doi: 10.1016/s0959-440x(99)00057-3. [DOI] [PubMed] [Google Scholar]
- 7.Grkovic S, Brown M H, Roberts N J, Paulsen I T, Skurray R A. QacR is a repressor protein that regulates expression of the Staphylococcus aureus multidrug efflux pump QacA. J Biol Chem. 1998;273:18665–18673. doi: 10.1074/jbc.273.29.18665. [DOI] [PubMed] [Google Scholar]
- 8.Harrison S C, Aggarwal A K. DNA recognition by proteins with the helix-turn-helix motif. Annu Rev Biochem. 1990;59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- 9.Hillen W, Berens C. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu Rev Microbiol. 1994;48:345–369. doi: 10.1146/annurev.mi.48.100194.002021. [DOI] [PubMed] [Google Scholar]
- 10.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 11.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 12.Lanzer M, Bujard H. Promoters largely determine the efficiency of repressor action. Proc Natl Acad Sci USA. 1988;85:8973–8977. doi: 10.1073/pnas.85.23.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis K. Multidrug resistance: versatile drug sensors of bacterial cells. Curr Biol. 1999;9:R403–R407. doi: 10.1016/s0960-9822(99)80254-1. [DOI] [PubMed] [Google Scholar]
- 14.Lewis M, Chang G, Horton N C, Kercher M A, Pace H C, Schumacher M A, Brennan R G, Lu P. Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. Science. 1996;271:1247–1254. doi: 10.1126/science.271.5253.1247. [DOI] [PubMed] [Google Scholar]
- 15.Littlejohn T G, Paulsen I T, Gillespie M T, Tennent J M, Midgley M, Jones I G, Purewal A S, Skurray R A. Substrate specificity and energetics of antiseptic and disinfectant resistance in Staphylococcus aureus. FEMS Microbiol Lett. 1992;95:259–266. doi: 10.1016/0378-1097(92)90439-u. [DOI] [PubMed] [Google Scholar]
- 16.Lomovskaya O, Lewis K, Matin A. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J Bacteriol. 1995;177:2328–2334. doi: 10.1128/jb.177.9.2328-2334.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lucas C E, Balthazar J T, Hagman K E, Shafer W M. The MtrR repressor binds the DNA sequence between the mtrR and mtrC genes of Neisseria gonorrhoeae. J Bacteriol. 1997;179:4123–4128. doi: 10.1128/jb.179.13.4123-4128.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Markham P N, Ahmed M, Neyfakh A A. The drug-binding activity of the multidrug-responding transcriptional regulator BmrR resides in its C-terminal domain. J Bacteriol. 1996;178:1473–1475. doi: 10.1128/jb.178.5.1473-1475.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Markham P N, LoGuidice J, Neyfakh A A. Broad ligand specificity of the transcriptional regulator of the Bacillus subtilis multidrug transporter, Bmr. Biochem Biophys Res Commun. 1997;239:269–272. doi: 10.1006/bbrc.1997.7467. [DOI] [PubMed] [Google Scholar]
- 20.Orth P, Schnappinger D, Hillen W, Saenger W, Hinrichs W. Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nat Struct Biol. 2000;7:215–219. doi: 10.1038/73324. [DOI] [PubMed] [Google Scholar]
- 21.Pabo C O, Sauer R T. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 22.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perry L J, Wetzel R. The role of cysteine oxidation in the thermal inactivation of T4 lysozyme. Protein Eng. 1987;1:101–105. doi: 10.1093/protein/1.2.101. [DOI] [PubMed] [Google Scholar]
- 24.Raumann B E, Rould M A, Pabo C O, Sauer R T. DNA recognition by beta-sheets in the Arc repressor-operator crystal structure. Nature (London) 1994;367:754–757. doi: 10.1038/367754a0. [DOI] [PubMed] [Google Scholar]
- 25.Rouch D A, Cram D S, DiBerardino D, Littlejohn T G, Skurray R A. Efflux-mediated antiseptic resistance gene qacA from Staphylococcus aureus: common ancestry with tetracycline- and sugar-transport proteins. Mol Microbiol. 1990;4:2051–2062. doi: 10.1111/j.1365-2958.1990.tb00565.x. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 27.Senear D F, Laue T M, Ross J B, Waxman E, Eaton S, Rusinova E. The primary self-assembly reaction of bacteriophage lambda cI repressor dimers is to octamer. Biochemistry. 1993;32:6179–6189. doi: 10.1021/bi00075a010. [DOI] [PubMed] [Google Scholar]
- 28.Shi W, Dong J, Scott R A, Ksenzenko M Y, Rosen B P. The role of arsenic-thiol interactions in metalloregulation of the ars operon. J Biol Chem. 1996;271:9291–9297. doi: 10.1074/jbc.271.16.9291. [DOI] [PubMed] [Google Scholar]
- 29.Solomon M J, Varshavsky A. Formaldehyde-mediated DNA-protein crosslinking: a probe for in vivo chromatin structures. Proc Natl Acad Sci USA. 1985;82:6470–6474. doi: 10.1073/pnas.82.19.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Somers W S, Phillips S E V. Crystal structure of the met repressor-operator complex at 2.8 Å resolution reveals DNA recognition by β-strands. Nature (London) 1992;359:387–393. doi: 10.1038/359387a0. [DOI] [PubMed] [Google Scholar]
- 31.Spott S, Dong F, Kisters-Woike B, Muller-Hill B. Dimerisation mutants of Lac repressor. II. A single amino acid substitution, D278L, changes the specificity of dimerisation. J Mol Biol. 2000;296:673–684. doi: 10.1006/jmbi.1999.3469. [DOI] [PubMed] [Google Scholar]
- 32.Stark M J. Multicopy expression vectors carrying the lac repressor gene for regulated high-level expression of genes in Escherichia coli. Gene. 1987;51:255–267. doi: 10.1016/0378-1119(87)90314-3. [DOI] [PubMed] [Google Scholar]
- 33.Tomazic S J, Klibanov A M. Mechanisms of irreversible thermal inactivation of Bacillus alpha-amylases. J Biol Chem. 1988;263:3086–3091. [PubMed] [Google Scholar]
- 34.Utschig L M, Bryson J W, O'Halloran T V. Mercury-199 NMR of the metal receptor site in MerR and its protein-DNA complex. Science. 1995;268:380–385. doi: 10.1126/science.7716541. [DOI] [PubMed] [Google Scholar]
- 35.Vázquez-Laslop N, Markham P N, Neyfakh A A. Mechanism of ligand recognition by BmrR, the multidrug-responding transcriptional regulator: Mutational analysis of the ligand-binding site. Biochemistry. 1999;38:16925–16931. doi: 10.1021/bi991988g. [DOI] [PubMed] [Google Scholar]
- 36.Volkin D B, Mach H, Middaugh C R. Degradative covalent reactions important to protein stability. In: Shirley B A, editor. Protein stability and folding: theory and practice. Vol. 40. Totowa, N.J: Humana Press Inc.; 1995. pp. 35–63. [DOI] [PubMed] [Google Scholar]
- 37.Wissmann A, Meier I, Hillen W. Saturation mutagenesis of the Tn10-encoded tet operator O1: identification of base-pairs involved in Tet repressor recognition. J Mol Biol. 1988;202:397–406. doi: 10.1016/0022-2836(88)90273-2. [DOI] [PubMed] [Google Scholar]
- 38.Zheleznova E E, Markham P N, Neyfakh A A, Brennan R G. Structural basis of multidrug recognition by BmrR, a transcriptional activator of a multidrug transporter. Cell. 1999;96:353–362. doi: 10.1016/s0092-8674(00)80548-6. [DOI] [PubMed] [Google Scholar]
- 39.Zheleznova Heldwein E E, Brennan R G. Crystal structure of the transcription activator BmrR bound to DNA and a drug. Nature (London) 2001;409:378–382. doi: 10.1038/35053138. [DOI] [PubMed] [Google Scholar]