Abstract
Background
Biological sex and the estrogen receptor alpha (ESR1) modulate human immunodeficiency virus (HIV) activity. Few women have enrolled in clinical trials of latency reversal agents (LRAs); their effectiveness in women is unknown. We hypothesized that ESR1 antagonism would augment induction of HIV expression by the LRA vorinostat.
Methods
AIDS Clinical Trials Group A5366 enrolled 31 virologically suppressed, postmenopausal women on antiretroviral therapy. Participants were randomized 2:1 to receive tamoxifen (arm A, TAMOX/VOR) or observation (arm B, VOR) for 5 weeks followed by 2 doses of vorinostat. Primary end points were safety and the difference between arms in HIV RNA induction after vorinostat. Secondary analyses included histone 4 acetylation, HIV DNA, and plasma viremia by single copy assay (SCA).
Results
No significant adverse events were attributed to study treatments. Tamoxifen did not enhance vorinostat-induced HIV transcription (between-arm ratio, 0.8; 95% confidence interval [CI], .2–2.4). Vorinostat-induced HIV transcription was higher in participants with increases in H4Ac (fold increase, 2.78; 95% CI, 1.34–5.79) vs those 9 who did not (fold increase, 1.04; 95% CI, .25–4.29). HIV DNA and SCA plasma viremia did not substantially change.
Conclusions
Tamoxifen did not augment vorinostat-induced HIV RNA expression in postmenopausal women. The modest latency reversal activity of vorinostat, postmenopausal status, and low level of HIV RNA expression near the limits of quantification limited assessment of the impact of tamoxifen. This study is the first HIV cure trial done exclusively in women and establishes both the feasibility and necessity of investigating novel HIV cure strategies in women living with HIV.
Clinical Trials Registration
Keywords: HIV cure, ESR1, latency reversal agent, biological sex
Combination therapy with tamoxifen and vorinostat did not augment human immunodeficiency virus (HIV) latency reversal over vorinostat alone in post-menopausal women. Histone acetylation changes did associate with level of HIV RNA induction. Enrollment of women in interventional trials of cure strategies is feasible.
To achieve the goal of human immunodeficiency virus (HIV) cure, latently infected cells that carry replication-competent virus must be eliminated. Inducing virus expression in this latent reservoir, which leads to the production of HIV RNA and proteins, is the foundation of the shock and kill strategy [1]. Proof of concept for latency reversal agents (LRAs) was first demonstrated with the histone deacetylase inhibitor (HDACi) vorinostat [2], and multiple trials evaluating a range of HDACi and other classes of LRAs have followed [3]. In vitro studies indicate that uninduced, replication-competent proviruses remain even after vigorous stimulation [4], highlighting the importance of identifying new strategies to augment HIV expression from all reservoir cells that harbor infectious HIV with in vivo treatments.
There has also been substantial heterogeneity in participants’ responses to LRAs across trials. Variable time to rebound in analytic treatment interruptions [5] and levels of virus induction following LRA treatment [6] or administration of immunomodulating agents such as anti-PD1 therapy [7] have been consistently observed. Notably, this diversity of response is present despite a relative homogeneity of trial participants, most of whom are male [8, 9]. Identifying sources of heterogeneity has value for both predicting probability of response to treatments in development and potentially for probing mechanistic pathways of latency maintenance and immune clearance.
Abundant evidence indicates that HIV expression is less robust in women compared with men [10]. In untreated infection, women have an approximately 0.35 log10 lower viral load (VL) early in disease [11], lower per cell production of HIV RNA in lymph nodes [12], and higher levels of T-cell activation for a given VL [13, 14]. This difference in VL excluded women at risk for progression to AIDS from treatment in early guidelines [15], underscoring the clinical implications of ignoring biological sex. In treated HIV, women have lower levels of detectable cell-associated HIV RNA [16, 17], lower levels of residual viremia [17, 18], and lower levels of inducible RNA [17] and lower levels of replication-competent virus in some but not all studies [19, 20].
The basis of these sex differences is still being defined [10], but data suggest that sex hormones play a role [10]. In vitro models of active infection reported inhibition of HIV expression by 17β-estradiol mediated through the estrogen receptor alpha (ESR1) [21], and studies of acute infection support an influence of estradiol on HIV VL in untreated disease [13]. ESR1 has also been implicated in HIV latency control. In an unbiased small hairpin RNA (shRNA) screen in an in vitro HIV latency model, ESR1 was a dominant regulator of HIV latency reversal [22]. In complementary studies, 17β-estradiol treatment blunted HIV transcription induced by T-cell activation in ex vivo assays in CD4+ T cells from women on suppressive antiretroviral therapy (ART). The selective estrogen receptor modulator tamoxifen enhanced HIV transcriptional activation by vorinostat [22]. Taken together, these data point to a role for 17β-estradiol and ESR1 in HIV transcriptional control during both active infection and latency reversal.
Although mounting evidence demonstrates sex-specific features of HIV latency, sparse data are available on the therapeutic strategies for HIV cure in women. Despite the fact that women constitute 51% of people with HIV, cure trials have overwhelming enrolled male participants, and the efficacy of LRAs in women is not known. In this study, we sought to determine whether antagonism of the estrogen receptor in vivo would augment HIV latency reversal by vorinostat in women. We hypothesized that combined tamoxifen and vorinostat would be safe and would lead to greater induction of HIV RNA compared with vorinostat alone.
METHODS
Study Design and Participants
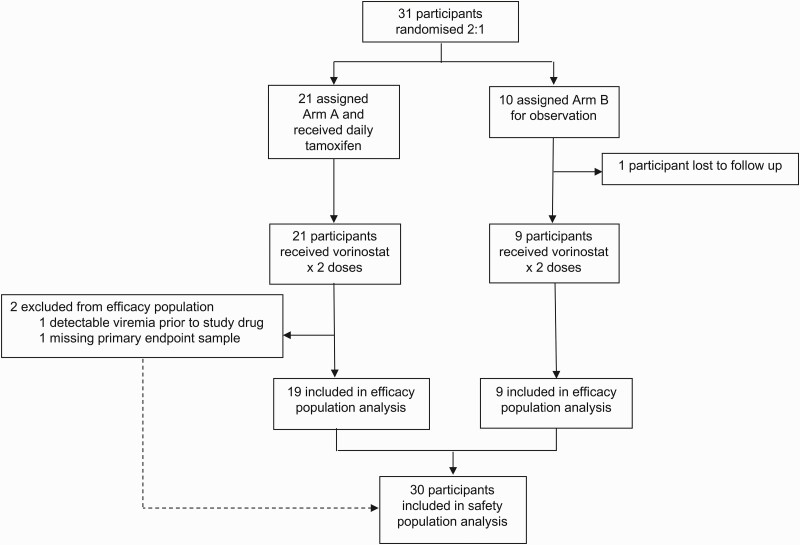
The AIDS Clinical Trials Group (ACTG) study A5366 (MOXIE: tamoxifen for enhancement of latency reversal) is a randomized, open-label, proof-of-concept study that enrolled postmenopausal women aged 18–65 years with HIV who were virologically suppressed on ART for >1 year at 15 sites. Postmenopausal women were enrolled due to potential genotoxicity of vorinostat [23] and symptoms from tamoxifen in premenopausal women. Participants had CD4+ T-cell counts >300 cells/μL and were on continuous ART for >1 year. Participants were randomized 2:1 to receive either tamoxifen at 20 mg daily for 38 days (arm A, TAMOX/VOR) or observation arm B (VOR) and then all received 2 doses of 400 mg vorinostat on days 35 and 38 (Figure 1). The study was approved by the local institutional review boards. Detailed methods are in the Supplementary Materials.
Figure 1.
Trial profile.
The primary end points were safety of tamoxifen and vorinostat and change from baseline in HIV type 1 (HIV-1) cell-associated RNA (caRNA) in CD4+ T cells following treatment with vorinostat and tamoxifen compared with vorinostat alone. Secondary analyses included change in proviral HIV DNA levels, proportion with low level viremia by single copy assay, and H4Ac levels.
Virologic, Hormonal, and Pharmacologic Assays
Baseline blood samples were obtained at the preentry and entry. Clinical HIV-1 RNA assays at entry and day 28 confirmed suppression prior to vorinostat dosing. The primary end point was 5 hours post the second dose of vorinostat based on prior studies that suggested this would yield maximal HIV RNA expression [2].
CD4+T cells were isolated by negative immunoselection from cryopreserved peripheral blood mononuclear cells (PBMCs); genomic DNA and total RNA (AllPrep, Qiagen) were isolated from approximately 5 × 106 total CD4+T cells. Total HIV-1 DNA and unspliced caRNA levels were quantified in triplicate using real-time polymerase chain reaction (PCR) with primers targeting the gag region [24]. Approximately 500 ng of genomic DNA were assessed per well, with cell input quantification by CCR5 gene DNA copy number and a limit of quantification of 1 copy per reaction [24]. HIV-1 caRNA was quantified with the same primers, and RNA integrity was confirmed by quantitative PCR (qPCR) of the human reference gene IPO8. The limit of quantification of the caRNA assay was 3 copies per reaction.
Quantification of spliced envelope RNA transcripts was performed using the EDITS assay [22]. CD4+T cells were isolated from cryopreserved PBMCs by immunoselection and RNA isolated (RNeasy Kit, Qiagen). The total input (approximately 1.25 × 106 CD4+cells/sample) was then used in a nested PCR with primers spanning the spliced region of Env and thereby excluding proviral amplification based on product length [22]. The sample was taken into library preparation and sequencing on the Ion Torrent platform. Mapped reads were quantified as the frequency of cells spontaneously producing spliced envelope RNA using a standard curve spanning a range of 1 to 300 primary memory CD4+cells infected with replication-competent GFP-tagged HIV-1 NL4-3 in a total pool of 1.25 × 106 uninfected cells [22].
Residual HIV-1 plasma viremia was quantified by a single copy assay using 4.5 mL of plasma. The assay uses primers specific for the integrase region of the pol gene and was performed as previously described; the limit of quantification was 0.38 copies/mL [25].
Histone acetylation was assessed using an H4K5/8/12/16 enzyme-linked immunosorbent assay on PBMC lysates as previously described [26, 27]. Estradiol levels were measured using a standard clinical liquid chromatography-mass spectrometry (LC-MS) assay. Tamoxifen concentration was measured from plasma samples using ultraperformance LC-MS (Waters, Milford, MA). Vorinostat concentrations were measured from serum using LC-MS/MS (SCIEX, Framingham, MA).
Statistical Analyses
Arms were compared using t tests of log-transformed virology measures, imputing half an analytic lower limit for results below limit (prespecified primary analysis, anticipating 10% of caRNA results would be below the assay lower limit). Sensitivity analyses used longitudinal censored-data methods [28] using result-specific lower limits in a mixed model with random intercept. For the initial approach, a lower limit was set for all study participants using a cutoff of the participant with the highest lower limit (eg, if linear range ended at 50 copies for participant X, measured values of 38 and 42 in participant Y would not be included but would be imputed at half the analytic lower limit; in this example, imputed at 25 copies). Using the result-specific lower limits, we included all values measured in the linear range for each participant and only imputed those without a measured value. Additional sensitivity analyses included negative binomial regression applied to the well-specific replicates [29].
RESULTS
Between June 2018 and September of 2018, 31 women with a median age of 57 years and a median CD4 count of 688 cells/mm3 enrolled in A5366; 58% were African American (Table 1). The median time since ART initiation was 7.5 years (first quartile [25th percentile], 2.9 years; third quartile [75th percentile], 13.9 years) with the majority (27 of 31) currently receiving regimens that included integrase inhibitors (Supplementary Table 1). Of these 31 women, 27 constituted the efficacy population; these women received intended doses of both study medications, maintained plasma viral load <75 copies/mL prior to study interventions, and had samples available for evaluation. Four women were not included in the efficacy population because they withdrew or were lost to follow-up (n = 2), did not have the primary end point blood sample drawn (n = 1), or had detectable viremia prior to the study interventions (n = 1). Participant-level data are provided in the Supplementary Materials.
Table 1.
Characteristics of the Study Population
| Characteristic | Overall | Arm A (Vorinostat + Tamoxifen) | Arm B (Vorinostat ) |
|---|---|---|---|
| Sex, number (%), female | 31 (100) | 21 (100) | 10 (100) |
| Age, median (Q1, Q3), years | 57 (53–60) | 57 (54–61) | 55 (51–59) |
| Race, number (%) | |||
| American Indian or Alaskan Native | 1 (3) | 1 (5) | 0 (0) |
| Black or African American | 18 (58) | 11 (52) | 7 (70) |
| White | 12 (39) | 8 (38) | 3 (30) |
| Ethnicity, number (%) | |||
| Hispanic/Latino | 6 (19) | 4 (19) | 2 (20) |
| Not Hispanic/Latino | 25 (81) | 17 (81) | 8 (80) |
| Years since antiretroviral therapy start, median Q1, Q3) | 7.5 (2.9–13.9) | 6.1 (2.4–13.9) | 9.4 (5.9–12.2) |
| Nadir CD4+ T-cell count, median (Q1, Q3), cells/mm3 | 232 (46–363) | 232 (10–363) | 261 (80–402) |
| Screening CD4+ T-cell count, median (Q1, Q3), cells/mm3 | 688 (536–854) | 688 (536–773) | 722 (566–1106) |
| Antiretroviral regimen, number (%) | |||
| Integrase inhibitor + NRTIs | 24 (77%) | 18 (86%) | 6 (60%) |
| NNRTI + NRTIs | 3 (10%) | 1 (5%) | 2 (20%) |
| Protease inhibitor + NRTIs | 1 (3%) | 1 (5%) | 0 (0%) |
| Other combination | 3 (10%) | 1 (5%) | 2 (20%) |
Abbreviations: NNRTI, nonnucleoside reverse transcriptase inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; Q1, first quartile (25th percentile); Q3, third quartile (75th percentile).
The primary end points of the study were safety and the difference in HIV RNA induction between the study arms. With respect to safety, the intervention was well tolerated and there were no serious adverse events attributed to the study medications. Only 2 participants had adverse events of any grade (Supplementary Table 2). Study withdrawal or loss to follow-up occurred in 1 participant in each arm.
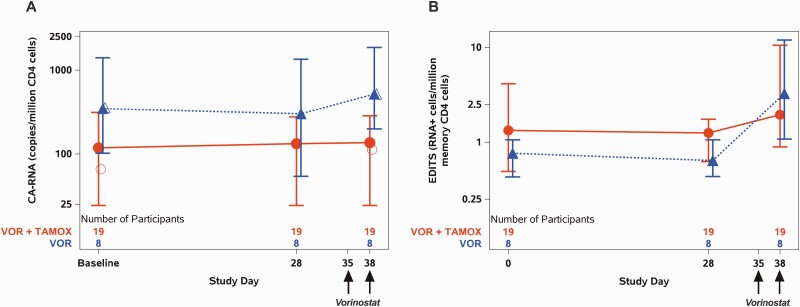
HIV caRNA induction was quantified using 2 approaches: qPCR measurement of unspliced HIV (usHIV) gag RNA transcripts in total CD4+T cells and the EDITS assay, a measure of spliced envelope transcripts from resting memory CD4+T cells (Figure 2, Supplementary Figures 1 and 2). The overall mean fold change for all participants for usHIV RNA was 1.2 (95% confidence interval [CI], .7–2.1); for spliced transcripts by EDITS, it was 2.0 (95% CI, 1.0–3.8). In the analysis by study arm, the mean fold change in usHIV RNA transcripts at 5 hours post the second dose of vorinostat was 1.2 (95% CI, .6–2.3) for arm A (VOR + TAMOX) vs 1.5 (95% CI, .7–3.2) for arm B (VOR; P = .6; Figure 2A, Table 2). Similar results were obtained when spliced envelope transcripts were analyzed using the EDITS assay, with a mean fold change of 1.5 (95% CI, .7–3.2) for arm A and 4.3 (95% CI, 1.2–15.0) for arm B (P = .12; Figure 2B). Tamoxifen treatment did not augment induced expression of HIV caRNA in the study population in either assay.
Figure 2.
Human immunodeficiency virus (HIV) RNA changes over time. A, Cell-associated unspliced HIV RNA measured in total CD4 cells at baseline (average of 2 measurements), at day 28 after either 4 weeks of tamoxifen (arm A) or observation (arm B), and 5 hours after the second dose of vorinostat (day 38). B, Spliced HIV RNA as measured with the EDITS assay in resting memory CD4 cells at the same time points. Symbols indicate geometric mean, bars Q1 (first quartile [25th percentile]), Q3 (third quartile [75th percentile]). Open symbols in (A) indicate geometric mean when using participant-specific lower levels of quantification. There was no significant difference between the study arms in the induction of HIV RNA by either assay. Abbreviations: caRNA, cell-associated RNA; TAMOX, tamoxifen; VOR, vorinostat.
Table 2.
Change in Unspliced Human Immunodeficiency Virus RNA Expression From Baseline to Post-Vorinostat Measurement
| Number of Participants | t Test With Imputation Below Overall LLQ | Longitudinal Censored Model With Participant-Specific LLQ | Negative Binomial Regression | |
|---|---|---|---|---|
| Arm A (vorinostat + tamoxifen) | 19 | 1.2 (0.6–2.3) | 1.7 (1.1–2.7) | 2.4 (1.3–4.6) |
| Arm B (vorinostat) | 8 | 1.5 (0.7–3.2) | 1.5 (0.7–2.9) | 2.6 (1.0–6.8) |
| Ratio between arms | 0.8 (0.2–2.4) | 1.2 (0.5–2.7) | 0.9 (0.3–2.9) |
Table shows the fold change and 95% confidence intervals (CIs) of unspliced human immunodeficiency virus RNA expression by study arm, measured as the ratio between expression at 5 hours after the second vorinostat dose compared with baseline, preintervention levels of expression (baseline = incorporates both preentry and entry visit values). The ratio in fold change between arms and 95% CIs are also shown. The fold change was assessed using 3 statistical approaches as detailed in the Methods and Results sections.
Abbreviation: LLQ, lower limit of quantification.
This usHIV RNA analysis was performed using a single lower limit of quantification (LLQ) of 49 copies per million total CD4+T cells across all samples. Thirty percent of the usHIV RNA values fell below this LLQ, and values were imputed. To assess the impact of imputation for values below the limit of detection, we performed planned secondary analyses of the usHIV RNA values using censored-data methods and result-specific LLQ (see Methods section) and using negative binomial regression including all measured values scaled to input [28, 29]. The majority of the imputed data points were in arm A participants; using a result-specific LLQ, this led to a lower baseline usHIV RNA estimate in arm A (Figure 2A, open circle at baseline). This alternative analysis correspondingly increased the estimate in arm A of usHIV RNA induction after vorinostat administration (Table 2).
To assess the usHIV RNA findings more comprehensively, we performed a negative binomial regression analysis. This approach allowed us to consider the values of each of the triplicate qPCR wells individually and permitted the inclusion of values below the LLQ without imputation and capturing the variability in replicate measurements as an additional measure of the uncertainty of the observations. When compared with the initial analysis, this method shifted the estimates of usHIV RNA induction. This effect was observed predominantly in arm A, which included the majority of the very low value observations (Table 2). Despite these shifts in estimates of induction, we again observed no augmentation of HIV caRNA by combined tamoxifen + vorinostat compared with vorinostat alone.
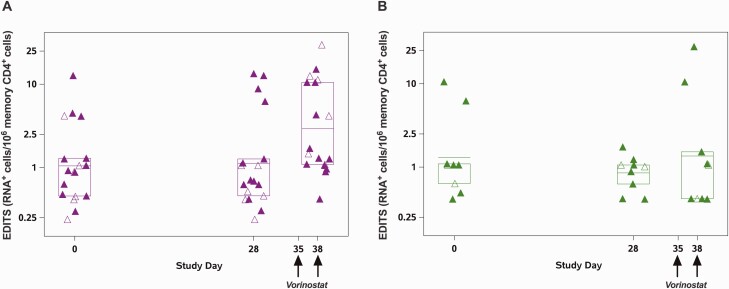
To assess the impact of vorinostat administration on histone 4 acetylation in PBMCs, we compared the level of acetylation prior to vorinostat exposure (day 28 specimen) with acetylation levels after the second vorinostat dose at the primary end point time point (day 38 post vorinostat specimen). Among the 27 women in the trial, 18 had evidence of an increase in histone acetylation consistent with the predicted biological effect of vorinostat, and 9 women demonstrated a decrease in histone acetylation (Table 3, Supplementary Figure 3). The median vorinostat concentration at day 38 was the same (75 ng/mL) for both of the histone acetylation groups (P = .5, Wilcoxon signed rank test; Supplementary Table 3). When stratified by change in histone acetylation, the women with evidence of increased histone acetylation showed a mean fold change of 2.78 (95% CI, 1.34–5.79 in spliced HIV transcripts by the EDITS assay), whereas those with decreased histone acetylation had a fold change in spliced HIV transcripts of 1.04 (95% CI, .25–4.29; Figure 3, Table 3, Supplementary Figure 4).
Table 3.
Change in Human Immunodeficiency Virus RNA Expression Based on Histone Acetylation
| Histone 4 Acetylation | ||
|---|---|---|
| Decrease N = 9 | Increase N = 18 | |
| Fold change (95% confidence interval) | 1.04 (0.25–4.29) | 2.78 (1.34–5.79) |
Table shows the fold change and 95% confidence intervals of human immunodeficiency virus RNA expression measured in resting memory CD4 cells using the EDITS assay 5 hours after the second vorinostat compared with a single baseline level of expression at entry.
Figure 3.
Human immunodeficiency virus (HIV) RNA changes using the EDITS assay stratified by histone acetylation changes. Data from all participants were pooled and stratified into two groups: those with an increase in H4 acetylation between day 28 and day 38 (n = 18, panel A) and those with a decrease in H4 acetylation (n = 9, panel B). HIV RNA+ cells per million resting memory CD4+ T cells quantified using EDITS is shown, box plots indicate mean and Q1 (first quartile [25th percentile]), Q3 (third quartile [75th percentile]). Closed symbols denote arm A participants and open symbols arm B participants.
Secondary end points that were analyzed included the level of detectable plasma viremia by single copy assay (lower limit of quantification, 0.47 copies HIV RNA/mL plasma) and cell-associated proviral HIV DNA. Neither of these values showed variation in response to vorinostat administration overall or when considered by study arm (Supplementary Tables 4 and 5, Supplementary Figure 5). Levels of 17β-estradiol were assessed in all participants at entry, day 28, day 38, and at the follow-up visits; tamoxifen concentrations were in the predicted range (Supplementary Table 6). Minimal variation in hormone levels was observed; only 4 participants (all in arm A) showed substantial variations in estradiol levels across the study period (Supplementary Figure 6).
DISCUSSION
ACTG A5366 (the MOXIE trial) is the first interventional trial to investigate potential HIV cure strategies conducted exclusively in women, a group that has been underrepresented in trials relevant to cure [8, 9]. The trial enrolled rapidly, the intervention was safe, and participants endorsed positive experiences with the trial, expressing willingness to participate in future HIV cure research [30]. Data suggesting that some features of the reservoir have sex specificity [16–18, 20, 31, 32] indicate that cure strategies should be tested in both sexes, and the successful enrollment of this trial supports the feasibility of including women in future trials of investigational cure strategies.
The trial did not demonstrate enhanced induction of HIV transcription by vorinostat in vivo after tamoxifen treatment. This is in contrast to a preclinical study that found that ex vivo exposure to 17β-estradiol decreases HIV transcription in response to latency reversal [22]. The reason for this difference may be related to study participant selection; the most pronounced suppressive effect of estradiol ex vivo was observed in cells from women of reproductive age, with more modest effects in cell cultures of men and older women [22]. Our study enrolled only postmenopausal women due to the potential for genotoxicity of vorinostat and/or adverse symptomatic effects of estrogen antagonism in premenopausal women [23]. In postmenopausal women, estrone is the dominant circulating estrogen; although it correlates with levels of 17β-estradiol [33], the effects on HIV latency and efficiency of blockade by tamoxifen are less clear. Our results demonstrated that only 4 of the 27 participants had substantial variations in 17β-estradiol levels over the course of the study. Low levels of circulating 17β-estradiol may have contributed to the lack of impact of tamoxifen in our study. Vorinostat had only a marginal effect on HIV transcriptional activity, which may also have limited the possibility of detecting an effect. Vorinostat was chosen based on preclinical studies of the combination with tamoxifen [22], safety of the combination (vorinostat plus tamoxifen) in women with breast cancer [34], and due to the high-affinity interaction of romidepsin with the estrogen receptor, rendering that HDACi unsuitable for a study of estrogen receptor antagonism [35].
Our study is the first to assess the efficacy of HDACi in women. In studies that assessed the impact of the HDACi vorinostat [2, 36–39], panobinostat [40], and romidepsin [41–43], only 14 of the 206 participants were women (7%). HDACi including vorinostat impacts estrogen receptor expression [44], and romidepsin interacts with the estrogen receptor [35]. Taken together with the impact of estrogen exposure during in vitro latency reversal treatments, the data suggest that latency reversal agents need direct assessment in women. Optimally, studies should enroll premenopausal women when safety concerns can be adequately addressed during informed consent. Our results are consistent with observations in prior studies that show a substantial degree of host variability in the response to latency reversal treatment. Some prior studies of HDACi have enriched for participants with a higher probability of response by using prescreening assays to identify participants with an ex vivo response to HDACi [2], but no clinical assay has emerged as a predictive correlate. In this study, stratification by H4Ac was associated with higher induction of spliced HIV RNA transcripts, suggesting that this may be a biomarker for responsiveness to HDACi response that could potentially be adapted as an ex vivo screening test of participants.
We observed a substantial frequency of usHIV RNA levels that were below the limit of detection. Data from prior studies [16, 18, 41] showed below-limit frequencies of 1.5% and 7.6%, respectively, which are substantially lower than our results. This finding may reflect generally lower levels of HIV transcriptional activity in females on suppressive ART [17], may be specific to the population enrolled in this trial, or could be related to a technical aspect of the assays we used (eg, direct measurements on total CD4+ T cells rather than on PBMCs used in prior ACTG studies). We used 3 statistical approaches to analyze the usHIV RNA dataset, as there is no consensus on the optimal method for analyzing data that are close to the analytic limits of detection [29]. All 3 approaches were consistent in showing no augmentation of latency reversal with tamoxifen. These data highlight the challenges of differentiating when a biological effect has been achieved and when that effect has a magnitude that is clinically significant. Prior studies have leveraged intensive sampling and assessment of multiple replicates and large numbers of cells to yield robust results [2]; however, leukapheresis and large blood volume sampling are not always feasible. Ideally, assays will be sensitive enough to validate or disprove effects shown with in vitro assays. Our results support the use of methods such as the EDITS assay to achieve better precision at lower values and the development of new techniques such as the ultrasensitive measurement of HIV proteins [45–47]. Improving sensitivity and precision of clinical trial end points will aid decisions on whether an intervention should be optimized or abandoned.
In conclusion, our study did not demonstrate an impact of estrogen antagonism on HIV latency reversal. The rapid enrollment and successful completion of the first HIV cure trial done exclusively in women support the feasibility and importance of enrolling women into future studies investigating curative interventions. The larger than anticipated fraction of women with very low values of caRNA in this trial emphasizes the potential for sex-based differences in HIV reservoir dynamics as initial predictions were based on prior studies with predominantly male participants. These findings highlight the need to adequately power proof-of-concept trials where a range of host responses are expected and underscore the need for sensitive and reliable assays of virologic outcomes.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. The full study team developed the study concept and design. E. P. S. and R. G. were the cochairs of the study, D. R. K. was the vice chair, A. T. was the protocol virologist, E. A. and R. J. B. were the protocol statisticians, S. S. was the protocol immunologist, Q. M. and G. M. were the protocol pharmacologists, K. S. was the community representative, K. E. S. was the industry representative, L. H. was the clinical trial specialist, F. G. was the laboratory technologist, and C. G. was the Division of AIDS representative. Study investigators included N. A., N. C., S. D., M. G., L. E., and E. C. Virological assays were done by K. C., A. T., C. D., and J. K. Histone acetylation assays were performed at Merck and Co under supervision of B. H. The manuscript was written by E. P. S. with input of all authors.
Acknowledgments. The study team thanks all of the study site staff, the data management team, and the AIDS Clinical Trials Group (ACTG) for the opportunity to perform this study. The team is grateful to all of the women who participated in this trial.
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health (NIH). The full protocol is available through the ACTG.
Financial support. This study was supported by the ACTG. The research reported here was supported by the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institute of Health under awards UM1 AI068634, UM1 AI068636, and UM1 AI106701. R. T. G. receives grant funding from the Harvard University Center for AIDS Research (NIH P30 AI060354) and the AIDS Clinical Trials Group (ACTG) (NIH/NIAID 2 UMAI069412-09). Pharmacology assay support was provided by the University of North Carolina at Chapel Hill Center for AIDS Research (NIH P30 AI050410).
Contributor Information
Eileen P Scully, Departement of Medicine, Johns Hopkins University, Baltimore, Maryland, USA.
Evgenia Aga, Department of Biostatistics, Harvard T.H. Chan School of Public Health, Boston, Massachusetts, USA.
Athe Tsibris, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Nancie Archin, University of North Carolina, Chapel Hill, North Carolina, USA.
Kate Starr, ACTG Clinical Research Site, Ohio State University, Hilliard, Ohio, USA.
Qing Ma, Translational Pharmacology Research Core, University at Buffalo, Buffalo, New York, USA.
Gene D Morse, Translational Pharmacology Research Core, University at Buffalo, Buffalo, New York, USA.
Kathleen E Squires, Merck Research Labs, Upper Gwynned, Pennsylvania, USA.
Bonnie J Howell, Department of Infectious Disease and Vaccines, Merck and Co, West Point, Pennsylvania, USA.
Guoxin Wu, Department of Infectious Disease and Vaccines, Merck and Co, West Point, Pennsylvania, USA.
Lara Hosey, ACTG Network Coordinating Center, Silver Spring, Maryland, USA.
Scott F Sieg, Department of Molecular Biology and Microbiology, Case Western Reserve University, Cleveland, Ohio, USA.
Lynsay Ehui, Whitman-Walker Health, Washington, D.C., USA.
Francoise Giguel, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Kendyll Coxen, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Curtis Dobrowolski, Department of Molecular Biology and Microbiology, Case Western Reserve University, Cleveland, Ohio, USA.
Monica Gandhi, Department of Medicine, University of California, San Francisco, California, USA.
Steve Deeks, Department of Medicine, University of California, San Francisco, California, USA.
Nicolas Chomont, Department of Microbiology, Infectiology and Immunology, Université de Montréal, Centre Hospitalier de l'Université de Montréal (CHUM), Montreal, Canada.
Elizabeth Connick, Department of Medicine, University of Arizona, Tucson, Arizona, USA.
Catherine Godfrey, Office of the Global AIDS Coordinator, Department of State, Washington D.C., USA.
Jonathan Karn, Department of Molecular Biology and Microbiology, Case Western Reserve University, Cleveland, Ohio, USA.
Daniel R Kuritzkes, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Ronald J Bosch, Department of Biostatistics, Harvard T.H. Chan School of Public Health, Boston, Massachusetts, USA.
Rajesh T Gandhi, Department of Medicine, Massachusetts General Hospital, Harvard University, Boston, Massachusetts, USA.
References
- 1. Deeks SG. HIV: shock and kill. Nature 2012; 487:439–40. [DOI] [PubMed] [Google Scholar]
- 2. Archin NM, Liberty AL, Kashuba AD, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 2012; 487:482–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Margolis DM, Garcia JV, Hazuda DJ, Haynes BF.. Latency reversal and viral clearance to cure HIV-1. Science 2016; 353:aaf6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ho YC, Shan L, Hosmane NN, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 2013; 155:540–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wen Y, Bar KJ, Li JZ.. Lessons learned from HIV antiretroviral treatment interruption trials. Curr Opin HIV AIDS 2018; 13:416–21. [DOI] [PubMed] [Google Scholar]
- 6. Ait-Ammar A, Kula A, Darcis G, et al. Current status of latency reversing agents facing the heterogeneity of HIV-1 cellular and tissue reservoirs. Front Microbiol 2019; 10:3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Abbar B, Baron M, Katlama C, et al. Immune checkpoint inhibitors in people living with HIV: what about anti-HIV effects? AIDS 2020; 34:167–75. [DOI] [PubMed] [Google Scholar]
- 8. Curno MJ, Rossi S, Hodges-Mameletzis I, Johnston R, Price MA, Heidari S.. A systematic review of the inclusion (or exclusion) of women in HIV research: from clinical studies of antiretrovirals and vaccines to cure strategies. J Acquir Immune Defic Syndr 2016; 71:181–8. [DOI] [PubMed] [Google Scholar]
- 9. Johnston RE, Heitzeg MM.. Sex, age, race and intervention type in clinical studies of HIV cure: a systematic review. AIDS Res Hum Retroviruses 2015; 31:85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scully EP. Sex differences in HIV infection. Curr HIV/AIDS Rep 2018; 15:136–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gandhi M, Bacchetti P, Miotti P, Quinn TC, Veronese F, Greenblatt RM.. Does patient sex affect human immunodeficiency virus levels?. Clin Infect Dis 2002; 35:313–22. [DOI] [PubMed] [Google Scholar]
- 12. Meditz AL, Folkvord JM, Lyle NH, et al. CCR5 expression is reduced in lymph nodes of HIV type 1-infected women, compared with men, but does not mediate sex-based differences in viral loads. J Infect Dis 2014; 209:922–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. El-Badry E, Macharia G, Claiborne D, et al. Better viral control despite higher CD4(+) T cell activation during acute HIV-1 infection in Zambian women is linked to the sex hormone estradiol. J Virol 2020; 94:e00758–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Meier A, Chang JJ, Chan ES, et al. Sex differences in the Toll-like receptor-mediated response of plasmacytoid dendritic cells to HIV-1. Nat Med 2009; 15:955–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sterling TR, Lyles CM, Vlahov D, Astemborski J, Margolick JB, Quinn TC.. Sex differences in longitudinal human immunodeficiency virus type 1 RNA levels among seroconverters. J Infect Dis 1999; 180:666–72. [DOI] [PubMed] [Google Scholar]
- 16. Gandhi RT, McMahon DK, Bosch RJ, et al. Levels of HIV-1 persistence on antiretroviral therapy are not associated with markers of inflammation or activation. PLoS Pathog 2017; 13:e1006285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Scully EP, Gandhi M, Johnston R, et al. Sex-based differences in human immunodeficiency virus type 1 reservoir activity and residual immune activation. J Infect Dis 2019; 219:1084–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cyktor JC, Bosch RJ, Mar H, et al. Male sex and obesity are associated with residual plasma HIV-1 viremia in persons on long-term antiretroviral therapy. J Infect Dis 2021; 223:462–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Falcinelli SD, Shook-Sa BE, Dewey MG, et al. Impact of biological sex on immune activation and frequency of the latent HIV reservoir during suppressive antiretroviral therapy. J Infect Dis 2020; 222:1843–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Prodger JL, Capoferri AA, Yu K, et al. Reduced HIV-1 latent reservoir outgrowth and distinct immune correlates among women in Rakai, Uganda. JCI Insight 2020; 5:e139287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Szotek EL, Narasipura SD, Al-Harthi L.. 17beta-estradiol inhibits HIV-1 by inducing a complex formation between beta-catenin and estrogen receptor alpha on the HIV promoter to suppress HIV transcription. Virology 2013; 443:375–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Das B, Dobrowolski C, Luttge B, et al. Estrogen receptor-1 is a key regulator of HIV-1 latency that imparts gender-specific restrictions on the latent reservoir. Proc Natl Acad Sci U S A 2018; 115:E7795–E804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Merck and Co. I. Vorinostat. Merck and Co, Whitehouse Station, NJ, 2006. Available at: https://www.accessdata.fda.gov/drugsatfda_docs/label/2006/021991lbl.pdf. [Google Scholar]
- 24. Malnati MS, Scarlatti G, Gatto F, et al. A universal real-time PCR assay for the quantification of group-M HIV-1 proviral load. Nat Protoc 2008; 3:1240–8. [DOI] [PubMed] [Google Scholar]
- 25. Cillo AR, Vagratian D, Bedison MA, et al. Improved single-copy assays for quantification of persistent HIV-1 viremia in patients on suppressive antiretroviral therapy. J Clin Microbiol 2014; 52:3944–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fidler S, Stohr W, Pace M, et al. Antiretroviral therapy alone versus antiretroviral therapy with a kick and kill approach, on measures of the HIV reservoir in participants with recent HIV infection (the RIVER trial): a phase 2, randomised trial. Lancet 2020; 395:888–98. [DOI] [PubMed] [Google Scholar]
- 27. Tsai P, Wu G, Baker CE, et al. In vivo analysis of the effect of panobinostat on cell-associated HIV RNA and DNA levels and latent HIV infection. Retrovirology 2016; 13:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vaida F, Liu L.. Fast implementation for normal mixed effects models with censored response. J Comput Graph Stat 2009; 18:797–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bacchetti P, Bosch RJ, Scully EP, et al. Statistical analysis of single-copy assays when some observations are zero. J Virus Erad 2019; 5:167–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dube K, Hosey L, Starr K, et al. Participant perspectives in an HIV cure-related trial conducted exclusively in women in the United States: results from AIDS Clinical Trials Group 5366. AIDS Res Hum Retroviruses 2020; 36:268–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cuzin L, Pugliese P, Saune K, et al. Levels of intracellular HIV-DNA in patients with suppressive antiretroviral therapy. AIDS 2015; 29:1665–71. [DOI] [PubMed] [Google Scholar]
- 32. Fourati S, Flandre P, Calin R, et al. Factors associated with a low HIV reservoir in patients with prolonged suppressive antiretroviral therapy. J Antimicrob Chemother 2014; 69:753–6. [DOI] [PubMed] [Google Scholar]
- 33. Henderson VW, St John JA, Hodis HN, et al. Cognition, mood, and physiological concentrations of sex hormones in the early and late postmenopause. Proc Natl Acad Sci U S A 2013; 110:20290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Munster PN, Thurn KT, Thomas S, et al. A phase II study of the histone deacetylase inhibitor vorinostat combined with tamoxifen for the treatment of patients with hormone therapy-resistant breast cancer. Br J Cancer 2011; 104:1828–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Celgene. Romidepsin. Celgene, Summit, NJ, 2009. Available at: https://www.accessdata.fda.gov/drugsatfda_docs/label/2009/022393lbl.pdf. [Google Scholar]
- 36. Archin NM, Bateson R, Tripathy MK, et al. HIV-1 expression within resting CD4+ T cells after multiple doses of vorinostat. J Infect Dis 2014; 210:728–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Elliott JH, Wightman F, Solomon A, et al. Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS Pathog 2014; 10:e1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gay CL, James KS, Tuyishime M, et al. Stable latent HIV infection and low-level viremia despite treatment with the broadly neutralizing antibody VRC07-523LS and the latency reversal agent vorinostat. J Infect Dis 2021:jiab487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kroon E, Ananworanich J, Pagliuzza A, et al. A randomized trial of vorinostat with treatment interruption after initiating antiretroviral therapy during acute HIV-1 infection. J Virus Erad 2020; 6:100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rasmussen TA, Tolstrup M, Brinkmann CR, et al. Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial. Lancet HIV 2014; 1:e13–21. [DOI] [PubMed] [Google Scholar]
- 41. McMahon DK, Zheng L, Cyktor JC, et al. A phase I/II randomized, placebo-controlled trial of romidepsin in persons with HIV-1 on suppressive antiretroviral therapy to assess safety and activation of HIV-1 expression (A5315). J Infect Dis 2020; 224:648–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mothe B, Rosas-Umbert M, Coll P, et al. HIVconsv vaccines and romidepsin in early-treated HIV-1-infected individuals: safety, immunogenicity and effect on the viral reservoir (study BCN02). Front Immunol 2020; 11:823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sogaard OS, Graversen ME, Leth S, et al. The depsipeptide romidepsin reverses HIV-1 latency in vivo. PLoS Pathog 2015; 11:e1005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yi X, Wei W, Wang SY, Du ZY, Xu YJ, Yu XD.. Histone deacetylase inhibitor SAHA induces ERalpha degradation in breast cancer MCF-7 cells by CHIP-mediated ubiquitin pathway and inhibits survival signaling. Biochem Pharmacol 2008; 75:1697–705. [DOI] [PubMed] [Google Scholar]
- 45. Puertas MC, Bayon-Gil A, Garcia-Guerrero MC, et al. VIP-SPOT: an innovative assay to quantify the productive HIV-1 reservoir in the monitoring of cure strategies. mBio 2021; 12:e0056021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stone M, Rosenbloom DIS, Bacchetti P, et al. Assessing the suitability of next-generation viral outgrowth assays to measure human immunodeficiency virus 1 latent reservoir size. J Infect Dis 2021; 224:1209–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu G, Swanson M, Talla A, et al. HDAC inhibition induces HIV-1 protein and enables immune-based clearance following latency reversal. JCI Insight 2017; 2:e92901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.